Abstract
We previously reported a protective association between single nucleotide polymorphisms (SNPs; rs4415345G and rs4610776A alleles) of Paneth cell α-defensin-5 against acute graft-versus-host disease (aGVHD). Because dysbiosis has been associated with aGVHD, we hypothesized that these SNPs may have a gut microbiota signature. In Lasso regression analysis of 248 healthy individuals, rs4415345G was associated with a higher abundance of Odoribacter splanchnicus, an anaerobic butyrogenic commensal. In multivariable analysis of data from 613 allogeneic hematopoietic cell transplant recipients, peri-engraftment presence of O. splanchnicus was associated with ~50% lower risk for grade II-IV aGVHD (hazard ratio 0.53, 95% confidence interval 0.28–1.00, P = 0.05). O. splanchnicus may protect rs4415345G individuals against aGVHD.
Keywords: Graft-versus-host disease, Paneth cell, Microbiota, Odoribacter, Polymorphism
Via the secretion of antimicrobial peptides (AMPs) such as defensins and regenerating islet-derived protein-3 alpha, small intestinal Paneth cells regulate the gut microbiota (Salzman et al, 2009). α-defensin-5 (HD5) accounts for 70% of the bactericidal peptide activity of Paneth cells (Ayabe et al, 2000). Interactions between the host and microbiota at the gut mucosal barrier regulate mucosal and systemic immune responses and, in pathological states, may contribute to disease states such as acute graft-versus-host disease (aGVHD) after allogeneic hematopoietic cell transplantation (allo-HCT) (Staffas et al, 2017). The hallmark of acute GVHD is alloimmune inflammatory damage to host epithelial cells, with the gut being a primal target tissue (Zeiser & Blazar, 2017).
We previously reported an association between two single nucleotide polymorphisms (SNPs rs4415345 [G allele] and rs4610776 [A allele]) in the gene for HD5 (DEFA5) and lower incidence of aGVHD (Rashidi et al, 2018). Although the functional consequences of these SNPs have not been studied, we suspected that these SNPs may influence HD5 protein levels. Therefore, considering that Paneth cell function is partly genetically regulated (Gulati et al, 2012) and fecal concentrations of α-defensins are surrogate markers for gut microbial homeostasis (Eriguchi et al, 2015), we hypothesized that rs4415345 and rs4610776 may be associated with specific gut microbiota signatures.
First, we used a linked host-microbiota genetic database to test our hypothesis in healthy individuals. Metagenomic data were generated by the Human Microbiome Project (HMP), which comprises samples from 300 healthy adult volunteers (age 18-40) recruited from two geographic locations in the US. We downloaded the available operational taxonomic unit (OTU) table comprising normalized relative abundance data from https://www.hmpdacc.org/hmsmcp2/. Taxonomic profiling was conducted with MetaPhlAn2 and strain-level characterization was performed with StrainPhlAn. Because some individuals had multiple SRA IDs, we averaged the OTU abundances across IDs. All data were submitted to the Data Repository for the University of Minnesota at https://doi.org/10.13020/9wn7-9142. Subject genetic variation was provided as part of dbGaP project #19406. We subsetted the available HMP subjects to those with samples in the OTU table and focused on our rs4415345 and rs4610776. We coded each genotype according to the presence of the allele in question (G for rs4415345 and A for rs4610776). Therefore, rs4415345 genotypes were coded as 0 (AA) vs. 1 (GG or GC), and rs4610776 genotypes were coded as 0 (TT) vs. 1 (AA vs AT). After matching metagenome samples to the genotyped individuals, we retained 248 subjects for the association study. Major allele frequencies at rs4610776 and rs4415345 were 0.73 and 0.67, consistent with the 1000 Genomes Project.
With the assumption that many microbial taxa have no association with a given SNP, we used least absolute shrinkage and selection operator (Lasso) regression as a test of association between host genotype and taxon abundance. This method permits variable selection by ‘shrinking away’ taxa that have no association. We used binary logistic regression to evaluate the association between taxonomic abundances (independent variable; arcsine square-root transformed) and each coded genotype (dependent variable). We implemented Lasso using the glmnet package (R Foundation for Statistical Computing, Vienna, Austria), with 10-fold cross validation across multiple values of the tuning parameter lambda, using area under the curve (AUC) as the measure of error. We identified the value of lambda associated with the maximum AUC value (AUCm), that is, the value yielding the best prediction. Because Lasso is sensitive to the training data used, we re-ran the regression with cross-validation 1000 times, each time retaining the AUCm value. To build a null distribution for AUCm, we again performed 1000 Lasso regressions with cross-validation, permuting genotype encodings before each test. Our significance test determined whether the mean AUCm from the 1000 Lasso regressions on our true data fell outside the AUCm distribution from permuted runs. As indicated in Supplementary Table S1, although the mean AUCm of un-permuted data was within 95% of the permuted AUCm distribution for both loci, the P value of 0.066 for rs4415345 was suggestive of an association. Therefore, we focused our additional analyses on this locus.
To test whether specific taxa were repeatedly retained as predictors for rs4415345G (repeatability analysis), we recorded the taxa list with the highest AUCm for each simulation. 1000 simulations were performed. We repeated the same analysis for permuted samples. If the regression performs better than random taxa selection, we expect the selected taxa to occur more frequently in the un-permuted dataset than in the permuted dataset. We observed very high repeatability; the frequency of selection of taxa as predictors fell almost entirely outside the distribution of the null distribution. Specifically, a single taxon (Odoribacter splanchnicus) was present in 99% of the simulations. Next, we performed stability selection analysis using R packages stabs and lars (cutoff 0.6, per-family error rate 2) to determine how frequently a taxon is predictive of rs4415345G in Lasso across 100 random subsamples of the data. O. splanchnicus was again the only taxon that exceeded the recommended frequency cutoff of 0.6 (Figure 1A), with a positive association. Therefore, a higher relative abundance of this species was associated with the G allele of rs4415345. Finally, because Lasso selects only one of multi-collinear predictors, we tested whether the relative abundance of O. splanchnicus correlated strongly with any other species. The Spearman’s correlation coefficient was consistently <0.50, confirming the absence of additional taxa in association with rs4415345.
Figure 1. Association between rs4415345, gut microbiota, and acute GVHD.
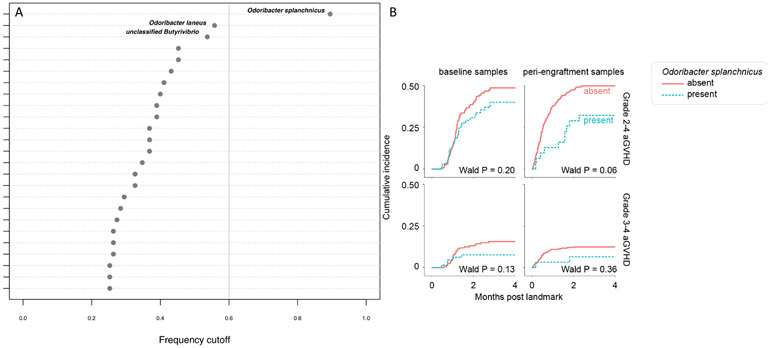
Stability selection analyses for the G allele of rs4415345. We chose the lower bound (0.6, represented by the vertical gray line) of the frequency cutoff recommended by the authors of the stabs R package. Each taxon is represented by a circle. Numbers along the x-axis show the frequency at which taxa were predictive in simulations across the 100 subsamples. The names of the top three taxa are shown. (B) Presence of O. splanchnicus in baseline and peri-engraftment fecal samples and the risk of aGVHD. The horizontal axis indicates months post the landmark. For the analysis of baseline samples (earliest sample collected per patient between day −30 and −6), the landmark was HCT day 0 (when cells were infused). Among 226 patients with evaluable baseline samples, O. splanchnicus was present in 65 and absent in 161 patients. For the analysis of peri-engraftment samples (median abundance in all samples collected per patient during days 7–21), the landmark was HCT day 21. Among 432 patients with evaluable peri-engraftment samples who had not been diagnosed with grade II-IV aGVHD as of the landmark, O. splanchnicus was present in 31 and absent in 401 patients. Among the 450 patients with evaluable peri-engraftment samples who had not been diagnosed with grade III-IV aGVHD as of the landmark, O. splanchnicus was present in 31 and absent in 419 patients.
So far, we have shown an association between rs4415345 G allele and less aGVHD in HCT recipients (Rashidi et al, 2018) and an association between the same allele and higher O. splanchnicus abundance in healthy individuals (current work), suggesting that this taxon may mediate the observed association in HCT recipients (protective effect). Next, we evaluated whether O. splanchnicus presence in the stool of HCT patients is associated with less aGVHD. The patients were admitted to the Memorial Sloan Kettering Cancer Center from 2009 to 2018 and enrolled in a biorepository protocol approved by the institutional review board (Supplementary Table S2). For the analysis of baseline samples (earliest sample collected per patient between day −30 and −6), the landmark was HCT day 0 (when cells were infused). For the analysis of peri-engraftment samples (median abundance in all samples collected per patient during days 7-21), the landmark was HCT day 21. The genomic 16S ribosomal RNA V4/V5 variable region was amplified and sequenced on the Illumina MiSeq (San Diego, CA) platform. Quality-filtered sequences with >97% identity were grouped into OTUs. Among 226 patients with evaluable baseline samples, O. splanchnicus was present in 65 and absent in 161 patients. Among 432 patients with evaluable peri-engraftment samples who had not been diagnosed with grade II-IV aGVHD as of the landmark, O. splanchnicus was present in 31 and absent in 401 patients. Among the 450 patients with evaluable peri-engraftment samples who had not been diagnosed with grade III-IV aGVHD as of the landmark, O. splanchnicus was present in 31 and absent in 419 patients.
The results of univariate cumulative incidence analysis with grade II-IV or III-IV aGVHD as the dependent variable and baseline or peri-engraftment presence of O. splanchnicus as the independent variable are shown in Figure 1B. Relapse and non-GVHD death were considered as competing risks. The strongest association in univariate analysis was between peri-engraftment presence of O. splanchnicus and grade II-IV aGVHD (hazard ratio [HR] 0.54, 95% confidence interval [95%CI] 0.29-1.02, P = 0.06). A pre-determined set of covariates were included in multivariable regression (Table 1). In multivariable analysis, peri-engraftment presence of O. splanchnicus was associated with ~50% lower risk for grade II-IV aGVHD (HR 0.53, 95%CI 0.28-1.00, P = 0.05). Although the P values for O. splanchnicus in all other models were >0.05, the results were consistently in the same direction, with O. splanchnicus presence showing a protective association (Figure 1B; Table 1).
Table 1:
Multivariable analysis of the association between O. splanchnicus and acute GVHD
| HR (95%CI) | P | HR (95%CI) | P | |
|---|---|---|---|---|
| Grade II-IV aGVHD | Grade III-IV aGVHD | |||
| Baseline O. splanchnicus | 0.23 | 0.12 | ||
| Absent | Reference | Reference | ||
| Present | 0.76 (0.48-1.19) | 0.46 (0.17-1.21) | ||
| Graft source | ||||
| Unmodified bone marrow | Reference | Reference | ||
| Unmodified peripheral blood | 1.14 (0.47-2.75) | 0.77 | 0.82 (0.17-3.99) | 0.81 |
| Cord blood | 1.38 (0.15-12.5) | 0.78 | 0.28 (0.02-4.23) | 0.36 |
| Conditioning intensity | ||||
| Myeloablative | Reference | Reference | ||
| Reduced intensity | 1.26 (0.72-2.22) | 0.42 | 1.34 (0.49-3.67) | 0.57 |
| Non-myeloablative | 0.57 (0.22-1.11) | 0.10 | 0.28 (0.06-1.38) | 0.12 |
| HCT-CI (continuous variable) | 0.94 (0.86-1.04) | 0.26 | 0.99 (0.83-1.19) | 0.94 |
| GVHD prophylaxis | ||||
| CNI+MMF-based | Reference | Reference | ||
| CNI+MTX-based | 0.58 (0.08-4.26) | 0.59 | 0.27 (0.03-2.11) | 0.21 |
| PT-Cy-based | 0.83 (0.10-7.00) | 0.86 | 0.24 (0.02-2.97) | 0.27 |
| Peri-engraftment O. splanchnicus | 0.05 | 0.34 | ||
| Absent | Reference | Reference | ||
| Present | 0.53 (0.28-1.00) | 0.50 (0.12-2.06) | ||
| Graft source | ||||
| Unmodified bone marrow | Reference | Reference | ||
| Unmodified peripheral blood | 1.19 (0.73-1.94) | 0.49 | 1.11 (0.43-2.88) | 0.83 |
| Cord blood | 1.70 (0.22-13.16) | 0.61 | 0.56 (0.06-5.28) | 0.61 |
| Conditioning intensity | ||||
| Myeloablative | Reference | Reference | ||
| Reduced intensity | 0.93 (0.64-1.35) | 0.71 | 1.08 (0.52-2.22) | 0.84 |
| Non-myeloablative | 0.38 (0.22-0.64) | <0.01 | 0.40 (0.13-1.26) | 0.12 |
| HCT-CI (continuous variable) | 0.97 (0.90-1.03) | 0.30 | 1.04 (0.91-1.18) | 0.57 |
| GVHD prophylaxis | ||||
| CNI+MMF-based | Reference | Reference | ||
| CNI+MTX-based | 0.72 (0.10-5.22) | 0.75 | 0.37 (0.05-2.74) | 0.33 |
| PT-Cy-based | 1.00 (0.13-7.70) | 1.00 | 0.25 (0.02-2.66) | 0.25 |
aGVHD: acute graft-versus-host disease; CI: Confidence interval; CNI: Calcineurin inhibitor; HCT-CI: Hematopoietic cell transplantation-specific comorbidity index; HR: Hazard ratio; MMF: Mycophenolate mofetil; MTX: Methotrexate; PT-Cy: Post-transplant cyclophosphamide
In summary, we found that the G allele of rs4415345, a SNP in the gene for Paneth cell α-defensin-5, is associated with the higher relative abundance of O. splanchnicus in healthy individuals and less aGVHD in HCT recipients. In an independent dataset, we found that the peri-engraftment presence of O. splanchnicus was associated with a 50% reduction in risk for grade II-IV aGVHD. O. splanchnicus presence in baseline samples was less powerfully predictive of GVHD risk. The peri-engraftment microbiota may be a stronger predictor of immunologic outcomes because the interaction between microbiota and the graft may be maximal in the interval surrounding engraftment. O. splanchnicus is an anaerobic bacteria in the phylum Bacteroidetes (Hardham et al, 2008) and present at low abundance in the gut of most individuals. O. splanchnicus has beneficial effects on the host, partly related to its butyrogenic activity (Werner et al, 1975). Butyrate is a short-chain fatty acid implicated in protection of the gut barrier and reduction of aGVHD (Toubai et al, 2016). In addition, O. splanchnicus produces indole from tryptophan (Göker et al, 2011); indole produced from the gut microbiota is known to limit GVHD (Swimm et al, 2018). O. splanchnicus presence is also correlated with the host’s lower inflammatory cytokine production capacity (Schirmer et al, 2016). O. splanchnicus is depleted in patients with inflammatory bowel disease (Morgan et al, 2012). Based on the results of the present study and our previous report (Rashidi et al, 2018), we propose that a higher abundance of O. splanchnicus in patients with the G allele of rs4415345 is protective against colitis and aGVHD. Host genetics-mediated regulation of the abundance of O. splanchnicus was suggested in a previous twin study (Lim et al, 2017), albeit with unknown mechanisms. Our findings suggest Paneth cell’s most abundant AMP as a key mediator, consistent with physiologic modulation of the gut microbiome by Paneth cell defensins (Ehmann et al, 2019). Future research should evaluate the effect of the SNP on HD5 gene expression, the effect of HD5 on O. splanchnicus, and the association between rs4415345 and O. splanchnicus in HCT patients.
Supplementary Material
Supplementary Table S1. Permuted and unpermuted AUCm distributions
Supplementary Table S2: Patient characteristics in the HCT cohort
Acknowledgements:
AR designed the study and wrote the manuscript. ALCG, JUP, DGB, and RRJ collected and analyzed data from the clinical cohort. AH analyzed HMP data. CS, BRB, and DJW critically reviewed the results and enhanced the manuscript. We thank Joshua Baller for help with dbGaP setup and methodological advice, as well as Evan Bollig for assistance with the Stratus computing environment required for protected data.
Funding: Armin Rashidi was supported by an American Cancer Society Institutional Research Grant from the University of Minnesota and an American Society of Blood and Marrow Transplantation Young Investigator Award. This research was also supported by NHLBI NIH Award K08HL143189, MSKCC Cancer Center Core Grant P30 CA008748, the Parker Institute for Cancer Immunotherapy at Memorial Sloan Kettering Cancer Center, the Sawiris Foundation, the Society of Memorial Sloan Kettering Cancer Center, MSKCC Cancer Systems Immunology Pilot Grant, Empire Clinical Research Investigator Program, and Seres Therapeutics.
Footnotes
Competing interests: The authors have no competing interests.
References
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME & Ouellette AJ (2000) Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nature Immunology, 1, 113–118 Available at: http://www.ncbi.nlm.nih.gov/pubmed/11248802 [Accessed April 8, 2018]. [DOI] [PubMed] [Google Scholar]
- Ehmann D, Wendler J, Koeninger L, Larsen IS, Klag T, Berger J, Marette A, Schaller M, Stange EF, Malek NP, Jensen BAH & Wehkamp J (2019) Paneth cell α-defensins HD-5 and HD-6 display differential degradation into active antimicrobial fragments. Proceedings of the National Academy of Sciences, 116, 3746–3751 Available at: https://www.pnas.org/content/116/9/3746 [Accessed February 27, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriguchi Y, Nakamura K, Hashimoto D, Shimoda S, Shimono N, Akashi K, Ayabe T & Teshima T (2015) Decreased secretion of Paneth cell α-defensins in graft-versus-host disease. Transplant Infectious Disease, 17, 702–706 Available at: http://www.ncbi.nlm.nih.gov/pubmed/26198302 [Accessed January 12, 2018]. [DOI] [PubMed] [Google Scholar]
- Göker M, Gronow S, Zeytun A, Nolan M, Lucas S, Lapidus A, Hammon N, Deshpande S, Cheng J-F, Pitluck S, Liolios K, Pagani I, Ivanova N, Mavromatis K, Ovchinikova G, Pati A, Tapia R, Han C, Goodwin L, Chen A, et al. (2011) Complete genome sequence of Odoribacter splanchnicus type strain (1651/6T). Standards in Genomic Sciences, 4, 200–209 Available at: http://www.standardsingenomics.org/index.php/sigen/article/view/sigs.1714269 [Accessed September 28, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati AS, Shanahan MT, Arthur JC, Grossniklaus E, von Furstenberg RJ, Kreuk L, Henning SJ, Jobin C & Sartor RB (2012) Mouse background strain profoundly influences Paneth cell function and intestinal microbial composition. PloS one, 7, e32403 Available at: http://dx.plos.org/10.1371/journal.pone.0032403 [Accessed August 10, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardham JM, King KW, Dreier K, Wong J, Strietzel C, Eversole RR, Sfintescu C & Evans RT (2008) Transfer of Bacteroides splanchnicus to Odoribacter gen. nov. as Odoribacter splanchnicus comb. nov., and description of Odoribacter denticanis sp. nov., isolated from the crevicular spaces of canine periodontitis patients. INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, 58, 103–109 Available at: http://www.ncbi.nlm.nih.gov/pubmed/18175692 [Accessed September 28, 2019]. [DOI] [PubMed] [Google Scholar]
- Lim MY, You HJ, Yoon HS, Kwon B, Lee JY, Lee S, Song Y-M, Lee K, Sung J & Ko G (2017) The effect of heritability and host genetics on the gut microbiota and metabolic syndrome. Gut, 66, 1031–1038 Available at: http://gut.bmj.com/lookup/doi/10.1136/gutjnl-2015-311326 [Accessed September 28, 2019]. [DOI] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, LeLeiko N, Snapper SB, Bousvaros A, Korzenik J, Sands BE, Xavier RJ & Huttenhower C (2012) Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biology, 13, R79 Available at: http://www.ncbi.nlm.nih.gov/pubmed/23013615 [Accessed September 28, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidi A, Shanley R, Yohe SL, Thyagarajan B, Curtsinger J, Anasetti C, Waller EK, Scott BL, Blazar BR & Weisdorf DJ (2018) Recipient single nucleotide polymorphisms in Paneth cell antimicrobial peptide genes and acute graft-versus-host disease: analysis of BMT CTN-0201 and −0901 samples. British Journal of Haematology, 182, 887–894 Available at: http://www.ncbi.nlm.nih.gov/pubmed/30004111 [Accessed February 14, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjöberg J, Amir E, Teggatz P, Barman M, Hayward M, Eastwood D, Stoel M, Zhou Y, Sodergren E, Weinstock GM, Bevins CL, Williams CB & Bos NA (2009) Enteric defensins are essential regulators of intestinal microbial ecology. Nature Immunology, 11, Available at: http://www.nature.com.ezp2.lib.umn.edu/ni/journal/v11/n1/pdf/ni.1825.pdf [Accessed March 26, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Jansen T, Jacobs L, Bonder MJ, Kurilshikov A, Fu J, Joosten LAB, Zhernakova A, Huttenhower C, Wijmenga C, Netea MG & Xavier RJ (2016) Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell, 167, 1125–1136.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staffas A, Burgos da Silva M & van den Brink MRM (2017) The intestinal microbiota in allogeneic hematopoietic cell transplant and graft-versus-host disease. Blood, 129, 927–933 Available at: http://www.ncbi.nlm.nih.gov/pubmed/27940475 [Accessed August 17, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swimm A, Giver CR, DeFilipp Z, Rangaraju S, Sharma A, Ulezko Antonova A, Sonowal R, Capaldo C, Powell D, Qayed M, Kalman D & Waller EK (2018) Indoles derived from intestinal microbiota act via type I interferon signaling to limit graft-versus-host disease. Blood, 132, 2506–2519 Available at: http://www.ncbi.nlm.nih.gov/pubmed/30257880 [Accessed December 23, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toubai T, Mathewson ND, Magenau J & Reddy P (2016) Danger Signals and Graft-versus-host Disease: Current Understanding and Future Perspectives. Frontiers in immunology, 7, 539 Available at: http://journal.frontiersin.org/article/10.3389/fimmu.2016.00539/full [Accessed March 25, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner H, Rintelen G & Kunstek-Santos H (1975) [A new butyric acid-producing bacteroides species: B. splanchnicus n. sp. (author’s transl)]. Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. Erste Abteilung Originale. Reihe A: Medizinische Mikrobiologie und Parasitologie, 231, 133–44 Available at: http://www.ncbi.nlm.nih.gov/pubmed/168701 [Accessed September 28, 2019]. [PubMed] [Google Scholar]
- Zeiser R & Blazar BR (2017) Acute Graft-versus-Host Disease — Biologic Process, Prevention, and Therapy. New England Journal of Medicine, 377, 2167–2179 Available at: http://www.ncbi.nlm.nih.gov/pubmed/29171820 [Accessed February 27, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. Permuted and unpermuted AUCm distributions
Supplementary Table S2: Patient characteristics in the HCT cohort


