Abstract
Purpose
Periodontitis is the leading cause of tooth loss. The role of long non-coding RNA (lncRNA) in periodontal inflammation remains unclear. The aim of this study was to investigate the role of lncRNA H19 in periodontitis and its possible regulation of autophagy in periodontitis.
Material and Methods
Inflammation level was determined by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and enzyme-linked immunosorbent assay (ELISA) in periodontal ligament cells (PDLCs). Western blotting, flow cytometric analysis, and immunofluorescence staining were used to detect the autophagy flux. Overexpression or knockdown of H19 was used to confirm its function. Ligature-induced periodontitis model in mice and periodontitis-affected human gingival tissue were used in vivo. RNA sequencing was performed to determine the differentially expressed genes.
Results
Autophagy was significantly increased in PDLCs after inflammatory stimulation as well as in a ligature-induced periodontitis model in mice and periodontitis-affected human gingival tissue. During the inflammatory process, H19 expression was also significantly upregulated. Further, the levels of autophagic markers were significantly upregulated after overexpressing H19 in PDLCs, and the increased autophagic activity induced by inflammatory stimulation was reversed by H19 knockdown. RNA sequencing showed that the expression profiles of mRNAs were significantly altered after H19 overexpression, and the differentially expressed genes were enriched in the PI3K/AKT signaling pathway, which was confirmed by the decreased p-AKT protein expression in the H19 overexpression group.
Conclusion
Periodontal inflammation activates autophagy flux, and H19 mediates the activation of autophagy via AKT pathway in periodontitis. This study expands our understanding of molecular regulation in periodontitis.
Keywords: autophagy, periodontitis, long non-coding RNA
Introduction
Periodontitis is a chronic inflammatory disease characterized by gingival swelling, bleeding, alveolar bone resorption, and tooth loosening.1 It is the leading cause of tooth loss in adults.2 Microbial infection is the main cause of periodontitis. The subsequent inflammation caused by microbial infection triggers the host response, and the excessive inflammation destroys the periodontal tissue.2 The periodontal ligament (PDL) is a connective tissue located between the alveolar bone and the tooth root that supports the teeth, buffers masticatory force, and contributes to tooth nutrition. Chronic inflammation leads to progressive damage of the PDL and gradually results in tooth loss. Thus, further investigation of the response of periodontal ligament cells (PDLCs) to inflammatory stimulation and the mechanisms involved is essential to develop effective therapeutic strategies for oral periodontitis.
Autophagy is an important mechanism of the conserved lysosomal degradation process, which maintains intracellular homeostasis and integrity.3 It is reported to be involved in starvation, hypoxia, infection, tumorigenesis, and stimulation.4 Appropriate autophagy removes damaged or useless organelles and maintains cellular energy and material levels. Previous studies have illustrated that autophagy is involved in inflammation and host immunity, and the interaction between autophagy and periodontitis has gradually attracted attention.5–7 Recent studies found that autophagy protects PDL stem cells from apoptosis and promotes angiogenesis in the inflammatory microenvironment.6,8 Autophagy helps eliminate periodontal pathogen infection and modulates the immune inflammatory response in periodontitis.9 However, the precise mechanism by which autophagy is regulated under inflammatory conditions remains unclear.
Long non-coding RNAs (lncRNAs), a class of non-protein-coding transcribed RNAs greater than 200 nucleotides in length, have been shown to regulate various cellular functions and processes.10 Many studies have demonstrated that lncRNAs participate in both biological and pathological control via regulation of autophagy-related DNA, RNA, and/or proteins.11–14 lncRNA H19 is a maternally expressed and paternally imprinted gene.15 Recently, H19 has been shown to play an important role in the regulation of autophagy in several kinds of diseases.16,17 One study reported that H19 promotes autophagy via the PI3K/AKT/mTOR pathway in insulin-like growth factor binding protein-associated protein-induced hepatic stellate cell activation,18 while another study showed that H19 inhibits autophagy by epigenetic silencing of DIRAS3 in diabetic cardiomyopathy.19 Hence, H19 shows dual effects on autophagy. At present, the role and function of H19 in periodontitis and whether H19 regulates autophagy in the inflammatory microenvironment remain unclear. Thus, this study aimed to investigate the role of H19 and its possible regulation of autophagy in periodontitis.
Materials and Methods
Cell Isolation and Culture
This study was approved by the Ethics Committee of the Peking University School of Stomatology (PKUSSIRB-2011007). First premolars extracted due to orthodontic reasons were collected from 3 individuals aged 20–30 years at the Department of Oral Maxilla-facial Surgery, Peking University School and Hospital of Stomatology. All three patients have provided written informed consents. The periodontal ligaments were gently scraped from the middle third surface of the root and digested in collagenase and trypsin (Gibco, Grand Island, NY, USA) for 60 min, then the isolated cells were cultured in α-modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Gibco) in a humidified atmosphere of 5% CO2 at 37°C. When the cell confluence reached 80% in 6-well culture dish, the cells were passaged and passage 4 were used for subsequent experiment.
Inflammatory Treatment
Tumor necrosis factor α (TNFα) and Porphyromonas gingivalis-derived lipopolysaccharide (LPS; Sigma-Aldrich, St. Louis, MO, USA) were used to create an inflammatory microenvironment in vitro. PDLCs were treated with TNFα (10 ng/mL and 50 ng/mL) or LPS (10 μg/mL) for 24 h according to previous study.5,6
Cell Transfection
The small interfering RNA (siRNA) against H19 (H19 siRNA: 5ʹ-CCC ACA ACA UGA AAG AAA UTT AUU UCU UUC AUG UUG UGG GTT-3ʹ) and the control siRNA (siNC: 5ʹ-ACG UGA CAC GUU CGG AGA ATT-3ʹ) were purchased from Gene Pharma (Suzhou, China). The full-length H19 (Gene Bank accession number: NR_002196.1) cDNA was cloned into the pcDNA3.1 vector as described previously.20 Lipofectamine 3000 was used as the transfection reagent. When the cell confluence reached 80%, the plasmids or siRNAs were transfected into PDLCs for 48 h according to the manufacturer’s instructions.
Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
RNA was extracted from cells and tissues using TRIzol reagent according to the manufacturer’s instructions. Then, the RNA was converted into cDNA using a cDNA Reverse Transcription Kit (Takara, Tokyo, Japan) under the following reaction conditions: 37°C for 15 min and 85°C for 5 s. qRT-PCR was performed using a SYBR Green PCR Master Mix in a quantitative PCR System (Applied Biosystems, Foster City, CA, USA) under the following conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous normalization control for mRNAs and lncRNAs. The primer sequences used in this study are listed in the Supplementary Table.
Enzyme-Linked Immunosorbent Assay (ELISA)
The protein level of TNFα and IL-6 in the cell-free supernatants were detected using ELISA kits (Tiangen Biotech Company, Beijing, China), according to the manufacturer’s instructions.
Western Blotting
Protein was extracted from PDLCs as described previously.21 The concentration of protein was determined using a Bicinchoninic Acid Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). The protein sample was separated via 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then electroblotted onto a polyvinylidene fluoride membrane. After blocking, the membranes were incubated with primary antibodies against LC3 (1:1000; Abcam, Cambridge, UK), Beclin-1 (1:1000; Abcam), phosphorylated-AKT (p-AKT; 1:1000; Abcam), AKT (AKT; 1:1000; Abcam), and GAPDH (1:2000; Abcam) at 4°C overnight. Then, membranes were washed and incubated with secondary antibodies (1:10,000; Zhongshan Golden Bridge, Beijing, China) at room temperature for 1 h. The protein strips were detected using an enhanced chemiluminescence kit (Applygen, Beijing, China). ImageJ software was used to quantify the strip intensities.
Flow Cytometric Analysis
The PDLCs were fixed with 4% formaldehyde for 10 min, permeabilized with 0.1% phosphate-buffered saline (PBS)-Triton X-100 for 15min, and incubated in 10% normal Goat serum followed by the antibody (Abcam), according to the manufacturer’s instructions. The Beclin-1 expression was detected using flow cytometric analysis as described previously.22
Immunofluorescence Staining
Cells seeded onto coverslips in 12-well plates were collected and fixed in 4% paraformaldehyde for 15 min. After blocking with goat serum, the fixed cells were incubated in anti-Beclin-1 primary antibody (1:200; Abcam) and anti-LC3 primary antibody (1:200; Abcam) at 4°C overnight, then further incubated in secondary antibody (1:10,000; Zhongshan Golden Bridge). The nuclei were stained with 4ʹ,6-diamidino-2-phenylindole. The fields in each group were randomly selected and captured using a confocal imaging system (Carl Zeiss, Jena, Germany).
RNA Sequencing
RNA (4 μg) extracted from PDLCs was treated with DNase as described previously.23 After removing the ribosomal RNA, the RNA library was prepared. RNA sequencing was performed on a HiSeq 2000 System (Illumina, San Diego, CA, USA). After the raw data were trimmed and filtered, the remaining data were then mapped to the human genome (hg19) using HISAT2. Differential gene expression was evaluated using the EBSeq package of the Bioconductor R program as previously described.24 Among the EBSeq results, the false discovery rate (FDR) was applied using the Benjamini–Hochberg algorithm based on the P-value. The differentially expressed genes were defined according to the following criteria: (1) fold change >1.5 or fold change <0.6667; and (2) FDR < 0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed using the Database for Annotation, Visualization and Integrated Discovery. The RNA sequence data have been uploaded into NCBI Sequence Read Archive with accession number GSE145112.
Ligature-Induced Periodontitis Model in Mice
All animal experimental protocols following the ARRIVE guidelines were approved by the Animal Care Committee of the Peking University School and Dental Hospital (LA2018305). Five BALB/c mice (male, 5–6 weeks old) used for the in vivo experiments were purchased from the Vital River Laboratory Animal Technology Company (Beijing, China). Mice were anesthetized and ligated with a 6–0 non-absorbable silk ligature around the neck of the left maxillary second molar. The right maxillary second molar was used as the control. The mice were sacrificed, and periodontal tissue samples around maxillary second molar from the inflammation side and the non-inflammation side were collected 7 days after ligation. For qRT-PCR analyses, the periodontal tissue samples were washed in sterile PBS and prepared using TissueLyser II for RNA extraction. For immunohistochemical staining, the tissues were fixed with 4% paraformaldehyde for 24 h and decalcified in 10% ethylenediaminetetraacetic acid buffer at room temperature for 5 weeks. All samples were embedded in paraffin blocks, and 4-μm sections were prepared.
Gingival Tissue with Periodontitis in Humans
The human study was approved by the Ethics Committee of the Peking University School of Stomatology (PKUSSIRB-201950166). All patients signed written informed consent forms. The gingival samples were collected from periodontal surgery performed in the Department of Periodontology, Peking University School and Hospital of Stomatology. The healthy periodontal tissue was obtained from crown-lengthening surgery in five patients without periodontitis (three females, two males; mean age: 33.2 years, standard deviation: 3.1). The inflammatory periodontal tissue was obtained from periodontal flap surgery in five patients with periodontitis (three females, two males; mean age: 35.7 years, standard deviation: 2.8). For patients with periodontitis, the periodontal flap surgery was performed when probing pocket depth > 5mm persisted 3 months after subgingival debridement. All subjects included in this study had no history of systematic disease, never smoke, and had no use of antibiotics or hormones in the previous 3 months. For qRT-PCR analyses, the gingival tissue samples were washed in sterile PBS, and prepared using TissueLyser II for RNA extraction. For immunohistochemical staining, the gingival tissue was fixed with 4% paraformaldehyde for 24 h, dehydrated, and cleared in xylene. Then, the samples were embedded in paraffin blocks, and 4-μm sections were prepared.
Immunohistochemistry
Immunohistochemical staining was performed as described previously.25 Briefly, the sections were deparaffinized and rehydrated. The sections were incubated with anti-LC3 antibody (1:200; Abcam) at 4°C overnight, then incubated with biotinylated goat immunoglobulin G for 30 min. Finally, the immunoreactivity was visualized using diaminobenzidine (Beyotime, Shanghai, China). The fields in mouse periodontitis model were located at maxillary second molar furcation. Regarding human gingival tissue, the fields representing the epithelium and connective tissue were randomly selected in each group. Images were taken using an Olympus BX light microscope with a photographic attachment (Olympus Optical, Tokyo, Japan). The integrated optical density (IOD) was measured to quantificationally analyze the LC3 staining using ImageJ software.
Statistics
All statistical analyses were performed using SPSS software (IBM, Armonk, NY, USA). All data are presented as the mean ± standard deviation. Student’s t-test was used to analyze the differences between two groups. One-way analysis of variance (ANOVA) was used for multiple group testing. When the ANOVA was significant, LSD Post Hoc test was further performed between groups. A P-value <0.05 was considered to indicate statistical significance.
Results
Inflammatory Stimulation Increased the Level of Autophagy in PDLCs
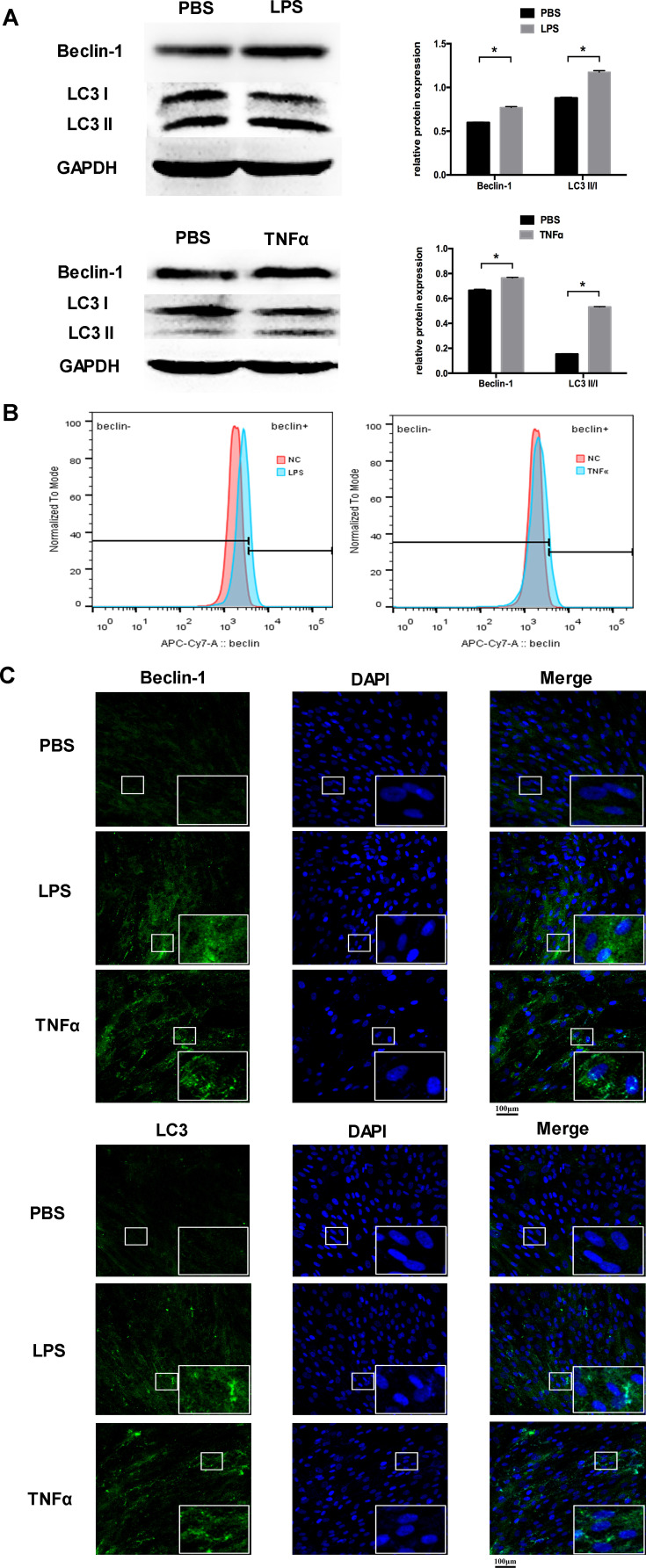
The level of autophagy in PDLCs was determined by Western blot analysis, flow cytometric analysis, and immunofluorescence staining. The expression of Beclin-1 and the ratio of LC3II/LC3I were selected as autophagic markers. In the both LPS and TNFα group, the protein expression of Beclin-1 and the ratio of LC3II/LC3I were both significantly upregulated compared to the control group (Figure 1A). These results were confirmed by flow cytometric analysis and immunofluorescence staining. The flow cytometric results of Beclin-1 demonstrated that inflammatory stimulation increased the level of autophagy in PDLCs (Figure 1B). After TNFα and LPS simulation, the staining of Beclin-1 was more extensive, and the LC3 punctate dots were significantly enhanced (Figure 1C).
Figure 1.
Inflammation increased the level of autophagy in PDLCs. Cells were treated with LPS or TNFα for 24 h. (A) Western blot analyses of Beclin-1 and LC3II/LC3I. Histograms show quantification of the band intensities. (B) The flow cytometric results of Beclin-1. (C) Images of Beclin-1 and LC3 immunofluorescence staining. Scale bar, 100μm. (Analysis of variance *P <0.05; n=9 replicates derived from three donors).
H19 Was Upregulated in an Inflammatory Microenvironment in PDLCs
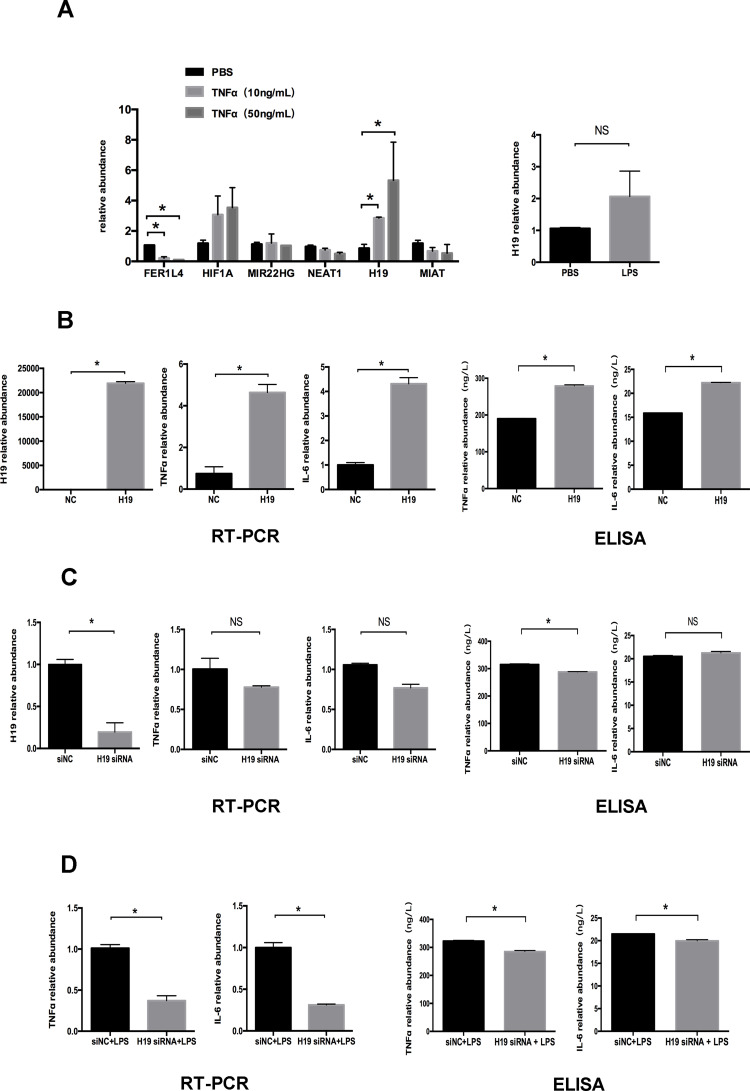
After 24 h treatment of PDLCs with 10 ng/mL and 50 ng/mL TNFα, the levels of six lncRNAs (FER1L4, HIF1A-AS1, MIR22HG, NEAT1, H19, and MIAT) supposed to be involved in the inflammatory process were detected by qRT-PCR. Inflammatory simulation distinctly increased H19 expression, which was more significant following treatment with 50 ng/mL TNFα. The increased expression of H19 under LPS simulation was further verified (Figure 2A). To further investigate the role of H19 in inflammation, we used the H19 plasmid to overexpress H19 in PDLCs, which increased H19 expression by ~22,000 fold. The mRNA expression of the inflammatory markers TNFα and IL-6 was increased significantly after H19 overexpression (Figure 2B). Furthermore, H19 siRNA was used to knock down the level of H19 in PDLCs. H19 expression decreased to ~20% after H19 knockdown. TNFα and IL-6 expression tended to decrease after H19 knockdown in non-inflammatory or inflammatory environment (Figure 2C and D). Then, we used ELISA to further determine the protein level of TNFα and IL-6 in the cell-free supernatants. The ELISA results were consistent with the qRT-PCR results (Figure 2B–D). The above results indicated a positive feedback loop between H19 and inflammation in PDLCs.
Figure 2.
H19 was upregulated under inflammation in PDLCs. Cells were transfected with H19 vector or H19 siRNA for 48 h to overexpress or knock down H19 expression. (A) Relative mRNA expression of six lncRNAs (FER1L4, HIF1A-AS1, MIR22HG, NEAT1, H19, and MIAT) after 24 h treatment of PDLCs with 10 ng/mL and 50 ng/mL TNFα, and relative RNA expression of H19 following 24 h treatment of PDLCs with LPS. (B) Overexpression efficiency of H19 vector and relative RNA and supernatant protein expression of TNFα, and IL-6 following overexpression of H19 in PDLCs. (C) Knockdown efficiency of H19 siRNA and relative RNA and supernatant protein expression of TNFα, and IL-6 after knocking down H19 in PDLCs. (D) Relative RNA and supernatant protein expression of TNFα and IL-6 in PDLCs under LPS stimulation with or without H19 knockdown (Analysis of variance *P <0.05; NS: non-significance, P >0.05; n=9 replicates derived from three donors).
H19 Promoted the Level of Autophagy in PDLCs
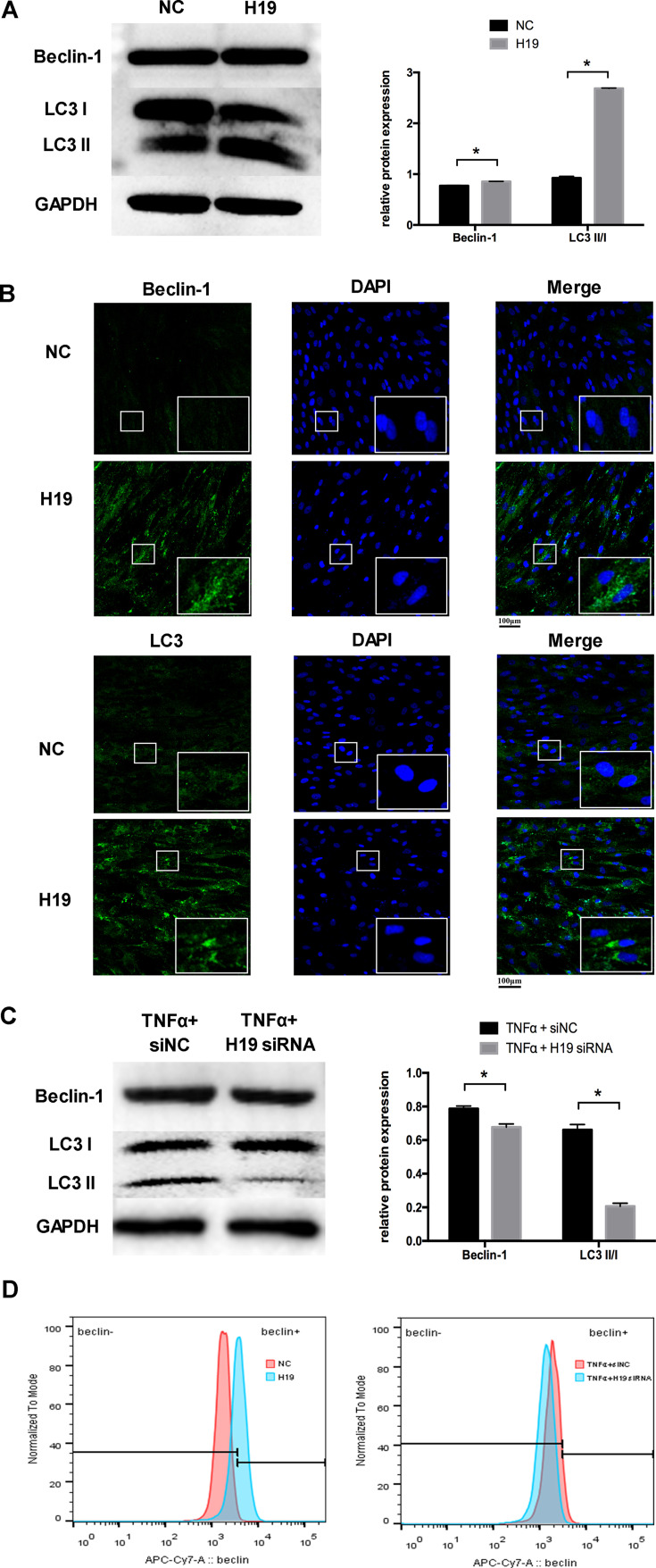
To investigate whether H19 regulates autophagy, we transfected the H19 plasmid into PDLCs. Beclin-1 and LC3II/LC3I were increased markedly in the H19 overexpression group (Figure 3A). Similar results were obtained using immunofluorescence staining. The staining of Beclin-1 and the formation of LC3 puncta were enhanced after overexpressing H19 in PDLCs (Figure 3B). To further investigate whether H19 mediates the increased autophagy induced by inflammation, we knocked down H19 expression in PDLCs under inflammatory stimulation. Western blot analysis showed that the increased ratio of LC3II/LC3I and Beclin-1 expression induced by inflammation were reversed by H19 knockdown (Figure 3C). The flow cytometric results of Beclin-1 further confirmed that H19 promoted the level of autophagy and H19 knockdown reversed the induced autophagy in PDLCs under inflammatory stimulation (Figure 3D).
Figure 3.
H19 promoted the level of autophagy in PDLCs. (A) Western blot analyses of Beclin-1 and LC3II/LC3I after PDLCs transfected with a vector expressing H19 for 48 h. Histograms show quantification of the band intensities. (B) Images of Beclin-1 and LC3 immunofluorescence staining after PDLCs transfected with a vector expressing H19 for 48 h. Scale bar, 100μm. (C) Western blot analyses of Beclin-1 and LC3II/LC3I in PDLCs treated with TNFα with or without H19 knockdown for 48 h. Histograms show quantification of the band intensities. (D) The flow cytometric results of Beclin-1 in H19 overexpression group and in H19 knockdown group with TNFα stimulation for 48 h. (Analysis of variance *P <0.05; n=9 replicates derived from three donors).
H19 Promoted the Level of Autophagy via the PI3K/AKT Signaling Pathway
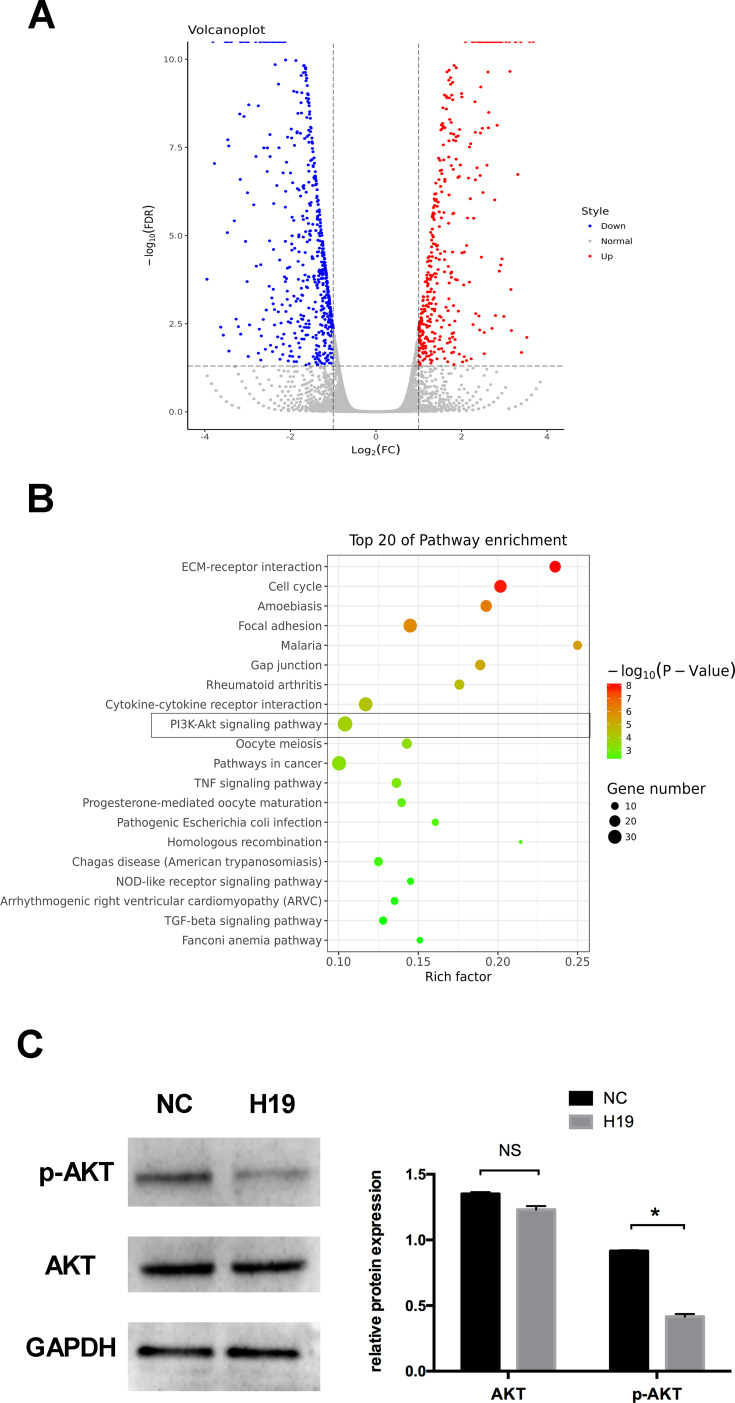
To preliminarily study the mechanism of H19 in the regulation of autophagy, RNA sequencing after transfection with the H19 vector was performed. The RNA sequencing data showed that 1120 mRNAs, including 431 upregulated and 689 downregulated mRNAs, were differentially expressed in the H19 overexpression group compared to the control group. The differential expression levels of mRNAs were visually expressed with a volcano plot (Figure 4A). To evaluate the function of differentially expressed genes, KEGG pathway analyses were performed. The data showed that the differentially expressed mRNAs were enriched in the PI3K/AKT signaling pathway, which is relevant to autophagy regulation (Figure 4B). Western blot analysis further showed that the p-AKT protein expression was significantly decreased in the H19 overexpression group (Figure 4C).
Figure 4.
RNA sequencing following transfection with H19 vector for 48 h. (A) Volcano plot of differentially expressed mRNAs in the control and H19 groups. Red points: upregulated mRNAs; blue points: downregulated mRNAs. (B) The top 20 enrichments in the KEGG pathway analysis of differentially expressed genes. (C) Western blot analyses of p-AKT and AKT after PDLCs transfected with H19 vector for 48 h. Histograms show quantification of the band intensities (Analysis of variance *P <0.05; NS: non-significance, P >0.05; n=9 replicates derived from three donors).
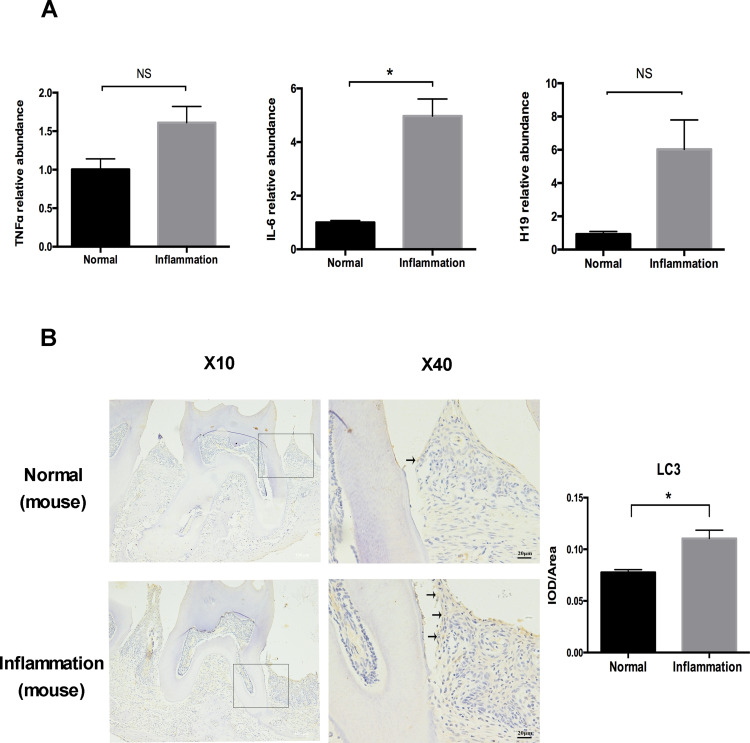
The Levels of Autophagy and H19 Were Increased in the Mouse Periodontitis Model
To further verify the participation of H19 in modulating autophagy under inflammation, we conducted a ligature-induced periodontitis model in mice. RNA was extracted from inflamed and healthy periodontal tissue. The levels of TNFα, IL-6, and H19 were increased in the inflammatory group compared to the healthy group (Figure 5A). The immunohistochemistry showed an increase of autophagy in the inflammatory group, as revealed by the enhanced staining of LC3 in the periodontal ligament tissue of the inflammatory group, whereas the area of staining was relatively narrow and the level of staining was mild in the healthy group. Quantitative analysis showed the LC3 staining in inflammatory group was significantly stronger than the healthy group (Figure 5B).
Figure 5.
The levels of autophagy and H19 were increased in a mouse periodontitis model. (A) Relative RNA expression of H19, TNFα, and IL-6 in the mouse periodontitis model. (B) Immunohistochemical staining of LC3 in the healthy and inflammatory groups. Black arrows identified the LC3 staining of periodontal ligament tissue of ligature-induced periodontitis mice. Quantitative analysis showed the LC3 staining in inflammation group was significantly stronger than the control group. (Analysis of variance *P <0.05; NS: non-significance, P >0.05; n=10 replicates derived from 5 mice in each group).
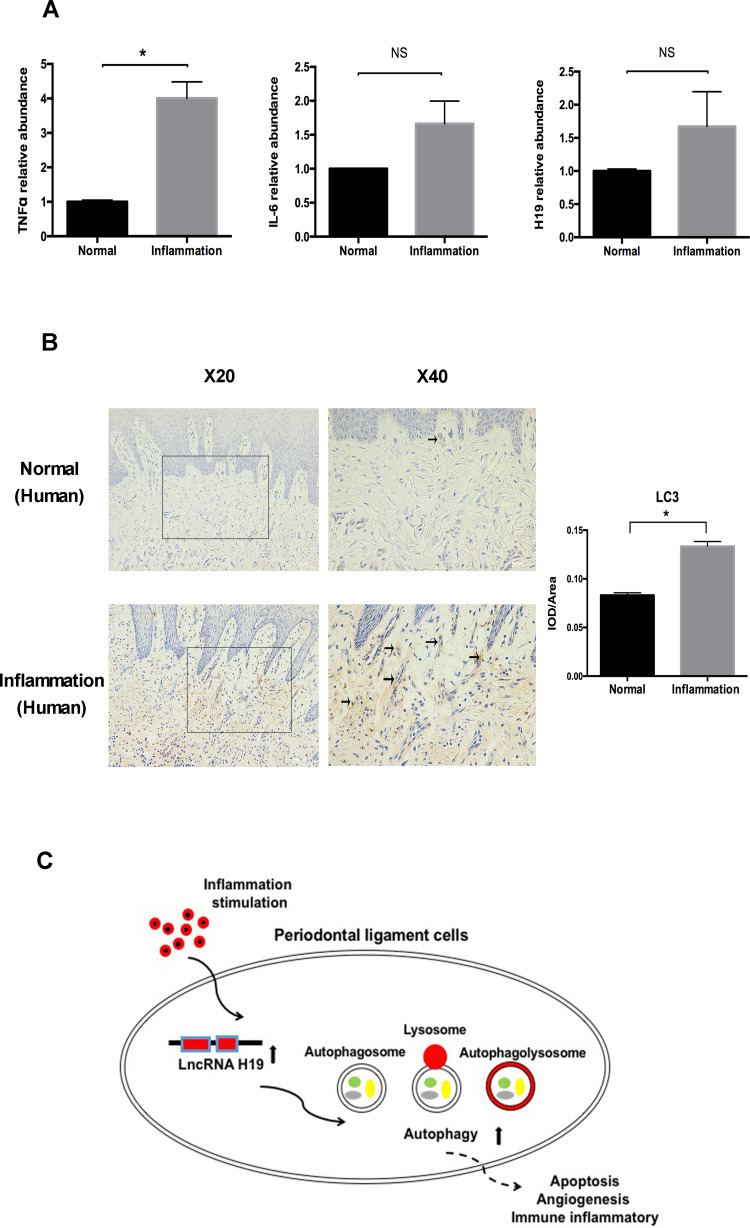
The Levels of Autophagy and H19 Were Increased in Periodontitis-Affected Human Gingival Tissue
The role of H19 was further determined in human gingival tissue with periodontitis. Similar to the results of the mouse model, the expression levels of TNFα, IL-6, and H19 were increased in the inflamed gingival tissue (Figure 6A). The level of autophagy in gingival tissue was tested by immunohistochemistry. Compared to the healthy group, the inflamed gingival tissue showed significantly enhanced staining of the LC3 in connective tissue (Figure 6B).
Figure 6.
The levels of autophagy and H19 were increased in periodontitis-affected human gingival tissue. (A) Relative RNA expression of H19, TNFα, and IL-6 in the human gingival tissue with or without periodontitis. (B) Immunohistochemical staining of LC3 in healthy and inflamed human gingival tissue. Black arrows identified the LC3 staining of gingival tissue of periodontitis patients. Quantitative analysis showed the LC3 staining in inflammation group was significantly stronger than the control group. (C) Schematic diagram showing the regulation of autophagy by H19 in PDLCs (Analysis of variance *P <0.05; NS: non-significance, P >0.05; n=10 replicates derived from 5 donors in each group).
Discussion
In this study, we found that autophagy was significantly increased under inflammatory conditions in periodontal cells and tissues. H19 mediated the autophagy induced by inflammatory stimulation, indicating the importance of H19 in the pathogenesis of periodontal disease.
Autophagy plays a distinct role in periodontitis.6,7 Autophagy maintains cellular homeostasis, but aberrant autophagy processes occur in certain diseases such as tumors and inflammatory diseases.26 In the present study, we selected Beclin-1 and LC3 as autophagy markers to monitor the autophagy process. Beclin-1 participates in autophagy as a major component of the initial formation of the autophagosomal membrane. Conversion of the soluble LC3 I to the vesicle-associated LC3 II is part of the late phase of autophagosomal membrane formation.27 We used two cytokines (TNFα and LPS) to induce inflammatory conditions in vitro, which were successfully and commonly applied to simulate the periodontal inflammatory microenvironment in previous studies.5,6 Interestingly, we found that autophagy was more sensitive to TNFα simulation compared to LPS. The highly expressed Beclin-1, the increased ratio of LC3 II/I in PDLCs under TNFα and LPS stimulation, and the enhanced staining of LC3 in the periodontitis model all indicated that the autophagy process was enhanced in periodontitis, which is consistent with previous studies.28,29 However, the exact function of autophagy in periodontitis remains controversial. In the early stage of periodontitis, autophagy might play a protective role, such as by protecting cells from apoptosis, promoting angiogenesis, and modulating the immune inflammatory response to suppress inflammation. However, this protective effect might function only in the early stage. One study found that autophagy was inhibited if inflammatory stimulation continued.6 In addition, autophagy could provide certain bacteria, such as P. gingivalis, with a route toward shelter from the immune system.30 Autophagy also induces the differentiation and proliferation of osteoclasts and modulates their function, resulting in alveolar bone resorption.31,32 The involvement of autophagy in periodontitis should be further studied.
lncRNAs modulate autophagy via a variety of mechanisms. To determine the role of lncRNAs in autophagy, we tested six candidate lncRNAs (FER1L4, HIF1A-AS1, MIR22HG, NEAT1, H19, and MIAT) supposed to be involved in the process of inflammation.19,33–37 Among them, H19, a conserved lncRNA, was the most differentially expressed. H19 has been reported to regulate autophagy in various types of cancer, and its aberrant expression was associated with tumorigenesis.38,39 The role of H19 in periodontitis has not been investigated previously. In the present study, we determined the expression of H19 in PDLCs under TNFα and LPS stimulation as well as in inflamed periodontal tissue. The expression of H19 was upregulated under inflammatory stimulation in vivo and in vitro, indicating that H19 is involved in the pathogenesis of periodontitis. While, the vivo experiment of human periodontal ligament would better connect with the cell culture data. Unfortunately, the periodontal ligament tissue of periodontitis patient is difficult to collect. Hence, we used the human gingival tissue in our study.
To further explore the possible effect of H19 on autophagy in PDLCs, we overexpressed and knocked down H19 in PDLCs under inflammatory conditions. Autophagy increased after overexpressing H19 in PDLCs, and the autophagy induced by inflammation was reversed by H19 knockdown. These results indicate that H19 participates in periodontitis by activating autophagy, consistent with several studies showing that H19 promotes the activation of autophagy in ischemic stroke, breast cancer, and liver fibrosis.18,40,41 Through mRNA sequencing, our data showed that the differentially expressed mRNAs were enriched in the PI3K/AKT signaling pathway after overexpressing H19 in PDLCs, which was further confirmed by the decreased p-AKT protein expression in the H19 overexpression group. The PI3K/AKT signaling pathway is a classic regulatory pathway for the autophagy process. A previous study demonstrated that H19 promotes autophagy in trophoblast cells via PI3K/AKT pathways.42 Our results indicated that H19 promote autophagy by inhibiting the PI3K/AKT signaling pathway in PDLCs. However, further studies are required to completely understand the interaction of H19 and PI3K/AKT signaling pathways in periodontitis.
Conclusion
Our study demonstrated that periodontal inflammation activated autophagy in vivo and in vitro, and H19 mediated the activation of autophagy via AKT signaling pathway in periodontitis (Figure 6C). These findings suggest a mechanism of autophagy regulation by H19 in the pathogenesis of periodontitis.
Acknowledgment
We thank Xiaowei Wu for his support in flow cytometric analysis.
Funding Statement
This work was supported by the National Natural Science Foundation of China, Beijing, China (No. 81670957, 81801010) and the Fund for Fostering Young Scholars of Peking University Health Science Center, Beijing, China (BMU2017PY022). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Abbreviations
lncRNA, long non-coding RNA; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; ELISA, enzyme-linked immunosorbent assay; PDLCs, periodontal ligament cells; PDL, periodontal ligament; TNFα, tumor necrosis factor α; LPS, Porphyromonas gingivalis-derived lipopolysaccharide; siRNA, small interfering RNA; p-AKT, phosphorylated-AKT; PBS, phosphate-buffered saline; FDR, false discovery rate; ANOVA, one-way analysis of variance.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Papapanou PN, Sanz M, Buduneli N, et al. Periodontitis: consensus report of workgroup 2 of the 2017 world workshop on the classification of periodontal and peri-Implant diseases and conditions. J Periodontol. 2018;89(Suppl 1):s173–s182. doi: 10.1002/JPER.17-0721 [DOI] [PubMed] [Google Scholar]
- 2.Könönen E, Gursoy M, Gursoy UK. Periodontitis: A multifaceted disease of tooth-supporting tissues. J Clin Med. 2019;8(8):8. doi: 10.3390/jcm8081135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Margeta M. Autophagy defects in skeletal myopathies. Annu Rev of Pathol. 2020;15:261–285. doi: 10.1146/annurev-pathmechdis-012419-032618 [DOI] [PubMed] [Google Scholar]
- 4.Tan YQ, Zhang J, Zhou G. Autophagy and its implication in human oral diseases. Autophagy. 2017;13(2):225–236. doi: 10.1080/15548627.2016.1234563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bullon P, Cordero MD, Quiles JL, et al. Autophagy in periodontitis patients and gingival fibroblasts: unraveling the link between chronic diseases and inflammation. BMC Med. 2012;10(1):122. doi: 10.1186/1741-7015-10-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.An Y, Liu W, Xue P, Zhang Y, Wang Q, Jin Y. Increased autophagy is required to protect periodontal ligament stem cells from apoptosis in inflammatory microenvironment. J Clin Periodontol. 2016;43(7):618–625. doi: 10.1111/jcpe.12549 [DOI] [PubMed] [Google Scholar]
- 7.Jiang M, Li Z, Zhu G. The role of autophagy in the pathogenesis of periodontal disease. Oral Dis. 2019;8(2):259–269. doi: 10.1111/odi.13045 [DOI] [PubMed] [Google Scholar]
- 8.Lorenzo-Pouso AI, Castelo-Baz P, Perez-Sayans M, Lim J, Leira Y. Autophagy in periodontal disease: evidence from a literature review. Arch Oral Biol. 2018;9:55–64. doi: 10.1016/j.archoralbio.2019.03.029 [DOI] [PubMed] [Google Scholar]
- 9.Rodrigues PH. Porphyromonas gingivalis and the autophagic pathway: an innate immune interaction? Frontiers in Bioscience. 2013;29(8):178. doi: 10.2741/2668 [DOI] [PubMed] [Google Scholar]
- 10.Chi Y, Wang D, Wang J, Yu W, Yang J. Long non-Coding RNA in the pathogenesis of cancers. Cells. 2019;8:1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barangi S, Hayes AW, Reiter R, Karimi G. The therapeutic role of long non-coding RNAs in human diseases: A focus on the recent insights into autophagy. Pharmacol Res. 2019;142:22–29. doi: 10.1016/j.phrs.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 12.Yin Q, Feng W, Shen X, Ju S. Regulatory effects of lncRNAs and miRNAs on autophagy in malignant tumorigenesis. Biosci Rep. 2018;38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang L, Wang H, Shen Q, Feng L, Jin H. Long non-coding RNAs involved in autophagy regulation. Cell Death Dis. 2017;8:e3073. doi: 10.1038/cddis.2017.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiong H, Ni Z, He J, et al. LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene. 2017;36(25):3528–3540. doi: 10.1038/onc.2016.521 [DOI] [PubMed] [Google Scholar]
- 15.Huang Y, Zheng Y, Jia L, Li W. Long Noncoding RNA H19 promotes osteoblast differentiation via TGF-β1/Smad3/HDAC signaling pathway by deriving miR-675. Stem Cells. 2015;33(12):3481–3492. doi: 10.1002/stem.2225 [DOI] [PubMed] [Google Scholar]
- 16.Wang M, Han D, Yuan Z, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9(12):1149. doi: 10.1038/s41419-018-1187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu TX, Chung HK, Xiao L, et al. Long noncoding RNA H19 impairs the intestinal barrier by suppressing autophagy and lowering paneth and goblet cell function. Cell Mol Gastroenterol Hepatol. 2019;9(4):611–625. doi: 10.1016/j.jcmgh.2019.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang TJ, Ren JJ, Zhang QQ, et al. IGFBPrP1 accelerates autophagy and activation of hepatic stellate cells via mutual regulation between H19 and PI3K/AKT/mTOR pathway. Biomed Pharmacother. 2019;116:109034. doi: 10.1016/j.biopha.2019.109034 [DOI] [PubMed] [Google Scholar]
- 19.Zhuo C, Jiang R, Lin X, Shao M. LncRNA H19 inhibits autophagy by epigenetically silencing of DIRAS3 in diabetic cardiomyopathy. Oncotarget. 2017;8(1):1429–1437. doi: 10.18632/oncotarget.13637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep. 2016;6(1):28897. doi: 10.1038/srep28897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiaobei L, Yan Z, Yan Z, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR-7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9(1):232. doi: 10.1186/s13287-018-0976-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y, Mo Z, Xue Z, Fang Y. Establish a flow cytometric method for quantitative detection of Beclin-1 expression. Cytotechnology. 2013;65(4):481–489. doi: 10.1007/s10616-012-9503-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng Y, Li X, Huang Y, Jia L, Li W. The Circular RNA landscape of periodontal ligament stem cells during osteogenesis. J Periodontol. 2017;1–17. [DOI] [PubMed] [Google Scholar]
- 24.Leng N, Dawson JA, Thomson JA, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–1043. doi: 10.1093/bioinformatics/btt087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schläfli AM, Berezowska S, Adams O, Langer R, Tschan MP. Reliable LC3 and p62 autophagy marker detection in formalin fixed paraffin embedded human tissue by immunohistochemistry. Eur J Histochem. 2015;59(2):2481. doi: 10.4081/ejh.2015.2481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie W, Zhou J. Aberrant regulation of autophagy in mammalian diseases. Biol Lett. 2018;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Deng X, Shi X, Dong X. Silencing H19 regulated proliferation, invasion, and autophagy in the placenta by targeting miR-18a-5p. J Cell Biochem. 2019;120(6):9006–9015. doi: 10.1002/jcb.28172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirasawa M, Kurita-Ochiai T. Porphyromonas gingivalis induces apoptosis and autophagy via ER stress in human umbilical vein endothelial cells. Mediators Inflamm. 2018;2018:1967506. doi: 10.1155/2018/1967506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park MH, Jeong SY, Na HS, Chung J. Porphyromonas gingivalis induces autophagy in THP-1-derived macrophages. Mol Oral Microbiol. 2017;32(1):48–59. doi: 10.1111/omi.12153 [DOI] [PubMed] [Google Scholar]
- 30.Belanger M, Rodrigues PH, Dunn WA Jr, Progulske-Fox A. Autophagy: a highway for Porphyromonas gingivalis in endothelial cells. Autophagy. 2006;2(3):165–170. doi: 10.4161/auto.2828 [DOI] [PubMed] [Google Scholar]
- 31.Chung YH, Jang Y, Choi B, et al. Beclin-1 is required for RANKL-induced osteoclast differentiation. J Cell Physiol. 2014;229(12):1963–1971. doi: 10.1002/jcp.24646 [DOI] [PubMed] [Google Scholar]
- 32.Chung YH, Yoon SY, Choi B, et al. Microtubule-associated protein light chain 3 regulates Cdc42-dependent actin ring formation in osteoclast. Int J Biochem Cell Biol. 2012;44(6):989–997. doi: 10.1016/j.biocel.2012.03.007 [DOI] [PubMed] [Google Scholar]
- 33.Huang Y, Han Y, Guo R, et al. Long non-coding RNA FER1L4 promotes osteogenic differentiation of human periodontal ligament stromal cells via miR-874-3p and vascular endothelial growth factor A. Stem Cell Res Ther. 2020;11(1):5. doi: 10.1186/s13287-019-1519-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruggi M, Layng FI, Lemos R Jr, et al. Absence of HIF1A leads to glycogen accumulation and an inflammatory response that enables pancreatic tumor growth. Cancer Res. 2019;79(22):5839–5848. doi: 10.1158/0008-5472.CAN-18-2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Ye S, Lu Y. Long non-coding RNA NEAT1 overexpression associates with increased exacerbation risk, severity, and inflammation, as well as decreased lung function through the interaction with microRNA-124 in asthma. J Clin Lab Ana. 2019;e23023. doi: 10.1002/jcla.23023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu X, Tan Z, Yang Y, Yang P. Long non-coding RNA MIR22HG inhibits cell proliferation and migration in cholangiocarcinoma by negatively regulating the Wnt/beta-catenin signaling pathway. J Gene Med. 2019;21(5):e3085. doi: 10.1002/jgm.3085 [DOI] [PubMed] [Google Scholar]
- 37.Sun G, Li Y, Ji Z. Up-regulation of MIAT aggravates the atherosclerotic damage in atherosclerosis mice through the activation of PI3K/Akt signaling pathway. Drug Deliv. 2019;26(1):641–649. doi: 10.1080/10717544.2019.1628116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Sun J, Yang F. The role of long non-coding RNA H19 in breast cancer. Oncol Lett. 2020;19:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bi Y, Fu Y, Wang S, Chen X, Schizandrin CX. A exerts anti-tumor effects on A375 cells by down-regulating H19. Braz J Med Biol Res. 2019;52(10):e8385. doi: 10.1590/1414-431X20198385 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Wang J, Cao B, Han D, Sun M, Feng J. Long non-coding RNA H19 induces cerebral ischemia reperfusion injury via activation of autophagy. Aging Dis. 2017;8(1):71–84. doi: 10.14336/AD.2016.0530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang J, Xie S, Yang J, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol. 2019;12(1):81. doi: 10.1186/s13045-019-0747-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu J, Xia Y, Zhang H, Guo H, Feng K, Zhang C. Overexpression of long non-coding RNA H19 promotes invasion and autophagy via the PI3K/AKT/mTOR pathways in trophoblast cells. Biomed Pharmacother. 2015;33:691–697. doi: 10.1016/j.biopha.2018.02.134 [DOI] [PubMed] [Google Scholar]