Summary
Congenital anomalies of the kidney and urinary tract (CAKUT) constitute one of the most frequent birth defects and represent the most common cause of chronic kidney disease in the first three decades of life. Despite the discovery of dozens of monogenic causes of CAKUT, most pathogenic pathways remain elusive. We performed whole-exome sequencing (WES) in 551 individuals with CAKUT and identified a heterozygous de novo stop-gain variant in ZMYM2 in two different families with CAKUT. Through collaboration, we identified in total 14 different heterozygous loss-of-function mutations in ZMYM2 in 15 unrelated families. Most mutations occurred de novo, indicating possible interference with reproductive function. Human disease features are replicated in X. tropicalis larvae with morpholino knockdowns, in which expression of truncated ZMYM2 proteins, based on individual mutations, failed to rescue renal and craniofacial defects. Moreover, heterozygous Zmym2-deficient mice recapitulated features of CAKUT with high penetrance. The ZMYM2 protein is a component of a transcriptional corepressor complex recently linked to the silencing of developmentally regulated endogenous retrovirus elements. Using protein-protein interaction assays, we show that ZMYM2 interacts with additional epigenetic silencing complexes, as well as confirming that it binds to FOXP1, a transcription factor that has also been linked to CAKUT. In summary, our findings establish that loss-of-function mutations of ZMYM2, and potentially that of other proteins in its interactome, as causes of human CAKUT, offering new routes for studying the pathogenesis of the disorder.
Keywords: congenital anomalies of the kidney and urinary tract, genetic kidney disease, syndromic CAKUT, extra-renal features, whole-exome sequencing, genomic analysis, transcription regulator, ZMYM2, FIM, ZNF198
Introduction
Congenital anomalies of the kidney and urinary tract (CAKUT) constitute one of the most frequent birth defects, causing almost 50% of all cases of end-stage kidney disease (ESKD) in the first three decades of life.1 In humans, the identification of 40 monogenic causes of isolated CAKUT and 153 monogenic causes of syndromic CAKUT has allowed delineation of multiple pathways of human CAKUT including those involving bone morphogenic protein signaling and retinoic acid signaling.2,3 However, the genes thus far implicated in monogenic forms of CAKUT account for only ∼14%–20% of cases.3, 4, 5 Thus, a significant proportion of CAKUT is still molecularly unidentified and many pathogenic pathways are still elusive.2 To gain further insight into the pathogenesis of human CAKUT, we performed whole-exome sequencing (WES) in a large international cohort of 551 individuals from different families with CAKUT. We identified 14 different heterozygous mutations in ZMYM2 in 15 unrelated families (19 affected individuals) with syndromic CAKUT who also had extrarenal features. ZMYM2 (MIM: 602221), previously known as FIM, ZNF198, or RAMP, is a nuclear zinc finger protein that localizes to the nucleus, specifically to the PML body.6 It forms part of a transcriptional complex acting as a corepressor by interacting with different nuclear receptors, and the LSD1-CoREST-HDAC1 complex on chromatin.7 The role of ZMYM2 in kidney and ureter development is largely unknown and ZMYM2 mutations have not previously been implicated in kidney disease.
Material and Methods
Subjects, Whole-Exome Sequencing, and Variant Evaluation
Approval for human subjects research was obtained from the Institutional Review Boards of the University of Michigan, Boston Children’s Hospital, and from other relevant local Ethics Review Boards. The procedures followed in this study were in accordance with the ethical standards of the responsible committee on human experimentation. Proper informed consent was obtained from all participants. Following informed consent, we obtained clinical data, pedigree data, and blood samples from individuals with CAKUT from worldwide sources using a standardized questionnaire. Informed consent was obtained from the individuals and/or the substitute decision maker, as appropriate. The diagnosis of CAKUT was made by nephrologists and/or urologists based on relevant imaging.
Whole-exome sequencing was performed as previously described.3 Briefly, DNA samples from affected individuals and unaffected family members were subjected to WES using Agilent SureSelect human exome capture arrays (Life Technologies) with next generation sequencing (NGS) on an Illumina sequencing platform. Sequence reads were mapped against the human reference genome (NCBI build 37/hg19) using CLC Genomics Workbench (v.6.5.1) software (CLC bio). Mutation analysis was performed under recessive, dominant, or de novo models, as previously published.3,8,9 Mutation analysis was performed by geneticists and cell biologists, who had knowledge regarding clinical phenotypes, pedigree structure, and genetic mapping, and was in line with proposed guidelines.10,11 Sequence variants remaining after WES evaluation were examined for segregation. Filtering was performed to retain only alleles with a minor allele frequency (MAF) < 0.1%, a widely accepted cutoff for autosomal-dominant disorders.12,13 MAF was estimated using combined datasets incorporating all available data from the 1000 Genomes Project, the Exome Variant Server (EVS) project, dbSNP145, the Exome Aggregation Consortium (ExAC), and gnomAD. We filtered to retain variants in genes with a PLI score of >0.3 based on a dominant hypothesis. To predict deleteriousness of variants, we used the University of Santa Cruz Human Genome Browser for the presence of paralogous genes, pseudogenes, or misalignments, then scrutinized all variants with MAF < 0.1% within the sequence alignments of the CLC Genomic Workbench software program and employed other web based programs (see Web Resources). Variants were confirmed by Sanger sequencing for segregation of phenotype with genotype.
When trios were available for analysis, data processing of FASTQs were performed by the Genomics Platform at the Broad Institute of Harvard and Massachusetts Institute of Technology (Broad Institute). Single-nucleotide polymorphism (SNPs) and insertions/deletions (indels) were jointly called across all samples using Genome Analysis Toolkit (GATK) HaplotypeCaller. Default filters were applied to SNP and indel calls using the GATK Variant Quality Score Recalibration (VQSR) approach. Lastly, the variants were annotated using Variant Effect Predictor (VEP).14 The variant call set was uploaded on to Seqr for analysis of the WES output.
Through collaboration with GeneDx, using genomic DNA from the proband or proband and parent(s), the exonic regions and flanking splice junctions of the genome were captured using the Clinical Research Exome kit (Agilent Technologies) or the IDT xGen Exome Research Panel v.1.0. Massively parallel (NextGen) sequencing was done on an Illumina system with 100 base pairs or greater paired-end reads. Reads were aligned to human genome build GRCh37/UCSC hg19 and analyzed for sequence variants using a custom-developed analysis tool. Additional sequencing technology and variant interpretation protocol has been previously described.15 The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission.
Control Cohorts
Variants were also tested for absence from in-house control populations. The control cohort consisted of 100 families with steroid-resistant nephrotic syndrome (SRNS) in whom a definitive underlying monogenic cause had already been established. An additional control cohort of 257 different families with a clinical diagnosis of nephronophthisis (NPHP) with no genetic cause identified was also used as previously described.16
cDNA Cloning
Full-length human ZMYM2 cDNA (cDNA clone HsCD00082148) was subcloned by PCR from full-length cDNA and cDNA clones. Expression vectors were generated using LR Clonase (Thermo Fisher) according to the manufacturer’s instructions. The following expression vectors were used in this study: pRK5-N-Myc and pcDNA6.2-N-GFP. Mutagenesis was performed using the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies) to generate clones with the ZMYM2 mutations identified in each family available at the time of analysis (Table S1). Each construct was sequenced to verify the correct frame as well as the proper sequence of any linker introduced during the cloning procedure.
Cell Culture and Transfections for cDNA Cloning
Experiments were performed in HEK293 cells purchased from the American Type Culture Collection (ATCC) biological resource center, unless otherwise stated. For transient transfections, HEK293 cells were seeded at 60%–70% confluency in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal calf serum and 1% penicillin/streptomycin and grown overnight. Transfections were carried out using Lipofectamine2000 (Thermo Fisher) and OptiMEM (Thermo Fisher) following the manufacturer’s instructions unless otherwise stated.
Immunofluorescence and Confocal Microscopy in Cell Lines
For immunostaining, HEK293 cells were seeded on fibronectin-coated coverslips in 6-well plates. After 16–24 h, cells were transiently transfected using Lipofectamine2000 (Thermo Fisher) according to the manufacturer’s instructions. Experiments were performed 24–48 h after transfection. Cells were fixed for 15 min using 4% paraformaldehyde and permeabilized for 15 min using 0.5% Triton X-100. After blocking with 10% donkey serum + BSA, cells were incubated with primary antibody overnight at 4°C. The following day, cells were incubated in secondary antibody for 60 min at room temperature and subsequently stained for 5 min with DAPI in PBS. Confocal imaging was performed using the Leica SP5X system with an upright DM6000 microscope, and images were processed with the Leica AF software suite. Immunofluorescence experiments were repeated at least two times in independent experiments. The following antibodies were used for immunostaining: mouse anti-Myc (sc-40, Santa Cruz Biotechnology), rabbit anti-ZMYM2 (ab106624, Abcam), and rabbit anti-ZMYM2 (PA5-28265, Thermo Fisher), all diluted 1:100. Donkey anti-mouse secondary antibodies conjugated to Alexa Fluor 488 (A-21202, Thermo Fisher) and donkey anti-rabbit secondary antibody conjugated to Alexa Fluor 594 (A-21207, Thermo Fisher) were used. We originally hypothesized that missense variants may be present in individuals with a milder phenotype. In total, we identified 12 missense variants in ZMYM2 (Table S2) in our CAKUT cohort. As such, along with testing loss-of-function variants, we tested the following missense variants in our immunoflurosence (IF) data: p.Val61del (c.181_183del), p.Glu126Ala (c.377A>C), p.Ile387Ala (c.1159A>G), p.Lys649Arg (c.1946A>G), p.Tyr763His (c.2287T>C), p.Tyr763Leu (c.2287_2288delinsTA>CT), p.Gly775Glu (c.2324G>A), p.Asp997del (c.2990_2992del), and p.Glu1031Lys (c.3091G>A) (Table S2, Figure S1). Please note that although variant p.Tyr763Glnfs∗6 (c. 2287_2288del) and p.Cys823∗ (c.2469T>A) were initially included in our IF data, these two variants were not included in the analysis of our clinical data as the families in question provided consent to study their variants but did not provide consent to include any clinical data.
Bioluminescence Resonance Energy Transfer (BRET) Assays
We employed Bioluminescence Resonance Energy Transfer (BRET) assays to test the interactions between both wild-type and mutant ZMYM2 and wild-type FOXP1. Briefly, wild-type and mutant ZMYM2 constructs were subcloned from the pRK5-N-Myc vector into YFP- and rLuc-vectors as previously described.17 Wild-type FOXP1, FOXP2, and NLS rLuc-constructs were generated in a prior study.17 HEK293T/17 cells were grown in 96-well plates and cultured for 24 h at 37°C with 5% CO2. Cells were transfected using GeneJuice Transfection Reagent (Merck Millipore) according to manufacturer’s instructions. At 36 h post-transfection, EnduRen luciferase substrate (Promega) was added at 60 μM and cells were incubated for 4 h. Emission values were measured using an Infinite 200Pro plate reader (Tecan) using the Blue1 and Green1 filter sets. Corrected BRET ratios were obtained using the following formula [Green1(experimental condition)/Blue1(experimental condition)] − [Green1(control condition)/Blue1(control condition)]. Further details of the BRET assay set-up are discussed by Deriziotis et al.17
Xenopus tropicalis Model
X. tropicalis were housed and cared for in the aquatics facility at Yale University School of Medicine according to established protocols approved by Yale Institutional Animal Care and Use Committee.
Expression Pattern of zmym2
Previous data have demonstrated RNA expression patterns for zmym2 in Xenopus (Figure S2).18 In addition, the Papalopulu lab has deposited images of zmym2 expression in Xenbase (see Web Resources), which are suggestive of expression in the pronephros. We further confirmed expression in a Xenopus model (Figure S2C) at stage 34 and although expression is somewhat ubiquitous, we did find some enrichment in the pronephros and tubule.
Selection of Variants Tested in Xenopus Modeling
We prioritized testing certain variants (p.Val61del [c.181_183del], p.Asp997del [c.2990_2992del], p.Gly257∗ [c.766_767dupGT], p.Gln398∗ [c.1192C>T], p.Cys536Leufs∗13 [c.1607delG], p.Arg540∗ [c.1618C>T], p.Lys812Aspfs∗13 [c.2434_2437delAAAG], p.Gly1045Argfs∗18 [c.3130_3131dupAA], and p.Gly1045Argfs∗33 [c.3130_3131dupAA]) since we hypothesize that these particular variants best represent the varying kinds of disruptions found widely distributed across the ZMYM2 gene locus and so testing these variants could lead to more informative results about dysfunction.
Microinjection of Morpholinos and mRNA in Xenopus Embryos
We induced ovulation and collected embryos by in vitro fertilization as previously described.19,20 Embryos were raised to stage 34 or 45 in 1/9MR + gentamycin. Staging of Xenopus tadpoles was performed according to Faber and Nieuwkoop.21 Antisense morpholino oligonucleotides (MO) or mRNAs were injected at either the one-cell stage or into one cell of the two-cell embryo as previously described.22 We employed a splice blocking MO which conceptually leads to exon 3 skipping. The following MOs were used: Control: 5′-CCTCTTACCTCAGTTACAATTTATA-3′ and zmym2 exon-3 intron-3 splice blocking 5′-TTGCTGTGGAGGCTGAAAAACCT-3′ (GeneTools in Web Resources). We performed PCR with primers that span this exon and expect a loss of ∼800 bp. To confirm efficient and specific knockdown of ZMYM2, we also tested for rescue of our MO phenotype using the human mRNA (and individual variants that do not rescue), confirming that our knockdown is specific for ZMYM2 in the experiments presented. We generated in vitro capped mRNA of wild-type and mutated human ZMYM2 from sequences cloned into in the pKR5-Myc backboned using the SP6 mMessage machine kit (Thermo Fisher) following the manufacturer’s instructions. MOs for knockdown experiments were injected with 5 ng in a 2 nanoliter volume into one cell of the two-cell embryo. This volume included the fluorescent tracer Mini-ruby (Thermo Fisher) in order to determine the injected side for later analysis via in situ hybridization. For rescue experiments, MOs were injected in the one-cell embryo with 10 ng in a 2 nanoliter volume. Subsequently, mRNA corresponding to either wild-type or mutated ZMYM2 sequences was injected into one cell of the two-cell embryo with 50 pg in a volume of 2 nanoliters including fluorescent tracer to determine the side injected with mRNA for subsequent analysis via in situ hybridization.
Whole-Mount In Situ Hybridization
We detected Xenopus atp1a1 expression by generating a digoxigenin-labeled antisense probe using the T7 High Yield RNA Synthesis kit (NEB, E2040S) and DIG-dUTP (Sigma) from clone number TTpA007m23. Atp1a1 expression serves as a well-established readout, allowing us to monitor a majority of the pronephric and tubule tissue.23 Embryos were collected at stage 34 and fixed in MEMFA (1:1:8 10× MEMFA salts, 37% formaldehyde, distilled water) (10× MEMFA salts: 1 M MOPS, 20 mM EGTA, 10 mM MgSO4) for 1–2 h at room temperature and dehydrated in 100% ethanol. Whole-mount in situ hybridization was done as previously described.22 We quantitatively assessed loss of function by measuring the area corresponding to the proximal and intermediate tubule24 in injected and control sides of each embryo. We qualitatively assessed posterior atp1a1 expression in the pronephric tubule. Rescue efficiency was assessed in the mRNA injected side of embryos in which the contralateral side had morphological abnormalities. Successful rescue was based on the comparison between mRNA injected and mRNA un-injected sides of the area of the proximal pronephros. Areas were determined by manually delineating the bounds of this region in ImageJ. In situ hybridization results were imaged with a Canon EOS 5d digital camera mounted on a Zeiss discovery V8 stereomicroscope.
Alcian Blue Staining
Stage 45 embryos were fixed in 100% ethanol for 48 h at room temperature and then washed briefly in acid alcohol (1.2% HCl in 70% ETOH). A 0.25% alcian blue solution in acid alcohol was used to stain the embryos over 48 h at room temperature. Specimens were then washed in acid alcohol several times, rehydrated into H2O, and bleached for 2 h in 1.2% hydrogen peroxide under a bright light. They were then washed several times in 2% KOH and left rocking overnight in 10% glycerol in 2% KOH. Samples were processed through 20%, 40%, 60%, and 80% glycerol in 2% KOH. Craniofacial cartilage was then imaged with a Canon EOS 5d digital camera mounted on a Zeiss discovery V8 stereomicroscope.
Statistical Analysis
All experiments in the Xenopus model were performed a minimum of three times and numbers stated in graphs are the composite of multiple experiments. Statistical significance of abnormalities and rescues with respect to proximal pronephric size were evaluated using unpaired t tests, while percentages of posterior loss of signal and abnormal craniofacial cartilage were evaluated using Fisher’s exact tests using GraphPad Prism v.8. In all figures, statistical significance was defined as p < 0.05. A single asterisk indicates p < 0.05, while double, triple, and quadruple asterisks indicate p < 0.005, p < 0.0005, and p < 0.0001, respectively. Bars in graphs indicate means and standard deviations.
Mice
Zmym2+/− mice were generated by the Transgenic Core Facility of the Goodman Cancer Research Centre in a C57BL/6 background using a CRISPR-Cas9 targeting approach. For this mouse model, we chose to replicate the truncating mutation in an early exon found in individual GM1-21 (p. Gly257∗ [c.766_767dupGT] Table 1), because it is associated with a strong human phenotype, and since any resulting mutant protein would be severely truncated, showing subcellular mislocalization according to cellular assays (Figure S1). To model this frameshift mutation, sgRNA 5′-GTTACAACCTTAGAAACAGG-3′ was designed against exon 3 found both in humans and mice. A Δ1 allele was selected and propagated in C57BL/6 background for three generations prior to phenotypical analyses. Genotyping was performed by PCR amplification using primers 5′-ACCTCCTCCATCTTCTGCAC-3′ and 5′-AAAAGGTCCAACTCCAGCCT-3′ amplicon sequencing. Animals and experiments were kept in accordance with the standards of the animal ethics committee of McGill University, and the guidelines of the Canadian Council of Animal Care.
Table 1.
Fourteen Heterozygous Mutations in ZMYM2 in 19 Individuals from 15 Families with Syndromic CAKUT
| Family, Individual | Nucleotide Change | Amino Acid Changea | Exon Segregation | Ethnicity, Gender | CAKUT (Sidednessb) | Extra-renal Manifestation | Neurologic Involvement |
|---|---|---|---|---|---|---|---|
| GM10-21 | c.622C>T | p.Arg208∗ | 3: 20% mosaic | Dutch, F | RUS normal | skeleton: downslanting palpebral fissures | hypotonia, ID, stereotypic movements |
| GM1-21 | c.766_767dupGT | p.Gly257∗ | 3: de novo | USA, M | UUT: renal agenesis LUT: hypospadias, cryptorchidism, chordee, Müllerian duct remnants |
heart: BAV skeleton: small hands & feet skin: facial dysmorphsims, convex dysplastic finger nails, hypoplastic toenails other: feeding problems, oral phase dysphagia, IUGR, growth delay |
microcephaly, DD, hypotonia, tethered cord |
| GM3-21 | c.1192C>T | p.Gln398∗ | 5: de novo | USA, M | UUT: RUS normal LUT: enuresis, incontinence |
heart: PDA skeleton: facial dysmorphsims other: dental caries |
DD, autistic spectrum |
| GM9-21 | c.1367dupA | p.Tyr456∗ | 6: de novo | white, F | UUT: hypoplastic pelvic kidney (R) detected by “reverse phenotyping” | skeleton: facial dysmorphisms (triangular face, broad neck, broad nasal bridge), scoliosis single palmar crease on left, tapered fingers, tapered lower extremities |
DD, ‾ auditory attention, startle reflex, motor stereotypies |
| SSC3-21 | c.1607delG | p.Cys536Leufs∗13 | 8: pat. NA, mat. NA | Italy, F | UUT: UPJO (L) | — | mild ID |
| A4730-21 | c.1618C>T | p.Arg540∗ | 8: de novo | Macedonia, M | UUT: pre-natal hydronephrosis LUT: BL VUR grade 3, urethral stricture, hydrocele testis |
skeleton: facial dysmorphism (wide interpupillary distance, mild epicanthal folds, long nose with a bulbous tip, farsightedness, low set posteriorly rotated ears with a simple helix and protuberant ears), hyper-extensibility of the joints | speech delay |
| A1204-21 | c.1618C>T | p.Arg540∗ | 8: pat. NA, mat. NA | Macedonia, F | UUT: renal agenesis (R) | other: hematocolpos, imperforate hymen | no data |
| GM11-21 | c.1623_1627delACAGT | p.Cys543Valfs∗3 | 8: de novo | Moroccan, M | RUS normal | skeleton: hypertelorsism, small ears, thick lips, high palate, facial dysmorphsims other: OSA |
DD, mild ID, seizures, autism, psychosis |
| GM17-21 | c.2165T>A | p.Leu722∗ | 12: pat. WT, mat. Het | white, M | mild hypospadias, distal chordee and dorsal hooding | skeleton: aplasia cutis other: acute lymphoblastic leukemia |
DD, autism spectrum disorder (mother has ADHD and learning disability) |
| GM19-21 | c.2338C>T | p.Arg780∗ | 13: de novo | Switzerland, M | RUS normal | — | seizure disorder, MRI normal, low IQ (85) |
| GM6-21 | c.2434_2437delAAAG | p.Lys812Aspfs∗18 | 13:de novo | white, F | RUS normal | heart: VSD, atrial septal defect, PDA skeleton: short stature −2 SD, short 5th digit with abnormal nails, BL epicanthi, abnormal palmar crease, upturned nasal tip and severe feeding problems |
microcephaly, DD |
| GM6-22 | c.2434_2437delAAAG | p.Lys812Aspfs∗18 | 13:de novo | white, F | RUS normal | BL epicanthus, abnormal palmar crease | speech delay |
| GM18-12 | c.2494−1G>A | IVS15-1G>A | Intron 14: pat. WT, mat. het | white, F | RUS NA | heart: atrial septal defect skeleton: epicanthal folds |
ADHD, autism, behavioral concerns |
| GM18-22 | c.2494−1G>A | IVS15-1G>A | white, M | brother: RUS NA | heart: atrial septal defect | ||
| GM18-12 | c.2494−1G>A | IVS15-1G>A; obligatory splice site | white, F | mother: RUS normal | heart: atrial septal defect | ||
| GM7-21 | c.3130_3131dupAA | p.Gly1045Argfs∗33 | 19: pat. WT, mat. NA | white, F | RUS normal | heart: ECHO normal skeleton: dysmorphic facial features, short 5th fingers & thumbs, broad big toes, 5th finger clinodactyly, mild short stature (9th percentile) |
microcephaly, DD, hypotonia, high hyperopia |
| GM13-21 | c.3176dupA | p.Asp1059Glufs∗2 | 20: de novo | white, F | RUS normal | heart: ECHO normal skeleton: short stature (3rd percentile), dysmorphic facial features (wide eyebrows, wide interpupillary and intercanthal distance, epicanthal folds, narrow downslanting palpebral fissures, nose with a wide tip, downturned corners of the mouth, small and low set ears with hypoplastic lobule), 5th finger clinodactyly |
microcephaly, DD, speech delay |
| GM12-21 | c.3246G>A | p.Trp1082∗ | 20: pat. NA, mat. het | white, M | RUS: bilateral malrotated kidneys and the right is low lying | skeleton: dysmorphic facial features (narrow palpebral fissures, epicanthi and telecanthus, small nose and a grooved single philtrum with mild hypoplastic nasal nares), short, thick fingers, ↑ range of motion joints | DD, speech delay, hypotonia, |
| GM12-12 | c.3246G>A | p.Trp1082∗ | white, F | mother RUS NA | intellectually disability |
Transcript accession number for ZMYM2: GenBank: NM_001190965. Mutations listed in this table were not present in gnomAD database. ADHD, attention deficit hyperactivity disorder; ASD, atrial septal defect; BAV, bicuspid aortic valve; BL, bilateral; DC, disease causing, DD; developmental delay; Del, deleterious; ECHO, echocardiogram; F, female; het, heterozygous; ID, intellectual disability; IUGR, intra-uterine growth retardation; L, left; LUT, lower urinary tract; mat., maternal; M, male; NA, not available; OSA, obstructive sleep apnoea; pat., paternal; PDA, patent ductus arteriosus; PNS, peripheral nervous system; PPH2 score, HumVar PolyPhen-2 prediction score; R, right; RUS, renal ultrasound; SIFT, sorting tolerant from intolerant; Tol., tolerated; UUT, upper urinary tract; UPJO; ureteropelvic junction obstruction; RUS, renal ultrasound; VACTERL, vertebral defects, anal atresia, cardiac defects, tracheo-esophageal fistula, renal anomalies, and limb abnormalities; VSD, ventricular septal defect; VUR, vesicoureteral reflux; WT, wild type.
Underline indicates Macedonian founder mutation.
Sidedness of CAKUT phenotype given in parentheses.
Vesicoureteral Reflux
Vesicoureteral reflux was assessed through methylene blue injection into the bladder as described.25 Briefly, the urinary tract of freshly sacrificed newborns was exposed and the bladder was injected using a 30-gauge needle connected to a reservoir filled with methylene blue dye (1 mg/mL). To increase hydrostatic pressure, the syringe was raised at 5 cm/s to 120 cm. Reflux and urethral voiding pressures were recorded as reservoir height (cm).
Histology
Whole urogenital tracts from E18.5 embryos and P0 neonates were dissected in PBS and fixed overnight in 4% PFA at 4°C. Samples were then processed for paraffin embedding and sectioning (4 μm thickness). Tissue sections from each sample were stained with Hematoxylin & Eosin for tissue analysis. Immunohistofluorescence analyses were performed as described.26 The following antibodies were used for immunostaining: anti-Pax2 (Covance cat# PRB-276P) at 1:200, anti-Podocalyxin (R&D systems cat# AF1556) at 1:200, anti-Cytokeratin 8/18 (Fitzgerald cat# 20R-CP004) at 1:300, and anti-Zmym2 (Origene cat# AP08258PU) at 1:100 dilution in PBS.
Proximity-Based Biotinylation (BioID) Analysis
Proximity-based biotinylation, or BioID, is a method developed for the characterization of protein-protein interactions in living cells.27 Briefly, Flp-In T-REx 293 cells were stably transfected with pcDNA5 FRT/TO Flag-BirA-R118G (FlagBirA∗) expression vectors, containing open reading frames for human ZMYM2 (wild type or deletion mutants) or ZMYM3. Cells at 80% confluence were incubated for 24 h in complete media supplemented with 1 μg/mL tetracycline (Sigma-Aldrich) and 50 μM biotin (BioShop). Cells were harvested, lysed (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 0.1% SDS, protease inhibitor cocktail, turbonuclease), sonicated twice for 10 s at 35% amplitude (Sonic Dismembrator 500; Fisher Scientific), and centrifuged at 16,000 rpm (35,000 × g) for 30 min at 4°C. Supernatants were passed through a Micro Bio-Spin Chromatography column (Bio-Rad 732-6204) and incubated with 30 μL of high performance streptavidin-Sepharose beads (GE Healthcare) for 3 h at 4°C on an end-over-end rotator. Beads were pelleted (2,000 rpm, 2 min) and washed six times with 50 mM ammonium bicarbonate (pH 8.3). Washed beads were treated with L-1-Tosylamide-2-phenylethyl chloromethyl ketone (TPCK)-treated trypsin (Promega) for 16 h at 37°C with end-over-end rotation. After 16 h, another 1 μL of TPCK-trypsin was added for 2 h and incubated in a water bath at 37°C. Supernatants were lyophilized and stored at 4°C. Two biological and two technical replicates were analyzed using mass spectrometry (below) to identify high confidence proximity interactors.
Mass Spectrometry Analysis
Liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) was conducted as previously described.27 Briefly, high performance liquid chromatography was conducted using a 2 cm pre-column (Acclaim PepMap 50 mm × 100 μm inner diameter [ID]) and 50 cm analytical column (Acclaim PepMap, 500 mm × 75 μm diameter; C18; 2 μm; 100 Å, Thermo Fisher Scientific), running a 120 min (35,000 × g) reversed-phase buffer gradient at 225 nL/min on a Proxeon EASY-nLC 1000 pump in-line with a Thermo Q-Exactive HF quadrupole-Orbitrap mass spectrometer. A parent ion scan was performed using a resolving power of 60,000, then up to the 20 most intense peaks were selected for MS/MS (minimum ion count of 1,000 for activation), using higher energy collision induced dissociation (HCD) fragmentation. Dynamic exclusion was activated such that MS/MS of the same m/z (within a range of 10 ppm; exclusion list size = 500) detected twice within 5 s were excluded from analysis for 15 s. For protein identification, Thermo.RAW files were converted to the .mzXML format using Proteowizard,28 then searched using X!Tandem29 and Comet30 against the human Human RefSeq Version 45 database (containing 36,113 entries). Search parameters specified a parent ion mass tolerance of 10 ppm and an MS/MS fragment ion tolerance of 0.4 Da, with up to 2 missed cleavages allowed for trypsin. Variable modifications of +16@M and W, +32@M and W, +42@N terminus, and +1@N and Q were allowed. Proteins identified with an iProphet cut-off of 0.9 (corresponding to ≤1% FDR) and at least two unique peptides were analyzed with SAINT Express v.3.6.1.31 Twenty control runs (consisting of BioID conducted on the same cell type stably expressing the FLAG-BirA∗ epitope tag alone) were collapsed to the four highest spectral counts for each prey and compared to the two technical replicates and two biological replicates of the baits. High confidence interactors were defined as those with FDR ≤ 0.01. All raw mass spectrometry files have been deposited at the MassIVE archive (see Data and Code Availability), with accession number ID MSV000085033.
Results
Mutations of ZMYM2 Cause CAKUT
In pursuit of additional monogenic causes for CAKUT, we performed WES in 551 individuals with CAKUT.32 We detected a heterozygous mutation (p.Arg540∗ [c.1618C>T]) in the gene Zinc Finger MYM-Type Containing 2 (ZMYM2, GenBank: NM_197968.2) in an affected individual with syndromic CAKUT in an outbred Macedonian family (A4730, Table 1). The mutation was de novo and was absent from control databases ExAC and gnomAD (Table 1, Figure S3). Direct inspection of sequence alignments from whole-exome data did not yield a mutation in any of the 40 known isolated human CAKUT genes, the 153 known human syndromic CAKUT genes, nor the 185 known murine CAKUT genes, as previously described.3 The individual, A4730_21, presented with pre-natal hydronephrosis due to a urethral stricture (Table 1). Post-natal ultrasound revealed bilateral grade three vesicoureteral reflux (VUR) with clinical evidence of a hydrocele. Post WES, extra-renal features were noted including facial dysmorphism, hyper-extensibility of the joints, and speech delay (Figure 1A).
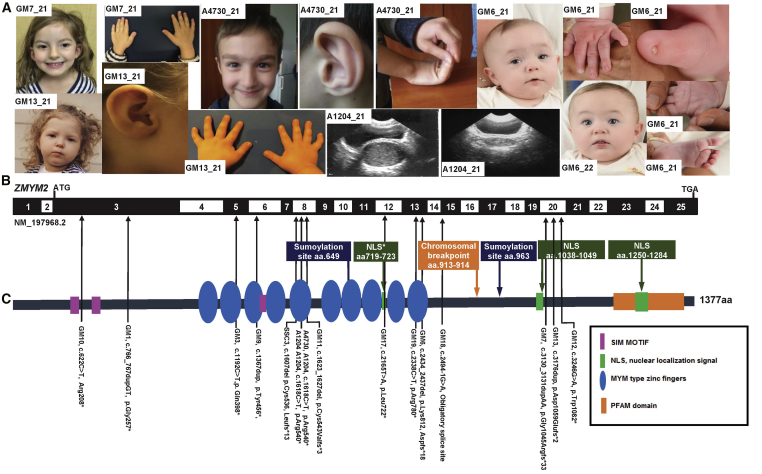
Figure 1.
Whole-Exome Sequencing Identifies 14 Heterozygous Loss-of-Function Mutations in ZMYM2 in 15 Families with 19 Affected Individuals
(A) Clinical features of individuals with ZMYM2 mutations (see Table 1); family number is shown in the white rectangle.
Family GM7: Hypertelorism; simple helix and protuberant ears; 5th fingers and thumbs; 5th finger clinodactyly.
Family A4730: Wide eyebrows, mild synophrys, short filtrum; long nose with a bulbous tip; auricle with hypoplastic lobule; hyperextensibility of joints.
Family GM13: Wide interpupillary distance and intercanthal distance; small auricle; clinodactyly.
Family A1204: hematocolpos pre- and post-drainage.
Family GM6: (GM6_21) dysmorphic facial features with epicanthi, short 5th digit with hypoplastic nails, abnormal palmar crease and sandal gap toe; (GM6_22) Dysmorphic features – epicanthi.
(B) Exon structure of human ZMYM2 cDNA (GenBank: NM_197968.2) and positions of mutations (arrowheads).
(C) Protein domain structure of human Zmym2 showing the positions of each of the 14 different heterozygous mutations identified in 15 families (position indicated by the arrows shafts).
aa, amino acid; ATG, start codon; NLS, nuclear localization site.
In an unrelated Macedonian family (A1204_21, Table 1), we detected the same variant as in the index family (p.Arg540∗ [c.1618C>T]) from our initial cohort of 551 individuals with CAKUT. This female individual had right renal agenesis. Similar to the index person (A4730_21), additional genitourinary tract pathologies were noted: hematocolpos secondary to imperforate hymen (Figure 1A). Due to loss of follow-up, additional extra-renal or neurological features could not be assessed.
Through collaboration with Columbia University, New York, we identified family SSC3 with a mutation in ZMYM2 (Table 1). The proband (SSC3_21) was a female with a truncating frameshift mutation (p.Cys536Leufs∗13 [c.1607del]) (Table 1). She had uretero-pelvic junction obstruction and evidence of intellectual disability (Table 1). Using Genematcher,33 we identified an additional 12 families who carried loss-of-function mutations in ZMYM2 (Table 1, Figure 1). All families had extra-renal features of disease or neurological involvement.
Phenotypic Features
In total, we detected 14 different heterozygous nonsense or frameshift mutations of ZMYM2 in 15 families with 19 affected individuals with CAKUT and/or syndromic extra-renal features. The phenotypic spectrum included CAKUT in 7 of 14 families, while all affected individuals displayed extra-renal features. Common extra-renal features included cardiac defects, facial dysmorphisms, small hands and feet with dysplastic/hypoplastic nails, clinodactyly, and neurological features. Neurological manifestations were noted in 14 families (16 affected individuals) and included microcephaly (4/14), developmental delay (9/14), intellectual disability (4/14), speech delay (4/14), and infantile hypotonia (3/14) (Table 1, Figure 1).
De Novo Pattern of Inheritance
In 8 of 15 families, DNA was available from both parents, and for four families a single parental DNA sample was available. For eight of these families segregation analysis was consistent with a de novo mutation (Table 1, Figure S3). Germline mosaicism was observed in 1 of the 14 families (GM10). In the two families (GM17 and GM18) where maternal DNA was available, we were able to confirm that the variant was inherited from an affected mother (Table 1). In both cases, the affected mother displayed neurological manifestations of disease (attention deficit hyperactivity disorder [ADHD] and learning disability), while the affected mother from family GM_18 also had evidence of cardiac involvement (Table 1).
Our findings regarding de novo and mosaic occurrence strongly suggest that heterozygous truncating mutations of ZMYM2 convey infertility or interfere with germline transmission. Allelic frequency data in gnomAD further support this hypothesis. All mutations observed in the affected individuals of this study are absent from control populations (ExAC and gnomAD) while other ZMYM2 loss-of-function variants are extremely rare, with only 31 such variants recorded, 27 of which only occurred once in approximately 270,000 indiviudals (Table S3).
We originally hypothesized that missense variants may be present in persons with a milder phenotype, as has previously been described for other genes implicated in CAKUT.34 In total, we identified 12 missense variants in ZMYM2 (Table S2) in our CAKUT cohort. As such, we tested the following missense variants in our IF data: p.Val61del (c.181_183del), p.Glu126Ala (c.377A>C), p.Ile387Val (c.1159A>G), p.Lys649Arg (c.1946A>G), p.Tyr763His (c.2287T>C), p.Tyr763Leu (c.2287_2288delinsTA>CT), p.Gly775Glu (c.2324G>A), p.Asp997del (c.2990_2992del), p.Glu1031Lys (c.3091G>A) (Table S2). However, the frequency of these variants was similar to the frequency of missense variants in a control steroid-resistant nephrotic syndrome and nephronopthisis cohorts (Table S4). In addition, these variants all retained their nuclear localization and transcriptional repression properties, making causality unlikely (Figures S1 and S4).
Zmym2 Knockdown in X. tropicalis Leads to Defects in Renal and Craniofacial Development
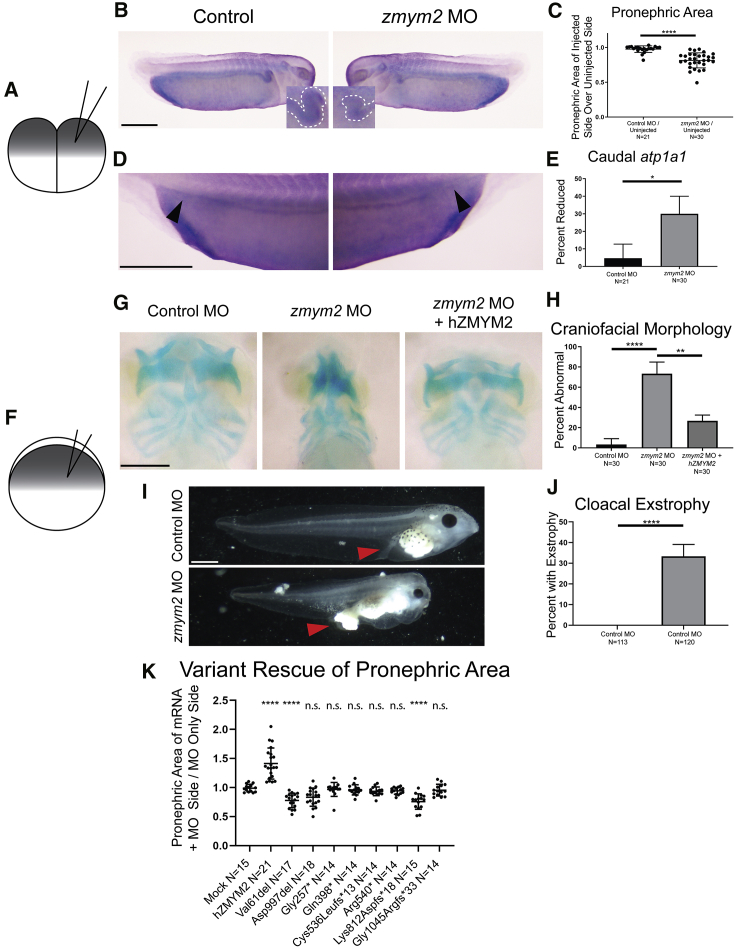
To evaluate the deleteriousness of the mutations observed in vivo, we generated an X. tropicalis model of zmym2 loss-of-function (Figures 2 and S5). At the two-cell stage, Xenopus embryos were injected with a morpholino oligo (MO) targeting zmym2. While one side of the embryo developed from the un-injected cell and served as an internal control, the contralateral side developed from the MO injected cell (Figures 2A–2E). At stage 34, in situ hybridization for the pronephric marker atp1a1 was employed to assess for defects in pronephros morphology in response to MO-mediated knockdown. Specifically, the posterior segment of the pronephric tubules were evaluated for the level of atp1a1 expression, which characteristically meets the proctodeum of the embryo at its caudal aspect. Additionally, the proximal region of the embryonic pronephri corresponding to the proximal and intermediate tubule in humans24 were quantified to determine zmym2 knockdown effects on these structures. Compared to control sides of the embryos, sides with MO knockdown of zmym2 demonstrated loss of caudal atp1a1 signal and decreased proximal pronephric area, suggesting that zmym2 has a specialized role in pronephric development in a subset of regions. Quantification of these experiments revealed a loss of posterior atp1a1 signal in 30% of embryos on the side representing zmym2 knockdown (Figure 2C).
Figure 2.
Xenopus tropicalis Model of Zmym2 Loss of Function
(A) Schematic of the experimental procedure for injection of morpholino into one cell of a two-cell embryo. One side of the embryo is subject to the knockdown, while the other serves as an internal control.
(B and C) Representative images and quantitation of decreased pronephric area in one-sided zmym2 morphants.
(D and E) Representative images and quantitation of decreased caudal atp1a1 signal in one-sided zmym2 morphants.
(F) Schematic of the experimental procedure for injection of morpholino into a one-cell stage embryo.
(G and H) Representative images and quantitation of craniofacial dysmorphology in zmym2 morphants, and frequency of rescue of this phenotype in zmym2 morphants co-injected with ZMYM2 mRNA.
(I and J) Representative images and quantitation of cloacal exstrophy in zmym2 morphants.
(K) Quantitation of proximal pronephric size abnormalities comparing the ratio of proximal pronephric size on the mRNA versus MO only side of an embryo between those injected with mock mRNA, control missense mutants, and those injected with the human ZMYM2 mRNA variants representing truncating mutants.
Scale bars depict 500 μm. ∗∗∗∗p < 0.0001, ∗∗p < 0.005, ∗p < 0.05 by unpaired t test (C, K) and Fisher’s exact test (E, H, J). Bars indicate mean and standard deviation.
To assess the functionality of ZMYM2 mutations discovered in individuals, we employed unilateral injection of zmym2 morphants with wild-type or variant ZMYM2 mRNA reflecting individual sequences (p.Gly257∗ [c.766_767dupGT], p.Gln398∗ [c.1192C>T], p.Cys536Leufs∗13[c.1607delG], p.Arg540∗ [c.1618C>T], p.Lys812Aspfs∗18 [c.2434_2437delAAAG], and p.Gly1045Argfs∗33 [c.3130_3131dupAA], in addition to two missense variants, p.Val61del [c.181_183del] and p.Asp997del [c.2990_2992del]). This approach revealed that only wild-type mRNA resulted in rescue of area size (Figures 2F–2K and S5). In contrast, unilateral injection of mRNA reflecting the truncating variants, that we identified in individuals, resulted in little to no restoration of proximal pronephric area (Figure 2K).
We then allowed zmym2 knockdown tadpoles to develop further in order to identify phenotypes that may not be visible early on. At these later stages, protrusion of tissue through the primitive cloaca was also apparent in 33% of zmym2 MO-injected embryos and none of the control MO-injected embryos. In addition, craniofacial abnormalities became readily apparent. To assess these abnormalities further, we used Alcian blue staining to delineate the cartilage morphology within stage 45 embryos. In 73% of zmym2 MO-injected embryos, gross morphological anomalies were observed as compared to 3% of control MO-injected embryos. The frequency of this phenotype in MO-injected embryos was reduced to 27% via reintroduction of wild-type ZMYM2 mRNA (Figure 2K). Noteworthy is that variant p.Lys812Aspfs∗18 actually worsens the phenotype when used in a rescue experiment, which is suggestive that this variant has the potential to function as dominant negative.These findings are consistent with a pathologic role for heterozygous ZMYM2 truncating mutations in urinary tract abnormalities and facial dysmorphisms observed in individuals with this disorder.
Mouse Model
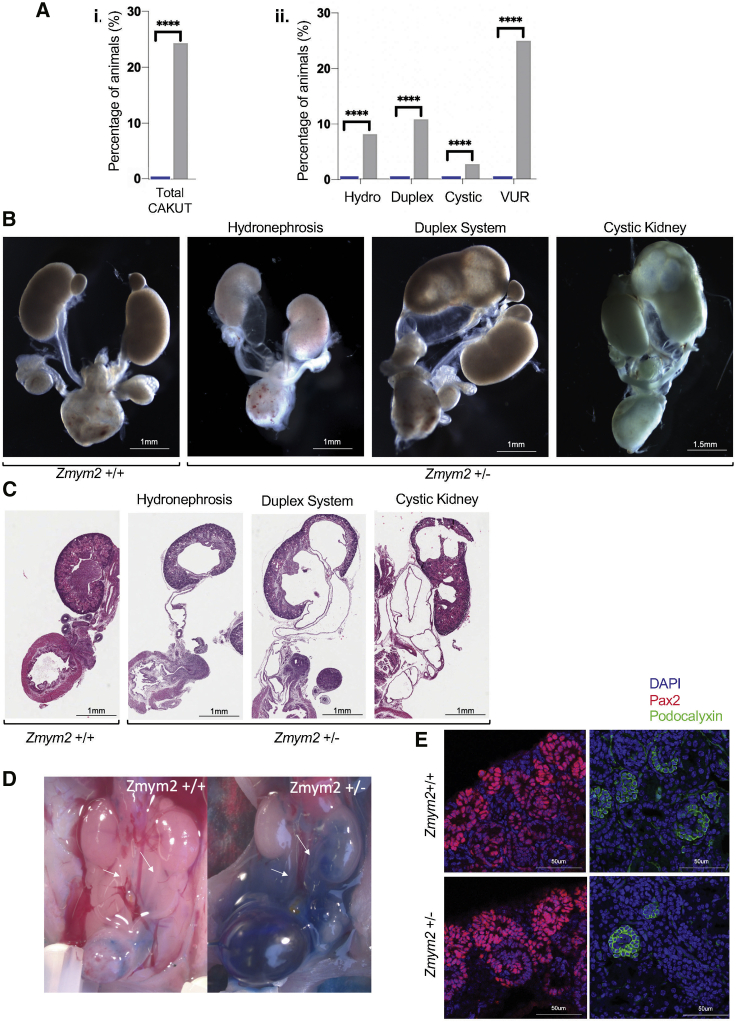
To further validate the causal role of ZMYM2 in CAKUT-related developmental defects, we generated a mouse model recapitulating the frameshift mutation found in exon 3 of individual GM1-21 using CRISPR-Cas9 gene targeting (Figure S6). Zmym2+/− mice showed a spectrum of CAKUT-like defects including hydroureter as well as duplex and cystic kidneys at E18.5 (Figures 3A–3C). When tested for vesicoureteral reflux, 25% of newborns had a reflux phenotype, including a majority at or below voiding pressure (Figures 3D and S6). None of those malformations were observed in wild-type animals. The presence of CAKUT-like phenotypes in Zmym2+/− animals is compatible with the low but widespread expression levels of Zmym2 in the developing kidney (Figure S7). Immunofluorescence analysis of E18.5 kidneys without overt malformations showed normal tissue architecture in Zmym2+/− animals (Figure 3E). No additional phenotypes were observed in Zmym2+/− animals.
Figure 3.
Array of CAKUT Phenotypes Observed in a Zmym2+/− Mutant Mouse Model
Zmym2+/− mice heterozygous for a frameshift mutation in exon 3 were analyzed at embryonic stage E18.5 and post-natal stage P0.
(A) Percentages of mice with given CAKUT phenotype observed in Zmym2+/− pups and their wild-type littermates. Statistical analysis was done using a binomial test. For CAKUT, hydroureter, duplex, and cystic kidneys phenotypes were compiled from Zmym2+/+ (n = 35) and Zmym2+/− (n = 37). Vesicoureteral reflux (VUR) was assessed from Zmym2+/+ (n = 25) and Zmym2+/− (n = 20). Note, some animals harbored more than one CAKUT phenotype.
(B) Dissected E18.5 and P0 urogenital system of Zmym2+/+and Zmym2+/− mice demonstrating gross CAKUT phenotypes including hydroureter, hydronephrosis, duplex systems, and cystic kidneys, respectively.
(C) Haemotoxylin and Eosin staining of tissue sections derived from Zmym2+/+and Zmym2+/− urogenital systems.
(D) Intravesical dye injection showing vesicoureteral reflux in Zmym2+/+ and Zmym2+/− P0 mice.
(E) Immunohistofluorescence analysis of E18.5 kidneys reveals no overt difference in cap mesenchyme and ureter tips (Pax2) nor in podocytes (podocalyxin) between Zmym2+/+and Zmym2+/− kidneys.
Intracellular Localization of Truncated ZMYM2 Proteins
ZMYM2 is a MYM type zinc finger protein that harbors two putative nuclear localization signals (NLS) and ten MYM type zinc fingers (Figures 1B–1G).35 The loss-of-function variants that we identified were frameshifts and/or stop-gains prior to the final exon, and therefore mutant mRNA transcripts are predicted to undergo nonsense-mediated decay (NMD), suggesting a haploinsufficiency as the most likely mechanism for the associated disorder. Nonetheless, we used cell-based assays to assess the functional properties of truncated protein that might result from escaping this NMD process, comparing them to wild-type ZMYM2 protein, as well as missense variants. Transfection of expression constructs in HEK293 cells revealed that, whereas the wild-type and missense ZMYM2 protein (p.Val61del [c.181_183del], p.Glu126Ala [c.377A>C], p.Ile387Val [c.1159A>G], p.Lys649Arg [c.1946A>G], p.Tyr763His [c.2287T>C], p.Tyr763Leu [c.2287_2288delinsTA>CT], p.Gly775Glu [c.2324G>A], p.Asp997del [c.2990_2992del], and p.Glu1031Lys [c.3091G>A]) was translocated to the nucleus, the ZMYM2 truncated proteins (p.Gly257∗ [c.766_767dupGT], p.Gln398∗ [c.1192C>T], p.Cys536Leufs∗13 [c.1607delG], p.Arg540∗ [c.1618C>T], p.Lys812Aspfs∗18 [c.2434_2437delAAAG], p.Gly1045Argfs∗33 [c.3130_3131dupAA]) remained primarily localized to the cytoplasm (Figures 4A and S1).
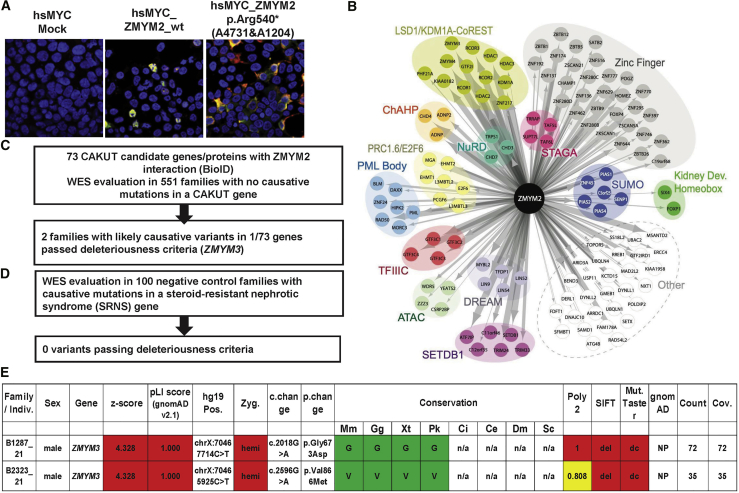
Figure 4.
Functional Characterization of ZMYM2 Variants and Identification of Protein-Protein Interaction Partners of ZMYM2 as Candidates for Monogenic Causes of CAKUT
(A) Representative immunofluoroscence images following overexpression of myc labeled cDNA constructs for mock, wild-type ZMYM2 (hsMYC_wtZMYM2), and cDNA representing mutation p.Arg540∗ (detected in A4730 and A1204) showing mislocalization of truncated protein to the cytoplasm rather than the nucleus.
(B) BioID of human wild-type ZMYM2 expressed in Flp-In T-REx 293 cells yields 123 proximity interaction partners. Interactors are grouped according to protein complex, intracellular localization, shared protein domain, or function. Edge size is proportional to total peptide counts.
(C) All 73 candidate genes resulting from the BioID experiments were evaluated for heterozygous mutations in 551 families with CAKUT using the American College of Medical Genetics criteria for deleteriousness.
(D) ZMYM3 variants in families B1287_21 and B2323_21 as a potential candidate gene in CAKUT pathogenesis.
CAKUT, congenital anomalies of the kidney and urinary tract; c. change, nucleotide change; Cov., coverage; gnomAD, genome aggregation; Miss., missense; Mut. Taster, Mutation Taster; NS, nephrotic syndrome; p. change, amino acid change, Poly2, Polymorphism Phenotyping v2; SIFT, Sorting Intolerant From Tolerant; WES, whole-exome sequencing; Zyg, zygosity; Mm, Mus musculus; Gg, Gallus gallus; Xt, Xenopus tropicalis; Pk, Paramormyrops kingsleyae; Ci, Ciona intestinalis; Ce, Caenorhabditis elegans; Dm, Drosophila melanogaster; Sc, Saccharomyces cerevisiae.
For three of the truncations (p.Gly257∗ [c.766_767dupGT], p.Gln398∗ [c.1192C>T], p.Arg540∗ [c.1618C>T]), the proteins showed exclusively cytoplasmic patterns in all cells tested. However, for the other four truncations that we tested (p.Try763Glnfs∗6 [c.2287_2288del], p.Cys812Aspfs∗18 [c.2434_2437delAAAG], p.Cys823∗ [c.2469T>A], and p.Gly1045Argfs∗33 [c.3130_3131dupAA]), protein localization was partially nuclear in a subset of cells, despite the loss of one or both known NLSs (p.1038–1049 and p.1250–1284) in the truncated proteins. We hypothesize that an additional functional NLS lies between p.540 and p.763, thereby accounting for the partial nuclear staining for these truncated proteins (see Supplemental Discussion and Figure S8 for further details).
ZMYM2 Interaction with FOXP1
In prior work, Bekheirnia et al. identified de novo mutations in FOXP1 (MIM: 613670) in six families with syndromic CAKUT,36 while Estruch et al. demonstrated that ZMYM2 is able to interact with different FOXP transcription factors.37 As noted above, transcripts with truncating ZMYM2 variants found in affected individuals of this study most likely undergo NMD. However, if such transcripts (partially) escape NMD, there remains the question of whether truncated ZMYM2 proteins would retain the ability to interact with FOXP transcription factors. To test this possibility, we used Bioluminescence Resonance Energy Transfer (BRET), a live-cell assay system for detecting putative protein-protein interactions.17 In these experiments, either wild-type FOXP1 or FOXP2 (MIM: 602081) was expressed as a renilla luciferase (RLuc) fusion protein, to function as a donor in the assay. Different variant ZMYM2 constructs were expressed as fusion proteins with YFP, to function as acceptors. If the donor and acceptor proteins come into close proximity of each other in co-transfected cells, energy transfer takes place from RLuc to YFP, which can be quantified by monitoring emission. Consistent with earlier studies, we were able to thereby demonstrate interactions between wild-type ZMYM2 and wild-type FOXP1, FOXP2, or ZMYM2 (homodimerization). All three truncated versions of ZMYM2 that we tested with BRET (p.Gly257∗ [c.766_767dupGT], p.Gln398∗ [c.1192C>T], p.Arg540∗ [c.1618C>T]). showed impaired interaction with FOXP1 and FOXP2 when compared with wild-type ZMYM2 (Figure S9).
Expanding the ZMYM2 Interactome
We employed proximity-dependent biotin identification (BioID: 22412018) to characterize the ZMYM2 protein interaction landscape. In total, 123 high-confidence (FDR ≤ 1%) ZMYM2 proximity interactors were identified (Table S5; all raw data available at MassIVE archive). The interactome is significantly enriched in DNA binding transcription factors (p = 8.3 × 10−22), transcriptional co-repressors (p = 3.5 × 10−7), and proteins linked to chromatin regulation (p = 9.26 × 10−14), chromatin organization (p = 5.91 × 10−14), and the SUMO system (p = 6.7 × 10−05). A number of previously reported ZMYM2 interactors were identified in our analysis:7 the LSD1(KDM1A)-CoREST (Corum complexes 633 and 1492),38 HDAC139 and HDAC2 and many of their known interacting partners (e.g., Corum 632: HDAC1, HDAC2, KDM1A/LSD1, GTF2I, GSE1/KIAA0182, PHF21A/BHC80, RCOR1, RCOR2, RCOR3, ZNF217, ZMYM2, and ZMYM3), and the transcription factors FOXP137 and SIX4 (Table S5). Consistent with a recent report linking ZMYM2 function to endogenous retrovirus silencing,40 BioID also identified components of the epigenetic repressor complex SETDB1-ATF7IP (SETDB1, ATF7IP, C11orf46), the non-canonical polycomb complex E2F6/PRC1.6 (PCGF6, E2F6, MGA, L3MBTL2, L3MBTL3, EHMT1, EHMT2, TFDP1), components of the DREAM (MYBL2, LIN52, LIN54, LIN9), ChAHP (CHD4, CBX1, ADNP), and TFIIIC (GTF3C1-4) complexes, and a large number of zinc finger-containing DNA binding proteins as high-confidence ZMYM2 interactors (Table S5). Consistent with a previous report,6 ZMYM2 BioID also identified a number of PML body components (PML, BLM, DAXX, HIPK2, MORC3, RAD50, ZNF24). Finally, consistent with recent reports identifying ZMYM2 as a SUMO binding protein,41,42 we also detected high-confidence interactions with a number of SUMO conjugation system components (PIAS1, PIAS2, PIAS4, ZNF451, SIMC1/C5orf25) (Table S5). Together, these data link ZMYM2 to transcriptional repression and the epigenetic regulation of heterochromatin and generate excellent candidates for additional genes potentially involved in monogenic forms of CAKUT (Figure S10).
Our ZMYM2 BioID identified two transcription factor proteins that have been linked to kidney development, SIX443, 44, 45 and FOXP1.36 SIX4 was shown to work together with the related SIX1 protein to regulate gene expression in metanephric mesenchyme,44 while SIX1 mutations cause CAKUT in humans.46 To characterize how the ZMYM2 protein interaction landscape is affected by CAKUT truncations, p.Gly257∗ (c.766_767dupGT), p.Gln398∗ (c.1192C>T), p.Cys536Leufs∗13 (c.1607delG), and p.Lys812Aspfs∗18 (c.2434_2437delAAAG) ZMYM2 variant proteins were also subjected to BioID (Table S5 and Figure S11). These mutants lost the vast bulk of interactions detected with the wild-type protein, including the interaction with FOXP1, consistent with the data from BRET assays.
ZMYM3 Variants as Potential Candidates in CAKUT Pathogenesis
We hypothesized that, like FOXP1, the genes encoding other ZMYM2 interacting partners could also represent candidate genes for involvement in CAKUT. Indeed, examination of WES data from our cohort of 551 individuals with CAKUT (Figure 4C, Table S6), revealed two male individuals with hemizygous variants in ZMYM3 (MIM: 300061, p.Gly673Asp [c.2018G>A] and p.Val866Met [c.2596G>A], Figures 4D and 4E). Consistent with our ZMYM2 data, BioID of the ZMYM3 protein yielded a reciprocal interaction with ZMYM2, and an interactome largely overlapping with that of ZMYM2 (68% overlap; Table S5), including components of the LSD1-CoREST, ChAHP, DREAM, and TFIIIC complexes. While further investigation of the functional role of these variants in ZMYM2 pathogenesis is necessary, these methods reveals strategies to identifiy variants in potentially novel target genes involved in the mechanism of disease development in CAKUT.
Discussion
In summary, here we describe the discovery of predominantly de novo loss-of-function mutations of ZMYM2 as an autosomal-dominant cause of human syndromic CAKUT. Consistent with its known nuclear function,6 we demonstrate that wild-type ZMYM2 is located in the nucleus, whereas truncated ZMYM2 proteins can mislocalize to the cytoplasm. By expression and morpholino knockdown experiments in Xenopus larvae, we confirm that ZMYM2 plays critical roles in kidney and craniofacial development. In addition, the renal phenotype of Xenopus morphants was rescued by wild-type but not mutant mRNA, consistent with pathogenicity for the alleles that we identified in individuals. Interestingly, one of the variants resulted in worsening the phenotype, suggesting the potential for it to yield dominant-negative effects, if the variant transcript escapes NMD in individual cells.
Furthermore, by generating a mouse model of heterozygous Zmym2 disruption, we recapitulated the human CAKUT phenotype, confirming the importance of ZMYM2 in renal development and as a cause of CAKUT in humans when mutated. Finally, we generate independent evidence confirming that mutant ZMYM2 leads to loss of interaction with FOXP1, a transcription factor already linked to CAKUT, as well as uncovering a potential interaction with ZMYM3, which we suggest may represent a method to identify candidate genes involved in this disorder.
ZMYM2, also known as FIM, ZNF198, or RAMP, is a nuclear zinc finger protein that contains 1,377 amino acids with a molecular mass of 150 kDa.47,48 ZMYM2 localizes to the nucleus, specifically the PML body,6 where it has been characterized as a corepressor of transcription by interacting with different nuclear receptors, and the LSD1-CoREST-HDAC1 complex on chromatin.7 A recent report has also linked ZMYM2 to silencing of endogenous retrovirus sequences.40 However, the importance of ZMYM2 for kidney and ureter development was largely unknown and despite a role in myeloproliferative disorder as a fusion protein, ZMYM2 mutations have not previously been implicated in human disease.
ZMYM2 has previously been shown to interact with FOXP transcription factors,37 and mutations in FOXP1 were recently identified in individuals with syndromic CAKUT.36 Given that they are all located prior to the final exon of the gene, the truncating and stop gain ZMYM2 variants found in our study will most likely undergo NMD in vivo, preventing their translation and leading to ZMYM2 haploinsufficiency. However, as demonstrated in our Xenopus model, some variants resulted in worsening the phenotype, which raises the possibility that some variants may in fact function as dominant negative rather than haploinsufficiency. Furthermore, our BRET assays indicate that if aberrant transcripts escape NMD, there would be expression of truncated versions of ZMYM2 that are unable to interact with either FOXP1 or FOXP2. These findings are also supported by our BioID data, again indicating a loss of FOXP1 interaction for truncated ZMYM2 proteins.
In addition, the BioID data identified multiple ZMYM2 interactors, including members of the LSD1-COREST-HDAC1 pathway, suggesting that the broader ZMYM2 interactome, which include DNA binding transcription factors, transcriptional co-repressors, and proteins linked to chromatin regulation and organization may represent potential candidate genes in urinary tract malformation.7 Given the observation that either FOXP1 or ZMYM2 loss-of-function mutations can cause CAKUT, the other genes in this interactome could also be considered as candidates for involvement in the disorder. Further work is now required to determine the role of these potential interactors in the pathogenesis of kidney malformation in individuals with ZMYM2 mutations. Mechanistically, further studies will help elucidate how ZMYM2 mutations lead to the development of CAKUT, giving novel insights into the biological basis of the disorder.
Published: September 4, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.013.
Web Resources
1000 Genomes Browser, https://www.internationalgenome.org/1000-genomes-browsers
Conifer software, http://conifer.sourceforge.net
Database of genomic variants, http://projects.tcag.ca/variation
Ensembl Genome Browser, http://www.ensembl.org
Exome Variant Server, https://evs.gs.washington.edu/EVS
Exome Aggregation Consortium, http://exac.broadinstitute.org
GeneDx ClinVar submission, https://www.ncbi.nlm.nih.gov/clinvar/submitters/26957/
Genematcher, https://genematcher.org/
GeneTools, https://www.gene-tools.com/
gnomAD browser beta, https://gnomad.broadinstitute.org/
MassIVE archive, https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp
MutationTaster, http://www.mutationtaster.org
OMIM, https://www.omim.org/
PolyPhen2, http://genetics.bwh.harvard.edu/pph2
Renal Genes, http://www.renalgenes.org
Sorting Intolerant From Tolerant (SIFT), http://sift.jcvi.org
UCSC Genome Browser, https://genome.ucsc.edu
Xenbase, http://www.xenbase.org
Data and Code Availability
All raw mass spectrometry files have been deposited at the MassIVE archive with accession number ID MSV000085033.
Human ZMYM2 full-length protein is GenBank: NM_197968.2.
Xenopus Zmym2 full-length protein is GenBank: NM_001123434.1.
Supplemental Data
References
- 1.Vivante A., Kohl S., Hwang D.Y., Dworschak G.C., Hildebrandt F. Single-gene causes of congenital anomalies of the kidney and urinary tract (CAKUT) in humans. Pediatr. Nephrol. 2014;29:695–704. doi: 10.1007/s00467-013-2684-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Ven A.T., Vivante A., Hildebrandt F. Novel Insights into the Pathogenesis of Monogenic Congenital Anomalies of the Kidney and Urinary Tract. J. Am. Soc. Nephrol. 2018;29:36–50. doi: 10.1681/ASN.2017050561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Ven A.T., Connaughton D.M., Ityel H., Mann N., Nakayama M., Chen J., Vivante A., Hwang D.Y., Schulz J., Braun D.A. Whole-Exome Sequencing Identifies Causative Mutations in Families with Congenital Anomalies of the Kidney and Urinary Tract. J. Am. Soc. Nephrol. 2018;29:2348–2361. doi: 10.1681/ASN.2017121265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber S., Moriniere V., Knüppel T., Charbit M., Dusek J., Ghiggeri G.M., Jankauskiené A., Mir S., Montini G., Peco-Antic A. Prevalence of mutations in renal developmental genes in children with renal hypodysplasia: results of the ESCAPE study. J. Am. Soc. Nephrol. 2006;17:2864–2870. doi: 10.1681/ASN.2006030277. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen M., Sunde L., Nielsen M.L., Ramsing M., Petersen A., Hjortshøj T.D., Olsen T.E., Tabor A., Hertz J.M., Johnsen I. Targeted gene sequencing and whole-exome sequencing in autopsied fetuses with prenatally diagnosed kidney anomalies. Clin. Genet. 2018;93:860–869. doi: 10.1111/cge.13185. [DOI] [PubMed] [Google Scholar]
- 6.Kunapuli P., Kasyapa C.S., Chin S.-F., Caldas C., Cowell J.K. ZNF198, a zinc finger protein rearranged in myeloproliferative disease, localizes to the PML nuclear bodies and interacts with SUMO-1 and PML. Exp. Cell Res. 2006;312:3739–3751. doi: 10.1016/j.yexcr.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 7.Gocke C.B., Yu H. ZNF198 stabilizes the LSD1-CoREST-HDAC1 complex on chromatin through its MYM-type zinc fingers. PLoS ONE. 2008;3:e3255. doi: 10.1371/journal.pone.0003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warejko J.K., Tan W., Daga A., Schapiro D., Lawson J.A., Shril S., Lovric S., Ashraf S., Rao J., Hermle T. Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome. Clin. J. Am. Soc. Nephrol. 2018;13:53–62. doi: 10.2215/CJN.04120417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vivante A., Hwang D.Y., Kohl S., Chen J., Shril S., Schulz J., van der Ven A., Daouk G., Soliman N.A., Kumar A.S. Exome Sequencing Discerns Syndromes in Patients from Consanguineous Families with Congenital Anomalies of the Kidneys and Urinary Tract. J. Am. Soc. Nephrol. 2017;28:69–75. doi: 10.1681/ASN.2015080962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacArthur D.G., Manolio T.A., Dimmock D.P., Rehm H.L., Shendure J., Abecasis G.R., Adams D.R., Altman R.B., Antonarakis S.E., Ashley E.A. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–476. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E., Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 12.Bamshad M.J., Ng S.B., Bigham A.W., Tabor H.K., Emond M.J., Nickerson D.A., Shendure J. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 13.Lee H., Deignan J.L., Dorrani N., Strom S.P., Kantarci S., Quintero-Rivera F., Das K., Toy T., Harry B., Yourshaw M. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Retterer K., Juusola J., Cho M.T., Vitazka P., Millan F., Gibellini F., Vertino-Bell A., Smaoui N., Neidich J., Monaghan K.G. Clinical application of whole-exome sequencing across clinical indications. Genet. Med. 2016;18:696–704. doi: 10.1038/gim.2015.148. [DOI] [PubMed] [Google Scholar]
- 16.Kitzler T.M., Schneider R., Kohl S., Kolvenbach C.M., Connaughton D.M., Dai R., Mann N., Nakayama M., Majmundar A.J., Wu C.W. COL4A1 mutations as a potential novel cause of autosomal dominant CAKUT in humans. Hum. Genet. 2019;138:1105–1115. doi: 10.1007/s00439-019-02042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deriziotis P., Graham S.A., Estruch S.B., Fisher S.E. Investigating protein-protein interactions in live cells using bioluminescence resonance energy transfer. J. Vis. Exp. 2014 doi: 10.3791/51438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neilson K.M., Pignoni F., Yan B., Moody S.A. Developmental expression patterns of candidate cofactors for vertebrate six family transcription factors. Dev. Dyn. 2010;239:3446–3466. doi: 10.1002/dvdy.22484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.del Viso F., Bhattacharya D., Kong Y., Gilchrist M.J., Khokha M.K. Exon capture and bulk segregant analysis: rapid discovery of causative mutations using high-throughput sequencing. BMC Genomics. 2012;13:649. doi: 10.1186/1471-2164-13-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.del Viso F., Khokha M. Generating diploid embryos from Xenopus tropicalis. Methods Mol. Biol. 2012;917:33–41. doi: 10.1007/978-1-61779-992-1_3. [DOI] [PubMed] [Google Scholar]
- 21.Faber J., Nieuwkoop P.D., editors. Normal Table of Xenopus laevis (Daudin) Garland Science; 1994. p. 418. [Google Scholar]
- 22.Khokha M.K., Chung C., Bustamante E.L., Gaw L.W., Trott K.A., Yeh J., Lim N., Lin J.C., Taverner N., Amaya E. Techniques and probes for the study of Xenopus tropicalis development. Dev. Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- 23.Miller R.K., Canny S.G., Jang C.W., Cho K., Ji H., Wagner D.S., Jones E.A., Habas R., McCrea P.D. Pronephric tubulogenesis requires Daam1-mediated planar cell polarity signaling. J. Am. Soc. Nephrol. 2011;22:1654–1664. doi: 10.1681/ASN.2010101086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raciti D., Reggiani L., Geffers L., Jiang Q., Bacchion F., Subrizi A.E., Clements D., Tindal C., Davidson D.R., Kaissling B., Brändli A.W. Organization of the pronephric kidney revealed by large-scale gene expression mapping. Genome Biol. 2008;9:R84. doi: 10.1186/gb-2008-9-5-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boualia S.K., Gaitan Y., Murawski I., Nadon R., Gupta I.R., Bouchard M. Vesicoureteral reflux and other urinary tract malformations in mice compound heterozygous for Pax2 and Emx2. PLoS ONE. 2011;6:e21529. doi: 10.1371/journal.pone.0021529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Narlis M., Grote D., Gaitan Y., Boualia S.K., Bouchard M. Pax2 and pax8 regulate branching morphogenesis and nephron differentiation in the developing kidney. J. Am. Soc. Nephrol. 2007;18:1121–1129. doi: 10.1681/ASN.2006070739. [DOI] [PubMed] [Google Scholar]
- 27.Coyaud E., Mis M., Laurent E.M., Dunham W.H., Couzens A.L., Robitaille M., Gingras A.C., Angers S., Raught B. BioID-based Identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 Ligase Substrates. Mol. Cell. Proteomics. 2015;14:1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Craig R., Beavis R.C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 30.Eng J.K., Jahan T.A., Hoopmann M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 31.Teo G., Liu G., Zhang J., Nesvizhskii A.I., Gingras A.C., Choi H. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics. 2014;100:37–43. doi: 10.1016/j.jprot.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otto E.A., Hurd T.W., Airik R., Chaki M., Zhou W., Stoetzel C., Patil S.B., Levy S., Ghosh A.K., Murga-Zamalloa C.A. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat. Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kohl S., Hwang D.Y., Dworschak G.C., Hilger A.C., Saisawat P., Vivante A., Stajic N., Bogdanovic R., Reutter H.M., Kehinde E.O. Mild recessive mutations in six Fraser syndrome-related genes cause isolated congenital anomalies of the kidney and urinary tract. J. Am. Soc. Nephrol. 2014;25:1917–1922. doi: 10.1681/ASN.2013101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ollendorff V., Guasch G., Isnardon D., Galindo R., Birnbaum D., Pébusque M.J. Characterization of FIM-FGFR1, the fusion product of the myeloproliferative disorder-associated t(8;13) translocation. J. Biol. Chem. 1999;274:26922–26930. doi: 10.1074/jbc.274.38.26922. [DOI] [PubMed] [Google Scholar]
- 36.Bekheirnia M.R., Bekheirnia N., Bainbridge M.N., Gu S., Coban Akdemir Z.H., Gambin T., Janzen N.K., Jhangiani S.N., Muzny D.M., Michael M. Whole-exome sequencing in the molecular diagnosis of individuals with congenital anomalies of the kidney and urinary tract and identification of a new causative gene. Genet. Med. 2017;19:412–420. doi: 10.1038/gim.2016.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estruch S.B., Graham S.A., Quevedo M., Vino A., Dekkers D.H.W., Deriziotis P., Sollis E., Demmers J., Poot R.A., Fisher S.E. Proteomic analysis of FOXP proteins reveals interactions between cortical transcription factors associated with neurodevelopmental disorders. Hum. Mol. Genet. 2018;27:1212–1227. doi: 10.1093/hmg/ddy035. [DOI] [PubMed] [Google Scholar]
- 38.Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Ruepp A. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47(D1):D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joshi P., Greco T.M., Guise A.J., Luo Y., Yu F., Nesvizhskii A.I., Cristea I.M. The functional interactome landscape of the human histone deacetylase family. Mol. Syst. Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F., Huang X., Zang R., Chen J., Fidalgo M., Sanchez-Priego C., Yang J., Caichen A., Ma F., Macfarlan T. DUX-miR-344-ZMYM2-Mediated Activation of MERVL LTRs Induces a Totipotent 2C-like State. Cell Stem Cell. 2020;26:234–250.e7. doi: 10.1016/j.stem.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guzzo C.M., Ringel A., Cox E., Uzoma I., Zhu H., Blackshaw S., Wolberger C., Matunis M.J. Characterization of the SUMO-binding activity of the myeloproliferative and mental retardation (MYM)-type zinc fingers in ZNF261 and ZNF198. PLoS ONE. 2014;9:e105271. doi: 10.1371/journal.pone.0105271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguilar-Martinez E., Chen X., Webber A., Mould A.P., Seifert A., Hay R.T., Sharrocks A.D. Screen for multi-SUMO-binding proteins reveals a multi-SIM-binding mechanism for recruitment of the transcriptional regulator ZMYM2 to chromatin. Proc. Natl. Acad. Sci. USA. 2015;112:E4854–E4863. doi: 10.1073/pnas.1509716112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J., Liu M., Zhao L., Li Y., Zhang M., Jin Y., Xiong Q., Liu X., Zhang L., Jiang H. Disabling of nephrogenesis in porcine embryos via CRISPR/Cas9-mediated SIX1 and SIX4 gene targeting. Xenotransplantation. 2019;26:e12484. doi: 10.1111/xen.12484. [DOI] [PubMed] [Google Scholar]
- 44.Kobayashi H., Kawakami K., Asashima M., Nishinakamura R. Six1 and Six4 are essential for Gdnf expression in the metanephric mesenchyme and ureteric bud formation, while Six1 deficiency alone causes mesonephric-tubule defects. Mech. Dev. 2007;124:290–303. doi: 10.1016/j.mod.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 45.Xu J., Xu P.X. Eya-six are necessary for survival of nephrogenic cord progenitors and inducing nephric duct development before ureteric bud formation. Dev. Dyn. 2015;244:866–873. doi: 10.1002/dvdy.24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ruf R.G., Xu P.X., Silvius D., Otto E.A., Beekmann F., Muerb U.T., Kumar S., Neuhaus T.J., Kemper M.J., Raymond R.M., Jr. SIX1 mutations cause branchio-oto-renal syndrome by disruption of EYA1-SIX1-DNA complexes. Proc. Natl. Acad. Sci. USA. 2004;101:8090–8095. doi: 10.1073/pnas.0308475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Still I.H., Cowell J.K. The t(8;13) atypical myeloproliferative disorder: further analysis of the ZNF198 gene and lack of evidence for multiple genes disrupted on chromosome 13. Blood. 1998;92:1456–1458. [PubMed] [Google Scholar]
- 48.Baumann H., Kunapuli P., Tracy E., Cowell J.K. The oncogenic fusion protein-tyrosine kinase ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. J. Biol. Chem. 2003;278:16198–16208. doi: 10.1074/jbc.M300018200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw mass spectrometry files have been deposited at the MassIVE archive with accession number ID MSV000085033.
Human ZMYM2 full-length protein is GenBank: NM_197968.2.
Xenopus Zmym2 full-length protein is GenBank: NM_001123434.1.