A transcript repressor is responsible for the normal development of spikelet and floral organs in rice and can be used in breeding design to improve yield.
Abstract
An understanding of flower and panicle development is crucial for improving yield and quality in majority of grass crops. In this study, we used mapping-based cloning to identify MULTI-FLORET SPIKELET2 (MFS2), which encodes a MYB transcription factor and regulates flower and spikelet development in rice (Oryza sativa). In the mfs2 mutant, specification of palea identity was severely disturbed and showed degradation or transformation into a lemma-like organ, and the number of all floral organs was increased to varying degrees. Due to the increase in the number of floral organs and development of extra transformed palea/marginal region of the palea-like organs, some mfs2 spikelets had a tendency to produce two florets. These defects implied that the mfs2 mutation caused abnormal specification of palea identity and partial loss of spikelet determination. We confirm that MFS2 is a transcriptional repressor that shows strong repression activity by means of two typical ethylene-responsive element binding factor-associated amphiphilic motifs, one of which locates at the C terminus and is capable of interaction with three rice TOPLESS and TOPLESS-related proteins. The results indicate that MFS2 acts as a repressor that regulates floral organ identities and spikelet meristem determinacy in rice by forming a repression complex with rice TOPLESS and TOPLESS-related proteins.
The inflorescence and flower, which are characteristic reproductive structures of angiosperms, have an important effect on crop yield. During the reproductive phase of angiosperms, the shoot apical meristem first differentiates into inflorescence meristem (IM), on which lateral and terminal flower meristem (FM) is formed and typically give rise to four whorls of floral organs (sepals, petal, stamen, and carpels; McSteen et al., 2000).
The fate of IM underlies FM production. With regard to the behavior of the apical meristems of the inflorescence, IM is categorized as indeterminate or determinate based on whether the meristem remains continuously active or is transformed into a terminal FM. An indeterminate IM, such as in Arabidopsis (Arabidopsis thaliana), continuously produces lateral FMs but is not transformed into a terminal FM; a determinate IM, such as in tobacco (Nicotiana tabacum), is finally transformed into a terminal FM after production of a fixed number of lateral FMs (Bradley et al., 1997; Ratcliffe et al., 1999; Sussex and Kerk, 2001; Chuck et al., 2008). In Arabidopsis, TERMINAL FLOWER (TFL) and LEAFY are expressed in IMs and antagonistically regulate IM identity (Prusinkiewicz et al., 2007). TFL inhibits FM fate acquisition in the top domain of the IM, and tfl mutants produce a determinate inflorescence with a terminal floret. By contrast, TFL promotes FM formation in the lateral domain of the IM, mutation of which results in the formation of secondary IMs instead of lateral FMs (Alvarez et al., 1992; Blázquez et al., 1997; Mimida et al., 2001).
The inflorescence in grasses is usually named panicle, comprising the main rachis, branch, spikelet, and floret, which were produced in an orderly manner after flower induction. In rice (Oryza sativa), branch meristems (BMs) are regulated by a similar mechanism to IMs of Arabidopsis, whereby the rice TFL1-orthologs RICE CENTRORADIALIS1 (RCN1) or RCN2, prolong IM fate. Overexpression of either RCN1 or RCN2 delays the transition of BM to spikelet meristems (SM) and ultimately results in the production of a higher number of branches and spikelets than in the wild type (Nakagawa et al., 2002; Rao et al., 2008). The spikelet is produced on the lateral or top of a branch, and usually considered to be a grass-specific basal inflorescence unit made up of a short branch bearing a pair of glumes and an uncertain number of florets. The SM also shows a determinate or indeterminate fate in different grass species (Coen and Nugent, 1994; Itoh et al., 2005; Kobayashi et al., 2010; Bartlett and Thompson, 2014). In species with an indeterminate SM, such as wheat (Triticum aestivum), SM fate is maintained and an indefinite number of FMs are produced. In species that produce a determinate spikelet, such as maize (Zea mays) and rice, the SM is ultimately transformed into a terminal FM after production of a fixed number of lateral FM(s), such as two in maize and one in rice, thus terminating the SM fate. Several genes have been reported to regulate the determination of SM. LEAFY HULL STERILE1 (LHS1)/OsMADS1 is a member of the E-class gene family of MADS-box transcription factors (TF) and is similar to Arabidopsis SEPALLATA genes. According to the “selecting gene hypothesis,” LHS1 and its orthologs might be required to specifically determine the production of a terminal meristem in determinate SM species (Prasad et al., 2001, 2005; Malcomber and Kellogg, 2004; Zahn et al., 2005; Khanday et al., 2013). SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET1 (OsIDS1) are APETALA2 (AP2)-like TFs that are essential for correct timing of the transition from the SM to the FM. Mutation of SNB causes differentiation of a number of rudimentary glumes, extra lemma/palea-like organs, and even extra florets, implying partial loss of SM determination. The osids1 single mutant shows a weak phenotype, whereas the snb+osids1 double mutant displays a more severe phenotype than that of the snb single mutant, which suggests that OsIDS1 functions synergistically with SNB in the regulation of SM determination (Lee et al., 2007; Lee and An, 2012). In addition, MULTI-FLORET SPIKELET1 (MFS1) encodes an AP2 protein involved in the determination of SM fate in rice. The mfs1 mutant exhibits a delayed SM-to-FM transition, which results in development of an additional lemma-like organ or an extra floret (Ren et al., 2013). Therefore, the regulation of SM fate differs from that of BMs in grasses and IMs in other species, and the mechanism remains unclear to a large degree.
On the basis of the genetic and molecular studies of floral homeotic mutants in eudicots, such as Arabidopsis and Antirrhinum majus, the ABC model and revised ABCE models were proposed to elucidate the molecular mechanism of floral organ specification, in which A-, B-, C-, and E-class genes cooperatively regulate the identities of four whorls of floral organs (Coen and Meyerowitz, 1991; Coen and Nugent, 1994; Ohmori et al., 2009). The ABCE model is also applicable to rice to a large degree. The rice genome contains four A-class genes, three B-class genes, two C-class genes, and at least five E-class genes, some of which have a conserved function with their ABCE orthologs in Arabidopsis, such as the A-class APE1-like gene OsMADS15, B-class AP3-like gene OsMADS16, B-class PISTILLATA-like genes OsMADS2 and OsMADS4, C-class AGAMOUS (AG)-like genes OsMADS3 and OsMADS58, and E-class gene LHS1/OsMADS1 (Jeon et al., 2000; Nagasawa et al., 2003; Yamaguchi et al., 2006; Yao et al., 2008; Wang et al., 2010; Dreni et al., 2011; Hu et al., 2011). However, in contrast to a eudicot such as Arabidopsis, the floret of rice develops a specialized outer whorl of floral organs (lemma and palea). In addition to the A- and E-class genes, such as OsMADS15 and LHS1/OsMADS1, many other genes have been determined to regulate development of the lemma and/or palea. STAMENLESS1, THIAMINE REQUIRING1 (TH1), and CONSERVED IN CILIATED SPECIES AND IN THE LAND PLANTS1 (CCP1)/DFO1 regulate the characteristic development of the palea and lemma simultaneously (Xiao et al., 2009; Li et al., 2012; Yan et al., 2015; Zheng et al., 2015). MOSAIC FLORAL ORGANS1 (MFO1)/OsMADS6 and CHIMERIC FLORAL ORGANS1 (CFO1)/OsMADS32 specifically regulate the marginal region of palea (mrp) development (Ohmori et al., 2009; Li et al., 2010; Sang et al., 2012). In the mfo1 and cfo1 mutants, mrp development is severely disrupted and results in lemma-like palea. In contrast, in the depressed palea1 (dp1), the body of palea (bop) is degraded so that only two mrps remain, which indicated that DP1 function in bop development (Luo et al., 2005; Jin et al., 2011).
Transcriptional repressors and their corepressors play crucial roles in plant flower development (Liu and Karmarkar, 2008; Krogan and Long, 2009). TPL/TPR, a conserved family of plant transcription corepressor proteins, inhibit expression of the target gene by interaction with TFs that contain ethylene-responsive element binding factor-associated amphiphilic (EAR) motifs (LxLxL/DLNxxP), after which histone deacetylases (HDACs) are recruited to the transcription complexes and the chromatin state is changed from active to inactive (Long et al., 2006; Gonzalez et al., 2007; Krogan and Long, 2009). In Arabidopsis, TPL and four TPR proteins (TPR1, TPR2, TPR3, and TPR4) act redundantly in Arabidopsis development. Silencing TPR2 in the quadruple mutant tpl tpr1 tpr3 tpr4 results in defective embryonic development similar to that of the tpl-1 mutant (Long et al., 2002, 2006). Three members of the TPL/TPR corepressor family are known in rice: OsTPR1, OsTPR2/ABERRANT SPIKELET AND PANICLE1 (ASP1), and OsTPR3. The ASP1 acts as a transcriptional corepressor to repress genes that regulate determinacy and maintenance of BM, IM, and SM (Yoshida et al., 2012), whereas the functions of the two OsTPRs have not been reported. The Arabidopsis A-function gene AP2 acts as a repressor and interacts with the corepressor complex TPL–HDAC19 to restrict expression of the C-function gene AG in whorls 1 and 2 (Ó’Maoiléidigh et al., 2014). In rice, NONSTOP GLUME1 (NSG1) interacts with OsTPRs via the EAR motif to repress expression of OsMADS1 in the rudimentary glume, sterile lemma, and mrp by increased acetylation of histone H3 on target region (Zhuang et al., 2020). However, the identity of the factors that interact with OsTPRs to regulate flower and inflorescence development in rice, and the mechanism involved, remain unclear.
The MYB family of TFs is present in almost all plants. Members of the family contain three types of conserved MYB DNA binding domains, namely R1, R2, and R3. Based on the number of MYB domain repeats, MYB proteins can be classified as R2R3-MYB, 1R-MYB, 3R-MYB, and 4R-MYB (Dubos et al., 2010). In rice, several MYB genes play important roles in organ development. The genes OsGAMYB, ANTHER INDEHISCENCE1, and CARBON STARVED ANTHER predominantly regulate pollen development (Kaneko et al., 2004; Zhu et al., 2004, 2015; Zhang et al., 2010), and SHALLOT-LIKE1 (SLL1) affects leaf and hull shape (Zhang et al., 2009; Ren et al., 2019). However, it remains unclear how MYB genes regulate rice development. In this study, we identified a MYB TF in rice named MFS2. The mfs2 mutation caused abnormal specification of palea identity and partial loss of spikelet determination. MFS2 was indicated to be a transcriptional repressor with strong activity owing to two typical EAR motifs, one of which was located at the C terminus and interacted with three rice TPL/TPR proteins. Therefore, our results indicated that MFS2 plays a crucial role in the regulation of floral organ identities and SM determinacy in rice by forming a repression complex with TPL/TPR proteins to restrict the expression of downstream genes.
RESULTS
Pleiotropic Defects in mfs2 Spikelet
In rice, inflorescence and spikelet development are categorized into the In1 to In9 and Sp1 to Sp8 stages, respectively (Ikeda et al., 2004). In this study, we identified a recessive mutant mfs2 that related to the development of the rice spikelet and floral organs. The floral morphologies of the mfs2 mutant and wild type were compared at the In9 stage (heading stage).
Generally, a typical wild-type spikelet consisted of two rudimentary glumes, two sterile lemmas, and one terminal floret that contained four whorls of floral organs: the lemma and palea in whorl 1, and two lodicules, six stamens, and one pistil arranged sequentially in the inner three whorls (Fig. 1, A1–A7). In addition to the slightly larger size of the lemma, the palea can be distinguished from the lemma by several features, such as the number of vascular bundles and distinct marginal region. The lemma contained five vascular bundles, whereas the palea contained three vascular bundles. The palea could be considered as a combinational organ of two parts: bop and mrp. Although the bop was anatomically similar to the lemma, with four cell layers discernible (silicified abaxial epidermis, fibrous sclerenchyma, spongy parenchymatous cells, and non-silicified adaxial epidermis), the mrp displayed a smooth non-silicified abaxial epidermis, a large amount of spongy parenchymatous tissue, and a small number of sclerenchyma cells (Figs. 1, A1 and A4–A7, and 2, A and B). The two lodicules were small semitransparent organs of a regular lung shape formed asymmetrically to the inner of the two lemmas (Fig. 1, A6 and A7), and consisted of parenchymatous cells and several interspersed tracheal elements (Fig. 2C).
Figure 1.
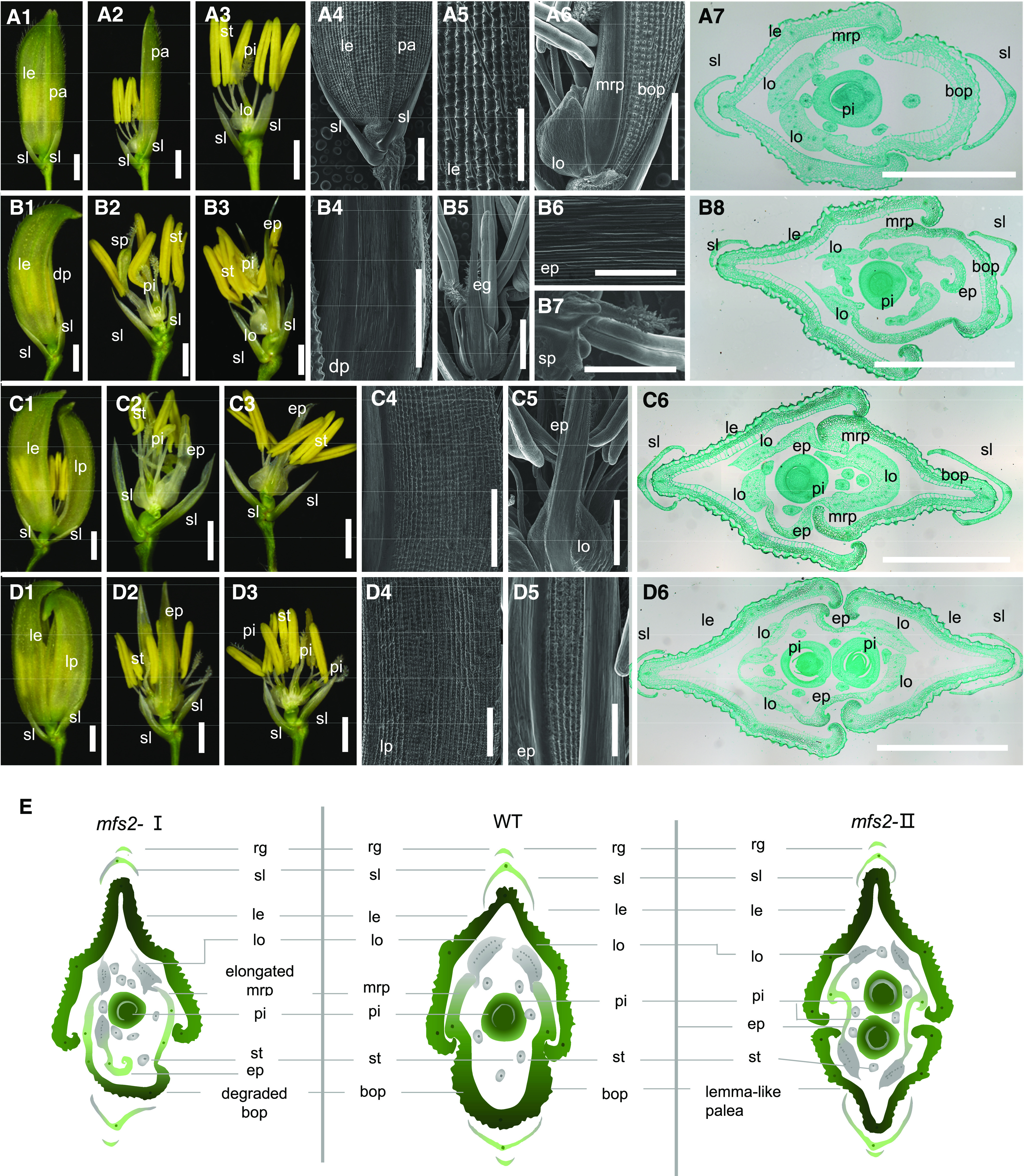
Phenotypes of spikelets of the wild-type and mfs2. A1, Complete spikelet of the wild type. A2, Spikelet from A1with lemma (le) removed. A3, Spikelet from A1 with lemma and palea (pa) removed. A4, Surface of a wild-type spikelet. A5 and A6, Surface characters of le, lodicule (lo), bop, and mrp, respectively. A7, Transverse sections of a wild-type spikelet. B1, mfs2 spikelet with degraded palea (dp). B2 and B3, Spikelet with lemma and palea removed. B4 and B5, Surfaces of the degraded palea and extra glume, respectively. B6, Magnification of B5. B7, Surface of the stamen-pistil fusion organ (sp). B8, Transverse sections of a mfs2 spikelet with degraded palea. C1, mfs2 spikelet with a partially lemma-like palea (lp). C2 and C3, Spikelet with lemma and palea removed. C4 and C5, Surfaces of the partially lemma-like palea and extra glume, respectively. C6, Transverse sections of a mfs2 spikelet with a partially lemma-like palea. D1, mfs2 spikelet with a lemma-like palea. D2, Spikelet from D1 with lemma and lemma-like palea removed. D3, Spikelet from D2 with the extra palea/mrp-like organ removed. D4 and D5, Surfaces of the lemma-like palea and extra glume, respectively. D6, Transverse sections of a mfs2 spikelet with lemma-like palea. E, Schematic diagrams of wild-type (WT) and mfs2 spikelet structure in transverse section. ep, Extra palea/mrp-like organ; pi, pistil; sl, sterile lemma; st, stamen. Scale bars = 1 mm (A1–A3, B1–B3, C1–C3, and D1– D3), 200 μm (A4–A6, B4– B7, C4, C5, D4, and D5), and 500 μm (A7, B8, C6, and D6).
Figure 2.
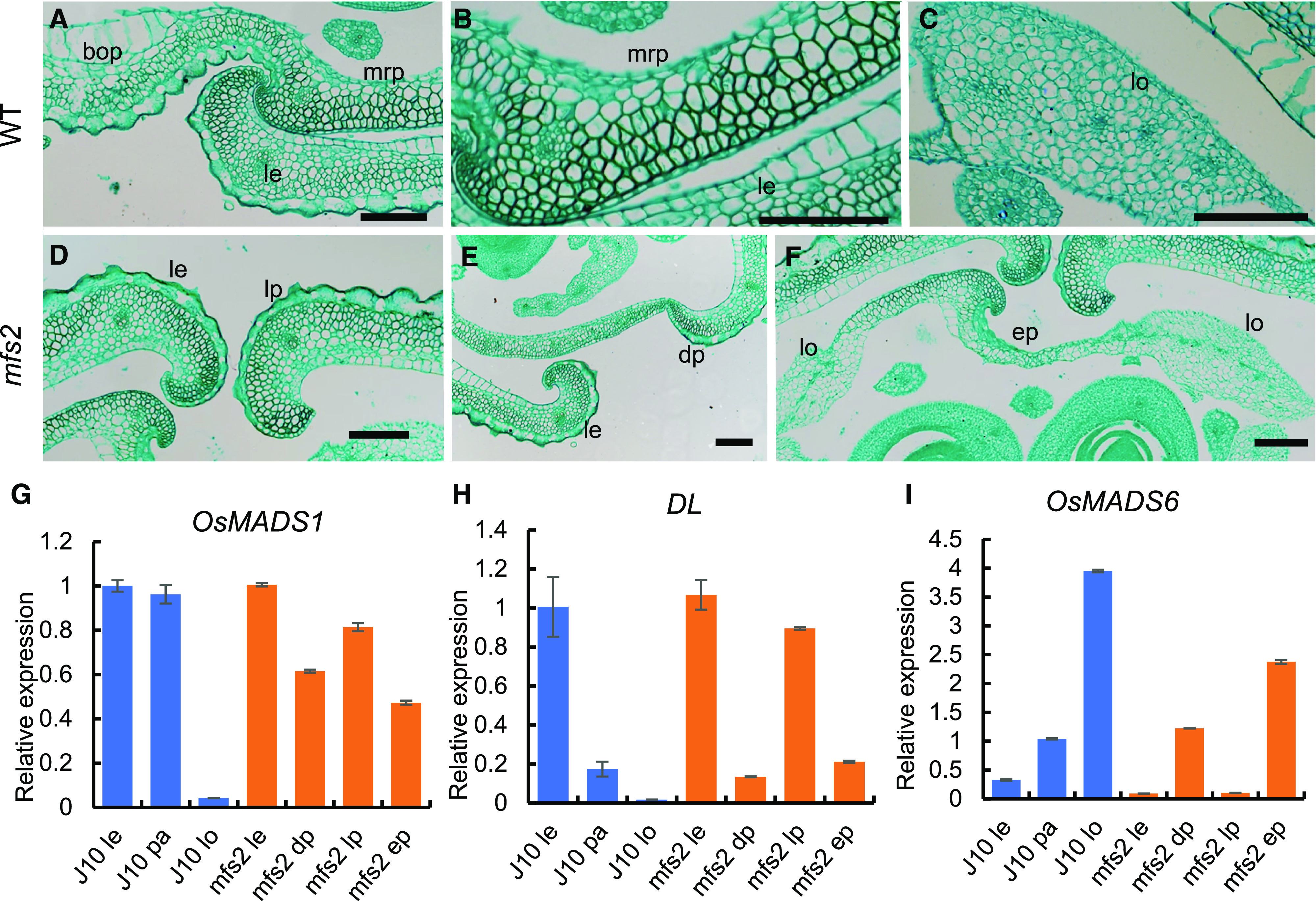
Histological and reverse transcription (RT)-qPCR analysis of organs in spikelets of the wild-type (WT) and mfs2 mutant. A, Transverse section of the joint of lemma (le) and palea (pa) in wild-type spikelets. B, Transverse section of the wild-type mrp. C, Magnification of the wild-type lodicule (lo). D, Transverse sections of the lemma and lemma-like palea (lp) in mfs2. E, Transverse section of the degraded palea (dp). F, Transverse section of the extra glume. G to I, Relative expression of OsMADS1, DL and OsMADS6 in organs of wild-type (blue) and mfs2 (orange) spikelets, respectively. ep, Extra palea/mrp-like organ. Scale bars = 100 μm.
The mfs2 spikelets displayed pleiotropic defects in the palea and inner floral organs. According to statistical analysis of 100 spikelets selected randomly from 10 mfs2 panicles, paleas were transformed into lemma-like organs in ∼57% of the spikelets, and degraded paleas were observed in 29% of the spikelets (Supplemental Fig. S1). Based on the palea phenotype, then mfs2 spikelets were classified into two types: spikelets with a degraded palea (type I; Fig. 1E) and spikelets with a lemma-like palea (type II; Fig. 1E).
In type-I spikelets, the degraded palea displayed a reduced bop, some of which remained only two mrp structures (Figs. 1, B1, B4, and B8, and 2E). These mrps were usually broader and included more vascular bundle-like elements than those in the wild type, and lost the hook structure locking with lemma (Figs. 1B8 and 2E). Stripping off the lemma and palea, extra palea/mrp-like organs and other defects of inner three whorls of organs could be observed. Of the type-I spikelets examined, ∼35% contained an extra palea/mrp-like organ in whorl 2. A majority of these organs were mrp-like in appearance with a smooth epidermis (Fig. 1, B3, B5, B6, and B8). In addition, nearly 35% of the type I displayed an increased number of stamens and ∼30% included an increased number of pistils (Supplemental Fig. S1). Furthermore, in ∼4% of the spikelets, stamen–pistil fusion organs were observed in whorl 3, which displayed a stigma developed on an anther (Fig. 1, B2 and B7; Supplemental Fig. S1).
The distinctive feature of the type-II spikelets was that the paleas were transformed into lemma-like organs and the number of inner floral organs were increased (Fig. 1, C1 and D1). In a portion of these spikelets, the paleas were larger and contained five vascular bundles similar to the lemma but retained an extremely narrow mrp with a smooth epidermis (Fig. 1, C4 and C6). In the remaining spikelets, the paleas were completely transformed into a lemma-like organ and developed four cell layers identical to those of the lemma (Figs. 1, D4 and D6, and 2D). Notably, ∼91% of the type-II spikelets developed two extra palea/mrp-like structures in whorl 2, each of which consisted of two lateral mrp-like structures with a smooth epidermis and one narrow medial bop-like tissue with a silicified adaxial epidermis bearing trichomes and protrusions, extremely similar to the degraded paleas in the type-I spikelets (Fig. 1, C2, C3, and D2; Supplemental Fig. S1). Unsurprisingly, four lodicules were always attached to the extra palea/mrp-like structures in whorl 2 (although they often displayed an irregular shape), with an increased number of stamens usually observed in whorl 3, and sometimes an increased number of pistils observed in whorl 4 (Figs. 1, C5, C6, D3, D5, and D6, and 2F). Therefore, type-II spikelets usually contained two lemmas (the original lemma and a lemma-like palea), two palea/mrp-like organs, two pairs of lodicules, and increased numbers of stamens and pistils, which largely resulted in the formation of “two-florets” and partial loss of SM determinacy in the mfs2 mutant (Fig. 1, C6, D6, and E).
We further detected the expression levels of the lemma and/or palea identity genes DROOPING LEAF (DL), OsMADS1, and OsMADS6 in related organs of the wild type and mfs2 mutant (Fig. 2, G–I). In lemma-like paleas, a high expression level of DL and OsMADS1, and a low expression level for OsMADS6 were detected, which were extremely similar to the expression patterns detected in the wild-type lemma. This result suggested that the lemma-like paleas had generally assumed a lemma-like identity. In degraded paleas, the transcript levels of DL and OsMADS6 were similar to those in wild-type paleas, whereas transcription of OsMADS1 was downregulated substantially, consistent with the phenotype of bop degradation. In the extra glumes, a high level of OsMADS1 expression was detected, although the level was slightly lower than that in wild-type paleas; the expression level of OsMADS6 was substantially higher than that detected in the wild-type paleas, and DL expression was detected at a low level similar to that in the wild-type paleas. These results implied that the extra glumes assumed a palea-like identity.
Abnormal Early Spikelet Development in the mfs2 Mutant
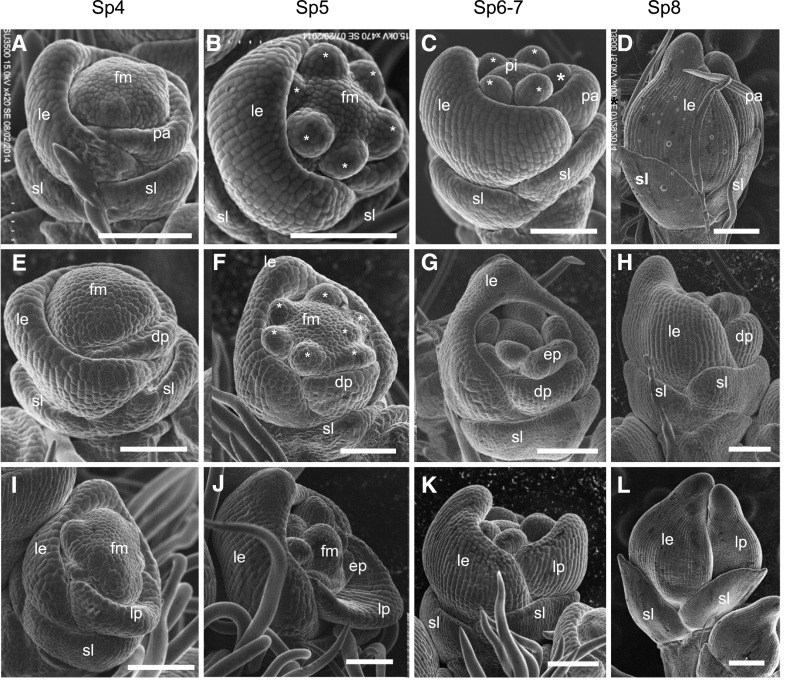
The early development of the rice spikelet was a complex but well-organized process. We observed notable differences in early spikelet development between the mfs2 mutant and wild type at the Sp4 to Sp8 developmental stages using scanning electron microscopy. In wild-type spikelets, the palea primordium was initiated in the opposite position to the lemma primordium during the Sp4 stage, and the lemma primordium was larger than the palea primordium (Fig. 3A). During Sp5 to Sp8 stages, primordia of lodicules, stamens, and the pistil developed in an orderly manner. During the Sp8 stage, the primordia of lemmas and paleas were hooked together and enclosed the inner floral organs (Fig. 3, B–D). In the mfs2 spikelets, morphological differences were observed since the Sp4 stage. Some spikelets developed smaller palea primordia than those of the wild type at the Sp4 stage (Fig. 3E). The development process of the palea primordium was slower than that of the wild-type palea primordium. The degraded palea lost the ability to hook with the lemma at the Sp8 stage (Fig. 3, F–H). It was often observed that some palea primordia were larger than those of the wild type at the same stage, and elongated rapidly after the Sp4 stage, which showed similarities to the development of the lemma primordia. During the Sp8 stage, the abnormal palea primordia were similar to the lemma primordia both in shape and size (Fig. 3, I–L). These results suggested that the palea identity is affected at an early developmental stage in the mfs2 mutant. In addition, extra palea/mrp-like primordia and increased stamen primordia were observed in some spikelets of the mfs2 mutant (Fig. 3, F, G, and J).
Figure 3.
Scanning electron micrographs of wild-type and mfs2 spikelets at early developmental stages. A to D, Wild-type spikelets at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages, respectively. E to H, mfs2 spikelets with degraded palea primordia at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages, respectively. I to L, mfs2 spikelets with lemma-like palea primordia at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages, respectively. ep, Extra palea/mrp-like organ; le, lemma; lp, lemma-like palea; pa, palea; pi, pistil; sl, sterile lemma; *, stamen. Scale bars = 200 μm.
Expression Patterns of Floral Organ Identity Genes during Early Stages of Floral Development
To confirm the abnormal development of floral organs in the mfs2 mutant, the expression levels of genes that regulate identities of palea and other floral organs were detected by quantitative PCR (qPCR; Supplemental Fig. S2), including DL, OsMADS1, OsMADS2, OsMADS6, OsMASD32, REP1, CCP1, TH1, AH2, and DP1. In the mfs2 mutant, expression levels of OsMADS6, REP1 and TH1 were suppressed sharply, whereas expression levels of OsMADS1, OsMADS2, DL, and DP1 were promoted substantially, being consistent with defects in the palea and other floral organs.
In situ hybridization was used to investigate the expression patterns of OsMADS1, OsMADS6, OsMADS2, and DL. During the Sp4 to Sp8 stages, in wild-type spikelets, OsMADS1 was mainly expressed in the whole lemma and bop, but not in the mrp (Fig. 4, G1–G3). In type-II mfs2 spikelets, except for the lemma, expression of OsMADS1 was detected in the whole lemma-like palea (both the mrp and bop), and in extra palea/mrp-like organs (Fig. 4, I1–I3). In type-I spikelets, in addition to the lemma, OsMADS1 signal was detected in the degraded bops (Fig. 4, H1–H3). In wild-type spikelets, OsMADS6 was expressed in the floral meristem during the Sp4 and Sp5 stages, and was subsequently expressed predominantly in the mrp and lodicule primordia during the Sp6 to Sp8 stages (Fig. 4, A1–A3). In type-I mfs2 spikelets, OsMADS6 expression was repressed to some degree in the mrps of degraded paleas, but no notable differences were detected in other domains compared with those of the wild-type spikelets (Fig. 4, B1–B3). As expected, in type-II mfs2 spikelets, almost no signal for OsMADS6 was detected in the whole of the lemma-like palea (Fig. 4, C1–C3), whereas a distinct signal was observed in the marginal regions of the extra palea-like organs. Expression of DL was first detected in the wild-type lemma during the Sp4 and Sp5 stages, and in the lemmas and pistil after the Sp6 stage when the pistil primordium was initiated (Fig. 4, D1–D3). In the mfs2 spikelets, no DL signal was detected in the reduced palea and extra palea-like organs (Fig. 4, E1–E3), whereas DL was ectopically expressed in the lemma-like palea from its initiation (Fig. 4, F1–F3). Thus, in the lemma-like paleas of the mfs2 mutant, the expression patterns of OsMADS1 and DL were similar to those of the wild-type lemma, whereas expression of OsMADS6 was substantially reduced, which confirmed that these organs had acquired a lemma-like identity. The expression of OsMADS1 and OsMADS6 in the extra palea/mrp-like organ also indicated that these were palea-like organs. In the wild type, OsMADS2 was predominantly expressed in the lodicules and stamens. In type-II mfs2 spikelet, strong expression of OsMADS2 was detected in the four lodicule-like organs that were usually connected to the margin of the extra palea/mrp-like organs (Supplemental Fig. S3), indicating that the former organs represented two pairs of lodicules.
Figure 4.
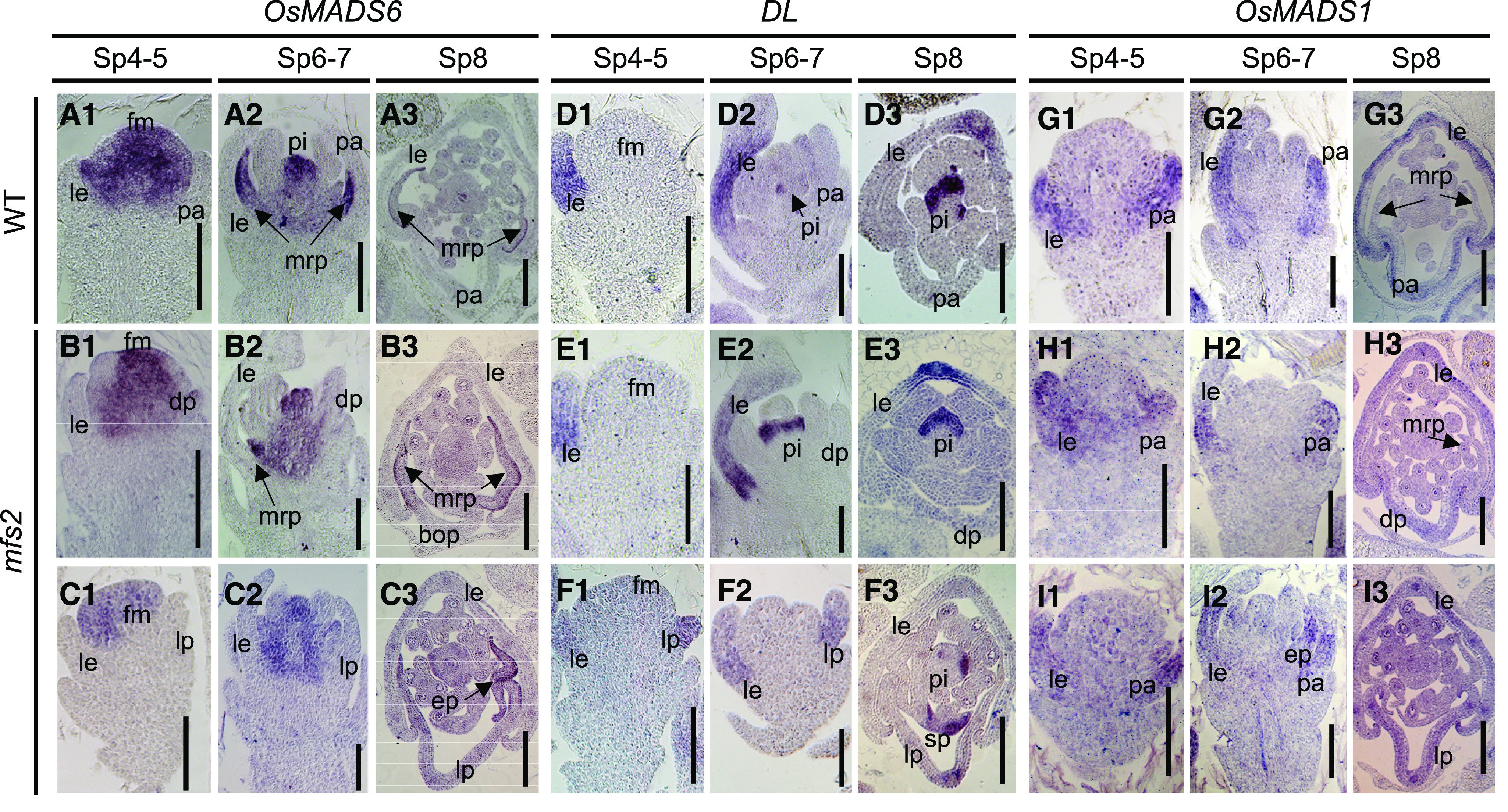
Expression pattern of floral-organ identity genes in spikelets of the wild-type and mfs2 mutant. A1 to A3, Expression of OsMADS6 in wild-type (WT) spikelets at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. B1 to B3, Expression of OsMADS6 in mfs2 spikelets with degraded palea (dp) at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. C1 to C3, Expression of OsMADS6 in mfs2 spikelets with lemma-like palea (lp) at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. D1 to D3, Expression of DL in wild-type spikelets at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. E1 to E3, Expression of DL in mfs2 with degraded spikelets at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. F1 to F3, Expression of DL in mfs2 spikelets with lemma-like palea at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. G1 to G3, Expression of OsMADS1 in wild-type spikelets at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. H1 to H3, Expression of OsMADS1 in mfs2 with degraded spikelets at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. I1 to I3, Expression of OsMADS1 in mfs2 spikelets with lemma-like palea at the Sp4, Sp5, Sp6, Sp7, and Sp8 developmental stages. ep, Extra palea/mrp-like organ; le, lemma; lo, lodicule; pa, palea; pi, pistil; sl, sterile lemma; sp, stamen-pistil fusion organ. Scale bars = 100 μm.
In addition, we detected expression of OsIDS1, SNB, and MFS1, which function in the regulation of SM determinacy during early developmental stages (Lee and An, 2012; Ren et al., 2013). While MFS1 was slightly upregulated, OsIDS1 and SNB were distinctly downregulated in young panicles of the mfs2 mutant (Supplemental Fig. S4). These results suggested that SM activity was prolonged and led to development of an additional floral organ and even florets in the mfs2 mutant, due to the mutation of MFS2 interfering with the transcription of OsIDS1 and SNB.
Map-Based Cloning of MFS2
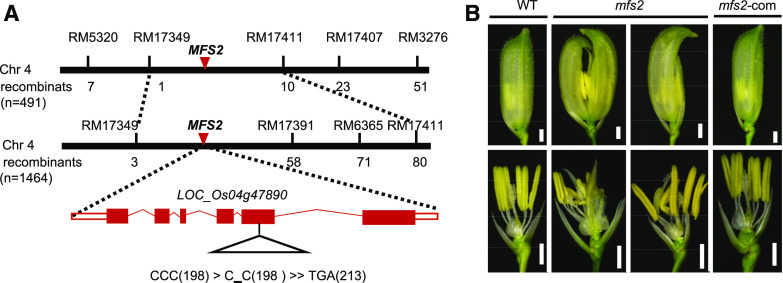
The mfs2 mutant was crossed with the sterile line cv Xinong 1A. All F1 hybrids displayed the wild-type phenotype, whereas the F2 progeny showed segregation of wild-type and mutant phenotypes in accordance with a 3:1 ratio (χ2 = 2.57 < χ20.05 = 3.84), which indicated that the mutant trait was controlled by a single recessive gene. A total of 1,464 individuals exhibiting the mutant phenotype in the F2 progeny was used as the mapping population. The MFS2 gene was mapped to the long arm of chromosome 4 within an ∼1.02-Mb region between the simple sequence repeat markers RM17349 and RM17391. In the mapped region, there was a MYB family TF (LOC_Os04g47890). Sequence analysis identified a single-nucleotide deletion (C) in the open reading frame of Os04g47890 in the mutant, which caused a frameshift mutation from the 198th codon and premature termination of translation at the 213th codon (Fig. 5A). To confirm these observations, the complementary vector that contained the LOC_Os04g47890 coding sequence (3,413 bp), 2,282-bp sequence upstream of the start codon, and 1,037-bp sequence downstream of the stop codon was transformed into the mfs2 mutant. In total, 17 transgenic plants were obtained, in nine of which the mutant phenotypes were rescued compared with that of the wild-type spikelets (Fig. 5B). In addition, we constructed an RNA interference (RNAi) vector using a 161-bp fragment of LOC_Os04g47890 complementary DNA (cDNA), and transformed the vector into the japonica cv ZH11 to silence LOC_Os04g47890 (Supplemental Fig. S5A). In 13 of the 15 transgenic plants obtained, qPCR analysis showed that the MFS2 transcript level was reduced to varying degrees (Supplemental Fig. S5C). These RNAi lines displayed similar pleiotropic spikelet defects to those of mfs2 mutant spikelets, containing lemma-like palea, degraded palea, and an extra palea-like organ (Supplemental Fig. S5B). Taken together, these results indicated that LOC_Os04g47890 was the MFS2 gene.
Figure 5.
Map-based cloning of MFS2. A, Fine mapping of MFS2. B, Genetic complementation of mfs2. WT, wild type. Scale bars = 1,000 μm.
Spatiotemporal Expression Pattern of MFS2
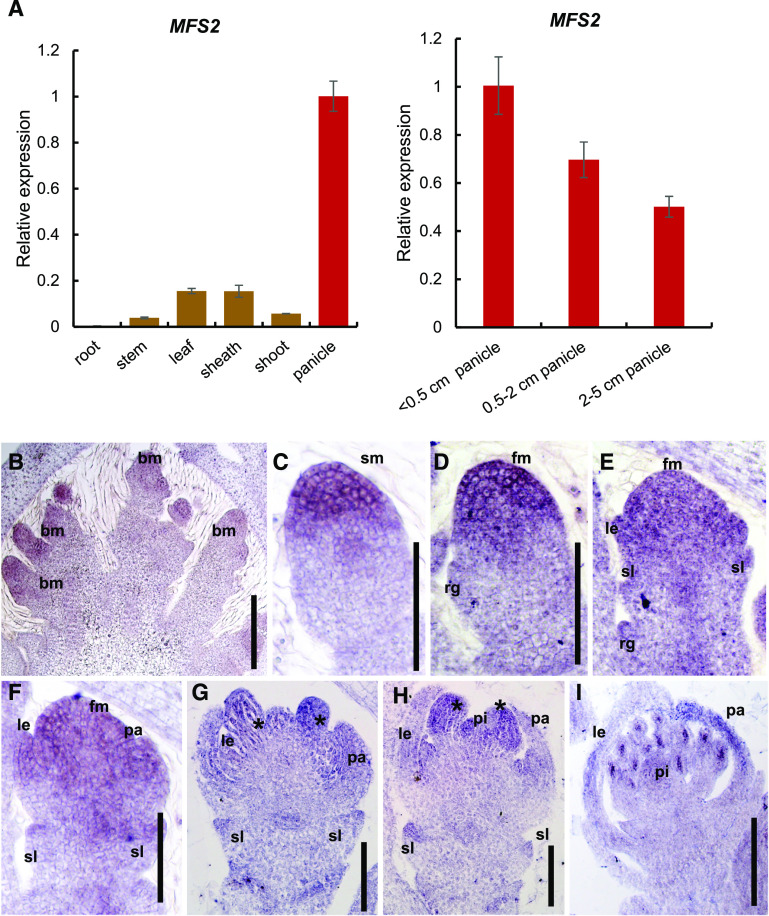
To determine the spatiotemporal expression pattern of MFS2, we employed qPCR and in situ hybridization to examine MFS2 expression in wild-type plants. The qPCR analysis showed that MFS2 was highly expressed in young panicles, whereas its transcripts were extremely low in the stem, leaf, sheath, and shoot, and not even detected in the root (Fig. 6A). In situ hybridization revealed that weak accumulation of MFS2 mRNA was first detected in the BM (Fig. 6B). Subsequently, strong MFS2 signals were detected in SM (Fig. 6C). With initiation of the floral organ primordia, MFS2 transcripts were detected in the FM, and the primordia of sterile lemma, lemma, and palea (Fig. 6, D–F). Subsequently, MFS2 was expressed in the lodicule, stamen, and pistil (Fig. 6, G–I). This expression pattern was consistent with the mfs2 phenotype.
Figure 6.
Expression pattern of MFS2. A, Expression of MFS2 in different tissues of rice (vegetative organs are marked in brown; reproductive organs are marked in red). B to I, In situ hybridization in wild-type panicles and spikelets, using a MFS2 antisense probe. B, A young panicle (<0.5-cm length). C and D, Spikelet at Sp1 to Sp2. E and F, Spikelet at Sp4. G and H, Spikelet at Sp6 and Sp7. I, Spikelet at Sp8. fm, Floral meristem; le, lemma; pa, palea; pi, pistil; sl, sterile lemma; *, stamen. Scale bars = 100 μm.
MFS2 Encodes a Nuclear-Localized MYB Transcriptional Repressor
MFS2 encodes a 1R-type MYB protein of 306 amino acids with one MYB repeat unit. MYB-related proteins are classified into five subfamilies: CCA1-like, I-box–like, TRF-like, CPC-like, and TBP-like (Chen et al., 2006). Using proteins from rice, Arabidopsis, and Amborella trichopoda, phylogenetic analysis revealed that a MFS2/SLL1-like clade was formed, containing MFS2, SLL1 (a known rice rolling leaf gene), several additional rice homeologs, and their orthologs in Arabidopsis and A. trichopoda, independent of the CCA1-like, I-box–like, TRF-like, CPC-like, and TBP-like clades (Fig. 7A). Furthermore, a phylogenetic tree for MFS2-like orthologs was constructed, containing 28 genes from 23 eudicots and monocots. The MFS2-like genes from eudicots and monocots consisted of a large branch respectively, and the genes from grass species constituted a subbranch independent of genes from monocots (Supplemental Fig. S6). Thus, evolution of MFS2-like genes was consistent with the evolution of flowering plants.
Figure 7.
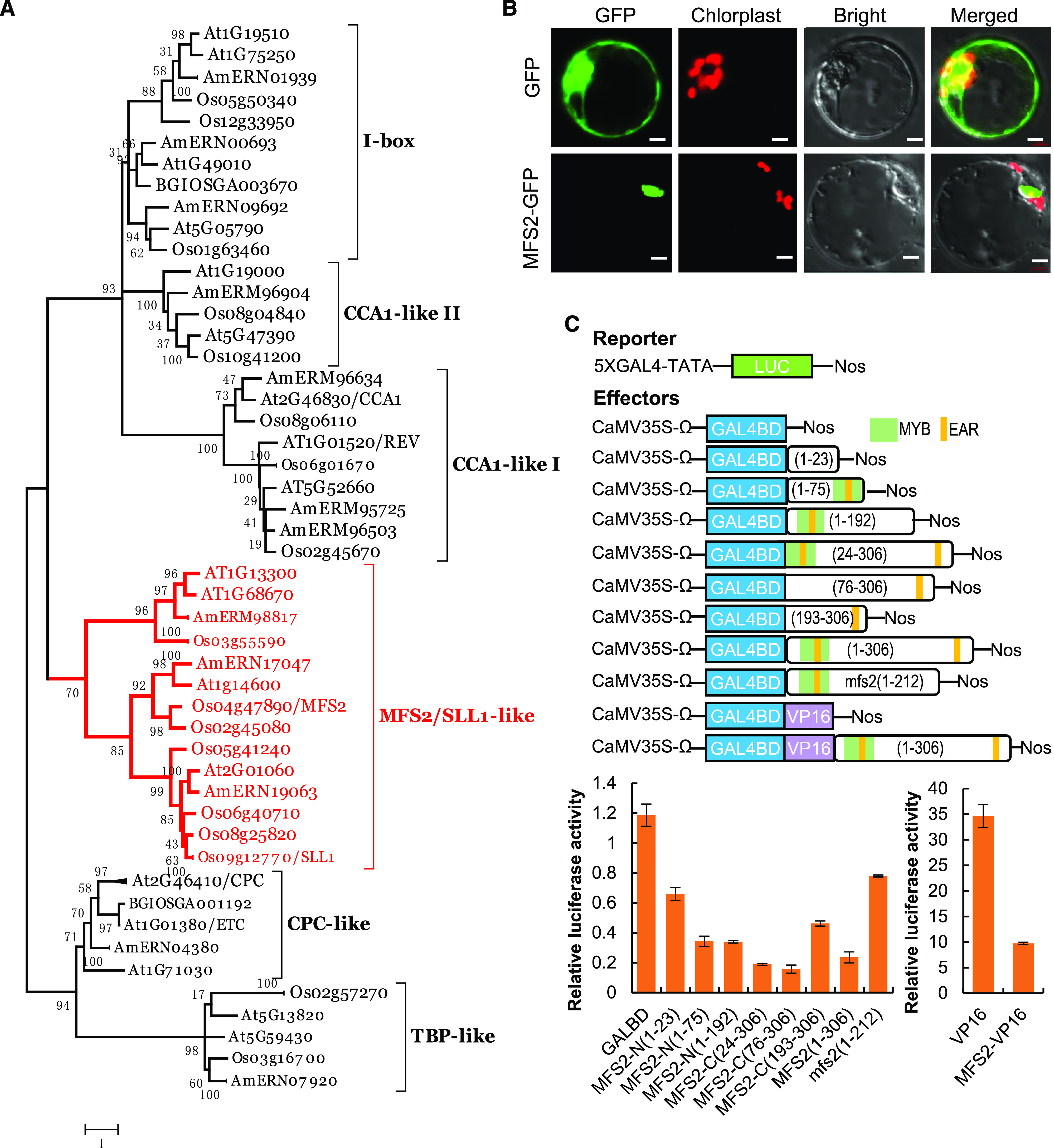
MFS2 encodes a MYB transcriptional repressor with two EAR motif. A, Phylogenetic tree analysis. The phylogenetic tree was performed using the neighbor-joining method, and bootstrap support values calculated from 1,000 replicates are given at the branch nodes; MFS2 protein and its homologous proteins are highlighted in red. Os, O. sativa; At, Arabidopsis; Am, A. trichopoda; BGIOSGA, the gene annotation of 9311 genome. B, Analysis of the subcellular localization of the MFS2 protein. Scale bars = 50 µm. C, Analysis of the transcriptional activation of MFS2 using the dual luciferase reporter assay system. VP16, a transcriptional activator, was used as a positive control and GAL4-BD was regarded as a negative control.
Sequence analysis of four MFS2-like proteins, three SLL1-like proteins, and four CCA1-like proteins showed that there was a highly conservative motif at the end of the MYB domain, which was manifested as the sequence SHLQMYR, SHLQKYR, and SHAQK(Y/F) in the three types of proteins, respectively (Supplemental Fig. S7). We fused MFS2 with the GFP reporter gene and expressed the fusion protein in rice protoplasts. The MFS2-GFP protein was localized in the nucleus, whereas the GFP protein alone was uniformly expressed in the cytoplasm and nucleus (Fig. 7B). This result indicated that MFS2 was a nuclear-localized protein.
In addition to the MYB domain, MFS2 contained two typical LxLxL-type EAR motifs, located within the MYB domain and at the C terminus, respectively (Supplemental Fig. S8), which was generally considered to be a hallmark of transcriptional repressors. In the mfs2 mutant, the single nucleotide “C” deletion led to a frameshift and prematurely truncated protein that lacked the C-terminal EAR motif (Supplemental Fig. S8). A dual-luciferase reporter assay was performed to investigate the transcriptional activity of MFS2. According to the positions of the MYB domain (24–76 amino acids), EAR motif1 (52–57 amino acids), and EAR motif2 (248–253 amino acids), we constructed dual-luciferase vectors for the truncated MFS2 proteins, comprising MFS2-N(1–23), MFS2-N(1–76), MFS2-N(1–192), MFS2-C(24–306), MFS2-C(76–306), MFS2-C(193–306), MFS2 (1–306), mutant mfs2 (1–212), and MFS2(1–306)-VP16 fusion proteins. The transcriptional activation activity of these proteins was measured in rice protoplasts. Almost all truncated and full-length MFS2 proteins containing the EAR motif1 and/or EAR motif2, except the mutant mfs2 (1–212) protein, showed substantially stronger repression activity than the empty vector and MFS2-N (1–23). However, when using the mutant mfs2 protein as an effector, the luciferase activity was substantially increased, compared with that of the full-length and all truncated MFS2 proteins except for MFS2-N (1–23). Furthermore, the MFS2(1–306)-VP16 fusion protein also showed distinctly reduced LUC single compared with VP16 protein (Fig. 7C). Therefore, these results demonstrated that MFS2 acted as a transcriptional repressor, and the mutated mfs2 protein had lost repression activity to large degree, probably reflecting the absence of the C-terminal EAR motif or an alternative change in the protein structure.
MFS2 Interacts with Rice TPL/TPR Proteins
It is well known that EAR-motif repressors usually interact with TPL/TPR transcriptional corepressors to regulate a variety of development processes in plants. Therefore, a yeast two-hybrid (Y2H) assay was performed to test interaction of the full-length MFS2, truncated proteins, and mutated mfs2 protein with the three TPL/TPR proteins in rice, namely OsTPR1, OsTPR2/ASP, and OsTPR3. As expected, the variable-sized MFS2 and mfs2 proteins showed no auto-activation (Fig. 8A). The MFS2 protein, as well as the C-terminal truncated protein MFS2-C(193–306) with EAR motif2, showed interaction with any one of the three OsTPRs, whereas the N-terminal truncated protein MFS2-C(1–192) with MYB and EAR motif1, and mfs2 proteins lacking EAR motif2, failed to interact with any one of the three OsTPRs (Fig. 8A). These results indicated that the MFS2 protein showed the ability to interact with TPL/TPR proteins and the C-terminal EAR motif was necessary for this interaction.
Figure 8.
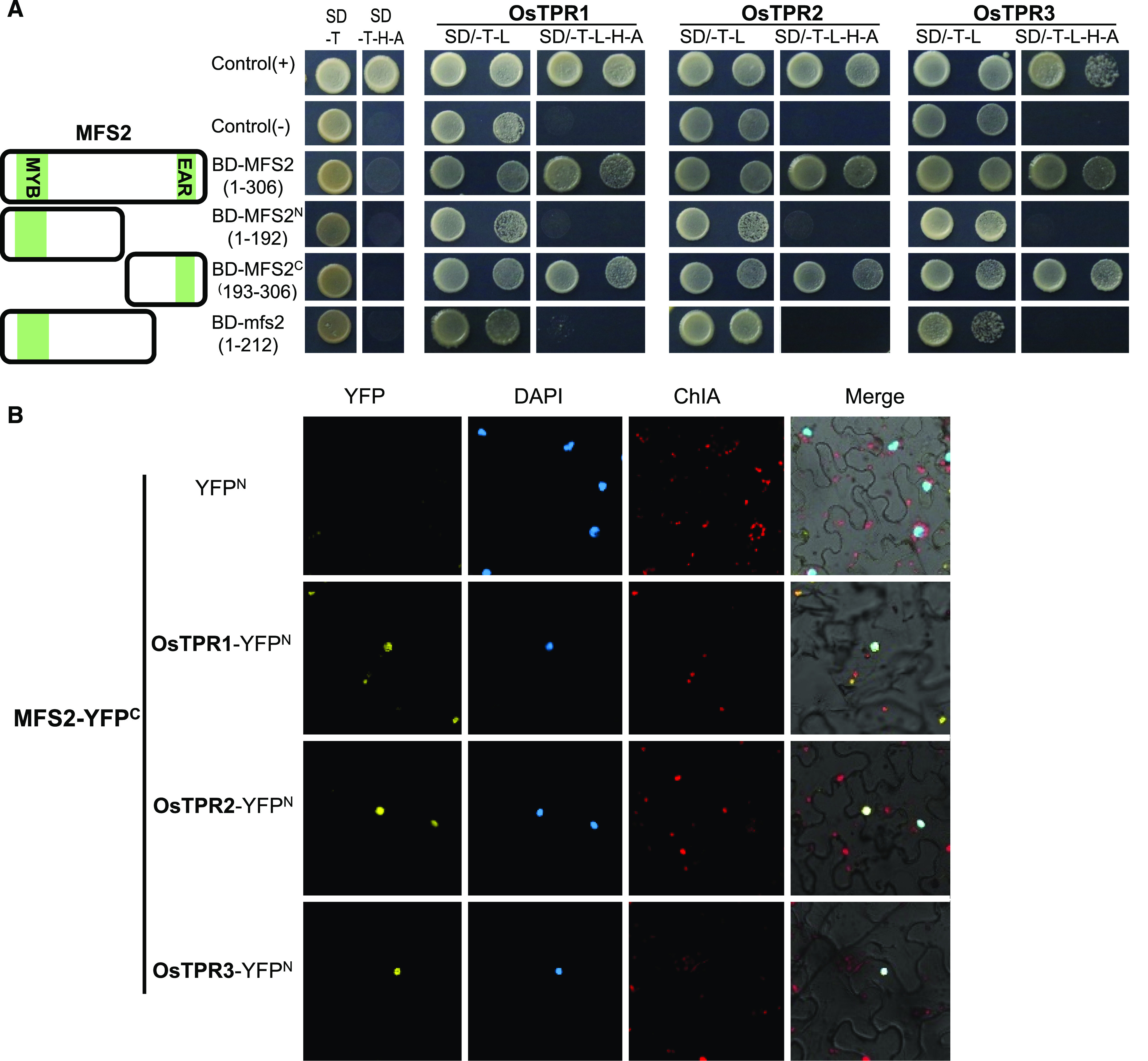
Analysis of MFS2 interaction with rice TPRs. A, Autoactivation and interaction with rice TPR1, TPR2, and TPR3 of MFS2 and its truncated proteins in a Y2H assay, respectively. B, Interaction analysis of MFS2 with TPR1, TPR2, and TPR3 in epidermal cells of N. benthamiana leaves in a BiFC assay, respectively.
We conducted a bimolecular fluorescence complementation (BiFC) assay to detect whether MSF2 and OsTPRs interacted in Nicotiana benthamiana leaves. Yellow fluorescent protein signal was detected in the nucleus of epidermal cells of N. benthamiana leaves that coexpressed the MFS2-YC fusion protein with YN-OsTPR1, YN-OsTPR2, or YN-OsTPR3 (Fig. 8B). This result further confirmed that MFS2 is capable of interaction with OsTPRs in plant cells, which implied that MFS2 acted as a transcriptional repressor and interacted with related corepressors.
DISCUSSION
MFS2 Regulates Palea Development
In the rice spikelet, the palea and the lemma together enclose the inner floral organs to shape and protect the grain, which is of considerable importance to the yield and quality of rice. The palea is considered to be distinct from the lemma in both identity and origin. The palea is believed to be a homolog of the prophyll (the first leaf produced by the axillary meristem), whereas the lemma corresponds to a bract (the leaf subtending the axillary meristem; Kellogg, 2001). The surface of the lemma is covered with silicified cells and burrs. Different from the lemma, the palea is a fused organ composed of the bop and the mrp. The bop is similar to the lemma in shape and structure, while the mrp is a smooth membranous structure, which can hook with the lemma to form a closed structure (Ohmori et al., 2009; Li et al., 2010).
In this study, mutation of MFS2 resulted in severely defective palea development. By analysis of phenotypes and expression of marker genes, we confirmed that, in type-I spikelets, the bop was reduced whereas the mrp was broader or more lemma-like to different degrees; in type-II spikelets, the bop was almost unaffected, and the mrp largely or entirely gained a lemma-like identity, which resulted in the whole palea resembling the lemma overall. Thus, loss of function of MFS2 affected either the bop or mrp, which suggests that MFS2 played a crucial role in both bop and mrp development. Previous studies have shown that several genes are involved in palea development. LHS1/OsMADS1 acts as an E-class gene and is responsible for lemma and palea specification, and mutation of it results in a leaf-like lemma and palea (Jeon et al., 2000; Prasad et al., 2005). The TH1 gene, which encodes a DUF640 protein, regulates the size of the lemma and palea, and the th1 mutant develops a smaller and triangular lemma and palea (Li et al., 2012). CCP1/DFO1, an EMBRYONIC FLOWER1 (EMF1)-like protein, plays an important role in palea development by maintaining H3K27me3-mediated epigenetic silence of OsMADS58, the ectopic expression of which causes pistil-like palea in the ccp1 mutant (Yan et al., 2015; Zheng et al., 2015). Several additional genes that specifically regulate only bop or mrp development have been isolated. DP1 encodes an AT-hook DNA binding protein and regulates bop growth. In the dp1 mutant, the bop is almost entirely lost, leaving two marginal leafy organs (Jin et al., 2011). REP1 belongs to the TCP gene family and is hypothesized to be a downstream gene of the DP1. In the rep1 mutant, the phenotype is weaker than that of the dp1 and the bop is strongly reduced, which results in a substantially smaller palea (Yuan et al., 2009). AH2 encodes a MYB domain protein and plays an important role in bop identity specification. In the ah2 mutant, the bop is degenerated to different degrees, but the mrp is not affected (Ren et al., 2019). In contrast to the aforementioned genes that regulate bop development, two MADS-box genes, MFO1/OsMADS6 and CFO1/OsMADS32, are required to specify mrp identity. In the mfo1 and cfo1 mutants, the mrp acquired entire features of the lemma, which results in transformation of the palea into a lemma (Ohmori et al., 2009; Li et al., 2010; Sang et al., 2012). Therefore, MFS2 mediation of bop development is similar to that of REP1, DP1, and AH2, whereas MFS2 specifies mrp identity in a similar manner to that of MFO1 or CFO1.
MFS2 Affects SM Determinacy
The fate of SM is associated with the number of lateral organs and florets produced in the spikelet. In wild-type rice, as one terminal FM is formed on the top of the SM, the determinate spikelet ultimately produces a fixed number of organs/florets, containing one pair of rudimentary glumes, two sterile lemmas, and one floret. In this study, the majority of type-II spikelets of the mfs2 spikelet displayed a lemma-like palea, four extra mrp-/palea-like structures, two pairs of lodicules, and an increased number of stamens and pistils. Therefore, this phenotype meant partial loss of SM determinacy, and formation of “two-florets” to a large degree. Similarly, in the mfs1 mutant, some spikelets contained an additional lemma-like organ, and in these spikelets the number of inner floral organs was often increased coincident with degradation of the palea (Ren et al., 2013). In the snb mutant, some spikelets develop supernumerary rudimentary glumes, additional lemma-like or palea-like structures, or lateral florets before the terminal floret emerges. The snb+ids1 double mutant produces a number of additional bract-like organs, including rudimentary glumes, lemmas, or paleas, compared with the corresponding single mutants (Lee et al., 2007; Lee and An, 2012). In this study, a substantial decrease in the expression level of SNB and OsIDS1 was observed in young panicles of the mfs2 mutant, but no distinct difference in mfs1 expression levels was detected between the mutant and the wild type. These results suggested that MFS2 might regulate SM fate to ensure the correct timing of the SM-to-FM transition by influencing the SNB and OsIDS1 pathways, but not the MFS1 pathway.
MFS2 Acts as a Transcription Repressor
Transcriptional repression complexes comprise a repressor and co-repressors, and play crucial roles in plant development. Plant TPL/TPR proteins are conserved transcription corepressors in the Groucho/Tup1 family, which commonly lack DNA-binding activity but can interact with the EAR motif of TFs. The TF–TPL complex then inhibits expression of the target gene by recruiting HDACs to transcription complexes and the chromatin state is changed from active to inactive (Long et al., 2006; Gonzalez et al., 2007; Liu and Karmarkar, 2008; Krogan and Long, 2009). In Arabidopsis, the A-function gene AP2 acts as a repressor, which interacts with the corepressor complex TPL-HDA19 to restrict expression of the C-function gene AG in whorls 1 and 2, and B-function genes AP3 and PISTILLATA and E-function gene SEPALLATA3 in sepals (Ó’Maoiléidigh et al., 2014). The rice genome contains three TPR proteins: OsTPR1, OsTPR2, and OsTPR3. However, little information is available on the mechanism of TF-TPR formation in rice. Mutation of OsTPR2/ASP1 caused a disorganized branching pattern, an elongated sterile lemma, and rudimentary glumes (Yoshida et al., 2012). Thus far, the functions of OsTPR1 and OsTPR3 have not been elucidated. Recently, NSG1 was identified as a transcriptional repressor, encoding a single C2H2 zinc finger domain and interacting with OsTPRs via its C-terminal EAR motif, to regulate spikelet development by restricting the expression of OsMADS1 in the rudimentary glumes and sterile lemma (Zhuang et al., 2020).
The MFS2 protein contained two typical EAR motifs, with EAR motif1 located within the MYB domain and EAR motif2 at the C terminus. A dual luciferase assay revealed that the MFS2 protein and all truncated proteins with one or both EAR motifs showed strong repression activity, and Y2H and BIFC assays indicated that the MFS2 protein and truncated proteins with the C-terminal EAR motif2 interacted with the three OsTPRs. Therefore, these data suggested that MFS2 might show a similar mechanism to that of NSG1, i.e. interacting with OsTPRs via the C-terminal EAR motif to repress the expression of downstream genes, thereby regulating spikelet development in rice. Interestingly, although it contained the EAR motif1, the repression activity of the mutant mfs2 protein was strongly decreased, compared with that of truncated proteins containing the same EAR motif1. We speculated that this difference was mainly because of the different protein structures. Owing to the presence of a longer C-terminal, EAR motif1 in the mfs2 protein might not be exposed to allow interaction with the TPR proteins and exertion of repression activity, whereas EAR motif1 might be exposed in truncated proteins MFS2-N (1–76) and MFS2-N (1–192) to exert distinct repression activity. Therefore, these results implied that in rice spikelet cells, as MYB domain harboring EAR motif1 is responsible for DNA binding, the EAR motif2 in the MFS2 protein may be responsible for interaction with OsTPRs to repress target genes. Identification of the direct targets of MFS2 will further improve our understanding of its function and mechanism in spikelet development in rice and other grass species.
CONCLUSION
In this research, we identified a multifloret mutant mfs2. The mfs2 spikelets displayed degraded/lemma-like palea, and an increased number of palea/mrp-like organs and other inner-whorl floral organs. The spikelet structure implied that mutation of MFS2 resulted in partial loss of SM determinacy and a tendency to produce two florets in one spikelet. MFS2 encoded a 1R-type MYB protein that contains a MYB domain and two EAR motifs, and displayed strong repression activity. MFS2 was capable of interaction with the three OsTPR corepressors via the C-terminal EAR motif. These data indicated that MFS2 acted as a repressor to regulate floral organ identities and SM determinacy in rice.
MATERIALS AND METHODS
Plant Materials
In this study, the mfs2 mutant was a natural mutant, and cv Jinhui 10 (cv J10; Oryza sativa ssp. indica) was used as the wild type. Both mfs2 mutant and cv J10 were all the offspring of cv Luhui 1089 (cv LH1089), a restorer line of hybrid rice. cv J10 is an oriented-genetic–improvement restorer line of cv LH1089 raised using backcross breeding by our group, and the mfs2 mutant was identified from an advanced backcross generation. Thus, the mfs2 mutant has a highly similar genetic background to that of cv J10. Therefore, we used cv J10 but not cv LH1089 as the wild type.
All plant materials were grown in experimental fields of the Rice Research Institute of Southwest University in Chongqing and Hainan, China.
Morphological and Microscopic Analysis of mfs2
The early spikelet development of mfs2 and wild-type plants was examined using a scanning electron microscope (SU3500; Hitachi) with a −20°C cool stage under a low-vacuum environment. At the flowering stage, spikelets from mfs2 and wild-type plants were observed using a scanning electron microscope (model no. SU3500; Hitachi) and a stereomicroscope (model no. SMZ1500; Nikon).
Paraffin Sectioning and Histological Analysis
Panicles at different developmental stages were collected and fixed in FAA (50% [v/v] ethanol, 0.9 m [m/v] glacial acetic acid, and 3.7% [v/v] formaldehyde) over 16 h at 4°C under vacuum. The fixed panicles were dehydrated with a graded ethanol series, infiltrated with xylene, and embedded in paraffin. Thin sections (8-mm thick) were prepared with a microtome (RM2245; Leica), then deparaffinized in xylene, and dehydrated through an ethanol series. The sections were stained sequentially with 1% (m/v) safranine O (Amresco) and 1% (m/v) fast green (Amresco), and a coverslip was mounted with neutral balsam. The sections were observed using a model no. E600 microscope (Nikon).
Map-Based Cloning of MFS2
The mfs2 mutant was crossed with cv Xinong 1A to generate F1 and F2 populations. The 1,464 F2 plants that exhibited a mutant phenotype were selected as a mapping population. Simple sequence repeats markers from publicly available rice databases, including Gramene (http://www.gramene.org) and the Rice Genomic Research Program (https://rgp.dna.affrc.go.jp/index.html.en) were used for fine-mapping of MFS2. The sequences of primers used in the mapping and candidate gene analysis are listed in Supplemental Table S1.
Vector Construction and Transformation
To construct a complementation plasmid, a 6,732-bp genomic fragment that contained the MFS2 coding sequence, coupled with the 2,282-bp upstream and 1,037-bp downstream sequences, was amplified using the primers MFS2-com-F and MFS2-com-R. The fragment was inserted into the binary vector pCAMBIA1300 using the pEASY-Uni Seamless Cloning and Assembly Kit (Transgene). The recombinant plasmids were transformed into the mfs2 mutant using the Agrobacterium tumefaciens-mediated transformation method as described in Xiao et al. (2009). To generate a construct for RNAi, we amplified a 161-bp fragment of MFS2 cDNA with the primers MFS2RiF (SacI and SpeI) and MFS2RiR (BamHI and KpnI). The resulting PCR products were ligated into the vector PTCK303. The recombinant plasmids were transformed into cv ZH11 plants using the A. tumefaciens-mediated transformation method. The primer sequences used are listed in Supplemental Table S1.
RNA Isolation and RT-qPCR Analysis
Total RNA from root, stem, leaf, sheath, panicle, and shoot of wild-type plants was isolated using the RNAprep Pure Plant Kit (Tiangen). The first-strand complementary cDNA was synthesized from 2 μg of total RNA using oligo(dT)18 primers in 20 μL of reaction volume using the PrimeScript Reagent Kit with gDNA Eraser (Takara). The RT-qPCR analysis was performed using the SYBR Premix Ex Taq II Kit (Takara) in an ABI 7500 Sequence Detection System (Applied Biosystems). At least three replicates were performed. ACTIN (OsRac1, LOC_Os01g12900) was used as an endogenous control.
In Situ Hybridization
Fresh young panicles from mfs2 and wild-type plants were fixed immediately in FAA solution, then dehydrated, infiltrated, embedded, and sectioned as described in Sang et al. (2012). The 426-bp gene-specific MFS2 probe was amplified with the primers MYBishF1 and MYBishR1 and labeled using the DIG RNA Labeling Kit (catalog no. SP6/T7; Roche). Other RNA probes for known floral-organ genes were generated in vitro using the same method. Images were captured with a model no. E600 microscope (Nikon). Sequences of primers used for in situ hybridization are listed in Supplemental Table S1.
Subcellular Localization
The full-length coding region (without the termination codon) of MFS2 was amplified using the primers OEMFS2-F1 and OEMFS2-R1 (Supplemental Table S1). The fragment was cloned into the expression cassette 35S-GFP -NOS (pCAMBIA1301) with appropriate modifications, which generated the MFS2-GFP fusion vector. The pA7-GFP and MFS2-GFP plasmids were transformed into rice protoplasts. After incubation for 12 to 16 h at 28°C, GFP fluorescence was detected using a confocal laser scanning microscope (model no. LSM710; Zeiss).
Protein Sequence and Phylogenetic Analysis
Protein sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/genbank/) using the MFS2 sequence as a query. A phylogenetic tree was constructed using the software MEGA 5.0 (Tamura et al., 2011). The tree was constructed using the maximum likelihood method based on the JTT matrix-based model with the lowest Bayesian information criterion scores (Jones et al., 1992; Tamura et al., 2011). Statistical support for the tree topology was assessed by means of a bootstrap analysis with 500 replicates.
Transcriptional Activity Analysis
Transcriptional activity of full-length and truncated MFS2 proteins were analyzed in rice protoplasts using the dual luciferase reporter assay system. A GloMax 20-20 Luminometer (Promega) was used to measure the relative luciferase activity. The primers used are listed in Supplemental Table S1.
Y2H Assays
The full-length coding region of MFS2, MFS21-192, MFS2193-306, and mfs2 were amplified and ligated into the yeast expression vector pGBKT7 to detect autoactivation of the MFS2 and mfs2 protein. The constructs pGBKT7-MFS2 and pGBKT7 (used as a blank control) were transformed separately into the yeast strain Y2HGold. Transformants were selected on synthetic dropout medium (SD)/-Trp or SD/-Ade/-His/-Trp medium. The primers used are listed in Supplemental Table S1.
The full-length coding sequences of OsTPR1, OsTPR2, and OsTPR3 were fused to the transcriptional activation domain of GAL4 (pGADT7) to serve as bait proteins and prey proteins, respectively. The bait and prey plasmids were transformed into the yeast strain Y2HGold. The Matchmaker Two-Hybrid System 3 (Clontech) was used to detect interactions. Each interaction was tested at least three times.
BiFC Assays
To conduct BiFC assays, the full-length coding region of MFS2 and OsTPRs were amplified by gene-specific primers and then ligated into BiFC vectors, including pSCYNE (SCN, modified) and pSCYCE (SCC, modified). The recombinant vector pairs were cotransformed into Nicotiana benthamiana leaves. The fluorescence signals were captured under a confocal laser scanning microscope (model no. LSM 800; Zeiss). The primers used are listed in Supplemental Table S1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers: SNB (ABD24033), OsIDS1 (NM_001058244), MFS1 (AK242393), DL (AB106553), OsMADS1 (NM_001055911), OsMADS6 (FJ666318), OsTPR1 (AP014957), OsTPR2 (AK111830), and OsTPR3 (AP014959).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Percentage of defective organs in mfs2 spikelets.
Supplemental Figure S2. Expression analysis of related floral organ development genes in wild-type and mfs2 plants.
Supplemental Figure S3. In situ hybridization of OsMADS2 in wild-type and mfs2 spikelets.
Supplemental Figure S4. Expression analysis of related spikelet development genes in wild-type and mfs2 plants.
Supplemental Figure S5. RNAi analysis of MFS2.
Supplemental Figure S6. Phylogenetic evolutionary analysis of the MFS2-like genes.
Supplemental Figure S7. Protein sequence analysis of the MFS2-like genes.
Supplemental Figure S8. Sequence alignment between normal and mutant MFS2 protein.
Supplemental Table S1. Primers used in the study.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31971919, 31730063, and 31271304), and the National Key Program for Research and Development (grant no. 2017YFD0100202).
References
- Alvarez J, Guli CL, Yu X-H, Smyth DR(1992) Terminal flower: A gene affecting inflorescence development in Arabidopsis thaliana. Plant J 2: 103–116 [Google Scholar]
- Bartlett ME, Thompson B(2014) Meristem identity and phyllotaxis in inflorescence development. Front Plant Sci 5: 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Soowal LN, Lee I, Weigel D(1997) LEAFY expression and flower initiation in Arabidopsis. Development 124: 3835–3844 [DOI] [PubMed] [Google Scholar]
- Bradley D, Ratcliffe O, Vincent C, Carpenter R, Coen E(1997) Inflorescence commitment and architecture in Arabidopsis. Science 275: 80–83 [DOI] [PubMed] [Google Scholar]
- Chen Y, Yang XY, He K, Liu M, Gao Z, Lin Z, Zhang Y, Wang X(2006) The MYB transcription factor superfamily of Arobidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60: 107–124 [DOI] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S(2008) Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135: 3013–3019 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM(1991) The war of the whorls: Genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Coen ES, Nugent JM(1994) Evolution of flowers and inflorescences. Development 1994: 107 [Google Scholar]
- Dreni L, Pilatone A, Yun D, Erreni S, Pajoro A, Caporali E, Zhang D, Kater MM(2011) Functional analysis of all AGAMOUS subfamily members in rice reveals their roles in reproductive organ identity determination and meristem determinacy. Plant Cell 23: 2850–2863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L(2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15: 573–581 [DOI] [PubMed] [Google Scholar]
- Gonzalez D, Bowen AJ, Carroll TS, Conlan RS(2007) The transcription corepressor LEUNIG interacts with the histone deacetylase HDA19 and mediator components MED14 (SWP) and CDK8 (HEN3) to repress transcription. Mol Cell Biol 27: 5306–5315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L, Liang W, Yin C, Cui X, Zong J, Wang X, Hu J, Zhang D(2011) Rice MADS3 regulates ROS homeostasis during late anther development. Plant Cell 23: 515–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Sunohara H, Nagato Y(2004) Developmental course of inflorescence and spikelet in rice. Japan J Breeding 54: 147–156 [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y(2005) Rice plant development: From zygote to spikelet. Plant Cell Physiol 46: 23–47 [DOI] [PubMed] [Google Scholar]
- Jeon J-S, Jang S, Lee S, Nam J, Kim C, Lee S-H, Chung Y-Y, Kim S-R, Lee YH, Cho Y-G, et al. (2000) leafy hull sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. Plant Cell 12: 871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Luo Q, Tong H, Wang A, Cheng Z, Tang J, Li D, Zhao X, Li X, Wan J, et al. (2011) An AT-hook gene is required for palea formation and floral organ number control in rice. Dev Biol 359: 277–288 [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM(1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8: 275–282 [DOI] [PubMed] [Google Scholar]
- Kaneko M, Inukai Y, Ueguchi-Tanaka M, Itoh H, Izawa T, Kobayashi Y, Hattori T, Miyao A, Hirochika H, Ashikari M, et al. (2004) Loss-of-function mutations of the rice GAMYB gene impair α-amylase expression in aleurone and flower development. Plant Cell 16: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg EA.(2001) Evolutionary history of the grasses. Plant Physiol 125: 1198–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanday I, Yadav SR, Vijayraghavan U(2013) Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiol 161: 1970–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J(2010) PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol 51: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NT, Long JA(2009) Why so repressed? Turning off transcription during plant growth and development. Curr Opin Plant Biol 12: 628–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D-Y, An G(2012) Two AP2 family genes, supernumerary bract (SNB) and Osindeterminate spikelet 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. Plant J 69: 445–461 [DOI] [PubMed] [Google Scholar]
- Lee D-Y, Lee J, Moon S, Park SY, An G(2007) The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. Plant J 49: 64–78 [DOI] [PubMed] [Google Scholar]
- Li H, Liang W, Jia R, Yin C, Zong J, Kong H, Zhang D(2010) The AGL6-like gene OsMADS6 regulates floral organ and meristem identities in rice. Cell Res 20: 299–313 [DOI] [PubMed] [Google Scholar]
- Li X, Sun L, Tan L, Liu F, Zhu Z, Fu Y, Sun X, Sun X, Xie D, Sun C(2012) TH1, a DUF640 domain-like gene controls lemma and palea development in rice. Plant Mol Biol 78: 351–359 [DOI] [PubMed] [Google Scholar]
- Liu Z, Karmarkar V(2008) Groucho/Tup1 family co-repressors in plant development. Trends Plant Sci 13: 137–144 [DOI] [PubMed] [Google Scholar]
- Long JA, Ohno C, Smith ZR, Meyerowitz EM(2006) TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312: 1520–1523 [DOI] [PubMed] [Google Scholar]
- Long JA, Woody S, Poethig S, Meyerowitz EM, Barton MK(2002) Transformation of shoots into roots in Arabidopsis embryos mutant at the TOPLESS locus. Development 129: 2797–2806 [DOI] [PubMed] [Google Scholar]
- Luo Q, Zhou K, Zhao X, Zeng Q, Xia H, Zhai W, Xu J, Wu X, Yang H, Zhu L(2005) Identification and fine mapping of a mutant gene for palealess spikelet in rice. Planta 221: 222–230 [DOI] [PubMed] [Google Scholar]
- Malcomber ST, Kellogg EA(2004) Heterogeneous expression patterns and separate roles of the SEPALLATA gene LEAFY HULL STERILE1 in grasses. Plant Cell 16: 1692–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J(2000) A floret by any other name: Control of meristem identity in maize. Trends Plant Sci 5: 61–66 [DOI] [PubMed] [Google Scholar]
- Mimida N, Goto K, Kobayashi Y, Araki T, Ahn JH, Weigel D, Murata M, Motoyoshi F, Sakamoto W(2001) Functional divergence of the TFL1-like gene family in Arabidopsis revealed by characterization of a novel homologue. Genes Cells 6: 327–336 [DOI] [PubMed] [Google Scholar]
- Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y(2003) SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice. Development 130: 705–718 [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Shimamoto K, Kyozuka J(2002) Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J 29: 743–750 [DOI] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H(2009) MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. Plant Cell 21: 3008–3025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó’Maoiléidigh DS, Graciet E, Wellmer F(2014) Gene networks controlling Arabidopsis thaliana flower development. New Phytol 201: 16–30 [DOI] [PubMed] [Google Scholar]
- Prasad K, Parameswaran S, Vijayraghavan U(2005) OsMADS1, a rice MADS-box factor, controls differentiation of specific cell types in the lemma and palea and is an early-acting regulator of inner floral organs. Plant J 43: 915–928 [DOI] [PubMed] [Google Scholar]
- Prasad K, Sriram P, Kumar CS, Kushalappa K, Vijayraghavan U(2001) Ectopic expression of rice OsMADS1 reveals a role in specifying the lemma and palea, grass floral organs analogous to sepals. Dev Genes Evol 211: 281–290 [DOI] [PubMed] [Google Scholar]
- Prusinkiewicz P, Erasmus Y, Lane B, Harder LD, Coen E(2007) Evolution and development of inflorescence architectures. Science 316: 1452–1456 [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U(2008) Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 105: 3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Bradley DJ, Coen ES(1999) Separation of shoot and floral identity in Arabidopsis. Development 126: 1109–1120 [DOI] [PubMed] [Google Scholar]
- Ren D, Cui Y, Hu H, Xu Q, Rao Y, Yu X, Zhang Y, Wang Y, Peng Y, Zeng D, et al. (2019) AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J 100: 813–824 [DOI] [PubMed] [Google Scholar]
- Ren D, Li Y, Zhao F, Sang X, Shi J, Wang N, Guo S, Ling Y, Zhang C, Yang Z, et al. (2013) MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol 162: 872–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang X, Li Y, Luo Z, Ren D, Fang L, Wang N, Zhao F, Ling Y, Yang Z, Liu Y, et al. (2012) CHIMERIC FLORAL ORGANS1, encoding a monocot-specific MADS box protein, regulates floral organ identity in rice. Plant Physiol 160: 788–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussex IM, Kerk NM(2001) The evolution of plant architecture. Curr Opin Plant Biol 4: 33–37 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S(2011) MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tang D, Hong L, Xu W, Huang J, Li M, Gu M, Xue Y, Cheng Z(2010) DEP and AFO regulate reproductive habit in rice. PLoS Genet 6: e1000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Tang J, Li Y, Wang W, Li X, Jin L, Xie R, Luo H, Zhao X, Meng Z, et al. (2009) STAMENLESS 1, encoding a single C2H2 zinc finger protein, regulates floral organ identity in rice. Plant J 59: 789–801 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano H-Y(2006) Functional diversification of the two C-class MADS box genes OSMADS3 and OSMADS58 in Oryza sativa. Plant Cell 18: 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Zhang X, Zhang L, Ye S, Zeng L, Liu J, Li Q, He Z(2015) Curved chimeric palea 1 encoding an EMF1-like protein maintains epigenetic repression of OsMADS58 in rice palea development. Plant J 82: 12–24 [DOI] [PubMed] [Google Scholar]
- Yao S-G, Ohmori S, Kimizu M, Yoshida H(2008) Unequal genetic redundancy of rice PISTILLATA orthologs, OsMADS2 and OsMADS4, in lodicule and stamen development. Plant Cell Physiol 49: 853–857 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Ohmori Y, Kitano H, Taguchi-Shiobara F, Hirano H-Y(2012) Aberrant spikelet and panicle1, encoding a TOPLESS-related transcriptional co-repressor, is involved in the regulation of meristem fate in rice. Plant J 70: 327–339 [DOI] [PubMed] [Google Scholar]
- Yuan Z, Gao S, Xue D-W, Luo D, Li L-T, Ding S-Y, Yao X, Wilson ZA, Qian Q, Zhang D-B(2009) RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol 149: 235–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn LM, Kong H, Leebens-Mack JH, Kim S, Soltis PS, Landherr LL, Soltis DE, Depamphilis CW, Ma H(2005) The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169: 2209–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G-H, Xu Q, Zhu X-D, Qian Q, Xue H-W(2009) SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 21: 719–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Liang W, Yang X, Luo X, Jiang N, Ma H, Zhang D(2010) Carbon starved anther encodes a MYB domain protein that regulates sugar partitioning required for rice pollen development. Plant Cell 22: 672–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Wang Y, Wang Y, Wang C, Ren Y, Lv J, Peng C, Wu T, Liu K, Zhao S, et al. (2015) DEFORMED FLORAL ORGAN1 (DFO1) regulates floral organ identity by epigenetically repressing the expression of OsMADS58 in rice (Oryza sativa). New Phytol 206: 1476–1490 [DOI] [PubMed] [Google Scholar]
- Zhu Q-H, Ramm K, Shivakkumar R, Dennis ES, Upadhyaya NM(2004) The ANTHER INDEHISCENCE1 gene encoding a single MYB domain protein is involved in anther development in rice. Plant Physiol 135: 1514–1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liang W, Cui X, Chen M, Yin C, Luo Z, Zhu J, Lucas WJ, Wang Z, Zhang D(2015) Brassinosteroids promote development of rice pollen grains and seeds by triggering expression of Carbon Starved Anther, a MYB domain protein. Plant J 82: 570–581 [DOI] [PubMed] [Google Scholar]
- Zhuang H, Wang H-L, Zhang T, Zeng X-Q, Chen H, Wang Z-W, Zhang J, Zheng H, Tang J, Ling Y-H, et al. (2020) NONSTOP GLUMES1 encodes a C2H2 zinc finger protein that regulates spikelet development in rice. Plant Cell 32: 392–413 [DOI] [PMC free article] [PubMed] [Google Scholar]