Summary
More than 100 genetic etiologies have been identified in developmental and epileptic encephalopathies (DEEs), but correlating genetic findings with clinical features at scale has remained a hurdle because of a lack of frameworks for analyzing heterogenous clinical data. Here, we analyzed 31,742 Human Phenotype Ontology (HPO) terms in 846 individuals with existing whole-exome trio data and assessed associated clinical features and phenotypic relatedness by using HPO-based semantic similarity analysis for individuals with de novo variants in the same gene. Gene-specific phenotypic signatures included associations of SCN1A with “complex febrile seizures” (HP: 0011172; p = 2.1 × 10−5) and “focal clonic seizures” (HP: 0002266; p = 8.9 × 10−6), STXBP1 with “absent speech” (HP: 0001344; p = 1.3 × 10−11), and SLC6A1 with “EEG with generalized slow activity” (HP: 0010845; p = 0.018). Of 41 genes with de novo variants in two or more individuals, 11 genes showed significant phenotypic similarity, including SCN1A (n = 16, p < 0.0001), STXBP1 (n = 14, p = 0.0021), and KCNB1 (n = 6, p = 0.011). Including genetic and phenotypic data of control subjects increased phenotypic similarity for all genetic etiologies, whereas the probability of observing de novo variants decreased, emphasizing the conceptual differences between semantic similarity analysis and approaches based on the expected number of de novo events. We demonstrate that HPO-based phenotype analysis captures unique profiles for distinct genetic etiologies, reflecting the breadth of the phenotypic spectrum in genetic epilepsies. Semantic similarity can be used to generate statistical evidence for disease causation analogous to the traditional approach of primarily defining disease entities through similar clinical features.
Keywords: electronic medical records, Human Phenotype Ontology, childhood epilepsies, neurogenetic disorders, developmental and epileptic encephalopathies, whole-exome sequencing, computational phenotypes
Introduction
In 1954, Dr. Andreas Rett, a pediatrician in Vienna, Austria, noticed two girls with unusual repetitive hand-washing motions in his waiting room. Rett concluded that these unusual features may be the presentation of a new disease entity and subsequently identified additional girls with similar features and related developmental trajectories. This initial observation laid the foundation for recognizing a neurodevelopmental disorder that came to bear Dr. Rett’s name.1,2 In 1999, MECP2 (MIM: 300005) was eventually discovered as the causative genetic etiology for Rett syndrome (MIM: 312750), which is thought to affect 1 in 10,000 girls worldwide.3, 4, 5 Similar observations on related clinical features led to discoveries of other genetic neurodevelopmental disorders and childhood developmental and epileptic encephalopathies, including Dravet syndrome (MIM: 607208) and epilepsy of infancy with migrating focal seizures (MIM: 614959).6,7
Although the syndrome-based approach is the time-proven, established method of defining disease entities in the epilepsies, it has several shortcomings that are particularly relevant in the era of large-scale genomics.8,9 First, the recognition of clinical symptoms is often fortuitous, depending on individuals with shared features to be seen by the same clinician or at the same center. Second, only a subset of clinical syndromes is linked to unique genetic etiologies, whereas many clinical entities, such as infantile spasms or Lennox-Gastaut syndrome, are associated with a wide range of underlying genetic causes.10, 11, 12, 13, 14, 15, 16 Third, the recognition, documentation, and comparison of clinical features is a manual, non-scalable process requiring significant human resources in contrast to the industrial scale of massive parallel sequencing that can be performed on DNA from tens of thousands of individuals.17
Large collaborative studies that are designed primarily for genetic discovery also collect descriptive clinical data, and these phenotypic data can be exploited for clinical discovery.18,19 Following the logic of primarily defining disease entities through shared clinical features, we reasoned that applying computational algorithms to available phenotype datasets might detect disease entities by identifying individuals with rare, overlapping phenotypic features that share the same genetic etiology. However, phenotype data is typically sparse and unstructured, which impedes the comparison of clinical features between individuals.
The Human Phenotype Ontology (HPO) is a standardized biomedical representation of the semantic relationships among over 14,000 phenotypic terms with defined relationships, enabling the mapping of heterogeneous clinical features to a common framework.20, 21, 22 Consequently, the value of a phenotypic feature can be weighted on the basis of its position in the ontological tree and frequency in the overall cohort. We and others have previously developed algorithms to identify individuals with significant phenotypic similarities on the basis of HPO terms within patient cohorts.18,23
Here, we translated clinical findings in 846 individuals with developmental and epileptic encephalopathies (DEEs) with available trio whole-exome data to 31,742 HPO terms. We then assessed whether individuals with de novo variants in the same genetic etiology had phenotypic features that were more similar than expected by chance and identified 11 genetic etiologies with significant phenotypic similarity. Our results demonstrate that phenotype data in HPO format represents a valuable resource in providing statistical evidence in gene-disease relationships and reconstructs meaningful disease patterns from sparse clinical data.
Material and Methods
Participant Recruitment
Clinical and phenotypic data included in this study were derived through local studies and data obtained through dbGaP (dbGaP Study Accession: phs000653.v1.p1, n = 335). For local cohorts, informed consent for participation in this study was obtained from parents of all probands in agreement with the Declaration of Helsinki and completed per protocol with local approval by the respective institutional review boards (IRBs). These cohorts included individuals from the EuroEPINOMICS-RES cohort (RES, n = 319), Epi4K cohort (EPGP, n = 335), and a cohort of individuals recruited through the Epilepsy Genetics Research Project at the Children’s Hospital of Philadelphia (EGRP, n = 192). A sub-cohort of 320 individuals from the RES and EGRP populations were included in a previous study.18 Phenotypes for these cohorts were collected through standardized phenotyping and questionnaires to physicians and healthcare providers. Description of the recruitment and phenotyping of the Epi4K dbGaP cohort (phs000653.v1.p1) has been reported previously.24,25
Translation to HPO Terms, Information Content (IC)
For the various phenotyping forms and databases provided for the individuals included in this project, we manually generated dictionaries to map phenotyping terms to HPO terms (HPO version 1.2; release format-version: 1.2; data-version: releases/2019-11-08; downloaded on 1/23/20). The phenotype of each individual from the EGRP dataset was manually coded by expert reviewers. Phenotypes were first extracted by research staff with clinical and biomedical knowledge and experience with the HPO by using all available clinical and research notes for an individual and by using the most specific HPO terms applicable. These assigned terms were then reviewed and verified by domain experts in the field of epilepsy, i.e., either epilepsy genetic counselors or specialist physicians. In cases of ambiguity and uncertainty, the higher level HPO term was coded (e.g., if autism spectrum disorder was not clearly diagnosed but mentioned, we assigned the higher level “autistic behavior” [HP: 0000729]).
For each individual, all higher-level (ancestral) HPO terms were derived, followed by de-duplication of HPO terms for each individual. We refer to this method as “propagation,” resulting in a base and propagated set of HPO terms for each individual. The propagated HPO dataset from the entire cohort was used to generate baseline frequencies f for all HPO terms. Information content (IC) of each term was defined as the −log2(f) with a higher IC value, reflecting a more specific and less frequently encountered HPO term in the cohort. In the current manuscript, we use a compact internationalized resource identifier (CURIE) to refer to HPO terms, i.e., “HP: 0001250” (“seizures”) abbreviates “https://hpo.jax.org/app/browse/term/HP:0001250” in accordance with the Open Biological and Biomedical Ontologies (OBO) Citation and Attribution Policy.
Genetic Analysis
Trio-based whole-exome sequencing was performed as previously described,18,26 including research sequencing within the EuroEPINOMICS-RES project (n = 335) performed at the Wellcome Trust Sanger Institute (Hinxton, UK) with the Illumina TruSeq DNA Sample Preparation Kit, the Agilent Technologies SureSelect Human All Exon 50 Mb Kit, and the Illumina HiSeq2000 per manufacturer’s protocols;11,26,27 research sequencing at the Institute of Clinical Molecular Biology at the University of Kiel and the Cologne Center for Genomics with NimbleGen SeqCap EZ Human Exome Library v2.0, Nextera Rapid Capture Exome, Nextera Rapid Capture Expanded Exome, Agilent SureSelect Human All Exon V5, and Agilent SureSelect Human All Exon 50 Mb; research sequencing at the Broad Institute with Nextera Rapid Capture Exome kit; sequencing in a diagnostic setting at GeneDx (n = 69) with SureSelect Human All Exon V4 (50Mb) kit; and sequencing at the Division of Genomic Diagnostics at the Children’s Hospital of Philadelphia (n = 49) with SureSelect Clinical Research Exome kits.
All genetic data on individuals included in the overall cohort were re-analyzed via a standardized pipeline as previously described.18,26 The Burrows Wheeler Alignment (v 0.7.12) MEM algorithm was used to align the raw data to the HS37d5 human reference genome, and Samblaster (v 0.1.20) was used to add mate tags (MC and MQ) to the paired-end lines. Base quality score recalibration (BQSR) was performed with GATK tools (v4.0.0.0), followed by SNP and indel calling via HaplotypeCaller with interval lists specific to the exome enrichment kit used for each sample. GVCF files for each trio were combined with PICARD tools (v2.0.1), and genotyping was performed with the GATK genotype GVCF tool. GATK tools was used for variant selection and filtration, and the PICARD tools MergeVcfs functionality was used to generate merged variant files (VCFs). A customized version of ANNOVAR was used to annotate the VCF file. De novo, homozygous, and compound heterozygous variants were derived from the annotated file. The following quality criteria were used for variant filtration: (1) read depth in proband and parents 310×; (2) genotype quality in proband and parents 320, (3) absent in all population databases including 1000G, EVS, and ExAC, (4) RVIS percentile <70, and (5) read ratio 30.25 and £0.75 of the alternate alleles in the proband. All de novo variants were visually inspected with the Integrative Genomics Viewer (IGV, 2.4.14), and a subset of genes were excluded due to inconsistency of calls. A subset of de novo variants was validated via Sanger sequencing in previous studies, confirmed clinically, or had been reported as causative genetic etiologies by diagnostic laboratories.10,11,18 The probability of n de novo variants in a given gene was determined with “denovolyzer.”28
Phenotypic Similarity Analysis
We used two similarity measures to determine phenotypic similarity (sim score): the previously reported simmax algorithm18 and a novel simcm algorithm (Figure S1). The simmax was used as the primary algorithm for this study. The basic concept of both phenotypic similarity algorithms is the generation of symmetric phenotypic similarity scores between two individuals on the basis of the similarity between the phenotypic concepts represented by their HPO terms. The greater the similarity score, the more similar the individuals’ phenotypes. This similarity score of a pair of individuals is derived from the summation of the IC of the most informative common ancestor (MICA) terms of all pairwise comparisons of the base HPO terms of the two individuals.29
| (Equation 1) |
A matrix is formed with the m base HPO terms, i of individual P1 as rows, and the n base HPO terms, j of individual P2 as columns. Each sij corresponds to the IC of the MICA of HPO terms i and j, that is the maximum information content within the set of propagated terms shared by i and j. In summary, the simmax algorithm sums over all rows and columns of a matrix that holds all base HPO terms in individual P1 (n terms as rows) and all HPO terms in individual P2 (m terms as columns; Equation 1).
The faster simcm algorithm operates on the propagated HPO terms of each individual and determines the intersect of propagated HPO terms between individual P1 and individual P2, summing up the IC of all ancestral HPO terms shared by both individuals. All computations were performed with the R Statistical Package.30
Although more computationally costly, this study used simmax as the primary similarity measure because this algorithm has been successfully utilized previously.18 However, results from both similarity measures are highly correlated (Figure S1).
Expected Phenotypic Similarity Score per Gene
All genetic etiologies with de novo variants in two or more individuals were included in the primary analysis. The expected phenotypic similarity per gene with n individuals was determined by comparing distribution of the median similarities of n individuals that were randomly selected with 100,000 permutations from the overall cohort, resulting in an exact p value via the comparison of observed versus expected phenotypic similarity. For example, only 10 out of 100,000 permutations of 16 randomly selected individuals showed a median sim score that was greater than or equal to the observed median phenotypic similarity in the 16 individuals with de novo variants in SCN1A (MIM: 182389), resulting in an exact p value of <1.0 × 10−5 for SCN1A (median sim score = 17.69).
Phenograms and Analysis of Gene-Specific Phenotypic Signals
For each genetic etiology, the frequency of all assigned and derived (propagated) HPO terms in patients was identified and compared to the frequency in individuals without the genetic etiology deriving a p value via Fisher’s tests. We refer to the display of these frequencies as “phenograms,” which provide a visual intuition of the phenotypic spectrum of each disease. Phenograms were generated for all genes included in the analysis. We compiled p values by comparing the observed versus expected contribution for all HPO terms across all genes.
Assessment of Positive Predictive Value of HPO Term Combinations
In order assess the predictive power of the combination of HPO terms for the presence of a specific genetic etiology, we selected HPO terms associated with each genetic etiology that were more frequent in gene-positive individuals compared to gene-negative individuals by using the propagated HPO dataset. “Gene-positive individuals” refers to individuals with de novo variants in a given genetic etiology, whereas “gene-negative individuals” refers to individuals without de novo variants in a given genetic etiology. We then selected HPOs present in at least 10% of individuals of gene-negative individuals to prevent the effect of very rare HPO terms. We then used HPO term frequency in gene-positive and gene-negative individuals to assess the combined frequency of n HPO terms. For example, if three HPO terms have a frequency of 0.9, 0.85, and 0.7, the combined frequency would be 0.9 × 0.85 × 0.7 = 0.54. Ranking HPO terms by strength of association with a given genetic etiology, we then assessed the positive predictive value (PPV) of the combination of HPO terms when successively including additional HPO terms. We used this method to determine the number of HPO terms needed for a PPV of 0.8.
Results
Phenotypic Information Translated to HPO Is Sparse with a Wide Range of Phenotypic Depth
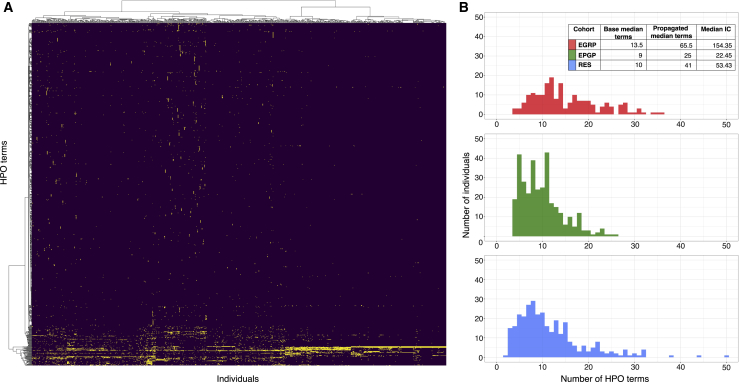
After translation to HPO terms, the 846 individuals included in the study were coded with a total of 31,742 HPO terms, including 1,616 unique HPO terms. The overall number of HPO terms differed widely between individuals, ranging from 12 terms to 181 terms with a median of 30 terms per individual. The cohorts included in the study showed significant differences: the Epi4K cohort (median of 22 terms) demonstrated a lower number of HPO terms per individual than the remaining cohort (median of 38 terms). The distribution of HPO terms in the cohort was sparse: only 29 terms were present in 100 or more individuals (Figure 1). “Seizures” (HP: 0001250), “infantile spasms” (HP: 0012469), and “hypsarrhythmia” (HP: 0002521) were the most common explicitly assigned HPO terms. Only 15% of all HPO terms were found in two or more individuals, and 50.1% of all HPO terms were only coded in a single individual.
Figure 1.
Heterogenous Distribution of HPO Terms
(A) Heatmap of all 846 individuals in the cohort with all 31,742 HPO terms. A yellow dot signifies that an HPO term is present in an individual. The heatmap displays the overall sparsity and heterogeneity of the cohort and indicates that only a small subset of the 1,616 unique HPO terms are shared between individuals.
(B) Distributions of number of HPO terms per patient in the three sub-cohorts (EGRP, EPGP/Epi4K, and EuroEPINOMICS-RES), indicating the varying depth of phenotyping across these cohorts. Base terms refer to the explicitly assigned terms in each cohort, and propagated terms refer to the assigned terms including all higher-level terms in the ontology.
Propagation of HPO Terms Enables an Accurate Assessment of Term Frequencies
Because HPO terms are interrelated within the tree-like structure of the HPO, assessing the baseline frequency of HPO terms provides a misleading estimate of the general frequencies of disease features in the cohort. For example, the higher-level term “neurodevelopmental abnormality” (HP: 0012759) was coded as an explicit term in only one individual. However, a much greater number of individuals had developmental differences consistent with “neurodevelopmental abnormality” (HP: 0012759) but had been assigned more specific terms. For example, “global developmental delay” (HP: 0001263) was coded in 272 individuals and “intellectual disability” (HP: 0001249) was coded in 62 individuals. We therefore generated the true frequency of all HPO terms by a process we referred to as “propagation.” In brief, for each individual, all higher-level HPO terms were added for the baseline HPO terms assigned to each individual, followed by de-duplication of HPO terms per individual. This method ensures that each individual coded with “global developmental delay” (HP: 0001263) was also coded with all higher-level, less specific ancestral terms, including “neurodevelopmental abnormality” (HP: 0012759) and “abnormality of the nervous system” (HP: 0012638). The propagated HPO terms allow for a meaningful estimate of the frequencies of clinical features in the cohort (Table S4). The frequencies of high-level HPO terms were particularly affected by the propagation (Figure S2), indicating that estimates derived from baseline HPO terms generally underestimate the frequency of higher-level, less specific terms for phenotypic features. In brief, when we used the propagated HPO terms, 803/846 individuals had seizures (HP: 0001250 and child terms), 227/846 had intellectual disability (HP: 0001249 and child terms), 254/846 had movement disorders or “abnormality of central motor function” (HP: 0011442 and child terms), and 97/846 individuals had autistic behavior (HP: 0000729 and child terms).
Genetic Analysis Identifies 41 Genetic Etiologies Shared by Two or More Individuals
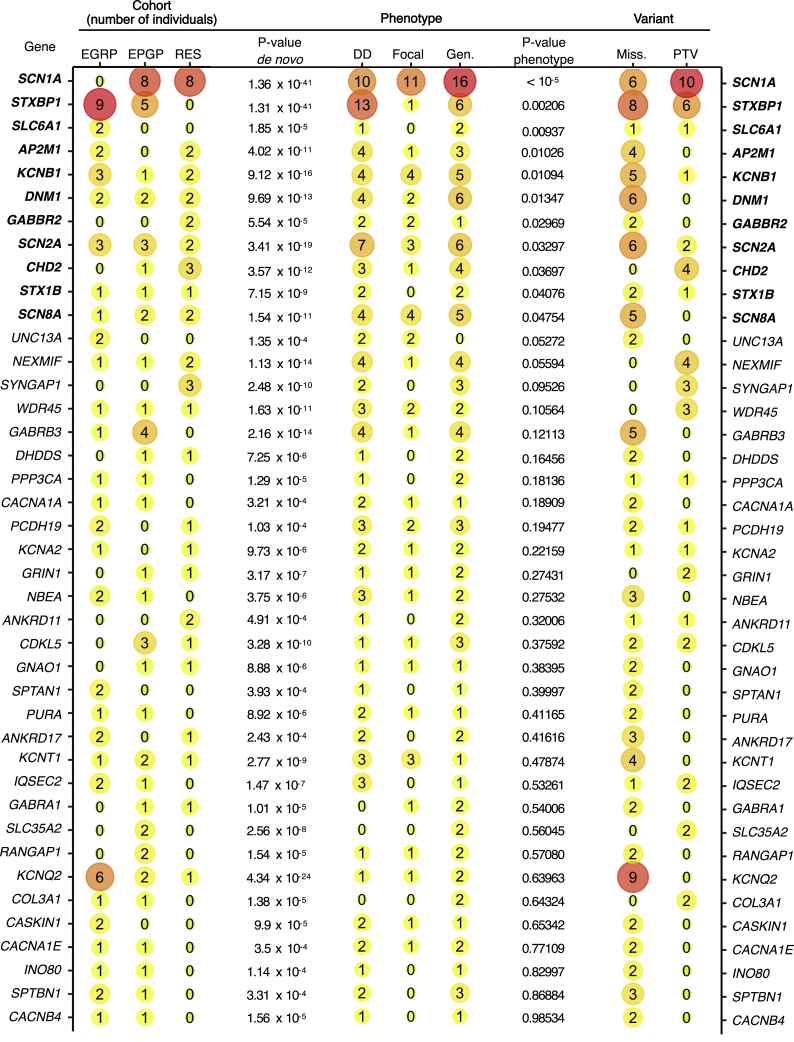
Using a standardized pipeline across all samples for variant calling, annotation, and inheritance models, we identified 41 genetic etiologies with de novo variants in two or more individuals (Table S6). The most common genetic etiologies in our cohort were SCN1A (n = 16), STXBP1 (MIM: 602926) (n = 14), KCNQ2 (MIM: 602235) (n = 9), SCN2A (MIM: 182390) (n = 8), and KCNB1 (MIM: 600397) (n = 6). When we used denovolyzer to estimate the probability of n de novo variants expected to occur in a given cohort of 846 individuals,28 19/41 genes with two or more de novo variants had a nominal p value of £0.05, suggesting that the observed number of de novo variants in these genes was higher than expected by chance (Figure 2).
Figure 2.
Overview of Genetic Etiologies and Associations in the Current Study
Overview of the genetic etiologies with de novo variants in the cohort of 846 individuals included in the current study, sorted by significance of phenotypic similarity (p value phenotype). The number of individuals per sub-cohort (cohort), variant type (variant), and broad phenotypes (phenotypes) is shown. The number reflects the number of individuals with a certain feature, and the size and color of a bubble reflects relative frequency within the specific column. The cohort columns list the number of individuals with de novo variants in the EGRP, EPGP/Epi4K, and RES cohorts. The variant column lists the total number of individuals with missense (miss.) and protein-truncating variants (PTV). Genotype p values were calculated with denovolyzer and reflect the probability of identifying the observed number of de novo variants in a given cohort. Phenotype p values were derived through a semantic similarity analysis via the simmax method. In the phenotype column, the total number of individuals with neurodevelopmental delay (DD; HP: 0012758), focal-onset seizures (focal; HP: 0007359), and generalized-onset seizures (gen.; HP: 0002197) are listed; these were derived from the harmonized and propagated HPO dataset.
Genetic Etiologies Implicated in DEE Have Distinct HPO Signatures
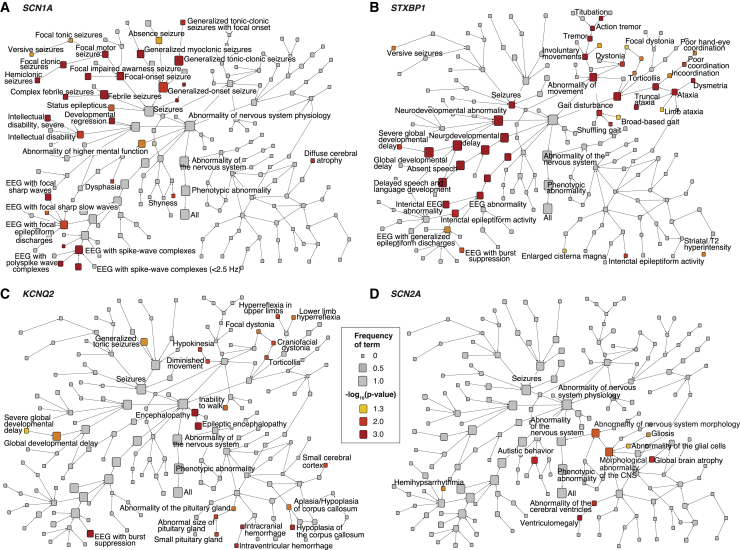
To determine the specific HPO terms driving phenotypic similarity for distinct genetic etiologies, we determined the relative contribution of specific HPO terms to each gene-specific similarity, comparing the observed and expected contribution of each HPO term (Figures 3 and 4). In summary, we identified 882 nominally significant gene-HPO associations (Table S3), and the comparison of observed and expected HPO terms resulted in gene-specific patterns (Figures 3, 4, and S7). The significant HPO terms reflect known phenotypic features associated with each genetic etiology, such as “febrile seizures” (HP: 0002373; p = 2.0 × 10−10) and “hemiclonic seizures” (HP: 0006813; p = 3.4 × 10−5) with SCN1A, “abnormality of central motor function” (HP: 0011442; p = 0.0015) with STXBP1, and “developmental regression” (HP: 0002376; p = 0.019) with SLC6A1 (MIM: 137165).
Figure 3.
Phenotype Association with Four Epilepsy Genes Shown as Phenotrees
(A–D) Each graph (phenotree) displays the branches of the Human Phenotype Ontology (HPO) beginning under the subbranch “abnormality of the nervous system” (HP: 0000707) for SCN1A, STXBP1, KCNQ2, and SCN2A. The size of each node indicates the frequency of each HPO term in the group of individuals with de novo variants with this gene, and the color indicates the level of statistical significance. The overall structure of the HPO tree is identical for each graph, which enables the visualization of phenotypic associations within the HPO tree. For example, for SCN1A, “generalized-onset seizure” (HP: 0002197) is present in 100% of individuals (n = 16) with a p value of 0.005. The more specific term “generalized tonic-clonic seizures” (HP: 0002069) is present in less individuals (n = 15, f = 0.94), but the association with the gene is stronger (p < 0.0001). The even more specific term “generalized tonic-clonic seizures with focal onset” (HP: 0007334) is less common (n = 4, f = 0.25) but is still associated with SCN1A (p = 0.01).
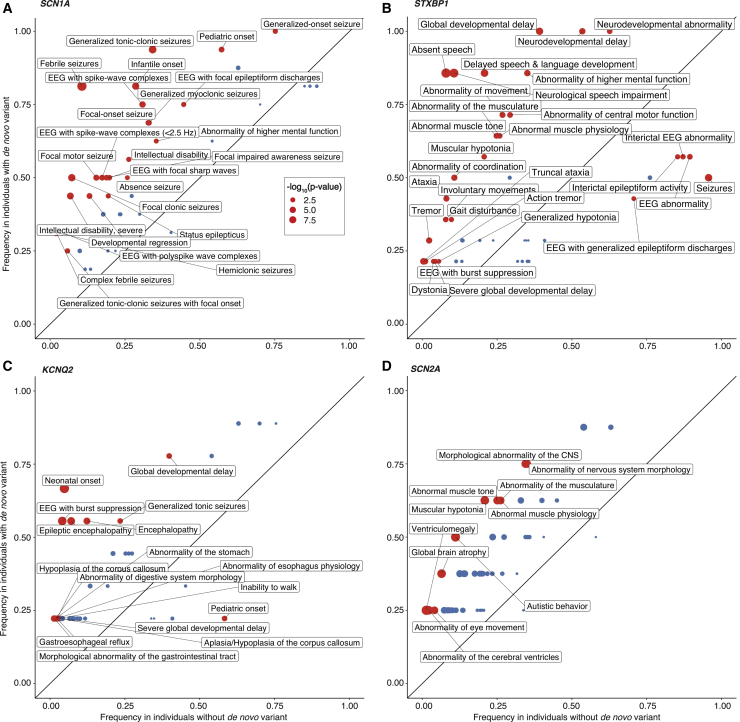
Figure 4.
Phenotype Association with Four Epilepsy Genes Shown as Phenograms
(A–D) Each graph (phenogram) displays the frequencies of HPO terms in SCN1A, STXBP1, KCNQ2, and SCN2A compared to the frequency in the overall cohort. The information contained reflects the associations shown in Figure 3 but allows for an alternative view of the gene-phenotype associations that includes the comparison to the wider cohort. Red dots indicate significant associations (p < 0.05) between HPO terms and specific genes. The size of the dot denotes the degree of significance displayed as −log10(p value). Because there are 1,616 unique HPO terms, rare and redundant terms were removed, e.g., “morphological abnormality of the central nervous system” (HP: 0002011) was removed when the more specific term “abnormality of brain morphology” (HP: 0012443) was present. For example, for SCN1A, “generalized tonic-clonic seizures” (HP: 0002069) are present in 94% of individuals with de novo variants compared to 34% in the remaining cohort. Accordingly, “generalized tonic-clonic seizures” (HP: 0002069) is located in the upper left corner of the phenogram and this association is significant (p = 1.5 × 10−6), as indicated by the color and size of the dot. In comparison, as can be seen from the relative positioning on the phenogram, “febrile seizures” (HP: 0002373) are less common in individuals with SCN1A than “generalized tonic-clonic seizures” (HP: 0002069). However, as indicated by the size of the dot, the association with SCN1A is stronger (p = 2 × 10−10) because the frequency in the overall cohort is very low.
Phenotypic Similarity Analysis Provides Statistical Evidence in 11 Genetic Etiologies
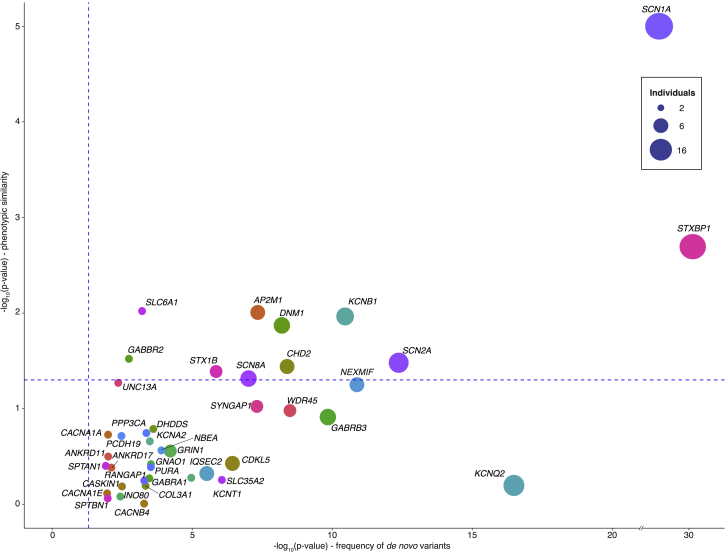
We next assessed whether genetic etiologies shared by two or more individuals have phenotypic similarities that were higher than expected by chance (Figures 2 and 5). We determined the median phenotypic similarity between individuals with each of the 41 genetic etiologies with two or more de novo variants and compared the observed median similarity score to the expected similarity score derived through 100,000 permutations. We identified 11 genetic etiologies with nominally significant phenotypic similarities (Figure 2). The significance for each of these genetic etiologies emerges consistently when adding individuals to the overall cohort and is not dependent on a single sub-cohort in this study (Figure 6). Comparing the statistical evidence for disease causation based on phenotypic evidence (phenotypic similarity) to genetic evidence (frequency of de novo variants) shows that the statistical evidence from the frequency of de novo variants is typically higher than the evidence derived from phenotypic similarity. However, both lines of evidence are independent. Some genetic etiologies with strong evidence based on the frequency of de novo variants are not significant based on phenotypic similarity, such as IQSEC2 (MIM: 300522) and PCDH19 (MIM: 300460). Other genetic etiologies, including DNM1 (MIM: 602377), SCN8A (MIM: 600702), and AP2M1 (MIM: 601024), have a relatively high phenotypic similarity compared to the significance based on the frequency of de novo variants. In addition, SCN1A and STXBP1 demonstrate a high degree of phenotypic similarity and statistical significance based on the frequency of de novo variants. The simcm and simmax algorithms showed some degree of variation between the statistical evidence for distinct genetic etiologies, but results from both algorithms were highly correlated (Figure S1).
Figure 5.
Comparison of Statistical Significance for the Frequency of Observed De Novo Variants and Phenotypic Similarity in 41 Genes
The graph compares the statistical significance for 41 genetic etiologies for genetic and phenotypic evidence. The point size indicates the number of individuals with de novo variants in each gene, and dashed blue lines represent −log10(0.05). Genetic evidence (x axis) reflects the significance, which was assessed with denovolyzer, for observed de novo variants. Phenotypic evidence reflects phenotypic similarity generated with simmax followed by permutation analysis (y axis). Contrasting genetic and phenotypic evidence allows for the comparison of both approaches and identification where one method deviates from the expected correlation. For example, de novo variants in KCNQ2 are present in nine individuals, but the phenotypic evidence is less than would be expected for genes with the same number of de novo variants. This discrepancy might be due to incomplete phenotyping or the inability of the HPO to capture the defining features of the disease correctly.
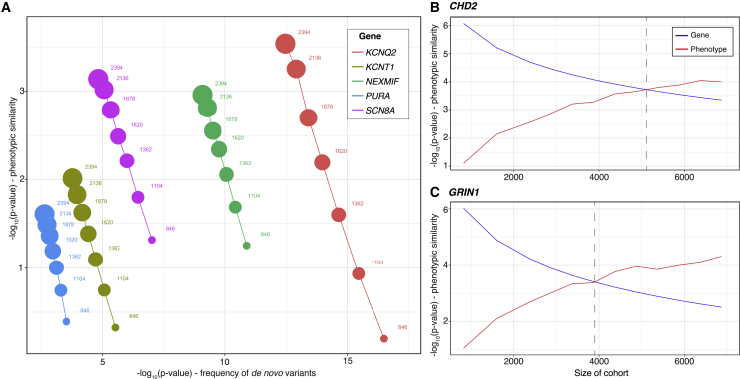
Figure 6.
Addition of Controls Results in Increased Phenotype-Based Significance and Reduced Genotype-Based Significance
(A) On the basis of the initial cohort of 846 individuals with DEE, subsequent addition of 1,548 population controls sequenced for de novo variants and without HPO terms results in a steady increase in the statistical significance of gene-based phenotypic similarity. Inversely, statistical significance based on the frequency of observed de novo variants steadily decreases with the addition of controls.
(B and C) With additional simulated controls, significance based on phenotypic similarity eventually exceeds significance based on frequency of de novo variants for CHD2 (B) and GRIN1 (C). The gray line indicates the critical cohort size when phenotypic significance becomes more significant than genotype-based significance.
The correlation between both algorithms is intriguing because both techniques emphasize slightly different aspects of the assigned phenotypes: the simmax generates a higher degree of similarity when multiple related phenotypes were assigned, e.g., “focal clonic seizures” (HP: 0002266) in addition to “focal aware seizure” (HP: 0002349), whereas the simcm algorithm would only assign similarity on the basis of the shared ancestral terms. In summary, the simmax algorithm is affected by the density of the assigned HPO terms within a specific sub-branch, whereas the simcm algorithm is dependent on the granularity of the HPO framework (Supplemental Notes).
In our study, we used a uniform bioinformatic pipeline for variant filtration. Because our pipeline processed heterogeneous exome data with varying quality, we decided to implement conservative thresholds for variant filtration, requiring at least 10 reads of the alternate allele to be present for the de novo analysis. We compared our results with the previously reported data from the Epi4K study and found that the threshold used in our study reliably identified all previously reported de novo variants.10 In addition, several individuals from the Epi4K cohort and EuroEPINOMICS-RES cohort had been found to carry de novo copy number variants, including known disease genes such as SCN1A, SCN2A, and GABRB3 (MIM: 137192).31 We subsequently repeated the phenotypic similarity analysis including the previously reported copy number variants (Tables S1 and S2). Neither analysis resulted in significant changes to the phenotypic similarities generated for each genetic etiology.
HPO Term Combinations Result in Unique Phenotype Profiles
In order to assess whether HPO terms can yield unique profiles that are predictive of the presence of a genetic etiology, we assessed the positive predictive value (PPV) of the combination of HPO terms that showed the strongest associations with genetic etiologies. As expected, we found that PPV increases with the addition of more HPO terms (Figure S3) but that the predicted frequency of individuals with the combination of HPO terms decreased. We then assessed the number of HPO terms per genetic etiology required to yield a PPV of 0.8 (Table 1). These term combinations, although only estimated to be present in a subset of individuals, have a probability of at least 80% for a de novo variant in the gene to be present. For some genetic etiologies, the combination of HPO terms required for a PPV of 0.8 is expected to be present in a significant number of individuals. For example, for DNM1, the combination of four HPO terms, including “brain atrophy” (HP: 0012444), “atrophy/degeneration affecting the central nervous system” (HP: 0007367), “aplasia/hypoplasia involving the central nervous system” (HP: 0002977), and “EEG with spike-wave complexes (<2.5 Hz)” (HP: 0010847), is expected in 41% of individuals with de novo variants in the gene, compared to 0.06% of individuals in the remainder of the cohort, resulting in a PPV of 0.8 for this combination of terms.
Table 1.
HPO Terms Required to Reach a PPV of at Least 80% for Genetic Etiologies in the Cohort
| Gene | PPV | Number of Terms | Cumulative Frequency | Individuals with Etiology | HPO ID | HPO Term | Frequency |
|---|---|---|---|---|---|---|---|
| DNM1 | 0.80 | 4 | 0.41 | 5 | HP: 0012444 | brain atrophy | 0.80 |
| HP: 0007367 | atrophy/degeneration affecting the CNS | 0.80 | |||||
| HP: 0002977 | aplasia/hypoplasia involving the CNS | 0.80 | |||||
| HP: 0010847 | EEG with spike-wave complexes (<2.5 Hz) | 0.80 | |||||
| KCNB1 | 0.81 | 5 | 0.070 | 6 | HP: 0011442 | abnormality of central motor function | 0.83 |
| HP: 0011443 | abnormality of coordination | 0.50 | |||||
| HP: 0000729 | autistic behavior | 0.50 | |||||
| HP: 0000708 | behavioral abnormality | 0.67 | |||||
| HP: 0000234 | abnormality of the head | 0.50 | |||||
| SCN1A | 0.90 | 5 | 0.23 | 16 | HP: 0002373 | febrile seizures | 0.81 |
| HP: 0002069 | generalized tonic-clonic seizures | 0.94 | |||||
| HP: 0003593 | infantile onset | 0.81 | |||||
| HP: 0010850 | EEG with spike-wave complexes | 0.75 | |||||
| HP: 0011153 | focal motor seizure | 0.50 | |||||
| STXBP1 | 0.87 | 5 | 0.63 | 14 | HP: 0002167 | neurological speech impairment | 0.86 |
| HP: 0000750 | delayed speech and language development | 0.86 | |||||
| HP: 0001263 | global developmental delay | 1.00 | |||||
| HP: 0011446 | abnormality of higher mental function | 0.86 | |||||
| HP: 0012758 | neurodevelopmental delay | 1.00 | |||||
| AP2M1 | 0.86 | 6 | 0.18 | 4 | HP: 0001252 | muscular hypotonia | 0.75 |
| HP: 0000750 | delayed speech and language development | 0.75 | |||||
| HP: 0011463 | childhood onset | 0.75 | |||||
| HP: 0010819 | atonic seizures | 0.75 | |||||
| HP: 0000708 | behavioral abnormality | 0.75 | |||||
| HP: 0003808 | abnormal muscle tone | 0.75 | |||||
| CHD2 | 0.84 | 6 | 0.12 | 4 | HP: 0002133 | status epilepticus | 0.75 |
| HP: 0011463 | childhood onset | 0.75 | |||||
| HP: 0000708 | behavioral abnormality | 0.75 | |||||
| HP: 0001249 | intellectual disability | 0.75 | |||||
| HP: 0002373 | febrile seizures | 0.50 | |||||
| HP: 0002123 | generalized myoclonic seizures | 0.75 |
For each genetic etiology in the cohort, the number of terms needed to reach a positive predictive value (PPV) of at least 80% was calculated. Displayed are all etiologies that required 6 terms or less to reach this threshold. HPO terms and their frequency within each genetic etiology are displayed.
Phenotypic Similarity Increases with the Inclusion of Unaffected Population Controls
In our current cohort of 846 individuals, the genetic evidence based on the probability of de novo variants in identified genetic etiologies was stronger than the statistical evidence derived from phenotypic similarity associated with that etiology. We reason that both parameters are driven by different factors in the overall cohort. The statistical significance of the frequency of de novo variants is greatest when the study cohort consists of a large number of affected individuals with a single underlying genetic etiology. Accordingly, inclusion of additional individuals with heterogeneous or unselected phenotypes will reduce the frequency of de novo variants for a specific genetic etiology in the larger cohort. In contrast, the phenotypic similarity associated with a given etiology is artificially diminished in cohorts of individuals with homogeneous phenotypes because the information content of terms depends upon its frequency in the cohort and, consequently, variation in phenotypic features is necessary for phenotypic similarity analysis to distinguish individuals who share a particular genetic etiology from those who do not. This is exemplified by the relatively low IC of “seizures” (HP: 0001250, IC = 0.075). In contrast to the diluting effect on the frequency of de novo variants, inclusion of individuals with heterogeneous phenotypes is likely to increase the phenotypic similarity of individuals with the same underlying genetic etiology because gene-related phenotypic features would become less frequent and therefore more informative. We tested this hypothesis by expanding our cohort to include 1,548 population controls that were sequenced for de novo variants and not assigned HPO terms (Figure 6).32
We observed the expected reduction in statistical significance for de novo variants, whereas the statistical evidence for phenotypic similarity increased. The trend continued when we subsequently added simulated population controls without de novo variants or phenotypic features. These results indicate that methods assessing phenotypic similarity may have an advantage in cohorts with heterogeneous phenotypes where genetic evidence based on the frequency of de novo variants may be insufficient to identify gene-disease associations. In these cohorts, the statistical evidence derived from phenotypic similarity may exceed the genetic evidence, particularly if future studies can exploit deeper phenotype data.
Discussion
In our study we assessed whether harmonization of sparse and heterogeneous phenotypic data via the HPO is capable of capturing associated clinical features and phenotypic similarities. Our aim was to model the cognitive process of recognizing gene-disease relationships through computational algorithms, providing a scalable method for phenotype analysis in large datasets. We reasoned that clinical features associated with distinct genetic etiologies may be prominent enough to stand out from the phenotypes in the larger cohort. We identified gene-specific phenotypic signatures and found that, for 11 genetic etiologies with de novo variants, the associated phenotypic similarities were greater than expected by chance.
Because of a lack of consistent frameworks and techniques, correlating clinical and genetic findings at scale remains a major hurdle in biomedical research33 and, despite attempts at standardization, phenotypic terminology remains heterogenous. Concepts to harmonize clinical phenotypic descriptions and to provide defined relationships between individual terms attempt to address this issue, and the HPO is one of the most frequently used frameworks. We demonstrate that the structure of the HPO can be used to harmonize phenotypic data across cohorts, including all major studies in the field of epilepsy research where trio exome data has been generated and where phenotypic features have been systematically captured. We further demonstrate that this conceptual framework can be used to operationalize previously vague concepts, such as phenotypic depth. For example, we find that the EPGP/Epi4K cohort only has a median of nine assigned phenotypic terms compared to the manually phenotyped EGRP cohort, which has a median of 13.5 phenotypic terms, translating into a median difference in IC of 131.9 (Figure 1, inset). Such concepts may help advance the understanding on how quality and quantity of phenotypic data associated with large genomic datasets can be measured and evaluated.
We find that the gene-phenotype associations identified in the harmonized clinical data correspond to the known phenotypic features in many of the genetic etiologies that are included in our study. For example, the most significant HPO terms associated with SCN1A accurately reflect the clinical spectrum of Dravet syndrome34, 35, 36 even though none of the individuals included in the study were primarily diagnosed with this condition given that the included data resources (EPGP/Epi4K and EuroEPINOMICS) were gene-discovery studies that excluded individuals with known genetic diagnoses.24 Likewise, the phenotypic spectrum linked to STXBP1 with “absent speech” (HP: 0001344; p = 1.31 × 10−11) and “truncal ataxia” (HP: 0002078; p = 7.03 × 10−5) reflects known phenotypic associations,37 as does the association of SCN2A with “autistic behavior” (HP: 0000729; p = 0.0079),38, 39, 40, 41 DNM1 with “obtundation status” (HP: 0011151; p = 0.00058),11,42 and KCNQ2 with “neonatal onset” (HP: 0003623; p = 1.39 × 10−6).43, 44, 45
We next evaluated whether the phenotypic terms linked to specific genetic etiologies were sufficiently strong for a gene-specific phenotypic signature to emerge. We applied two algorithms based on the MICA concept, assessing pairwise phenotypic similarities between individuals through the combination of the most specific terms shared by both individuals.29 Although both our algorithms are based on slightly different strategies, we find convergence for both concepts—both our simmax and simcm measures identify at least ten distinct genes associated with phenotype features more similar than expected by chance. Given that all individuals included in our study had epilepsy or neurodevelopmental disorders, we conclude that phenotypic features associated with genetic etiologies, including SCN1A, STXBP1, SLC6A1, AP2M1, and KCNB1, are not only similar per se, but they are also sufficiently similar to be identified within a cohort of individuals with related phenotypes.
Given that we used the example of Rett Syndrome as an introduction to the conceptual framework of phenotypic similarity, we performed a simulation to test whether the phenotypic similarity between six individuals with Rett Syndrome would appear significant if they were added to our existing dataset (Figure S3 and Supplemental Notes). In our simulation, although these four individuals with hypothetical MECP2 de novo variants would not be significantly similar if only a single term is added (“stereotypical hand wringing” [HP: 001217]), these individuals will have significant phenotypic similarity when two phenotypic terms are assigned (“stereotypical hand wringing” [HP: 0012171] and “developmental regression” [HP: 0002376], p = 0.05) or when four phenotypic terms are assigned (“stereotypical hand wringing” [HP: 0012171], “developmental regression” [HP: 0002376], “absent speech” [HP:0001344], and “apraxia” [HP:0002186], p = 0.002). This hypothetical example highlights that our approach can recapitulate the clinical recognition of specific phenotypes, such as Rett Syndrome.
To demonstrate the role of phenotypic homogeneity on the results of our study, we assessed how the inclusion of actual and simulated control individuals would affect the results of our study. We find that the phenotypic distinctiveness of all genetic etiologies increases with the inclusion of controls, whereas the probability of n de novo variants decreases. We further demonstrate that, with sufficient numbers of controls, the significance derived from our phenotypic similarity analysis will surpass the significance derived on the basis of the probability of de novo variants, even when using sparse phenotypic features. This emphasizes the utility of methods based on phenotypic similarity when assessing the causative role of rare genetic changes in large cohorts. Such methods may be useful for identifying individuals with extremely rare monogenic causes when analyzing population-based studies or entire healthcare systems.
Our phenotypic similarity analysis also showed several unexpected findings. Several genetic etiologies with relatively homogeneous phenotypes did not demonstrate the degree of phenotypic similarity that would have been expected. Most prominently, individuals carrying de novo variants in KCNQ2 did not show more phenotypic similarity than expected by chance. This finding is surprising given that clinical features in individuals with KCNQ2-related disorders are strikingly similar given the almost universal seizure onset in the neonatal period. A total of 45 HPO terms, including “neonatal onset” (HP: 0003623), “EEG with burst suppression” (HP: 0010851), “epileptic encephalopathy” (HP: 0200134), “encephalopathy” (HP: 0001298), and “gastroesophageal reflux” (HP: 0002020), were nominally associated with KCNQ2. We reviewed the phenotypic terms contributing to the nine individuals with KCNQ2-related disorders and found that the ten most strongly associated phenotypic terms were absent in three individuals (EIEE49, EPGP011188, and EPGP015469). In two of these individuals, we observed a very low depth of phenotyping. Individuals EIEE49 and EPGP015469 only had four and six phenotypic terms assigned, respectively, whereas the seven other individuals with de novo variants in KCNQ2 had a median of 11 assigned HPO terms. This observation suggests that the lack of similarity in individuals with KCNQ2 may be due to incomplete phenotyping rather than true phenotypic variation. As expected, when we added missing phenotypic terms to the three individuals, the overall phenotypic similarity became significant. The phenotypic similarity for all nine individuals reached p = 0.008 when adding the top three terms and p = 5.0 × 10−5 when adding the top ten terms associated with KCNQ2 missing in individuals EIEE49, EPGP011188, and EPGP015469.
The ability to pinpoint the lack of phenotypic similarity to individual factors, such as incomplete phenotyping, may highlight a strength of our approach—by harmonizing phenotypic information into a common format, it becomes possible to dissect phenotypes in individual genetic etiologies and identify sets of clinical features that drive the observed phenotypic similarity. However, the KCNQ2 example also highlights the need for methods that ensure that phenotypes are encoded uniformly and in an exhaustive manner. Although the overall framework of the HPO allows for both detailed and shallow datasets to be merged and analyzed jointly, it is a conceptual weakness of the HPO that phenotype quality and certainty cannot be encoded. Because it will remain conceptually challenging to distinguish incomplete phenotyping from truly absent phenotypes, quality measures and standard operation procedures for phenotypes will be required to ensure that the already heterogeneous phenotype data is not confounded as a result of low-quality phenotyping data.
Our study had several limitations. We observed a range of phenotypic terms assigned to the individuals and a significant difference among the different cohorts included: the EGRP cohort was significantly more deeply phenotyped compared to the EPGP/Epi4K or EuroEPINOMICS cohorts. Given the difference in phenotyping depth within and between cohorts, key aspects of the clinical presentation in some individuals may be incomplete, thus limiting the capacity of the similarity algorithms to identify individuals with shared features. Furthermore, the phenotypic features captured for an individual may only capture those manifesting by the age at last data collection. For example, individuals with loss-of-function variants in SCN2A typically present with developmental delay and autism, and seizures are frequently observed only after the age of two. Consequently, for younger individuals, seizures may not be recorded. In the EGRP sub-cohort in which age of recruitment was systematically recorded in 151/192 individuals, 30/151 individuals were recruited and phenotyped before the age of two. Accordingly, phenotypic similarities due to clinical features with later onset would not be able to be detected in this cohort. However, this limitation in recruitment strategy and data collection applies to traditional phenotypic analyses. We expect that more thorough longitudinal phenotypic details will be made available in the future through improved methods of extracting clinical information from electronic medical records, including advanced natural language processing and corrections for age-dependent phenotypic features.
A further limitation of our study was our reliance on retrospective data and that there may have been bias on how clinicians assigned HPO terms on the basis of their knowledge or assumption of the underlying genetic cause. Although we cannot exclude such an effect in the EGRP cohort, both the EPGP and RES cohorts were phenotyped prior to sequencing and HPO term assignment was not performed knowing the individuals’ genotypes. Despite this blinding, we cannot exclude that clinicians may have been biased toward an assumed underlying genetic diagnosis.
In summary, we demonstrate that an HPO-based framework is capable of bridging and harmonizing phenotypic data across various clinical datasets that were captured alongside large sequencing projects in the epilepsies. Although clinical data is heterogeneous and sparse, the mapping of features to a common ontology allows for the detection of frequently associated clinical features. The subsequent use of phenotypic similarity algorithms enables the detection of significant clinical similarities between individuals with shared genetic etiologies. These methods provide independent statistical evidence for disease causation and can be viewed as an extension of the clinical-genetic approach of defining disease entities through phenotypic resemblance. Given the increasing amounts of deep phenotypic data available for systematic analysis, methods that use computational phenotypes have the potential to identify novel genetic etiologies, particularly in situations when individuals have distinct phenotypic features and when the causative genetic etiology is rare.
Consortia
The members of the Non-Classical Epileptic Encephalopathy (NCEE) Study Group are Ralf Berkenfeld, Ingo Borggräfe, Andrea Dieckmann, Milda Endziniene, Andreas Faber, Andre Franke, Helge Gallwitz, Markus Gschwind, Christian M. Korff, Gerd Kurlemann, Sebastien Lebon, Johannes R. Lemke, Frank Maier, Thomas Mayer, Rikke Möller, Susanne Schubert-Bast, Niklas Schwarz, Simone Seifert, Bernhard J. Steinhoff, Inga Talvik, Shan Tang, and Holger Thiele.
The EPGP Investigators are Bassel Abou-Khalil, Brian Alldredge, Dina Amrom, Eva Andermann, Jocelyn Bautista, Sam Berkovic, Judith Bluvstein, Alex Boro, Gregory Cascino, Damian Consalvo, Sabrina Cristofaro, Patricia Crumrine, Orrin Devinsky, Dennis Dlugos, Michael Epstein, Robyn Fahlstrom, Miguel Fiol, Nathan Fountain, Kristen Fox, Jacqueline French, Catharine Freyer, Daniel Friedman, Eric Geller, Tracy Glauser, Simon Glynn, Kevin Haas, Sheryl Haut, Jean Hayward, Sucheta Joshi, Andres Kanner, Heidi Kirsch, Robert Knowlton, Eric Kossoff, Rachel Kuperman, Ruben Kuzniecky, Daniel Lowenstein, Shannon McGuire, Paul Motika, Gerard Nesbitt, Edward Novotny, Ruth Ottman, Juliann Paolicchi, Jack Parent, Kristen Park, Annapurna Poduri, Neil Risch, Lynette Sadleir, Ingrid Scheffer, Renee Shellhaas, Elliott Sherr, Jerry J. Shih, Shlomo Shinnar, Rani Singh, Joseph Sirven, Michael Smith, Michael R. Sperling, Joe Sullivan, Liu Lin Thio, Anu Venkat, Eileen Vining, Gretchen Von Allmen, Judith Weisenberg, Peter Widdess-Walsh, and Melodie Winawer.
The members of the EuroEPINOMICS-RES consortium are Rudi Balling, Nina Barisic, Stéphanie Baulac, Hande Caglayan, Dana Craiu, Peter De Jonghe, Christel Depienne, Renzo Guerrini, Helle Hjalgrim, Dorota Hoffman-Zacharska, Johanna Jähn, Karl Martin Klein, Bobby P. C. Koeleman, Vladimir Komarek, Eric Leguern, Anna-Elina Lehesjoki, Johannes R. Lemke, Holger Lerche, Tarja Linnankivi, Carla Marini, Patrick May, Rikke S. Møller, Deb K. Pal, Aarno Palotie, Felix Rosenow, Kaja Selmer, Jose M. Serratosa, Sanjay Sisodiya, Ulrich Stephani, Katalin Štěrbová, Pasquale Striano, Arvid Suls, Tiina Talvik, Sarah Weckhuysen, and Federico Zara.
The members of the Genomics Research and Innovation Network are Paul Avillach, Anna Bartels, Alan H. Beggs, Sawona Biswas, Florence T. Bourgeois, Jeremy Corsmo, Andrew Dauber, Batsal Devkota, Gary R. Fleisher, Tracy Glauser, Adda Grimberg, Tiffiney Hartman, Colin Hawkes, Allison P. Heath, Ingo Helbig, Joel N. Hirschhorn, Judson Kilbourn, Susan Kornetsky, Ian D. Krantz, Joseph A. Majzoub, Kenneth D. Mandl, Eric Marsh, Keith Marsolo, Lisa J. Martin, Jeremy Nix, Amy Schwarzhoff, Jason Stedman, Arnold Strauss, Kristen L. Sund, Deanne M. Taylor, Peter S. White, and Sek Won Kong.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
We thank the participants and their family members for taking part in the study. We also thank Margaret O’Brien, Mahgenn Cosico, Priya Vaidiswaran, and Eryn Fitch for support in enrolling research participants. I.H. was supported by The Hartwell Foundation through an Individual Biomedical Research Award. This work was also supported by NINDS (K02 NS112600), including the Channelopathy-associated Research Center (U54 NS108874), NICHD through the Intellectual and Developmental Disabilities Research Center (IDDRC) at Children’s Hospital of Philadelphia and the University of Pennsylvania (U54 HD086984), and NCATS (UL1 TR001878). The content is the responsibility of the authors and does not necessarily represent the official views of the NIH. This project was also supported in part by the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics at the Perelman School of Medicine of the University of Pennsylvania and by the Children's Hospital of Philadelphia through the Epilepsy NeuroGenetics Initiative (ENGIN). The study also received support through the German Research Foundation (DFG; HE5415/3-1, HE5415/5-1, and HE5415/6-1 to I.H. and WE4896/3-1 to Y.W.) and the DFG/FNR INTER Research Unit FOR2715 (We4896/4-1 and He5415/7-1 to I.H. and Y.W. and INTER/DFG/17/11583046 to R.K.). I.H. also received support through the International League Against Epilepsy (ILAE) and the Genomics Research and Innovation Network. C.A.E. was supported an NIH Ruth L. Kirschstein National Research Service Award (NRSA) Institutional Research Training Grant (T32 NS091008). D.L.S. was supported by the Wellcome Trust 4ward North Clinical PhD Academy.
Published: August 26, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.003.
Web Resources
Human Phenotype Ontology, https://hpo.jax.org/app/
OBO Citation and Attribution Policy, http://www.obofoundry.org/docs/Citation.html
Online Mendelian Inheritance in Man, https://www.omim.org/
Data and Code Availability
The code generated during this study are available through GitHub (https://github.com/galerp/helbig_lab_hpo_sim). The data from EuroEPINOMICS-RES (EGA), accession numbers EGAS00001000190, EGAS00001000386, and EGAS00001000048, and Epi4k (dbgap), accession number phs000653.v2.p1, are publicly available. A subset of the datasets supporting the current study have not been deposited in a public repository because of local consent and IRB restrictions but are available from the corresponding author on request.
Supplemental Data
Figures S1–S7, Tables S7–S12, and Supplemental Notes
References
- 1.Hagberg B., Aicardi J., Dias K., Ramos O. A progressive syndrome of autism, dementia, ataxia, and loss of purposeful hand use in girls: Rett’s syndrome: report of 35 cases. Ann. Neurol. 1983;14:471–479. doi: 10.1002/ana.410140412. [DOI] [PubMed] [Google Scholar]
- 2.Rett A. Uber ein eigenartiges hirnatrophisches Syndrom bei Hyperammonämie im Kindersalter. Wien. Med. Wochenschr. 1966;116:723–726. [PubMed] [Google Scholar]
- 3.Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 4.Kozinetz C.A., Skender M.L., MacNaughton N., Almes M.J., Schultz R.J., Percy A.K., Glaze D.G. Epidemiology of Rett syndrome: a population-based registry. Pediatrics. 1993;91:445–450. [PubMed] [Google Scholar]
- 5.Laurvick C.L., de Klerk N., Bower C., Christodoulou J., Ravine D., Ellaway C., Williamson S., Leonard H. Rett syndrome in Australia: a review of the epidemiology. J. Pediatr. 2006;148:347–352. doi: 10.1016/j.jpeds.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Claes L., Del-Favero J., Ceulemans B., Lagae L., Van Broeckhoven C., De Jonghe P. De novo mutations in the sodium-channel gene SCN1A cause severe myoclonic epilepsy of infancy. Am. J. Hum. Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barcia G., Fleming M.R., Deligniere A., Gazula V.R., Brown M.R., Langouet M., Chen H., Kronengold J., Abhyankar A., Cilio R. De novo gain-of-function KCNT1 channel mutations cause malignant migrating partial seizures of infancy. Nat. Genet. 2012;44:1255–1259. doi: 10.1038/ng.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helbig I., Lindhout D. Advancing the phenome alongside the genome in epilepsy studies. Neurology. 2017;89:14–15. doi: 10.1212/WNL.0000000000004053. [DOI] [PubMed] [Google Scholar]
- 9.Helbig I., Riggs E.R., Barry C.A., Klein K.M., Dyment D., Thaxton C., Sadikovic B., Sands T.T., Wagnon J.L., Liaquat K. The ClinGen Epilepsy Gene Curation Expert Panel-Bridging the divide between clinical domain knowledge and formal gene curation criteria. Hum. Mutat. 2018;39:1476–1484. doi: 10.1002/humu.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epi4K Consortium. Epilepsy Phenome/Genome Project. Allen A.S., Berkovic S.F., Cossette P., Delanty N., Dlugos D., Eichler E.E., Epstein M.P., Glauser T. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–221. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.EuroEPINOMICS-RES Consortium. Epilepsy Phenome/Genome Project. Epi4K Consortium De Novo Mutations in Synaptic Transmission Genes Including DNM1 Cause Epileptic Encephalopathies. Am. J. Hum. Genet. 2017;100:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Epi25 Collaborative Ultra-Rare Genetic Variation in the Epilepsies: A Whole-Exome Sequencing Study of 17,606 Individuals. Am. J. Hum. Genet. 2019;105:267–282. doi: 10.1016/j.ajhg.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindy A.S., Stosser M.B., Butler E., Downtain-Pickersgill C., Shanmugham A., Retterer K., Brandt T., Richard G., McKnight D.A. Diagnostic outcomes for genetic testing of 70 genes in 8565 patients with epilepsy and neurodevelopmental disorders. Epilepsia. 2018;59:1062–1071. doi: 10.1111/epi.14074. [DOI] [PubMed] [Google Scholar]
- 14.Truty R., Patil N., Sankar R., Sullivan J., Millichap J., Carvill G., Entezam A., Esplin E.D., Fuller A., Hogue M. Possible precision medicine implications from genetic testing using combined detection of sequence and intragenic copy number variants in a large cohort with childhood epilepsy. Epilepsia Open. 2019;4:397–408. doi: 10.1002/epi4.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyne H.O., Artomov M., Battke F., Bianchini C., Smith D.R., Liebmann N., Tadigotla V., Stanley C.M., Lal D., Rehm H. Targeted gene sequencing in 6994 individuals with neurodevelopmental disorder with epilepsy. Genet. Med. 2019;21:2496–2503. doi: 10.1038/s41436-019-0531-0. [DOI] [PubMed] [Google Scholar]
- 16.Heyne H.O., Singh T., Stamberger H., Abou Jamra R., Caglayan H., Craiu D., De Jonghe P., Guerrini R., Helbig K.L., Koeleman B.P.C., EuroEPINOMICS RES Consortium De novo variants in neurodevelopmental disorders with epilepsy. Nat. Genet. 2018;50:1048–1053. doi: 10.1038/s41588-018-0143-7. [DOI] [PubMed] [Google Scholar]
- 17.Helbig I., Ellis C.A. Personalized medicine in genetic epilepsies - possibilities, challenges, and new frontiers. Neuropharmacology. 2020;172:107970. doi: 10.1016/j.neuropharm.2020.107970. [DOI] [PubMed] [Google Scholar]
- 18.Helbig I., Lopez-Hernandez T., Shor O., Galer P., Ganesan S., Pendziwiat M., Rademacher A., Ellis C.A., Hümpfer N., Schwarz N., EuroEPINOMICS-RES Consortium. GRIN Consortium A Recurrent Missense Variant in AP2M1 Impairs Clathrin-Mediated Endocytosis and Causes Developmental and Epileptic Encephalopathy. Am. J. Hum. Genet. 2019;104:1060–1072. doi: 10.1016/j.ajhg.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastarache L., Hughey J.J., Hebbring S., Marlo J., Zhao W., Ho W.T., Van Driest S.L., McGregor T.L., Mosley J.D., Wells Q.S. Phenotype risk scores identify patients with unrecognized Mendelian disease patterns. Science. 2018;359:1233–1239. doi: 10.1126/science.aal4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson P.N., Köhler S., Bauer S., Seelow D., Horn D., Mundlos S. The Human Phenotype Ontology: a tool for annotating and analyzing human hereditary disease. Am. J. Hum. Genet. 2008;83:610–615. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köhler S., Vasilevsky N.A., Engelstad M., Foster E., McMurry J., Aymé S., Baynam G., Bello S.M., Boerkoel C.F., Boycott K.M. The Human Phenotype Ontology in 2017. Nucleic Acids Res. 2017;45(D1):D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akawi N., McRae J., Ansari M., Balasubramanian M., Blyth M., Brady A.F., Clayton S., Cole T., Deshpande C., Fitzgerald T.W., DDD study Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nat. Genet. 2015;47:1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epi4K Consortium Epi4K: gene discovery in 4,000 genomes. Epilepsia. 2012;53:1457–1467. doi: 10.1111/j.1528-1167.2012.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Epi4K Consortium Phenotypic analysis of 303 multiplex families with common epilepsies. Brain. 2017;140:2144–2156. doi: 10.1093/brain/awx129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vögtle F.N., Brändl B., Larson A., Pendziwiat M., Friederich M.W., White S.M., Basinger A., Kücükköse C., Muhle H., Jähn J.A. Mutations in PMPCB Encoding the Catalytic Subunit of the Mitochondrial Presequence Protease Cause Neurodegeneration in Early Childhood. Am. J. Hum. Genet. 2018;102:557–573. doi: 10.1016/j.ajhg.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Suls A., Jaehn J.A., Kecskés A., Weber Y., Weckhuysen S., Craiu D.C., Siekierska A., Djémié T., Afrikanova T., Gormley P., EuroEPINOMICS RES Consortium De novo loss-of-function mutations in CHD2 cause a fever-sensitive myoclonic epileptic encephalopathy sharing features with Dravet syndrome. Am. J. Hum. Genet. 2013;93:967–975. doi: 10.1016/j.ajhg.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ware J.S., Samocha K.E., Homsy J., Daly M.J. Interpreting de novo Variation in Human Disease Using denovolyzeR. Curr. Protoc. Hum. Genet. 2015;87:7.25.21–27.25.15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Resnik P. 14th International Joint Conference on Artificial Intelligence. Montreal, Morgan Kaufmann Publishers Inc.; San Francisco, CA, USA: 1995. Using information content to evaluate semantic similarity in a taxonomy; pp. 448–453. [Google Scholar]
- 30.R Core Team . R Foundation for Statistical Computing; 2013. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 31.Epilepsy Phenome/Genome Project Epi4K Consortium Copy number variant analysis from exome data in 349 patients with epileptic encephalopathy. Ann. Neurol. 2015;78:323–328. doi: 10.1002/ana.24457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jónsson H., Sulem P., Kehr B., Kristmundsdottir S., Zink F., Hjartarson E., Hardarson M.T., Hjorleifsson K.E., Eggertsson H.P., Gudjonsson S.A. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature. 2017;549:519–522. doi: 10.1038/nature24018. [DOI] [PubMed] [Google Scholar]
- 33.Haendel M.A., Chute C.G., Robinson P.N. Classification, Ontology, and Precision Medicine. N. Engl. J. Med. 2018;379:1452–1462. doi: 10.1056/NEJMra1615014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harkin L.A., McMahon J.M., Iona X., Dibbens L., Pelekanos J.T., Zuberi S.M., Sadleir L.G., Andermann E., Gill D., Farrell K., Infantile Epileptic Encephalopathy Referral Consortium The spectrum of SCN1A-related infantile epileptic encephalopathies. Brain. 2007;130:843–852. doi: 10.1093/brain/awm002. [DOI] [PubMed] [Google Scholar]
- 35.de Lange I.M., Gunning B., Sonsma A.C.M., van Gemert L., van Kempen M., Verbeek N.E., Sinoo C., Nicolai J., Knoers N.V.A.M., Koeleman B.P.C., Brilstra E.H. Outcomes and comorbidities of SCN1A-related seizure disorders. Epilepsy Behav. 2019;90:252–259. doi: 10.1016/j.yebeh.2018.09.041. [DOI] [PubMed] [Google Scholar]
- 36.Myers K.A., Burgess R., Afawi Z., Damiano J.A., Berkovic S.F., Hildebrand M.S., Scheffer I.E. De novo SCN1A pathogenic variants in the GEFS+ spectrum: Not always a familial syndrome. Epilepsia. 2017;58:e26–e30. doi: 10.1111/epi.13649. [DOI] [PubMed] [Google Scholar]
- 37.Stamberger H., Nikanorova M., Willemsen M.H., Accorsi P., Angriman M., Baier H., Benkel-Herrenbrueck I., Benoit V., Budetta M., Caliebe A. STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–962. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]
- 38.Sanders S.J., Murtha M.T., Gupta A.R., Murdoch J.D., Raubeson M.J., Willsey A.J., Ercan-Sencicek A.G., DiLullo N.M., Parikshak N.N., Stein J.L. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y.H., Yuen R.K., Jin X., Wang M., Chen N., Wu X., Ju J., Mei J., Shi Y., He M. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am. J. Hum. Genet. 2013;93:249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolff M., Johannesen K.M., Hedrich U.B.S., Masnada S., Rubboli G., Gardella E., Lesca G., Ville D., Milh M., Villard L. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140:1316–1336. doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 41.Lim E.T., Uddin M., De Rubeis S., Chan Y., Kamumbu A.S., Zhang X., D’Gama A.M., Kim S.N., Hill R.S., Goldberg A.P., Autism Sequencing Consortium Rates, distribution and implications of postzygotic mosaic mutations in autism spectrum disorder. Nat. Neurosci. 2017;20:1217–1224. doi: 10.1038/nn.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.von Spiczak S., Helbig K.L., Shinde D.N., Huether R., Pendziwiat M., Lourenço C., Nunes M.E., Sarco D.P., Kaplan R.A., Dlugos D.J., Epi4K Consortium. EuroEPINOMICS-RES NLES Working Group DNM1 encephalopathy: A new disease of vesicle fission. Neurology. 2017;89:385–394. doi: 10.1212/WNL.0000000000004152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh N.A., Charlier C., Stauffer D., DuPont B.R., Leach R.J., Melis R., Ronen G.M., Bjerre I., Quattlebaum T., Murphy J.V. A novel potassium channel gene, KCNQ2, is mutated in an inherited epilepsy of newborns. Nat. Genet. 1998;18:25–29. doi: 10.1038/ng0198-25. [DOI] [PubMed] [Google Scholar]
- 44.Charlier C., Singh N.A., Ryan S.G., Lewis T.B., Reus B.E., Leach R.J., Leppert M. A pore mutation in a novel KQT-like potassium channel gene in an idiopathic epilepsy family. Nat. Genet. 1998;18:53–55. doi: 10.1038/ng0198-53. [DOI] [PubMed] [Google Scholar]
- 45.Weckhuysen S., Mandelstam S., Suls A., Audenaert D., Deconinck T., Claes L.R., Deprez L., Smets K., Hristova D., Yordanova I. KCNQ2 encephalopathy: emerging phenotype of a neonatal epileptic encephalopathy. Ann. Neurol. 2012;71:15–25. doi: 10.1002/ana.22644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figures S1–S7, Tables S7–S12, and Supplemental Notes
Data Availability Statement
The code generated during this study are available through GitHub (https://github.com/galerp/helbig_lab_hpo_sim). The data from EuroEPINOMICS-RES (EGA), accession numbers EGAS00001000190, EGAS00001000386, and EGAS00001000048, and Epi4k (dbgap), accession number phs000653.v2.p1, are publicly available. A subset of the datasets supporting the current study have not been deposited in a public repository because of local consent and IRB restrictions but are available from the corresponding author on request.