Abstract
The actin cytoskeleton has the particularity of being assembled into many functionally distinct filamentous networks from a common reservoir of monomeric actin. Each of these networks has its own geometrical, dynamical and mechanical properties, because they are capable of recruiting specific families of actin-binding proteins (ABPs), while excluding the others. This review discusses our current understanding of the underlying molecular mechanisms that cells have developed over the course of evolution to segregate ABPs to appropriate actin networks. Segregation of ABPs requires the ability to distinguish actin networks as different substrates for ABPs, which is regulated in three different ways: (1) by the geometrical organization of actin filaments within networks, which promotes or inhibits the accumulation of ABPs; (2) by the identity of the networks' filaments, which results from the decoration of actin filaments with additional proteins such as tropomyosin, from the use of different actin isoforms or from covalent modifications of actin; (3) by the existence of collaborative or competitive binding to actin filaments between two or multiple ABPs. This review highlights that all these effects need to be taken into account to understand the proper localization of ABPs in cells, and discusses what remains to be understood in this field of research.
Keywords: actin isoforms, tropomyosin, post-translational modifications, network architecture, actin-binding protein segregation, actin filament identity
1. Introduction
Actin plays a major role in many different biological processes such as cytokinesis, migration, vesicular trafficking and infection [1,2]. For each of these functions, actin filaments are organized into networks of optimized architectures, dynamics and mechanical properties. The main types of organizations include (but are not limited to) branched and linear networks of actin filaments [3,4]. Branched actin networks are generated by the association of a seven-subunit complex called the Arp2/3 complex, which nucleates short actin filament branches. Linear networks, where polar actin filaments are parallel or randomly organized, are generated from the de novo nucleation of actin filaments by factors such as formins, or from the debranching and reorganization of branched networks.
Actin networks are regulated by the association of different families of actin-binding proteins (ABPs). It is important to note that although all these proteins coexist in the cell cytoplasm, only a specific subset of ABPs interacts with each actin network while being excluded from the others [5–7]. Such an observation is surprising since it would be natural to assume that all actin filaments in the cell represent equivalent substrates for ABPs. On the contrary, these observations reveal the existence of complex mechanisms capable of precisely addressing the cell's ABPs, and research conducted in recent years has revealed a much more complex picture than anticipated. This work will review the multifarious strategies that cells use to guide ABPs to the appropriate actin networks, the molecular mechanisms behind these processes, and discuss future directions for research in this area.
2. Cellular strategies for distinguishing actin networks as different substrates for actin-binding protein binding
There are multiple arguments to assert that the binding of most ABPs to specific actin subnetworks does not rely solely on their transport or on their local activation. First of all, actin networks in cells are often very close to each other. Sometimes an actin subnetwork can even form from a pre-existing one. This is the case, for example, of filopodia, which emerge through the elongation of actin filaments assembled in lamellipodia. In this context, the fast diffusion of proteins in the cytoplasm of cells (typical diffusion rates measured for globular proteins of 10 to 100 kDa range from around 10 to 100 µm2 s−1) would prevent a precise and efficient targeting of ABPs [8,9]. Second, further evidence comes from the fact that local activation of specific actin assembly pathways in cells is often sufficient to induce the formation of functional actin networks. For example, the recruitment by optogenetic tools of specific RhoGTPases is sufficient to trigger actin assembly, and to initiate actin-dependent processes such as cell migration [10] or cytokinesis [11]. Similarly, triggering actin assembly from cellular extracts by specific factors such as WASp (which is an activator of the Arp2/3 complex) or formins, leads to the formation of actin filament networks with a composition of ABPs comparable to branched and linear actin networks, respectively [12–14].
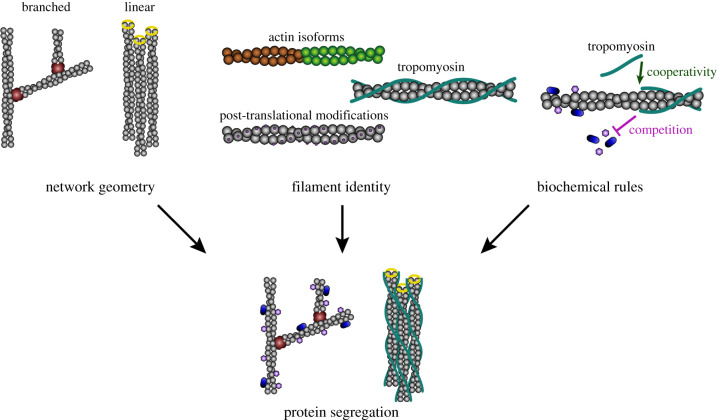
All of these observations unambiguously indicate that, to a large extent, actin networks themselves represent different substrates for downstream protein interactions. This has led the community to ask what specific features could allow actin networks to distinguish themselves from each other, in order to be identified as different substrates for the cell's ABPs [15]. To date, two main hypotheses, not mutually exclusive, are guiding this field towards a better understanding of these principles. The first hypothesis is that the geometrical organization of filaments within actin networks is itself a sufficient characteristic to make these substrates distinct for ABPs (figure 1). The second hypothesis is that the actin filaments themselves within the actin networks could present different biochemical signatures, which could differentiate them for the different ABPs of the cell (figure 1).
Figure 1.
Three main mechanisms that account for the segregation of ABPs to different actin networks in cells. Schematic of the different molecular mechanisms described in this review. Actin networks are distinguished by different geometries. For example, the Arp2/3 complex generates branched networks and formins generate linear arrays. Actin filaments have different molecular identities based on the use of various actin isoforms, post-translational modifications and/or the presence of tropomyosin. Additionally, ABPs can compete or cooperate to restrict or promote their binding to actin filaments.
3. The geometry of actin networks as an intrinsic feature of actin-binding proteins segregation
Actin filaments that are polymerized in vitro for standard actin-binding assays are generally between 1 and 100 µm in length, corresponding to approximately 360–36 000 actin subunits [16]. Since actin filaments are semi-flexible polymers with a persistence length of about 17 µm, this size range is of interest for studying certain biophysical aspects of actin filaments and their interactions with ABPs [17]. However, since most actin filaments in vivo are shorter than 1 µm, the results of these studies may bias our interpretation of how ABPs interact with actin filaments in cells.
Furthermore, actin filaments also differ significantly from one actin network to another. Primarily, they vary in length and relative orientation to each other. For example, the branched actin networks of the lamellipodium are composed of short actin filaments of 7–18 actin subunits [18], which are connected to each other by the Arp2/3 complex at an angle of 70°. At endocytic sites, filaments are also branched but appear to be longer, between 18 and 68 actin subunits [19–22]. At these scales, actin filaments are short enough to be considered totally rigid. Linear arrays, on the contrary, are often composed of longer filaments with up to 300 actin subunits [23,24]. The actin filaments are approximately parallel to each other, and may all have similar (e.g. in the case of filopodia) or random (e.g. in the case of cytokinetic rings or stress fibres) orientations.
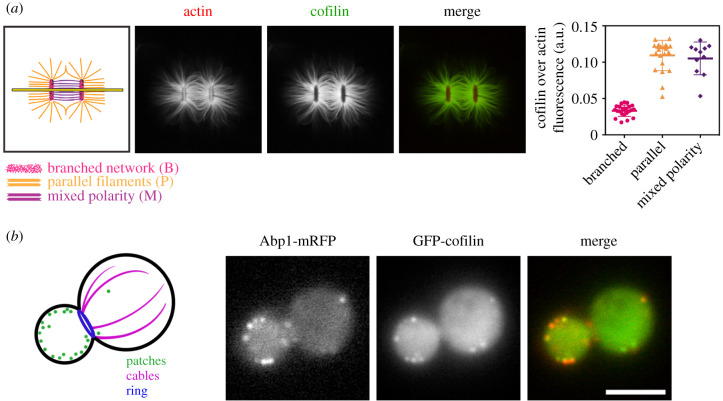
Studying the relationship between the geometrical organization of actin networks and the apparent affinity of ABPs has required the development of more complex biomimetic systems than previously envisaged. The best description to date of the impact of actin network organization on the binding and activity of an ABP concerns myosins, particularly contractile myosins with multiple motor domains [25,26]. Several studies have shown that their recruitment and activity strongly depend on whether the actin networks are disorganized, branched by the Arp2/3 complex, parallel or antiparallel [27–29]. More particularly, these motors are capable of strong contractile activity, even to the point of disassembling actin networks, when actin filaments do not have the same orientation and polarity. These observations are very consistent with the effects of these molecular motors in vivo. This is the case, for example, at the sarcomeres of muscle fibres or at cell–cell junctions in tissues, where the contractile activity of the motors is exerted on actin filaments of opposite polarity [30]. This is also the case for many disorganized actin networks where myosin activity is capable of driving actin flows or pulsatile phenomena [31–33]. On the contrary, actin filament structures such as filopodia, where the actin filaments all have the same orientation, are not contractile structures [4,34]. On these structures, molecular motors are generally used for trafficking. It is important to note that the sensitivity of ABPs to different actin filament organizations does not seem to be limited to the case of molecular motors, but seems, on the contrary, to be quite general. For example, crosslinkers such as α-actinin bind preferentially when the spacing between two actin filaments is favourable [35]. Other proteins such as ADF/cofilin, which is involved in the disassembly of actin networks, accumulate on linear networks of actin filaments, but are not efficiently recruited on branched actin networks in vitro [36] (figure 2a).
Figure 2.
Actin network architecture as an important property to consider in explaining the accumulation of ABPs. (a) Profile of accumulation of ADF/cofilin on different actin network architectures reconstituted in vitro (adapted from [36]). Left: Schematic of the different actin network architectures assembled on a micropattern coated with an activator of the Arp2/3 complex (two vertical bars), in the presence of soluble actin and Arp2/3. Branched actin networks are assembled on the patterns, whereas linear actin networks (parallel or mixed polarity) are assembled from the elongation of the filaments away from the patterns. Middle: Localization of ADF/cofilin (in green) after its addition to polymerized actin network. Cofilin accumulates preferentially to linear actin networks. Right: Quantification of ADF/cofilin over actin intensities for each architecture. (b) Accumulation of ADF/cofilin on different actin network architectures in vivo (adapted from [38]). Left: Schematic of the different actin networks found in S. cerevisiae cells: branched networks in actin patches (dots), linear networks in actin cables (intra-cellular lines) and the cytokinetic ring (at the yeast bud neck). Right: ADF/cofilin (in green) co-localizes preferentially with the actin patch protein Abp1 (in red), indicating that other principles than network architecture are at play to account for the cellular localization of ADF/cofilin.
The geometrical organization of actin networks is, therefore, a key parameter to consider when evaluating the affinity and activity of ABPs. However, other observations made at the cellular level show that the geometric organization of actin networks alone is not sufficient to fully address how ABP recruitment is carried out in vivo. Indeed, some proteins, such as ADF/cofilin mentioned above, are not found in cells on actin networks for which their affinity should be the strongest. While ADF/cofilin is clearly localized on branched actin networks such as endocytic actin patches, ADF/cofilin is hardly detectable on linear networks such as actin cables [37–39] (figure 2b). These observations indicate that additional principles, beyond the actin filament network architecture, need to be taken into account to obtain a global picture of how ABPs are addressed in cells.
4. Biochemical opportunities for generating different actin substrates
Although attractive, we saw that the segregation of ABPs observed in cells could not be explained solely by the geometric organization of actin filaments. The necessity to discriminate different populations of actin filaments in cells must lead us to consider other possibilities, including that small differences in structure and surface properties of the actin filaments themselves could modulate their affinity for certain ABPs. Our community has been working on a number of additional hypotheses to explain how actin filaments could be functionally different and acquire identities of their own. A first hypothesis is that cells could possibly use different actins, either through the expression of different actin isoforms or through post-translational modifications (PTMs) [40,41]. A second hypothesis is that specific ABPs could progressively decorate actin filaments to give them a specific identity, reinforcing or limiting the binding of other ABPs to the same filaments by steric effects or by stabilizing particular conformations of the filaments [5]. These two hypotheses are not mutually exclusive, meaning that cells could also use several strategies simultaneously to create the greatest possible diversity of actin substrates.
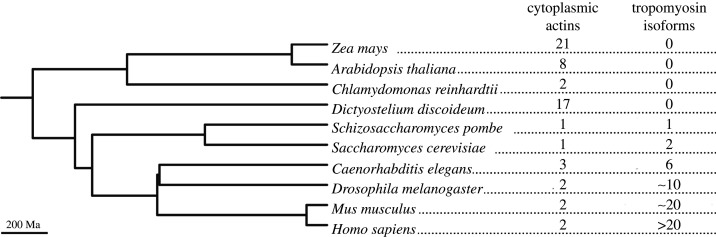
Proof that both of these strategies exist in cells is evident from a careful analysis of genomes of all eukaryotes (figure 3). Many species express a variable number of actin isoforms, which can be either cell-specific or expressed simultaneously in the same cell types. An extreme example is plants, which express multiple actin isoforms, that originated from genome duplications (figure 3). The number of actins varies for each plant species and can reach, for example, 21 isoforms in Zea mays. A potential limitation of this strategy is that the actin sequence must remain highly conserved in order to maintain its assembly properties and its ability to interact with the most essential ABPs. For example, even between two distant eukaryotes such as budding yeast and human, which diverged more than a billion years ago, actin sequences still retain around 90% identity. Actin mutations are generally rare, and usually lead in humans to serious diseases such as Baraitser–Winter syndrome [42]. Overall, while the existence of many actins in eukaryotes supports the possibility that cells can use a variety of actins to generate different actin-related functions, the difficulty of bringing mutations and generating variety also questions the effectiveness of such a mechanism to generate diversity of functions.
Figure 3.
Phylogenetic tree showing how the number of cytoplasmic actin and tropomyosin isoforms changed over the course of evolution in eukaryotes. These numbers are strongly anti-correlated, suggesting that different lineages used different strategies to generate a complex actin cytoskeleton. For clarity, muscle actins which are specific to Metazoa and which are highly specialized were excluded.
An alternative strategy to differentiate actin filaments without the need to mutate the actin itself is through the use of specialized ABPs. This hypothesis has gained considerable credibility with the comparison of eukaryotic genomes. Indeed, whereas some species, such as plants and amoebas, generally express dozens of different actin isoforms, other eukaryotes, for example, those from the kingdoms Animalia or Fungi, express only one or a very limited number of cytoplasmic actin isoforms (figure 3). Conversely, species expressing one or few actin isoforms express a multitude of tropomyosins, which are specific ABPs that wrap around actin filaments, whereas plants or amoebas do not express any (figure 3) [43]. This very strong anticorrelation is a signature that these two phenomena are probably related to each other. It suggests that while some species use multiple actin isoforms, representing different substrates for ABPs, in order to create different actin-related functions, other species that had gained tropomyosins in the course of evolution could use a limited number of actins, decorated by different tropomyosins, to generate functional diversity.
5. Generating a diversity of actin substrates by expressing a variety of actin isoforms
The question that will now be addressed is whether very similar eukaryotic actins are nevertheless able (1) to assemble separately within a common cytoplasm and (2) to form filaments of sufficiently specific molecular identity to interact differently with ABPs and carry specific cellular functions. The answer to this question is far from being clear today and is the subject of intense research.
5.1. Plant actins localization and functions
Plant actins were originally studied from the model organism Arabidopsis thaliana, which has 10 actin genes. Eight of these genes have been demonstrated to code for functional actin isoforms, grouped in two classes according to their sequence similarities and their tissue-specific expressions: vegetative (ACT2, 7 and 8) and reproductive (ACT1, 3, 4, 11 and 12) [40]. Vegetative and reproductive actins are involved in different cellular processes [44], and plant actin isoforms that are expressed in the same tissue can also assemble into isoform-specific structures. GFP-fusion proteins of ACT2 and ACT7, the main vegetative actin isoforms, co-localize only partially at the surface of chloroplasts, where ACT2 is mainly found in thinner and longer bundles, whereas ACT7 is organized into thick bundles [45]. Besides their differential expression and their spatial segregation, these isoforms are not functionally equivalent. Expression of the reproductive actin ACT1 in vegetative tissues causes dwarfing and altered morphology in most organs, showing that expression of ACT1 in these tissues is affecting the dynamics of actin and its associated proteins [46].
Another well-studied organism is the unicellular green algae Chlamydomonas reinhardtii, which carries two actin genes. The main isoform IDA5 is a conventional actin that is expressed in normal conditions. The second actin, called NAP1 (for Novel Actin-like Protein 1), is highly divergent as it shares only 65% sequence identity with IDA5 [47]. The expression of NAP1 in wild-type cells is negligible, but it is highly upregulated in certain conditions, for example, when IDA5 is absent or after addition of the actin monomer sequestering drug latrunculin B [48,49]. This drug can prevent IDA5 polymerization, but surprisingly NAP1 generates latrunculin B-resistant structures. Despite being so different, essential actin functions can be performed by either of these actins, and cells lacking any of the actin genes can grow and divide normally. However, they also seem to maintain some specialized functions. The conventional actin IDA5 has a function in mating since it is involved in the elongation of the fertilization tube, a function that NAP1 cannot substitute [50–52]. Both IDA5 and NAP1 are found in the axoneme of the flagella but apparently in different structures. While IDA5 seems to be part of the inner dynein arms, NAP1 plays a role in flagellar formation independently of axonemal dyneins [52,53].
5.2. β- and γ-actin localization and functions
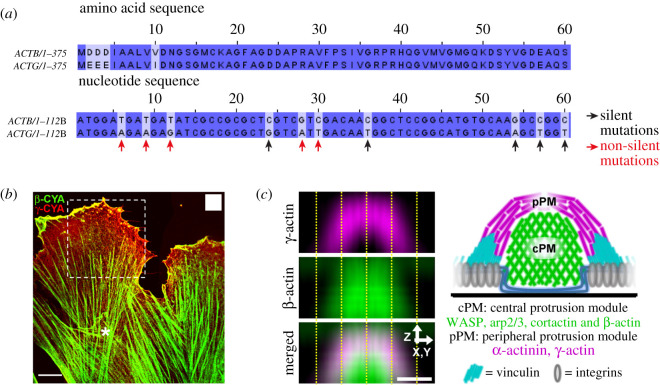
Generating actin structures from different actin isoforms is also possible in higher eukaryotes, including mammals. Mammalian organisms have six different actin isoforms: four muscle actins and two cytoplasmic actins. The latter, called β- and γ-actins, differ only in four amino acids at the N-terminal end (figure 4a) and are simultaneously expressed in cells. Due to their extreme similarities, determining the cellular localization of β- and γ-actin is challenging. Specific monoclonal antibodies, recognizing specifically the different N-terminal regions, are now available to visualize the localization of both isoforms in different cell types [54,55].
Figure 4.
Evidence that highly similar cytoplasmic actins can nevertheless assemble into functionally different actin networks in cells. (a) Beginning of the nucleotide and amino acid sequence of β- and γ-actins. These proteins only differ in four amino acids located at the N-terminal end, although their nucleotide sequences have a much higher number of silent mutations (e.g. black arrows). (b) Example of the differential localization of β- (green) and γ-(red) actins at the cell scale, in migrating human subcutaneous fibroblasts (adapted from [54], scale bar, 10 µm) (c) Another example of the differential localization of β- (green) and γ-actins (magenta) within a same structure, podosomes (adapted from [57], scale bar: 0.5 µm). A linear network of γ-actin is surrounding a branched β-actin core.
β-actin was originally found mainly in actin bundles of basal stress fibres, filopodia, at cell–cell contacts and in contractile rings, whereas γ-actin is present mainly in lamellar and dorsal cell regions (figure 4b) [54,55]. In epithelial cells, β-actin has been shown to play a role in adherens junction maintenance, and γ-actin in tight junction integrity [56]. In podosomes, which are actin-rich adhesive structures involved in migration and invasion, the use of better super-resolution microscopy techniques allowed a differential localization between actin isoforms to be distinguished. Actin filaments in podosomes are organized into two distinct networks, consisting of a β-actin core, composed of branched actin filaments nucleated with WASp, Arp2/3 and cortactin, and surrounded by a γ-actin envelope, composed of linear actin filaments bound to α-actinin and connected with myosins (figure 4c) [57].
These differences in localization suggest that these two isoforms, despite being so similar, could be associated with different cellular functions. During wound closure, cells assemble significantly more β-actin beneath the plasma membrane, which suggests a role for this actin in cell motility [58]. β-actin implication in cell motility was confirmed in fibroblasts where decreased β-actin protein levels lead to reduced motility [59,60]. The implication of γ-actin in cell migration is less clear. While γ-actin knocked-down cells are shown to migrate less in some studies [54,61], loss of γ-actin can also induce epithelial-to-mesenchymal transitions in another model [62]. In agreement with β-actin localization in the cytokinetic ring, β-actin knocked-down cells also show reduced proliferation and can be multinucleated [59,60,63,64]. In breast cancer cells, cycle entry and proliferation seem to be regulated by γ-actin, particularly in G1, while β-actin plays a role in later mitotic stages, especially in telophase for cytokinesis [64]. β-Actin implication in cell motility and proliferation can be explained by the direct activity of β-actin in the filaments of the structures controlling these processes, but also by the role that this specific actin plays in the regulation of transcription. β-Actin binds directly to chromatin remodelling proteins as well as RNA polymerases, a first indication of its role in nuclear processes [65]. This role is also confirmed as β-actin was shown to regulate the expression of cell cycle and actin dynamics related genes, as well as its own expression [59,66].
Cellular localization of these actins does not suggest any particular preference for a certain type of architecture. For instance, β-actin is localized at the linear structure of the stress fibres and contractile ring but it is also localized at the branched core of the podosomes. This lack of a general obvious rule complicates our understanding of the molecular mechanisms implicated in the assembly of these two actin isoforms into distinct networks. Moreover, this understanding is further complicated by functional tests at the whole organism level. Despite their amino acid sequences being so similar, the nucleotide sequences of beta and γ-actin genes possess silent mutations that affect 40% of the codons (figure 4a). By taking the β-actin gene, and changing only four codons to express γ-actin from this gene, it is possible to generate viable mice that are not expressing the β-actin protein [67]. This result is surprising, since β-actin knock-out mice are reported to be embryonic lethal [68,69]. Therefore, essential functions of β-actin may not be related primarily to its amino acid sequence, but may also rely heavily on its nucleotide sequence. This difference in nucleotide sequence results in different translation speeds [70], which could lead to protein regulation at different levels: differential expression levels, alternative splicing and differential co- and PTMs.
5.3. Biochemical similarities and differences between actins
Differences in cell localization described above suggest that different actins, although very similar, must still have significantly different biochemical properties. However, while divergent actins expressed by prokaryotes have clearly distinguishable assembly properties, actins expressed in eukaryotes seem to have much more subtle biochemical differences [71,72]. The search for these subtleties has long suffered from the difficulty of purifying a variety of actin isoforms in order to study them independently. Most of our knowledge is based on studies using the same mammalian actin muscle isoform. A more limited number of studies have used the yeast actin S. cerevisiae, and only a few studies used mixtures of γ- and β-actin or actins from other species.
Actins from budding yeast and rabbit muscle are 87% identical, which indicates that these two actins are quite different comparatively to all actins expressed in eukaryotes. Budding yeast and rabbit muscle actins can nevertheless copolymerize [73]. Surprisingly, this is less clear for beta and γ-actin, despite being 99% identical. While these two isoforms were shown to copolymerize in some studies, other studies reported their ability to assemble into independent filaments [55,74,75]. Yeast and rabbit muscle actins show differences in flexibility, with a persistence length of rabbit muscle actin two-to-threefold higher than yeast actin [76,77]. In the presence of magnesium, yeast actin polymerizes faster than muscle actin, which is due to a faster trimer nucleus formation rather than a faster elongation of the filaments [78–81]. This difference in rates of polymerization is also observed in plants, as the two vegetative actins ACT2 and ACT7 polymerize faster than the reproductive actins ACT1 and ACT11 [82]. Nucleotide hydrolysis, nucleotide exchange and Pi release are also faster for yeast actin compared to muscle actin filaments [80,81,83–85]. This correlates with the fact that the nucleotide-binding cleft of S. cerevisiae's actin appears more open than for muscle actin [86]. In summary, we can hypothesize from few well-characterized actins, that many biochemical and biophysical subtleties might overall account for important functional differences in cells.
Differences among actins are also sufficient to modulate some interactions with ABPs. Actin nucleators, which play an important role in architecture formation, are reported in few studies to favour specific actin isoforms. The formin DIAPH3, for example, has a preference for β-actin compared to γ-actin, suggesting that actin cables assembled from DIAPH3 could be enriched with β-actin [55]. The VCA domain of N-WASP, an activator of Arp2/3, does not show specificity for β- or γ-actin [87], but S. pombe's Arp2/3 is reported to be a better nucleator of S. pombe's actin than rabbit muscle actin [80]. In Arabidopsis thaliana, the binding affinity of profilins for actin monomers seems lower for a specific isoform, ACT2 [82]. Since profilin enhances formin-linear actin cable assembly, at the expense of Arp2/3-branched network assembly, it is tempting to speculate whether ACT2 would assemble more specifically within branched networks. Moreover, to achieve different actin functions, it appears that plant actins and ABPs have co-evolved to generate class-specific protein–protein interactions. The expression of the reproductive ACT1 isoform in vegetative tissues leads to aberrant cell and tissue morphology, a phenotype that is rescued by co-expression of the reproductive profilin (PRF4) and cofilin (ADF7) [88]. Evidence for coevolution of actin with ABPs can be also found by studying proteins from different species. For example, in both yeasts S. cerevisiae and S. pombe, profilin inhibits endogenous actin polymerization but has little effect on rabbit muscle actin polymerization [81,89,90]. Another example is vertebrate cofilin, which can bind to S. cerevisiae's actin but does not increase their flexibility nor promote severing [77,91].
These studies indicate to the scientific community that highly similar actin isoforms have subtle but significantly different properties to display preferential binding to a variety of ABPs. We are just beginning to identify the molecular mechanisms by which actin isoforms could assemble into distinct actin networks of specialized properties. However, we still do not have a satisfying overview of the variety of possible differences among all actin isoforms expressed in eukaryotes. Recently, new protocols have been developed [92–94], allowing for a wider variety of actin isoforms to be purified. It is likely that future comparisons of a greater diversity of actin isoforms, purified from similar protocols, will strengthen our knowledge of these mechanisms.
6. Actin's post-translational modifications
Co- and PTMs are covalent modifications to one or several amino acids of a protein, a process that is usually mediated by specific enzymes. These modifications can affect the interactions of the protein with its partners by changing its surface charge density, its structure, or by steric hindrance.
First, actin can be arginylated, which is the addition of an arginine residue at the N-terminal end. This modification is mediated by the arginyl-tRNA-protein transferase Ate1, a protein that has been identified in several organisms, including mammals, plants and budding yeast [95]. In Dictostellium discoideum, an organism expressing a large number of cytoplasmic actin isoforms, several actins (Act3, Act10, Act17, Act22, Act23 and the most abundant one Act8) are arginylated, and impairing Ate1 activity affects cell migration and substrate adhesion [96]. In mammals, arginylation is possible for β- and γ-actins, but the latter is specifically degraded when it is arginylated [70,97]. As this PTM does not affect both actin isoforms equally, arginylation could be an important PTM to regulate specifically β-actin-dependent cellular processes. For example, arginylated β-actin, which corresponds to around 1% of total β-actin, is likely to be involved in lamella formation, as downregulation of Ate1 reduces the formation of this structure [97–99].
Studies in Ate1 knocked-out cells indicate that actin arginylation is responsible for a decreased interaction with gelsolin, but for an increased recruitment of capping protein (CP) and twinfilin [100]. Arginylation adds positive charges to normally negative charged surfaces, a change that logically affects actin's interaction with binding partners such as gelsolin, whose binding relies on the first 10 amino acids of actin [101,102]. On the contrary, CP and twinfilin are not shown to bind to this area, but their increased binding could be explained by an absence of gelsolin which would leave excessive free actin filament barbed ends for these two proteins to bind to.
The most abundant PTM for β- and γ-actin is N-terminal acetylation, which is the addition of an acetyl group [103]. In animals, this PTM occurs after cleavage of the first one or two amino acids, and is modifying an important fraction of the actin [104–107]. It is mediated by the acetyltransferase NAA80, which is specific to actin and acetylates preferentially the monomeric actin-profilin complex [108,109]. Interestingly, plants and fungi do not express NAA80, but do express the general acetylase NatB, which acetylates many other proteins. In yeast, actin is co-translationally acetylated by NatB [110,111], but in plants, the role of NatB is less clear. Even though the lack of NatB affects plant growth, actin is not identified as a substrate for this protein [112]. As NatB targets the N-terminal part of proteins starting with Met-Glu-, Met-Asp-, or Met-Asn-, plant actins, which start with Met-Ala-, may not be modified or may be modified by a mechanism not yet identified [111]. Since plants like Arabidopsis thaliana already express several actin isoforms, we can also speculate that actin acetylation might be less important to generate different actin-based functions in this organism. In HeLa cells, actin acetylation affects cell motility and cytoskeletal organization [103]. Acetylated actin has a faster polymerization rate, including formin-induced polymerization, and a faster depolymerization rate, so filaments composed of acetylated actin are shorter lived [103]. The N-terminal residue of actin is not the only amino acid that can be acetylated. A complex of lysine-acetylated actin and cyclase-associated protein (CAP) was shown to promote the inhibition of the formin INF2 [113]. This proves that PTMs can not only regulate actin properties and its binding to other proteins, but also the activity of the other proteins themselves.
Arginylation and acetylation are two main PTMs of actin. Other PTMs, including phosphorylation and methylation, can also modify the chemistry of the actin molecule. For more details, we refer readers to a more detailed review [114].
7. Tropomyosins and the biogenesis of new actin substrates
7.1. Generating diversity from a limited number of actin isoforms: tropomyosin as the missing link
We will now study the more complex case where a cell is able to generate distinct actin networks from identical (or nearly identical) actin molecules. The distinction between actin networks can no longer be made on the basis of biochemical differences between the actin composing different networks, but on the basis of biochemical particularities of the ABPs composing each of the networks. For greater clarity, our discussion will distinguish two different cases: the first case corresponds to the de novo generation of new actin networks from determined actin nucleation factors; the second case corresponds to the reorganization of pre-existing networks into networks with different properties.
In the first case, the assembly of new actin filament networks suggests that filaments acquire particular identities at the moment when they are generated by nucleation factors. The idea that this function is carried by factors such as the Arp2/3 complex or formins is a priori tempting. However, the coincubation of the Arp2/3 complex, its VCA activator and a formin (FMNL2) leads to the formation of mixed actin networks (i.e. having both Arp2/3 branches and formin-bound filaments) and not of distinct actin networks [115]. It should be noted that in the experiment described, the actin filament branches are much longer than the branches present in the cells, and that we could not exclude the possibility that formin cannot bind to very short branches. It is also possible that formin FMNL2 is a peculiar isoform that can bind to branched networks [115,116]. Nevertheless, this experiment rather suggests that another regulator is needed to effectively segregate formins and the Arp2/3 complex on separate networks. We have already seen in paragraph 4 that careful genomic analysis strongly suggests that proteins of the tropomyosin family are responsible for functional diversity of the actin cytoskeleton in higher eukaryotes [43]. We shall see that genetics, cell biology and biochemistry have also provided additional evidence for the importance of tropomyosins.
The second case corresponds to situations where actin networks undergo major dynamic reorganizations, independently of any nucleation of new actin filaments. For instance, linear actin structures found beneath the lamellipodia are not exclusively generated by formins, but also emerge to a large extent from pre-existing lamellipodial actin networks [117–120]. Interestingly, in this case, where actin filaments are not generated de novo, tropomyosin recruitment correlates also very well with filament debranching and the re-organization of actin filaments into new linear actin structures [121,122]. These observations suggest that regardless of the mechanism by which actin networks are formed, tropomyosins are consistently of key importance in giving actin filaments a new identity.
7.2. Tropomyosins localization and functions
Most cells express multiple tropomyosin isoforms and splicing variants, and those proteins have been proposed to provide actin filaments specific identities [5,123–125]. This concept has been reinforced by the observation of cellular localization of tropomyosins, which is highly dependent on the type of tropomyosin isoform [126,127] (figure 5a). Among the tens of isoforms that exist in metazoans, individual actin structures usually interact with a subset of tropomyosins. Most actin networks in cells are decorated by specific families of tropomyosins, including filopodia, lamella and stress fibres, with the exception of branched networks such as lamellipodia or endocytic actin patches which do not recruit tropomyosins (figure 5b) [124,125,127,128]. Some tropomyosins do not form copolymers (figure 5c), indicating that they are therefore involved in many different cellular functions [126,129], and modulation of the expression of tropomyosins triggers specific cellular responses. For example, some cancer cell lines can remarkably recover rigidity sensing and rigidity-dependent growth, when a single tropomyosin isoform (Tpm2.1) is over-expressed [127,130]. However, structures such as stress fibres are highly sensitive to the expression level of any isoform of tropomyosin [127,131]. Recent data also suggest that modulation of any tropomyosin isoform impacts the whole myosin organization in cells, thus acting on both tension and traction forces driven by focal adhesions [132]. Therefore, some actin networks might bind to multiple families of tropomyosins simultaneously, which is coherent with the high concentration of tropomyosin present in cells, and with the fact that some tropomyosin isoforms have the ability to copolymerize as demonstrated in vitro (figure 5c) [133,134].
Figure 5.
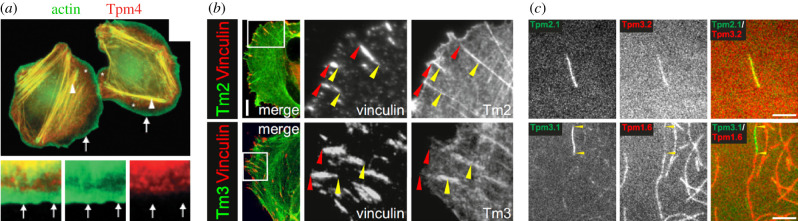
Functional differentiation of actin networks by tropomyosins. (a) Example of the specific localization of tropomyosin 4 (in red) in MTLn3 cells. Tropomyosin localizes with actin (in green) in stress fibres and in lamellar structures (arrowheads), while it is absent from the branched actin structures at the leading edge (arrows) (adapted from [128]). (b) Example of the differential localization of tropomyosin isoforms (in green) in U2OS cells. While Tpm2 shows a strong colocalization with the focal adhesion-specific protein vinculin (in red), Tpm3 localizes proximally to focal adhesions (adapted from [131]). (c) In vitro single-filament scale imaging reveals that while some tropomyosin isoforms (top images: Tpm2.1 in green and Tpm3.2 in red) can copolymerize with actin filaments, others cannot (bottom images: Tpm3.1 in green and Tpm1.6 in red) (adapted from [134], scale bar, 5 µm).
The presence of tropomyosins on linear actin networks suggests a preference for formin-generated filaments. Indeed, incubation of the pan-formin inhibitor SMIFH2 in oocytes decreases the cortical tropomyosin level [135]. Tropomyosin depletion promotes the expansion of lamellipodia while its overexpression inhibits this branched structure while promoting linear networks [136–138]. Moreover, it seems that some formins assemble actin filaments bound to specific tropomyosins. This is beautifully illustrated in fission yeast, where an exchange of the localization of the fission yeast formins For3 and Cdc12 results in an exchange in localizations of the tropomyosin forms on the corresponding actin networks [139]. Also, the absence of CP in fission yeast cells induces simultaneously ectopic recruitment of the tropomyosin Cdc8 and of both formins Fus1 and Cdc12 [140]. However, specific downregulation of some formins (mDia1 and mDia3) does not affect the localization of tropomyosins, indicating that some formins may not share this specificity for tropomyosins [141].
7.3. Impact of tropomyosins on actin filament nucleation, debranching and the binding of other actin-binding proteins
Tropomyosins are dimers of α-helices forming parallel coiled-coils that span several actin subunits [123]. A biochemical link between formins and tropomyosins has been described in vitro, and cooperativity between these proteins is established [136,142,143]. In budding yeast, the presence of tropomyosin can specifically increase the nucleation rate of a formin. Conversely, tropomyosins are generally strong inhibitors of Arp2/3-induced actin nucleation and branch formation [144,145]. Debranching and re-organization of actin networks into linear arrays is also favourable to tropomyosins, as this process generates more actin pointed ends from where tropomyosins can bind [121,122]. These observations agree well with the localization of tropomyosin in cells.
The binding of tropomyosin around actin filaments contributes directly to the recruitment of particular families of ABPs, and the dissociation of others. Tropomyosins regulate the activity of the different families of myosins by modifying their binding to actin filaments and their enzymatic kinetics [146,147]. This mechanism is important because it allows cargoes to be directed to appropriate locations and regulates contractility. Elegant in vitro studies confirm at the level of single actin filaments that tropomyosin excludes other ABPs, such as fimbrin or ADF/cofilin, therefore preventing filaments from disassembly [129,134,148,149]. Interestingly, tropomyosin is not required per se to assemble cables in vitro in the absence of disassembling factors but it becomes necessary to maintain cable assembly in biomimetic assays where treadmilling has been reconstituted [136,150]. Tropomyosins are hence major biochemical regulators that define the identity of actin filaments and regulate the binding of many families of ABPs, thereby leading to the segregation of these proteins to different actin networks.
8. Regulation of actin networks protein composition by competition between ABPs
Numerous lines of evidence indicate that not only tropomyosins, but most ABPs, display cooperative or competitive binding effects to actin filaments, and that these effects need to be taken into account to understand globally how an appropriate ABP composition of actin networks is reached [5]. A number of cellular biology studies demonstrate unambiguously that the removal of a given ABP from one actin network may trigger a global relocation of ABPs from other actin networks [6,140]. As a consequence, phenotypes observed in cells are not only due to the absence of the ABP of interest, but also to the mislocalization of other ABPs. Several hypotheses could explain this phenomenon. First, it is possible that the absence of a protein in a network may open a binding site for other proteins, or allow the binding of competing proteins. For instance, in yeast, removal of fimbrin from actin patches, which are Arp2/3-branched networks, leads to an ectopic localization of tropomyosin to those networks [6]. Second, it is possible that ectopic protein localization triggers the cooperative binding of additional proteins. For instance, loss of CP from actin patches creates free actin filament barbed ends, where formins can bind, which in turn favours the ectopic binding of tropomyosin [140]. Finally, the absence of an ABP could also have consequences on the geometry of the network, which would consequently impact its ABP composition. Overall, these results indicate that although tropomyosins are key regulators for addressing ABPs to appropriate networks, proper segregation of ABPs on specific actin networks in cells also relies on a global and complex biochemical equilibrium, involving many different families of ABPs. Addressing these questions in the future will require to integrate all these parameters into a comprehensive model.
9. Conclusion
The aim of this review was to describe our current knowledge of the different molecular mechanisms involved in the definition of the identity of actin filaments and networks for a proper segregation of ABPs in cells. We conclude this work by emphasizing that these mechanisms are often not purely distinct from each other, but interrelated. A clear example is the fact that a protein like tropomyosin gives an identity to actin filaments, but is also involved in competitive binding with other ABPs. As many different protein–protein interactions and molecular mechanisms are simultaneously involved, a comprehensive understanding of these complex systems requires non-superficial analysis.
Acknowledgements
As the topic of this review is quite broad, we apologize to our colleagues whose work may have been accidentally missed while writing the manuscript. The authors thank David Drubin, Voytek Okreglak, Christine Chaponnier, Vera Dugina, Alessandra Cambi, Koen van den Dries, Vera DesMarais, John Condeelis, Pekka Lappalainen and Sari Tojkander for allowing us to use some of their published material.
Data accessibility
This article has no additional data.
Authors' contributions
This review was written collectively by all authors.
Competing interests
We declare we have no competing interests.
Funding
This work has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 638376/Segregactin) and from the Labex INFORM (ANR-11-LABX-0054, funded by the ‘Investissements d'Avenir French Government program').
References
- 1.Chhabra ES, Higgs HN. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110–1121. ( 10.1038/ncb1007-1110) [DOI] [PubMed] [Google Scholar]
- 2.Skau CT, Waterman CM. 2015. Specification of architecture and function of actin structures by actin nucleation factors. Annu. Rev. Biophys. 44, 285–310. ( 10.1146/annurev-biophys-060414-034308) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard TD, Cooper JA. 2009. Actin, a central player in cell shape and movement. Science 326, 1208–1212. ( 10.1126/science.1175862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. 2014. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev. 94, 235–263. ( 10.1152/physrev.00018.2013) [DOI] [PubMed] [Google Scholar]
- 5.Michelot A, Drubin DG. 2011. Building distinct actin filament networks in a common cytoplasm. Curr. Biol. 21, R560–R569. ( 10.1016/j.cub.2011.06.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skau CT, Kovar DR. 2010. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr. Biol. 20, 1415–1422. ( 10.1016/j.cub.2010.06.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kovar DR, Sirotkin V, Lord M. 2011. Three's company: the fission yeast actin cytoskeleton. Trends Cell Biol. 21, 177–187. ( 10.1016/j.tcb.2010.11.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrio-Dupont M, Foucault G, Vacher M, Devaux PF, Cribier S. 2000. Translational diffusion of globular proteins in the cytoplasm of cultured muscle cells. Biophys. J. 78, 901–907. ( 10.1016/S0006-3495(00)76647-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Milo R, Phillips R. 2016. Cell biology by the numbers. New York, NY: Garland Science. See https://www.taylorfrancis.com/books/e/9780429258770.
- 10.Levskaya A, Weiner OD, Lim WA, Voigt CA. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. ( 10.1038/nature08446) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wagner E, Glotzer M. 2016. Local RhoA activation induces cytokinetic furrows independent of spindle position and cell cycle stage. J. Cell Biol. 213, 641–649. ( 10.1083/jcb.201603025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michelot A, Costanzo M, Sarkeshik A, Boone C, Yates JR, Drubin DG. 2010. Reconstitution and protein composition analysis of endocytic actin patches. Curr. Biol. 20, 1890–1899. ( 10.1016/j.cub.2010.10.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao Y, Wong CCL, Mennella V, Michelot A, Agard DA, Holt LJ, Yates JR, Drubin DG. 2013. Cell-cycle regulation of formin-mediated actin cable assembly. Proc. Natl Acad. Sci. USA 110, E4446–E4455. ( 10.1073/pnas.1314000110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee K, Gallop JL, Rambani K, Kirschner MW. 2010. Self-assembly of filopodia-like structures on supported lipid bilayers. Science 329, 1341–1345. ( 10.1126/science.1191710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jégou A, Romet-Lemonne G. 2016. Single filaments to reveal the multiple flavors of actin. Biophys. J. 110, 2138–2146. ( 10.1016/j.bpj.2016.04.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.von der Ecken J, Müller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. 2015. Structure of the F-actin–tropomyosin complex. Nature 519, 114–117. ( 10.1038/nature14033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ott A, Magnasco M, Simon A, Libchaber A. 1993. Measurement of the persistence length of polymerized actin using fluorescence microscopy. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 48, R1642–R1645. ( 10.1103/physreve.48.r1642) [DOI] [PubMed] [Google Scholar]
- 18.Svitkina TM, Borisy GG. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145, 1009–1026. ( 10.1083/jcb.145.5.1009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young ME, Cooper JA, Bridgman PC. 2004. Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166, 629–635. ( 10.1083/jcb.200404159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodal AA, Kozubowski L, Goode BL, Drubin DG, Hartwig JH. 2005. Actin and septin ultrastructures at the budding yeast cell cortex. Mol. Biol. Cell 16, 372–384. ( 10.1091/mbc.e04-08-0734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sirotkin V, Berro J, Macmillan K, Zhao L, Pollard TD. 2010. Quantitative analysis of the mechanism of endocytic actin patch assembly and disassembly in fission yeast. Mol. Biol. Cell 21, 2894–2904. ( 10.1091/mbc.E10-02-0157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akamatsu M, Vasan R, Serwas D, Ferrin MA, Rangamani P, Drubin DG. 2020. Principles of self-organization and load adaptation by the actin cytoskeleton during clathrin-mediated endocytosis. eLife 9, e49840 ( 10.7554/eLife.49840) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160, 409–421. ( 10.1083/jcb.200210174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamasaki T, Arai R, Osumi M, Mabuchi I. 2005. Directionality of F-actin cables changes during the fission yeast cell cycle. Nat. Cell Biol. 7, 916–917. ( 10.1038/ncb1295) [DOI] [PubMed] [Google Scholar]
- 25.Nagy S, Ricca BL, Norstrom MF, Courson DS, Brawley CM, Smithback PA, Rock RS. 2008. A myosin motor that selects bundled actin for motility. Proc. Natl Acad. Sci. USA 105, 9616–9620. ( 10.1073/pnas.0802592105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagy S, Rock RS. 2010. Structured post-IQ domain governs selectivity of myosin X for fascin–actin bundles. J. Biol. Chem. 285, 26 608–26 617. ( 10.1074/jbc.M110.104661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachowiak MR, McCall PM, Thoresen T, Balcioglu HE, Kasiewicz L, Gardel ML, O'Shaughnessy B. 2012. Self-organization of myosin II in reconstituted actomyosin bundles. Biophys. J. 103, 1265–1274. ( 10.1016/j.bpj.2012.08.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reymann A-C, Boujemaa-Paterski R, Martiel J-L, Guérin C, Cao W, Chin HF, De La Cruz EM, Théry M, Blanchoin L.. 2012. Actin network architecture can determine myosin motor activity. Science 336, 1310–1314. ( 10.1126/science.1221708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ennomani H, Letort G, Guérin C, Martiel J-L, Cao W, Nédélec F, De La Cruz EM, Théry M, Blanchoin L.. 2016. Architecture and connectivity govern actin network contractility. Curr. Biol. 26, 616–626. ( 10.1016/j.cub.2015.12.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svitkina T. 2018. The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 10, a018267 ( 10.1101/cshperspect.a018267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori M, Monnier N, Daigle N, Bathe M, Ellenberg J, Lénárt P. 2011. Intracellular transport by an anchored homogeneously contracting F-actin meshwork. Curr. Biol. 21, 606–611. ( 10.1016/j.cub.2011.03.002) [DOI] [PubMed] [Google Scholar]
- 32.Azoury J, Lee KW, Georget V, Rassinier P, Leader B, Verlhac M-H. 2008. Spindle positioning in mouse oocytes relies on a dynamic meshwork of actin filaments. Curr. Biol. 18, 1514–1519. ( 10.1016/j.cub.2008.08.044) [DOI] [PubMed] [Google Scholar]
- 33.Dehapiot B, Clément R, Alégot H, Gazsó-Gerhát G, Philippe J-M, Lecuit T. 2020. Assembly of a persistent apical actin network by the formin Frl/Fmnl tunes epithelial cell deformability. Nat. Cell Biol. 22, 791–802. ( 10.1038/s41556-020-0524-x) [DOI] [PubMed] [Google Scholar]
- 34.Koenderink GH, Paluch EK. 2018. Architecture shapes contractility in actomyosin networks. Curr. Opin. Cell Biol. 50, 79–85. ( 10.1016/j.ceb.2018.01.015) [DOI] [PubMed] [Google Scholar]
- 35.Winkelman JD, Suarez C, Hocky GM, Harker AJ, Morganthaler AN, Christensen JR, Voth GA, Bartles JR, Kovar DR. 2016. Fascin- and α-actinin-bundled networks contain intrinsic structural features that drive protein sorting. Curr. Biol. 26, 2697–2706. ( 10.1016/j.cub.2016.07.080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gressin L, Guillotin A, Guérin C, Blanchoin L, Michelot A. 2015. Architecture dependence of actin filament network disassembly. Curr. Biol. 25, 1437–1447. ( 10.1016/j.cub.2015.04.011) [DOI] [PubMed] [Google Scholar]
- 37.Lappalainen P, Drubin DG. 1997. Cofilin promotes rapid actin filament turnover in vivo. Nature 388, 78–82. ( 10.1038/40418) [DOI] [PubMed] [Google Scholar]
- 38.Okreglak V, Drubin DG. 2007. Cofilin recruitment and function during actin-mediated endocytosis dictated by actin nucleotide state. J. Cell Biol. 178, 1251–1264. ( 10.1083/jcb.200703092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dawe HR, Minamide LS, Bamburg JR, Cramer LP. 2003. ADF/cofilin controls cell polarity during fibroblast migration. Curr. Biol. 13, 252–257. ( 10.1016/S0960-9822(03)00040-X) [DOI] [PubMed] [Google Scholar]
- 40.Slajcherová K, Fišerová J, Fischer L, Schwarzerová K. 2012. Multiple actin isotypes in plants: diverse genes for diverse roles? Front. Plant Sci. 3, 226 ( 10.3389/fpls.2012.00226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrin BJ, Ervasti JM. 2010. The actin gene family: function follows isoform. Cytoskeleton (Hoboken) 67, 630–634. ( 10.1002/cm.20475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rivière J-B, et al. 2012. De novo mutations in the actin genes ACTB and ACTG1 cause Baraitser–Winter syndrome. Nat. Genet. 44, 440–444. ( 10.1038/ng.1091) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. 2015. The evolution of compositionally and functionally distinct actin filaments. J. Cell. Sci. 128, 2009–2019. ( 10.1242/jcs.165563) [DOI] [PubMed] [Google Scholar]
- 44.McDowell JM, Huang S, McKinney EC, An YQ, Meagher RB. 1996. Structure and evolution of the actin gene family in Arabidopsis thaliana. Genetics 142, 587–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kijima ST, Staiger CJ, Katoh K, Nagasaki A, Ito K, Uyeda TQP. 2018. Arabidopsis vegetative actin isoforms, AtACT2 and AtACT7, generate distinct filament arrays in living plant cells. Sci. Rep. 8, 4381 ( 10.1038/s41598-018-22707-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kandasamy MK, McKinney EC, Meagher RB. 2002. Functional nonequivalency of actin isovariants in Arabidopsis. Mol. Biol. Cell 13, 251–261. ( 10.1091/mbc.01-07-0342) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee VD, Finstad SL, Huang B. 1997. Cloning and characterization of a gene encoding an actin-related protein in Chlamydomonas. Gene 197, 153–159. ( 10.1016/s0378-1119(97)00254-0) [DOI] [PubMed] [Google Scholar]
- 48.Kato-Minoura T, Uryu S, Hirono M, Kamiya R. 1998. Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem. Biophys. Res. Commun. 251, 71–76. ( 10.1006/bbrc.1998.9373) [DOI] [PubMed] [Google Scholar]
- 49.Onishi M, Pringle JR, Cross FR. 2016. Evidence that an unconventional actin can provide essential F-actin function and that a surveillance system monitors F-actin integrity in Chlamydomonas. Genetics 202, 977–996. ( 10.1534/genetics.115.184663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Detmers PA, Goodenough UW, Condeelis J. 1983. Elongation of the fertilization tubule in Chlamydomonas: new observations on the core microfilaments and the effect of transient intracellular signals on their structural integrity. J. Cell Biol. 97, 522–532. ( 10.1083/jcb.97.2.522) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Detmers PA, Carboni JM, Condeelis J. 1985. Localization of actin in Chlamydomonas using antiactin and NBD-phallacidin. Cell Motil. 5, 415–430. ( 10.1002/cm.970050505) [DOI] [PubMed] [Google Scholar]
- 52.Kato-Minoura T, Hirono M, Kamiya R. 1997. Chlamydomonas inner-arm dynein mutant, ida5, has a mutation in an actin-encoding gene. J. Cell Biol. 137, 649–656. ( 10.1083/jcb.137.3.649) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hirono M, Uryu S, Ohara A, Kato-Minoura T, Kamiya R. 2003. Expression of conventional and unconventional actins in Chlamydomonas reinhardtii upon deflagellation and sexual adhesion. Eukaryot. Cell 2, 486–493. ( 10.1128/ec.2.3.486-493.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dugina V, Zwaenepoel I, Gabbiani G, Clément S, Chaponnier C. 2009. Beta and gamma-cytoplasmic actins display distinct distribution and functional diversity. J. Cell. Sci. 122, 2980–2988. ( 10.1242/jcs.041970) [DOI] [PubMed] [Google Scholar]
- 55.Chen A, Arora PD, McCulloch CA, Wilde A. 2017. Cytokinesis requires localized β-actin filament production by an actin isoform specific nucleator. Nat. Commun. 8, 1530 ( 10.1038/s41467-017-01231-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Baranwal S, Naydenov NG, Harris G, Dugina V, Morgan KG, Chaponnier C, Ivanov AI. 2012. Nonredundant roles of cytoplasmic β- and γ-actin isoforms in regulation of epithelial apical junctions. Mol. Biol. Cell 23, 3542–3553. ( 10.1091/mbc.E12-02-0162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van den Dries K, et al. 2019. Modular actin nano-architecture enables podosome protrusion and mechanosensing. Nat. Commun. 10, 5171 ( 10.1038/s41467-019-13123-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoock TC, Newcomb PM, Herman IM. 1991. Beta actin and its mRNA are localized at the plasma membrane and the regions of moving cytoplasm during the cellular response to injury. J. Cell Biol. 112, 653–664. ( 10.1083/jcb.112.4.653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bunnell TM, Burbach BJ, Shimizu Y, Ervasti JM. 2011. β-Actin specifically controls cell growth, migration, and the G-actin pool. Mol. Biol. Cell 22, 4047–4058. ( 10.1091/mbc.E11-06-0582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joseph R, Srivastava OP, Pfister RR. 2014. Downregulation of β-actin and its regulatory gene HuR affect cell migration of human corneal fibroblasts. Mol. Vis. 20, 593–605. [PMC free article] [PubMed] [Google Scholar]
- 61.Shum MSY, Pasquier E, Po'uha ST, O'Neill GM, Chaponnier C, Gunning PW, Kavallaris M. 2011. γ-Actin regulates cell migration and modulates the ROCK signaling pathway. FASEB J. 25, 4423–4433. ( 10.1096/fj.11-185447) [DOI] [PubMed] [Google Scholar]
- 62.Lechuga S, Baranwal S, Li C, Naydenov NG, Kuemmerle JF, Dugina V, Chaponnier C, Ivanov AI. 2014. Loss of γ-cytoplasmic actin triggers myofibroblast transition of human epithelial cells. Mol. Biol. Cell 25, 3133–3146. ( 10.1091/mbc.E14-03-0815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Patrinostro X, O'Rourke AR, Chamberlain CM, Moriarity BS, Perrin BJ, Ervasti JM. 2017. Relative importance of βcyto- and γcyto-actin in primary mouse embryonic fibroblasts. Mol. Biol. Cell 28, 771–782. ( 10.1091/mbc.E16-07-0503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dugina V, Shagieva G, Khromova N, Kopnin P. 2018. Divergent impact of actin isoforms on cell cycle regulation. Cell Cycle 17, 2610–2621. ( 10.1080/15384101.2018.1553337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zheng B, Han M, Bernier M, Wen J. 2009. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 276, 2669–2685. ( 10.1111/j.1742-4658.2009.06986.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalo A, Kanter I, Shraga A, Sheinberger J, Tzemach H, Kinor N, Singer RH, Lionnet T, Shav-Tal Y. 2015. Cellular levels of signaling factors are sensed by β-actin alleles to modulate transcriptional pulse intensity. Cell Rep. 11, 419–432. ( 10.1016/j.celrep.2015.03.039) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vedula P, Kurosaka S, Leu NA, Wolf YI, Shabalina SA, Wang J, Sterling S, Dong DW, Kashina A. 2017. Diverse functions of homologous actin isoforms are defined by their nucleotide, rather than their amino acid sequence. Elife 6, e31661 ( 10.7554/eLife.31661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shawlot W, Deng JM, Fohn LE, Behringer RR. 1998. Restricted beta-galactosidase expression of a hygromycin-lacZ gene targeted to the beta-actin locus and embryonic lethality of beta-actin mutant mice. Transgenic Res. 7, 95–103. ( 10.1023/a:1008816308171) [DOI] [PubMed] [Google Scholar]
- 69.Shmerling D, et al. 2005. Strong and ubiquitous expression of transgenes targeted into the beta-actin locus by Cre/lox cassette replacement. Genesis 42, 229–235. ( 10.1002/gene.20135) [DOI] [PubMed] [Google Scholar]
- 70.Zhang F, Saha S, Shabalina SA, Kashina A. 2010. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science 329, 1534–1537. ( 10.1126/science.1191701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Popp D, Robinson RC. 2011. Many ways to build an actin filament. Mol. Microbiol. 80, 300–308. ( 10.1111/j.1365-2958.2011.07599.x) [DOI] [PubMed] [Google Scholar]
- 72.Stairs CW, Ettema TJG. 2020. The archaeal roots of the eukaryotic dynamic actin cytoskeleton. Curr. Biol. 30, R521–R526. ( 10.1016/j.cub.2020.02.074) [DOI] [PubMed] [Google Scholar]
- 73.McKane M, Wen K-K, Meyer A, Rubenstein PA. 2006. Effect of the substitution of muscle actin-specific subdomain 1 and 2 residues in yeast actin on actin function. J. Biol. Chem. 281, 29 916–29 928. ( 10.1074/jbc.M602251200) [DOI] [PubMed] [Google Scholar]
- 74.Bergeron SE, Zhu M, Thiem SM, Friderici KH, Rubenstein PA. 2010. Ion-dependent polymerization differences between mammalian beta- and gamma-nonmuscle actin isoforms. J. Biol. Chem. 285, 16 087–16 095. ( 10.1074/jbc.M110.110130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Müller M, Diensthuber RP, Chizhov I, Claus P, Heissler SM, Preller M, Taft MH, Manstein DJ. 2013. Distinct functional interactions between actin isoforms and nonsarcomeric myosins. PLoS ONE 8, e70636 ( 10.1371/journal.pone.0070636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Orlova A, Galkin VE, VanLoock MS, Kim E, Shvetsov A, Reisler E, Egelman EH. 2001. Probing the structure of F-actin: cross-links constrain atomic models and modify actin dynamics. J. Mol. Biol. 312, 95–106. ( 10.1006/jmbi.2001.4945) [DOI] [PubMed] [Google Scholar]
- 77.McCullough BR, et al. 2011. Cofilin-linked changes in actin filament flexibility promote severing. Biophys. J. 101, 151–159. ( 10.1016/j.bpj.2011.05.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kim E, Miller CJ, Reisler E. 1996. Polymerization and in vitro motility properties of yeast actin: a comparison with rabbit skeletal alpha-actin. Biochemistry 35, 16 566–16 572. ( 10.1021/bi9623892) [DOI] [PubMed] [Google Scholar]
- 79.Buzan JM, Frieden C. 1996. Yeast actin: polymerization kinetic studies of wild type and a poorly polymerizing mutant. Proc. Natl Acad. Sci. USA 93, 91–95. ( 10.1073/pnas.93.1.91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ti S-C, Pollard TD. 2011. Purification of actin from fission yeast Schizosaccharomyces pombe and characterization of functional differences from muscle actin. J. Biol. Chem. 286, 5784–5792. ( 10.1074/jbc.M110.199794) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Takaine M, Mabuchi I. 2007. Properties of actin from the fission yeast Schizosaccharomyces pombe and interaction with fission yeast profilin. J. Biol. Chem. 282, 21 683–21 694. ( 10.1074/jbc.M611371200) [DOI] [PubMed] [Google Scholar]
- 82.Kijima ST, Hirose K, Kong S-G, Wada M, Uyeda TQP. 2016. distinct biochemical properties of Arabidopsis thaliana actin isoforms. Plant Cell Physiol. 57, 46–56. ( 10.1093/pcp/pcv176) [DOI] [PubMed] [Google Scholar]
- 83.Yao X, Rubenstein PA. 2001. F-actin-like ATPase activity in a polymerization-defective mutant yeast actin (V266G/L267G). J. Biol. Chem. 276, 25 598–25 604. ( 10.1074/jbc.M011797200) [DOI] [PubMed] [Google Scholar]
- 84.Bryan KE, Rubenstein PA. 2005. An intermediate form of ADP-F-actin. J. Biol. Chem. 280, 1696–1703. ( 10.1074/jbc.M410180200) [DOI] [PubMed] [Google Scholar]
- 85.Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. 1998. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry 37, 11 171–11 181. ( 10.1021/bi9720033) [DOI] [PubMed] [Google Scholar]
- 86.Orlova A, Chen X, Rubenstein PA, Egelman EH. 1997. Modulation of yeast F-actin structure by a mutation in the nucleotide-binding cleft. J. Mol. Biol. 271, 235–243. ( 10.1006/jmbi.1997.1163) [DOI] [PubMed] [Google Scholar]
- 87.Marzook NB, Latham SL, Lynn H, Mckenzie C, Chaponnier C, Grau GE, Newsome TP. 2017. Divergent roles of β- and γ-actin isoforms during spread of vaccinia virus. Cytoskeleton (Hoboken) 74, 170–183. ( 10.1002/cm.21356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kandasamy MK, Burgos-Rivera B, McKinney EC, Ruzicka DR, Meagher RB. 2007. Class-specific interaction of profilin and ADF isovariants with actin in the regulation of plant development. Plant Cell 19, 3111–3126. ( 10.1105/tpc.107.052621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ezezika OC, Younger NS, Lu J, Kaiser DA, Corbin ZA, Nolen BJ, Kovar DR, Pollard TD. 2009. Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: insights from crystal structures of fission yeast profilin. J. Biol. Chem. 284, 2088–2097. ( 10.1074/jbc.M807073200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nefsky B, Bretscher A. 1992. Yeast actin is relatively well behaved. Eur. J. Biochem. 206, 949–955. ( 10.1111/j.1432-1033.1992.tb17005.x) [DOI] [PubMed] [Google Scholar]
- 91.Kang H, Bradley MJ, Cao W, Zhou K, Grintsevich EE, Michelot A, Sindelar CV, Hochstrasser M, De La Cruz EM.. 2014. Site-specific cation release drives actin filament severing by vertebrate cofilin. Proc. Natl Acad. Sci. USA 111, 17 821–17 826. ( 10.1073/pnas.1413397111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Noguchi TQP, Kanzaki N, Ueno H, Hirose K, Uyeda TQP. 2007. A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J. Biol. Chem. 282, 27 721–27 727. ( 10.1074/jbc.M703165200) [DOI] [PubMed] [Google Scholar]
- 93.Hatano T, et al. 2018. Rapid production of pure recombinant actin isoforms in Pichia pastoris. J. Cell. Sci. 131, jcs.213827 ( 10.1242/jcs.213827) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hatano T, Sivashanmugam L, Suchenko A, Hussain H, Balasubramanian MK. 2020. Pick-ya actin—a method to purify actin isoforms with bespoke key post-translational modifications. J. Cell. Sci. 133, jcs.241406 ( 10.1242/jcs.241406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kashina A. 2014. Protein arginylation, a global biological regulator that targets actin cytoskeleton and the muscle: protein arginylation, a global biological regulator. Anat. Rec. 297, 1630–1636. ( 10.1002/ar.22969) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Batsios P, Ishikawa-Ankerhold HC, Roth H, Schleicher M, Wong CCL, Müller-Taubenberger A. 2019. Ate1-mediated posttranslational arginylation affects substrate adhesion and cell migration in Dictyostelium discoideum. Mol. Biol. Cell 30, 453–466. ( 10.1091/mbc.E18-02-0132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karakozova M, Kozak M, Wong CCL, Bailey AO, Yates JR, Mogilner A, Zebroski H, Kashina A. 2006. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196. ( 10.1126/science.1129344) [DOI] [PubMed] [Google Scholar]
- 98.Pavlyk I, Leu NA, Vedula P, Kurosaka S, Kashina A. 2018. Rapid and dynamic arginylation of the leading edge β-actin is required for cell migration. Traffic 19, 263–272. ( 10.1111/tra.12551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen L, Kashina A. 2019. Quantification of intracellular N-terminal β-actin arginylation. Sci Rep 9, 16669 ( 10.1038/s41598-019-52848-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saha S, Mundia MM, Zhang F, Demers RW, Korobova F, Svitkina T, Perieteanu AA, Dawson JF, Kashina A. 2010. Arginylation regulates intracellular actin polymer level by modulating actin properties and binding of capping and severing proteins. Mol. Biol. Cell 21, 1350–1361. ( 10.1091/mbc.e09-09-0829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Feinberg J, Benyamin Y, Roustan C. 1995. Definition of an interface implicated in gelsolin binding to the sides of actin filaments. Biochem. Biophys. Res. Commun. 209, 426–432. ( 10.1006/bbrc.1995.1520) [DOI] [PubMed] [Google Scholar]
- 102.Burtnick LD, Koepf EK, Grimes J, Jones EY, Stuart DI, McLaughlin PJ, Robinson RC. 1997. The crystal structure of plasma gelsolin: implications for actin severing, capping, and nucleation. Cell 90, 661–670. ( 10.1016/s0092-8674(00)80527-9) [DOI] [PubMed] [Google Scholar]
- 103.Drazic A, et al. 2018. NAA80 is actin's N-terminal acetyltransferase and regulates cytoskeleton assembly and cell motility. Proc. Natl Acad. Sci. USA 115, 4399–4404. ( 10.1073/pnas.1718336115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Redman K, Rubenstein PA. 1981. NH2-terminal processing of Dictyostelium discoideum actin in vitro. J. Biol. Chem. 256, 13 226–13 229. [PubMed] [Google Scholar]
- 105.Rubenstein PA, Martin DJ. 1983. NH2-terminal processing of Drosophila melanogaster actin. Sequential removal of two amino acids. J. Biol. Chem. 258, 11 354–11 360. [PubMed] [Google Scholar]
- 106.Rubenstein PA, Martin DJ. 1983. NH2-terminal processing of actin in mouse L-cells in vivo. J. Biol. Chem. 258, 3961–3966. [PubMed] [Google Scholar]
- 107.Solomon LR, Rubenstein PA. 1985. Correct NH2-terminal processing of cardiac muscle alpha-isoactin (class II) in a nonmuscle mouse cell. J. Biol. Chem. 260, 7659–7664. [PubMed] [Google Scholar]
- 108.Goris M, et al. 2018. Structural determinants and cellular environment define processed actin as the sole substrate of the N-terminal acetyltransferase NAA80. Proc. Natl Acad. Sci. USA 115, 4405–4410. ( 10.1073/pnas.1719251115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rebowski G, Boczkowska M, Drazic A, Ree R, Goris M, Arnesen T, Dominguez R. 2020. Mechanism of actin N-terminal acetylation. Sci. Adv. 6, eaay8793 ( 10.1126/sciadv.aay8793) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Polevoda B, Cardillo TS, Doyle TC, Bedi GS, Sherman F. 2003. Nat3p and Mdm20p are required for function of yeast NatB Nalpha-terminal acetyltransferase and of actin and tropomyosin. J. Biol. Chem. 278, 30 686–30 697. ( 10.1074/jbc.M304690200) [DOI] [PubMed] [Google Scholar]
- 111.Van Damme P, et al. 2012. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc. Natl Acad. Sci. USA 109, 12 449–12 454. ( 10.1073/pnas.1210303109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huber M, et al. 2020. NatB-mediated N-terminal acetylation affects growth and biotic stress responses. Plant Physiol. 182, 792–806. ( 10.1104/pp.19.00792) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mu A, Fung TS, Kettenbach AN, Chakrabarti R, Higgs HN. 2019. A complex containing lysine-acetylated actin inhibits the formin INF2. Nat. Cell Biol. 21, 592–602. ( 10.1038/s41556-019-0307-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Terman JR, Kashina A. 2013. Post-translational modification and regulation of actin. Curr. Opin. Cell Biol. 25, 30–38. ( 10.1016/j.ceb.2012.10.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Block J, et al. 2012. FMNL2 drives actin-based protrusion and migration downstream of Cdc42. Curr. Biol. 22, 1005–1012. ( 10.1016/j.cub.2012.03.064) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kage F, et al. 2017. FMNL formins boost lamellipodial force generation. Nat. Commun. 8, 14 832 ( 10.1038/ncomms14832) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ponti A. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science 305, 1782–1786. ( 10.1126/science.1100533) [DOI] [PubMed] [Google Scholar]
- 118.Hotulainen P, Lappalainen P. 2006. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J. Cell Biol. 173, 383–394. ( 10.1083/jcb.200511093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Delorme V, Machacek M, DerMardirossian C, Anderson KL, Wittmann T, Hanein D, Waterman-Storer C, Danuser G, Bokoch GM. 2007. Cofilin activity downstream of Pak1 regulates cell protrusion efficiency by organizing lamellipodium and lamella actin networks. Dev. Cell 13, 646–662. ( 10.1016/j.devcel.2007.08.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Burnette DT, Manley S, Sengupta P, Sougrat R, Davidson MW, Kachar B, Lippincott-Schwartz J. 2011. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 13, 371–382. ( 10.1038/ncb2205) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bugyi B, Didry D, Carlier M-F. 2010. How tropomyosin regulates lamellipodial actin-based motility: a combined biochemical and reconstituted motility approach. EMBO J. 29, 14–26. ( 10.1038/emboj.2009.316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hsiao JY, Goins LM, Petek NA, Mullins RD. 2015. Arp2/3 complex and cofilin modulate binding of tropomyosin to branched actin networks. Curr. Biol. 25, 1573–1582. ( 10.1016/j.cub.2015.04.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gunning PW, Schevzov G, Kee AJ, Hardeman EC. 2005. Tropomyosin isoforms: divining rods for actin cytoskeleton function. Trends Cell Biol. 15, 333–341. ( 10.1016/j.tcb.2005.04.007) [DOI] [PubMed] [Google Scholar]
- 124.Vindin H, Gunning P. 2013. Cytoskeletal tropomyosins: choreographers of actin filament functional diversity. J. Muscle Res. Cell. Motil. 34, 261–274. ( 10.1007/s10974-013-9355-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gunning PW, Hardeman EC. 2017. Tropomyosins. Curr. Biol. 27, R8–R13. ( 10.1016/j.cub.2016.11.033) [DOI] [PubMed] [Google Scholar]
- 126.Manstein DJ, Meiring JCM, Hardeman EC, Gunning PW. 2019. Actin–tropomyosin distribution in non-muscle cells. J. Muscle Res. Cell. Motil. 41, 11–22. ( 10.1007/s10974-019-09514-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hardeman EC, Bryce NS, Gunning PW. 2019. Impact of the actin cytoskeleton on cell development and function mediated via tropomyosin isoforms. Semin. Cell Dev. Biol. 102, 122–131. ( 10.1016/j.semcdb.2019.10.004) [DOI] [PubMed] [Google Scholar]
- 128.DesMarais V. 2002. Spatial regulation of actin dynamics: a tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115, 4649–4660. ( 10.1242/jcs.00147) [DOI] [PubMed] [Google Scholar]
- 129.Jansen S, Goode BL. 2019. Tropomyosin isoforms differentially tune actin filament length and disassembly. Mol. Biol. Cell 30, 671–679. ( 10.1091/mbc.E18-12-0815) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang B, Wolfenson H, Chung VY, Nakazawa N, Liu S, Hu J, Huang RY-J, Sheetz MP. 2020. Stopping transformed cancer cell growth by rigidity sensing. Nat. Mater. 19, 239–250. ( 10.1038/s41563-019-0507-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. 2011. A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21, 539–550. ( 10.1016/j.cub.2011.03.007) [DOI] [PubMed] [Google Scholar]
- 132.Hu S, Grobe H, Guo Z, Wang Y-H, Doss BL, Pan M, Ladoux B, Bershadsky AD, Zaidel-Bar R. 2019. Reciprocal regulation of actomyosin organization and contractility in nonmuscle cells by tropomyosins and alpha-actinins. Mol. Biol. Cell 30, 2025–2036. ( 10.1091/mbc.E19-02-0082) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meiring JCM, Bryce NS, Wang Y, Taft MH, Manstein DJ, Liu Lau S, Stear J, Hardeman EC, Gunning PW. 2018. Co-polymers of actin and tropomyosin account for a major fraction of the human actin cytoskeleton. Curr. Biol. 28, 2331–2337. ( 10.1016/j.cub.2018.05.053) [DOI] [PubMed] [Google Scholar]
- 134.Gateva G, et al. 2017. Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr. Biol. 27, 705–713. ( 10.1016/j.cub.2017.01.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kim H-C, Jo Y-J, Kim N-H, Namgoong S. 2015. Small molecule inhibitor of formin homology 2 domains (SMIFH2) reveals the roles of the formin family of proteins in spindle assembly and asymmetric division in mouse oocytes. PLoS ONE 10, e0123438 ( 10.1371/journal.pone.0123438) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Antkowiak A, Guillotin A, Boiero Sanders M, Colombo J, Vincentelli R, Michelot A. 2019. Sizes of actin networks sharing a common environment are determined by the relative rates of assembly. PLoS Biol. 17, e3000317 ( 10.1371/journal.pbio.3000317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Iwasa JH, Mullins RD. 2007. Spatial and temporal relationships between actin-filament nucleation, capping, and disassembly. Curr. Biol. 17, 395–406. ( 10.1016/j.cub.2007.02.012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gupton SL, et al. 2005. Cell migration without a lamellipodium. J. Cell Biol. 168, 619–631. ( 10.1083/jcb.200406063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Johnson M, East DA, Mulvihill DP. 2014. Formins determine the functional properties of actin filaments in yeast. Curr. Biol. 24, 1525–1530. ( 10.1016/j.cub.2014.05.034) [DOI] [PubMed] [Google Scholar]
- 140.Billault-Chaumartin I, Martin SG. 2019. Capping protein insulates Arp2/3-assembled actin patches from formins. Curr. Biol. 29, 3165–3176. ( 10.1016/j.cub.2019.07.088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meiring JCM, Bryce NS, Niño JLG, Gabriel A, Tay SS, Hardeman EC, Biro M, Gunning PW. 2019. Tropomyosin concentration but not formin nucleators mDia1 and mDia3 determines the level of tropomyosin incorporation into actin filaments. Sci. Rep. 9, 6504 ( 10.1038/s41598-019-42977-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wawro B, Greenfield NJ, Wear MA, Cooper JA, Higgs HN, Hitchcock-DeGregori SE. 2007. Tropomyosin regulates elongation by formin at the fast-growing end of the actin filament. Biochemistry 46, 8146–8155. ( 10.1021/bi700686p) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Skau CT, Neidt EM, Kovar DR. 2009. Role of tropomyosin in formin-mediated contractile ring assembly in fission yeast. Mol. Biol. Cell 20, 2160–2173. ( 10.1091/mbc.E08-12-1201) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Blanchoin L, Pollard TD, Hitchcock-DeGregori SE. 2001. Inhibition of the Arp2/3 complex-nucleated actin polymerization and branch formation by tropomyosin. Curr. Biol. 11, 1300–1304. ( 10.1016/S0960-9822(01)00395-5) [DOI] [PubMed] [Google Scholar]
- 145.Alioto SL, Garabedian MV, Bellavance DR, Goode BL. 2016. Tropomyosin and profilin cooperate to promote formin-mediated actin nucleation and drive yeast actin cable assembly. Curr. Biol. 26, 3230–3237. ( 10.1016/j.cub.2016.09.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. 2010. Differential regulation of unconventional fission yeast myosins via the actin track. Curr. Biol. 20, 1423–1431. ( 10.1016/j.cub.2010.07.026) [DOI] [PubMed] [Google Scholar]
- 147.Ostap EM. 2008. Tropomyosins as discriminators of myosin function. In Tropomyosin (ed. Gunning P.), pp. 273–282. New York, NY: Springer; ( 10.1007/978-0-387-85766-4_20) [DOI] [PubMed] [Google Scholar]
- 148.Christensen JR, Hocky GM, Homa KE, Morganthaler AN, Hitchcock-DeGregori SE, Voth GA, Kovar DR. 2017. Competition between tropomyosin, fimbrin, and ADF/cofilin drives their sorting to distinct actin filament networks. eLife 6, e23152 ( 10.7554/eLife.23152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Christensen JR, Homa KE, Morganthaler AN, Brown RR, Suarez C, Harker AJ, O'Connell ME, Kovar DR. 2019. Cooperation between tropomyosin and α-Actinin inhibits fimbrin association with actin filament networks in fission yeast. Elife 8, e47279 ( 10.7554/eLife.47279) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pollard LW, Garabedian MV, Alioto SL, Shekhar S, Goode BL. 2020. Genetically-inspired in vitro reconstitution of S. cerevisiae actin cables from seven purified proteins. Mol. Biol. Cell. 31, 319–396. ( 10.1091/mbc.E19-10-0576) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.