An Arabidopsis phosphatidylinositol kinase regulates plant growth and disease resistance through interacting with and stabilizing an E3 ligase, providing clues how E3 is regulated at post-translational level.
Abstract
Root development is important for normal plant growth and nutrient absorption. Studies have revealed the involvement of various factors in this complex process, improving our understanding of the relevant regulatory mechanisms. Here, we functionally characterize the role of Arabidopsis (Arabidopsis thaliana) phosphatidylinositol 4-kinase γ2 (PI4Kγ2) in root elongation regulation, which functions to modulate stability of the RING-type E3 ligase MYB30-INTERACTING E3 LIGASE1 (MIEL1) and auxin metabolism. Mutant plants deficient in PI4Kγ2 (pi4kγ2) exhibited a shortened root length and elongation zone due to reduced auxin level. PI4Kγ2 was shown to interact with MIEL1, regulating its degradation and furthering the stability of transcription factor MYB30 (which suppresses auxin metabolism by directly binding to promoter regions of GH3.2 and GH3.6). Interestingly, pi4kγ2 plants presented altered hypersensitive response, indicating that PI4Kγ2 regulates synergetic growth and defense of plants through modulating auxin metabolism. These results reveal the importance of protein interaction in regulating ubiquitin-mediated protein degradation in eukaryotic cells, and illustrate a mechanism coordinating plant growth and biotic stress response.
The phosphatidylinositol (PI) signaling pathway is a vital regulator of biological function in eukaryotes. Phosphoinositides have long been recognized as major components of eukaryotic membranes essential for many functions in eukaryotes, including cell signaling, membrane dynamics, and trafficking (Ischebeck et al., 2010). The functions of the PI signaling pathway are less understood in plants compared with mammals and yeast.
Phosphatidylinositol-4-kinases (PI4Ks) phosphorylate PIs at the 4′ position of inositol ring to produce phosphatidylinositol-4-phosphate. A BLAST program analysis showed that there are 12 distinct loci of PI4K homologs in Arabidopsis (Arabidopsis thaliana): eight belonging to the type-II group (PI4Kγ1–8; none have been demonstrated as a functional PI4K) and four belonging to the type-III group (PI4KIIIα1, PI4KIIIα2, PI4KIIIβ1, and PI4KIIIβ2; Mueller-Roeber and Pical, 2002). PI4Ks are essential for many aspects of developmental processes including root hair growth (Preuss et al., 2006), chloroplast division (Okazaki et al., 2015), and pollen growth (Chapman and Goring, 2011). Moreover, a previous study showed that RabA4b and PI4Kβ1 localize to budding secretory vesicles in the trans-Golgi network and regulate secretory vesicle size (Kang et al., 2011). Phosphatidylinositol 4-phosphate interacts specifically with PLASTID DIVISION1 and PLASTID DIVISION2 proteins to regulate chloroplast division (Okazaki et al., 2015).
Although physiological roles of type-III PI4Ks have been reported, most of the eight type-II PI4Ks in Arabidopsis have not been functionally characterized. Type-II PI4Ks contain PI3/PI4 kinase domains and variable numbers (none, one, or two) of ubiquitin-like domains, whereas they lack the PI-binding domains such as pleckstrin homolog (in PI4Kα) or novel homology domains (in PI4Kβ; Mueller-Roeber and Pical, 2002). Lipid kinase activity has not previously been detected in type-II PI4Ks, whereas Arabidopsis PI4Kγ3, PI4Kγ4, and PI4Kγ7 were shown to possess autophosphorylation activity. AtPI4Kγ4 phosphorylates Ser/Thr residues of ubiquitin fusion degradation1 and regulatory particle non-ATPase10 (Galvão et al., 2008). TaPI4KIIγ has been confirmed as a stress-inducible gene in wheat (Triticum aestivum), which interacts with and phosphorylates wheat ubiquitin fusion degradation protein (Liu et al., 2013); however, the effects of ubiquitin fusion degradation1 and regulatory particle non-ATPase10 phosphorylation remain unknown. AtPI4Kγ3 selectively binds to a few PIs and is important for reinforcing plant responses to environmental stresses and in the delay of floral transition (Akhter et al., 2016). AtPI4Kγ5 interacts with the membrane-bound transcription factor ANAC078 to regulate auxin biosynthesis and leaf margin development (Tang et al., 2016). These studies indicate that type-II PI4Ks participate in the regulation of specific physiological processes through different mechanisms.
The functionality of proteins is tightly regulated by various kinds of posttranslational modifications. Ubiquitination is an essential regulatory mechanism that alters protein stability by mediating the proteasomal degradation of target proteins (Schwechheimer, 2018). Ubiquitination involves E1 ubiquitin-activating enzymes, E2 ubiquitin-conjugating enzymes, and E3 ligases (Rape, 2018). E3 ligases recognize and bind specifically to the substrate (Vierstra, 2009), and thus play key roles in protein degradation. Although an increasing number of reports show the crucial roles of E3 ligases in plants, only a few reports have focused on the regulation of E3 ligases at the post-translational level. Indeed, in addition to transcriptional regulation, E3 ligases are subjected to various post-translational modifications. Tyr phosphorylation of the E3 ligase GARU at Tyr-321 inhibits the GARU–GID1A interaction and hence degradation of GID1 (Nemoto et al., 2017). Cop9 Signalosome, which mediates the de-neddylation of cullins, is required to inactivate Cullin-RING E3 ubiquitin–Ligase complexes, and then deter substrate recognition and prevent Cullin-RING E3 ubiquitin–Ligase subunit instability by counteracting their autoubiquitination and subsequent degradation (Merlet et al., 2009). Pirh2 protein is a p53-induced E3 ligase regulated by different proteins: CaMK II and Cdk9 phosphorylate Pirh2, then abrogate its E3 ligase activity toward p53 (Duan et al., 2007; Bagashev et al., 2013); Tat-interactive protein of 60 kD interacts with Pirh2 to alter the subcellular localization of Pirh2 (Logan et al., 2004); and PLAGL2 interacts with Pirh2 to prevent the proteasomal degradation of Pirh2 (Zheng et al., 2007). These findings indicate that posttranslational modification is critical in E3 functions.
Many signaling pathways control root growth and development, particularly the phytohormone auxin, which regulates both cell proliferation and elongation of root cells (Street et al., 2016). Previous studies showed that AUX1 mediates the inhibitory effects of cytokinin on root cell elongation (Péret et al., 2012; Street et al., 2016). YDK1, a GH3 gene, might function as a negative regulator of root cell elongation by controlling the level of the active form of auxin (Takase et al., 2004). In rice (Oryza sativa), OsARF12, a transcription activator of auxin-response genes, mediates root cell elongation by inhibiting most OsYUCCAs auxin-synthesis genes (Qi et al., 2012). In addition, phospholipase d-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN2 (Lin et al., 2020). Phosphatidylinositol monophosphate 5-kinase2 and phosphatidylinositol monophosphate 5-kinase1 are involved in root development regulation through regulation of PIN proteins (Mei et al., 2012) and PIN-mediated auxin efflux (Barbosa et al., 2016).
Recent genetic studies have revealed important roles of distinct E3 ligases in phytohormone signaling pathways (Kelley, 2018), operating via the degradation of various components, including receptors, positive or negative regulators, or those related to hormone metabolisms. Through systematic biochemical and genetic studies, we here report the functional characterization of an Arabidopsis type-II PI4K, PI4Kγ2, that regulates root growth through its interaction with the ubiquitin E3 ligase MYB30-INTERACTING E3 LIGASE1 (MIEL1). MIEL1 is a negative regulator of hypersensitive response (HR) cell death by promoting MYB30 turnover in Arabidopsis (Marino et al., 2013). MIEL1 protein level is significantly reduced in pi4kγ2 mutant plants and MYB30 directly binds to promoter regions of GH3.2 and GH3.6, thereby suppressing auxin metabolism and hence root growth. Our data demonstrate that PI4Kγ2 is critical for MIEL1 stability and MYB30 turnover, and elucidate how MIEL1 coordinates auxin-dependent plant growth and defensive HR in Arabidopsis.
RESULTS
Deficiency of PI4Kγ2 Results in Short Roots
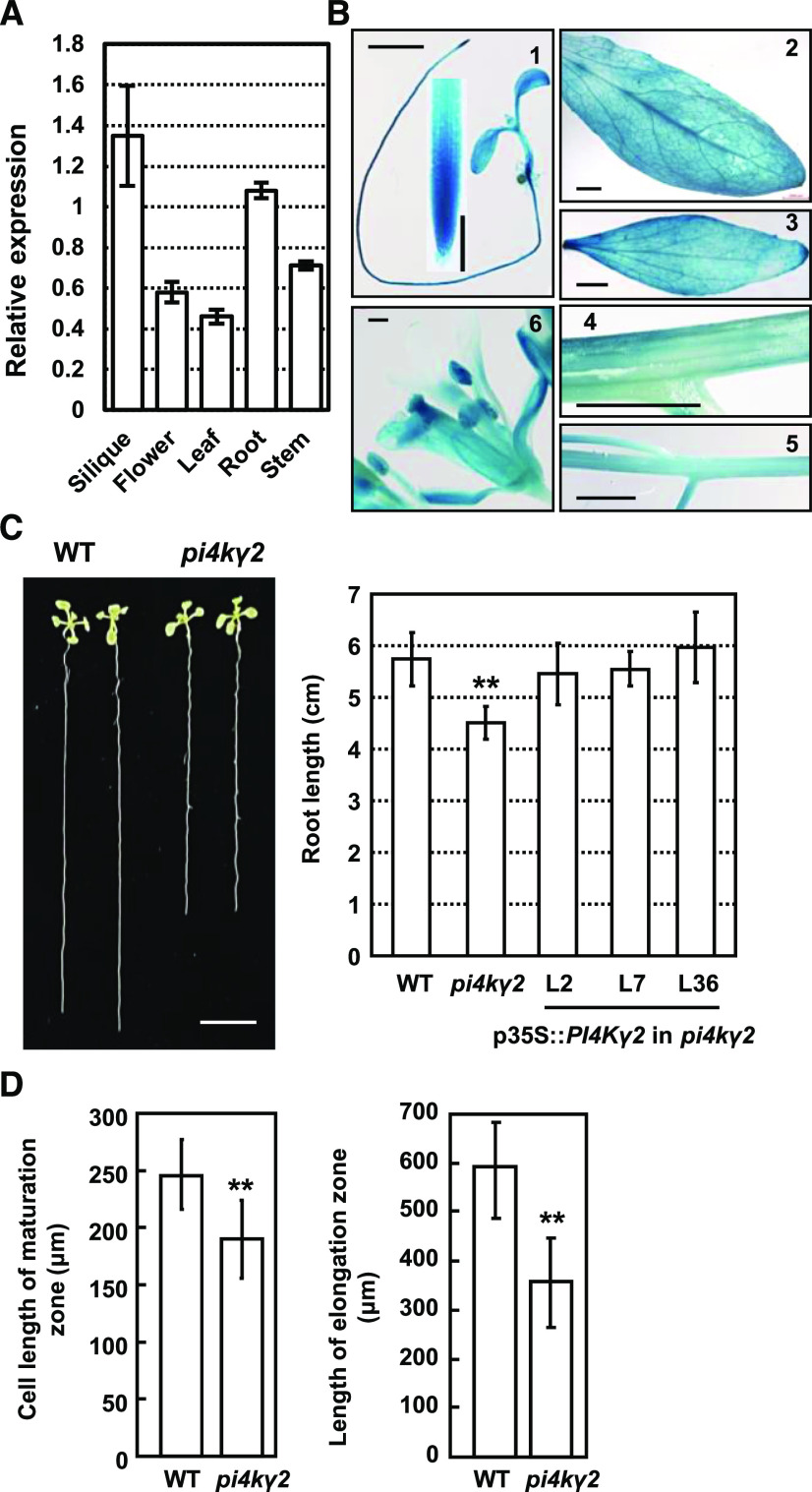
Type-II PI4Ks, which lack the PI-binding domains, play roles in plant development via protein–protein interaction or protein phosphorylation (Mueller-Roeber and Pical, 2002; Galvão et al., 2008; Liu et al., 2013). Our previous study showed that Arabidopsis PI4Kγ5 regulates auxin biosynthesis and leaf margin development by interacting with and regulating the cleavage of the membrane-bound transcription factor ANAC078 (Tang et al., 2016). To expand our understanding of type-II PI4K functions, we further functionally characterized AtPI4Kγ2. Analysis by reverse-transcription quantitative PCR (RT-qPCR) revealed PI4Kγ2 expression in various tissues (Fig. 1A). Histochemical analysis of transgenic lines harboring a PI4Kγ2 promoter-reporter construct (Glucuronidase, pPI4Kγ2::GUS) showed that PI4Kγ2 is expressed in young seedlings, floral tissue, rosette leaf, cauline leaf, stem, and inflorescence (Fig. 1B). GUS signals were predominantly observed in the root apical region of young seedlings (Fig. 1B, 1), suggesting a potential role of PI4Kγ2 in root development.
Figure 1.
Deficiency of PI4Kγ2 results in shortened roots. A, RT-qPCR analysis revealed PI4Kγ2 expression in various tissues including leaf, stem, flower, root, and silique. ACTIN7 gene was used as an internal reference. Analysis were biologically repeated three times and data are presented as means ± se (n = 3). B, Promoter–GUS fusion analysis revealed that PI4Kγ2 is expressed in young seedlings (1), rosette leaf (2), cauline leaf (3), stem (4), inflorescence (5), and floral tissue (6). Scale bars = 200 μm (1 and 6) and 2 mm (2–5). C, Observation (left) and measurement (right) of root growth of 10-d–old wild-type (WT), pi4kγ2, and pi4kγ2 seedlings expressing p35S::PI4Kγ2-mCherry. Scale bar = 1 cm. Experiments were repeated three times and data are presented as means ± sd (n > 30). Statistical analysis revealed significant difference (**P < 0.01). D, pi4kγ2 plants exhibited a decreased root elongation zone length and root epidermal cell length in the maturation zone compared to those of wild type. Roots of 7-d–old seedlings were measured and statistically analyzed (**P < 0.01). Experiments were repeated three times and data are presented as means ± sd (n = 30).
A transfer DNA (T-DNA) insertional mutant pi4kγ2 (Alonso et al., 2003) was identified, which we used to investigate the physiological role of PI4Kγ2. T-DNA was inserted in the first exon of PI4Kγ2 (Supplemental Fig. S1, A and B) and RT-qPCR analysis confirmed the deficient transcription of PI4Kγ2 in homozygous mutant plant (Supplemental Fig. S1C), indicating that pi4kγ2 is a knockout mutant.
Phenotypic observation showed that growth of the pi4kγ2 mutant is similar to wild type; however, interestingly, it presents short roots (Fig. 1C). Considering that primary root growth is dependent on both cell proliferation and cell elongation, detailed analysis revealed a decreased length of root elongation zone and decreased root epidermal cell length in the maturation zone for pi4kγ2 (Fig. 1D). Considering that there was no change of length of meristematic zone of pi4kγ2 roots (Supplemental Fig. S1D), these results suggest that PI4Kγ2 regulates root growth by modulating cell length. As expected, complementation studies demonstrated that reinstated PI4Kγ2 expression (Supplemental Fig. S2) resulted in recovered root growth, confirming the role of PI4Kγ2 in root development.
PI4Kγ2 Interacts with MIEL1
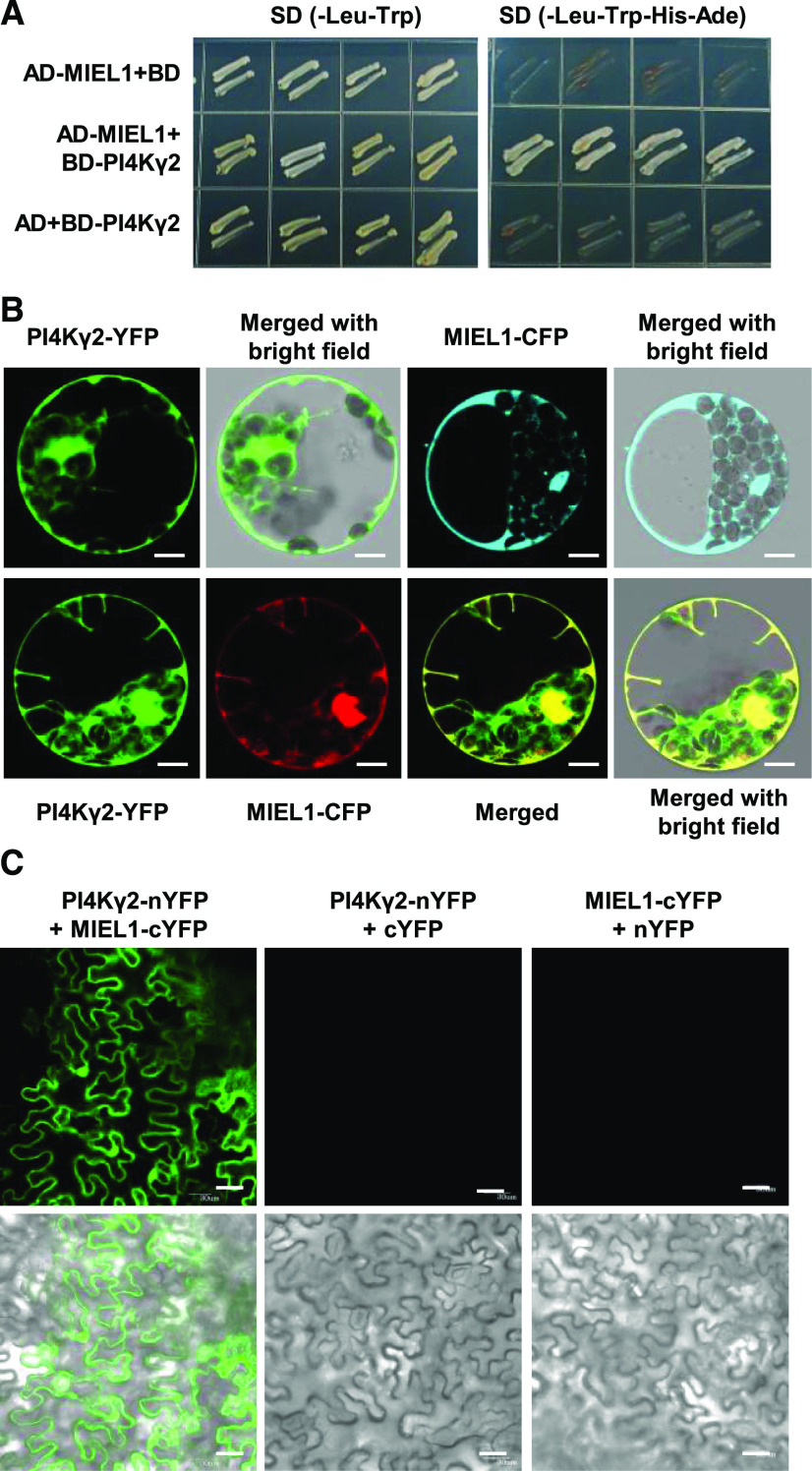
To investigate the functional mechanism of how PI4Kγ2 regulates root cell elongation, yeast two-hybrid screening was performed using whole PI4Kγ2 protein as bait to search for PI4Kγ2-interacting partners. Interestingly, MIEL1, a zinc finger RING-type protein localized to the cytoplasm and nucleus (Marino et al., 2013), was identified (Fig. 2A). Subcellular localization studies by observing fluorescence showed the distribution of yellow fluorescent protein (YFP)-PI4Kγ2 fusion protein in cytoplasm and nucleus of Arabidopsis cells, which is similar to that of MIEL1 (Fig. 2B, upper). Transient expression of YFP-PI4Kγ2 and cyan fluorescent protein (CFP)-MIEL1 fusion proteins in Arabidopsis protoplasts confirmed that PI4Kγ2 colocalizes with MIEL1 (Fig. 2B, lower), further suggesting a possible interaction between them, which was investigated by using bimolecular fluorescence complementation (BiFC) assays. Indeed, strong fluorescence was detected in the nucleus and cytoplasm of cells co-expressing cYFP-MIEL1 and nYFP-PI4Kγ2 (Fig. 2C), confirming the PI4Kγ2-MIEL1 interaction in vivo.
Figure 2.
PI4Kγ2 interacts with E3 ligase MIEL1. A, Yeast two-hybrid assays revealed a direct interaction between PI4Kγ2 and MIEL1. Transformed yeast cells were grown on SD-Leu-Trp or SD-Leu-Trp-His-Ade. AD, Activation domain; BD, binding domain. B, Observation of transiently expressed fusion proteins in Arabidopsis protoplasts showed that PI4Kγ2 colocalizes with MIEL1. Scale bars = 10 μm. C, BiFC assays confirmed the MIEL1-PI4Kγ2 interaction in planta. A model no. FV1000 confocal microscope (Olympus) was used and collection wavelength was set at 510 to 550 nm to strictly filter out the autofluorescence (details in “Materials and Methods” section), resulting in two control images (upper, middle and right) that are blank. Scale bars = 30 μm.
Deficiency of MIEL1 and MYB30 Results in Shorter or Longer Roots, Respectively
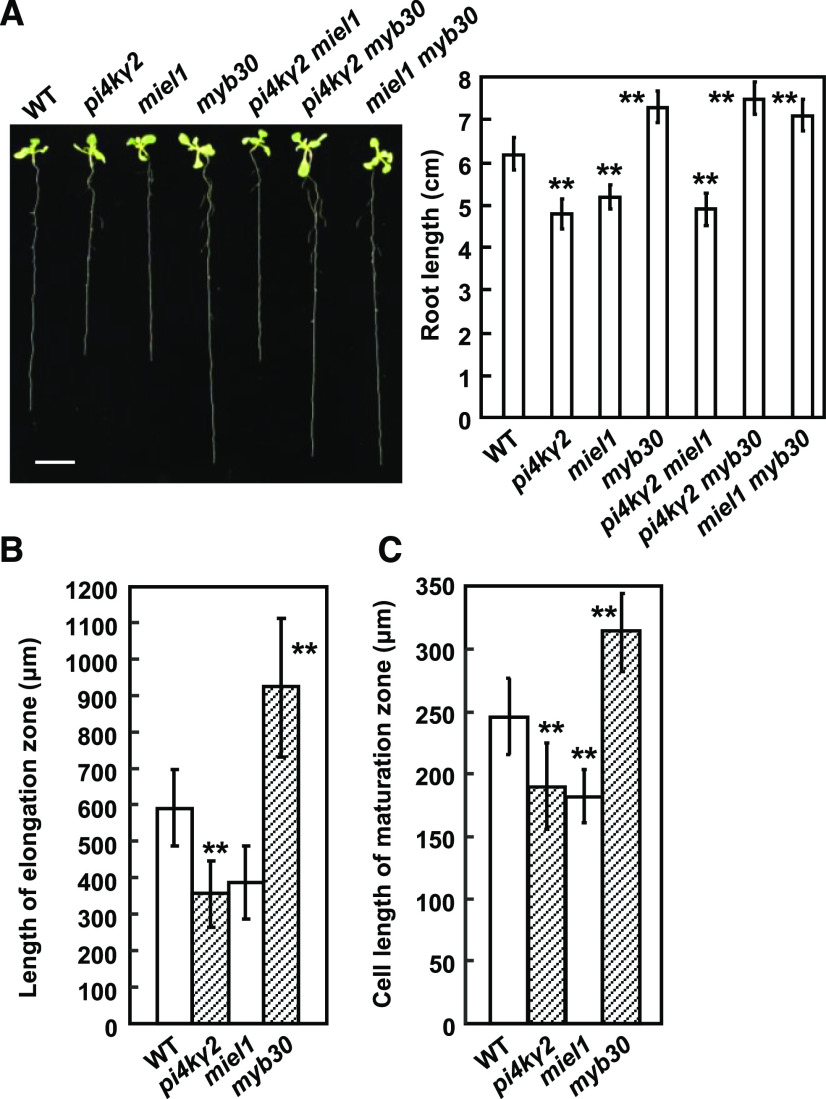
Previous studies showed that MIEL1 interacts with and ubiquitinates MYB30, leading to MYB30 proteasomal degradation and suppressed plant defense responses (Marino et al., 2013). Considering that PI4Kγ2 interacts with MIEL1, we thus investigated the function of MIEL1 and MYB30 in root growth. Observation of growth of miel1 (Marino et al., 2013) and myb30 knockout lines (Liu et al., 2014) showed that, similar to pi4kγ2 mutant, miel1 exhibited shortened roots and decreased length of root elongation zone and epidermal cells of the root maturation zone, whereas myb30 showed longer root length and increased length of root elongation zone and epidermal cells of root maturation zone (Fig. 3), which is consistent with MIEL1-facilitated MYB30 degradation and indicates a specific role for MIEL1 and MYB30 in regulating root growth.
Figure 3.
miel1 and myb30 mutants have shortened or elongated roots. A, Root length of 12-d-old wild type (WT), pi4kγ2, miel1, myb30, pi4kγ2 miel1, pi4kγ2 myb30, and miel1 myb30 seedlings were observed (left) and measured (right). Scale bar = 1 cm. B, Length of root elongation zone of 7-d-old wild type, pi4kγ2, miel1, and myb30 seedlings. C, Length of epidermal cell of root maturation zone of 7-d-old wild type, pi4kγ2, miel1, and myb30 seedlings. Experiments were repeated three times and data are presented as means ± sd (n > 30). Statistical analysis was performed using a two-tailed Student’s t test compared with wild type (**P < 0.01).
Genetic analysis showed that miel1 pi4kγ2 double mutants displayed similar root growth as miel1 and pi4kγ2, whereas pi4kγ2 myb30 or miel1 myb30 double mutants presented root growth similar to myb30 (longer roots), indicating the genetic epistasis of MYB30 and that root growth regulation by PI4Kγ2 and MIEL1 is achieved via the effect of MIEL1 on MYB30.
PI4Kγ2 Regulates MIEL1 Stability and Accelerates MYB30 Turnover In Planta
Many reports in animals have shown that functions of E3 ubiquitin ligase can be regulated at the posttranslational level, including de-neddylation, phosphorylation, and interaction with other proteins. To test whether MIEL1 is regulated by PI4Kγ2 through direct interaction in planta, MIEL1 fused with mCherry was expressed in wild type and pi4kγ2 to study the effects of PI4Kγ2 on MIEL1 in vivo.
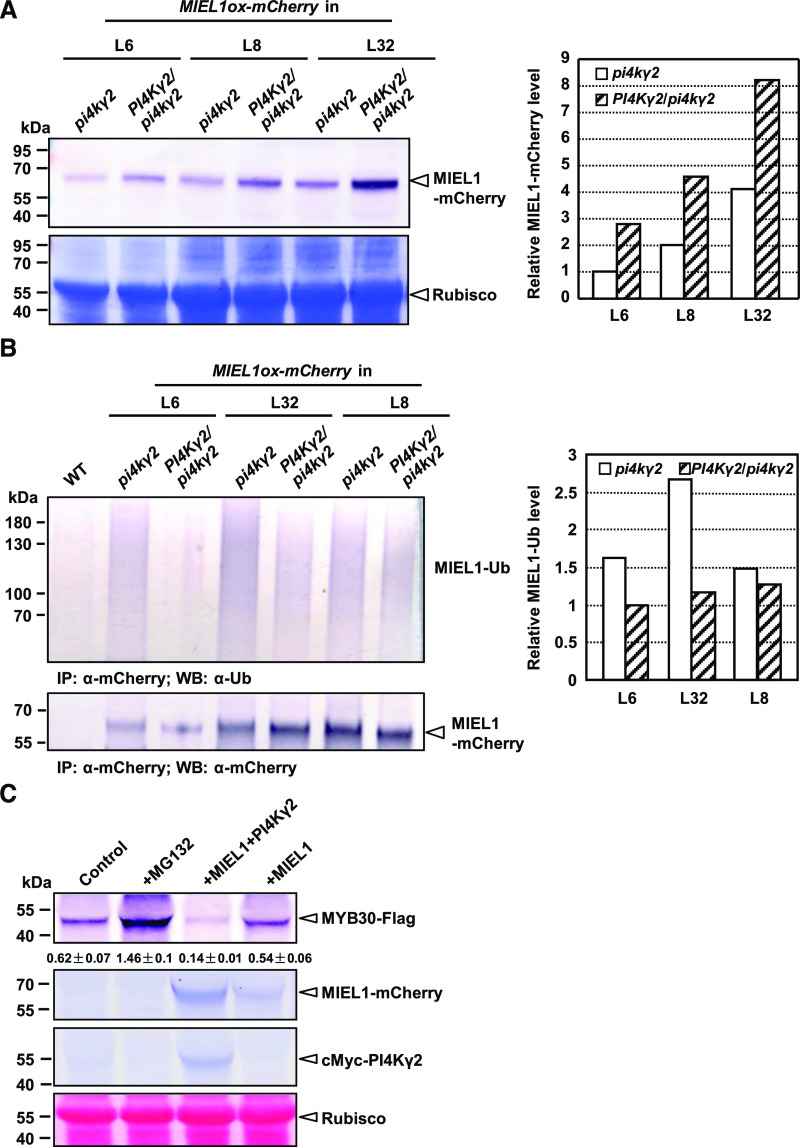
A quick cross was performed to obtain the isogenic MIEL1-mCherry plants. Pollens from MIEL1-mCherry (in pi4kγ2) homozygous lines were respectively spread on the stigmas of pi4kγ2 and wild type, and analysis of F1 plants with comparable MIEL1 expression levels by immunoblot analysis showed that MIEL1-mCherry accumulated less in pi4kγ2 compared to that in PI4Kγ2/pi4kγ2 (Fig. 4A), suggesting that PI4Kγ2 functions in regulating MIEL1 stability. Consistently, transient expression of FLAG-tagged MIEL1 alone or with Myc-tagged PI4Kγ2 in Nicotiana benthamiana leaves further showed greater accumulation of MIEL1 protein after the co-expression of PI4Kγ2 (Supplemental Fig. S3A). These results indicate that PI4Kγ2 positively regulates MIEL1 stability through direct interaction.
Figure 4.
PI4Kγ2 regulates MIEL1 stability and accelerates MYB30 turnover in planta. A, Immunoblot analysis showed that deficiency of PI4Kγ2 leads to reduced accumulation of MIEL1-mCherry. Wild-type (WT) lines expressing MIEL1 (PI4Kγ2/pi4kγ2) were generated by crossing homozygous pi4kγ2 lines expressing MIEL1. Seven-day-old seedlings were used to extract proteins and analyzed with an mCherry antibody (leaf upper). Coomassie Brilliant Blue staining showed the protein loading (leaf bottom). Band density of MIEL1-mCherry was measured using the software ImageJ (https://imagej.en.softonic.com/) by setting band density in pi4kγ2 (L6) as “1” (right). B, PI4Kγ2 regulates the ubiquitination of MIEL1. Immunoprecipitation (IP) assays showed that deficiency of PI4Kγ2 leads to increased ubiquitination of MIEL1. Wild-type lines expressing MIEL1 (PI4Kγ2/pi4kγ2) were generated by crossing homozygous pi4kγ2 lines expressing MIEL1. Seven-day-old seedlings were used to extract proteins and analyzed using the α-Ubiquitin (α-UB) antibody (left upper). Loading proteins were quantified by using an mCherry antibody (left bottom). Band density of ubiquitinated MIEL1 was measured using the software ImageJ by setting band density in PI4Kγ2/pi4kγ2 (L6) as “1” (right). C, PI4Kγ2 accelerated the degradation of MYB30. FLAG-tagged MYB30 was expressed in N. benthamiana in the absence or presence of mCherry-tagged MIEL1 or Myc-tagged PI4Kγ2, or treated with proteasome inhibitor MG132. Levels of MYB30, MIEL1, and PI4Kγ2 were analyzed by immunoblotting with relevant antibodies. Ponceau-S staining confirmed the equal loading of proteins. Band density is measured by the software ImageJ and relative density of MYB30-FLAG was calculated by dividing the density of MYB30-Flag by that of Rubisco. Data are presented as means ± sd (n = 3).
MIEL1 is degraded by the ubiquitin-26S proteasome pathway (Lee and Seo, 2016). Considering the PI4Kγ2 effects on MIEL1 stability, whether PI4Kγ2 affects the ubiquitination of MIEL1 in vivo and hence, degradation, was investigated by immunoblot analysis. Total protein extracted from 35S::MIEL1-mCherry (pi4kγ2) and 35S::MIEL1-mCherry (PI4Kγ2/pi4kγ2) seedlings were immunoprecipitated with an anti-mCherry antibody, and the resultant precipitates were subsequently analyzed by immunoblot assays with anti-Ub or anti-mCherry antibody (Fig. 4B, left bottom). Indeed, analysis revealed increased ubiquitination of MIEL1 in the pi4kγ2 mutant (Fig. 4B, upper; Supplemental Fig. S3B), confirming that PI4Kγ2 affects MIEL1 ubiquitination via their interaction.
Several type-II PI4K members (PI4Kγ1, PI4Kγ4, PI4Kγ5, and PI4Kγ7) are Ser/Thr protein kinases (Galvão et al., 2008; Tang et al., 2016); however, in vitro phosphorylation assay by autoradiograph showed that there was no autophosphorylation of PI4Kγ2 and that PI4Kγ2 does not phosphorylate MIEL1 (Supplemental Fig. S3C), further indicating that PI4Kγ2 affects MIEL1 ubiquitination and stability via their interaction.
MIEL1 mediates the proteasomal degradation of MYB30 (Marino et al., 2013). We thus examined whether PI4Kγ2-MIEL1 interaction influences the degradation and turnover of MYB30. Analysis of N. benthamiana leaves transiently expressing FLAG-tagged MYB30 showed that MYB30 protein accumulated after treatment with proteasome inhibitor MG132 and was reduced after the co-expression of MIEL1 (Fig. 4C), which is consistent with the previous reports (Marino et al., 2013). Interestingly, MYB30 protein level was significantly reduced when co-expressed with PI4Kγ2 and MIEL1 (Fig. 4C), indicating that PI4Kγ2 regulates the turnover of MYB30 through interacting with and stabilizing MIEL1.
PI4Kγ2 Regulates Auxin Metabolism through MYB30
Root growth is a complex process and regulated by various developmental signals and environmental factors, particularly phytohormones. Auxin level, signaling, and polar transport are involved in root growth regulation. Considering that free indole-3-acetic acid (IAA) plays main roles in auxin effects, we first investigated whether IAA level is altered in pi4kγ2 by liquid chromatography-mass spectrometry (LC-MS) analysis and results revealed decreased free IAA content in pi4kγ2 roots (Fig. 5A). Consistently, the short-root phenotype of pi4kγ2 and miel1 was recused by exogenous IAA and both mutants presented similar root length under 0.01 μm of IAA (Fig. 5B), confirming that reduced IAA level leads to the short-root phenotype of pi4kγ2 and miel1.
Figure 5.
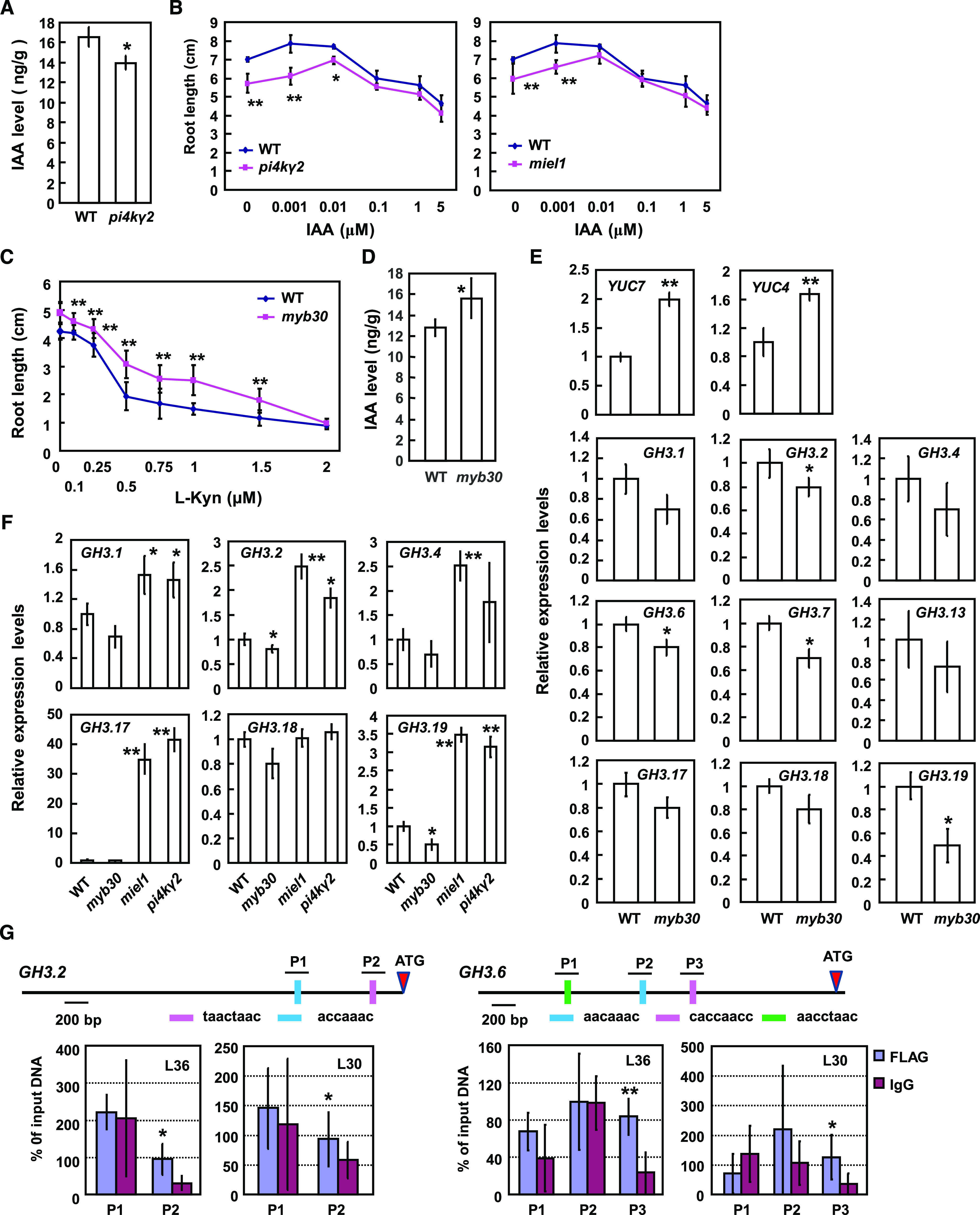
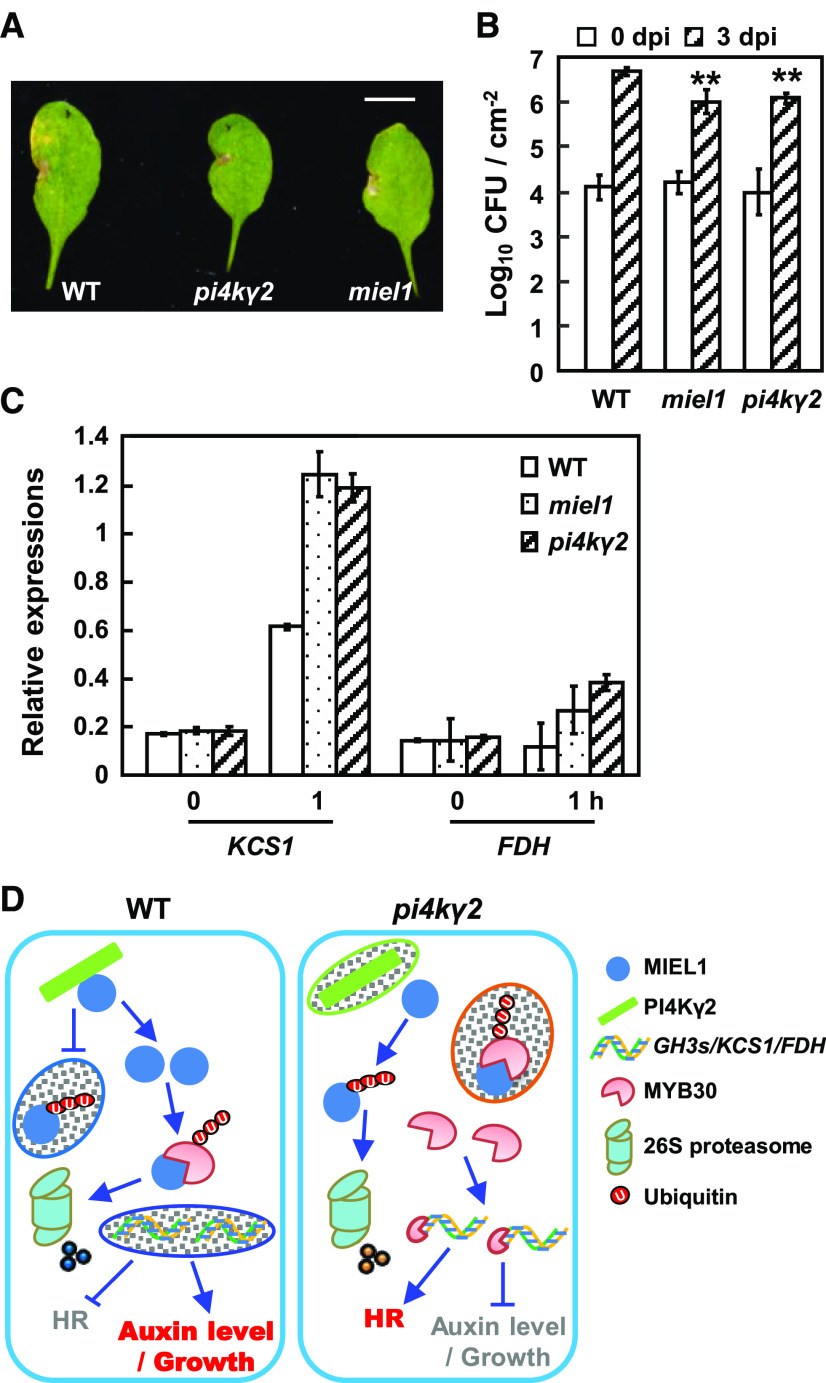
PI4Kγ2 deficiency results in reduced auxin level and MYB30 deficiency results in altered expression levels of GH3 or YUC genes in planta. A, Quantification of free IAA content by LC-MS revealed a reduced IAA level in pi4kγ2. Roots of 5-d-old wild-type (WT) and pi4kγ2 seedlings were used for analysis and data are presented as means ± sd (n = 5). Statistical analysis was performed using a two-tailed Student’s t test (*P < 0.05, compared to wild type). B, Wild-type, miel1, and pi4kγ2 seedlings were grown on Murashige-and-Skoog medium supplemented with IAA for 15 d, and root lengths were measured. Experiments were repeated three times and data are presented as means ± sd (n > 30). Statistical analysis revealed significant differences compared to wild type (*P < 0.05 and **P < 0.01). C, Wild-type and myb30 seedlings were grown on Murashige-and-Skoog medium supplemented with IAA biosynthesis inhibitor l-Kynurenine (l-Kyn) for 10 d, and root lengths were measured. Experiments were repeated three times and data are presented as means ± sd (n > 30). Statistical analysis revealed the significant differences compared to wild type (*P < 0.05 and **P < 0.01). D, Quantification of free IAA content by LC-MS revealed the increased IAA level in myb30. Roots of 5-d-old wild type and myb30 seedlings were used for analysis and data are presented as means ± sd (n = 5). Statistical analysis was performed using a two-tailed Student’s t test compared to wild type (*P < 0.05). E, Relative expression levels of YUCCA and GH3 genes in myb30 and wild type (expression of examined genes in wild type was set as “1”). Roots of 7-d–old seedlings were used for RNA extraction and RT-qPCR analysis. ACTIN7 gene was used as an internal reference. Experiments were repeated three times and data are presented as means ± se (n = 3). Statistical analysis was performed using a two-tailed Student’s t test compared to wild type (*P < 0.05 and **P < 0.01). F, Relative expression levels of GH3 genes in wild type, pi4kγ2, miel1, and myb30 (expression of examined genes in wild type was set as “1”). Roots of 7-d-old seedlings were used for RNA extraction and RT-qPCR analysis. ACTIN7 gene was used as an internal reference. Experiments were repeated three times and data are presented as means ± se (n = 3). Statistical analysis was performed using a two-tailed Student’s t test compared to wild type (*P < 0.05 and **P < 0.01). G, ChIP-qPCR analysis showed that MYB30 directly binds the promoter regions of GH3.2 and GH3.6 to regulate their expression. Two independent lines expressing MYB30-FLAG (L30 and L36) were used for analysis. Input DNA (from the un-immunoprecipitated DNA) was used as a positive control (100%). Immunoprecipitated DNA from IgG chromatin was used as a negative control. P1, P2, and P3 are the putative MYB30 binding sequences in promoters of GH3.2 and GH3.6. Statistical analysis was performed using a two-tailed Student’s t test compared to Input DNA (*P < 0.05 and **P < 0.01).
Considering the genetic epistasis of MYB30 on PI4Kγ2 and MILE1, and that MIEL1 mediates the proteasomal degradation of MYB30, it was hypothesized that MYB30 negatively regulates auxin metabolism and hence, cell elongation. The chemical compound l-Kynurenine (l-Kyn, an inhibitor of TAA1/TAR to suppress auxin synthesis; He et al., 2011) was utilized to test whether the phenotype of myb30 was the result of increased auxin accumulation. Indeed, the longer root of myb30 was suppressed under l-Kyn treatment (similar root length was observed at 2 μm of l-Kyn; Fig. 5C), suggesting the longer roots of myb30 might be due to increased auxin content. This was confirmed by increased free IAA level of myb30 roots demonstrated by LC-MS (Fig. 5D). As expected, RT-qPCR analysis of expression of some IAA metabolism-related genes showed that GH3 members (GH3.1, GH3,2, GH3.4, GH3.6, GH3.7, GH3.13, GH3.17, GH3.18, and GH3.19, encoding auxin-metabolism enzymes) presented decreased expression levels, whereas YUC4 and YUC7 (encoding key enzymes in auxin biosynthesis) presented increased expression levels in myb30 roots (Fig. 5E). Further analysis showed that expression levels of GH3 members (GH3.1, GH3.2, GH3.4, GH3.17, GH3.18, and GH3.19) were increased in pi4kγ2 and miel1 roots (Fig. 5F). GH3 proteins synthesize IAA-amino acid conjugates, either the intermediate destined for IAA metabolism or the inactive storage form of IAA, to reduce the free IAA level. Increased (or decreased) expression levels of GH3s are consistent with the reduced auxin contents in pi4kγ2 and miel1 (or myb30), and confirmed that PI4Kγ2 regulates auxin metabolism and root growth by interacting with MILE1 to modulate MYB30.
MYB30 is a R2R3-MYB transcriptional activation factor involved in the very first step of HR (Vailleau et al., 2002). Interestingly, prediction analysis (The Arabidopsis Gene Regulatory Information Server, http://arabidopsis.med.ohio-state.edu/) revealed the presence of putative binding sequences of MYB members (Supplemental Table S1). A chromatin immunoprecipitation (ChIP)-qPCR analysis with an anti-Flag antibody was subsequently performed to investigate whether MYB30 directly binds the promoter of these genes. A MYB30-Flag fusion protein (driven by CaMV35S promoter) was expressed in wild-type seedlings and two transgenic lines with comparable protein expression levels were selected for a ChIP assay. qPCR analysis revealed that distinct DNA fragments of GH3.2 (P2; Fig. 5G) and GH3.6 (P3; Fig. 5G) promoters were more enriched, indicating that MYB30 regulates the expression levels of GH3.2 and GH3.6 genes by directly binding their promoters.
PI4Kγ2 Negatively Regulates Resistance and HR After Bacterial Inoculation
MIEL1 attenuates HR to bacterial infection, consistent with the enhanced or reduced responses of myb30 or MYB30-overexpression lines (Marino et al., 2013). Observation showed that compared to chlorosis symptoms of wild type with Pseudomonas syringae pv. tomato AvrRpm1 (Pst AvrRpm1), miel1 plants showed clear HR cell death symptoms (Fig. 6A). Similar to miel1, pi4kγ2 presented HR cell death symptoms as well at 64-h post infection (hpi) with Pst AvrRpm1 (Fig. 6A), indicating the important function of PI4Kγ2 in plant response to bacterial inoculation. Consistent with the faster HR, pi4kγ2 showed increased resistance in response to inoculation with Pst AvrRpm1 compared to wild-type plants (Fig. 6B), confirming that PI4Kγ2 functions as a negative regulator of plant defense.
Figure 6.
PI4Kγ2 is a negative regulator of HR responses in response to bacterial inoculation. A, Symptoms developed by pi4kγ2, miel1, and wild-type (WT) 64 hpi with Pst AvrRpm1 (2 × 106 cfu mL−1). Experiments were repeated three times (>5 seedlings each time) and representative images were shown. Scale bar = 1 cm. B, Growth of Pst AvrRpm1 in pi4kγ2, miel1, and wild type. Bacterial growth at 3 d was measured after inoculation (5 × 105 cfu mL−1). Experiments were repeated three times and data are presented as means ± sd (n > 5). Statistical analysis revealed significant differences compared to wild type (**P < 0.01). C, Relative expression levels of KCS1 and FDH, the target genes for MYB30. Wild-type, pi4kγ2, and miel1 seedlings were used for extracting RNA after inoculation with Pst AvrRpm1 (5 × 107 cfu mL−1) for 1 h. ACTIN7 gene was used as an internal reference. Experiments were repeated three times and data are presented as means ± se (n = 3). D, Model illustrating the regulation of PI4Kγ2-MIEL1-MYB30 and the effects on plant growth and HR. In wild type, PI4Kγ2 interacts with MIEL1 to suppress its ubiquitination and degradation, leading to the proteasomal degradation of MYB30 by MIEL1. Reduced MYB30 protein results in increased auxin content/signaling, and in turn promoted growth and suppressed HR to avoid superfluous activation. Under PI4Kγ2 deficiency, enhanced MIEL1 ubiquitination and degradation results in the suppressed ubiquitination and degradation of MYB30. Accumulated MYB30 thereby activates the expression of target genes, resulting in decreased auxin level, suppressed growth, and stimulated HR.
RT-qPCR analysis of expression of MYB30 very long chain fatty acid-related target genes, specifically KCS1 and FDH (Marino et al., 2013), showed their increased expression in pi4kγ2 and miel1 plants compared to wild-type plants after bacterial inoculation (Fig. 6C), further indicating that PI4Kγ2 negatively mediates Arabidopsis defense response, possibly by regulating MYB30 stability.
DISCUSSION
Large numbers of E3 ubiquitin ligases (∼1,500 E3-encoding genes are predicted in the Arabidopsis genome; Guzmán, 2014) regulate multiple cellular and biological processes by interacting with and degrading distinct target proteins (Merlet et al., 2009). Many reports have shown that E3 ligases can be regulated through posttranslational modifications in animals, which remain relatively undocumented in plants. Our studies functionally characterized the role of PI4Kγ2, an Arabidopsis atypical type-II PI4K, in root elongation and indicated that PI4Kγ2 is an E3 ligase mediator that inhibits ubiquitination of E3 (MIEL1) through protein–protein interaction. These findings provide further insights into Arabidopsis type-II PI4Ks and help to elucidate the regulatory mechanism of plant growth/development control, particularly the balance between plant resistance and development, at a posttranslational level (Fig. 6D).
MIEL1 is a C3H2C3 canonical RING-type E3 ligase and plays important roles in abscisic acid signaling and plant defense responses; however, the upstream regulation of MIEL1 remains poorly understood. A previous study showed that MIEL1 can be ubiquitinated (Lee and Seo, 2016). We found that PI4Kγ2 ensures the reduced ubiquitination and enhanced stability of MIEL1 (Fig. 4) through their interactions, leading to stimulated MYB30 degradation, providing additional evidence as to how type-II PI4Ks confer their functions in plants. Similarly, in mammalian cells, interaction between PLAGL2 and Pirh2 (Pirh2 encodes a RING-H2 domain-containing protein with intrinsic ubiquitin–protein ligase activity) prevents the proteasomal degradation of Pirh2 (Zheng et al., 2007). These results suggest that protein–protein interaction is a general regulatory mechanism for ubiquitination and stability of E3 ligases; however, the exact function of ubiquitin-like domains of type-II PI4Ks and the detailed mechanisms behind this regulation require further investigation.
MYB30 is a pleiotropic mediator that regulates a variety of physiological processes and signaling responses, including pathogen-induced HR, flowering time, and brassinosteroid and abscisic acid signaling (Raffaele et al., 2006, 2008; Li et al., 2009; Zheng et al., 2012; Raffaele and Rivas, 2013; Liu et al., 2014). However, its function in root cell elongation is unclear. Our studies show that MYB30 deficiency results in the promoted cell elongation of roots by altering auxin metabolism, which is consistent with the decreased expression of many GH3s and reveals a further function of MYB30 in regulating auxin metabolism.
Understanding control of the balance between plant resistance and development is fundamental, and it was recently shown that GA–abscisic acid and auxin–jasmonic acid synergistically regulate resistance and growth (Yuan et al., 2018). Interestingly, we found that PI4Kγ2-MIEL1-MYB30 regulates auxin-mediated growth and HR, and MYB30 may act in crosstalk of growth and defense signaling. MYB30 positively regulates HR cell death in response to pathogen attack (Vailleau et al., 2002) while inhibiting growth by decreasing auxin level. Indeed, auxin is an important plant growth regulator and also influences plant–pathogen interactions, and previous studies showed that elevated IAA biosynthesis in plants led to enhanced susceptibility to DC3000 (Mutka et al., 2013). Considering its myriad of regulatory targets and differential expression of PI4Kγ2-MIEL1-MYB30 after inoculation with Pst AvrRpm1, i.e. PI4Kγ2 and MYB30 expression was specifically induced at 4 and 1 hpi, respectively, whereas MIEL1 expression was repressed after inoculation at 1 hpi (Supplemental Fig. S4), it would seem that MYB30 acts on upstream effectors of auxin signaling and HR progress.
In sum, PI4Kγ2 interacts with MIEL1, thus inhibiting its turnover and leading to increased proteasomal degradation of MYB30, which attenuates auxin metabolism, and hence increases auxin level to avoid superfluous activation of HR. Under PI4Kγ2 deficiency, MIEL1 is ubiquitinated and degraded, thereby resulting in accumulated MYB30 protein, and hence activated HR and decreased auxin level/suppressed growth (Fig. 6D). Increased auxin enhances plant growth and auxin-mediated susceptibility, whereas decreased auxin results in growth inhibition and enhanced disease resistance. Considering the positive effect of PI4Kγ2 on MIEL1 stability and negative effect of MIEL1 on MYB30 activity, these results manifest that PI4Kγ2 may contribute to synergistic regulation of plant defense and growth.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
All Arabidopsis (Arabidopsis thaliana) plants used in this study were in the Columbia-0 (Col-0) background. The T-DNA insertion mutants, pi4kγ2 (SAIL_11_B03), miel1 (SALK_097638), and myb30 (SALK_027644C) were obtained from the Arabidopsis Biological Resource Center (Li et al., 2009; Marino et al., 2013).
Seeds were surface-sterilized and sown on plates containing Murashige-and-Skoog medium (Duchefe Biochemie). After stratification at 4°C for 3 d, seedlings were grown in a phytotron with a 16-h light/8-h dark cycle (22°C) for normal growth and seed harvesting.
Identification of T-DNA Mutants
Mutant pi4kγ2 carrying a T-DNA insertion in the first exon was confirmed by PCR amplification using the primers PI4Kγ2-l and PI4Kγ2-R. Transcription level of PI4Kγ2 was examined by RT-qPCR using the primers PI4Kγ2-3 and PI4Kγ2-4. T-DNA insertion of mutant miel1 was confirmed by PCR amplification using the primers MIEL1-1 and MIEL1-2. T-DNA insertion of mutant myb30 was confirmed by PCR amplification using the primers MYB30-1 and MYB30-2. All primers used in this study are listed in Supplemental Table S2.
Constructs and Plant Transformation
For expression of PI4Kγ2 in wild type or pi4kγ2, the coding sequence of PI4Kγ2 (1–903 bp) was amplified with primers (PI4Kγ2-5 and PI4Kγ2-6) and subcloned into the mCherry vector with C-terminal fusion. For expression of cMyc-PI4Kγ2 in Nicotiana benthamiana, the coding sequence of PI4Kγ2 was amplified (primers PI4Kγ2-7 and PI4Kγ2-8) and subcloned into a modified pEGAD-4XcMyc vector with N-terminal fusion. For expression of MIEL1-Flag in N. benthamiana, the coding sequence of MIEL1 was amplified by PCR (primers MIEL1-3 and MIEL1-4) and subcloned into 1306-Flag vector with C-terminal fusion. For expression of MIEL1-mCherry in wild type, pi4kγ2, or N. benthamiana, the coding sequence of MIEL1 was amplified by PCR (primers MIEL1-5 and MIEL1-6) and subcloned into an mCherry vector with C-terminal fusion. For expression of MYB30-Flag in N. benthamiana, the coding sequence of MYB30 was amplified by PCR (primers MYB30-3 and MYB30-4) and subcloned into the 1306-Flag vector with C-terminal fusion.
Arabidopsis transformation was performed by the floral-dip method (Clough and Bent, 1998).
Promoter-Reporter Gene Fusion Studies
The promoter of PI4Kγ2 (−2,491 bp upstream of ATG) was amplified (primers PI4Kγ2-P1/PI4Kγ2–P2), then cloned into a modified pCAMBIA1300 vector including a GUS reporter. A histochemical assay of GUS activities was performed according to Tang et al. (2016). Arabidopsis transformation samples were observed using differential interference contrast microscopy (SMZ1500; Nikon).
RT-qPCR Analysis
RT-qPCR analysis was performed to examine the PI4Kγ2 transcription in various tissues, expression of MIEL1 in N. benthamiana (primers MIEL1–7 and MIEL1–8), and expression of MYB30, GH3.1, GH3.2, GH3.4, GH3.6, GH3.7, GH3.13, GH3.17, GH3.18, GH3.19, YUC4, YUC7, GH3.2-P1, GH3.2-P2, GH3.6-P1, GH3.6-P2, GH3.6-P3, KCS1, and FDH in Arabidopsis (primers are listed in Supplemental Table S2). Total RNA was extracted from various tissues and used to synthesize complementary DNA by reverse transcription. The ACTIN7 (AT5G09810) and Elongation Factor1a (EF1a) genes were amplified (primers ACTIN7-1, ACTIN7-2, EF1a-1, and EF1a-2; Geng et al., 2017) and used as internal reference.
Measurement of Free IAA Contents by LC-MS
Approximately 200 mg of roots from 10-d-old seedlings was frozen in liquid nitrogen and ground to a fine powder for free IAA content measurement using a TSQ Quantum Ultra LC-MS-MS system (Thermo Fisher Scientific) according to the description in Tang et al. (2016).
Immunoblot Analysis
Isogenic MIEL1-mCherry plants were first prepared by crossing MIEL1-mCherry (in pi4kγ2) homozygous lines (as the male parent) with wild type and pi4kγ2, respectively. Protein extracted from transgenic plants was resuspended in extraction buffer (20 mm of Tris-HCl at pH 7.5, 150 mm of NaCl, 0.5% [v/v] TWEEN 20, 1 mm of EDTA, and 1 mm of dithiothreitol) containing a protease inhibitor cocktail (Roche). After addition of an equal volume of 2× SDS buffer, the samples were boiled for 5 min, fractionated by 10% SDS-PAGE, and transferred to a polyvinylidene fluoride membrane by semidry blotting. The blots were incubated with a mouse anti-mCherry antibody (Abcam) and then with a bovine anti-mouse IgG alkaline phosphatase-conjugated secondary antibody (Santa Cruz Biotechnology). Alkaline phosphatase activity was detected by BCIP/NBT Detection Reagents (Invitrogen).
For mCherry immunoprecipitation assays, the de-ubiquitination inhibitor PR-619 (Sigma-Aldrich) was added into the extraction buffer. Supernatants were pre-cleared for 1 h at 4°C with Dynabeads (Life Technology). Then samples were incubated with Anti-mCherry Dynabeads (Life Technology) overnight. All steps were performed at 4°C. Pulled-down proteins were eluted by 2× SDS buffer, then boiled for 5 min. MIEL1-immunoprecipitated fractions and ubiquitination of MIEL1 were analyzed by using anti-mCherry or anti-Ubiquitin antibody, respectively.
Yeast Two-Hybrid Analysis
The coding sequence of PI4Kγ2 was amplified (primers PI4Kγ2-9 and PI4Kγ2-10) and subcloned into pGBKT7 or pGADT7 vectors. For yeast two-hybrid screening, BD-PI4Kγ2 was used as bait to screen the candidate interacting proteins on synthetic dropout (SD) medium (-Leu/-Trp/-His/-Ade; Clontech) plates. The pGADT7-MIEL1 construct was generated using primers MIEL1-9 and MIEL1-10.
For auxotroph assays, candidate clones were streaked onto SD medium (-Leu/-Trp/-His/-Ade) and grown at 30°C for 4 d.
BiFC Assays
The coding regions of PI4Kγ2 (primers PI4Kγ2-11 and PI4Kγ2-12) and MIEL1 (primers MIEL1-11 and MIEL1-12) were fused with nYFP or cYFP, respectively. Resultant constructs PI4Kγ2-nYFP and MIEL1-cYFP were transformed into Agrobacterium tumefaciens and observed after 48 h according to Fang and Spector (2010). Considering YFP was excited at 488 nm and emission at 510 to 580 nm, and chlorophyll autofluorescence was emission at 644 to 714 nm (Vermeer et al., 2006; Body et al., 2019), BiFC results were imaged with a model no. FV1000 confocal microscope (Olympus) using the 488-nm argon laser and a dichroic filter to visualize YFP. The laser power was set at 5% for the 488-nm lasers. All images were collected using a 20× aperture objective. Collection wavelength was set at 510 to 550 nm.
Colocalization Studies of PI4Kγ2 and MIEL1
The coding regions of PI4Kγ2 (primers PI4Kγ2-13 and PI4Kγ2-14) and MIEL1 (primers MIEL1-13 and MIEL1-14) were amplified and subcloned into vector pA7 (N terminus fusion with YFP or CFP, respectively), resulting in the YFP-PI4Kγ2 and CFP-MIEL1 fusion constructs, which were transiently expressed in Arabidopsis protoplasts by PEG/CaCl2 methods (Yoo et al., 2007). The fluorescence was observed by confocal laser scanning microscopy (model no. FV1000; Olympus). For YFP/CFP, we used excitation/emission combinations of 514 nm/530 to 580 nm for YFP and 458 nm/470 to 500 nm for CFP. The laser power was set at 20% for the 458-nm laser and 5% for the 514-nm lasers. All images were collected using a 20× aperture objective.
Recombinant Expression of PI4Kγ2 and MIEL1, and In Vitro Kinase Assay
The coding region of PI4Kγ2 was amplified (primers PI4Kγ2-15 and PI4Kγ2-16) and subcloned into the pGEX-4T-1 vector (Novagen). The coding region of MIEL1 was amplified (primers MIEL1-15 and MIEL1-16) and subcloned into the vector pET28a (Novagen). All constructs were confirmed by sequencing.
PI4Kγ2 and MIEL1 proteins were expressed in Escherichia coli (strain BL21) supplemented with 1 mm of isopropyl-β-d-thiogalactoside (28°C, 3 h), or expressed in E. coli (strain Rosetta) supplemented with 0.1 mm of isopropyl-β-d-thiogalactoside (16°C, 15 h). Proteins were then purified using Ni-NTA His-binding resin (Novagen) or Glutathione Sepharose (Novagen) according to the manufacturer protocols.
The assay of kinase activity was performed according to a description in Tan et al. (2013) with a few modifications.
Bacterial Materials
Arabidopsis 4-week-old plants were kept at high humidity for 12 h before inoculation and injected with a bacterial suspension of Pst AvrRpm1 at the indicated bacterial densities using a blunt syringe on the abaxial side of the leaves. For measure of in planta bacterial growth, injected leaves samples were harvested 0 and 3 d. A predetermined dilution for each sample was plated on King’s B medium and incubated at 28°C for 2 d. Data were submitted to a statistical analysis (Marino et al., 2013).
ChIP-qPCR Assay
Ten-day-old wild-type seedlings expressing 35S::MYB30-FLAG were used for ChIP analysis. The ChIP assay was performed according to the manufacturer’s protocol of the EpiQuik Plant ChIP Kit (catalog no. P-2014; Epigenetics). qPCR analysis using Input DNA, immunoprecipitated DNA from IgG antibody, and Flag antibody as templates, respectively, was performed to examine the fraction of DNA fragments including MYB30 binding sequence in GH3.2 and GH3.6 promoters. Input DNA from un-immunoprecipitated DNA was used as a positive control (100%). The negative control was immunoprecipitated DNA from IgG antibody that binds with a nonspecific target and the associated DNA fragments were immunoprecipitated.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers listed in Supplemental Tables S1 and S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Characterization of pi4kγ2 mutant.
Supplemental Figure S2. Analysis of pi4kγ2 lines expressing PI4Kγ2.
Supplemental Figure S3. PI4Kγ2 regulates the stability and ubiquitination of MIEL1.
Supplemental Figure S4. PI4Kγ2, MYB30, and MIEL1 genes are spatially and temporally accumulated under inoculation.
Supplemental Table S1. Candidate binding sequences of MYB proteins in promoter of GH3 and YUC genes.
Supplemental Table S2. Primers used for mutant genotyping and plasmid construction.
Acknowledgments
We thank Lang-Tao Xiao (Hunan Agricultural University) for assistance with quantification of IAA contents by LC-MS, and Xiao-Shu Gao (Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences) for microscopy observation.
Footnotes
The work was supported by the National Science Foundation of China (grant no. 91954206), the China Postdoctoral Science Foundation (grant no. 2018M642002), and the Ten-Thousand Talent Program.
Articles can be viewed without a subscription.
References
- Akhter S, Uddin MN, Jeong IS, Kim DW, Liu XM, Bahk JD(2016) Role of Arabidopsis AtPI4Kγ3, a type II phosphoinositide 4-kinase, in abiotic stress responses and floral transition. Plant Biotechnol J 14: 215–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bagashev A, Fan S, Mukerjee R, Claudio PP, Chabrashvili T, Leng RP, Benchimol S, Sawaya BE(2013) Cdk9 phosphorylates Pirh2 protein and prevents degradation of p53 protein. Cell Cycle 12: 1569–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa IC, Shikata H, Zourelidou M, Heilmann M, Heilmann I, Schwechheimer C(2016) Phospholipid composition and a polybasic motif determine D6 PROTEIN KINASE polar association with the plasma membrane and tropic responses. Development 143: 4687–4700 [DOI] [PubMed] [Google Scholar]
- Body MJA, Dave DF, Coffman CM, Paret TY, Koo AJ, Cocroft RB, Appel HM(2019) Use of yellow fluorescent protein fluorescence to track OPR3 expression in Arabidopsis thaliana responses to insect herbivory. Front Plant Sci 10: 1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman LA, Goring DR(2011) Misregulation of phosphoinositides in Arabidopsis thaliana decreases pollen hydration and maternal fertility. Sex Plant Reprod 24: 319–326 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Duan S, Yao Z, Hou D, Wu Z, Zhu WG, Wu M(2007) Phosphorylation of Pirh2 by calmodulin-dependent kinase II impairs its ability to ubiquitinate p53. EMBO J 26: 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Spector DL(2010) BiFC imaging assay for plant protein-protein interactions. Cold Spring Harb Protoc 2: pdb prot5380. [DOI] [PubMed] [Google Scholar]
- Galvão RM, Kota U, Soderblom EJ, Goshe MB, Boss WF(2008) Characterization of a new family of protein kinases from Arabidopsis containing phosphoinositide 3/4-kinase and ubiquitin-like domains. Biochem J 409: 117–127 [DOI] [PubMed] [Google Scholar]
- Geng C, Wang HY, Liu J, Yan ZY, Tian YP, Yuan XF, Gao R, Li XD(2017) Transcriptomic changes in Nicotiana benthamiana plants inoculated with the wild-type or an attenuated mutant of Tobacco vein banding mosaic virus. Mol Plant Pathol 18: 1175–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzmán P.(2014) ATLs and BTLs, plant-specific and general eukaryotic structurally-related E3 ubiquitin ligases. Plant Sci 215-216: 69–75 [DOI] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, et al. (2011) A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ischebeck T, Seiler S, Heilmann I(2010) At the poles across kingdoms: Phosphoinositides and polar tip growth. Protoplasma 240: 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Nielsen E, Preuss ML, Mastronarde D, Staehelin LA(2011) Electron tomography of RabA4b- and PI-4Kβ1-labeled trans Golgi network compartments in Arabidopsis. Traffic 12: 313–329 [DOI] [PubMed] [Google Scholar]
- Kelley DR.(2018) E3 ubiquitin ligases: Key regulators of hormone signaling in plants. Mol Cell Proteomics 17: 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HG, Seo PJ(2016) The Arabidopsis MIEL1 E3 ligase negatively regulates ABA signalling by promoting protein turnover of MYB96. Nat Commun 7: 12525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y(2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DL, Yao HY, Jia LH, Tan JF, Xu ZH, Zheng WM, Xue HW(2020) Phospholipase d-derived phosphatidic acid promotes root hair development under phosphorus deficiency by suppressing vacuolar degradation of PIN-FORMED2. New Phytol 226: 142–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang J, Adrian J, Gissot L, Coupland G, Yu D, Turck F(2014) Elevated levels of MYB30 in the phloem accelerate flowering in Arabidopsis through the regulation of FLOWERING LOCUS T. PLoS One 9: e89799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Xu ZS, Pan-Pan L, Hu D, Chen M, Li LC, Ma YZ(2013) A wheat PI4K gene whose product possesses threonine autophophorylation activity confers tolerance to drought and salt in Arabidopsis. J Exp Bot 64: 2915–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan IR, Sapountzi V, Gaughan L, Neal DE, Robson CN(2004) Control of human PIRH2 protein stability: involvement of TIP60 and the proteasome. J Biol Chem 279: 11696–11704 [DOI] [PubMed] [Google Scholar]
- Marino D, Froidure S, Canonne J, Ben Khaled S, Khafif M, Pouzet C, Jauneau A, Roby D, Rivas S(2013) Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat Commun 4: 1476. [DOI] [PubMed] [Google Scholar]
- Mei Y, Jia WJ, Chu YJ, Xue HW(2012) Arabidopsis phosphatidylinositol monophosphate 5-kinase 2 is involved in root gravitropism through regulation of polar auxin transport by affecting the cycling of PIN proteins. Cell Res 22: 581–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlet J, Burger J, Gomes JE, Pintard L(2009) Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66: 1924–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller-Roeber B, Pical C(2002) Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutka AM, Fawley S, Tsao T, Kunkel BN(2013) Auxin promotes susceptibility to Pseudomonas syringae via a mechanism independent of suppression of salicylic acid-mediated defenses. Plant J 74: 746–754 [DOI] [PubMed] [Google Scholar]
- Nemoto K, Ramadan A, Arimura GI, Imai K, Tomii K, Shinozaki K, Sawasaki T(2017) Tyrosine phosphorylation of the GARU E3 ubiquitin ligase promotes gibberellin signalling by preventing GID1 degradation. Nat Commun 8: 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki K, Miyagishima SY, Wada H(2015) Phosphatidylinositol 4-phosphate negatively regulates chloroplast division in Arabidopsis. Plant Cell 27: 663–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, Li G, Zhao J, Band LR, Voß U, Postaire O, Luu DT, Da Ines O, Casimiro I, Lucas M, et al. (2012) Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol 14: 991–998 [DOI] [PubMed] [Google Scholar]
- Preuss ML, Schmitz AJ, Thole JM, Bonner HK, Otegui MS, Nielsen E(2006) A role for the RabA4b effector protein PI-4Kbeta1 in polarized expansion of root hair cells in Arabidopsis thaliana. J Cell Biol 172: 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Wang S, Shen C, Zhang S, Chen Y, Xu Y, Liu Y, Wu Y, Jiang D(2012) OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol 193: 109–120 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Rivas S(2013) Regulate and be regulated: Integration of defense and other signals by the AtMYB30 transcription factor. Front Plant Sci 4: 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Rivas S, Roby D(2006) An essential role for salicylic acid in AtMYB30-mediated control of the hypersensitive cell death program in Arabidopsis. FEBS Lett 580: 3498–3504 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D(2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M.(2018) Ubiquitylation at the crossroads of development and disease. Nat Rev Mol Cell Biol 19: 59–70 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C.(2018) NEDD8—its role in the regulation of Cullin-RING ligases. Curr Opin Plant Biol 45(Pt A): 112–119 [DOI] [PubMed] [Google Scholar]
- Street IH, Mathews DE, Yamburkenko MV, Sorooshzadeh A, John RT, Swarup R, Bennett MJ, Kieber JJ, Schaller GE(2016) Cytokinin acts through the auxin influx carrier AUX1 to regulate cell elongation in the root. Development 143: 3982–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M(2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37: 471–483 [DOI] [PubMed] [Google Scholar]
- Tan ST, Dai C, Liu HT, Xue HW(2013) Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling. Plant Cell 25: 2618–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Zhao CY, Tan ST, Xue HW(2016) Arabidopsis type II phosphatidylinositol 4-kinase PI4Kγ5 regulates auxin biosynthesis and leaf margin development through interacting with membrane-bound transcription factor ANAC078. PLoS Genet 12: e1006252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylidès C, Roby D(2002) A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proc Natl Acad Sci USA 99: 10179–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr., Munnik T(2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Vierstra RD.(2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J(2007) Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yuan P, Du L, Poovaiah BW(2018) Ca2+/calmodulin-dependent AtSR1/CAMTA3 plays critical roles in balancing plant growth and immunity. Int J Mol Sci 19: E1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Ning J, Yang YC(2007) PLAGL2 controls the stability of Pirh2, an E3 ubiquitin ligase for p53. Biochem Biophys Res Commun 364: 344–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Schumaker KS, Guo Y(2012) Sumoylation of transcription factor MYB30 by the small ubiquitin-like modifier E3 ligase SIZ1 mediates abscisic acid response in Arabidopsis thaliana. Proc Natl Acad Sci USA 109: 12822–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]