Highlights
-
•
SARS-CoV-2 serology offers limited clinical utility for individuals.
-
•
SARS-CoV-2 serology may be useful for seroprevalence studies.
-
•
Claims regarding protective immunity or infectivity cannot be made using serology.
-
•
Assay clinical sensitivity and specificity should approach 100%.
-
•
Harmonized reporting of SARS-CoV-2 serology minimizes result misinterpretation.
Keywords: SARS-CoV-2, COVID-19, Serology, Antibody, Clinical performance, Guidance document
Abstract
Clinical laboratories across the world are working to validate and perform testing for SARS-CoV-2 antibodies. Herein, we present interim consensus guidance for Canadian clinical laboratories testing and reporting SARS-CoV-2 serology, with emphasis on the capabilities and limitations of these tests and recommendations for interpretative comments in an effort to achieve harmonized laboratory practices. The consensus document provides a broad overview of topics including sample type and contamination risk; kinetics of antibody response to COVID-19 and the impact on serology testing; clinical utility of SARS-CoV-2 serology testing; clinical performance of commercial laboratory-based assays commonly deployed in North America; recommendations for interim reporting; utility of SARS-CoV-2 antibody testing for pediatric patients; and utility of point-of-care testing. The information is based on the current literature and is subject to change as additional information becomes available.
1. Background & scope
SARS-CoV-2 is a novel betacoronavirus responsible for coronavirus disease 19 (COVID-19), first identified in Wuhan, China in December of 2019. Current evidence suggests human-to-human transmission occurs via droplets and aerosols, with a wide array of symptoms ranging from asymptomatic to severe acute respiratory distress syndrome (SARDS) [1]. Although molecular assays serve as the cornerstone of acute COVID-19 diagnosis, antibody, or serological, tests are expected to play an important role in identifying persons with prior infection of SARS-CoV-2 and assessing the extent of COVID-19 exposure in the general population.
This interim guidance document has been developed to aid Canadian clinical laboratories considering validating and performing SARS-CoV-2 serology testing. This document focuses on the appropriate testing and reporting of SARS-CoV-2 serology, with emphasis on the capabilities and limitations of these tests, and provides recommendations to guide harmonized laboratory practices. It has been developed based on current understanding of the humoral immune response to SARS-CoV-2 and is subject to change as additional information becomes available through basic and clinical investigations. Laboratories should work with local clinicians as well as regional and provincial/territorial public health departments to ensure appropriate utilization of SARS-CoV-2 serology testing.
2. Abbreviations
| COVID-19 | Coronavirus Disease 2019; disease caused by SARS-CoV-2 |
| IFU | Instructions for Use |
| IgA | Immunoglobulin isotype A |
| IgG | Immunoglobulin isotype G |
| IgM | Immunoglobulin isotype M |
| LOINC | Logical Observation Identifiers Names and Codes |
| NAAT | Nucleic acid amplification tests |
| NPV | Negative predictive value |
| MIS-C | Multisystem inflammatory syndrome in children |
| PPV | Positive predictive value |
| RT-PCR | Real-time Polymerase Chain Reaction |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 of the genus Betacoronavirus |
3. SARS-CoV-2 serology specimens and specimen contamination risk
Laboratory personnel must use appropriate personal protective equipment when collecting, handling, or analyzing patient specimens. When handling and processing samples for SARS-CoV-2 antibody testing, local guidelines for processing of potentially infectious material should be followed, based on institutional risk assessment and standard precautions [2], [3]. This includes minimizing the exposure to aerosols and droplets created during technical procedures, and appropriate personal protective equipment when collecting, handling, or analyzing patient specimens [4]. Clinical laboratories should follow the instructions provided in the Manufacturer’s Instructions for Use (IFU) including specimen collection and storage procedures, or thoroughly validate alternate conditions. Validated sample types for commercial in vitro diagnostic tests of SARS-CoV-2 antibodies typically specify whole blood, serum, or plasma matrices. The value of alternative matrices requires thorough validation and demonstration of equivalency to venipuncture specimens before implementation. For antibody testing, it is best-practice to not use specimens that are heat-inactivated, pooled, hemolyzed, contaminated with microbial or fungal growth, or poorly separated (if serum or plasma). Although sample pooling is of increasing interest to reduce cost, turn-around time, and to manage supply chain issues for SARS-CoV-2 molecular testing, inadequate data exists to evaluate the impact on serology testing. Several Manufacturer’s IFU state that pooled specimens should not be used.
To minimize potential analytical false-positive results, the necessary protocols must be in place to mitigate sample-to-sample contamination. This includes appropriate glove-hygiene when manually handling specimens (uncapping, aliquoting, pipetting, washing, etc.), adequate decontamination protocols on automated sample handing equipment (decappers, pipettors, recappers, etc.), assessment and mitigation of sample carry-over on automated instruments (e.g. use of pipet tips or stringent wash protocols), and others as per equipment manufacturer recommendations.
3.1. Recommendations
-
i)
Clinical laboratories should follow the instructions provided in the Manufacturer’s Instructions For Use (IFU) regarding acceptable sample type, sample collection and storage procedures, or thoroughly validate alternate conditions.
-
ii)
Clinical laboratories should consider performing a contamination risk assessment prior to implementing SARS-CoV-2 antibody testing to mitigate risk for potential specimen cross- contamination.
4. Kinetics of antibody response to SARS-CoV-2 and impact on serology testing
Much remains unknown regarding the extent and duration of antibody response after SARS-CoV-2 infection. In most reports, antibody detection is most reliable three weeks post-symptom onset or post-exposure, particularly in the case of IgG [5], [6], [7], [8]. In some mild and asymptomatic cases, antibodies were not detected during the timeframe of the reported studies (i.e. up to 46 days) [8], [9], [10], [11], [12]. Evidence suggests that IgM and IgG antibody levels are higher in severe cases compared to mild/asymptomatic cases [13]. At present, the dynamics of the IgM and IgA antibody response in COVID-19 are not well understood, and therefore, their utility in discriminating between recent and past infection remains questionable. To some extent, SARS-CoV-2 IgA, IgM, and IgG isotypes appear to be concomitantly expressed during convalescence [14] and at this time, there is no apparent clinical advantage of IgA or IgM versus IgG assays for SARS-CoV-2. Furthermore, the current performance characteristics of SARS-CoV-2 IgA and IgM assays (Table 2) suggest they are inadequate for clinical use and that results from these assays be interpreted with significant caution.
Table 2.
Clinical performance of common commercially available laboratory-based SARS-CoV-2 antibody assays.
| Manufacturer | Type | Target | Sens (95% CI)a N |
Spec (95% CI) N |
1% prevalence (95% CI) |
5% prevalence (95% CI) |
||
|---|---|---|---|---|---|---|---|---|
| PPV | NPV | PPV | NPV | |||||
| Abbott Architect and Alinity | IgG | N protein | 100% (95.89, 100.00) N = 88/88 |
99.63% (99.05, 99.90) N = 1066/1070 |
72.99% (50.40, 87.78) | 100% | 93.37% (84.11, 97.40) | 100% |
| Beckman | IgG | S1 RBD | 99.41% (96.77, 99.99) N = 169/170b |
99.64% (99.17, 99.88) N = 1395/1400 |
73.76% (53.96, 87.09) | 99.99% (99.96, 100.00) | 93.61% (85.93, 97.23) | 99.97% (99.78, 100.00) |
| Bio-Rad Platelia | Total | N protein | 97.50% (86.84, 99.94) N = 39/40c |
99.56% (98.73, 99.91) N = 684/687 |
69.28% (42.14, 87.47) | 99.97% (99.82, 100.00) | 92.16% (79.15, 97.33) | 99.87% (99.09, 99.98) |
| Diasorin Liaison | IgG | Spike, S1 and S2 | 97.56% (87.40, 99.47) N = 40/41 |
99.3% (98.6, 99.6) N = 1082/1090 |
57.31% (40.19, 72.85) | 99.98% (98.55, 99.67) | 87.49% (77.79, 93.32) | 99.87% (99.11, 99.98) |
| Euroimmun | IgG | Spike, S1 | 94.44% (86.38, 98.47) N = 68/72d |
99.63% (99.13, 99.88) N = 1339/1344 |
71.94% (51.62, 86.04) | 99.94% (99.85, 99.98) | 93.04% (84.76, 96.98) | 99.71% (99.25, 99.89) |
| Euroimmun | IgA | Spike, S1 | 100% (47.82, 100.00) N = 5/5d |
92.5% (87.93, 95.74) N = 185/200 |
11.87% (7.65, 17.97) | 100% | 41.24% (30.13, 53.31) | 100% |
| Ortho | Total | Spike | 100% (92.75, 100.00) N = 49/49e |
100% (99.08, 100.00) N = 400/400 |
100% | 100% | 100% | 100% |
| Ortho | IgG | Spike | 85.71% (69.71, 95.19) N = 30/35e |
100% (99.10, 100.00) N = 407/407 |
100% | 99.86% (99.68, 99.94) | 100% | 99.25% (98.34, 99.67) |
| Roche cobas | Total | N protein | 100% (88.06, 100.00) N = 29/29 |
99.81% (99.65,99.91) N = 5262/5272 |
84.19% (74.14, 90.82) | 100% | 96.52% (93.73, 98.10) | 100% |
| Siemens Atellica and Centaur | Total | S1 RBD | 100% (91.59, 100.00) N = 42/42 |
99.82% (99.34, 99.98) N = 1089/1091 |
84.64% (57.98, 95.65) | 100% | 96.63% (87.79, 99.14) | 100% |
Assay performance reflects manufacturer-generated validation data as of July 11, 2020. Refer to IFU for Manufacturer updates. EuroImmun and BioRad assays are Enzyme-Linked Immunosorbent Assays (ELISAs), and the others are Chemiluminescent Immunoassays (CLIAs).
Unless stated otherwise, ≥14 days post onset of symptoms or positive RT- PCR is presented. Assays demonstrate reduced clinical sensitivity prior to approximately 13 days.
>14 days post onset of symptoms or positive SARS-CoV-2 RT-PCR.
>8 days post onset of symptoms or positive SARS-CoV-2 RT-PCR.
>10 days post onset of symptoms or positive SARS-CoV-2 RT-PCR.
≥6 days post onset of symptoms or positive SARS-CoV-2 RT-PCR.
Over time, antibodies to other human coronaviruses diminish [15], [16], and evidence is emerging that neutralizing antibodies to SARS-CoV2 decrease but are still detectable in convalescent patients two to three months after infection [12].
4.1. Recommendations
-
i)
Clinical laboratories should advise healthcare providers that the presence of SARS-CoV-2 IgA, IgM, and IgG may co-occur during the initial phases of the humoral immune response.
-
ii)
Clinical laboratories should communicate that serological testing may be negative during early infection (e.g. within 0–14 days of initial presentation). Some patients (e.g. immunocompromised, asymptomatic or with mild disease) may fail to illicit an immune response detectable by serological tests. Antibody levels may wane over time and become undetectable by serological tests.
5. Clinical utility of SARS-CoV-2 serological testing
SARS-CoV-2 serology is anticipated to be particularly valuable for prevalence surveys because serological testing can potentially detect prior infection, regardless of symptom or hospitalization history. There are, however, several circumstances where evidence does not yet exist to support serological testing. Table 1 contrasts the types of applications where SARS-CoV-2 serology testing may or may not be appropriate. Support for these recommendations can be found through various organization guidance documents [17], [18] and reviews [19], [20], [21].
Table 1.
Potential Utility of SARS-CoV-2 antibody testing.
| Existing Evidence Supports the Application | Existing Evidence is Lacking to Support the Application |
|---|---|
|
|
Critical to evaluating the utility of SARS-CoV-2 antibodies is an understanding of SARS-CoV-2 antibody kinetics as well as the relationship between SARS-CoV-2 binding and neutralizing antibodies. As described above, the robustness and kinetics of the SARS-CoV-2 antibody response (i.e. that levels may wane over time) influences the positivity rate and may lead to an underestimation of SARS-CoV-2 exposure, particularly with seroprevalence studies. Furthermore, a positive antibody binding result does not necessarily equate to a non-infectious state, particularly in the case of non-neutralizing antibodies wherein seropositive individuals may continue to shed virus [12], [22], [23]. All currently available commercial serological assays indicate antibody binding to SARS-CoV-2 antigens. This is in comparison to virus neutralization tests (VNT), currently the gold-standard tests used to assess whether antibodies prevent viral entry and replication [24]. Not all SARS-CoV-2 binding antibodies have viral neutralizing functions, with several studies demonstrating a wide range of correlations between binding and neutralizing activity [25], [26], [27]. Although VNTs are highly specific, they provide low throughput and turn-around time, require specialized equipment and laboratories with biosafety level 3 clearance [28], thereby limiting their utility. Further studies are required to understand how binding assays correlate with neutralizing antibody titers and whether neutralizing antibodies are the primary mechanism of immune protection [29], [30], [31]. Until the relationship between SARS-CoV-2 antibodies, protective immunity, and viral infectivity are elucidated, serological testing should not be used as a sole indicator to guide decisions regarding physical distancing or need for personal protective equipment. Once established, titres of neutralizing antibodies, measured either through VNT or proxy binding assays, may be used as correlates of protection in screening of convalescent plasma and in assessing efficacy of vaccines to SARS-CoV-2.
5.1. Recommendations
-
i)
Clinical laboratories must recognize the differences between “active infection”, “prior infection”, and “immunity” and be able to communicate these differences.
-
ii)Clinical laboratories must make clear that serological testing of SARS-CoV-2 can be used to assess prior infection but should not be used in the diagnosis of active infection.
- a.
- b.
-
iii)
Clinical laboratories should communicate that a positive SARS-CoV-2 serology result supports prior infection, but not necessarily immunity to nor clearance of SARS-CoV-2 infection. Clinical laboratories should not promote so-called “immunity passports” given the current lack of evidence supporting this application.
6. Targets of serological assays and impact on SARS-CoV-2 serology testing
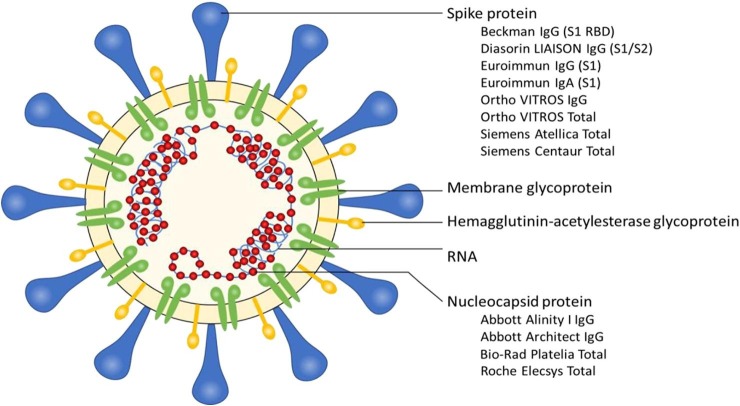
The SARS-CoV-2 virus contains four major structural proteins to which antibodies may develop: the surface or spike glycoprotein (S), which is comprised of an N-terminal S1 subunit and a C-terminal S2 subunit, envelope (E), membrane (M), and nucleocapsid (N) [35]. Major commercial assays target antibodies against either the S protein or N protein epitopes, with several assays focusing on the S1 subunit, which is thought to offer improved specificity and correlation with neutralizing titers [26], [27], [36], [37] (Fig. 1 ). Additional studies are needed to determine how commercially available antibody binding assays correlate with neutralization assays for SARS-CoV-2. At the current time, there is no clear evidence refuting the equivalency of the antigenic targets.
Fig. 1.
Schematic of the antigenic targets of commercial SARS-CoV-2 antibody assays. The list of assays is not exhaustive and reflects the instruments commonly used in Canadian clinical laboratories. At this time, not all assays listed have received Health Canada in vitro diagnostic device licensing.
7. Sensitivity, specificity, positive predictive value, and negative predictive value
To avoid false-positive and false-negative results, it is critical that both clinical sensitivity and specificity of SARS-CoV-2 antibody assays are as high as possible. Estimates of clinical performance provided by the commercial vendors are limited by the small number of positive samples included in the validation studies (see N values in Table 2), as well as non-standardized timing of serology testing in relation to symptom onset and/or positive RT-PCR result. Regarding sensitivity, individuals with reduced immune response (e.g. immunocompromised, elderly, etc.) and those with mild disease may have antibody titers that are too low to be detected by all, or any, assays. Regarding specificity, assays may be impacted by common immunoassay interferences (e.g. rheumatoid factor, antibodies to other viruses including endemic coronaviruses, human anti-animal antibodies, monoclonal antibodies, particulate matter, etc.) as well as assay-specific interferences. There is a growing number of studies aimed to better characterize test performance at different windows of time following viral exposure and to understand factors leading to false positive and negative results in asymptomatic, mild, and severe COVID-19 [26], [27], [38], [39], [40], [41].
In addition to test accuracy, the impact of disease prevalence on the positive and negative predictive values must be considered. As of August 20, 2020, with over 123,000 confirmed cases in Canada, the prevalence of reported SARS-CoV-2 infections in Canada is estimated to be approximately 2.5% [42]. As outlined in Table 2, the low prevalence has a substantial impact on the positive predictive value (PPV) of the tests. With this in mind, strategies to improve the PPV, have been proposed including: a) implementing assays with high specificity (>99.5%); b) restricting testing to high-risk populations with increased pre-test probability of disease; and, c) performing orthogonal testing in which initial positives are retested by a second method [4]. At this time, the performance of orthogonal testing strategies has not yet been systematically evaluated in a real-world setting.
7.1. Recommendations
-
i)
Tests performed by clinical laboratories should have clinical sensitivity and specificity approaching 100% as validated using well-characterized specimen set(s) with known positive cases obtained ≥14 days post-onset of symptoms or positive PCR result. Health Canada currently advises that assays exceed a sensitivity target of at least 95% for IgG or total antibodies, and a specificity target of at least 98% in pre-market studies [43]. Sensitivity and specificity information should be made available to clinicians upon request.
-
ii)
Laboratories are encouraged to report all confirmed false-positive and false-negative results to the Manufacturer and/or Health Canada (https://www.canada.ca/en/health-canada/services/drugs-health-products/medeffect-canada/adverse-reaction-reporting/medical-device.html).
-
iii)
Clinical laboratories must be familiar with known limitations associated with the SARS-CoV-2 antibody assay(s) performed and should make this information available to clinicians upon request.
-
iv)
Clinical laboratories must ensure assay performance specifications and limitations are up to date as communicated by the assay manufacturer.
8. Laboratory reporting of SARS-CoV-2 serology results
It is possible that testing for SARS-CoV-2 antibodies will be performed by all laboratory types, whether rural or urban hospitals, private reference laboratories, or Public Health laboratories. It is similarly expected that various assays will be implemented across laboratories based on performance, instrument availability, workflow, cost, and supply chain availability. It is reasonable to speculate, then, that the same patient may be tested by different clinical laboratories at different times during their clinical management, recovery and follow up. To ensure consistent messaging and avoid result misinterpretation, harmonized reporting and application of interpretation comments among laboratories is encouraged. A recent UK report [44] showed variability in the clinical interpretation of SARS-CoV-2 serology results especially with respect to inferring immunity and infectivity. This may have serious implications for ongoing public health efforts in mitigating COVID-19 and for individual patient management. At minimum, laboratories must comply with local regulations and directives in accordance with regional health authorities.
8.1. Recommendations
-
a.Test name and mapping
-
i)When mapping SARS-CoV-2 serology assays, select the appropriate LOINC. Mappings for SARS-CoV-2 tests can be found through the LOINC website.
-
ii)Specify one of the following, as appropriate:
-
•SARS-CoV-2 Total antibody
-
•SARS-CoV-2 IgG antibody
-
•SARS-CoV-2 IgM antibody1
-
•SARS-CoV-2 IgA antibody1
-
•
-
i)
1Note: Results from SARS-CoV-2 IgM and IgA antibody tests should be interpreted with significant caution until more robust performance characterization studies are available.
-
b.
Reporting negative results
-
i)
Report as negative or non-reactive, as specified in the Manufacturer’s IFU.
-
ii)
Specify the vendor and method used and the target antigen(s).
-
iii)Include as appended comments:
-
•Absence of SARS-CoV-2 antibodies does not rule out acute SARS-CoV-2 infection. False-negative results may occur in immunocompromised individuals, elderly or those with mild illness.
-
•Absence of SARS-CoV-2 antibodies should not be used to infer infectious status of an individual.
-
•
-
c.
Reporting indeterminate/inconclusive results
-
i)
Report as indeterminate or inconclusive, if specified in the Manufacturer’s IFU.
-
ii)
Specify the vendor and method used and the target antigen(s).
-
iii)Include as appended comments:
-
•An indeterminate/inconclusive result neither rules-in nor rules-out SARS-CoV-2 infection.
-
•Consider sampling time post-onset of symptoms or post-positive SARS-CoV-2 PCR result and repeat serology testing in seven to 14 days if indicated.
-
•
-
d.
Reporting positive results
-
i)
Report as positive or reactive, as specified in the Manufacturer’s IFU.
-
ii)
Specify the vendor and method used and the target antigen(s).
-
iii)Include as appended comments:
-
•Presence of SARS-CoV-2 antibodies indicates current or previous infection. False-positive results may occur due to cross-reacting antibodies or other causes. At this time, it is unknown whether antibodies indicate protective or long-term immunity.
-
•Presence of SARS-CoV-2 antibodies should not be used to infer infectious status of an individual.
-
•
9. Utility of SARS-CoV-2 serology testing in pediatric patients
Although SARS-CoV-2 infections are thought to be symptomatically mild in children, there is growing evidence of a temporal relationship between SARS-CoV-2 exposure and inflammatory presentations resembling Kawasaki disease, termed multisystem inflammatory syndrome in children (MIS-C; alternatively reported as Pediatric Inflammatory Multisystem Syndrome temporally associated with SARS-CoV-2 (PIMS-TS)). This syndrome appears several weeks after SARS-CoV-2 exposure. Compared with Kawasaki disease, MIS-C cases present with atypical Kawasaki disease symptoms and at an older age of onset [45]. In most MIS-C case series, evidence of SARS-CoV-2 exposure has been ascertained by positive RT-PCR results, presence of antibodies against SARS-CoV-2, and/or history of contact with a COVID-19-positive patient. From these studies, between 80 and 100% of patients have positive SARS-CoV-2 serology results, with lower rates of PCR positivity [46], [47], [48]. Case definitions for MIS-C have been outlined by several organizations including the WHO, CDC, and Royal College of Paediatrics and Child Health in the UK [34], [48], [49] with serological testing included as one source of evidence of SARS-CoV-2 exposure. Several Canadian provinces are in the process of adopting these case criteria and have identified MIS-C as a notifiable disease, requiring reporting of suspected cases to the local public health office [50], [51], [52].
Despite the inclusion of serological testing in MIS-C case definitions, the performance of serological testing in children and in MIS-C patients remains poorly characterized. Cases reported in the literature have used a variety of serological tests for which clinical performance characteristics are inconsistently reported or absent. Vendor performance claims appear limited to adult blood donors, while the age of COVID-19 patients are not usually described.
9.1. Recommendations
-
i)
Clinical laboratories performing SARS-CoV-2 serology testing should be prepared to support MIS-C investigations.
-
ii)
Clinical laboratories should strive to include pediatric samples in their validation studies of SARS-CoV-2 antibody assays and acknowledge to healthcare providers that the performance in children remains largely unknown. Serial serology testing may be advisable (i.e. looking for evidence of seroconversion) given possible differences in pediatric versus adult humoral immune responses.
10. SARS-CoV-2 serology point of care testing (POCT)
Although the role of SARS-CoV-2 serological point-of-care testing (POCT) remains unclear, POCT may provide several benefits when centralized laboratory testing cannot meet turnaround time needs or when access to testing is limited. At the time of writing, no SARS-CoV-2 serological POCT have received Health Canada authorization. Most SARS-CoV-2 serological POCT devices are qualitative tests based on immunochromatographic (lateral-flow) assays which typically lack the precision and accuracy of laboratory-based systems [53], [54], [55]. The devices are simple to use and require a small amount of sample. While the same performance specifications as described above apply, additional considerations must be undertaken in order to have confidence in POCT results including: proper training of the healthcare workers (typically non-laboratory professionals) who perform the test; adequate measures to prevent the spread of infectious diseases (both blood borne and respiratory); and ongoing comparison of test results against a reference method, peer group, and/or laboratory based-method [56]. Importantly, the IFU must be followed as recommended by the manufacturer in order to mitigate against potential error.
Technical validations for SARS-CoV-2 serological POCT present additional challenges beyond laboratory-based testing. Verification against a laboratory-based serological assay may yield discrepant results due to differences in the antigen substrate(s) or antibody(ies) detected by each method. Although a true reference method is unlikely to be developed for SARS-CoV-2 serology testing, the availability of well characterized sera-sets could provide an accurate basis for detection of antibody subtypes and seroconversion. Where visual interpretation of a colour change is required to obtain a POCT result, the sensitivity of detection may be influenced by background colour and/or band granularity. The type of specimen (e.g. capillary whole blood versus venous plasma) must be strictly followed as specified as any substitution may invalidate a test result. This may pose additional challenges for comparison against laboratory results. Finally, the validation data should be derived from end-users performing the POCT.
11. Recommendation
-
i)
At this time, there is insufficient evidence to support use of POCT devices for SARS-CoV-2 serology as an alternative to traditional laboratory-based systems.
12. Conclusions
The clinical laboratory has a key role to play in the health care response to the ongoing evolution of the COVID-19 pandemic. Through this interim guidance document, the recommendations provided will allow Canadian laboratories and beyond to appropriately mobilize SARS-CoV-2 serology testing in a robust and harmonized fashion. While the ultimate role of SARS-CoV-2 serology testing will continue to evolve as new data becomes available, at this time, this framework assists laboratories to support clinical and public health efforts in understanding and ongoing management of COVID-19.
Acknowledgements
We would like to thank all the Clinical Chemists and Medical Biochemists who have worked tirelessly to review the literature surrounding SARS-CoV-2 serology testing as well as the many iterations of this manuscript including: Adil Khan, Robert Kozak, Amy Lou, Cristiana Stefan, Curtis Oleschuk, Ihssan Bouhtiauy, Ivan Stevic, Kika Veljkovic, Lusia Sepiashvili, Maria Pasic, Mari DeMarco, Michael J. Knauer, Saranya Arnoldo, Sophia L. Wong, Tony Chetty, and Vincent De Guire. This document reflects the contributions of many laboratory scientists. We would additionally like to thank all the medical laboratory staff who bravely and tirelessly collect and perform SARS-CoV-2 molecular and serology testing and for the personal sacrifices they have made during the 2020 pandemic.
References
- 1.Garg S. Hospitalization rates and characteristics of patients hospitalized with laboratory-confirmed coronavirus disease 2019 – COVID-NET, 14 States, March 1–30, 2020. MMWR Morb. Mortal. Wkly Rep. 2020;69:458–464. doi: 10.15585/mmwr.mm6915e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Coronavirus Disease 2019 (COVID-19): Biosafety for Specimen Handling. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/lab-biosafety-guidelines.html.
- 3.World Health Organization. Laboratory biosafety guidance related to coronavirus disease (COVID-19). 2020. https://www.who.int/publications/i/item/laboratory-biosafety-guidance-related-to-coronavirus-disease-2019-(covid-19).
- 4.Respiratory Virus Infections Working Group Canadian Public Health Laboratory Network best practices for COVID-19. Can. Commun. Dis. Rep. 2020;46:112–118. doi: 10.14745/ccdr.v46i05a02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun B. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg. Microbes Infect. 2020;9:940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G., Nie S., Zhang Z. Longitudinal change of severe acute respiratory syndrome coronavirus 2 antibodies in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:183–188. doi: 10.1093/infdis/jiaa229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryan A. Performance characteristics of the abbott architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yongchen Z. Different longitudinal patterns of nucleic acid and serology testing results based on disease severity of COVID-19 patients. Emerg. Microbes Infect. 2020;9:833–836. doi: 10.1080/22221751.2020.1756699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burbelo P.D., Riedo F.X., Morishima C. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis. 2020;222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z.L. Antibody profiles in mild and severe cases of COVID-19. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long Q.X. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 12.Long Q.X. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020 doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 13.Lynch K.L., Whitman J.D., Lacanienta N.P. Magnitude and kinetics of anti-SARS-CoV-2 antibody responses and their relationship to disease severity. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma H. Serum IgA, IgM, and IgG responses in COVID-19. Cell. Mol. Immunol. 2020;17:773–775. doi: 10.1038/s41423-020-0474-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020 doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Q., Zhu L., Ni Z., Meng H., You L. Duration of serum neutralizing antibodies for SARS-CoV-2: Lessons from SARS-CoV infection. J. Microbiol. Immunol. Infect. 2020 doi: 10.1016/j.jmii.2020.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Centers for Disease Control and Prevention. Interim guidelines for COVID-19 antibody testing in clinical and public health settings. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html (2020).
- 18.Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19: Serologic Testing: https://www.idsociety.org/COVID19guidelines/serology.
- 19.Van Caeseele P., Bailey D., Forgie S.E., Dingle T.C., Krajden M. SARS-CoV-2 (COVID-19) serology: implications for clinical practice, laboratory medicine and public health. CMAJ. 2020 doi: 10.1503/cmaj.201588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng M.P., Yansouni C.P., Basta N.E. Serodiagnostics for severe acute respiratory syndrome-related coronavirus-2: a narrative review. Ann. Intern. Med. 2020 doi: 10.7326/M20-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant J.E., Azman A.S., Ferrari M.J. Serology for SARS-CoV-2: Apprehensions, opportunities, and the path forward. Sci. Immunol. 2020;5(47) doi: 10.1126/sciimmunol.abc6347. [DOI] [PubMed] [Google Scholar]
- 22.Wölfel R. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 23.Atkinson B., Petersen E. SARS-CoV-2 shedding and infectivity. Lancet. 2020;395:1339–1340. doi: 10.1016/S0140-6736(20)30868-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kilic T., Weissleder R., Lee H. Molecular and immunological diagnostic tests of COVID-19: current status and challenges. iScience. 2020;23(8) doi: 10.1016/j.isci.2020.101406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gattinger P., Borochova K., Dorofeeva Y. Antibodies in serum of convalescent patients following mild COVID-19 do not always prevent virus receptor binding. Allergy. 2020 doi: 10.1111/all.14523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jääskeläinen A.J., Kuivanen S., Kekäläinen E. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meschi S., Colavita F., Bordi L. Performance evaluation of Abbott ARCHITECT SARS-CoV-2 IgG immunoassay in comparison with indirect immunofluorescence and virus microneutralization test. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K., Long Q.X., Deng H.J. Longitudinal dynamics of the neutralizing antibody response to SARS-CoV-2 infection. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amanat F. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020 doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao Y. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients' B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandrashekar A. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020 doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo L. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao R. Early detection of SARS-CoV-2 antibodies in COVID-19 patients as a serologic marker of infection. Clin. Infect. Dis. 2020 [Google Scholar]
- 34.World Health Organization. Multisystem inflammatory syndrome in children and adolescents temporally related to COVID-19. https://www.who.int/news-room/commentaries/detail/multisystem-inflammatory-syndrome-in-children-and-adolescents-with-covid-19. (2020).
- 35.Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okba N.M.A. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weidner L., Gänsdorfer S., Unterweger S. Quantification of SARS-CoV-2 antibodies with eight commercially available immunoassays. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theel E.S., Harring J., Hilgart H., Granger D. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin. Microbiol. 2020;58(8) doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perkmann T., Perkmann-Nagele N., Breyer M.K. Side by side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Charlton C.L., Kanji J.N., Johal K. Evaluation of six commercial mid to high volume antibody and six point of care lateral flow assays for detection of SARS-CoV-2 antibodies. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.01361-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johns Hopkins University and Medicine Coronavirus Resource Center. COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (JHU). https://coronavirus.jhu.edu/map.html. (2020).
- 43.Government of Canada. COVID-19 serological testing devices: Notice on sensitivity and specificity values. https://www.canada.ca/en/health-canada/services/drugs-health-products/covid19-industry/medical-devices/testing/serological/notice-sensitivity-specificity-values.html (2020).
- 44.Bermingham W.H., Wilding T., Beck S., Huissoon A. SARS-CoV-2 serology: Test, test, test, but interpret with caution! Clin. Med. (Lond.) 2020 doi: 10.7861/clinmed.2020-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCrindle B.W., Manlhiot C. SARS-CoV-2-related inflammatory multisystem syndrome in children: different or shared etiology and pathophysiology as Kawasaki disease? JAMA. 2020 doi: 10.1001/jama.2020.10370. [DOI] [PubMed] [Google Scholar]
- 46.Toubiana J. Kawasaki-like multisystem inflammatory syndrome in children during the covid-19 pandemic in Paris, France: prospective observational study. BMJ. 2020;369 doi: 10.1136/bmj.m2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Belhadjer Z. Acute heart failure in multisystem inflammatory syndrome in children (MIS-C) in the context of global SARS-CoV-2 pandemic. Circulation. 2020 doi: 10.1161/CIRCULATIONAHA.120.048360. [DOI] [PubMed] [Google Scholar]
- 48.Centers for Disease Control and Prevention. Case definition for MIS-C. https://www.cdc.gov/mis-c/hcp/ (2020).
- 49.Whittaker E. Clinical characteristics of 58 children with a pediatric inflammatory multisystem syndrome temporally associated with SARS-CoV-2. JAMA. 2020 doi: 10.1001/jama.2020.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ministry of Health Government of Alberta. Public health disease management guidelines: Multisystem Inflammatory Syndrome in Children (MIS-C). https://open.alberta.ca/dataset/7332b06a-515e-4194-8ffa-a80e4fc0ea3b/resource/d8a1b57f-3117-4f6d-afa8-c355e260a55b/download/health-misc-c-public-health-disease-management-guidelines.pdf (2020).
- 51.Public Health Ontario. COVID-19 – What we know so far about...Kawasaki disease-like illness. https://www.publichealthontario.ca/-/media/documents/ncov/covid-wwksf/2020/05/what-we-know-kawasaki-disease-like-illness.pdf?la=en (Queens’s Printer for Ontario, Toronto, Ontario 2020).
- 52.BC Centre for Disease Control BC Ministry of Health. Multisystem Inflammatory Syndrome in Children (MIS-C) Temporally Associated with COVID-19: Guidance for Clinicians in B.C. http://www.bccdc.ca/Health-Professionals-Site/Documents/COVID19_MIS-C_ClinicianGuidance.pdf (2020).
- 53.Lisboa Bastos M. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370 doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deeks J.J. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst. Rev. 2020;6 doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espejo A.P. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020 doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yip P.M., Venner A.A., Shea J. Point-of-care testing: A position statement from the Canadian Society of Clinical Chemists. Clin. Biochem. 2018;53:156–159. doi: 10.1016/j.clinbiochem.2018.01.015. [DOI] [PubMed] [Google Scholar]