Dear Editor,
We read with interest the paper by Azzi and colleagues who report on the reliability of saliva testing for SARS-CoV2 infection.1 We have carried out a study to analyze the efficiency of saliva testing in monitoring the viral load of confirmed patients and get a similar conclusion. Saliva testing has been widely used in diagnosing and screening suspected COVID-19 patients due to it being easy to collect and noninvasiveness and having a high positive rate.2 , 3 For inpatients, the current standard for discharge is a negative RT-qPCR result from two sets of nasopharyngeal and throat swab specimens. Multiple throat swab specimens from each patient are needed to monitor the viral load, which will not only inevitably increase the risk of cross-infection but also increase the discomfort of the patient and cause possible complications such as bleeding.4 There is no doubt that saliva testing can greatly improve patient comfort and reduce the risk of medical staff contracting the virus.
In this study, inpatients with a diagnosis of COVID-19 provided by real-time reverse-transcriptase polymerase chain reaction (rRT-PCR) on oropharyngeal swabs in Beijing Ditan Hospital, Capital Medical University from July 10, 2020, to July 20, 2020, were included. Saliva was collected and one-step rRT-PCR was performed using the Da'an Gene 2019-nCoV Detection Kit (fluorescent PCR method, batch number: 2020032). Ct values of the ORF1a gene and N gene were also tested simultaneously. The results were considered ‘positive’ when the cycle threshold (Ct) values of FAM and VIC channels were less than 40, and there were obvious amplification curves. SPSS 24.0 and Prism 8.0 were used for statistical analyses, the difference between groups was analyzed by ANOVA and Student's t-test, P < 0.05 was considered to be statistically significant.
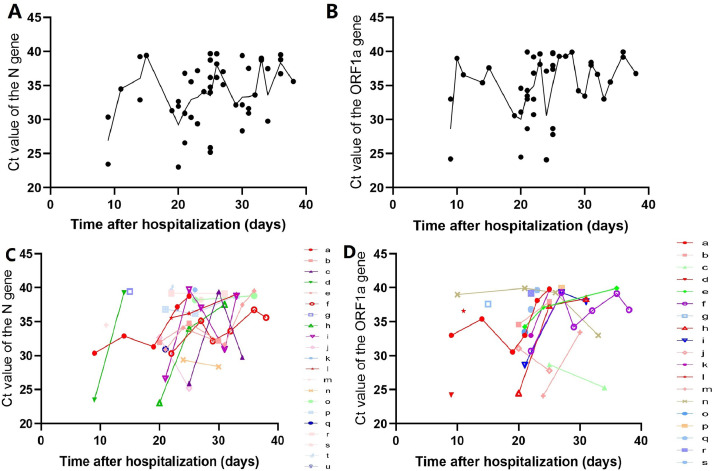
A total of 34 patients were included (Table 1 ), and 709 nucleic acid tests, consisting of 150 saliva tests (average of 4.41±1.89 times per patient), 326 oropharyngeal swab tests (average of 9.59±2.63 times per patient), and 232 sputum tests (average of 6.82±2.61 times per patient) were performed. The Ct value of 91 saliva tests was recorded; the median Ct value of the ORF1a gene was 36.64 (range 24.10–39.90), and the median Ct value of the N gene was 33.99 (range 23.03–39.67). According to the number of weeks after hospitalization, the median Ct value of the two genes gradually increased, and the amplitude gradually decreased (Fig. 1 A, B) (see Appendix Table A1). The Ct value of most patients increased with time. However, in some patients, the Ct value first decreased with increasing time and finally increased and became negative (Fig. 1C, D). Univariate analysis found that the reduction in red blood cells significantly affected the peak value of the ORF1a gene (p = 0.027), while for the N gene, there was no significant difference (p = 0.059). In multivariate analysis, no related factors that significantly affected the Ct peak were found (see Appendix Table A2).
Table 1.
Patient characteristics by severity of disease.
| Asymptomatic disease (n = 6) | Mild disease (n = 6) | Moderate disease (n = 22) | p value | |
|---|---|---|---|---|
| Age, years | 37 (28–48) | 38.7 (21–57) | 44.4 (21–64) | 0.354 |
| Sex | ||||
| Female | 2 (33.3%) | 0 | 12 (54.5%) | 0.764 |
| Male | 4 (66.7%) | 6 (100%) | 10 (45.5%) | 0.821 |
| Presenting symptoms | ||||
| Fever | 0 | 5 (83.3%) | 21 (95.5%) | 0.683 |
| Chills | 0 | 1 (16.7%) | 3 (13.6%) | 0.898 |
| Dyspnea | 0 | 0 | 4 (18.2%) | 0.853 |
| Cough | 0 | 4 (66.7%) | 11 (50%) | 0.638 |
| Runny nose | 0 | 0 | 1 (4.5%) | 0.792 |
| Blocked nose | 0 | 2 (33.3%) | 3 (13.6%) | 0.483 |
| Sore throat | 0 | 1 (16.7%) | 5 (22.7%) | 0.606 |
| Chest discomfort | 0 | 0 | 0 | — |
| Nausea | 0 | 0 | 0 | — |
| Diarrhea | 0 | 1 (16.7%) | 1 (4.5%) | 0.64 |
| Myalgia | 0 | 0 | 2 (9.1%) | 0.898 |
| Malaise | 0 | 0 | 2 (9.1%) | 0.443 |
| Loss of taste | 0 | 1 (16.7%) | 4 (18.2%) | 0.81 |
| Loss of smell | 0 | 2 (33.3%) | 4 (18.2%) | 0.316 |
| Antibody | ||||
| IgM | 0 | 2 (33.3%) | 10 (45.5%) | 0.947 |
| IgG | 0 | 1 (16.7%) | 9 (40.9%) | 0.537 |
| Blood tests on admission | ||||
| Total white blood cell count, × 10⁹ per L | 4.26 (3.39–5.33) | 5.85 (3.16–8.91) | 4.91 (2.83–10.98) | 0.21 |
| Total white blood cells <4 × 10⁹ per L | 2 (33.3%) | 1 (16.7%) | 5 (22.7%) | 0.777 |
| Lymphocyte count, × 10⁹ per L | 1.69 (1.26–2.27) | 1.87 (1.23–3.21) | 1.65 (0.58–3.38) | 0.091 |
| Lymphocytes <1•0 × 10⁹ per L | 0 | 0 | 4 (18.2) | 0.762 |
| red blood cell count, × 10⁹ per L | 4.45 (3.95–5.07) | 4.54 (2.54–5.28) | 4.89 (3.90–5.87) | 0.184 |
| red blood cell count, 4 × 10⁹ per L | 1 (16. 7%) | 1 (16.7%) | 1 (4.5%) | 0.251 |
| Platelet count, × 10⁹ per L | 202.67 (162–282) | 221.67 (147–364) | 190.74 (118–296) | 0.536 |
| Platelets <100 × 10⁹ per L | 0 | 0 | 0 | — |
Data are n (%) or median (range), unless otherwise stated. For statistical analyses, ANOVA was performed for continuous variables, and chi-squared test was performed for categorical variables.
Fig. 1.
Ct value from serial semiquantitative detection of SARS-CoV-2 for all 34 patients(A-B); Fig. 1A shows the N gene, and Fig. 1B shows the ORF1a gene. Datapoints denote the Ct value, and the curve indicates the median value.
Ct value of each patient after hospitalization(C-D). Fig. 1C shows the N gene, and Fig. 1D shows the ORF1a gene.
The total positive rate of nucleic acid detection from sputum was the highest (67.2%), followed by oropharyngeal swabs (53.1%) and saliva (36%). According to the number of weeks after hospitalization, the positive rate of nucleic acid detection from the three sample types gradually decreased, the positive rate of nucleic acid detection from saliva was 83.33% in the second week, 48% in the third week, and 0% in the seventh week (see Appendix Fig. A1). While the positive rates of nucleic acid detection from saliva, sputum, and oropharyngeal swab samples were significantly different at 3 and 6 weeks (see Appendix Table A3).
The average time for nucleic acid detection results to become negative was 27.29±7.73 days for sputum samples, 27.82±12.09 days for oropharyngeal swab samples, and 24.53±13.59 days for saliva samples (see Appendix Table A4). Univariate analysis revealed that the clinical classification had a significant impact on both the time of the positive to negative conversion of sputum, oropharyngeal swab and saliva samples (p = 0.001, p = 0.001, p = 0.012), while only red blood cell reduction had a significant effect on the positive to negative conversion time of saliva samples (p = 0.032). Multivariate analysis found that clinical classification had a significant impact on the time of sputum and oropharyngeal swab samples to become negative (p = 0.007, p = 0.002) (see Appendix Table A5). Taking sputum specimens as an example, the average time for test results to become negative in asymptomatic patients was 14 days, while the average times for patients with mild and moderate disease were 25 days and 32 days, respectively.
Using the sputum-oropharyngeal swab test results as a reference, that is, a negative result was when the nucleic acid results of both specimen types were negative, and if one of the samples had a positive test result, it is considered a positive result. The efficiency of saliva single detection method and saliva-sputum combined detection method was tested. The results showed that the total sensitivity, efficiency and specificity of saliva single detection method were 74.10%, 83.90% and 94.40%, respectively. The overall sensitivity, efficiency and specificity of saliva-sputum combined detection method were 93.40%, 94.00% and 95.20%, respectively (see Appendix Table A6). Studies have conducted research on the effectiveness of saliva to diagnosis COVID-19, and the overall efficiency rate differs, ranging from 30.7% to 100%.1 , 5, 6, 7, 8, 9 total efficiency and specificity of the saliva detection method in this study were higher than those of the sputum and oropharyngeal swab detection methods (83.90% and 94.40%, respectively). The saliva-sputum combined diagnosis is more effective, with a total efficiency and specificity of 94.00% and 95.20%, respectively. In addition, to verify the specificity of saliva testing, the saliva and oropharyngeal swab samples of 50 patients were tested, and the results of all of these patients were negative.
However, only 34 patients were included and it was not possible to collect all three sample types from every patient at the same time. We also fails to obtain the true copy of the virus, that is, the viral copies per ml of sample. Nonetheless, our results show that combined sputum-saliva detection is a reliable method for monitoring the viral load of patients recovering from COVID-19.
Funding
None.
Role of the funding source
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Data availability
Data directly supporting the study results can be found in Beijing Ditan Hospital in paper form.
Declaration of Competing Interest
None.
Acknowledgements
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2020.10.002.
Appendix. Supplementary materials
References
- 1.Azzi L., Carcano G., Gianfagna F., Grossi P., Gasperina D.D., Genoni A. Saliva is a reliable tool to detect SARS-CoV-2. J Infect. 2020;81:e45–e50. doi: 10.1016/j.jinf.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.To K.K., Lu L., Yip C.C., Poon R.W., Fung A.M. Cheng A., et al. Additional molecular testing of saliva specimens improves the detection of respiratory viruses. Emerg Microbes Infect. 2017;6:e49. doi: 10.1038/emi.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To K.K.W., Yip C.C.Y., Lai C.Y.W., Wong C.K.H., Ho D.T.Y., Pang P.K.P. Saliva as a diagnostic specimen for testing respiratory virus by a point-of-care molecular assay: a diagnostic validity study. Clin Microbiol Infect. 2019;25:372–378. doi: 10.1016/j.cmi.2018.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Chan J.F.W., Yuan S., Kok K.H., To K.K.W., Chu H., Yang J. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.To K.K.W., Tsang O.T.Y., Yip C.C.Y., Chan K.H., Wu T.C., Chan J.M.C. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis. 2020;71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azzi L., Carcano G., Dalla Gasperina D., Sessa F., Maurino V., Baj A. Two cases of COVID-19 with positive salivary and negative pharyngeal or respiratory swabs at hospital discharge: a rising concern. Oral Dis. 2020:1–3. doi: 10.1111/odi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen L., Zhao J., Peng J., Li X., Deng X., Geng Z., et al. Detection of 2019-nCoV in saliva and characterization of oral symptoms in COVID-19 patients. 2020. https://ssrn.com/abstract=3557140. [DOI] [PMC free article] [PubMed]
- 8.To K.K.W., Tsang O.T.Y., Leung W.S., Tam A.R., Wu T.C., Lung D.C. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams E., Bond K., Zhang B., Putland M., Williamson D.A. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol. 2020;58:e00776. doi: 10.1128/JCM.00776-20. -20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data directly supporting the study results can be found in Beijing Ditan Hospital in paper form.