Characterization of the TCP4-WRI1 complex regulating Arabidopsis seed oil biosynthesis expands our understanding of WRI1-interacting factors and describes a further role for TCP transcription factors.
Abstract
Cross-family transcription factor (TF) interactions play critical roles in the regulation of plant developmental and metabolic pathways. WRINKLED1 (WRI1) is a key TF governing oil biosynthesis in plants. However, little is known about WRI1-interacting factors and their roles in oil biosynthesis. We screened a TF library using Arabidopsis (Arabidopsis thaliana) WRI1 (AtWRI1) as bait in yeast two-hybrid assays and identified three TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP) family TFs, namely TCP4, TCP10, and TCP24, as AtWRI1-interacting partners. The physical interaction between AtWRI1 and TCPs was further validated using bimolecular fluorescence complementation assays. TCPs play important roles in various plant developmental processes; however, their involvement in fatty acid biosynthesis was not previously known. Coexpression of TCP4, but not TCP10 or TCP24, with AtWRI1 reduced AtWRI1-mediated oil biosynthesis in Nicotiana benthamiana leaves. Transcriptomic analysis in transgenic Arabidopsis plants with enhanced TCP4 activity engineered by expressing rTCP4 (i.e. miR319-resistant TCP4) revealed that AtWRI1 target genes were significantly repressed. TCP4 expression is strongly correlated with AtWRI1 during embryo development. A tcp4 loss-of-function mutant, the jaw-D mutant with a strong reduction of TCP4 expression, and a tcp2 tcp4 tcp10 triple mutant accumulated more seed oil than wild-type Arabidopsis. In addition, TCP4 repressed the AtWRI1-mediated transactivation of the promoters of fatty acid biosynthetic genes. Collectively, our findings suggest that TCP4 represses fatty acid biosynthetic gene expression through interaction with AtWRI1, leading to a reduction of AtWRI1-mediated seed oil accumulation.
Plant oils are important for both human diet and industry as renewable feedstocks. Many plants synthesize triacylglycerol (TAG) in seeds as a crucial mechanism for storing and providing energy for seedling development (Chapman and Ohlrogge, 2012). The APETALA2 (AP2) transcription factor (TF), WRINKLED1 (WRI1), is a key regulator of plant oil biosynthesis (Cernac and Benning, 2004; Masaki et al., 2005; Kong and Ma, 2018). The Arabidopsis (Arabidopsis thaliana) loss-of-function mutant wri1-1 has ∼80% reduced seed oil content compared with the wild-type plant (Focks and Benning, 1998). Microarray analysis revealed that the repressed genes in the wri1-1 mutant mainly encode fatty acid biosynthetic and late glycolytic enzymes (Ruuska et al., 2002). Subsequent studies validated that the genes encoding proteins of late glycolysis and fatty acid biosynthesis are targets of AtWRI1 (Baud et al., 2007; Maeo et al., 2009). WRI1 orthologs have been identified from many plant species, such as Brassica napus (Liu et al., 2010), Zea mays (Shen et al., 2010), Elaeis guineensis (Ma et al., 2013), Brachypodium distachyon (Yang et al., 2015), Glycine max (Chen et al., 2020), and rice (Oryza sativa; Mano et al., 2019). In addition, transgenic plants overexpressing AtWRI1 or other WRI1 orthologs produce higher oil in seeds and leaves (Cernac and Benning, 2004; Shen et al., 2010; Yang et al., 2015). AtWRI1 is regulated at the posttranslational level and subject to 26S proteasome-mediated degradation (Chen et al., 2013). A PEST motif in the C-terminal intrinsically disordered region of AtWRI1 mediates the protein stability, which, in turn, affects oil biosynthesis (Ma et al., 2015). The 14-3-3 proteins interact with AtWRI1 in the mediation of AtWRI1 stability, transcriptional activity, and, consequently, oil production (Ma et al., 2016). In addition, phosphorylation of AtWRI1 by the Suc nonfermentation-related kinase KIN10 was recently reported to be crucial for AtWRI1 degradation (Zhai et al., 2017). A subsequent study showed that trehalose 6-phosphate stabilizes AtWRI1, resulting in increased fatty acid production through KIN10 activity suppression (Zhai et al., 2018).
Protein interactions between TFs from the same family or different families play crucial roles in regulating metabolic or developmental pathways. In Arabidopsis, about half of more than 2,000 TF–TF interactions are cross-family interactions (Bemer et al., 2017). Transcriptional networks are frequently formed by cross-family TF interaction, which enables the integration of various signals that dictate the outcome of a plant metabolic or developmental pathway. However, the potential interactions of AtWRI1 with other TFs in oil biosynthesis are not known. TFs frequently involved in cross-family interaction form transcriptional hubs (Bemer et al., 2017). One such TF family is TEOSINTE BRANCHED1/CYCLOIDEA/PROLIFERATING CELL FACTOR (TCP). TCPs are plant-specific TFs that play important roles in diverse biological processes, such as shoot apical meristem and leaf development, phytohormone biosynthesis, regulation of circadian clock rhythm, and immunity (Palatnik et al., 2003; Schommer et al., 2008; Giraud et al., 2010; Li et al., 2012; Kim et al., 2014a). In Arabidopsis, 24 TCP TFs are divided into class I and class II subfamilies (Cubas et al., 1999; Palatnik et al., 2003). Of note for our study, the Arabidopsis jaw-D mutant overproducing the microRNA319 (miRNA319) leads to the repression of several class II TCPs, including TCP2, TCP3, TCP4, TCP10, and TCP24 (Palatnik et al., 2003). The miRNA319-regulated TCPs interact with the TF ASYMMETRIC LEAVES2, thus repressing class I KNOX genes (Li et al., 2012). Arabidopsis TCPs show unique and redundant functions (Danisman et al., 2013). TCP3 interacts with MYB12 or MYB111, thus regulating the expression of flavonoid biosynthetic genes (Li and Zachgo, 2013). Six TCPs have been identified as interacting partners of SUPPRESSOR OF rps4-RLD1 (SRFR1), and the interaction between SRFR1 and TCPs is a key mechanism to balance plant development and immune responses (Kim et al., 2014a). TCPs also interact with key circadian regulators (e.g. CCA1 and LHY; Giraud et al., 2010). Therefore, TCPs are known to control plant development, defense, and redox regulation. However, no report thus far indicates the involvement of TCPs in regulating oil biosynthesis.
In this work, we found that AtWRI1 interacted with TCPs in the regulation of fatty acid biosynthesis. We discovered that expression of TCP4 displayed strong correlation with AtWRI1 during embryo development and that TCP4 negatively affected AtWRI1-stimulated oil biosynthesis. Arabidopsis loss-of-function mutants, including tcp4, tcp2 tcp4 tcp10, and jaw-D (overproducing miRNA319, resulting in reduced expression of TCPs), were found to have higher seed oil content compared with the wild type. Our findings suggest that TCP4 acts as a transcriptional repressor of AtWRI1 through protein-protein interactions. This interaction possibly plays a role in the fine-tuning of plant oil biosynthesis.
RESULTS
TCPs Physically Interact with AtWRI1 in Yeast and Plant Cells
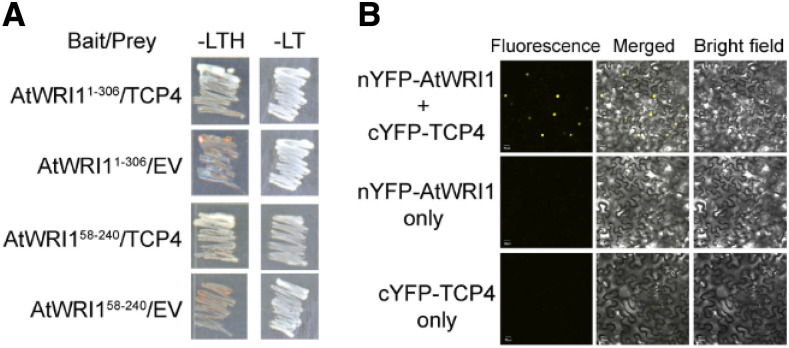
TFs function in the nucleus. We demonstrated that a nuclear localization signal at the N terminus (AtWRI133-41) enables AtWRI1 to localize to the nucleus, where it likely interacts with other factors in the regulation of target gene expression (Supplemental Fig. S1). In order to identify AtWRI1-interacting factors, we employed the yeast two-hybrid (Y2H) assay to screen an Arabidopsis TF library (Kim et al., 2014b). We used two truncated AtWRI1 variants (AtWRI11-306 and AtWRI158-240) as baits for the screening, as the AtWRI1 C-terminal part of the protein shows high autoactivation in yeast cells (Ma et al., 2015). Our initial assay identified three TCP family TFs, namely TCP4, TCP10, and TCP24, as interacting partners of AtWRI1. We next performed independent Y2H assays to confirm the interaction between the TCPs and AtWRI1. Our results showed that TCP4, TCP10, and TCP24 interacted with AtWRI1 in yeast cells (Fig. 1A; Supplemental Fig. S2A). To further validate the physical AtWRI1-TCP interaction in plant cells, we performed bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana leaves. For this purpose, the coding sequences for the TCPs (TCP4, TCP10, and TCP24) were individually fused at their N-terminal ends to the C-terminal half of YFP (cYFP), and the N-terminal half of YFP (nYFP) was fused to the N terminus of AtWRI1. Reconstituted YFP fluorescence was detected in the nucleus when nYFP-AtWRI1 and cYFP-TCPs were transiently coexpressed in N. benthamiana leaves (Fig. 1B; Supplemental Fig. S2B). YFP signal was not detected when only one of the fusion proteins, meaning nYFP-AtWRI1 or one of the cYFP-TCP fusions, was present (Fig. 1B; Supplemental Fig. S2B). Collectively, our results suggest that AtWRI1 physically interacts with TCPs.
Figure 1.
The TCP4 TF physically interacts with AtWRI1. A, TCP4 shows physical interaction with AtWRI1 variants in the Y2H assay. Yeast growth on either stringent selective (-Leu/-Trp/-His [-LTH]) or permissive (-Leu/-Trp [-LT]) medium is shown. Empty vector (EV) was used as a negative control. B, AtWRI1 interacts with the TCP4 TF in planta. A BiFC assay was performed to validate the interaction between AtWRI1 and TCP4. N. benthamiana epidermal cells were transiently cotransformed with plasmids encoding nYFP-AtWRI1 and cYFP-TCP4 as indicated. Bars = 20 μm.
Expression of TCP4 Strongly Correlates with AtWRI1 during Seed Development
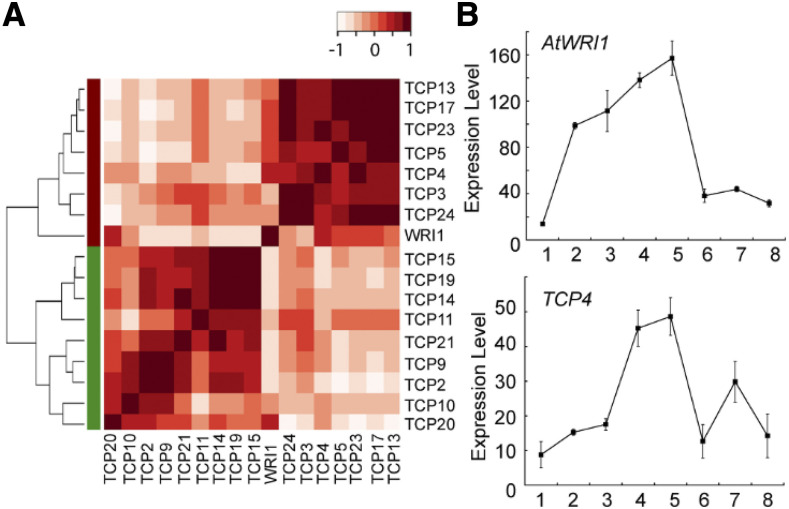
The Arabidopsis genome harbors 24 TCPs, which are broadly divided into two major groups, namely class I and class II (Supplemental Fig. S3). The class II TCPs are further divided into two subgroups: The CINCINNATA clade-containing genes are involved in lateral organ development and the CYC/TB1/ECE clade-containing genes are involved in axillary meristem development. TCP4, TCP10 and TCP24 belong to the CINCINNATA subclade. To determine the temporal expression correlation of TCPs with AtWRI1, we conducted coexpression analysis of AtWRI1 and TCPs at various stages of Arabidopsis embryo development (Le et al., 2010). Expression data of Arabidopsis embryos at five developmental stages (24 h, globular, cotyledon, mature-green, and post-mature-green stages) were used to generate a heat map. Expression of 16 of 24 TCPs was detected during embryo development. Hierarchical clustering analysis showed that TCPs form two major clusters, as indicated in Figure 2A by red and green bars, and AtWRI1 is grouped together with TCP4 and TCP24, whereas TCP10 is found in the other cluster. TCP4 displayed strong correlation with the expression of AtWRI1 during embryo development (Fig. 2A). In addition, using the eFP browser (Winter et al., 2007), we also found that TCP4 expression during embryo development mirrors that of AtWRI1 (Fig. 2B). Although TCP10 and TCP24 are expressed during embryo development, their expression patterns are different from that of WRI1 (Supplemental Fig. S4). Taken together, the spatial and temporal expression relationship of their respective genes suggests that AtWRI1 and TCP4 meet the criteria for possible interaction in the regulation of seed oil biosynthesis.
Figure 2.
In silico analysis of AtWRI1 and TCP expression. A, Coexpression of AtWRI1 with TCPs in Arabidopsis embryo tissues. Hierarchical clustering and heat map shows that AtWRI1 strongly correlates with and clusters with TCP4. B, Expression profiles of AtWRI1 and TCP4 during various stages of embryo development. Expression data are taken from the Arabidopsis eFP browser. Points on the graphs are as follows: (1) seed stage 3 with siliques; (2) seed stage 4 with siliques; (3) seed stage 5 with siliques; (4) seed stage 6 without siliques; (5) seed stage 7 without siliques; (6) seed stage 8 without siliques; (7) seed stage 9 without siliques; and (8) seed stage 10 without siliques. Results are shown as means ± sd (n = 3). Experimental details were described for the eFP browser (Winter et al., 2007).
TCP4 Represses AtWRI1-Regulated Oil Biosynthesis
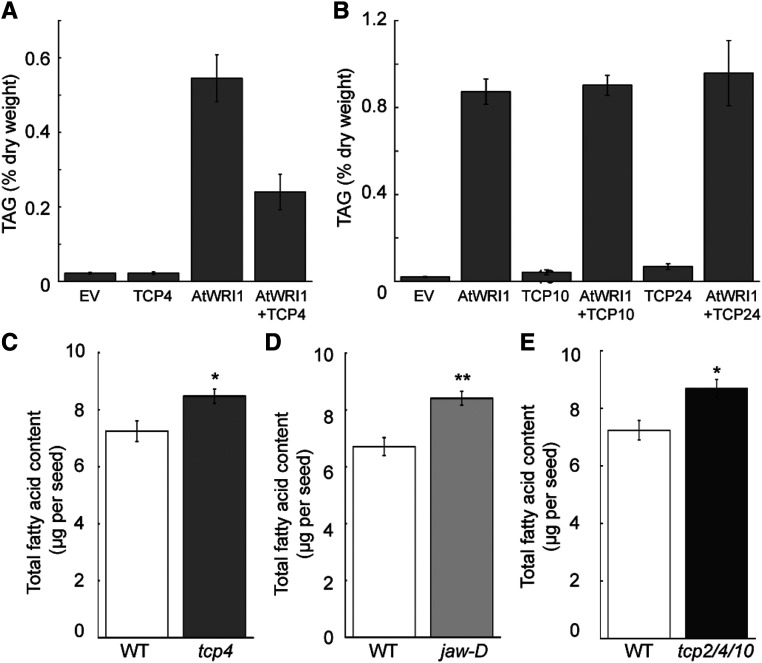
Recent studies have validated that the transient expression of coding sequences for plant oil regulators in N. benthamiana leaves followed by TAG analysis is a robust and fast approach to study the regulation of plant oil biosynthesis (Vanhercke et al., 2013; Ma et al., 2015). We therefore evaluated the effects of the TCPs on AtWRI1-regulated oil biosynthesis using an established N. benthamiana transient expression system (Ma et al., 2015). As demonstrated previously, expression of AtWRI1 induced oil accumulation in N. benthamiana leaves; however, coexpression of TCP4 with AtWRI1 significantly reduced oil production compared with expression of AtWRI1 alone (Fig. 3A). By contrast, coexpression of TCP10 or TCP24 with AtWRI1 had no effect on AtWRI1-mediated oil biosynthesis. Expression of TCP4, TCP10, or TCP24 alone did not lead to oil production (Fig. 3, A and B).
Figure 3.
TCP effects on AtWRI1-mediated plant oil biosynthesis. A, TAG content in N. benthamiana leaves transiently coproducing AtWRI1 and TCP4. Results are shown as means ± se (n = 3). B, TAG content in N. benthamiana leaves transiently coproducing AtWRI1 and TCP10 or TCP24. Results are shown as means ± se (n = 3–4). Empty vector (EV) was used as a control. C, Fatty acid content of seeds of the wild type (WT) and tcp4. Results are shown as means ± se (n = 3). D, Fatty acid content of seeds of the wild type and jaw-D. Results are shown as means ± se (n = 3–4). E, Fatty acid content of seeds of the wild type and tcp2 tcp4 tcp10. Results are shown as means ± se (n = 3). Asterisks indicate significant differences between the mutant and the wild type by one-way ANOVA (*P < 0.05 and **P < 0.01).
To gain further insight into the function of TCP4 in AtWRI1-regulated oil biosynthesis, we searched publicly available transcriptome data of TCP4 transgenic plants. We examined the previously published microarray data of rTCP4 transgenic Arabidopsis, in which the miR319-resistant TCP4 is more abundantly expressed (Schommer et al., 2008). We discovered that the expression of AtWRI1 target genes in the oil biosynthetic pathway (e.g. BCCP2, ACP1, and PKP-β1) was down-regulated in rTCP4 transgenic plants compared with that in lines producing the native form of TCP4 (Supplemental Fig. S5). We hence hypothesized that TCP4 plays a role in repressing oil biosynthesis through interaction with AtWRI1.
The tcp4 Mutant Accumulates Higher Seed Oil Compared with the Wild Type
We subsequently measured the seed oil content in the tcp4 loss-of-function mutant and found a significant increase (17%) compared with the wild type (Fig. 3C). The activation-tagging mutant jaw-D exhibited increased levels of miRNA319, which consequently led to reduced expression of several miRNA319-targeted TCPs, including TCP2, TCP3, TCP4, TCP10, and TCP24 (Palatnik et al., 2003). Among the several miRNA319a-targeted TCPs, expression of TCP4 displayed the strongest reduction in the jaw-D mutant (Palatnik et al., 2003). When we measured the seed oil content of the jaw-D mutant, it was significantly increased (23%) compared with the wild type (Fig. 3D). We also measured the seed oil content of the tcp2 tcp4 tcp10 triple mutant. Notably, the tcp4 allele in this triple mutant is another allele (SAIL_1174_02), different from the tcp4 single mutant in Figure 3C. The seed oil content of the triple mutant was 20% higher compared with the wild type (Fig. 3E).
TCP4 Represses AtWRI1 Transactivation Activity
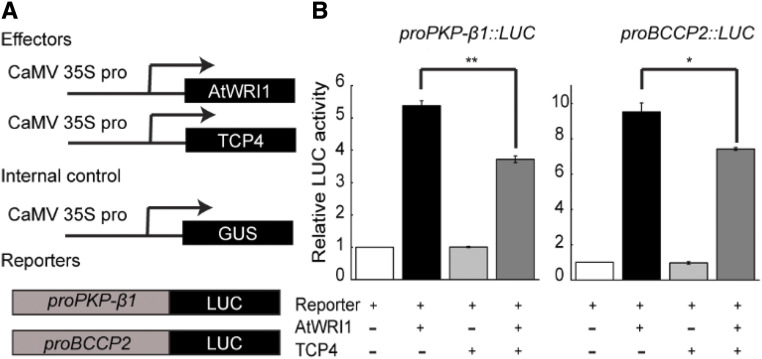
We next examined whether the interaction of TCP4 with AtWRI1 affects AtWRI1 transactivation activity in plant cells. We used a transient expression system (Pattanaik et al., 2010) in which the promoters of two AtWRI1 target genes (proBCCP2 and proPKP-β1) were fused to the firefly Luciferase (LUC) reporter gene. Vectors expressing AtWRI1 and TCP4, driven by the cauliflower mosaic virus (CaMV) 35S promoter, served as effectors. The reporters were introduced into Nicotiana tabacum protoplasts, alone or in combination with the effectors (Fig. 4A). Cointroduction of AtWRI1 with proBCCP2-LUC or proPKP-β1-LUC led to a significant increase in LUC activity, indicating that AtWRI1 activated the promoters of BCCP2 and PKP-β1. However, coexpression of TCP4 with AtWRI1 significantly reduced the activation by AtWRI1 (Fig. 4B). TCP4 by itself did not affect the promoter-driven LUC activity (Fig. 4B). These results indicate that the interaction between TCP4 and AtWRI1 represses the transactivation activity of AtWRI1.
Figure 4.
TCP4 repressed the transactivation activity of AtWRI1. A, Schematic representation of constructs used in a transient expression assay in protoplasts. B, Coexpression of TCP4 with AtWRI1 repressed the transactivation activity of AtWRI1 on the promoters of PKP-β1 and BCCP2 in protoplasts. Results are shown as means ± se (n = 3). Asterisks indicate significant differences by one-way ANOVA (*P < 0.05 and **P < 0.01) between AtWRI1 alone and coproduction of TCP4 as indicated.
DISCUSSION
TFs often work together in a complex. Interactions between TFs of the same or different families, evident in numerous developmental or metabolic pathways, dictate the ultimate outcome of a biological process in plants. Accumulating evidence suggests that TCPs are transcriptional hubs, as they interact with various regulatory proteins to control many biological processes (Bemer et al., 2017). However, TCPs have not been implicated in seed oil biosynthesis until now. Here, we provide evidence that TCP4 is involved in AtWRI1-mediated oil biosynthesis in Arabidopsis.
We identified TCP4, TCP10, and TCP24 in an Arabidopsis TF library as interacting partners of AtWRI1 using a Y2H assay and further validated the interactions by BiFC (Fig. 1). AtWRI1 is a major regulator of seed-specific acyl lipid biosynthesis in Arabidopsis. Coexpression analysis of AtWRI1 and TCP genes during embryo development showed a strong correlation between AtWRI1 and TCP4 expression (Fig. 2), suggesting a potential role of TCP4 in seed oil biosynthesis. Although TCP10 and TCP24 are expressed during embryo development, the expression patterns are different from that of AtWRI1 (Supplemental Fig. S4), suggesting that TCP10 and TCP24 are possibly involved in other biological processes during embryogenesis. Using an N. benthamiana leaf infiltration assay, we demonstrated that TCP4, but not TCP10 and TCP24, repressed AtWRI1-induced oil biosynthesis (Fig. 3, A and B). Furthermore, oil accumulation is moderately but significantly higher in the tcp4 and tcp2 tcp4 tcp10 loss-of-function mutants (Fig. 3, C and E). The class II TCPs, including TCP4, are posttranscriptionally regulated by miRNA319 (Palatnik et al., 2003). We showed that seed oil accumulation was significantly higher in the jaw-D mutant, in which the expression of up to five class II TCP genes (TCP2, TCP3, TCP4, TCP10, and TCP24) was altered due to the elevated expression of miRNA319 (Fig. 3D). We reasoned that the increased seed oil content in jaw-D is due to altered expression of TCP4 because (1) TCP4 expression showed the strongest reduction compared with the other four TCPs, (2) TCP10 and TCP24 had no effect on AtWRI1-mediated oil biosynthesis (Fig. 3B), (3) TCP2 and TCP3 are not known to regulate fatty acid biosynthesis thus far, and (4) the seed oil content of the jaw-D mutant is comparable to that of the tcp4 single mutant and the tcp2 tcp4 tcp10 triple mutant. Collectively, these findings indicate that TCP4 is a negative regulator of seed oil biosynthesis in Arabidopsis.
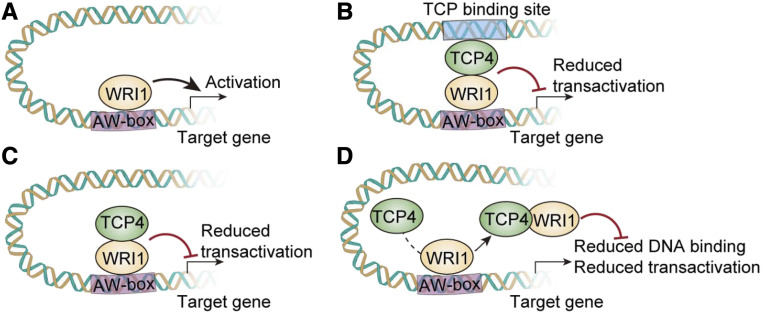
Synergistic or antagonistic interactions between TFs are known to regulate metabolic or developmental pathways. Synergistic interaction between R2R3 MYB, bHLH, and WD40 proteins form the MYB-bHLH-WD40 complex that regulates anthocyanin biosynthesis in a wide range of plant species (Patra et al., 2013). Antagonistic interactions between HLH and bHLH TFs regulate brassinosteroid-mediated cell elongation in rice and Arabidopsis (Zhang et al., 2009). Similarly, interactions between the bHLH TF MYC2 and the basic Leu zipper proteins GBFs regulate hypocotyl elongation in Arabidopsis (Maurya et al., 2015) and terpenoid indole alkaloid biosynthesis in Catharanthus roseus (Sui et al., 2018). Interaction between the TCP TF OsTB1 and the MADS domain protein OsMADS57 regulates tillering in rice (Guo et al., 2013). Here, we demonstrated that the interaction between TCP4 and AtWRI1 repressed the transcriptional activity of AtWRI1 acting on key fatty acid pathway promoters (Fig. 4). We envision three possible mechanisms by which TCP4 decreases AtWRI1 activity toward its target promoters (Fig. 5): (1) the TCP4-AtWRI1 complex occupies a target gene promoter through binding to the TCP- and WRI1-binding cis-elements (Fig. 5B); (2) the complex binds to the promoter only through the WRI1-binding cis-element (Fig. 5C); and (3) TCP4 interaction with AtWRI1 reduces AtWRI1 binding to its binding cis-element (Fig. 5D). TCP proteins bind to specific consensus DNA motifs (Kosugi and Ohashi, 2002). We hence searched the promoter regions of the known AtWRI1 target genes, including BCCP2 and PKP-β1, for potential TCP-binding sites. We found TCP consensus-binding motifs in the promoter of PKP-β1 but not in the BCCP2 promoter (Supplemental Fig. S6). TCP2, TCP3, and TCP10 have been found to bind to the promoters of BP and KNAT2, although the classical TCP binding cis-elements are not present in these promoters (Li et al., 2012). Therefore, it is possible that TCP4 binds to the BCCP2 promoter at an unidentified cis-element or is recruited to the promoter by AtWRI1. TCP4 alone has no activity on the BCCP2 and PKP-β1 promoters (Fig. 4B), suggesting that TCP4 acts as a repressor of AtWRI1 activity. As interaction between TFs affects the promoter-binding specificity, it is possible that the interaction between AtWRI1 and TCP4 affects the AtWRI1-binding affinity to the promoters of its target genes.
Figure 5.
Model of the proposed TCP4-mediated repression of AtWRI1 transactivation. A, AtWRI1 activates the expression of target genes for oil biosynthesis by its binding to the AW-box. B to D, The interaction of TCP4 and AtWRI1 results in acting as a repressor of AtWRI1 (binding to promoters of AtWRI1 target genes; B), acting as a repressor of AtWRI1 (not binding to promoters of AtWRI1 target genes; C), or interference of AtWRI1 binding to the promoters of AtWRI1 target genes (D). In any case, the AtWRI1-TCP4 protein complex displays decreased AtWRI1 target transactivation activity as a result of the AtWRI1-TCP4 interaction.
Fatty acids are essential components of plant cells. A question thus arose about the biological significance of a negative regulator of the fatty acid biosynthetic pathway. AtWRI1 and its orthologs are key regulators of seed-specific acyl lipid biosynthesis in Arabidopsis and other plant species. Seed oil accumulation during embryo development changes slightly during the initial and final stages but changes dramatically during the maturation phase. In addition, partitioning of fatty acids, proteins, and carbohydrates during embryo development is well balanced. Oxidation of fatty acids releases more energy than oxidation of carbohydrates or proteins, and acyl lipid biosynthesis is a high-energy-demanding process. Therefore, fatty acid biosynthesis must be tightly regulated. One mechanism to efficiently fine-tune fatty acid biosynthesis is activity modulation of the key transcriptional regulator. Posttranslational regulation of AtWRI1 by phosphorylation and proteasomal degradation has been previously reported (Chen et al., 2013; Ma et al., 2015; Zhai et al., 2017; Kong et al., 2020). Here, we propose an additional regulatory mechanism by which TCP4 and possibly other regulators act in concert to modulate acyl lipid accumulation in seeds. Whereas multiple TFs are likely involved in regulating the basal fatty acid biosynthetic gene expression in embryonic development, AtWRI1 is a major activator that enables high-level oil accumulation. Nevertheless, AtWRI1 activity must be spatiotemporally regulated in order to respond to developmental cues and to prevent hyperactivation that leads to metabolic imbalance. Such higher level regulation can be effectively achieved by combinatorial action of both transcriptional activators and repressors. Simultaneous expression or coordinated induction of activators and repressors in response to stimuli is evident in many metabolic pathways, likely serving to modulate the amplitudes of target gene expression (Memelink and Gantet, 2007; Sui et al., 2018). Our study suggests that the AtWRI1-TCP4 interaction is one of the mechanisms that allow fine-tuning of the oil biosynthetic pathway. A recent study showed that TCP4, similar to AtWRI1, is a target of posttranslational modification. Mass spectrometry analysis identified nine phosphorylated residues in TCP4 (Kubota et al., 2017). Our in silico analysis using different phosphorylation analysis tools (P3DB, Musite, PhosPhAt 4.0, and DISPHOS 1.3) identified more than 30 potential phosphorylation residues in TCP4. Among the nine phosphorylated residues in TCP4 identified by mass spectrometry (Kubota et al., 2017), eight residues are found in our in silico analyses (Supplemental Fig. S7; Supplemental Table S1). It is well known that phosphorylation affects the stability, transcriptional activity, protein-protein interaction, and subcellular localization of proteins (Ptacek and Snyder, 2006). As TCP4 interacts with and represses WRI1 activity, we hypothesize that reversible phosphorylation in response to stimuli affects the interaction with WRI1 and, ultimately, the expression of fatty acid biosynthetic genes. Identification of candidate kinases and elucidation of the molecular mechanism governing phosphorylation of the AtWRI1-TCP4 module to fine-tune oil production remain to be explored. In summary, our findings uncover a previously uncharacterized role for TCP TFs and open new doors to investigate the complicated regulatory network centered on AtWRI1-regulated gene expression.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) and Nicotiana benthamiana plants were grown in a growth chamber at 22°C with a photoperiod of 16 h of light (100–150 μmol m−2 s−1 illumination)/8 h of dark (Ma et al., 2013, 2015). Arabidopsis wild-type ecotype Columbia was used in this study. The jaw-D mutant (Palatnik et al., 2003) and the tcp2 tcp4 tcp10 triple mutant were described previously (Schommer et al., 2008). Seeds of the tcp4 mutant (SALK_125700) were obtained from the Arabidopsis Biological Resource Center. Seed sterilization and germination were performed as previously described (Ma et al., 2013).
Bioinformatics Analysis
The nuclear localization signal was predicted using the PSORT II program (Nakai and Horton, 1999). Publicly available transcriptome data for Arabidopsis seeds at different stages of development were used to extrapolate the gene expression correlation among TCPs and WRI1 (Gene Expression Omnibus accession no. GSE680; Le et al., 2010). The correlation between every sample pair was calculated as the Euclidean distance and presented as a heat map using the heatmap.2 function with the gplots R package (http://CRAN.R-project.org/package=gplots). The full-length protein sequences of Arabidopsis TCPs were aligned using ClustalW (Larkin et al., 2007) with default settings. MEGA software (version X; Kumar et al., 2018) was used to construct the phylogenetic tree using the neighbor-joining method with bootstrap values set at 1,000 replicates. The expression of TCP4 and other oil biosynthesis-related genes was analyzed in the microRNA-resistant (rTCP4-GFP) and -sensitive (TCP4-GFP) transgenic Arabidopsis lines as previously reported (ArrayExpression accession no. E-MEXP-469; Schommer et al., 2008). TCP binding site analysis was performed using AthaMap (Steffens et al., 2005). Phosphorylation site analysis was performed using P3DB (Gao et al., 2009), Musite (Gao et al., 2010), PhosPhAt 4.0 (Zulawski et al., 2013), and DISPHOS 1.3 (Iakoucheva et al., 2004).
Plasmid Construction
The coding sequences of AtWRI1 (including truncated AtWRI1 variants) and TCPs were amplified by PCR and introduced into the pENTR4 vector (Life Technologies). Entry constructs were combined with destination vectors (Y2H vectors, BiFC vectors, and pEarlygate binary vectors) through Gateway LR reactions (Life Technologies) as previously described (Ma et al., 2015). Entry constructs of TCP4 and TCP24 were obtained from the Arabidopsis Biological Resource Center. The construct for TCP4-GFP was previously described (Schommer et al., 2008). A list of the primers used for plasmid construction in this study is provided in Supplemental Table S2.
Y2H Assays
The Arabidopsis TF library was a gift from Dr. Michael Thomashow. For library screening, TFs were cloned into the pDEST22 vector (prey) and introduced into yeast strain Y187 (Clontech). AtWRI1 variants were cloned into pDEST32 vector (bait) and transformed into yeast strain AH109. The prey and bait were mated and spotted on -Trp-Leu medium. After 3 d of growth, the colonies were streaked onto -Trp-Leu-His medium for screening of positive interactions. TCPs were cloned into pDEST22 as the prey and AtWRI1 variants were cloned into pDEST32 as the bait. The Y2H assay was done as previously described (Ma et al., 2015).
Transient Expression in N. benthamiana, BiFC, and Confocal Microscopy
Agrobacterium tumefaciens-mediated transient expression in N. benthamiana leaves, BiFC, and confocal microscopy were performed as previously described (Ma et al., 2015, 2016). The plasmid pEAQ HT expressing the P19 protein was coinfiltrated with other constructs to ensure high-level expression in N. benthamiana (Sainsbury et al., 2009).
Transient Expression Assay in Protoplasts
The reporter plasmids used in the protoplast assay contained the firefly LUC coding sequence driven by BCCP2 or PKP-β1 promoters and the rbcS terminator. The effector plasmids were generated by cloning the full-length coding sequences of AtWRI1 and TCP4 into a pBluescript vector containing the CaMV 35S promoter and rbcS terminator. The primers for these constructs are listed in Supplemental Table S2. Isolation of protoplasts from tobacco cell suspension cultures and electroporation of tobacco protoplasts with plasmid DNA were performed as previously described (Pattanaik et al., 2010). A plasmid containing the GUS gene under the control of the CaMV 35S promoter and rbcS terminator was coelectroporated with each promoter construct as an internal control. After 20 to 22 h, protoplasts were harvested for GUS and LUC activity assays. All constructs were tested in at least three independent experiments. The GUS activity was normalized against LUC activity.
Fatty Acid Analysis
Lipid analysis in seeds and leaves was done as previously described (Ma et al., 2013, 2015).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource database (www.arabidopsis.org) under accession numbers: WRI1 (AT3G54320), TCP4 (AT3G15030), TCP10 (AT2G31070), TCP24 (AT1G30210), BCCP2 (AT5G15530), PKP-β1 (AT5G52920), and ACP1 (AT3G05020).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Identification of the nuclear localization signal in AtWRI1.
Supplemental Figure S2. Assays of physical interaction between AtWRI1 and TCPs.
Supplemental Figure S3. Phylogenetic analysis of the Arabidopsis TCP gene family.
Supplemental Figure S4. Expression profiles of TCP10 and TCP24 during various stages of embryo development.
Supplemental Figure S5. Expression analysis of TCP4, WRI1, and WRI1 target genes in TCP4 transgenic plants.
Supplemental Figure S6. In silico analysis of TCP-binding sites in the promoter region of AtWRI1 target genes.
Supplemental Figure S7. Phosphorylation site analysis of TCP4.
Supplemental Table S1. TCP4 phosphorylation sites identified in this study and a previous study.
Supplemental Table S2. Primers used for plasmid construction in this study.
Acknowledgments
We thank Dr. Detlef Weigel (Max Planck Institute for Developmental Biology) for the seeds of the jaw-D and tcp2 tcp4 tcp10 mutants and TCP4 constructs, Dr. Michael Thomashow (Michigan State University) for the Arabidopsis TF library, Dr. Melinda Frame (Michigan State University Center for Advanced Microscopy) for confocal microscopy experiments, and Dr. Eva Farre (Michigan State University) for helpful discussion.
Footnotes
This work was supported by a Department of Energy-Great Lakes Bioenergy Research Center Cooperative Agreement (grant no. DE–FC02–07ER64494 to C.B.), the Harold R. Burton Endowed Professorship (to L.Y.), a Nanyang Technological University Startup Grant (to W.M.), and the Ministry of Education of Singapore Tier 1 (grant no. 2018–T1–002–019 to W.M.).
Articles can be viewed without a subscription.
References
- Baud S, Mendoza MS, To A, Harscoët E, Lepiniec L, Dubreucq B(2007) WRINKLED1 specifies the regulatory action of LEAFY COTYLEDON2 towards fatty acid metabolism during seed maturation in Arabidopsis. Plant J 50: 825–838 [DOI] [PubMed] [Google Scholar]
- Bemer M, van Dijk ADJ, Immink RGH, Angenent GC(2017) Cross-family transcription factor interactions: An additional layer of gene regulation. Trends Plant Sci 22: 66–80 [DOI] [PubMed] [Google Scholar]
- Cernac A, Benning C(2004) WRINKLED1 encodes an AP2/EREB domain protein involved in the control of storage compound biosynthesis in Arabidopsis. Plant J 40: 575–585 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Ohlrogge JB(2012) Compartmentation of triacylglycerol accumulation in plants. J Biol Chem 287: 2288–2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Zhang G, Li P, Yang J, Guo L, Benning C, Wang X, Zhao J(2020) Multiple GmWRI1s are redundantly involved in seed filling and nodulation by regulating plastidic glycolysis, lipid biosynthesis and hormone signalling in soybean (Glycine max). Plant Biotechnol J 18: 155–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Lee JH, Weber H, Tohge T, Witt S, Roje S, Fernie AR, Hellmann H(2013) Arabidopsis BPM proteins function as substrate adaptors to a cullin3-based E3 ligase to affect fatty acid metabolism in plants. Plant Cell 25: 2253–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E(1999) The TCP domain: A motif found in proteins regulating plant growth and development. Plant J 18: 215–222 [DOI] [PubMed] [Google Scholar]
- Danisman S, van Dijk AD, Bimbo A, van der Wal F, Hennig L, de Folter S, Angenent GC, Immink RG(2013) Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J Exp Bot 64: 5673–5685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focks N, Benning C(1998) wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol 118: 91–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Agrawal GK, Thelen JJ, Xu D(2009) P3DB: A plant protein phosphorylation database. Nucleic Acids Res 37: D960–D962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Thelen JJ, Dunker AK, Xu D(2010) Musite, a tool for global prediction of general and kinase-specific phosphorylation sites. Mol Cell Proteomics 9: 2586–2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ng S, Carrie C, Duncan O, Low J, Lee CP, Van Aken O, Millar AH, Murcha M, Whelan J(2010) TCP transcription factors link the regulation of genes encoding mitochondrial proteins with the circadian clock in Arabidopsis thaliana. Plant Cell 22: 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xu Y, Liu H, Mao Z, Zhang C, Ma Y, Zhang Q, Meng Z, Chong K(2013) The interaction between OsMADS57 and OsTB1 modulates rice tillering via DWARF14. Nat Commun 4: 1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK(2004) The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res 32: 1037–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Son GH, Bhattacharjee S, Kim HJ, Nam JC, Nguyen PD, Hong JC, Gassmann W(2014a) The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J 78: 978–989 [DOI] [PubMed] [Google Scholar]
- Kim WC, Reca IB, Kim Y, Park S, Thomashow MF, Keegstra K, Han KH(2014b) Transcription factors that directly regulate the expression of CSLA9 encoding mannan synthase in Arabidopsis thaliana. Plant Mol Biol 84: 577–587 [DOI] [PubMed] [Google Scholar]
- Kong Q, Ma W(2018) WRINKLED1 transcription factor: How much do we know about its regulatory mechanism? Plant Sci 272: 153–156 [DOI] [PubMed] [Google Scholar]
- Kong Q, Yang Y, Guo L, Yuan L, Ma W(2020) Molecular basis of plant oil biosynthesis: Insights gained from studying the WRINKLED1 transcription factor. Front Plant Sci 11: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y(2002) DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J 30: 337–348 [DOI] [PubMed] [Google Scholar]
- Kubota A, Ito S, Shim JS, Johnson RS, Song YH, Breton G, Goralogia GS, Kwon MS, Laboy Cintrón D, Koyama T, et al. (2017) TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet 13: e1006856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K(2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35: 1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948 [DOI] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Zachgo S(2013) TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J 76: 901–913 [DOI] [PubMed] [Google Scholar]
- Li Z, Li B, Shen WH, Huang H, Dong A(2012) TCP transcription factors interact with AS2 in the repression of class-I KNOX genes in Arabidopsis thaliana. Plant J 71: 99–107 [DOI] [PubMed] [Google Scholar]
- Liu J, Hua W, Zhan G, Wei F, Wang X, Liu G, Wang H(2010) Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Biochem 48: 9–15 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Arondel V, Kilaru A, Bates PD, Thrower NA, Benning C, Ohlrogge JB(2013) Wrinkled1, a ubiquitous regulator in oil accumulating tissues from Arabidopsis embryos to oil palm mesocarp. PLoS ONE 8: e68887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Kong Q, Grix M, Mantyla JJ, Yang Y, Benning C, Ohlrogge JB(2015) Deletion of a C-terminal intrinsically disordered region of WRINKLED1 affects its stability and enhances oil accumulation in Arabidopsis. Plant J 83: 864–874 [DOI] [PubMed] [Google Scholar]
- Ma W, Kong Q, Mantyla JJ, Yang Y, Ohlrogge JB, Benning C(2016) 14-3-3 protein mediates plant seed oil biosynthesis through interaction with AtWRI1. Plant J 88: 228–235 [DOI] [PubMed] [Google Scholar]
- Maeo K, Tokuda T, Ayame A, Mitsui N, Kawai T, Tsukagoshi H, Ishiguro S, Nakamura K(2009) An AP2-type transcription factor, WRINKLED1, of Arabidopsis thaliana binds to the AW-box sequence conserved among proximal upstream regions of genes involved in fatty acid synthesis. Plant J 60: 476–487 [DOI] [PubMed] [Google Scholar]
- Mano F, Aoyanagi T, Kozaki A(2019) Atypical splicing accompanied by skipping conserved micro-exons produces unique WRINKLED1, an AP2 domain transcription factor in rice plants. Plants (Basel) 8: E207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki T, Mitsui N, Tsukagoshi H, Nishii T, Morikami A, Nakamura K(2005) ACTIVATOR of Spomin:LUC1/WRINKLED1 of Arabidopsis thaliana transactivates sugar-inducible promoters. Plant Cell Physiol 46: 547–556 [DOI] [PubMed] [Google Scholar]
- Maurya JP, Sethi V, Gangappa SN, Gupta N, Chattopadhyay S(2015) Interaction of MYC2 and GBF1 results in functional antagonism in blue light-mediated Arabidopsis seedling development. Plant J 83: 439–450 [DOI] [PubMed] [Google Scholar]
- Memelink J, Gantet P(2007) Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem Rev 6: 353–362 [Google Scholar]
- Nakai K, Horton P(1999) PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24: 34–36 [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D(2003) Control of leaf morphogenesis by microRNAs. Nature 425: 257–263 [DOI] [PubMed] [Google Scholar]
- Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L(2013) Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim Biophys Acta 1829: 1236–1247 [DOI] [PubMed] [Google Scholar]
- Pattanaik S, Werkman JR, Kong Q, Yuan L(2010) Site-directed mutagenesis and saturation mutagenesis for the functional study of transcription factors involved in plant secondary metabolite biosynthesis. Methods Mol Biol 643: 47–57 [DOI] [PubMed] [Google Scholar]
- Ptacek J, Snyder M(2006) Charging it up: Global analysis of protein phosphorylation. Trends Genet 22: 545–554 [DOI] [PubMed] [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB(2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14: 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP(2009) pEAQ: Versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7: 682–693 [DOI] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chételat A, Cubas P, Farmer EE, Nath U, Weigel D(2008) Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol 6: e230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B, Allen WB, Zheng P, Li C, Glassman K, Ranch J, Nubel D, Tarczynski MC(2010) Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol 153: 980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens NO, Galuschka C, Schindler M, Bülow L, Hehl R(2005) AthaMap web tools for database-assisted identification of combinatorial cis-regulatory elements and the display of highly conserved transcription factor binding sites in Arabidopsis thaliana. Nucleic Acids Res 33: W397–W402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Singh SK, Patra B, Schluttenhofer C, Guo W, Pattanaik S, Yuan L(2018) Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J Exp Bot 69: 4267–4281 [DOI] [PubMed] [Google Scholar]
- Vanhercke T, El Tahchy A, Shrestha P, Zhou XR, Singh SP, Petrie JR(2013) Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett 587: 364–369 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ(2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Munz J, Cass C, Zienkiewicz A, Kong Q, Ma W, Sanjaya, Sedbrook J, Benning C(2015) Ectopic expression of WRINKLED1 affects fatty acid homeostasis in Brachypodium distachyon vegetative tissues. Plant Physiol 169: 1836–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Keereetaweep J, Liu H, Feil R, Lunn JE, Shanklin J(2018) Trehalose 6-phosphate positively regulates fatty acid synthesis by stabilizing WRINKLED1. Plant Cell 30: 2616–2627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Z, Liu H, Shanklin J(2017) Phosphorylation of WRINKLED1 by KIN10 results in its proteasomal degradation, providing a link between energy homeostasis and lipid biosynthesis. Plant Cell 29: 871–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X, et al. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21: 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulawski M, Braginets R, Schulze WX(2013) PhosPhAt goes kinases: Searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res 41: D1176–D1184 [DOI] [PMC free article] [PubMed] [Google Scholar]