Abstract
Background and purpose
Hypoxia Positron-Emission-Tomography (PET) as well as Computed Tomography (CT) radiomics have been shown to be prognostic for radiotherapy outcome. Here, we investigate the stratification potential of CT-radiomics in head and neck cancer (HNC) patients and test if CT-radiomics is a surrogate predictor for hypoxia as identified by PET.
Materials and methods
Two independent cohorts of HNC patients were used for model development and validation, HN1 (n = 149) and HN2 (n = 47). The training set HN1 consisted of native planning CT data whereas for the validation cohort HN2 also hypoxia PET/CT data was acquired using [18F]-Fluoromisonidazole (FMISO). Machine learning algorithms including feature engineering and classifier selection were trained for two-year loco-regional control (LRC) to create optimal CT-radiomics signatures.
Secondly, a pre-defined [18F]FMISO-PET tumour-to-muscle-ratio (TMRpeak ≥ 1.6) was used for LRC prediction. Comparison between risk groups identified by CT-radiomics or [18F]FMISO-PET was performed using area-under–the-curve (AUC) and Kaplan-Meier analysis including log-rank test.
Results
The best performing CT-radiomics signature included two features with nearest-neighbour classification (AUC = 0.76 ± 0.09), whereas AUC was 0.59 for external validation. In contrast, [18F]FMISO TMRpeak reached an AUC of 0.66 in HN2. Kaplan-Meier analysis of the independent validation cohort HN2 did not confirm the prognostic value of CT-radiomics (p = 0.18), whereas for [18F]FMISO-PET significant differences were observed (p = 0.02).
Conclusions
No direct correlation of patient stratification using [18F]FMISO-PET or CT-radiomics was found in this study. Risk groups identified by CT-radiomics or hypoxia PET showed only poor overlap. Direct assessment of tumour hypoxia using PET seems to be more powerful to stratify HNC patients.
Keywords: Radiomics, PET-Imaging, Quantitative Imaging, CT-Imaging, Machine Learning, Imaging biomarkers
1. Introduction
Tumour hypoxia has been shown to be prognostic for poor outcome after chemoradiotherapy (CRT) in head and neck squamous cell carcinoma (HNSCC) by several studies [1], [2], [3], [4], [5]. In addition, also biological heterogeneity as identified by radiomics analyses based on computed tomography (CT) and others factors have also been linked to poor outcome after chemoradiation therapy [6], [7], [8]. Hypoxia can be measured invasively using probes, or assessed non-invasively using specific radiotracers in positron emission tomography (PET) imaging [9]. In clinical research, the most commonly used hypoxia PET tracer is [18F]FMISO [10]. Tumour-to-muscle ratio (TMR) is a simple but very robust metric that has been used in many studies to derive the hypoxic status from PET data, though other methods exist as well [9], [10], [11]. Different studies have shown that TMR assessed 2 to 4 h after tracer injection enables differentiation between hypoxic and normoxic tumours based on a threshold value (e.g. TMRpeak ≥ 1.4) [3], [12], and consequently to distinguish patients at increased risk of loco-regional failure (LRF) at different time points of CRT [3], [4], [5].
Radiomics, which is a technique for quantitative analysis of medical images, hypothesises that imaging features capture anatomical or functional tumour heterogeneity in solid tumours without the need for additional diagnostic interventions such as biopsies [6]. Different research teams have shown not only a significant prognostic power of radiomics features and signatures in the task of patient stratification for LRF in patients after CRT but also correlations with gene expression in different forms of cancer [7], [13], [14], [15], [16], [17]. Therefore, some authors hypothesised that CT radiomics captures tissue heterogeneity caused by tumour hypoxia [6], [7], [18], [19].
Consequently, CT radiomics might be able to identify similar risk groups of patients compared to [18F]FMISO PET. Since hypoxia PET requires non-standard tracers, long examination times and complex data post-processing it is only available at a small number of academic institutions. It might therefore by very attractive to identify high-risk patient subgroups using CT radiomics instead of [18F]FMISO PET imaging for patient stratification and outcome prediction after CRT.
Therefore, the hypothesis of the current study was that an independently trained CT radiomics model might serve as surrogate for hypoxia PET imaging to stratify patients into risk groups according to outcome after RCT of HNSCC. Ideally, a CT radiomics signature might be able to capture similar risk profiles as hypoxia imaging using [18F]FMISO PET. To investigate this hypothesis, the aim of this study was to first develop a CT radiomics model based on n = 149 HNSCC and subsequent validation with an independent, bi-institutional data set of n = 47 patients for whom [18F]FMISO PET data were also available to compare the potential of CT radiomics versus [18F]FMISO PET imaging for patient stratification.
2. Material and methods
2.1. Patient data
The data set consisted of 196 patients in total with HNSCC in advanced stages scheduled for definitive CRT who had been recruited in a period of 10 years (from 2005 to 2015) at the University Hospital Tübingen (UHT, n = 171) and the University Hospital Dresden (UHD, n = 25). This study represents a secondary analysis of data collected within two different clinical trials, approved by the respective local ethics committees (NCT00180180, NCT02552792).
The patient cohort consisted of two distinct groups: HN1 and HN2. For HN1 (n = 149 all from UHT) only native radiotherapy (RT) planning CT data with delineations of gross tumour volumes (GTV) by an experienced radiation oncologist were available. For HN2 (n = 47), in addition to native planning CT images and GTV delineations also [18F]FMISO PET/CT data were available at baseline before the start of treatment [1], [4]. At both hospitals, patients were treated with definitive CRT with a radiation dose of 70 Gy, in addition to fluorouracil (5-FU) and mitomycin (MMC) or concomitant weekly cisplatin. After the end of CRT, follow-up examinations were done every six months. LRF was defined as CT- or PET/CT-proven local recurrence. For the current analysis, LRF two years after CRT was used as an endpoint. For further patient details refer to table 1.
Table 1.
Patient characteristics.
| HN1 | HN2 | |
|---|---|---|
| Number of patients | 149* | 47† |
| Age (mean, range) | 62 (39–87) years | 58 (45–76) years |
| GTV volume (mean, range) | 61.6 (1.4 – 326.7) cm3 | 62.7 (10.4–238.8) cm3 |
| Gender (female/male) | 25/124 (16.8%/83.2%) | 7/40 (14.9%/85.1%) |
| Number of loco-regional failures | 50 (34%) | 15 (32%) |
| Median follow-up-time (median, range) | 12 (0–82) months | 17 (1–75) months |
| Distant metastases | 26 (17%) | 7 (15%) |
| T-stage (Tis/T1/T2/T3/T4) | 1/1/17/46/84 | 0/0/2/19/26 |
| N-stage (N0/N1/N2a/N2b/N2c/N3) | 20/14/46/3/55/11 | 5/4/7/16/13/2 |
| Radiation dose (mean, range) | 70 (66–72) Gy | 71 (69–72) Gy |
| Chemotherapy | ||
| 5-FU/MMC | 116 (77.8%) | 25 (53.2%) |
| Cisplatin | 16 (10.7%) | 1 (2.1%) |
| Cisplatin/5-Fu | 3 (2.0%) | 21 (44.7%) |
| Other | 14 (9.4%) | 0 |
*from UHT only, †n = 23 from UHT and n = 25 from UHD.
2.2. Imaging data
For all patients of HN1, native planning CT scans were acquired using a Somatom Sensation Open (Siemens Healthineers, Erlangen, Germany). In subgroup HN2, patients also received a planning CT. In addition, [18F]FMISO PET/CT scans were acquired using a Siemens Biograph 16 (UHD, UHT) or a Siemens Biograph mCT (UHT). PET data were reconstructed using OSEM 3D (four iterations, eight subsets) with a 5-mm 3D Gaussian filtering. The [18F]FMISO PET/CT acquisition protocol consisted of static scans acquired four hours post injection with injected activities of 250 – 444 MBq.
For further details of CT and PET image acquisition see Table 2.
Table 2.
Details of CT and PET imaging parameters.
| Modality | Scanning parameters | HN1 | HN2 |
|---|---|---|---|
| CT | Scanners | Siemens Somatom Sensation Open | Siemens Biograph (n = 36), Siemens Biograph mCT (n = 11) |
| Slice thickness [43] | 3 | 3 (n = 22), 5 (n = 25) | |
| In-plane resolution [mm] | 1.27 | 1.27 (n = 22), 1.38 (n = 25) | |
| Tube Voltage [kVP] | 120 | 120 | |
| Tube Current [mA] | 40 | 40 (n = 22), 100 (n = 25) | |
| Reconstruction Kernel | Convolution kernel B40S filtered back projection | Convolution kernel B40S filtered back projection | |
| PET | Scanners | Siemens Biograph (n = 36),Siemens Biograph mCT (n = 11) | |
| Slice thickness [mm] | 5 | ||
| In-plane resolution [mm] | 1.38 (n = 25), 2.42 (n = 22) | ||
| Administrated [18F]FMISO activity [MBq] | 250 – 300 (n = 25), 315 – 444 (n = 22) |
||
| Reconstruction kernel | 5-mm Gaussian filter OSEM3D 4 integration 8 subsets | ||
| Scan duration time | 15 min (n = 22), 12 min (n = 25) |
||
| Attenuation correction | Based on CT | ||
| Standard Uptake Value (SUV) normalisation | Body weight |
2.3. CT radiomics
2.3.1. Imaging pre-processing and feature extraction
For the radiomics analysis, CT images were used without voxel resizing, in order to avoid inclusion of artificial information that might cause noise at the moment of feature calculations. In an internal preliminary analysis (data not shown) radiomics feature calculations in intensity, shape and texture families did not showed major difference with or without voxel resizing. Only soft tissue voxels with values between −250 and 120 HU were considered in order to make sure that only tissue regions were included into the analysis. Dental artefacts [20] were present in both of the cohorts; however they were treated as noise data in the feature pre-processing strategy described in the following section. A total of 64 bins were used to group voxel values for texture feature calculations.
Feature definitions were obtained from the Imaging Biomarker Standardisation Initiative (IBSI) [21], cf. Appendix 1. For the texture features, we used the grey-level co-occurrence (GLCM), grey-level run length (GRLM), neighbourhood grey tone difference (NGTDM), grey-level size zone (GLZSM) and grey-level distance zone (GLDZM) matrix. They were computed in three dimensions regardless of differences between in-plane and in-slice voxel dimensions. One level undecimated wavelet features were obtained as follows. Firstly, the original images were filtered using high (H) or low-pass (L) “Coiflet 1” filter in every image (x, y, z) direction. Different filter combinations resulted in eight filtered images (cf. Appendix, fig A1). Subsequently, intensity and texture features were computed for each filtered image [21]. In total, we extracted 1150 radiomics features from GTV regions contoured in the planning CT scans. All filtering and feature computations were implemented in-house in Python 3.6.
2.3.2. Feature pre-processing
Several of the radiomics features described by the IBSI [21] are highly correlated and therefore redundant. Hence, in the training phase, we clustered correlated features (Pearson correlation coefficient > 95%), in order to optimise the feature selection process. To do so, features were first scaled according to the interquartile range (IQR), which ranges between the first quartile (25% quantile) and the third quartile (75% quantile). Then, they were clustered hierarchically according to Pearson correlation coefficient [22]. Finally, every cluster was reduced to one single feature using principal component analysis (PCA) to conserve the maximum possible variance inside the cluster [23]. Moreover, all features with variance lower than 0.3 were excluded from the final feature set.
2.3.3. Feature selection and model tuning
According to Leger et al. [11] feature selection methods play a more important role in predicting outcomes than the models themselves. Therefore, a four-step feature selection method was implemented as follows:
Step 1: The HN1 training cohort was randomly subsampled with replacement in a balanced fashion so that each subsample contained 50 patients with and without LRF, respectively. This was repeated 100 times, thus creating a set of 100 subsamples.
Step 2: Within each subsample, variable importance was determined using:
-
•
correlation measures (Pearson [24], Kendall [25], Spearman [26]),
-
•
mutual information (mutual information maximisation [27]),
-
•
univariate significance test scores (Fischer, χ2 [28]),
-
•
multivariate forward selection using classification models (decision trees (DT), k-nearest neighbours (KNN), logistic regression (LogR), random forest (RF), naïve-Bayes (GNB), support vector machines (SVM) [29])
based on the model Area under the curve of the Receiver Operating Characteristic Curve (AUC-ROC) score [30].
For all methods above, up to twenty most important features were kept, and the remaining features were discarded. These feature subsets were then aggregated across the different methods to form a final subset of the five most commonly occurring features for each subsample.
Step 3: The features in the final subset of each of the 100 subsamples were then aggregated and heuristically ranked using the following scoring:
| (1) |
The rank score RS favours the number of appearances of a feature in the 100 subsets, and penalises its mean rank μr together with the standard deviation of its rank σr in the different subsets. The five most highly ranked features were subsequently selected.
Step 4: Finally, we determined a CT radiomics signature for each of the classifiers using a sequential forward feature selection method [31]. For this purpose, we performed 5-fold cross-validation using the HN1 data set. For each classifier, the set of features that produced the model with the highest average AUC on the validation folds was used as a signature.
After feature selection, model hyperparameters such as the number of neighbours for KNN were optimised using grid search (cf. Table 4) and 5-fold cross validation. All methods and algorithms were implemented in-house in Python 3.6 using the packages Pandas, Scikit-learn and mlxtend for machine learning. Fig. 2 presents a schematic overview of the algorithmic workflow used in this study.
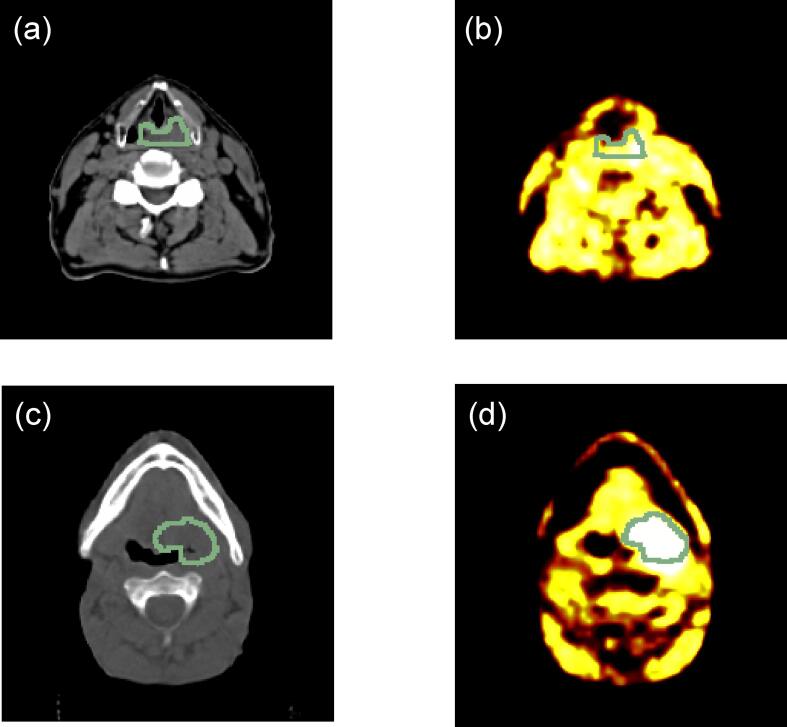
Fig. 2.
Image (a) is a planning CT scan with (b) the [18F]FMISO PET scan after 4 h post injection and their ROIs of a patient who did not recur after CRT. [18F]FMISO TMRpeak was determined as 1.44 and CT radiomics model probability for LRF was 0.18. Images (c) and (d) are the planning CT scan and the [18F]FMISO PET scan with tumour ROIs of a patient who had a recurring tumour after CRT. Here, a TMRpeak of 1.96 and a radiomics model probability for recurrence of 0.54 were observed.
2.3.4. Model validation for CT radiomics signature
Finally, we tested the models created using the signature obtained in our training cohort (HN1) for each classifier in the HN2 cohort. Subsequently the model in our training phase was used to stratify patients into high and low risk groups at a 0.5 risk threshold probability (cf. Supplementary Fig. S1 for a schematic overview).
2.4. [18F]FMISO PET/CT imaging
2.4.1. Tumour-to-muscle ratio extraction
TMRpeak values were extracted from [18F]FMISO PET/CT scans according to:
| (2) |
Peak values of FMISO standardised uptake values (SUVpeak) in the GTV were determined by averaging voxels represented in a 0.5 cm3 sphere of highest tumour uptake as described in previous studies [3], [4], [12]. The mean muscle standardised intensity uptake value (SUVmuscle) was obtained from manually contoured regions of deep neck muscles.
2.4.2. Model validation for TMRpeak
Model validation was performed in HN2 where the TMRpeak was used to classify tumours into high and low risk groups based on the 1.6 threshold obtained in an earlier study [3], [4].
2.5. Model comparison
In order to assess whether the patients at risk classified by the best CT radiomics signature are similar to the classified patients at risk based on TMRpeak, the following simple matching score (MS) was used:
| (3) |
MS measures the ratio between the number of patients that both models predict either as patients at high risk (true positives, TP) or as low risk patients (true negatives, TN) divided by the total number of patients (NT) in HN2. If MS equals 1, it means that both modalities predict the same treatment outcome for a patient, whereas 0 indicates complete disagreement.
2.6. Statistical analysis
Stratification of patients into risk groups for LRC was assessed using Kaplan-Meier curves and the log-rank test. The endpoint of this study was defined as a binary information about LRC as available at last patient follow-up. All statistical analyses were performed using the lifelines package implemented in Python 3.6. A p-value < 0.05 was considered as significant.
3. Results
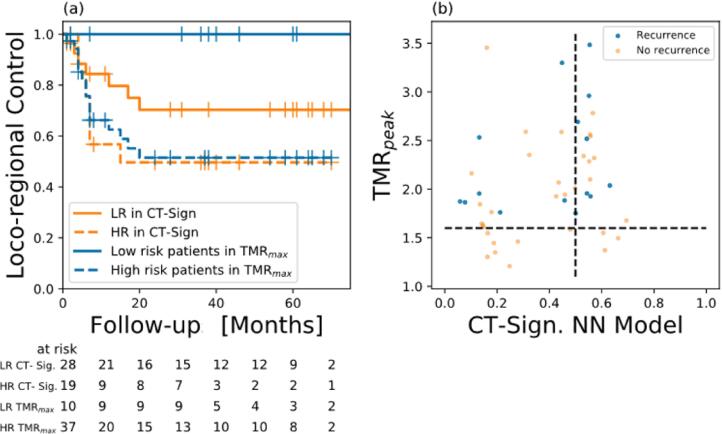
Following model training in the HN1 cohort, the six best models had AUC-ROC values ranging between 0.70 ± 0.09 and 0.76 ± 0.09. The best performing CT radiomics model was a 25-Nearest Neighbours model based on two radiomics meta-features associated according to the first principle component to ‘LLL Size Zone (SZ): Large Zone High Grey Level Emphasis’ and ‘LHH Minimum histogram gradient’ (cf. Table 3). However, in the external validation using the HN2 cohort the AUC of the models decreased to a range between 0.52 and 0.59, using a 0.5 threshold for risk classification. Stratification of the validation cohort HN2 into high and low risk patients failed according to this CT radiomics model, underlined by a p-value = 0.18 in the log-rank test (cf. Fig. 1a).
Table 3.
Best performing CT radiomics signatures and models.
| Feature selection criteria | Model | # of meta features | Name of the associated features in clusters | Hyperparameters | AUC in training cohort HN1 | AUC in validation cohort HN2 |
|---|---|---|---|---|---|---|
| RF | KNN | 2 | LLL SZ: LZHGLE LHH Minimum Histogramm Gradient |
number of neighbors: 25 weights: distance |
0.76 ± 0.09 | 0.59 |
| RF | RF | 4 | LLH Area under IVH curve HHH RL: LGLRE HHL Intensity histogram median LLH SZ: LZLGLE |
Class weight: {0: 0.5} Criterion: Entropy Max depth: 10 Number of estimators: 9 |
0.75 ± 0.07 | 0.56 |
| DT | RF | 3 | LLH NGTD: Busyness NGTD: Busyness LHL Median |
Bootstrap: False Class weight: None Criterion: Gini Max depth: None Number of estimators: 10 |
0.75 ± 0.10 | 0.59 |
| χ2 | KNN | 5 | HLH Energy LLH Energy LLL SZ: LZHGLE LLL SZ: LZE LHH SZ: ZS non-uniformity |
number of neighbors: 23 weights: distance |
0.74 ± 0.10 | 0.52 |
| KNN | LR | 4 | LLL LZE HHL Intensity histogram median LLH Range HLH Energy |
C: 1000 Class weight: {0: 0.5} |
0.71 ± 0.10 | 0.53 |
| DT | LR | 3 | HHL Intensity histogram median LLL SZ: LZHGLE LHL DZ: ZD non-uniformity |
C: 1.0 Class weight: None |
0.70 ± 0.09 | 0.52 |
Abbreviations for classifiers; Random Forest (RF), Decision Trees (DT), k-nearest neighbours (KNN), Logistic Regression (LR). Abbreviations for features obtained after applying filters to CT scans in directions ×, y, z follow the rule of appearance in the direction of application, for instance LLL means the Low-pass filter was applied in x-, y- and z-direction. For more details, please refer to the Supplementary Material.
Fig. 1.
(a) Kaplan-Maier curves for loco-regional control stratified by TMRpeak > 1.6 (p = 0.02) in comparison to the best-performing CT radiomics signature using the 0.5 threshold to stratify patients at risk (p = 0.18). (b) Patient classification according to CT radiomics signature (AUC = 0.59, x-axis) and TMRpeak (AUC = 0.66, y-axis), yielding a matching score of 0.553.
On the contrary, in the same HN2 cohort, the [18F]FMISO PET TMRpeak imaging marker resulted in an AUC score of 0.66 using the threshold of 1.6, as identified earlier for an exploratory cohort. Likewise, in the same cohort a better stratification was achieved by TMRpeak using the log-rank test (p = 0.02, cf. Fig. 1a).
A matching score of MS = 55% was obtained between the two models, which suggests that there is only a weak correlation between the CT radiomics signature classification and the risk classification of patients by [18F]FMISO TMRpeak (cf. Fig. 1b). Consequently, the CT radiomics model does not perform better than TMRpeak in stratifying the HN2 cohort according to LRF.
The most relevant CT radiomics features identified in this study are associated with the quantification of pattern-variation-values with respect to image heterogeneity in a LLL and LHH frequency filtered tumour in a volumetric image. As an example, two patient image sets representing low and high risk groups for LRF are visualized in Fig. 2. In Fig. 2c-d a patient presenting with an irregular CT pattern-structure variation distributed homogeneously across the tumour region of interest (ROI) is shown, the probability for LRF estimated by the radiomics model is p = 0.18 confirmed by a low FMISO TMRpeak of 1.44. In contrast, Fig. 2a-b visualizes an image data set of a patient with a low pattern-structure variation, but equally distributed within the ROI, leading to high risk of LRF predicted by the radiomics model (p = 0.54). Similarly, for this patient high levels of tumour hypoxia were identified (TMRpeak = 1.96).
4. Discussion
In this study, two features, out of 550 radiomics meta-features, along with the KNN model were identified as the best-performing CT radiomics signature from the training cohort (HN1), yielding an AUC value of 0.76 ± 0.09. To validate this signature, an independent validation cohort was used with n = 47 data sets consisting not only of planning CT data but also of FMISO PET images. Validation of the best CT radiomics model resulted in an AUC of 0.59 (log rank p = n.s.), whereas a previously trained simple FMISO TMRpeak threshold reached an AUC-value of 0.66 yielding significant stratification potential (p = 0.02). The matching score was 55% indicating only a low correlation of the CT radiomics and the FMISO PET model, respectively. We assumed the two most important radiomics features to be associated to phenotypical expressions of heterogeneity in tumours, since these features are defined to quantify pattern-variations of grey-levels in medical images [7], [16], [21], [32]. As we had a retrospective data set and therefore could not access genetic information of the tumours, a direct proof of this assumption is lacking.
The aim of the current study was to investigate if a CT radiomics model stratified the same patient risk groups compared to hypoxia PET information. This study design seems unconventional, but it was explicitly chosen because of the low number of patients with both, CT and hypoxia PET images available. In contrast, other groups trained a radiomics model to predict hypoxia information directly. This approach is advantageous in terms of the desired model output, whereas is appears challenging with respect to the required number of imaging data to get a robust model [33], [34], [35].
Several recent studies published CT radiomics models for predicting local control or overall survival in HNC patients following CRT [36], [37], [38], [39]. Similar to our findings, those studies identified features related to CT value homogeneity as most relevant for outcome prognosis. However, in our study the AUC value determined for the validation cohort was still 0.59 but the significance of the CT radiomics model to stratify patient risk groups could not be confirmed in this cohort in contrast to other published studies [37], [38], [39]. In one study by Bogowicz et al. [36] a CT radiomics model based on the primary tumour volume could not be validated in contrast to a model, which was applied to primary tumour and lymph node volumes. According to these findings, the fact that in our study CT radiomics was assessed for the primary tumour only whereas loco-regional failure was used as a prediction variable might be a further limitation. There are a few other reasons that may have led to the non-significant validation of out CT radiomics model. As part of the validation data set was acquired in a different centre, differences in this data set such as the different tube voltage used for CT acquisition or the different slice thicknesses may have introduced too large variation. Especially as we did not perform voxel reformatting, this may be a major limitation of the study. Most previous studies were single centre evaluations [36], [37], [38], [39]. Furthermore, in our study native CT data were used, whereas other studies often based their model training of contrast-enhanced CT images [36], [37], [39].
As previously indicated [4], [5], the results of our study show that pre-treatment [18F]FMISO PET TMRpeak has a significant prognostic power to discriminate between patients with high and low risk of LRF following CRT. This is aligned to the study of Zips et al. [4] which found a significant prognostic power of the TMRmedian feature in the baseline and at the second week after the start of the CRT treatment. The approach in our study is based on the results of Löck et al. [3]. A better discriminative power of the TMRpeak threshold may however be reached in second-week images after the start of CRT or by using dynamic [18F]FMISO PET data [5]. We did not explore time-dependency, because we were limited by the data. This study was performed retrospectively and we did not have neither dynamic data nor weekly [18F]FMISO PET and CT scans for all patients. Also CT radiomics, features extracted from imaging during treatment have been shown to result in a higher prognostic power compared to features acquired before the start of CRT, as shown by Leger et al. [19].
In the publication of Löck et al. [3], TMRpeak was not found to be significantly related to LRF for their exploratory cohort (n = 25). However, in Mönnich et al. [12] TMRpeak was found significant for 22 patients out of the HN2 cohort. The two studies showed some methodological differences. The first approach [3] might be a more robust method because it used an exploratory cohort for assessing TBRpeak thresholds at different time points during the course of CRT in addition to an independent validation cohort for testing. Whereas in the study of Mönnich et al. [12], the derivation of a TMRpeak threshold consisted of the median-value in the exploratory cohort, which was not independently validated [40]. The AUC was lowered from 0.77 in Mönnich et al. to 0.66 in this study. A possible explanation for these results might be the lack of standardisation for the determination of SUVmuscle and SUVpeak in 0.5 cm3 of tumour or muscle tissue, which depends strongly on manual delineation procedures. Moreover, the difference in AUC results may be an effect of the increased sample size.
Larger, more heterogeneous solid tumours often develop hypoxia and have therefore increased risk of LRF [7], [13], [16], [41]. The hypothesis of this study was that CT radiomics, which is assumed to quantify heterogeneity in tumours, could be used to provide a prognostic model that significantly correlates with LRF after CRT and thus also up to a significant extent with an imaging metric for hypoxia, such as TMRpeak. However, hypoxia may not be the only cause of LRF. Different factors such as patient characteristics, tumour biology and also treatment related issues contribute to the observed outcome which may not be captured entirely by both approaches used in this study. A more robust approach to test our hypothesis would be directly targeting hypoxia gene expressions, hypoxia imaging biomarkers [42] or potentially generate [18F]FMISO image distributions via a deep learning architecture such as a Convolutional Neural Network (CNN) based approach, instead of targeting loco-regional outcomes of tumours. However, these approaches were not possible in the context of the current study because of the small cohort size to train and test findings.
In this study, model training is performed using LRF data. The binary nature of the response variable introduces limitations to this study as censored events as well as the time to recurrence is neglected. Other studies have presented radiomics models which include time-to-event data [19] and might thus be considered more accurate in terms of event modelling.
Another possible limitation of our study is the application of wavelet filters in the 3D image space. Voxel lengths were not interpolated and thus no equal voxel spacing was used leading to larger voxel dimensions in slice direction compared to in-plane voxel spacing. This may affect the generation of new filtered images and subsequently the corresponding feature values. However, interpolation operations might also introduce additional artefacts to the data. To date, it is unknown to which extend this might affect the process of feature selection and machine learning modelling.
The chosen CT radiomics signature is based on the best performing signature inside the training phase. This is not always the safest choice according to Leger et al. [11]. As a result, we tested the six best CT radiomics signatures from the training phase in our validation cohort, where similar results were obtained (cf. Table 3). We therefore did not see any impediment to compare simply the best signature from our training phase with the results obtained for TMRpeak as a matter of consistency.
In this study, no direct correlation between [18F]FMISO PET TMRpeak and the best performing CT radiomics model was found. This finding might potentially also be compromised by the low sample size in the validation data set (n = 47). Larger cohorts of coherently acquired hypoxia PET data would be needed to assess this in more detail. The basic processes leading to image formation in CT and hypoxia PET are very different and therefore capture complementary biological tissue characteristics. CT radiomics may pick tumour phenotypic heterogeneity from CT data which might be linked to tumour hypoxia, but indirectly. However, direct assessment of tumour hypoxia with specific imaging techniques and radiotracers is suggested to have a more powerful prediction power.
Filter-based features
In this study we applied to the original image coiflet1 based filter to original images and decomposed as shown in Fig. S2. In the eight final images we computed the set of features from Table S1. For more details, please refer to the IBSI collaboration publication [21].
Classifiers
In this study, different classifiers were used for model generation. Details about the hyperparameters are summarized in Table S2.
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: DT and DZ declare institutional collaborations including financial support with the companies Siemens Healthineers (2014–2019), Elekta AB, Philips and PTW Freiburg without any direct relation to this study.
In the past 5 years, MB attended an advisory board meeting of MERCK KGaA (Darmstadt), for which the University of Dresden received a travel grant. He further received funding for his research projects and for educational grants to the University of Dresden by Teutopharma GmbH (2011–2015), IBA (2016), Bayer AG (2016–2018), Merck KGaA (2014–2030), Medipan GmbH (2014–2018). For the German Cancer Research Center (DKFZ, Heidelberg) MB is on the supervisory boards of HI-STEM gGmbH (Heidelberg). MB, as former chair of OncoRay (Dresden) and present CEO and Scientific Chair of the German Cancer Research Center (DKFZ, Heidelberg), signed/signs contracts for his institute(s) and for the staff for research funding and/or collaborations with a multitude of companies worldwide. MB confirms that none of the above funding sources were involved in the design of this study, the preparation of this paper, the materials used, or the collection, analysis, and interpretation of data.
Acknowledgment
This project has in parts received funding from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (FP7/2007-2013), grant agreement no. ERC StG 335367.
Footnotes
"Daniela Thorwarth, a co-author of this paper, is an Editor-in-Chief of Physics & Imaging in Radiation Oncology. The editorial process for this manuscript was managed independently from Dr. Thorwarth and the manuscript was subject to the Journal's usual peer-review process."
A detailed list of features extracted in this study is given in table S1. For details about the construction of the texture matrices, please refer to the IBSI collaboration document [21]. For the sake of the document extension, the construction was not included in this appendix. Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2020.07.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Welz S., Monnich D., Pfannenberg C., Nikolaou K., Reimold M., La Fougere C. Prognostic value of dynamic hypoxia PET in head and neck cancer: Results from a planned interim analysis of a randomized phase II hypoxia-image guided dose escalation trial. Radiother Oncol. 2017;124:526–532. doi: 10.1016/j.radonc.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 2.Zegers C.M., van Elmpt W., Reymen B., Even A.J., Troost E.G., Ollers M.C. In vivo quantification of hypoxic and metabolic status of NSCLC tumors using [18F]HX4 and [18F]FDG-PET/CT imaging. Clin Cancer Res. 2014;20:6389–6397. doi: 10.1158/1078-0432.CCR-14-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lock S., Perrin R., Seidlitz A., Bandurska-Luque A., Zschaeck S., Zophel K. Residual tumour hypoxia in head-and-neck cancer patients undergoing primary radiochemotherapy, final results of a prospective trial on repeat FMISO-PET imaging. Radiother Oncol. 2017;124:533–540. doi: 10.1016/j.radonc.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Zips D., Zophel K., Abolmaali N., Perrin R., Abramyuk A., Haase R. Exploratory prospective trial of hypoxia-specific PET imaging during radiochemotherapy in patients with locally advanced head-and-neck cancer. Radiother Oncol. 2012;105:21–28. doi: 10.1016/j.radonc.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 5.Thorwarth D., Welz S., Monnich D., Pfannenberg C., Nikolaou K., Reimold M. Prospective Evaluation of a Tumor Control Probability Model Based on Dynamic (18)F-FMISO PET for Head and Neck Cancer Radiotherapy. J Nucl Med. 2019;60:1698–1704. doi: 10.2967/jnumed.119.227744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lambin P., Rios-Velazquez E., Leijenaar R., Carvalho S., van Stiphout R.G., Granton P. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z., Tang L.H., Klimstra D.S. Effect of tumor heterogeneity on the assessment of Ki67 labeling index in well-differentiated neuroendocrine tumors metastatic to the liver: implications for prognostic stratification. Am J Surg Pathol. 2011;35:853–860. doi: 10.1097/PAS.0b013e31821a0696. [DOI] [PubMed] [Google Scholar]
- 9.Thorwarth D., Alber M. Implementation of hypoxia imaging into treatment planning and delivery. Radiother Oncol. 2010;97:172–175. doi: 10.1016/j.radonc.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 10.Horsman M.R., Mortensen L.S., Petersen J.B., Busk M., Overgaard J. Imaging hypoxia to improve radiotherapy outcome. Nat Rev Clin Oncol. 2012;9:674–687. doi: 10.1038/nrclinonc.2012.171. [DOI] [PubMed] [Google Scholar]
- 11.Leger S., Zwanenburg A., Pilz K., Lohaus F., Linge A., Zophel K. A comparative study of machine learning methods for time-to-event survival data for radiomics risk modelling. Sci Rep. 2017;7:13206. doi: 10.1038/s41598-017-13448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Monnich D., Welz S., Thorwarth D., Pfannenberg C., Reischl G., Mauz P.S. Robustness of quantitative hypoxia PET image analysis for predicting local tumor control. Acta Oncol. 2015;54:1364–1369. doi: 10.3109/0284186X.2015.1071496. [DOI] [PubMed] [Google Scholar]
- 13.Karlo C.A., Di Paolo P.L., Chaim J., Hakimi A.A., Ostrovnaya I., Russo P. Radiogenomics of clear cell renal cell carcinoma: associations between CT imaging features and mutations. Radiology. 2014;270:464–471. doi: 10.1148/radiol.13130663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kickingereder P., Gotz M., Muschelli J., Wick A., Neuberger U., Shinohara R.T. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin Cancer Res. 2016;22:5765–5771. doi: 10.1158/1078-0432.CCR-16-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Zhu Y., Burnside E.S., Drukker K., Hoadley K.A., Fan C. MR Imaging Radiomics Signatures for Predicting the Risk of Breast Cancer Recurrence as Given by Research Versions of MammaPrint, Oncotype DX, and PAM50 Gene Assays. Radiology. 2016;281:382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gevaert O., Echegaray S., Khuong A., Hoang C.D., Shrager J.B., Jensen K.C. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci Rep. 2017;7:41674. doi: 10.1038/srep41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bogowicz M., Riesterer O., Ikenberg K., Stieb S., Moch H., Studer G. Computed Tomography Radiomics Predicts HPV Status and Local Tumor Control After Definitive Radiochemotherapy in Head and Neck Squamous Cell Carcinoma. Int J Radiat Oncol Biol Phys. 2017;99:921–928. doi: 10.1016/j.ijrobp.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Gevaert O., Mitchell L.A., Achrol A.S., Xu J., Echegaray S., Steinberg G.K. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leger S., Zwanenburg A., Pilz K., Zschaeck S., Zophel K., Kotzerke J. CT imaging during treatment improves radiomic models for patients with locally advanced head and neck cancer. Radiother Oncol. 2019;130:10–17. doi: 10.1016/j.radonc.2018.07.020. [DOI] [PubMed] [Google Scholar]
- 20.Wei L., Rosen B., Vallieres M., Chotchutipan T., Mierzwa M., Eisbruch A. Automatic recognition and analysis of metal streak artifacts in head and nekc computed tomography for radiomics modeling. Phys Imaging Radiother Oncol. 2019;10:49–54. doi: 10.1016/j.phro.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwanenburg A., Vallieres M., Abdalah M.A., Aerts H., Andrearczyk V., Apte A. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;191145 doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker M., Kublin J.G., Zunt J.R. Fast R functions for rubust correlations and hierarchical clustering. J Stat Softw. 2009;42:115–125. [PMC free article] [PubMed] [Google Scholar]
- 23.Kambhatla M., Leen T.K. Dimension reduction by local principal component analysis. Neural Comput. 1997;9:1493–1516. [Google Scholar]
- 24.Benestly J., Cheng J., Huang Y., Cohen I. Springer; 2009. Pearson correlation coefficient. [Google Scholar]
- 25.Abdi H. The Kendall rank correlation coefficient.: Encycl Meas Stat Sage, Thousand Oaks, CA.; 2007.
- 26.Dodge Y. Spearman Rank Correlation Coefficient.: Springer; 2008.
- 27.Peng H., Long F., Ding C. Feature selection based on mutual information: criteria of max-dependency, max-relevance, and min-redundancy. IEEE Trans Pattern Anal Mach Intell. 2005;27:1226–1238. doi: 10.1109/TPAMI.2005.159. [DOI] [PubMed] [Google Scholar]
- 28.Huberty C.J., Morris J.D. Multivariate analysis versus multiple univariate analyses. Psychol Bull. 1989;105:302. [Google Scholar]
- 29.Bishop C.M. Springer; 2006. Pattern recognition and machine learning. [Google Scholar]
- 30.Huang J., Ling C.X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans Knowl Data Eng. 2005;17:299–310. [Google Scholar]
- 31.Pudil P., Novovicova J., Kittler J. Floating search methods in feature selection. Pattern Recognit Lett. 1994;15:1119–1125. [Google Scholar]
- 32.Lambin P., Leijenaar R.T.H., Deist T.M., Peerlings J., de Jong E.E.C., van Timmeren J. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 33.Crispin-Ortuzar M., Apte A., Grkovski M., Oh J.H., Lee N.Y., Schoder H. Predicting hypoxia status using a combination of contrast-enhanced computed tomography and [(18)F]-Fluorodeoxyglucose positron emission tomography radiomics features. Radiother Oncol. 2018;127:36–42. doi: 10.1016/j.radonc.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorensen A., Carles M., Bunea H., Majerus L., Stoykow C., Nicolay N.H. Textural features of hypoxia PET predict survival in head and neck cancer during chemoradiotherapy. Eur J Nucl Med Mol Imaging. 2020;47:1056–1064. doi: 10.1007/s00259-019-04609-9. [DOI] [PubMed] [Google Scholar]
- 35.Even A.J.G., Reymen B., La Fontaine M.D., Das M., Jochems A., Mottaghy F.M. Predicting tumor hypoxia in non-small cell lung cancer by combining CT, FDG PET and dynamic contrast-enhanced CT. Acta Oncol. 2017;56:1591–1596. doi: 10.1080/0284186X.2017.1349332. [DOI] [PubMed] [Google Scholar]
- 36.Bogowicz M., Tanadini-Lang S., Guckenberger M., Riesterer O. Combined CT radiomics of primary tumor and metastatic lymph nodes improves prediction of loco-regional control in head and neck cancer. Sci Rep. 2019;9:15198. doi: 10.1038/s41598-019-51599-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogowicz M., Riesterer O., Stark L.S., Studer G., Unkelbach J., Guckenberger M. Comparison of PET and CT radiomics for prediction of local tumor control in head and neck squamous cell carcinoma. Acta Oncol. 2017;56:1531–1536. doi: 10.1080/0284186X.2017.1346382. [DOI] [PubMed] [Google Scholar]
- 38.Cozzi L., Franzese C., Fogliata A., Franceschini D., Navarria P., Tomatis S. Predicting survival and local control after radiochemotherapy in locally advanced head and neck cancer by means of computed tomography based radiomics. Strahlenther Onkol. 2019;195:805–818. doi: 10.1007/s00066-019-01483-0. [DOI] [PubMed] [Google Scholar]
- 39.Head M.D.A.C.C. Neck Quantitative Imaging Working G. Investigation of radiomic signatures for local recurrence using primary tumor texture analysis in oropharyngeal head and neck cancer patients. Sci Rep. 2018;8:1524. doi: 10.1038/s41598-017-14687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwanenburg A., Lock S. Why validation of prognostic models matters? Radiother Oncol. 2018;127:370–373. doi: 10.1016/j.radonc.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 41.Mroz E.A., Tward A.D., Pickering C.R., Myers J.N., Ferris R.L., Rocco J.W. High intratumor genetic heterogeneity is related to worse outcome in patients with head and neck squamous cell carcinoma. Cancer. 2013;119:3034–3042. doi: 10.1002/cncr.28150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lock S., Linge A., Seidlitz A., Bandurska-Luque A., Nowak A., Gudziol V. Repeat FMISO-PET imaging weakly correlates with hypoxia-associated gene expressions for locally advanced HNSCC treated by primary radiochemotherapy. Radiother Oncol. 2019;135:43–50. doi: 10.1016/j.radonc.2019.02.020. [DOI] [PubMed] [Google Scholar]
- 43.Ouyang L., Folkerts M., Zhang Y., Hrycushko B., Lamphier R., Lee P. Volumetric modulated arc therapy based total body irradiation: Workflow and clinical experience with an indexed rotational immobilization system. Phys Imag Radiat Oncol. 2017;4:22–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.