Abstract
Proanthocyanidins are the second most abundant plant phenolic polymer, but, despite intensive investigation, several aspects of their biosynthesis and functions remain unclear.
Proanthocyanidins (PAs, also known as condensed tannins) are polymers of flavan-3-ols that bind to proteins and have been ascribed functions as herbivore feeding deterrents and antimicrobial compounds. They provide astringency to fruits and beverages, positively impact human health, and benefit ruminant livestock by improving nitrogen nutrition and providing protection from pasture bloat (McMahon et al., 2000; Dixon et al., 2013; Rauf et al., 2019). Much progress has been made in recent years in understanding the molecular genetic basis of PA biosynthesis. However, there remain difficulties in resolving the chemical labeling pattern of PAs with their proposed biosynthetic pathway, and defining the subcellular sites of biosynthesis. There is also no model that fully explains the cell biological phenotypes of mutations that interrupt the pathway and disturb the accumulation of PAs in the central vacuole.
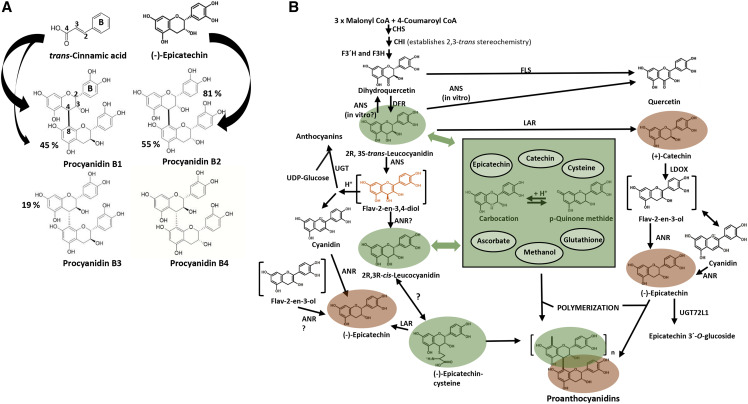
Pioneering studies in the 1970s (Jacques and Haslam, 1974; Haslam et al., 1977) established that dimeric PAs can be assembled by attack of a carbocation “extension unit” derived from a flavan-3-4-diol (leucocyanidin in the case of PAs derived from catechin and/or epicatecin) primarily at the nucleophilic C8 position of a flavan-3-ol [(epi)catechin] “starter unit.” The diversity of dimeric PAs depends on the 2,3-sterochemistry of the starter and extension units (Fig. 1A). The C8 position of the upper unit of the dimer can be attacked by a second extension unit, a process resulting in chains of 4 to 8-linked units that become insoluble as chain length increases. Based on this model, PA assembly in planta could be nonenzymatic, contrary to the assembly of other major plant polymers such as cellulose, hemicelluloses, and lignin. The facile assembly of PAs according to simple thermodynamic control may necessitate physical and possibly temporal separation of PA precursor units to protect plant cells from reactive pathway intermediates, in addition to protection of proteins from the final oligomeric PAs.
Figure 1.
Biosynthetic pathways to the starter and extension units of proanthocyanidins. A, Structures and labeling patterns of 4 to 8-linked procyanidin dimers. Procyanidins B1 to B4 represent the four possible combinations arising from dimerization of 2,3-cis-(−)-epicatechin and 2,3-trans(+)-catechin, with procyanidins B2 and B3 being the homodimers of epicatechin and catechin, respectively. The percentage values represent the approximate percent of known structures with that particular unit as starter (lower) or extension (upper) unit. The large black arrows signify radiolabel incorporation from trans-cinnamic acid; the upper units incorporate 3 to 5 times more label than the lower units, and labeled epicatechin only labels lower units in procyanidin B2. B, Scheme for separate origins of starter and extension units in plants with the LDOX/LAR pathway. Green highlighting indicates potential extension units, and the central green box shows the reactive species thus derived (carbocation and quinone methide) and the nucleophiles that can trap them (light blue ovals). Flav-2-en-3,4-diol is proposed as a potential substrate for generation of 2,3-cis-leucocyanidin for epicatechin extension units. In species that possess LAR, expression of this enzyme can determine chain length by converting Epi-cys (extension unit) to epicatchin (starter unit). Brown highlighting indicates starter units. Enzymes are as follows: CHS, chalcone synthase; CHI, chalcone isomerase; F3′H, flavonoid 3′-hydroxylase; F3H, flavanone 3-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol reductase; ANS, anthocyanidin synthase; ANR, anthocyanidin reductase; UGT, uridine diphosphate glycosyltransferase; LAR, leucoanthocyanidin reductase; LDOX, leucoanthocyandin dioxygenase. Not all species (e.g. Arabidopsis) possess the LAR/LDOX route to epicatechin starter units.
The discovery of transcription factors (TFs) that control PA biosynthesis in crop plants (Mellway et al., 2009; Koyama et al., 2014; James et al., 2017) will facilitate the engineering of these compounds for commercial applications, and recent discoveries of lignin-flavonoid interactions during cell wall biosynthesis open up the possibility of developing novel biomaterials. In addition, the ability to manipulate PA levels in plants is helping our understanding of the ecological roles of PAs in plant–herbivore and plant–pathogen interactions, and some of these results are surprising. We discuss key questions and opportunities in this expanding but complex field.
BIOSYNTHESIS OF PA STARTER AND EXTENSION UNITS—DO WE HAVE THE WHOLE STORY?
The enzymes responsible for formation of the flavan-ol skeleton and the hydroxylation pattern of the flavanol B-ring have been known for some time (Dixon et al., 2013). Most commonly, PAs are of the procyanidin type, in which the B-ring possesses 3′-4´-dihydroxy substitution (Fig. 1A). Lack of activity of a flavonoid 3′-hydroxylase results in the less common propelargonidin type, with 4´-hydroxy B-ring substitution, whereas activity of flavonoid 3′, 5′-hydroxylase results in the prodelphinidin type with 3′, 4´, 5′-trihydroxy B-ring substitution, as commonly found in the PAs and flavanols of tea (Camellia sinensis; Wang et al., 2014). One feature of PA biosynthesis that is proving more difficult to explain is the lack of equivalence of the starter and extension units. Paradoxically, 14C-cinnamic acid is incorporated into the upper extension unit of procyanidin B2 [(−)-epicatechin-(4β→8)-(−)-epicatechin] at around 3 to 5 times the level of incorporation into the lower starter unit (Fig. 1A; Haslam et al., 1977), whereas labeled epicatechin exclusively labels the starter unit. A second problem is that the 2,3-trans-stereochemistry of flavan 3,4-diol, fixed through the chalcone isomerase reaction (Fig. 1B), must switch to allow for formation of the 2,3-cis-epicatechin units that account for ∼55% of PA starter units and 80% of extension units across plant species (Fig. 1A). Stafford (1983) proposed the involvement of an epimerase in this stereochemical switch, but the discovery of anthocyanidin reductase (ANR) provided a pathway to 2,3-cis-epicatechin from 2,3-trans-leucocyanidin via achiral cyanidin (Xie et al., 2003; Fig. 1B). However, ANRs from different species can produce different cis- and trans-stereoisomers in vitro irrespective of the stereochemistry of the PAs in that species (Xie and Dixon, 2005; Gargouri et al., 2009; Pang et al., 2013), and the biological significance of this remains to be explained. The discovery of leucocyanidin reductase provided a pathway to 2,3-trans-catechin by direct reduction of 2,3-trans-leucocyanidin (Tanner et al., 2003).
It now seems likely that 2,3-cis extension units can arise through promiscuous reactions of four late pathway enzymes. Anthocyanidin synthase (ANS, also known as leucoanthocyanidin dioxygenase [LDOX)]) catalyzes, among other reactions (Fig. 1B), the 2-oxoglutarate-dependent oxidation of 2,3-trans-leucocyanidin to cyanidin (Turnbull et al., 2004). A hypothetical intermediate, a 4S-flav-2-en-3,4-diol, can be captured by a 3-O-glucosyltransferase to form a stable anthocyanin (Turnbull et al., 2004), accounting for the biosynthesis of this class of plant pigment, but could theoretically also provide an alternative substrate for reduction (by ANR?) to 2,3-cis-leucocyanidin (Fig. 1B). In Medicago truncatula, a second LDOX can convert catechin to cyanidin via a flav-2-en-3-ol intermediate, and this pathway has been shown both genetically and by precursor labeling studies to generate the starter unit but not the extension unit of PAs (Jun et al., 2018). Finally, leucoanthocyanidin reductase (LAR) forms (+)-catechin from 2,3,-trans-leucocyanidin and also generates (−)-epicatechin (starter unit) from its Cys adduct (Epi-cys). Epi-cys is present in planta, overaccumulates in lar mutants, and acts as an extension unit in vitro (Liu et al., 2016). Loss of function of LAR results in increased levels of insoluble higher-molecular-weight PAs in M. truncatula (Liu et al., 2016), and single nucleotide polymorphism (SNP) analysis in a grapevine diversity panel links LAR to PA polymer chain length (Huang et al., 2012; Liu et al., 2016), suggesting that the in vitro conversion of Epi-cys to (−)-epicatechin by LAR has physiological significance for the conversion of extension units to starter units in planta.
Figure 1B presents a model for the generation of PA starter and extension units consistent with the available (bio)chemical and genetic evidence. Unlike other pathways of specialized metabolism, this is neither a linear pathway nor a metabolic grid. The model proposes 2,3-cis-leucocyanidin generated by ANR as an extension unit, consistent with the presence of epicatechin-only PAs in Arabidopsis (Arabidopsis thaliana), a species that lacks an LAR gene (Jun et al., 2018). Leucocyanidins are very difficult to purify and structurally characterize from plant tissues; they form highly reactive intermediates that can be trapped by reaction with either a nucleophile, such as positions C8 or C6 of a flavan-3-ol or upper unit of a PA, or a thiol, such as Cys or glutathione. The presence in planta of the leucocyanidin carbocation was recently inferred from experiments in which it was trapped as the 4-methyl derivative by extraction in acidic methanol (Wang et al., 2020). If formation of Epi-cys turns out to be enzymatic, its formation could be seen as an example of genetically controlled metabolic damage pre-emption (de Crécy-Lagard et al., 2018). For the pathways in Figure 1B to form oligomeric PAs, some form of temporal or physical separation of the shared reactions leading to starter and extension units is required. Haslam et al. (1977) described a gradual switch from starter to extension unit synthesis associated with increasing PA chain length in some species, but the in vivo presence of trapped carbocations in the form of Epi-cys in model plants examined to date suggests the absence of tight, stoichiometric coupling of starter and extension unit generation.
TOXICITY OF PA STARTER AND EXTENSION UNITS
PA extension units are highly reactive and therefore potentially toxic to the cell; managing this toxicity appears to require complex subcellular compartmentation as proposed below. However, the stable flavan-3-ols that comprise the starter units of PAs can also exhibit cellular toxicity. In this regard, there has been much controversy surrounding reports that (+/−)-catechin exhibits allelopathic activity (Duke et al., 2009). Indirect genetic evidence for catechin toxicity was obtained in studies of the ldox mutant of Arabidopsis that accumulates (+)-catechin (Jun et al., 2018). Independent homozygous null mutants exhibit a strong developmental phenotype in which more than ∼80% to 90% of the seeds show developmental arrest at 2 to 4 d after pollination, and cannot be recovered. Generally only 1 or 2 seeds per pod develop normally (Jun et al., 2018). By contrast, the lar ldox double mutant, in which catechin does not accumulate, does not exhibit this seed development phenotype.
Plants generally detoxify polyphenolic compounds by sugar conjugation and transport to the vacuole (Wan and Hou, 2009). Epicatechin 3′-O-glucoside has been detected in both Arabidopsis and M. truncatula (Pang et al., 2008, 2013; Kitamura et al., 2010), and is formed by a glucosyltransferase (UGT72L1) that is regulated by the same TFs that control expression of ANR and LAR (Pang et al., 2008). Epicatechin 3′-O-glucoside is substrate for the vacuolar MATE1 transporter from M. truncatula and the TT12 MATE transporter from Arabidopsis (Zhao and Dixon, 2009). Because PAs have been assumed to be assembled in the vacuole, it has been tempting to assume that glucosylation and subsequent transport of epicatechin to the central vacuole is a critical step in PA biosynthesis. However, testing this hypothesis is problematic because no ortholog of UGT72L1 has been found in Arabidopsis and exhaustive screening of transparent testa (tt) mutants has not revealed a flavanol-specific UGT. UGT72L1 does not act on (+)-catechin, although this molecule can be a starter unit for PAs in M. truncatula ldox mutants (Jun et al., 2018). It therefore remains unclear whether glycosylation of flavan-3-ols is essential for PA biosynthesis or represents a mechanism for detoxification and storage of excess PA starter units; both could potentially be true if synthesis of starter and extension units is temporally or spatially separated.
RECONCILING CELL BIOLOGY WITH BIOCHEMISTRY
Phenotypes of tt Mutants
Genetic mutations that block accumulation of oxidized PAs result in a tt phenotype, sometimes with dramatic changes to cellular ultrastructure. TT19 is a cytosolic GST of somewhat unclear function. In the tt19 mutant, PAs are found in small vesicle structures localized around small vacuoles outlined by the MATE transporter TT12 (Kitamura et al., 2010). tt19 has higher levels of insoluble PAs than wild type, and these are reduced in the tt19/tt12 double mutant (Kitamura et al., 2010). To reconcile these findings with the pathways in Figure 1B, Figure 2 presents a model in which reactive PA extension units are stabilized through interaction with TT19, either in soluble complexes or vesicles, and then delivered to prevacuole-like vesicles that contain stable starter units, possibly, although perhaps not exclusively, loaded as glycosylated flavan-3-ols by TT12. Sequestration of starter units is required because they themselves can be phytotoxic (Jun et al., 2018). In this model, fusion of the TT19- and TT12-containing structures results in mixing of starter and extension units to initiate oligomerization; the resulting PA dimers and higher oligomers are finally delivered, perhaps by vesicle fusion, to the central vacuole. TT19 is also present in the tonoplast, where it may be associated with transporters for anthocyanins (Sun et al., 2012) or PAs. Consistent with the model in Figure 2, loss of function of LAR results in an increased proportion of higher molecular weight PAs through increasing the ratio of Epi-cys extension units to epicatechin starter units (Liu et al., 2016), and loss of function of TT19 results in higher-molecular-weight PAs formed in the cytosol through interaction of excess “unprotected” extension units with cytosolic epicatechin; this is reversed in the tt19/tt12 double mutant because of the elevated levels of cytosolic starter units (Kitamura et al., 2010). TT13 encodes a tonoplast ATPase necessary for generation of a proton gradient for transport of PA starter units by TT12 (Appelhagen et al., 2015). In both tt12 and tt13 mutants, PAs accumulate on the outside of TT12-containing small vacuoles, suggesting that the TT19 complex is targeting the extension units to these vacuoles where the starter units are now backed up; glycosylated epicatechin, presumably cytosolic, accumulates in the tt12 mutant (Kitamura et al., 2010; Appelhagen et al., 2015).
Figure 2.
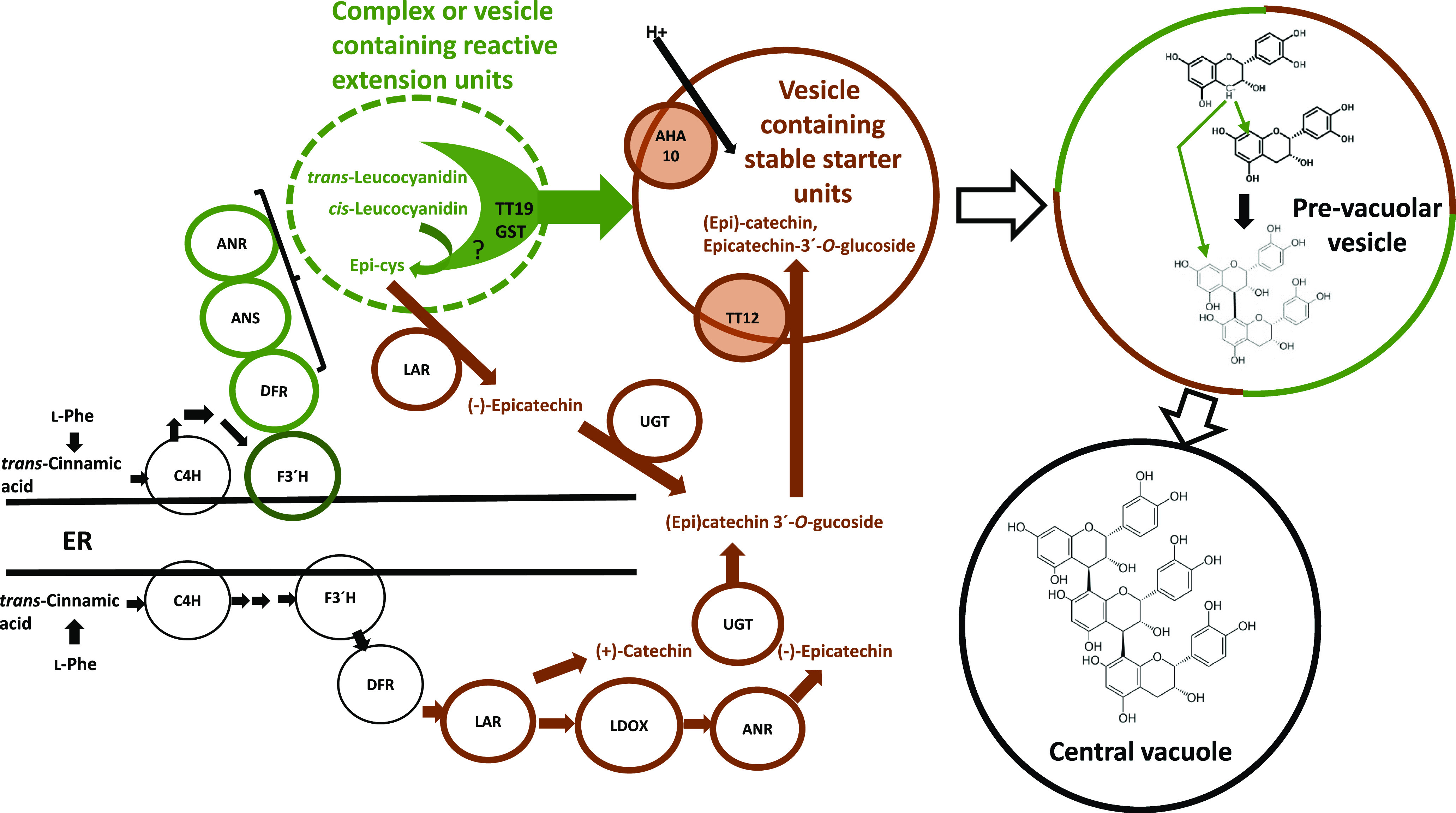
Hypothetical scheme for the sequestration of starter and extension units during PA biosynthesis in M. truncatula. The model proposes that reactions specific for starter unit formation from LAR occur on freely soluble enzymes, whereas those associated with extension unit formation occur through a hypothetical metabolon associated with a subdomain of the endoplasmic reticulum (ER) with tethering through the membrane anchor of the F3′H cytochrome P450 enzyme. The products of these reactions (leucoanthocyanidins) are “captured” and protected by the TT19 GST, including through formation of Epi-cys from 2,3,-cis-leucocyanidin. The TT19 complex interacts with vesicles loaded with starter units through the combined activities of a UGT, the MATE transporter TT12, and the proton APTase TT13 (also known as AHA10). Localization of TT19 to the tonoplast may indicate a tight association with PA extension units until they are safely loaded into vesicles harboring starter units. Fusion of the structures containing starter and extension units allows nonenzymatic condensation to form PA dimers and higher oligomers during migration of the prevacuolar vesicles to ultimately fuse with the central vacuole, where they are finally deposited as tannin accretions. Enzymes, represented by small circles, are as in the legend to Figure 1B. C4H, cinnamate 4-hydroxylase. Enzymes and structures circled in green are associated with extension unit formation, in brown with starter unit formation. This model assumes the existence of soluble and insoluble forms of DFR, but other spatial or even temporal controls could allow for separation of the pathways.
TT10 encodes a laccase enzyme (AtLAC15 in Arabidopsis) that was originally ascribed a role in PA polymerization, although the nonspecific linkage pattern of TT10-catalyzed oligomerization products has led to the hypothesis that the function of the enzyme is more likely the oxidation of preformed PAs in the cell wall of the seed coat (Pourcel et al., 2005). TT10 can also catalyze lignin polymerization (Liang et al., 2006), and could potentially catalyze the formation of cross-links between PAs and other cell wall polymers. Whether oxidation by TT10 is important for formation of insoluble PAs or whether these simply reflect higher molecular weight forms (Liu et al., 2016) remains unclear. In fact, insoluble PAs, although often accounting for more than 50% of the total PA species, remain somewhat poorly characterized. They are easy to quantify by conversion to anthocyanidins by heating in acidic butanol, but this results in the loss of structural information. New methods for analysis of insoluble PAs are clearly required. Although NMR approaches are now being applied to PA analysis (Fryganas et al., 2018), they have yet to provide the structural resolution of similar approaches for the phenylpropanoid polymer lignin (Sette et al., 2011). Increasingly sophisticated mass spectrometry approaches are being applied to PA characterization (Salminen, 2018), but detailed structural features of the internal portions of polymers remain elusive.
The role of metabolons in reactions specific for PA biosynthesis, as suggested in Figure 2, requires further investigation. It is now well established that the earlier reactions in the flavonoid pathway are organized in metabolons anchored to the endoplasmic reticulum through association with the cytochrome P450 enzymes of the pathway (Nakayama et al., 2019; Waki et al., 2020), but physical interactions between the later enzymes of the pathway have yet to be demonstrated. The need for such physical organization of the pathway would appear necessary to explain the differentiation of starter unit from extension unit synthesis using shared enzymes (Jun et al., 2018).
The Tannosome Model
A very different model to describe the subcellular sites of PA biosynthesis has been proposed based on microscopy of tannin-producing cells from across the plant kingdom (Brillouet 2014, 2015; Brillouet et al., 2013,2014). In this model, PA precursors are synthesized in chloroplasts and polymerized in an organelle termed the tannosome, which is derived from thylakoids, and protected during their intracellular journey to the vacuole in “shuttles” bounded by membranes derived from both inner and outer chloroplast envelopes that have budded from the chloroplast (Brillouet et al., 2014). The shuttles are then incorporated into the vacuole as tannin accretions by invagination of the tonoplast, thus protecting the cell contents from the protein-binding activity of polymerized PAs (Brillouet et al., 2014). It appears hard at first sight to reconcile the tannosome model with our current understanding of the biochemistry and genetics of PA biosynthesis (Box 1), unless different pathways of PA synthesis and trafficking occur in different species and/or cell types. It is also hard to reconcile a model in which all reactions of PA biosynthesis occur together in the same subcellular compartment with the asymmetric labeling of PA subunits (Haslam et al., 1977).

Testing Intracellular Routes of PA Biosynthesis
One impediment to a reconciliation of the models derived from cellular, biochemical, and genetic examinations of PA biosynthesis is the lack of an optimal experimental model system. The descriptive studies on the tannosome have used species in which the biochemistry and genetics of PA biosynthesis are less well defined, and which are poorly amenable to genetic manipulation. Furthermore, histological observations provide varying pictures of tannin deposition and vesicles in different cell types (Vio-Michaelis et al., 2020). The species with the best developed genetic tools (Arabidopsis and Medicago spp.) have significant differences in the biochemical pathways to starter units (Arabidopsis possesses neither LAR nor a functional ortholog of Medicago spp. LDOX), and furthermore only accumulate PAs at significant levels during early seed coat development. Poplar (Populus spp.) may be a better model for future studies. In poplar, PAs are produced naturally in leaves, providing large amounts of material for analysis, and the pathway is also inducible by a number of stresses and chemicals (Mellway et al., 2009; Ullah et al., 2019a). Moreover, poplar is genetically transformable and amenable to gene editing (Bewg et al., 2018), and a very large collection of fully sequenced natural variants is available for genome-wide association studies that are already providing new information on multiple traits including the biosynthesis of the phenolic polymer lignin (Chhetri et al., 2019).
Other emerging model species for PA biosynthesis are tea, in which genome sequences of cultivated and wild accessions have been mined to inform the biosynthesis of both PAs and health-promoting PA monomers such as epigallocatechin gallate (Zhang et al., 2020), and grapevine (Vitis vinifera), in which transcriptomic and association genetic approaches have been applied to study the PA pathway (Huang et al., 2012; Carrier et al., 2013). All the above systems produce the classical 4 to 8-linked B-type PAs. Cranberry (Vaccinium macrocarpon), also with a sequenced genome, accumulates A-type PAs that possess additional C2-O-C7 or C2-O-C5 bonds. Transcriptomic studies have addressed the core genes of PA biosynthesis in this species (Sun et al., 2015), but the exact mechanism for formation of the additional A-type linkages remains to be determined.
With a suitable model selected, the key approach to solving the problem of coupled PA synthesis and intracellular trafficking will be the proteomic and metabolomic characterization of the vesicles seen in the tannosome model and in the various mutants described above. Methods are available for labeling cellular organelles/compartments to allow for visualization of colocalization within the cell (Geldner et al., 2009) and affinity purification for biochemical analysis (Bayraktar et al., 2019; Xiong et al., 2019), and the sensitivity of both proteomics and metabolomics has now improved to the point where analysis of plant subcellular compartments is possible (Fürtauer et al., 2019). The vesicles purported to contain PAs in the tannosome model sedimented to the bottom of the ultracentrifuge tube during purification (Brillouet et al., 2014) and could therefore be contaminated with precipitated PAs. It is critical to prove directly whether PAs or their precursors are sequestered in vesicles showing chloroplast membrane origin and, if so, whether proteins such as TT12, TT13, and TT19 colocalize with these vesicles. These studies will require the initial generation of a number of engineered lines in which various membrane protein markers are introduced into different genetic backgrounds in which the PA pathway is perturbed, as well as marker lines designed for tracking the cellular pathways temporally after suitable induction. These materials could also be subjected to labeling with 13C-Phe or 13C-cinnamic acid to track the distribution of label in starter and extension units as assembling PAs are trafficked to the central vacuole.
An alternative approach is to apply new tools to existing models. Autophagy of chloroplasts can occur through multiple routes, including the process of microchlorophagy in which more than one type of small vesicle can be targeted to the main vacuole (Zhuang and Jiang, 2019). Monitoring this process during the early developmental stages of the Arabidopsis seed coat endothelium, where PA synthesis occurs, could provide a means of testing the tannosome model in a genetically tractable system.
EXAMINING THE PROPERTIES OF PAS FROM A FUNCTIONAL PERSPECTIVE
Roles in Biotic and Abiotic Stress Protection
PAs have been suggested to possess protective functions against oxidative stress, herbivory (insect and animal), and pathogen attack. Studies to address such functions benefit from the use of a plant that produces PAs in major organs and tissues, that possesses well-studied ecology and extensive genetic variation, and that is suitable for genetic manipulation to alter PA profiles. Poplar has emerged as a model species that fits these criteria well. Extracts from poplar bark contain higher concentrations of PAs and greater antioxidant capacity than similar extracts from fir, beech, pine, and oak (Hamad et al., 2019). Manipulation of the expression of two MYB TFs, MYB115 and MYB134, makes it possible to generate poplar lines with widely differing levels of leaf catechin and PAs (James et al., 2017), and such lines have been exploited to address various potential ecological functions of PAs (Box 2).
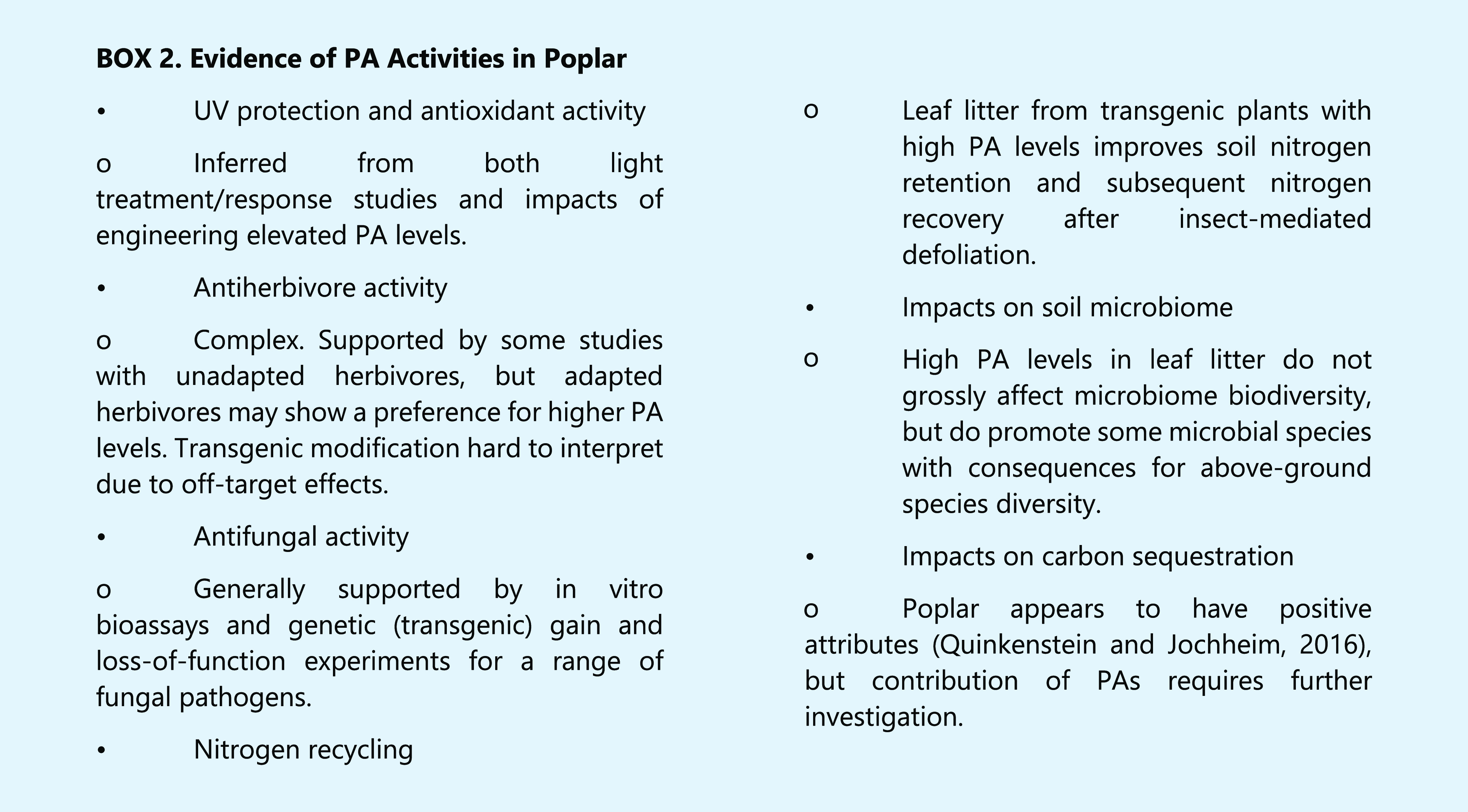
High light stress and nitrogen deficiency both generate reactive oxygen species in hybrid poplar (Populus tremula × Populus tremuloides); exposure to natural sunlight for 2 weeks caused a 14-fold increase in foliar PA levels, whereas subjection to soil nitrogen deficiency led to a 4- to 5-fold increase. In both cases, the antioxidant capacity of the poplar extracts paralleled the increase in PA levels (Gourlay and Constabel 2019). Moreover, when MYB overexpressing transgenic poplar with elevated PA levels were treated with the reactive oxygen species–generator methyl viologen, they retained greater chlorophyll fluorescence and produced less hydrogen peroxide and superoxide (Gourlay and Constabel, 2019). High cytosolic flavonol concentrations may be detrimental by acting as pro-oxidants and thereby damaging DNA in the presence of hydrogen peroxide (Krych and Gebicka, 2013; Harding, 2019), but overexpression of MYB134/115 induces PAs without elevating cytosolic flavonol levels (Gourlay and Constabel, 2019).
Many phenolic compounds such as PAs are bitter and thus predicted to act as feeding deterrents, and PAs have additional specific antinutritional effects. For example, mountain hares (Lepus timidus) fed birch (Betula spp.) bark showed high sodium output through urine, rapid loss of body weight, and did not survive on the diet (Palo, 1984). High PA levels can cause oxidative stress in the insect gut, and intestinal damage in vertebrate herbivores (Barbehenn and Peter Constabel, 2011). These detrimental effects are predicted to reduce herbivore food preference for plants with higher concentrations of PAs, and in a classic study with Quercus spp., foliar PA concentration was negatively correlated with herbivore community density (Forkner et al., 2004). However, moving beyond correlations has proven difficult. PAs are induced when poplar is subject to natural herbivory, for example by the white satin moth Leucoma salicis, or when mechanically wounded (Peters and Constabel, 2002; Tsai et al., 2006), and this induction may protect the trees against unadapted species. However, PAs are often not effective against adapted herbivore species (Barbehenn and Peter Constabel, 2011). In fact, coevolution may cause CTs to become feeding stimulants for some herbivores (Hjältén and Axelsson, 2015). Further research is therefore necessary to elucidate the roles of PAs in herbivore defense, but the use of transgenic models requires careful examination. For example, after a chance outbreak of thrips, transgenic poplar lines expressing high PA levels were found to be more damaged than their wild-type counterparts. This appears to result from reduction in the levels of phenolic glycosides as a result of the up-regulation of the PA pathway (Mellway et al., 2009). In a separate study, high-PA poplar was likewise preferred by the forest tent caterpillar (Malacosoma disstria) and the gypsy moth (Lymantria dispar) because of the reduced phenolic glycoside levels (Boeckler et al., 2014). Future transgenic manipulation studies will need to consider ways of limiting changes in polyphenols to PAs alone.
An interesting concept that requires further evaluation is that PAs are mediators of insect herbivore tolerance rather than resistance (Madritch and Lindroth, 2015). The protein-binding capacity of PAs can enhance nitrogen retention and subsequent recycling from the soil, and genetically engineered PA levels in P. tremuloides were shown to correlate with increased nitrogen recovery after defoliation through insect herbivory (Madritch and Lindroth, 2015). PAs may therefore exert their protective functions as much or more after leaves are shed than when they are still on the plant.
Evidence of a role for PAs in pathogen defense is less disputed than for insect defense. Infection by fungal biotrophs such as Melampsora medusae, Marssonina brunnea f.sp. multigermtubi, and Plectosphaerella populi induces transcriptional activation of the PA biosynthetic pathway in poplar leaves (Mellway et al., 2009; Yuan et al., 2012; Ullah et al., 2019b), and PAs and monomeric catechin reduce mycelial growth of P. populi (Ullah et al., 2017, 2019b). Overexpression of MYB115 in poplar resulted in a 50% reduction in lesions caused by Dothiorella gregaria, the causative agent of branch canker, whereas lesion numbers were increased by 137.5% in CRISPR/Cas9-generated myb115 mutant plants (Wang et al., 2017). Both D. gregaria and M. brunnea f. sp. multigermtubi exhibited reduced mycelial growth, shorter hyphae, swollen tips, and fewer hyphal branches on exposure to extracts from MYB115-overexpressing plants as compared with control plants (Yuan et al., 2012; Wang et al., 2017), and the improved response to fungal attack was directly attributed to PA accumulation, as MYB115 overexpression did not appear to enhance other defense pathways involving genes such as PR5, JAZ10, MYB44, and NPR1 (Wang et al., 2017).
Most PAs in the soil probably originate from leaf litter. Levels in roots tend to be lower, and not correlated with above-ground levels (Dettlaff et al., 2018). Although tannin-rich leaf litter from MYB134-overexpressing poplar did not result in changes in microbial biodiversity, the leaf litter was found to promote the growth of Eocronartium muscicola, a parasite of mosses, which reduced moss proliferation in soil microcosms (Winder et al., 2013). Use of short-term coppiced plants such as poplar for carbon sequestration has been considered (Quinkenstein and Jochheim, 2016). Leaf PAs represent a potentially large sink of carbon for greenhouse gas sequestration, but, before attempting to implement increasing PAs as a carbon-reduction strategy, it will be important to consider further their impacts on nutrient recycling, microbial communities, and greenhouse gas emissions from soil. Furthermore, because of the increasingly realized importance of root PAs for soil carbon sequestration (Adamczyk et al., 2020), more studies are required on the biosynthesis of PAs in roots and the effects of their structural modification on the stabilization of organic matter.
PAs in Pastures and Animal Feed
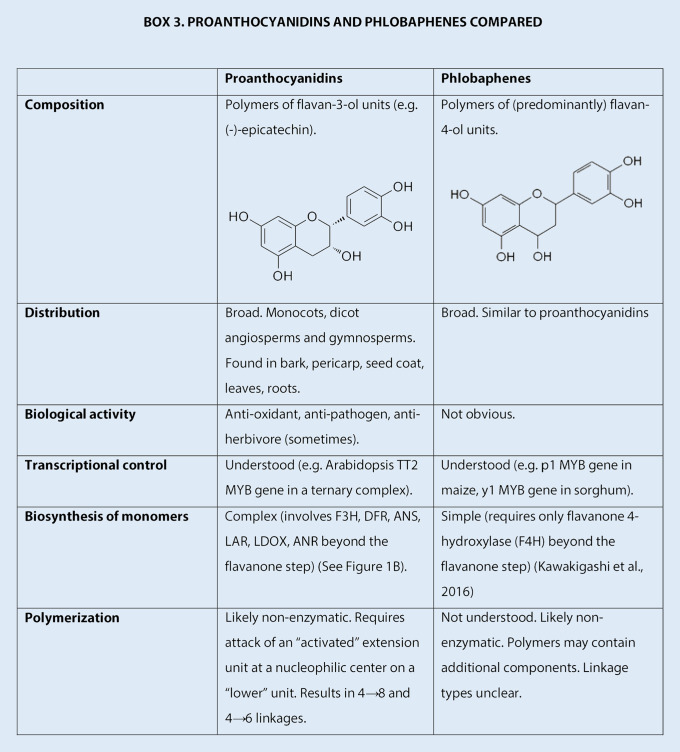
The protein-binding activity of PAs accounts for their ability to protect ruminants from pasture bloat and shield proteins in the ruminant diet and in silage from precocious degradation (McMahon et al., 2000). This has been the stimulus for attempts to engineer PAs in forage crops that lack PAs in vegetative tissues (Hancock et al., 2012), and long-term proprietary studies using structural and regulatory genes are underway to this end, although as yet unpublished. Because PAs have multiple effects on animal health and performance, it is important to understand exactly what PA structures we need to engineer. This will require collaboration between analytical chemists, ruminant nutritionists, and molecular biologists. Useful approaches will be the engineering of PAs of broadly defined composition and chain length in planta facilitated by our knowledge of how LAR and LDOX help define starter/extension unit composition and degree of polymerization (Liu et al., 2016; Jun et al., 2018), and the development of more versatile analytical methods for PAs that can be predictive of PA function rather than structure alone (Marsh et al., 2020). There is also a need for further investigation of the biosynthesis of PAs in cereal crops, and the development of the best model organism for such studies. Many grasses and grains, such as tall fescue (Festuca arundinacea), perennial ryegrass (Lolium perenne), and wild rice (Zizania spp.), possess PAs in the seeds (Fraser et al., 2016; Hosoda et al., 2018), but few have them in vegetative tissues. Protective flavonoid synthesis in infected leaves of both sorghum (Sorghum bicolor) and maize (Zea mays) involves the formation of 3-deoxyanthocyanidins, generated via flavan-4-ols through the activity of a flavanone 4-reductase (Kawahigashi et al., 2016). Flavan-4-ols are also believed to be the precursors of the still poorly defined red phlobaphene pigments found in maize seeds. There is strong interest in engineering PAs in maize; however, despite a number of early genetic studies, much more needs to be understood about phlobaphene biosynthesis, which appears, at least on paper, to parallel and potentially compete with flavan-3-ol-derived PA biosynthesis (Box 3). Paradoxically, although several studies have shown that flavonoid-pathway TFs from maize can induce PA accumulation in dicot species (Li et al., 2007), maize does not itself appear to accumulate PAs to levels that allow rigorous identification. Although the maize genome contains genes with sequence similarity to ANR, the functions of these genes are yet to be determined.
PAs as Structural Components: The Lignin Connection Revisited
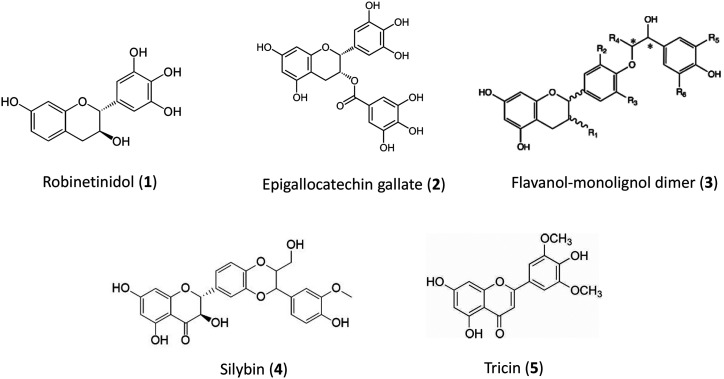
In a classic review entitled “Proanthocyanidins and the lignin connection,” Stafford (1988) discussed similarities between lignin and PAs, concluding that, although both polymers often occur together in plants, PAs are unlikely to play major structural roles. This conclusion still appears generally valid, although it was hypothesized that the helicoidal tridimensional structures of PAs in cell walls of some African “resuscitation plants” might provide protection from cell wall cracking under intense desiccation, allowing the plants to recover rapidly following reinstated water availability (Pizzi and Cameron, 1986). Whatever the natural functions of PAs in cell walls, it is interesting to consider whether it may be possible to generate, in planta, novel lignin-PA copolymers as biomaterials (Grishechko et al., 2013). This concept appears feasible both chemically and biologically, in view of recent progress in the use of PAs as core molecules for biomaterial design (Garcıa et al., 2016), and the newly realized plasticity of lignin structure with resulting potential for designer lignins (Mottiar et al., 2016). Lignin-PA aerogels have been synthesized chemically from lignins and wattle tannin (Grishechko et al., 2013), a polymer comprised primarily of units derived from robinetinidol (a 5-deoxy-flavan-3-ol with a 3′-,4´-,5′-hydroxy-substituted B-ring; Fig. 3, compound 1). Epigallocatechin and epigallocatechin gallate (Fig. 3, compound 2) have the same B-ring substitution pattern as robinetinidol, and have been shown to incorporate into lignin when fed, along with monolignols, to isolated maize primary cell walls (Elumalai et al., 2012; Grabber et al., 2012). Such incorporation enhances sugar release from lignocellulosic biomass (Elumalai et al., 2012), and density functional theory calculations confirm that incorporation of lignin-flavonoid bonds in the lignin polymer (Fig. 3, compound 3) will lead to linkage properties conducive to more facile lignin depolymerization (Berstis et al., 2020).
Figure 3.
Flavonoids that form linkages to monolignols or lignin. See the “PAs as Structural Components: The Lignin Connection Revisited’ section for details.
Lignin-flavonoid linkages occur in nature. Flavonolignans are dimers or higher oligomers comprising at least one monolignol linked to at least one flavonoid, with the best-known example being silybin, a popular dietary supplement from milk thistle (Silybum marianum; Fig. 3, compound 4), in which the flavonoid component (taxifolin) may be synthesized in flowers and transported to the seed coat where it is polymerized with coniferyl alcohol (Lv et al., 2017). In some cases, molecules of this type include flavan-3-ol units (Kinyok et al., 2017). Importantly, the flavone tricin (Fig. 3, compound 5) is a natural component of lignin in grasses, where it serves as a nucleation site for lignin polymerization (Lan et al., 2015). In maize defective in the CHALCONE SYNTHASE2 gene, near total loss of tricin results in increased levels of Klason lignin with the enhancement of dimer linkages (Eloy et al., 2017), whereas loss of function of FLAVONE SYNTHASE2 in rice (Oryza sativa) results in partial replacement of lignin-associated tricin with its precursor naringenin, decreased lignin syringyl:guaiacyl unit ratio, and enhanced sugar release efficiency (Lam et al., 2017). These results indicate that flavonoids are made in or transported to cells undergoing lignification, and that they can cross into the apoplast to initiate or further contribute to lignification. Although levels of PAs are much lower in stems and developing xylem than in leaves and root tips of poplar (Tsai et al., 2006), it should be possible to increase these levels through overexpression of the TFs described above.
CONCLUDING REMARKS
Despite an apparently near-complete understanding of the biochemical machinery required for PA biosynthesis, how these molecules are assembled is still perplexing (see Outstanding Questions). The complex pathways for metabolic elaboration and sequestration in PA biosynthesis may be necessary for protecting the plant cell against reactive/toxic intermediates, and providing order to a polymerization mechanism that relies on thermodynamic rather than enzymatic control. Furthermore, the spatial and temporal separation of starter and extension unit biosynthesis and accumulation provides an explanation for the difference in labeling of the upper and lower units in PAs, despite their largely shared biochemical pathways. The potential toxicity to the plant of the intermediates and final products of the PA pathway presents challenges to successful metabolic engineering of the pathway. It is well worth attempting to overcome these challenges in view of the potential advances in agriculture, chemical ecology, carbon sequestration, and biomaterials science that this will facilitate.
Footnotes
This work was supported by the Hagler Institute for Advanced Study, Texas A&M University, by research grants from Forage Genetics International and GrasslaNZ Research Inc (to R.A.D.), and the Texas Academy of Mathematics and Science Summer Student Scholarship (to S.S.).
Articles can be viewed without a subscription.
References
- Adamczyk B, Heinonsalo J, Simon J(2020) Mechanisms of carbon sequestration in highly organic ecosystems – importance of chemical ecology. ChemistryOpen 9: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhagen I, Nordholt N, Seidel T, Spelt K, Koes R, Quattrochio F, Sagasser M, Weisshaar B(2015) TRANSPARENT TESTA 13 is a tonoplast P3A -ATPase required for vacuolar deposition of proanthocyanidins in Arabidopsis thaliana seeds. Plant J 82: 840–849 [DOI] [PubMed] [Google Scholar]
- Barbehenn RV, Peter Constabel C(2011) Tannins in plant-herbivore interactions. Phytochemistry 72: 1551–1565 [DOI] [PubMed] [Google Scholar]
- Bayraktar EC, Baudrier L, Özerdem C, Lewis CA, Chan SH, Kunchok T, Abu-Remaileh M, Cangelosi AL, Sabatini DM, Birsoy K, et al. (2019) MITO-Tag Mice enable rapid isolation and multimodal profiling of mitochondria from specific cell types in vivo. Proc Natl Acad Sci USA 116: 303–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berstis L, Elder T, Dixon RA, Crowley M, Beckham GT(2020) Coupling of flavonoid nucleation sites with monolignols studied by density functional theory. ACS Green Chem 8: 11033–11045 [Google Scholar]
- Bewg WP, Ci D, Tsai C-J(2018) Genome editing in trees: From multiple repair pathways to long-term stability. Front Plant Sci 9: 1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckler GA, Towns M, Unsicker SB, Mellway RD, Yip L, Hilke I, Gershenzon J, Constabel CP(2014) Transgenic upregulation of the condensed tannin pathway in poplar leads to a dramatic shift in leaf palatability for two tree-feeding Lepidoptera. J Chem Ecol 40: 150–158 [DOI] [PubMed] [Google Scholar]
- Brillouet J-M.(2014) Plasticity of the tannosome ontogenesis in the Tracheophyta. J Plant Sci 2: 317–323 [Google Scholar]
- Brillouet J-M.(2015) On the role of chloroplasts in the polymerization of tannins in Tracheophyta: A monograph. Am J Plant Sci 6: 1401–1409 [Google Scholar]
- Brillouet J-M, Romieu C, Lartaud M, Jublanc E, Torregrosa L, Cazevieille C(2014) Formation of vacuolar tannin deposits in the chlorophyllous organs of Tracheophyta: From shuttles to accretions. Protoplasma 251: 1387–1393 [DOI] [PubMed] [Google Scholar]
- Brillouet J-M, Romieu C, Schoefs B, Solymosi K, Cheynier V, Fulcrand H, Verdeil J-L, Cone’je’ro G(2013) The tannosome is an organelle forming condensed tannins in the chlorophyllous organs of Tracheophyta. Ann Bot 112: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrier G, Huang Y-F, Le Cunff L, Fournier-Level A, Vialet S, Souquet J-M, Cheynier V, Terrier N, This P(2013) Selection of candidate genes for grape proanthocyanidin pathway by an integrative approach. Plant Physiol Biochem 72: 87–95 [DOI] [PubMed] [Google Scholar]
- Chhetri HB, Macaya-Sanz D, Kainer D, Biswal AK, Evans LM, Chen JG, Collins C, Hunt K, Mohanty SS, Rosenstiel T, et al. (2019) Multitrait genome-wide association analysis of Populus trichocarpa identifies key polymorphisms controlling morphological and physiological traits. New Phytol 223: 293–309 [DOI] [PubMed] [Google Scholar]
- de Crécy-Lagard V, Haas D, Hanson AD(2018) Newly-discovered enzymes that function in metabolite damage-control. Curr Opin Chem Biol 47: 101–108 [DOI] [PubMed] [Google Scholar]
- Dettlaff MA, Marshall V, Erbilgin N, Cahill JF Jr.(2018) Root condensed tannins vary over time, but are unrelated to leaf tannins. AoB Plants 10: ply044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RA, Liu C, Jun JH(2013) Metabolic engineering of anthocyanins and condensed tannins in plants. Curr Opin Biotechnol 24: 329–335 [DOI] [PubMed] [Google Scholar]
- Duke SO, Blair AC, Dayan FE, Johnson RD, Meepagala KMM, Cook D, Bajsa J(2009) Is (-)-catechin a novel weapon of spotted knapweed (Centaurea stoebe)? J Chem Ecol 35: 141–153 [DOI] [PubMed] [Google Scholar]
- Eloy NB, Voorend W, Lan W, Saleme ML, Cesarino I, Vanholme R, Smith RA, Goeminne G, Pallidis A, Morreel K, et al. (2017) Silencing CHALCONE SYNTHASE in maize impedes the incorporation of tricin into lignin and increases lignin content. Plant Physiol 173: 998–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elumalai S, Tobimatsu Y, Grabber JH, Pan X, Ralph J(2012) Epigallocatechin gallate incorporation into lignin enhances the alkaline delignification and enzymatic saccharification of cell walls. Biotechnol Biofuels 5: 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forkner R, Marquis R, Lill J(2004) Feeny revisited: Condensed tannins as anti-herbivore defences in leaf-chewing herbivore communities of Quercus. Environ Entomol 29: 174–187 [Google Scholar]
- Fraser K, Collette V, Hancock KR(2016) Characterization of proanthocyanidins from seeds of perennial ryegrass (Lolium perenne L.) and tall fescue (Festuca arundinacea) by liquid chromatography–mass spectrometry. J Agric Food Chem 64: 6676–6684 [DOI] [PubMed] [Google Scholar]
- Fryganas C, Drake C, Ropiak HM, Mora-Ortiz M, Smith LMJ, Mueller-Harvey I, Kowalczyk RM(2018) Carbon-13 cross-polarization magic-angle spinning nuclear magnetic resonance for measuring proanthocyanidin content and procyanidin to prodelphinidin ratio in sainfoin (Onobrychis viciifolia) issues. J Agric Food Chem 66: 4073–4081 [DOI] [PubMed] [Google Scholar]
- Fürtauer L, Küstner L, Weckwerth W, Heyer AG, Nägele T(2019) Resolving subcellular plant metabolism. Plant J 100: 438–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcıa DE, Glasser WG, Pizzi A, Paczkowski SP, Marie-Pierre Laborie M-P(2016) Modification of condensed tannins: From polyphenol chemistry to materials engineering. New J Chem 40: 36 [Google Scholar]
- Gargouri M, Manigand C, Maugé C, Granier T, Langlois d’Estaintot B, Cala O, Pianet I, Bathany K, Chaudière J, Gallois B(2009) Structure and epimerase activity of anthocyanidin reductase from Vitis vinifera. Acta Crystallogr D Biol Crystallogr 65: 989–1000 [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J(2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J 59: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourlay G, Constabel CP(2019) Condensed tannins are inducible antioxidants and protect hybrid poplar against oxidative stress. Tree Physiol 39: 345–355 [DOI] [PubMed] [Google Scholar]
- Grabber JH, Ress D, Ralph J(2012) Identifying new lignin bioengineering targets: Impact of epicatechin, quercetin glycoside, and gallate derivatives on the lignification and fermentation of maize cell walls. J Agric Food Chem 60: 5152–5160 [DOI] [PubMed] [Google Scholar]
- Grishechko LI, Amaral-Labat G, Szczurek A, Fierro V, Kuznetsov BN, Pizzi A, Celzard A(2013) New tannin–lignin aerogels. Ind Crops Prod 41: 347–355 [Google Scholar]
- Hancock KR, Collette V, Fraser K, Greig M, Xue H, Richardson K, Jones C, Rasmussen S(2012) Expression of the R2R3-MYB transcription factor TaMYB14 from Trifolium arvense activates proanthocyanidin biosynthesis in the legumes Trifolium repens and Medicago sativa. Plant Physiol 159: 1204–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamad AMA, Ates S, Olgun C, Gür M(2019) Chemical composition and antioxidant properties of some industrial tree bark extracts. BioRes 14: 5657–5671 [Google Scholar]
- Harding SA.(2019) Condensed tannins: Arbiters of abiotic stress tolerance? Tree Physiol 39: 341–344 [DOI] [PubMed] [Google Scholar]
- Haslam E, Opie CT, Porter LJ(1977) Procyanidin metabolism -a hypothesis. Phytochemistry 16: 99–102 [Google Scholar]
- Hjältén J, Axelsson EP(2015) GM trees with increased resistance to herbivores: Trait efficiency and their potential to promote tree growth. Front Plant Sci 6: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda K, Sasahara H, Matsushita K, Tamura Y, Miyaji M, Matsuyama H(2018) Anthocyanin and proanthocyanidin contents, antioxidant activity, and in situ degradability of black and red rice grains. Asian-Australas J Anim Sci 31: 1213–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YF, Doligez A, Fournier-Level A, Le Cunff L, Bertrand Y, Canaguier A, Morel C, Miralles V, Veran F, Souquet JM, et al. (2012) Dissecting genetic architecture of grape proanthocyanidin composition through quantitative trait locus mapping. BMC Plant Biol 12: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques D, Haslam E(1974) Biosynthesis of plant proanthocyanidins. J Chem Soc Chem Commun 1974: 231–232 [Google Scholar]
- James AM, Ma D, Mellway R, Gesell A, Yoshida K, Walker V, Tran L, Stewart D, Reichelt M, Suvanto J, et al. (2017) Poplar MYB115 and MYB134 transcription factors regulate proanthocyanidin synthesis and structure. Plant Physiol 174: 154–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Xiao X, Rao X, Dixon RA(2018) Proanthocyanidin subunit composition determined by functionally diverged dioxygenases. Nat Plants 4: 1034–1043 [DOI] [PubMed] [Google Scholar]
- Kawahigashi H, Kasuga S, Sawada Y, Yonemaru J-i, Ando A, Kanamori H, Wu J, Mizuno H, Momma M, Fujimoto Z, et al. (2016) Thes for leaf color changes upon wounding (P) encodes a flavanone 4-reductase in the 3-deoxyanthocyanidin biosynthesis pathway. G3 (Bethesda) 6: 1439–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinyok MJ, Bonnet S, Noté OP, Ngo Mbing J, Kamto EL, Van der Westhuizen JH, Pegnyemb DE(2017) A new flavanolignan and a new alkane from the Stem bark of Newtonia griffoniana. Nat Prod Res 31: 2233–2238 [DOI] [PubMed] [Google Scholar]
- Kitamura S, Matsuda F, Tohge T, Yonekura-Sakakibara K, Yamazaki M, Saito K, Narumi I(2010) Metabolic profiling and cytological analysis of proanthocyanidins in immature seeds of Arabidopsis thaliana flavonoid accumulation mutants. Plant J 62: 549–559 [DOI] [PubMed] [Google Scholar]
- Koyama K, Numata M, Nakajima I, Goto-Yamamoto N, Matsumura H, Tanaka N(2014) Functional characterization of a new grapevine MYB transcription factor and regulation of proanthocyanidin biosynthesis in grapes. J Exp Bot 65: 4433–4449 [DOI] [PubMed] [Google Scholar]
- Krych J, Gebicka L(2013) Catalase is inhibited by flavonoids. Int J Biol Macromol 58: 148–153 [DOI] [PubMed] [Google Scholar]
- Lam PY, Tobimatsu Y, Takeda Y, Suzuki S, Yamamura M, Umezawa T, Lo C(2017) Disrupting flavone synthase II alters lignin and improves biomass digestibility. Plant Physiol 174: 972–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan W, Lu F, Regner M, Zhu Y, Rencoret J, Ralph SA, Zakai UI, Morreel K, Boerjan W, Ralph J(2015) Tricin, a flavonoid monomer in monocot lignification. Plant Physiol 167: 1284–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Flachowsky H, Fischer TC, Hanke MV, Forkmann G, Treutter D, Schwab W, Hoffmann T, Szankowski I(2007) Maize Lc transcription factor enhances biosynthesis of anthocyanins, distinct proanthocyanidins and phenylpropanoids in apple (Malus domestica Borkh.). Planta 226: 1243–1254 [DOI] [PubMed] [Google Scholar]
- Liang M, Davis E, Gardner D, Cai X, Wu Y(2006) Involvement of AtLAC15 in lignin synthesis in seeds and in root elongation of Arabidopsis. Planta 224: 1185–1196 [DOI] [PubMed] [Google Scholar]
- Liu C, Wang X, Shulaev V, Dixon RA(2016) A role for leucoanthocyanidin reductase in the extension of proanthocyanidins. Nat Plants 2: 16182. [DOI] [PubMed] [Google Scholar]
- Lv Y, Gao S, Xu S, Du G, Zhou J, Chen J(2017) Spatial organization of silybin biosynthesis in milk thistle [Silybum marianum (L.) Gaertn]. Plant J 92: 995–1004 [DOI] [PubMed] [Google Scholar]
- Madritch MD, Lindroth RL(2015) Condensed tannins increase nitrogen recovery by trees following insect defoliation. New Phytol 208: 410–420 [DOI] [PubMed] [Google Scholar]
- Marsh KJ, Wallis IR, Kulheim C, Clark R, Nicolle D, Foley WJ, Salminen JP(2020) New approaches to tannin analysis of leaves can be used to explain in vitro biological activities associated with herbivore defence. New Phytol 225: 488–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellway RD, Tran LT, Prouse MB, Campbell MM, Constabel CP(2009) The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol 150: 924–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon LR, McAllister TA, Berg BP, Majak W, Acharya SN, Popp JD, Coulman BE, Wang Y, Cheng K-J(2000) A review of the effects of forage condensed tannins on ruminal fermentation and bloat in grazing cattle. Can J Plant Sci 80: 469–485 [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD(2016) Designer lignins: Harnessing the plasticity of lignification. Curr Opin Biotechnol 37: 190–200 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Takahashi S, Waki T(2019) Formation of flavonoid metabolons: Functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front Plant Sci 10: 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palo RT.(1984) Distribution of birch (Betula SPP.), willow (Salix SPP.), and poplar (Populus SPP.) secondary metabolites and their potential role as chemical defense against herbivores. J Chem Ecol 10: 499–520 [DOI] [PubMed] [Google Scholar]
- Pang Y, Abeysinghe ISB, He J, He X, Huhman D, Mewan KM, Sumner LW, Yun J, Dixon RA(2013) Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol 161: 1103–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang Y, Peel GJ, Sharma SB, Tang Y, Dixon RA(2008) A transcript profiling approach reveals an epicatechin-specific glucosyltransferase expressed in the seed coat of Medicago truncatula. Proc Natl Acad Sci USA 105: 14210–14215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP(2002) Molecular analysis of herbivore-induced condensed tannin synthesis: Cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701–712 [DOI] [PubMed] [Google Scholar]
- Pizzi A, Cameron FA(1986) Flavonoid tannins - structural wood components for drought-resistance mechanisms of plants. Wood Sci Technol 20: 119–124 [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I(2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinkenstein A, Jochheim H(2016) Assessing the carbon sequestration potential of poplar and black locust short rotation coppices on mine reclamation sites in Eastern Germany - Model development and application. J Environ Manage 168: 53–66 [DOI] [PubMed] [Google Scholar]
- Rauf A, Imran M, Abu-Izneid T, Iahtisham-Ul-Haq, Patel S, Pan X, Naz S, Sanches Silva A, Saeed F, Rasul, et al. (2019) Proanthocyanidins: A comprehensive review. Biomed Pharmacother 116: 108999. [DOI] [PubMed] [Google Scholar]
- Sette M, Wechselberger R, Crestini C(2011) Elucidation of lignin structure by quantitative 2D NMR. Chemistry 17: 9529–9535 [DOI] [PubMed] [Google Scholar]
- Salminen J-P.(2018) Two-dimensional tannin fingerprints by liquid chromatography tandem mass spectrometry offer a new dimension to plant tannin analyses and help to visualize the tannin diversity in plants. J Agric Food Chem 66: 9162–9171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stafford HA.(1983) Enzymic regulation of procyanidin biosynthesis; lack of a flav-3-en-3-ol intermediate. Phytochemistry 22: 2643–2646 [Google Scholar]
- Stafford HA.(1988) Proanthocyanidins and the lignin connection. Phytochemistry 27: 1–6 [Google Scholar]
- Sun H, Liu Y, Gai Y, Geng J, Chen L, Liu H, Kang L, Tian Y, Li Y(2015) De novo sequencing and analysis of the cranberry fruit transcriptome to identify putative genes involved in flavonoid biosynthesis, transport and regulation. BMC Genomics 16: 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li H, Huang J-R(2012) Arabidopsis TT19 functions as a carrier to transport anthocyanin from the cytosol to tonoplasts. Mol Plant 5: 387–400 [DOI] [PubMed] [Google Scholar]
- Tanner GJ, Francki KT, Abrahams S, Watson JM, Larkin PJ, Ashton AR(2003) Proanthocyanidin biosynthesis in plants. Purification of legume leucoanthocyanidin reductase and molecular cloning of its cDNA. J Biol Chem 278: 31647–31656 [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Harding SA, Tschaplinski TJ, Lindroth RL, Yuan Y(2006) Genome-wide analysis of the structural genes regulating defense phenylpropanoid metabolism in Populus. New Phytol 172: 47–62 [DOI] [PubMed] [Google Scholar]
- Turnbull JJ, Nakajima J, Welford RWD, Yamazaki M, Saito K, Schofield CJ(2004) Mechanistic studies on three 2-oxoglutarate-dependent oxygenases of flavonoid biosynthesis: anthocyanidin synthase, flavonol synthase, and flavanone 3beta-hydroxylase. J Biol Chem 279: 1206–1216 [DOI] [PubMed] [Google Scholar]
- Ullah C, Tsai C-J, Unsicker SB, Xue L, Reichelt M, Gershenzon J, Hammerbacher A(2019a) Salicylic acid activates poplar defense against the biotrophic rust fungus Melampsora larici-populina via increased biosynthesis of catechin and proanthocyanidins. New Phytol 221: 960–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C, Unsicker SB, Fellenberg C, Constabel CP, Schmidt A, Gershenzon J, Hammerbacher A(2017) Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol 175: 1560–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah C, Unsicker SB, Reichelt M, Gershenzon J, Hammerbacher A(2019b) Accumulation of catechin and proanthocyanidins in black poplar stems after infection by Plectosphaerella populi: Hormonal regulation, biosynthesis and antifungal activity. Front Plant Sci 10: 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vio-Michaelis S, Feucht W, Gómez M, Hadersdorfer J, Treutter D, Schwab W(2020) Histochemical analysis of anthocyanins, carotenoids, and flavan-3-ols/proanthocyanidins in Prunus domestica L. fruits during ripening. J Agric Food Chem 68: 2880–2890 [DOI] [PubMed] [Google Scholar]
- Waki T, Mameda R, Nakano T, Yamada S, Terashita M, Ito K, Tenma N, Li Y, Fujino N, Uno K, et al. (2020) A conserved strategy of chalcone isomerase-like protein to rectify promiscuous chalcone synthase specificity. Nat Commun 11: 870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Hou B(2009) Glycosyltransferases: key players involved in the modification of plant secondary metabolites. Front Biol China 4: 39–46 [Google Scholar]
- Wang L, Ran L, Hou Y, Tian Q, Li C, Liu R, Fan D, Luo K(2017) The transcription factor MYB115 contributes to the regulation of proanthocyanidin biosynthesis and enhances fungal resistance in poplar. New Phytol 215: 351–367 [DOI] [PubMed] [Google Scholar]
- Wang P, Liu Y, Zhang L, Wang W, Hou H, Zhao Y, Jiang X, Yu J, Tan H, Wang Y, et al. (2020) Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J 101: 18–36 [DOI] [PubMed] [Google Scholar]
- Wang YS, Xu YJ, Gao LP, Yu O, Wang XZ, He XJ, Jiang XL, Liu YJ, Xia T(2014) Functional analysis of flavonoid 3′,5′-hydroxylase from tea plant (Camellia sinensis): critical role in the accumulation of catechins. BMC Plant Biol 14: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder RS, Lamarche J, Constabel CP, Hamelin RC(2013) The effects of high-tannin leaf litter from transgenic poplars on microbial communities in microcosm soils. Front Microbiol 4: 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D-Y, Dixon RA(2005) Proanthocyanidin biosynthesis—still more questions than answers? Phytochemistry 66: 2127–2144 [DOI] [PubMed] [Google Scholar]
- Xie D-Y, Sharma SB, Paiva NL, Ferreira D, Dixon RA(2003) Role of anthocyanidin reductase, encoded by BANYULS in plant flavonoid biosynthesis. Science 299: 396–399 [DOI] [PubMed] [Google Scholar]
- Xiong J, He J, Xie WP, Hinojosa E, Ambati CSR, Putluri N, Kim H-E, Zhu MX, Du G(2019) Rapid affinity purification of intracellular organelles using a twin strep tag. J Cell Sci 132: jcs235390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Wang L, Han Z, Jiang Y, Zhao L, Liu H, Yang L, Luo K(2012) Molecular cloning and characterization of PtrLAR3, a gene encoding leucoanthocyanidin reductase from Populus trichocarpa, and its constitutive expression enhances fungal resistance in transgenic plants. J Exp Bot 63: 2513–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhang Y, Qiu H, Guo Y, Wan H, Zhang X, Scossa F, Alseekh S, Zhang Q, Wang P, et al. (2020) Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat Commun 11: 3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dixon RA(2009) MATE transporters facilitate vacuolar uptake of epicatechin 3′-O-glucoside for proanthocyanidin biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21: 2323–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Jiang L(2019) Chloroplast degradation: Multiple routes into the vacuole. Front Plant Sci 10: 359. [DOI] [PMC free article] [PubMed] [Google Scholar]