Abstract
Objective
To detect the expression of CEA-related cell adhesion molecule 5 (CEACAM5) in non-small-cell lung cancer (NSCLC) and explore its function in the progression and development of NSCLC.
Methods
qRT-PCR and immunohistochemistry were performed to detect CEACAM5 expression in human NSCLC tissues and cell lines. The correlation between CEACAM5 expression and the clinicopathological features of patients with NSCLC was also investigated. MTT, colony formation, wound healing, and immunoblot assays were performed to detect the functions of CEACAM5 in NSCLC cells in vitro, and immunoblotting was used to detect the effects of CEACAM5 on p38–Smad2/3 signaling.
Results
CEACAM5 expression was elevated in human NSCLC tissues and cells. We further found that CEACAM expression was correlated with clinicopathological features including T division, lymph invasion, and histological grade in patients with NSCLC. The in vitro assays confirmed that CEACAM5 depletion inhibited the proliferation and migration of NSCLC cells by activating p38–Smad2/3 signaling. We verified the involvement of CEACAM5 in the suppression of NSCLC tumor growth in mice.
Conclusion
CEACAM5 stimulated the progression of NSCLC by promoting cell proliferation and migration in vitro and in vivo. CEACAM5 may serve as a potential therapeutic target for the treatment of NSCLC.
Keywords: Non-small-cell lung cancer, CEA-related cell adhesion molecule 5, proliferation, migration, therapeutic target, p38–Smad2/3 signaling, tumorigenesis
Introduction
Lung cancer is the leading cause of cancer-related mortality globally. Non-small-cell lung cancer (NSCLC), as a major type of lung cancer, can progress quickly, including with multiple metastases in its later stages. NSCLC caused an estimated 228,150 new cases and 142,670 deaths in the USA in 2018.1 The 5-year survival rate of metastatic NSCLC is only 2%.1 Currently, mortality caused by distant metastases remains the major obstacle in the treatment of NSCLC.2 Thus, exploring the mechanisms of NSCLC progression is particularly urgent.
CEA-related cell adhesion molecule 5 (CEACAM5) belongs to the CEACAM family of highly glycosylated proteins with a typical N-terminal variable Ig-like domain followed by zero to six constant Ig-like domains, as well as a hydrophobic transmembrane domain with a cytoplasmic tail (CEACAM1 to CEACAM4) or a glycosylphosphatidylinositol lipid moiety (CEACAM5 to CEACAM8).3,4 CEACAM5 is involved in intercellular contact via both homophilic and heterophilic binding (with CEACAM1 or CEACAM6).5 In addition to its functions in cell adhesion and migration, CEACAM5 also inhibits anoikis.6 Because resistance to anoikis is a characteristic of cancer cells, the inhibitory effect of CEACAM5 on anoikis suggests its role in facilitating tumorigenesis and metastasis. The tumorigenic functions of CEACAM5 have been depicted using 3D cultures and transgenic mice.7–9
The members of the CEACAM family were reported to participate in cancerous growth and invasion by acting as either tumor suppressors or poor prognostic markers for the progression of malignancies.10–12 CEACAM5 is upregulated in approximately 90% of gastrointestinal, colorectal, and pancreatic cancers and 50% of breast cancers.13 CEACAM5 has been applied in the clinical detection of liver metastasis, colorectal cancer, and colon cancer relapse.14 Notably, CEACAM5 is commonly used as an accepted tumor biomarker and indicator of recurrence in patients with cancer, especially those with colorectal cancer.15 However, the utility of CEACAM5 in NSCLC has been less investigated.
In the present study, we investigated the role of CEACAM5 in NSCLC progression. We found that CEACAM5 was upregulated in NSCLC compared with its levels in adjacent non-tumor tissue. Notably, high CEACAM5 levels were closely related to the clinical features of patients with NSCLC. We also found that CEACAM5 depletion dramatically inhibited NSCLC cell proliferation and invasion by regulating p38–Smad2/3 signaling, which was also confirmed in mice. Therefore, CEACAM5 may serve as a potential therapeutic target for the treatment of NSCLC.
Materials and methods
Bioinformatic analysis
We performed bioinformatic analysis using GEPIA (http://gepia.cancer-pku.cn/detail.php?gene=CEACAM5/) to analyze The Cancer Genome Atlas data with thresholds of P < 0.05 and LogFC > 1 or < −1 for differentially expressed genes.
Clinical sample collection and preparation
The use of clinical specimens in this research was reviewed and approved by the Ethics Committee of Linyi Central Hospital on January 23, 2018, and the approval number was YKRL (26). NSCLC tissue specimens and adjacent normal tissue specimens were collected from patients at Linyi Central Hospital. This study was approved by the Ethics Committee and Scientific Medical Board of our hospital according to the Declaration of Helsinki of 2008. No patients had received radiotherapy or chemotherapy prior to tissue collection. Lung tissue was examined using qRT-PCR and immunohistochemical analysis. Written informed consent was obtained from all patients.
Immunohistochemistry (IHC)
Tumor samples were freshly isolated, fixed with paraformaldehyde (PFA), embedded in paraffin, and cut into 5-μm sections. Sections were deparaffinized with xylene, rehydrated via an ethanol gradient, and incubated with H2O2 for 10 minutes. Sections were blocked using 1% normal goat serum and treated with CEACAM5 antibody (1:500, Abcam, Cambridge, UK) for 2 hours at room temperature. Then, sections were incubated with secondary antibody and counterstained with hematoxylin before coverslip mounting.
According to the results of IHC, CEACAM5 was mainly localized to the cytoplasm of NSCLC cells. The proportion of positively stained cells was graded as follows: 0, 0% positive staining; 1, 1% to 30% positive staining; 2, 31% to 80% positive staining; and 3, ≥81% positive staining.
The staining intensity was evaluated on a three-point scale as follows: 0, no or weak staining; 1, moderate staining; and 2, intense staining. CEACAM5 expression was categorized using the staining index, which was calculated by summing the scores for positive staining and staining intensity. Staining indices of <3 indicated low expression, whereas scores of ≥3 indicated high expression. For CEACAM5, the cutoff was defined as the median number of positive cells.
Cell culture and transfection
H1299, H358, HCC827, and A549 human NSCLC cells and HBE normal human bronchial epithelial cells were obtained from American Type Culture Collection (Manassas, VA, USA). H358, HCC827, H1299, and A549 cells were maintained and grown in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Waltham, MA, USA), and HBE cells were maintained in Dulbecco's modified Eagle’s medium (DMEM; Gibco; Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (HyClone; GE Healthcare, Chicago, IL, USA) at 37°C in an atmosphere of 5% CO2 in a humidified incubator.
Cells were seeded in six-well plates, incubated for 12 hours, then transfected with a plasmid encoding shRNA targeting CEACAM5 using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific). The sequence for shCEACAM5 was CCGGGCAGTATTCTTGGCGTATCAACTCGAGTTGATACGCCAAGAATACTGCTTTTTG. In total, 5 µL of transfection reagent and 1 µg of the plasmid were mixed in 200 µL of serum-free medium, incubated for 10 minutes, then mixed again. The mixture was left at room temperature for 15 minutes, and serum-starved cells were then incubated with the mixture for 6 hours. Stable depletion clones were screened via lentivirus infection in the presence of 5 mg/mL Polybrene (Sigma-Aldrich, St. Louis, MO, USA). Infected cells were selected for 14 days in the presence of 1 mg/mL puromycin (Sigma-Aldrich) and used for the animal assays.
qRT-PCR assays
Total RNA was extracted using TRIzol reagent (Invitrogen, Thermo Fisher Scientific) and reverse-transcribed using a Transcriptor First Strand cDNA Synthesis Kit (Takara, Kusatsu, Japan). Gene expression was normalized to that of GAPDH. The mRNA levels of CEACAM5 were detected in tumor and normal tissues from patients. Power analyses in the study were conducted using G power. The number of required patients was 87 (97.8% [up 95% G-power]). The primers used in this assay were as follows: CEACAM5 forward, 5ʹ-AGGCCAATAACTCAGCCAGT-3ʹ; CEACAM5 reverse, 5ʹ-GGGTTTGGAGTTGTTGCTGG-3ʹ; β-actin forward, 5ʹ-CTCCATCCTGGCCTCGCTGT-3ʹ; and β-actin reverse, 5ʹ-GCTGTCACCTTCACCGTTCC-3ʹ.
Immunoblotting
After extraction, proteins were separated via SDS-PAGE and transferred to polyvinylidene difluoride membranes (MilliporeSigma, Burlington, MA, USA). The membrane was blocked in 5% BSA and incubated with specific primary antibodies against CEACAM5 (1:1000, 1:1000, 11077-R327, Sino Biological, Beijing, China Sino Biological, Beijing, China), anti-Ki67, anti-PCNA (1:1000, Cell Signaling Technology, Danvers, MA, USA), anti-MMP2 (1:1500, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-MMP9 (1:1500, Santa Cruz Biotechnology), anti-cyclin D1 (1:1500 Santa Cruz Biotechnology), and β-actin (1:5000, ab8227, Abcam) at 4°C overnight. After washing, the membrane was incubated with HRP-conjugated secondary antibodies, and signals were detected using a Novex™ ECL Chemiluminescent Substrate Reagent kit (Thermo Fisher Scientific). The relative protein levels were quantified using ImageJ (US National Institutes of Health, Bethesda, MD, USA).
Cell viability and colony formation assays
For cell viability assays, cells were incubated in 96-well plates at a density of 2000 cells per well 1 day before the experiment. Then, 10 μL of Cell Counting Kit 8 reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan) in 100 μL of RPMI 1640 medium were added to each well, and cells were cultured for 2 hours at 37°C. The absorbance was measured at 450 nm. The experiment was replicated five times, including three independent replicates.
For colony formation assays, cells were seeded and incubated for 2 weeks to permit colony formation. To visualize colonies, cells were fixed in PFA for 10 minutes, stained with crystal violet, and photographed. For the control group, the shRNA targeting sequence did not target any intracellular RNAs. The numbers of colonies were counted manually.
Wound healing assay
To assess cell migration, the wound healing assay was performed. Briefly, cells transfected with the indicated plasmids were wounded via scraping with a tip, followed by washing. Subsequently, the cells were cultured with complete culture medium to stimulate wound healing. Photographs were taken at immediately and 24 hours after wounding to evaluate the migration of cancer cells.
Animal experiments
The animal experiments performed in this research were reviewed and approved by the Ethics Committee of Medical Experimental Animals of Linyi Central Hospital on February 12, 2019, and the approval number was YKDL (52). Eight-week-old female BALB/c (nu/nu) nude mice were obtained from the Shanghai Experimental Animal Center (Shanghai, China).
To measure tumor growth in vivo, BALB/c (nu/nu) nude mice were divided into two groups randomly (n = 6 for each group). A549 cells stably transfected with CEACAM5 shRNA or negative-control shRNA lentivirus cells were suspended in 50 μL of 50% Matrigel in DMEM medium. Cells were subcutaneously injected into each mouse. After 14 days, tumors had formed, and we measured the tumor volume weekly. After 49 days, all tumors were isolated, and representative photographs were taken. All mice were given adequate food and water, and none died of natural causes. Mice were sacrificed via cervical dislocation before tumor tissue was removed, and their heartbeats were checked to confirm death. Adequate humanitarian care was given. The tumor volume (length × width2 × 0.5236) was calculated every week.
Statistical analysis
Statistical analysis was performed using GraphPad (GraphPad, La Jolla, CA, USA) in this study. All data are presented as the mean ± SEM for at least three independent experiments. Statistically significant correlations between CEACAM5 expression and clinical features in patients with NSCLC were determined using the χ2 test. A paired t-test was used to detect significant differences in CEACAM5 mRNA levels between tumor and normal tissues. Student’s t-test was used for statistical analysis in the in vitro and in vivo assays. P < 0.05 indicated statistical significance.
Results
CEACAM5 is abnormal high expression in human NSCLC tissues
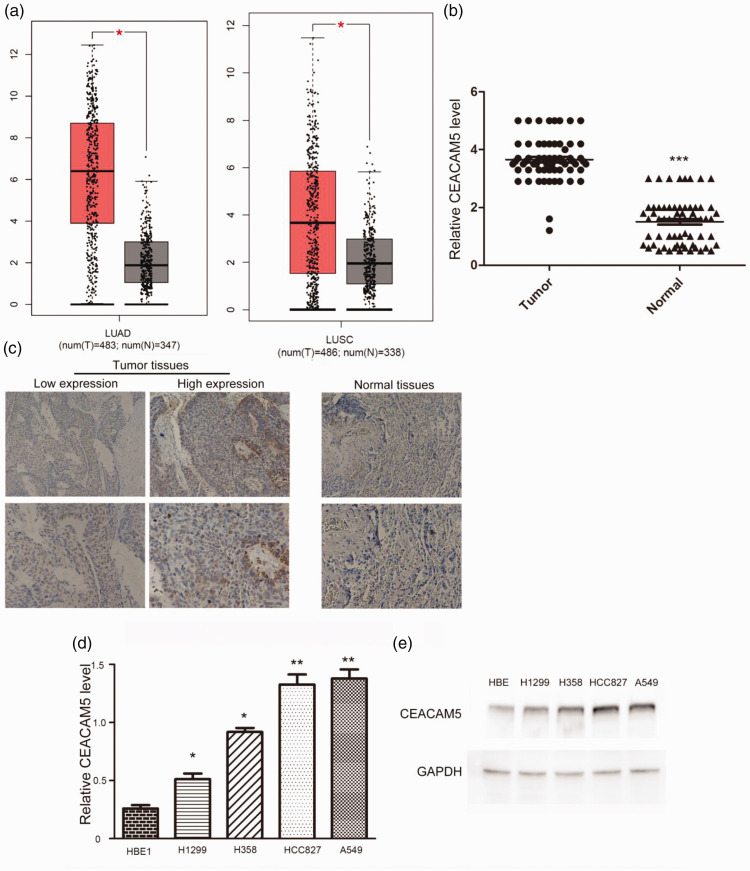
To assess the potential involvement of CEACAM5 in the development of lung cancer, we first conducted bioinformatic analysis of CEACAM5 expression in 483 human lung adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) tissues, as well as 347 normal tissues. CEACAM5 expression was abnormally high in LUAD and LUSC tumor tissues compared with that in normal tissues (http://gepia.cancer-pku.cn/, Figure 1a). To further confirm the result, we performed IHC to assess CEACAM5 expression in tumor tissues and corresponding normal tissues collected from 87 patients (53 men and 34 women; age range, 47 to 73 years) in our hospital (Figure 1b). Similarly, CEACAM5 levels were higher in tumor tissues than in adjacent non-tumor tissues (Figure 1c).
Figure 1.
CEA-related cell adhesion molecule 5 (CEACAM5) expression was enhanced in human non-small-cell lung cancer (NSCLC) tissues and cells. (a) Bioinformatic analysis revealed enhanced CEACAM5 expression in patients with NSCLC compared with that in normal tissues according to The Cancer Gene Atlas database. (b) qRT-PCR assays depicted the mRNA levels of CEACAM5 in 87 pairs of NSCLC tissues and corresponding normal tissues (c) Immunohistochemical assays revealing higher CEACAM5 expression in NSCLC tissues than in the corresponding normal tissues. Representative photograph are presented (×100 and ×200 magnification, respectively). (d) qRT-PCR and (e) immunoblot assays revealed the expression of CEACAM5 in HBE, H1299, H358, HCC827, and A549 cells.
Subsequently, we detected the mRNA levels of CEACAM5 in H1299, H358, HCC827, A549, and HBE cells. CEACAM5 levels were dramatically enhanced in NSCLC cells compared with those in normal cells, consistent with the previous results (Figure 1d). Immunoblot assays also revealed the upregulation of CEACAM5 in NSCLC cells (Figure 1e). We therefore conclude that CEACAM5 is elevated in human NSCLC cells and tissues.
CEACAM5 expression is correlated with the clinical pathological features of patients with NSCLC
To further explore the involvement of CEACAM5 in the progression of NSCLC, we assessed the correlations of CEACAM5 expression with the clinicopathological features of 87 patients with NSCLC. Patients were divided into two groups according to CEACAM5 expression. Fifty-six patients exhibited high CEACAM5 expression, and the remaining patients displayed low expression (Table 1). No obvious correlations were found between CEACAM5 expression and clinicopathological features including patient gender (P = 0.5263), age (P = 0.1444), smoking history (P = 0.3437), and histology (P = 0.4001, Table 1). Importantly, CEACAM5 expression was significantly correlated with T division (P = 0.0268), lymph invasion (P = 0.0052), and histological grade (P = 0.0144, Table 1) in patients with NSCLC. We therefore demonstrated that CEACAM5 expression was correlated with the clinicopathological features of patients with NSCLC.
Table 1.
Relationship between CEACAM5 expression and clinical pathological characteristics of NSCLC patients (N = 87).
| N |
CEACAM5 |
Chi-squared value | P | ||
|---|---|---|---|---|---|
| Low expression | High expression | ||||
| Gender | |||||
| Female | 34 | 13 | 21 | 0.165 | 0.685 |
| Male | 53 | 18 | 35 | ||
| Age (years) | |||||
| <60 <60 | 35 | 15 | 20 | 1.333 | 0.248 |
| ≥60 ≥60 | 52 | 16 | 36 | ||
| Smoking history | |||||
| Non-smoker | 28 | 13 | 15 | 2.098 | 0.147 |
| Smoker | 59 | 18 | 41 | ||
| T stage | |||||
| T1/2 | 48 | 12 | 36 | 5.278 | 0.022 |
| T3/4 | 39 | 19 | 20 | ||
| Lymph invasion | |||||
| N0 | 45 | 9 | 36 | 9.931 | 0.002 |
| N1 or N2 | 42 | 22 | 20 | ||
| Histology | |||||
| Squamous cancer | 32 | 9 | 23 | 1.256 | 0.534 |
| Adenocarcinoma | 27 | 11 | 16 | ||
| other | 28 | 11 | 17 | ||
| Histological grade | |||||
| Well | 15 | 11 | 4 | 12.916 | 0.002 |
| Moderate | 41 | 14 | 27 | ||
| Poor | 31 | 6 | 25 | ||
CEACAM5, CEA-related cell adhesion molecule 5.
Ablation of CEACAM5 blocks NSCLC cell proliferation and migration in vitro
To further explore the role of CEACAM5 in the progression of NSCLC, we transfected A549 and HCC827 cells with CEACAM5 shRNA plasmids to suppress CEACAM expression in these cells. As detected by qRT-PCR, CEACAM5 mRNA levels were obviously reduced after CEACAM5 shRNA transfection in A549 and HCC827 cells (Figure 2a). Similarly, immunoblot assays demonstrated that CEACAM5 expression was decreased after transfection of its shRNA plasmids (Figure 2b). Therefore, the efficiency of CEACAM5 shRNA plasmids was validated.
Figure 2.
CEA-related cell adhesion molecule 5 (CEACAM5) expression was downregulated in A549 and HCC827 cells after CEACAM5 shRNA transfection. qRT-PCR (a) and immunoblot analyses (b) revealed the decrease in CEACAM5 levels in A549 and HCC827 cells transfected with CEACAM5 shRNA plasmids. β-actin was used as an internal control. *P < 0.05.
To assess cell proliferation after CEACAM5 depletion, we performed colony formation assays. We observed reduced cell proliferation after CEACAM5 depletion (Figure 3a). We next performed wound healing assays to evaluate the effects of CEACAM5 on the migration of NSCLC cells and observed that CEACAM5 depletion dramatically inhibited wound closure in A549 and HCC827 cells (Figure 3b). Subsequently, we performed immunoblot assays to detect the expression of Ki67 and PCNA, two proliferation cell markers, in A549 and HCC827 cells. As expected, the ablation of CEACAM5 dramatically suppressed the expression of Ki67 and PCNA (Figure 3c). Because MMP2 and MMP9 are necessary for tumor migration, we also examined their expression after CEACAM5 depletion. We observed decreased MMP2 and MMP9 expression after CEACAM5 depletion (Figure 3d). Collectively, our results indicated that CEACAM5 contributes to the proliferation and migration of NSCLC cells in vitro.
Figure 3.
CEA-related cell adhesion molecule 5 (CEACAM5) promoted the proliferation and migration of non-small-cell lung cancer cells in vitro. (a) Colony formation assays revealed the inhibitory effect of CEACAM5 ablation on cell proliferation. (b) Wound healing assays were conducted to evaluate cell migration using A549 and HCC827 cells transfected with control or CEACAM5 shRNA plasmids. Representative photographs are presented. (c) Immunoblot assays revealed the reduced expression of PCNA and Ki67 in A549 and HCC827 cells after CEACAM5 ablation. (d) Immunoblot assays indicated the decreased expression of MMP2 and MMP9 in A549 and HCC827 cells following CEACAM5 ablation. Results are presented as the mean ± SEM, *P < 0.05, **P < 0.01.
CEACAM5 promotes NSCLC cell proliferation and migration via the p38–Smad2/3 signaling pathway
The p38 kinases are involved in many complex biologic processes, such as cell proliferation, differentiation, death, migration, and invasion. Dysregulation of p38 mitogen-activated protein kinase (MAPK) levels are associated with advanced stages and short survival in patients with cancer. CEACAM5 has been reported to inhibit p38 activity and promote the growth of metastatic lesions.16 Thus, we examined the potential association of p38 activity with the CEACAM5-mediated enhancement of cell proliferation. Importantly, CEACAM5 depletion increased the TGF-β-mediated phosphorylation of Smads and p38 in A549 cells (Figure 4a). To explore the involvement of p38 in CEACAM5-mediated cell proliferation and migration, we depleted CEACAM5 and overexpressed p38 in A549 cells to examine for MMP2 and PCNA levels. We observed significant decreases in MMP2 and PCNA levels after CEACAM5 depletion (P < 0.05), and p38 overexpression blocked these effects (Figure 4b). Therefore, we deduced that p38 might be involved in CEACAM5-regulated proliferation and migration in NSCLC cells.
Figure 4.
CEA-related cell adhesion molecule 5 (CEACAM5) promoted the proliferation and migration of non-small-cell lung cancer cells via TGF-β signaling. (a) Immunoblot assays revealed the levels of p38 and Smad3 and the phosphorylation status of p38 and Smad2/3 in A549 cells transfected with control or CEACAM5 shRNA plasmids and stimulated in the presence of 1 ng/mL TGF-β for 2 hours. (b) Immunoblot assays indicated the levels of MMP2 and PCNA and the phosphorylation of p38 in A549 cells transfected with CEACAM5 shRNA plasmids or both CEACAM5 shRNA and pcDNA3.1-p38 plasmids. Results are presented as the mean ± SEM, *P < 0.05, **P < 0.01.
CEACAM5 induces NSCLC tumor growth via the p38/SMAD2/3 pathway in vivo
To further assess the function of CEACAM5 in vivo, we then evaluated whether CEACAM5 promoted NSCLC tumor growth in mice. The tumor volume and tumor growth curve are displayed in Figure 4a. Consistent with our expectation, the tumor volume in the CEACAM5 depletion group was significantly smaller than that in the control group (P < 0.05, Figure 5a, left). Similar results were denoted by the tumor growth curve (Figure 5a, right).
Figure 5.
CEA-related cell adhesion molecule 5 (CEACAM5) depletion impaired non-small-cell lung cancer tumor growth in vivo. (a) A549 cells infected with CEACAM5 or control shRNA lentivirus were implanted into nude mice. The tumor volume was monitored every week from the third week. After 49 days, tumors were isolated (n = 6 for each group). Representative images of tumors are presented (left), and tumor growth curves and mouse weight were calculated (right). (b) Immunohistochemical assays revealed the expression of CEACAM5 in control or CEACAM5-depleted tumor tissues. (c) Immunoblot assays further confirmed the expression of CEACAM5, MMP2, and PCNA and the phosphorylation of p38 and SMAD2/3 in tumor tissues from control or CEACAM5-depleted mice. Results are presented as the mean ± SEM, *P < 0.05, **P < 0.01.
We further conducted IHC to detect CEACAM5 expression in tumor tissues from both groups. As expected, CEACAM5 expression was obviously downregulated in CEACAM5-depleted tumor tissues, confirming the effective depletion of CEACAM5 (Figure 5b). Similarly, we detected lower MMP2 and PCNA expression following CEACAM5 depletion as well as higher phosphorylated p38 and Smad2/3 expression (Figure 5c). We thus revealed that CEACAM5 might contribute to NSCLC tumor growth via the p38/SMAD2/3 pathway in mice.
Discussion
NSCLC is a primary type of lung cancer and one of the most common and deadly malignant tumors on a global scale.17 Because of the strong invasiveness of cancer, liposomal drug delivery systems have achieved good results,18,19 and many patients are diagnosed with advanced disease and distant metastasis.20 Although great progresses have been made in past years, 5-year overall survival for NSCLC remains unsatisfactory.21 Identifying strategies for improving survival is thus necessary. Targeted therapy has been identified as an effective strategy with potential efficacy against lung cancer.22 Given the intratumoral heterogeneity in lung cancer, novel therapeutic targets remain urgently needed. In this study, we revealed that CEACAM5, a highly glycosylated protein, could serve as a novel molecular target for NSCLC.
In this study, we explored the possible involvement of CEACAM5 in NSCLC. Using qRT-PCR and IHC assays, we detected enhanced CEACAM5 expression in tumor tissues compared with that in normal tissues in patients with NSCLC, indicating the critical role of CEACAM5 in the pathogenesis of NSCLC. Concurrently, we observed high CEACAM5 expression in NSCLC cells compared with that in normal cells, which was consistent with the clinical data. Furthermore, we investigated the link between clinicopathological features and CEACAM5 expression in patients with NSCLC. We observed that clinical characteristics, including T division, lymph invasion, and histological grade, were associated with CEACAM5 expression. Similar to our findings, high CEACAM5 levels have also been implicated with enhanced growth in malignancy.14,23–25 In breast tumors, high CEACAM5 expression is correlated with reduced patient survival.24 CEACAM5 is also associated with the progression of colorectal and pancreatic cancers.26,27 These findings all indicated the important regulatory role of CEACAM5 in tumor progression.
CEACAM5 is overexpressed in multiple cancer types, including most gastrointestinal, colorectal, and pancreatic cancers.23–25 CEACAM5 could serve as a biomarker for these cancers.23–25 Recently, CEACAM5 expression has been detected using immunoassays and related techniques.25 However, its effects on lung cancer were unclear. In this study, we confirmed the high expression of CEACAM5 in NSCLC cancers, further suggesting that CEACAM5 could act as a biomarker for NSCLC.
We further explored the role of CEACAM5 in the proliferation and migration of NSCLC cells using colony formation and wound healing assays. We demonstrated that CEACAM5 promotes the proliferation and invasion of NSCLC cells. Mechanically, CEACAM5 promotes cell proliferation and invasion via p38/Smad2/3 signaling in NSCLC, and we further verified this finding in an animal model. Consistently, overproduction of CEACAM5 delayed TGF-β–induced epithelial–mesenchymal transition (EMT). CEACAM5 inhibited p38 activity and promoted invasion and metastasis, in line with previous findings that the pharmacologic inhibition of p38 promoted the outgrowth of micrometastatic tumor cells.28 Another study declared that CEACAM5 was also associated with the inhibition of cell differentiation, anoikis, and apoptosis in colon cells through its co-localization and activation of α5β1 integrin signal transduction, promoting PI3K/AKT activity.29 These studies demonstrated that CEACAM5 plays a vital role in promoting tumor progression.
TGF-β kinase is activated when TGF-β binds to TGF-βRII, resulting in Smad2/3 phosphorylation. In addition, the stress-activated protein kinase p38 plays fundamental roles in TGF-β signaling in a variety of systems.30 The p38–Smad2/3 pathway mediates multiple cellular processes.31 In this study, we noticed that CEACAM5 promoted the progression of NSCLC via this pathway and confirmed that the p38–Smad2/3 axis could serve as a therapeutic target for NSCLC treatment.
TGF-β signaling is considered a key driver of cancer cell proliferation and invasion through a variety of Smad-dependent and Smad-independent pathways, including the p38 MAPK pathway.30 In progressive cancer tissues, TGF-β promotes tumor formation, and the upregulation of TGF-β is often correlated with cancer malignancy.31 TGF-β1 induces EMT in pancreatic ductal adenocarcinoma cells along with the upregulation of various mesenchymal markers such as the small leucine-rich proteoglycan biglycan.32 These studies, together with our findings in this study, confirmed the key involvement of the TGF-β signaling pathway in cancer progression. Therefore, developing inhibitors targeting this pathway is imperative for developing anti-tumor drugs.
Conclusions
We observed the enhanced expression of CEACAM5 in human NSCLC tissues and cells and its correlations with the clinicopathological features of patients with NSCLC. We further verified the role of CEACAM5 in the progression of NSCLC in vitro and in vivo via the p38–Smad2/3 signaling pathway. Therefore, CEACAM5 could act as a potential therapeutic target for the treatment of NSCLC.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This work was supported by the Key Research & Development Program of Shandong Province (2018GGX109006).
ORCID iD
Hongbang Xu https://orcid.org/0000-0002-6788-0714
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 2.Jung G, Kim SH, Kim TG, et al. Demographic and Socioeconomic Factors for Renouncing Further Active Therapy for Patients with Brain Metastasis of Non-Small Cell Lung Cancer. Brain Tumor Res Treat 2019; 7: 112–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol 2006; 6: 433–446. [DOI] [PubMed] [Google Scholar]
- 4.Fahlgren A, Baranov V, Frangsmyr L, et al. Interferon-gamma tempers the expression of carcinoembryonic antigen family molecules in human colon cells: a possible role in innate mucosal defence. Scand J Immunol 2003; 58: 628–641. [DOI] [PubMed] [Google Scholar]
- 5.Taheri M, Saragovi U, Fuks A, et al. Self recognition in the Ig superfamily. Identification of precise subdomains in carcinoembryonic antigen required for intercellular adhesion. J Biol Chem 2000; 275: 26935–26943. [DOI] [PubMed] [Google Scholar]
- 6.Ordonez C, Screaton RA, Ilantzis C, et al. Human carcinoembryonic antigen functions as a general inhibitor of anoikis. Cancer Res 2000; 60: 3419–3424. [PubMed] [Google Scholar]
- 7.Ilantzis C, DeMarte L, Screaton RA, et al. Deregulated expression of the human tumor marker CEA and CEA family member CEACAM6 disrupts tissue architecture and blocks colonocyte differentiation. Neoplasia 2002; 4: 151–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan CH, Cook D, Stanners CP. Increased colon tumor susceptibility in azoxymethane treated CEABAC transgenic mice. Carcinogenesis 2006; 27: 1909–1916. [DOI] [PubMed] [Google Scholar]
- 9.Chan CH, Camacho-Leal P, Stanners CP. Colorectal hyperplasia and dysplasia due to human carcinoembryonic antigen (CEA) family member expression in transgenic mice. PLoS One 2007; 2: e1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Obrink B. On the role of CEACAM1 in cancer. Lung Cancer 2008; 60: 309–312. [DOI] [PubMed] [Google Scholar]
- 11.Litkouhi B, Litkouhi B, Fleming E, et al. Overexpression of CEACAM6 in borderline and invasive mucinous ovarian neoplasms. Gynecol Oncol 2008; 109: 234–239. [DOI] [PubMed] [Google Scholar]
- 12.Scheffrahn I, Singer BB, Sigmundsson K, et al. Control of density-dependent, cell state-specific signal transduction by the cell adhesion molecule CEACAM1, and its influence on cell cycle regulation. Exp Cell Res 2005; 307: 427–435. [DOI] [PubMed] [Google Scholar]
- 13.Thompson JA, Grunert F, Zimmermann W. Carcinoembryonic antigen gene family: molecular biology and clinical perspectives. J Clin Lab Anal 1991; 5: 344–366. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Fan X, Chen N, et al. Identification of CEACAM5 as a Biomarker for Prewarning and Prognosis in Gastric Cancer. J Histochem Cytochem 2015; 63: 922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Minton JP, Hoehn JL, Gerber DM, et al. Results of a 400-patient carcinoembryonic antigen second-look colorectal cancer study. Cancer 1985; 55: 1284–1290. [DOI] [PubMed] [Google Scholar]
- 16.Powell E, Shao J, Picon HM, et al. A functional genomic screen in vivo identifies CEACAM5 as a clinically relevant driver of breast cancer metastasis. NPJ Breast Cancer 2018; 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu ZT, Liu B, Li CZ. A case report of metastatic bilateral ovarian cancer due to non-small cell lung cancer with ALK gene rearrangement. Eur J Gynaecol Oncol 2019; 40: 151–153. [Google Scholar]
- 18.Yang Y, Zhao Z, Xie CW, et al. Dual-targeting liposome modified by glutamic hexapeptide and folic acid for bone metastatic breast cancer. Chem Phys Lipids 2020; 228: 104882. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Z, Zhao Y, Xie CW, et al. Dual-active targeting liposomes drug delivery system for bone metastatic breast cancer: synthesis and biological evaluation. Chem Phys Lipids 2019; 223: 104785. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Jing W, An N, et al. Prognostic significance of peripheral CD8+CD28+ and CD8+CD28- T cells in advanced non-small cell lung cancer patients treated with chemo(radio)therapy. J Transl Med 2019; 17: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.An B, Pan T, Hu J, et al. The discovery of a potent and selective third-generation EGFR kinase inhibitor as a therapy for EGFR L858R/T790M double mutant non-small cell lung cancer. Eur J Med Chem 2019; 183: 111709. [DOI] [PubMed] [Google Scholar]
- 22.Chen XY, Yang F. [ Progress of immune checkpoint inhibitors in neoadjuvant therapy of non-small cell lung cancer]. Zhonghua Wai Ke Za Zhi 2019; 57: 872–877. [DOI] [PubMed] [Google Scholar]
- 23.Zheng C, Feng J, Lu D, et al. A novel anti-CEACAM5 monoclonal antibody, CC4, suppresses colorectal tumor growth and enhances NK cells-mediated tumor immunity. PLoS One 2011; 6: e21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang XM, Zhang Z, Pan LH, et al. KRT19 and CEACAM5 mRNA-marked circulated tumor cells indicate unfavorable prognosis of breast cancer patients. Breast Cancer Res Treat 2019; 174: 375–385. [DOI] [PubMed] [Google Scholar]
- 25.Blumenthal RD, Leon E, Hansen HJ, et al. Expression patterns of CEACAM5 and CEACAM6 in primary and metastatic cancers. BMC Cancer 2007; 7: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baczynska D, Wietrzyk J, Madej J, et al. The tumorigenic potential of human CX-1 colon adenocarcinoma cells depends on carcinoembryonic antigen (CEACAM5) expression. Cell Mol Biol Lett 2003; 8: 471–486. [PubMed] [Google Scholar]
- 27.Govindan SV, Cardillo TM, Moon SJ, et al. CEACAM5-targeted therapy of human colonic and pancreatic cancer xenografts with potent labetuzumab-SN-38 immunoconjugates. Clin Cancer Res 2009; 15: 6052–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han SU, Kwak TH, Her KH, et al. CEACAM5 and CEACAM6 are major target genes for Smad3-mediated TGF-beta signaling. Oncogene 2008; 27: 675–683. [DOI] [PubMed] [Google Scholar]
- 29.Li Q, Li Y, Li J, et al. FBW7 suppresses metastasis of colorectal cancer by inhibiting HIF1alpha/CEACAM5 functional axis. Int J Biol Sci 2018; 14: 726–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Y, Wang X, Li Y, et al. PAK4 enhances TGF-beta1-induced epithelial-mesenchymal transition through activating beta-catenin signaling pathway in renal tubular epithelial cells. Int J Clin Exp Pathol 2018; 11: 3026–3035. [PMC free article] [PubMed] [Google Scholar]
- 31.Peng L, Hu C, Zhang C, et al. Anti-cancer activity of Conyza blinii saponin against cervical carcinoma through MAPK/TGF-beta/Nrf2 signaling pathways. J Ethnopharmacol 2019; 251: 112503. [DOI] [PubMed] [Google Scholar]
- 32.Yan W, Wang X, Pei Y, et al. TGF-beta1 suppresses syndecan-2 expression through the ERK signaling pathway in nucleus pulposus cells. Int J Clin Exp Pathol 2018; 11: 2017–2024. [PMC free article] [PubMed] [Google Scholar]