Genetic, biochemical, and molecular characterization of phloretin glycosyltransferase2 reveals its importance to the production of sweet-tasting trilobatin (phloretin-4′-o-glucoside) in wild apple.
Abstract
Epidemics of obesity and type 2 diabetes drive strong consumer interest in plant-based low-calorie sweeteners. Trilobatin is a sweetener found at high concentrations in the leaves of a range of crabapple (Malus) species, but not in domesticated apple (Malus × domestica) leaves, which contain trilobatin’s bitter positional isomer phloridzin. Variation in trilobatin content was mapped to the Trilobatin locus on LG 7 in a segregating population developed from a cross between domesticated apples and crabapples. Phloretin glycosyltransferase2 (PGT2) was identified by activity-directed protein purification and differential gene expression analysis in samples high in trilobatin but low in phloridzin. Markers developed for PGT2 cosegregated strictly with the Trilobatin locus. Biochemical analysis showed PGT2 efficiently catalyzed 4′-o-glycosylation of phloretin to trilobatin as well as 3-hydroxyphloretin to sieboldin. Transient expression of double bond reductase, chalcone synthase, and PGT2 genes reconstituted the apple pathway for trilobatin production in Nicotiana benthamiana. Transgenic M. × domestica plants overexpressing PGT2 produced high concentrations of trilobatin in young leaves. Transgenic plants were phenotypically normal, and no differences in disease susceptibility were observed compared to wild-type plants grown under simulated field conditions. Sensory analysis indicated that apple leaf teas from PGT2 transgenics were readily discriminated from control leaf teas and were perceived as significantly sweeter. Identification of PGT2 allows marker-aided selection to be developed to breed apples containing trilobatin, and for high amounts of this natural low-calorie sweetener to be produced via biopharming and metabolic engineering in yeast.
Diabetes is a major public health problem that is approaching epidemic proportions globally. The key risk factor linked with type 2 (adult-onset) diabetes is obesity, which is associated with physical inactivity and overeating (Bray and Popkin, 2014). One response to this health crisis by the food and beverage industry has been to develop products utilizing a range of non-sugar sweeteners to reduce calorie intake. Artificial sweeteners such as aspartame, sucralose, and saccharin are the most widely used (Kroger et al., 2006); however, there has been increasing consumer awareness and demand for the use of natural low-calorie sweeteners. Natural compounds with intense sweetening capacity can be found in numerous chemical families, including proteins, terpenoids, and flavonoids. Examples of plant-based sweeteners include steviol glycosides from stevia (Stevia rebaudiana) leaves, mogrosides from the juice of monkfruit (Siraitia grosvenorii), and glycyrrhizin from liquorice (Glycyrrhiza glabra) roots (Kim and Kinghorn, 2002).
Trilobatin is a plant-based sweetener that is reported to be ∼100× sweeter than Suc (Jia et al., 2008). It is found at high concentrations in the leaves of a range of crabapple species, including Malus trilobata, Malus sieboldii, and Malus toringoides (Williams, 1982; Gutierrez et al., 2018b). It is not found in the domesticated apple (Malus × domestica), but has been reported in low amounts in the leaves of wild grape (Vitis spp.) species (Tanaka et al., 1983). Some Lithocarpus species also contain trilobatin and their leaves are used to prepare sweet tea in China (Sun et al., 2015). The potential utility of trilobatin as a sweetener is recognized in many food and beverage formulations (e.g. Jia et al., 2008; Walton et al., 2013); however, its usefulness is limited by its scarcity. Methods for extraction have been documented from a range of tissues (Sun and Zhang, 2015; Sun, 2015) and after biotransformation of citrus waste (Lei et al., 2018). Biosynthesis of trilobatin in yeast has also been achieved (Eichenberger et al., 2017), but efficient production has been hampered by lack of knowledge of all the enzymes in the biosynthetic pathway.
Sweet-tasting trilobatin (phloretin-4′-o-glucoside [Ph4′-oGT]) and bitter-tasting phloridzin (phloretin-2′-o-glucoside [Ph2′-oGT]) are positional isomers of the dihydrochalcone (DHC) phloretin, which is produced on a side branch of the phenylpropanoid pathway. The first committed step in the biosynthesis of DHCs can be catalyzed by a double bond reductase (DBR) that converts p-coumaryl-CoA to p-dihydrocoumaryl-CoA (Gosch et al., 2009; Dare et al., 2013b; Ibdah et al., 2014). The next step involves decarboxylative condensation and cyclization of p-dihydrocoumaryl-CoA and three units of malonyl-CoA by chalcone synthase (CHS) to produce phloretin (Gosch et al., 2009; Yahyaa et al., 2017). The final step in the pathway requires the action of UDP-glycosyltransferases (UGTs) to attach Glc at either the 2′ or 4′ positions of the chalcone A-ring. Another apple DHC, sieboldin (3-hydroxyphloretin-4′-o-glucoside), is also glycosylated at the 4′ position either after the conversion of phloretin to hydroxyphloretin or by conversion of trilobatin directly to sieboldin.
UGTs are typically encoded by large gene families, with over 100 genes having been described in Arabidopsis (Arabidopsis thaliana; Ross et al., 2001) and over 200 genes in the M. × domestica genome (Caputi et al., 2012). All UGTs contain a conserved Plant Secondary Product Glycosyltransferase motif that binds the UDP moiety of the activated sugar (Li et al., 2001). Although some UGTs can utilize a broad range of acceptor substrates (Yauk et al., 2014; Hsu et al., 2017), others have been shown to be highly specific (Fukuchi-Mizutani et al., 2003; Jugdé et al., 2008). Systematic classification can facilitate the identification of some UGT activities; however, functionality is generally difficult to ascribe through phylogenetic relatedness alone. In apple, multiple UGTs have been identified that catalyze the 2′-o-glycosylation of phloretin to phloridzin: UGT88F1/MdPGT1 (Jugdé et al., 2008), UGT88F8 (Elejalde-Palmett et al., 2019), UGT88F4, UGT71K1 (Gosch et al., 2010), and UGT71A15 (Gosch et al., 2012).
DHC levels have been manipulated in transgenic apples by downregulation or overexpression of UGTs and other genes in the phenylpropanoid pathway. UGT88F1 knockdown lines showed significantly reduced phloridzin accumulation (30% of wild type) and were severely dwarfed, with greatly reduced internode lengths and narrow lanceolate leaves (Dare et al., 2017). UGT88F1 knockdown lines also showed increased resistance to Valsa spp. canker infection (Zhou et al., 2019). MdCHS knockout lines with 3% to 5% of wild-type foliar phloridzin levels also showed a severely dwarfed growth pattern (Dare et al., 2013b). Overexpression of UGT71A15 in transgenic apples increased the molar ratio of phloridzin to phloretin, but did not affect plant morphology or susceptibility to the fire-blight–causing bacterium Erwinia amylovora (Gosch et al., 2012). Overexpression of a chalcone 3′-hydroxylase in transgenic apple seedlings increased concentrations of 3-hydroxyphloridzin and reduced susceptibility to both fire blight and apple scab (Hutabarat et al., 2016).
Two apple enzymes, UGT71A15 and UGT75L17 (MdPh-4′-oGT), which glycosylate phloretin at the 4′ position in vitro, have been reported (Gosch et al., 2012; Yahyaa et al., 2016). However, these proteins are expressed in the leaves and fruit of domesticated apples, which do not accumulate trilobatin. To identify the glycosyltransferase leading to the production of trilobatin in planta, we focused on tissue from a range of wild apple accessions that produced trilobatin and sieboldin, and compared this with material from cultivated and wild apples producing only phloridzin. Using a range of genetic, biochemical, molecular, and transgenic tools, we demonstrate that phloretin glycosyltransferase2 (PGT2) is responsible for trilobatin production in Malus spp., and that its overexpression increases perception of sweetness in apple leaf teas.
RESULTS
Candidate Glycosyltransferases Identified by Activity-Directed Protein Purification
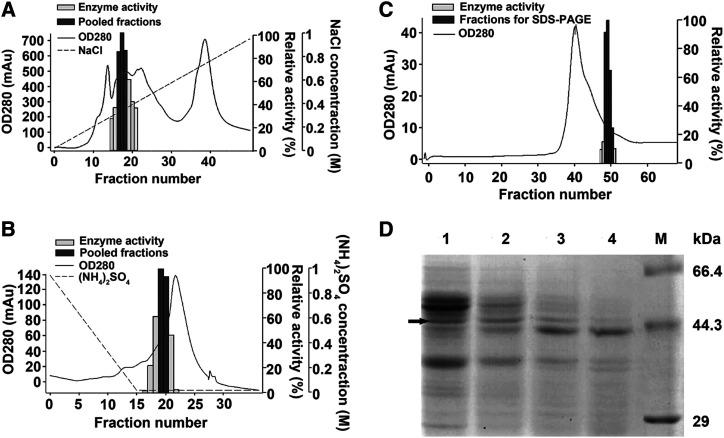
Tissues high in Ph4′-oGT activity required for trilobatin production, but containing very low Ph2′-oGT activity for phloridzin synthesis, were used to identify candidate Ph4′-oGTs by activity-directed protein purification. Flower petals of the crabapple hybrid ‘Adams’ were identified as suitable experimental material as they have low amounts of Rubisco compared with leaves, but higher Ph4′-oGT activity compared with fruit. Purification involved sequential chromatographic steps (Q-sepharose, phenyl sepharose, and Superdex 75; Fig. 1, A–C), after which fractions with high Ph4′-oGT activity were pooled and used for further purification. In the final step, after size exclusion chromatography, four protein fractions with different Ph4′-oGT enzyme activities were analyzed by SDS-PAGE. The abundance of a single band 44 to 66 kD (the size expected of a typical UGT) changed in a similar pattern (Fig. 1D) to that of the Ph4′-oGT activities (Fig. 1C). This band was subjected to liquid chromatography tandem mass spectroscopy (LC-MS/MS) analysis and peptides corresponding to 50 proteins were identified in the M. × domestica ‘Golden Delicious’ v1.0 genome assembly (Velasco et al., 2010; Supplemental Table S1). Of the five most abundant proteins, peptides corresponding to gene models MDP0000836043/MDP0000318032, encoding predicted UGT 88A1-like proteins, were observed at highest abundance (26% of total peptides; Table 1).
Figure 1.
Activity-directed purification of Ph4′-oGT activity from flowers of the crabapple hybrid ‘Adams’. Active fractions are shown as dark gray bars. A, Purification by Q-sepharose chromatography. B, Purification by phenyl sepharose chromatography using pooled fractions from Q-sepharose. C, Purification by Superdex 75 chromatography using pooled fractions from phenyl sepharose. Protein concentration (280 nm), enzyme activity, pooled fractions, and NaCl or (NH4)2SO4 gradient in the elution buffer are indicated. D, SDS-PAGE analysis of the four active fractions after purification by Superdex 75 chromatography are shown in lanes 1 to 4. M, Premixed broad protein marker (Takara). Arrow indicates the band sent for LC-MS/MS analysis.
Table 1. Proteins identified after activity-directed purification.
Abundant proteins identified by LC-MS/MS analysis of bands isolated after activity-directed purification of Ph4′-oGT activity from flowers of the crabapple hybrid ‘Adams’ (containing trilobatin and not phloridzin). The analysis was performed twice and the most abundant proteins found in both analyses are given as R1 and R2. iBAQ, sum of all peptide intensities divided by the number of observable peptides of a protein.
| Protein | Description | iBAQ R1 | iBAQ R2 |
|---|---|---|---|
| MDP0000836043/MDP0000318032 | M.× domestica UGT 88A1-like | 3786800000 | 3146000000 |
| MDP0000155691 | M. × domestica pentatricopeptide repeat-containing protein At4g14190, chloroplastic | 96169000 | 33639000 |
| MDP0000267350 | M. × domestica monodehydroascorbate reductase-like | 85148000 | 273050000 |
| MDP0000705244 | M. × domestica UGT 76B1-like | 75047000 | 628300 |
| MDP0000234480 | M. × domestica transaldolase-like | 31272000 | 246460000 |
Candidate Glycosyltransferases Identified by Differential Gene Expression Analysis
A second approach using differential gene expression (DGE) analysis was used to identify candidate Ph4′-oGTs in tissues high in trilobatin but low in phloridzin. A cross was produced between the ornamental crabapple hybrid ‘Radiant’ (containing both trilobatin and phloridzin) and M. × domestica ‘Fuji’ (containing only phloridzin). The F1 progeny were separated into two phenotypes with or without trilobatin for RNA extraction and transcriptome analysis. Expression levels of 109 genes were upregulated at least 16-fold (log2-fold change > 4) in progeny producing trilobatin (Supplemental Table S2). Of the five genes showing the greatest log-fold change, the expression level of the predicted UGT 88A1-like protein MDP0000836043 exhibited the largest differential expression, and was upregulated over 128-fold (log2-fold change > 7) in plants with trilobatin compared with those without (Table 2).
Table 2. Differentially expressed genes.
The five most differentially expressed genes identified after transcriptome analysis of pooled leaf samples of an F1 population between the crabapple hybrid ‘Radiant’ (containing both trilobatin and phloridzin) and M. × domestica ‘Fuji’ (containing only phloridzin).
| Gene | Description | Log2-Fold Change |
|---|---|---|
| MDP0000836043 | M. × domestica UGT 88A1-like (LOC103410306), mRNA | 7.54 |
| MDP0000204525 | M. × domestica cinnamoyl-CoA reductase 1 (LOC103427062), mRNA | 6.89 |
| MDP0000206483 | M. × domestica cytokinin hydroxylase-like (LOC114826167), mRNA | 6.77 |
| MDP0000219066 | M. × domestica cytochrome P450 CYP72A219-like (LOC103427349), mRNA | 6.68 |
| MDP0000737403 | M. × domestica probable mannitol dehydrogenase (LOC103446373), mRNA | 6.63 |
Mapping Concentrations of Trilobatin and Candidate Genes in a Segregating Population
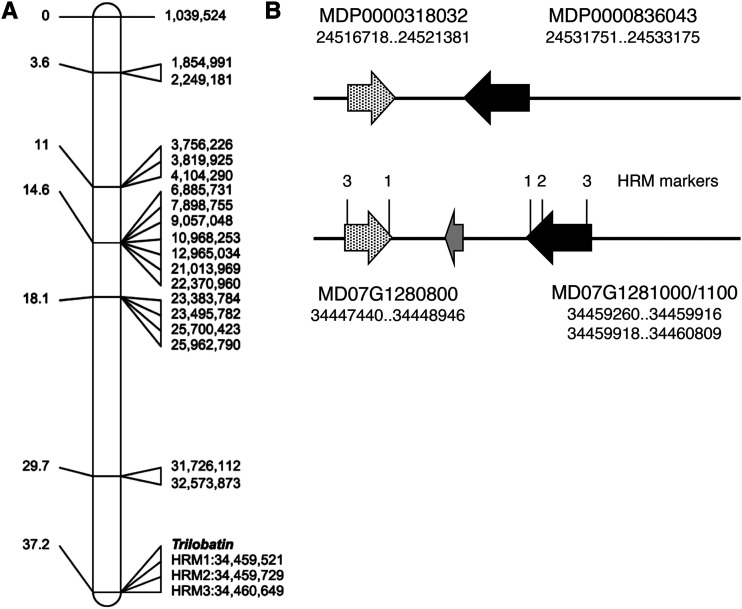
Trilobatin levels were mapped in a segregating population developed from a cross between domesticated and wild apples (‘Royal Gala’ × Y3). The female parent ‘Royal Gala’ (M. × domestica) produced only phloridzin, while the male parent Y3 (derived from M. sieboldii) produced both trilobatin and phloridzin. Of the 51 plants phenotyped, 30 contained trilobatin and phloridzin, and 21 phloridzin alone. The segregation ratio (1:0.7; χ2 = 1.59) suggested trilobatin content was segregating as a qualitative trait controlled by a single gene. Data obtained by screening leaf DNA with the International RosBREED SNP Consortium (IRSC) 8K SNP array (Chagné et al., 2012) were analyzed using the software JoinMap v.4.0 (van Ooijen, 2006). A single locus for control of trilobatin biosynthesis was identified on the lower arm of linkage group (LG)7, distal to a single-nucleotide polymorphism (SNP) marker located at position 32,527,873 bp (Fig. 2A) on the cv Golden Delicious doubled-haploid genome assembly (GDDH13 v1.1; Daccord et al., 2017). The locus was further defined using high-resolution melting (HRM) SNP markers developed from two candidate PGT genes at 34,460,000 to 34,461,000 bp (Fig. 2, A and B) close to the base of LG7 (total length of LG7 is 36,691,129 in GDDH13 v1.1). The position mapped on LG7 was consistent with one of the three independently segregating loci for DHC content reported recently in Malus spp., using linkage and association analysis (Gutierrez et al., 2018a).
Figure 2.
Genetic mapping of trilobatin production in a cv Royal Gala × cv Y3 segregating population. A, The Trilobatin locus was mapped near the base of LG7 of cv Y3 using the IRSC 8K SNP array (Chagné et al., 2012). Genetic locations in centimorgans are shown on the left and physical location in basepairs on the right (based on the cv Golden Delicious doubled-haploid assembly GDDH13 v1.1). The physical locations of three HRM-SNP markers (Supplemental Fig. S1; Supplemental Table S3) are indicated. B, The genomic region of the Trilobatin locus in the cv Golden Delicious v1.0p assembly (top) and the doubled-haploid assembly GDDH13 v1.1 (bottom). The physical positions of two UGT genes identified at the locus in each assembly are shown below the gene model. The black arrow corresponds to PGT2, the speckled arrow to PGT3, and the gray arrow to MD07G1280900 (annotated as a suppressor of auxin resistance). Note: PGT2 in the doubled-haploid assembly was incorrectly annotated as two truncated gene models MD07G1281000 and MD07G1281100. N.B. HRM1 and 3 amplified on both genes.
Genetic mapping by HRM marker analysis of gene model MDP0000836043, the candidate UGT 88A1-like protein identified by activity-directed protein purification and DEG analysis, demonstrates that it colocates with the locus identified for trilobatin production on LG7 (Fig. 2A). In the cv Golden Delicious v1.0p assembly (Velasco et al., 2010), MDP0000836043 is located at ∼24,531,751 bp and corresponds to gene models MD07G1281000/1100 (located at 34,459,260 bp) in the doubled-haploid assembly GDDH13 v1.1 (Fig. 2B). The second UGT 88A1-like protein identified by activity-directed protein purification encoded by MDP0000318032 (MD07G1280800) is located ∼10.3 kb away in both assemblies (Fig. 2B). Four SNP variants identified in the region of these two UGT gene models were used to develop markers for HRM analysis (Supplemental Fig. S1; HRM primer sequences in Supplemental Table S3). The mapping results validated the position of the MDP0000836043 and MDP0000318032 on LG7 (Fig. 2A), and three of the markers (HRM1, HRM2, and HRM3) showed precise concordance with presence/absence of trilobatin in the segregating progeny (Supplemental Table S4). HRM1 and HRM3 amplified both MDP0000836043 (MD07G1281000/1100) and MDP0000318032 (MD07G1280800), while HRM2 was specific for MDP0000836043 (MD07G1281000/1100). HRM4 was not specific to the locus and was not used further.
Biochemical Characterization of PGT2 and PGT3
Complete open reading frames (ORFs) corresponding to MDP0000836043 (hereafter termed PGT2, UGT88A32) were amplified from the leaves of six Malus spp. accessions. The PGT2 ORFs from five accessions synthesizing trilobatin showed 91% to 94% amino acid identity to the MDP0000836043 gene model from M. × domestica ‘Golden Delicious’ (Supplemental Fig. S2). A complete ORF for PGT2 obtained from the leaves of M. × domestica ‘Fuji’ was identical to that obtained from M. toringoides, although PGT2 was difficult to obtain from cv Fuji because of very low expression levels. The complete ORF corresponding to MDP0000318032 (termed PGT3, UGT88A33) was also amplified from five Malus spp. accessions and exhibited 97% to 99% identity to the MDP0000318032 gene model from cv Golden Delicious, and 85% to 87% identity to PGT2 (Supplemental Fig. S3).
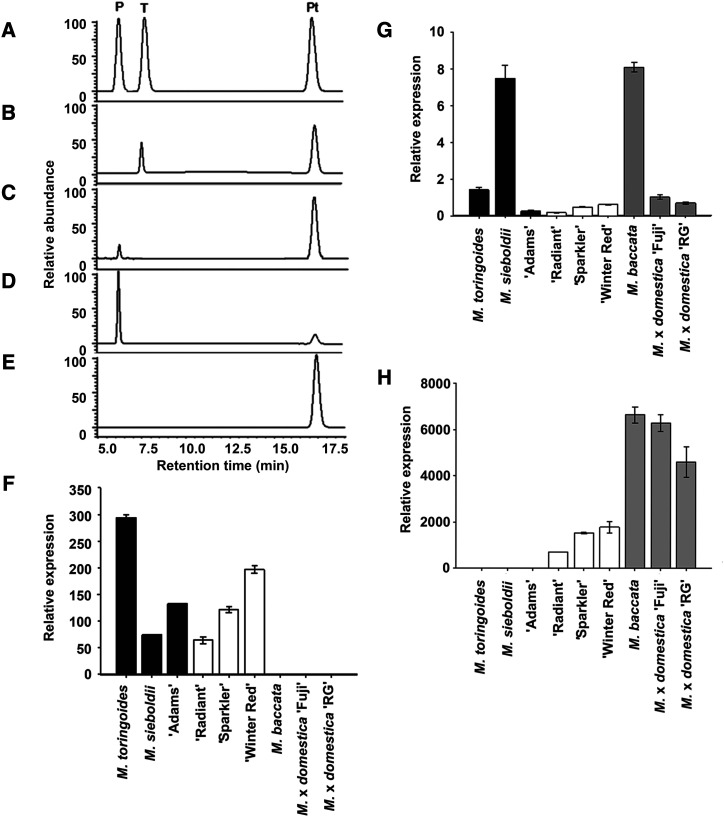
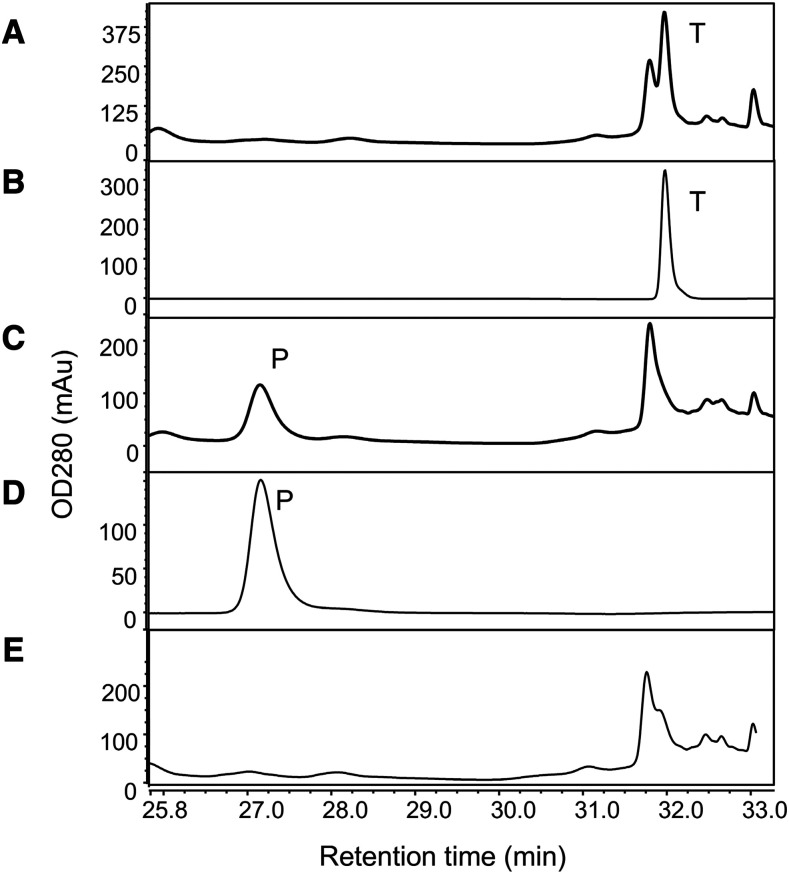
PGT2 and PGT3 from five Malus spp. accessions and PGT1 from cv Fuji were expressed in Escherichia coli and the products formed using phloretin and UDP-glucoside as substrates were determined by high performance liquid chromatography (HPLC). All purified recombinant PGT2 enzymes produced a single peak at 7.5 min that ran at the same retention time as the trilobatin standard (Fig. 3A). A representative HPLC trace for the product produced by PGT2 from M. toringoides is shown in Figure 3B. All purified recombinant PGT3 enzymes (Fig. 3C) and PGT1 from cv Fuji (Fig. 3D) produced a peak at 6.0 min with the same retention time as phloridzin (Fig. 3A), but no peak for trilobatin. No phloridzin or trilobatin were produced by the empty vector control (Fig. 3E).
Figure 3.
Biochemical and expression analysis of PGT1 to PGT3. A to E, Authentic standards of phloridzin (P), trilobatin (T), and phloretin (Pt) compared with the products formed by recombinant PGT2 from M. toringoides (B), PGT3 from M. sieboldii (C), PGT1 from M. × domestica ‘Fuji’ (D), and an empty vector control (E) in the presence phloretin and UDP-Glc. Experiments were performed in triplicate and a single representative trace is shown. F to H, Expression of PGT2 (F), PGT3 (G), and PGT1 (H) were analyzed by RT-qPCR using gene-specific primers (Supplemental Table S3) in three Malus spp. accessions containing only trilobatin (black bars), three containing trilobatin and phloridzin (white bars), and three containing only phloridzin (gray bars). MdACTIN was used as the reference gene. RG, cv Royal Gala. Data are means (±se) of three biological replicates from young leaves. Expression is presented relative to M. × domestica ‘Fuji’ in F and G and to M. toringoides in H (values set as 1).
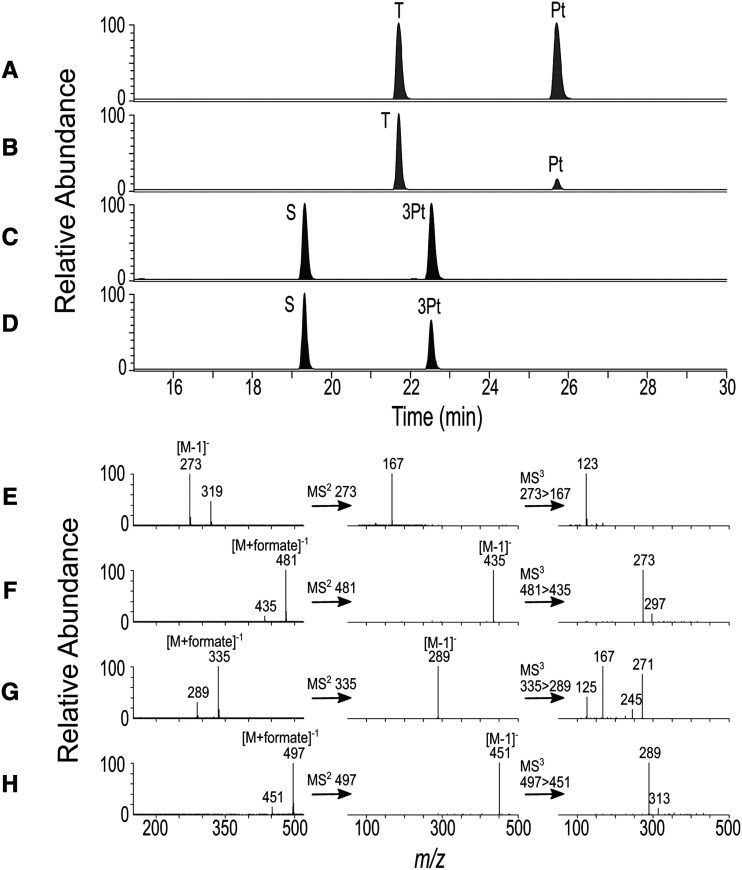
The substrate specificity of recombinant PGT2 from M. toringoides was further characterized using UDP-glucoside as the sugar donor and 12 substrates typically found in apple or with structural homology to phloretin (Table 3). The products of each reaction were determined by LC-MS/MS. Phloretin was the best acceptor for PGT2 and base peak plots indicated that a single peak at 21.5 min was formed that coeluted with the trilobatin standard (Fig. 4, A and B). PGT2 also catalyzed glycosylation of 3-OH phloretin to produce sieboldin (Fig. 4, C and D) with a relatively high conversion rate of ∼60% (Table 3). Quercetin-3-o-glucoside was detected as a reaction product using quercetin (Supplemental Fig. S4, A–F) with a lower conversion rate of 9.1% (Table 3). This result is surprising, as the 4′ position of DHCs corresponds to the 7 position of quercetin, but no quercetin-7-o-glucoside was observed. No products were detected in reactions with the other acceptor substrates; however, trace activity (1.4% compared with UDP-Glc) was detected with PGT2 using phloretin and UDP-Gal as the activated sugar donor. Full-scan and MS/MS data were used to further characterize the products of the PGT2 reactions. Phloretin (Fig. 4E) was detected as its pseudo-molecular ion mass-to-charge ratio (m/z) 273 [M−1]−), whereas trilobatin (Fig. 4F), 3-OH phloretin (Fig. 4G), and sieboldin (Fig. 4H) were detected predominantly as the corresponding formate adducts [M+formate])−1. MS/MS on the formate adducts identified the expected pseudo-molecular ion at m/z 435 and 451 [M−1]−) for the trilobatin and sieboldin glucosides. Triple-stage MS data on the m/z 435 and 451 [M−1]−) glucoside ions identified the m/z 273 and m/z 289 [M−1]−) ions of the phloretin and 3-OH phloretin aglycones, respectively.
Table 3. Substrate specificity of recombinant PGT2 cloned from M. toringoides.
The products of reactions using UDP-glucoside as the sugar donor and the 12 substrates shown were determined by LC-MS/MS. Conversion percent is the amount of product formed relative to the conversion of phloretin to trilobatin, which was set at 100%. All reactions were performed in triplicate. Nd, No products detected.
| Substrate | Product | Conversion |
|---|---|---|
| % | ||
| Phloretin | Trilobatin | 100.0 ± 6.1 |
| 3-OH Phloretin | Sieboldin | 58.7 ± 2.3 |
| Quercetin | Quercetin 3-o-glucoside | 9.1 ± 0.1 |
| Phloridzin | Nd | 0 |
| Trilobatin | Nd | 0 |
| Sieboldin | Nd | 0 |
| Naringenin | Nd | 0 |
| Cyanidin | Nd | 0 |
| Caffeic acid | Nd | 0 |
| 4-Coumaric acid | Nd | 0 |
| Neohesperidin | Nd | 0 |
| Chlorogenic acid | Nd | 0 |
| Boiled protein | Nd | 0 |
Figure 4.
LC-MS/MS analysis of products formed by PGT2. A to D, Base peak plots. A, Mixed standard of phloretin (Pt) and trilobatin (T). B, PGT2 + phloretin + UDP-Glc. C, Mixed standard of 3-OH phloretin (3Pt) + sieboldin (S). D, PGT2 + 3-OH phloretin + UDP-Glc. E to H, Mass spectra for reaction products and standards. E, Full-scan, MS/MS, and triple-stage MS data for phloretin. F, Full-scan, MS/MS, and triple-stage MS data for trilobatin. G, Full-scan, MS/MS, and triple-stage data for 3-OH phloretin. H, Full-scan, MS/MS, and triple-stage MS data for sieboldin.
PGT2 and PGT1 enzyme activities were compared over a pH range of 4 to 12 and with temperatures from 15°C to 60°C. The pH optimum of both enzymes was between pH 8 to 9 (Supplemental Fig. S5, A and B), while the optimum temperature was ∼40°C (Supplemental Fig. S5, C and D). The Michaelis constant (Km) values of PGT2 for phloretin were 18.0 ± 6.7 µm (the maximum velocity of enzyme-catlyzed reaction [Vmax] = 1.85 ± 0.17 nmol min−1) and for UDP-Glc were 103.6 ± 23.0 µm: (Vmax = 2.07 ± 0.17 nmol min−1; Supplemental Fig. S5, E and F). These Km values are comparable to those obtained for PGT1 for phloretin of 4.1 ± 1.2 µm (Vmax = 1.54 ± 0.08 nmol min−1) and for UDP-Glc of 491 ± 41 μm (Vmax = 8.84 ± 0.35 nmol min−1) under the same purification conditions (Supplemental Fig. S5, E and F).
Expression of PGT2 and PGT3 in Malus spp.
The relative expression of PGT2 and PGT3 were determined by reverse-transcription quantitative PCR (RT-qPCR) in the leaves of nine Malus spp. accessions (Fig. 3). Three accessions produced predominantly trilobatin, three both trilobatin and phloridzin, and three only phloridzin. PGT2 was highly expressed in all six Malus spp. accessions producing trilobatin; however, expression was essentially absent in the three accessions that did not synthesize trilobatin (Fig. 3F). Conversely, the expression of PGT1 was high in the six Malus spp. accessions producing phloridzin (Fig. 3G). Expression of PGT3 was observed in all nine accessions, and did not correspond with the presence/absence of trilobatin or phloridzin in the samples (Fig. 3H).
Structural Comparison of the Active Sites in PGT1 to PGT3
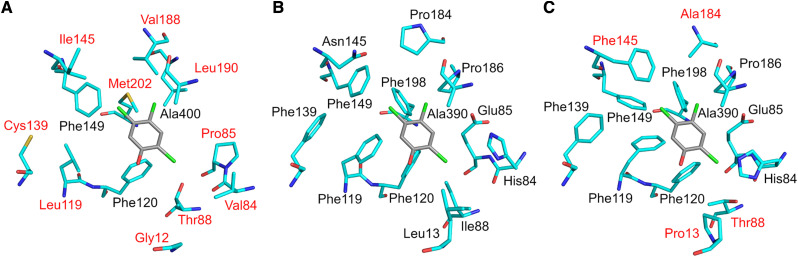
To investigate the structural basis for the difference in positional specificity for glycosylation on phloretin, structural homology models were independently obtained for PGT1 to PGT3 using the iTASSER server (Yang and Zhang, 2015). The models were superimposed and compared with the crystal structure of UGT72B1 bound with UDP-Glc (donor) and 2,4,5-trichlorophenol (acceptor; Brazier-Hicks et al., 2007; PDB: 2VCE). Overall, all structures were very similar, with root mean square deviations of ∼1 Å between each other (Supplemental Fig. S6). Sequence identities among PGT2/PGT1, PGT2/PGT3, and PGT1/PGT3 were 48%, 86%, and 47%, respectively. Around the predicted UDP binding site, however, the amino acid conservation between the three enzymes was much higher (>95% identity; Supplemental Table S5), consistent with the ability of these enzymes to bind the same donor molecule. Furthermore, the positions of the catalytic dyad residues in the models (His-16/Asp-118 in PGT2 and PGT3, His-15/Asp-118 in PGT1) were in excellent agreement with the crystal structure of UGT72B1. In contrast, conservation between PGT2 and PGT1 was considerably lower among the amino acids shaping the acceptor binding pocket (23% identity; 3 of 13 residues; Fig. 5, A and B). Similarly, although less pronounced, conservation between PGT2 and PGT3 around the acceptor binding pocket dropped to 69% (9 of 13 residues; Fig. 5, B and C).
Figure 5.
Structural comparison of the acceptor binding pockets of PGT1 to PGT3 models. The models were constructed by the iTASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). PGT1 (A) and PGT3 (C) residues labeled in red are different from PGT2 (B). The 2,4,5-trichlorophenol acceptor (shown in gray) from the UGT72B1 cocrystal structure (PDB: 2VCE) is superimposed onto the PGT models to highlight the approximate position of the acceptor binding pocket.
Metabolic Engineering of Trilobatin Production in Nicotiana benthamiana
To reconstitute the full apple pathway for trilobatin and phloridzin production in N. benthamiana, MdMyb10 and two biosynthetic genes MdDBR and MdCHS were transiently expressed together to catalyze the synthesis of phloretin substrate for glycosylation. Leaves infiltrated with MdMyb10, MdDBR, MdCHS, and PGT2 were analyzed by Dionex-HPLC and exhibited a peak at 32 min that corresponded to the trilobatin standard (Fig. 6, A and B), while those infiltrated with PGT1 exhibited a peak at 27.2 min corresponding to phloridzin (Fig. 6, C and D). Concentrations of trilobatin produced with PGT2 and phloridzin produced with PGT1 were similar (45 versus 87 ug g−1, respectively; Supplemental Table S6). Neither phloridzin nor trilobatin was detected in leaves inoculated with the GUS control vector (Fig. 6E). Interestingly, no phloretin was detected in the reactions, suggesting that glycosylation is needed to stabilize the reactive phloretin in N. benthamiana leaves.
Figure 6.
Engineering of trilobatin and phloridzin production in N. benthamiana. N. benthamiana leaves were infiltrated with Agrobacterium tumefaciens suspensions containing pHEX2_PGT2, pHEX2_PGT1, or the negative control pHEX2_GUS (each in combination with pHEX2_MdMyb10, pHEX2_MdCHS, and pHEX2_MdDBR + pBIN61-p19). Production of trilobatin and phloridzin were analyzed by Dionex-HPLC analysis (Thermo Fisher Scientific) at 7-d post-infiltration. Experiments were performed in triplicate and a single representative trace is shown. A, pHEX2_PGT2. B, Trilobatin (T) standard. C, pHEX2_PGT1. D, Phloridzin (P) standard. E, Negative control pHEX2_GUS.
MdDBR and MdCHS1 were required for synthesis of trilobatin in coinfiltrations with PGT2 (Supplemental Table S6). Substitution of MdDBR for MdENRL3 (Dare et al., 2013a) abolished trilobatin production. The MdMyb10 transcription factor increased trilobatin formation presumably via increased substrate flux through the phenylpropanoid pathway (Supplemental Table S6). These results indicate that three Malus spp. biosynthetic genes and a transcription factor are sufficient to reconstitute the pathway to trilobatin and phloridzin production in N. benthamiana (and likely any other plant) for biotechnological applications.
Overexpression of PGT2 in M. × domestica
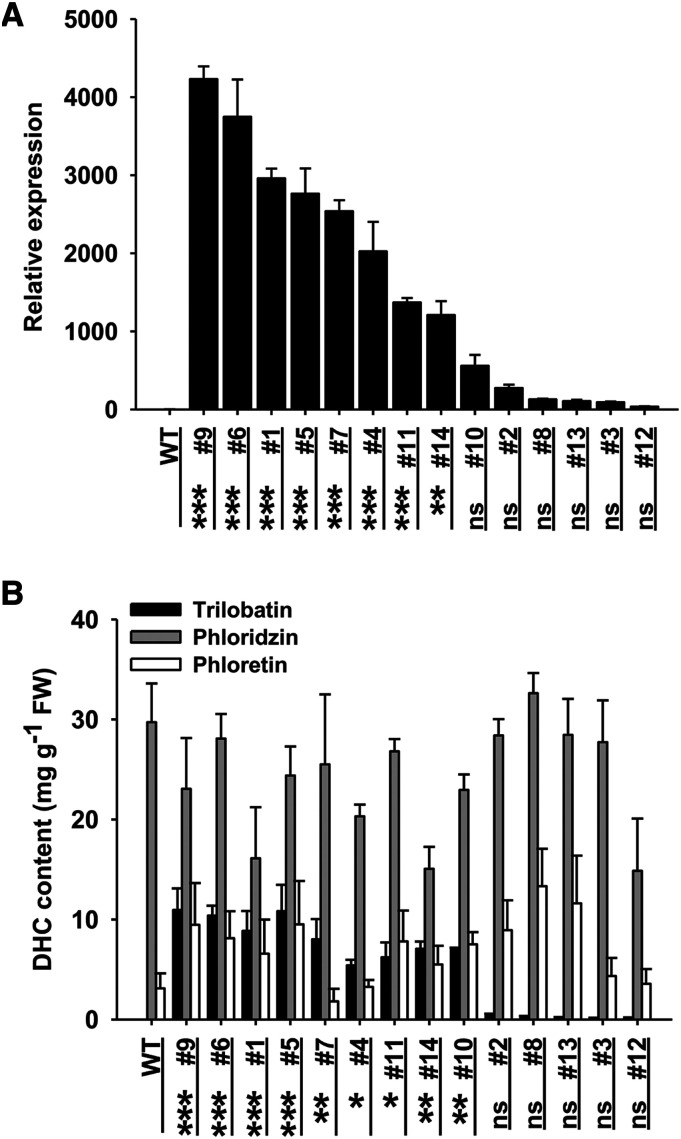
PGT2 was overexpressed in M. × domestica backgrounds GL3 and ‘Royal Gala’. Fourteen transgenic GL3 lines were obtained and PGT2 expression was significantly increased in the leaves of 4-week-old plants from eight lines (1, 4, 5, 6, 7, 9, 11, and 14) compared with wild type (Fig. 7A). Amounts of trilobatin were significantly increased in the same eight lines + line 10 compared with wild type, with concentrations ranging from ∼5 to 11 mg g−1 fresh weight (Fig. 7B). No significant differences were observed in phloridzin, phloretin (Fig. 7B), or total content of trilobatin + phloridzin (Supplemental Fig. S7A) among the GL3 transgenic lines. Eleven transgenic cv Royal Gala lines overexpressing PGT2 were also regenerated. An initial screen of shoots in tissue culture showed that all transgenic lines contained increased concentrations of trilobatin (Supplemental Fig. S8A), but similar total DHC content (Supplemental Fig. S8B) compared with wild types. Further analysis of the young leaves from six of the cv Royal Gala lines grown in a containment glasshouse confirmed these results (Supplemental Fig. S8, C and D).
Figure 7.
PGT2 expression levels and DHC content in transgenic GL3 APPLE LINES. A, Relative expression of PGT2 in 14 transgenic GL3 lines was determined by RT-qPCR using RNA extracted from young leaves. Expression was corrected against MdACTIN and is given relative to the wild-type (WT) GL3 control (value = 1). Primers and product sizes are given in Supplemental Table S3. B, Phenolic compounds were extracted from young leaves and individual DHC content determined by HPLC. A and B, Data are presented as mean ± se, n = 3 biological replicates. Statistical analysis was performed in the software GraphPad Prism (www.graphpad.com): one-way ANOVA using Dunnett’s Multiple Comparison Test versus wild type. No significant differences in phloridzin or phloretin content were observed. Significantly higher PGT2 expression and trilobatin content versus control are shown at ***P < 0.001, **P < 0.01, and *P < 0.05. FW, Fresh weight, ns, not significant.
The relative expression of PGT1, MdCHS, and PGT3 were also analyzed by RT-qPCR in the GL3 transgenic PGT2 overexpression lines. The expression levels of PGT1 (Supplemental Fig. S7B) and MdCHS (Supplemental Fig. S7C) were not significantly altered in the 14 transgenic apple lines. Interestingly, the relative expression of PGT3 in all 13 of 14 transgenic GL3 lines decreased significantly (Supplemental Fig. S7D). Strongest suppression was observed in lines expressing PGT2 and trilobatin at the lowest levels, suggesting co-suppression of the endogenous PGT3 gene by the introduced PGT2 transgene.
Physiology of PGT2 Overexpression Lines Grown under Simulated Field Conditions
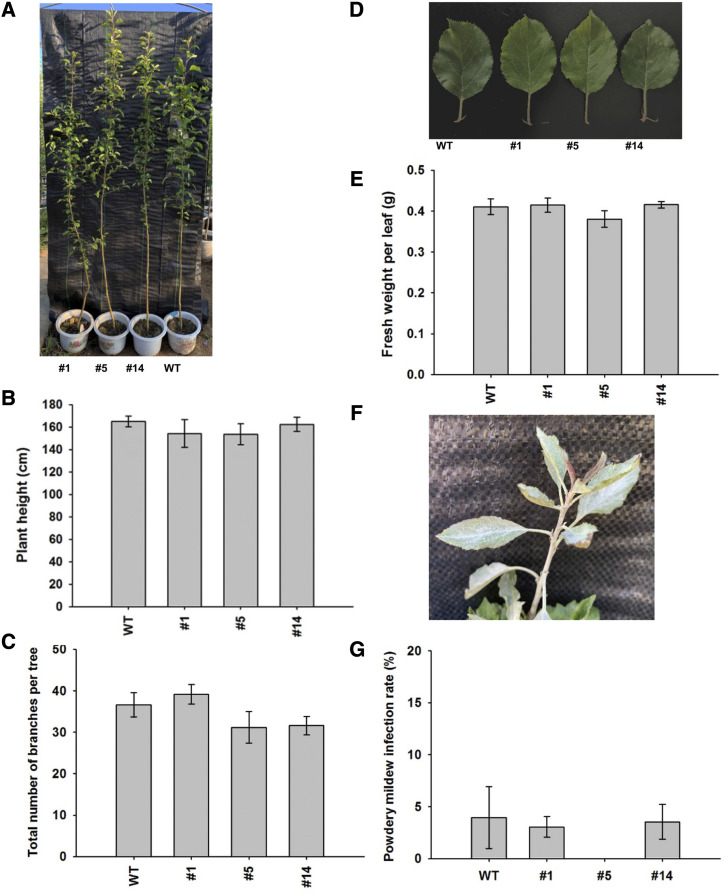
Manipulation of DHC levels in some transgenic apple lines has been shown to severely affect plant morphology (Dare et al., 2013b, 2017; Zhou et al., 2019). Multiple seedlings of three PGT2 overexpression lines (1, 5. and 14) and matching wild types were assessed for changes in morphology at 18 months of age when grown under simulated field conditions. No significant differences were observed in plant height (Fig. 8, A and B), total number of branches per tree (Fig. 8C), leaf morphology (Fig. 8D), or fresh weight per leaf (Fig. 8E).
Figure 8.
Physiology of PGT2 transgenic lines grown under simulated field conditions. A, Phenotype at age 18 months of wild-type (WT) and PGT2 transgenic lines 1, 5, and 14. B, Plant height, n = 3 trees per line. C, Total number of branches per tree, n = 5 trees per line. D, Leaf phenotype. E, Fresh weight per leaf, n = 5 trees per line, three leaves per tree. F, Powdery mildew infection phenotype in the young leaves of PGT2 line 1. G, Powdery mildew infection rate, n = 5 trees per line, three leaves per tree. Data are presented as mean ± se. Statistical analysis was performed in the software GraphPad Prism: one-way ANOVA using Dunnett’s Multiple Comparison Test versus wild type. No significant differences in plant height, branch number, leaf weight, or infection rate were observed.
Changes in DHC levels have also been implicated in pathogen susceptibility in some transgenic apple lines (Hutabarat et al., 2016; Zhou et al., 2019), but not in others (Gosch et al., 2012). PGT2 overexpression lines and matching wild types grown under simulated field conditions were subject to natural pathogen infection; no disease or pest management schemes were imposed on the plants. Under these conditions, no differences in disease susceptibility were observed between the PGT2 transgenics and wild types. The most common infection on all plants was by powdery mildew (Fig. 8F), but no significant differences in powdery mildew infection rate were observed (Fig. 8G).
Sensory Evaluation of Apple Leaf Teas from PGT2 Transgenic Plants
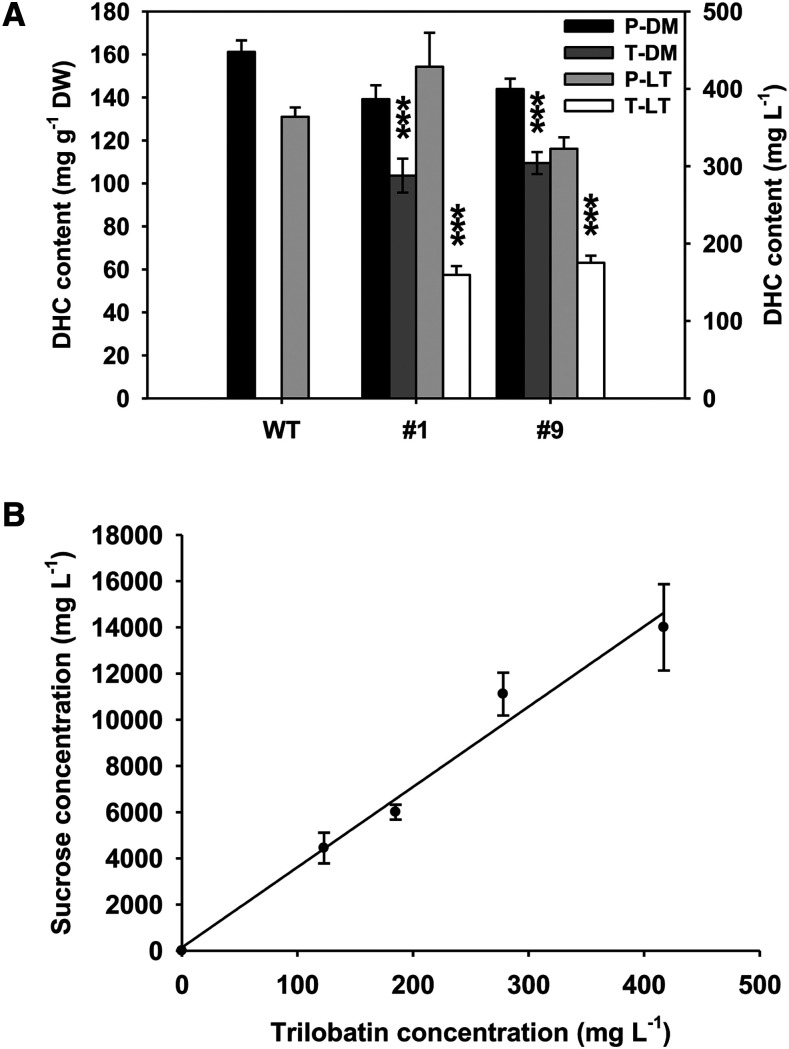
Sensory analysis was used to investigate the impact of PGT2 overexpression on the taste of apple leaf tea. Leaves were harvested from 4-month–old wild-type GL3 plants and two transgenic lines (1 and 9). After drying, the phloridzin contents in the wild-type and transgenic leaves were similar (∼150 mg g−1 dry weight). The transgenic lines also contained trilobatin (∼100 mg g−1 dry weight), while wild type contained none (Fig. 9A). After steeping, ∼27% of the phloridzin and 16% of the trilobatin was extracted into the tea (Fig. 9A).
Figure 9.
Analysis of apple leaf teas and trilobatin iso-sweetness. A, Phenolic compounds were extracted from dried leaf material and apple leaf tea prepared from wild-type (WT) apple and two transgenic GL3 lines overexpressing PGT2 (lines 1 and 9). Individual phloridzin (P) and trilobatin (T) content was determined by HPLC. Data are presented as mean ± se, n ≥ 7 for dried leaf material (DM) and n = 3 for apple leaf teas (LT). Statistical analysis was performed in the software GraphPad Prism: one-way ANOVA using Dunnett’s Multiple Comparison Test versus wild type. Significantly higher than wild type at ***P < 0.001. B, Iso-sweetness comparison test between trilobatin and Suc. Each participant was given one trilobatin solution and eight Suc solutions at different concentrations to taste. Iso-sweetness was established as 35.2 ± 1.66 (R2 = 0.98). Data presented are mean ± se, n = 5 participants.
In triangle tests, panelists were clearly able to distinguish the flavor of tea produced from the transgenic PGT2 leaves compared with tea produced from wild type (P < 0.01, 38 correct observations out of n = 70). To determine the basis for this discrimination, panelists were then asked to rate the sweetness of each sample on a scale from 1 to 10. The average sweetness of the two transgenic lines was rated significantly (P < 0.05, n = 23) higher at 4.8 and 4.6, respectively, compared with that of wild type, rated at 3.2.
DISCUSSION
Glycosyltransferases are encoded by large gene families, and identifying enzymes with specific activities based on homology is difficult. Two enzymes capable of 4′-o-glycosylation of phloretin in vitro have been reported (Gosch et al., 2012; Yahyaa et al., 2016), but these genes are expressed in tissues that produce only phloridzin. In this study, we used multiple approaches to show that PGT2 is responsible for production of trilobatin in apple. The genetic locus for trilobatin production colocated with the PGT2 gene, and HRM markers developed to PGT2 segregated strictly with trilobatin production. Molecular and biochemical analysis demonstrated that PGT2 was only expressed in accessions where trilobatin (or sieboldin) was produced and that the enzyme showed Ph4′-oGT activity in vitro. Finally, overexpression of PGT2 in domesticated apple confirmed that PGT2 leads to the production of trilobatin in planta.
The modeling results for PGT1 to PGT3 showed strong amino acid conservation among the three enzymes around the predicted UDP binding site, a feature common to the UGT superfamily (Li et al., 2001). Although both are members of the UGT88 family, PGT2 (UGT88A32) and PGT1 (UGT88F1) showed large variations in the amino acid composition of their respective acceptor binding pockets, which is consistent with these two enzymes having different activities and generating different products. More surprisingly, PGT2 and PGT3 (UGT88A32), which are putative paralogs and share high amino acid identity, also show strong differences in overall activity and specificity. In the case of PGT3, the 3D model analysis suggests that the low enzymatic activity may be due to one (or to a combination) of the four amino acids differing with PGT2 in the acceptor binding pocket. Among these, the substitution at position 145 of an Asn in PGT2 for a bulkier Phe in PGT3 may restrict the size of the pocket and impair the binding of the acceptor. However, site-directed mutagenesis and/or crystallographic work is required to confirm this hypothesis and to further understand the Ph2′-oGT activity of PGT3, compared with the Ph4′-oGT activity of PGT2.
UGTs with the ability to glycosylate phloretin have been described in M. × domestica from multiple UGT families (Jugdé et al., 2008; Gosch et al., 2010; Yahyaa et al., 2016; Zhou et al., 2017; Elejalde-Palmett et al., 2019). However, the majority, including the PGT2 and PGT3, belong to the UGT88 family (for a phylogeny, see Supplemental Fig. S9). The absence of trilobatin biosynthesis in M. × domestica is not due to mutations in the PGT2 gene, as a complete ORF was amplified from cv Fuji and shown to be identical to the PGT2 ORF from M. toringoides (a species producing trilobatin). Instead, DGE and RT-qPCR analyses indicated that the mutation was at the transcriptional level, with M. × domestica expressing PGT2 at very low levels. M. × domestica and all other accessions tested do express the putative PGT3 paralog located within ∼10 kb of PGT2. However, despite high homology, PGT3 does not exhibit Ph4′-oGT activity, but weak Ph2′-oGT activity. The Pyrus spp. (pear) genome (Linsmith et al., 2019) contains a close homolog of the PGT3 gene (XP_009368718.2), but not PGT2, suggesting that the PGT2 gene may have evolved recently only in the Malus spp. lineage, or have been lost from the Pyrus spp. lineage. The absence of trilobatin production in M. × domestica does not appear to be driven by domestication, as a number of wild apple species (e.g. Malus baccata; Fig. 3F) produce only phloridzin and lack expression of PGT2. Most wild Malus species that express PGT2 accumulate sieboldin, or trilobatin in combination with phloridzin. Only one species expressing PGT2, M. trilobata, accumulates trilobatin alone.
Manipulation of DHC levels in transgenic apples has been associated with changes in plant physiology, notably in plant morphology (Dare et al., 2017), and susceptibility to pathogen infection (Gosch et al., 2012; Hutabarat et al., 2016; Zhou et al., 2019). Transgenic PGT2 overexpressing lines appeared phenotypically normal, and no differences in disease susceptibility were observed between the PGT2 transgenics and wild types grown under simulated field conditions. In transgenic apple seedlings overexpressing chalcone 3′-hydroxylase, total DHC content was not altered, but reduced susceptibility to both fire blight and apple scab was associated with an increase (up to 11.5%) in 3-hydroxyphloridzin accumulation (Hutabarat et al., 2016). In the PGT2 transgenics, total DHC content was similarly not altered, but trilobatin accumulation (up to 38%) did not alter disease susceptibility. This difference may suggest that sieboldin is more effective in promoting disease resistance. However, in Hutabarat et al., (2016), pathogen sensitivity assays were performed using artificial shoot inoculations, and it was not reported if increased sieboldin concentrations altered susceptibility under field conditions. Conversely, our observations do not preclude the possibility that disease susceptibility in the PGT2 transgenics will change as the plants age, or that the plants were not exposed to pathogens sensitive to trilobatin under simulated field conditions.
Sensory analysis of apple leaf teas made from transgenic plants overexpressing PGT2 demonstrated that they could be clearly distinguished from teas made from wild-type apple leaves. The concentrations of trilobatin extracted into tea (Fig. 9A, ∼150 mg L−1) were above the sweetness detection threshold reported for trilobatin (3–200 mg L−1; Jia et al., 2008). The perception of increased sweetness in the transgenic leaf teas was consistent with increased production of trilobatin and not a decrease in concentrations of bitter-tasting phloridzin. However, a more detailed analysis by trained panelists would be required to understand the sensory properties fully. An iso-sweetness comparison test between trilobatin purified from leaves of the crabapple hybrid ‘Adams’ and Suc indicated that trilobatin was ∼35-fold sweeter than Suc (Fig. 9B). This number is slightly lower than figures reported in Jia et al. (2008), which may relate to the purity of the trilobatin tested, the delivery system, or variation in panelist sensitivity to Suc or trilobatin.
Identification of the Ph4′-oGT for trilobatin production will allow us to investigate whether PGT2 overexpression in the fruit affects consumer perception of sweetness. Transgenic ‘Royal Gala’ plants overexpressing the PGT2 gene have been produced and these contain both trilobatin and phloridzin in the leaves. This result indicates that PGT2 is reasonably competitive with PGT1 for the pool of phloretin substrate available in leaves and that PGT2 should be competitive with PGT1 for the smaller pool of phloretin produced in fruit. This hypothesis will be tested when the transgenic cv Royal Gala plants reach maturity in several years’ time. An alternative approach to increasing trilobatin production in M. × domestica would be to use molecular markers to accelerate the introgression of PGT2 into elite breeding material. However, for this approach to be successful, parental material in which PGT2 is well-expressed in fruit (rather than leaves and flowers) needs to be identified.
Identification of the Ph4′-oGT for trilobatin production and reconstitution of the apple pathway to trilobatin and phloridzin production in N. benthamiana may also allow high amounts of trilobatin to be produced via biotechnological means, such as biopharming and metabolic engineering in yeast. The utility of this approach has already been demonstrated for PGT1 in yeast (Eichenberger et al., 2017), but not in planta. The ability to produce large quantities of trilobatin would allow it to be tested not only as a natural sweetener in the food and beverage industry, but also for its potential health benefits (Fan et al., 2015; Xiao et al., 2017).
MATERIALS AND METHODS
Plant Material
Trilobatin production was mapped in an F1 seedling population between cv Royal Gala and cv Y3 grown in a greenhouse at The New Zealand Institute for Plant and Food Research, Auckland, New Zealand. Y3 is derived from the crabapple hybrid ‘Aotea’ × Malus × domestica ‘M9’. ‘Aotea’ is an open-pollinated Malus sieboldii (which produces sieboldin) selection. Malus trilobata and ‘Aotea’ were grown at the PFR research orchard in Havelock North, New Zealand.
Malus micromalus ‘Makino’ and the F1 population for DGE analysis between the crabapple hybrid ‘Radiant’ and M. × domestica ‘Fuji’ were grown at the Luochuan Apple Experimental Station, Northwest A&F University, Shaanxi, China. All other material was grown in an experimental orchard at Northwest A&F University, Yangling, Shaanxi, China. All trees were grown on their own roots and managed using standard horticultural growth practices and management for disease and pest control.
For physiological experiments on transgenic lines, GL3 and wild-type plants were grown in a plastic tunnel house in a randomized layout at Northwest A&F University. To meet transgenic containment restrictions, plants were grown in pots (height 22 cm, diameter 32 cm). To simulate field conditions, plants were grown under ambient temperature and environmental conditions. As the tunnel house was open-ended, plants were subject to natural pathogen infection. No disease- or pest-management schemes were imposed.
Chemicals
Trilobatin was purified from the crabapple hybrid ‘Adams’ (Xiao et al., 2017). Sieboldin, 3-OH phloretin, and quercetin glycosides were purchased from PlantMetaChem (www.PlantMetaChem.com) and cyanidin from Extrasynthese (www.extrasynthese.com). All other chemicals, including phloridzin and phloretin, were obtained from Sigma-Aldrich.
Mapping the Trilobatin Locus
Leaf tissue from seedlings in the cv Royal Gala × cv Y3 population were harvested and weighed before snap-freezing in liquid nitrogen. Phenolics were extracted from 100 to 250 mg of leaf tissue as described by Dare et al., (2017) and polyphenols quantified by Dionex-HPLC on an Ultimate 3000 system (Thermo Fisher Scientific) equipped with a diode array detector at 280 nm as described by Andre et al., (2012). Seedling DNA was extracted using the DNAeasy Plant Mini Kit (Qiagen) and genotypes determined using the IRSC 8K SNP array (Chagné et al., 2012). The SNP array data were analyzed using the Genotyping Module in the data analysis software GenomeStudio (Illumina). The genetic map was constructed using the software JoinMap v.4.0 (www.kyazma.nl) and the position of the Trilobatin locus on LG7 of Y3 was identified after conversion of SNP physical coordinates in the cv Golden Delicious v1.0 genome assembly (upon which the IRSC 8K SNP array was designed) to the latest version of the apple reference genome (GDDH13 v1.1). The position of PGT2 was then defined using HRM primers designed within the PGT2 candidate genes (Fig. 1; Supplemental Table S3) and PCR conditions as in Chagné et al., (2008).
Activity-Directed Protein Purification
A detailed protocol for activity-directed purification of Ph4′-oGT activity from apple flower petals is given in the legend to Supplemental Table S1. Purified protein fractions were separated on 12% (w/v) SDS-PAGE gels and visualized by Coomassie Blue R-250 staining. Target bands were cut and digested in gel with trypsin according to the method of Gao et al. (2017). The peptide mixture was then loaded onto a reverse-phase trap column (100 μm × 2 cm, Acclaim PepMap 100 column; Thermo Fisher Scientific) connected to the C18-reversed phase analytical column (10-cm long, 75 μm internal diameter, 3-μm resin Easy Column; Thermo Fisher Scientific) in buffer A (0.1% [v/v] formic acid) and separated with a linear gradient of buffer B (84% [v/v] acetonitrile and 0.1% [v/v] formic acid) at a flow rate of 300 nL min−1 controlled by IntelliFlow (Applied Flow Technology). LC-MS/MS analysis was performed on a Q-Exactive Mass Spectrometer (Thermo Fisher Scientific) that was coupled to Easy nLC (Proxeon Biosystems, now Thermo Fisher Scientific) for 60 min, and the mass spectrometer was operated in positive ion mode. Details of MS/MS spectra analysis are given in the legend to Supplemental Table S1.
DGE Analysis
The F1 population developed from a cross between the crabapple hybrid ‘Radiant’ and M. × domestica ‘Fuji’ was screened for trilobatin and phloridzin by HPLC. Eighty-one plants containing trilobatin + phloridzin (T+P) and 81 plants containing only phloridzin (P) were identified. One expanding leaf was collected from each seedling (∼20 cm tall) and three pooled replicate samples for T+P and P were prepared (each replicate containing leaves from 27 plants). Total RNA was extracted from frozen ground powder using Trizol Reagent (Life Technologies) following the manufacturer’s instructions and checked for RNA integrity on a model no. 2100 Bioanalyzer (Agilent). Sequencing libraries were generated from 3 μg of RNA per sample using the NEBNext Ultra RNA Library Prep Kit for Illumina (New England Biolabs) following the manufacturer’s recommendations and index codes were added to attribute sequences to each sample. RNA was sequenced by Novogene using the HiSeq4000 (Illumina) platform. Details of sequence alignment and DGE analysis are given in the legend to Supplemental Table S2.
RT-qPCR Analysis
Total RNA was extracted from young leaves as described by Malnoy et al., (2001). First-strand complementary DNA was synthesized from 1 μg of total RNA using the PrimeScript RT Reagent Kit (Takara), according to the manufacturer’s instructions. RT-qPCR was performed with a model no. CFX96 system (Bio-Rad) using the TB Green Premix Ex Taq (Takara). MdACTIN was used as the reference gene. The relative expression levels were calculated according to the 2ΔΔCT method (Livak and Schmittgen, 2001). Three biological replicates each with three technical repeats were used for RT-qPCR analysis. Gene-specific primers are listed in Supplemental Table S3.
Biochemical Characterization of PGT1 to PGT3 in Escherichia coli
The ORFs of PGT1 to PGT3 were amplified using primers in Supplemental Table S3 and ligated into pET28a(+) using the One Step Cloning Kit (www.vazyme.com). Recombinant proteins were expressed in E. coli BL21 (DE3) cells with 0.5 mm of isopropyl-1-thio-β-galactopyranoside at 16°C for 24 h at 80 rpm. Purification of recombinant proteins was performed using Ni-NTA agarose (EMD Millipore). Eluted fractions were used for determining enzyme activity and for SDS-PAGE analysis (Supplemental Fig. S10). Active fractions were concentrated using Vivaspin 2 concentrators (Sartorius).
GT activity assays were performed in triplicate in 200-µL reactions containing 50 mm of Tris-HCl (pH 9.0), 1 mm of DTT, 0.5 mm of phloretin, 0.5 mm of UDP-Glc, and 30 to 80 ng enzyme. Reaction mixtures were incubated for 10 min at 40°C and reactions stopped by adding 40 µL of 1 m HCl. NaOH (1 m) was used to adjust the pH to neutral for HPLC analysis of the products at 280 nm.
HPLC and LC-MS/MS Analysis of Phenolic Compounds
Leaf tissue (500 mg) for HPLC was snap-frozen in liquid nitrogen, and extracted with 1.5 mL of a solution containing 50% (v/v) methanol and 2% (v/v) formic acid at 0°C to 4°C. The homogenate was centrifuged at 10,000g for 10 min, and the supernatant used for HPLC after filtering with a 0.45-µm syringe (Li et al., 2013). Polyphenols were quantified on a model no. LC-20A Liquid Chromatograph equipped with a Diode Array Detector (Shimadzu) at 280 nm, as described in Zhang et al. (2018).
For LC-MS/MS analysis, scaled-up reactions were performed containing ∼10 µg of enzyme, 10 μm of substrate, and UDP-Glc at a final concentration of 250 μm. Reactions were performed in triplicate, and stopped after 1 h by addition of 10 μL of 10% (v/v) glacial acetic acid and blown down to dryness under a gentle stream of N2. The reaction products were reconstituted in 100 μL 5:95 (v/v) acetonitrile: water + 0.1% (v/v) formic acid, and then a 10-μL aliquot taken, which was diluted 10-fold in the same solvent. LC-MS/MS employed an LTQ Linear Ion Trap Mass Spectrometer fitted with an electrospray ionization interface (Thermo Fisher Scientific) coupled to an Ultimate 3000 UHPLC and PDA detector (Dionex).
Phenolic compound separation was achieved using a Hypersil Gold aQ Analytical Column (1.9-μm, 150- × 2.1-mm, maintained at 45°C; Thermo Fisher Scientific). Solvents were (A) water + 0.1% (v/v) formic acid and (B) acetonitrile + 0.1% (v/v) formic acid and the flow rate was 200 μL min−1. The initial mobile phase, 5% B/95% A, was held for 2 min, then ramped linearly to 15% B at 10 min and held for 3.75 min, before ramping linearly to 25% B at 18 min, 33% B at 25 min, 50% B at 28 min, and 100% B between 29 and 32 min before resetting to the original conditions. The sample injection volume was 2 μL. MS data were acquired in the negative mode using a data‐dependent LC-MS/MS method. This method isolates and fragments the most intense parent ion to give triple-stage MS data, then isolates and fragments the most intense daughter ion (triple-stage MS data). The electrospary ionization voltage, capillary temperature, sheath gas pressure, and sweep gas were set at −10 V, 275°C, 35 psi, and 5 psi, respectively.
Molecular Modeling
The sequences for PGT1 to PGT3 were independently submitted to the iTASSER server (Yang and Zhang, 2015). C-scores of the best models used for structural analysis were −0.38, 0.94, and 1.52 for PGT1, PGT2, and PGT3, respectively. Superimposition, structural analysis, and figures were performed using the PyMOL Molecular Graphics System, v.2.0 (Schrödinger).
Transient Expression in Nicotiana benthamiana
PGT2 was amplified from M. trilobata, pHEX2-MdCHS, and MdDBR (Yahyaa et al., 2016) from ‘Royal Gala’ using the primers in Supplemental Table S3. Genes were cloned into pHEX2 to generate the binary vectors pHEX2-PGT2, pHEX2-CHS, and pHEX2-DBR, respectively. Construction of pHEX2-Myb10, pGreen0029-ENRL3, pBIN61-p19 (containing the suppressor of gene silencing p19), and the control construct pHEX2-GUS have been reported in Espley et al. (2007), Dare et al. (2013a), and Nieuwenhuizen et al. (2013). All constructs were electroporated in Agrobacterium tumefaciens strain GV3101. Freshly grown cultures were mixed in equal ratio and infiltrated into N. benthamiana leaves as described in Hellens et al., (2005). After 7 d, leaves were harvested and phenolic compounds extracted for Dionex-HPLC analysis (Thermo Fisher Scientific).
Generation of Transgenic Apple Plants
The coding region of PGT2 was amplified from Malus toringoides using the primers in Supplemental Table S3 and cloned into pCAMBIA2300 using the One Step Cloning Kit (www.vazyme.com). The PGT2:pCAMBIA plasmid was then transformed into A. tumefaciens (strain GV3101) cells. Transgenic GL3 apple plants were generated by A. tumefaciens -mediated transformation according to Dai et al. (2013) and Sun et al. (2018). Transgeniccv Royal Gala plants were transformed with pHEX2-PGT2 and plants regenerated as described in Yao et al. (1995, 2013).
Sensory Panel Analysis
Apple leaves from wild type and two PGT2 transgenic GL3 lines were washed with water and dried at room temperature. Leaves were held at 200°C for 1 min to inactivate enzymes, then dried at 80°C in an oven for 60 min. Apple leaf tea was made using 5 g of dried leaves with the ratio of leaves/water being 1:100 (g mL−1). Water at ∼80°C was added to the leaves for 15 min, then all leaves were removed to stop further extraction. The tea was then kept at 50°C in water bath for sensory analysis. The sensory panel consisted of 23 individuals and included 14 females and nine males (all 20–30 y). Participation was voluntary and all participants gave their written consent before participation in the study. For the triangle tests, participants were given three trays; each tray had three cups (2 mL tea in each cup) with transgenic and wild-type leaf tea in a random design, either two transgenic and one wild type or two wild type and one transgenic. Participants were asked to sequentially taste the three samples on each tray and select which sample was different. To assess the relative sweetness of wild-type versus transgenic apple leaf teas, two samples (one transgenic and one wild type) were presented and the 23 panelists were asked to score the two samples on a sweetness scale from 1 to 10. For all the tasting tests, participants kept the samples in their mouths for 1 to 2 s, then spat them out into a waste container. Participants rinsed their mouths between samples with water and a dry biscuit was provided between each sample set.
Five participants with high acuity for trilobatin in the triangle test were selected to perform the iso-sweetness comparison test between trilobatin and Suc. Each participant was given one trilobatin solution and eight Suc solutions at different concentrations to taste. Solutions were prepared as described above for the apple leaf teas. The trilobatin solutions were presented at 12.3, 18.5, 27.8, and 41.7 mg per 100 mL, while the Suc solutions were presented at 296.3, 444.4, 592.6, 666.7, 888.9, 1,000, 1,333.3, and 2,000 mg per 100 mL.
Accession Numbers
Nucleotide sequences for genes characterized as part of this study were deposited in GenBank (www.ncbi.nlm.nih.gov) and received the accession numbers MN38099 to MN381012.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. HRM profiles of the four SNP markers located at the Trilobatin locus.
Supplemental Figure S2. Amino acid alignment of PGT2 sequences from Malus spp. accessions.
Supplemental Figure S3. Amino acid alignment of PGT3 sequences from Malus spp. accessions.
Supplemental Figure S4. LC-MS/MS analysis of reactions containing PGT2, quercetin, and UDP-Glc.
Supplemental Figure S5. Biochemical properties of recombinant PGT2 and PGT1.
Supplemental Figure S6. 3D-superimposition of PGT1, PGT2, and PGT3 models with the UGT71B1 crystal structure.
Supplemental Figure S7. HPLC and RT-qPCR analysis of transgenic GL3 plants overexpressing PGT2.
Supplemental Figure S8. HPLC analysis of transgenic cv Royal Gala plants overexpressing PGT2.
Supplemental Figure S9. Phylogeny of UGTs.
Supplemental Figure S10. SDS-PAGE of recombinant PGT2 and PGT1 proteins.
Supplemental Table S1. Proteins identified after activity-directed purification of Ph4′-oGT activity.
Supplemental Table S2. DGE analysis in tissues high in trilobatin but low in phloridzin.
Supplemental Table S3. Primer sequences for HRM analysis, RT-qPCR, and cloning.
Supplemental Table S4. Phenotype-to-genotype comparisons for individuals used to construct the genetic map and in HRM assays.
Supplemental Table S5. Amino acids surrounding the donor and acceptor binding sites in PGT1 to PGT3, identified from the respective 3D models.
Supplemental Table S6. Transient production of DHCs in N. benthamiana.
Acknowledgments
We thank Monica Dragulescu and her team for plant care at the New Zealand Institute for Plant and Food Research; Shanshan Zhao, Xiaohui Cui, and Ruijia Yang (Northwest A&F University) for help running the sensory trial; and Andrew Dare, Cecilia Deng, and Sue Gardiner (New Zealand Institute for Plant and Food Research) for reviewing the article.
Footnotes
This work was supported by the National Key R&D Program of China (grant no. 2018YFD1000200), the New Zealand Ministry of Business, Innovation and Employment, the New Zealand Institute for Plant and Food Research Ltd internal funding derived in part from apple variety and royalty income; the Innovation Capability Support Plan (grant no. 2018GHJD–11 to Y.W.), and the Key Research & Development Program of Shaanxi Province, China (grant no. S2019–YF–GHMS–0050).
References
- Andre CM, Greenwood JM, Walker EG, Rassam M, Sullivan M, Evers D, Perry NB, Laing WA(2012) Anti-inflammatory procyanidins and triterpenes in 109 apple varieties. J Agric Food Chem 60: 10546–10554 [DOI] [PubMed] [Google Scholar]
- Bray GA, Popkin BM(2014) Dietary sugar and body weight: Have we reached a crisis in the epidemic of obesity and diabetes?: Health be damned! Pour on the sugar. Diabetes Care 37: 950–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazier-Hicks M, Offen WA, Gershater MC, Revett TJ, Lim E-K, Bowles DJ, Davies GJ, Edwards R(2007) Characterization and engineering of the bifunctional n- and o-glucosyltransferase involved in xenobiotic metabolism in plants. Proc Natl Acad Sci USA 104: 20238–20243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi L, Malnoy M, Goremykin V, Nikiforova S, Martens S(2012) A genome-wide phylogenetic reconstruction of family 1 UDP-glycosyltransferases revealed the expansion of the family during the adaptation of plants to life on land. Plant J 69: 1030–1042 [DOI] [PubMed] [Google Scholar]
- Chagné D, Crowhurst RN, Troggio M, Davey MW, Gilmore B, Lawley C, Vanderzande S, Hellens RP, Kumar S, Cestaro A, et al. (2012) Genome-wide SNP detection, validation, and development of an 8K SNP array for apple. PLoS One 7: e31745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DR, Lawrence TJ, Rikkerink EH, Gardiner SE, Korban SS(2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92: 353–358 [DOI] [PubMed] [Google Scholar]
- Daccord N, Celton JM, Linsmith G, Becker C, Choisne N, Schijlen E, van de Geest H, Bianco L, Micheletti D, Velasco R, et al. (2017) High-quality de novo assembly of the apple genome and methylome dynamics of early fruit development. Nat Genet 49: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Dai H, Li W, Han G, Yang Y, Ma Y, Li H, Zhang Z(2013) Development of a seedling clone with high regeneration capacity and susceptibility to Agrobacterium in apple. Sci Hortic (Amsterdam) 164: 202–208 [Google Scholar]
- Dare AP, Tomes S, Cooney JM, Greenwood DR, Hellens RP(2013a) The role of enoyl reductase genes in phloridzin biosynthesis in apple. Plant Physiol Biochem 72: 54–61 [DOI] [PubMed] [Google Scholar]
- Dare AP, Tomes S, Jones M, McGhie TK, Stevenson DE, Johnson RA, Greenwood DR, Hellens RP(2013b) Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus × domestica). Plant J 74: 398–410 [DOI] [PubMed] [Google Scholar]
- Dare AP, Yauk Y-K, Tomes S, McGhie TK, Rebstock RS, Cooney JM, Atkinson RG(2017) Silencing a phloretin-specific glycosyltransferase perturbs both general phenylpropanoid biosynthesis and plant development. Plant J 91: 237–250 [DOI] [PubMed] [Google Scholar]
- Eichenberger M, Lehka BJ, Folly C, Fischer D, Martens S, Simón E, Naesby M(2017) Metabolic engineering of Saccharomyces cerevisiae for de novo production of dihydrochalcones with known antioxidant, antidiabetic, and sweet tasting properties. Metab Eng 39: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elejalde-Palmett C, Billet K, Lanoue A, De Craene JO, Glévarec G, Pichon O, Clastre M, Courdavault V, St-Pierre B, Giglioli-Guivarc’h N, et al. (2019) Genome-wide identification and biochemical characterization of the UGT88F subfamily in Malus x domestica Borkh. Phytochemistry 157: 135–144 [DOI] [PubMed] [Google Scholar]
- Espley RV, Hellens RP, Putterill J, Stevenson DE, Kutty-Amma S, Allan AC(2007) Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J 49: 414–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan X, Zhang Y, Dong H, Wang B, Ji H, Liu X(2015) Trilobatin attenuates the LPS-mediated inflammatory response by suppressing the NF-κB signaling pathway. Food Chem 166: 609–615 [DOI] [PubMed] [Google Scholar]
- Fukuchi-Mizutani M, Okuhara H, Fukui Y, Nakao M, Katsumoto Y, Yonekura-Sakakibara K, Kusumi T, Hase T, Tanaka Y(2003) Biochemical and molecular characterization of a novel UDP-glucose:anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol 132: 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Li Z, Xia C, Qu Y, Liu M, Yang P, Yu L, Song X(2017) Combining manipulation of transcription factors and overexpression of the target genes to enhance lignocellulolytic enzyme production in Penicillium oxalicum. Biotechnol Biofuels 10: 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosch C, Flachowsky H, Halbwirth H, Thill J, Mjka-Wittmann R, Treutter D, Richter K, Hanke M-V, Stich K(2012) Substrate specificity and contribution of the glycosyltransferase UGT71A15 to phloridzin biosynthesis. Trees (Berl) 26: 259–271 [Google Scholar]
- Gosch C, Halbwirth H, Kuhn J, Miosic S, Stich K(2009) Biosynthesis of phloridzin in apple (Malus domestica Borkh.). Plant Sci 176: 223–231 [Google Scholar]
- Gosch C, Halbwirth H, Schneider B, Holscher D, Stich K(2010) Cloning and heterologous expression of glycosyltransferases from Malus × domestica and Pyrus communis, which convert phloretin to phloretin 2′-o-glucoside (phloridzin). Plant Sci 178: 299–306 [Google Scholar]
- Gutierrez BL, Arro J, Zhong G-Y, Brown SK(2018a) Linkage and association analysis of dihydrochalcones phloridzin, sieboldin, and trilobatin in Malus. Tree Genet Genomes 14: 91 [Google Scholar]
- Gutierrez BL, Zhong G-Y, Brown SK(2018b) Genetic diversity of dihydrochalcone content in Malus germplasm. Genet Resour Crop Evol 65: 1485–1502 [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA(2005) Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu YH, Tagami T, Matsunaga K, Okuyama M, Suzuki T, Noda N, Suzuki M, Shimura H(2017) Functional characterization of UDP-rhamnose-dependent rhamnosyltransferase involved in anthocyanin modification, a key enzyme determining blue coloration in Lobelia erinus. Plant J 89: 325–337 [DOI] [PubMed] [Google Scholar]
- Hutabarat OS, Flachowsky H, Regos I, Miosic S, Kaufmann C, Faramarzi S, Alam MZ, Gosch C, Peil A, Richter K, et al. (2016) Transgenic apple plants overexpressing the chalcone 3-hydroxylase gene of Cosmos sulphureus show increased levels of 3-hydroxyphloridzin and reduced susceptibility to apple scab and fire blight. Planta 243: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibdah M, Berim A, Martens S, Valderrama AL, Palmieri L, Lewinsohn E, Gang DR(2014) Identification and cloning of an NADPH-dependent hydroxycinnamoyl-CoA double bond reductase involved in dihydrochalcone formation in Malus × domestica Borkh. Phytochemistry 107: 24–31 [DOI] [PubMed] [Google Scholar]
- Jia ZM, Yang X, Hansen CA, Naman CB, Simons CT, Slack J P, Gray K, inventors (December 11, 2008) Consumables. International Patent Application No. WO2008/148239A1.
- Jugdé H, Nguy D, Moller I, Cooney JM, Atkinson RG(2008) Isolation and characterization of a novel glycosyltransferase that converts phloretin to phlorizin, a potent antioxidant in apple. FEBS J 275: 3804–3814 [DOI] [PubMed] [Google Scholar]
- Kim NC, Kinghorn AD(2002) Highly sweet compounds of plant origin. Arch Pharm Res 25: 725–746 [DOI] [PubMed] [Google Scholar]
- Kroger M, Meister K, Kava R(2006) Low-calorie sweeteners and other sugar substitutes: A review of the safety issues. Compr Rev Food Sci Food Saf 5: 35–47 [Google Scholar]
- Lei L, Huang B, Liu A, Lu Y-J, Zhou J-L, Zhang J, Wong W-L(2018) Enzymatic production of natural sweetener trilobatin from citrus flavanone naringin using immobilised α-L-rhamnosidase as the catalyst. Int J Food Sci Technol 53: 2097–2103 [Google Scholar]
- Li P, Ma F, Cheng L(2013) Primary and secondary metabolism in the sun-exposed peel and the shaded peel of apple fruit. Physiol Plant 148: 9–24 [DOI] [PubMed] [Google Scholar]
- Li Y, Baldauf S, Lim EK, Bowles DJ(2001) Phylogenetic analysis of the UDP-glycosyltransferase multigene family of Arabidopsis thaliana. J Biol Chem 276: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Linsmith G, Rombauts S, Montanari S, Deng CH, Celton J-M, Guérif P, Liu C, Lohaus R, Zurn JD, Cestaro A, et al. (2019) Pseudo-chromosome length genome assembly of a double haploid ‘Bartlett’ pear (Pyrus communis L.). Gigascience 8: giz138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malnoy M, Reynoird JP, Mourgues F, Chevreau E, Simoneau P(2001) A method for isolating total RNA from pear leaves. Plant Mol Biol Report 19: 69a–69f [Google Scholar]
- Nieuwenhuizen NJ, Green SA, Chen X, Bailleul EJD, Matich AJ, Wang MY, Atkinson RG(2013) Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol 161: 787–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Li Y, Lim E, Bowles DJ(2001) Higher plant glycosyltransferases. Genome Biol 2: 3004.3001-3004.3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Wang P, Jia X, Huo L, Che R, Ma F(2018) Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol J 16: 545–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y-S, inventor (December 15, 2015) A method of sweetening natural bulk separation trilobatin. Chinese Patent Application No. CN104974201B.
- Sun Y, Li W, Liu Z(2015) Preparative isolation, quantification and antioxidant activity of dihydrochalcones from Sweet Tea (Lithocarpus polystachyus Rehd.). J Chromatogr B Analyt Technol Biomed Life Sci 1002: 372–378 [DOI] [PubMed] [Google Scholar]
- Sun Y-S, Zhang YW, inventors (June 25, 2015) Preparation of isolated natural sweetener trilobatin by crushing trilobatin, adding alcohol, heating to reflux, separating, filtering, separating filter residue and filter liquor, separating filter residue, and combining filtered liquors. Chinese Patent Application No. CN104974201A
- Tanaka T, Tanaka O, Kohda H, Chou W-H, Chen F-H(1983) Isolation of trilobatin, a sweet dihydrochalcone-glucoside from leaves of Vitis piasezkii Maxim and Vitis saccharifera Makino. Agric Biol Chem 47: 2403–2404 [Google Scholar]
- van Ooijen J.(2006) JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. Kyazma B.V., Wageningen, Netherlands [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. (2010) The genome of the domesticated apple (Malus × domestica Borkh.). Nat Genet 42: 833–839 [DOI] [PubMed] [Google Scholar]
- Walton SK, Denardo T, Zanno PR, Topalovic M, inventors (May 23, 2013) Taste modifiers. International Patent Application No. WO2013074811A1.
- Williams AH.(1982) Chemical evidence from the flavonoids relevant to the classification of Malus species. Bot J Linn Soc 84: 31–39 [Google Scholar]
- Xiao Z, Zhang Y, Chen X, Wang Y, Chen W, Xu Q, Li P, Ma F(2017) Extraction, identification, and antioxidant and anticancer tests of seven dihydrochalcones from Malus ‘Red Splendor’ fruit. Food Chem 231: 324–331 [DOI] [PubMed] [Google Scholar]
- Yahyaa M, Ali S, Davidovich-Rikanati R, Ibdah M, Shachtier A, Eyal Y, Lewinsohn E, Ibdah M(2017) Characterization of three chalcone synthase-like genes from apple (Malus × domestica Borkh.). Phytochemistry 140: 125–133 [DOI] [PubMed] [Google Scholar]
- Yahyaa M, Davidovich-Rikanati R, Eyal Y, Sheachter A, Marzouk S, Lewinsohn E, Ibdah M(2016) Identification and characterization of UDP-glucose:phloretin 4′-o-glycosyltransferase from Malus × domestica Borkh. Phytochemistry 130: 47–55 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Y(2015) I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res 43(W1): W174–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao JL, Cohen D, Atkinson R, Richardson K, Morris B(1995) Regeneration of transgenic plants from the commercial apple cultivar ‘Royal Gala’. Plant Cell Rep 14: 407–412 [DOI] [PubMed] [Google Scholar]
- Yao JL, Tomes S, Gleave AP(2013) Transformation of apple (Malus × domestica) using mutants of apple acetolactate synthase as a selectable marker and analysis of the T-DNA integration sites. Plant Cell Rep 32: 703–714 [DOI] [PubMed] [Google Scholar]
- Yauk Y-K, Ged C, Wang MY, Matich AJ, Tessarotto L, Cooney JM, Chervin C, Atkinson RG(2014) Manipulation of flavour and aroma compound sequestration and release using a glycosyltransferase with specificity for terpene alcohols. Plant J 80: 317–330 [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu Q, You Y, Chen W, Xiao Z, Li P, Ma F(2018) Characterization of quercetin and its glycoside derivatives in Malus germplasm. Hortic Environ Biotechnol 59: 909–917 [Google Scholar]
- Zhou K, Hu L, Li P, Gong X, Ma F(2017) Genome-wide identification of glycosyltransferases converting phloretin to phloridzin in Malus species. Plant Sci 265: 131–145 [DOI] [PubMed] [Google Scholar]
- Zhou K, Hu L, Li Y, Chen X, Zhang Z, Liu B, Li P, Gong X, Ma F(2019) MdUGT88F1-mediated phloridzin biosynthesis regulates apple development and Valsa canker resistance. Plant Physiol 180: 2290–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]