Abstract
5-fluorouracil (5-FU) has been shown to have suffered from resistance which demands a solution entailing the development of 5-FU analogues. Our study aims to design a number of analogues of 5-FU and evaluate their effectiveness against thymidylate synthase (TS) in silico compared to parent 5-FU with an effort to obtain better hit(s). All the molecules were optimized by molecular mechanics method utilizing MM2 forcefield parameters. Molecular docking of these molecules against TS was performed to catch on the binding strength of these molecules, determination of binding energy & interaction types of each ligand-target complex as well as subsequent analysis. PRA10 showed highest binding affinity (-9.1 Kcal/mol). Although binding energy of PRA6 & PRA14 are slightly lower than PRA10, they can be of special interest since they interact with crucial amino acids for binding and exhibit substantial non-bonded interactions. Residue analysis revealed that Arg50A, Arg175B, Arg215A, Ser216A, Arg176B and Asn226A of TS active site were crucial for binding/interaction. The best scored drug candidates demonstrated considerable pharmacokinetic as well as druglike properties. The present study also revealed that PRA6, PRA10 and PRA14 can be potential anticancer drugs for further development.
Keywords: Pharmaceutical chemistry, Theoretical chemistry, 5-Fluorouracil, Thymidylate synthase, Energy optimization, Docking, ADME
Pharmaceutical Chemistry, Theoretical Chemistry, 5-fluorouracil; Thymidylate synthase; Energy Optimization; Docking; ADME.
1. Introduction
Since the introduction of 5-Fluorouracil (5-FU) in 1957, it continues to be extensively utilized in the management of several common malignancies including cancer of the colon, breast and skin [1]. From the viewpoint of usage of chemotherapeutics in the treatment of solid tumors across the globe 5-FU holds the third position [2]. It is an organic aromatic heterocyclic compound resembling the construction similar to the pyrimidine nitrogen bases found in DNA and RNA [3].
5-Fluorouracil (5-FU) is a prodrug and hence requires metabolic activation through enzymatic reactions [1]. The most prominent mechanism of action suggests that 5-Fluorouracil (5-FU) is converted to 5-fluoro-2′-deoxyuridine-5′-monophosphate (FdUMP) which then forms a complex with the enzyme thymidylate synthase (TS). Due to the presence of a fluorine atom in the molecule, FdUMP binds more strongly to TS active site than dUMP. For the de novo synthesis of thymidylate, a crucial component of replication process, TS is essential. Such binding diminishes the DNA synthesis through a so-called “thymineless death” (Figure 1). Another mechanism suggests that 5-FU is transformed to 5-fluorouridine-5′- triphosphate (FUTP) which then incorporates itself into RNA culminating in interference with RNA processing and mRNA translation. An alternative mode of action proposes that 5-FU can integrates itself into the DNA by converting itself to 5-fluorodeoxyuridine-5′-triphosphate (FdUTP). Such incorporation knocks down DNA synthesis and function. In summary, the anticancer effects of 5-FU stems from the inhibition of thymidylate synthase (TS) and integration of its metabolites into RNA and DNA [4, 5, 6]
Figure 1.
A brief mechanism of the cytotoxic activity of 5-FU.
Throughout the past 20 years, a comprehensive understanding of the 5-FU mode of action has steered the development of strategies leading to its increased anticancer activity. Despite these advances, resistance to 5-FU remains a substantial impediment to its clinical use [7]. Many researchers have reported on the incidence and possible mechanism of 5-fu resistance. Causes of anti-cancer drug resistance include change in drug transportation through the cell membranes in either ways, augmentation of drug inactivation, mutation of the drug target leading to anomalous conformation and so on. Excessive expression of TS, amplified activity of deoxyuridine triphosphatase, MLH1 gene methylation, and overproduction of Bcl-2, Bcl-XL, and Mcl-1 proteins have all been linked to 5-FU resistance, pointing towards the involvement of multiple factors in 5-FU resistance rather than a single one [8].
Like all anticancer drugs, 5-FU leads to several untoward effects. The major toxic effect of 5-FU reported so far is myelotoxicity, particularly in patients who are given bolus doses whereas other toxicities like Hand-foot syndrome, stomatitis as well as neuro- and cardiotoxicities were found to be associated with continuous infusions. Other commonly observed adverse effects accompanying the 5-FU therapy include nausea, vomiting, diarrhoea, alopecia, and dermatitis. All these limitations clearly explain the rising demand for more effective and less toxic fluoropyrimidines. Such demand has pushed the recent research to focus on improving the therapeutic effectiveness of this long – used drug employed in the treatment of solid malignancies through biomodulation [9, 10]
In one study, researchers investigated the potentiality of anti-cancer activities of some novel co-crystals of 5-FU against human thymidylate synthase. As part of the in-silico investigation they have prepared those novel co-crystals and performed docking studies using the CDOCKER protocol in Discovery Studio Version 2.5 on human thymidylate synthase whose PDB ID was 1HVY. Their control ligand was dUMP, which is present as the natural ligand in the structure of the deposited protein model, 1HVY. The results of co-crystals were compared with dUMP [8]. Co-crystal 4 (5-Fluorouracil and 4, 4′-bipyridine in 1:1 stoichiometry) showed the strongest binding energy among other co-crystals. It was also highlighted that co-crystals mainly formed hydrogen bonds to stabilize the complex of co-crystal and TS.
To our knowledge, no extensive study was performed computationally in order to find out better novel 5-fluorouracil analogues with improved pharmacokinetic properties. Limitations of previous studies include using blind docking rather than targeted docking (where the active site is defined as the binding site), describing amino acid interactions involved in binding with TS and absence of pharmacokinetic studies in silico. Keeping these limitations in mind, the study was undertaken to address these issues and delve out the crucial interactions governing the action of these molecules while forming complexes with TS. The findings of this theoretical study will provide a basement for the ensuing experimental works towards obtaining a better drug candidate with better efficacy and lesser side effects.
2. Methodology
2.1. Preparation of ligands
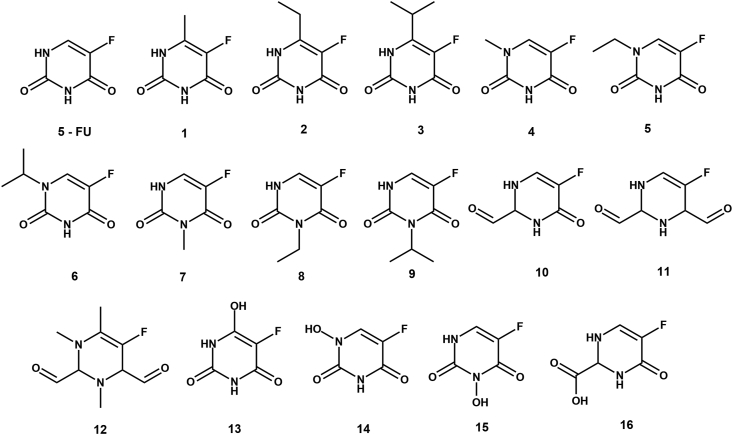
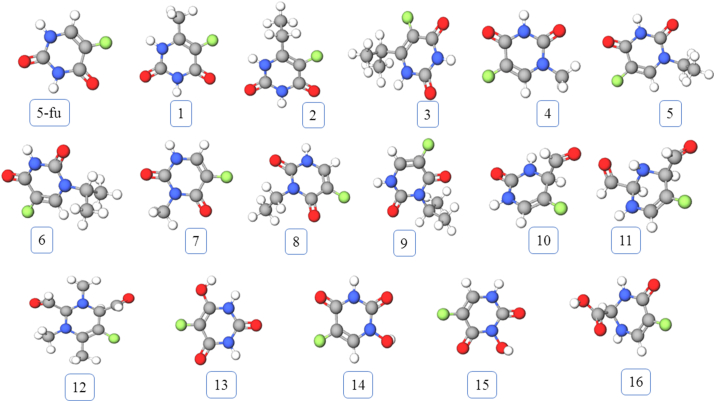
The 3D structures of 5-FU and its analogues were drawn using Chem3D Pro 12.0 software package at first (Figure 1). For our purpose we have constructed 16 analogues (Figure 2). According to the mechanism of the pharmacological action of 5-FU, it needs to be converted to 5-FdUMP where a monophosphorylated ribose gets attached to 5-FU. Keeping that in mind, we have fused each of our analogues and parent 5-FU with monophosphorylated ribose unit which we have termed as phosphoribosyl analogues (PRA) and 5-FdUMP respectively. Then the 3D structures of 5-FdUMP and all PRAs were subjected to energy minimization by molecular mechanics method using MM2 force field parameters, in order to get the most stable conformation (Figure 3). All energy minimizations were done using Chem3D Pro 12.0 software package. Such energy minimization treatment is essential to remove the impact of any possible unfavorable non-bonded interactions, bond lengths, bond angles, or torsional rotations.
Figure 2.
2D structures of 5-FU and it's analogues. All structures were drawn using ChemDraw 12.0 drawing tool.
Figure 3.
3D structures of 5-FU and it's analogues. All structures were generated using Molview, an open-Source web-application for virtual construction of molecular model.
2.2. Preparation of target
The crystal structure of human thymidylate synthase (PDB ID: 1HVY) was fetched from Protein Data Bank PDB. The rationale behind selecting this particular protein was the presence of dUMP in its structure which was found to be bound in its active site. The protein was prepared by expunging all the ligands and water molecules from the crystal structure. Only chain A & chain B were kept for further docking purpose. PyMOL (version 2.3) software package was utilized for preparing the target [11].
2.3. Docking
All the candidates were docked to thymidylate synthase utilizing AutoDock Vina platform [12, 13]. This program uses grids to pre-calculate the binding interactions at different positions within the binding site where values are stored in look-up tables and accessed automatically. Thus, binding energies for each pose can be obtained reasonably fast by adding up table entries. The grid box co-ordinate was set in a manner where the center of the box was X: -2.5397, Y: 3.3367, Z: 15.1541 and the dimension was X: 23.4558, Y: 25.0000, Z: 23.5748 to cover the binding pocket. AutoDock Vina employs flexible docking keeping the protein rigid while allowing torsional rotation for all rotatable bonds of optimized drug structures. PyMOL and Accelrys Discovery Studio 4.1 were used to investigate the pose of the conformer having the lowest binding free energy with the respective protein. This was done for confirming that ligands were docked at the proper binding pocket after docking [14].
2.4. Pharmacokinetics and druglikeness investigation
Pharmacokinetics and druglikeness parameters of the modified drug candidates were analyzed using online SwissADME server [15, 16]. Assessment of pharmacokinetic properties usually takes place earlier in the drug discovery process when a large library of compounds is at hand to be considered but entree to the physical samples is limited. This web tool computes important ADME-Tox and drug properties in order to assist hit selection prior to chemical synthesis.
3. Result & discussion
3.1. Binding energy of 5-FU analogues
In our study, 5-FdUMP was considered as the standard. Hence, the binding energy of 5-FdUMP was standard against which the binding affinity of all PRAs were compared. The binding energy is tabulated in Table 1. The binding energy difference of all PRAs from that of standard were calculated using the following formula:
| Binding energy difference between standard and PRA (BEDSA) = Binding energy of standard - Binding energy of PRA |
Table 1.
Binding affinity and Percent of resemblance of all 5-FU analogues compared to standard 5-FU.
| Candidate | Binding affinity | BEDSA | Percentage of resemblance |
|---|---|---|---|
| 5-FU | -8.1 | 0 | 100% |
| PRA1 | -8.6 | 0.5 | 88.89% |
| PRA2 | -8.5 | 0.4 | 88.89% |
| PRA3 | -8.2 | 0.1 | 66.67% |
| PRA4 | -8.8 | 0.7 | 88.89% |
| PRA5 | -8.8 | 0.7 | 88.89% |
| PRA6 | -8.8 | 0.7 | 88.89% |
| PRA7 | -8.4 | 0.3 | 88.89% |
| PRA8 | -8.4 | 0.3 | 100.00% |
| PRA9 | -8.8 | 0.7 | 88.89% |
| PRA10 | -9.1 | 1 | 66.67% |
| PRA11 | -7.7 | -0.4 | 66.67% |
| PRA12 | -8.4 | 0.3 | 66.67% |
| PRA13 | -8.3 | 0.2 | 66.67% |
| PRA14 | -8.9 | 0.8 | 88.89% |
| PRA15 | -8.3 | 0.2 | 100.00% |
| PRA16 | -8.1 | 0 | 66.67% |
After calculating binding energy differences of all PRAs, the values were utilized to generate a scatter plot (Figure 4). In brief, greater the value of BEDSA better is the binding affinity of PRA than standard. Strikingly, all the PRAs were found to be better bound to the target than the parent compound as revealed by the targeted docking except PRA11. Since, a positive value of BEDSA indicates better interaction than the standard we selected all PRAs other than PRA11 for further analysis. Since, PRA10 shows the highest BEDSA value, it was our automatic choice.
Figure 4.
Deviation of binding energy of phosphoribosyl analogues from that of 5-fdUMP. A downward curve from the baseline indicates poorer binding whereas an upward curve from the baseline indicates stronger binding.
In an attempt to find out which PRA(s) among these 14 chosen PRAs provide better activity, a binding interaction analysis was undertaken (Table 1). In order to facilitate the interaction study, another new formula was devised which is as follows.
| Percentage of resemblance = (no. of interacting amino acids in analogue that matches with the interacting amino acids in standard/no. of interacting amino acids in standard) × 100% |
Percent of resemblance indicates the relative similarity of binding of an analogue with respect to standard. It gives a measure of non-bonded interaction pattern. Higher value of resemblance indicates that PRA binds with similar type of amino acids which interacts with standard. Lower value of resemblance indicates that the analogue binds with different type of amino acids interacting with standard.
3.2. Binding interaction of 5-FU analogues
Phan et. al. reported that the binding site of thymidylate synthase within a 4 Å region of dUMP is composed of Arg50, Leu192, Cys195, His196, Gln214, Arg215, Ser216, Asn226, His256, and Tyr258 of chain A [17]. The crystal structure of thymidylate synthase retrieved from PDB reveals that residues that are important for binding are Arg50A, Arg175B, Arg215A, Ser216A, Arg176B and Asn226A. Our study revealed that most of our PRAs along with the standard interacted with these amino acids which indicates that these amino acids are also important in interacting with 5-FU analogues (Table 2 & Figure 5).
Table 2.
Interacting amino acids in TS found after docking of 5-FU PRAs.
| fdUMP | PRA1 | PRA2 | PRA3 | PRA4 | PRA5 | PRA6 | PRA7 | PRA8 | PRA9 | PRA10 | PRA11 | PRA12 | PRA13 | PRA14 | PRA15 | PRA16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARG50 | ARG175 | ARG50 | ARG50 | ARG50 | ARG50 | ARG50 | ARG175 | ARG175 | ARG176 | ARG175 | ARG215 | ARG50 | ALA312 | ARG175 | HIS256 | ARG175 |
| ARG175 | ARG176 | ARG175 | ARG175 | ARG175 | ARG175 | ARG175 | ARG176 | ARG176 | ARG215 | ARG176 | ARG50 | ARG175 | ARG175 | ARG176 | ARG175 | ARG176 |
| ARG176 | ARG215 | ARG176 | ARG176 | ARG176 | ARG176 | ARG215 | ARG215 | ARG215 | ARG50 | ARG215 | ASN226 | ARG176 | ARG176 | ARG215 | ARG176 | ARG215 |
| ARG215 | ARG50 | ARG215 | ARG215 | ARG215 | ARG215 | ASN226 | ASN226 | ARG50 | ASN226 | ASN226 | ASP218 | ARG215 | ARG215 | ARG50 | ARG215 | ARG50 |
| ASN226 | ASN226 | ASN226 | ASN226 | ASN226 | ASN226 | ASP218 | CYS195 | ASN226 | CYS195 | ASP218 | CYS195 | ASN226 | ARG50 | ASN226 | ARG50 | ASN226 |
| CYS195 | CYS195 | CYS195 | CYS195 | ASP218 | CYS195 | CYS195 | GLN214 | CYS195 | HIS196 | GLN214 | GLY222 | ASP218 | ASP218 | ASP218 | ASN226 | ASP218 |
| HIS196 | LEU192 | MET311 | MET311 | CYS195 | GLN214 | HIS196 | HIS196 | HIS196 | ILE108 | HIS256 | SER216 | GLN214 | MET311 | CYS195 | CYS195 | GLY222 |
| SER216 | SER216 | SER216 | TRP109 | SER216 | HIS196 | LEU192 | HIS256 | ILE108 | PHE225 | SER216 | TYR258 | MET311 | SER216 | SER216 | GLN214 | HIS256 |
| TYR258 | TRP109 | TRP109 | TYR258 | SER216 | SER216 | SER216 | SER216 | SER216 | TYR258 | TYR258 | TYR258 | TYR258 | HIS196 | SER216 | ||
| TYR258 | TYR258 | TRP109 | TYR258 | TRP109 | TRP109 | SER216 | ||||||||||
| TYR135 | TYR258 | TYR258 | TYR258 | |||||||||||||
| TYR258 |
Figure 5.
(A) Important amino acids for interaction with 5-FU PRAs. Note that Arg215 interacted with all 5FU analogues. (B), (C) & (D) Binding pose of selected hits PRA14, PRA10 and PRA6. PRAs are shown in ball and stick model while residues were shown in stick representation. Green dashed lines represent hydrogen bonds while others represent hydrophobic interactions except the red line which represents unfavourable interaction.
It is clearly visible from Figure 6 that PRAs interacted with polar residues predominantly. This is quite expected since the ribose and three consecutive phosphates build into the structures of PRAs made them polar in general. Furthermore, in terms of acidity/basicity, interacting residues were either neutral or basic. Acidic residues were involved in binding rarely which could be explained by presence of negatively charged oxygen atoms in phosphate moieties. Polar residues can be further considered to be of two types based on the charge. Figure 6 also shows us clearly that polar charged residues were mainly involved in binding. Contribution of alkyl- and sulfur containing residues were negligible, in general.
Figure 6.
Relative distribution of interacting residues (A, B, C) and bond types (D) are shown in this figure. Section A depicts the frequency of interacting residues based on their polarity for each ligand. Section B & C describes the occurrence of residues based on their pH and side chain respectively. Section D picturizes an overall contribution of different types of non-bonded interactions for all ligands.
PRA6 can be of special interest since it's binding affinity is -8.8 Kcal/mol and percentage of resemblance is 88.89%. Besides that, it interacts with 12 amino acid residues while most of other PRAs didn't cross 11 residues. PRA6 exhibits 12 hydrogen bonds, 3 hydrophobic bonds, 1 halogen bond and one electrostatic interaction (Table 3). In addition, it interacted with Arg50A, Arg175B, Arg215A and Asn226A which were important amino acids for interaction.
Table 3.
Distribution of bond types. All blank spaces correspond to “0”.
| Drug candidate | Hydrogen bond | Hydrophobic | Halogen | Others |
|---|---|---|---|---|
| FdUMP | 8 | 2 | 1 | |
| PRA 1 | 10 | 4 | ||
| PRA 2 | 13 | 3 | ||
| PRA 3 | 10 | 4 | ||
| PRA 4 | 14 | 1 | ||
| PRA 5 | 11 | 1 | ||
| PRA 6 | 12 | 3 | 2 | 1 |
| PRA 7 | 12 | 1 | 3 | |
| PRA 8 | 9 | 4 | 2 | |
| PRA 9 | 8 | 5 | 2 | |
| PRA 10 | 13 | |||
| PRA 11 | 10 | 1 | ||
| PRA 12 | 10 | 1 | ||
| PRA 13 | 11 | 1 | 1 | |
| PRA 14 | 14 | 1 | ||
| PRA 15 | 12 | 1 | 3 | |
| PRA 16 | 11 |
Another candidate, PRA14 can also be a potential hit since its binding affinity holds the second position (-8.9 Kcal/mol) and percentage of resemblance is 88.89%. Besides that, it interacted with 11 amino acid residues and exhibits 12 hydrogen bonds, 1 hydrophobic bond and 3 halogen bonds (Table 3).
3.3. Analysis of non-bonded interaction types
Among all non-covalent interactions, hydrogen bond is considered to be the strongest although its binding strength may vary from 01-40 Kcal/mol. Hydrogen bond plays crucial in case of drug-receptor interaction in many instances. Increased number of hydrogen bonds corresponds to better and strong binding, especially for binding sites composed of polar residues. The active site of TS is composed of amino acid residues which are mostly polar in nature. Therefore, we expected that our designed PRAs will exhibit a substantial number of hydrogen bonds after binding with TS active site. According to our anticipation, hydrogen bonds were formed to a greater proportion for all PRAs and FdUMP when compared with other types of non-bonded interactions (Table 3). This explains why most PRAs exhibited greater binding strength than the parent one. Most PRAs presented greater number of hydrogen bonds than FdUMP which explains the reason of enhanced activity conferred by our substitutions. Besides hydrogen bond, other types of non – covalent interactions were not significant in stabilizing PRA – TS complexes.
3.4. Pharmacokinetic and druglikeness analysis
The ADME study of 5-FU and its analogues was performed using SwissADME web service. GI absorption of drug becomes extremely important when the drug is intended to be administered via oral route. According to this study, all analogues were expected to be rapidly absorbed from GI tract. However, since these molecules won't be able to distinguish between normal cell and cancer cell, they should be reserved for parenteral administration unless modified suitably. BBB permeation determines whether a molecule will exert its action, either beneficial or detrimental, on brain. Crossing of conventional small anticancer molecules through the BBB can cause significant destruction of the neurons in brain producing serious neuro-consequence [18]. It is noteworthy that after administration, all PRAs, except PRA6 and PRA9, are expected to produce no neurotoxicity, at least not in brain, since they will not cross the BBB (Figure 7 and Table – S1). The degree of skin permeation can be evaluated from Log Kp value. A higher value Log Kp indicates better skin permeation and vice versa (Table 4 and Table – S1). All molecules under the study will not penetrate the skin layer very easily which means that these molecules will produce little or no reduced skin toxicity.
Figure 7.
BOILED-egg representation of chosen hits along with the parent drug. Molecules that lie within the egg yolk are predicted to cross blood brain barrier while molecules lying in the egg white indicates intestinal absorption. Red circle means that the drug is not a substrate for P-gp.
Table 4.
Physicochemical, Pharmacokinetic & druglike properties of selected hits. MW = molecular weight, HA = No. of heavy atoms, RB = No. of rotatable bonds, HBA = No. of hydrogen bond acceptor, HBD = No. of hydrogen bond donor, TPSA = Total polar surface area.
| PRA6 | PRA10 | PRA14 | ||
|---|---|---|---|---|
| Physicochemical parameter | Formula | C7H9FN2O2 | C5H5FN2O2 | C4H3FN2O3 |
| MW | 172.16 | 144.1 | 146.08 | |
| HA | 6 | 0 | 6 | |
| RB | 1 | 1 | 0 | |
| HBA | 3 | 3 | 4 | |
| HBD | 1 | 2 | 2 | |
| TPSA(Å2) | 54.86 | 58.2 | 75.09 | |
| Pharmacokinetic parameter | Lipophilicity | -0.93 | -0.51 | -0.15 |
| GI absorption | High | High | High | |
| BBB permeant | Yes | No | No | |
| P-gp substrate | No | No | No | |
| Skin permeation (logKp) | -7.12 | -7.53 | -7.88 | |
| Druglike property | Lipinski | Yes | Yes | Yes |
| Bioavailability Score | 0.55 | 0.55 | 0.55 | |
| Synthetic accessibility | 1.88 | 3.32 | 2.27 |
P-gp, a crucial member of ABC transporter family, is involved in the active efflux of many drug molecules from the cell. Overexpression of P-gp occurs in some tumor cells leading to multidrug-resistant cancers [19]. No molecules were found to be a substrate of P-gp indicating that these molecules are not susceptible to such resistance. All candidates passed Lipinski's filter [20] which indicates that, qualitatively, they bear the chance to become an oral drug with respect to bioavailability. Martin et. al. described a bioavailability scoring system to forecast the likelihood of a molecule to exhibit at least 10% oral bioavailability in rat or measurable Caco-2 permeability [21]. According to this scoring system, all candidates were moderately orally bioavailable. Lastly, the chemical synthesis of these compounds was graded as “easy” meaning that they will not be very difficult to synthesize in the laboratory as signposted by Synthetic Accessibility Score (Table 4 and Table – S1).
4. Conclusion
In this work the interaction of some anticancer drug candidates with thymidylate synthase has been investigated using molecular docking method. Modified derivatives PRA6, PRA10 and PRA14 exhibited stronger binding affinity against the target protein primarily through hydrogen bonding along with other non-bonding interactions. Important amino acid residues for interaction with modified drug candidates were Arg50A, Arg175B, Arg215A, Ser216A, Arg176B and Asn226A among which Arg215A was the most important. According to this study, all analogues were expected to be rapidly absorbed from GI tract, cross BBB, escape P-gp (except PRA6 and PRA9) and produce no skin toxicity. The present study exposes that PRA10 and PRA14 can be potential anticancer drugs for further development. Therefore, these analogues should be selected for subsequent synthesis. Synthetic Accessibility Score pointed out the relative ease of synthesis of the chosen analogues. Simple reactions (e.g. aromatic alkylation) are required to prepare the analogues while using 5 – FU as the starting material which is quite available and cheap.
The idea of computational design of drugs came into being with the aim to substantially reduce the experimental time and cost. Our study shared the same theme where the aim was to find a better candidate than the existing one i.e. 5 – FU. Due to resource limitation, advanced computational analysis (e.g. observing the mobile behaviour of drug – protein complex, membrane permeability etc.) were not possible to be introduced in our work. The computational findings of our results will be a guide for medicinal chemists interested in the experimental validation of our findings.
Declarations
Author contribution statement
Surid Mohammad Chowdhury: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Md. Rajdoula Rafe: Analyzed and interpreted the data; Wrote the paper.
Md. Nuruzzaman Hossain: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Diasio R.B., Harris B.E. Clinical pharmacology of 5-fluorouracil. Clin. Pharmacokinet. 1989;16:215–237. doi: 10.2165/00003088-198916040-00002. [DOI] [PubMed] [Google Scholar]
- 2.Sorrentino M.F., Kim J., Foderaro A.E., Truesdell A.G. 5-fluorouracil induced cardiotoxicity: review of the literature. Cardiol. J. 2012;19:453–457. doi: 10.5603/cj.2012.0084. [DOI] [PubMed] [Google Scholar]
- 3.Rutman R.J., Cantarow A., Paschkis K.E. Studies in 2-acetylaminofluorene carcinogenesis: III. The utilization of uracil-2-C14 by preneoplastic rat liver and rat hepatoma. Canc. Res. 1954;14:119–123. [PubMed] [Google Scholar]
- 4.Thomas D.M., Zalcberg J.R. 5-Fluorouracil: a pharmacological paradigm in the use of cytotoxics. Clin. Exp. Pharmacol. Physiol. 1998;25:887–895. doi: 10.1111/j.1440-1681.1998.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 5.Chernyshev A., Fleischmann T., Kohen A. Thymidyl biosynthesis enzymes as antibiotic targets. Appl. Microbiol. Biotechnol. 2007;74:282–289. doi: 10.1007/s00253-006-0763-1. [DOI] [PubMed] [Google Scholar]
- 6.Ceilley R.I. Mechanisms of action of topical 5-fluorouracil: review and implications for the treatment of dermatological disorders. J. Dermatol. Treat. 2012;23:83–89. doi: 10.3109/09546634.2010.507704. [DOI] [PubMed] [Google Scholar]
- 7.Longley D.B., Harkin D.P., Johnston P.G. 5-fluorouracil: mechanisms of action and clinical strategies. Nat. Rev. Canc. 2003;3:330. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 8.Nadzri N.I., Sabri N.H., Lee V.S., Halim S.N.A. 5-fluorouracil co-crystals and their potential anti-cancer activities calculated by molecular docking studies. J. Chem. Crystallogr. 2016;46:144–154. [Google Scholar]
- 9.Malet-Martino M., Martino R. Oncologist clinical studies of three oral prodrugs of 5-fluorouracil. Oncol. 2002;7:288–323. doi: 10.1634/theoncologist.7-4-288. [DOI] [PubMed] [Google Scholar]
- 10.Focaccetti C., Bruno A., Magnani E., Bartolini D., Principi E., Dallaglio K., Bucci E.O., Finzi G., Sessa F., Noonan D.M. Effects of 5-fluorouracil on morphology, cell cycle, proliferation, apoptosis, autophagy and ROS production in endothelial cells and cardiomyocytes. PloS One. 2015;10 doi: 10.1371/journal.pone.0115686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrodinger L.L.C. 2008. The PyMOL Molecular Graphics System Version 1.2 R3pre. [Google Scholar]
- 12.Trott O., Olson A.J., Vina AutoDock. Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallakyan S., Olson A.J. Chem. Biol. Springer; 2015. Small-molecule library screening by docking with PyRx; pp. 243–250. [DOI] [PubMed] [Google Scholar]
- 14.Biovia D.S. 2017. Discovery Studio Visualizer, San Diego, CA, USA. [Google Scholar]
- 15.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017;7:42717. doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daina A., Zoete V. A boiled-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11:1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phan J., Koli S., Minor W., Dunlap R.B., Berger S.H., Lebioda L. Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry. 2001;40:1897–1902. doi: 10.1021/bi002413i. [DOI] [PubMed] [Google Scholar]
- 18.Pardridge W.M. The blood-brain barrier: bottleneck in brain drug development. NeuroRx. 2005;2:3–14. doi: 10.1602/neurorx.2.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharom F.J. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9(1):105–127. doi: 10.2217/14622416.9.1.105. [DOI] [PubMed] [Google Scholar]
- 20.Lipinski C.A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Martin Y.C. A bioavailability score. J. Med. Chem. 2005;48(9):3164–3170. doi: 10.1021/jm0492002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.