Summary
There is growing recognition that epivariations, most often recognized as promoter hypermethylation events that lead to gene silencing, are associated with a number of human diseases. However, little information exists on the prevalence and distribution of rare epigenetic variation in the human population. In order to address this, we performed a survey of methylation profiles from 23,116 individuals using the Illumina 450k array. Using a robust outlier approach, we identified 4,452 unique autosomal epivariations, including potentially inactivating promoter methylation events at 384 genes linked to human disease. For example, we observed promoter hypermethylation of BRCA1 and LDLR at population frequencies of ∼1 in 3,000 and ∼1 in 6,000, respectively, suggesting that epivariations may underlie a fraction of human disease which would be missed by purely sequence-based approaches. Using expression data, we confirmed that many epivariations are associated with outlier gene expression. Analysis of variation data and monozygous twin pairs suggests that approximately two-thirds of epivariations segregate in the population secondary to underlying sequence mutations, while one-third are likely sporadic events that occur post-zygotically. We identified 25 loci where rare hypermethylation coincided with the presence of an unstable CGG tandem repeat, validated the presence of CGG expansions at several loci, and identified the putative molecular defect underlying most of the known folate-sensitive fragile sites in the genome. Our study provides a catalog of rare epigenetic changes in the human genome, gives insight into the underlying origins and consequences of epivariations, and identifies many hypermethylated CGG repeat expansions.
Keywords: epimutation, epivariation, DNA methylation, repeat expansion, folate-sensitive fragile site
Introduction
The main focus of the field of human genetics over the past few decades has been the investigation of sequence variation as a driver of human phenotypic variation. Projects such as the HapMap, 1000 Genomes, and the Exome Aggregation Consortium1, 2, 3, 4, 5 have provided deep surveys of genetic variation in both coding and non-coding regions, facilitating many novel insights into genotype-phenotype relationships in both common and rare diseases.
However, a number of recent studies have also demonstrated that rare epigenetic variation, sometimes termed epivariations or epimutations, can also underlie human disease. For example, between 5% and 15% of patients with hereditary nonpolyposis colorectal cancer who are negative for pathogenic coding variants present with constitutional MLH1 (MIM: 120436) promoter methylation.6 Similarly, allelic methylation of the BRCA1 (MIM: 113705) promoter has been identified in several pedigrees with familial breast/ovarian cancer,7,8 and inborn errors of vitamin B12 metabolism have been shown to result from an epivariation that silences MMACHC9 (MIM: 609831). Other studies have shown a significant increase of de novo epivariations in individuals with congenital disorders compared to control subjects10 and provided evidence that epivariations contribute to the mutational spectra underlying autism and schizophrenia.11
Epivariations can be subdivided based on their apparent etiology.12 Primary epivariations are thought to be caused by stochastic errors in the establishment or maintenance of the epigenome, such as certain types of imprinting anomalies.13 In contrast, secondary epivariations occur as a result of an underlying change in local DNA sequence and include mutations that disrupt regulatory elements9,10,14 and expansions of CpG-rich tandem repeats.15 Large hypermethylated expansions of CGG repeats have been identified at a number of folate-sensitive fragile sites in the human genome,16 including several that are associated with neurodevelopmental anomalies, such as the CGG expansions that occur at FMR1 (MIM: 309550), AFF2 (MIM: 300806), DIP2B (MIM: 611379), and AFF3 (MIM: 601464).17, 18, 19, 20
Originally the term “epimutation” was used in the literature to refer specifically to purely epigenetic changes that occur without a change in DNA sequence.21 However, over the past two decades many reports have applied this term to a variety of epigenetic changes, some of which apparently result from nearby sequence alterations,13 but often their etiology was undetermined.22 Here we use the term “epivariation” to refer to any rare alteration in DNA methylation, irrespective of their underlying cause, because in the majority of cases this is difficult to unambiguously determine.
Despite this growing evidence that epigenetic defects contribute to a wide variety of human diseases, currently little information exists on the prevalence and distribution of rare epigenetic variation in the human population. As a result, the potential contribution of epivariations to human disease is unclear. In order to address this, here we have analyzed data from >23,000 individuals that were originally generated for use in epigenome-wide association studies, representing the largest cohort of methylomes assembled to date. Utilizing a robust outlier analysis, we identified >4,000 epivariation loci that each span multiple CpGs in these samples, including several hundred that occur at the promoters of known Mendelian disease genes, thus implicating epivariations as a potentially causative factor in many human disorders. Using hundreds of monozygous (MZ) twin pairs and available variation and expression data in thousands of samples, we investigated the causes and consequences of epivariations. Furthermore, by applying long-read sequencing, we validated the presence of CGG expansions as the cause of some epivariations, identifying the molecular defect underlying most of the known folate-sensitive fragile sites in the human genome. Our study provides a catalog of rare epigenetic changes in the human genome and identifies many hypermethylated CGG repeat expansions. These data suggest that epivariations mark or represent a subset of pathogenic alleles at some disease loci, which would likely be missed by purely sequence-based approaches.
Material and Methods
Datasets
For the identification of epivariations, we accessed methylation data from a total of 24,985 individuals from 22 cohorts, listed in Table S1. Each cohort comprised DNA methylation profiles from at least 300 individuals generated using the Illumina 450k HumanMethylation BeadChip (450k array). Eighteen studies utilized DNA extracted from peripheral whole blood, while the remaining four studies utilized DNA extracted from newborn cord blood, dried neonatal blood spots, purified monocytes, or adipose tissue. Seventeen of the cohorts represented samples drawn from the general population without ascertainment for any specific condition, while five of the cohorts included some samples ascertained due to a diagnosis of ischemic stroke, asthma, Parkinson disease, facial clefts, or rheumatoid arthritis. Four of the cohorts were comprised partially or wholly of pairs of monozygous and dizygous twins. For additional studies of rare sequence variants associated with epivariations, we utilized data from 457 Parkinson disease and control individuals from the Parkinson’s Progression Markers Initiative (PPMI) cohort, where peripheral whole-blood DNA methylation profiles generated with the Illumina Infinium MethylationEPIC BeadChip (850k array) are available.23 This study was approved by, and the procedures followed were in accordance with, the ethical standards of the Institutional Review Board of the Icahn School of Medicine under HS# 18-01169.
Quality Control and Data Processing
Within each cohort, we performed a number of quality-control steps to identify samples for exclusion, as follows. (1) We removed any sample with >1% of autosomal probes with detection p value > 0.01. (2) We performed principal component analysis (PCA) based on β-values of all probes located on chr1. Based on scatterplots of the first two principal components, we removed samples judged to be outliers. (3) We utilized the array data to infer the likely sex of each sample, based on scatterplots of mean β-value of probes located on chrX versus the fraction of probes located on chrY with detection p > 0.01. We compared these predictions against self-reported gender for each sample where available and removed any samples with a potential sex mismatch. Furthermore, outlier samples and samples with potential sex chromosome aneuploidies were also removed. Samples then underwent normalization, as described previously.10,11 Briefly, raw signal intensities were subjected to color correction, background correction, and quantile normalization using the Lumi package in R,24 and the normalized intensities converted into β-values, which range between 0 and 1, representing the methylation ratio at each measured CpG. In order to correct for inherent differences in the distribution of β-values reported by Infinium I and Infinium II probes, we applied BMIQ.25 Each cohort was normalized independently, and data for probes located on chrX in males were normalized separately from autosomal data. After normalization, we estimated the major cellular fractions comprising each blood sample directly from β-values using the method described by Houseman et al.26 and removed outlier samples, defined as those that showed cellular fractions either ≥99th percentile +2% or ≤1st percentile −2% of any cell type. After all quality-control and filtering steps, 23,173 samples assayed with the 450k array were processed to identify epivariations.
Identification of Rare Epigenetic Variants
In order to identify rare epigenetic variants, also termed differentially methylated regions (DMRs), we utilized a sliding window approach to compare individual methylation profiles of a single sample against all other samples from the same cohort. We chose this approach of testing for DMRs within each cohort in order to minimize batch effects that might result if we performed comparisons across different cohorts. We defined DMRs as regions of outlier methylation represented by multiple independent probes using the following parameters:
-
•
Hypermethylated DMR: any 1 kb region with at least three or more probes with β-values ≥ 99.5th percentile plus 0.15 and containing at least three consecutive probes with β-values ≥ 99.5th percentile. In addition, we required that the minimum distance spanned by probes that were ≥99.5th percentile was ≥100 bp.
-
•
Hypomethylated DMR: any 1 kb region with at least three or more probes with β-values ≤ 0.5th percentile minus 0.15 and containing at least three consecutive probes with β-values ≤ 0.5th percentile. In addition, we required that the minimum distance spanned by probes that were ≤0.5th percentile was ≥100 bp.
As the presence of an underlying homozygous deletion at a probe binding site can result in spurious β-values,11 we removed any DMR call in which the carrier individual reported one or more probes within the DMR with failed detection p value (p > 0.01). Finally, we removed 57 samples which each reported an unusually high number (n > 20) of autosomal DMRs, leaving a final cohort of 23,116 samples that were used in downstream analysis of autosomal loci. We performed manual curation of epivariation calls by visual inspection of plots, identifying 102 loci that showed clear technical effects and were removed.
For analysis of DMRs on the X chromosome, due to the confounder of X chromosome inactivation that can result in highly variable β-values at many X-linked loci in females, we only considered male samples in our analysis. Furthermore, to ensure statistical robustness for detecting outlier events, we utilized chrX data only from the ten cohorts that each contained at least 300 males after performing all QC steps (total n = 8,027 males analyzed). Furthermore, due to hemizygosity for the X chromosome in males, which will result in stronger signals compared to heterozygous events on the autosomes, we increased thresholds for identifying DMRs on chrX to require three probes within a 1 kb window with a β-value difference to ≥99.5th percentile plus 0.4 for hypermethylated DMRs, and ≤0.5th percentile minus 0.4 for hypomethylated DMRs. Before summarizing (Tables S3 and S4), overlapping DMRs identified in different individuals, but which showed methylation changes in the same direction, were merged.
We annotated DMRs using the following data sources: (1) overlap with Refseq gene bodies and promoter regions (defined here as the region ± 2 kb of transcription start sites); (2) overlap with imprinted loci that exhibit significant parental bias in DNA methylation;27,28 (3) overlap with repetitive elements identified by RepeatMasker and Tandem Repeats Finder (RepeatMasker and Simple Repeats tracks downloaded from the UCSC Genome Browser); and (4) OMIM disease genes based on overlap with Refseq gene promoters. All enrichment analyses were performed using a background list of 38,646 1kb windows on the 450k array that contained three or more probes, which overlap 68.8% of the 457,201 autosomal probes on the 450k array.
Identification of Candidate Unstable Tandem Repeats
We utilized hipSTR29 to profile genome-wide variation of short tandem repeats (motif sizes 2–6 bp) in a cohort of 600 individuals who had undergone whole-genome sequencing using Illumina 150 bp paired-end reads, representing the parents of individuals with congenital heart defects (dbGaP: phs001138.v3.p2).
Validation of Rare Epigenetic Variants using Targeted Bisulfite Sequencing
We selected four epivariations located at the promoters of OMIM genes for secondary validation (LDLR, CCT5, PNPO, and PIK3R1). DNA samples from a carrier of each of these epivariations were bisulfite converted using the Epitect kit (QIAGEN), and PCR amplification of each locus was performed in all samples (Table S2). Amplicons were then barcoded, pooled in equimolar amounts, and sequenced with paired-end 150 bp reads using a Nano flowcell on an Illumina MiSeq instrument. Reads were mapped to the amplified regions ±2 kb of additional flanking sequence using BisMark30 (v.0.18.2) with default parameters. For each target region, we estimated percent methylation per CpG site by calculating the relative number of T (unmethylated) and C (methylated) nucleotides at each CpG position within the amplicon using samtools mpileup.31
Analysis of Monozygotic Twins
For concordance analysis of epivariations found in MZ twins, we generated β-value plots of each epivariation identified in any MZ twin and used these to manually categorize each locus as fully concordant, partially concordant, or discordant within each MZ twin pair.
Analysis of Gene Expression Data
Four of the cohorts utilized in this study had available gene expression data, as follows.
-
1.
BIOS study: We downloaded gene-level RNA-seq read counts for 3,560 samples made using HTSeq (EGA: EGAD00010001420).32 Read counts were normalized using DESeq2.33 We only considered autosomal genes with mean expression value in the top half of all genes assayed.
-
2.
MuTHER study: We used normalized expression values for 825 samples with expression in subcutaneous fat generated using the Illumina HumanHT-12 v3.0 Expression BeadChip (ArrayExpress: E-TABM-1140). Probe sequences were mapped using BWA, and only uniquely aligned probes were retained. We removed any probe that overlapped with single nucleotide variations (SNVs) identified by the 1000 Genomes Project that had minor allele frequency (MAF) > 0.01 in European populations and only considered autosomal genes with mean expression value in the top half of all genes assayed.
-
3.
MESA study: We used normalized expression values for 1,202 samples generated using the Illumina HumanHT-12 v4.0 expression beadchip (GEO: GSE56045). We removed any expression value with detection p > 0.01, removed probes with more than 10% missing values, and considered only autosomal genes with mean expression value in the top half of all genes assayed.
-
4.
Framingham Heart Study: We used normalized expression values for 2,198 samples generated using the Affymetrix GeneChip Human Exon 1.0 ST Array (dbGaP: phs000363.v5.p7). We considered only autosomal genes with mean expression value in the top half of all genes assayed.
In each cohort, we linked epivariations to corresponding expression data based on the overlap of epivariations with RefSeq gene promoters (as defined above), retaining only those genes that showed a unique mapping position with a single gene promoter. Normalized gene expression values were converted to both z-scores and ranks, and we compared expression data for samples carrying hypomethylated epivariations or hypermethylated epivariations against the entire population. p values were generated by randomly permuting expression values 10,000 times among samples and comparing the mean gene expression of these permuted values with the observed means of genes associated with epivariations.
cis-Association Analysis of Epivariations with SNVs
We used available SNV array data from 933 samples from the WHI cohort genotyped with the Illumina Multi-Ethnic Genotyping Array for whom methylation data were also available.
We performed pre-imputation quality control on the raw SNV array data which included removing multi-allelic sites, indels, resolving strand inconsistencies, and converting coordinates from hg18 to hg19, where applicable, using PLINK (v.1.07 and 2b3.43),34,35 vcftools (v.0.1.15),36 and Beagle utilities. We performed imputation and phasing in each of the datasets separately using Beagle 4.037 and the 1000 Genomes Project (1KGP) Phase3 reference panel downloaded from the Beagle website. For efficiency, genotype data from each chromosome were divided into segments of 5,000 SNVs for imputation, processed separately, and subsequently merged together for downstream analysis. We performed quality control on imputed and phased genotypes, removing SNVs with imputed R2 < 0.95, Hardy-Weinberg equilibrium p < 10−4, and multiallelic sites.
We selected 97 epivariations that were present in 2 or more individuals in the WHI cohort and performed a χ-square test using SNVs located within ±1 Mb around each epivariation, comparing allele frequencies between epivariation carriers and all other samples who did not carry that epivariation. We considered SNVs as significantly associated at 1% FDR.
Identification of Rare SNVs and CNVs Associated with Epivariations
In order to study the relationship of rare sequence variants with epivariations, we utilized samples from the PPMI cohort, which includes 457 Parkinson disease and control individuals in whom both PCR-free Illumina whole-genome sequencing data and DNA methylation data generated using the Illumina Infinium MethylationEPIC BeadChip (850k array) are available.23
We utilized SNV calls downloaded from the PPMI, considering rare SNVs (MAF < 0.1%) located within ±5 kb of the midpoint of each epivariation. To add specificity for potential regulatory sequences, we intersected these rare SNVs with transcription factor binding sites based on ChIPseq data generated by the ENCODE project,38 downloaded from the track “Transcription Factor ChIP-seq Peaks (338 factors in 130 cell types) from ENCODE 3” in the hg19 UCSC Genome Browser.
In order to identify rare copy number variations (CNVs) that were potentially associated with epivariations, we performed CNVnator analysis with a bin size of 100 bp and performed CNV calling using default parameters.39 Putative CNVs of length <2 kb or >1 Mb were removed. To avoid artifactual fragmentation of large CNVs into multiple smaller events, we merged multiple CNV calls in the same individual that shared the same direction of copy number change and were separated by <3 kb. We then focused on rare CNVs located within ±50 kb of each DMR that were observed in only a single individual in the PPMI cohort. For both rare SNVs and rare CNVs, we tested for a global enrichment of rare variants in epivariation carriers versus control subjects by considering all loci at which an epivariation was identified using a two-sided χ-square test, where controls were defined as any other PPMI sample which did not have an epivariation at the loci in question.
Validation of Repeat Expansions via Long-Read Sequencing
Pacific Biosciences long-insert libraries with the addition of barcodes were prepared for samples with epivariations at ABCD3 and PCMTD2, the two samples mixed at equimolar amounts, and sequenced on a single 8M SMRT cell with the Pacific Biosciences Sequel II system. Mean coverage was 12.5× and 9.1×, mean polymerase read lengths were 35.8 kb and 34.2 kb, and mean subread lengths were 10.7 kb and 10.0 kb for samples with epivariations of PCMTD2 and ABCD3, respectively. Subreads were aligned to the hg19 human reference genome using pbmm2 v1.0.040 with default parameters. Subreads were extracted from hg19 coordinates chr1:94,883,969–94,884,008 and chr20:62,887,069–62,887,108, and the number of CGG motifs were detected using the TR-specific dynamic programming algorithm PacmonSTR41 from the extracted subreads. We sequenced samples with epivariations at LINGO3 and FZD6 using Oxford Nanopore Technology, generating mean coverage of 3× and 27×, respectively. Reads were mapped to the hg19 human reference genome using minimap2 (v.2.7),40 and bam files for samples sequenced in multiple runs were merged, sorted, and indexed using samtools (v.1.7).31 To estimate methylation levels on normal and expanded CGG repeat alleles separately, we first separated reads in each sample based on the presence or absence of a CGG expansion. Using nanopolish (v.0.10.2),42 we created index files to link reads with their signal level data in FAST5 files, followed by estimation of DNA methylation status at each CpG located within 2 kb of CGG TRs, requiring a minimum log likelihood ratio ≥2.5 at each site.
Southern Blot, Repeat-Primed PCR, Methylation, and Expression Analysis in a Carrier of FRA22A
A Southern blot was created by digesting 8 μg DNA extracted from peripheral blood, using restriction enzymes HindIII and XbaI. The digested DNA was then separated by electrophoresis on a 0.7% agarose gel, and after denaturation and neutralization, transferred to Hybond N+ membranes. Hybridization was performed at 65°C using a specific probe generated by PCR (forward primer 5′-GCTGGAGAGGGAGGGAAGG-3′ and reverse primer 5′-ATAGAAACGAAGGCAAAGGAGACC-3′).
Repeat-primed PCR was performed to interrogate the number of CGG repeats in CSNK1E with the Asuragen CGG Repeat Primed PCR system designed for detection of fragile X expanded alleles. Samples were PCR-amplified using 2 μL of DNA sample (20 ng/μL), 11.45 μL of GC-rich AMP buffer, 0.25 μL of FAM-labeled CSNK1E forward primer F1 (5′-AGGCTGGGGAACTGCGTCT-3′) or FAM-labeled CSNK1E forward primer F2 (5′-GAGAGCCCAGAGCCAGAGC-3′), 0.25 μL of CSNK1E reverse primer R3 (5′-CAAAAACAAAGAGGCTGAGGGAG-3′), 0.5 μL of CGG primer (5′-TACGCATCCCAGTTTGAGACGGCCGCCGCCGCCGCC-3′), 0.5 μL of nuclease-free water, and 0.05 μL of GC-rich polymerase mix from Asuragen Inc. Samples were amplified with an initial heat denaturation step of 95°C for 5 min, followed by 10 cycles of 97°C for 35 s, 62°C for 35 s, and 68°C for 4 min, and then 20 cycles of 97°C for 35 s, 62°C for 35 s, and 68°C for 4 min, with a 20 s auto extension at each cycle. The final extension step was 72°C for 10 min. After PCR, 2 μL of the PCR product was added to a mix with 11 μL formamide and 2 μL Rox 1000 size standard (Abbott). After a brief denaturation step, samples were analyzed using an ABI Prism 3130 Genetic Analyzer (Applied Biosystems).
DNA methylation analysis was performed using bisulphite treatment with the Epitect bisulfite kit (QIAGEN). Primers specific for the methylated bisulphite-converted DNA (5′-GAGGAGGAGGGGGTTTGTTAT-3′ and 5′-AAATCAATAACCTAATAACCACACAC-3′) were designed using Methyl Primer Express (Applied Biosystems). After PCR amplification, the CGG surrounding area was sequenced using the forward primer on an ABI Prism 3130 (Applied Biosystems). We performed pyrosequencing to quantify the methylation using the CSNK1E_001 PyroMark CpG assay and analyzed the results on a PyroMark Q24 (QIAGEN). Methylation threshold values used were 10%.
Quantitative RT-PCR analysis was used to assess expression levels of CSNK1E. After homogenizing cultured lymphoblastoid cells from the FRA22A-expressing individual in triplicate and from nine control individuals, total cellular RNA was isolated using Trizol (Invitrogen) according to the manufacturer’s instructions, with RNase-free DNase treatment (Ambion). Subsequently, cDNA was reverse transcribed from total patient and control RNA samples using random hexamers primers from the SuperScript III First-Strand Synthesis System for RT-PCR kit (Invitrogen) according to manufacturer’s guidelines. Genomic contamination of the cDNA was checked with 2 control primers (5′-ATAGTCACCCCATTCAAACTCAAG-3′ and 5′-ATTCATAGCAGCAGCATTTGTTTTA-3′), spanning a large intron. First-strand cDNA was diluted in TE buffer to a concentration of 20 ng/μL. Primers were designed to span the exon-exon junction between protein coding exons 6 and 7 of CSNK1E (5′-TCAGCGAGAAGAAGATGTCAAC-3′ and 5′-GTAGGTAAGAGTAGTCGGGC-3′), and mRNA expression assayed with a two-step real-time quantitative PCR assay with qPCR MasterMix Plus w/o UNG with SYBR Green I No Rox (Eurogentec S.A) using a Lightcycler 480 Instrument (Roche Applied Science). Cycling conditions were as follows: an UNG step of 2 min 50°C, 10 min 95°C, and 45 cycles at 95°C for 15 s and 60°C for 1 min. Subsequently, specificity of the amplification was checked using a melting curve analysis by rapid heating to 97°C to denature the DNA (11°C/s), followed by cooling to 65°C (0.4°C/s). The protocol was terminated with a cooling step of 10 s at 40°C. All samples were assayed in triplicate. Expression of CSNK1E was normalized against the geometric mean of three stably expressed reference genes (B2M, GAPDH, and YWHAZ), and a Mann Whitney U test was used to assess statistical significance.
Results
Using a sliding window approach to identify regions containing ≥3 CpGs on the 450k array with outlier methylation levels (see Material and Methods), we identified 13,879 curated autosomal epivariations in 7,653 individuals, and 26 chrX epivariations in 26 males. Overall, 33.1% of the 23,116 samples tested carried one or more epivariations, corresponding to 4,452 unique autosomal loci and 18 unique chrX loci (Tables S3 and S4). Table S5 shows the underlying probe-level data per sample for each epivariation we identified, while the distributions of methylation levels and differences versus the population average across all epivariations are summarized in Figure S1. We observed an ∼2.3-fold excess of hypermethylated compared to hypomethylated epivariations: of the autosomal loci, 3,095 epivariations were gains of methylation, 1,329 epivariations were losses, while 28 autosomal epivariations were bidirectional, exhibiting either hyper- or hypomethylation in different samples.
Given the size of our cohort, we were able to estimate the population frequency of each epivariation (Tables S3 and S4), including several that have been described previously and/or are associated with disease. For example, the second most frequent epivariation we observed was hypermethylation of the promoter region of FRA10AC1 (MIM: 608866), with a population frequency of ∼1 per 325 individuals. This epivariation is known to be caused by expansion of an underlying CGG repeat which causes silencing of FRA10AC1, and in heterozygous form is thought to be a benign variant.43 Similarly, we observed gains of methylation at DIP2B with a frequency of ∼1 per 1,050 samples, and XYLT1 (MIM: 608124) in ∼1 per 2,100 samples. Both of these events are also caused by underlying expansions of CGG repeats and have been associated with intellectual disability19 and recessive Desbuquois dysplasia 2, respectively.15 Other known epivariations we observed include promoter methylation of MMACHC, which can cause recessive inborn errors of vitamin B12 metabolism9 and which we observed at a population frequency of ∼1 in 950. The frequency distribution of hyper- and hypomethylated autosomal epivariations is shown in Figure S2.
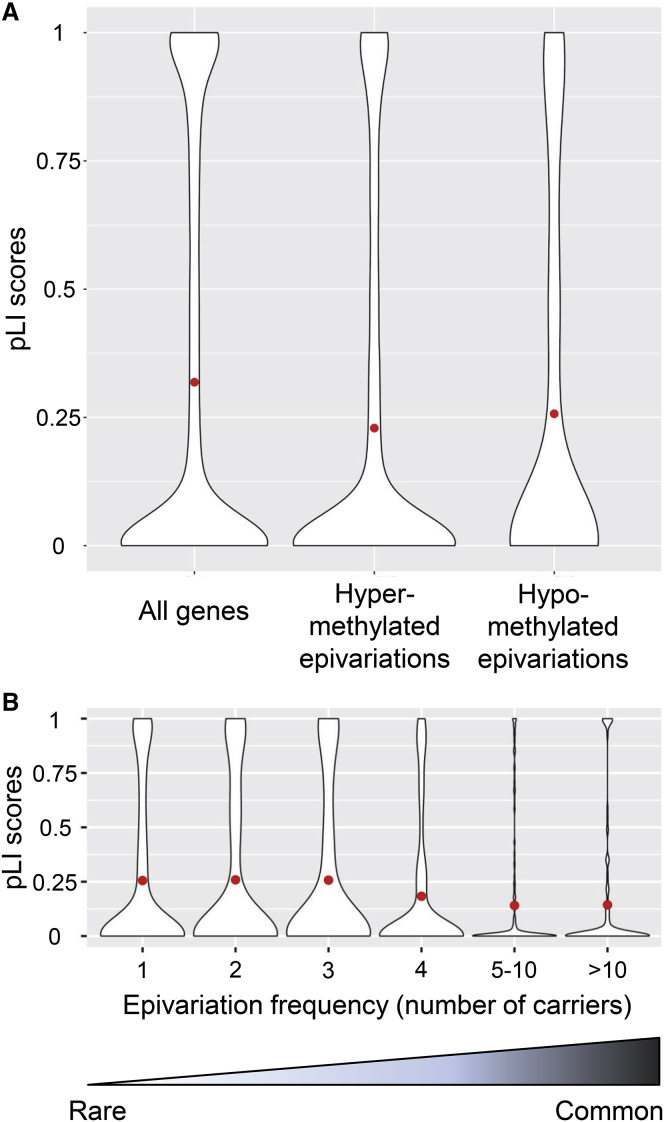
2,723 (61.2%) of 4,452 epivariations overlapped broad gene promoter regions (±2 kb of transcription start site), including 499 (402 hypermethylated, 91 hypomethylated, and 6 bidirectional epivariations) that overlapped promoter regions of OMIM disease genes (Tables S3 and S4). We observed evidence suggestive of purifying selection operating on promoter-associated epivariations (Figure 1). Using pLI scores generated by the Exome Aggregation Consortium (ExAC),5 hypermethylated promoter epivariations were biased away from the promoters of genes under selective constraint (permutation p < 10−7). Similarly, hypomethylated epivariations also showed bias away from constrained genes (permutation p = 1.6 × 10−3), but to a lesser degree than hypermethylated epivariations. We also observed a weak but significant inverse relationship between the population frequency of hypermethylated promoter epivariations and selective constraint of the associated genes (Pearson r = −0.11, p = 1.8 × 10−6) (Figure 1B).
Figure 1.
Evidence of Purifying Selection Operating on Promoter Epivariations
(A) Using pLI scores generated by the ExAC,5 we observed that hypermethylated promoter epivariations were preferentially associated with genes showing reduced selective constraint (permutation p < 10−7). Similarly, hypomethylated epivariations were also biased away from genes with high pLI scores, but to a lesser degree (permutation p = 1.6 × 10−3).
(B) We observed an inverse relationship between the population frequency of hypermethylated promoter epivariations and selective constraint of the associated gene (Pearson p = 1.8 × 10−6). These distributions are consistent with many promoter epivariations undergoing purifying selection which acts to reduce their frequency in the population. The red dot shows the mean pLI score in each distribution.
Epivariations Are Frequently Associated with cis-Linked Changes in Gene Expression
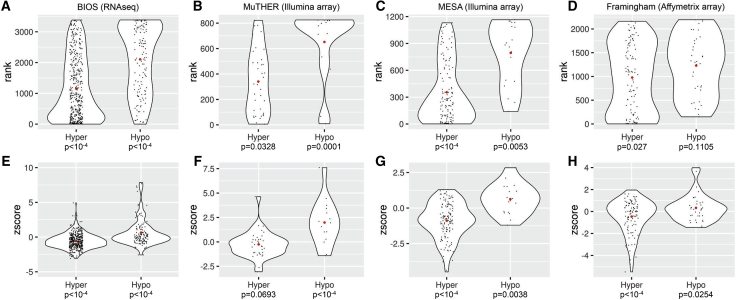
To determine the functional effect of epivariations on local gene expression, we analyzed available gene expression data in four different cohorts, comprising a total of 7,786 samples, analyzed with three different expression platforms. Focusing on epivariations that occurred at the promoter regions of genes, we observed significantly altered gene expression levels associated with epivariations in every cohort (Figure 2). Consistent with the known repressive effects of promoter methylation,44 promoter hypomethylation was associated with increased expression in all four cohorts tested, while promoter hypermethylation was associated with reduced repression (Tables S6, S7, S8, and S9).
Figure 2.
Promoter Epivariations Are Associated with Altered Gene Expression
Rank distributions of expression for genes with promoter epivariations (A–D) and z-scores of expression for genes with promoter epivariations (E–H). Using available gene expression data in four different cohorts, we observed that epivariations located at promoter regions were associated with altered gene expression levels. Consistent with the known repressive effects of promoter DNA methylation, promoter hypermethylation was associated with transcriptional repression and promoter hypomethylation with increased expression. In each violin plot, the red dot shows the mean expression value of each distribution, with individual genes shown as black dots. Above each plot is shown the cohort name and expression platform. All p values were calculated by permutation testing.
We also performed similar tests of the effect on expression of epivariations that either overlapped gene bodies (excluding promoter regions) and for the effects of intergenic epivariations on the closest gene. Using RNA-seq data from the BIOS cohort, we observed small but nominally significant effects of gene body methylation (p < 0.05), which showed a weak positive correlation with expression (Figure S3). However, using closest gene annotations, we observed no significant associations.
Epivariations and Known Disease Genes
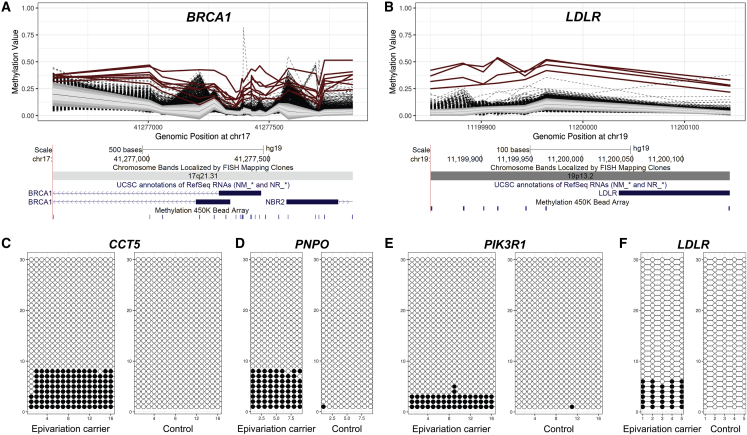
To gain insight into the potential contribution of epivariations to human disease, we utilized OMIM disease gene annotations, identifying 384 autosomal OMIM genes with hypermethylated epivariations at their promoter regions that may result in allelic silencing (Table S3). This includes 7 of 59 genes in which pathogenic mutations are considered medically actionable by the American College of Medical Genetics.45 For example, we detected seven individuals with promoter methylation of BRCA1, which has previously been reported in pedigrees with hereditary breast/ovarian cancer who lack pathogenic coding mutations in BRCA1,7,8 and four individuals with promoter methylation of LDLR (MIM: 606945), haploinsufficiency of which is associated with familial hypercholesterolemia (Figure 3). We selected four loci where we observed gains of methylation at the promoter regions of OMIM disease genes for secondary validation using amplicon bisulfite sequencing, obtaining between 59,732 and 170,388 reads per locus in each sample. At all four loci tested, the individual identified from array data as carrying a putative epivariation showed an elevated methylation level compared to control subjects, therefore confirming our predictions of gains of methylation at these loci (Figure 3, Table S2).
Figure 3.
Multiple Individuals with Epivariations at the Promoters of BRCA1 and LDLR and Secondary Validation by Bisulfite Sequencing
Coding mutations of BRCA1 and LDLR are associated with familial breast/ovarian cancer and with familial hypercholesterolemia, respectively. Loss-of-function mutations in both genes are considered highly penetrant and medically actionable and are recommended for return to patients when identified as incidental or secondary findings in clinical sequencing under current guidelines published by the American College of Medical Genetics and Genomics.45
(A and B) We identified (A) promoter hypermethylation of BRCA1 in seven individuals and (B) promoter hypermethylation of LDLR in four individuals, yielding population estimates for these epivariations of approximately 1 per 3,300 and 1 per 5,800 individuals, respectively. In each methylation plot, individuals with epivariations are shown as bold red lines, while the other ∼23,000 samples tested are shown as dashed black lines. Grey shaded regions indicate 1, 1.5, and 2 standard deviations from the mean of the distribution, while the solid black line shows the mean β-value of the entire population. Below each methylation plot are screenshots from the UCSC Genome Browser showing the relevant genomic region, genes, and the location of CpGs assayed by probes on the Illumina 450k array.
(C–F) Validation of array results of promoter hypermethylation events at four OMIM disease genes. Each plot shows 30 randomly selected reads from bisulfite PCR and sequencing of the promoter regions of CCT5 (MIM: 610150), PNPO (MIM: 603287), PIK3R1 (MIM: 171833), and LDLR. Filled circles show methylated CpG sites, while open circles show unmethylated CpGs. At each locus, the individual with the epivariation identified by array profiling shows a fraction of methylated reads that are absent in control subjects, which is consistent with methylation on one of their two alleles. However, we note that at all four loci the methylation fraction reported by bisulfite PCR/sequencing was lower than that predicted by array, which likely indicates a consistent PCR bias against amplification of the methylated allele.
Segregation of Epivariations with Local Sequence Variants
We hypothesized that some epivariations might represent secondary events caused by underlying genetic variation. We performed two complementary analyses to study this, investigating both common and rare variation.
First, we asked whether some epivariations segregate within the population on specific haplotype backgrounds.7,9 Using data from 933 individuals from the Women’s Health Initiative in whom both methylation and single-nucleotide variation (SNV) data derived from bead arrays were available, we identified 97 epivariations that were present in at least two individuals, and performed association analysis of these with local SNVs. Overall, using a stringent statistical threshold (1% FDR), 68 of the 97 epivariations tested (70%) showed at least one significantly associated SNV (Figure 4, Table S10). There was a trend for significantly associated variants to be located in close proximity to the epivariation, and in many cases the region of significantly associated SNVs directly overlapped the epivariation. These results indicate that many epivariations result from genetic variants located within their immediate vicinity.10 However, in a few cases, significant associations occurred with SNVs located >500 kb away, suggesting that some genetic variants can disrupt epigenetic regulation over large distances in cis (Figure S4).
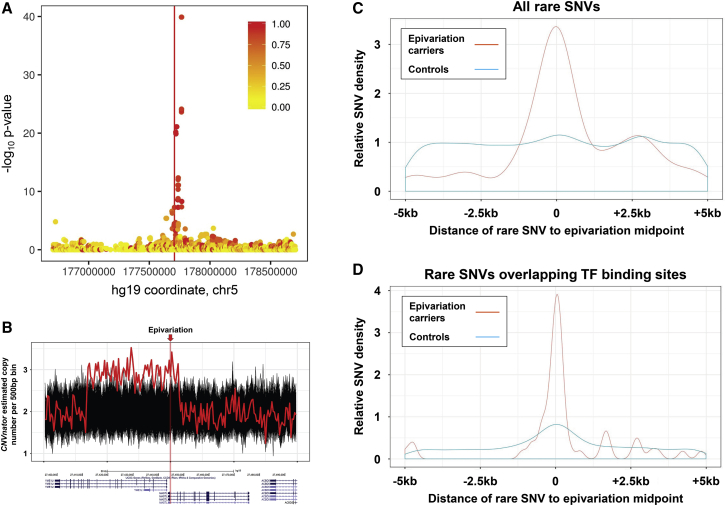
Figure 4.
Genetic Association and Rare Variant Analyses Provide Insight into the Origins of Epivariations
(A) Many epivariations are associated with local sequence variation, indicating an underlying genetic cause. Using data from 933 individuals from the Women’s Health Initiative in whom both methylation and SNV data were available, we performed association analysis with local SNVs for each recurrent epivariation. Shown are results for an epivariation located intronic within COL23A1 (MIM: 610043) (chr5:177,707,036–177,707,227, hg19) that showed a strong association with local SNVs. Eight of the nine epivariation carriers were heterozygous for a cluster of 12 SNVs (lead SNV rs73346815, p = 1.3 × 10−40, χ-square test), each of which had minor allele frequency of only 1.3% in non-carriers. The region of significant association directly overlaps the epivariation, indicating that this epivariation is likely a secondary event that occurs as a result of genetic variation segregating on a local haplotype. The color of each point indicates the fraction of epivariation carriers that carry the associated allele, while the location of the epivariation is indicated by the vertical red line. Examples of other epivariations with significantly associated SNVs are shown in Figure S4.
(B) Using CNVnator, we identified rare CNVs located adjacent to or overlapping the epivariation in ten carrier individuals, representing a 37.4-fold enrichment for rare CNVs to co-occur with an epivariation (p = 2 × 10−71, Χ-square test). The plot shows one of these loci, a ±50 kb region around an epivariation located at the bidirectional promoter region of YME1L1 (MIM: 607472) and MASTL (MIM: 608221). Each line shows estimated diploid copy number per 500 bp interval in 457 individuals from the PPMI cohort, with the epivariation carrier, who has a 36.4 kb duplication, shown in red, and all other individuals who do not carry the epivariation in black. The location of the epivariation is indicated by a vertical red bar. Below the plot is a screenshot from the UCSC Genome Browser showing gene annotations in the region. Other rare CNVs found in association with epivariations are shown in Figure S5.
(C) We observed a strong enrichment for rare SNVs to co-occur with rare epivariations, with 33 of 371 epivariations (8.9%) containing one or more rare SNVs within ±500 bp of the epivariation midpoint (10.7-fold enrichment, p = 2.6 × 10−63, χ-square test).
(D) This enrichment was even stronger when considering rare SNVs that overlapped transcription factor binding sites (12.3-fold enrichment, p = 2.3 × 10−70, Χ-square test). Full details of all rare SNVs identified in the PPMI cohort within ±500 bp of epivariation midpoints are shown in Table S12.
Second, as association analysis using array-based genotypes typically gives limited insight into the effect of rare variants, we investigated whether some epivariations might be attributable to rare SNVs or CNVs that disrupt local regulatory elements.10 Using data from 457 individuals from the PPMI cohort in whom both methylation and WGS data were available, we first identified 371 unique epivariations within this cohort (Table S11), and then related these to the presence of rare SNVs located within ±5 kb, and rare CNVs located within ±50 kb. We observed a clear enrichment for rare SNVs to co-occur in the immediate vicinity of rare epivariations (Figure 4), with 33 of 371 epivariations (8.9%) containing one or more rare SNVs within ±500 bp of the epivariation midpoint. This represents a 10.7-fold enrichment for rare SNVs in epivariation carriers compared to the background frequency of rare variants at these same loci in other individuals from the PPMI cohort (p = 2.6 × 10−63, χ-square test). Furthermore, this enrichment was even stronger when considering rare SNVs that overlapped transcription factor binding sites (12.3-fold enrichment, p = 2.3 × 10−70) (Table S12). Similarly we also observed a significant enrichment for rare CNVs to co-occur with the presence of an epivariation. We identified ten epivariation carriers that had rare deletions or duplications located within ±50 kb of the epivariation (Figure S5, Table S13). Compared to the background frequency of rare CNVs at these same loci in other samples from the PPMI cohort who did not carry an epivariation, this represents a 37.4-fold enrichment for rare CNVs to co-occur with an epivariation (p = 2 × 10−71). Overall, these data suggest that ∼8% of epivariations result from the presence of an underlying rare SNV, and ∼3% result from rare CNVs that occur within the immediate vicinity.
Epivariations Are Frequently Discordant in Monozygous Twins
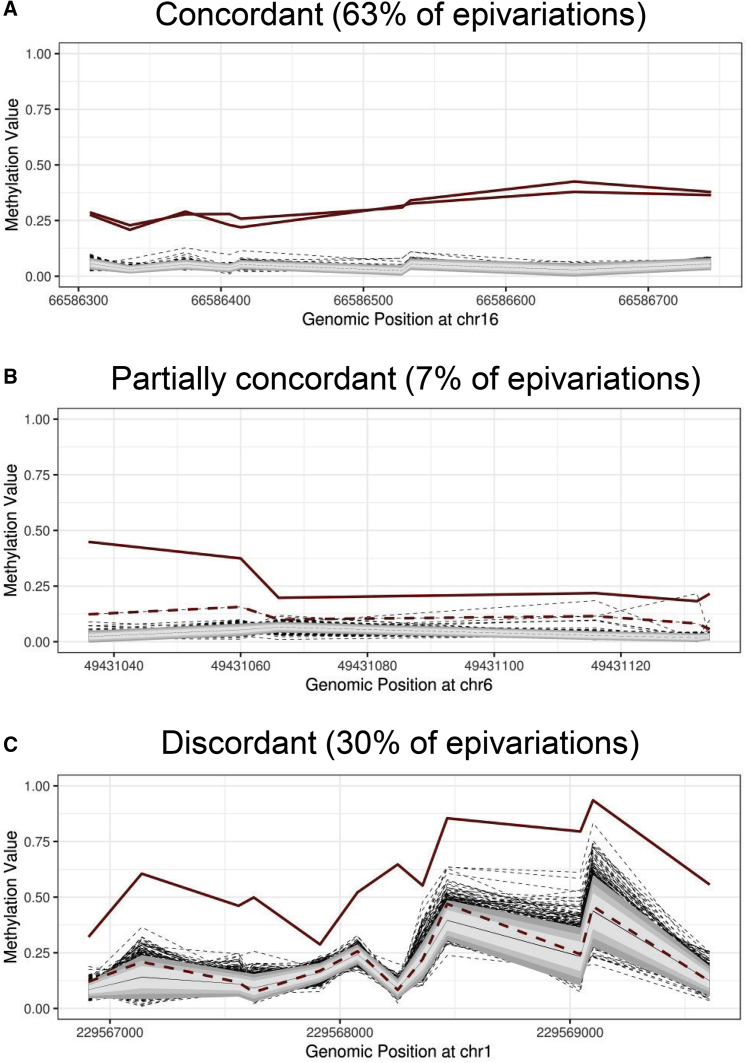
As MZ twins arise from the splitting of a single embryo post-fertilization, they provide a unique opportunity to gain insights into the developmental origins of epivariations. Our study population included 700 pairs of MZ twins derived from four different cohorts, and we identified a total of 333 loci where epivariations were identified in one or both of these MZ twins. Manual curation of these events showed that while 63% were concordant (i.e., both members of the MZ twin pair carried the same epivariation), 30% showed complete discordance (where one twin carried the epivariation and the second twin showed a normal methylation pattern at that locus), and 7% were scored as partially concordant (where one twin carried the epivariation and the second twin showed an outlier methylation profile at that locus, but of reduced magnitude) (Table S14). Examples of these three categories are shown in Figure 5. Overall, these observations indicate that approximately one third of epivariations are somatic events that occur post-zygotically.
Figure 5.
Twin Studies Suggest Many Epivariations Have a Post-zygotic Origin
One third of epivariations are discordant in monozygotic twin pairs, suggesting a high frequency of somatic epivariation. We identified 333 epivariations in 700 pairs of MZ twins. Of these, 63% were concordant with both members of the twin pair carrying the same epivariation, 7% were partially concordant, with one twin carrying the epivariation and the second twin being a weak outlier, and 30% were discordant with only one of the two twins carrying the epivariation.
(A) chr16:66,586,308–66,586,746, located at the promoter of CKLF (MIM: 616074).
(B) chr6:49,431,037–49,431,136, located at the bidirectional promoter of CENPQ (MIM: 611506) and MMUT (MIM: 609058), the latter of which is associated with methylmalonic aciduria.
(C) chr1:229,566,907–229,569,609, overlapping ACTA1 (MIM: 102610), which is associated with a number of congenital myopathies. In each methylation plot, the two members of an MZ twin pair are shown as bold red lines. Individuals formally identified as carrying an epivariation by our sliding window analysis are shown as solid red lines, while a member of the twin pair who was not formally identified as carrying this epivariation are shown as dashed red lines. All other samples from the cohort are shown as dashed black lines, with the gray shaded regions indicating 1, 1.5, and 2 standard deviations from the mean of the distribution, while the solid black line shows the mean β-value of the entire population.
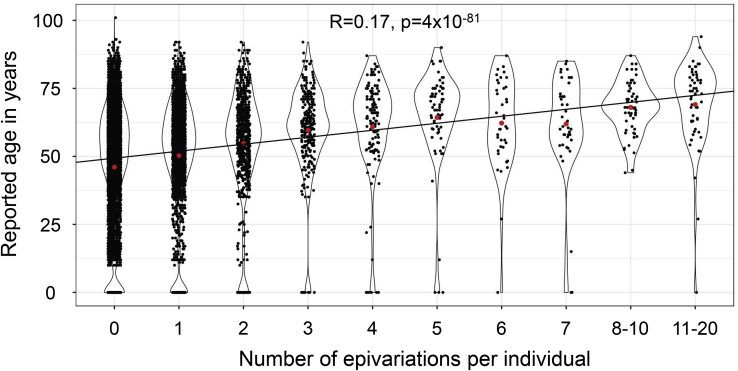
Epivariations Are More Common with Age
Using 11,690 samples with reported age at sampling, we observed a significant trend for the number of epivariations identified per individual to increase with age (Spearman r = 0.17, p = 4 × 10−81) (Figure 6). Consistent with this, MZ twin pairs who were fully or partially discordant for an epivariation (mean age 36.5 years) were significantly older than MZ twins with concordant epivariations (mean age 26.2 years) (p = 0.002, two-sided t test). These observations suggest that some epivariations are sporadic somatic events that accumulate with age.
Figure 6.
Epivariations Are More Common with Age
Using 11,690 individuals with reported age at sampling and methylation profiled in blood, we observed a significant trend for the number of epivariations per individual to increase with age (Spearman r = 0.17, p = 4 × 10−81). The red dot in each violin shows the group mean.
Epivariations at Imprinted Loci
Epivariations at imprinted loci exhibited a frequency profile that differed from the overall genomic distribution of epivariations in several ways. (1) Imprinted loci were more prone to epivariations, showing a 4.2-fold increase in epivariations compared to non-imprinted loci (p = 8.5 × 10−22, hypergeometric test). (2) Loss of methylation defects predominate over gains of methylation at imprinted loci: 60% of epivariations at imprinted loci were hypomethylation events, representing a 2.3-fold increase compared to the rest of the genome (p = 6.3 × 10−45, hypergeometric test) (Figure S6). (3) Consistent with their hemi-methylated nature, epivariations at imprinted loci were 85-fold enriched for bi-directional changes compared to the entire genome (p = 1.2 × 10−24, hypergeometric test) (Figure S7).
Prediction and Validation of CGG Expansions at Hypermethylated Epivariations
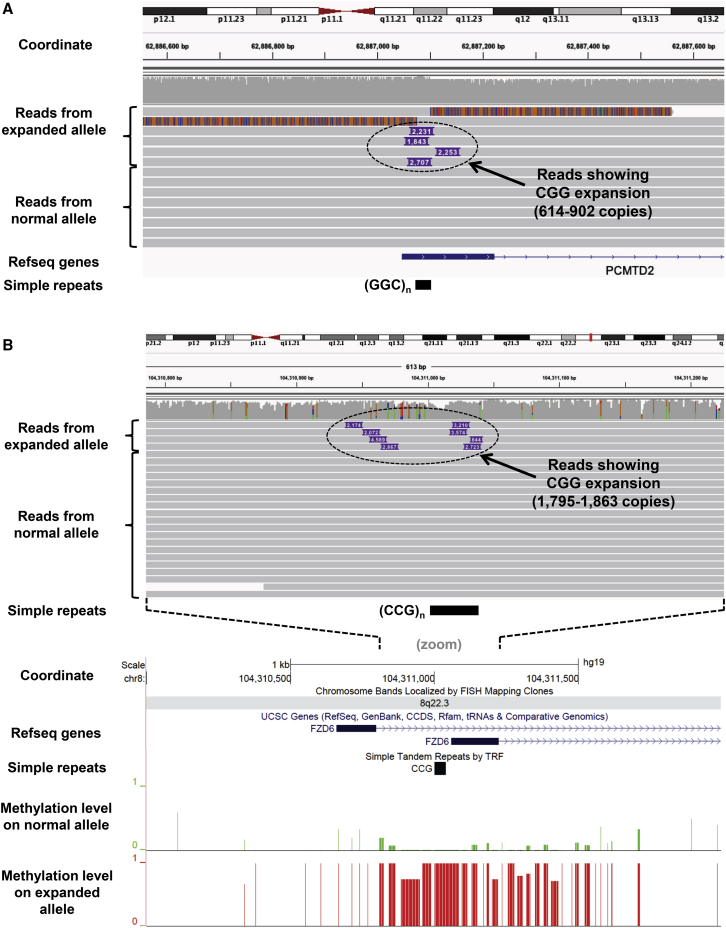
Using tandem repeat (TR) genotypes generated by hipSTR in 600 unrelated individuals who had undergone Illumina WGS, we observed that TRs that are known to undergo occasional expansion in human disease tend to show extremely high levels of polymorphism in the general population (Figure S8). For example, nearly all known pathogenic TRs had ≥10 different alleles in the 600 genotyped samples, placing them in the top 3% of the most polymorphic TRs in the genome. Thus, we hypothesized that we could use high levels of population variability to predict unstable TRs in the human genome that are prone to occasional expansion. Based on this approach, we identified 180 TRs with motif size of 3–6 bp and 100% GC-content that each showed ≥10 different alleles in our cohort of 600 sequenced individuals. Intersection of these potentially unstable GC-rich TRs with hypermethylated epivariations yielded 25 overlaps. This included six TRs that were already known to undergo rare expansion and hypermethylation (FRA10AC1, C11orf80 [MIM: 616109], CBL [MIM: 165360], C9orf72 [MIM: 614260], DIP2B, and TMEM185A [MIM: 300031])15,18,19,43,46, 47, 48 and highlighted 19 additional epivariations that we hypothesized might be caused by previously unidentified TR expansions (Table S15). Plots of all methylation profiles of epivariations overlapping putatively unstable CG-rich TRs are shown in Figure S9.
To investigate whether these epivariations were attributable to expansions of an underlying TR, we obtained DNA samples from four individuals in whom we had identified hypermethylated epivariations overlapping putatively unstable CGG repeats and performed long-read WGS using either Pacific Biosciences SMRT sequencing or Oxford Nanopore Technology (ONT). In all four samples tested, we confirmed the presence of a heterozygous TR expansion comprising several hundred copies of CGG at the epivariation (Figure 7, Table S15), thus identifying hypermethylated CGG expansions within the promoter/5′ UTR regions of ABCD3 (MIM: 170995), FZD6/LOC105369147 (MIM: 603409), LINGO3 (MIM: 609792), and PCMTD2. Furthermore, by analyzing the signal profiles of phased ONT reads, we demonstrated that in an individual with hypermethylation of FZD6, the expanded TR allele was highly methylated, while the normal TR allele was largely unmethylated (Figure 7B), thus showing that this epivariation represents monoallelic hypermethylation associated with a CGG expansion.
Figure 7.
Validation of CGG Expansions Underlying Epivariations using Long Read Sequencing
We obtained DNA samples from individuals with hypermethylated epivariations at the promoter/5′ UTR regions of PCMTD2 and FZD6, both of which contained putatively unstable CGG repeats.
(A) Pacific Biosciences SMRT sequencing confirmed the presence of a heterozygous repeat expansion at PCMTD2 composed of 614–902 CGG motifs, suggesting considerable somatic variation in the size of the expanded allele.
(B) Sequencing with Oxford Nanopore Technology confirmed the presence of a heterozygous repeat expansion at FZD6, composed of 1,795–1,863 CGG motifs. By analyzing the signal profiles of the reads after phasing based on presence/absence of the expansion, we demonstrated that the expanded allele was highly methylated, while the normal TR allele was largely unmethylated. We consistently observed <50% of reads coming from the expanded allele at each locus, which we believe likely represents an inherent difficulty in sequencing across expanded CGG repeats. In support of this, we observed multiple reads that returned low-quality sequence after traversing the CGG tract, visible as mis-aligned segments of reads shown at top of IGV screenshot of PCMTD2 locus.
Multiple folate-sensitive fragile sites (FSFS) in the human genome are known to be caused by underlying CGG expansions, including FRA2A (AFF3), FRA7A (ZNF713 [MIM: 616181]), FRA10A (FRA10AC1), FRA11A (C11orf80), FRA11B (CBL), FRA12A (DIP2B), FRA16A (XYLT1), FRAXA (FMR1), FRAXE (AFF2), and FRAXF (TMEM185A).16, 17, 18, 19, 20,43,46,47,49, 50, 51 We thus hypothesized that CGG expansions might underlie other FSFS. Consistent with this, 8 of the 25 putative or validated CGG repeat expansions we identified coincide with the cytogenetic location of other rare FSFS that to date have not been molecularly mapped,16 strongly suggesting that these epivariations likely represent the unstable CGG repeats that are responsible for the FSFS FRA1M (ABCD3), FRA2B (BCL2L11 [MIM: 603827]), FRA5G (FAM193B [MIM: 615813]), FRA8A (FZD6), FRA9A (C9orf72), FRA19B (LINGO3), FRA20A (RALGAPA2), and FRA22A (CSNK1E [MIM: 600863]) (Table S15).
To formally test whether this approach accurately identifies CGG expansions underlying FSFS, we obtained DNA from an individual who expressed the FSFS FRA22A but who was not part of our discovery cohort. Our epivariation analysis had identified six individuals with a gain of methylation overlapping the 5′ UTR of CSNK1E, a region that includes a highly polymorphic CGG repeat and lies within 22q13.1, the cytogenetic band to which FRA22A has been mapped. Thus, based on our epivariation and TR data, we predicted that expansions of this CGG repeat within CSNK1E likely underlie the FRA22A fragile site, which was subsequently confirmed by several complementary experiments. (1) Repeat primed-PCR of the CGG repeat52 in the individual with the FRA22A fragile site showed a characteristic saw-tooth pattern on the fluorescence trace, with periodicity of 3 bp, indicative of a triplet repeat expansion. (2) Subsequent Southern blot in the FRA22A carrier identified a novel smeared fragment of approximately 3.2 kb, in addition to the expected fragment of 2.2 kb, which, together with the PCR result, indicate the presence of an expanded CGG tract of approximately 340 repeats. (3) Analysis of CpGs in the promoter of CSNK1E using both bisulfite sequencing and pyrosequencing showed methylation levels of 40%–50% in the FRA22A carrier, while control samples were unmethylated. (4) Finally, using real-time RT-PCR in lymphoblastoid cells, we observed that in the FRA22A carrier, expression of CSNK1E was reduced to ∼37% of the levels observed in control subjects (Figure S10). Overall, these results indicated that expansion of a CGG repeat in the 5′ UTR of CSNK1E results in allelic methylation and silencing of the gene and represents the molecular defect underlying the FRA22A FSFS.
Discussion
Our large-scale survey of epivariations in >23,000 individuals represents the largest cohort of methylomes assembled to date, providing a comprehensive catalog of epivariations that are found in the human population. While a handful of previous studies have identified epivariations as causative factors in some human genetic diseases, here we identified promoter epivariations at hundreds of genes that are known to cause genetic disease, suggesting that epivariations may contribute to the mutational spectra underlying many Mendelian disorders. Using available expression data, we show that many of these epivariations exert functional effects on the genome, with promoter epivariations in particular being associated with significant alterations in gene expression. In previous work, we have shown that hypermethylated promoter epivariations are often associated with monoallelic expression, and thus can have an impact comparable to that of loss-of-function coding mutations.10 Based on this observation, we anticipate that epigenetic profiling in patients with overt genetic disease, but who lack pathogenic sequence mutations in the gene(s) relevant to their phenotype, will lead to the identification of epivariants as a causative factor in some conditions, and potentially providing additional diagnostic yield compared to purely sequence-based approaches.11
Through genetic association, rare variant analysis, and by studying patterns of epivariation in twin pairs and samples of different ages, we gained insights into the underlying mechanisms of epivariations. Association analysis using epivariations observed in multiple individuals suggests that ∼70% of epivariations segregate on specific haplotype backgrounds, indicating that the majority of epivariations are secondary events that occur downstream of stably inherited genetic variants. Analysis of rare SNVs and CNVs ascertained from WGS data indicated that ∼11% of epivariations are likely caused by rare variants within the immediate region of altered methylation. Our data therefore indicate that the majority of epivariations are secondary events resulting from underlying sequence variants that disrupt either the establishment and/or maintenance of the normal epigenetic state, such as mutations of regulatory elements and transcription factor binding sites.10,53 It is possible that some mutations might be low-penetrance events that predispose to gradual gain or loss of methylation during development, and might therefore result in somatic mosaicism. Contrastingly, analysis of MZ twins found that approximately one third of epivariations are discordant between genetically identical twins, indicating that a significant fraction of epivariations occur post-zygotically. This conclusion is further supported by the observations that (1) the incidence of epivariations increases with age, and (2) in MZ twins, discordant epivariations are observed more frequently in older versus younger twins. This suggests that many epivariations will likely exhibit somatic mosaicism and therefore, depending on their tissue distribution, might show attenuated or absent phenotypic effects, and/or reduced heritability between generations. In support of this latter prediction, we previously observed a significant reduction in heritability of epivariations between parents and offspring.10 This observation of reduced heritability is consistent either with some epivariations being mosaic events that are confined to somatic tissues and absent from the germline, and/or that some epivariations are primary events, i.e., sporadic errors that arise as a result of the epigenetic remodeling that occurs during cellular differentiation, and that undergo epigenetic reprogramming back to the default state during gametogenesis/early embryogenesis. In contrast, secondary epivariations that occur downstream of a sequence change will likely exhibit Mendelian inheritance.
We postulate that post-zygotic epivariations may represent either (1) primary epivariations or (2) secondary epivariations resulting from somatically acquired sequence mutations. Further work will be needed to distinguish these possibilities. However, even with twin studies and extensive analysis of rare and common sequence variation, as most epivariations are rare, and we only had access to variation data in a small fraction of our cohort, we emphasize that for the majority of epivariations that we describe it is difficult to determine their underlying etiology. Thus, although our studies do provide reasonable estimates of the relative proportion of epivariations that are attributable to local sequence variation or are somatic in origin, in most cases we are unable to state which specific epivariations might be primary events (i.e., purely sporadic defects unlinked to a change in DNA sequence) and we are only able to infer a small number that are almost certainly secondary events that occur downstream of an underlying sequence mutation.
While dysregulation of several different imprinted genes is associated with a number of developmental disorders,54 we found that epivariations at some imprinted loci were relatively common events (Table S3, Figures S6 and S7). Indeed, we observed that epivariations were enriched at imprinted loci in general and specifically for hypomethylation events. For example, the most frequent epivariation we observed in this study was at the HM13 imprinted locus, where our results indicate bi-allelic methylation occurring in ∼1 per 350 individuals and hypomethylation (loss of imprinting) occurring in ∼1 per 3,300. Relatively frequent imprinting defects (>1 per 1,000 individuals) were also observed at several other imprinted loci, such as FAM50B, L3MBTL1, SNU13, VTRNA2-1, and KCNQ1OT1, although in some cases these epivariations covered only part of the imprinted region. These data indicate that parent-of-origin specific methylation at some imprinted loci may be relatively labile.
We also identified CGG repeat expansions as the causative factor underlying a small subset of epivariations. Large expansions of CGG repeats are known to be associated with local DNA hypermethylation of the expanded allele, and have been found to underlie multiple rare folate-sensitive fragile sites in the genome.16 By combining our map of outlier hypermethylation events with predictions of unstable TRs, we identified 25 epivariations that we predicted as being caused by underlying CGG repeat expansions. Six of these loci represent previously identified TR expansions, thus both validating our approach and providing population estimates of the prevalence of these events, some of which are surprisingly frequent. For example, our data indicate that hypermethylated expansions at FRA10AC1 occur with a prevalence of ∼1 per 325 individuals. In order to assess the validity of our predictions for the 19 other loci containing CGG repeats, we obtained DNA samples from five individuals in whom we identified hypermethylation of the candidate loci and validated the presence of a heterozygous expanded repeat at all five of these loci in carrier individuals. Although we were unable to obtain DNA samples with putative expansions at the 14 other putatively unstable CGG TRs we identify, we suggest that these represent strong candidates for TR expansions. In support of this, several of these candidate loci coincide with the approximate location of rare FSFSs that have been cytogenetically mapped, suggesting that these candidate repeats represent the molecular defect underlying these FSFSs. While several hypermethylated CGG expansions are known to be associated with neurodevelopmental disorders,17, 18, 19, 20,49,51 the possible phenotypic consequences of the CGG expansions we identified will require further study. Given that many of these occur within the 5′ UTRs of genes, one intriguing possibility is that unmethylated premutation-sized alleles might predispose to late-onset neurodegenerative disease, similar to the fragile X tremor/ataxia syndrome that occurs in some carriers of FMR1 premutations.55 In direct support of this hypothesis, one of the candidate hypermethylated repeat expansions we identified was a CGG repeat located within the 5′ UTR of GIPC1 (MIM: 605072). A recent study reported heterozygous unmethylated moderate expansions (73–161 copies) of this same CGG repeat in patients with the adult-onset neuromuscular disorder oculopharyngodistal myopathy.56 Thus, although we have not yet shown that hypermethylation of GIPC1 is caused by large expansions of this CGG repeat, it seems likely that this locus behaves similarly to the CGG repeat in FMR1, in that intermediate “premutation” alleles can cause late-onset disease through a gain of function via overexpression of the expanded CGG repeat, while larger expansions become hypermethylated and inactive.
In an era where genome sequencing is being applied to millions of individuals, our results show that the study of epigenetic variation can provide additional insights into genome function.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by NIH grant NS105781 to A.J.S., NIH predoctoral fellowship NS108797 to O.R., and American Heart Association Postdoctoral Fellowship 18POST34080396 to A.M.T. R.F.K. acknowledges support of the Research Fund of the University of Antwerp (Methusalem-OEC grant – “GENOMED”). Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD018522. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Data used in the preparation of this article were obtained from the Parkinson’s Progression Markers Initiative (PPMI) database (see Web Resources). PPMI, a public-private partnership, is funded by the Michael J. Fox Foundation for Parkinson’s Research and funding partners, a full list of which can be found online.
The Biobank-Based Integrative Omics Studies (BIOS) Consortium is funded by BBMRI-NL, a research infrastructure financed by the Dutch government (NWO 184.021.007). The Parkinson disease patient and control study was funded by NIEHS grants ES024356, R01ES10544, and P01ES016732. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195 and HHSN268201500001I). Additional funding for SABRe was provided by Division of Intramural Research, NHLBI, and Center for Population Studies, NHLBI. The Women’s Health Initiative (WHI) program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study or WHI and does not necessarily reflect the opinions or views of the Framingham Heart Study, WHI investigators, Boston University, or NHLBI.
Published: September 15, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.019.
Web Resources
Array Express, https://www.ebi.ac.uk/arrayexpress/
Beagle, http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a/
Database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap/
Epivariation scripts, https://github.com/AndyMSSMLab/Epivariation-in-23K-samples
European Genome-phenome Archive, https://www.ebi.ac.uk/ega/home
Gene Expression Omnibus (GEO), https://www.ncbi.nlm.nih.gov/geo/
OMIM, https://www.omim.org/
UCSC Genome Browser, http://genome.ucsc.edu
Data and Code Availability
The code generated during this study is available at github (see Epivariation scripts in Web Resources). Original source data utilized in this study are listed in Table S1.
Supplemental Data
22 Cohorts of Illumina 450k Methylation Data Used in This Study
PCR Amplicons and Results of Targeted Bisulfite Sequencing of Four Epivariations
All Autosomal Epivariations Identified
All chrX Epivariations Identified
Individual-Level Data for All Probes within Epivariations
Gene Expression Data in BIOS Population
Gene Expression Data in MuTHER Population
Gene Expression Data in MESA Population
Gene Expression Data in Framingham Heart Study Population
Association of Epivariations with Local SNVs in the WHI Cohort
Autosomal Epivariations Identified in the PPMI Cohort from Analysis of 850k Methylation Array Data
Rare SNVs in DMR Carriers Located within ± 500 bp of DMR Midpoint in the PPMI Cohort
Rare CNVs Identified within ± 50 kb of DMRs in the PPMI Cohort
Concordance of Epivariations in MZ Twin Pairs
25 Epivariations Overlapping Putative Unstable CG-Rich Repeats
References
- 1.Belmont J.W., Boudreau A., Leal S.M., Hardenbol P., Pasternak S., Wheeler D.A., Willis T.D., Yu F., Yang H., Gao Y., International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., Donnelly P., Egholm M., 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H.Y., 1000 Genomes Project Consortium An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castillejo A., Hernández-Illán E., Rodriguez-Soler M., Pérez-Carbonell L., Egoavil C., Barberá V.M., Castillejo M.I., Guarinos C., Martínez-de-Dueñas E., Juan M.J. Prevalence of MLH1 constitutional epimutations as a cause of Lynch syndrome in unselected versus selected consecutive series of patients with colorectal cancer. J. Med. Genet. 2015;52:498–502. doi: 10.1136/jmedgenet-2015-103076. [DOI] [PubMed] [Google Scholar]
- 7.Evans D.G.R., van Veen E.M., Byers H.J., Wallace A.J., Ellingford J.M., Beaman G., Santoyo-Lopez J., Aitman T.J., Eccles D.M., Lalloo F.I. A dominantly inherited 5¢ UTR variant causing methylation-associated silencing of BRCA1 as a cause of breast and ovarian cancer. Am. J. Hum. Genet. 2018;103:213–220. doi: 10.1016/j.ajhg.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansmann T., Pliushch G., Leubner M., Kroll P., Endt D., Gehrig A., Preisler-Adams S., Wieacker P., Haaf T. Constitutive promoter methylation of BRCA1 and RAD51C in patients with familial ovarian cancer and early-onset sporadic breast cancer. Hum. Mol. Genet. 2012;21:4669–4679. doi: 10.1093/hmg/dds308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guéant J.L., Chéry C., Oussalah A., Nadaf J., Coelho D., Josse T., Flayac J., Robert A., Koscinski I., Gastin I. Publisher Correction: A PRDX1 mutant allele causes a MMACHC secondary epimutation in cblC patients. Nat. Commun. 2018;9:554. doi: 10.1038/s41467-018-03054-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barbosa M., Joshi R.S., Garg P., Martin-Trujillo A., Patel N., Jadhav B., Watson C.T., Gibson W., Chetnik K., Tessereau C. Identification of rare de novo epigenetic variations in congenital disorders. Nat. Commun. 2018;9:2064. doi: 10.1038/s41467-018-04540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garg P., Sharp A.J. Screening for rare epigenetic variations in autism and schizophrenia. Hum. Mutat. 2019;40:952–961. doi: 10.1002/humu.23740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horsthemke B. Epimutations in human disease. Curr. Top. Microbiol. Immunol. 2006;310:45–59. doi: 10.1007/3-540-31181-5_4. [DOI] [PubMed] [Google Scholar]
- 13.Buiting K., Gross S., Lich C., Gillessen-Kaesbach G., el-Maarri O., Horsthemke B. Epimutations in Prader-Willi and Angelman syndromes: a molecular study of 136 patients with an imprinting defect. Am. J. Hum. Genet. 2003;72:571–577. doi: 10.1086/367926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ligtenberg M.J.L., Kuiper R.P., Chan T.L., Goossens M., Hebeda K.M., Voorendt M., Lee T.Y.H., Bodmer D., Hoenselaar E., Hendriks-Cornelissen S.J.B. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3¢ exons of TACSTD1. Nat. Genet. 2009;41:112–117. doi: 10.1038/ng.283. [DOI] [PubMed] [Google Scholar]
- 15.LaCroix A.J., Stabley D., Sahraoui R., Adam M.P., Mehaffey M., Kernan K., Myers C.T., Fagerstrom C., Anadiotis G., Akkari Y.M., University of Washington Center for Mendelian Genomics GGC repeat expansion and exon 1 methylation of XYLT1 Is a common pathogenic variant in Baratela-Scott syndrome. Am. J. Hum. Genet. 2019;104:35–44. doi: 10.1016/j.ajhg.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debacker K., Kooy R.F. Fragile sites and human disease. Hum. Mol. Genet. 2007;16 Spec No. 2:R150–R158. doi: 10.1093/hmg/ddm136. [DOI] [PubMed] [Google Scholar]
- 17.Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S.T., Schlessinger D., Sutherland G.R., Richards R.I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 18.Knight S.J.L., Flannery A.V., Hirst M.C., Campbell L., Christodoulou Z., Phelps S.R., Pointon J., Middleton-Price H.R., Barnicoat A., Pembrey M.E. Trinucleotide repeat amplification and hypermethylation of a CpG island in FRAXE mental retardation. Cell. 1993;74:127–134. doi: 10.1016/0092-8674(93)90300-f. [DOI] [PubMed] [Google Scholar]
- 19.Winnepenninckx B., Debacker K., Ramsay J., Smeets D., Smits A., FitzPatrick D.R., Kooy R.F. CGG-repeat expansion in the DIP2B gene is associated with the fragile site FRA12A on chromosome 12q13.1. Am. J. Hum. Genet. 2007;80:221–231. doi: 10.1086/510800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metsu S., Rooms L., Rainger J., Taylor M.S., Bengani H., Wilson D.I., Chilamakuri C.S.R., Morrison H., Vandeweyer G., Reyniers E. FRA2A is a CGG repeat expansion associated with silencing of AFF3. PLoS Genet. 2014;10:e1004242. doi: 10.1371/journal.pgen.1004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holliday R. Mutations and epimutations in mammalian cells. Mutat. Res. 1991;250:351–363. doi: 10.1016/0027-5107(91)90192-q. [DOI] [PubMed] [Google Scholar]
- 22.Gicquel C., Rossignol S., Cabrol S., Houang M., Steunou V., Barbu V., Danton F., Thibaud N., Le Merrer M., Burglen L. Epimutation of the telomeric imprinting center region on chromosome 11p15 in Silver-Russell syndrome. Nat. Genet. 2005;37:1003–1007. doi: 10.1038/ng1629. [DOI] [PubMed] [Google Scholar]
- 23.Marek K., Jennings D., Lasch S., Siderowf A., Tanner C., Simuni T., Coffey C., Kieburtz K., Flagg E., Chowdhury S., Parkinson Progression Marker Initiative The Parkinson Progression Marker Initiative (PPMI) Prog. Neurobiol. 2011;95:629–635. doi: 10.1016/j.pneurobio.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 25.Teschendorff A.E., Marabita F., Lechner M., Bartlett T., Tegner J., Gomez-Cabrero D., Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Houseman E.A., Kelsey K.T., Wiencke J.K., Marsit C.J. Cell-composition effects in the analysis of DNA methylation array data: a mathematical perspective. BMC Bioinformatics. 2015;16:95. doi: 10.1186/s12859-015-0527-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Court F., Tayama C., Romanelli V., Martin-Trujillo A., Iglesias-Platas I., Okamura K., Sugahara N., Simón C., Moore H., Harness J.V. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zink F., Magnusdottir D.N., Magnusson O.T., Walker N.J., Morris T.J., Sigurdsson A., Halldorsson G.H., Gudjonsson S.A., Melsted P., Ingimundardottir H. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat. Genet. 2018;50:1542–1552. doi: 10.1038/s41588-018-0232-7. [DOI] [PubMed] [Google Scholar]
- 29.Willems T., Zielinski D., Yuan J., Gordon A., Gymrek M., Erlich Y. Genome-wide profiling of heritable and de novo STR variations. Nat. Methods. 2017;14:590–592. doi: 10.1038/nmeth.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krueger F., Andrews S.R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhernakova D.V., Deelen P., Vermaat M., van Iterson M., van Galen M., Arindrarto W., van ’t Hof P., Mei H., van Dijk F., Westra H.J. Identification of context-dependent expression quantitative trait loci in whole blood. Nat. Genet. 2017;49:139–145. doi: 10.1038/ng.3737. [DOI] [PubMed] [Google Scholar]
- 33.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang C.C., Chow C.C., Tellier L.C.A.M., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunham I., Kundaje A., Aldred S.F., Collins P.J., Davis C.A., Doyle F., Epstein C.B., Frietze S., Harrow J., Kaul R., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abyzov A., Urban A.E., Snyder M., Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–984. doi: 10.1101/gr.114876.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ummat A., Bashir A. Resolving complex tandem repeats with long reads. Bioinformatics. 2014;30:3491–3498. doi: 10.1093/bioinformatics/btu437. [DOI] [PubMed] [Google Scholar]
- 42.Simpson J.T., Workman R.E., Zuzarte P.C., David M., Dursi L.J., Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat. Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 43.Sarafidou T., Kahl C., Martinez-Garay I., Mangelsdorf M., Gesk S., Baker E., Kokkinaki M., Talley P., Maltby E.L., French L., European Collaborative Consortium for the Study of ADLTE Folate-sensitive fragile site FRA10A is due to an expansion of a CGG repeat in a novel gene, FRA10AC1, encoding a nuclear protein. Genomics. 2004;84:69–81. doi: 10.1016/j.ygeno.2003.12.017. [DOI] [PubMed] [Google Scholar]
- 44.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 45.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 46.Debacker K., Winnepenninckx B., Longman C., Colgan J., Tolmie J., Murray R., van Luijk R., Scheers S., Fitzpatrick D., Kooy F. The molecular basis of the folate-sensitive fragile site FRA11A at 11q13. Cytogenet. Genome Res. 2007;119:9–14. doi: 10.1159/000109612. [DOI] [PubMed] [Google Scholar]
- 47.Jones C., Slijepcevic P., Marsh S., Baker E., Langdon W.Y., Richards R.I., Tunnacliffe A. Physical linkage of the fragile site FRA11B and a Jacobsen syndrome chromosome deletion breakpoint in 11q23.3. Hum. Mol. Genet. 1994;3:2123–2130. doi: 10.1093/hmg/3.12.2123. [DOI] [PubMed] [Google Scholar]
- 48.Xi Z., Zinman L., Moreno D., Schymick J., Liang Y., Sato C., Zheng Y., Ghani M., Dib S., Keith J. Hypermethylation of the CpG island near the G4C2 repeat in ALS with a C9orf72 expansion. Am. J. Hum. Genet. 2013;92:981–989. doi: 10.1016/j.ajhg.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metsu S., Rainger J.K., Debacker K., Bernhard B., Rooms L., Grafodatskaya D., Weksberg R., Fombonne E., Taylor M.S., Scherer S.W. A CGG-repeat expansion mutation in ZNF713 causes FRA7A: association with autistic spectrum disorder in two families. Hum. Mutat. 2014;35:1295–1300. doi: 10.1002/humu.22683. [DOI] [PubMed] [Google Scholar]
- 50.Nancarrow J.K., Kremer E., Holman K., Eyre H., Doggett N.A., Le Paslier D., Callen D.F., Sutherland G.R., Richards R.I. Implications of FRA16A structure for the mechanism of chromosomal fragile site genesis. Science. 1994;264:1938–1941. doi: 10.1126/science.8009225. [DOI] [PubMed] [Google Scholar]
- 51.Hirst M.C., Barnicoat A., Flynn G., Wang Q., Daker M., Buckle V.J., Davies K.E., Bobrow M. The identification of a third fragile site, FRAXF, in Xq27--q28 distal to both FRAXA and FRAXE. Hum. Mol. Genet. 1993;2:197–200. doi: 10.1093/hmg/2.2.197. [DOI] [PubMed] [Google Scholar]
- 52.Filipovic-Sadic S., Sah S., Chen L., Krosting J., Sekinger E., Zhang W., Hagerman P.J., Stenzel T.T., Hadd A.G., Latham G.J., Tassone F. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin. Chem. 2010;56:399–408. doi: 10.1373/clinchem.2009.136101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onuchic V., Lurie E., Carrero I., Pawliczek P., Patel R.Y., Rozowsky J., Galeev T., Huang Z., Altshuler R.C., Zhang Z. Allele-specific epigenome maps reveal sequence-dependent stochastic switching at regulatory loci. Science. 2018;361:361. doi: 10.1126/science.aar3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eggermann T., Perez de Nanclares G., Maher E.R., Temple I.K., Tümer Z., Monk D., Mackay D.J.G., Grønskov K., Riccio A., Linglart A., Netchine I. Imprinting disorders: a group of congenital disorders with overlapping patterns of molecular changes affecting imprinted loci. Clin. Epigenetics. 2015;7:123. doi: 10.1186/s13148-015-0143-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hagerman R.J., Leehey M., Heinrichs W., Tassone F., Wilson R., Hills J., Grigsby J., Gage B., Hagerman P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 56.Deng J., Yu J., Li P., Luan X., Cao L., Zhao J., Yu M., Zhang W., Lv H., Xie Z. Expansion of GGC repeat in GIPC1 Is associated with oculopharyngodistal myopathy. Am. J. Hum. Genet. 2020;106:793–804. doi: 10.1016/j.ajhg.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
22 Cohorts of Illumina 450k Methylation Data Used in This Study
PCR Amplicons and Results of Targeted Bisulfite Sequencing of Four Epivariations
All Autosomal Epivariations Identified
All chrX Epivariations Identified
Individual-Level Data for All Probes within Epivariations
Gene Expression Data in BIOS Population
Gene Expression Data in MuTHER Population
Gene Expression Data in MESA Population
Gene Expression Data in Framingham Heart Study Population
Association of Epivariations with Local SNVs in the WHI Cohort
Autosomal Epivariations Identified in the PPMI Cohort from Analysis of 850k Methylation Array Data
Rare SNVs in DMR Carriers Located within ± 500 bp of DMR Midpoint in the PPMI Cohort
Rare CNVs Identified within ± 50 kb of DMRs in the PPMI Cohort
Concordance of Epivariations in MZ Twin Pairs
25 Epivariations Overlapping Putative Unstable CG-Rich Repeats
Data Availability Statement
The code generated during this study is available at github (see Epivariation scripts in Web Resources). Original source data utilized in this study are listed in Table S1.