Abstract
Background
Although miR-758-3p has been reported to be associated with multiple cancers, including hepatocellular carcinoma, bladder cancer, gastric cancer and papillary thyroid cancer, its role in clear cell renal cell carcinoma (ccRCC) remains unclear.
Methods
The expression levels of miR-758-3p in ccRCC tissues and cell lines were examined using qRT-PCR. Survival analysis was performed using Kaplan–Meier, while the prognostic significance of miR-758-3p was evaluated by Cox regression analysis. The effects of miR-758-3p on cell proliferation, migration and invasion were analyzed with CCK-8, crystal violet and transwell assays. Luciferase reporter assays were performed to determine the effect of miR-758-3p on MDM2. Western blot was applied to measure the expression of MDM2.
Results
The expression levels of miR-758-3p were down-regulated in human ccRCC tissues and cell lines. Moreover, the expression of miR-758-3p was closely associated with histological grade, TNM stage and vascular invasion. High expression of miR-758-3p was found to be capable of predicting favorable clinical prognosis in ccRCC patients. Additionally, whilst the proliferation, migration and invasion of ccRCC cells were inhibited upon overexpression of miR-758-3p, the effects were reversed upon miR-758-3p knockdown. Moreover, miR-758-3p was a modulator of MDM2 in ccRCC.
Conclusion
This study demonstrated that miR-758-3p is a potential prognostic biomarker for ccRCC patients. Data from this study showed that miR-758-3p exhibits different biological functions that inhibit the progression of ccRCC cells, hence it can be a potential therapeutic target candidate for treating ccRCC.
Keywords: ccRCC, miR-758-3p, progression, prognosis, biomarker
Introduction
Renal cell carcinoma (RCC) is a commonly diagnosed human kidney cancer that accounts for approximately 3.8% of all adult malignancies and 2.5% of all cancer-related mortality.1 Recently, the annual incidence of RCC has been reported to be increased by 0.6% globally.2 Clear cell renal cell carcinoma (ccRCC) is the most common RCC subtype that constitutes 70–80% of all five histological subtypes.3 However, approximately 20–30% of firstly diagnosed RCC patients have developed metastatic diseases, including locally invasive or metastatic renal cell carcinoma.4 Moreover, it has been shown that nearly a third of RCC patients may develop recurrence even after resection of localized disease.5 Despite considerable progress in the development of therapeutic strategies, the prognosis of ccRCC patients at advanced stages remains poor with a median survival of only 13 months.6
The identification of molecular biomarkers to date has led to new insights into the biology of ccRCC and the development of novel targeted therapies.7 Emerging data have demonstrated the associations between the prognosis of ccRCC patients with different cellular and molecular factors, including somatic mutations, gene expression profiles, gene methylations, germline variations and immune biomarkers, such as CD8 and PD-L1.8–10 Some of these markers, when used in combination especially, have improved the discrimination of current prognostic models, however, none of the markers were extensively validated, so their underlying mechanisms remain poorly understood. As such, their routine use in clinical practice is currently not recommended. Therefore, it is of vital importance to investigate the molecular mechanisms of the markers during the pathogenesis of ccRCC in order to facilitate the development of novel prevention strategies for these patients.
MicroRNAs (miRNAs) are endogenous small non-coding RNAs with a length of 19 to 25 nucleotides. Recently, the expression of miRNAs and their functions in tumor progression have been extensively elucidated. In particular, abnormal miRNA expression has been found to be highly involved in some critical cellular processes of multiple cancers, including proliferation, differentiation, development and apoptosis.11–13 Recently, increasing evidence has indicated that miRNAs can act as oncogenes or tumor suppressors in the regulation of ccRCC development and progression.14 It has been widely reported in recent years, that miR-758-3p plays critical roles in different cancer types, including hepatocellular carcinoma,15 bladder cancer,16 gastric cancer,17 and papillary thyroid cancer.18 Nevertheless, the roles of miR-758-3p in ccRCC remain unclear.
In this study, we explored the prognostic and biological functions of miR-758-3p in the progression of ccRCC. We found that miR-758-3p was down-regulated in human primary ccRCC tissues and cell lines. Moreover, we demonstrated that the high expression of miR-758-3p was associated with favorable prognosis. Additionally, our data revealed that miR-758-3p inhibited the proliferation, migration and invasion of ccRCC cells in vitro via targeting MDM2.
Materials and Methods
Clinical Samples
The tumor tissues and paired adjacent normal tissues were obtained from 68 ccRCC patients at the First Affiliated Hospital of Jinzhou Medical University (Liaoning, China) between January 2016 and December 2016. Patients included in this study fulfilled the following selection criteria: (1) Must have undergone primary surgery; (2) Must not have received pre-surgery anticancer treatments like chemotherapy or radiation treatment; (3) All collected tumor samples must be pathologically confirmed as clear cell renal cell carcinoma; and (4) Must have complete follow-up information. All collected samples were immediately frozen in liquid nitrogen for further analysis. Written informed consent was obtained from all patients. This study was approved by the ethics committee of the First Affiliated Hospital of Jinzhou Medical University (JMU-20200010), and was conducted in accordance with the Declaration of Helsinki.
Cell Culture
Human ccRCC cell lines (ACHN, Caki-1 and 786-O) and normal renal epithelial (HK-2) were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences (Shanghai, China). ACHN, Caki-1 and 786-O cells were cultured in RPMI-1640 medium (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA), 100 U/mL penicillin and 100 g/mL streptomycin (Sangong Biotech) in a 37 ◦C incubator with 5% CO2. HK-2 cells were cultured in keratinocyte serum-free medium (K-SFM; Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA).
RNA Extraction and Quantitative Reverse Transcription‑polymerase Chain Reaction (RT-qPCR)
Total RNA from the clinical tissues and cell lines samples was extracted using RNAisoPlus reagent (Takara, Bio, Inc., Otsu, Japan). A total of 1 µg RNA was reverse transcribed to cDNA using TaqMan microRNA Reverse Transcription kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) in accordance with the manufacturer’s protocol. RT-qPCR was conducted using SYBR-Green Kit (Takara Biotechnology Co., Ltd.) on an iCycler IQ multicolor Detection System (Applied Biosystems). The thermal cycling conditions were as follows: 95°C for 3 min; 40 cycles of 95°C for 20 sec, 60°C for 20 sec, and 72°C for 40 sec; 72°C for 3 min; 4°C for the remaining period. The relative expression of miR-758-3p was normalized to that of U6 using the 2−∆∆Ct method.19 The following oligonucleotides (GenePharma, Shanghai, China) were used in RT-qPCR reactions: miR-758-3p-forward (5ʹ-ACACTCCAGCTGGGTTTGTGACCTGGTCCA-3ʹ), miR-758-3p-reverse (5ʹ-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGTTAGTG-3ʹ), U6-forward (5ʹ-CTCGCTTCGGCAGCACA-3ʹ), U6-reverse (5ʹ-AACGCTTCACGAATTTGCGT −3ʹ), MDM2-forward (5ʹ- ACCCATCTACCCTGACCACA-3ʹ), MDM2-reverse (5ʹ- AGAATGCTTTAGTCCACCTAACCT-3ʹ), GAPDH-forward (5ʹ- GCACCGTCAAGGCTGAGAAC −3ʹ), GAPDH – reverse (5ʹ- TGGTGAAGACGCCAGTGGA −3ʹ).
Western Blotting
786-O and ACHN cells were lysed in RIPA solution (Thermo Fisher Scientific, Inc.). Protein concentration was determined using BCA Assay kit (KeyGEN Biotech, China) according to the manufacturer’s instructions. Proteins samples were separated by 10% SDS-PAGE and transferred onto PVDF membranes (Millipore, MA, USA). Then, membranes were blocked with 5% fat-free milk, and incubated with anti-MDM2 (dilution, 1:1000; cat. no. ab16895; Abcam, CA, USA) and anti-GAPDH (dilution, 1:1000; cat. no. 60,004-1-Ig; ProteinTech Group, Inc.) primary antibodies at 4°C overnight. Membranes were then incubated with appropriate horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The immunoreactive protein bands were visualized by chemiluminescence.
Cell Transfection
The miR-758-3p mimics, miR-758-3p inhibitor and their corresponding negative controls (miR-NCs) were all designed and purchased from GenePharma (Shanghai, China). ccRCC cells were seeded in 6-well plates (5×105 cells per well) and cultured to ~60‑70% confluency before transfection using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Transfected cells were cultured for 48 h at 37°C in accordance with the manufacturer’s instructions before subsequent experiments.
Cell Counting Kit-8 (CCK-8) Proliferation Assay
Three replicates of transfected ccRCC cells (3×103 cells in 100 μL culture medium per well) were seeded into 96-well plates. After that, 10 μL of CCK-8 solution (Dojindo, Kumamoto, Japan) was added to the medium at 24, 48, 72 and 96 h, and then incubated at 37°C for 2 h before the optical density (OD) measurement at a wavelength of 450nm using an automatic microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Crystal Violet Assay
Three replicates of transfected ccRCC cells were seeded into 6-well plates at a density of 1×103 cells per well. The cells were cultured in culture medium containing 10% FBS, and the culture medium was changed every 3 days for a duration of 2 weeks. The culture medium was then removed and the cells were stained with crystal violet solution for 15 min at room temperature. Ten minutes later, the fixed cells were washed with phosphate-buffered saline (PBS) before optical density (OD) measurement at a wavelength of 570 nm using an automatic microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Transwell Assay
The migration and invasion ability of transfected ccRCC cells were examined using a Transwell assay with polyethylene terephthalate membranes (24-well inserts; 8.0 μm; Corning Inc.). A total of 100 μL cell suspension (5 × 104 cells) was seeded in the upper chamber with serum-free medium. The lower chamber contained 500 μL medium supplemented with 10% FBS. For invasion assay, the membrane precoated with 50 µL Matrigel at 1:6 dilution (BD Biosciences) was incubated at 37°C for 3 h. The cells were maintained in a humidified atmosphere with 5% CO2 at 37°C. After 48 h, cells that have migrated or invaded the bottom part of the membrane were fixed with polyoxymethylene at room temperature for 30 min before staining with 0.5% crystal violet for 15min at room temperature. Cells in five randomly selected fields were counted under a light microscope (Olympus Corporation) at ×400 magnification.
Luciferase Reporter Assay
The amplified 3′-UTR of MDM2 was subcloned into the firefly plasmids in pmirGLO luciferase vector (GeneChem, Shanghai, China). The wild type of MDM2 3′-UTR (MDM2-WT) was constructed. The site-directed mutation of miR-758-3p binding sites in MDM2 3′-UTR (MDM2-MUT) was generated using the GeneTailor™ Site-Directed Mutagenesis System (Invitrogen, Carlsbad, CA, USA). 786-O cell lines were co-transfected with aforementioned plasmids and miRNAs (miR-758-3p mimic or mi-NC) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h of incubation, the cells were lysed, and reporter activities were determined using a dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Statistical Analysis
All the data are expressed as mean ± standard deviation. Statistical evaluations were performed with GraphPad Prism 5.0 (GraphPad Software Inc., CA, USA) and SPSS 20.0 (IBM SPSS Inc., Chicago, IL, USA). The difference between two groups was evaluated using Student’s t-test. Categorical data were compared using chi-square test. Overall survival (OS) and recurrence-free survival (RFS) of patients with ccRCC were assessed using Kaplan–Meier curves. Prognostic factors were examined using Cox regression proportional hazards analysis. Differences were considered to be significant when P< 0.05.
Results
miR-758-3p Was Down-Regulated in ccRCC Tissues and Cell Lines
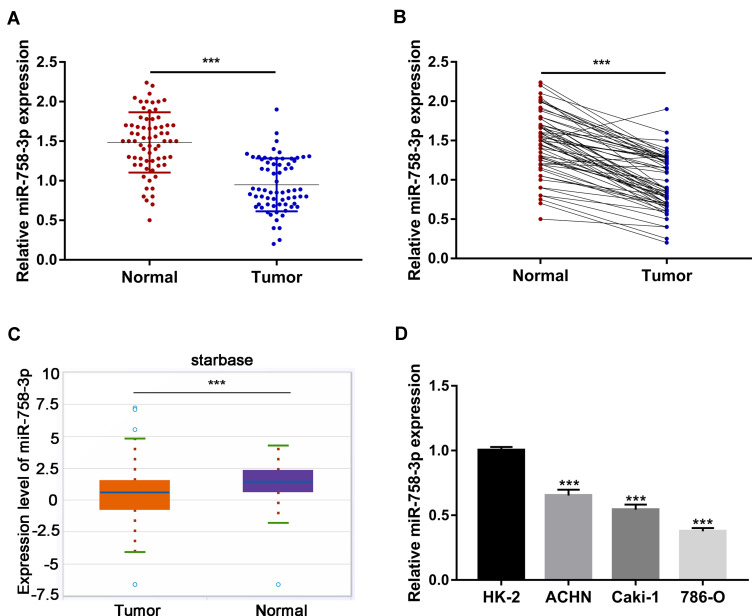
To primarily explore whether the miR-758-3p expression is associated with ccRCC progression, the expression levels of miR-758-3p in 68 pairs of ccRCC tissues and corresponding normal tissues were evaluated using RT-qPCR. Our results showed that the expression of miR-758-3p was significantly down-regulated in the primary ccRCC tissues compared to that of the normal tissues (all P< 0.001, Figure 1A and B). Besides, the starbase data analysis from 517 ccRCC tissues and 71 normal tissues showed a significant decrease of miR-758-3p expression in the primary ccRCC tissues compared to that of the normal tissues (P< 0.001, Figure 1C). Subsequently, the expression levels of miR-758-3p were analyzed in several ccRCC cell lines (ACHN, Caki-1 and 786-O) and normal renal epithelial cell line (HK-2). As shown in Figure 1D, the expression of miR-758-3p was markedly lower in the ccRCC cell lines, especially in 786-O cells, than that in HK-2 cells (all P< 0.001).
Figure 1.
Down-regulation of miR-758-3p expression in ccRCC tissues and cell lines. (A-B) Relative expression of miR-758-3p in 68 pairs of ccRCC tissues and adjacent normal tissues determined by RT-qPCR. Scatter plot view (A) and paired view (B) are shown. (C) starbase data analysis of miR-758-3p expression in 517 ccRCC tissues and 71 normal tissues. (D) Relative expression of miR-758-3p in three ccRCC cell lines (ACHN, Caki-1 and 786-O) and normal renal epithelial cell line (HK-2) determined by RT-qPCR. ***P <0.001.
Abbreviations: ccRCC, clear cell renal cell carcinoma; RT-qPCR, quantitative reverse transcription‑polymerase chain reaction.
Association Between Clinicopathological Characteristics and miR-758-3p Expression in ccRCC Patients
Next, with the median value of miR-758-3p expression in tumor tissues as the cut-off value, the 68 ccRCC patients were divided into two groups: low miR-758-3p expression group (below the median, 34 patients) and high miR-758-3p expression group (above the median, 34 patients). Then, the association between the expression of serum miR-758-3p and clinicopathologic parameters was analyzed in the 68 ccRCC patients. As shown in Table 1, a higher proportion of G3-G4 histological grade was observed in the low miR-758-3p expression group than that in the high miR-758-3p expression group (P = 0.026). There was a higher number of patients with TNM stage III-IV in the low miR-758-3p expression group than that in the high miR-758-3p expression group (P = 0.027). In addition, vascular invasion was more common in patients in the miR-758-3p low expression group than that in the high expression group (P = 0.028). However, no significant difference was found between miR-758-3p expression and other characteristics, such as age, gender, tumor size and lymph nodes metastasis (all P > 0.05).
Table 1.
The Relationship Between miR-758-3p Expression and Clinical Features in Patients with Clear Cell Renal Cell Carcinoma
| Parameters | Cases (n=68) |
miR-758-3p, n (%) | P-value | |
|---|---|---|---|---|
| High Expression (n=34) |
Low Expression (n=34) |
|||
| Age, year | 0.625 | |||
| < 60 | 30 (44.1) | 14 (41.2) | 16 (47.1) | |
| ≥ 60 | 38 (55.9) | 20 (58.8) | 18 (52.9) | |
| Gender | 0.612 | |||
| Male | 44 (64.7) | 23 (67.6) | 21 (61.8) | |
| Female | 24 (35.3) | 11 (32.4) | 13 (38.2) | |
| Tumor size, cm | 0.331 | |||
| < 7 | 36 (52.9) | 20 (58.8) | 16 (47.1) | |
| ≥ 7 | 32 (47.1) | 14 (41.2) | 18 (52.9) | |
| Histological grade | 0.026* | |||
| G1-G2 | 29 (42.6) | 18 (52.9) | 9 (26.5) | |
| G3-G4 | 39 (57.4) | 16 (47.1) | 25 (73.5) | |
| TNM stage | 0.027* | |||
| I-II | 29 (42.6) | 19 (55.9) | 10 (29.4) | |
| III-IV | 39 (57.4) | 15 (44.1) | 24 (70.6) | |
| Lymph nodes metastasis | 0.088 | |||
| Positive | 31 (45.6) | 12 (35.3) | 19 (55.9) | |
| Negative | 37 (54.4) | 22 (64.7) | 15 (44.1) | |
| Vascular invasion | 0.028* | |||
| Yes | 18 (26.5) | 5 (14.7) | 13 (38.2) | |
| No | 50 (73.5) | 29 (85.3) | 21 (61.8) | |
Note: *P <0.05.
The Prognostic Value of miR-758-3p in ccRCC Patients
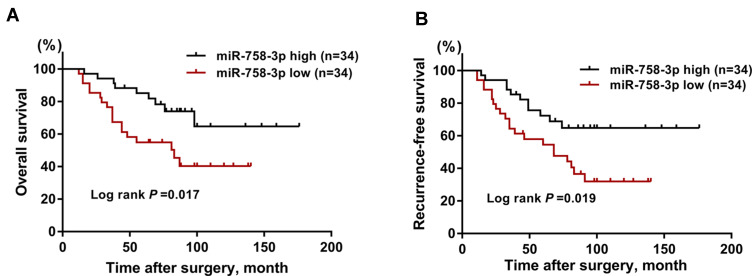
Next, the role of miR-758-3p in the prognosis of ccRCC patients was investigated using Kaplan–Meier curves. In ccRCC patients, high miR-758-3p expression was found to be associated with better OS (Figure 2A; P = 0.017) and RFS (Figure 2B; P = 0.019) compared to those with low miR-758-3p expression. To explore the prognostic factors for OS and RFS, multivariate Cox regression analysis was applied. For prognostic factors for OS (Table 2), the following results were obtained: TNM stage (hazard ratio (HR): 1.71, 95% confidence interval (CI): 1.23–2.02, P =0.011); vascular invasion (HR: 2.12, 95% CI: 1.65–3.01, P= 0.001); miR-758-3p expression (HR: 0.53, 95% CI: 0.46–0.88, P= 0.013). Meanwhile, for prognostic factors for RFS (Table 3), the following data were obtained: TNM stage (HR: 1.82, 95% CI: 1.31–2.13, P= 0.001); vascular invasion (HR: 2.12, 95% CI: 1.65–3.01, P =0.003); miR-758-3p expression (HR: 0.57, 95% CI: 0.44–0.84, P =0.033). Taken together, our results showed that high expression of miR-758-3p was correlated with favorable prognosis in ccRCC patients.
Figure 2.
Prognostic value assessment of miR-758-3p in ccRCC patients. (A-B) Kaplan–Meier curves for overall survival (A) and recurrence-free survival (B) according to serum levels of miR-758-3p in ccRCC patients.
Abbreviation: ccRCC, clear cell renal cell carcinoma.
Table 2.
Univariate and Multivariate Analyses of Prognostic Factors Associated with Overall Survival
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (≥60 vs <60) | 1.11 (0.53–1.67) | 0.512 | ||
| Gender (male vs female) | 1.09 (0.54–1.86) | 0.345 | ||
| Tumor size (≥ 7 vs <7) | 1.43 (0.95–2.12) | 0.078 | ||
| Histological grade (G3-4 vs G1-2) | 2.25 (1.17–3.56) | 0.010* | 2.01 (0.89–3.23) | 0.089 |
| TNM stage (III-IV vs I-II) | 1.78 (1.34–2.13) | 0.001* | 1.71 (1.23–2.02) | 0.011* |
| Lymph nodes metastasis (positive vs negative) | 1.45 (0.78–2.87) | 0.098 | ||
| Vascular invasion (yes vs no) | 2.56 (1.78–3.13) | <0.001* | 2.12 (1.65–3.01) | 0.001* |
| miR-758-3p expression (high vs low) | 0.57 (0.48–0.93) | 0.001* | 0.53 (0.46–0.88) | 0.013* |
Note: *P <0.05.
Abbreviations: HR, hazard ratio; CI, confidence interval.
Table 3.
Univariate and Multivariate Analyses of Prognostic Factors Associated with Recurrence-Free Survival
| Variables | Univariate | Multivariate | ||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age (≥60 vs <60) | 1.13 (0.49–1.56) | 0.425 | ||
| Gender (male vs female) | 1.11 (0.64–1.93) | 0.278 | ||
| Tumor size (≥ 7 vs <7) | 1.33 (0.87–2.08) | 0.112 | ||
| Histological grade (G3-4 vs G1-2) | 2.11 (0.91–3.32) | 0.093 | ||
| TNM stage (III-IV vs I-II) | 1.98 (1.43–2.34) | <0.001* | 1.82 (1.31–2.13) | 0.001* |
| Lymph nodes metastasis (positive vs negative) | 1.56(0.68–2.98) | 0.113 | ||
| Vascular invasion (yes vs no) | 2.78 (1.69–3.24) | 0.001* | 2.12 (1.65–3.01) | 0.003* |
| miR-758-3p expression (high vs low) | 0.53 (0.41–0.79) | 0.002* | 0.57 (0.44–0.84) | 0.033* |
Note: *P <0.05.
Abbreviations: HR, hazard ratio; CI, confidence interval.
Overexpression of miR-758-3p Inhibited the Proliferation, Migration and Invasion of ccRCC Cells
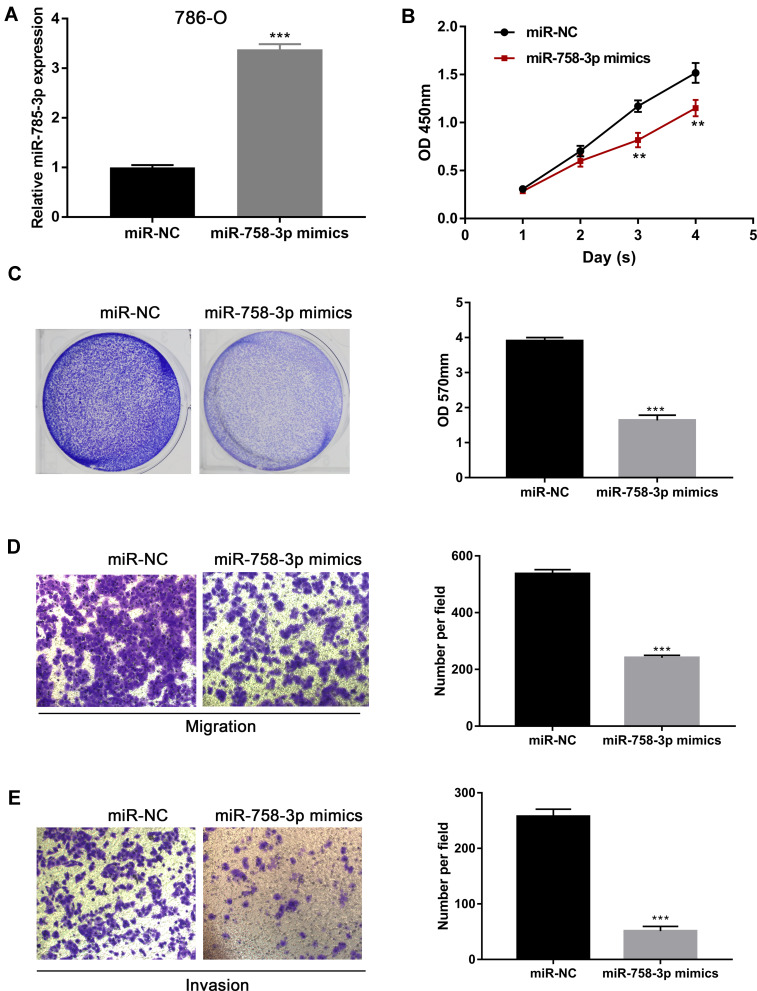
Based on the findings above, we further investigated the biological functions of miR-758-3p in the progression of ccRCC cells. Firstly, miR-758-3p was overexpressed using miR-758-3p mimics in 786-O cells with relatively low miR-758-3p expression. As shown in Figure 3A, qRT-PCR analysis indicated that miR-758-3p mimics successfully induced efficient miR-758-3p overexpression in 786-O cells (P < 0.001). Results from CCK-8 assays as shown in Figure 3B revealed that the absorbance values of 786-O cells at 3 and 4 days after transfection with miR-758-3p mimics were significantly lower than those of the untreated cells (all P < 0.01). Moreover, data from crystal violet assays (Figure 3C) indicated that overexpression of miR-758-3p significantly inhibited the proliferation of 786-O cells (P < 0.001). In addition, to explore the migratory and invasive capacity of 786-O cells transwell assays were performed. Results as shown in Figure 3D and E demonstrated that the migration and invasion of 786-O cells were notably inhibited by miR-758-3p overexpression (all P < 0.001).
Figure 3.
Inhibition of ccRCC cell proliferation, migration and invasion by miR-758-3p overexpression. (A) Expression of miR-758-3p in 786-O cells after transfection with miR-758-3p mimics and negative controls. (B and C) Effects of miR-758-3p overexpression on the proliferation of 786-O cells assessed by CCK-8 (B) and crystal violet assays (C). (D and E) Effects of miR-758-3p overexpression on the migration (D) and invasion (E) of 786-O cells assessed by transwell assays (magnification: 400×). **P< 0.01, ***P< 0.001.
Abbreviation: ccRCC, clear cell renal cell carcinoma.
Knockdown of miR-758-3 Promoted the Proliferation, Migration and Invasion of ccRCC Cells
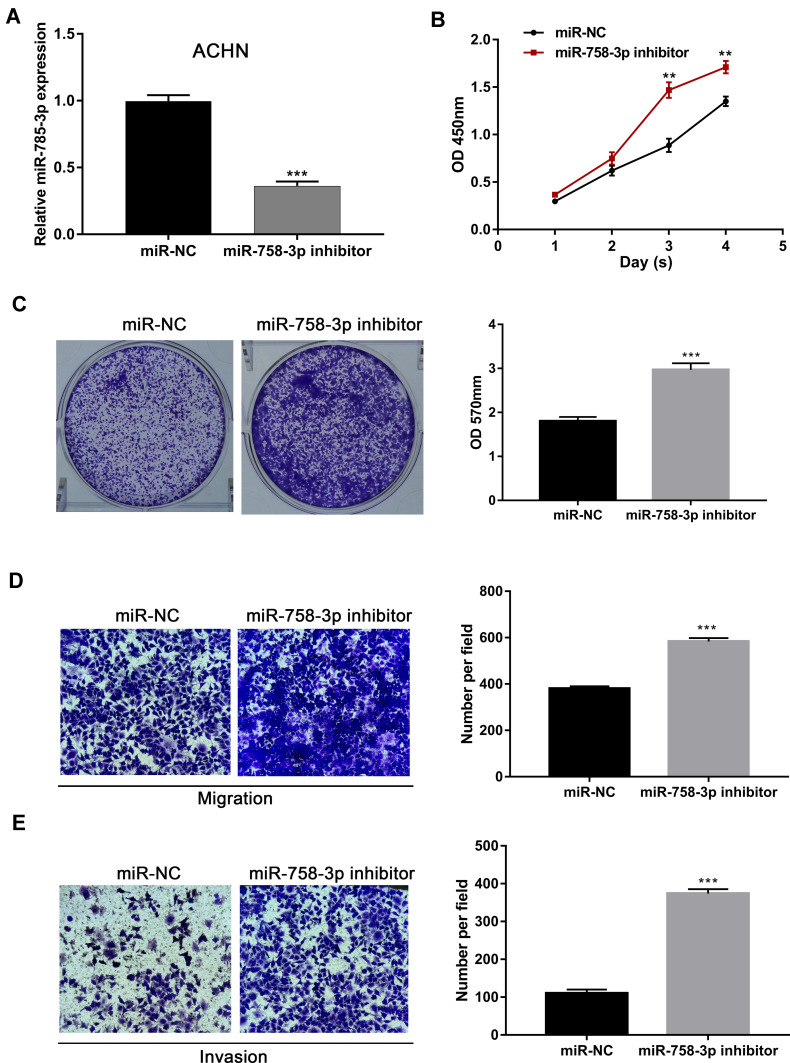
To down-regulate the expression of miR-758-3p, miR-758-3p inhibitor was used in ACHN cell lines with relatively high miR-758-3p expression and the knockdown efficacy was confirmed by qRT-PCR (P < 0.001, Figure 4A). Results from CCK-8 assays and crystal violet assays as shown in Figure 4B and C revealed that knockdown of miR-758-3p significantly promoted the proliferation ability of ACHN cells (all P< 0.01). In addition, transwell assays indicated that ACHN cells transfected with miR-758-3p inhibitor exhibited a significantly increased cell migratory and invasive capacity compared with that of the negative control (all P < 0.001, Figure 4D and E).
Figure 4.
Promotion of ccRCC cell proliferation, migration and invasion through miR-758-3p knockdown. (A) Expression of miR-758-3p in ACHN cells after transfection with miR-758-3p inhibitor and negative controls. (B and C). Effects of miR-758-3p knockdown on the proliferation of ACHN cells assessed by CCK-8 (B) and crystal violet assays (C). (D and E) Effects of miR-758-3p knockdown on the migration (D) and invasion (E) of ACHN cells assessed by transwell assays (magnification: 400×). **P< 0.01, ***P< 0.001.
Abbreviation: ccRCC, clear cell renal cell carcinoma.
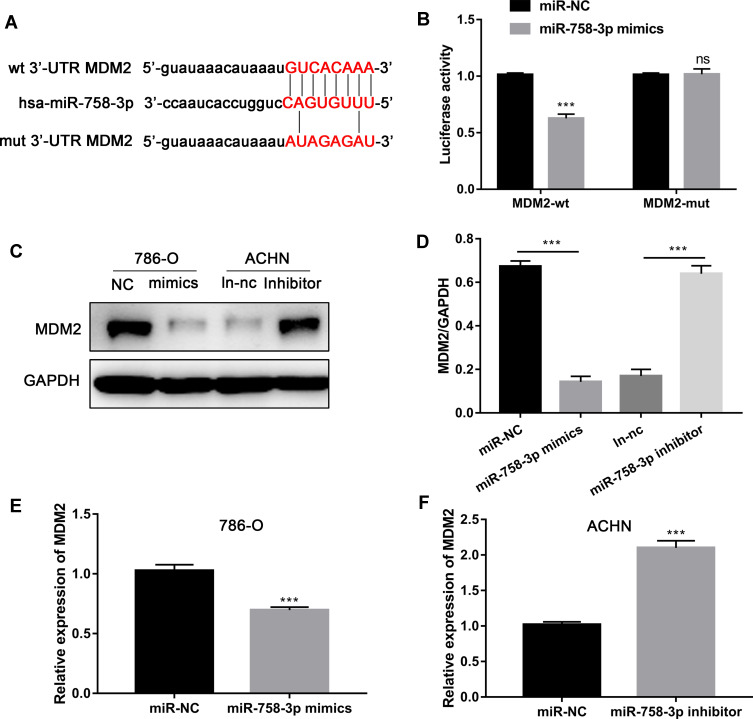
MDM2 is the Direct Target of miR-758-3p
We screened that MDM2 may act as the potential target gene of miR-758-3p via bioinformatics prediction using Targetscan tool. At first, the binding sequence of miR-758-3p and MDM2 3′ UTR was predicted and obtained (Figure 5A). Then, we examined the luciferase activity of reporter containing wild-type MDM2 (MDM2-WT) or mutant-type MDM2 (MDM2-MUT) in cells transfected with miR-758-3p mimics. The results suggested that the luciferase activity of MDM2-WT vector but not MDM2-MUT vector was decreased by miR-758-3p mimics (Figure 5B). Moreover, the protein levels of MDM2 were decreased in 786-O cells transfected with miR-758-3p mimics and increased in ACHN cells transfected with miR-758-3p inhibitors (Figure 5C and D). In addition, the mRNA levels of MDM2 were decreased in 786-O cells transfected with miR-758-3p mimics and increased in ACHN cells transfected with miR-758-3p inhibitors (Figure 5E and F). Taken together, above data demonstrated that miR-758-3p was a modulator of MDM2 in ccRCC.
Figure 5.
MDM2 is direct targets of miR-758-3p. (A) Diagram of the potential associating site in the 3ʹ-UTR region of MDM2 with miR-758-3p. (B) miR-758-3p overexpression inhibited the luciferase activity in 786-O cells. (C and D) MiR-758-3p inhibits MDM2 expression at protein level. (E and F) MiR-758-3p inhibits MDM2 and mRNA expression. ***P <0.001.
Abbreviation: ns, not significant.
Discussion
Recently, an increasing number of miRNAs with fundamental biological functions in the progression of ccRCC has been identified as diagnostic or prognostic biomarkers.19–21 For instance, Luo and colleagues have developed a novel three-miRNA signature (miR130b, miR-18a and miR-223) with reliable prognostic for the survival of patients with ccRCC, and hence may facilitate the development of individualized management of ccRCC patients.22 Based on a discovery cohort of 18 ccRCC patients, Zhang and colleagues have identified multiple dysregulated miRNAs in ccRCC tissues, among which miR-155-5p and miR-210-3p were capable of predicting ccRCC recurrence.23 In addition, Jingushi and colleagues have reported that miR-629 can accelerate the cell mortality and invasion of ccRCC via the regulation of TGFβ/Smad signaling.24
In this study, we documented that miR-758-3p was down-regulated in human ccRCC tissues and cell lines compared with that of adjacent normal tissues and normal renal epithelial cells. Moreover, the expression of miR-758-3p was found to be closely associated with histological grade, TNM stage and vascular invasion. Notably, high expression of miR-758-3p was shown to be able to predict favorable clinical prognosis in ccRCC patients. These findings indicated the prognostic capability of miR-758-3p in ccRCC patients. In addition, our functional assays indicated that, whilst the proliferation, migration and invasion of ccRCC cells were inhibited when miR-758-3p was overexpressed; opposite effects were observed upon knockdown of miR-758-3p. Through binding partner predication, we found the possible interaction between miR-758-3p and the 3ʹUTR of MDM2 and validated their binding through mutation analysis. miR-758-3p overexpression likely led to a decrease in MDM2 expression at both of transcriptional and translational levels. Thus, MDM2 might be a functional target of miR-758-3p in ccRCC.
Previous studies have demonstrated that miR-758-3p plays a tumor suppressor role in multiple human cancers. For example, in hepatocellular carcinoma, overexpression of miR-758-3p has been reported to play a critical role in the inhibition of proliferation, migration and invasion of hepatocellular carcinoma cells via MDM2 and mTOR.15 In addition, Wu and colleagues have demonstrated that miR-758-3p can suppress the proliferation, migration and invasion of bladder cancer cells by targeting NOTCH2.16 Meanwhile, Guo and colleagues have reported that miR-758-3p can serve as a tumor suppressor, playing a crucial inhibitory role in the proliferation, migration and invasion of gastric cancer via chromobox 5.17 Moreover, miR-758-3p has also been shown to act as a tumor suppressor in the initiation of papillary thyroid cancer via inhibition of cell proliferation and invasion, and promotion of cell apoptosis.18 Overall, for the first time, this study determined the prognostic and tumor suppressor role of miR-758-3p in the progression of ccRCC.
In conclusion, this study demonstrated that miR-758-3p is associated with clinical prognosis of ccRCC patients, and that miR-758-3p inhibits the progression of ccRCC cells. Thus, miR-758-3p may be a potential prognostic and therapeutic target for ccRCC patients.
Funding
There is no funding to report.
Disclosure
No conflict of interests was declared in the study.
References
- 1.Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906. [DOI] [PubMed] [Google Scholar]
- 2.Petejova N, Martinek A. Renal cell carcinoma: review of etiology, pathophysiology and risk factors. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2016;160(2):183–194. [DOI] [PubMed] [Google Scholar]
- 3.Hancock SB, Georgiades CS. Kidney Cancer. Cancer j. 2016;22(6):387–392. [DOI] [PubMed] [Google Scholar]
- 4.Lam JS, Shvarts O, Pantuck AJ. Changing concepts in the surgical management of renal cell carcinoma. Eur Urol. 2004;45(6):692–705. [DOI] [PubMed] [Google Scholar]
- 5.Russo P. Renal cell carcinoma: presentation, staging, and surgical treatment. Semin Oncol. 2000;27(2):160–176. [PubMed] [Google Scholar]
- 6.Lam JS, Belldegrun AS, Pantuck AJ. Long-term outcomes of the surgical management of renal cell carcinoma. World J Urol. 2006;24(3):255–266. [DOI] [PubMed] [Google Scholar]
- 7.Majer W, Kluzek K, Bluyssen H, Wesoły J. Potential approaches and recent advances in biomarker discovery in clear-cell renal cell carcinoma. J Cancer. 2015;6(11):1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao D, Margolis CA, Gao W, et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science. 2018;359(6377):801–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joseph RW, Kapur P, Serie DJ, et al. Loss of BAP1 protein expression is an independent marker of poor prognosis in patients with low-risk clear cell renal cell carcinoma. Cancer. 2014;120(7):1059–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rydzanicz M, Wrzesiński T, Bluyssen HA, Wesoły J. Genomics and epigenomics of clear cell renal cell carcinoma: recent developments and potential applications. Cancer Lett. 2013;341(2):111–126. [DOI] [PubMed] [Google Scholar]
- 11.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. [DOI] [PubMed] [Google Scholar]
- 12.Qadir MI, Faheem A. miRNA: a diagnostic and therapeutic tool for pancreatic cancer. Crit Rev Eukaryot Gene Expr. 2017;27(3):197–204. [DOI] [PubMed] [Google Scholar]
- 13.Sun Z, Shi K, Yang S, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braga EA, Fridman MV, Loginov VI, Dmitriev AA, Morozov SG. Molecular mechanisms in clear cell renal cell carcinoma: role of miRNAs and hypermethylated miRNA genes in crucial oncogenic pathways and processes. Front Genet. 2019;10:320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang D, Cho W, Li Z, et al. MiR-758-3p suppresses proliferation, migration and invasion of hepatocellular carcinoma cells via targeting MDM2 and mTOR. Biomed Pharmacotherapy. 2017;96:535–544. [DOI] [PubMed] [Google Scholar]
- 16.Wu X, Chen B, Shi H, et al. miR-758-3p suppresses human bladder cancer cell proliferation, migration and invasion by targeting NOTCH2. Exp Ther Med. 2019;17(5):4273–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J, Zhang Z, Pan L, Zhou Y. Identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer. Technol Cancer Res Treat. 2018;17:1533033818816061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Chen J, Xu Z, Yu C, et al. MiR-758-3p regulates papillary thyroid cancer cell proliferation and migration by targeting TAB1. Die Pharmazie. 2019;74(4):235–238. [DOI] [PubMed] [Google Scholar]
- 19.Szabó Z, Szegedi K, Gombos K, et al. Expression of miRNA-21 and miRNA-221 in clear cell renal cell carcinoma (ccRCC) and their possible role in the development of ccRCC. Urol Oncol. 2016;34(12):533.e21-.e27. [DOI] [PubMed] [Google Scholar]
- 20.Xie M, Lv Y, Liu Z, et al. Identification and validation of a four-miRNA (miRNA-21-5p, miRNA-9-5p, miR-149-5p, and miRNA-30b-5p) prognosis signature in clear cell renal cell carcinoma. Cancer Manag Res. 2018;10:5759–5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sage AP, Minatel BC, Marshall EA, et al. Expanding the miRNA transcriptome of human kidney and renal cell carcinoma. Int J Genomics. 2018;2018:6972397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Y, Chen L, Wang G, Xiao Y, Ju L, Wang X. Identification of a three-miRNA signature as a novel potential prognostic biomarker in patients with clear cell renal cell carcinoma. J Cell Biochem. 2019;120(8):13751–13764. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Ye Y, Chang DW, et al. Global and targeted miRNA expression profiling in clear cell renal cell carcinoma tissues potentially links miR-155-5p and miR-210-3p to both tumorigenesis and recurrence. Am J Pathol. 2018;188(11):2487–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jingushi K, Ueda Y, Kitae K, et al. miR-629 targets TRIM33 to promote TGFβ/Smad signaling and metastatic phenotypes in ccRCC. Mol Cancer Res. 2015;13(3):565–574. [DOI] [PubMed] [Google Scholar]