Abstract
Background
Hypomyces is a large genus of fungicolous fungi, parasitising the fruiting bodies of Agaricales, Boletales, Helotiales, Pezizales and Polyporales. Hypomyces currently comprises of 147 species widely distributed in Australia, China, France, Germany, Italy, Japan, North America, Sri Lanka, Thailand and UK. Amongst them, 28 species have been recorded in China.
New information
Hypomyces pseudolactifluorum sp. nov., growing on the fruiting bodies of Russula sp. in subsect. Lactarioideae and collected from Yunnan, China, is described with illustrations and molecular phylogenetic data (combined ITS, LSU, TEF1-α and RPB2 sequence dataset). The new species is characterised by semi-immersed to immersed perithecia and fusiform, apiculate and verrucose ascospores. We also review the species diversity of the genus Hypomyces in China.
Keywords: Mycoparasite, species diversity, muti-gene phylogeny
Introduction
Fungicolous fungi are a large and diverse ecological group, currently containing more than 1500 taxa distributed in many lineages across the fungal kingdom (Põldmaa 2011, Sun et al. 2019a). Hypomyces (Fr.) Tul. & C. Tul. is an important genus of fungicolous fungi and placed in the family Hypocreaceae (Hypocreales, Sordariomycetes, Ascomycota) (Hyde et al. 2020). Hypomyces was originally introduced as a subgenus of Hypocrea Fr. (Fries 1825) and then Tulasne and Tulasne (1860) revised it to a genus and designated H. lactifluorum (Schwein.) Tul. & C. Tul. from the USA as its type. Hypomyces parasitises the fruiting bodies of Agaricales, Boletales, Helotiales, Pezizales and Polyporales (Rossman et al. 1999, Tamm and Põldmaa 2013, Sun et al. 2019a). Hypomyces is characterised by: superficial or immersed, spherical to ovate, pyriform, papillate and yellow, orange, tawny red or green perithecia in a subiculum; 8-spored, subcylindrical to cylindrical and with a thickened apical asci; and ellipsoid, lanceolate, fusiform to navicular, 0-1-septate or rarely 3-septate, hyaline, spinulose or verrucose and smooth-walled ascospores (Rossman et al. 1999, Zeng and Zhuang 2015). Its allied genera include Cladobotryum Nees, Mycogone Link, Sepedonium Link and Stephanoma Wallr (Wijayawardene et al. 2017) and its asexual morphs are Acremonium-, Dactylaria-, Papulaspora-, Trichothecium- or Verticillium-like (Jaklitsch et al. 2006, Hyde et al. 2020). Hypomyces currently comprises of 147 species in Species Fungorum (http://www.speciesfungorum.org/, accessed in April 2020) and is widely distributed in Australia, China, France, Germany, Italy, Japan, North America, Sri Lanka, Thailand and UK (Zhuang et al. 2012, Rossman et al. 2013, Zeng and Zhuang 2016, Zare and Gams 2016, Lechat et al. 2017, Wei and Kirschner 2017, Sun et al. 2019a, Sun et al. 2019b, Zeng and Zhuang 2019). Amongst them, 28 species have been reported in China (Table 1).
Table 1.
Species diversity of the genus Hypomyces in China (29 species in total).
| Taxa names | Hosts | Distribution | References |
| Hypomyces amaniticola | Amanita sp. | China (Yunnan) | Zeng and Zhuang 2016 |
| H. aurantius | Agaricus bisporus, Polyporales (Cymatoderma sp., Laetiporus sulphureus, Panellus sp., Polyporus picipes), Stereum sp. | China (Anhui, Fujian, Guangxi, Hainan, Hebei, Hunan, Jiangsu, Jiangxi, Shanghai, Sichuan, Zhejiang), New Zealand, USA | Chen and Fu 1989, Põldmaa 2011, Luo and Zhuang 2012 |
| H. aureonitens | Phlebia tremellosa, Polyporus sp. | China (Fujian, Guangxi), Europe | Teng 1963, Sun et al. 2019a |
| H. chlorinigenus | Agaricaceae, Boletaceae | Belgium, China (Taiwan), Guyana; Indonesia, New Zealand, USA | Rogerson and Samuels 1989, Zeng and Zhuang 2016 |
| H. chrysospermus | Boletus sp., Hemileccinum impolitum, Suillus americanus, Russula sp. | China (Fujian, Jiling, Nanjing), Russia | Ma 2008, Luo and Zhuang 2012 |
| H. completiopsis | Boletus sp. | China (Yunnan) | Zeng and Zhuang 2016 |
| H. fistulina | Fistulina sp. | China (Guangxi) | Sun et al. 2019b |
| H. hubeiensis | Agaricus sp. | China (Hubei) | Zeng and Zhuang 2019 |
| H. hyalinus | Agaricales (Amanita sp.), Polyporales | Canada, China (Jiangsu), Japan, USA | Teng 1934, Teng 1963, Rogerson and Samuels 1994 |
| H. lateritius | Lactarius camphoratus, L. chelidonium, L. controversus, L. deliciosus, L. sanguifluus, L. thejogalus, L. trivialis, Lactarius sp. | Canada, China (Tibet), Europe, Japan, Mexico, New Zealand, USA | Rogerson and Samuels 1994, Luo and Zhuang 2012 |
| H. luteovirens | Russula atropurpurea, R. rosea, R. sanguinaria, Russula sp. | Canada, China (Inner Mongolia), Europe, Japan, Russia, USA | Rogerson and Samuels 1994, Ma 2008 |
| H. macrosporus | Russulaceae | China (Hubei), Mexico, USA | Rogerson and Samuels 1994, Luo and Zhuang 2012 |
| H. microspermus | Boletaceae, Boletus sp., Imleria badia, Xanthoconium affine, Xerocomellus chrysenteron, Xerocomus sp. | Canada, China (Fujian, Guizhou, Hainan, Hubei, Jilin, Taiwan, Yunan), Indonesia, New Zealand, USA | Rogerson and Samuels 1989, Zeng and Zhuang 2016 |
| H. mycophilus | Auricularia sp., Bulgari sp., Marasmius sp., Polyporus sp., Trametes versicolor | China (Guangdong), USA | Rogerson and Samuels 1993, Zeng et al. 2017 |
| H. ochraceus | Decaying leaves, wood and fungi (e.g. Russula sp.) | China (Guangxi, Yunnan), Europe, USA | Teng 1963, Sun et al. 2019a |
| H. orthosporus | Polyporales | China (Tibet), Estonia, Finland, The Netherlands | Põldmaa 1996, Zeng and Zhuang 2019 |
| H. papulasporae | Geoglossum difforme, G. fallax, G. glabrum, G. nigritum, G. simile, Glutinoglossum glutinosum, Trichoglossum hirsutum, T. walteri | China, USA, New Zealand | Rogerson and Samuels 1985, Sun et al. 2019a |
| H. polyporinus | Auricularia auricula-judae, Polyporales, Trametes versicolor, T. pubescens, Polyporus sp. | Canada, China (Guangxi), USA | Teng 1963, Rogerson and Samuels 1993 |
| H. pseudolactifluorum sp. nov. | Russula sp. | China (Yunnan) | This study |
| H. rosellus | Agaricus bisporus, Armillaria sp., Hydnellum sp., Hyphoderma sp., Mycena sp., Polyporus sp., Russula sp., Trichaptum sp. | China (Gansu), Europe, Iran, Japan, Korea, USA | Tamm and Põldmaa 2013, Sun et al. 2019b |
| H. semicircularis | Ganoderma sichuanense, Microporus xanthopus | Cuba, China | Wei and Kirschner 2017, Sun et al. 2019a |
| H. sibirinae | Aphyllophorales, Boletus sp., Polyporales | China (Hunan), Indonesia, USA | Samuels et al. 1990, Zeng et al. 2017, Sun et al. 2019a |
| H. sinicus | Schizophyllum sp. | China (Anhui) | Zhuang et al. 2012 |
| H. stephanomatis | Humaria hemisphaerica, Humaria sp. | Canada, China (Hubei), Germany, USA | Rogerson and Samuels 1985,Zeng and Zhuang 2016 |
| H. subiculosus | Polyporaceae (Microporus affinis, Trametes versicolo) | China (Anhui, Beijing, Guangxi, Zhejiang), Cuba, Japan | Rogerson and Samuels 1993, Luo and Zhuang 2012 |
| H. succineus | Pholiota sp. | China (Taiwan), USA | Rogerson and Samuels 1994, Zeng and Zhuang 2016 |
| H. tegillum | Aphyllophorales, Polyporales | Brazil, China (Guangxi, Yunnan), Panama, USA | Rogerson and Samuels 1993, Luo and Zhuang 2012 |
| H. triseptatus | Bark or associated with an ascomycete; Pyrenomycete | China (Hunan, Guangdong), Gabon | Rossman and Rogersson 1981, Zeng et al. 2017 |
| H. yunnanensis | Boletus sp. | China (Yunnan) | Zeng and Zhuang 2016 |
Fungicolous fungi play important roles in the processes of the growths and degradations of their hosts. With the rapid development of mushroom industries, the fungicolous fungi on mushrooms have received more and more attention (Hyde et al. 2019). In this paper, we introduce a new member of fungicolous fungi, Hypomyces pseudolactifluorum sp. nov., on the fruiting bodies of Russula sp., collected from Yunnan Province, China. At the same time, we review the species diversity of the genus Hypomyces in China.
Materials and methods
Collections and Morphology
Hypomyces specimens, including their host mushrooms, were collected in an evergreen broad-leaved forest in Baihualing, Baoshan, Yunnan Province, China. The specimens, as well as collected host mushrooms, were placed on a piece of aluminium foil at first, then rolled the paper into a cylinder, twisted at the ends for sealing and lastly taken back to the laboratory for study (McKnight and McKnight 1997). Colour codes were recorded following those of Kornerup and Wanscher (1978). A Nikon Coolpix P510 camera was used to take photos in the wild. Dried specimens were observed and photographed using an Olympus SZ61 stereomicroscope and a Nikon ECLIPSE Ni compound microscope fitted with a Canon EOS 600D digital camera. Measurements were made using the Tarosoft® Image Frame Work programme v.0.9.7. The colour change of the perithecial wall was tested using 5% potassium hydroxide (KOH). Type specimens are deposited at the Herbarium of Mae Fah Luang University, Thailand (MFLU) and the Herbarium of Cryptogams Kunming Institute of Botany, Chinese Academy of Sciences, PR China (HKAS).
DNA extraction, PCR amplification and sequencing
The genomic DNA was extracted from the dried materials using the CTAB method (Doyle 1987). Tissues from the ascocarps of parasitic fungi and fruiting bodies of the host mushrooms were used to extract DNA, respectively. Primer pairs ITS1F/ITS4 (White et al. 1990), LR0R/LR5 (Rehner and Samuels 1994, Vilgalys and Hester 1990), TEF1-α 983f/TEF1-α 2218r (Carbone and Kohn 1999, Rehner and Buckley 2005) and RPB2-5f/RPB2-7cR (Liu et al. 1999) were used for amplification of the ITS, LSU, TEF1-α and RPB2 gene regions.
PCR was performed in a 25 μl reaction volume: 12.5 μl Taq PCR Master Mix (Abmgood, Richmond, BC, Canada), 1 μl forward primer, 1 μl reverse primer, 1 μl DNA template and 9.5 μl ddH2O. For ITS and LSU, PCR reaction conditions are: 8 min at 94ºC, followed by 30 s at 94ºC, 30 s at 52ºC and 1 min at 72ºC for 35 cycles and a final extension of 10 min at 72ºC. PCR reaction conditions of TEF1-α and RPB2 are: 8 min at 94ºC, followed by 1 min at 95ºC, 45 s at 59ºC for RPB2/55ºC for TEF1-α and 1 min at 72ºC for 35 cycles and a final extension of 10 min at 72ºC. The PCR products were detected using agarose gel electrophoresis and, in the gel documentation system, clear bands were observed. Sequencing was performed by Sangon Biotech (Shanghai) Co. Ltd., PR China; partial impure products were purified using the Cycle-pure-kit (Omega, America) and then cloned into pClone007 Simple vector (TSV-007S from Beijing TsingKe Biotech). Twenty clones of PCR products of each gene were sequenced using the universal primer pairs M13-47/M13-48.
Sequence alignment and phylogenetic analyses
The parasitic fungus: Hypomyces pseudolactifluorum sp. nov.
Molecular phylogenetic trees were constructed using our sequencing results of H. pseudolactifluorum sp. nov. and the voucher sequences of their allies obtained from NCBI GenBank (Table 2). Two species of Trichoderma, T. hamatum (DAOM 167057) and T. viride (CBS 119325) were used as outgroup taxa. All sequences were assembled and aligned using MAFFT v6.8 (Katoh et al. 2005) and manually edited via BioEdit version 7.0.9 (Hall 1999). Four sequence matrices of ITS, LSU, TEF1-α and RPB2 genes, respectively, were compiled. The optimal substitution model for each gene dataset was determined using jModelTest2 under the Akaike information criterion (AIC) (Darriba et al. 2012). The results indicated that the GTR+I+G model (-lnL = 8658.2624) is optimal for the ITS dataset, as well as the TIM1+I+G model (-lnL = 4392.5417) for LSU, the TrN+I+G model (-lnL = 5751.4959) for TEF1-α and the model SYM+I+G (-lnL = 6419.6669) for RPB2, respectively. Using the aligned sequence matrices, a combined gene sequence dataset (ITS, LSU, TEF1-α and RPB2, orderly) was assembled and aligned and was finally deposited in TreeBASE database (http://purl.org/phylo/treebase/phylows/study/TB2:S26593?x-access-code=152eadfc2292343af7627cfad5c2946c&format=html).
Table 2.
Voucher information and GenBank accession numbers for samples appearing in the Hypomyces phylogenetic tree. Our sequencing results are displayed in bold. (Label T indicate the sequences from ex-type strains.)
Maximum Likelihood (ML) analysis was performed using IQ-Tree (Nguyen et al. 2014, Chernomor et al. 2016) with the computing models listed above and a bootstrap test of 1000 replicates. Bayesian Inference (BI) analysis was carried out using MrBayes v3.2.6 (Ronquist et al. 2012). The TIM1 and TrN substitution models were replaced by the GTR model (Huelsenbeck and Rannala 2004). Four simultaneous Markov Chain Monte Carlo (MCMC) chains were run for random trees of 10,000,000 generations and were sampled by every 100 generations. The computing was stopped when the standard deviation of the split frequencies fell below 0.01 and ESS values > 200. Subsequently, phylogenetic trees were summarised and posterior probabilities (PP) were performed using MCMC by discarding the first 25% generations as “burn-in” (Huelsenbeck and Ronquist 2001). Gaps were treated as missing data. Phylogenetic trees were viewed in FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree).
The host mushroom: Russula sp.
Voucher sequences (ITS gene) for phylogenetic analyses of the host mushroom and its allies were obtained from our sequencing results and GenBank databases (Li et al. 2020) (Table 3). Five species of Russula subg. Compactae, R. acrifolia, R. adusta, R. eccentrica, R. nigricans and R. subnigricans were selected as the outgroup taxa. Sequence alignment and phylogenetic analyses followed those of the parasitic fungus above. ML analysis was performed using IQ-Tree with TVM+I+G model (-lnL = 5298.7964) (Nguyen et al. 2014, Chernomor et al. 2016). The ITS sequence matrix of the host mushroom and its allies were deposited in the TreeBASE database (http://purl.org/phylo/treebase/phylows/study/TB2:S26693?x-access-code=2e445b17aebe1f93266051a8920ae62f&format=html).
Table 3.
Voucher information and GenBank accession numbers for samples appearing in the Russula phylogenetic tree. Our sequencing results are displayed in bold.
| Taxa names | Specimen/Strain number | GenBank accession | References |
| Russula acrifolia | TUB UE12.09.2003-3 | DQ421998 | Eberhardt 2002 |
| R. adusta | PC 547RUS27 | AY061652 | Miller and Buyck 2002 |
| R. aff. chloroides | FH 12273 | KT934015 | Looney et al. 2016 |
| R. brevipes | SMI329 | FJ845429 | Kranabetter et al. 2009 |
| R. brevipes | JS160927-01 | MG407682 | GenBank |
| R. brevipes | TENN 070667 | KY848511 | Looney et al. 2018 |
| R. brevipes var. acrior | JMP 0058 | EU819422 | Palmer et al. 2008 |
| R. byssina | HGAS-MF 009907 | MN648951 | Li et al. 2020 |
| R. byssina | HGAS-MF009921 | MN648949 | Li et al. 2020 |
| R. byssina | HGAS-MF 009913 | MN648950 | Li et al. 2020 |
| R. cascadensis | UBC F30189 | KX812838 | Bazzicalupo 2018 |
| R. cascadensis | UBC F19691 | HM240541 | Buyck et al. 2017 |
| R. cf. angustispora | PC BB2004-252 | EU598152 | GenBank |
| R. cf. brevipes | F 28785 | MH718203 | GenBank |
| R. cf. brevipes | F CDW47 | GQ166868 | GenBank |
| R. cf. brevipes | GO 2009-276 | KC152212 | GenBank |
| R. cf. delica | UBC F30260 | KX812852 | Bazzicalupo 2018 |
| R. chloroides | PC 205RUS24 | AY061663 | Miller and Buyck 2002 |
| R. chloroides | UBC F20353 | KC581331 | GenBank |
| R. chloroides | RUS-12091401 | KF432954 | Wisitrassameewong et al. 2014 |
| R. cremicolor | HGAS-MF 009901 | MN648955 | Li et al. 2020 |
| R. cremicolor | HGAS-MF 009908 | MN648952 | Li et al. 2020 |
| R. cremicolor | HGAS-MF 009912 | MN648953 | Li et al. 2020 |
| R. cremicolor | HGAS-MF 009919 | MN648954 | Li et al. 2020 |
| R. delica | hue22 (TUB) | AF418605 | Eberhardt 2002 |
| R. delica | FH 12-272 | KF432955 | Wisitrassameewong et al. 2014 |
| R. delica | HA 2015-004 | KX263000 | Aghajani et al. 2017 |
| R. delica | PC 496RUS26 | AY061671 | Miller and Buyck 2002 |
| R. delica | TUB hue22 | AF418605 | Eberhardt 2002 |
| R. delica | UBC F30263 | KX812842 | Bazzicalupo 2018 |
| R. delica | RMUKK 37 | KX267630 | GenBank |
| R. delica | KA 12-1327 | KR673555 | Kim et al. 2015 |
| R. delica | HMJAU 32182 | KX094989 | Liu et al. 2017 |
| R. eccentrica | HCCN 23685 | KC699778 | Park et al. 2014 |
| R. japonica | MHHNU 31049 | MK167414 | Chen and Zhang 2019 |
| R. japonica | HGAS-MF 009923 | MN648957 | Li et al. 2020 |
| R. japonica | HGAS-MF 009915 | MN648956 | Li et al. 2020 |
| R. leucocarpa | HGAS-MF 009910 | MN648948 | Li et al. 2020 |
| R. leucocarpa | HGAS-MF 009916 | MN648947 | Li et al. 2020 |
| R. littoralis | PC 1222IS87 | AY061702 | Miller and Buyck 2002 |
| R. marangania | MEL 2293694 | EU019930 | Lebel and Tonkin 2007 |
| R. nigricans | TUB fo46761 | AF418607 | Eberhardt 2002 |
| R. pallidospora | PC 2-1221IS85 | AY061701 | Miller and Buyck 2002 |
| R. pumicoidea | MEL T-14771 | EU019931 | Lebel and Tonkin 2007 |
| R. sinuata | MEL H4755 | EU019943 | Lebel and Tonkin 2007 |
| R. subnigricans | MHHNU ZP6932 | EF534351 | Yin et al. 2008 |
| R. vesicatoria | PC 0124666 | KY800359 | Buyck et al. 2017 |
| Russula sp. | MFLU 20-0265 (host) | MT755627 | In this study |
Taxon treatments
Hypomyces pseudolactifluorum
F. M. Yu, Q. Zhao & K. D. Hyde sp. nov.
1119A7CE-9897-55CC-94D7-5CF3C180F567
Materials
Type status: Holotype. Occurrence: catalogNumber: MFLU 20-0265; recordedBy: Jian-Wei Liu; lifeStage: Telemorph; Taxon: scientificName: Hypomyces pseudolactifluorum; Location: country: China; stateProvince: Yunnan; locality: Baoshan, Longyang, Baihualing; verbatimElevation: 2094m; locationRemarks: label transliteration: "Yunnan, Baoshan, Longyang, Baihualing, on Russula sp., 20 July 2018, Jian-Wei Liu; [云南保山百花岭 2094 m, 2018.07.20, 刘建伟]; verbatimCoordinates: 25°17.931’N, 98°47.0718’E; decimalLatitude: 25.2989; decimalLongitude: 98.7845; georeferenceProtocol: label; Identification: identifiedBy: Feng-Ming Yu; dateIdentified: 2019
Type status: Paratype. Occurrence: catalogNumber: MFLU 20-0266; recordedBy: Jian-Wei Liu; lifeStage: Telemorph; Taxon: scientificName: Hypomyces pseudolactifluorum; Location: country: China; stateProvince: Yunnan; locality: Baoshan, Longyang, Baihualing; verbatimElevation: 2094m; locationRemarks: label transliteration: "Yunnan, Baoshan, Longyang, Baihualing, on Russula sp., 20 July 2018, Jian-Wei Liu; [云南保山百花岭 2094 m, 2018.07.20, 刘建伟]; verbatimCoordinates: 25°17.931’N, 98°47.0718’E; decimalLatitude: 25.2989; decimalLongitude: 98.7845; georeferenceProtocol: label; Identification: identifiedBy: Feng-Ming Yu; dateIdentified: 2019
Type status: Isotype. Occurrence: catalogNumber: HKAS 107300; recordedBy: Jian-Wei Liu; lifeStage: Telemorph; Taxon: scientificName: Hypomyces pseudolactifluorum; Location: country: China; stateProvince: Yunnan; locality: Baoshan, Longyang, Baihualing; verbatimElevation: 2094m; locationRemarks: label transliteration: "Yunnan, Baoshan, Longyang, Baihualing, on Russula sp., 20 July 2018, Jian-Wei Liu; [云南保山百花岭 2094 m, 2018.07.20, 刘建伟]; verbatimCoordinates: 25°17.931’N, 98°47.0718’E; decimalLatitude: 25.2989; decimalLongitude: 98.7845; georeferenceProtocol: label; Identification: identifiedBy: Feng-Ming Yu; dateIdentified: 2019
Description
Index Fungorum number: IF557817
Sexual morph. Subiculum light yellow (4A4–5) when fresh and pale orange, light orange to brownish-orange (5A3–4, 5C4, 6C6) after being dried, usually covering the pileus, stipe and deformed gills of the host mushroom. Perithecia aggregated, semi-immersed to immersed in subiculum, except for their erumpent papilla, yellowish-brown to dark brown (5E6, 6E6, 6F6–8), pyriform to subglobose, 262–484 × 136–284 μm; perithecial wall 12–25 μm thick, single-layer, cells 9–22 × 4–8 μm. Papilla prominent, 129–177 μm high, at base 135–284 μm wide. Asci 8-spored, cylindrical, 147–222 × 4–9 μm; apex thickened, 4.9–6.0 wide and 2.5-3.0 μm high. Ascospores uniseriate and with ends overlapping, fusiform, 30–38 × 6–8 μm, single-septate, septum median and with dense verrucae and prominently apiculate, apiculi 4.5–8.0 μm long, straight or curved. Asexual morph: unknown. (Fig. 1)
Figure 1.
Hypomyces pseudolactifluorum sp. nov.. a: The host mushroom (Russula sp.); b-e: Perithecia embedded in subiculum effused over the substratum; d-e: Median sections of an ascoma; f: Section of peridium; g-l: Asci with ascospores; m-t: Ascospores. Scale bars: a = 5 cm; b = 1 mm; c = 200 μm; d, e = 100 μm; f, g= 50 μm; h - l = 20 μm; m - t = 10 μm.
Diagnosis
The new species is similar to Hypomyces lactifluorum on Russula and Lactarius spp. from North America (Rogerson and Samuels 1994), but has smaller perithecia and shorter asci. The main differences of the two species are compared in Table 4.
Table 4.
Main differences between Hypomyces lactifluorum and H. pseudolactifluorum sp. nov..
|
H.
lactifluorum (Rogerson and Samuels 1994) |
H. pseudolactifluorum | |
| Subiculum | Pale yellowish-orange to bright orange (young), in age becoming deep red, reddish-purple to very dark purple (old), occasionally fading to pink, turning purple in 3% KOH. | Light yellow (4A4–5) when fresh, and pale orange to light orange to brownish-orange (5A3–4, 5C4, 6C6) after being dried, KOH (-). |
| Perithecia | Ovate to obpyriform, deep orange to reddish-purple, 400–600 × 200–450 μm | Pyriform to subglobose, yellowish-brown to dark brown (5E6, 6E6, 6F6–8), 262–484 × 136–284 μm |
| Embedded type | Immersed except for papilla | Semi-immersed to immersed except for papilla |
| Papilla | Averaging 120 μm high, 120 μm wide | 129–177 μm high and 135 –284 μm wide at base |
| Asci | Long cylindrical, 200–260 × 5–10 μm | Cylindrical, 147–222 × 4–8.5 μm |
| Ascospores | Fusiform, 1-septate, 35–40 × 4.5–7 μm | Fusiform, 1-septate, 30–38 × 5.5–8 μm |
| Apiculi | 4.5–7.5 μm long | 4–6 μm long |
| Hosts | Russula and Lactarius spp. | Russula sp. |
| Distribution | North America | P.R. China (Yunnan) |
Etymology
Referring to the most closely-related species, Hypomyces lactifluorum.
Distribution
PR CHINA (Yunnan).
Host
On the fruiting bodies of Russula sp. that grew on the humus layer in an evergreen broad-leaved forest of a rainforest. The host mushrooms: basidiocarps medium-sized and infundibuliform, pilei 63−77 mm in diameter. As serious degradation has occurred, the colour and other characters of the host mushrooms cannot be determined. Molecular phylogenetic evidence indicates it is a Russula species.
Notes
Only sexual morph had been discovered on the hosts (Russula sp.) of the new species.
Analysis
Phylogenetic analyses
Parasitic fungus: Hypomyces pseudolactifluorum sp. nov
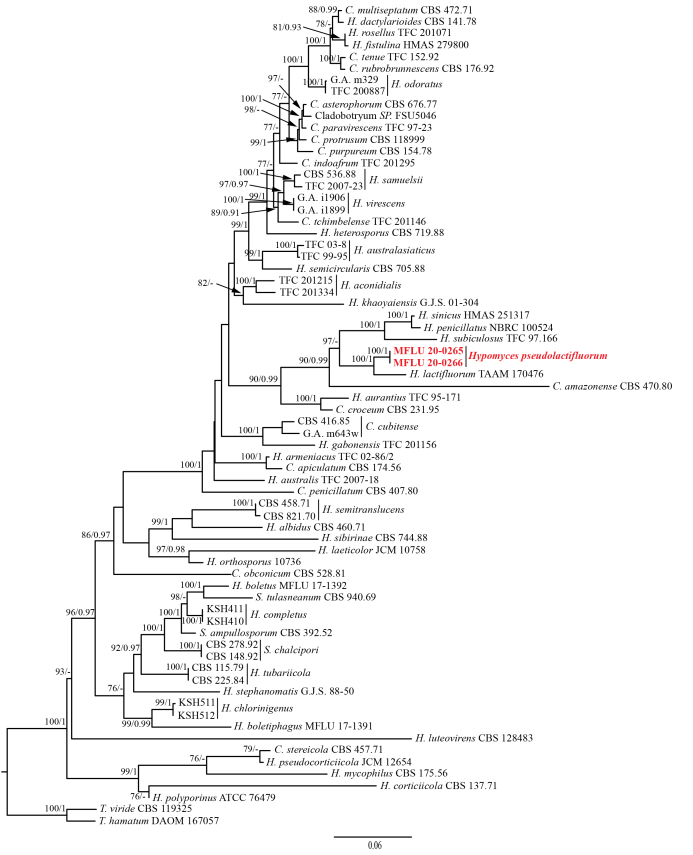
The combined ITS+LSU+TEF1-α+RPB2 sequence dataset (excluding the outgroup taxa) contains 3,262 characters (709 for ITS, 893 for LSU, 921 for TEF1-α and 739 for RPB2) from 56 Hypomyces species and two Trichoderma species. Amongst them, 2,246 characters are constant, 209 variable characters are parsimony-uninformative and 807 characters are parsimony-informative. The ML and BI analyses resulted in trees with similar topology and support values and the ML tree is shown in Fig. 2.
Figure 2.
ML tree of Hypomyces pseudolactifluorum sp. nov. and its allies generated from a combined ITS, LSU,TEF1-α and RPB2 gene sequence dataset. Supporting values of MLBP (left, greater than 75%) and BIBP (right, greater than 0.9) are shown at the nodes, respectively. The new species is marked in red.
In the phylogenetic tree, the parasitic fungi MFLU 20-0265 and MFLU 20-0266 are clustered together and formed a distinct lineage with the same branch length and strong supportive values (MLBP = 100%, BIPP = 1), which support them to be conspecific. The parasitic fungi are closely related H. lactifluorum and they form a sister clade also with strong supportive values (MLBP = 100%, BIPP = 1). Comparing the gene sequences of the two species, there are 25 bp (4.3%) differences across 582 bp in ITS, 28 bp (3.2%) differences across 870 bp in LSU, 24 bp (2.6%) differences across 921 bp in TEF1-α and 24 bp (3.2%) differences across 739 bp in RPB2 (Suppl. material 1). Following the recommendations from Jeewon and Hyde (2016), we assign the parasitic fungi as H. pseudolactifluorum sp. nov.
The host mushroom: Russula sp.
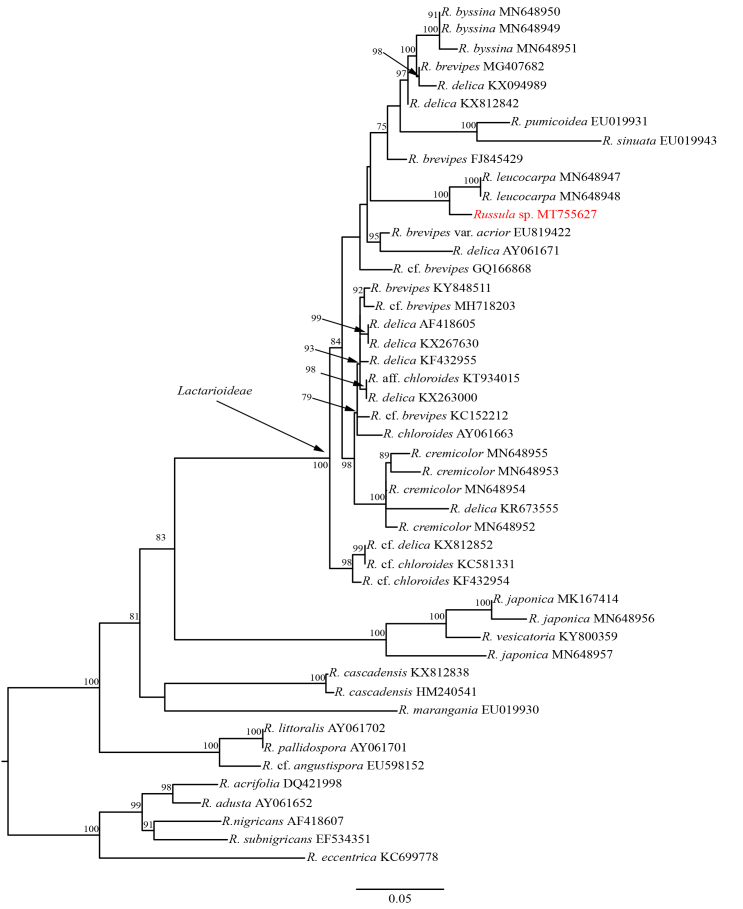
According to the ITS phylogenetic tree of the host mushroom and its allies, the host mushroom (MFLU 20-0265) is clustered together with Russula leucocarpa (HGAS-MF 009910 and HGAS-MF 009916) (MLBP = 100%) in subsect. Lactarioideae. However, their ITS sequences have 24 bp (3.5%) differences across 694 bp, which indicated they may be two distinct species. Due to lack of sufficient morphological evidence, the host mushroom was temporarily identified as Russula sp. (Fig. 3).
Figure 3.
ML tree of Russula sp. (in red) and it allies inferred from the ITS sequence dataset. Five species of Russula subg. Compactae were used as the outgroup taxa. Supporting values of MLBP (greater than 75%) are shown at the nodes.
Discussion
Zeng and Zhuang (2016) described H. amaniticola on Amanita sp. and H. completiopsis and H. yunnanensis on Boletus sp., also from China. Though with similar colour and shapes of perithecia, the host of H. pseudolactifluorum sp. nov. is decidedly different from those of these three species. Furthermore, H. pseudolactifluorum sp. nov. (KOH-) has smaller perithecia and larger ascospores than those of H. completiopsis (KOH+) and H. pseudolactifluorum sp. nov. has larger perithecia, asci and ascospores than those of H. amaniticola (KOH+) and H. yunnanensis (KOH-). Unfortunately, these three species all lack molecular data.
With the rapid development of mushroom industries, fungal pathogens on mushrooms have received more and more attention (Hyde et al. 2019). The fungicolous fungi Hypomyces is an important group of mushroom pathogens. Many Hypomyces species, for example, H. aurantius, H. perniciosus, H. rosellus, H. odoratus etc., have all been recorded as the causes of Cobweb or Web bubble disease which seriously influence mushroom industries (Fletcher and Gaze 2007, Carrasco et al. 2017, Zhang et al. 2017, Zhang et al. 2017). Russula is the largest subgenus in agaric with approximately 800 species (Li et al. 2020) and many Russula species are important edible mushrooms. Since growing on Russula sp., H. pseudolactifluorum sp. nov., as well as H. lactifluorum from North America (Rogerson and Samuels 1994), could be one of the potential pathogens of some Russula species in Asia.
Supplementary Material
Hypomyces pseudolactifluorum sp. nov. (Hypocreales: Hypocreaceae) on Russula sp. from Yunnan, PR China
FENG-MING YU, RUVISHIKA S. JAYAWARDENA, JIAN-WEI LIU, KEVIN D. HYDE, QI ZHAO
Data type
word
Brief description
Sequence differences of ITS, LSU, TEF1-α and RPB2 genes between H. lactifluorum (TAAM 170476) and H. pseudolactifluorum sp. nov.. The locus’ numbers refer to the nucleotide positions of the gene sequences of H. lactifluorum from GenBank. Gap is replaced by ‘-’.
File: oo_438866.docx
Acknowledgements
The research is supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) Program (Grant No. 2019QZKK0503); the Open Research Project of “Cross-Cooperative Team” of the Germplasm Bank of Wild Species, Kunming Institute of Botany, Chinese Academy of Sciences (Grant No. 292019312511043); Science and Technology Service Network Initiative of the Chinese Academy of Sciences (KFJ-STS-QYZD-171); the Biodiversity Survey and Assessment Project of the Ministry of Ecology and Environment, PR China (2019HJ2096001006) and Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (Grant No. RDG6130001).
References
- Aghajani Hamed, Hojjati Seyed Mohammad, Tajick-Ghanbari Mohammad Ali, Puormajidian Mohammad Reza, Borhani Ali. Molecular Identification of Ectomycorrhizal Fungal Communities Associated with Oriental Beech Trees (Fagus orientalis Lipsky) in Hyrcanian Forest of Iran. Iranian Journal of Science and Technology, Transactions A: Science. 2017;43(1):25–32. doi: 10.1007/s40995-017-0435-2. [DOI] [Google Scholar]
- Bazzicalupo A. L. Evaluating morphology and geographic range extent of genetically delimited species of mushrooms. The University of British Columbia; Vancouver: 2018. 1-247 [Google Scholar]
- Buyck B., Duhem B., Das K., Jayawardena R. S., Niveiro N., Pereira O. L., Prasher I. B., Adhikari S., Albertó E. O., Bulgakov T. S., Castañeda-Ruiz R. F., Hembrom M. E., Hyde K. D., Lewis D. P., Michlig A., Nuytinck J., Parihar A., Popoff O. F., Ramirez N. A., da Silva M., Verma R. K., Hofstetter V. Fungal biodiversity profiles 21-30. Cryptogamie Mycologie. 2017;38(1):101–146. doi: 10.7872/crym/v38.iss1.2017.101. [DOI] [Google Scholar]
- Carbone Ignazio, Kohn Linda M. A method for designing primer sets for speciation studies in filamentous Ascomycetes. Mycologia. 1999;91:553–556. doi: 10.2307/3761358. [DOI] [Google Scholar]
- Carrasco Jaime, Navarro María-Jesús, Gea Francisco J. Cobweb, a serious pathology in mushroom crops: A review. Spanish Journal of Agricultural Research. 2017;15(2) doi: 10.5424/sjar/2017152-10143. [DOI] [Google Scholar]
- Chen JD, Fu XH. The verticillium-like genera parasitic on mushrooms. Acta Mycologica Sinica. 1989;8:123–132. Chinese. [Google Scholar]
- Chen Z. H., Zhang P. Atlas of macrofungi in Hunan. Normal University Press; Changsha: 2019. 1-426. Chinese. [Google Scholar]
- Chernomor Olga, von Haeseler Arndt, Minh Bui Quang. Terrace Aware Data Structure for Phylogenomic Inference from Supermatrices. Systematic Biology. 2016;65(6):997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie Cameron R, Wong Bess, Stuart Alison E, Schultz Ted R, Rehner Stephen A, Mueller Ulrich G, Sung Gi-Ho, Spatafora Joseph W, Straus Neil A. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299:386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- Darriba Diego, Taboada Guillermo L, Doallo Ramón, Posada David. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9(8):772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]
- Eberhardt Ursula. Molecular kinship analyses of the agaricoid Russulaceae: Correspondence with mycorrhizal anatomy and sporocarp features in the genus Russula. Mycological Progress. 2002;1(2):201–223. doi: 10.1007/s11557-006-0019-6. [DOI] [Google Scholar]
- Fletcher J. T., Gaze R. H. Mushroom Pest and Disease Control: A Colour Handbook. Academic Press, San Diego. 2007 doi: 10.1201/b15139. [DOI]
- Fries EM. Systema Orbis Vegetabilis. Primas lineas novæ constructionis periclitatur EF Pars I. Plantæ Homonemeæ. Lundae : Typographia Academica; Lund: 1825. 369 [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Hoyos-Carvajal Lilliana, Orduz Sergio, Bissett John. Genetic and metabolic biodiversity of Trichoderma from Colombia and adjacent neotropic regions. Fungal Genetics and Biology. 2009;46:615–631. doi: 10.1016/j.fgb.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck John P., Rannala Bruce. Frequentist Properties of Bayesian Posterior Probabilities of Phylogenetic Trees Under Simple and Complex Substitution Models. Systematic Biology. 2004;53(6):904–913. doi: 10.1080/10635150490522629. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J. P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Hyde Kevin D., Xu Jianchu, Rapior Sylvie, Jeewon Rajesh, Lumyong Saisamorn, Niego Allen Grace T., Abeywickrama Pranami D., Aluthmuhandiram Janith V. S., Brahamanage Rashika S., Brooks Siraprapa, Chaiyasen Amornrat, Chethana K. W. Thilini, Chomnunti Putarak, Chepkirui Clara, Chuankid Boontiya, de Silva Nimali I., Doilom Mingkwan, Faulds Craig, Gentekaki Eleni, Gopalan Venkat, Kakumyan Pattana, Harishchandra Dulanjalee, Hemachandran Hridya, Hongsanan Sinang, Karunarathna Anuruddha, Karunarathna Samantha C., Khan Sehroon, Kumla Jaturong, Jayawardena Ruvishika S., Liu Jian-Kui, Liu Ningguo, Luangharn Thatsanee, Macabeo Allan Patrick G., Marasinghe Diana S., Meeks Dan, Mortimer Peter E., Mueller Peter, Nadir Sadia, Nataraja Karaba N., Nontachaiyapoom Sureeporn, O’Brien Meghan, Penkhrue Watsana, Phukhamsakda Chayanard, Ramanan Uma Shaanker, Rathnayaka Achala R., Sadaba Resurreccion B., Sandargo Birthe, Samarakoon Binu C., Tennakoon Danushka S., Siva Ramamoorthy, Sriprom Wasan, Suryanarayanan T. S., Sujarit Kanaporn, Suwannarach Nakarin, Suwunwong Thitipone, Thongbai Benjarong, Thongklang Naritsada, Wei Deping, Wijesinghe S. Nuwanthika, Winiski Jake, Yan Jiye, Yasanthika Erandi, Stadler Marc. The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Diversity. 2019;97:1–136. doi: 10.1007/s13225-019-00430-9. [DOI] [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, Bhat DJ, Jones EBG, Bundhun D, Chen YJ, Bao DF, Boonmee S, Calabon MS, Chaiwan N, Chethana KWT, Dai DQ, Dayarathne MC, Devadatha B, Dissanayake AJ, Dissanayake LS, Doilom M, Dong W, Fan XL, Goonasekara ID, Hongsanan S, Huang SK, Jayawardena RS, Jeewon R, Karunarathna A, Konta S, Kumar V, Lin CG, Liu JK, Liu NG, Luangsa-ard J, Lumyong S, Luo ZL, Marasinghe DS, McKenzie EHC, Niego AGT, Niranjan M, Perera RH, Phukhamsakda C, Rathnayaka AR, Samarakoon MC, Samarakoon SMBC, Sarma VV, Senanayake IC, Shang QJ, Stadler M, Tibpromma S, Wanasinghe DN, Wei DP, Wijayawardene NN, Xiao YP, Yang J, Zeng XY, Zhang SN, Xiang MM. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- Jaklitsch Walter M., Samuels Gary J., Dodd Sarah L., Lu Bing-Sheng, Druzhinina Irina S. Hypocrea rufa/Trichoderma viride: a reassessment, and description of five closely related species with and without warted conidia. Studies in Mycology. 2006;56:135–177. doi: 10.3114/sim.2006.56.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeewon R, Hyde K. D. Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere. 2016;7(11):1669–1677. doi: 10.5943/mycosphere/7/11/4. [DOI] [Google Scholar]
- Katoh Kazutaka, Kuma Kei-ichi, Toh Hiroyuki, Miyata Takashi. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic acids research. 2005;33:511–8. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Chang Sun, Jo Jong Won, Kwag Young-Nam, Sung Gi-Ho, Lee Sle-gee, Kim Sang-Yong, Shin Chang-Ho, Han Sang-Kuk. Mushroom Flora of Ulleung-gun and a Newly Recorded Bovista Species in the Republic of Korea. Mycobiology. 2015;43(3):239–257. doi: 10.5941/myco.2015.43.3.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornerup A., Wanscher J. H. Methuen Handbook of Colour, 3rd edn. London: Eyre Methuen; 1978. [Google Scholar]
- Kranabetter J. M., Friesen J., Gamiet S., Kroeger P. Epigeous fruiting bodies of ectomycorrhizal fungi as indicators of soil fertility and associated nitrogen status of boreal forests. Mycorrhiza. 2009;19(8):535–548. doi: 10.1007/s00572-009-0255-0. [DOI] [PubMed] [Google Scholar]
- Lebel Teresa, Tonkin Jennifer E. Australasian species of Macowanites are sequestrate species of Russula (Russulaceae, Basidiomycota) Australian Systematic Botany. 2007;20(4):355–381. doi: 10.1071/sb07007. [DOI] [Google Scholar]
- Lechat C, Gardiennet A, Fournier J. First report of a lichenicolous species of Hypomyces (Hypocreaceae), H. peltigericola sp. nov. Ascomycete. org. 2017;9:23–26. [Google Scholar]
- Li G. J., Deng C. Y., Shi L. Y., Wang J., Meng Q. F., Li S. M. Three new species of Russula subsect. Lactarioideae from China. Mycosystema. 2020;39(4):618–636. [Google Scholar]
- Liu X. L., Bau T., Wang X. H. Species diversity of Russula from the Greater and Lesser Hinggan Mountains in Northeast China. Mycosystema. 2017;36(10):1355–1368. [Google Scholar]
- Liu Y. J., Whelen S., Hall B. D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerse II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Looney Brian P., Ryberg Martin, Hampe Felix, Sánchez-García Marisol, Matheny P. Brandon. Into and out of the tropics: global diversification patterns in a hyperdiverse clade of ectomycorrhizal fungi. Molecular Ecology. 2016;25(2):630–647. doi: 10.1111/mec.13506. [DOI] [PubMed] [Google Scholar]
- Looney Brian P., Meidl Peter, Piatek Marek J., Miettinen Otto, Martin Francis M., Matheny P. Brandon, Labbé Jessy L. Russulaceae: a new genomic dataset to study ecosystem function and evolutionary diversification of ectomycorrhizal fungi with their tree associates. New Phytologist. 2018;218(1):54–65. doi: 10.1111/nph.15001. [DOI] [PubMed] [Google Scholar]
- Luo J, Zhuang WY. Re-examinations of the Hypomyces specimens on deposit in HMAS. Mycosystema. 2012;31:784–788. [Google Scholar]
- Ma J. Study on morphology and taxonomy of mycoparasite fungi. Jilin Agricultural University; Changchun: 2008. 73. Chinese. [Google Scholar]
- McKnight K. H., McKnight V. B. A field guide to mushrooms: North America. Vol. 34. Houghton Mifflin Harcourt.; 1997. [Google Scholar]
- Miller Steven L., Buyck Bart. Molecular phylogeny of the genus Russula in Europe with a comparison of modern infrageneric classifications. Mycological Research. 2002;106(3):259–276. doi: 10.1017/s0953756202005610. [DOI] [Google Scholar]
- Nguyen Lam-Tung, Schmidt Heiko A., von Haeseler Arndt, Minh Bui Quang. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Molecular Biology and Evolution. 2014;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto Alexander, Laub Annegret, Wendt Lucile, Porzel Andrea, Schmidt Jürgen, Palfner Götz, Becerra José, Krüger Dirk, Stadler Marc, Wessjohann Ludger, Westermann Bernhard, Arnold Norbert. Chilenopeptins A and B, peptaibols from the Chilean Sepedonium aff. chalcipori KSH 883. Journal of Natural Products. 2016;79:929–938. doi: 10.1021/acs.jnatprod.5b01018. [DOI] [PubMed] [Google Scholar]
- Palmer Jonathan M., Lindner Daniel L., Volk Thomas J. Ectomycorrhizal characterization of an American chestnut (Castanea dentata)-dominated community in Western Wisconsin. Mycorrhiza. 2008;19(1):27–36. doi: 10.1007/s00572-008-0200-7. [DOI] [PubMed] [Google Scholar]
- Park Myung Soo, Lee Hyun, Oh Seung-Yoon, Jung Paul Eunil, Seok Soon Ja, Fong Jonathan J., Lim Young Woon. Species delimitation of three species within the Russula subgenus Compacta in Korea: R. eccentrica, R. nigricans, and R. subnigricans. Journal of Microbiology. 2014;52(8):631–638. doi: 10.1007/s12275-014-4168-z. [DOI] [PubMed] [Google Scholar]
- Põldmaa K. A new species of Hypomyces and three of Cladobotryum from Estonia, with a discussion of their taxonomic position. Mycotaxon. 1996;59:389–405. [Google Scholar]
- Põldmaa Kadri, Larsson Ellen, Kõljalg Urmas. Phylogenetic relationships in Hypomyces and allied genera, with emphasis on species growing on wood-decaying homobasidiomycetes. Canadian Journal of Botany. 2000;77(12):1756–1768. doi: 10.1139/b99-148. [DOI] [Google Scholar]
- Põldmaa Kadri. Tropical species of Cladobotryum and Hypomyces producing red pigments. Studies in Mycology. 2011;68:1–34. doi: 10.3114/sim.2011.68.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehner Stephen A., Samuels Gary J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycological Research. 1994;98(6):625–634. doi: 10.1016/s0953-7562(09)80409-7. [DOI] [Google Scholar]
- Rehner S. A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1- sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97(1):84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- Rogerson Clark T., Samuels Gary J. Species of Hypomyces and Nectria occurring on Discomycetes. Mycologia. 1985;77(5) doi: 10.2307/3793285. [DOI] [Google Scholar]
- Rogerson Clark T., Samuels Gary J. Boleticolous species of Hypomyces. Mycologia. 1989;81(3) doi: 10.2307/3760079. [DOI] [Google Scholar]
- Rogerson Clark T., Samuels Gary J. Polyporicolous species of Hypomyces. Mycologia. 1993;85(2) doi: 10.2307/3760461. [DOI] [Google Scholar]
- Rogerson Clark T., Samuels Gary J. Agaricicolous species of Hypomyces. Mycologia. 1994;86(6) doi: 10.2307/3760597. [DOI] [Google Scholar]
- Ronquist Fredrik, Teslenko Maxim, van der Mark Paul, Ayres Daniel L., Darling Aaron, Höhna Sebastian, Larget Bret, Liu Liang, Suchard Marc A., Huelsenbeck John P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman Amy Y., Rogersson Clark T. A new species of Hypomyces (Hypocreaceae) with phragmosporous ascospores. Brittonia. 1981;33(3) doi: 10.2307/2806428. [DOI] [Google Scholar]
- Rossman AY, Samuels GJ, Rogerson CT, Lowen R. Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes) Studies in Mycology. 1999;42:1–260. [Google Scholar]
- Rossman Amy Y., Seifert Keith A., Samuels Gary J., Minnis Andrew M., Schroers Hans-Josef, Lombard Lorenzo, Crous Pedro W., Põldmaa Kadri, Cannon Paul F., Summerbell Richard C., Geiser David M., Zhuang Wen-ying, Hirooka Yuuri, Herrera Cesar, Salgado-Salazar Catalina, Chaverri Priscila. Genera in Bionectriaceae, Hypocreaceae, and Nectriaceae (Hypocreales) proposed for acceptance or rejection. IMA Fungus. 2013;4(1):41–51. doi: 10.5598/imafungus.2013.04.01.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels GJ, Doi Y, Rogerson CT. Hypocreales . Memoirs of the New York Botanical Garden. 1990;59:6–108. [Google Scholar]
- Sun Jing-Zu, Liu Xing-Zhong, McKenzie Eric H. C., Jeewon Rajesh, Liu Jian-Kui (Jack), Zhang Xiao-Ling, Zhao Qi, Hyde Kevin D. Fungicolous fungi: terminology, diversity, distribution, evolution, and species checklist. Fungal Diversity. 2019;95(1):337–430. doi: 10.1007/s13225-019-00422-9. [DOI] [Google Scholar]
- Sun JZ, Liu XZ, Jeewon R, Lin CG, Tian Q, Zhao Q, Xiao XP, Hyde KD, Nilthong S. Fifteen fungicolous Ascomycetes on edible and medicinal mushrooms in China and Thailand. Asian Journal of Mycology. 2019;2(1):129–169. doi: 10.5943/ajom/2/1/7. [DOI] [Google Scholar]
- Tamm Heidi, Põldmaa Kadri. Diversity, host associations, and phylogeography of temperate aurofusarin-producing Hypomyces / Cladobotryum including causal agents of cobweb disease of cultivated mushrooms. Fungal Biology. 2013;117(5):348–367. doi: 10.1016/j.funbio.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Teng SC. Notes on Hypocreales from China. Sinensia. 1934;4:269–298. [Google Scholar]
- Teng SC. Fungi of China. 1st Edition. Science Press; Beijing: 1963. 808. Chinese. [Google Scholar]
- Tulasne LR, Tulasne C. De quelques sphéries fongicoles, à propos d’un mémoire de M. Antoine de Bary sur les Nyctalis. Annales des Sciences Naturelles Botanique. 1860;13:5–19. [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology. 1990;172(8):4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D., Groenewald M., de Vries M., Gehrmann T., Stielow B., Eberhardt U., Al-Hatmi A., Groenewald J. Z., Cardinali G., Houbraken J., Boekhout T., Crous P. W., Robert V., Verkley G. J.M. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Studies in Mycology. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei IC, Kirschner R. Two fungicolous anamorphic species of Hypomyces s. lat. from Taiwan. Fungal Science. 2017;32:15–25. [Google Scholar]
- White T. J., Bruns T., Lee S., Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols. 1990:315–322. doi: 10.1016/b978-0-12-372180-8.50042-1. [DOI]
- Wijayawardene NN, Hyde KD, Tibpromma S, Wanasinghe DN, Thambugala KM, Tian Q, Wang Y. Towards incorporating asexual fungi in a natural classification: checklist and notes 2012–2016. Mycosphere. 2017;8:1457–1555. doi: 10.5943/mycosphere/8/9/10. [DOI] [Google Scholar]
- Wisitrassameewong Komsit, NUYTINCK JORINDE, HYDE KEVIN D, VERBEKEN ANNEMIEKE. Lactarius subgenus Russularia (Russulaceae) in Southeast Asia: 1. Species with very distant gills. Phytotaxa. 2014;158(1):23–42. doi: 10.11646/phytotaxa.158.1.2. [DOI] [Google Scholar]
- Yin J. H., Zhang P., Gong Q. F., Chen Z. H. Sequence analysis of the internal transcribed spacer of gene coding for rDNA in Russula subnigricans and R. nigricans. Mycosystema. 2008;27(2):237–242. In Chinese. [Google Scholar]
- Zare Rasoul, Gams Walter. More white verticillium-like anamorphs with erect conidiophores. Mycological Progress. 2016;15:993–1030. doi: 10.1007/s11557-016-1214-8. [DOI] [Google Scholar]
- Zeng ZQ, Zhuang WY. Current understanding of the genus Hypomyces (Hypocreales) Mycosystema. 2015;34:809–816. Chinese. [Google Scholar]
- Zeng ZQ, Zhuang WY. Three new species and two new Chinese records of Hypomyces (Hypocreales) Mycosystema. 2016;35:1048–1055. Chinese. [Google Scholar]
- Zeng ZQ, Wing XC, Zhuang WY. Three new Chinese records of Hypomyces (Hypocreales) Mycosystema. 2017;36:522–527. [Google Scholar]
- Zeng Zhao Qing, Zhuang Wen Ying. Two new species and a new Chinese record of Hypocreaceae as evidenced by morphological and molecular data. Mycobiology. 2019;47(3):280–291. doi: 10.1080/12298093.2019.1641062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Chunlan, Kakishima Makoto, Xu Jize, Wang Qi, Li Yu. The effect of Hypomyces perniciosus on the mycelia and basidiomes of Agaricus bisporus. Microbiology. 2017;163(9):1273–1282. doi: 10.1099/mic.0.000521. [DOI] [PubMed] [Google Scholar]
- Zhang C. L., Xu J. Z., Kakishima M., Li Y. First Report of Wet Bubble Disease Caused by Hypomyces perniciosus on Pleurotus citrinopileatus in China. Plant Disease. 2017;101(7):1321–1321. doi: 10.1094/pdis-02-17-0179-pdn. [DOI] [Google Scholar]
- Zhuang WY, Chen SL, Zeng ZQ, Zheng HD. A new species of Hypomyces (Hypocreales) on Schizophyllum sp. from China. Mycosystema. 2012;31:821–826. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hypomyces pseudolactifluorum sp. nov. (Hypocreales: Hypocreaceae) on Russula sp. from Yunnan, PR China
FENG-MING YU, RUVISHIKA S. JAYAWARDENA, JIAN-WEI LIU, KEVIN D. HYDE, QI ZHAO
Data type
word
Brief description
Sequence differences of ITS, LSU, TEF1-α and RPB2 genes between H. lactifluorum (TAAM 170476) and H. pseudolactifluorum sp. nov.. The locus’ numbers refer to the nucleotide positions of the gene sequences of H. lactifluorum from GenBank. Gap is replaced by ‘-’.
File: oo_438866.docx