Abstract
Simulation plays a central role in population genomics studies. Recent years have seen rapid improvements in software efficiency that make it possible to simulate large genomic regions for many individuals sampled from large numbers of populations. As the complexity of the demographic models we study grows, however, there is an ever-increasing opportunity to introduce bugs in their implementation. Here, we describe two errors made in defining population genetic models using the msprime coalescent simulator that have found their way into the published record. We discuss how these errors have affected downstream analyses and give recommendations for software developers and users to reduce the risk of such errors.
Simulation plays a central role in population genomics studies. Recent years have seen rapid improvements in software efficiency that make it possible to simulate large genomic regions for many individuals sampled from large numbers of populations. As the complexity of the demographic models we study grows, however, there is an ever-increasing opportunity to introduce bugs in their implementation. Here, we describe two errors made in defining population genetic models using the msprime coalescent simulator that have found their way into the published record. We discuss how these errors have affected downstream analyses and give recommendations for software developers and users to reduce the risk of such errors.
Main Text
In the effort to build more realistic simulations of genetic diversity, scientific software developers often focus on computational speed and biological realism. As the models simulated become more realistic, however, they also become more complex and difficult to specify. The interface through which users define their models is therefore increasingly important. Without an intuitive and thoroughly documented interface, it is very difficult to simulate complex population models correctly.
The msprime coalescent simulator1, 2, 3 is now widely used in genetics studies. Much of its appeal is the large increase in efficiency over the classical ms program,4 which makes it feasible to simulate large samples of whole chromosomes for the first time. Another distinct advantage of msprime is its Python application programming interface (API), which greatly increases the flexibility and ease of use over the standard approach of text-based command line interfaces. In particular, programs like ms require users to specify cryptic command line options to describe demographic models. For example, the Gutenkunst et al.5 demographic model (which is the subject of this note), as written in ms syntax, is shown in Figure 1A. This model is relatively simple, and models with many more populations and parameters are increasingly common. Descriptions of such models in ms syntax are not easy to comprehend. The Python interface for msprime, by contrast, allows the user to state models in a more human-readable and programmatic manner and has many advantages over ms’ command line interface.
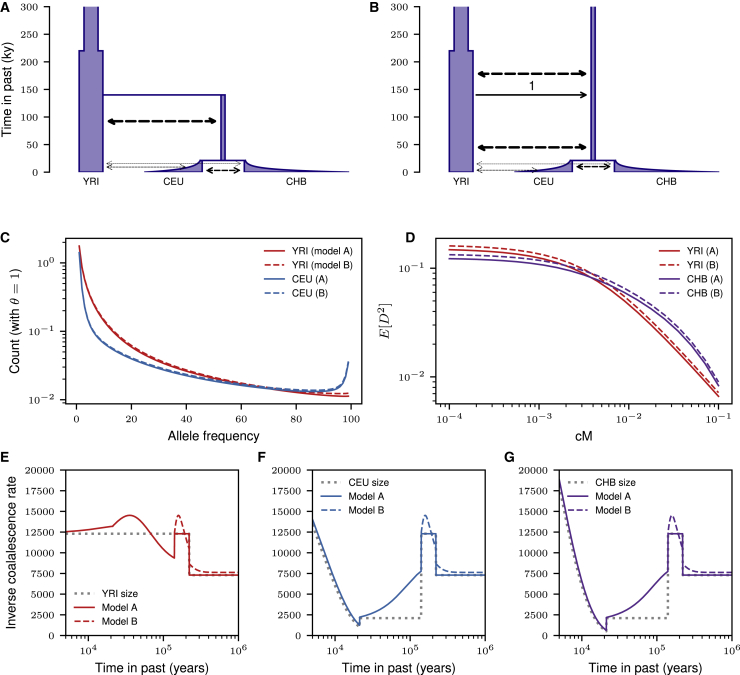
Figure 1.
Implementations of Demography in Coalescent Simulations
(A) The Gutenkunst et al. (2009)5 human expansion model as written in ms syntax.
(B) The same human expansion model with demographic events written in msprime syntax. In earlier versions of the msprime documentation, the highlighted line 17 was missing, so population structure erroneously continued into the past.
(C) A simulation of this model in Martin et al. (2017)6 omitted demographic events in the simulation function (highlight). This omission meant that populations remained distinct in the distant past and had excess .
(D) In msprime version 1.0, models will be specified so that population configurations and demographic events are coupled, reducing the possibility of making such errors of omission.
Even when using a high-level programming language like Python, however, implementing multi-population models of demographic history is difficult and prone to error. In this note, we discuss two implementation errors that arose through unfortunate design decisions in msprime’s demography API and that then found their way into the scientific record. The first error has relatively mild effects on genetic diversity but was used in many publications, whereas the second error was used only once but had a large impact on the simulation results. In light of these implementation errors, we discuss improvements to msprime’s API motivated by these discoveries and, more generally, best practices for implementing and simulating complex multi-population demography.
Case 1: A Misspecified Model in msprime’s Documentation
To illustrate the demography API, msprime included a description of a widely used three-population Out-of-Africa model5 as part of its tutorial documentation. In this model (Figure 2A), Eurasian (CEU and CHB) and African (YRI) populations split from each other in the deep past, followed by a more recent split of European and Asian populations with variable rates of continuous migration between each of the populations. Regrettably, the implementation in the msprime tutorial was incorrect. Before the time of the split of African and Eurasian populations, when there should have been just a single randomly mating population, migration was allowed to occur between the ancestral population and a second population with size equal to the Eurasian bottleneck size for all time into the past (Figure 2B). This incorrect model was introduced into the tutorial for msprime version 0.3.0 and remained in the documentation for around 4 years.
Figure 2.
Expected Diversity Statistics under the Gutenkunst et al. Model
(A) The correctly implemented model. Dashed arrows depict continuous migration.
(B) The incorrectly implemented model from the msprime tutorial with migration continuing into the past beyond the mass migration event with proportion 1 from the ancestral population to the bottleneck population.
(C) Marginal allele frequency spectra under the two models. Heterozygosity in the incorrect model is inflated by , although the general shapes of the distributions are qualitatively similar.
(D) Similarly, the increased heterozygosity leads to excess , although the LD-decay is qualitatively similar between models.
(E–G) We compared the true size history for each population under this model to their expected inverse coalescent rates, which are often interpreted as . The correct and incorrect models (A and B) are equivalent in the recent history, so the inverse coalescence rates only differ in the more distant past. For multi-population demographic history, the inverse coalescence rates are not expected to match the “true” historical size along each branch because population structure and migration between populations change coalescent probabilities over time.
Fortunately, the effects of this error are subtle. Population sizes and structure since the time of the earliest split are unaffected, so differences in expected are negligible between the correct and incorrect models. However, the ancient structure distorts the distribution of early TMRCAs (times to most recent common ancestor) (Figures 2E–2G). The extraneous ancient population increases the long-term effective population size, resulting in roughly excess heterozygosity in contemporary populations compared to the intended model, but the overall effects on patterns of diversity are minimal (Figures 2C and 2D).
Even though the error has a limited effect on simulated data, the tutorial code has been copied many times and used in publications. By searching for some identifying strings from the model definition on GitHub, we found 32 repositories containing either direct copies of the erroneous model code or code that was obviously derived from it (we have opened issues on each of these repositories to alert the authors). In most cases, the publications used simulations to test a non-demographic inference method, such as inferring gene genealogies from sequencing data,7 determining the age of a genetic variant,8 or studying the power of variant association tests.9 In each of these studies, this model was used as an example of a complex population history. Zhou et al.10 used the incorrect model as an example of how their method for visualizing demographic models can support msprime input. Finally, Pfaffelhuber et al.11 used simulations of the incorrect model demography to evaluate their method for choosing ancestry-informative markers. Given the very subtle effect of the incorrect model on demography (and the fact the method was evaluated with other simulations and real data), it seems unlikely that the model details had any qualitative effect on their conclusions.
This long-standing error could have been prevented by better API design. To model a population split currently in msprime, a user must specify a “MassMigration” event that moves lineages from one population to another and then must also remember to turn off migration between those populations at the same time. The demographic events for the correct model are given in Figure 1B. Note that the migration rate change on line 17 was missing in the incorrect model. The release of msprime version 1.0 will introduce a “PopulationSplit” event, which more intuitively links the movement of lineages with appropriate changes in migration rates at the time of the split.
Case 2: Incorrect Model Parameters in an Analysis Pipeline
In another publication using this model,6 a separate error was introduced: the model itself was defined as suggested in the documentation (with updated parameters from Gravel et al.12) and inspected with the msprime debugging tools. Despite these initial checks, the simulation was performed without passing the list of demographic events, such as historical changes in size or population splits, so that the three populations never merged and remained separated with low levels of migration (Figure 3A). This leads to a vast overestimate of the divergence across human populations. Although the correct model predicts a mean of across the three populations, the simulated model generated ranging between , depending on the populations considered. Overall diversity was also strongly affected: heterozygosity was more than doubled in African populations and reduced by more than half in Eurasian populations relative to the correct model.
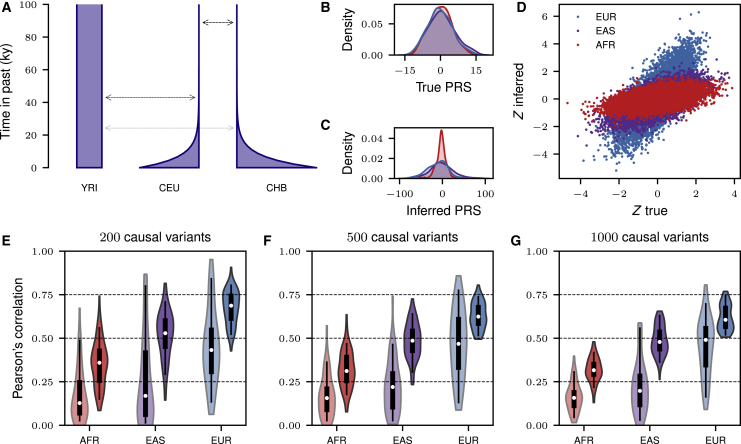
Figure 3.
The Transferability of Polygenic Risk Scores (PRSs) under Neutrality
(A) In Martin et al.,6 the simulated demographic model did not apply demographic events in the past, so continental populations were simulated as isolated with low levels of migration for all time. The correct model has historical events as shown in Figure 2A.
(B–G) We repeated the simulation experiment in Martin et al.6 by using the correct demographic model. GWAS summary statistics were computed from 10,000 case and control subjects in the European population, and the distribution of inferred PRSs was compared across the three simulated populations. Correlations between true and inferred polygenic scores were computed over 100 simulation replicates. Unlike the original study, we do not observe large differences in mean inferred PRS across the three populations (C and D), and although risk prediction in the African and East Asian populations is still reduced compared to the European population, the reduction in prediction accuracy is not as large as reported in the original study (E–G). (B) and (C) show the distribution of true and inferred PRSs after standardizing PRSs across all individuals in the simulation (this is in contrast to Figure 5 in Martin et al., which plotted unstandardized PRS6). Because the polygenic trait was simulated as a threshold trait, it is the relative distribution of PRSs across individuals that determined cases and controls instead of absolute true or inferred PRS. (D) shows a comparison of PRS distributions for a simulation replicate with 1,000 causal variants. For each population in the violin plots, the original correlations from Martin et al.6 are shown on the left, and correlations via the correct model are shown on the right. For direct comparison to the original study, see Figure 5 in Martin et al.6 Box plots show median, first and third quartiles, and 95% intervals.
This simulation was performed to assess the transferability of polygenic risk scores across human populations. In other words, it sought to explore how human demographic history and population structure affect our ability to predict genetic risk in diverse populations given the well-documented unequal representation in medical genetic studies.13 The resulting publication has been influential in the discussion of health inequalities and genomics: it has been cited over 350 times since 2017. The large excess in divergence under the incorrect model here exaggerated the role of demography and genetic drift in limiting the transferability of genetic risk scores across populations.
Difficulties in transferability remain in the corrected model (Figure 3), although risk prediction in each population is significantly improved (compare to Figure 5 in Martin et al.6). Simulations under the correct model indicate that the accuracy of genetic risk scores is still substantially reduced in understudied populations (Figures 3E–3G), supporting one of the main conclusions of Martin et al.6 However, the reduction is much less pronounced than reported. In particular, we do not observe large differences in mean predicted risk across populations (Figures 3C and 3D) that were present in data and simulations from Martin et al.6 Thus, a model with continental-scale demographic structure, highly polygenic architecture, and neutral evolution does not appear to explain the large directional biases in mean predicted risk identified in Martin et al.6
From a software perspective, this error was easy to make and could have been prevented by better API design. The original msprime API requires the user to pass three separate parameters to specify a demographic model (see example in Figure 1C). Note that the same three parameters must be passed to both the debugger and the simulate function. To help prevent such errors, msprime version 1.0 will introduce a demography class. The above snippet would then be rewritten (approximately) as shown in Figure 1D. This simple change to the interface makes it much less likely that different models are passed to the debugger and simulator, reducing the potential for error.
Conclusions
The implementation of complex demographic models is error prone, and such errors can have a large impact on downstream analyses and interpretation. The discovery and correction of the demographic models discussed here underscore how API design choice can lead to the propagation of mistakes that are difficult to notice. There are myriad other ways to get models slightly or very wrong. Migration rates can be set in the wrong direction, for example, and parameters can be rescaled incorrectly when a new mutation rate is used. We therefore recommend the following steps to ensure more robust simulations.
First, if possible, we recommend using the stdpopsim14 library, which removes the need to re-implement demographic models from scratch. This is a “standard library” of quality-controlled models and simulation resources for a growing number of commonly studied species. The resource is built around an open-source community development model with rigorous code review and quality control (QC) procedures. For example, when a contributor adds a new model, it is not fully integrated into the catalog until a second, entirely independent, implementation of the model is provided by another developer. If the original and “QC” versions of the model are precisely equal, then we can have a reasonable degree of confidence in correctness. It is through this QC process that we discovered the misspecified model in msprime’s documentation. However, stdpopsim has QC-verified models available for only a handful of published models and species, so it might not meet the needs of a particular simulation study. If the required model is not present in stdpopsim, then extra care should be taken to validate the implementation. This could include verification through code review or comparison with an independent implementation of the model. If the model is of wider interest to the community, we would encourage developers to contribute it to stdpopsim; at the very least, the QC procedures in place provide a strong reassurance of correctness.
Second, regardless of whether the model has been implemented locally or obtained from a resource like stdpopsim, basic statistical validation should always be performed on the results. Errors are all too easy to make, and an analysis of the basic statistical properties of the simulations is essential due diligence. For example, a brief analysis of values most likely would have prevented the error in Martin et al.6 It is understandable that such analyses were not undertaken in this case because processing such a huge dataset (600,000 samples) was highly challenging. Recent progress has made statistical analysis at this scale much easier15 so that , heterozygosity, the allele frequency spectrum, or the distribution of coalescence times can be rapidly computed. However, the problems would have been apparent if pilot simulations with smaller sample sizes were undertaken and analyzed because simulations and analysis of small samples sizes are very efficient. Graphical inspection of demographic models can also help identify issues, especially if visualization can be automated. The demography debugging tool in msprime summarizes demographic events occurring over time, and various efforts are underway to facilitate the specification and visualization of population genetics models, such as Zhou et al.10 and the demography package used in this paper.
Finally, openness is essential to the self-correcting nature of science. We only know about these errors because of open code and open-source development processes. By making their entire pipeline available, Martin et al.6 not only enabled other research teams to build upon their findings, but they also made it possible for errors to be found and corrected. There must be many, many more mistakes out there, and we need both pre- and post-publication vigilance from users and developers to ensure the soundness of the large body of simulation-based analyses.
Acknowledgments
We would like to thank Alicia Martin for comments on the manuscript and for providing the simulation data from their study reproduced in Figure 3. We would also like to thank Chris Gignoux, Ilan Gronau, and John Novembre for helpful comments on the manuscript. J.K. would like to apologize to those affected by the error in the example Out-of-Africa model and by the poor design decisions in msprime’s demography API. This research is funded in part by the Canada Research Chair Program, CIHR project grant MOP 136855, and an NSERC Discovery grant to S.G. J.K. is supported by the Robertson Foundation.
Contributor Information
Simon Gravel, Email: simon.gravel@mcGill.ca.
Jerome Kelleher, Email: jerome.kelleher@bdi.ox.ac.uk.
Data and Code Availability
We computed the expected allele frequency spectrum (AFS) by using moments version 1.0.316 and LD-decay curves by using moments.LD.17 and other diversity statistics were computed from the expected AFS and verified with branch statistics from the output of msprime simulations using tskit version 0.2.3.15 Demographic models were plotted with the demography package written for Python (https://github.com/apragsdale/demography, version 0.0.3). The demography package allows users to define a demographic model as a directed acyclic graph using networkx,18 which can be plotted as in Figures 2A, 2B, and 3A. Demography also can translate the graph-based demography into msprime input commands or simulate the AFS or two-locus statistics by using moments16,17 or dadi.5 For the analysis of polygenic risk, we used the original pipeline from Martin et al.6 available from https://github.com/armartin/ancestry_pipeline/blob/master/simulate_prs.py. We updated the pipeline to run with the correct demographic parameters and more recent versions of msprime and tskit, and the updated pipeline is available at https://github.com/apragsdale/PRS. Data and Python scripts to recreate Figures 2 and 3 can be found at https://github.com/jeromekelleher/msprime-model-errors. A full list of the GitHub repositories containing copies of the erroneous model are also given here.
References
- 1.Kelleher J., Etheridge A.M., McVean G. Efficient coalescent simulation and genealogical analysis for large sample sizes. PLoS Comput. Biol. 2016;12:e1004842. doi: 10.1371/journal.pcbi.1004842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nelson D., Kelleher J., Ragsdale A.P., Moreau C., McVean G., Gravel S. Accounting for long-range correlations in genome-wide simulations of large cohorts. PLoS Genet. 2020;16:e1008619. doi: 10.1371/journal.pgen.1008619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelleher J., Lohse K. Coalescent simulation with msprime. In: Dutheil J.Y., editor. Statistical Population Genomics. Springer US; New York, NY: 2020. pp. 191–230. [Google Scholar]
- 4.Hudson R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 5.Gutenkunst R.N., Hernandez R.D., Williamson S.H., Bustamante C.D. Inferring the joint demographic history of multiple populations from multidimensional snp frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelleher J., Wong Y., Wohns A.W., Fadil C., Albers P.K., McVean G. Inferring whole-genome histories in large population datasets. Nat. Genet. 2019;51:1330–1338. doi: 10.1038/s41588-019-0483-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Albers P.K., McVean G. Dating genomic variants and shared ancestry in population-scale sequencing data. PLoS Biol. 2020;18:e3000586. doi: 10.1371/journal.pbio.3000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong D.M.H., Hernandez R.D. Population genetic simulation study of power in association testing across genetic architectures and study designs. Genet. Epidemiol. 2020;44:90–103. doi: 10.1002/gepi.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y., Tian X., Browning B.L., Browning S.R. POPdemog: visualizing population demographic history from simulation scripts. Bioinformatics. 2018;34:2854–2855. doi: 10.1093/bioinformatics/bty184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaffelhuber P., Grundner-Culemann F., Lipphardt V., Baumdicker F. How to choose sets of ancestry informative markers: A supervised feature selection approach. Forensic Sci. Int. Genet. 2020;46:102259. doi: 10.1016/j.fsigen.2020.102259. [DOI] [PubMed] [Google Scholar]
- 12.Gravel S., Henn B.M., Gutenkunst R.N., Indap A.R., Marth G.T., Clark A.G., Yu F., Gibbs R.A., Bustamante C.D. 1000 Genomes Project. Demographic history and rare allele sharing among human populations. Proc. Natl. Acad. Sci. USA. 2011;108:11983–11988. doi: 10.1073/pnas.1019276108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adrion J.R., Cole C.B., Dukler N., Galloway J.G., Gladstein A.L., Gower G., Kyriazis C.C., Ragsdale A.P., Tsambos G., Baumdicker F. A community-maintained standard library of population genetic models. eLife. 2020;9:e54967. doi: 10.7554/eLife.54967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ralph P., Thornton K., Kelleher J. Efficiently summarizing relationships in large samples: a general duality between statistics of genealogies and genomes. Genetics. 2020;215:779–797. doi: 10.1534/genetics.120.303253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouganous J., Long W., Ragsdale A.P., Gravel S. Inferring the joint demographic history of multiple populations: beyond the diffusion approximation. Genetics. 2017;206:1549–1567. doi: 10.1534/genetics.117.200493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ragsdale A.P., Gravel S. Models of archaic admixture and recent history from two-locus statistics. PLoS Genet. 2019;15:e1008204. doi: 10.1371/journal.pgen.1008204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagberg A., Swart P., Schult D. Los Alamos National Lab; 2008. Exploring network structure, dynamics, and function using networkx. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We computed the expected allele frequency spectrum (AFS) by using moments version 1.0.316 and LD-decay curves by using moments.LD.17 and other diversity statistics were computed from the expected AFS and verified with branch statistics from the output of msprime simulations using tskit version 0.2.3.15 Demographic models were plotted with the demography package written for Python (https://github.com/apragsdale/demography, version 0.0.3). The demography package allows users to define a demographic model as a directed acyclic graph using networkx,18 which can be plotted as in Figures 2A, 2B, and 3A. Demography also can translate the graph-based demography into msprime input commands or simulate the AFS or two-locus statistics by using moments16,17 or dadi.5 For the analysis of polygenic risk, we used the original pipeline from Martin et al.6 available from https://github.com/armartin/ancestry_pipeline/blob/master/simulate_prs.py. We updated the pipeline to run with the correct demographic parameters and more recent versions of msprime and tskit, and the updated pipeline is available at https://github.com/apragsdale/PRS. Data and Python scripts to recreate Figures 2 and 3 can be found at https://github.com/jeromekelleher/msprime-model-errors. A full list of the GitHub repositories containing copies of the erroneous model are also given here.