A resource of resistance gene analogs within Brassicaceae species identifies functional resistance genes to improve plant breeding programs.
Abstract
The Brassicaceae consists of a wide range of species, including important Brassica crop species and the model plant Arabidopsis (Arabidopsis thaliana). Brassica spp. crop diseases impose significant yield losses annually. A major way to reduce susceptibility to disease is the selection in breeding for resistance gene analogs (RGAs). Nucleotide binding site-leucine rich repeats (NLRs), receptor-like kinases (RLKs), and receptor-like proteins (RLPs) are the main types of RGAs; they contain conserved domains and motifs and play specific roles in resistance to pathogens. Here, all classes of RGAs have been identified using annotation and assembly-based pipelines in all available genome annotations from the Brassicaceae, including multiple genome assemblies of the same species where available (total of 32 genomes). The number of RGAs, based on genome annotations, varies within and between species. In total 34,065 RGAs were identified, with the majority being RLKs (21,691), then NLRs (8,588) and RLPs (3,786). Analysis of the RGA protein sequences revealed a high level of sequence identity, whereby 99.43% of RGAs fell into several orthogroups. This study establishes a resource for the identification and characterization of RGAs in the Brassicaceae and provides a framework for further studies of RGAs for an ultimate goal of assisting breeders in improving resistance to plant disease.
The Brassicaceae family encompasses ∼340 genera and 3,350 species, including important crop species from the genus Brassica, such as Brassica napus (canola), Brassica oleracea (including kale [Brassica oleracea var. acephala], broccoli [Brassica oleracea var. italica], cabbage [Brassica oleracea var. capitata], and brussels sprouts [Brassica oleracea var. gemmifera]), and Brassica rapa (bok choy [Brassica rapa ssp. chinensis] and turnip [Brassica rapa ssp. rapa]; Schmidt et al., 2001). Several members of this family are also employed as model species, including Arabidopsis (Arabidopsis thaliana), which is the most widely used plant in research; Arabidopsis halleri, a model species for heavy metal tolerance and hyperaccumulation (Palmer et al., 2001); Lepidium meyenii, a model species for floral structure (Lee et al., 2002); Eutrema salsugineum, a model species for salinity stress (Wu et al., 2012); and Barbarea vulgaris, a model species for insect resistance (Nielsen et al., 2010). Huang et al. (2016) grouped tribes from the Brassicaceae into six major clades (A to F) using nuclear markers from newly sequenced transcriptomes of 32 Brassicaceae species. Clade A includes plants from the genera Lepidium, Arabidopsis, Capsella, and Boechera; the genera Brassica, Eutrema, Raphanus, Arabis, and Thlaspi are examples of clade B; the tribes Cochlearieae, Anastaticeae, and Biscutelleae are from clade C; clade D includes the tribe Alysseae; clade E includes species from four tribes, including Chorisporeae and Hesperideae; and clade F includes the Atheionemeae tribe (Huang et al., 2016). Guo et al. (2017) also reported a similar phylogeny of Brassicaceae using the plastome of 53 species of Brassicales. However, clade D was not identified as a result of the limited taxon sampling (Guo et al., 2017).
Crop species from the Brassicaceae are often affected by several diseases, including blackleg (Leptosphaeria maculans), Sclerotinia stem rot (Sclerotinia sclerotiorum), downy mildew (Hyaloperonospora parasitica), clubroot (Plasmodiophora brassicae), and Turnip mosaic virus (Rimmer et al., 2007; Neik et al., 2017). Plant resistance gene analogs (RGAs) play a role in host resistance to disease and consist of genes with conserved domains and motifs and diverse structure, function, and evolution (Sekhwal et al., 2015). The nucleotide-binding-site leucine-rich repeat (NLR) gene family is the best-known family of RGAs, with a major role in plant disease resistance (Meyers et al., 1999; McHale et al., 2006). NLR genes are known as Resistance genes (R genes). R gene-induced resistance occurs in a gene-for-gene manner whereby an R gene in the host plant has a corresponding avirulence gene in the pathogen (Flor, 1971). In a typical NLR gene, the nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains are located in the middle and in the C terminus of the gene, respectively (Meyers et al., 1999; Xiao et al., 2001; Shao et al., 2014). The remaining structure of NLR proteins consists of three main domains at the N terminus, which are also used to classify R genes: the TIR-NBS-LRR (TNL) class is characterized by a Toll/IL-1 receptor (TIR) domain, the CC-NBS-LRR (CNL) class contains the coiled-coil domain, and the RPW8-NBS-LRR (RNL) class contains the RESISTANCE TO POWDERY MILDEW8 (RPW8) domain.
Receptor-like protein kinases (RLKs) or membrane-associated receptor-like proteins (RLPs), known as pattern recognition receptors (PRRs), are another class of RGA and the main component of the first line of plant immunity (Zipfel, 2014; Sekhwal et al., 2015). In plants, RLKs are the most abundant RGAs, and their structure is very similar to that of RLPs. They have an extracellular domain at the beginning of their N terminus involved in the perception of the microbial pattern and a transmembrane helix domain that can anchor the RLP and RLK in the plasma membrane. However, RLKs differ from RLPs by having a cytoplasmic kinase domain such as Ser/Thr protein kinases and Tyr kinase (STTK) at their C termini (Walker, 1994; Shiu and Bleecker, 2003; Sekhwal et al., 2015). Plant PRRs are classified according to their extracellular N-terminal domain. The major PRR subclasses involved in pathogen recognition carry a Lys motif or an LRR domain (Couto and Zipfel, 2016). RLPs do not possess any signaling domain in their intracellular region, and their domain structure is similar to extracellular domains of RLKs, suggesting that they might function in conjunction with one or several RLKs (Shiu and Bleecker, 2003; Zipfel, 2014). Along with defense mechanisms, RLKs and RLPs are also involved in developmental processes (Sekhwal et al., 2015), including meristem and stomatal development (Jeong et al., 1999; Nadeau and Sack, 2002).
RGAs play an important role in plant defense and have been widely used in breeding programs to improve crop disease resistance. A better understanding of their function, structure, and distribution is extremely valuable for breeders to enhance crop disease resistance. Several studies have analyzed the diversity and evolution of RGAs among the Brassicaceae (Fritz-Laylin et al., 2005; Wang et al., 2008; Shao et al., 2016; Yu et al., 2016; Zhang et al., 2016b); however, most focus on one or a few members of the Brassicaceae, and no study includes all sequenced species from this family. Moreover, the analyses have been undertaken using different methodologies, making the results between studies harder to compare. In this research, we used a unified methodology for RGA identification, phylogeny, and distribution analysis in all currently available genome annotations for 25 species of the Brassicaceae, including both wild and domesticated species, and one species from the Cleomaceae, the Brassicaceae sister family (Cheng et al., 2013). The results provide a valuable resource of candidate R genes in the Brassicaceae, which can be employed by breeders to improve disease resistance in Brassica spp. crops.
RESULTS
Identification, Classification, and Distribution of RGAs in the Brassicaceae Family Using RGAugury
We used the RGAugury pipeline to identify three main classes of RGAs (RLKs, RLPs, and NLRs) in the Brassicaceae. A total of 34,065 candidate RGAs were identified in all genomes, with the most common being RLK (21,691 genes), followed by NLR (8,588 genes) and RLP (3,786 genes) classes. Among all genomes, B. napus ‘ZS11’ (NLRs, 566; RLKs, 1,517; and RLPs, 260), B. napus ‘Darmor-bzh V4’ (NLRs, 621; RLKs, 1,497; and RLPs, 273), and Camelina sativa (NLRs, 504; RLKs, 1,469; and RLPs, 280) contained the highest number of NLRs, RLKs, and RLPs (Supplemental Figs. S1 and S2; Supplemental Table S1).
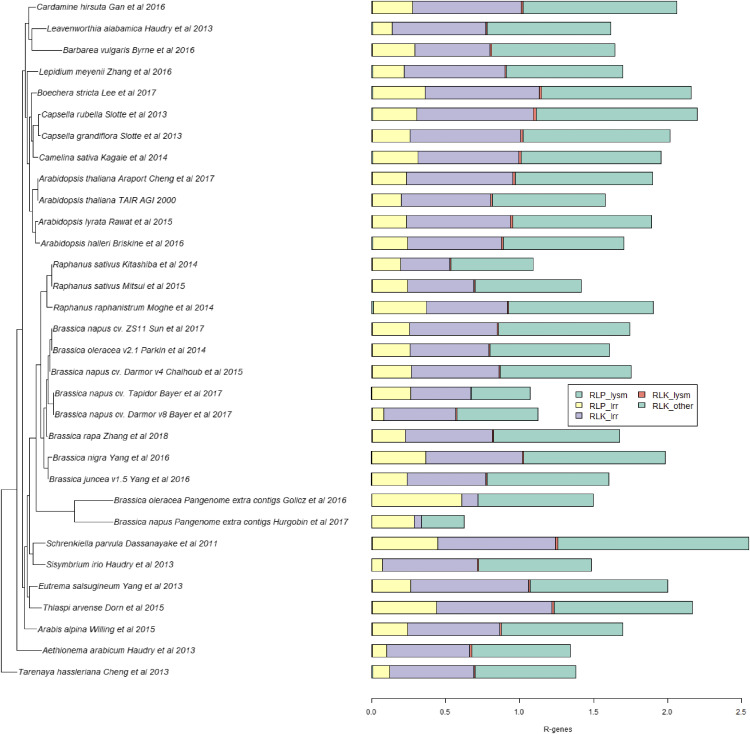
RLKs and RLPs were subdivided into three classes based on their N-terminal domain, namely LRR, LysM, and Other. RLKs were further divided into RD and non-RD classes, where RD refers to the kinases that are involved in phosphorylation with positively charged Arg (R) and negatively charged Asp (D) amino acid residues at the activation site and non-RD refers to kinases that are lacking these amino acids (Dardick et al., 2012). In total, the majority of candidate RLKs and RLPs were of class LRR, 9,066 and 3,702, respectively, whereas only 157 LysM-RLKs and 84 LysM-RLPs were identified. There were fewer than 10 LysM-RLPs identified in all the Brassicaceae genomes. C. sativa and L. meyenii contained the highest number of LysM-RLKs, 15 and 10, respectively. The additional contigs of the B. napus and B. oleracea pangenomes were not found to contain any LysM-RLK and LysM-RLP RGAs (Supplemental Table S1).
Among RLK subclasses, the majority of LysM-RLKs and other-RLKs were classified as RD; however, almost half of LRR-RLKs (4,068 out of 9,066) were non-RD. L. meyenii (total, 655; non-RD, 311), C. sativa (total, 609; non-RD, 263), and B. napus ‘ZS11’ (total, 604; non-RD, 269) were found to harbor the highest number of LRR-RLKs and non-RD LRR-RLKs. However, the highest proportion of both LRR-RLK types was found in Schrenkiella parvula (0.79%), E. salsugineum (0.79%), and Capsella rubella (0.78%). The highest number of LRR-RLPs was identified in C. sativa (271), B. napus ‘Darmor-bzh V4’ (268), and B. napus ‘ZS11’ (254). The highest proportion of LRR-RLPs was found in S. parvula (0.43%), Thlaspi arvense (0.43%), and Brassica nigra (0.36%; Fig. 1; Supplemental Table S1).
Figure 1.
Phylogeny of Brassicaceae genomes based on R genes. The colored bars show the percentages of RLKs and RLPs in each genome. Genes were identified using RGAugury.
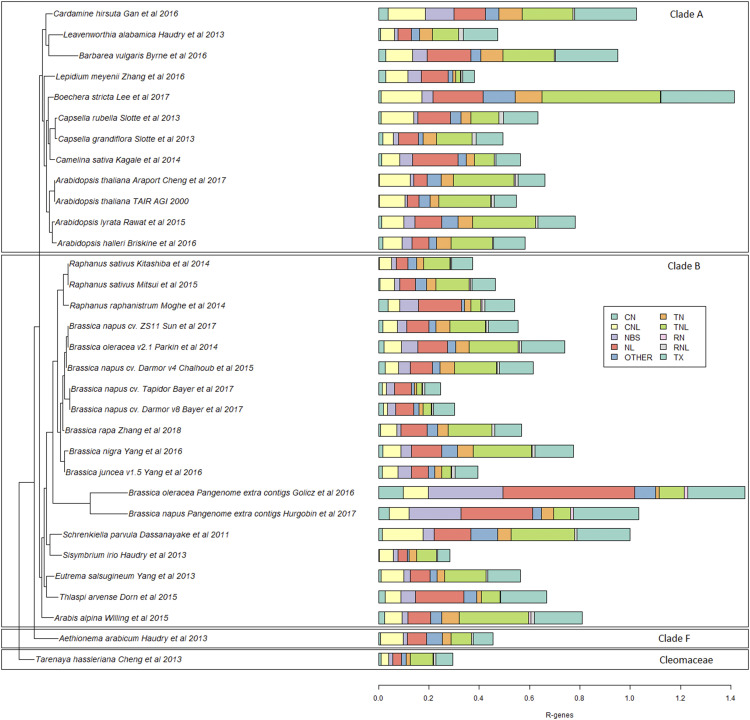
Out of the total 8,588 NLR genes, 3,146 were typical NLRs (NLR genes with all three domains) and 5,442 were atypical (genes with partial or disordered domains). TX (TIR with unclassified domains; 1,822 genes), NL (NBS-LRR; 1,532), NBS (631), and TN (TIR-NBS; 618) were the most commonly identified atypical NLR genes, whereas RN (RPW8-NBS) was the least frequent (30). The number of atypical NLRs was higher than typical NLRs in all genomes except for three: B. rapa (typical, 130; atypical, 120), Arabidopsis Araport reannotation (typical, 117; atypical, 88), and ArabidopsisTAIR (typical, 119; atypical, 86; Supplemental Fig. S1; Supplemental Table S1). The total numbers of the three typical classes of NLR genes identified were as follows: TNLs, 1,954; CNLs, 1,055; and RNLs, 137 (Supplemental Table S1). The B. napus ‘Darmor-bzh V4’ genome was revealed to have the highest number of TNLs (169 genes), L. meyenii contained the highest number of CNLs (86 genes), and Brassica juncea contained the highest number of RNLs (11 genes). The largest numbers of NLR genes were identified in B. napus ‘Darmor-bzh V4’ (621), B. napus ‘ZS11’ (566), and C. sativa (504). The highest percentages of NLR genes were found in Boechera stricta (1.41%), Cardamine hirsute (1.02%), and S. parvula (0.99%). B. stricta and S. parvula also had the highest percentages of typical NLR genes, 0.63% and 0.42%, respectively (Fig. 2; Supplemental Table S1).
Figure 2.
Phylogeny of Brassicaceae genomes based on R genes. The colored bars show the percentage of NLRs in each genome. Genes were identified using RGAugury. Boxes show the agreement of the presented results with the previously reported phylogenetic relationship of Brassicaceae by Huang et al. (2016). All species from clade A grouped and formed a separate clade, similar to species from clade B and F. Species from the Cleomaceae family formed a separate clade from all Brassicaceae family species.
Despite the greater overall total number of TNLs in the Brassicaceae, six of the genomes were found to contain more CNLs than TNLs, including B. juncea (TNLs, 30; CNLs, 49), L. meyenii (TNLs, 18; CNLs, 86), and Raphanus raphanistrum (TNLs, 15; CNLs, 18). The number of RNLs and RNs identified was lower than that of TNLs and CNLs, with 14 genomes not containing any RNLs. Furthermore, no RNLs were found in the additional contigs of B. napus and B. oleracea pangenomes, even though these extra contigs contained all other classes of RGAs (Supplemental Table S1).
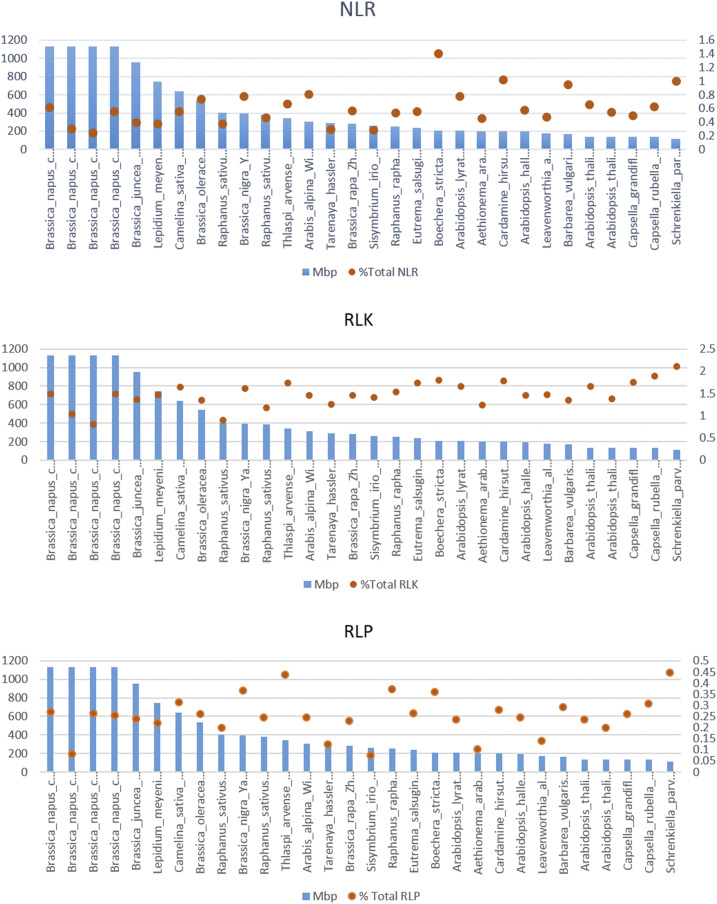
Comparing the ratios of RGAs to the assembled sizes of the genomes shows that species with larger genome sizes do not necessarily harbor higher percentages of RGAs (Fig. 3). For example, S. parvula (NLRs, 0.99%; RLKs, 2.1%; and RLPs, 0.44%) and C. rubella (NLRs, 0.63%; RLKs, 1.89%; and RLPs, 0.3%) contained higher percentages of RGAs compared with the species with larger genomes, such as B. napus ‘Darmor-bzh V4’ (NLRs, 0.61%; RLKs, 1.48%; and RLPs 0.27%) and B. juncea (NLRs, 0.39%; RLKs, 1.36%; and RLPs, 0.23%; Fig. 3; Supplemental Table S1).
Figure 3.
Comparing the NLR, RLK, and RLP ratios with assembled sizes of the genomes, illustrating no clear link between the genome size and the ratio of RGAs.
The percentage of atypical NLRs within whole NLR genes indicates that genomes with larger genome sizes have higher percentages of atypical NLRs. For example, C. sativa (72.22%), L. meyenii (69.67%), B. napus ‘Darmor-bzh V8’ (82.16%), R. raphanistrum (82.03%), and B. napus ‘Tapidor’ (81.73%) have higher percentages of atypical NLRs compared with species with small genome sizes, such as Arabidopsis TAIR (41.95%) and S. parvula (57.43%; Supplemental Table S1).
We analyzed and compared the numbers of candidate RGAs among different assembly versions of B. napus, Raphanus sativus, and Arabidopsis. The total percentages of RLK genes among different assembly versions of these species show the percentage of RLKs to be comparable between B. napus ‘Darmor-bzh V4’ (1.48%) and ‘Darmor-bzh V8’ (1.04%). However, the percentage of NLR genes in B. napus 'Darmor-bzh V4' (0.61%) is almost twice that of B. napus ‘Darmor-bzh V8’ (0.3%). Similarly, the percentages of RLPs and total RGAs were higher in B. napus ‘Darmor-bzh V4’ (RLPs, 0.27%; total, 2.36%) in comparison with B. napus ‘Darmor-bzh V8’ (RLPs, 0.08%; total, 1.42%). The percentages of NLR genes and total RGAs between the two assembly versions of Arabidopsis from TAIR (NLRs, 0.54%; total RGAs, 2.12%; RLKs, 1.37%; and RLPs, 0.19%) and Araport (NLRs, 0.66%; total RGAs, 2.56%; RLKs, 1.66%; and RLPs, 0.23%), R. sativus 2014 (NLRs, 0.37%; total RGAs, 1.46%; RLKs, 0.89%; and RLPs, 0.19%), and R. sativus 2015 (NLRs, 0.46%; total RGAs, 1.884%; RLKs, 1.17%; and RLPs, 0.24%) were not significantly different (Fig. 2; Supplemental Table S1).
Identification of NLRs in the Brassicaceae Using NLR-Annotator
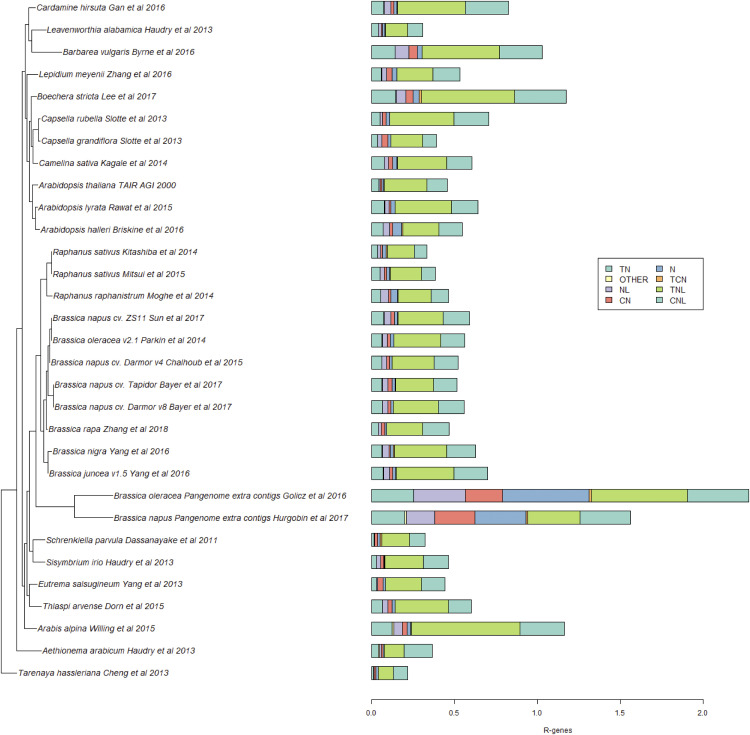
To evaluate the impact of different assembly and annotation methods on RGA prediction, NLR genes were also identified using the assembly-based NLR-Annotator, in contrast to the annotation-based RGAugury. The total numbers of NLRs for both methods were comparable: RGAugury (8,588 NLR genes) and NLR-Annotator (8,889 NLR genes; Fig. 4; Supplemental Table S1).
Figure 4.
Phylogeny of Brassicaceae genomes based on R genes. The colored bars show the percentage of NLRs in each genome. Genes were identified using NLR-Annotator.
RGA Distribution on Chromosomes/Pseudochromosomes and Subgenomes
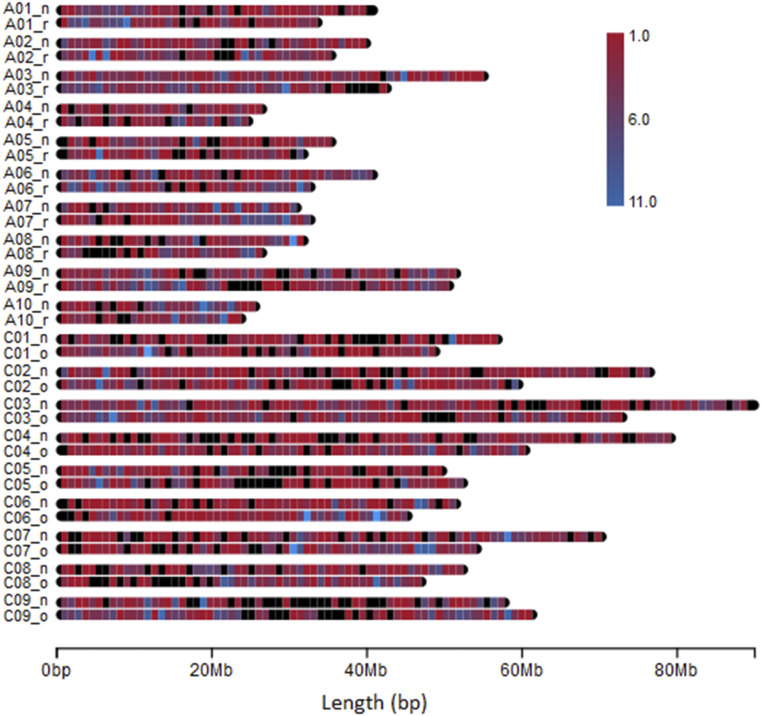
We investigated the distribution and density of RGAs in the annotations of the closely related genomes B. oleracea, B. rapa, and B. napus. Overall, RGAs were unevenly distributed along the chromosomes. The RGA distribution pattern in the C subgenome (B. oleracea) and the A subgenome (B. rapa) is comparable with their counterpart subgenome in B. napus (Fig. 5). In general, a similar distribution and density pattern of RLKs was found between subgenome A in B. napus and B. rapa. However, some regions, such as the end of chromosome A3 and the beginning of chromosome A8 in B. rapa, do not contain any RGAs, which is not the case in A3 and A8 of B. napus. Similarly, in the C subgenome, there are some similarities and differences between B. napus and B. oleracea. For example, both species do not contain any RGAs in the middle of chromosome C9, but chromosome C7 has a similar distribution and density pattern in both species (Fig. 5).
Figure 5.
Distribution and density of RGAs in genomes of Brassica spp. RGAs are presented from B. oleracea (ending in o), B. rapa (ending in r), and B. napus (ending in n) genomes.
RGA analyses were carried out for the A and C subgenomes in B. napus ‘Darmor-bzh V8’, ‘Tapidor’, ‘Darmor-bzh V4’, and ‘ZS11’. The total number of RLKs was found to be similar between the A and C subgenomes in each of the cultivars and assembly versions of B. napus (Supplemental Table S1). However, the total number of NLRs was found to be noticeably higher in the C subgenome of all cultivars and assembly versions of B. napus than in subgenome A (Supplemental Table S1). For example, subgenomes A and C in B. napus ‘Darmor-bzh V8’ contained 479 and 486 RLKs and 119 and 157 NLRs, respectively. For RLPs, there was no specific pattern: whereas both subgenomes contained similar numbers of RLPs in B. napus ‘Darmor-bzh V8’ and ‘Darmor-bzh V4’, in B. napus ‘Tapidor’ and ‘ZS11’, the C subgenomes have noticeably more RLPs than the A subgenome (Supplemental Table S1).
Orthogroup Clustering and Phylogenetic Analysis
We investigated the protein sequence similarity of all candidate RGAs across all genomes using OrthoFinder. All RGAs were grouped into 1,396 multigene clusters (orthogroups). The cluster with the largest number of RGAs contained 2,969 genes, including 917 non-RGAs, 2,028 NLRs, zero RLKs, and 24 RLPs. Of the 2,029 NLR-RGAs, the majority were of class TNL (1,098), followed by 428 NLs and 141 TNs.
CNLs and RNLs, similar to TNLs, mostly grouped together, and their clusters also contained high numbers of their respective atypical genes. For instance, the two clusters with the largest number of CNLs (572 and 168) contained the highest number of CNs, 108 and 56, respectively (Supplemental Table S2). The number of non-RGAs in clusters varied from zero to 2,195. Out of 1,396 clusters, 12 contained at least 1,000 non-RGAs and 256 contained at least 100 non-RGAs. The non-RGA domain structure was examined among 20 clusters with the highest number of candidate RGAs, and the results showed that most of the non-RGAs in each cluster contained some conserved domains of the dominant RGAs in the cluster (Supplemental Table S2).
To gain insight into the dynamic evolution of NLR genes, phylogenetic analysis based on the genome annotation was performed for all 30 genomes and two extra contigs of pangenomes. The tree formed two main clades: one clade included only species from the Brassicaceae family and the other clade included the Tarenaya genus from the Cleomaceae family. All the genomes from the same species grouped together: all species from the Brassica genus were in one clade and all species from the Arabidopsis genus grouped together (Figs. 1 and 2).
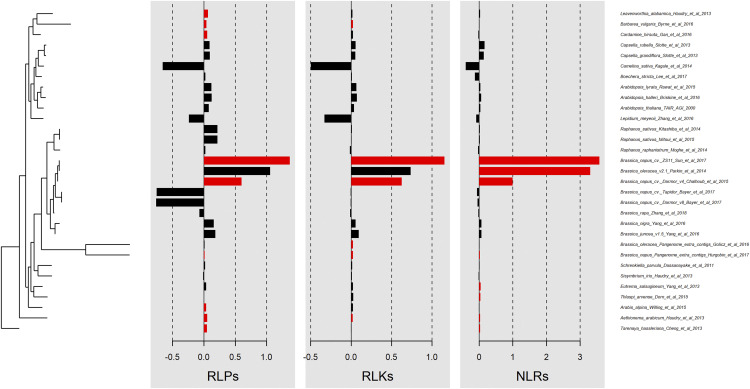
The phylogenetic signal was also tested for RLKs, RLPs, and NLRs across the Brassicaceae using phylosignal (Keck et al., 2016). Significant positive autocorrelation (P < 0.05) was detected in clade B of the phylogeny tree for RLKs and NLRs and in clades A and B for RLPs. There was a strong phylogenetic signal for RLPs among three species (Leavenworthia alabamica, B. vulgaris, and C. hirsute) in clade A that was not present in species that are distanced phylogenetically. For all three subclasses of RGAs there were strong positive phylogenetic signals for B. napus ‘Darmor-bzh V4’ and B. napus ‘ZS11’; however, there were no positive phylogenetic signals for other B. napus cultivars and their other surrounding species in the clade (Fig. 6).
Figure 6.
Significance test of phylogenetic signals for RLKs, RLPs, and NLRs across the Brassicaceae using autocorrection (local Moran’s I) metrics. Red bars represent genomes with significant phylogenetic autocorrelation (P < 0.05).
DISCUSSION
Distribution of RGAs in the Brassicaceae Family
In this study, the whole-genome distribution of RGAs was studied among different members of the Brassicaceae. The results revealed variation in RLK, RLP, and NLR gene numbers among different species of this family. The large number of RLKs compared with the other RGAs has been previously reported in several species, including Arabidopsis (Shiu and Bleecker, 2001; Meyers et al., 2003), rice (Oryza sativa; Shiu et al., 2004; Zhou et al., 2004; Fritz-Laylin et al., 2005), and Fragaria vesca (Li et al., 2018b). This is likely to be due to RLKs also having a signaling function and their involvement in different plant processes, such as growth and development, as well as defense (Shiu et al., 2004), whereas NLR genes are primarily involved with plant resistance responses (McHale et al., 2006).
The largest class of PRRs have an extracellular LRR domain (RLK/RLP) that has been reported in many plant species, including Arabidopsis (LRR-RLK:216; Shiu and Bleecker, 2001), potato (Solanum tuberosum; LRR-RLK:246; Li et al., 2018a), cotton (Gossypium hirsutum; LRR-RLK:543; Yuan et al., 2018), wheat (Triticum aestivum; LRR-RLK:531; Shumayla et al., 2016), rice (LRR-RLK:309; Sun and Wang, 2011), and poplar (Populus trichocarpa; LRR-RLP:82; Petre et al., 2014). Similarly, in this study, the majority of RLKs and RLPs were classified as LRR-RLK/RLP. Here, a large number of non-RD RLKs were identified across the Brassicaceae, and it has been suggested that non-RD RLKs are mainly involved in immunity responses (Dardick et al., 2012; Rameneni et al., 2015).
Different numbers of NLR genes have also been reported within and between plant species, including Arachis duranensis (Song et al., 2017), Manihot esculenta (Lozano et al., 2015), Arabidopsis and B. rapa (Mun et al., 2009), B. rapa, C. rubella, and Arabidopsis lyrata (Zhang et al., 2016b), Arabidopsis (Meyers et al., 2003; Kong et al., 2018), B. napus, B. rapa, and B. oleracea (Alamery et al., 2018), the B. oleracea pangenome (Golicz et al., 2016), tomato (Solanum lycopersicum), pepper (Capsicum annuum), and potato (Seo et al., 2016), Fragaria × ananassa, Fragaria iinumae, Fragaria nipponica, Fragaria nubicola, and Fragaria orientalis (Zhong et al., 2018), and wheat (Gu et al., 2015). Using different pipelines for gene prediction and different versions of a genome assembly causes the contradiction of reported numbers of NLR genes among different studies, including our study. It has been confirmed that different approaches in genome annotation and masking of repetitive elements have a major impact on gene prediction (Bayer et al., 2018; Slotkin, 2018). In our study, the quality of different assembly versions of the genera Arabidopsis and Raphanus was comparable, and no significant differences were observed between the percentages of candidate RGAs. However, there were noticeable differences among different assembly versions of B. napus, potentially due to different repeat-masking approaches.
The higher number of TNLs than CNLs and RNLs observed in our study is similar to the findings in B. rapa and B. napus (Alamery et al., 2018) and Arabidopsis (Meyers et al., 2003). However, some of the genomes contain more CNLs than TNLs, such as Aethionema arabicum and R. raphanistrum, which was also reported in potato (Seo et al., 2016) and Medicago truncatula (Ameline-Torregrosa et al., 2008). The reports of RNLs being the least prevalent among different classes of NLRs in 25 species of angiosperms (Shao et al., 2014; Zhang et al., 2016b) are consistent with our findings. RNLs are not directly involved in pathogen recognition, and they assist other NLR genes to accomplish a resistance response (Xiao et al., 2001). Therefore, plants do not need a high number of RNLs, and thus the wide range of TNLs and CNLs is necessary for plants to expand their ability for pathogen recognition to counteract rapid pathogen evolution. The observed variation of identified RGAs and their uneven distribution along the genomes indicate that the genome evolutionary events, namely whole-genome duplication (WGD), whole-genome triplication (WGT), transposon-mediated gene duplication, and tandem and segmental gene duplication, have an important role in generating different numbers of RGAs (Walker et al., 1995; Mun et al., 2009; Franzke et al., 2011; Lisch, 2013). Loss of function, subfunctionalization, and neofunctionalization play major roles in the evolution of duplicated genes (Lynch and Conery, 2000) by increasing the retention rate of these genes (Rastogi and Liberles, 2005). The involvement of these processes in the evolution of RGAs has been also reported in other plant families such as Rosaceae (Zhong et al., 2015, 2018).
NLR-Annotator and RGAugury Comparison
Different annotation approaches can affect the prediction of NLR genes (Bayer et al., 2018). To evaluate the results from RGAugury, a pipeline for RGA identification using all predicted proteins, we used NLR-Annotator, a tool for de novo genome annotation of NLR loci. The total number of candidate NLRs was very similar between the RGAugury and NLR-Annotator methods. Most of the assemblies that have been used in this study are Illumina based; however, a few assemblies are based on other sequencing methods; for example, B. rapa and B. napus ‘Darmor-bzh’ are PacBio and Illumina based (Supplemental Table S1).
Only one assembly showed substantial differences between the two methods. The results from RGAugury show that B. napus ‘Darmor-bzh V8’ contains substantially fewer RGAs in comparison with B. napus ‘Darmor-bzh V4’; however, using NLR-Annotator, B. napus ‘Darmor-bzh V8’ and ‘Darmor-bzh V4’ contain exactly the same number of RGAs. Of note, B. napus ‘Darmor-bzh V8’ and ‘Darmor-bzh V4’ are different in both annotation and assembly. As NLR-Annotator is only annotation independent, this observation suggests that the differences between B. napus ‘Darmor-bzh V4’ and ‘Darmor-bzh V8’ as a result of using different annotation methods and RGA prediction are not affected by assembly methods. The stringent repeat-masking method used for both B. napus ‘Darmor-bzh V8’ and ‘Tapidor’ (Bayer et al., 2017, 2018) can explain the lower number of RGAs identified by RGAugury in comparison with that identified for other B. napus cultivars and assembly versions. Using NLR-Annotator, the number of RGAs in cv Tapidor is comparable with other B. napus cultivars. These observations show that the results have not been significantly affected by sequencing and assembly methods.
RGA Distribution on Pseudomolecules
The distribution of RGAs in and within the pseudomolecules of B. napus and its progenitor B. rapa and B. oleracea genomes was not even, which is similar to observations made in other studies (Chalhoub et al., 2014; Yu et al., 2014; Golicz et al., 2016; Zhang et al., 2016b). The high similarity of distribution and density pattern of RGAs in the C subgenome (B. oleracea) and the A subgenome (B. rapa) with their counterpart subgenome in B. napus shows that RGAs have been conserved during genome evolutionary events. The results also show that a chromosome might be rich in one class of RGAs whereas it has only a few or no genes of another class. Different NBS-encoding gene numbers were also reported in C. rubella, Thellungiella salsuginea, A. lyrata, Arabidopsis, and B. rapa (Zhang et al., 2016b). The observed differences of distribution and density pattern of RGAs between different assembly versions of B. napus and B. rapa reflect the importance of the genome assembly quality, whereby the accuracy of gene prediction mainly depends on the quality of genome assembly.
Orthogroup Clustering and Phylogenetic Analysis
Based on Orthogroup clustering, most of the genes from the same class of RGAs, across all genomes, grouped together. This observation confirms a significant homology between RGA protein sequences among all species. Non-RGAs in a cluster carry some of the key domains of the dominant RGA class in the cluster. Non-RGAs in RLK-dominated clusters consisted of different types of kinase domain, such as STTK, and other types of domains, including PAN/Apple domain, a type of RLK receptor (Shiu and Bleecker, 2001); LRR; legume lectin domain; and Gnk2-homologous domain. Non-RGAs in NBS-dominated clusters mostly include a winged helix-like DNA-binding domain, which is a subdomain of the NBS domain (McHale et al., 2006), and LRR and TIR domains. The non-RGAs might be an incomplete form of the RGAs that have not been identified with the applied pipeline (RGAugury).
Our phylogenetic analysis is consistent with the proposed Brassicaceae phylogeny and evolutionary history presented by Guo et al. (2017) and Huang et al. (2016). Species from the Brassicaceae family formed a separate clade from the Cleomaceae family. The clade of the Brassicaceae family also divided into three subclades: one subclade includes all species from clade A, the second one includes all species from clade B, and the third one includes the Aethionema genus from clade F.
NLR distribution among the species shows that species with a larger genome size (such as Brassica spp.) contain a higher number of NLR genes; however, they contain a lower percentage of typical NLR genes and a higher percentage of atypical NLR genes compared with species with a small genome size (such as Arabidopsis). Additionally, the difference between the percentages of typical and atypical NLRs is more significant in species with a larger genome size. This suggests that the large number of R genes must be costly, so that in the Brassicaceae family, despite the increased genome size during genome evolutionary events, the R gene number has not increased proportionally with genome size. The biological cost of containing a high copy number of R genes is increasing energy consumption under stress conditions, when plant adaptation relies on energy-saving responses (Tomé et al., 2014), in order to balance the transcription and translation of R genes where the high expression of R genes can be lethal for plants (Li et al., 2012). The lower percentage of NLRs among Brassica spp., those species with larger genome sizes, could also be a consequence of extensive gene loss, which occurred during WGD, WGT, and polyploidization events in these species. Gene loss is reported to be common during polyploidization (Town et al., 2006), and extensive gene and segmental loss have been frequently reported in B. oleracea (Town et al., 2006; Liu et al., 2014; Alamery et al., 2018), B. napus (Parkin et al., 2005; Chalhoub et al., 2014; Alamery et al., 2018), B. rapa (Alamery et al., 2018), and B. juncea (Yang et al., 2016). Consequently, both genome evolutionary events and the biological cost of the high copy number of R genes lead plants to keep the number of R genes at a definite range, regardless of genome size.
Phylogenetic signals were observed among species with close phylogenetic distance for all three classes of RGA (i.e. RLKs, RLPs, and NLRs). However, the strong positive phylogenetic signals that were detected for B. napus ‘Darmor-bzh V4’ and B. napus ‘ZS11’ were not detected for other B. napus cultivars and their other surrounding species in the phylogeny tree, which suggests that the observed signals for B. napus ‘Darmor-bzh V4’ and B. napus ‘ZS11’ might be false-positive signals due to technological artifacts. This is more likely linked to the different annotation methods, as we show that RGA prediction is not affected by the sequencing technologies and assembly approaches in this study.
In summary, we identified more than 34,000 RGAs across Brassicaceae wild and domesticated species. In Brassica spp., despite their large genome size and WGD, WGT, and polyploidization events, the number of R genes has not expanded widely. Comparative analysis indicates that the number and distribution of RGAs greatly vary among species, whereas orthogroup clustering confirmed a high homology of the RGA proteome across all genomes. Despite many studies that have identified RGAs across different plant species, only a few R genes have been cloned due to the complexity of fine-mapping of R genes, which is partially due to the lack of information about their genomic structure and distribution. These complications are further intensified in plants that experience WGD and WGT, like Brassica spp. that harbor many copy numbers of RGAs. Different methods of RGA identification make further fine-mapping and comparative genomic analysis between species more complicated. Here, by using a unified methodology, we performed a comparative analysis of RGAs among all sequenced wild and cultivated species in addition to two sets of extra contigs from two Brassica spp. pangenomes. These comparative analyses provide a better insight into the genomic distribution and variation of RGAs across this plant family, which can be used to assist the identification and cloning of RGAs from previously untapped sources and their subsequent application in breeding programs for producing resistant cultivars.
MATERIALS AND METHODS
Genomic Resources
Whole-genome RGA identification was performed on 32 sequenced and annotated genomes, including 30 genomes from the Brassicaceae in addition to two sets of extra contigs of Brassica napus and Brassica oleracea pangenomes and one genome from Cleomaceae, included as an outlier. To minimize the assembly impact on RGA prediction, different assembly versions for identical species were included (Table 1).
Table 1. List of the genomes used in this study, their assembly size, and number of predicted proteins.
N/A, Not available.
| Species | Total Assembled Size | No. of Proteins | Reference |
|---|---|---|---|
| Aethionema arabicum | 199,432,295 | 37,839 | Haudry et al. (2013) |
| Arabidopsis halleri | 127,615,339 | 26,911 | Briskine et al. (2017) |
| Arabidopsis lyrata | 206,667,935 | 33,132 | Rawat et al. (2015) |
| Arabidopsis Araport | N/A | 48,359 | Arabidopsis Genome Initiative (2000) |
| Arabidopsis TAIR | 119,667,750 | 37,513 | Cheng et al. (2017) |
| Arabis alpina | 336,719,365 | 34,220 | Willing et al. (2015) |
| Barbarea vulgaris | 167,749,783 | 25,350 | Byrne et al. (2017) |
| Boechera stricta | 189,344,188 | 29,812 | Lee et al. (2017) |
| Brassica juncea | 937,030,072 | 79,644 | Yang et al. (2016) |
| Brassica napus ‘Darmor-bzh V4’ | 850,288,203 | 101,040 | Chalhoub et al. (2014) |
| Brassica napus ‘Tapidor’ | 636,323,668 | 84,347 | Bayer et al. (2017) |
| Brassica napus ‘ZS11’ | 976,245,903 | 101,942 | Sun et al. (2017) |
| Brassica napus pangenome | 1,048,086,857 | 111,283 | Hurgobin et al. (2018) |
| Additional contigs | 197,794,754 | 16,518 | Hurgobin et al. (2018) |
| Darmor-bzh V8.1 | 850,292,103 | 94,765 | Bayer et al. (2017) |
| Brassica nigra | 402,096,365 | 47,953 | Yang et al. (2016) |
| Brassica oleracea pangenome | 587,191,128 | 66,298 | Golicz et al. (2016) |
| Additional contigs | 98,568,621 | 7,078 | Golicz et al. (2016) |
| V2.1 | 488,622,507 | 59,220 | Parkin et al. (2014) |
| Brassica rapa | N/A | 45,985 | Zhang et al. (2018) |
| Camelina sativa | 641,452,262 | 94,495 | Kagale et al. (2014) |
| Capsella grandiflora | 105,346,052 | 26,561 | Slotte et al. (2013) |
| Capsella rubella | 134,834,574 | 28,447 | Slotte et al. (2013) |
| Cardamine hirsuta | 198,654,690 | 38,094 | Gan et al. (2016) |
| Eutrema salsugineum | 243,117,811 | 26,351 | Yang et al. (2013) |
| Leavenworthia alabamica | 174,200,922 | 38,676 | Haudry et al. (2013) |
| Lepidium meyenii | 743,171,196 | 96,417 | Zhang et al. (2016a) |
| Raphanus raphanistrum | 254,586,761 | 50,972 | Moghe et al. (2014) |
| Raphanus sativus | 402,330,269 | 80,521 | Kitashiba et al. (2014) |
| Raphanus sativus | 383,104,752 | 64,657 | Mitsui et al. (2015) |
| Sisymbrium irio | 259,494,581 | 49,956 | Haudry et al. (2013) |
| Tarenaya hassleriana | 249,929,577 | 40,802 | Cheng et al. (2013) |
| Schrenkiella parvula | 137,251,937 | 28,178 | Dassanayake et al. (2011) |
| Thlaspi arvense | 343,012,389 | 27,390 | Dorn et al. (2015) |
Identification and Classification of RGA Genes in the Brassicaceae Family
RGAs were identified using RGAugury, a pipeline for genome-wide RGA prediction (Li et al., 2016). Three main classes of RGAs were identified and classified: RLK, RLP, and NLR genes. The RGAugury pipeline also divided the NLR gene family members into several subgroups according to their domain architecture, namely NBS, CNL, TNL, TN, CN, NL, TX (TIR with unclassified domains), and Other. Genes carrying RPW8 domains were manually reassigned based on their original domains: genes classified as NBS with an RPW8 were reclassified as RN, genes classified as NL with an additional RPW8 domain were reclassified as RNL, and all other remaining genes (TNL, CNL, CN, TN, and TX) carrying an additional RPW8 domain were reclassified as Other. Among NLRs, TNL, CNL, and RNL subgroups were named as typical NLRs and the rest of the subgroups that contain partial or disordered domains was named as atypical NLRs. Using a Python script, RLKs and RLPs were divided into three subclasses: PRRs containing an LRR domain (LRR-RLK/RLP), PRRs containing Lys motifs (LysM-RLK/RLP), and PRRs with any other domain (other-RLK/RLP). RLKs were further divided into RD and non-RD classes by performing BLAST between RLK candidates and previously published RD and non-RD RLK candidates in Brassica rapa (Rameneni et al., 2015). To evaluate the effect of annotation on gene prediction, we also used NLR-Annotator (Steuernagel et al., 2018) for the identification of NLR genes based on motifs present in the genome assembly.
Phylogenetic Analysis of RGAs
All proteomes were clustered using OrthoFinder v1.1.8 (Emms and Kelly, 2015). The species-level tree obtained from OrthoFinder was compared with the phylogenies stored for the Brassicaceae in the Open Tree Of Life (Wang et al., 2009; Parfrey et al., 2011; Soltis et al., 2011; Ruhfel et al., 2014; Magallón et al., 2015; Sun et al., 2016; Foster et al., 2017). The expansion of RGA classes for different branches of the tree was carried out using the R package phytools v0.6 (Revell, 2012). Phylogenetic signal was tested with the function phyloSignal in the R package phylosignal v.1.2 (Keck et al., 2016); its significance was estimated with 999 iterations.
Gene Density Plots
ChromoMap v0.2 (Anand, 2019) was used to calculate and then plot a histogram of RGA candidate density and distribution for B. oleracea, B. rapa, and B. napus by plotting the density of NLR candidates for B. napus ‘ZS11’, B. oleracea V2.1, and B. rapa V3.0.
Accession Numbers
All candidate sequences are available at https://doi.org/10.26182/5dbf848ca28c3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogeny of Brassicaceae genomes based on NLR R genes.
Supplemental Figure S2. Phylogeny of Brassicaceae genomes based on RLP and RLK R genes.
Supplemental Table S1. RGA classification and clustering analysis.
Supplemental Table S2. Orthogroup clustering of RGAs
Footnotes
This work was supported by the Australian Research Council (grant nos. FT130100604, DP1601004497, LP140100537, and LP160100030), the University of Western Australia, the Grains Research and Development Corporation (grant no. 9175960 to S.T.), and a Forrest Research Fellowship.
Articles can be viewed without a subscription.
References
- Alamery S, Tirnaz S, Bayer P, Tollenaere R, Chaloub B, Edwards D, Batley J(2018) Genome-wide identification and comparative analysis of NBS-LRR resistance genes in Brassica napus. Crop Pasture Sci 69: 72–93 [Google Scholar]
- Ameline-Torregrosa C, Wang BB, O’Bleness MS, Deshpande S, Zhu H, Roe B, Young ND, Cannon SB(2008) Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol 146: 5–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand L.(2019) ChromoMap: An R package for interactive visualization and annotation of chromosomes. bioRxiv 605600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bayer PE, Edwards D, Batley J(2018) Bias in resistance gene prediction due to repeat masking. Nat Plants 4: 762–765 [DOI] [PubMed] [Google Scholar]
- Bayer PE, Hurgobin B, Golicz AA, Chan CKK, Yuan Y, Lee H, Renton M, Meng J, Li R, Long Y, et al. (2017) Assembly and comparison of two closely related Brassica napus genomes. Plant Biotechnol J 15: 1602–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briskine RV, Paape T, Shimizu‐Inatsugi R, Nishiyama T, Akama S, Sese J, Shimizu KK(2017) Genome assembly and annotation of Arabidopsis halleri, a model for heavy metal hyperaccumulation and evolutionary ecology. Mol Ecol Resour 17: 1025–1036 [DOI] [PubMed] [Google Scholar]
- Byrne SL, Erthmann PØ, Agerbirk N, Bak S, Hauser TP, Nagy I, Paina C, Asp T(2017) The genome sequence of Barbarea vulgaris facilitates the study of ecological biochemistry. Sci Rep 7: 40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345: 950–953 [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD(2017) Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J 89: 789–804 [DOI] [PubMed] [Google Scholar]
- Cheng S, van den Bergh E, Zeng P, Zhong X, Xu J, Liu X, Hofberger J, de Bruijn S, Bhide AS, Kuelahoglu C, et al. (2013) The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25: 2813–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, Zipfel C(2016) Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol 16: 537–552 [DOI] [PubMed] [Google Scholar]
- Dardick C, Schwessinger B, Ronald P(2012) Non-arginine-aspartate (non-RD) kinases are associated with innate immune receptors that recognize conserved microbial signatures. Curr Opin Plant Biol 15: 358–366 [DOI] [PubMed] [Google Scholar]
- Dassanayake M, Oh DH, Haas JS, Hernandez A, Hong H, Ali S, Yun DJ, Bressan RA, Zhu JK, Bohnert HJ, et al. (2011) The genome of the extremophile crucifer Thellungiella parvula. Nat Genet 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn KM, Fankhauser JD, Wyse DL, Marks MD(2015) A draft genome of field pennycress (Thlaspi arvense) provides tools for the domestication of a new winter biofuel crop. DNA Res 22: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S(2015) OrthoFinder: Solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH.(1971) Current status of the gene-for-gene concept. Annu Rev Phytopathol 9: 275–296 [Google Scholar]
- Foster CSP, Sauquet H, van der Merwe M, McPherson H, Rossetto M, Ho SYW(2017) Evaluating the impact of genomic data and priors on Bayesian estimates of the angiosperm evolutionary timescale. Syst Biol 66: 338–351 [DOI] [PubMed] [Google Scholar]
- Franzke A, Lysak MA, Al-Shehbaz IA, Koch MA, Mummenhoff K(2011) Cabbage family affairs: The evolutionary history of Brassicaceae. Trends Plant Sci 16: 108–116 [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Krishnamurthy N, Tör M, Sjölander KV, Jones JD(2005) Phylogenomic analysis of the receptor-like proteins of rice and Arabidopsis. Plant Physiol 138: 611–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Hay A, Kwantes M, Haberer G, Hallab A, Ioio RD, Hofhuis H, Pieper B, Cartolano M, Neumann U, et al. (2016) The Cardamine hirsuta genome offers insight into the evolution of morphological diversity. Nat Plants 2: 16167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golicz AA, Bayer PE, Barker GC, Edger PP, Kim H, Martinez PA, Chan CKK, Severn-Ellis A, McCombie WR, Parkin IA, et al. (2016) The pangenome of an agronomically important crop plant Brassica oleracea. Nat Commun 7: 13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Si W, Zhao L, Yang S, Zhang X(2015) Dynamic evolution of NBS-LRR genes in bread wheat and its progenitors. Mol Genet Genomics 290: 727–738 [DOI] [PubMed] [Google Scholar]
- Guo X, Liu J, Hao G, Zhang L, Mao K, Wang X, Zhang D, Ma T, Hu Q, Al-Shehbaz IA, et al. (2017) Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics 18: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A, Platts AE, Vello E, Hoen DR, Leclercq M, Williamson RJ, Forczek E, Joly-Lopez Z, Steffen JG, Hazzouri KM, et al. (2013) An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat Genet 45: 891–898 [DOI] [PubMed] [Google Scholar]
- Huang CH, Sun R, Hu Y, Zeng L, Zhang N, Cai L, Zhang Q, Koch MA, Al-Shehbaz I, Edger PP, et al. (2016) Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol Biol Evol 33: 394–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurgobin B, Golicz AA, Bayer PE, Chan CKK, Tirnaz S, Dolatabadian A, Schiessl SV, Samans B, Montenegro JD, Parkin IAP, et al. (2018) Homoeologous exchange is a major cause of gene presence/absence variation in the amphidiploid Brassica napus. Plant Biotechnol J 16: 1265–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong S, Trotochaud AE, Clark SE(1999) The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11: 1925–1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, Clarke C, et al. (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5: 3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck F, Rimet F, Bouchez A, Franc A(2016) phylosignal: An R package to measure, test, and explore the phylogenetic signal. Ecol Evol 6: 2774–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitashiba H, Li F, Hirakawa H, Kawanabe T, Zou Z, Hasegawa Y, Tonosaki K, Shirasawa S, Fukushima A, Yokoi S, et al. (2014) Draft sequences of the radish (Raphanus sativus L.) genome. DNA Res 21: 481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Li B, Wang Q, Wang B, Duan X, Ding L, Lu Y, Liu LW, La H(2018) Analysis of the DNA methylation patterns and transcriptional regulation of the NB-LRR-encoding gene family in Arabidopsis thaliana. Plant Mol Biol 96: 563–575 [DOI] [PubMed] [Google Scholar]
- Lee CR, Wang B, Mojica JP, Mandáková T, Prasad KV, Goicoechea JL, Perera N, Hellsten U, Hundley HN, Johnson J(2017) Young inversion with multiple linked QTLs under selection in a hybrid zone. Nat Ecol Evol 1: 0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Mummenhoff K, Bowman JL(2002) Allopolyploidization and evolution of species with reduced floral structures in Lepidium L. (Brassicaceae). Proc Natl Acad Sci USA 99: 16835–16840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Pignatta D, Bendix C, Brunkard JO, Cohn MM, Tung J, Sun H, Kumar P, Baker B(2012) MicroRNA regulation of plant innate immune receptors. Proc Natl Acad Sci USA 109: 1790–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Quan X, Jia G, Xiao J, Cloutier S, You FM(2016) RGAugury: A pipeline for genome-wide prediction of resistance gene analogs (RGAs) in plants. BMC Genomics 17: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Salman A, Guo C, Yu J, Cao S, Gao X, Li W, Li H, Guo Y(2018a) Identification and characterization of LRR-RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells 7: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wei W, Feng J, Luo H, Pi M, Liu Z, Kang C(2018b) Genome re-annotation of the wild strawberry Fragaria vesca using extensive Illumina- and SMRT-based RNA-seq datasets. DNA Res 25: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D.(2013) How important are transposons for plant evolution? Nat Rev Genet 14: 49–61 [DOI] [PubMed] [Google Scholar]
- Liu S, Liu Y, Yang X, Tong C, Edwards D, Parkin IA, Zhao M, Ma J, Yu J, Huang S, et al. (2014) The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat Commun 5: 3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Hamblin MT, Prochnik S, Jannink JL(2015) Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genomics 16: 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery JS(2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T(2015) A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207: 437–453 [DOI] [PubMed] [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW(2006) Plant NBS-LRR proteins: Adaptable guards. Genome Biol 7: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND(1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20: 317–332 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW(2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15: 809–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui Y, Shimomura M, Komatsu K, Namiki N, Shibata-Hatta M, Imai M, Katayose Y, Mukai Y, Kanamori H, Kurita K, et al. (2015) The radish genome and comprehensive gene expression profile of tuberous root formation and development. Sci Rep 5: 10835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe GD, Hufnagel DE, Tang H, Xiao Y, Dworkin I, Town CD, Conner JK, Shiu SH(2014) Consequences of whole-genome triplication as revealed by comparative genomic analyses of the wild radish Raphanus raphanistrum and three other Brassicaceae species. Plant Cell 26: 1925–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Yu HJ, Park S, Park BS(2009) Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Genet Genomics 282: 617–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JA, Sack FD(2002) Control of stomatal distribution on the Arabidopsis leaf surface. Science 296: 1697–1700 [DOI] [PubMed] [Google Scholar]
- Neik TX, Barbetti MJ, Batley J(2017) Current status and challenges in identifying disease resistance genes in Brassica napus. Front Plant Sci 8: 1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen NJ, Nielsen J, Staerk D(2010) New resistance-correlated saponins from the insect-resistant crucifer Barbarea vulgaris. J Agric Food Chem 58: 5509–5514 [DOI] [PubMed] [Google Scholar]
- Palmer CE, Warwick S, Keller W(2001) Brassicaceae (Cruciferae) family, plant biotechnology, and phytoremediation. Int J Phytoremediation 3: 245–287 [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA(2011) Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA 108: 13624–13629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ(2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171: 765–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IA, Koh C, Tang H, Robinson SJ, Kagale S, Clarke WE, Town CD, Nixon J, Krishnakumar V, Bidwell SL, et al. (2014) Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol 15: R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre B, Hacquard S, Duplessis S, Rouhier N(2014) Genome analysis of poplar LRR-RLP gene clusters reveals RISP, a defense-related gene coding a candidate endogenous peptide elicitor. Front Plant Sci 5: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rameneni JJ, Lee Y, Dhandapani V, Yu X, Choi SR, Oh MH, Lim YP(2015) Genomic and post-translational modification analysis of leucine-rich-repeat receptor-like kinases in Brassica rapa. PLoS ONE 10: e0142255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi S, Liberles DA(2005) Subfunctionalization of duplicated genes as a transition state to neofunctionalization. BMC Evol Biol 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat V, Abdelsamad A, Pietzenuk B, Seymour DK, Koenig D, Weigel D, Pecinka A, Schneeberger K(2015) Improving the annotation of Arabidopsis lyrata using RNA-Seq data. PLoS ONE 10: e0137391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ.(2012) Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol Evol 3: 217–223 [Google Scholar]
- Rimmer SR, Shattuck VI, Buchwaldt L(2007) Compendium of Brassica Diseases. APS Press, St. Paul, MN [Google Scholar]
- Ruhfel BR, Gitzendanner MA, Soltis PS, Soltis DE, Burleigh JG(2014) From algae to angiosperms: Inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol Biol 14: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R, Acarkan A, Boivin K(2001) Comparative structural genomics in the Brassicaceae family. Plant Physiol Biochem 39: 253–262 [Google Scholar]
- Sekhwal MK, Li P, Lam I, Wang X, Cloutier S, You FM(2015) Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci 16: 19248–19290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo E, Kim S, Yeom SI, Choi D(2016) Genome-wide comparative analyses reveal the dynamic evolution of nucleotide-binding leucine-rich repeat gene family among Solanaceae plants. Front Plant Sci 7: 1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Xue JY, Wu P, Zhang YM, Wu Y, Hang YY, Wang B, Chen JQ(2016) Large-scale analyses of angiosperm nucleotide-binding site-leucine-rich repeat genes reveal three anciently diverged classes with distinct evolutionary patterns. Plant Physiol 170: 2095–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao ZQ, Zhang YM, Hang YY, Xue JY, Zhou GC, Wu P, Wu XY, Wu XZ, Wang Q, Wang B, et al. (2014) Long-term evolution of nucleotide-binding site-leucine-rich repeat genes: Understanding gained from and beyond the legume family. Plant Physiol 166: 217–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB(2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Bleecker AB(2003) Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu SH, Karlowski WM, Pan R, Tzeng YH, Mayer KF, Li WH(2004) Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16: 1220–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumayla, Sharma S, Kumar R, Mendu V, Singh K, Upadhyay SK(2016) Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front Plant Sci 7: 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK.(2018) The case for not masking away repetitive DNA. Mob DNA 9: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T, Hazzouri KM, Ågren JA, Koenig D, Maumus F, Guo YL, Steige K, Platts AE, Escobar JS, Newman LK, et al. (2013) The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat Genet 45: 831–835 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Smith SA, Cellinese N, Wurdack KJ, Tank DC, Brockington SF, Refulio-Rodriguez NF, Walker JB, Moore MJ, Carlsward BS, et al. (2011) Angiosperm phylogeny: 17 genes, 640 taxa. Am J Bot 98: 704–730 [DOI] [PubMed] [Google Scholar]
- Song H, Wang P, Li C, Han S, Zhao C, Xia H, Bi Y, Guo B, Zhang X, Wang X(2017) Comparative analysis of NBS-LRR genes and their response to Aspergillus flavus in Arachis. PLoS ONE 12: e0171181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuernagel B, Witek K, Krattinger SG, Ramirez-Gonzalez RH, Schoonbeek H, Yu G, Baggs E, Witek A, Yadav I, Krasileva KV(2018) Physical and transcriptional organisation of the bread wheat intracellular immune receptor repertoire. bioRxiv 339424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Fan G, Hu Q, Zhou Y, Guan M, Tong C, Li J, Du D, Qi C, Jiang L(2017) The high-quality genome of Brassica napus cultivar ‘ZS11’reveals the introgression history in semi-winter morphotype. Plant J 92: 452–468 [DOI] [PubMed] [Google Scholar]
- Sun X, Wang GL(2011) Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PLoS ONE 6: e16079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Moore MJ, Zhang S, Soltis PS, Soltis DE, Zhao T, Meng A, Li X, Li J, Wang H(2016) Phylogenomic and structural analyses of 18 complete plastomes across nearly all families of early-diverging eudicots, including an angiosperm-wide analysis of IR gene content evolution. Mol Phylogenet Evol 96: 93–101 [DOI] [PubMed] [Google Scholar]
- Tomé F, Nägele T, Adamo M, Garg A, Marco-Llorca C, Nukarinen E, Pedrotti L, Peviani A, Simeunovic A, Tatkiewicz A, et al. (2014) The low energy signaling network. Front Plant Sci 5: 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, et al. (2006) Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EL, Robbins TP, Bureau TE, Kermicle J, Dellaporta SL(1995) Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J 14: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JC.(1994) Structure and function of the receptor-like protein kinases of higher plants. Plant Mol Biol 26: 1599–1609 [DOI] [PubMed] [Google Scholar]
- Wang G, Ellendorff U, Kemp B, Mansfield JW, Forsyth A, Mitchell K, Bastas K, Liu CM, Woods-Tör A, Zipfel C, et al. (2008) A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol 147: 503–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE(2009) Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA 106: 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing EM, Rawat V, Mandáková T, Maumus F, James GV, Nordström KJ, Becker C, Warthmann N, Chica C, Szarzynska B(2015) Genome expansion of Arabis alpina linked with retrotransposition and reduced symmetric DNA methylation. Nat Plants 1: nplants201423. [DOI] [PubMed] [Google Scholar]
- Wu HJ, Zhang Z, Wang JY, Oh DH, Dassanayake M, Liu B, Huang Q, Sun HX, Xia R, Wu Y, et al. (2012) Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc Natl Acad Sci USA 109: 12219–12224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG(2001) Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 291: 118–120 [DOI] [PubMed] [Google Scholar]
- Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, Hu Z, Chen S, Pental D, Ju Y, et al. (2016) The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet 48: 1225–1232 [DOI] [PubMed] [Google Scholar]
- Yang R, Jarvis DE, Chen H, Beilstein MA, Grimwood J, Jenkins J, Shu S, Prochnik S, Xin M, Ma C, et al. (2013) The reference genome of the halophytic plant Eutrema salsugineum. Front Plant Sci 4: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Zhang X, Huang Z, Chu M, Song T, Falk KC, Deora A, Chen Q, Zhang Y, McGregor L, et al. (2016) Identification of genome-wide variants and discovery of variants associated with Brassica rapa clubroot Resistance gene Rcr1 through bulked segregant RNA sequencing. PLoS ONE 11: e0153218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Tehrim S, Zhang F, Tong C, Huang J, Cheng X, Dong C, Zhou Y, Qin R, Hua W, et al. (2014) Genome-wide comparative analysis of NBS-encoding genes between Brassica species and Arabidopsis thaliana. BMC Genomics 15: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan N, Rai KM, Balasubramanian VK, Upadhyay SK, Luo H, Mendu V(2018) Genome-wide identification and characterization of LRR-RLKs reveal functional conservation of the SIF subfamily in cotton (Gossypium hirsutum). BMC Plant Biol 18: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Tian Y, Yan L, Zhang G, Wang X, Zeng Y, Zhang J, Ma X, Tan Y, Long N, et al. (2016a) Genome of plant maca (Lepidium meyenii) illuminates genomic basis for high-altitude adaptation in the central Andes. Mol Plant 9: 1066–1077 [DOI] [PubMed] [Google Scholar]
- Zhang L, Cai X, Wu J, Liu M, Grob S, Cheng F, Liang J, Cai C, Liu Z, Liu B, et al. (2018) Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic Res 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YM, Shao ZQ, Wang Q, Hang YY, Xue JY, Wang B, Chen JQ(2016b) Uncovering the dynamic `evolution of nucleotide-binding site-leucine-rich repeat (NBS-LRR) genes in Brassicaceae. J Integr Plant Biol 58: 165–177 [DOI] [PubMed] [Google Scholar]
- Zhong Y, Yin H, Sargent DJ, Malnoy M, Cheng ZMM(2015) Species-specific duplications driving the recent expansion of NBS-LRR genes in five Rosaceae species. BMC Genomics 16: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Zhang X, Cheng ZM(2018) Lineage-specific duplications of NBS-LRR genes occurring before the divergence of six Fragaria species. BMC Genomics 19: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wang Y, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D(2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics 271: 402–415 [DOI] [PubMed] [Google Scholar]
- Zipfel C.(2014) Plant pattern-recognition receptors. Trends Immunol 35: 345–351 [DOI] [PubMed] [Google Scholar]