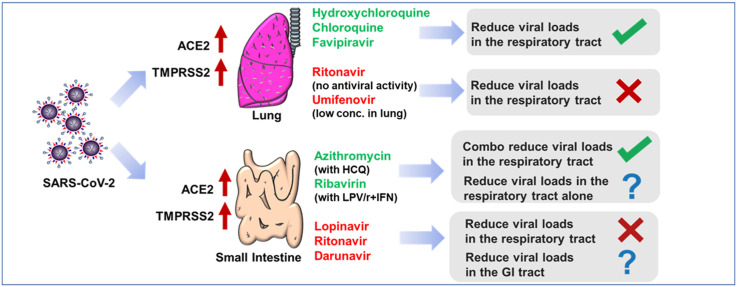
Abstract
Repurposing of approved antiviral drugs against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a promising strategy to treat Coronavirus disease 2019 (COVID-19) patients. Previously we reported our hypothesis that the antiviral drugs with high lung distributions might benefit COVID-19 patients by reducing viral loads. So far, chloroquine, lopinavir, hydroxychloroquine, azithromycin, favipiravir, ribavirin, darunavir, remdesivir, and umifenovir have been tested in COVID-19 clinical trials. Here we validated our hypothesis by comparing the pharmacokinetics profiles of these drugs and their capabilities of reducing viral load in clinical trials. According to bulk RNA and single cell RNA sequencing analysis, we found that high expression of both angiotensin converting enzyme 2 (ACE2) and transmembrane Serine Protease 2 (TMPRSS2) makes the lung and intestine vulnerable to SARS-CoV-2. Hydroxychloroquine, chloroquine, and favipiravir, which were highly distributed to the lung, were reported to reduce viral loads in respiratory tract of COVID-19 patients. Conversely, drugs with poor lung distributions, including lopinavir/ritonavir, umifenovir and remdesivir, were insufficient to inhibit viral replication. Lopinavir/ritonavir might inhibit SARS-CoV-2 in the GI tract according to their distribution profiles. We concluded here that the antiviral drugs should be distributed straight to the lung tissue for reducing viral loads in respiratory tract of COVID-19 patients. Additionally, to better evaluate antiviral effects of drugs that target the intestine, the stool samples should also be collected for viral RNA test in the future.
Keywords: Coronavirus disease 2019, Tissue distribution, Antiviral drugs, Hydroxychloroquine, Chloroquine, Favipiravir
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes Coronavirus disease 2019 (COVID-19) and killed more than 881,000 patients (as of Sep 7, 2020, World Health Organization, WHO). Previously we reported our hypothesis that the high distributions of antiviral drugs in the lung is a key factor that results in reducing viral loads in COVID-19 patients (Wang and Chen, 2020). We speculated that the disappointing clinical outcome of lopinavir might result from its low lung tissue distribution, whereas the high concentration of chloroquine in the lung might help to promote viral clearance in COVID-19 patients (Wang and Chen, 2020). So far, many antiviral drugs, including hydroxychloroquine, azithromycin, favipiravir, ribavirin, darunavir, umifenovir and remdesivir, have been tested in COVID-19 clinical trials and the results have been released. Here we validated our hypothesis by comparing the pharmacokinetics profiles of these drugs and their capabilities of reducing viral loads in clinical trials.
2. Materials and methods
2.1. Viral receptors expression landscape profile
Bulk RNA-seq data of mouse tissues are presented as mean ± S.E.M. (n = 4 biological replicates per tissue). The data was downloaded from public database National Center for Biotechnology Information (NCBI, Source: PRJNA375882 (Yan et al., 2017)). Bulk RNA-seq data of human tissues was downloaded from THE HUMAN PROTEIN ATLAS (Uhlen et al., 2015), available from https://www.proteinatlas.org. SI: Small intestine; LI: Large intestine (colon); NX: Normalized eXpression; pTPM: protein-coding transcripts per million.
2.2. Single-cell sequencing analysis
The single-cell RNA-Seq (scRNA-seq) data of human lung (Ziegler et al., 2020) and ileum (Ziegler et al., 2020) are visualized by Single Cell Portal - Broad Institute, available from https://singlecell.broadinstitute.org. The scRNA-seq data of human kidney (Liao et al., 2020) (GSE131685) was downloaded from the public Gene Expression Omnibus (GEO) database and re-analyzed and visualized by Seurat (Butler et al., 2018; Stuart et al., 2019).
2.3. Tissue/plasma ratio calculation
The tissue drug mean concentrations and the plasma drug mean concentrations were compiled from publications. The tissue drug area-under-curve (AUC) and the plasma drug AUC were compiled from publications. The tissue/plasma ratio was determined by dividing the tissue concentrations or AUC of drugs by the plasma concentrations or AUC of drugs.
3. Results
3.1. High expression of viral receptor makes tissues vulnerable to SARS-CoV-2
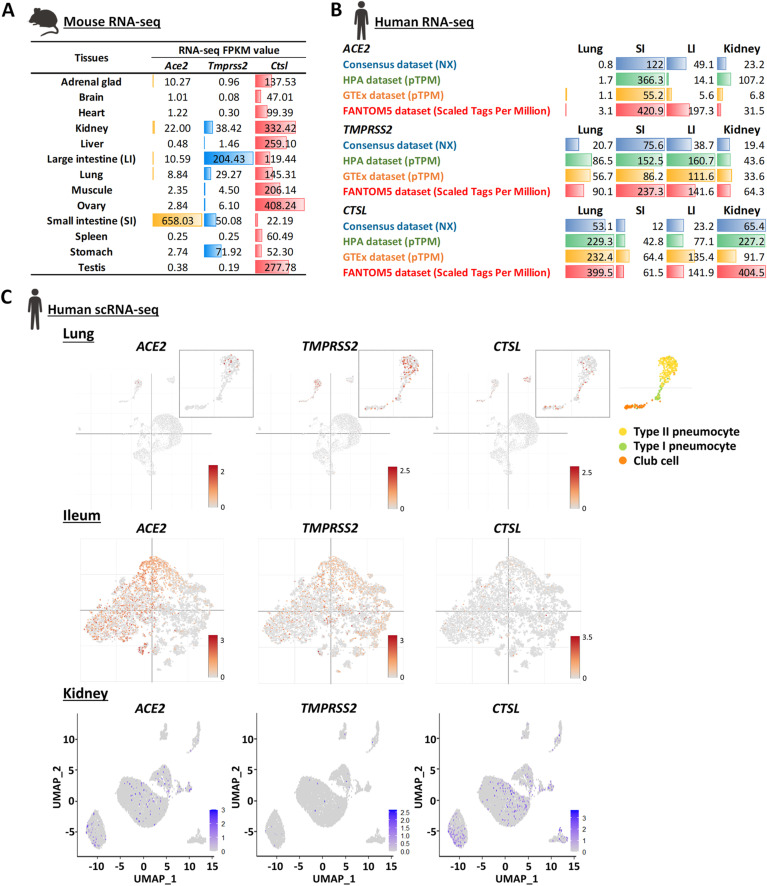
Angiotensin converting enzyme 2 (ACE2) is required for SARS-CoV-2 cell entry. The spike protein (S protein) of SARS-CoV-2 directly binds to ACE2 and is primed by transmembrane Serine Protease 2 (TMPRSS2), therefore allowing the fusion of viral membrane with the plasma membrane (Hoffmann et al., 2020). Additionally, SARS-CoV-2 was reported to enter cell through endocytosis pathway where the endosomal cysteine protease cathepsin L (CTSL) primes S protein to make the membrane fusion (Hoffmann et al., 2020). In this way, tissues that highly co-express ACE2, TMPRSS2 or CTLS are more likely to be attacked by SARS-CoV-2. We re-analyzed mouse and human bulk RNA-seq data and found these genes are highly expressed in the lung, kidney and intestine, indicating that these tissues are vulnerable for SARS-CoV-2 (Fig. 1 A and B). Consistently, COVID-19 patients exhibit cough symptoms and abnormal lung findings on CT, as well as the high viral loads in their bronchoalveolar lavage fluid samples (Liu et al., 2020). Therefore, the lung is believed to be a major target tissue of SARS-CoV-2. Diarrhea is another reported COVID-19 symptom, and the SARS-CoV-2 was also detected in the stools of COVID-19 patients (Hindson, 2020). It indicates that the intestine is another target tissue of SARS-CoV-2.
Fig. 1.
RNA-seq analysis reveals transcript levels of ACE2, TMPRSS2 and CTSL genes in lung, intestine and kidney tissues. (A) Bulk RNA-seq data of mouse tissues are presented as mean ± S.E.M. (n = 4 biological replicates per tissue). Data source: PRJNA375882 (Yan et al., 2017). (B) Bulk RNA-seq data of human tissues. Data Sources: THE HUMAN PROTEIN ATLAS (Uhlen et al., 2015), available from https://www.proteinatlas.org. SI: Small intestine; LI: Large intestine (colon); NX: Normalized eXpression; pTPM: protein-coding transcripts per million. (C) The scRNA-seq data of human lung (Ziegler et al., 2020) and ileum (Ziegler et al., 2020) are visualized by Single Cell Portal - Broad Institute, available from https://singlecell.broadinstitute.org. The scRNA-seq data of human kidney (Liao et al., 2020) (GSE131685) are re-analyzed and visualized by Seurat (Butler et al., 2018; Stuart et al., 2019).
Considering the decent expression level of ACE2 and TMPRSS2 in the kidney, we would think that SARS-CoV-2 should also attack the kidney. However, kidney involvement is not frequent in COVID-19. In most cases, SARS-CoV-2 was not observed in urine samples of COVID-19 patients, and only 0.5%–9% COVID-19 patients had the acute kidney injury (Lescure et al., 2020). We then re-analyzed the public single cell RNA-seq data to visualize the distribution of ACE2 + TMPRSS2 + and ACE2 + CTSL + cells in the human lung, intestine (ileum) and kidney. ACE2 + TMPRSS2 + cells were widely distributed in type II pneumocyte (AT2) cells of lung (Fig 1C, top panel) and ileum (Fig. 1C, middle panel), as reported by others. Conversely, massive ACE2 + CTSL + cells but not ACE2 + TMPRSS2 + were observed in the kidney (Fig. 1C, bottom panel), indicating that if SARS-CoV-2 indeed enters kidney, the virus is more likely to use CTSL for S protein priming. The CTSL-mediated priming was reported to make coronaviruses less transmissible and toxic (Shirato et al., 2017, 2018), which might partially explain why the kidney damage is barely observed in the COVID-19 patients. Another possible reason is that lung and intestine could be exposed to SARS-CoV-2 directly, and SARS-CoV-2 might be difficult to reach kidney, as in the most cases, the plasma virus was undetectable (Wolfel et al., 2020). This makes kidney reduce potential exposures to SARS-CoV-2. Taken together, the relatively direct exposure and high expression of both ACE2 and TMPRSS2 make lung and intestine vulnerable to SARS-CoV-2, suggesting that the antiviral drugs should accumulate in lung and intestine tissues to reduce viral loads.
3.2. Distribution of antiviral drugs in susceptible tissues impacts on their capabilities of reducing viral loads
Currently, many antiviral drugs have been tested in COVID-19 clinical trials and their clinical outcomes have been reported. Here we compiled the AUC or mean concentrations of these antiviral drugs in different tissues from various animal studies and calculated tissue/plasma ratio. We then ranked these ratios and identified the major distribution tissues of these antiviral drugs (Table 1 ).
Table 1.
Summary of tissue distributions of antiviral drugs in COVID-19 treatment.
| Tissues | Tissue/Plasma Ratio (AUC* or Mean Concentration) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Azithromycin* | Hydroxychloroquine | Chloroquine | Favipiravir* | Ribavirin | Lopinavir* | Ritonavir | Darunavir | Umifenovir | |
| Adrenal glad | 56.4 | 4.6 | 1.6 | 12.8 | |||||
| Brain | 2.1 | 4.3 | 0.2 | 7.1 | 0.0 | 1.7 | 4.6 | 0.1 | |
| Heart | 63.9 | 12.4 | 14.1 | 17.5 | 0.5 | 8.2 | 0.4 | 0.1 | |
| Kidney | 100.0 | 28.3 | 43.3 | 0.2 | 3.4 | 0.9 | 20.7 | 4.0 | 0.1 |
| Liver | 51.5 | 97.2 | 3.8 | 22.2 | 76.7 | 20.0 | 0.5 | ||
| Large intestine | 30.8 | 20.4 | 83.0 | ND | 6.5 | ||||
| Lung | 13.6 | 50.6 | 83.1 | 0.2 | 2.2 | 0.5 | 18.4 | 0.8 | 1.7 |
| Muscule | 91.4 | 0.2 | 3.9 | 0.1 | |||||
| Ovary | 11.2 | 0.2 | |||||||
| Small intestine | 137.0 | 18.4 | 40.8 | ND | 10.0 | 1.1 | |||
| Spleen | 34.9 | 0.2 | 2.9 | 0.4 | 10.3 | 1.4 | 0.3 | ||
| Stomach | 3.6 | 84.8 | ND | 10.0 | 10.6 | ||||
| Testis | 3.8 | 0.2 | 6.4 | 0.0 | |||||
Given that SARS-CoV-2 mainly attacks the lung and intestine, the antiviral drugs might be more effective if they could be distributed straight to the lung and intestine tissues. How potent the drugs are in vitro and how the drugs work might also impact on their capabilities of reducing viral loads in COVID-19 patients, therefore we included cell-based antiviral EC50 results as well as mechanism of action (MOA) of antiviral drugs for our discussion (Table 2 ).
Table 2.
Summary of Antiviral activities of antiviral drugs in COVID-19 treatment.
| Antiviral Drugs | MOA | EC50 [μM] | Major distribution organs | Does drug reduce viral loads in COVID-19 patients |
|---|---|---|---|---|
| Azithromycin | Block endocytosis | 2.12 | Intestine, Heart, Kidney | Reduced viral loads in nasopharyngeal swabs with hydroxychloroquines |
| Hydroxychloroquine | Block endocytosis | 4.51 | AG, Muscule, Liver, Lung | Reduced viral loads in nasopharyngeal swabs |
| Chloroquine | Block endocytosis | 1.13–2.17 | Liver, Lung, Kidney | Reduced viral loads in patients |
| Favipiravir | Inhibit RdRp | 61.88–100 | Lung, Kidney, Spleen, Brain | Reduced viral loads in nasopharyngeal swabs |
| Ribavirin | Inhibit RdRp | 109.5–500 | Intestine, Heart | Reduced viral loads in nasopharyngeal swabs with LPV/r and interferon |
| Lopinavir | Inhibit 3CLpro | 26.1 | Stomach, Intestine | Failed to reduce viral loads in oropharyngeal swab |
| Ritonavir | Inhibit 3CLpro | >100 | Liver, Kidney, Lung | Failed to reduce viral loads in oropharyngeal swab |
| Darunavir | Inhibit 3CLpro | >100 | Liver, Intestine, Stomach | Failed to reduce viral loads in nasopharyngeal swab |
| Umifenovir | Unknown | 10.7 | Stomach, Intestine, Lung | Failed to reduce viral loads in pharyngeal swab |
| Remdesivir | Inhibit RdRp | 0.11–0.77 | Unknown but not lung | Failed to reduce viral loads in nasopharyngeal and oropharyngeal swab |
Lung is one of the major distribution tissues for chloroquine (Ono et al., 2003), hydroxychloroquine (Wei et al., 1995), favipiravir (Gowen et al., 2015), ritonavir (Denissen et al., 1997) and umifenovir (Liu et al., 2013). Chloroquine showed strong inhibitory effects on SARS-CoV-2 replication in vitro (Wang et al., 2020a) and was the first reported drug that can reduce viral loads and benefit COVID-19 patients (Wang et al., 2020a). Hydroxychloroquine, the analog of chloroquine, inhibited this coronavirus in vitro with EC50 at 4.51 μM (Wang et al., 2020a), and significantly reduced viral loads in clinical trials (Gautret et al., 2020). Favipiravir, a mild RdRp inhibitor of SARS-CoV-2 (Wang et al., 2020a), was reported to accelerate viral clearance in an open-label control study (Cai et al., 2020). Ritonavir was highly distributed to lung (Denissen et al., 1997), but it did not show anti-SARS-CoV-2 activity in vitro (Choy et al., 2020). Therefore, it is not surprising that ritonavir failed to promote viral clearance in the clinical trial. Umifenovir acts as a potent viral inhibitor in vitro with EC50 at 4.11 μM (Choy et al., 2020), and the lung is one of its major distribution organs (Liu et al., 2013), but still failed to reduce viral loads in the clinical trial (Choy et al., 2020). We speculated that although the drug distribution of umifenovir in lung is relatively high, its absolute concentration in lung is not adequate to clear the virus. Only 0.833 μg/g of umifenovir was detected in lung after 54 mg/kg P.O. in rats (Liu et al., 2013), whereas 13.439 μg/g of hydroxychloroquine was probed in lung after 30 mg/kg P.O. in rats (Wei et al., 1995).
Both azithromycin combo (with hydroxychloroquine) (Gautret et al., 2020) and ribavirin combo (with lopinavir/ritonavir and interferon) (Hung et al., 2020) were reported to benefit COVID-19 patients by reducing viral loads. However, whether these two drugs can help to kill virus alone requires further validation. Lopinavir/ritonavir (LPV/r), the drug combination for HIV treatment, significantly reduced SARS-CoV viral loads in the patients with SARS 17 years ago (Stockman et al., 2006). Currently, lopinavir showed a mild inhibitory effect on SARS-CoV-2 replication in vitro with EC50 at 26.1 μM (Choy et al., 2020) but LPV/r failed to promote viral clearance in the COVID-19 patients (Cao et al., 2020). Lopinavir was not mainly distributed to the lung and its concentration in lung is only 1.18 μg equiv/ml (Kumar et al., 2004). On the other hand, the concentration of ritonavir in lung is high (Denissen et al., 1997). But ritonavir is an inhibitor of P450 3A4, which is not active in the antiviral screening (Choy et al., 2020). Considering that viral loads of SARS-CoV-2 might be much higher than viral loads of SARS-CoV in lung, the low concentration of lopinavir in lung limited its capability of reducing viral loads in the respiratory tract of COVID-19 patients (Wang and Chen, 2020). Intestine is another susceptible tissue of SARS-CoV-2, and the viral RNA has been detected in the stools (Wolfel et al., 2020). Lopinavir is mainly distributed to gastrointestinal tract (GI tract), including small intestine, large intestine and stomach (Kumar et al., 2004). In this context, lopinavir might reduce viral loads in GI tract rather than respiratory tract.
Currently, remdesivir showed the most potent inhibitory effect on SARS-CoV-2 replication in vitro with EC50 at 0.11–0.77 μM (De Meyer et al., 2020; Wang et al., 2020a). Notably, as compared with placebo, remdesivir did not accelerate the viral clearance in the COVID-19 patients (Wang et al., 2020b). Because of the poor distribution profiles in the lung, it was believed that remdesivir and its active metabolites might not be adequate to inhibit SARS-CoV-2 in the lung (Sun, 2020).
4. Discussion
We conclude here that the antiviral drugs should be distributed straight to or accumulate in the lung for reducing viral loads in respiratory tract of COVID-19 patients. Some antiviral drugs, like LPV/r, might inhibit SARS-CoV-2 in the GI tract according to their distribution profiles. However, most of samples for viral RNA test were collected from nasopharyngeal swabs and oropharyngeal saliva in the clinical trials. To better evaluate antiviral drugs that target GI tract, the stool samples should also be collected for viral RNA test in the future.
CRediT authorship contribution statement
Yan Wang: Conceptualization, Methodology, Data curation, Investigation, Writing - review & editing, writing, editing. Lei Chen: Conceptualization, Methodology, Data curation, Investigation, Writing - review & editing, writing, editing.
Declaration of competing interest
All authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment Statement
This study was supported by the National Natural Science Foundation of China (81903875).
References
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., Liao X., Gu Y., Cai Q., Yang Y., Shen C., Li X., Peng L., Huang D., Zhang J., Zhang S., Wang F., Liu J., Chen L., Chen S., Wang Z., Zhang Z., Cao R., Zhong W., Liu Y., Liu L. 2020. Experimental Treatment with Favipiravir for COVID-19: an Open-Label Control Study. Engineering. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antivir. Res. 2020;178:104786. doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer S., Bojkova D., Cinatl J., Van Damme E., Meng C.B., Van Loock M., Woodfall B., Ciesek S. Lack of antiviral activity of darunavir against SARS-CoV-2. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.1005.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissen J.F., Grabowski B.A., Johnson M.K., Buko A.M., Kempf D.J., Thomas S.B., Surber B.W. Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug Metabol. Dispos.: Biol. Fate Chem. 1997;25:489–501. [PubMed] [Google Scholar]
- Gautret P., Lagier J.C., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honore S., Colson P., Chabriere E., La Scola B., Rolain J.M., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020:105949. doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gowen B.B., Sefing E.J., Westover J.B., Smee D.F., Hagloch J., Furuta Y., Hall J.O. Alterations in favipiravir (T-705) pharmacokinetics and biodistribution in a hamster model of viral hemorrhagic fever. Antivir. Res. 2015;121:132–137. doi: 10.1016/j.antiviral.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson J. COVID-19: faecal-oral transmission? Nature reviews. Gastroenterol. Hepatol. 2020;17:259. doi: 10.1038/s41575-020-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically Proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung I.F., Lung K.C., Tso E.Y., Liu R., Chung T.W., Chu M.Y., Ng Y.Y., Lo J., Chan J., Tam A.R., Shum H.P., Chan V., Wu A.K., Sin K.M., Leung W.S., Law W.L., Lung D.C., Sin S., Yeung P., Yip C.C., Zhang R.R., Fung A.Y., Yan E.Y., Leung K.H., Ip J.D., Chu A.W., Chan W.M., Ng A.C., Lee R., Fung K., Yeung A., Wu T.C., Chan J.W., Yan W.W., Chan W.M., Chan J.F., Lie A.K., Tsang O.T., Cheng V.C., Que T.L., Lau C.S., Chan K.H., To K.K., Yuen K.Y. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G.N., Jayanti V.K., Johnson M.K., Uchic J., Thomas S., Lee R.D., Grabowski B.A., Sham H.L., Kempf D.J., Denissen J.F., Marsh K.C., Sun E., Roberts S.A. Metabolism and disposition of the HIV-1 protease inhibitor lopinavir (ABT-378) given in combination with ritonavir in rats, dogs, and humans. Pharmaceut. Res. 2004;21:1622–1630. doi: 10.1023/b:pham.0000041457.64638.8d. [DOI] [PubMed] [Google Scholar]
- Lescure F.X., Bouadma L., Nguyen D., Parisey M., Wicky P.H., Behillil S., Gaymard A., Bouscambert-Duchamp M., Donati F., Le Hingrat Q., Enouf V., Houhou-Fidouh N., Valette M., Mailles A., Lucet J.C., Mentre F., Duval X., Descamps D., Malvy D., Timsit J.F., Lina B., van-der-Werf S., Yazdanpanah Y. Infectious diseases; 2020. Clinical and Virological Data of the First Cases of COVID-19 in Europe: a Case Series. The Lancet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Yu Z., Chen Y., Bao M., Zou C., Zhang H., Liu D., Li T., Zhang Q., Li J., Cheng J., Mo Z. Single-cell RNA sequencing of human kidney. Scientific data. 2020;7:4. doi: 10.1038/s41597-019-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Pei K., Chen X.H., Bi K.S. Distribution and excretion of arbidol hydrochloride in rats. Chin. J. New Drugs. 2013;22:829–833. [Google Scholar]
- Liu Y., Yang Y., Zhang C., Huang F., Wang F., Yuan J., Wang Z., Li J., Li J., Feng C., Zhang Z., Wang L., Peng L., Chen L., Qin Y., Zhao D., Tan S., Yin L., Xu J., Zhou C., Jiang C., Liu L. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono C., Yamada M., Tanaka M. Absorption, distribution and excretion of 14C-chloroquine after single oral administration in albino and pigmented rats: binding characteristics of chloroquine-related radioactivity to melanin in-vivo. J. Pharm. Pharmacol. 2003;55:1647–1654. doi: 10.1211/0022357022340. [DOI] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J. Virol. 2017;91 doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockman L.J., Bellamy R., Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D. Remdesivir for treatment of COVID-19: combination of Pulmonary and IV administration may offer aditional benefit. AAPS J. 2020;22:77. doi: 10.1208/s12248-020-00459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C.A., Odeberg J., Djureinovic D., Takanen J.O., Hober S., Alm T., Edqvist P.H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J.M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen L. Lung tissue distribution of drugs as a key factor for COVID-19 treatment. Br. J. Pharmacol. 2020 doi: 10.1111/bph.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Nygard G.A., Ellertson S.L., Khalil S.K. Stereoselective disposition of hydroxychloroquine and its metabolite in rats. Chirality. 1995;7:598–604. doi: 10.1002/chir.530070807. [DOI] [PubMed] [Google Scholar]
- Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brunink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Yan K.S., Janda C.Y., Chang J., Zheng G.X.Y., Larkin K.A., Luca V.C., Chia L.A., Mah A.T., Han A., Terry J.M., Ootani A., Roelf K., Lee M., Yuan J., Li X., Bolen C.R., Wilhelmy J., Davies P.S., Ueno H., von Furstenberg R.J., Belgrader P., Ziraldo S.B., Ordonez H., Henning S.J., Wong M.H., Snyder M.P., Weissman I.L., Hsueh A.J., Mikkelsen T.S., Garcia K.C., Kuo C.J. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., Feldman J., Muus C., Wadsworth II Marc H., II, Kazer S.W., Hughes T.K., Doran B., Gatter G.J., Vukovic M., Taliaferro F., Mead B.E., Guo Z., Wang J.P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J.M.S., Taylor C.J., Lin B., Waghray A., Mitsialis V., Dwyer D.F., Buchheit K.M., Boyce J.A., Barrett N.A., Laidlaw T.M., Carroll S.L., Colonna L., Tkachev V., Peterson C.W., Yu A., Zheng H.B., Gideon H.P., Winchell C.G., Lin P.L., Bingle C.D., Snapper S.B., Kropski J.A., Theis F.J., Schiller H.B., Zaragosi L.-E., Barbry P., Leslie A., Kiem H.-P., Flynn J.L., Fortune S.M., Berger B., Finberg R.W., Kean L.S., Garber M., Schmidt A.G., Lingwood D., Shalek A.K., Ordovas-Montanes J. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020 doi: 10.1016/j.cell.2020.1004.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]