Abstract
The emergence of a new coronavirus, in around late December 2019 which had first been reported in Wuhan, China has now developed into a massive threat to global public health. The World Health Organization (WHO) has named the disease caused by the virus as COVID-19 and the virus which is the culprit was renamed from the initial novel respiratory 2019 coronavirus to SARS-CoV-2. The person-to-person transmission of this virus is ongoing despite drastic public health mitigation measures such as social distancing and movement restrictions implemented in most countries. Understanding the source of such an infectious pathogen is crucial to develop a means of avoiding transmission and further to develop therapeutic drugs and vaccines. To identify the etiological source of a novel human pathogen is a dynamic process that needs comprehensive and extensive scientific validations, such as observed in the Middle East respiratory syndrome (MERS), severe acute respiratory syndrome (SARS), and human immunodeficiency virus (HIV) cases. In this context, this review is devoted to understanding the taxonomic characteristics of SARS-CoV-2 and HIV. Herein, we discuss the emergence and molecular mechanisms of both viral infections. Nevertheless, no vaccine or therapeutic drug is yet to be approved for the treatment of SARS-CoV-2, although it is highly likely that new effective medications that target the virus specifically will take years to establish. Therefore, this review reflects the latest repurpose of existing antiviral therapeutic drug choices available to combat SARS-CoV-2.
Keywords: COVID-19, HIV, Origin and taxonomy, Molecular mechanisms, Potential therapeutics
List of abbreviation
| CoVs | Coronaviruses |
| CNS | Central nervous system |
| SARS-CoV-2 MERS-CoV |
Severe acute respiratory syndrome coronavirus 2 Middle East respiratory syndrome coronavirus |
| ARDS | Acute respiratory distress syndrome |
| WHO | World Health Organization |
| HIVs | Human immunodeficiency viruses |
| SIV | Immunodeficiency virus |
| GP41 | Glycoprotein-41 |
| CCR5 | C-C chemokine receptor-5 |
| CXCR4 | C-C chemokine receptor-4 |
| (IFI)-16 | Interferon-gamma induced protein-16 |
| IL-1β | Interleukin-1β |
| ACE2 | Angiotensin-converting enzyme-2 |
| FGF2 | Basic fibroblast growth factor 2 |
| GCSF | Granulocyte-colony stimulating factor |
| IFNγ | Interferon-gamma |
| IP10 | IFN-γ-inducible protein-10 |
| MCP1 | Monocyte chemoattractant protein 1 |
| MIP1A | Macrophage inflammatory proteins |
| TNF-α | Tumor necrosis factor-alpha |
| TGF-β | Transforming growth factor-beta |
| NCBD | National Center for Biotechnology Development |
| FDA | Food and Drug Administration |
| LPV | Lopinavir |
| RTV | Ritonavir |
| PWH | People with HIV |
1. Introduction
Coronaviruses (CoVs) are a group of enveloped and positive single-stranded RNA genome viruses that infect both animals and humans (Smith and Denison, 2013; Xu et al., 2020). They contain the largest known RNA genomes ranging from 27 to 32 kilobases in length (Smith and Denison, 2013). Coronavirus-related infections are known to affect the liver, central nervous system (CNS), respiratory and gastrointestinal tract of various birds and mammals, including humans (Xu et al., 2020). They are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus (Fung et al., 2020). The novel 2019 coronavirus (2019-nCoV) also belongs to the same group of CoVs. It was named ‘severe acute respiratory syndrome coronavirus 2’ (SARS-CoV-2) by the International Committee on Taxonomy of Viruses (ICTV) (Fung et al., 2020). The transmission pattern of SARS-CoV-2 primarily involves direct close contact with the infected person via nose and mouth secretions, however it can also be transmitted indirectly through contaminated objects or surfaces (World Health Organization). It was discovered in late December 2019 in Wuhan city of China in patients with acute respiratory distress syndrome (ARDS) symptoms, such as fever, cough, and dyspnea (Q.Li et al., 2020b; F.Zhu et al., 2020a) after isolated from the respiratory epithelium (Xu et al., 2020). The genome sequence analysis of SARS-CoV-2 showed that it belongs to the Betacoronavirus genus (β-CoV) (Lu et al., 2020b). The SARS-CoV-2-related disease was recently named as a novel respiratory 2019 coronavirus disease (COVID-19) by the World Health Organization (WHO) (Xu et al., 2020). Up to date, it is unclear how SARS-CoV-2 interacts with the host antiviral immunity, hence lessons can be learned from previous studies of other members of the coronavirus family and also human pathogenic viruses, such as human immunodeficiency viruses (HIVs) (Fung et al., 2020).
HIVs are the most studied viruses and the best models for understanding the interplay between host antiviral defense and viruses (Fung et al., 2020), contributing to a comprehensive understanding of the viral biology and pathogenesis (Schwetz and Fauci, 2018). The Joint United Nations Program (UNAIDS) data from 2018 reported that more than 77 million people had been diagnosed with HIV, 35 million of whom died as a result of severe disease condition, and currently, approximately 40 million people live with HIV (Schwetz and Fauci, 2018). Besides, HIVs may provide the basic framework for understanding the pathogenicity and cross-species transmission of SARS-CoV-2. In general, many similarities are present between SARS-CoV-2 and HIVs in terms of cross-species transmission (Fung et al., 2020). Based on its genetic diversity, HIVs are classified into two types, namely HIV-1 and HIV-2. The most common and well-studied form is HIV-1 (Pham and Mesplède, 2018). Fung and co-authors made an in-depth comparison between SARS-CoV-2 and HIVs (Fung et al., 2020). According to them, what is learned from HIV is very instructive and highly relevant to SARS-CoV-2 due to the following reasons.
-
1.
SARS-CoV-2 and HIVs are recognized as a potentially zoonotic origin.
-
2.
There are no or mild symptoms when parental viruses of SARS-CoV-2 and HIV infect the reservoir hosts, but severe symptoms are developed when they infect humans. However, this claim is somewhat controversial with the recent study conducted by Liu et al. (2020b) on Malayan pangolins proposed as intermediate hosts of SARS-CoV-2, suffered from serious respiratory disease and failed to be rescued (Liu et al., 2020b).
-
3.
Both SARS-CoV-2 and HIV are derived from their natural reservoirs in animals and spread to humans through cross-species transmission.
Up to date, there is no specific therapeutic drug available against the novel COVID-19. The current steps taken for the management of COVID-19 include isolation of COVID-19 patients, social distancing, provision of supportive medical care, enforcement of travel restrictions, and globally enforced lockdown (Zhang et al., 2020). Many pharmaceutical sectors have already started developing or are in the midst of conducting clinical trials on various potential drugs useful for the treatment of COVID-19 but more insights into the underlying pathobiology are required (Zhang et al., 2020; G.Li and De Clercq, 2020). Thus, this review will comprehensively discuss the taxonomic characteristics of SARS-CoV-2 and HIVs, together with their molecular mechanisms of viral infections and the latest repurpose of existing antiviral drug choices to combat COVID-19.
2. Taxonomy of SARS-CoV-2 and HIV
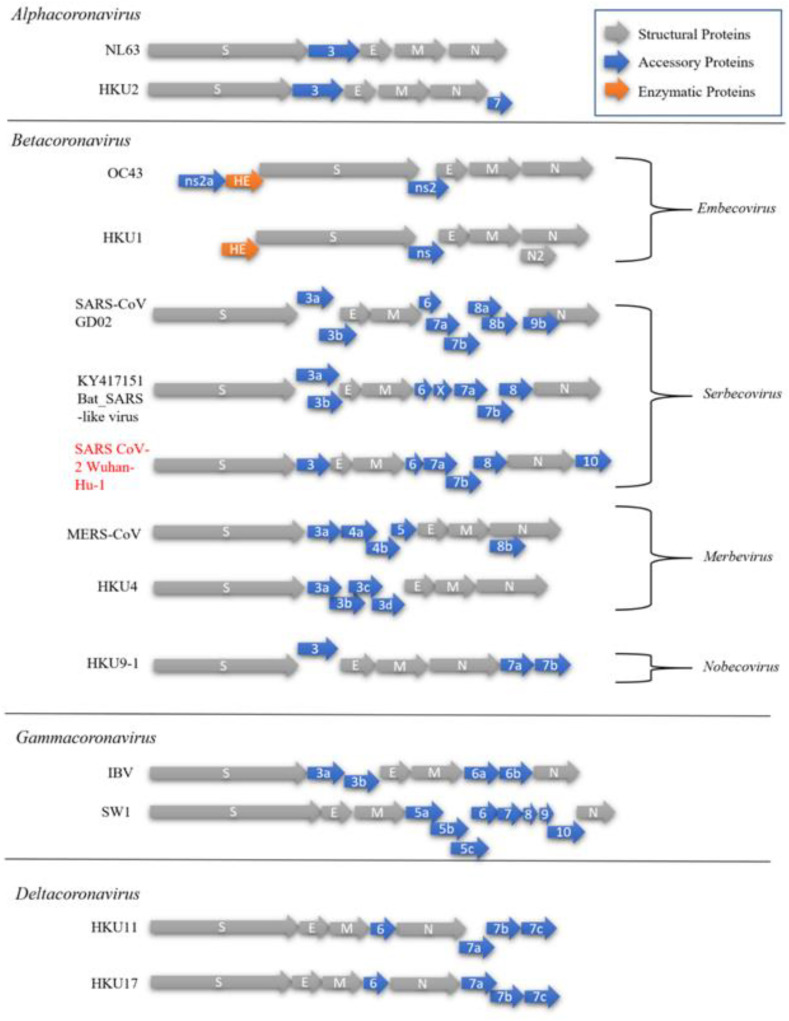
With the onset of a global pandemic looming over civilization, efforts to correctly classify COVID-19, and definitively study it is ever more pressing. Initially, the outbreak was identified back in December 2019 as a novel coronavirus (2019-nCov) and was later recognized as highly contagious with the impacts of massive global proportions. Pursuant to that, studies have begun to establish the origin and taxonomy of the virus which was then named SARS-CoV-2 by the Coronavirus study group of the International Committee on Taxonomy of Viruses. The disease caused by SARS-CoV-2 was later renamed by WHO to COVID-19 on the 11th of February 2020 (Gorbalenya, 2020). As mentioned, SARS-CoV-2 is a member of the order Nidovirales, suborder Coronavirinae divided into family Coronaviridae, subfamily Orthocoronavirinae and further classified into four genera such as Alpha-, Beta-, Gamma- and Delta-coronavirus. It is the latest addition to an already existing list of 6 other viruses, including 229E, OC43, NL63, HKU1, Middle East respiratory syndrome (MERS) CoV and severe acute respiratory syndrome (SARS) CoV known as human CoVs (HCoVs) due to their ability to cause human infections (Andersen et al., 2020). Aside from NL63 and 229E; which belong to Alphacoronavirus and the remaining five viruses come under the Betacoronavirus genus. However, SARS-CoV, MERS-CoV, and SARS-CoV-2 are known to cause severe disease leading to death, whereas 229E, OC43, NL63, and HKU1 have only been associated with mild symptoms in infected persons although they are recognized as a potentially zoonotic origins (Andersen et al., 2020; Corman et al., 2018). All members of the Coronaviridae contain enveloped, positive-stranded spherical RNA virions with a length of 120–160 nm and are usually decorated with large visible petal-shaped surface projections known as ‘spikes’ when viewed under an electron microscope (Singh et al., 2020). In addition, members of the Coronaviridae share common characteristics, which are outlined in (Table 1 ). Within the family, there is also another subfamily such as Torovirinae comprising two other genera Torovirus and Bafinivirus. They are different from Coronaviridae mainly due to their different tubular virion shape. The morphology of virion, such as Coronavirinae has been unraveled, as illustrated in (Fig. 1a). Moreover, the classification of various genera within the subfamily is a little more complicated with high CoV recombination frequency, thus necessitating the development of precise criteria proposed over the years (Corman et al., 2018). Currently, the primary means of establishing their phylogenetic relationship is through the genomic structure, as shown in (Fig. 2 ). It is based on these differences that a particular host may be infected by specific genus of the virus, for instance, Alpha- and Beta-coronavirus infects only mammals and Gamma- and Delta-coronavirus infects primarily birds and some studies suggest the possibility of the latter genera infecting mammals as well (Cui et al., 2019; King et al., 2012; Woo et al., 2012). The specific distinctions between four genera are closely associated with (1) the unique type of nsp1 known as a non-structural RNA-binding protein within their genome which may differ in size and sequence and (2) the presence or absence of a commonly shared accessory gene (Sheikh et al., 2020). Several examples of viruses within the genera with their unique accessory proteins and genome arrangements can be found in (Fig. 3 ).
Table 1.
Common Characteristics shared across the members of Coronaviridae.
| Characteristics | Description | References |
|---|---|---|
| Virion | Enveloped and perforated with large petal-shaped ‘spiky’ surface projections | (Schoeman and Fielding, 2019) |
| Nucleocapsid | Helical shaped comprising of the genome and multiple copies of a single phosphoprotein (N) | (Grunewald et al., 2018) |
| Envelope | Made up of 2 membrane protein
|
(Neuman et al., 2011; Song et al., 2004) |
| Genome | Linear, unimolecular, positive-sense RNA, 26-32 kb long, capped, polyadenylated and structurally polycistronic | (Cui et al., 2019) |
| Genome organization | Generally, 5’-UTR-replicase-S-M-N-UTR-3′ | (Fung et al., 2020; Lu et al., 2020b) |
| Replicase gene | Overlapped with ORFs 1a and 1b that codes two huge polyproteins, pp1a and pp1ab that are processed autoproteolytically into 16 non-structural proteins involved in genome transcription and replication. | (Sheikh et al., 2020; Singh et al., 2020; Chan et al., 2020; Clavel et al., 1986; Zahoor et al., 2014) |
| Morphogenesis | Assembly of virion takes place at the smooth intracellular membranes of the endoplasmic reticulum/early Golgi compartments. | (Masters, 2019) |
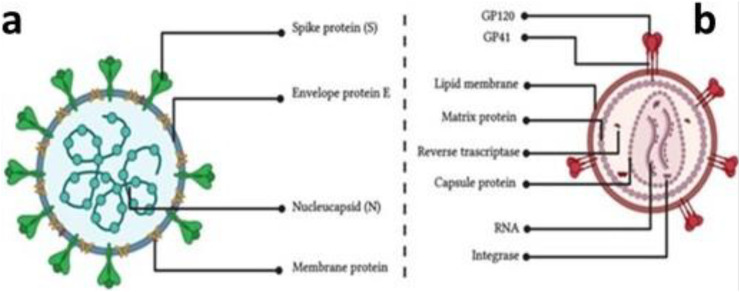
Fig. 1.
Schematic illustration of the virion morphology of coronavirus (a) and HIV (b).
Fig. 2.
The general genome structure of Coronaviridae shared across the family, consisting of a positive-sense ssRNA genome of 27–32 kb in size. Starting from the 5′-terminal with approximately two-thirds of the genome encoding for polyprotein, pp1ab, which is cleaved into 16 non-structural proteins that are involved in genome transcription and replication. The 3′-terminal end possesses the primary structural genes encoding for ‘spike’ (S) protein, envelope (E) protein, membrane (M) protein and nucleocapsid (N) protein.
Fig. 3.
Comparison of various coronavirus members of their respective genera: NL63 associated with lower respiratory tract disease, Rhinolophus bat coronavirus HKU2, Human coronavirus OC43, HKU1 possessing the enzyme haemagglutinin-esterase, severe acute respiratory syndrome coronavirus (SARS-CoV) strain GD02, Bat (SARS)-like Virus KY417151, severe acute respiratory syndrome coronavirus (SARS-CoV-2) isolated from Wuhan in December 2019 Genbank Accession number: MN908947 (Wu et al. 2020), Middle East respiratory syndrome coronavirus (MERS-CoV), Bat coronavirus HKU 14–1, Bat coronavirus HKU9-1, Infectious bronchitis virus (IBV), Beluga whale coronavirus SW1, Bulbul coronavirus HKU11 and Sparrow coronavirus HKU17.
The complete SARS-CoV-2 genome sequencing data had begun to emerge as the real gravity of the situation started to loom over the medical personnels who first encountered this novel coronavirus back in December 2019. Their findings have since been published and provided us the information necessary to classify SARS-CoV-2 into its respective group under the Serbecovirus (β-B Group) subgenus (Lu et al., 2020b; Chan et al., 2020). The sequence is publicly available on GenBank with the accession number MN908947 possessing 30,474 nucleotides with close relations to bat SARS-like coronavirus (CoV) isolate bat SL-CoVZC45 (Genbank accession number MG772933) with 89.1% nucleotide identity similarities (Lu et al., 2020b).
In contrast, the origins of HIV have been a subject of debate since its discovery back in 1980s with many theories floating about and evidences not sufficiently conclusive. What is known is that infected individuals were at risk of developing acquired immune deficiency syndrome (AIDS), which at that time resulted in opportunistic pathogens infecting the individual and due to a pronounced weakened immune system, the individual would succumb eventually to the infection. Current data suggest that over the past three decades, HIV has caused more than 35 million deaths worldwide with 35 million living with the virus (Pharr et al., 2017). HIV is classified with other known immunodeficiency viruses, which belong to phylum Artveviricota, order Ortervirales, and family Retroviridae. Further, it has been categorized into subfamily Orthoretrovirinae and genus Lentivirus including 10 species of viruses, such as feline immunodeficiency virus (FIV), bovine immunodeficiency virus (BIV), simian immunodeficiency virus (SIV) and dog immunodeficiency virus (DIV) (Vemuri et al., 2020). Besides, three other retroviruses, such as Human T-cell Lymphotropic Virus type 1, 2, 3 (HTLV 1, 2, 3) exist within the same subfamily but genus deltaretrovirus also affects the immune system in humans (Calattini et al., 2005).
As mentioned earlier, HIV has been categorized into two types; HIV-1 and HIV-2, the former being known as a more pathogenic than the latter, making up the majority of AIDS cases around the globe, whereas HIV-2 mainly infects individuals in West Africa (Cloyd et al., 2000). In general, the most plausible route of evolution can be established by genomic sequence of HIV, while its origin is yet to be understood. In 1986, the genomic analysis found that HIV-2 virus was morphologically similar but genetically different from type 1 and caused more serious AIDS disease in western Africa (Jaffar et al., 2004). Another genomic study showed that HIV-2 is firmly associated with the simian immunodeficiency virus (SIV) isolated from macaque monkeys (Nakayama and Shioda, 2015). Since then, the simian close relatives have been further expanded with more and more species of primates in Sub-Saharan Africa found to house similar viruses collectively named SIV with special suffixes denoting their species of origin. In general, SIV-derived host species have been found to be non-pathogenic, but can become pathogenic once they cross the species (Meyerson et al., 2018); resulting in the discovery of current pandemic HIV1 strain (group M or main) that is similar to SIVcpz in Chimpanzees (Pan troglodytes) after closer investigation. In addition, the natural host of viruses was identified in Cameroon (Hahn et al., 2000; Keele et al., 2006).
The morphological structure of HIV is usually spherical, enveloped with 80–100 nm in diameter, and 8 nm surface glycoprotein projections. The internal structure of the virus nucleocapsid, such as a rod or truncated cone-shaped has been shown in (Fig. 1b). The viral genome consists of a linear dimer of positive-sense ssRNA with each monomer at approximately 7–13 kb in size. Each monomer is polyadenylated at the 3′ end and contains a cap structure at the 5′ end. Within the virus, proteins make up about 60% of the biomolecular content, including two envelope proteins, SU (surface) and TM (Transmembrane), 3–6 internal and non-glycosylated structural proteins comprising of CA (capsid), NC (nucleocapsid), MA (matrix) and some unspecified functional proteins. Meanwhile, other proteins such as PR (protease), IN (integrase), and RT (reverse transcriptase) also exist inside the virus.
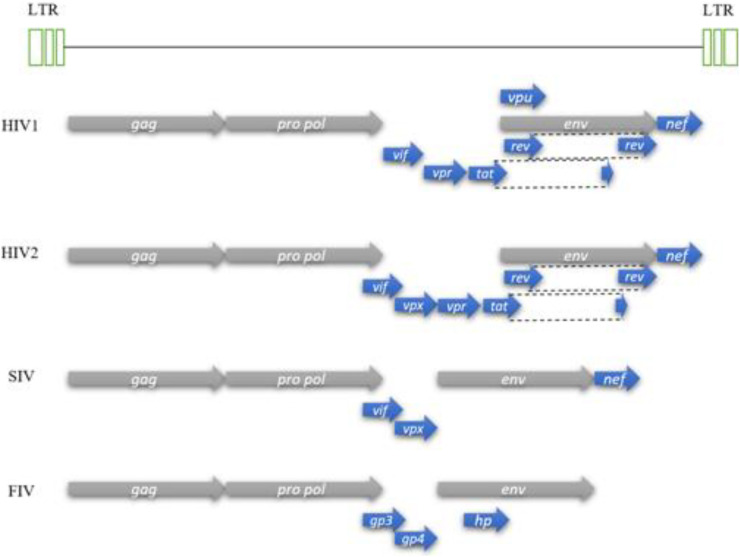
HIV viral genome follows the common pattern within the subfamily Orthoretrovirinae, which carries two copies with the arrangement order: 5′-gag-pro-pol-env-3′, each at approximately 9.3 kb in size. A schematic illustration of the HIV genome is shown in (Fig. 4 ) with comparisons made with other species within the Lentivirus genus. Upon successful infection and incorporation of retroviral DNA into the chromosomal DNA of the host, cellular RNA polymerase II transcribes DNA into virion RNA and mRNA which serves as a template for the translation of gag, pro and pol genes into the resulting protein from the 5′-half end. As a result, polyprotein precursors are formed and cleaved to produce the structural proteins such as PR, IN, and RT. The other half of mRNA 3′ end is translated into viral envelope precursors with other accessory proteins. In general, the unique sequences of viruses give each genus of a specific viral family its distinguishing characteristics allow its classification into the respective genus, which is Lentivirus in case of HIV. In addition to the primary structural genes reported, members of the Lentivirus genus also have additional genes, for instance, accessory protein genes vif, vpr, vpu, nef, and regulatory protein genes tat and rev. Several additional activities have been ascribed to the resulting proteins transcribed by these genes (secondary activities) which suggest the multifunctional role they play but generally their primary activities are highly conserved (Faust et al., 2017). A summary of these accessory proteins and their role has been shown in (Table 2 ).
Fig. 4.
Comparison between various members of the Lentivirus genus reference genome. Human Immunodeficiency Virus 1 (HIV1), Human Immunodeficiency Virus 2 (HIV2), Simian Immunodeficiency Virus (SIV) and Feline Immunodeficiency Virus (FIV).
Table 2.
List of accessory genes within the HIV genome and their associated functions.
| Gene | Primary function | Secondary Function | References |
|---|---|---|---|
| vif |
|
|
(LaRue et al., 2010; Greenwood et al., 2016; Kim et al., 2013) |
| vpr |
|
|
(Lai et al., 2005; Roshal et al., 2003; Ziebuhr, 2005; Zimmerman et al., 2006) |
| vpu |
|
|
(Galão et al., 2012; Roy et al., 2014) |
| nef |
|
|
(Geyer et al., 2001; Sauter et al., 2015) |
| tat |
|
|
(Feinberg et al., 1991; Izmailova et al., 2003) |
| rev |
|
|
(DiMattia et al., 2010; Felber et al., 1990) |
3. The emergence and transmission
SARS-CoV-2 was first reported in December 2019 and reportedly originated from the Huanan sea wet market in Wuhan city (Andersen et al., 2020; Zhu et al., 2020b; Singhal, 2020). Its unprecedented spread leads to a thorough investigation by Chinese researchers, who noticed that the cluster of atypical pneumonia cases emerged from the zoonotic transmission and showed >95% homology with bat coronaviruses and > 70% similarity with the previous SARS-CoV (Lu et al., 2020b; McIntosh, 2020). Thus, Chinese Health Authorities identified the outbreak as a novel coronavirus (COVID-19) (Lu et al., 2020a). Although environmental samples taken from the market tested positive, researchers considered the possibility of human-to-human transmission in other geographical areas after people returned from Wuhan celebrating the Chinese New Year (CNY) with their families (Wang et al., 2020; Chen et al., 2020). In this context, one study showed a significant correlation between domestic train transportation and the number of cases imported to other provinces in China. Besides, non-stop passenger flights were also linked to the number of cases reported abroad, subsequently the cause of this pandemic (Rodríguez-Morales et al., 2020). Evidence suggests that virus is transmitted by respiratory droplets, direct contact, and airborne transmission. While analysis of confirmed and suspected cases of COVID-19 suggested that novel virus could go beyond the respiratory tract (Peng et al., 2020). More interestingly, the study also found that virus can enter the body through eye exposure (J.P.O. Li et al., 2020a).
Notably, several asymptomatic cases have emerged across the globe, arguably the cause of such widespread transmission (Bai et al., 2020). Earlier this year, Italy declared a state of emergency after registering two cases from Wuhan tourists with suspected symptoms of coronavirus after they arrived in Rome (Giovanetti et al., 2020). Similarly, the USA confirmed the first positive case of SARS-CoV-2 identified in a woman in her 60's, who had returned from China with good health in mid-January. Later, her husband was also tested positive and admitted to the hospital. Although he did not travel but was considered to be in close contact with his wife (Ghinai et al., 2020). Similarly, a 50-year-old female entered the UK from Hubei province without previous medical history and no regular medications. After arrival, she developed symptoms of malaise and fever, accompanied by a sore throat and dry cough within three days (Lillie et al., 2020).
As some cases show delayed symptoms, the possibility of human-to-human transmission could occur during the asymptomatic incubation period (X.Li et al., 2020c; Backer et al., 2020). Until an appropriate diagnostic cure is found, researchers have alerted countries to maintain a distance of one meter from infected persons to reduce the risk of transmission (Mehraeen et al., 2020). Subsequently, many countries have now introduced movement control orders and lockdowns as preventative measures to curb the spread of the virus (Hamzelou, 1981), urging their citizens to maintain social distance to minimize the spread of the virus.
In recent history, another deadly virus called the human immunodeficiency virus (HIV) had emerged, which is the retrovirus responsible for the acquired immunodeficiency syndrome (AIDS). As a common disease in Asia, the concurrence of these diseases could present an alarming problem globally. Recently, Zhu and colleagues noted that “HIV infected patients need to be regarded as a vulnerable group,” after co-infection of COVID-19 and HIV was reported in a patient in Wuhan, China (Zhu et al., 2020a). Previously, HIV-1 emerged in young homosexual men in 1981 (Brodie et al., 2004). Five years later, HIV-2 virus was discovered with similar morphology to HIV-1, but antigenically distinct, which increased AIDS cases reported in patients from western Africa in 1986 (Clavel et al., 1986). Nonetheless, HIV-2 viral strain was distantly related to HIV-1 and both strains were acquired by humans from non-human primates infected with simian immunodeficiency virus (SIV) (Heeney et al., 2006).
Scientists believed that SIV was transmitted to humans after contact with the blood of infected chimpanzees during bushmeat handling. Notably, the virus mutated into HIV from its origins and gradually spread from Africa to other parts of the world (Sharp and Hahn, 2011; Ayouba et al., 2013; Sousa et al., 2017). Thus, confirming the occurrence of both viruses (HIV-1 and HIV-2) as a result of zoonotic transmission from infected primates. In general, human-to-human spread is known to be transmitted from mother to child, and during unprotected sexual intercourse between couples (Kassa, 2018). When HIV infected body fluids, including blood, breast milk, semen, vaginal, and rectal secretion come into contact with mucous membrane in the mouth, rectum, penis, and vagina, it destroys the tissue or injects directly into the bloodstream by a needle or syringe (Hladik and McElrath, 2008; Shaw and Hunter, 2012).
In summary, it is still unknown whether any identified interrelationship presents between HIVs and SARS-CoV-2. Despite the remarkable number of patients infected with HIV and novel COVID-19 separately, which extends to cover a wide zone in Asia and worldwide. There are very few reports on the SARS-CoV-2 infection among patients infected with HIV (Joob and Wiwanitkit, 2020).
4. Mechanism of infection
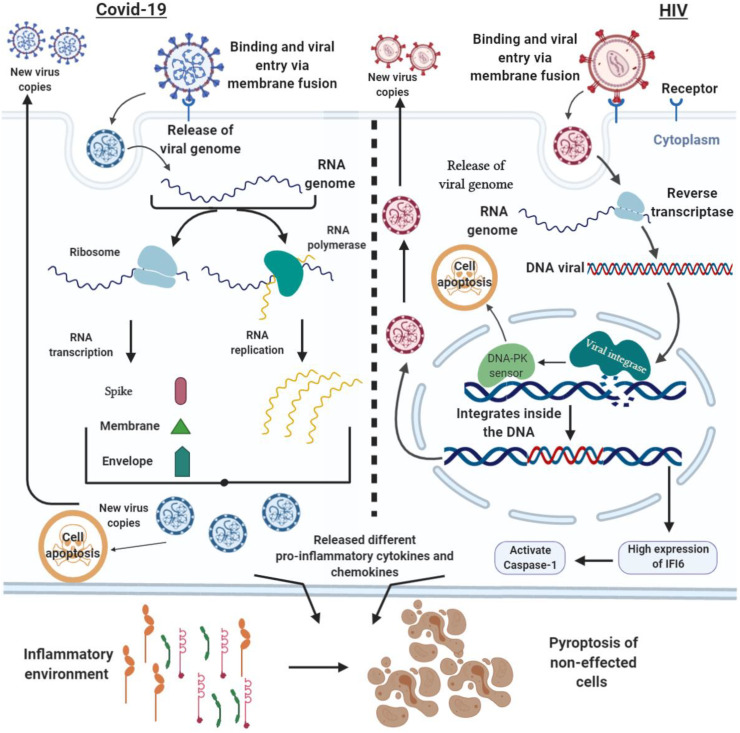
It has been observed that HIV targets the specific CD4+ immune cells, including T cells, macrophages, and dendritic cells, resulting in a significant reduction in the number of these immune cells in HIV-infected patients (Lane, 2010). These CD4+ T cells (known as helper cells) are the backbone of the immune system where they activate B cells, macrophage, and cytotoxic T cells to secrete antibodies, destroy ingested microbes and kill the infected cells, respectively (Laidlaw, 2016). The HIV membrane contains a transmembrane glycoprotein called glycoprotein-41 (GP41) and surface glycoprotein, namely glycoprotein-120 (GP120), which binds to CD4 receptors on the surface of CD4+ cells (Pancera et al., 2014). There are two other chemokine co-receptors known as C—C chemokine receptor 5 (CCR5) and C—C chemokine receptor 4 (CXCR4), which facilitate HIV binding and infusion into CD4+ cells (Didigu et al., 2014). CXCR4 co-receptor is expressed in many peripheral T-cells lymphoma found in the lymphatic sides, including lymph nodes, bone marrow, and spleen (Machado et al., 2009). However, CCR5 is highly expressed on the surface of T cells, macrophages, dendritic cells, eosinophils, and microglia (Moore et al., 2004). The binding of GP120 to CD4 receptor leads to a change in the envelope structure of virus, which allows the co-receptor, either CCR5 or CXCR4 (CXCR4 for T-tropic HIV strains, and CCR5 for M-tropic HIV strains) bind to a specific domain in the gp120. After binding, the N-terminal fusion peptide (GP41) penetrates in the host cell membrane, followed by entry of viral capsid into the cytosol of T cell. After penetration, virus removes and exposes its capsid, which contains the RNA genome and its associated enzymes (reverse transcriptase and integrase) into the host cell (Naif, 2013). The virus produces complementary DNA (cDNA) by transcribing its single-standard RNA based on reverse transcriptase activity for producing duplicate strands of viral DNA. However, complete viral DNA migrates to the nucleus and integrates with DNA inside the host cell and unfortunately affects the cellular activity. The integrated DNA is used to generate mRNA, thus synthesizing the viral proteins and allowing the new virus copy to develop. It has been observed that virus sabotages the CD4+ immune T cells for replication, which leads to increase virus load in the blood and a significant decrease in CD4+ T cells (Goodsell, 2015). Interestingly, some researchers found another mechanism of HIV destruction of CD4+ T cells (Bolton et al., 2002; Yue et al., 2005). They observed that depletion of CD4+ T cells by apoptosis pathway because of direct HIV infection was only 5–10% of the total CD4+ T cell pool. It has been found that HIV enzyme (integrase) plays a crucial role in the activation of DNA-PK sensor of host cell, which activates the apoptotic cascade inside the cell. However, another HIV enzyme (protease) activates the caspase-8 that further triggers the apoptosis of infected cells. Besides, specific HIV proteins display on the surface of infected CD4+ T cells, which alarm CD8+ T cells to destroy the infected CD4+ T cells. The immune system reacts by producing antibodies, such as anti-HIV antibodies, which stick to the infected CD4+ T cells and mark their destruction by other immune cells (Selliah and Finkel, 2001). However, it has been observed that indirect effect of HIV infection results in majority of CD4+ T cells death, which causes immune deficiency. The direct effect of HIV to CD4+ T cells triggers the expression of interferon-gamma induced protein-16 (IFI-16), followed by the activation of inflammatory cascade inside the infected cell, which ends up with cellular self-destruction called pyroptosis (Doitsh et al., 2014; Monroe et al., 2014). The caspase-1 activated by high expression of IFI-16 triggers the production of cytokines, especially interleukin-1 beta (IL-1β) inside the infected cells (Lupfer et al., 2015). The high production of IL1-β leads to the creation of inflammatory environment inside the cell, resulting in the initiation of apoptosis (Feria et al., 2018). The infected cells lead to release of interleukin-1 beta (IL-1β) to the outside cellular environment, which contributes to the activation of pyroptosis in the non-affected cells, causing a considerable loss of CD4+ T cells (Doitsh et al., 2014).
Unlike HIV, the exact mechanism how COVID-19 induces immune defect is still unknown; however, it could be similar to the previous coronaviruses, including the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV). These viruses induce low level of lymphocytes abnormality in blood, or it is known as lymphopenia related to HIV infection. The binding of coronavirus to the host cell required a specific class I viral glycol- transmembrane-protein, such as S protein on the surface of the virus envelope. It needs a specific receptor on the host cells called angiotensin-converting enzyme 2 (ACE2) receptor (Andersen et al., 2020; Dhama et al., 2020). ACE2 is an enzyme spread in many cell membranes of different organs, including lungs, arteries, heart, kidney, and intestines; it plays a significant role in blood pressure regulation. The dramatic reduction in the peripheral CD4+ and CD8+ T cells is one of the symptoms in COVID-19 cases related to HIV infection (Liu et al., 2020a; Diao et al., 2020). Besides, over circulation of proinflammatory cytokines and chemokines, including interleukins (IL-1β, IL1RA, IL2, IL4, IL5, IL6, IL7, IL8, IL9, IL10, IL12p70, IL13, IL15, IL17A), eotaxin, basic fibroblast growth factor (FGF2), granulocyte-colony stimulating factor (GCSF), granulocyte-macrophage colony-stimulating factor (GMCSF), Interferon-gamma (IFNγ), IFN-γ-inducible protein-10 (IP10), chemokines (Monocyte chemoattractant protein-1 (MCP1)), macrophage inflammatory proteins-1A (MIP1A), macrophage inflammatory proteins-1B (MIP1B), platelet-derived growth factor subunit B (PDGFB), C—C motif chemokine ligand 5 (CCL5)), and tumor necrosis factor (TNFα) was observed in the acute stage of positive COVID-19 patients (Rothan and Byrareddy, 2020; Huang et al., 2020). Moreover, it has been observed that level of NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome was high in patients infected with COVID-19, which triggers the release of IL-1β and IL-18 (Freeman and Swartz, 2020; Tang et al., 2020; Chen et al., 2019). However, a hyper-immune response was also reported during the clinical course of SARS-CoV infection that is responsible for the high morbidity of SARS-CoV. The high production of C—C motif chemokine ligand 2 (CCL2), C-X-C Motif Chemokine Ligand 9 (CXCL9), C-X-C Motif Chemokine Ligand 10 (CXCL10), interleukins (IL-1, IL-6, IL-8) with interferon-gamma (IFN-γ), and transforming growth factor-beta (TGF-β) occurred in the acute stage of SARS-CoV infection triggering the cytokine storm (Cameron et al., 2008; Huang et al., 2005). In addition, T cell lymphopenia has been reported in several SARS-COVID patients, where the total number of CD4+ and CD8+ T cells drastically decreased within two weeks of infection (Cui et al., 2003). The investigation by Jiang et al. (2019) proves the activation of pyroptosis during MERS-CoV infection. Like HIV infection, MERS-CoV activates caspase-1 in the spleen with high circulating amount of TNF-α, IL-1β, IFN-γ and IL-6, and upward stimulation of macrophages (Jiang et al., 2019). Taken together, the pro-inflammatory molecules released from the immune cells during the onset of COVID-19 infection create an acute inflammatory environment similar to that recorded in HIV exposure, which consecutively activates the pyroptotic cascade of immune cells. Notably, the immune deficiency is more severe in case of coronaviruses because it occurs at the early stage of infection (within a few days), while it takes a longer time for HIV after ten years. Fig. 5 explains the molecular mechanism and cell death of COVID-19 and HIV.
Fig. 5.
Crosstalk of the infection mechanism and cell death of COVID-19 and HIV.
5. Latest therapeutic potential of antiviral agents against COVID-19
At present, there is no effective antiviral therapy available for treating the latest respiratory 2019 coronavirus infection. Therefore, the development of an active antiviral agent and vaccine is crucial to stop the COVID-19 pandemic. The economic and social challenges produced by this outbreak also demand urgent interventions worldwide. An earlier study on MERS- and SARS-CoV has allowed expediting the discovery of viable drugs to combat the new outbreak of SARS-CoV-2. However, there is no therapeutic drug that targets MERS- and SARS-CoV which has progressed beyond phase-1 studies at present.
Chloroquine is a primary therapeutic drug used to prevent and treat patients infected with malaria. Fig. 6a shows the chemical structure of chloroquine. Recently, the National Center for Biotechnology Development (NCBD) in China stated that chloroquine is an effective drug that could be used to treat patients infected with novel COVID-19. Notably, the chloroquine therapeutic drug repurposing has been studied in hospitals in Beijing, Guangdong Province in South China, and Hunan Province in central China. The earlier studies by Gao et al. (2020) conducted in China proposed that approximately a hundred patients infected with SARS-CoV-2 were treated with drug chloroquine; as a result, a rapid fall in fever and improved computed tomography (CT) scan images of the lung were observed, and patients also needed a short recovery time compared to the control group, without any side effects. Therefore, the Chinese Medical Board (CMB) has suggested to include chloroquine as a therapeutic drug in the guidelines for treating the SARS-CoV-2. Probably, chloroquine is the first therapeutic drug to be used for the patients infected with SARS-CoV in China and worldwide. However, the more extended use of chloroquine could produce severe side effects, such as cardiomyopathy (Bernstein, 1991; Ratliff et al., 1987) and the presence of some reports on macular retinopathy as a minor risk caused by this drug (Cubero et al., 1993). A survey of the patients infected with SARS-CoV-2 for adverse side effects of chloroquine treatment is yet to be conducted. Chloroquine was considered the best therapeutic drug choice available for treating SARS-CoV-2 infected patients in hospitals. Recently, the WHO has advocated a solidarity clinical trial for COVID-19 treatments. The solidarity clinical trial will compare four treatment choices, including chloroquine/hydroxychloroquine, remdesivir, lopinavir/ritonavir, and lopinavir/ritonavir plus interferon beta-1a against standard therapy and determine their relative efficacy against COVID-19 (World Health Organization, 2020).
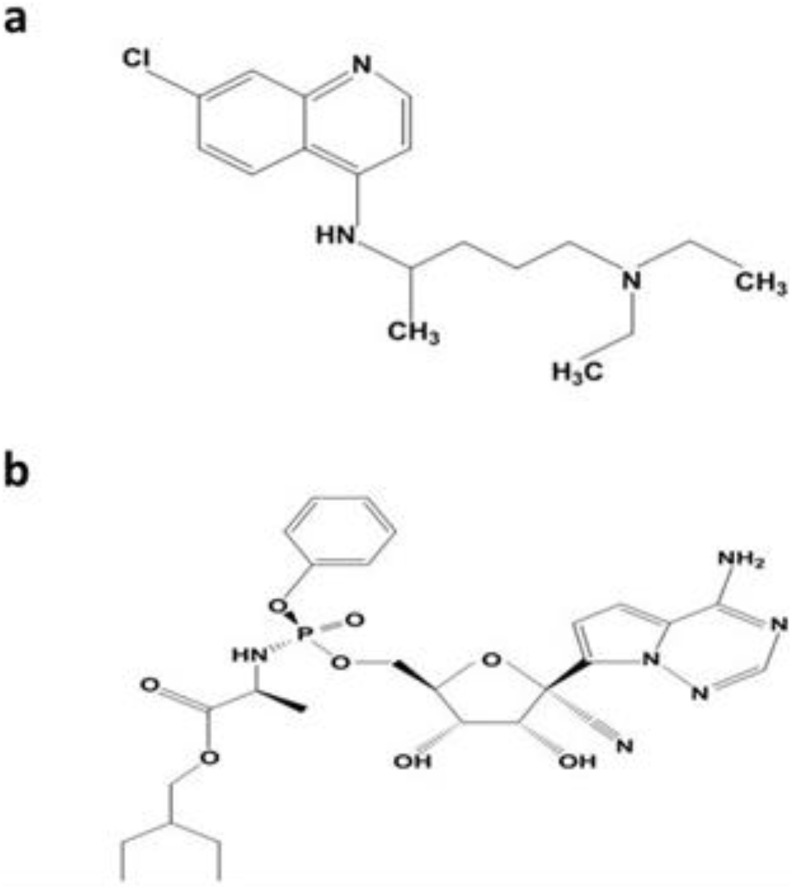
Fig. 6.
Chemical structure of (a) chloroquine and (b) remdesivir.
Remdesivir (GS-5734) is the chief antiviral therapeutic drug that could be used for combating SARS-CoV-2. Remdesivir is a class of adenosine nucleotide analogue drug with a wide range of antiviral activity towards pneumo-viruses, paramyxoviruses, filoviruses, and pathogenic strains of coronaviruses, such as MERS- and SARS-CoV (Sheahan et al., 2017). Fig. 6b shows the chemical structure of remdesivir. The pharmacokinetic trials have been accomplished, and clinical studies are being conducted for testing the effectiveness of remdesivir in the treatment of Ebola virus (Mulangu et al., 2019). In general, earlier studies have found that adenosine nucleotide analogues are less effective against coronaviruses because of the proofreading exonuclease of the virus. However, the drug remdesivir was effective against MERS- and SARS-CoV, and also many strains of bat-CoV (Sheahan et al., 2017). A study reported that remdesivir showed an effective half-maximum concentration (EC50s) of 0.074 μM for MERS-CoV and 0.069 μM for SARS-CoV (Brown et al., 2019). Notably, in-vitro studies have found that therapeutic drug remdesivir is also useful in the EC50 submicromolar scale against a wide range of significantly divergent coronaviruses, such as human endemic CoVs, 229E (HCoV-229E) and OC43 (HCoV-OC43). Hence, the remdesivir has a broad-spectrum activity against CoVs (Brown et al., 2019). The prophylactic administration of remdesivir significantly decreased the viral titers <2 on the lung in SARS-CoV mouse model. In addition, remdesivir improved respiratory function and the clinical symptoms of illness compared to control animals (Sheahan et al., 2017). The authors reported that comparable findings with MERS-CoV were attained in prophylactic studies conducted with MERS-CoV transgenic mouse model (Cockrell et al., 2016). They found that the enzyme carboxylesterase 1c (Ces1c) was deleted and receptor (dipeptidyl peptidase 4, hDPP4) of humanized MERS-CoV was expressed in order to increase the pharmacokinetics of nucleotide prodrugs. Another study conducted by Agostini et al. (2018) reported that two mutations were recognized, such as V553L and F276L in the RNA-dependent RNA polymerase gene of the virus after 23 passages of remdesivir drug (Agostini et al., 2018). They also observed that these changes in amino acid reduced the burden of virus and lessened the pathogenesis of SARS-CoV in mice. In recent months, the prophylactic efficacy of remdesivir was also tested in a rhesus macaque (non-human primate) MERS-CoV infection model (de Wit et al., 2020). When the treatment with prophylactic remdesivir was started 24 h before inoculation, MERS-CoV was found to be inhibited from replicating in the respiratory tissues and prevented from causing clinical disease, which inhibited the development of lung lesions. The same findings were obtained when treatment with remdesivir drug was initiated 12 h after the virus inoculation (de Wit et al., 2020). The remdesivir therapeutic drug safety data are readily available for humans (Mulangu et al., 2019); hence, human studies can be conducted to assess the effectiveness of this compound towards novel coronaviruses.
The Food and Drug Administration (FDA) approved the therapeutics against MERS- and SARS-CoV that have been assessed for antiviral activity. For instance, lopinavir (LPV), a protease inhibitor of HIV-1, was tested to combine with ritonavir (RTV) in order to enhance the half-life of LPV. The combination of these two drugs (LPV/RTV) was found to be more effective in patients against SARS-CoV and in-vitro study. The calculated EC50 was 4 μg/ml for the fetal rhesus kidney-4 cells (Chu et al., 2004). Meanwhile, Chan and coworkers reported that LPV/RTV also declined clinical sores, weight loss, and progression of disease in marmosets infected with MERS-CoV (Chan et al., 2015). However, the antiviral activity of LPV towards MERS-CoV in the in-vitro study remains controversial. Another study conducted by Chan et al. (2013) found no optimal EC50 in the Vero cells, whereas EC50 was reported to be 8 μM in the Huh7 cells in a different study (de Wilde et al., 2014).
It was found that infections with MERS-CoV were mediated by replication of the virus and host inflammatory responses after clinical observations in both humans and animals. Those studies had suggested the use of combination therapies including interferons I and II. However, interferon-beta (IFNb) reduced the replication of MERS-CoV in the tissue culture and showed the best efficacy with 1.37–17 IU/ml EC50s (Chan et al., 2013; Hart et al., 2014). From the study on marmosets infected with MERS-CoV, Chan et al. (2015) reported the clinical improvement with the use of LPV/RTV with IFNb. Also, the clinical randomized control trials were started in Saudi Arabia in order to determine the efficacy of combinations of therapies, such as IFNb and LPV/RTV in improving the clinical outcomes against MERS-CoV (Arabi et al., 2018). Notably, China has launched another controlled trial to test the effectiveness of IFNα-2b and LPV/RTV in hospitalized SARS-CoV-2 patients (ChiCTR2000029308).
In addition, the therapeutic and prophylactic activities of LPV/RTV-IFNb and remdesivir were compared in a transgenic mouse model infected with MERS-CoV (Sheahan et al., 2020). It was found that remdesivir reduced lung viral titers, virus replication, and improved the pulmonary function. At the same time, prophylactic combined therapeutics, such as LPV/RTV-IFNb, only resulted in a moderate decline in viral titers and did not produce any effects on other parameters of disease. The combined therapy boosted pulmonary function but did not influence viral replication (Sheahan et al., 2020). Thus, it was observed that remdesivir has shown to be effective for treating MERS-CoV infections compared to LPV/RTV-IFNb.
Another antiviral prodrug, Ribavirin is a guanosine analogue used to treat many infections caused by viruses, such as hepatitis C respiratory syncytial virus, and patients coinfected with HIV and hepatitis B. In 1980, it was launched to the marketplace for treating the respiratory syncytial virus, especially in children. In most cases, it is used in combination with interferon (IFN). Falzarano and co-authors found promising results when ribavirin was combined with IFNα-2b against the rhesus macaque model infected with MERS-CoV (Falzarano et al., 2013), but there have been some contradictory data on the MERS-CoV infected patients treated with IFNα2a or IFNβ1 and ribavirin (Arabi et al., 2017). Besides, ribavirin also produces adverse side effect such as decreasing haemoglobin concentrations in patient with respiratory disorder that could not be appropriate for use against SAR-CoV-2. In addition, various other antiviral therapeutic drugs/cocktails, including favipiravir, nitazoxanide, ganciclovir, acyclovir/penciclovir, and the latest FDA-approved ivermectin are listed in Table 3 .
Table 3.
Listed repurposed antiviral drugs in clinical development against COVID-19.
| Drug/cocktail | Mode of action | Status and anti-infective mechanisms | Target diseases | References |
|---|---|---|---|---|
| Chloroquine | 9-aminoquinolin | Status: approved, vet-approved, investigational; Mechanisms: the drug increasing endosomal pH, inhibitors of autophagy, and immunomodulating | Malaria, and autoimmune disease | (Savarino et al., 2003; Golden et al., 2015) |
| Remdesivir (GS-5734) | The prodrug of Nucleotide analogue | Status: experimental; Mechanisms: the drug interfering with the post-entry of virus | MERS, SARS, and Ebola | (Agostini et al., 2018; Tchesnokov et al., 2019; Lo et al., 2019) |
| Lopinavir/ Ritonavir | Protease inhibitors | Status: approved; Mechanisms: inhibiting the protease of HIV-1 for protein cleavage; as a result, immature or non-infectious viral particles | HIV/AIDS, MERS, and SARS | (Chu et al., 2004; Oldfield et al., 2005; Arabi et al., 2020) |
| Lopinavir/ritonavir, ASC09/ritonavir, with and without umifenovir | Protease inhibitors | Status: experimental, approved; Mechanisms: lopinavir/ritonavir are protease inhibitors; ASC09 is a protease inhibitor of HIV-1; Umifenovir is an entry inhibitor for influenza | HIV/AIDs, and influenza | (Harrison, 2020) |
| Ribavirin | Synthetic guanosine Nucleoside | Status: approved; Mechanisms: the drug interfering with the viral mRNA synthesis, also a broad-spectrum antiviral activity for both RNA and DNA | HCV, MERS, and SARS | (Chung et al., 2018; Arabi et al., 2019) |
| Different combinations of lopinavir/ritonavir and baloxavirmarboxil/favipiravir | Favipiravir is a guanine analog RNA-dependent RNA polymerase inhibitor, and baloxavirmarboxil is a Cap-dependent endonuclease inhibitor | Status: approved; Mechanisms: favipiravir is a guanine analog RNA-dependent RNA polymerase inhibitor, and baloxavirmarboxil is a Cap-dependent endonuclease inhibitor approved for influenza A and B | Influenza A, and B | (Harrison, 2020) |
| Oseltamivir | Neuraminidase inhibitor | Status: approved; Mechanisms: the drug inhibiting the viral neuraminidase activity, and also inhibiting the infectivity and viral replication | Influenza (A) viruses | (McQuade and Blair, 2015; Jefferson et al., 2014) |
| Oseltamivir, ritonavir/oseltamivir, ASC09/oseltamivir | Oseltamivir is a sialidase inhibitor | Status: approved; Mechanisms: oseltamivir is a sialidase inhibitor approved for influenza | Influenza | (Harrison, 2020) |
| Interferon α-2b alone or together with ribavirin and lopinavir/ritonavir | Interferon α-2b is a recombinant cytokine and ribavirin is a guanine derivative | Status: experimental, approved; Mechanisms: interferon α-2b is a recombinant cytokine protein with antiviral activity and ribavirin is a guanine derivative approved for viral infections | Cancer, hepatitis B, and C | (Harrison, 2020) |
| Ganciclovir | Nucleoside analog | Status: investigational, approved; Mechanisms: a potent inhibitor of family Herpesvirus, such as cytomegalovirus | HIV/AIDS-related cytomegalovirus infections | (Al-Badr and Ajarim, 2018) |
| Acyclovir/Penciclovir | Nucleoside analog | Status: approved; Mechanisms: it is a synthetic derivative of acyclic guanine, as a result of chain termination | VZV, HSV | (Chung et al., 2018) |
| Favipiravir (T-705) | Nucleoside analog: polymerase inhibitor of viral RNA | Status: investigational; Mechanisms: inhibiting viral reproduction by acting on its genetic copying without influencing on host cellular nucleic acid | Influenza A(H1N1), Ebola | (Shiraki, 2018; Cardile et al., 2017) |
| Nafamostat | Synthetic inhibitor of serine protease | Status: investigational; Mechanisms: inhibiting the membrane fusion through preventing the release of cathepsin B; role as an anticoagulant activity | MERS, Ebola, MERS, and influenza | (Hsieh and Hsu, 2007; Nishimura and Yamaya, 2015) |
| Azvudine | Inhibitor of reverse transcriptase | Status: experimental; Mechanisms: inhibitor of reverse transcriptase against AIDS/HIV-1 | HIV-1/AIDS | (Harrison, 2020) |
| Nitazoxanide | Antiprotozoal agent | Status: approved, vet-approved, investigational; Mechanisms: modulating the growth, survival, and proliferation of intracellular and extracellular viruses, protozoa, bacteria (microaerophilic and anaerobic), and helminths | Human/animal coronaviruses | (Rossignol, 2014; Rossignol, 2016) |
| Ivermectin | Antiparasitic agent | Status: FDA-approved, vet-approved, investigational; Mechanisms: targeting the glycine receptor subunit α-3 and gamma-aminobutyric acid receptor subunit β-3 in human | Onchocerciasis, strongyloidiasis, and scabies | (Caly et al., 2020) |
In summary, the repurposing of preexisting antiviral therapeutic drugs is finite, so it is almost inevitable that both new and repurposed drugs will be required to treat COVID-19. Therapeutic drug choices in response to COVID-19 are urgently required and fortunately, some of the preexisting antiviral therapeutics are already progressing into human clinical trials.
6. Impact of SARS-CoV-2 on HIV therapy
SARS-CoV-2 has dramatically altered the daily lives of people worldwide. This virus has already enforced social distancing, travel restrictions, and provision of supportive medical care, resulting in significant disruptions to routine functioning. With regard to the social nature of individuals, people are trying to adjust their lives with SARS-CoV-2 for the foreseeable future. Moreover, it is also crucial for the healthcare professionals (e.g., physicians, psychologists, etc.) to comprehend about the provision of healthcare therapy across time and how the biological impacts of SARS-CoV-2 can affect people with HIV (PWH). Besides, people with HIV (PWH) are placed at a higher risk for contracting and developing complications related to COVID-19. Given the long-term HIV prognosis, it is important for PWH to regularly attend their healthcare providers and adhere to therapy (National Institute of Allergy and Infectious Disease, 2020). The treatment of PWH can be affected due to stay at home orders enforced by countries worldwide. For instance, several medical practitioners canceled their appointments with HIV suspected people in the USA and switched to telehealth appointments to follow the WHO guidelines (Ives, 2020). However, telehealth programs are restricted in the wide range of resources that can be given to the customers (Siwicki, 2020), therefore, PWH may not be able to completely access the services needed for HIV therapy. In addition, many PWH patients may not be able to get access to telehealth services for many reasons, such as limited access to technology and lack of knowledge about telehealth that could hinder their therapy progression.
PWH are more vulnerable group to contract opportunistic infections, such as tuberculosis, toxoplasmosis, pneumonia, etc. (Department of veteran affairs, 2020), than those without immunocompromised systems. HIV-infected people who have suffered from any other disorders may undergo delayed treatment because of COVID-19. This can happen in already taxed healthcare system due to hospital overcrowding. Moreover, PWH who seek out emergency treatment can face a high risk of experiencing COVID-19 among other disorders in the healthcare systems (Collins, 2008).
7. Conclusion and outlook
The occurrence of person-to-person transmission for COVID-19 is increasing, which means that large numbers of cases with second COVID-19 wave of infection are likely to be recorded in the near future. Notably, it has created instability in healthcare systems and more economic losses worldwide (DiMattia et al., 2010). The preventive measurements are enforced globally, and more investigations are underway to identify the origin of infection and to learn more about the characteristics of the virus, route of transmission, severity of the disease, and mechanisms of infection. Since the infection caused by HIV is more prevalent in Asian countries. The rivalry between human immunodeficiency syndrome virus (HIV) and many other illnesses is an appealing problem. Zhu and colleagues reported the first study on the coinfection of HIV and SARS-CoV (Zhu et al., 2020b). They propositioned that persons infected with HIV need to be considered as a vulnerable group for COVID-19. Nonetheless, no robust evidence of any interrelationship between these two viral infections is yet to be identified. Despite the high number of patients infected with HIV as well as the remarkable number of patients with the novel COVID-19 disease, which extends to cover a wide zone in Asia and worldwide. There are very few reports on the co-infection of HIV and SARS-CoV. In addition, antiviral therapeutic drugs are broadly used for treating HIV-infected patients, and these drugs have potential to be used against SARS-CoV-2 (Felber et al., 1990; Savarino et al., 2003). Thus, the patients infected with HIV who are receiving anti-HIV therapeutic drugs might be at a lower risk for COVID-19 compared to the general population. Some antiviral therapeutic drugs, including chloroquine, remdesivir, ribavirin, lopinavir/ritonavir, and now FDA-approved ivermectin and others (summarized Table 3) have the potential to treat patients infected with SARS-CoV-2. At the same time, more clinical trials are required in order to obtain robust data. Also, further studies are urgently required to explore the pathogenicity, mechanisms, and transmission of novel coronavirus.
To obtain a better insight of the novel virus, countries should strive to provide more reliable and accurate data by transparency and sharing of data, and more research studies need to be conducted on the reported cases. Therefore, countries should continue to work towards developing preventive measures to minimize both the transmission and number of infected patients. In addition to unravelling the uncertainty of the mechanisms of viral replication and host cell entry, which will provide the fundamental knowledge for future research into the development of targeted vaccines and antiviral therapeutic drugs. With continuing efforts to curb the widespread transmission of COVID-19 globally, we hope that the novel coronavirus pandemic may alleviate after a few months. In summary, there is an urgent need to develop a new broad-spectrum antiviral therapeutic agent that will be effective to fight against not only the novel respiratory 2019 coronavirus, but also to prepare for a possibly similar virus outbreak in the future. For “If there is any message coming out from the latest outbreaks, it is almost certain that it will happen again.”
Author contributions
Mansab Ali Saleemi, Bilal Ahmad, Khaled Benchoula, Muhammad Sufyan Vohra, and Hing Jian Mea performed the literature search, planned, and wrote the manuscript draft. Pei Pei Chong, Navindra Kumari Palanisamy, and Eng Hwa Wong edited the manuscript and contributed to structure and design. All the authors reviewed and approved the final manuscript draft before submission.
Funding
This study was financially supported by Taylor's University Flagship Research Grant - Scheme Project No: TUFR/2017/001/05; TUFR/2017/003/05; TRGS/MFS/1/2017/SBS/003. The funders had no role in the study design, collection, and analysis of data, manuscript draft preparation, or decision to publish.
Declaration of Competing Interest
The authors declare no conflicts of interest.
Acknowledgements
This review was supported by Taylor's University Flagship Research Grant - Scheme Project No: TUFR/2017/001/05; TUFR/2017/003/05; TRGS/MFS/1/2017/SBS/003.
References
- Agostini M.L., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. MBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Badr A.A., Ajarim T.D.S. Ganciclovir. Profiles Drug Subst Excipients Relat Methodol. 2018;43:1–208. doi: 10.1016/bs.podrm.2017.12.001. [DOI] [PubMed] [Google Scholar]
- Andersen K.G., et al. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., et al. C63 VIRAL Respir Infect American Thoracic Society; 2017. Effect of Ribavirin and Interferon on the Outcome of Critically Ill patients with MERS; p. A6067. [Google Scholar]
- Arabi Y.M., et al. Treatment of Middle East Respiratory Syndrome with a combination of lopinavir-ritonavir and interferon-β1b (MIRACLE trial): study protocol for a randomized controlled trial. Trials. 2018;19:81. doi: 10.1186/s13063-017-2427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., et al. Ribavirin and interferon therapy for critically ill patients with Middle East respiratory syndrome: a Multicenter observational study. Clin. Infect. Dis. 2019;70(9):1837–1844. doi: 10.1093/cid/ciz544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y.M., et al. Treatment of Middle East respiratory syndrome with a combination of lopinavir/ritonavir and interferon-β1b (MIRACLE trial): statistical analysis plan for a recursive two-stage group sequential randomized controlled trial. Trials. 2020;21(1):8. doi: 10.1186/s13063-019-3846-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayouba A., Akoua-Koffi C., Calvignac-Spencer S., Esteban A., Locatelli S., Li H., Li Y., Hahn B.H., Delaporte E., Leendertz F.H., et al. Evidence for continuing cross-species transmission of SIVsmm to humans: characterization of a new HIV-2 lineage in rural Côte d’Ivoire. AIDS. 2013;27(15) doi: 10.1097/01.aids.0000432443.22684.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travellers from Wuhan, China, 20–28 January 2020. Eurosurveillance. 2020;25:2000062. doi: 10.2807/1560-7917.ES.2020.25.5.2000062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., et al. Presumed asymptomatic carrier transmission of COVID-19. JAMA. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H.N. Ocular safety of hydroxychloroquine. Ann. Ophthalmol. 1991;23:292–296. [PubMed] [Google Scholar]
- Bolton D.L., et al. Death of CD4+ T-cell lines caused by human immunodeficiency virus type 1 does not depend on caspases or apoptosis. J. Virol. 2002;76:5094–5107. doi: 10.1128/JVI.76.10.5094-5107.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie M., et al. AIDS at 21: Media coverage of the HIV epidemic 1981–2002. The Nation. 2004;49:68. [Google Scholar]
- Brown A.J., et al. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antivir. Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calattini S., et al. Discovery of a new human T-cell lymphotropic virus (HTLV-3) in Central Africa. Retrovirology. 2005;2:30. doi: 10.1186/1742-4690-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caly L., et al. The FDA-approved drug Ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020 doi: 10.1016/j.antiviral.2020.104787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron M.J., et al. Human immunopathogenesis of severe acute respiratory syndrome (SARS) Virus Res. 2008;133:13–19. doi: 10.1016/j.virusres.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardile A.P., et al. Will there be a cure for Ebola? Annu. Rev. Pharmacol. Toxicol. 2017;57:329–348. doi: 10.1146/annurev-pharmtox-010716-105055. [DOI] [PubMed] [Google Scholar]
- Chan J.F.W., et al. Broad-spectrum antivirals for the emerging Middle East respiratory syndrome coronavirus. J. Inf. Secur. 2013;67:606–616. doi: 10.1016/j.jinf.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., et al. Treatment with lopinavir/ritonavir or interferon-β1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J. Infect. Dis. 2015;212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.F.W., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen I.Y., et al. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M., et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung R.T., et al. Hepatitis C guidance 2018 update: AASLD-IDSA recommendations for testing, managing, and treating hepatitis C virus infection. Clin. Infect. Dis. 2018;67(10):1477–1492. doi: 10.1093/cid/ciy585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavel F., et al. Isolation of a new human retrovirus from West African patients with AIDS. Science. 1986;233:343–346. doi: 10.1126/science.2425430. [DOI] [PubMed] [Google Scholar]
- Cloyd M.W., Chen J.J.Y., Wang L. How does HIV cause AIDS? The homing theory. Mol Med Today. 2000;6:108–111. doi: 10.1016/S1357-4310(99)01663-9. [DOI] [PubMed] [Google Scholar]
- Cockrell A.S., et al. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat. Microbiol. 2016;2:1–11. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins A. In: In-Patient Safety and Quality: An Evidence-Based Handbook for Nurses. Hughes R.G., editor. Agency for Healthcare Research and Quality (US); Rockville, MD, USA: 2008. Preventing Health Care-Associated Infections; pp. 547–570. [PubMed] [Google Scholar]
- Corman V.M., et al. Hosts and sources of endemic human coronaviruses. Adv Viral Res. 2018;100:163–188. doi: 10.1016/bs.aivir.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubero G.I., Reguero J.J.R., Ortega J.N.R. Restrictive cardiomyopathy caused by chloroquine. Heart. 1993;69:451–452. doi: 10.1136/hrt.69.5.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui W., et al. Expression of lymphocytes and lymphocyte subsets in patients with severe acute respiratory syndrome. Clin. Infect. Dis. 2003;37:857–859. doi: 10.1086/378587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Veteran Affairs, Preventing Opportunistic Infections (OIs) 2020. https://www.hiv.va.gov/patient/diagnosis/OI-prevention.asp Available online.
- Dhama K., et al. 2020. Coronavirus disease 2019–COVID-19. Preprints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao B., et al. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didigu C.A., et al. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood J Am Soc Hematol. 2014;123:61–69. doi: 10.1182/blood-2013-08-521229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMattia M.A., et al. Implications of the HIV-1 Rev dimer structure at 3.2 Å resolution for multimeric binding to the Rev response element. Proc. Natl. Acad. Sci. 2010;107:5810–5814. doi: 10.1073/pnas.0914946107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doitsh G., et al. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature. 2014;505:509–514. doi: 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falzarano D., et al. Treatment with interferon-α2b and ribavirin improves outcome in MERS-CoV–infected rhesus macaques. Nat. Med. 2013;19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust T.B., et al. Making sense of multifunctional proteins: human immunodeficiency virus type 1 accessory and regulatory proteins and connections to transcription. Annu Rev Virol. 2017;4:241–260. doi: 10.1146/annurev-virology-101416-041654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg M.B., Baltimore D., Frankel A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. 1991;88:4045–4049. doi: 10.1073/pnas.88.9.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber B.K., Drysdale C.M., Pavlakis G.N. Feedback regulation of human immunodeficiency virus type 1 expression by the rev protein. J. Virol. 1990;64:3734–3741. doi: 10.1128/jvi.64.8.3734-3741.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feria M.G., et al. HIV replication is associated to inflammasomes activation, IL-1β, IL-18 and caspase-1 expression in GALT and peripheral blood. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0192845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman T.L., Swartz T.H. Targeting the NLRP3 inflammasome in severe COVID-19. Front. Immunol. 2020;11:1–12. doi: 10.3389/fimmu.2020.01518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S.Y., et al. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg Microbes Infect. 2020;9:558–570. doi: 10.1080/22221751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galão R.P., et al. Innate sensing of HIV-1 assembly by Tetherin induces NFκB-dependent proinflammatory responses. Cell Host Microbe. 2012;12:633–644. doi: 10.1016/j.chom.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- Geyer M., Fackler O.T., Peterlin B.M. Structure–function relationships in HIV-1 Nef. EMBO Rep. 2001;2:580–585. doi: 10.1093/embo-reports/kve141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghinai I., et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. 2020;395:1137–1144. doi: 10.1016/S0140-6736(20)30607-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovanetti M., et al. The first two cases of 2019-nCoV in Italy: where they come from? J. Med. Virol. 2020;92:518–521. doi: 10.1002/jmv.25699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden E.B., et al. Quinoline-based antimalarial drugs: a novel class of autophagy inhibitors. Neurosurg. Focus. 2015;38:E12. doi: 10.3171/2014.12.FOCUS14748. [DOI] [PubMed] [Google Scholar]
- Goodsell D.S. Illustrations of the HIV life cycle. Futur. HIV-1 Ther. 2015:243–252. doi: 10.1007/82_2015_437. [DOI] [PubMed] [Google Scholar]
- Gorbalenya A.E. Severe acute respiratory syndrome-related coronavirus–the species and its viruses, a statement of the Coronavirus Study Group. BioRxiv. 2020 doi: 10.1101/2020.02.07.937862. [DOI] [Google Scholar]
- Greenwood E.J.D., et al. Temporal proteomic analysis of HIV infection reveals remodelling of the host phosphoproteome by lentiviral Vif variants. Elife. 2016;5:e18296. doi: 10.7554/eLife.18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald M.E., et al. The coronavirus nucleocapsid protein is ADP-ribosylated. Virology. 2018;517:62–68. doi: 10.1016/j.virol.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn B.H., et al. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–614. doi: 10.1126/science.287.5453.607. [DOI] [PubMed] [Google Scholar]
- Hamzelou J. Kaposi’s sarcoma and Pneumocystis pneumonia among homosexual men--New York City and California. MMWR Morb. Mortal. Wkly Rep. 1981;30(25):305–308. [PubMed] [Google Scholar]
- Harrison C. Coronavirus puts drug repurposing on the fast track. Nat. Biotechnol. 2020;38(4):379–381. doi: 10.1038/d41587-020-00003-1. [DOI] [PubMed] [Google Scholar]
- Hart B.J., et al. Interferon-β and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J Gen Virol. 2014;95:571. doi: 10.1099/vir.0.061911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeney J.L., Dalgleish A.G., Weiss R.A. Origins of HIV and the evolution of resistance to AIDS. Science. 2006;313:462–466. doi: 10.1126/science.1123016. [DOI] [PubMed] [Google Scholar]
- Hladik F., McElrath M.J. Setting the stage: host invasion by HIV. Nat Rev Immunol. 2008;8:447–457. doi: 10.1038/nri2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh H.P., Hsu J.T.A. Strategies of development of antiviral agents directed against influenza virus replication. Curr. Pharm. Des. 2007;13:3531–3542. doi: 10.2174/138161207782794248. [DOI] [PubMed] [Google Scholar]
- Huang K., et al. An interferon-γ-related cytokine storm in SARS patients. J. Med. Virol. 2005;75:185–194. doi: 10.1002/jmv.20255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- J. Ives, Telemedicine surges, Fueled by coronavirus fears and shift in payment rules. News Medical Life Sciences (2020). Available online: https://www.news-medical.net/news/20200327/Telemedicine-surges-fueledby-coronavirus-fears-and-shift-in payment-rules.aspx ((accessed on 23 April 2020)).
- Izmailova E., et al. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 2003;9:191–197. doi: 10.1038/nm822. [DOI] [PubMed] [Google Scholar]
- Jaffar S., et al. The natural history of HIV-1 and HIV-2 infections in adults in Africa: a literature review. Bull. World Health Organ. 2004;82:462–469. [PMC free article] [PubMed] [Google Scholar]
- Jefferson T., et al. Oseltamivir for influenza in adults and children: systematic review of clinical study reports and summary of regulatory comments. Bmj. 2014;348:g2545. doi: 10.1136/bmj.g2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., et al. Complement receptor C5aR1 inhibition reduces pyroptosis in hDPP4-transgenic mice infected with MERS-CoV. Viruses. 2019;11:39. doi: 10.3390/v11010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joob B., Wiwanitkit V. SARS-CoV-2 and HIV. J. Med. Virol. 2020;1 doi: 10.1002/jmv.25782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassa G.M. Mother-to-child transmission of HIV infection and its associated factors in Ethiopia: a systematic review and meta-analysis. BMC Infect. Dis. 2018;18:216. doi: 10.1186/s12879-018-3126-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., et al. CBFβ stabilizes HIV Vif to counteract APOBEC3 at the expense of RUNX1 target gene expression. Mol. Cell. 2013;49:632–644. doi: 10.1016/j.molcel.2012.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.M.Q., et al. Virus taxonomy. Ninth Rep Int Comm Taxon Viruses. 2012:486–487. [Google Scholar]
- Lai M., et al. Activation of the ATR pathway by human immunodeficiency virus type 1 Vpr involves its direct binding to chromatin in vivo. J. Virol. 2005;79:15443–15451. doi: 10.1128/JVI.79.24.15443-15451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw B.J., Craft J.E., Kaech S.M. The multifaceted role of CD4+ T cells in CD8+ T cell memory. Nat Rev Immunol. 2016;16:102. doi: 10.1038/nri.2015.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane H.C. Pathogenesis of HIV infection: total CD4+ T-cell pool, immune activation, and inflammation. Top HIV Med a Publ Int AIDS Soc USA. 2010;18(1):2–6. [PubMed] [Google Scholar]
- LaRue R.S., et al. Lentiviral Vif degrades the APOBEC3Z3/APOBEC3H protein of its mammalian host and is capable of cross-species activity. J. Virol. 2010;84:8193–8201. doi: 10.1128/JVI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Li J.P.O., et al. Novel Coronavirus disease 2019 (COVID-19): the importance of recognising possible early ocular manifestation and using protective eyewear. Br. J. Ophthalmol. 2020;104:297–298. doi: 10.1136/bjophthalmol-2020-315994. [DOI] [PubMed] [Google Scholar]
- Li Q., et al. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., et al. Molecular immune pathogenesis and diagnosis of COVID-19. J Pharm Anal. 2020;47:777–780. doi: 10.1016/j.jpha.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie P.J., et al. Novel coronavirus disease (Covid-19): the first two patients in the UK with person to person transmission. J. Inf. Secur. 2020;80(5):578–606. doi: 10.1016/j.jinf.2020.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., et al. Decreased T cell populations contribute to the increased severity of COVID-19. Clin. Chim. Acta. 2020;508:110–114. doi: 10.1016/j.cca.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P., et al. Are pangolins the intermediate host of the 2019 novel coronavirus (SARS-CoV-2)? PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1008421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.K., et al. Remdesivir (GS-5734) protects African green monkeys from Nipah virus challenge. Sci. Transl. Med. 2019;11:eaau9242. doi: 10.1126/scitranslmed.aau9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Stratton C.W., Tang Y. Outbreak of pneumonia of unknown Etiology in Wuhan China: the mystery and the Miracle. J. Med. Virol. 2020;92:401–402. doi: 10.1002/jmv.25678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupfer C., Malik A., Kanneganti T.D. Inflammasome control of viral infection. Curr Opin Virol. 2015;12:38–46. doi: 10.1016/j.coviro.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado L., et al. Expression and function of T cell homing molecules in Hodgkin’s lymphoma. Cancer Immunol. Immunother. 2009;58:85–94. doi: 10.1007/s00262-008-0528-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters P.S. Coronavirus genomic RNA packaging. Virology. 2019;537:198–207. doi: 10.1016/j.virol.2019.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh K. 2020. Coronavirus disease 2019 (COVID-19) UpToDate. [Google Scholar]
- McQuade B., Blair M. Influenza treatment with oseltamivir outside of labeled recommendations. Am J Heal Pharm. 2015;72:112–116. doi: 10.2146/ajhp140390. [DOI] [PubMed] [Google Scholar]
- Mehraeen E., et al. Self-care instructions for people not requiring hospitalization for coronavirus disease 2019 (COVID-19) Arch Clin Infect Dis. 2020;15 doi: 10.5812/archcid.102978. [DOI] [Google Scholar]
- Meyerson N.R., et al. Species-specific vulnerability of RanBP2 shaped the evolution of SIV as it transmitted in African apes. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monroe K.M., et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343:428–432. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.P., et al. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- Mulangu S., et al. A randomized, controlled trial of Ebola virus disease therapeutics. N. Engl. J. Med. 2019;381:2293–2303. doi: 10.1056/NEJMoa1910993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naif H.M. Pathogenesis of HIV infection. Infect Dis Rep. 2013;5:e6. doi: 10.4081/idr.2013.s1.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama E.E., Shioda T. Impact of TRIM5α in vivo. AIDS. 2015;29:1733. doi: 10.1097/QAD.0000000000000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Allergy and Infectious Disease. Seeing Your Healthcare Provider. HIV Gov 2020. https://www.hiv.gov/hiv-basics/staying-in-hiv-care/provider-visits-and-lab-test/seeing-your-healthcare-provider Available online.
- Neuman B.W., et al. A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol. 2011;174:11–22. doi: 10.1016/j.jsb.2010.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura H., Yamaya M. A synthetic serine protease inhibitor, Nafamostat Mesilate, is a drug potentially applicable to the treatment of Ebola virus disease. Tohoku J. Exp. Med. 2015;237:45–50. doi: 10.1620/tjem.237.45. [DOI] [PubMed] [Google Scholar]
- Oldfield V., Keating G.M., Plosker G. Enfuvirtide: a review of its use in the management of HIV infection. Drugs. 2005;65:1139–1160. doi: 10.2165/00003495-200565080-00007. [DOI] [PubMed] [Google Scholar]
- Pancera M., et al. Structure and immune recognition of trimeric pre-fusion HIV-1 Env. Nature. 2014;514:455–461. doi: 10.1038/nature13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X., et al. Transmission routes of 2019-nCoV and controls in dental practice. Int J Oral Sci. 2020;12:1–6. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H.T., Mesplède T. The latest evidence for possible HIV-1 curative strategies. Drugs Context. 2018;7 doi: 10.7573/dic.212522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pharr J.R., et al. A cross-sectional study of the role of HIV/AIDS knowledge in risky sexual behaviors of adolescents in Nigeria. Int J High Risk Behav Addict. 2017;6(4):e63203. doi: 10.5812/ijhrba.63203. [DOI] [Google Scholar]
- Ratliff N.B., et al. Diagnosis of chloroquine cardiomyopathy by endomyocardial biopsy. N. Engl. J. Med. 1987;316:191–193. doi: 10.1056/NEJM198701223160405. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Morales A.J., et al. Going global–travel and the 2019 novel coronavirus. Travel Med. Infect. Dis. 2020;33 doi: 10.1016/j.tmaid.2020.101578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshal M., et al. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J. Biol. Chem. 2003;278:25879–25886. doi: 10.1074/jbc.M303948200. [DOI] [PubMed] [Google Scholar]
- Rossignol J.F. Nitazoxanide: a first-in-class broad-spectrum antiviral agent. Antivir. Res. 2014;110:94–103. doi: 10.1016/j.antiviral.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol J.F. Nitazoxanide, a new drug candidate for the treatment of Middle East respiratory syndrome coronavirus. J Infect Public Health. 2016;9:227–230. doi: 10.1016/j.jiph.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J. Autoimmun. 2020;109 doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., et al. Mechanisms underlying HIV-1 Vpu-mediated viral egress. Front. Microbiol. 2014;5:177. doi: 10.3389/fmicb.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter D., et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 2015;10:586–599. doi: 10.1016/j.celrep.2014.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savarino A., et al. Effects of chloroquine on viral infections: an old drug against today’s diseases. Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwetz T.A., Fauci A.S. The extended impact of human immunodeficiency virus/AIDS research. J. Infect. Dis. 2018;219:6–9. doi: 10.1093/infdis/jiy441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selliah N., Finkel T.H. Biochemical mechanisms of HIV induced T cell apoptosis. Cell Death Differ. 2001;8:127–136. doi: 10.1038/sj.cdd.4400822. [DOI] [PubMed] [Google Scholar]
- Sharp P.M., Hahn B.H. Origins of HIV and the AIDS pandemic. Cold Spring Harb Perspect Med. 2011;1:a006841. doi: 10.1101/cshperspect.a006841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw G.M., Hunter E. HIV transmission. Cold Spring Harb Perspect Med. 2012;2:a006965. doi: 10.1101/cshperspect.a006965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., et al. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh J.A., et al. Emerging genetic diversity among clinical isolates of SARS-CoV-2: lessons for today. Infect. Genet. Evol. 2020 doi: 10.1016/j.meegid.2020.104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki K. Antiviral drugs against alphaherpesvirus. Adv. Exp. Med. Biol. 2018;1045:103–122. doi: 10.1007/978-981-10-7230-7_6. [DOI] [PubMed] [Google Scholar]
- Singh H., et al. Mapping the genomic landscape & diversity of COVID-19 based on > 3950 clinical isolates of SARS-CoV-2: likely origin & transmission dynamics of isolates sequenced in India. Indian J. Med. Res. 2020;151(5):474. doi: 10.4103/ijmr.IJMR_1253_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:1–6. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siwicki B. Health Care News. 2020. Telemedicine during COVID-19: Benefits, limitations, burdens, adaptation.https://www.healthcareitnews.com/news/telemedicine-during-covid-19-benefitslimitations-burdens-adaptation Available online. (accessed on 23 April 2020) [Google Scholar]
- Smith E.C., Denison M.R. Coronaviruses as DNA wannabes: a new model for the regulation of RNA virus replication fidelity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.C., et al. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 2004;78:10328–10335. doi: 10.1128/JVI.78.19.10328-10335.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa J.D., Müller V., Vandamme A.M. The epidemic emergence of HIV: what novel enabling factors were involved? Future Virol. 2017;12:685–707. doi: 10.2217/fvl-2017-0042. [DOI] [Google Scholar]
- Tang Y., et al. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1–13. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchesnokov E.P., et al. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses. 2019;11:326. doi: 10.3390/v11040326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemuri H.S., Challa S., Neelapu N.R.R. Evolution, distribution, and diversity of immunodeficiency viruses. Dyn Immune Act Viral Dis. 2020:187–203. doi: 10.1007/978-981-15-1045-8_13. [DOI] [Google Scholar]
- Wang C., et al. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wilde A.H., et al. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of Middle East respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., et al. Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc. Natl. Acad. Sci. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo P.C.Y., et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavi. J. Virol. 2012;86:3995–4008. doi: 10.1128/JVI.06540-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Monitored Emergency Use of Unregistered and Experimental Interventions (MEURI) 2020. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/global-research-on-novel-coronavirus-2019-ncov/solidarity-clinical-trial-for-covid-19-treatments
- Xu J., et al. Systematic comparison of two animal-to-human transmitted human coronaviruses: SARS-CoV-2 and SARS-CoV. Viruses. 2020;12(2):244. doi: 10.3390/v12020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue F.Y., et al. Preferential apoptosis of HIV-1-specific CD4+ T cells. J. Immunol. 2005;174:2196–2204. doi: 10.4049/jimmunol.174.4.2196. [DOI] [PubMed] [Google Scholar]
- Zahoor M.A., et al. HIV-1 Vpr induces interferon-stimulated genes in human monocyte-derived macrophages. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., et al. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F., et al. Reply to Comments on’Co-infection of SARS-CoV-2 and HIV in a patient in Wuhan city, China. J. Med. Virol. 2020 doi: 10.1002/jmv.25838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Coronavirus Replication Reverse Genet. 2005:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman E.S., et al. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J. Virol. 2006;80:10407–10418. doi: 10.1128/JVI.01212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]