Summary
Quantifying the functional effects of complex disease risk variants can provide insights into mechanisms underlying disease biology. Genome-wide association studies have identified 39 regions associated with risk of epithelial ovarian cancer (EOC). The vast majority of these variants lie in the non-coding genome, where they likely function through interaction with gene regulatory elements. In this study we first estimated the heritability explained by known common low penetrance risk alleles for EOC. The narrow sense heritability () of EOC overall and high-grade serous ovarian cancer (HGSOCs) were estimated to be 5%–6%. Partitioned SNP heritability across broad functional categories indicated a significant contribution of regulatory elements to EOC heritability. We collated epigenomic profiling data for 77 cell and tissue types from Roadmap Epigenomics and ENCODE, and from H3K27Ac ChIP-seq data generated in 26 ovarian cancer and precursor-related cell and tissue types. We identified significant enrichment of risk single-nucleotide polymorphisms (SNPs) in active regulatory elements marked by H3K27Ac in HGSOCs. To further investigate how risk SNPs in active regulatory elements influence predisposition to ovarian cancer, we used motifbreakR to predict the disruption of transcription factor binding sites. We identified 469 candidate causal risk variants in H3K27Ac peaks that are predicted to significantly break transcription factor (TF) motifs. The most frequently broken motif was REST (p value = 0.0028), which has been reported as both a tumor suppressor and an oncogene. Overall, these systematic functional annotations with epigenomic data improve interpretation of EOC risk variants and shed light on likely cells of origin.
Keywords: genome-wide association studies, epithelial ovarian cancer, cells of origin, heritability, functional enrichment, epigenomics, motif
Introduction
Epithelial ovarian cancer (EOC [MIM: 167000]) describes a diverse group of tumors that are often diagnosed at a late stage, have a poor prognosis, and develop resistance to standard chemotherapeutic treatments.1, 2, 3 There are five main histological subtypes of invasive disease: high-grade serous (HGSOC), low-grade serous (LGSOC), mucinous (MOC), endometrioid (EnOC), and clear cell (CCOC) ovarian cancer. Ovarian tumors of low malignant potential (LMP) ovarian cancer comprise ∼20% of case subjects and only a small minority will progress to invasive disease. The histotypes show differences in underlying biology, genetic drivers, and to some extent different epidemiological and lifestyle risk factors. They may also derive from different cell types, with fallopian tube secretory epithelial cells the likely cells of origin for most HGSOCs4,5 and endometriosis the putative precursor of CCOC and EnOC.6, 7, 8 Thus, uncovering the underlying genetic architecture of different EOC histotypes is an urgent need and may be the most effective approach to reduce mortality due to EOC.9
Less than 40% of the estimated heritability of ovarian cancer is explained by known coding pathogenic mutations in susceptibility genes including BRCA1 (MIM: 113705), BRCA2 (MIM: 600185), BRIP1 (MIM: 605882), RAD51C (MIM: 602774), and RAD51D (MIM: 602954).10 More recently, genome-wide association studies (GWASs) have identified 39 independent regions associated with EOC risk.11 Some regions are associated with specific histotypes, while others appear pleiotropic across different EOC histotypes11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 or other phenotypes (e.g., breast cancer).21,22 Combined, these risk alleles explain a fraction of narrow sense heritability for ovarian cancer. Heritability estimates are complicated by linkage disequilibrium, which often results in the identification of tens to hundreds of tightly correlated SNPs at each susceptibility locus.23
The vast majority of genetic risk alleles for common complex traits identified by GWASs lie in non-protein coding DNA regions, with their mechanisms of function largely unknown.24 Previous studies for other complex phenotypes have shown that risk variants are enriched in regulatory elements, indicating that they function through the differential regulation of gene expression.25, 26, 27, 28 Many regulatory elements can be identified by epigenomic modifications.29, 30, 31, 32 Publicly available resources such as the Encyclopedia of DNA Elements (ENCODE) and the Roadmap Epigenome Mapping Consortium have characterized the epigenomic architecture of a multitude of cell types, showing that the epigenome and transcriptional program are highly tissue specific.29,33 Analyses of acetylated lysine 27 of histone H3 protein (H3K27Ac) activity in primary tumors shows that >80% of cell-type-specific regulatory elements lie in putative enhancers, reinforcing previous observations that cell-type-specific enhancers drive the spatial and temporal diversity of gene expression.29,34
We hypothesize that common ovarian cancer risk SNPs are located within tissue-specific regulatory elements and are likely to function by altering the activity of host enhancers active in ovarian cancers and cell types that represent precursors of the different EOC histotypes. We applied systematic, computational approaches to identify regulatory elements that are potentially disrupted at EOC GWAS risk loci. We first estimated the heritability for each EOC histotype using common SNPs, taking into account linkage disequilibrium; and then partitioning narrow sense heritability analysis across general broad functional categories. We focused our analyses on 39 germline genetic risk loci previously reported GWASs for one or more EOC histotypes with the aim of identifying putative regulatory targets and transcription factors associated with EOC risk variants and the initiation and early-stage EOC development.
Material and Methods
Genotyping Datasets for Ovarian Cancer
Summary statistics were available from the largest published meta-analysis of 25,509 EOC case subjects and 40,941 control subjects from Phelan et al.11 EOC cases comprised the five major histotypes of invasive disease—high-grade serous (HGSOC; n = 13,037), low-grade serous (LGSOC, n = 1,012), mucinous (MOC, n = 1,149), endometrioid (EnOC, n = 2,764), and clear cell carcinoma (CCOC, n = 1,366)—and additionally, borderline serous (n = 1,954) and other uncategorized EOC (n = 2,749) were available. Following Phelan et al.,11 we also included the meta-cohort analysis (OCAC+CIMBA), combining genotypes from EOC case subjects and control subjects in Ovarian Cancer Association Consortium (OCAC) and carriers of BRCA1/2 mutations from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA; 31,448 BRCA1/2 mutation carriers, of which 3,887 mutation carriers have EOC). Summary statistics for genetic associations were generated after imputing genotypes to the 1000 Genomes Project reference panel of 11,403,952 common variants (minor allele frequency [MAF] > 1%). These summary statistics were used for partitioning heritability. We further curated a list of all previously reported genome-wide significant risk regions (p value < 5.0 × 10−8) for EOC (including all histotypes) and the credible causal set of SNPs reported at each locus for all invasive ovarian cancer and for each histotype where there was evidence of a risk association from prior publications.11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21 This identified 39 risk regions for different histotypes at genome-wide significance (Table S1). Variant position and rsid for each variant were validated in dbSNP146 with hg19/GRCh37 coordinates.
Epigenomic Datasets for Ovarian Cancers and Their Precursor Tissues
Publicly available epigenomic profiling datasets were collected from the Roadmap Epigenomics Mapping Consortium34 and the ENCODE project29 (labeled ENCODE2012 in this study, Table S2). Additionally, a collection of chromatin immunoprecipitation-sequencing (ChIP-seq) for H3K27Ac in ovarian cancer-related cell and tissue types that were generated in house was compiled. This includes precursor normal and ovarian cancer cell lines from previously published studies and newly generated H3K27Ac ChIP-seq in additional cell lines and primary tumors (Table S3). Briefly, we have generated H3K27Ac-ChIP-seq data for: 20 primary EOC tumors, 5 each for the different histotypes of invasive ovarian cancer (HGSOC, CCOC, EnOC, and MOC) (Table S3); 12 established EOC cell lines that model undifferentiated EOC (HEYA8), HGSOC (CaOV3, UWB1.289, Kuramochi, OVCA429), LGSOC (VOA1056, OAW42), CCOC (JHOC5, ES2, and RMG-II), and MOC (GFTR230, MCAS, EFO27); and 3 ovarian cancer precursor cell types—fallopian tube secretory epithelial cells ((FTSECs), FT246, FT33), ovarian surface epithelial cells ([OSECs] IOSE4 and IOSE11), and endometrioid epithelial cells (EEC16)35 (Table S3). Methods for H3K27Ac-ChIP-seq and peak calling that was previously published have been described.11,29,36, 37, 38, 39, 40, 41
H3K27Ac ChIP-seq for six new cell lines (EFO27, VOA1056, HEYA8, Kuramochi, ES2, RMG-II) was performed according to previously published methods.40 Peak calling was performed using the AQUAS pipeline.42 Reads were aligned against the reference human genome hg38. Quality control metrics were computed for each individual replicate, including number of reads, percentage of duplicated reads, normalized strand coefficient, relative strand correlation, and fraction of reads in called peaks. Two biological replicates were available for EFO27, VOA1056, and Kuramochi. Peak calling was performed with macs2 with pooled replicate peaks that overlap 50% or more of each individual replicate selected for the final peak set. When replicates were not available (HEYA8, ES2, and RMG-II), pseudo replicates were formed and pooled peaks selected in the same manner from these pseudo replicates. To create consensus peak sets across a single histotype for enrichment analyses, peaks with least 50% overlap with at least one other peak in two or more samples from a histotype group were retained, with the boundaries stretched to the edge of each peak in the overlap. Files were then concatenated and peak co-ordinates merged such that if records within the concatenated file were overlapping, they were combined into a single peak.
We generated chromatin state calls in Roadmap Epigenomics and ENCODE2012 samples using StatepaintR43 (Table S2). This approach uses human expert rule-based segmentations, which allows the user to designate combinations of epigenomic marks to represent functional chromatin states. StatepaintR annotates chromatin states based upon available epigenomic marks, accommodating for the practical situation that not all histone marks are available for all samples. These chromatin state annotations are also released in the StateHub Model Repository under TrackHub ID 5813b67f46e0fb06b493ceb0 (see Web Resources). We note that the “active” state delineates regions overlapping with active marks, i.e., H3K27Ac, but when the marks required to call the more specific “active Enhancer” or “active promoter” states are not present in inputs.
Estimation of SNP Heritability
We estimated the variance explained by known SNP effects, or SNP heritability, by using linkage disequilibrium score regression (LDSC),44,45 v.1.0.0. LDSC models the expected statistics from a GWAS of SNP j as
where N is the number of individuals; M is the number of SNPs, such that is the average heritability explained per SNP; is a constant measuring the contribution of confounding biases, such as cryptic relatedness and population stratification; is the LD score of SNP j defined as , where is the Pearson correlation between SNP j and SNP k; and k denotes other SNPs within the LD region. The LD scores were pre-calculated from phased European ancestry individuals from the 1000 Genomes Project reference panel v.3.44
Partitioning SNP Heritability into Functional Categories
To examine the importance of specific functional categories in SNP heritability, we applied stratified LD score regression45 to EOC and HGSOC GWAS summary statistics.11 The goal was to partition SNP heritability into functional categories by combining SNPs in the same LD region together and quantify their overlaps with regions of interest. The stratified LDSC model was adapted from the above-mentioned regular LDSC model:
where C represents the functional categories; denotes the per-SNP contribution to heritability of category C; is the LD score of SNP j falling in category C, calculated as ; all the other parameters are the same as in LDSC. The category-specific enrichment was defined as the proportion of SNP heritability in the category divided by the proportion of SNPs in the same category.
The partitioned-heritability analyses were performed with two different sets of functional categories. The first is a full baseline model with 24 general broad functional annotations from public datasets, which were inclusive of all publicly available cell types and post-processed in Gusev et al.46 The 24 annotations include coding, 3′ UTR, 5′ UTR, promoter, and intron regions from UCSC Genome Browser;46,47 regions conserved in mammals;48,49 combined chromHMM and Segway predictions comprising CTCF-bound regions, promoter-flanking, transcribed, transcription start site (TSS), strong enhancer, weak enhancer, repressed annotations;50 digital genomic footprint (DGF) and transcription factor binding sites (TFBS) from ENCODE;46 open chromatin regions as reflected by DNase I hypersensitivity sites (DHSs) from a union of all cell types and a union of only fetal cell types on ENCODE and Roadmap Epigenomics;51 FANTOM5 enhancer;52 H3K27Ac,36,53 H3K4me1,29 and H3K4me329 histone marks from a union over cell types on Roadmap Epigenomics; and super-enhancers obtained from Hnisz et al.36
The second set contains 15 cell-type-specific annotations for H3K27Ac marks, which represent precursor normal and ovarian cancer cell lines (see the Epigenomic Datasets for Ovarian Cancers and Their Precursor Tissues section for details). We added these cell-type-specific annotations individually to the full baseline model, which resulted in 15 models for EOC and 15 models for HGSOC. This cell-type-specific analysis helped measure how much more the annotation contributes on top of the rest of the full baseline model and to justify which cell type is more enriched than the others.
Enrichment of Credible Causal SNPs in Biofeatures
EOC credible causal risk variants were combined to create the full credible set (n = 1,432) and then split to represent sets of risk variants associated with each EOC histotype. The background set of variants used in functional annotation and enrichment analysis were generated by aggregating SNPs within 2 Mb (±1 Mb) of the credible causal set, in an attempt to maintain similar genetic architecture (e.g., linkage disequilibrium, regulatory activity, and transcriptional program) as credible causal risk variants. Functional annotation of credible causal SNPs was performed with SNPnexus54 using SIFT55 and PolyPhen56 for protein effect, ENCODE,29 Roadmap Epigenomics,34 and Ensembl Regulatory Build57 for regulatory elements, and CADD,58 DeepSEA,59 and FunSeq260 for non-coding variation scoring. The difference between the average FunSeq2 functional score for the foreground and background SNP lists was determined with a two-tailed t test.
Enrichment analysis was performed with the FunciVar package (see Web Resources), a tool for annotation and functional enrichment of variant sets. In principle, FunciVar first takes two lists of variants as inputs: (1) a list of target variants, in this analysis the credible causal set of risk SNPs, which act as the foreground, and (2) a list of control variants, which act as the background. The background SNP lists from each locus were combined as necessary to ensure the local background set of variants for each locus was included in the histotype-specific enrichment. FunciVar then intersects each variant with biofeatures, which were provided as bed files. The likelihood of true enrichment for each variant list is modeled under the beta-binomial distribution,
where S is the number of observed overlaps with biofeature, N is the number of total variants, and subscripts and denote background and foreground, respectively. FunciVar uses an uninformative Jeffreys prior, which set and . To estimate the true enrichment, FunciVar by default simulates 10,000 times to obtain a distribution of foreground enrichment probability, , and a distribution of background enrichment probability, . The two sets of simulated probabilities were next directly subtracted to obtain a distribution of differences. FunciVar calculates a 95% credible interval for the range of enrichment probability differences between the two lists of variants. Enrichment is reported as the median of this credible interval, within the range of −1 to 1, where 1 means strong enrichment and −1 means strong depletion. The significance of results is reported as probability that foreground SNPs have more overlaps with the biofeature than background SNPs, within the range of 0 to 1, the higher the more confident. Results are plotted with significantly enriched biofeatures shown in color, and non-significantly enriched biofeatures shown in gray.
Identifying Transcription Factor Binding Consequences of EOC Credible Causal Variants in Enhancers
To identify the potential consequences of EOC risk variants in EOC enhancers, we used MotifBreakR61 to predict the transcription factor binding sites that a variant disrupts and the extent of disruptiveness. MotifBreakR uses a position weight matrix to score the difference of binding between reference and alternative alleles for every possible window that includes the variant, and then categorizes the normalized difference score as effect of the target variant (strong, weak, or neutral). We used seven TFBS motif databases: ENCODE motifs,62 Factorbook,63 Hocomoco,64 Homer,65 Transfac,66 Jaspar,67 and MotifDb.68
To identify significant TFs that were predicted to be impacted by the alternate allele at credible causal variants, we applied FunciVar package again. We curated two lists: (1) the foreground list, which are credible causal variants that intersect H3K27Ac peaks in any EOC cell type, and (2) the background list, credible causal variants that did not intersect H3K27Ac peaks in any EOC cell type. Significant differences in likelihood of the alternate allele of a credible causal variant disrupting a TFBS are reported for each TF.
Results
Regulatory Elements Significantly Account for Ovarian Cancer Heritability
The aim was to evaluate the functional significance of common, genetic variants associated with epithelial ovarian cancer (EOC) risk identified by GWASs, and the contribution of different functional states to EOC heritability. We utilized genotype data pooled from multiple GWASs comprising 25,509 EOC case subjects and 40,941 control subjects stratified into five major histotypes of invasive or low-grade/borderline disease: high-grade serous (HGSOC), low-grade serous (LGSOC), mucinous (MOC), endometrioid (EnOC), and clear cell (CCOC) ovarian cancers (see Material and Methods).11 We estimated the variance explained by known SNP effects, or SNP heritability, using linkage disequilibrium score regression (LDSC).44,45 LDSC measures narrow-sense heritability (, “SNP heritability” henceforth) using GWAS summary statistics to explicitly model linkage disequilibrium. Estimates of SNP heritability ranged from nearly 0% to 6% for the different EOC histotypes (Figure 1). We identified 5.1% heritability in our analyses for the All EOC GWAS, in keeping with a prior report of heritability performed with the same publicly available summary statistics for All EOC.69 The highest heritability identified was explained by risk variants associated with the HGSOC histotype and the lowest heritability for risk variants associated with LGSOC.
Figure 1.
Estimates of SNP Heritability () Explained by Common SNPs
Overall SNP heritability calculated based on GWAS summary statistics for each EOC histotype. The GWAS included 40,941 control subjects, and the number of case subjects by histotype are shown in parentheses. EOC, epithelial ovarian cancer; HGSOC, high-grade serous ovarian cancer; MOC, mucinous ovarian cancer; EnOC, endometrioid ovarian cancer; CCOC, clear cell ovarian cancer; LGSOC, low-grade serous ovarian cancer; LMP, low malignant potential.
Next, we partitioned SNP heritability across 24 broad non-cell-type-specific “functional” categories (see Material and Methods)70 in two analyses, EOC and HGSOC, as these groups were suitably powered for further analysis based on their observed heritability z-score. We observed a significant contribution of four functional categories (represented by five biofeatures) to the heritability of EOC, and two functional categories to the heritability of HGSOC (Table 1). The most significant enrichment was in regions of the genome marked by H3K27Ac (p value = 0.01–0.03). Up to 27% of the 1,432 candidate causal risk variants coincided with the histone modification H3K27Ac, accounting for as much as 97% of the estimated SNP heritability for EOC (3.6-fold enrichment, p value = 0.006). Other significant functional elements included three prime untranslated regions (3′ UTR) (17.3-fold enrichment, p value = 0.015), promoters (8.7-fold enrichment, p value = 0.016), and super-enhancers (2.1-fold enrichment, p value = 0.02) (Table 1). HGSOC heritability was most significantly enriched in 3′ UTRs (18.4-fold enrichment, p value = 0.009) and H3K27Ac marks (1.8-fold enrichment, p value = 0.033).
Table 1.
Enrichment Estimates for 24 Non-Cell-Type-Specific Functional Categories for EOC and HGSOC
| Functional Category |
All EOC |
HGSOC |
||
|---|---|---|---|---|
| Enrichment | p Value | Enrichment | p Value | |
| 3′ UTR | 17.29 | 0.02 | 18.40 | 0.01 |
| 5′ UTR | −0.62 | 0.89 | 5.12 | 0.71 |
| Coding | 3.55 | 0.75 | 8.05 | 0.30 |
| Conserved | 24.94 | 0.06 | 21.82 | 0.07 |
| CTCF | −9.62 | 0.11 | −3.86 | 0.47 |
| DGF | 1.71 | 0.82 | −0.92 | 0.52 |
| DHS | 3.42 | 0.35 | 0.50 | 0.83 |
| Enhancer | 2.66 | 0.69 | 2.56 | 0.71 |
| FANTOM5 enhancer | −2.46 | 0.86 | 12.42 | 0.51 |
| Fetal DHS | 0.41 | 0.87 | −1.52 | 0.50 |
| H3K27Ac (Hnisz et al.36) | 1.96 | 0.01 | 1.77 | 0.03 |
| H3K27Ac (PGC2) | 3.61 | 0.01 | 2.42 | 0.16 |
| H3Kme1 | 2.02 | 0.16 | 1.82 | 0.28 |
| H3Kme3 | 3.91 | 0.07 | 1.01 | 0.99 |
| H3K9ac | 3.25 | 0.33 | 0.90 | 0.96 |
| Intron | 1.46 | 0.12 | 1.24 | 0.35 |
| Promoter | 8.69 | 0.02 | 6.99 | 0.06 |
| Promoter flanking | 10.38 | 0.49 | −2.90 | 0.76 |
| Repressed | −0.32 | 0.07 | 0.67 | 0.64 |
| Superenhancer | 2.09 | 0.02 | 1.83 | 0.12 |
| Transcription factor binding site | 4.93 | 0.16 | 2.21 | 0.66 |
| Transcribed | 1.87 | 0.24 | 1.05 | 0.95 |
| TSS | 5.28 | 0.55 | 0.56 | 0.96 |
| Weak enhancer | 9.06 | 0.35 | 4.69 | 0.68 |
Enrichment was calculated as , which shows the proportion of estimated SNP heritability explained by the proportion of SNPs in the functional category. Statistically significant associations (p values < 0.05) are marked in bold.
Enrichment of EOC Risk Variants with Different Chromatin States by Cell Type
We collated a set of 1,423 credible causal SNPs, previously reported from 39 confirmed genome-wide significant risk regions (p value < 5.0 × 10−8) for all EOC histotypes (Table S1). We annotated the full credible set of EOC risk SNPs with SNPnexus71 to map each variant to intergenic, intronic, 3′ or 5′ UTR, or exonic regions (Figure S1A). The 1,432 credible causal risk variants were integrated with epigenomic data to evaluate enrichment of EOC risk variants in different chromatin states by cell type. The majority of credible causal SNPs (96%) fall into non-protein coding DNA regions; 71% of SNPs lie in intergenic regions; and 25% of SNPs lie in intronic regions. We obtained a functional impact score for each variant through FunSeq2 scoring algorithms.60 The average functional impact score of EOC risk variants was 0.404, which is significantly higher than regional, matched background SNPs (0.2404; p value = 2.02 × 10−9; Figure S1B).
We performed enrichment analyses to test whether EOC risk SNPs are enriched within specific classes of biofeatures. We used StatePaintR43 to combine epigenomic marks into chromatin state calls that represent functional elements, including active, poised, silenced, and weak states of enhancers and promoters. We first evaluated enrichment of EOC risk SNPs with chromatin states from Roadmap Epigenomics and ENCODE for publicly available tissues.29,34 Enrichment tests were performed using FunciVar (see Material and Methods). Overall, we observed the greatest enrichment of EOC risk SNPs in active regulatory regions in digestive, immune, epithelial, liver, thymus, smooth muscle, and stem cell types and each of the cancer-associated ENCODE2012 cell lines, which are all closely related cell types (Figure 2, Table S4). In contrast, we observed a depletion of EOC risk SNPs in heterochromatin in 68 cell types and an enrichment in polycomb repressed silenced regions in 48 cell types. Overall, these analyses indicate that the enrichment of EOC risk SNPs in active regulatory regions is typically more cell type restricted than in silenced regions.
Figure 2.
EOC Risk Variants Are Enriched in Active Regulatory Elements
Enrichment analyses were performed in different chromatin states in REMC and ENCODE tissues and cell lines. Enriched biofeatures are shown in red, depleted biofeatures in blue, and non-significantly enriched biofeatures in white. The size of the circle indicates the probability of enrichment, circles outlined met significance.
We observed the strongest enrichment in an active regulatory chromatin state in stimulated primary T helper cells (E041) and primary T helper memory cells (E037), where 165 and 128 of 1,432 EOC risk SNPs, respectively, overlapped active regions (Figure 2, Table S5). There was also enrichment in active regulatory regions in all digestive tissue types (sigmoid colon, rectal mucosa, small intestine, and stomach). By contrast, we found no evidence of enrichment for EOC risk SNPs in active regulatory regions in brain, heart, or lung tissues, but instead observed enrichment for silenced regions in these tissue types.
Enrichment of EOC Risk Variants in Regions Marked by H3K27Ac Peaks in Ovarian and Non-ovarian Cancer Tissues
Given the tissue-specific patterns of enrichment in active regulatory states, we restricted these analyses to regions only marked by H3K27Ac, the most widely profiled marks in Roadmap Epigenomics and ENCODE tissues. We also included in these analyses data we have generated through H3K27Ac-ChIP-seq profiling of primary tissues or cell lines for 26 ovarian cancers representing the different histotypes of invasive disease, and 6 normal cell lines representing putative cells of origin of the different ovarian cancer histotypes (see Material and Methods; Table S3).
We observed enrichment of EOC risk SNPs in H3K27Ac peaks in 38 of the 98 cell types from in Roadmap Epigenomics/ENCODE and depletion in only 10 cell types (track 1 of Figure 3 and Tables S6 and S7). EOC risk SNPs were most enriched in H3K27Ac in blood and T cell tissues and were significantly depleted in all seven brain cell types. After stratifying EOC risk SNPs by histological subtype, we found the strongest enrichment for risk variants at the 17q12 risk locus for the CCOC histotype; all 8 candidate causal SNPs at this locus lie in intronic regions of the HNF1B gene (hepatocyte nuclear factor 1 homeobox B [MIM: 189907]) (Figure 3 and Table S6), with the greatest enrichment in digestive (E106, E102, E101, E092, E085, E084) and liver (E080) tissues.
Figure 3.
Histotype-Specific Credible Causal Variants Show Different Patterns of Enrichment
Enrichment analyses were performed for each EOC histotype in active regulatory regions marked by H3K27Ac in Roadmap Epigenomics and ENCODE tissues and cell lines. Enriched biofeatures are shown in red, depleted biofeatures in blue, and non-significantly enriched biofeatures in white. The size of the circle indicates the probability of enrichment, circles outlined met significance.
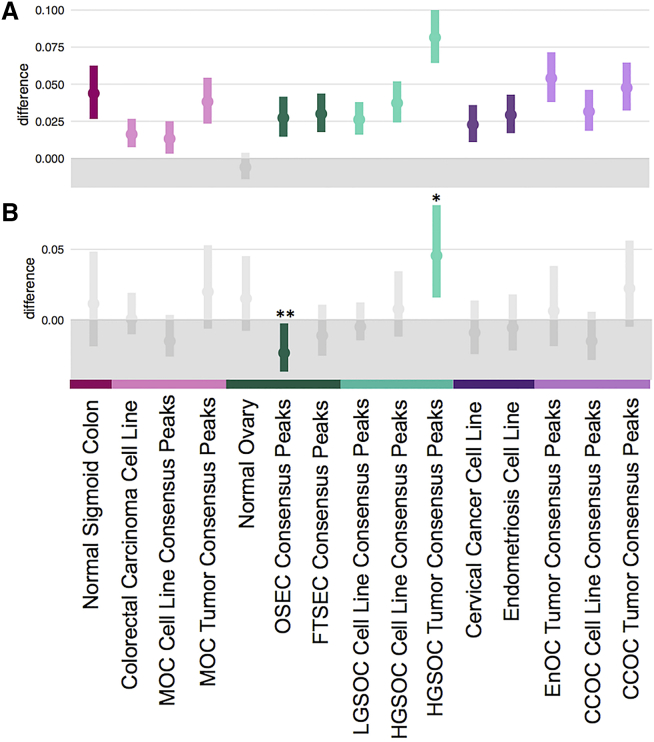
We next performed the same analysis for H3K27Ac marks profiled in 38 ovarian cancer-related tissues, including ovarian tumors for different histotypes, normal ovarian cancer precursor cell types and data from profiling of whole ovary specimens.36 We also compared these data to enrichment for other tissue types from Roadmap Epigenomics/ENCODE which may indicate other tissues of origin for ovarian cancers. We included the sigmoid colon as a potential cell type of origin for MOC, which currently has an undetermined origin72 but shows histological features similar to the gastrointestinal tract. The colorectal cancer cell line HCT116 was included as an additional model of cancers that arise from the gastrointestinal tract. The cervical cancer cell line HeLaS3 was included as a model of carcinoma of the cervix, which is a candidate cell type of origin for CCOC, and there are currently no suitable cell lines available to model normal cervix.72,73 We observed enrichment of EOC risk SNPs across all ovarian tissues except for whole ovary. The strongest enrichment was observed in H3K27Ac peaks in primary HGSOCs in which 197/1,432 SNPs (13.75%) overlapped H3K27Ac peaks, compared to 5.6% of the background (control SNPs) (p value < 0.001) (Figure 4A, Tables S8 and S9). In parallel, we also estimated enrichment of heritability in these H3K27Ac marks based on common SNPs with similar findings (Figure S3 and Supplemental Material and Methods, Enrichment of Common SNPs in Ovarian Cancer-Related H3K27Ac Peaks Based on Partitioned Heritability paragraph).
Figure 4.
Enrichment of EOC Risk Variants in Ovarian Cancer-Associated Tissues and Cell Lines
(A) EOC credible causal SNPs are significantly enriched in precursor (dark colors) and cell line models of EOC, and primary EOC tumors (light colors).
(B) Credible causal SNPs associated with HGSOC are enriched in active regulatory regions in primary HGSOCs (∗) and significantly depleted in ovarian surface epithelial cells (OSEC consensus peaks) (∗∗).
We repeated these analyses after stratifying the panel of candidate causal EOC risk SNPs by histotype. In total there were 315 candidate causal risk SNPs specific to HGSOC, 353 SNPs specific to LGSOC, 8 SNPs specific to CCOC, 8 SNPs specific to EnOC, 296 SNPs specific to MOC, and 47 SNPs specific to LMP histotypes. Risk SNPs for HGSOC were most significantly enriched in H3K27Ac marks in primary HGSOC tumors; 31/315 (9.8%) risk variants for HGSOC intersect H3K27Ac marks in primary HGSOCs, compared to local background SNPs (difference = 0.045, p value = 0.001; Figure 4B). We observed little or no enrichment for HGSOC risk SNPs in H3K27Ac marks generated in HGSOC cell lines, nor in normal FTSECs which are the reported precursors of HGSOC (Figure 4B). HGSOC risk SNPs were also significantly depleted in normal ovarian surface epithelial cells (OSECs). We also observed significant enrichment of risk variants associated with the LMP histotype in H3K27Ac marks in OSECs (Tables S10 and S11; Figure S2), but no tissue-specific enrichments for risk SNPs for other histotypes, which could largely be due to the lack of statistical power to detect enrichment.
In Silico Analysis of EOC Risk SNPs Intersecting Transcription Factor Binding Site (TFBS) Motifs
We evaluated the putative effects of the 590 EOC risk SNPs intersecting H3K27Ac marks on binding to TFBS motifs using the statistical tool motifbreakR.61 The 590 EOC risk SNPs were selected by intersecting with at least one H3K27Ac peak in any of the precursor normal or ovarian cancer cell lines or tumors. 469 out of 590 SNPs were predicted to significantly disrupt at least one TFBS (p value < 1 × 10−5; Table S12), compared to a background SNP set which was drawn from credible causal SNPs that did not intersect any EOC-related H3K27Ac marks. Eighty-two SNPs were predicted to break a single TFBS; the remaining SNPs break two or more (on average four) motifs with 5 SNPs predicted to break more than 20 motifs (Figure 5A). At the 18q11.2 locus, which confers risk of HGSOC, rs9955681 located in an intron of LAMA3 (MIM: 600805) was predicted to break 67 different motifs; and at the 4q26 EOC locus, rs7671665, which is located in intron 2 of SYNPO2, was predicted to break 31 different motifs (Table S12, Figure 5A). We analyzed eQTL association from in The Cancer Genome Atlas (TCGA) eQTL databases for HGSOC and breast cancer, and GTEx normal ovary dataset (Table S12). Out of 490 SNPs, 99 were reported as eQTLs in at least one of the datasets implicating 18 genes at 8 EOC loci as candidate susceptibility genes. Of these 99 SNPs, 32 (32%) are associated with PAX8 (MIM: 167415) and/or a PAX8-antisence transcript LOC654433, which is a well-established transcription factor overexpressed in ovarian cancer.40,41,74,75
Figure 5.
EOC Risk SNPs Disrupt TF Motifs at Risk Loci
(A) Number of motifs disrupted by credible causal SNPs intersecting EOC-related H3K27Ac peaks.
(B) Number of times motif is broken by credible causal SNPs that overlap EOC-related H3K27Ac peaks.
(C) REST motif logo from motifbreakR.
The most frequently disrupted TFBS motifs were for REST (repressor element-1 silencing transcription factor [MIM: 600571]) disrupted by 19 SNPs across 12 loci (p value = 0.0028); TCF3 (Transcription factor 3 [MIM: 147141]) disrupted by 11 SNPs (p value = 0.0075); ID4 (DNA-binding protein inhibitor [MIM: 600581]), which was disrupted by 8 SNPs (p value = 0.0025); and EHF (epithelial-specific transcription factor [MIM: 605439]) broken by 6 SNPs (p value = 0.0001) (Figure 5B and Table S13). The six SNPs disrupting EHF binding locate at five EOC risk loci associated with serous and mucinous histotypes (1p36, 2q13, 2q31, 8q24, 19p13).
Discussion
Identifying the functional effects of common susceptibility variants identified by GWASs is an important step in delineating the biological mechanisms underlying disease and in understanding the earliest stages of disease pathogenesis. In this study, we examined the heritability for risk variants associated across ovarian cancer and for each of the different histotypes of disease. Moreover, we partitioned heritability into broad functional categories to identify the functional drivers of neoplastic initiation and progression. Partitioning heritability has enabled a powerful integration of GWAS findings and functional annotation of risk-associated variants, but its utility is often restricted to datasets with a large sample size to enable a calculation of SNP heritability with sufficient confidence. In this study, our analyses were limited to all invasive EOC and HGSOC case subjects where samples sizes were large enough. Samples sizes were relatively small for other, rarer EOC histotypes such that there was insufficient statistical power to perform SNP-heritability analyses. We might expect the heritability estimates, and the functional elements that this heritability partitions to, to improve as GWASs are performed in increasingly large sample sizes for these and other histotypes.
We identified enrichment of EOC credible causal SNPs into active regulatory elements marked by H3K27Ac in ENCODE and Roadmap Epigenomics public datasets. This indicated germline risk variants that contribute to disease biology via disruption of enhancer activity in cell- and tissue-specific active regulatory regions, rather than regulatory elements that are active across a broad range of cell types. We further identified strong enrichment of the full credible causal variant list in 14 of the 15 EOC-relevant cell types included. We observed clear patterns of enrichment of HGSOC germline risk SNPs in HGSOC tumors, and depletion of these variants in H3K27Ac from precursor normal cells. These findings suggest that HGSOC germline risk variants affect cancer progression or development rather than initiation and underscore the need for variant annotation using cell types relevant to disease. It has been recognized that human primary tissue specimens serve as better representation of tumors than immortalized normal and cancer cell lines. Our lack of enrichment of risk variants in cell lines may be due in part to the lack of suitability of these models in evaluating the role of germline risk variants in tumor development. The significant enrichment for HGSOC credible causal SNPs in primary HGSOCs (n = 5) likely represents an underestimate of the true enrichment due to the small numbers of tumors; we anticipate a stronger signal if we use larger numbers of tumors in these analyses.
The cells of origin for the different histotypes of ovarian cancers are not precisely known. Fallopian tube epithelial cells are the most like precursors of HGSOCs and CCOC and EnOC are suspected to arise from endometriosis.4, 5, 6, 7, 8 Our comprehensive H3K27Ac ChIP-seq data in ovarian and non-ovarian cancer tissues makes it possible to identify the putative cells of origin of disease. The significant depletion of HGSOC credible causal variants we observed in H3K27Ac from OSECs active regions (Figure 4B) is consistent with an emerging consensus that HGSOC is less likely to arise from ovarian surface epithelial cells.4,5,76 The significant enrichment of LMP risk variants in OSECs active regions supports a role for this cell type in this histotype (Table S10 and Figure S2).77,78 It has been hypothesized, with supporting data from pathology examination,78 that ovarian surface epithelium invaginates into the underlying stroma of the ovary to form inclusion cysts that undergo transformation to become malignant.78 LMP and LGSOC are likely to arise from transformed OSECs trapped within inclusion cysts77 and the significant enrichment of six SNPs at two LMP rick loci (4q32.2 and 5p15) in OSECs (Table S11) supports an OSEC origin for these tumor types.
CCOCs are strongly associated with endometriosis and may derive from ciliated epithelial cells in ovarian endometriosis lesions.79,80 Only one locus has been confirmed to be associated with CCOC risk (the HNF1B 17q12 locus), which makes it challenging to investigate the likely cells of origin in the current study. This locus is pleiotropic for both HGSOC and CCOC, but we observed significant enrichment in H3K27Ac marks only for CCOC and MOC tumors and cell lines (Figure S4A). Here all eight candidate causal SNPs at 17q12 lie in intronic regions of HNF1B. HNF1B has been reported as a susceptibility gene and is highly expressed in CCOCs but largely absent in HGSOCs.7,81 We further investigated gene expression of HNF1B across our previously generated ovarian cancer tumor RNA-seq data.41 We found HNF1B is expressed in MOC, EnOC, and CCOC, but not in HGSOC (Figure S4B), which is consistent with the difference in H3K27Ac enrichment between histotypes.
We present here an approach to annotate risk SNPs that may influence transcriptional regulation by interacting with the epigenomic landscape to disrupt TF binding and alter gene regulation and expression. Functional validation would be the most convincing approach to confirm our in silico predictions of TFBS disrupted by risk variants within active regulatory regions. This would likely require genome editing assays to be developed in human cell line models to create isogenic cell lines for each SNP and to test their effects on binding of the relevant TFs, but this is beyond the scope of the current study. As an alternative, we have evaluated the evidence for these candidate TFs and risk variants through literature-based evidence searches and by confirming that several of the predicted impacted variants are expression QTLs in HGSOCs and breast cancer samples from TCGA, or in normal ovarian tissues profiled by GTEx (Table S12). For example, SNPs rs7671665 and rs9955681 were predicted to break the greatest number of motifs. We identified SNP rs7671665 that breaks 31 motifs within a regulatory element present in a wide range of Roadmap Epigenomics and ENCODE cell types and most of our panel of EOC-related cell types. This SNP is an eQTL located within intron 2 of SYNPO2 and is reported to loop to the promoter of SYNPO282 and METTL14 (MIM: 616504),83 a component of N6-methyladenosine (m6A) methyltransferase complex. This complex controls post translational modification of m6A RNA and has been implicated in cancer and cell differentiation and proliferation in development pathways.84 Interestingly, m6A is reported to be enriched in the 3′ UTR,85 which was the most significantly enriched biofeature in our partitioning of heritability analysis. Another example is SNP rs9955681, which is predicted to break 67 TF motifs in EOC tumor active regions. This SNP is located in an intron of LAMA3, a known enhancer in breast and cervical cancer cell lines and gastrointestinal tissues.29,33 This SNP is also a known eQTL in previous HGSOC susceptibility gene analyses.86
Our own study and other previous studies have identified a convergence of EOC risk variants on the PAX8 cistrome, with an enrichment of risk variants for serous ovarian cancer at PAX8 target genes.75 This is similar to findings for non-coding somatic variation in ovarian tumors, which are also enriched for PAX8 transcription factor binding sites and PAX8 targets.87 Identifying an increased burden of risk variants in TFBS motifs across the genome has the potential to uncover previously unknown biology and may explain how many risk variants across many loci act together to impact disease. We identified three TFs with motifs across the genome that were strongly enriched for risk variants. The results were supported by the published literatures: REST and ID4 both appear to have dual roles as oncogenic and antioncogenic neoplastic drivers.88,89 ID4 is associated with endometriosis risk and overexpressed in EOC tumors but not in normal tissues.90, 91, 92 EHF is also overexpressed in EOC tumors and induces apoptosis and impairs cell adhesion and invasion after knockdown in EOC cell lines.93
In conclusion, we have applied enrichment approaches to identify overrepresentations of risk SNPs within specific biofeatures. By intersecting risk SNPs with a catalog of regulatory elements, we identify putative enhancers impacted by risk variants that help explain the underlying functional mechanisms mediating genetic risk as ovarian cancer susceptibility loci. In additional we have shown the power of these approaches to elucidate the putative cells of origin of the different ovarian cancer histotypes, providing support for previously known cell types, and identifying other cell types associated with other histotypes. Finally, these studies have defined sets of putative causal variants at ovarian cancer risk loci that warrant further functional analyses to identify the genetic and regulatory mechanisms that drive initiation and early-stage development of ovarian cancers.
Consortia
We acknowledge the following investigators that contributed to OCAC: Joellen Schildkraut, Linda Kelemen, Georgia Chenevix-Trench, Penelope Webb, Peter Fasching, Diether Lambrechts, Anthony Swerdlow, Francisco Candido dos Reis, Digna Velez Edwards, James Brenton, Kexin Chen, Yan Li, Kang Shan, Javier Benítez, Cristina Rodriguez-Antona, María García, Larry Maxwell, Chad Hamilton, Cheryl Thompson, Jana Soukupova, Andrew Berchuck, Jennifer Doherty, Holly Harris, Antoinette Hollestelle, Ingrid Boere, Renée Fortner, Elio Riboli, Renée Fortner, Drakoulis Yannoukakos, Marc Goodman, Thilo Dörk-Bousset, Matthias Duerst, Natalia Bogdanova, Ralf Butzow, Heli Nevanlinna, Francesmary Modugno, Kirsten Moysich, Florian Heitz, Akira Hirasawa, Keitaro Matsuo, Sue Park, Beth Karlan, Håkan Olsson, Ellen Goode, Susanne Kruger Kjaer, Soo-Hwang Teo, Yin Ling Woo, Roger Milne, Michelle Hildebrandt, Loic Le Marchand, V. Wendy Setiawan, Alvaro Monteiro, Douglas Levine, Daniel Cramer, Kathryn Terry, Shelley Tworoger, Meir Stampfer, Walter Willett, Elisa Bandera, Line Bjorge, Lambertus Kiemeney, Tanja Pejovic, Linda Cook, Nhu Le, Nicolas Wentzensen, Jacek Gronwald, Estrid Høgdall, Claus Høgdall, Anita Koushik, Paul Pharoah, Dale P. Sandler, Alicja Wolk, Ian Campbell, Diana Eccles, Harvey Risch, Iain McNeish, Ros Glasspool, Nadeem Siddiqui, Weiva Sieh, Karin Sundfeldt, Björg Kristjansdottir, Wei Zheng, Rebecca Sutphen, Steven Narod, Andreas Hartkopf, Stefan Kommoss, Hoda Anton-Culver, Taymaa May, Simon Gayther, Usha Menon, Leigh Pearce, Anna Wu, David Huntsman, Emily White, Riki Peters, Anna DeFazio, and Jolanta Kupryjanczyk.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
This work was supported by NIH 7R01CA211574 (K.L. and S.A.G.), 5R0120745604 (K.L. and S.A.G.), R01CA204954 (M.L.F. and S.A.G.), 5R01CA211707 (S.A.G, M.L.F., and P.P.), R01CA193910 (M.L.F.), U01CA184826 (B.P.B. and S.A.G), and R01CA178535 (P.P. and S.A.G.), and Samuel Oschin Comprehensive Cancer Institute Cedars-Sinai Medical Center Developmental Funds (B.P.B.). K.L. is also supported by Ovarian Cancer Research Alliance Liz Tilberis Early Career Award (599175), an Ovarian Cancer Research Alliance Program Project Development Award (373356) and a Research Scholar’s Grant from the American Cancer Society (134005). M.L.F. is also supported by H.L. Snyder Medical Research Foundation.
The expression quantitative locus (eQTL) results published here are in part based upon data generated by The Cancer Genome Atlas (TCGA) Research Network.
Published: September 17, 2020
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2020.08.021.
Web Resources
FunciVar R package, https://github.com/Simon-Coetzee/funcivar
OMIM, https://www.omim.org/
StateHub-StatePaintR, http://www.statehub.org/
Data and Code Availability
Data availability for H3K27Ac ChIP-seq is detailed in Table S3. H3K27Ac for primary tumors is available in the Gene Expression Omnibus (GEO) under project GSE121103. H3K27Ac for OSEC, FTSEC, and EEC16 are available in the GEO under project GSE68104. H3K27Ac for VOA1056, GTFR230, EFO27, Kuramochi, JHOC5, MCAS, UWB1.289, OAW42, HeyA8, ES2, CaOV3, and RMG-II are available at GEO under GSE156275.
Supplemental Data
Tables S1–S13
References
- 1.Liu J., Cristea M.C., Frankel P., Neuhausen S.L., Steele L., Engelstaedter V., Matulonis U., Sand S., Tung N., Garber J.E., Weitzel J.N. Clinical characteristics and outcomes of BRCA-associated ovarian cancer: genotype and survival. Cancer Genet. 2012;205:34–41. doi: 10.1016/j.cancergen.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolton K.L., Chenevix-Trench G., Goh C., Sadetzki S., Ramus S.J., Karlan B.Y., Lambrechts D., Despierre E., Barrowdale D., McGuffog L., EMBRACE. kConFab Investigators. Cancer Genome Atlas Research Network Association between BRCA1 and BRCA2 mutations and survival in women with invasive epithelial ovarian cancer. JAMA. 2012;307:382–390. doi: 10.1001/jama.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alsop K., Fereday S., Meldrum C., deFazio A., Emmanuel C., George J., Dobrovic A., Birrer M.J., Webb P.M., Stewart C. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J. Clin. Oncol. 2012;30:2654–2663. doi: 10.1200/JCO.2011.39.8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klotz D.M., Wimberger P. Cells of origin of ovarian cancer: ovarian surface epithelium or fallopian tube? Arch. Gynecol. Obstet. 2017;296:1055–1062. doi: 10.1007/s00404-017-4529-z. [DOI] [PubMed] [Google Scholar]
- 5.Kim J., Park E.Y., Kim O., Schilder J.M., Coffey D.M., Cho C.-H., Bast R.C., Jr. Cell Origins of High-Grade Serous Ovarian Cancer. Cancers (Basel) 2018;10:10. doi: 10.3390/cancers10110433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kommoss F., Gilks C.B. Pathology of ovarian cancer: recent insights unveiling opportunities in prevention. Clin. Obstet. Gynecol. 2017;60:686–696. doi: 10.1097/GRF.0000000000000314. [DOI] [PubMed] [Google Scholar]
- 7.Kar S.P., Berchuck A., Gayther S.A., Goode E.L., Moysich K.B., Pearce C.L., Ramus S.J., Schildkraut J.M., Sellers T.A., Pharoah P.D.P. Common genetic variation and susceptibility to ovarian cancer: current insights and future directions. Cancer Epidemiol. Biomarkers Prev. 2018;27:395–404. doi: 10.1158/1055-9965.EPI-17-0315. [DOI] [PubMed] [Google Scholar]
- 8.Matulonis U.A., Sood A.K., Fallowfield L., Howitt B.E., Sehouli J., Karlan B.Y. Ovarian cancer. Nat. Rev. Dis. Primers. 2016;2:16061. doi: 10.1038/nrdp.2016.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones M.R., Kamara D., Karlan B.Y., Pharoah P.D.P., Gayther S.A. Genetic epidemiology of ovarian cancer and prospects for polygenic risk prediction. Gynecol. Oncol. 2017;147:705–713. doi: 10.1016/j.ygyno.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 10.Cuellar-Partida G., Lu Y., Dixon S.C., Fasching P.A., Hein A., Burghaus S., Beckmann M.W., Lambrechts D., Van Nieuwenhuysen E., Vergote I., Australian Ovarian Cancer Study Assessing the genetic architecture of epithelial ovarian cancer histological subtypes. Hum. Genet. 2016;135:741–756. doi: 10.1007/s00439-016-1663-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phelan C.M., Kuchenbaecker K.B., Tyrer J.P., Kar S.P., Lawrenson K., Winham S.J., Dennis J., Pirie A., Riggan M.J., Chornokur G., AOCS study group. EMBRACE Study. GEMO Study Collaborators. HEBON Study. KConFab Investigators. OPAL study group Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat. Genet. 2017;49:680–691. doi: 10.1038/ng.3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Permuth-Wey J., Lawrenson K., Shen H.C., Velkova A., Tyrer J.P., Chen Z., Lin H.-Y., Chen Y.A., Tsai Y.-Y., Qu X., Australian Cancer Study. Australian Ovarian Cancer Study. Consortium of Investigators of Modifiers of BRCA1/2 Identification and molecular characterization of a new ovarian cancer susceptibility locus at 17q21.31. Nat. Commun. 2013;4:1627. doi: 10.1038/ncomms2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolton K.L., Tyrer J., Song H., Ramus S.J., Notaridou M., Jones C., Sher T., Gentry-Maharaj A., Wozniak E., Tsai Y.-Y., Australian Ovarian Cancer Study Group. Australian Cancer Study (Ovarian Cancer) Ovarian Cancer Association Consortium Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat. Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchenbaecker K.B., Ramus S.J., Tyrer J., Lee A., Shen H.C., Beesley J., Lawrenson K., McGuffog L., Healey S., Lee J.M., EMBRACE. GEMO Study Collaborators. Breast Cancer Family Registry. HEBON. KConFab Investigators. Australian Cancer Study (Ovarian Cancer Investigators) Australian Ovarian Cancer Study Group. Consortium of Investigators of Modifiers of BRCA1 and BRCA2 Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat. Genet. 2015;47:164–171. doi: 10.1038/ng.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song H., Ramus S.J., Tyrer J., Bolton K.L., Gentry-Maharaj A., Wozniak E., Anton-Culver H., Chang-Claude J., Cramer D.W., DiCioccio R., Australian Cancer (Ovarian) Study. Australian Ovarian Cancer Study Group. Ovarian Cancer Association Consortium A genome-wide association study identifies a new ovarian cancer susceptibility locus on 9p22.2. Nat. Genet. 2009;41:996–1000. doi: 10.1038/ng.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kelemen L.E., Lawrenson K., Tyrer J., Li Q., Lee J.M., Seo J.-H., Phelan C.M., Beesley J., Chen X., Spindler T.J., Australian Cancer Study. Australian Ovarian Cancer Study Group. Ovarian Cancer Association Consortium Genome-wide significant risk associations for mucinous ovarian carcinoma. Nat. Genet. 2015;47:888–897. doi: 10.1038/ng.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goode E.L., Chenevix-Trench G., Song H., Ramus S.J., Notaridou M., Lawrenson K., Widschwendter M., Vierkant R.A., Larson M.C., Kjaer S.K., Wellcome Trust Case-Control Consortium. Australian Cancer Study (Ovarian Cancer) Australian Ovarian Cancer Study Group. Ovarian Cancer Association Consortium (OCAC) Ovarian Cancer Association Consortium (OCAC) A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat. Genet. 2010;42:874–879. doi: 10.1038/ng.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Couch F.J., Wang X., McGuffog L., Lee A., Olswold C., Kuchenbaecker K.B., Soucy P., Fredericksen Z., Barrowdale D., Dennis J., kConFab Investigators. SWE-BRCA. Ontario Cancer Genetics Network. HEBON. EMBRACE. GEMO Study Collaborators. BCFR. CIMBA Genome-wide association study in BRCA1 mutation carriers identifies novel loci associated with breast and ovarian cancer risk. PLoS Genet. 2013;9:e1003212. doi: 10.1371/journal.pgen.1003212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., Australian Cancer Study. Australian Ovarian Cancer Study. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) Gene Environment Interaction and Breast Cancer (GENICA) Swedish Breast Cancer Study (SWE-BRCA) Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) Epidemiological study of BRCA1 & BRCA2 Mutation Carriers (EMBRACE) Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers (GEMO) Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384, e1–e2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pharoah P.D.P., Tsai Y.-Y., Ramus S.J., Phelan C.M., Goode E.L., Lawrenson K., Buckley M., Fridley B.L., Tyrer J.P., Shen H., Australian Cancer Study. Australian Ovarian Cancer Study Group GWAS meta-analysis and replication identifies three new susceptibility loci for ovarian cancer. Nat. Genet. 2013;45:362–370, e1–e2. doi: 10.1038/ng.2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kar S.P., Beesley J., Amin Al Olama A., Michailidou K., Tyrer J., Kote-Jarai Z., Lawrenson K., Lindstrom S., Ramus S.J., Thompson D.J., ABCTB Investigators. AOCS Study Group & Australian Cancer Study (Ovarian Cancer) APCB BioResource. kConFab Investigators. NBCS Investigators. GENICA Network. PRACTICAL consortium Genome-Wide Meta-Analyses of Breast, Ovarian, and Prostate Cancer Association Studies Identify Multiple New Susceptibility Loci Shared by at Least Two Cancer Types. Cancer Discov. 2016;6:1052–1067. doi: 10.1158/2159-8290.CD-15-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawrenson K., Kar S., McCue K., Kuchenbaeker K., Michailidou K., Tyrer J., Beesley J., Ramus S.J., Li Q., Delgado M.K., GEMO Study Collaborators. EMBRACE. Hereditary Breast and Ovarian Cancer Research Group Netherlands (HEBON) KConFab Investigators. Australian Ovarian Cancer Study Group Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat. Commun. 2016;7:12675. doi: 10.1038/ncomms12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishizaki S.S., Boyle A.P. Mining the unknown: assigning function to noncoding single nucleotide polymorphisms. Trends Genet. 2017;33:34–45. doi: 10.1016/j.tig.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spielmann M., Mundlos S. Looking beyond the genes: the role of non-coding variants in human disease. Hum. Mol. Genet. 2016;25(R2):R157–R165. doi: 10.1093/hmg/ddw205. [DOI] [PubMed] [Google Scholar]
- 25.Hazelett D.J., Rhie S.K., Gaddis M., Yan C., Lakeland D.L., Coetzee S.G., Henderson B.E., Noushmehr H., Cozen W., Kote-Jarai Z., Ellipse/GAME-ON consortium. Practical consortium Comprehensive functional annotation of 77 prostate cancer risk loci. PLoS Genet. 2014;10:e1004102. doi: 10.1371/journal.pgen.1004102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smemo S., Tena J.J., Kim K.-H., Gamazon E.R., Sakabe N.J., Gómez-Marín C., Aneas I., Credidio F.L., Sobreira D.R., Wasserman N.F. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 32.Holwerda S.J.B., de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368:20120369. doi: 10.1098/rstb.2012.0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein B.E., Stamatoyannopoulos J.A., Costello J.F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M.A., Beaudet A.L., Ecker J.R. The NIH roadmap epigenomics mapping consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hernandez L., Kim M.K., Lyle L.T., Bunch K.P., House C.D., Ning F., Noonan A.M., Annunziata C.M. Characterization of ovarian cancer cell lines as in vivo models for preclinical studies. Gynecol. Oncol. 2016;142:332–340. doi: 10.1016/j.ygyno.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrenson K., Sproul D., Grun B., Notaridou M., Benjamin E., Jacobs I.J., Dafou D., Sims A.H., Gayther S.A. Modelling genetic and clinical heterogeneity in epithelial ovarian cancers. Carcinogenesis. 2011;32:1540–1549. doi: 10.1093/carcin/bgr140. [DOI] [PubMed] [Google Scholar]
- 38.Coetzee S.G., Shen H.C., Hazelett D.J., Lawrenson K., Kuchenbaecker K., Tyrer J., Rhie S.K., Levanon K., Karst A., Drapkin R., Ovarian Cancer Association Consortium, The Consortium of Investigators of Modifiers of BRCA1/2. Ovarian Cancer Association Consortium The Consortium of Investigators of Modifiers of BRCA1/2 Cell-type-specific enrichment of risk-associated regulatory elements at ovarian cancer susceptibility loci. Hum. Mol. Genet. 2015;24:3595–3607. doi: 10.1093/hmg/ddv101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrenson K., Notaridou M., Lee N., Benjamin E., Jacobs I.J., Jones C., Gayther S.A. In vitro three-dimensional modeling of fallopian tube secretory epithelial cells. BMC Cell Biol. 2013;14:43. doi: 10.1186/1471-2121-14-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adler E.K., Corona R.I., Lee J.M., Rodriguez-Malave N., Mhawech-Fauceglia P., Sowter H., Hazelett D.J., Lawrenson K., Gayther S.A. The PAX8 cistrome in epithelial ovarian cancer. Oncotarget. 2017;8:108316–108332. doi: 10.18632/oncotarget.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corona R.I., Seo J.-H., Lin X., Hazelett D.J., Reddy J., Abassi F., Lin Y.G., Mhawech-Fauceglia P.Y., Lester J., Shah S.P. Non-coding Somatic Mutations Converge on the PAX8 Pathway in Epithelial Ovarian Cancer. Nat. Commun. 2019 doi: 10.1038/s41467-020-15951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., Bernstein B.E., Bickel P., Brown J.B., Cayting P. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coetzee S.G., Ramjan Z., Dinh H.Q., Berman B.P., Hazelett D.J. StateHub-StatePaintR: rapid and reproducible chromatin state evaluation for custom genome annotation. F1000Res. 2017;7:214. [Google Scholar]
- 44.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finucane H.K., Bulik-Sullivan B., Gusev A., Trynka G., Reshef Y., Loh P.-R., Anttila V., Xu H., Zang C., Farh K., ReproGen Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. RACI Consortium Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium. Schizophrenia Working Group of the Psychiatric Genomics Consortium. SWE-SCZ Consortium Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lindblad-Toh K., Garber M., Zuk O., Lin M.F., Parker B.J., Washietl S., Kheradpour P., Ernst J., Jordan G., Mauceli E., Broad Institute Sequencing Platform and Whole Genome Assembly Team. Baylor College of Medicine Human Genome Sequencing Center Sequencing Team. Genome Institute at Washington University A high-resolution map of human evolutionary constraint using 29 mammals. Nature. 2011;478:476–482. doi: 10.1038/nature10530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ward L.D., Kellis M. Evidence of abundant purifying selection in humans for recently acquired regulatory functions. Science. 2012;337:1675–1678. doi: 10.1126/science.1225057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013;41:827–841. doi: 10.1093/nar/gks1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trynka G., Sandor C., Han B., Xu H., Stranger B.E., Liu X.S., Raychaudhuri S. Chromatin marks identify critical cell types for fine mapping complex trait variants. Nat. Genet. 2013;45:124–130. doi: 10.1038/ng.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dayem Ullah A.Z., Lemoine N.R., Chelala C. A practical guide for the functional annotation of genetic variations using SNPnexus. Brief. Bioinform. 2013;14:437–447. doi: 10.1093/bib/bbt004. [DOI] [PubMed] [Google Scholar]
- 55.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 56.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zerbino D.R., Wilder S.P., Johnson N., Juettemann T., Flicek P.R. The ensembl regulatory build. Genome Biol. 2015;16:56. doi: 10.1186/s13059-015-0621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fu Y., Liu Z., Lou S., Bedford J., Mu X.J., Yip K.Y., Khurana E., Gerstein M. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014;15:480. doi: 10.1186/s13059-014-0480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coetzee S.G., Coetzee G.A., Hazelett D.J. motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinformatics. 2015;31:3847–3849. doi: 10.1093/bioinformatics/btv470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kheradpour P., Kellis M. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014;42:2976–2987. doi: 10.1093/nar/gkt1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J., Zhuang J., Iyer S., Lin X., Whitfield T.W., Greven M.C., Pierce B.G., Dong X., Kundaje A., Cheng Y. Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res. 2012;22:1798–1812. doi: 10.1101/gr.139105.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulakovskiy I.V., Medvedeva Y.A., Schaefer U., Kasianov A.S., Vorontsov I.E., Bajic V.B., Makeev V.J. HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic Acids Res. 2013;41:D195–D202. doi: 10.1093/nar/gks1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matys V., Kel-Margoulis O.V., Fricke E., Liebich I., Land S., Barre-Dirrie A., Reuter I., Chekmenev D., Krull M., Hornischer K. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stormo G.D. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 68.Shannon P., Richards M. R Package Version; 2014. MotifDb: An annotated collection of protein-DNA binding sequence motifs. [Google Scholar]
- 69.Jiang X., Finucane H.K., Schumacher F.R., Schmit S.L., Tyrer J.P., Han Y., Michailidou K., Lesseur C., Kuchenbaecker K.B., Dennis J. Shared heritability and functional enrichment across six solid cancers. Nat. Commun. 2019;10:431. doi: 10.1038/s41467-018-08054-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gazal S., Finucane H.K., Furlotte N.A., Loh P.-R., Palamara P.F., Liu X., Schoech A., Bulik-Sullivan B., Neale B.M., Gusev A., Price A.L. Linkage disequilibrium-dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dayem Ullah A.Z., Oscanoa J., Wang J., Nagano A., Lemoine N.R., Chelala C. SNPnexus: assessing the functional relevance of genetic variation to facilitate the promise of precision medicine. Nucleic Acids Res. 2018;46(W1):W109–W113. doi: 10.1093/nar/gky399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cheasley D., Wakefield M.J., Ryland G.L., Allan P.E., Alsop K., Amarasinghe K.C., Ananda S., Anglesio M.S., Au-Yeung G., Böhm M. The molecular origin and taxonomy of mucinous ovarian carcinoma. Nat. Commun. 2019;10:3935. doi: 10.1038/s41467-019-11862-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kurman R.J., Shih IeM. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am. J. Pathol. 2016;186:733–747. doi: 10.1016/j.ajpath.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Soriano A.A., de Cristofaro T., Di Palma T., Dotolo S., Gokulnath P., Izzo A., Calì G., Facchiano A., Zannini M. PAX8 expression in high-grade serous ovarian cancer positively regulates attachment to ECM via Integrin β3. Cancer Cell Int. 2019;19:303. doi: 10.1186/s12935-019-1022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kar S.P., Adler E., Tyrer J., Hazelett D., Anton-Culver H., Bandera E.V., Beckmann M.W., Berchuck A., Bogdanova N., Brinton L. Enrichment of putative PAX8 target genes at serous epithelial ovarian cancer susceptibility loci. Br. J. Cancer. 2017;116:524–535. doi: 10.1038/bjc.2016.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lawrenson K., Fonseca M., Segato F., Lee J., Corona R., Seo J.-H., Coetzee S., Lin Y., Pejovic T., Mhawech-Fauceglia P. Integrated Molecular Profiling Studies to Characterize the Cellular Origins of High-Grade Serous Ovarian Cancer. bioRxiv. 2018 doi: 10.1101/330597. [DOI] [Google Scholar]
- 77.Hauptmann S., Friedrich K., Redline R., Avril S. Ovarian borderline tumors in the 2014 WHO classification: evolving concepts and diagnostic criteria. Virchows Arch. 2017;470:125–142. doi: 10.1007/s00428-016-2040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurman R.J., Shih IeM. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am. J. Surg. Pathol. 2010;34:433–443. doi: 10.1097/PAS.0b013e3181cf3d79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kolin D.L., Dinulescu D.M., Crum C.P. Origin of clear cell carcinoma: nature or nurture? J. Pathol. 2018;244:131–134. doi: 10.1002/path.5009. [DOI] [PubMed] [Google Scholar]
- 80.Cochrane D.R., Tessier-Cloutier B., Lawrence K.M., Nazeran T., Karnezis A.N., Salamanca C., Cheng A.S., McAlpine J.N., Hoang L.N., Gilks C.B., Huntsman D.G. Clear cell and endometrioid carcinomas: are their differences attributable to distinct cells of origin? J. Pathol. 2017;243:26–36. doi: 10.1002/path.4934. [DOI] [PubMed] [Google Scholar]
- 81.Shen H., Fridley B.L., Song H., Lawrenson K., Cunningham J.M., Ramus S.J., Cicek M.S., Tyrer J., Stram D., Larson M.C., PRACTICAL Consortium. Australian Ovarian Cancer Study Group. Australian Cancer Study Epigenetic analysis leads to identification of HNF1B as a subtype-specific susceptibility gene for ovarian cancer. Nat. Commun. 2013;4:1628. doi: 10.1038/ncomms2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fishilevich S., Nudel R., Rappaport N., Hadar R., Plaschkes I., Iny Stein T., Rosen N., Kohn A., Twik M., Safran M. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford) 2017;2017:2017. doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liu J., Eckert M.A., Harada B.T., Liu S.-M., Lu Z., Yu K., Tienda S.M., Chryplewicz A., Zhu A.C., Yang Y. m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol. 2018;20:1074–1083. doi: 10.1038/s41556-018-0174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H. VIRMA mediates preferential m6A mRNA methylation in 3'UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4:10. doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lawrenson K., Li Q., Kar S., Seo J.-H., Tyrer J., Spindler T.J., Lee J., Chen Y., Karst A., Drapkin R., Australian Ovarian Cancer Study Group Cis-eQTL analysis and functional validation of candidate susceptibility genes for high-grade serous ovarian cancer. Nat. Commun. 2015;6:8234. doi: 10.1038/ncomms9234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Corona R.I., Seo J.-H., Lin X., Hazelett D.J., Reddy J., Fonseca M.A.S., Abassi F., Lin Y.G., Mhawech-Fauceglia P.Y., Shah S.P. Non-coding somatic mutations converge on the PAX8 pathway in ovarian cancer. Nat. Commun. 2020;11:2020. doi: 10.1038/s41467-020-15951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Negrini S., Prada I., D’Alessandro R., Meldolesi J. REST: an oncogene or a tumor suppressor? Trends Cell Biol. 2013;23:289–295. doi: 10.1016/j.tcb.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 89.Best S.A., Hutt K.J., Fu N.Y., Vaillant F., Liew S.H., Hartley L., Scott C.L., Lindeman G.J., Visvader J.E. Dual roles for Id4 in the regulation of estrogen signaling in the mammary gland and ovary. Development. 2014;141:3159–3164. doi: 10.1242/dev.108498. [DOI] [PubMed] [Google Scholar]
- 90.Fung J.N., Rogers P.A.W., Montgomery G.W. Identifying the biological basis of GWAS hits for endometriosis. Biol. Reprod. 2015;92:87. doi: 10.1095/biolreprod.114.126458. [DOI] [PubMed] [Google Scholar]
- 91.Beger C., Pierce L.N., Kruger M., Marcusson E.G., Robbins J.M., Welcsh P., Welch P.J., Welte K., King M.C., Barber J.R., Wong-Staal F. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl. Acad. Sci. USA. 2001;98:130–135. doi: 10.1073/pnas.98.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ren Y., Cheung H.W., von Maltzhan G., Agrawal A., Cowley G.S., Weir B.A., Boehm J.S., Tamayo P., Karst A.M., Liu J.F. Targeted tumor-penetrating siRNA nanocomplexes for credentialing the ovarian cancer oncogene ID4. Sci. Transl. Med. 2012;4:147ra112. doi: 10.1126/scitranslmed.3003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Potapov P.P. [Activity of NADP-dependent cytoplasmic dehydrogenases in the liver and adipose tissue of rats in the restorative period after hypokinesia] Kosm. Biol. Aviakosm. Med. 1989;23:89–90. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S13
Data Availability Statement
Data availability for H3K27Ac ChIP-seq is detailed in Table S3. H3K27Ac for primary tumors is available in the Gene Expression Omnibus (GEO) under project GSE121103. H3K27Ac for OSEC, FTSEC, and EEC16 are available in the GEO under project GSE68104. H3K27Ac for VOA1056, GTFR230, EFO27, Kuramochi, JHOC5, MCAS, UWB1.289, OAW42, HeyA8, ES2, CaOV3, and RMG-II are available at GEO under GSE156275.