Candidatus Liberibacter asiaticus targets citrus CsACD2 to promote devastating Huanglongbing disease.
Abstract
Citrus Huanglongbing (HLB), caused by Candidatus Liberibacter asiaticus (Las), is one of the most destructive citrus diseases worldwide, yet how Las causes HLB is poorly understood. Here we show that a Las-secreted protein, SDE15 (CLIBASIA_04025), suppresses plant immunity and promotes Las multiplication. Transgenic expression of SDE15 in Duncan grapefruit (Citrus × paradisi) suppresses the hypersensitive response induced by Xanthomonas citri ssp. citri (Xcc) and reduces the expression of immunity-related genes. SDE15 also suppresses the hypersensitive response triggered by the Xanthomonas vesicatoria effector protein AvrBsT in Nicotiana benthamiana, suggesting that it may be a broad-spectrum suppressor of plant immunity. SDE15 interacts with the citrus protein CsACD2, a homolog of Arabidopsis (Arabidopsis thaliana) ACCELERATED CELL DEATH 2 (ACD2). SDE15 suppression of plant immunity is dependent on CsACD2, and overexpression of CsACD2 in citrus suppresses plant immunity and promotes Las multiplication, phenocopying overexpression of SDE15. Identification of CsACD2 as a susceptibility target has implications in genome editing for novel plant resistance against devastating HLB.
Global citrus industry is facing an unprecedented disease challenge caused by citrus Huanglongbing (HLB; also called citrus greening), which is presently the most devastating citrus disease worldwide (Bove, 2006; Gottwald, 2010; Wang et al., 2017). HLB is caused by phloem-limited, unculturable α-Proteobacteria Candidatus Liberibacter spp., with Candidatus Liberibacter asiaticus (Las) being the most widespread species (Bove, 2006). Citrus is one of the top tree crops worldwide and planted in approximately 140 countries. Las and its insect vector Asian citrus psyllid (Diaphorina citri) have been spreading from Asia to other continents, with 53 countries reporting HLB so far (https://www.cabi.org/isc/datasheet/16567). Billions of dollars of annual economic loss is related to yield, quality and tree loss, and costs for control and regulation measures of HLB (Bove, 2006; Gottwald, 2010; Wang 2020).
Suppression or avoidance of plant immunity is critical for pathogens to successfully infect their hosts. Las has been observed to activate nonself recognition immune responses in planta, including callose deposition, increased level of salicylic acid, and induction of pathogenesis-related (PR) genes. Las-triggered immune responses are generally slow (Kim et al., 2009; Folimonova and Achor, 2010; Li et al., 2017; Shi et al., 2018). Las does not cause a considerable induction of immune responses until 5 to 9 weeks after inoculation (Albrecht and Bowman, 2008), whereas extracellular bacterial pathogens induce immune responses within minutes to hours (Melotto et al., 2014). However, it remains poorly understood how Las suppresses plant immunity to promote its survival in planta. Many extracellular plant pathogenic bacteria have been shown to suppress plant immunity via translocation of virulence-associated effector proteins by the type-III secretion system (T3SS; Jones and Dangl, 2006; Toruño et al., 2016). Intracellular Las does not contain a T3SS. Instead, Las contains a complete Sec-dependent secretion apparatus (Duan et al., 2009). Previous studies showed that Las secretes Sec-dependent effectors (SDEs) into phloem sieve cells and neighboring companion cells (Pitino et al., 2016; Prasad et al., 2016; Pagliaccia et al., 2017). The Sec-dependent pathway has been shown to be critical for secreting virulence factors for other phloem-limited bacteria such as phytoplasma (Hogenhout et al., 2008). Importantly, one Las-secreted effector, SDE1, was reported to suppress plant immunity by inhibiting the activity of papain-like Cys proteases (PLCPs) in citrus (Clark et al., 2018).
The unculturability of Las and difficulties with genetic manipulation of Las and its host plant, namely citrus, present major challenges to using many well-developed methodologies in the study of citrus–Las interactions. Nevertheless, we have identified 86 putative SDE proteins with functional Sec-dependent secretion signal peptides (Prasad et al., 2016). Here, we conducted a detailed study of the Las effector SDE15 (CLIBASIA_04025). Our results showed that SDE15 targets ACCELERATED CELL DEATH2 (ACD2), a red chlorophyll catabolite reductase that functions as a susceptibility (S) target gene to promote HLB.
RESULTS
SDE15 Suppresses Plant Immune Responses and Promotes Las Growth in Planta
To determine whether SDE15 (CLIBASIA_04025, 11.17 kD) is involved in suppressing plant defenses and/or promoting Las growth, we overexpressed SDE15 in Duncan grapefruit (Citrus × paradisi) via stable Agrobacterium tumefaciens-mediated transformation. The resulting SDE15-transgenic plants were validated by western blotting analysis using an antibody raised against antigenic peptides of SDE15 (Supplemental Fig. S1). SDE15 suppressed the hypersensitive response (HR), a type of plant immune response characterized by rapid cell death, in Duncan grapefruit induced by Xanthomonas citri ssp. citri strain Aw (XccAw; Brunings and Gabriel, 2003). Specifically, XccAw triggered an HR in leaves of wild-type, but not SDE15-transgenic Duncan grapefruit (Fig. 1, A–C). We next examined the expression of immunity-associated genes in SDE15-transgenic plants after inoculation with XccAw (Asai et al., 2002; Fu and Dong, 2013; Zhang et al., 2015; Li et al., 2017). The expression of pattern-triggered immunity (PTI) marker genes FRK1, GST1, and WRKY22 was induced significantly after XccAw inoculation in nontransgenic Duncan grapefruit, but was repressed in SDE15-transgenic citrus. Similarly, expression of PR genes, PR1, PR3, and PR5, was markedly reduced in the SDE15-transgenic compared with that in the nontransgenic Duncan grapefruit (Fig. 1D). Taken together, these results suggest that SDE15 suppresses plant immunity in citrus.
Figure 1.
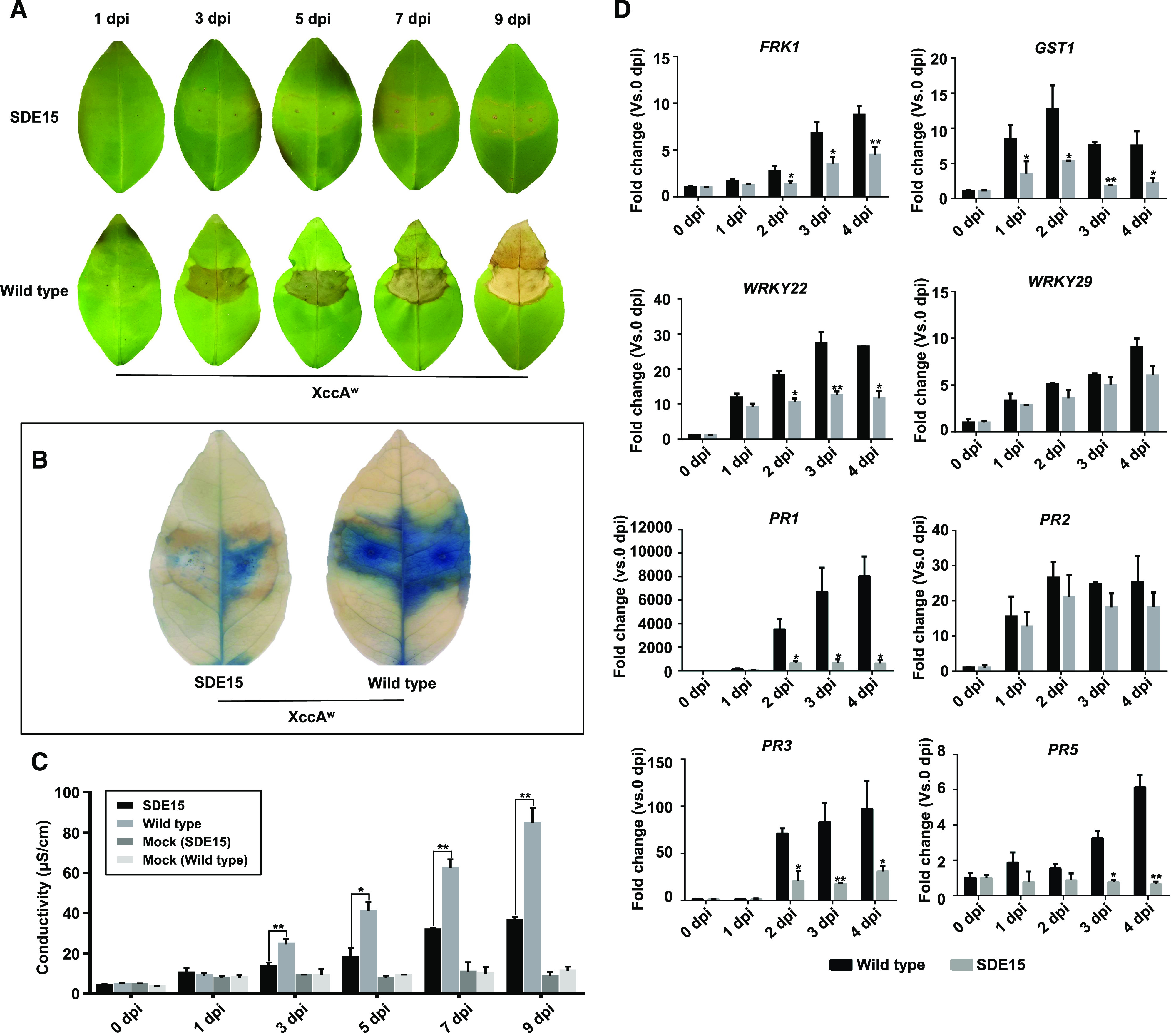
HR was repressed in SDE15-transgenic citrus. A, HR was observed in the wild-type but not in the SDE15-transgenic Duncan grapefruit inoculated with XccAW. B, Trypan-blue staining of the leaves that were inoculated with XccAW at 3 d post inoculation (dpi) indicated HR was repressed in SDE15-transgenic Duncan grapefruit. Whole leaf inoculated with XccAW was collected at 3 dpi and immersed with trypan-blue staining solution for 6 h with gently shaking, then de-stained with 98% to 100% ethanol. C, Dynamic electrolyte leakage assay demonstrated that the HR induced by XccAW was repressed in the SDE15-transgenic Duncan grapefruit. Leaf discs were floated on deionized water with shaking. The conductivity of the solution was measured after 4 h. Error bars indicate se (n = 3). Asterisks represent significant differences between SDE15-transgenic Duncan grapefruit and nontransgenic control by one-way ANOVA with posthoc test (*P < 0.05 and **Pe < 0.01). XccAW cells were infiltrated into citrus leaves at a concentration of 108 CFU mL−1. D, reverse transcription quantitative PCR (RT-qPCR) analyses of defense-related genes. Expression of FRK1, GST1, WRKY22, WRKY29, PR1, PR3, and PR5 was repressed in the SDE15-transgenic compared with that in wild-type Duncan after HR induction by XccAW. The housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase-C (GAPDH-C) was used as an endogenous control. Error bars indicate se (n = 4 biological replicates). Asterisks represent significant differences by one-way ANOVA with post hoc test (*P < 0.05 and **P < 0.01). All experiments were repeated three times with the similar results.
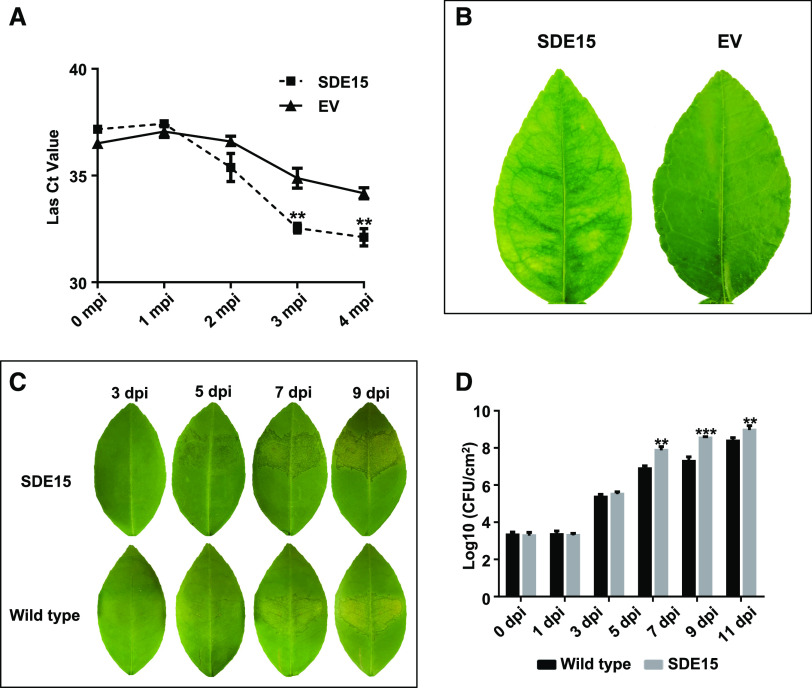
The ability of SDE15 to suppress plant immunity prompted us to examine whether the SDE15-transgenic plants were more susceptible to Las. For this purpose, we graft-inoculated SDE15-transgenic Duncan grapefruit plants with Las. The empty vector (EV)-transformed Duncan grapefruit plants were infected with Las by graft-inoculation as control. A significant increase of Las titers (indicated by the reduced Ct value of qPCR) was observed over a 4-month period in the SDE15-transgenic citrus compared with the EV-transgenic Duncan grapefruit plants (Fig. 2A). In accordance, visible HLB symptoms were observed earlier on leaves of SDE15-transgenic citrus than those of control plants after Las inoculation (Fig. 2B).
Figure 2.
SDE15-transgenic citrus plants are more susceptible to HLB and canker. A, The Las titers in the SDE15-transgenic citrus and EV-transgenic control citrus were determined by TaqMan qPCR at 0, 1, 2, 3, and 4 months post inoculation (mpi) with Las. Each Ct value was represented by means ± se (nSDE15 = 7, nEV = 5). Asterisks represent significant differences in the Las titer between SDE15-transgenic citrus and EV-transgenic control by one-way ANOVA with post hoc test (**P < 0.01). B, Leaf images taken at 3 mpi with Las via budding grafting. C, Canker symptoms were more severe on the SDE15-transgenic citrus than the wild-type citrus at 5 d post inoculation (dpi) with XccA 306. D, Bacterial growth analyses of XccA 306 in the SDE15-transgenic and wild-type Duncan grapefruit. Bacterial cells were infiltrated into citrus leaves at a concentration of 106 CFU mL−1. Error bars indicate se (n = 4). Asterisks represent significant differences in the bacterial population between the SDE15-transgenic citrus and nontransgenic control by one-way ANOVA with post hoc test (**P < 0.01 and ***P < 0.001).
Because SDE15 suppressed expression of defense-related genes in response to XccAw, we tested whether SDE15-transgenic plants were more susceptible to XccA. Indeed, the canker symptom caused by XccA 306 appeared earlier in SDE15-transgenic citrus compared with EV-transgenic plants. In addition, higher XccA 306 titers were detected in the SDE15-transgenic grapefruit than that in the EV-transgenic Duncan grapefruit (Fig. 2, C and D). Thus, SDE15 promotes citrus susceptibility to both HLB and canker, two severe citrus diseases worldwide.
SDE15 Is Detected in the Phloem Sap
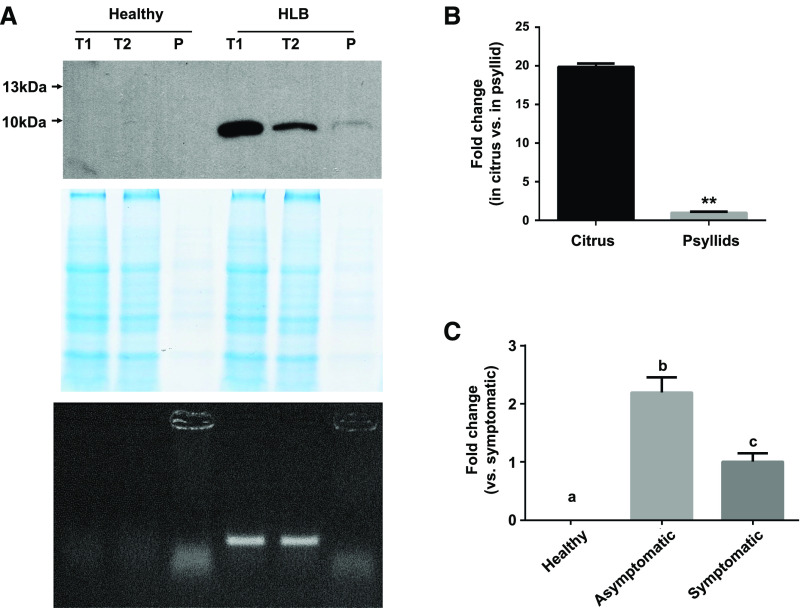
Sequence analysis of SDE15 did not reveal insights regarding its function except for the presence of a typical signal peptide (amino acids 1–22) predicted by SignalP v4.1 (http://www.cbs.dtu.dk/services/SignalP-4.1/). To determine whether SDE15 is secreted into the cytosol of phloem cells, and may be detected in the phloem sap of Las-infected plants, we used the SDE15-specific antibody to examine the presence of SDE15 in infected citrus plants. As shown in Figure 3A, SDE15 is clearly detected in the phloem sap extract of Las-infected citrus bark but not in that of healthy citrus by western blot analysis. PCR analysis using Las-specific primers showed the presence of Las in the bark samples, but not in the phloem sap extract samples (Fig. 3A), suggesting that SDE15 detected in the phloem sap is not directly associated with Las cells per se, but is secreted into phloem cells.
Figure 3.
Characterization of SDE15. A, SDE15 detection in phloem sap. Phloem sap was isolated from the bark of both healthy and HLB-diseased citrus (top). SDE15 was detected by western blot using SDE15-specific antibody generated by antigenic peptide ‘DDSHNQKPTEKKPN’. Coomassie brilliant blue was used as a loading control (middle), and PCR analysis using Las-specific primers was performed to detect Las in different samples (bottom). T1, Total bark proteins; T2, total bark proteins after phloem sap extraction; P, phloem sap. B and C, RT-qPCR analysis of SDE15 expression in different Las hosts (B) and in different stages of Las infection (C). Relative transcript abundances were determined using gyrase subunit A of Las (CLIBASIA_00325) as an endogenous control. Bars represent the mean of eight replicates for different hosts and six replicates for different stages of Las infection. Asterisks in B represent significant differences in the transcript abundance between citrus and psyllids by one-way ANOVA with post hoc test (**P < 0.01). Lowercase letters in C represent significant differences in samples of different Las infection stages by one-way ANOVA followed by lsd post hoc test (P < 0.05). All experiments were repeated three times with the similar results.
We next analyzed the expression pattern of SDE15 in Las-infected citrus and psyllids using RT-qPCR. SDE15 showed a higher expression level in citrus than in psyllids (Fig. 3B). Furthermore, SDE15 expression level was higher in the early stage (asymptomatic) than the later stage (symptomatic) of Las infection (Fig. 3C), indicating that SDE15 likely functions at a relatively early stage of Las infection in citrus before the appearance of visible HLB symptoms.
SDE15 Interacts with CsACD2
Because SDE15 does not contain obvious sequence domains that suggest enzymatic and DNA binding activity, we hypothesized that SDE15 might exert its suppressive effect on plant immunity via protein–protein interactions. To identify the targets of SDE15, we performed yeast two-hybrid (Y2H) screening with SDE15 without its signal peptide using complementary DNA (cDNA) libraries generated with mRNAs isolated from the Valencia sweet orange (Citrus × sinensis) at different Las infection stages (healthy [H], asymptomatic [AS], and symptomatic [S]). We identified 20 SDE15-interacting proteins in the H library, 60 in the AS library, and 6 in the S library (Supplementary Tables S1–S3). Among the SDE15-interacting proteins identified by Y2H, proteins encoded by Cs1g22670 and Cs8g20660 are potentially involved in plant immunity. Cs1g22670 encodes a red chlorophyll catabolite reductase (ACD2, hereinafter referred to as CsACD2) that represses programmed cell death in plants (Yao and Greenberg, 2006), whereas Cs8g20660 encodes a ubiquitin-activating enzyme E1 playing roles in innate immunity (Goritschnig et al., 2007). However, further experiments showed that the putative interaction between Cs8g20660 and SDE15 could not be validated by using pairwise Y2H assay. On the other hand, the interaction between CsACD2 and SDE15 was further confirmed using pairwise Y2H assay (Fig. 4A), a glutathione-S-transferase (GST) pull-down assay in vitro (Fig. 4B) and coimmunoprecipitation assays (CoIP) assay in vivo (Fig. 4C).
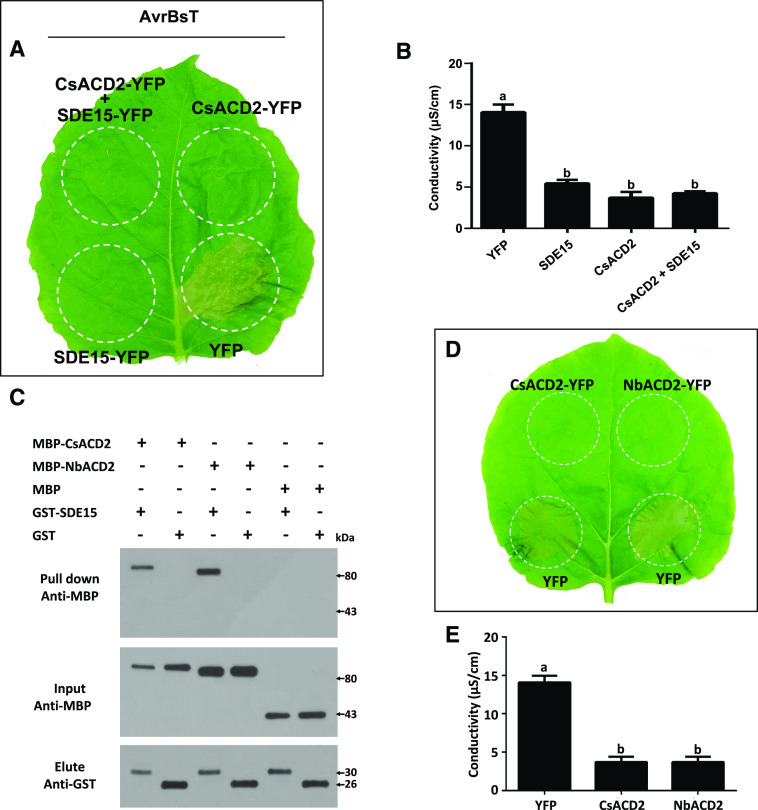
Figure 4.
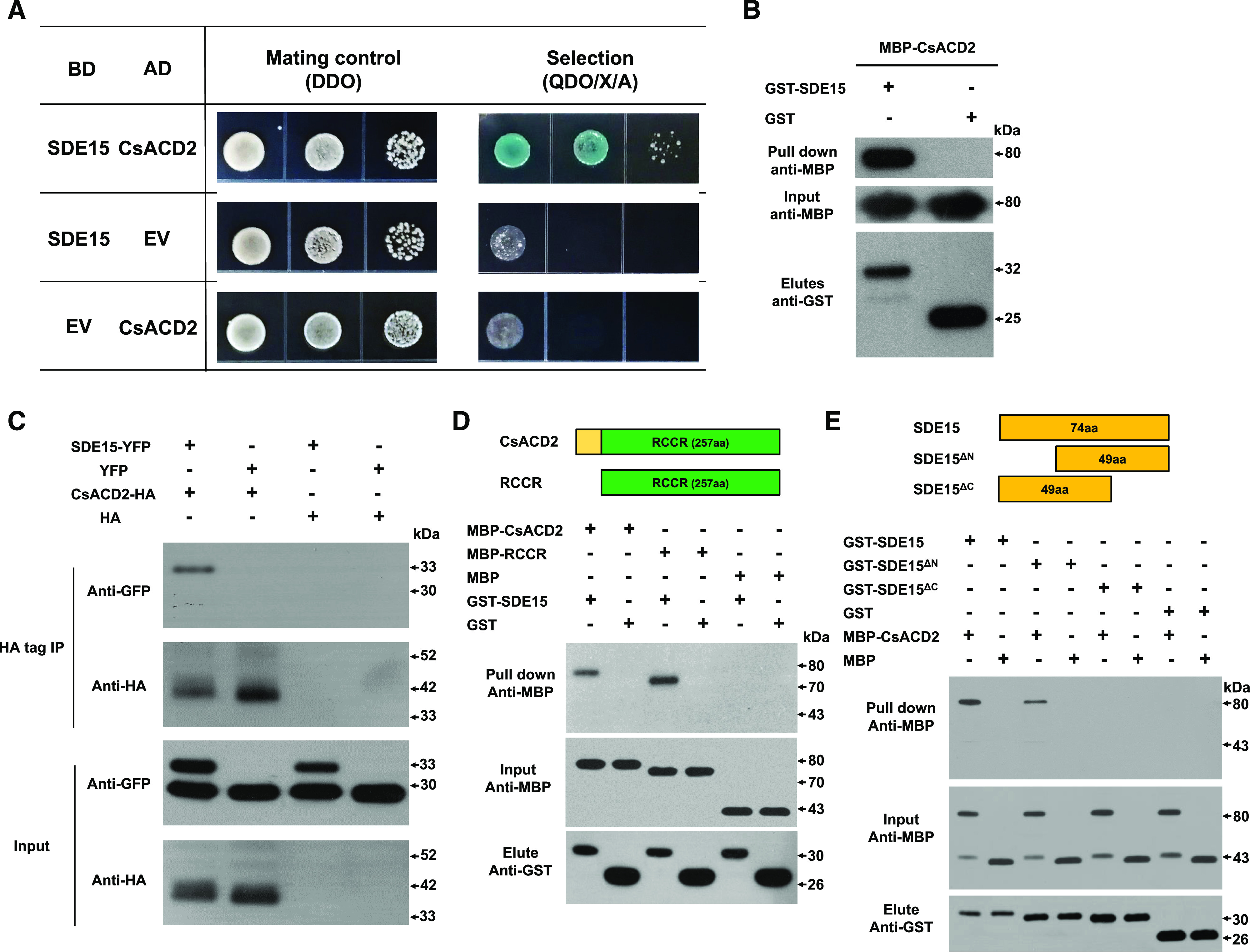
SDE15 interacts with CsACD2. A, Y2H assays using SDE15 without the signal peptide as the bait and full-length CsACD2 protein as the prey. SDE15 fused to the GAL4 DNA BD was expressed in combination with CsACD2 fused to the GAL4 AD in the yeast strain Y2HGold. Strains were grown on double dropout medium (DDO) with -Trp and -Leu and screened on quadruple dropout medium (QDO) with -Trp, -Leu, -Ade, and -His supplemented with X-α-Gal and Aureobasidin A (QDO/X/A). The empty BD and AD vectors were used as the negative controls. B, GST pull-down assay. GST-SDE15 and GST EV were expressed in E. coli, immobilized on glutathione sepharose beads, and incubated with E. coli lysate containing MBP-CsACD2. Total cell extract (Input) and eluted protein (Elute) were immunoblotted using the anti-MBP and anti-GST antibody. C, Coimmunoprecipitation (Co-IP) assay. HA-tagged CsACD2 and eYFP-tagged SDE15 were coexpressed in the leaves of N. benthamiana through agroinfiltration. HA tag and eYFP proteins were also expressed in the leaves of N. benthamiana as controls. Co-IP assays were performed using anti-HA and anti-GFP antibodies to determine associations. D, The RCCR domain of CsACD2 interacts with the SDE15 in E. coli lysate. E, The C-terminal of SDE15 interacts with CsACD2. GST-SDE15, GST-SDE15∆N, GST-SDE15∆C, and GST EV were expressed in E. coli, immobilized on glutathione sepharose beads, and incubated with E. coli lysate containing MBP-CsACD2, MBP-RCCR, and free MBP proteins. Total cell extract (Input) and eluted protein (Elute) were immunoblotted using the anti-MBP and anti-GST antibody.
To define the regions of SDE15 and CsACD2 involved in the interaction, truncated proteins GST-SDE15∆N (SDE15 protein without the putative signal peptide and additional 25 amio acides following the signal peptide at the N terminus), GST-SDE15∆C (SDE15 protein without the putative signal peptide and 25 amino acids from the C terminus), and MBP-RCCR (maltose-binding protein tag fused with red chlorophyll catabolite reductase; the RCCR domain of CsACD2 with 71-amino acid deletion at the N terminus of CsACD2) were expressed in Escherichia coli, purified and used in GST pull-down assays (Fig. 4, D and E). We found that the RCCR domain of CsACD2 was sufficient to interact with GST-SDE15 (Fig. 4D). In addition, deletion of the C terminus, but not the N terminus of SDE15 abolished its interaction with CsACD2 (Fig. 4E).
SDE15 Repression of Plant Immunity Is Dependent on CsACD2
To determine whether CsACD2 is involved in the SDE15-mediated suppression of plant immunity, we infiltrated the leaves of Nicotiana benthamiana with A. tumefaciens harboring the binary vector that expresses CsACD2-YFP fusion protein and/or SDE15, followed by infiltration with A. tumefaciens containing a binary vector that expresses AvrBsT at 2 d post-CsACD2 infiltration (Fig. 5, A and B; Supplemental Fig. S2; Kim et al., 2010). As expected, AvrBsT-triggered HR was repressed in the presence of CsACD2 or SDE15 plus CsACD2 (Fig. 5, A and B; Supplemental Fig. S2).
Figure 5.
SDE15 and CsACD2 repress the hypersensitive response in N. benthamiana. A, HR assay. A. tumefaciens strain GV2260 harboring binary vectors that are designed to express SDE15, CsACD2 were infiltrated or coinfiltrated into leaves of N. benthamiana at the concentration of 108 CFU mL–1. Two days later, another A. tumefaciens strain GV2260 harboring the binary vector expressing the AvrBsT protein, which can trigger an HR, was infiltrated in the same area of the leaves. HR induction was observed and photographed at 2 to 3 d post inoculation. All experiments were repeated three times with similar results. B, Electrolyte leakage associated with HR induced by AvrBsT at 2 d post infiltration. Leaf discs were floated on deionized water with shaking. The conductivity of the solution was measured after 4 h shaking. Error bars indicate se (n = 3). Lowercase letters represent significant differences in different types of samples by one-way ANOVA followed by lsd post hoc test (P < 0.05). C, GST pull-down assay demonstrates that SDE15 interacts with NbACD2 of N. benthamiana. GST-SDE15 and GST empty vectors were expressed in E. coli, immobilized on glutathione sepharose beads, and incubated with E. coli lysate containing MBP-CsACD2, MBP-NbACD2, and MBP. Total cell extract (Input) and eluted protein (Elute) were immunoblotted using the anti-MBP and anti-GST antibodies. D, HR assay. A. tumefaciens strain GV2260 harboring binary vectors that are designed to express CsACD2 and NbACD2 were infiltrated into leaves of N. benthamiana at the concentration of 108 CFU mL–1. Two days later, another A. tumefaciens strain GV2260 harboring the binary vector expressing the AvrBsT protein was infiltrated in the same area of the leaves. HR induction was observed and photographed at 2 to 3 d post inoculation. All experiments were repeated three times with similar results. E, Electrolyte leakage associated with the HR induced by AvrBsT at 2 d post infiltration. Leaf discs were floated on deionized water with shaking. The conductivity of the solution was measured after 4 h shaking. Error bars indicate se (n = 3). Lowercase letters represent significant differences in different types of samples by one-way ANOVA followed by lsd post hoc test (Pe < 0.05). The experiments were repeated twice with similar results.
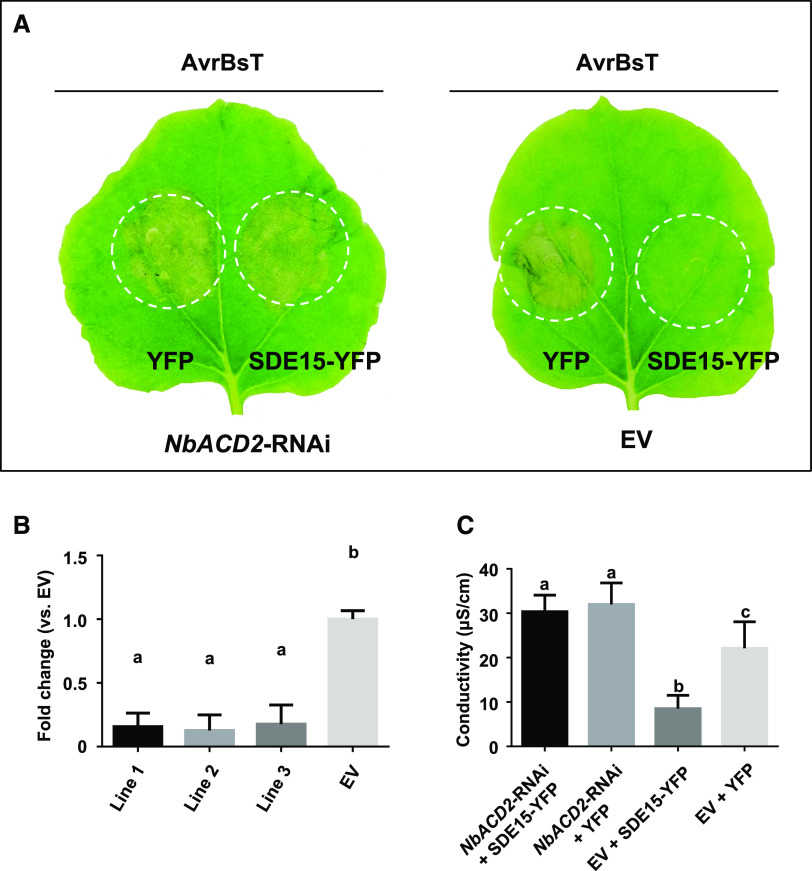
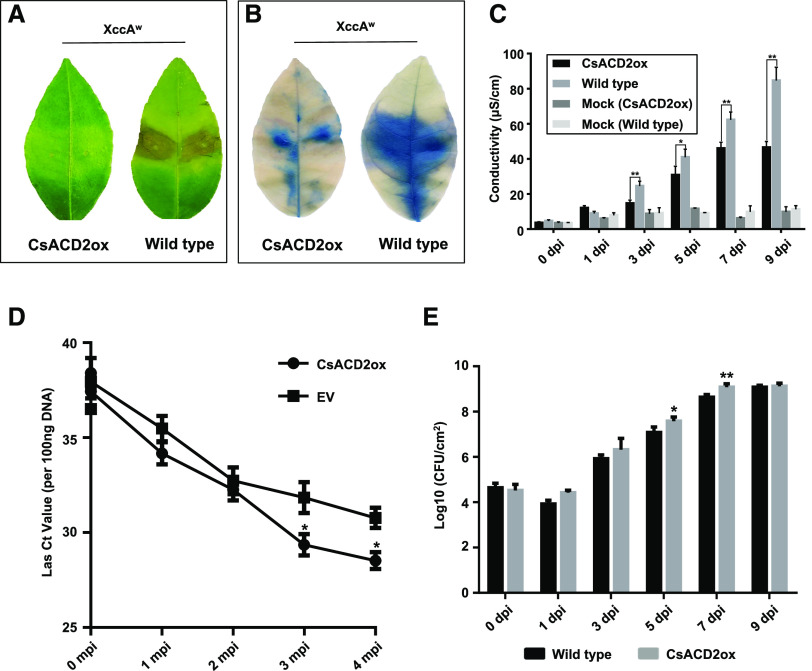
Intriguingly, however, we found that transient overexpression of SDE15-YFP protein alone was also sufficient to suppress AvrBsT-elicited HR in N. benthamiana (Fig. 5A), suggesting two possibilities: (1) SDE15 interacts with the endogenous homolog of ACD2 in N. benthamiana (NbACD2 hereinafter); or (2) SDE15 directly interferes with plant immunity without involving ACD2. In support of the former scenario, GST pull-down assays showed that SDE15 interacted strongly with NbACD2 (Fig. 5C). As in the case of CsACD2, the NbACD2-YFP fusion protein alone, when transiently overexpressed, was sufficient to suppress HR induction by AvrBsT (Fig. 5, D and E; Supplemental Fig. S2). Furthermore, silencing of NbACD2 in N. benthamiana plants abolished the SDE15 suppression of AvrBsT-triggered HR (Fig. 6), suggesting that SDE15 suppression of plant immunity is ACD2-dependent. In addition, overexpression of CsACD2 in Duncan grapefruit had a similar effect as overexpression of SDE15 in suppressing the HR induced by XccAw (Fig. 7, A–C; Supplemental Fig. S3). Overexpression of CsACD2 suppressed the expression of defense-related genes (Supplemental Fig. S4), promoted Las growth (Fig. 7D), and facilitated the multiplication of XccA 306 in the CsACD2ox plants than the EV-transgenic Duncan grapefruit plants (Fig. 7E). Thus, CsACD2 acts as a genuine susceptibility target gene for Las.
Figure 6.
Silencing of ACD2 inhibits HR suppression by SDE15. A, HR assay. A. tumefaciens strain GV2260 harboring TRV vectors that are designed to silence the expression of NbACD2 was infiltrated into leaves of N. benthamiana to generate the NbACD2-RNAi plants. Leaves of NbACD2-silenced plants were infiltrated with the A. tumefaciens strain GV2260 harboring the binary vector expressing SDE15. Two days later, another A. tumefaciens strain GV2260 harboring the binary vector expressing the AvrBsT protein, which can trigger HR, was infiltrated in the same area of the leaves. GV2260 harboring the EV was used as a control. HR induction was observed and photographed at 2 to 3 d post inoculation. All experiments were repeated twice with similar results. B, RT-qPCR analysis of NbACD2. Expression of NbACD2 was repressed in the NbACD2-silenced N. benthamiana plants compared with that in EV transformants. The house keeping gene encoding actin was used as an endogenous control. Error bars indicate se (n = 4). Lowercase letters represent significant differences in different types of samples by one-way ANOVA followed by lsd post hoc test (P < 0.05). C, Electrolyte leakage associated with the HR induced by AvrBsT at 2 d post infiltration. Leaf discs were floated on deionized water with shaking. The conductivity of the solution was measured after 4 h shaking. Error bars indicate se (n = 3). Lowercase letters represent significant differences in different types of samples by one-way ANOVA followed by lsd post hoc test (P < 0.05).
Figure 7.
Constitutively expressing CsACD2 in grapefruit suppresses the HR and promotes disease susceptibility. A, HR was observed in the wild-type but not the CsACD2ox-transgenic Duncan grapefruit at 5 d post inoculation (dpi) with XccAW. XccAW cells were infiltrated into citrus leaves at a concentration of 108 CFU mL−1. Photos were taken at 5 dpi with XccAW. B, Trypan-blue staining of the leaves inoculated with XccAW at 3 dpi indicated HR was repressed in CsACD2ox transgenic Duncan grapefruit. Whole leaf inoculated with XccAW was collected at 3 dpi and immersed with trypan-blue staining solution for 6 h with gently shaking, then de-stained with 98% to 100% ethanol. C, Dynamic electrolyte leakage assay associated with the HR induced by XccAW. Leaf discs were floated on deionized water with shaking. The conductivity of the solution was measured after 4 h shaking. Error bars indicate se (n = 3). Asterisks represent significant differences between CsACD2ox transgenic citrus and nontransgenic control by one-way ANOVA with posthoc test (*P < 0.05 and **P < 0.01). D, Las titers in the CsACD2ox transgenic citrus and EV-transgenic control citrus were determined by TaqMan qPCR at 0, 1, 2, and 3 months post infection (mpi) with Las. Each Ct value was represented by means ± se (n = 6). Asterisks represent significant differences in the Las titer between CsACD2-transgenic citrus and nontransgenic control by one-way ANOVA with posthoc test (*P < 0.05). E, Bacterial titers of XccA 306 in the CsACD2ox transgenic and nontransgenic Duncan grapefruit. Bacterial cells were infiltrated into citrus leaves at a concentration of 106 CFU mL−1. Error bars indicate se (n = 4). Asterisks represent significant differences in the bacterial population between the CsACD2ox-transgenic citrus and nontransgenic control by one-way ANOVA with post hoc test (*P < 0.05 and **P < 0.01).
Auto-immune Response was Observed on CsACD2-RNAi Transgenic Citrus
Next, we went on to produce CsACD2-silenced transgenic citrus plants to determine whether such plants would be resistant to Las. A fragment targeting the second exon and 3ʹ-UTR regions was designed to construct a sense-loop-antisense RNA interference (RNAi) vector and transferred into Duncan grapefruit via stable A. tumefaciens-mediated transformation. Of five independent shootings with endoplasmic reticulum GFP (erGFP)-specific fluorescence obtained in total, chlorosis was observed on four of them (lines 1, 2, 3, and 5), and spontaneous cell death was first observed on lines 3 and 5, then lines 1 and 2 (Fig. 8A). Each of these four transgenic shootings eventually died with the lesions spread over the whole plant.
Figure 8.
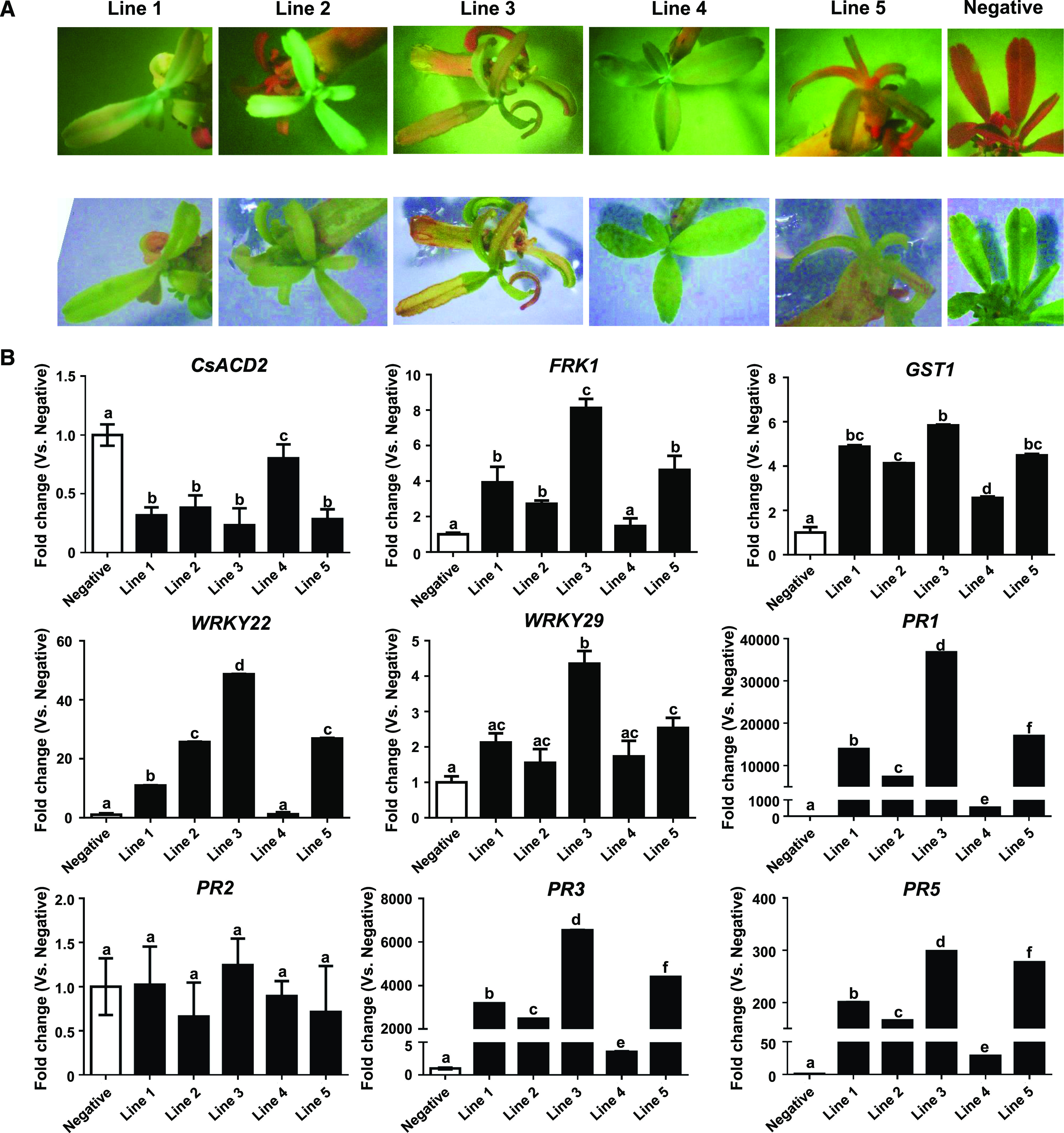
Auto-immune response was triggered on the CsACD2-RNAi transgenic Duncan grapefruit. A, Chlorosis and spontaneous cell death were observed on the CsACD2-RNAi positive transgenic citrus. Top, erGFP fluorescence screening on transgenic CsACD2-RNAi plant. Bottom, photos of transgenic plants in the bright field. Negative-control plant is wild-type Duncan plant which did not show erGFP fluorescence signal. B, RT-qPCR analysis of defense-related genes. Expression of FRK1, GST1, WRKY22, WRKY29, PR1, PR3, and PR5 in the CsACD2-RNAi transgenic citrus was compared with the wild-type Duncan grapefruit. The housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase-C (GAPDH-C) was used as an endogenous control. Error bars indicate se (n = 4). Lowercase letters represent significant differences in different types of samples by one-way ANOVA followed by lsd post hoc test (P < 0.05). All experiments were repeated three times with the similar results.
RNA samples from the transgenic shootings were collected before they died to detect the expression of CsACD2 and defense-related genes (Fig. 8B). The expression level of CsACD2 was significantly reduced in lines 1, 2, 3, and 5 but not in line 4. Interestingly, line 4 did not show spontaneous cell death, although it had erGFP fluorescence. This result indicates the spontaneous cell death triggered in lines 1, 2, 3, and 5 was caused by silencing of CsACD2. In addition, immunity-related genes FRK1, GST1, WRKY22, PR1, PR3, and PR5 were highly expressed in lines 1, 2, 3, and 5. The immature plant death precluded us from testing the resistance of RNAi lines against Las. Overall, the data demonstrate that silencing of CsACD2 triggers auto-immune responses and runaway cell death.
DISCUSSION
In this study, we showed that a Las-secreted protein SDE15 functions as a virulence effector by suppressing plant immunity. SDE15 is a conserved effector among Las strains and contains a typical Sec-dependent secretion signal peptide (Prasad et al., 2016). SDE15 promotes Las multiplication by targeting the citrus CsACD2 protein, a regulator of programmed cell death. Transgenic expression of SDE15 suppresses not only plant immunity induced by XccAw in citrus, but also that induced by Xanthomonas vesicatoria effector protein AvrBsT in N. benthamiana, suggesting that SDE15 may be a broad-spectrum suppressor of plant immunity. Importantly, transgenic expression of SDE15 in citrus facilitated Las multiplication and HLB symptom development, consistent with its role in suppressing plant defense responses.
Suppression of plant immunity is a common virulence strategy by pathogens. Many bacterial pathogens and fungal/oomycete pathogens live extracellularly in the leaf apoplast and produce effectors that can suppress HR (Jones and Dangl, 2006; Schornack et al., 2009; Toruño et al., 2016; Ramachandran et al., 2017). Liberibacter is one of the few bacteria (Liberibacter, Serratia marcescens [Zhang et al., 2005], Candidatus Phytoplasma, Spiroplasma, and Candidatus Phlomobacter fragariae [Zreik et al., 1998]) that live inside plant cells, mainly inside the sieve cells of the phloem, a highly specialized vascular tissue in plants. Our results, along with that of PLCP-inhibiting activity of another Las effector SDE1, suggest that phloem-colonizing bacteria have adopted virulence strategies similar to extracellular plant pathogens to suppress plant immune responses. Of note, most effectors of Phytoplasma spp. are smaller than 40 kD, allowing them to potentially move out of phloem sieve cells, probably through plasmodesmata, which have size exclusion limits ranging between 10 and 40 kD (Imlau et al., 1999; Bai et al., 2009). SDE15 without the putative signal peptide has a Mr of 8.68 kD, which could enable it to move out of phloem sieve cells and target CsACD2 in both sieve cells and the surrounding cell types, such as phloem companion cells.
Mechanistically, we found that SDE15 exerts its effect on plant immunity via interacting with CsACD2, a homolog of Arabidopsis ACD2 (Yao and Greenberg, 2006). CsACD2 is hypothesized to act as a S gene for HLB. S genes (van Schie and Takken, 2014) facilitate compatibility of pathogens (e.g. promoting pathogen growth or suppressing immune responses, or disease development; Wang et al., 2017). This hypothesis is supported by the following evidence: specific interaction between SDE15 and CsACD2 at the protein level, SDE15 suppression of HR in N. benthamiana dependent on ACD2, and constitutive expression of CsACD2 in citrus suppressing plant immunity and promoting Las multiplication similar as the phenotypes of overexpressing SDE15. It is noteworthy that the evidence for ACD2-dependent SDE15 suppression of plant immunity is obtained from transient gene silencing of NbACD2, because gene silencing of CsACD2 based on stable expression leads to immature plant death, which prevented us from performing immunity suppression assays. Arabidopsis ACD2 encodes a RCCR that in vitro can break down red chlorophyll catabolite (Yao and Greenberg, 2006). Interestingly, loss of the ACD2 gene also leads to spontaneous cell death in Arabidopsis, similar to our results in CsACD2-silenced citrus explants, implicating a conserved role of ACD2 family genes in regulating cell death in diverse plants (Greenberg et al., 1994). How the SDE15 and CsACD2 interaction leads to suppression of cell death and other immune responses in citrus needs further characterization.
Mutations of S genes or modulation of their expression have been successfully used to enhance disease resistance in agriculture (Li et al., 2012; Wang et al., 2014; Blanvillain-Baufumé et al., 2017; Oliva et al., 2019). We anticipate that future precision genome-editing efforts aimed at the CsACD2 promoter or coding sequences, via CRISPR technology (Jia et al., 2017; Wang 2019), to fine tune CsACD2 gene expression or interaction with SDE15 without causing spontaneous plant cell death could provide new possibilities to generate citrus resistance against HLB.
CONCLUSIONS
Our data enrich the current understanding of the interaction between Las and citrus. We identified a Las effector, SDE15, that negatively regulates citrus defense. Transgenic citrus constitutively expressing SDE15 has the function to repress HR and became more susceptible to HLB. By Y2H screening, CsACD2, a homologue of Arabidopsis ACD2, was identified as a downstream target of SDE15. SDE15 suppression of plant immunity is dependent on CsACD2. Overexpression of CsACD2 in citrus suppresses plant immunity and promotes Las multiplication.
MATERIALS AND METHODS
Vector Construction
To generate the SDE15-expressing construct for plant transformation, Las genomic DNA was isolated from HLB diseased citrus leaves by the cetyltrimethyl ammonium bromide method (Allen et al., 2006). The coding sequence of SDE15 without the signal peptide (222 bp) was PCR-amplified using gene-specific primers (Supplemental Table S4). A BamHI recognition sequence and a KpnI recognition site with two protecting nucleotides were added to the 5ʹ end of primers. The PCR product was purified and cloned into the pGEM-T Easy vector (Promega) and then cloned into the binary vector erGFP-1380N with the C-terminal 3×HA protein tag at the BamHI and KpnI sites to generate the SDE15-overexpression vector. The 996-bp coding sequence of CsACD2 was amplified from citrus leaf cDNA and cloned into the binary vector erGFP-1380N and fused with the C-terminal 3×HA protein tag at the BamHI and KpnI sites to generate the CsACD2-overexpression vector. To generate the vector of CsACD2-RNAi to silence CsACD2 in citrus, a 624-bp sequence encompassing the exon 2 and the 3ʹ-UTR sequence of CsACD2 was amplified from citrus leaf cDNA, assembled with the 199-bp sequence of the 1st intron of potato GA20 oxidase in an inverted repeat orientation by using NEBuilder HiFi DNA Assembly Cloning Kit (New England Biolab). The assembled sense-loop-antisense construct was cloned into the binary vector erGFP-1380N under the control of the CaMV 35S promoter. The resulting binary vectors were transferred into Agrobacterium tumefaciens strain EHA105 for citrus transformation. The EV was used in citrus transformation as a negative control.
For Y2H, the coding region of SDE15 (minus the putative signal peptide) was amplified and cloned in-frame with the GAL4 DNA-binding domain (BD) of the bait vector pGBKT7 to generate BD-SDE15 for Y2H library screening and cotransformation in yeast. The 996-bp coding sequence of CsACD2 was PCR-amplified from citrus leaf cDNA and cloned in-frame with the GAL4 DNA-activating domain (AD) of the prey vector pGADT7 to generate AD-CsACD2 for cotransformation with bait vector in yeast to confirm the interaction. BD and AD vectors were constructed by using the In-Fusion cloning kit (Clontech).
To generate recombinant protein constructs for GST pull-down assay, the coding region of SDE15 (without the predicted signal peptide) was inserted between EcoRI and XhoI sites of the pGEX-4T-1 vector (GE Healthcare) to generate the GST-SDE15 fusion protein vector as the bait. The coding sequence of CsACD2 was inserted between BamHI and EcoRI sites of the pMAL-c5X vector (NEB) to generate the MBP-CsACD2 fusion protein vector as the prey. The truncated sequences of SDE15 fragments (SDE15∆N and SDE15∆C) were inserted into the EcoRI-digested pGEX-4T-1 vector by using the In-Fusion cloning kit to generate the GST-SDE15∆N and GST-SDE15∆C fusion protein vectors as the bait. The coding sequence of RCCR domain of CsACD2 was amplified and inserted between BamHI and EcoRI sites of the pMAL-c5X vector by using the In-Fusion cloning kit to generate the MBP-RCCR fusion protein vector as the prey.
To generate constructs for agro-infiltration assay in Nicotiana benthamiana, modified pCambia1380 vectors were constructed by inserting the cauliflower mosaic virus promoter (CaMV 35S) and EYFP coding sequence to create the transient expression vector with the C-terminal EYFP reporter protein (pCambia1380-35S-EYFP). The coding sequence of SDE15 without signal peptide was PCR-amplified and inserted between BamHI and KpnI sites of pCambia1380-35S-EYFP to generate 35S-SDE15-EYFP. The coding sequence of CsACD2 was inserted between BamHI and KpnI sites of pCambia1380-35S-EYFP to generate 35S-CsACD2-EYFP. The coding sequence of NbACD2 was inserted between BamHI and KpnI sites of pCambia1380-35S-EYFP by using the In-Fusion cloning kit to generate 35S-NbACD2-EYFP. All the vectors were then transferred into A. tumefaciens strain GV2260 for agro-infiltration assays.
To generate constructs using the tobacco rattle virus (TRV) vector to silence the expression of NbACD2 in N. benthamiana through virus-induced gene silencing (VIGS), the 451-bp exon 2 sequence of NbACD2 was amplified and inserted at the KpnI site of the pTRV2 vector (CD3-1040 from Arabidopsis Biological Resource Center) by using the In-Fusion cloning kit to generate the NbACD2-TRV2 vector. This vector was transferred into A. tumefaciens strain GV2260 for coinfiltration with another A. tumefaciens strain GV2260 harboring the pTRV1 vector (CD3-1039 from Arabidopsis Biological Resource Center) to make NbACD2 silencing plants. The pTRV2 EV was transferred into GV2260 as the EV control. All the primers used for vector construction were listed in Supplemental Table S4.
Plant Transformation and Pathogen Inoculation
A. tumefaciens-mediated transformation of etiolated epicotyl segments of Duncan grapefruit (Citrus × paradisi) was carried out as described previously (Orbović and Grosser, 2015). A. tumefaciens EHA105 harboring the recombinant plasmid was used for citrus transformation. Transgenic lines showing kanamycin-resistance and erGFP-specific fluorescence were selected and then micrografted in vitro onto 1-month-old Carrizo citrange (Citrus sinensis × Poncirus trifoliata) nucellar rootstock seedlings. After a month of growth in vitro, the grafted shoots were potted into a peat-based commercial potting medium and acclimated under greenhouse conditions. The EV-transformed citrus were also generated as control for further pathogenicity assay.
For the HLB-pathogenicity assay, citrus plants, such as SDE15-transgenic plants (∼4-year-old), were graft-inoculated with Las as previously reported (Li et al., 2009). The CsACD2ox trees (1-year-old) were inoculated with Las via psyllid transmission. EV transgenic trees were used as the control. Midrib DNA was isolated from the graft-inoculated trees monthly after grafting up to 4 months post inoculation, whereas leaf punch DNA was isolated from the psyllid-feeding trees monthly to 4 months after inoculation. To guarantee the psyllids were Las positive, DNA of psyllids was extracted after infection for Las detection. Isolated DNA was used to quantify Las by Taqman qPCR with Primer/probe combination (CQULA04F-CQULAP10-CQULA04R) as described previously (Wang et al., 2006; Trivedi et al., 2009). The Ct value of each amplicon represents the Las genomic copy numbers in 100 ng citrus midrib DNA. The test was repeated three times.
For the Xanthomonas citri ssp. citri pathogenicity and HR assays in citrus, SDE15-transgenic and nontransgenic Duncan grapefruit plants were used for inoculation in a quarantine greenhouse. The wild-type strain Xcc306 causes disease on grapefruit whereas the Xcc Aw strain triggers HR in grapefruit leaves (Brunings and Gabriel, 2003; Rybak et al., 2009). Xcc strains were cultured with shaking overnight at 28°C in NB, pelleted, and suspended in sterile tap water, and the concentrations were adjusted to 106 CFU mL−1 (for Xcc 306) and 108 CFU mL−1 (for Xcc Aw). Bacterial solutions were infiltrated into fully expanded, immature leaves with needleless syringes (Yan and Wang, 2012; Teper et al., 2019). The tests were repeated three times with similar results. HR was characterized by the trypan-blue staining and the electrolyte-leakage assay. Citrus leaves were collected at 3 d after Xcc Aw infiltration to perform trypan-blue staining (Fernández-Bautista et al., 2016). For the electrolyte-leakage assay, Xcc Aw-infiltrated leaf discs were floated on deionized water with shaking. The conductivity of the solution was measured 4 h later using an Oakton Conductivity Benchtop Meter (Thermo Fisher). Disease symptoms and HR phenotype were photographed at 3, 5, 7, and 9 d postinoculation. Growth curve assays of Xcc 306 were conducted at 0, 1, 3, 5, 7, and 9 d postinoculation.
Agro-infiltration Assay in N. benthamiana
A. tumefaciens strain GV2260 cells containing binary vectors were cultured overnight in Luria–Bertani liquid medium with 50 μg mL−1 of rifampicin and 50 μg mL−1 kanamycin and resuspended in induction medium (10 mm MgCl2, 10 mm MES pH 5.6, 200 μm acetosyringone), and incubated at 25°C with shaking for 4 h. The cultures were diluted to OD600 of 0.1 or 0.2. For each vector, three leaves of young N. benthamiana plants were infiltrated with diluted A. tumefaciens suspension in triplicates.
For the HR assay, young leaves of N. benthamiana were first infiltrated with A. tumefaciens cells containing binary vectors for SDE15-EYFP and/or CsACD2-EYFP by using a needleless syringe, kept in a greenhouse for 2 d and then infiltrated with another A. tumefaciens strain harboring the binary vector carrying the AvrBsT protein, which can trigger HR as reported previously (Kim et al., 2010). Agro-infiltrated plants were kept in a greenhouse and HR was examined and photographed at 3 d post AvrBsT inoculation. For the electrolyte leakage assay, leaf discs of AvrBsT infiltrated plants at 2 d postinfiltration were floated on deionized water with shaking. The conductivity of the solution was measured 4 h later using an Oakton Conductivity Benchtop Meters (Thermo Fisher). The A. tumefaciens transformant cells harboring an EV were infiltrated into the leaves of N. benthamiana as controls.
Extraction of Phloem Sap Proteins
An optimized method of protein extraction from the phloem sap was performed by combining two methods reported before (Hijaz and Killiny, 2014; O’Leary et al., 2014). Briefly, 10 to 20 cm (0.5-cm diameter) stems from Las infected and uninfected trees were collected. The bark area was stripped into two pieces and was manually removed from the twig. The inner part of the bark was rinsed with deionized water and dried with Kim wipes. Then the bark strips were cut into about 1-cm pieces using a sterile razor blade and placed in a 60-mL syringe filled with distilled water. Vacuum was applied for 5 to 15 s repeatedly to let water penetrate the bark. Then the bark pieces were dried with Kim wipes and placed in a 20-mL syringe, centrifuged in 50-mL falcon tube for 10 min at 4,000 g at 4°C. The collected phloem sap was centrifuged at 15,000g for 5 min. The supernatant was heated for 5 min at 95°C in the SDS gel-loading buffer for SDE15 detection with specific antibody.
RNA Isolation and Gene Expression Analysis by RT-qPCR
RT-qPCR was performed to examine the expression of SDE15, CsACD2, NbACD2, and defense-related genes in different types of plants and psyllids samples (Andrade et al., 2020). Total RNA of transgenic citrus, transgenic N. benthamiana, and psyllids were extracted by Trizol reagent (Thermo Fisher) and digested with DNase I (Promega) following the manufacturers’ instructions. First-strand cDNA was synthesized from purified RNA with the ImProm-II Reverse Transcription System (Promega) and diluted 10 times for RT-qPCR to detect related genes with specific primers (Supplemental Table S5). Twenty microliters of qPCR reaction consisted of 10 µL of 2×KiCqStart SYBR Green qPCR ReadyMix (Sigma-Aldrich), 1 µL of each primer (5 µm), 2 µL of diluted cDNA template, and 6 µL of DNase/RNase free water. The PCR cycling consisted of an initial activation step at 95°C for 3 min, followed by 40 cycles of 95°C for 15 s and 60°C for 40 s. All cDNA samples were run in triplicates. The citrus glyceraldehyde-3-phosphate dehydrogenase gene, N. benthamiana actin gene, and Las gyrA gene were used as endogenous controls wherever appropriate. The qPCR primer sequences of specific genes and endogenous control genes are listed in Supplemental Table S5.
Y2H Screening and Protein-Protein Interaction Analysis
Total RNA was extracted from leaves of Valencia sweet orange (Citrus × sinensis), HLB S (Las Ct value 25–26 per 100 ng DNA), HLB AS (Las Ct value 28–30 per 100 ng DNA), and H (Las free) by Trizol reagent (Thermo Fisher), digested by DNase I (Promega). mRNA samples were purified using the NucleoSpin RNA kit (Clontech). ALL three types of mRNA samples were used to construct Y2H libraries in the pGADT7-Rec vector using the Make Your Own “mate and plate” library system (Clontech) following the manufacturer’s instructions and transformed into the yeast strain Y187 by using the Yeastmaker Yeast Transformation System 2 (Clontech). The titer of each constructed library is more than 3 × 108 indicating good transformation efficiency. The BD:SDE15 construct was transformed into Y2HGOLD yeast strain (Clontech). Library screening was performed according to the Matchmaker Gold Y2H system protocol (Clontech). Standard positive controls (pGBKT7-53 and pGADT7-T; Clontech) and standard negative controls (pGBKT7-Lam and pGADT7-T) were included. After mating between the Gold strain transformed with BD:SDE15 and the Y187 libraries, diploid yeasts were plated on synthetic dropout (SD)/−Leu/−Trp (DDO), SD/−Leu/−Trp/−Ade/−His (QDO) and SD/−Leu/-Trp/-Ade/-His plus X-α-gal and Aureobasidin A (AbA; QDO/A/X) agar plates to detect the activation of reporter genes HIS3, ADE2, MEL1 (for α-galactosidase activity), and AbAr (for Aureobasidin A resistance). The fragments of positive diploid yeast were amplified by colony PCR with Matchmaker Insert Check PCR Mix 2 (Clontech) and analyzed by electrophoresis on a 0.8% TAE Agarose/EtBr gel. The PCR products with a single band were purified and sent for sequencing. The PCR products with multiple bands indicate the presence of more than one prey plasmid in a heterozygote cell. For this situation, plasmids were isolated from the heterozygote cells with multiple plasmids with Easy Yeast Plasmid Isolation Kit (Clontech) and transferred into E. coli for sequencing. BLAST was used to compare the insert nucleotide sequences to the genome of sweet orange (Citrus × sinensis) to identify corresponding proteins that interact with SDE15.
Recombinant Protein Expression and GST Pull-Down Assays
Escherichia coli cells expressing GST or GST fusion proteins were washed in PBS buffer and suspended with CelLytic B Cell Lysis Reagent (Sigma-Aldrich) to generate the cell lysates. After centrifugation, the cell lysates were incubated with glutathione agarose beads in accordance with the GST Protein Interaction Pull-Down Kit instructions (Thermo Scientific). The beads were washed to remove the unbound proteins and incubated with E. coli cell lysates expressing either MBP or MBP fusion protein for 1 to 2 h at 4°C. After washing four times, the beads were eluted with 10 mm glutathione, and the eluates were collected and immunoblotted using anti-MBP (NEB) and anti-GST (Abcam) antibodies.
Co-IP Assay
CsACD2-HA was transiently coexpressed with SDE15-EYFP in the leaf of N. benthamiana. Total proteins were extracted and applied for immunoprecipitation with magnetic beads on which the anti-HA antibody is covalently immobilized in accordance with the HA-Tag IP/Co-IP Kit instructions (Thermo Scientific). The beads were washed with the IP Lysis/Wash buffer to remove the unbound proteins and incubated with extracted proteins expressing either HA or HA fusion protein overnight. After washing four times, the beads were eluted with the low pH Elution buffer and neutralized with the Neutralization buffer. The eluted proteins were boiled with nonreducing Sample buffer (5×) and detected by anti-HA (Sigma-Aldrich) and anti-GFP (Sigma-Aldrich) immunoblots.
TRV-based VIGS in N. benthamiana
TRV based VIGS was used to down-regulate the expression of NbACD2 in N. benthamiana as reported (Senthil-Kumar and Mysore, 2014). Briefly, TRV1 and NbACD2-TRV2 carrying GV2260 strains were coinoculated into 3-week-old N. benthamiana leaves by a needleless syringe. TRV1 and TRV2-EV carrying GV2260 strains were coinoculated into N. benthamiana leaves as controls. At 2 to 3 weeks later, the newly developed noninoculated leaves were collected to extract RNA for NbACD2 expression test. The NbACD2-RNAi positive plants were used to perform the subsequent HR test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CLIBASIA_04025 (SDE15), Cs1g22670 (CsACD2), Niben101Scf02681g01015.1 (NbACD2), Cs5g32800 (GST1), Cs1g16040 (FRK1), Cs5g03010 (WRKY22), orange1.1t00419 (WRKY29), Cs8g03430 (PR1), orange1.1t00647 (PR2), Cs1g26330 (PR3), and Cs3g24410 (PR5).
SUPPLEMENTAL DATA
The following supplemental materials are available.
Supplemental Figure S1. Generation of SDE15-transgenic citrus.
Supplemental Figure S2. Western blot was used to detect the expression of YFP fusion proteins during the HR induction in N. benthamiana.
Supplemental Figure S3. CsACD2ox transgenic citrus validation by western blot using the HA tag-specific antibody.
Supplemental Figure S4. RT-qPCR analysis of defense-related genes in the CsACD2ox transgenic citrus during HR induction.
Supplemental Table S1. List of citrus proteins interacting with SDE15 identified in healthy Valencia sweet orange using the Y2Hd assay.
Supplemental Table S2. List of citrus proteins interacting with SDE15 identified in asymptomatic Valencia sweet orange infected by Las using the Y2H.
Supplemental Table S3. List of citrus proteins interacting with SDE15 identified in symptomatic Valencia sweet orange infected by Las using the Y2H assay.
Supplemental Table S4. Primers used for vector construction.
Supplemental Table S5. Primers used for RT-qPCR and Taqman probe qPCR analysis.
Acknowledgments
The authors thank the transformation lab of Citrus Research and Education Center, University of Florida, for supplying transgenic citrus service.
Footnotes
The work was supported by USDA National Institute of Food and Agriculture (grant nos. 2018–70016–27412, 2016–70016–24833, and 2019–70016–29796), the Florida Citrus Initiative, and the Florida Citrus Research and Development Foundation.
Articles can be viewed without a subscription.
References
- Albrecht U, Bowman K(2008) Gene expression in Citrus sinensis (L.) Osbeck following infection with the bacterial pathogen Candidatus Liberibacter asiaticus causing Huanglongbing in Florida. Plant Sci 175: 291–306 [Google Scholar]
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF(2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1: 2320–2325 [DOI] [PubMed] [Google Scholar]
- Andrade MO, Pang Z, Achor DS, Wang H, Yao T, Singer BH, Wang N(2020) The flagella of ‘Candidatus Liberibacter asiaticus’ and its movement in planta. Mol Plant Pathol 21: 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J(2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Bai X, Correa VR, Toruño TY, Ammar D, Kamoun S, Hogenhout SA(2009) AY-WB phytoplasma secretes a protein that targets plant cell nuclei. Mol Plant Microbe Interact 22: 18–30 [DOI] [PubMed] [Google Scholar]
- Blanvillain-Baufumé S, Reschke M, Solé M, Auguy F, Doucoure H, Szurek B, Meynard D, Portefaix M, Cunnac S, Guiderdoni E, et al. (2017) Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol J 15: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove JM.(2006) Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J Plant Pathol 88: 7–37 [Google Scholar]
- Brunings AM, Gabriel DW(2003) Xanthomonas citri: Breaking the surface. Mol Plant Pathol 4: 141–157 [DOI] [PubMed] [Google Scholar]
- Clark K, Franco JY, Schwizer S, Pang Z, Hawara E, Liebrand TWH, Pagliaccia D, Zeng L, Gurung FB, Wang P, et al. (2018) An effector from the Huanglongbing-associated pathogen targets citrus proteases. Nat Commun 9: 1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Y, Zhou L, Hall DG, Li W, Doddapaneni H, Lin H, Liu L, Vahling CM, Gabriel DW, Williams KP, et al. (2009) Complete genome sequence of citrus huanglongbing bacterium, ‘Candidatus Liberibacter asiaticus’ obtained through metagenomics. Mol Plant Microbe Interact 22: 1011–1020 [DOI] [PubMed] [Google Scholar]
- Fernández-Bautista N, Domínguez-Núñez JA, Moreno MMC, Berrocal-Lobo M(2016) Plant tissue trypan blue staining during phytopathogen infection. Bio Protoc 6: e2078 [Google Scholar]
- Folimonova SY, Achor DS(2010) Early events of citrus greening (Huanglongbing) disease development at the ultrastructural level. Phytopathology 100: 949–958 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Dong XN(2013) Systemic acquired resistance: Turning local infection into global defense. Annu Rev Plant Biol 64: 839–863 [DOI] [PubMed] [Google Scholar]
- Goritschnig S, Zhang Y, Li X(2007) The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J 49: 540–551 [DOI] [PubMed] [Google Scholar]
- Gottwald TR.(2010) Current epidemiological understanding of citrus Huanglongbing. Annu Rev Phytopathol 48: 119–139 [DOI] [PubMed] [Google Scholar]
- Greenberg JT, Guo A, Klessig DF, Ausubel FM(1994) Programmed cell death in plants: A pathogen-triggered response activated coordinately with multiple defense functions. Cell 77: 551–563 [DOI] [PubMed] [Google Scholar]
- Hijaz F, Killiny N(2014) Collection and chemical composition of phloem sap from Citrus sinensis L. Osbeck (sweet orange). PLoS One 9: e101830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout SA, Oshima K, Ammar D, Kakizawa S, Kingdom HN, Namba S(2008) Phytoplasmas: Bacteria that manipulate plants and insects. Mol Plant Pathol 9: 403–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlau A, Truernit E, Sauer N(1999) Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11: 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H, Zhang Y, Orbović V, Xu J, White FF, Jones JB, Wang N(2017) Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol J 15: 817–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL(2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim JS, Sagaram US, Burns JK, Li JL, Wang N(2009) Response of sweet orange (Citrus sinensis) to ‘Candidatus Liberibacter asiaticus’ infection: Microscopy and microarray analyses. Phytopathology 99: 50–57 [DOI] [PubMed] [Google Scholar]
- Kim NH, Choi HW, Hwang BK(2010) Xanthomonas campestris pv. vesicatoria effector AvrBsT induces cell death in pepper, but suppresses defense responses in tomato. Mol Plant Microbe Interact 23: 1069–1082 [DOI] [PubMed] [Google Scholar]
- Li J, Pang Z, Trivedi P, Zhou X, Ying X, Jia H, Wang N(2017) ‘Candidatus Liberibacter asiaticus’ encodes a functional salicylic acid (SA) hydroxylase that degrades SA to suppress plant defenses. Mol Plant Microbe Interact 30: 620–630 [DOI] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B(2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30: 390–392 [DOI] [PubMed] [Google Scholar]
- Li W, Levy L, Hartung JS(2009) Quantitative distribution of ‘Candidatus Liberibacter asiaticus’ in citrus plants with citrus huanglongbing. Phytopathology 99: 139–144 [DOI] [PubMed] [Google Scholar]
- Melotto M, Panchal S, Roy D(2014) Plant innate immunity against human bacterial pathogens. Front Microbiol 5: 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary BM, Rico A, McCraw S, Fones HN, Preston GM(2014) The infiltration-centrifugation technique for extraction of apoplastic fluid from plant leaves using Phaseolus vulgaris as an example. J Vis Exp 94: 52113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, Eom JS, Li C, Nguyen H, Liu B, et al. (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37: 1344–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orbović V, Grosser JW(2015) Citrus transformation using juvenile tissue explants. Methods Mol Biol 1224: 245–257 [DOI] [PubMed] [Google Scholar]
- Pagliaccia D, Shi J, Pang Z, Hawara E, Clark K, Thapa SP, De Francesco AD, Liu J, Tran TT, Bodaghi S, et al. (2017) A pathogen secreted protein as a detection marker for Citrus Huanglongbing. Front Microbiol 8: 2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitino M, Armstrong CM, Cano LM, Duan Y(2016) Transient expression of Candidatus Liberibacter asiaticus effector induces cell death in Nicotiana benthamiana. Front Plant Sci 7: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad S, Xu J, Zhang Y, Wang N(2016) SEC-translocon dependent extracytoplasmic proteins of Candidatus Liberibacter asiaticus. Front Microbiol 7: 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran SR, Yin C, Kud J, Tanaka K, Mahoney AK, Xiao F, Hulbert SH(2017) Effectors from wheat rust fungi suppress multiple plant defense responses. Phytopathology 107: 75–83 [DOI] [PubMed] [Google Scholar]
- Rybak M, Minsavage GV, Stall RE, Jones JB(2009) Identification of Xanthomonas citri ssp. citri host specificity genes in a heterologous expression host. Mol Plant Pathol 10: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornack S, Huitema E, Cano LM, Bozkurt TO, Oliva R, Van Damme M, Schwizer S, Raffaele S, Chaparro-Garcia A, Farrer R, et al. (2009) Ten things to know about oomycete effectors. Mol Plant Pathol 10: 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS(2014) Tobacco rattle virus-based virus-induced gene silencing in Nicotiana benthamiana. Nat Protoc 9: 1549–1562 [DOI] [PubMed] [Google Scholar]
- Shi Q, Febres VJ, Zhang S, Yu F, McCollum G, Hall DG, Moore GA, Stover E(2018) Identification of gene candidates associated with Huanglongbing tolerance, using ‘Candidatus Liberibacter asiaticus’ Flagellin 22 as a proxy to challenge citrus. Mol Plant Microbe Interact 31: 200–211 [DOI] [PubMed] [Google Scholar]
- Teper D, Zhang Y, Wang N(2019) TfmR, a novel TetR-family transcriptional regulator, modulates the virulence of Xanthomonas citri in response to fatty acids. Mol Plant Pathol 20: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toruño TY, Stergiopoulos I, Coaker G(2016) Plant-pathogen effectors: Cellular probes interfering with plant defenses in spatial and temporal manners. Annu Rev Phytopathol 54: 419–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi P, Sagaram US, Kim J, Brlansky RH, Rogers ME, Stelinski LL, Oswalt C, Wang N(2009) Quantification of viable Candidatus Liberibacter asiaticus in hosts using quantitative PCR with the aid of ethidium monoazide (EMA). Eur J Plant Pathol 124: 553–563 [Google Scholar]
- van Schie CC, Takken FL(2014) Susceptibility genes 101: How to be a good host. Annu Rev Phytopathol 52: 551–581 [DOI] [PubMed] [Google Scholar]
- Wang N. (2020) A perspective of citrus Huanglongbing in the context of the Mediterranean Basin. J Plant Pathol 102: 635–640 [Google Scholar]
- Wang N.(2019) The citrus Huanglongbing crisis and potential solutions. Mol Plant 12: 607–609 [DOI] [PubMed] [Google Scholar]
- Wang N, Pierson EA, Setubal JC, Xu J, Levy JG, Zhang Y, Li J, Rangel LT, Martins J Jr.(2017) The Candidatus Liberibacter-host interface: Insights into pathogenesis mechanisms and disease control. Annu Rev Phytopathol 55: 451–482 [DOI] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL(2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947–951 [DOI] [PubMed] [Google Scholar]
- Wang Z, Yin Y, Hu H, Yuan Q, Peng G, Xia Y(2006) Development and application of molecular-based diagnosis for ‘Candidatus Liberibacter asiaticus’, the causal pathogen of citrus huanglongbing. Plant Pathol 55: 630–638 [Google Scholar]
- Yan Q, Wang N(2012) High-throughput screening and analysis of genes of Xanthomonas citri subsp. citri involved in citrus canker symptom development. Mol Plant Microbe Interact 25: 69–84 [DOI] [PubMed] [Google Scholar]
- Yao N, Greenberg JT(2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell death. Plant Cell 18: 397–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Melcher U, Zhou L, Najar FZ, Roe BA, Fletcher J(2005) Genomic comparison of plant pathogenic and nonpathogenic Serratia marcescens strains by suppressive subtractive hybridization. Appl Environ Microbiol 71: 7716–7723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jalan N, Zhou X, Goss E, Jones JB, Setubal JC, Deng X, Wang N(2015) Positive selection is the main driving force for evolution of citrus canker-causing Xanthomonas. ISME J 9: 2128–2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zreik L, Bové JM, Garnier M(1998) Phylogenetic characterization of the bacterium-like organism associated with marginal chlorosis of strawberry and proposition of a Candidatus taxon for the organism, ‘Candidatus phlomobacter fragariae’. Int J Syst Bacteriol 48: 257–261 [DOI] [PubMed] [Google Scholar]