Despite a tradeoff between vigor and the effect of salt stress, this relationship can be modulated by plasticity in root traits, leaf K and Na content, and an alpha-mannosidase gene.
Abstract
With rising food demands, crop production on salinized lands is increasingly necessary. Sunflower (Helianthus annuus), a moderately salt-tolerant crop, exhibits a tradeoff where more vigorous, high-performing genotypes have a greater proportional decline in biomass under salinity stress. Prior research has found deviations from this relationship across genotypes. Here, we identified the traits and genomic regions underlying variation in this expectation-deviation tolerance (the magnitude and direction of deviations from the expected effect of salinity). We grew a sunflower diversity panel under control and salt-stressed conditions and measured a suite of morphological (growth, mass allocation, plant and leaf morphology) and leaf ionomic traits. The genetic basis of variation and plasticity in these traits was investigated via genome-wide association, which also enabled the identification of genomic regions (i.e. haplotypic blocks) influencing multiple traits. We found that the magnitude and direction of plasticity in whole-root mass fraction, fine root mass fraction, and chlorophyll content, as well as leaf sodium and potassium content under saline conditions, were most strongly correlated with expectation-deviation tolerance. We identified multiple genomic regions underlying these traits as well as a single alpha-mannosidase gene directly associated with this tolerance metric. Our results show that, by taking the vigor-salinity effect tradeoff into account, we can identify unique traits and genes associated with salinity tolerance. Since these traits and genomic regions are distinct from those associated with high vigor (i.e. growth in benign conditions), they provide an avenue for increasing salinity tolerance in high-performing sunflower genotypes without compromising vigor.
The rapid rise of global population levels has increased strain on our food production systems (Ramankutty et al., 2018). With demands projected to nearly double by the middle of this century, expanded efforts to improve crop productivity are warranted. One factor limiting crop productivity is high soil salinity (generally NaCl) caused by poor irrigation practices, salt water encroachment, and/or drought. With over 20% of the world’s irrigated agricultural land being impacted by salinity and expansion needing to occur on less favorable lands, developing crop varieties more suitable for salinized soils is vital (Munns, 2005, FAO, 2005; Munns et al., 2020a). However, mitigating the physiological problems imposed on plants by high soil NaCl concentrations remains a challenging task (Munns et al., 2020a).
High soil salinity imposes two types of stress on plants. First, as a solute, dissolved NaCl imposes an osmotic stress that limits leaf expansion (Rawson and Munns, 1984) and photosynthesis through reduced transpiration, similar to drought (Munns, 2002; Munns and Tester, 2008). Second, either to combat the imposed osmotic stress or through unavoidable net leakage into the roots, plants take up NaCl from the soil (Munns et al., 2020a). Accumulated sodium (Na) poses a risk of ion toxicity (Munns and Tester, 2008) and must either be excreted (a trait limited to salt-adapted species; Cheeseman, 2015) or sequestered in roots and stems (Cuin et al., 2011; Munns et al., 2012; Guan et al., 2014) or vacuoles (Mansour et al., 2003; Hasegawa, 2013; Bassil et al., 2019; Shabala et al., 2019). An improved understanding of the genetic basis of the traits underlying salt tolerance would accelerate the development of increasingly resilient cultivars (Zhu et al., 2016; Morton et al., 2019). Given the large set of traits and mechanisms related to salinity tolerance (Munns and Tester, 2008; Munns et al., 2020a, 2020b), however, the ability to maintain productivity under saline conditions is likely to be genetically complex (Flowers, 2004; Munns, 2005).
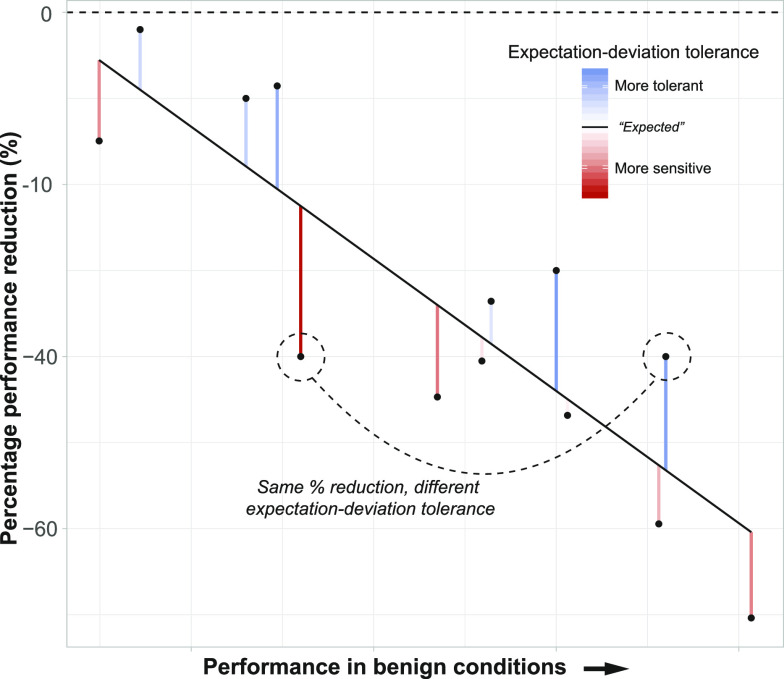
Numerous metrics for stress tolerance exist, leading to potentially conflicting conclusions depending on which metric is employed (Zhu et al., 2016; Morton et al., 2019). For example, a high performing genotype under saline conditions (suggesting tolerance) could also experience a large percentage performance decrease in salt stress (suggesting sensitivity). Furthermore, if there is a tradeoff (or negative correlation; Agrawal, 2020) between performance and performance under stress (Mayrose et al., 2011; Koziol et al., 2012), then optimizing for yield under ideal conditions could favor genotypes that are more negatively impacted by stress. Conversely, optimizing for genotypes that are least impacted by stress could result in poor overall performance. An alternative approach is to decouple the metric of tolerance from performance (Fig. 1). Given an observed relationship between performance under benign conditions and the impact of stress across genotypes, the deviation of a given genotype from this overall relationship can be viewed as a measure of tolerance/sensitivity. In other words, if a genotype outperforms the expectation given its ideal performance, it exhibits evidence of stress tolerance relative to other genotypes. Similarly, if a genotype underperforms relative to the expectation, it exhibits evidence of stress sensitivity.
Figure 1.
Conceptual diagram of expectation-deviation tolerance. Given a negative relationship between performance (estimated using any relevant measure) and the proportional effect of stress, along with variation in both, it can be difficult to define tolerance versus sensitivity. Two different genotypes (highlighted with dashed circles/line) could have the same percentage reduction in performance due to stress while differing greatly when compared to their expected performance reduction when accounting for inherent differences in performance under benign conditions (red/blue shading). The magnitude of this deviation (red/blue lines) from the best-fit line (solid line) provides an estimate of tolerance/sensitivity that is independent of differences in performance under benign conditions. Residuals from this best-fit line thus reflect genotypes that are more sensitive (below the line, highlighted red) or tolerant than expected (above the line, highlighted blue; see the introduction for more details).
In cultivated sunflower (Helianthus annuus), prior work has found a tradeoff between vigor and the effect of salinity, with the more vigorous genotypes (higher growth under ideal conditions) exhibiting a greater decrease in biomass under stress (Temme et al., 2019a, 2019b). In these small sets of genotypes, it was possible to take this tradeoff into account and quantify tolerance using a genotype’s deviation from the expected response (Fig. 1). This expectation-deviation tolerance was weakly correlated with Na sequestration in the stem, and this differed from trait associations with a more common proportional-reduction tolerance metric (Temme et al., 2019a). By selecting on traits associated with better-than-expected performance, it could be possible to modulate this vigor/stress response tradeoff. A clearer picture of the traits underlying vigor as well as the extent to which multivariate trait expression is driven by genetic correlations (i.e. pleiotropy or close linkage; Auge et al., 2019) will inform on the feasibility of this approach to modulate this negative relationship.
Given the central role of Na in salinity stress, extensive research has been done on its effect on plant processes (Mäser et al., 2002; Broadley et al., 2012; Hasegawa, 2013). Due to its ionic size, charge, and processes involved in sequestration, Na uptake affects the accumulation of other essential macro- and micronutrients. Most notably, potassium (K) uptake is greatly affected when Na passes through K channels and disrupts the K:Na balance across membranes during Na sequestration (Shabala and Cuin, 2008; Shabala et al., 2019; Munns et al., 2020a). However, other elements are also likely to play a role in salinity tolerance. Decreases in the cost of inductively coupled plasma mass spectroscopy have enabled the measurement of a range of elements, collectively referred to as the ionome (Salt et al., 2008) and how they relate to salinity tolerance (Wu et al., 2018; Temme et al., 2019a, 2019b; Munns et al., 2020b).
In stressful environments, plants exhibit different trait values than under benign conditions. Explicit consideration of this trait plasticity has gained momentum as a useful tool in understanding trait variation across environments (Nicotra et al., 2010; Kusmec et al., 2017; Arnold et al., 2019; Laitinen and Nikoloski, 2019). Moreover, plasticity in some traits can be required for robustness in others (e.g. performance or yield; Laitinen and Nikoloski, 2019). For example, in salt marshes, the success of invasive Japanese knotweed (Fallopia japonica) is linked to plasticity in leaf succulence (Richards et al., 2008). Similarly, in cultivated sunflower, changes in leaf sulfur (S) content are associated with the maintenance of growth under salinity stress (Temme et al., 2019b). Examples such as these highlight the potential utility of trait plasticity for improving salt tolerance. The finding of distinct genomic regions underlying trait variation and plasticity in maize (Zea mays; Kusmec et al., 2017) and sunflower (Mangin et al., 2017) further suggests that trait expression can be decoupled from trait adjustment under stress.
Here, we examine the suite of traits and genomic regions underlying salt tolerance, independent from vigor, in cultivated sunflower. Sunflower is a globally important oilseed crop that exhibits moderate, but genetically variable, salt tolerance (Katerji et al., 2000; Shi and Sheng, 2005; Temme et al., 2019a, 2019b). As demands for seed oil are expected to increase 70% by 2050 (Ramankutty et al., 2018), improvements need to be made in cultivars under production. Improving our understanding of the genetic basis of variation in the physiological mechanisms conferring tolerance to salinity stress will allow for rapid selection on these traits (Flexas and Gago, 2018; York, 2019). Here, we use detailed phenotypic and ionomic characterization and genome-wide association studies (GWASs) to address this issue by answering the following questions: (1) What is the effect of salinity on morphological traits (vegetative growth, biomass allocation, plant and leaf morphology) and leaf ionomic traits in cultivated sunflower? (2) What is the link among trait expression, trait plasticity, and salinity expectation-deviation tolerance? (3) How heritable are the traits underlying expectation-deviation tolerance? (4) Which genomic regions influence vigor, expectation-deviation tolerance, and associated traits, and to what extent are these traits genetically correlated?
RESULTS
Sunflower Traits Are Affected by Salinity Stress and Linked with Salinity Tolerance
Salinity stress greatly affected growth, morphology, and the leaf ionome across our cultivated sunflower diversity panel, which included 239 genotypes representing major heterotic group/market-type combinations as well as landraces, open-pollinated varieties, and cultivated genotypes carrying wild species introgressions (Mandel et al., 2011, 2013). While there was substantial variation among genotypes, the median response to salt stress across genotypes was a 56.3% reduction in biomass and substantially greater biomass allocation to roots (50.4% increase in root mass fraction [RMF]) at the expense of stem allocation (35.2% decrease in stem mass fraction [SMF]). Leaf mass per area increased by 46.4%, indicating thicker or denser leaves. Changes to the leaf ionome were substantial, with a median increase of 101.4% in manganese (Mn) content and 4395.2% in Na content (Table 1; Supplemental Fig. S1). For all growth and morphological traits (i.e. all nonelemental traits) analyzed, genotypes (G) differed significantly in their trait values (P < 0.001) as well as in their response to salinity (genotype-by-treatment [G×T] interaction; P < 0.001). Due to the presence of this interaction, the main effect of treatment was nonsignificant for chlorophyll content and several root traits. This indicates substantial spread in the response to salinity, with some genotypes increasing in these trait values and others decreasing, highlighting significant differences in the magnitude and direction of the salinity response between sunflower genotypes.
Table 1. Effect of salt stress on sunflower traits.
Traits measured in our cultivated sunflower diversity panel with their median value (based on genotype means) and range (in parentheses), when grown under control (0 mm NaCl) and salt-stressed (100 mm NaCl) conditions, as well as an estimate of their plasticity (trait adjustment) between treatments. Plasticity was calculated as the difference in natural log transformed values (control – salt) but converted here to Δ% versus control (via eΔln(trait) – 1) for ease of interpretation. For all nonelemental traits, stars indicate Wald’s χ2 significance (with four replicate plants per genotype) of the main effects of genotype (G), treatment (T), and their interaction (G×T). As tissue was bulked for elemental content (i.e., no replication per genotype). For elemental traits stars indicate significance of a t test between control and salt-stressed groups (*P < 0.05, **P < 0.01, and ***P < 0.001). LMF, Leaf mass fraction; MF, mass fraction; ns, nonsignificant; RF, root fraction.
| Trait | 0 mm NaCl | 100 mm NaCl | Plasticity (Δ%) | T | G | G×T | |||
|---|---|---|---|---|---|---|---|---|---|
| Plant mass (g) | 4.31 | (1.01–10.65) | 1.86 | (0.43–4.82) | −56.3 | (−85.29–47.7) | ** | *** | *** |
| Leaf mass (g) | 2.2 | (0.62–5.54) | 0.98 | (0.27–2.48) | −53.1 | (−83.89–43.36) | * | *** | *** |
| LMF (gleaves gplant−1) | 0.5 | (0.4–0.73) | 0.54 | (0.39–0.68) | 7 | (−14.62–42.75) | * | *** | *** |
| LMA (g m−2) | 22.62 | (14.12–32.69) | 32.87 | (26.91–41.65) | 46.4 | (7.12–123.92) | *** | *** | *** |
| Height (cm) | 57.75 | (9.5–80.63) | 23.88 | (6.63–43.38) | −57.7 | (−77.2 to −21.24) | *** | *** | *** |
| Stem mass (g) | 1.52 | (0.05–3.9) | 0.42 | (0.06–1.35) | −70.9 | (−94.92–4.15) | *** | *** | *** |
| SMF (gstem gplant−1) | 0.36 | (0.11–0.45) | 0.23 | (0.1–0.35) | −35.2 | (−61.36 to −4.4) | *** | *** | *** |
| Stem diameter (mm) | 8.7 | (5.2–14.61) | 5.12 | (2.71–7.55) | −39.9 | (−65.56 to −8.82) | *** | *** | *** |
| Shoot mass (g) | 3.78 | (0.8–9.45) | 1.48 | (0.33–3.83) | −59.5 | (−85.65 to −27.85) | ** | *** | *** |
| All root mass (g) | 0.56 | (0.12–1.57) | 0.36 | (0.03–0.89) | −36.7 | (−94.47–242.13) | ns | *** | *** |
| Tap root mass (g) | 0.23 | (0.05–0.51) | 0.14 | (0.02–0.35) | −36.3 | (−91.21–150.67) | ns | *** | *** |
| Lateral root mass (g) | 0.33 | (0.07–1.13) | 0.21 | (0.01–0.69) | −35.6 | (−97.34–298.95) | ns | *** | *** |
| RMF (groots gplant−1) | 0.13 | (0.08–0.22) | 0.2 | (0.11–0.28) | 50.4 | (−34.79–134.02) | ns | *** | *** |
| Tap root MF (gtap root gplant−1) | 0.05 | (0.02–0.14) | 0.08 | (0.03–0.17) | 49.5 | (−56.86–229.86) | * | *** | *** |
| Tap root RF (gtap root groots−1) | 0.41 | (0.2–0.71) | 0.41 | (0.17–0.63) | −1.6 | (−56.97–156.03) | ns | *** | *** |
| Fine root MF (gfine root gplant−1) | 0.08 | (0.04–0.14) | 0.12 | (0.06–0.22) | 52.4 | (−28.88–194.57) | ns | *** | *** |
| Fine root RF (gfine root groots−1) | 0.59 | (0.29–0.8) | 0.59 | (0.37–0.83) | 1 | (−38.43–46.88) | ns | *** | *** |
| Chlorophyll content (index) | 14.71 | (8.86–26.15) | 17.79 | (5.64–32.27) | 20.3 | (−47.6–127.69) | ns | *** | *** |
| B (µg g−1) | 100 | (55–162) | 109 | (52–213) | 9.3 | (−38.82–98.77) | *** | – | – |
| Ca (mass %) | 1.23 | (0.79–2.17) | 1.28 | (0.7–2.42) | 1.4 | (−47.18–130.48) | ns | – | – |
| Cu (µg g−1) | 27 | (15–49) | 36 | (21–65) | 37.5 | (−23.4–125) | *** | – | – |
| Fe (µg g−1) | 139 | (95–535) | 127.5 | (74–934) | −9.1 | (−64.11–661.67) | ** | – | – |
| Mg (mass %) | 0.41 | (0.26–0.64) | 0.41 | (0.23–0.76) | −2.5 | (−41.51–94.87) | ns | – | – |
| Mn (µg g−1) | 670 | (350–1363) | 1358.5 | (429–3663) | 101.4 | (−2.51–439.47) | *** | – | – |
| N (mass %) | 7.19 | (5.94–8.72) | 5.97 | (4.65–7.1) | −16.2 | (−34.94–3.24) | *** | – | – |
| P (mass %) | 0.44 | (0.25–0.84) | 0.3 | (0.17–0.46) | −30.6 | (−65.48–20) | *** | – | – |
| K (mass %) | 5.54 | (3.93–7.6) | 5 | (2.15–7) | −9.3 | (−52.51–41.34) | *** | – | – |
| Na (mass %) | 0.02 | (0.01–0.1) | 1.14 | (0.13–4.66) | 4395.2 | (288.33–48,916.67) | *** | – | – |
| S (mass %) | 0.71 | (0.52–1.54) | 0.52 | (0.36–0.95) | −27.5 | (−52.13–3.28) | *** | – | – |
| Zn (µg g−1) | 84 | (47–238) | 108 | (55–471) | 27.5 | (−60.14–324.32) | *** | – | – |
| K/Na ratio | 225.2 | (60.93–973.33) | 4.41 | (0.67–49.06) | −98 | (−99.87 to −75.19) | *** | – | – |
Due to our bulking of leaf tissue across replicates for each genotype, we could not contrast genotypes for elemental content. However, by using genotypes as the unit of replication, we could still assess the impact of salinity on the diversity panel as a whole. Of the 12 elements analyzed, all but calcium (Ca) and magnesium (Mg) were significantly affected by salinity. This lack of overall effect for these two elements is likely due to the large variance in response with content halving in some genotypes and doubling in others. Under salt stress, foliar concentrations significantly increased for boron (B; median 9.3%), copper (Cu; median 37.5%), Mn (median 101.4%), Na (median 4395.2%), and zinc (Zn; median 27.5%). In contrast, foliar concentrations decreased for iron (Fe; median 9.1%), nitrogen (N; median 16.2%), phosphorus (P; median 30.6%), K (median 9.3%), and S (median 27.5%; Table 1; Supplemental Fig. S1). These results indicate the substantial effect of excess NaCl on the accumulation and concentration of other elements.
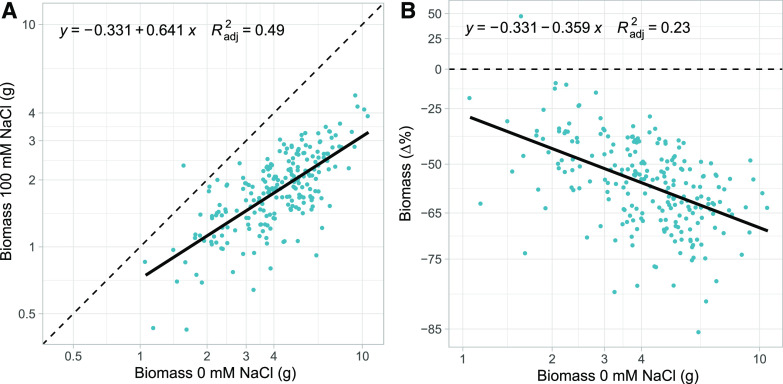
While all but one genotype decreased in biomass accumulation under salinity stress, there was a strong positive relationship between biomass in control conditions and biomass in saline conditions. More vigorous genotypes (i.e. higher biomass in control conditions) tended to accumulate the most biomass under salt-stressed conditions as well (P < 0.001, R2=0.49; Fig. 2A). However, these more vigorous genotypes also tended to have a greater proportional decrease in biomass under saline conditions (P < 0.001, R2 = 0.23; Fig. 2B). Given this tradeoff between vigor and the effect of salinity, we quantified tolerance as the deviation of each genotype from this expected response. Thus, residuals from the fitted line (Fig. 2B) served as estimates of these expectation-deviation tolerance values (Fig. 1). By decoupling tolerance from vigor, we can isolate those traits that are tied to better-than-expected performance, independent of traits conferring vigor.
Figure 2.
Effects of salt stress on whole-plant biomass. A, Whole-plant biomass of across our cultivated sunflower diversity panel under control (0 mm NaCl) and salt-stressed (100 mm NaCl) conditions. Dotted line indicates a 1:1 relationship, while the solid line is the fitted regression. B, Plasticity in biomass under salt stress versus biomass at control conditions. Plasticity was calculated as the difference in natural log transformed values (control-salt) but converted here to Δ% (via eΔln(trait) − 1) change from control for ease of interpretation. Dotted line indicates zero difference (no salt effect), while the solid line is the fitted regression. Blue dots indicate estimated marginal means of four replicate plants per genotype per treatment. Note the natural log scaling of the axes.
Across the measured traits, we observed numerous significant trait correlations in both the control and salt-stressed environments (Fig. 3A). Some notable relationships in the control treatment were that vigor (biomass) was negatively correlated with overall root mass fraction (⍴ = −0.27, P < 0.001) but positively correlated with the proportion of root mass made up by fine roots (⍴ = 0.36, P < 0.001). While vigor was negatively correlated with leaf N (⍴ = −0.26, P < 0.001) and P (⍴ = −0.49, P < 0.001), it was positively correlated with Mg (⍴ = 0.18, P < 0.05) and Mn (⍴ = 0.38, P < 0.001). The trait correlations under saline conditions differed from those of the control treatment. For example, biomass was not correlated with Mg concentration but was positively correlated with S (⍴ = 0.146, P < 0.05) and negatively correlated with Na (⍴ = −0.261, P < 0.001). Additionally, under saline conditions, Na was negatively correlated with K (⍴ = −0.520, P < 0.001), S (⍴ = −0.310, P < 0.001), and N (⍴ = −0.199, P < 0.01) but positively correlated with P (⍴ = 0.316, P < 0.001). All traits were positively correlated between treatments, though there was substantial variation in correlation strength across traits. While these results illustrate the high level of connectedness of trait expression in sunflower, it should be noted that the causality in these bivariate trait relationships cannot be undetermined.
Figure 3.
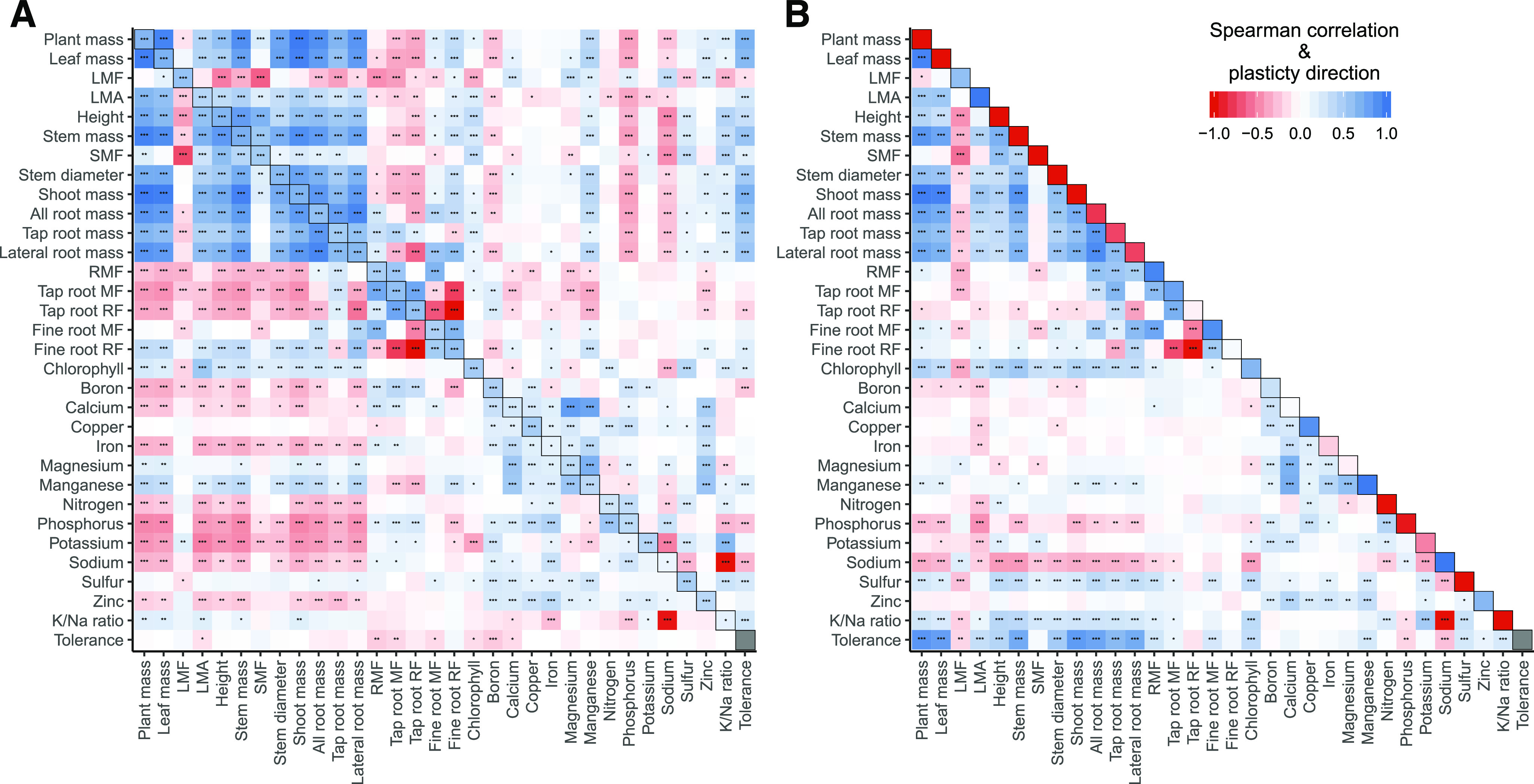
Sunflower trait correlations. A, Spearman correlation matrix of phenotypic and elemental traits in our cultivated sunflower diversity panel under control (0 mm NaCl; lower diagonal) and salt-stressed (100 mm NaCl; upper diagonal) conditions. Within-trait correlations between the control and stressed conditions are on the diagonal. B, Spearman correlation matrix of plasticity in trait value, Δln(trait) = ln(control) − ln(stress), between all phenotypic and elemental traits. The average direction and uniformity of the shift in trait value due to salinity stress is on the diagonal (−1 = all genotypes decrease in trait value, +1 = all genotypes increase in trait value). For tolerance, we used our expectation-deviation metric. As expectation-deviation tolerance is a compound trait based on both the control and salt treatments, comparisons across treatments do not apply, and genotype tolerance values were reused for each correlation matrix. Correlation values range from −1 (red) to +1 (blue). Stars in tiles indicate the significance of correlations (*P < 0.05, **P < 0.01, and ***P < 0.001).
Across genotypes, plasticity in response to salinity was also highly correlated among traits, with shifts in one trait being highly correlated with shifts in other traits (Fig. 3B). Note that, when interpreting the sign of the correlation between trait shifts, it is important to consider the sign of the change for each of the two correlated traits. For example, a positive correlation could result from two traits that both increase or both decrease with salinity but, perhaps counterintuitively, also between one trait that increases and another that decreases with salinity when a smaller decrease in one is coupled with a larger increase in the other. Changes in biomass traits were all positively correlated, such that a greater decrease in any biomass component in response to salinity was coupled with a greater decrease in other components. However, in terms of biomass allocation changes, the leaf, stem, and root mass fractions were all negatively correlated, indicating variability in how genotypes adjust their biomass allocation. For leaf elemental content, changes in leaf elemental concentration for Mg, Mn, and Ca were positively correlated. Sodium accumulation was correlated with multiple other elements, such that genotypes with a smaller increase in Na under salt stress had a smaller decrease in N (⍴ = −0.21, P < 0.001), S (⍴ = −0.29, P < 0.001), and K (⍴ = −0.36, P < 0.001). For whole-plant biomass, genotypes with a greater proportional increase in leaf Na content had a greater decrease in biomass accumulation (⍴ = −0.30, P < 0.001). These results show that trait adjustments occurred in a concerted manner across genotypes but that genotypes differed in the extent to which trait adjustments occurred in response to salinity stress.
Given the substantial correlation among traits within treatments and among proportional changes across treatments, a multivariate approach incorporating trait correlations was used to provide an integrated view of the relationship of traits with both vigor and tolerance (Table 2). After separating out our putatively size-independent traits (i.e. biomass ratios, leaf mass per area [LMA], and chlorophyll content) and our leaf ionome traits, regression analysis of vigor on the major principal component (PC) axes revealed that high vigor under control conditions was associated most strongly with the first principal component of putatively size-independent traits (P < 0.001, R2 = 0.27) and the second principal component of the leaf ionome (P < 0.001, R2 = 0.22). The top traits loading on these principal components were variation in root mass allocation for the putatively size-independent principal component analysis (PCA) and a suite of concentrations including Mn, Mg, P, and N for the leaf element PCA (Tables 3 and 4; Supplemental Fig. S2). Thus, high vigor in control conditions appears to be linked with nutrient uptake (root mass allocation) and leaf content of key macronutrients (N and P).
Table 2. Relationship between sunflower vigor under control conditions, salinity tolerance, and correlated suites of size-independent and ionomic traits.
R2 and significance of the ordinary least -squares regression of the PC1 and PC2 values of the size-independent and leaf ionomic traits (chlorophyll content, fine root allocation [mass fraction and root fraction], LMA, leaf mass fraction [LMF], RMF, SMF, tap root allocation [mass fraction and root fraction]) and elemental traits (B, Ca, Cu, Fe, Mg, Mn, N, P, K, Na, S, Zn, K/Na ratio) in control (0 mm NaCl) and salt-stressed (100 mm NaCl) conditions, as well as the plasticity in trait values between both environments (see "Materials and Methods" for details). Highlighted in bold are the top two regressions with the highest explanatory power for vigor and tolerance For elemental traits stars indicate significance of a t test between control and salt-stressed groups (*P < 0.05, **P < 0.01, and ***P < 0.001).
| Trait Set | Treatment | PC Axis | Vigor | Tolerance |
|---|---|---|---|---|
| R2adj. | R2adj. | |||
| Size-independent traits | Control | PC1 | 0.27*** | 0.02* |
| PC2 | 0.03** | – | ||
| Salt | PC1 | 0.08*** | 0.02* | |
| PC2 | – | 0.04** | ||
| Plasticity | PC1 | 0.02* | – | |
| PC2 | – | 0.17*** | ||
| Ionomic traits | Control | PC1 | 0.1*** | – |
| PC2 | 0.22*** | – | ||
| Salt | PC1 | 0.02* | – | |
| PC2 | – | 0.14*** | ||
| Plasticity | PC1 | 0.06*** | 0.04** | |
| PC2 | – | 0.04** |
Table 3. Principal component loadings of leaf elements.
Loadings (fraction of variance in trait explained by principal component) based on observed trait variation in our cultivated sunflower diversity panel for the first and second PCs under control (0 mm NaCl) and salt-stressed (100 mm NaCl) conditions as well as the plasticity in trait values between treatments. Bolded are the top three size-independent traits/elements per PC with the rank of the loading in parentheses.
| Element | Control | Salt | Plasticity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | |||||||
| B | 0.21 | (8) | 0.02 | (9) | 0.01 | (11) | 0.03 | (9) | 0.18 | (7) | 0.02 | (10) |
| Ca | 0.47 | (1) | 0.16 | (6) | 0.78 | (2) | >0.01 | (12) | 0.68 | (1) | 0.10 | (7) |
| Cu | 0.25 | (5) | >0.01 | (12) | 0.14 | (6) | >0.01 | (11) | 0.17 | (8) | >0.01 | (13) |
| Fe | 0.36 | (2) | >0.01 | (13) | 0.23 | (5) | 0.06 | (7) | 0.29 | (4) | 0.02 | (11) |
| K/NaRatio | 0.20 | (9) | 0.13 | (7) | 0.07 | (7) | 0.56 | (2) | 0.10 | (10) | 0.81 | (1) |
| Mg | 0.24 | (6) | 0.44 | (2) | 0.81 | (1) | >0.01 | (13) | 0.46 | (3) | 0.12 | (5) |
| Mn | 0.20 | (10) | 0.49 | (1) | 0.70 | (3) | 0.11 | (6) | 0.65 | (2) | 0.05 | (8) |
| N | 0.07 | (12) | 0.21 | (4) | 0.04 | (8) | 0.05 | (8) | 0.01 | (13) | 0.19 | (4) |
| P | 0.33 | (3) | 0.20 | (5) | 0.02 | (10) | 0.15 | (5) | 0.02 | (12) | >0.01 | (12) |
| K | 0.04 | (13) | 0.11 | (8) | >0.01 | (13) | 0.48 | (3) | 0.22 | (6) | 0.28 | (3) |
| Na | 0.23 | (7) | 0.26 | (3) | 0.04 | (9) | 0.74 | (1) | 0.06 | (11) | 0.77 | (2) |
| S | 0.13 | (11) | 0.02 | (10) | >0.01 | (12) | 0.25 | (4) | 0.13 | (9) | 0.10 | (6) |
| Zn | 0.25 | (4) | 0.01 | (11) | 0.36 | (4) | 0.03 | (10) | 0.26 | (5) | 0.02 | (9) |
Table 4. Principal component loadings of size-independent traits.
Loadings (fraction of variance in trait explained by principal component) based on observed trait variation in our cultivated sunflower diversity panel for the first and second PCs under control (0 mm NaCl) and salt-stressed (100 mm NaCl) conditions as well as the plasticity in trait values between treatments. Bolded are the top three size-independent traits/elements per PC with the rank of the loading in parentheses. LMF, Leaf mass fraction.
| Trait | Control | Salt | Plasticity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | PC1 | PC2 | |||||||
| Chlorophyll | 0.04 | (7) | 0.11 | (8) | 0.02 | (8) | 0.19 | (4) | 0.01 | (8) | 0.23 | (3) |
| fineMF | 0.04 | (8) | 0.36 | (3) | 0.34 | (4) | 0.52 | (3) | 0.09 | (5) | 0.79 | (1) |
| fineRF | 0.83 | (2) | 0.12 | (7) | 0.93 | (1) | 0.04 | (7) | 0.88 | (1) | 0.06 | (8) |
| LMA | 0.24 | (5) | 0.14 | (5) | 0.05 | (7) | 0.01 | (9) | 0.05 | (7) | 0.07 | (7) |
| LMF | >0.01 | (9) | 0.42 | (2) | 0.15 | (5) | 0.55 | (2) | 0.08 | (6) | 0.22 | (4) |
| RMF | 0.35 | (4) | 0.14 | (4) | 0.02 | (9) | 0.72 | (1) | 0.12 | (4) | 0.75 | (2) |
| SMF | 0.12 | (6) | 0.68 | (1) | 0.05 | (6) | 0.06 | (6) | 0.01 | (9) | 0.01 | (9) |
| tapMF | 0.86 | (1) | >0.01 | (9) | 0.75 | (3) | 0.11 | (5) | 0.77 | (3) | 0.14 | (5) |
| tapRF | 0.83 | (3) | 0.12 | (6) | 0.93 | (2) | 0.04 | (8) | 0.86 | (2) | 0.08 | (6) |
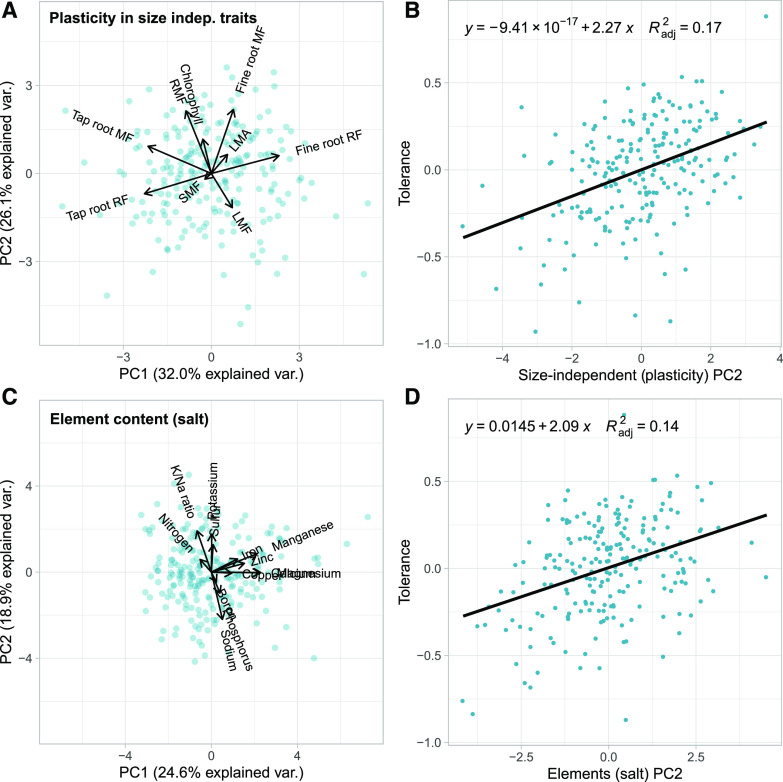
Tolerance to salinity, estimated as the residual from the fitted vigor versus effect-of-salinity tradeoff (i.e. expectation-deviation tolerance; Fig. 2B), was most strongly associated with principal components that differed from those associated with vigor. Expectation-deviation tolerance was most strongly associated with the second principal component of the PCA of plasticity in size-independent traits (P < 0.001, R2 = 0.17; Fig. 4, A and B) and the second principal component of the leaf ionomic PCA under salinity stress (P < 0.001, R2 = 0.14; Fig. 4, C and D). The top traits loading onto these principal components were, respectively, plasticity in root mass fraction, fine root mass fraction, and chlorophyll content; and Na content, K content, and K/Na ratio under saline conditions (Fig. 4, A and C; Tables 3 and 4). Thus, genotypes that had a greater increase in root mass and fine root mass fraction as well as a lower Na content, and higher K/Na ratio under saline conditions had a greater estimated tolerance, resulting in a lower than expected (based on their vigor) decrease in performance.
Figure 4.
Cultivated sunflower salinity tolerance and associated trait sets. A, Principal component analysis in our cultivated sunflower diversity panel (blue dots) plasticity in size-independent traits (chlorophyll content, fine root allocation [mass fraction and root fraction], LMA, leaf mass fraction (LMF), RMF, SMF, tap root allocation [mass fraction and root fraction]). B, Relationship between salinity tolerance (estimated as expectation-deviation tolerance; the deviation from the expected decrease in biomass due to salinity stress based on vigor in control conditions) and the second principal component of plasticity in size-independent traits. Negative tolerance values indicate more sensitive than expected and positive tolerance values indicate more tolerant than expected. C, Principal component analysis of leaf elemental traits (B, Ca, Cu, Fe, Mg, Mn, N, P, K, Na, S, Zn, and K/Na ratio) of plants grown under salt-stressed (100 mm NaCl) conditions. As above, dots reflect values for individual genotypes. D, Relationship between salinity tolerance and the second principal component of leaf ionomic traits under salt stress. Negative tolerance values indicate more sensitive than expected and positive tolerance values indicate more tolerant than expected.
Heritability Varies and GWAS Reveals Multiple Associations for Sunflower Traits and Plasticity
Estimates of narrow sense heritability (h2) revealed a broad range of additive genetic variance across traits in control and salt treatment with only modest differences between treatments (Table 5). A notable exception was that h2 of leaf Na concentration decreased from 0.5 to 0.22 under salt stress. It should be noted, however, that heritability values for the ionomic traits in control and salt-stressed treatment and the plasticity of all traits were based on genotypic averages, which increases uncertainty in the estimates. For the growth and morphological traits, h2 was generally high, with whole-plant biomass having an estimate of 0.47 in control conditions and 0.44 under saline conditions. Estimates for above-ground biomass traits were greater than for below-ground traits. Expectation-deviation tolerance had low h2 at just 0.05, although it should be noted again that this was necessarily estimated using genotypic averages.
Table 5. Heritability and genome-wide associations for sunflower traits in control (0 mm NaCl) and salt-stressed (100 mm NaCl) conditions and the plasticity between them.
h2, number of independent genomic regions with significant (GEMMA GWAS algorithm Wald test −log10(P) > 5.6) genome-wide associations, and the sum of the relative effect sizes (RES). Narrow sense heritability was calculated using individual plants for nonelemental traits and genotype means for elemental traits and trait plasticity values (h2 confidence interval in parentheses). Relative effect sizes (%) were calculated as (2*β)/(trait range), with β being the estimated effect of the minor allele in the region of interest, and then summed across significant regions for a given trait. Instances with RES > 100 likely reflect an overestimation of the number of independent regions, potentially due to inconsistencies in the underlying genome assembly. LMF, Leaf mass fraction; MF, mass fraction; RF, root fraction.
| Trait | 0 mm NaCl | 100 mm NaCl | Plasticity | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| h2 | No. Reg. | RES | h2 | No. Reg. | RES | h2 | No. Reg. | RES | ||||
| Plant mass (g) | 0.47 | (0.38–0.56) | 6 | 84.6 | 0.44 | (0.35–0.54) | 1 | 15.3 | 0.01 | (−0.03–0.05) | 1 | 10.7 |
| Leaf mass (g) | 0.44 | (0.35–0.54) | 4 | 52.7 | 0.44 | (0.35–0.53) | 3 | 56.8 | 0.01 | (−0.04–0.06) | – | – |
| LMF (gleaves gplant−1) | 0.45 | (0.37–0.53) | 15 | 310.1 | 0.32 | (0.23–0.41) | 1 | 22 | 0.08 | (0–0.15) | – | – |
| LMA (g m−2) | 0.28 | (0.19–0.37) | 1 | 11.7 | 0.25 | (0.16–0.34) | 2 | 49.7 | 0.03 | (−0.02–0.07) | 3 | 55.8 |
| Height (cm) | 0.54 | (0.47–0.62) | 5 | 100.8 | 0.64 | (0.57–0.7) | 2 | 43 | 0.13 | (0.03–0.24) | – | – |
| Stem mass (g) | 0.55 | (0.47–0.63) | 3 | 41.2 | 0.5 | (0.42–0.58) | 3 | 68.5 | 0.09 | (−0.01–0.18) | 1 | 14.3 |
| SMF (gstem gplant−1) | 0.53 | (0.45–0.6) | 6 | 125.8 | 0.49 | (0.4–0.57) | 2 | 39.1 | 0.05 | (−0.01–0.11) | 2 | 34.1 |
| Stem diameter (mm) | 0.45 | (0.36–0.54) | 1 | 20.9 | 0.3 | (0.21–0.38) | 2 | 29.8 | 0.11 | (0.01–0.2) | – | – |
| Shoot mass (g) | 0.49 | (0.4–0.58) | 3 | 41.3 | 0.47 | (0.38–0.56) | – | – | 0.01 | (−0.03–0.05) | 2 | 26.2 |
| All root mass (g) | 0.33 | (0.23–0.42) | 4 | 69.7 | 0.33 | (0.24–0.42) | 1 | 15.9 | 0.04 | (−0.02–0.1) | 4 | 43.2 |
| Tap root mass (g) | 0.3 | (0.21–0.39) | – | – | 0.3 | (0.21–0.39) | 2 | 27.4 | 0.07 | (−0.01–0.15) | 4 | 59.9 |
| Lateral root mass (g) | 0.35 | (0.26–0.44) | 5 | 83.6 | 0.31 | (0.22–0.39) | – | – | 0.03 | (−0.02–0.08) | 4 | 38.5 |
| RMF (groots gplant−1) | 0.19 | (0.12–0.27) | – | – | 0.14 | (0.07–0.21) | 1 | 17.7 | 0.06 | (0–0.12) | – | – |
| Tap root MF (gtap root gplant−1) | 0.07 | (0.04–0.11) | 1 | 16.3 | 0.09 | (0.04–0.14) | – | – | 0.09 | (0–0.17) | – | – |
| Tap root RF (gtap root groots−1) | 0.37 | (0.29–0.45) | – | – | 0.28 | (0.19–0.37) | – | – | 0.04 | (−0.02–0.09) | 1 | 10 |
| Fine root MF (gfine root gplant−1) | 0.07 | (0.02–0.12) | 1 | 19 | 0.12 | (0.05–0.19) | 4 | 72.3 | 0.03 | (−0.01–0.07) | – | – |
| Fine root RF (gfine root groots−1) | 0.37 | (0.29–0.45) | – | – | 0.28 | (0.19–0.37) | – | – | 0.03 | (−0.02–0.08) | 1 | 14.5 |
| Chlorophyll content (index) | 0.59 | (0.52–0.67) | 4 | 116.3 | 0.54 | (0.46–0.62) | 2 | 36.6 | 0.12 | (0.03–0.21) | – | – |
| B (µg g−1) | 0.29 | (0.13–0.45) | – | – | 0.18 | (0.05–0.31) | 4 | 84.1 | 0.09 | (0.01–0.18) | – | – |
| Ca (mass %) | 0.12 | (0.03–0.22) | 1 | 10.3 | 0.18 | (0.07–0.3) | 2 | 46.7 | 0.23 | (0.1–0.35) | 2 | 50.8 |
| Cu (µg g−1) | 0.19 | (0.07–0.31) | 3 | 85.7 | 0.23 | (0.1–0.37) | 2 | 48.2 | 0.14 | (0.03–0.25) | – | – |
| Fe (µg g−1) | 0.02 | (−0.02–0.07) | 19 | 294.9 | 0.03 | (−0.01 to −0.07) | 3 | 59.7 | 0.04 | (0–0.09) | – | – |
| Mg (mass %) | 0.25 | (0.1–0.4) | – | – | 0.28 | (0.13–0.43) | 2 | 51 | 0.31 | (0.15–0.46) | 1 | 18.6 |
| Mn (µg g−1) | 0.07 | (−0.01–0.15) | – | – | 0.15 | (0.06–0.25) | 4 | 65.9 | 0.13 | (0.03–0.22) | 6 | 104.4 |
| N (mass %) | 0.16 | (0.04–0.27) | – | – | 0.12 | (0.02–0.23) | 1 | 24 | 0.07 | (−0.01–0.16) | 3 | 48.1 |
| P (mass %) | 0.11 | (0.02–0.19) | – | – | 0.1 | (0.02–0.18) | 1 | 15.6 | 0.01 | (−0.02–0.04) | – | – |
| K (mass %) | 0.11 | (0.01–0.2) | 1 | 31.3 | 0.28 | (0.14–0.43) | 2 | 49.6 | 0.25 | (0.09–0.41) | 1 | 11.9 |
| Na (mass %) | 0.5 | (0.4-0.6) | 1 | 21.6 | 0.22 | (0.09–0.35) | 6 | 133.5 | 0.16 | (0.05–0.28) | 2 | 42.2 |
| S (mass %) | 0.58 | (0.39–0.77) | 3 | 55.9 | 0.38 | (0.2–0.55) | 4 | 68.5 | 0.19 | (0.06–0.33) | 2 | 29.8 |
| Zn (µg g−1) | 0.01 | (−0.03–0.05) | 1 | 14.5 | 0.06 | (0.02–0.11) | 8 | 76.6 | 0.06 | (0–0.11) | 4 | 81.8 |
| K/Na ratio | 0.01 | (−0.03–0.05) | 2 | 48.5 | 0.17 | (0.06–0.27) | 20 | 416.2 | 0.18 | (0.06–0.3) | 3 | 56.8 |
| Tolerance | – | – | – | – | 0.05 | (−0.01 to −0.11) | 1 | 19.9 | – | – | – | – |
Our haplotype block analysis divided the genome into blocks of single-nucleotide polymorphisms (SNPs) based on patterns of linkage disequilibrium (LD). This resulted in the identification of 19,918 haplotypic blocks consisting of multiple SNPs in strong LD along with 9,179 singleton SNPs (Fig. 5B; Supplemental Fig. S8). We identified 2,739 SNPs significantly associated with variation in one or more traits in the control and salt stress treatments or with variation in the proportional shift between treatments. Based on the haplotype map, these significant SNPs represented 242 haplotype blocks plus 17 singletons (Supplemental Figs. S3 and S8). Further clustering of these significant SNPs allowed for the combining of haplotype blocks (likely due to localized misordering in the underlying genome assembly), resulting in 169 genomic regions (158 haplotypic blocks and 11 singletons) significantly associated with variation in trait values (Supplemental Fig. S4). Of these 169 regions, 29 were significantly associated with variation in multiple trait and/or treatment combinations (up to 11 for a single region spanning 93,336 kb on chromosome 10), and 143 regions were significant for at least one trait and suggestive (top 0.1% of SNPs) for at least one other. Relative effect sizes of regions ranged from fairly high, 36% for chlorophyll content in control conditions at region 04-09, to fairly modest, 6% for Zn content under salt treatment for region 10-02 (Supplemental Fig. S5; Supplemental Table S1). Only 26 regions had a significant association with just a single trait and no significant or suggestive association with other traits (Supplemental Fig. S5; Supplemental Table S2). Across all traits, only four instances were identified where a region was significantly associated with a trait in more than one of either control conditions, the salt treatment, or the plasticity between them (Supplemental Table S2). While these results could be due to the stringent multiple comparison correction, the relatively few instances of colocalization of a trait across treatments suggests strong environmental dependence of allelic effects (i.e. strong G×T interaction).
Figure 5.
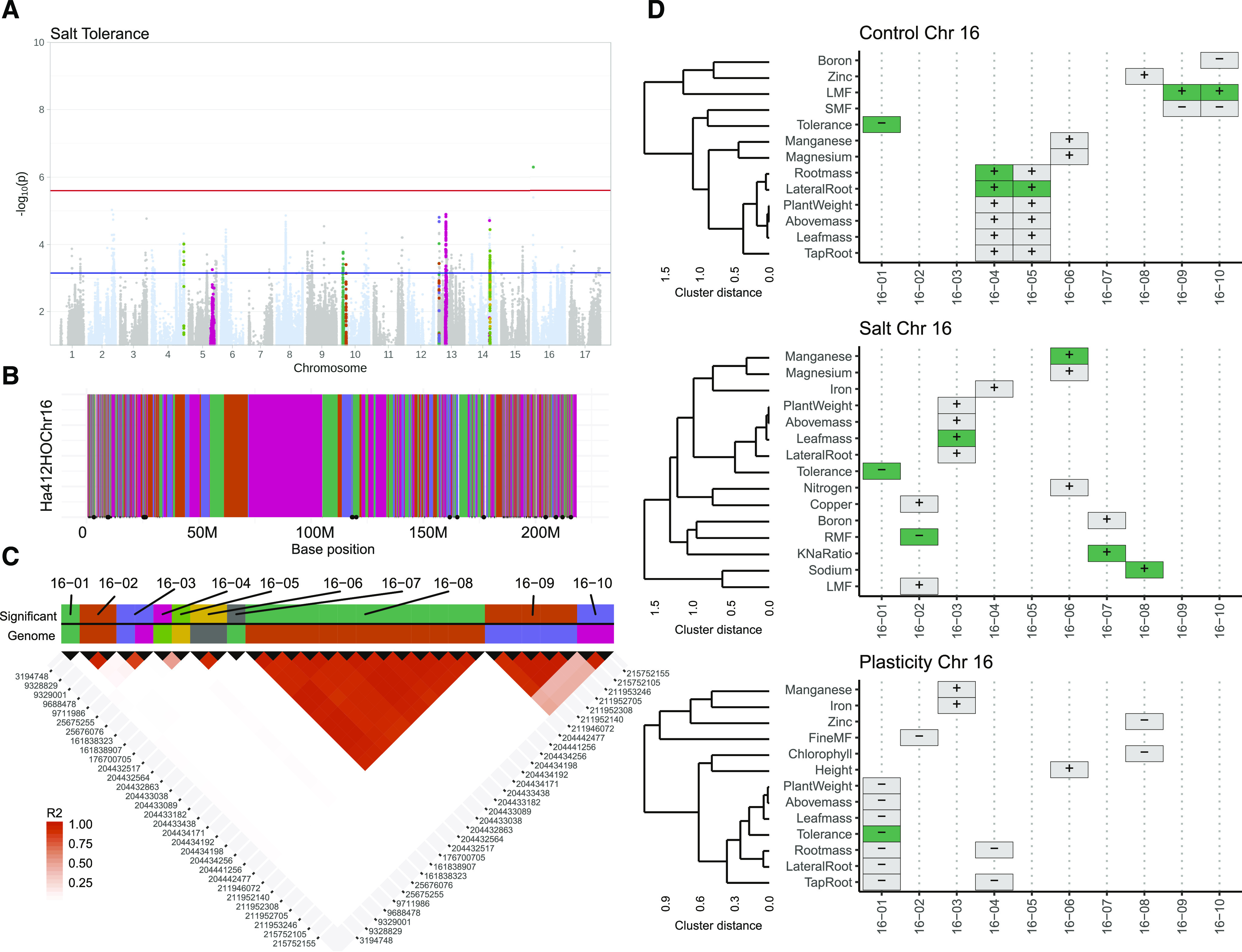
Overview of sunflower GWAS and trait colocalization analyses. Association testing for ∼1.48 m SNPs was performed on trait values from our cultivated sunflower diversity panel under control and salt-stressed conditions, as well as for the plasticity in trait value between treatments. A, Manhattan plot of GWAS results for salt tolerance (estimated as expectation-deviation tolerance; the deviation of the expected decrease in biomass based on vigor) showing a single significantly associated region on chromosome 16 (above the red line). An additional 12 regions harbored suggestive associations (i.e. SNPs in the top 0.1% of likelihood values that were also significant for other traits; above the blue line) for tolerance. B, Zooming in on chromosome 16, the haplotype block analysis clustered 103,607 SNPs into 1,732 blocks plus 856 individual SNPs. Blocks that contain significant SNPs for at least one trait are marked by a black dot on the x axis. Adjacent blocks are colored in an alternating fashion repeating the same four colors C, Evaluating all significant SNPs across all traits and treatments (plus plasticity) on chromosome 16 reveals that these SNPs correspond to 11 unique haplotype blocks. However, after rerunning the blocking algorithm on just the significant SNPs (see “Materials and Methods” for details), two blocks were further collapsed based on observed patterns of LD. Adjacent blocks are colored in an alternating fashion repeating the same seven colors. D, Visualizing trait colocalization along chromosome 16 reveals relatively few instances of genomic overlap in traits with significant SNPs (green tiles) but substantial overlap in regions that are suggestive for a trait (gray tiles). Plus or minus symbols in the tiles indicate the sign of β (effect of minor allele on trait). Traits and trait dendrogram are ordered using complete-linkage hierarchical clustering based on Spearman rank correlations.
Surprisingly, many highly correlated traits did not share significant regions (Supplemental Fig. S5). For example, Na concentration, K concentration, and leaf K/Na ratio (Fig. 3A) shared no significant regions for control and salt treatment. However, suggestive regions did overlap, highlighting the issue of stringent multiple comparison correction. Within traits, there were also few significant shared regions for trait values across environments or trait plasticity (Supplemental Fig. S6), although suggestive regions for control and salt treatment did overlap more frequently. Regions associated with trait plasticity were generally more distinct with even fewer significant and suggestive regions overlapping with control and salt treatment, indicating unique regions associated with shifts in trait value.
Based on the haplotype map, genome annotation, and our GWAS results, we determined the genes contained in each significant region. When a region consisted of multiple haplotype blocks that could be combined after a second LD analysis (Fig. 5C; Supplemental Fig. S3), we combined the genes contained in each individual haplotype block. Across the 169 genomic regions, we identified 4,167 genes potentially associated with variation in traits and trait plasticity (Supplemental Table S3; Supplemental Fig. S7). Of these genes, 1,586 were of unknown function, but the remaining 2,753 genes consisted of 720 unique proteins and 397 that were present two or more times (Supplemental Table S3). Contained in the gene list are several candidates that are of interest due to potential involvement with known salt tolerance mechanisms. It should be noted, however, that with over 38% genes of unknown function and 135 regions containing multiple genes, identifying the causal genes underlying trait variation will be challenging.
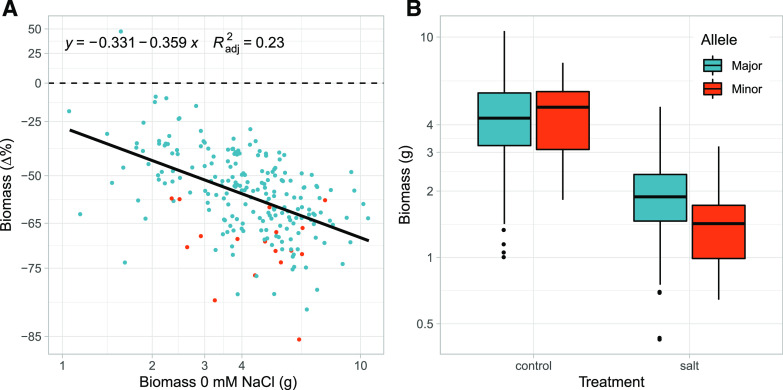
For salinity expectation-deviation tolerance, we identified a single, small, region (containing only one SNP; adjacent SNPs were not in strong LD) on chromosome 16 with a relative effect size of 20% (Fig. 5A). Given its short span, this region (16-01) contained only a single gene—putatively coding for alpha-mannosidase (gene ID Ha412HOChr16g0747981). While individuals carrying the major versus minor allele at this locus did not differ in vigor (Fig. 6B), genotypes carrying the minor allele had a greater-than-expected (based on their vigor) decline in biomass under salt stress (Fig. 6A). Surprisingly, no traits measured (except biomass plasticity, as could be expected) colocated to this region (Fig. 5D), suggesting an unmeasured mechanism or trait involved in the lower tolerance of these genotypes.
Figure 6.
Allelic effects of chromosomal region 16-1 on sunflower salinity tolerance. A, Percent reduction in biomass versus biomass under control conditions (i.e. vigor) for genotypes with the major (224 blue dots) versus minor (15 orange dots) allele for chromosomal region 16-1. The percent reduction was estimated as the difference in natural log transformed values (control – salt) but converted here to Δ% (via eΔln(trait) − 1) change from control for ease of interpretation. B, The relationship between the treatment, allele (major versus minor), and biomass accumulation. Note the natural log scaling of the axes.
DISCUSSION
Improving food security requires the development of crops that can withstand environmental stresses such as high salinity. Given the osmotic as well as ionic challenges presented by salt stress, the physiological and genetic basis of salinity tolerance is likely to be complex (Negrão et al., 2017; Morton et al., 2019). Here, we investigated the suites of traits underlying variation in salinity tolerance in sunflower and examined their underlying genetic basis. Genotypes differed in terms of morphological, growth/biomass allocation, and leaf ionomic responses to salt stress. Morphological and ionomic traits were highly correlated among themselves under both control and saline conditions, as well as in their plasticity to salt stress. Given an observed tradeoff between vigor (biomass accumulation in benign conditions) and the effect of salt stress, we decoupled our metric of tolerance from this expected response (Fig. 1). Consistent with prior findings, the most vigorous genotypes in control conditions tended to remain the best performers (most biomass) under salt stress (Fig. 2A; Temme et al., 2019a, 2019b). In addition, similar to previous work, these high-vigor genotypes had the greatest proportional reduction in biomass in response to stress (Temme et al., 2019a, 2019b). The low vigor of the genotypes least affected by salinity stress might be attributable to the maintenance of costly mechanisms related to salinity tolerance (Munns and Tester, 2008; Shabala et al., 2019). However, by taking into account the relationship between vigor and the proportional decline in performance, we were able to focus on the traits associated with better-/worse-than-expected performance, independent from vigor (compare Fig. 1 with Fig. 2B).
Using this metric of expectation-deviation tolerance, we found that genotypes that had a greater increase in root mass fraction, fine root mass fraction, and chlorophyll content had a lower-than-expected decline in biomass (i.e. they had a higher expectation-deviation tolerance; Fig. 4). As measured, greater chlorophyll content could be the result of more chlorophyll per area and/or thicker leaves. Comparing this to the response in LMA, which also increased, our results could indicate greater leaf succulence under salinity stress, thereby diluting accumulated Na (Munns et al., 2016). For root traits, rather than variation in the traits themselves, it was the magnitude and direction of root trait plasticity (i.e. strong G×T interaction across genotypes; Supplemental Fig. S1) that was most clearly linked to variation in expectation-deviation tolerance. Functionally, there are some known mechanisms that could play a role in these responses. For example, higher root mass fractions could allow for increased storage of Na in the roots, as has been found for soy (Guan et al., 2014). Alternatively, increased root branching (resulting in greater fine root mass fraction) has been suggested to reduce the energetic cost of Na exclusion (Zolla et al., 2010; Munns et al., 2020a). Notably, this could come at the cost of reduced water uptake during the day (Arsova et al., 2020), suggesting a possible interaction between the osmotic and ionic components of salt stress through root branching. Since our experimental design (well-watered greenhouse plants and a delay in root processing due to the scale of the experiment) is an abstraction of the real world, more field work, imaging of root traits, etc. is needed to determine if these results hold in real-world applications.
Besides a substantial increase in Na content, the concentration of a suite of elements (N, P, K, S, Ca, Mg, Mn, Fe, Cu, Zn, and B) was affected by salinity stress. We previously found a connection between leaf S content and salinity tolerance in sunflower (Temme et al., 2019b), possibly related to root sulpholipid content (Erdei et al., 1980). Additionally, Mn content has been linked to salt tolerance in Brassica spp. (Wang et al., 2010; Chakraborty et al., 2016). However, here we found that lower leaf Na content, higher leaf K content, and higher K/Na ratio were associated most strongly with greater expectation-deviation tolerance. This is not surprising, given that these traits are well known to be involved in salinity tolerance. Excluding Na from the leaf while maintaining K uptake and adequate levels in the cytosol are key to plants functioning under saline conditions (Munns and Tester, 2008; Munns et al., 2020a).
High vigor was correlated with a suite of traits, including a relatively high fine root mass fraction (yet low overall root mass fraction) and high leaf Mn and Mg content (Supplemental Fig. S2). Taken together, these traits suggest nutrient uptake and/or allometric scaling of root traits with vigor (Wang et al., 2019) as key components in vegetative growth. In a field setting, increased fertilization indeed produces increased growth in sunflower, though this did not ultimately translate into a yield increase; instead, it resulted in increased lodging and reduced yields (Schultz et al., 2018). This finding highlights the complexity of translating vegetative growth to yield.
In terms of GWAS results, we identified at least one genomic region with a significant association with the majority of traits either in the control treatment, salt treatment, or plasticity of the trait across treatments (Table 5). Heritability estimates for biomass, biomass fractions, and morphological traits were generally high, whereas estimates for ionomic traits were much lower, potentially owing to the lack of replicates for the ionomic traits (Kruijer et al., 2015). Relative effect sizes of alleles ranged from 6% for Zn under salt stress to 36% for chlorophyll content under control conditions (Supplemental Table S1). The identification of several potential regions of interest for the majority of traits combined with these sometimes considerable relative effect sizes of individual loci showcases the potential for adjusting trait expression through targeted breeding or even genome editing once the underlying genes have been identified.
Considering trait expression across both treatments, the infrequent occurrence of genomic regions with a significant association for a trait in both treatments (Supplemental Table S2) was consistent with the occurrence of substantial G×T interaction (Table 1) for the majority of traits (Des Marais et al., 2013). More commonly, regions were at least suggestively (or significant and suggestively) associated with traits in both environments (Supplemental Fig. S6). This highlights a potential consequence of stringent multiple comparison corrections: i.e. that false negatives may occur and give the appearance of environment specificity. Interestingly, there were far fewer significant and suggestive regions overlapping with plasticity in trait values. This suggests independent genomic control of the expression of a trait and the plasticity in that trait (Kusmec et al., 2017).
Significant colocalizations across traits were also infrequent, with only 39 out of 189 regions being significantly associated with multiple traits. However, if we include suggestive colocalizations (i.e. regions that were significant for one trait and in the top 0.1% for at least one other trait), this number rises to 161 regions being significant for at least one trait and suggestive for at least one other. Thus, it may be that the apparent paucity of regions influencing multiple traits was due, at least in part, to the highly stringent significance thresholds. The colocalization of multiple traits, either significant or suggestive (Supplemental Fig. S5; Supplemental Table S2), in either control, salt stressed, or the plasticity of trait responses between treatments, demonstrates the multivariate nature of the genetic controls of trait expression (Wagner et al., 2007; Wagner and Zhang, 2011; Sella and Barton, 2019). This multivariate nature represents a constraint on the development of new trait combinations and needs to be taken into account when attempting to improve salt tolerance. Indeed, adjustments in the expression of one trait will likely influence others (Pailles et al., 2019; Temme et al., 2019a), thereby restricting the response to selection for particular trait combinations. Nevertheless, since our metric of expectation-deviation tolerance is independent of vigor and correlated with plasticity in trait values (which are generally independent of the trait values themselves), it should be possible to improve tolerance while minimizing undesired tradeoffs.
Based on the annotations of genes in the significantly associated regions, we were able to identify several candidates for genes underlying observed trait variation (Supplemental Tables S2 and S3). Here, we highlight a few with potential impacts on the traits most closely related to salinity tolerance. In region 8-4 (53 genes, 6,161 kb), which was associated with Na content under salt stress, we found hypersensitive-induced reaction1 protein (gene identifier Ha412HOChr08g0366591), which has been linked to K efflux (impacting K/Na ratio) in plant cells (Jung and Hwang, 2007). Similarly, in region 12-14 (29 genes, 827 kb), which was likewise associated with Na content under salt stress, we found two genes related to vacuolar function (vacuolar (H+)-ATPase G subunit [Ha412HOChr12g0557101] and vacuolar protein sorting-associate protein Vta1/callose synthase [Ha412HOChr12g0557261]) that could play a role in vacuolar sequestration of Na, which is known to be an important tolerance mechanism (Gaxiola et al., 2016; Schilling et al., 2017; Munns et al., 2020a). While these gene descriptions are consistent with putative mechanisms, they are often not the only genes in an independent block. For example, region 10-2 contains a calcium permeable stress-gated cation channel1 transmembrane domain-containing protein (Ha412HOChr10g0441661) and has a significant association with K/Na ratio under salinity stress, yet this is also one of the larger regions (93,336 kb) and is associated with 11 traits (some in multiple environments; Supplemental Table S2) and contains 1,397 genes. Future work on gene expression changes under salinity stress along with functional analyses of the most promising candidates will allow us to develop a more complete understanding of the molecular basis of observed trait variation.
We identified a single small region on chromosome 16, containing a single gene, associated with expectation-deviation tolerance. Interestingly, aside from plasticity in biomass-related traits (Fig. 5D), none of the measured traits colocalized with that same region. This suggests the existence of a potentially unmeasured mechanism conferring salinity tolerance that is influenced by variation in this region. Genotypes carrying the minor allele at this locus were found to be less tolerant to salinity stress but did not show any difference in performance under control conditions (Fig. 6), indicating strong G×T interaction for this allele, along with a lack of substantial performance tradeoffs. The gene of interest in this region is a putative alpha-mannosidase (Ha412HOChr16g0747981; Supplemental Table S1). In Arabidopsis (Arabidopsis thaliana), α-mannosidases are involved in a complex pathway involved in regulating glycoprotein abundance during salt stress (Liu et al., 2018), and they also play a role in lateral root formation (Veit et al., 2018). Given that we found that plasticity in root mass allocation was associated with expectation-deviation tolerance (though it did not colocalize with this region) and that alpha-mannosidase is related to aspects of root formation in the literature, this gene appears to be a worthwhile candidate for future mechanistic work on sunflower root development and salinity tolerance.
Taken together, our results indicate that salinity tolerance can be improved in sunflower while minimizing tradeoffs with performance. Indeed, we found evidence that trait variation (Na and K content) and plasticity (particularly in root traits) influence salt tolerance independent of performance under benign conditions and that these traits and vigor tend to map to distinct genomic regions (though trait plasticity exhibits low heritability). Since the traits of interest and their underlying genomic regions are distinct from those associated with high vigor (i.e. growth in benign conditions), this provides an avenue for increasing salt tolerance by fine tuning this trait variation/plasticity in high-performing genotypes without incurring costly tradeoffs.
MATERIALS AND METHODS
Plant Materials and Experimental Design
This work used the sunflower association mapping (SAM) population (Mandel et al., 2011, 2013). This population includes representatives from all four cultivated sunflower (Helianthus annuus) heterotic group/market-type combinations (HA-Oil, HA-NonOil, RHA-Oil, RHA-NonOil) as well as landraces, open-pollinated varieties, and cultivated genotypes carrying wild species introgressions. These lines were previously advanced via single-seed descent to minimize residual heterozygosity. All lines in this population have been subjected to whole-genome shotgun resequencing (Hübner et al., 2019), and these data were used to identify SNPs and extract genotypic information for each line.
Due to greenhouse space constraints, we limited this study to the 239 genotypes from the SAM population with the least residual heterozygosity. With two treatments and four replicates/treatment, this resulted in a total 1,912 individual plants. The experiment was initiated by planting up to 16 achenes from each genotype in seedling germination trays filled with a 3:1 mixture of coarse sand and turface (Turface Athletics, PROFILE Products). The trays were treated with a 0.45 g L−1 solution of a broad-spectrum fungicide to inhibit fungal growth (Banrot, Everris) and watered twice daily for 5 d to allow for seedling emergence. At that point, eight representative seedlings per genotype were transplanted into individual, 36-cm-tall, 2.83-L pots filled with the same 3:1 sand/turface mixture plus 40 g Osmocote Plus (15-12-9 NPK; ScottsMiracle-Gro) and supplemental Ca2+ in the form of 5 mL each of gypsum (Performance Minerals Corporation) and lime powder (Austinville Limestone). Pots were assigned to one of two treatments (i.e. control [0 mm NaCl] and salt stressed [100 mm NaCl]) and arranged in a split-plot design with four replicate plots split between treatments.
Plots were arranged orthogonally to the direction of airflow within the greenhouse and alternated control and stress from east to west. Each subplot consisted of one pot per genotype randomly arranged. The differential treatments were implemented 2 d after transplanting with pots being placed in shallow ponds, with the bottom 10 cm standing in either fresh water (control) or a 100 mm NaCl solution (stress). Three times per week, these ponds were drained and refilled with new solutions. At that time, all pots were top-watered with ∼300 mL of the solution from their pond (both in control and stressed treatments) to stimulate nutrient release from the Osmocote pellets and to prevent buildup of NaCl at the soil surface in the stress treatment. Given this frequent top watering and fairly low water holding capacity of the sand/turface mix, water logging of the pot as well as anoxic conditions in the lower portion of the pot were minimal. After 3 weeks (while the plants were still in the vegetative growth stage), all plants were harvested over a 4-d period, with one entire plot (478 plants, control and stress treatment) being harvested each day. This experiment was carried out during the summer of 2017 in the Botany Greenhouses at the University of Georgia (Athens, Georgia).
Phenotypic and Leaf Elemental Traits
At harvest (28 d postgermination, 21 d post-treatment-initiation), all plants were measured for height (from the base of the stem to the apical meristem), stem diameter (at the base of the stem), and chlorophyll content (MC-100, Apogee Instruments) of the most recently fully expanded leaf (MRFEL). Plant biomass was separated into the MRFEL pair, all other leaves, stem (plus immature flower bud, if present), and roots. The MRFELs (both leaves in the pair) were scanned at 300 dpi on a flatbed scanner (Canon CanoScan LiDE120), and leaf area was determined using ImageJ (National Institutes of Health, http://rsb.info.nih.gov/ij/). Prior to scanning, half of the lamina of one member of the MRFEL pair was separated and preserved for use in a separate study on leaf anatomy; the amount of missing biomass was therefore estimated using the mass/area relationship (i.e. LMA) from the remaining leaf lamina. All aboveground biomass samples were bagged and oven dried at 60°C for 48 h. Pots containing roots were placed in cold storage to prevent degradation prior to processing. Given the large number of pots, all roots were gently washed to remove soil, bagged, and oven dried at 60°C for 48 h over the space of 2 weeks. Pots were washed on a first-in first-out principle, so any decay was evenly spread across replicates. All dried plant samples were placed into storage outside the oven and then redried prior to any further analysis.
After drying, any immature flower buds that might have been present were separated from the stem tissue, and root tissue was separated by dividing the taproot (only up to the point at which the taproot and lateral roots had similar widths) from the lateral fine roots. After weighing all biomass fractions (i.e. lateral root, tap root, stem, leaves, MRFEL, buds), all intact MRFELs (excluding the petiole) for each genotype were pooled across replicates (separately by treatment) and ground to a coarse powder using a Wiley Mill (Thomas Scientific). After mixing evenly, a 2-mL subsample of the coarse powder was ground to a fine powder using a Qiagen TissueLyser (Qiagen). Finely powdered leaf tissue was then sent to Midwest Laboratories for inductively coupled plasma mass spectroscopy to quantify the presence of the following elements: P, K, Ca, Na, S, Fe, Zn, Cu, Mg, Mn, and B. N content was determined via the Dumas method.
Data Analysis
All statistical analyses were performed using R (v3.6.2; R Core Team, 2019). Genotype mean values and the effects of salinity stress for all nonelemental traits were estimated by fitting a linear mixed model to the data with genotype and salinity treatment as fixed factors and pond within plot as a random factor using the R package lme4 (Bates et al., 2015). Genotype means were calculated by taking the estimated marginal means of genotype × treatment without the random pond effect using emmeans (Lenth, 2018). Significance of the fixed effects and their interaction was determined via Wald’s χ2 test using the package car (Fox and Weisberg, 2011). The effect of the salinity treatment on leaf elemental content was tested using two-sample t tests with genotype means as the unit of replication. All further analyses were carried out on the genotype means with genotype as the unit of replication. We quantified trait plasticity in response to salinity as the difference in natural log-transformed trait values between salt and control treatments. This plasticity metric has the benefit of being proportional to the trait value (in either the control or stressed treatments) and is symmetric to which treatment is set as the control, changing only in sign, not magnitude.
Correlational analyses were performed using corrr (https://github.com/tidymodels/corrr) with Spearman correlations and visualized using ggplot2 (Wickham, 2009). To relate trait expression and plasticity in traits to performance and tolerance, we first divided measured traits into three sets: mass/size related traits, (putatively) size-independent traits (i.e. mass fractions, LMA, chlorophyll content), and elemental traits. The principal components of the size-independent traits and elemental traits were then regressed against vigor and expectation-deviation tolerance. Principal component biplots were visualized using a modified version of ggbiplot (https://github.com/vqv/ggbiplot).
h2 was calculated using the package heritability (Kruijer et al., 2015) and a kinship matrix constructed from the SNP data (see GWAS below). For all nonelemental traits, individual plants could be included in the heritability analysis. However, for the elemental traits and plasticity metric, we only had genotype average values; this required us to use only genotypes’ mean values, resulting in reduced precision in the heritability estimate.
Genotypic Data Analysis and GWAS
Genome-wide association analyses were conducted using SNPs derived from whole-genome shotgun resequencing of the SAM population (Hübner et al., 2019) using a SNP set developed by Todesco et al. (2019). In brief, this involved aligning the whole-genome shotgun reads to the XRQv1 reference genome (Badouin et al., 2017) using NextGenMap v0.5 (Sedlazeck et al., 2013), calling SNPs using GATK4 (Van der Auwera et al., 2013) and processing the resulting SNPs using GATK’s variant quality score recalibration (Poplin et al., 2017). This initial dataset was filtered to retain SNPs in the 90% tranche with < 10% missing data and a minor allele frequency ≥ 1%. Missing genotype calls were then imputed using BEAGLE v5.0 (Browning and Browning, 2016). This set of ∼2.16 m high-quality SNPs was then reordered based on the more recent, improved assembly of the HA412-HO genome (Todesco et al., 2019) because it has better localized ordering of chromosomal segments. Finally, this dataset was filtered based on the 239 genotypes used in this study to remove SNPs with minor allele frequency < 5% and heterozygosity > 10%, resulting in ∼1.48 m SNPs.
To better visualize the haplotypic structure present within the sunflower gene pool and to estimate the effective number of independent markers in the genome, we used the ordered SNPs to construct a haplotype map based on observed patterns of LD using PLINK v1.9 (Purcell et al., 2007). Haplotypic blocks were identified based on D′ following Gabriel et al. (2002) with a standard confidence interval of 0.7005 to 0.98, an informative fraction of 0.9 instead of 0.95 to be modestly more permissive of misaligned SNPs within blocks, and a maximum block span of 100 Mb to accommodate the largest observed block in the genome (Supplemental Fig. S8; Gabriel et al., 2002).
For all traits, association analyses were performed using GEMMA v0.98.1 (Zhou and Stephens, 2012). These analyses included corrections for both kinship (calculated using GEMMA) and population structure (using the first four PC axes from an analysis using a pruned set of approximately 24,000 independent SNPs [i.e., D′ < 0.8 using a sliding window approach] in the R package SNPRelate; Zheng et al., 2012). To correct our significance thresholds for multiple comparisons, we used a modified Bonferroni correction with the number of independent tests set to the number of multi-SNP haplotype blocks identified in our haplotypic analysis. While this procedure helps to account for nonindependence among linked markers, it still results in a highly conservative significance threshold, since no genome assembly is perfect with respect to sequence ordering and orientation, thereby inflating the inferred number of independent regions.
Following the identification of significant marker-trait associations, associated genomic regions were identified on the basis of our haplotypic analysis. More specifically, significantly associated SNPs that occurred within a single haplotypic block were assumed to mark a single genomic region. To protect against the possibility of haplotype blocks being broken up by localized genotyping/ordering errors, thereby resulting in an overestimate of the number of independent regions influencing a given trait, we repeated the LD-based haplotypic analysis using only the markers on each chromosome that were found to be significant for one or more traits; if previously independent markers/haplotypes collapsed into a single haplotype in this reanalysis, they were assumed to mark the same region.
For each associated region, the magnitude of its effect on the trait of interest was set as the maximum beta (i.e. effect) value across all SNPs in the region that were significantly associated with that trait. The relative effect size was calculated as (2*β)/(trait range) (Masalia et al., 2018), with the relative effect size of a haplotype block set as the maximum of the significant SNPs in that block. Relative effect size values were summed across all regions per trait to calculate the total phenotypic range that was explained by the significantly associated genomic regions.
In cases where SNPs within an associated region were significantly associated with multiple traits, we considered those traits to colocalize, either due to close linkage among the causal factors or through pleiotropy. Because of our highly stringent significance thresholds, false negatives are possible and could give the erroneous appearance of independence across traits. Thus, we also searched for cases of suggestive colocalization, defined as instances where SNPs within a haplotypic block were significantly associated with a given trait and in the top 0.1% of SNPs for one or more other traits, even if they did not reach statistical significance for those traits.
Following the identification of significantly associated regions for all traits of interest, a list of the genes contained within each region was extracted from the annotation of the HA412-HOv2 genome (Todesco et al., 2019) based on the positions of the first and last SNPs in each region. When singleton SNPs exhibited significant associations, we extracted either the gene in which it was contained or the two flanking genes if it was in an intergenic region. Code for the GWAS pipeline including the trait colocalization steps and gene extraction is available online (at https://github.com/aatemme/Sunflower-GWAS-v2).
Accession Numbers
All raw data from the resequencing of the SAM population are stored in the Sequence Read Archive under Bioproject PRJNA353001. SNP set used and genome assembly as in Todesco et al. (2020).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Trait reaction norms to salinity stress.
Supplemental Figure S2. Vigor and associated trait complexes.
Supplemental Figure S3. LD heatmaps for significant SNPs on each chromosome.
Supplemental Figure S4. Manhattan plots of all traits.
Supplemental Figure S5. Trait colocalization under control and salt-stressed conditions as well as their plasticity.
Supplemental Figure S6. Colocalization of traits across environments.
Supplemental Figure S7. Number of genes per significant region.
Supplemental Figure S8. Haplotype block map of the sunflower genome.
Supplemental Table S1. Detailed information on significant and suggestive regions per trait/treatment combination.
Supplemental Table S2. List of significant genomic regions and associated significant/suggestive traits.
Supplemental Table S3. List of genes per region.
Acknowledgments
We thank Kelly Bettinger, Mike Boyd, Kevin Tarner, the rest of the greenhouse staff, the sunflower undergraduate army, and numerous members of the Donovan and Burke labs for help during the experiment. Special thanks to Emily Dittmar for instigating the expectation-deviation tolerance idea and to Greg Owens, Jean-Sébastien Légaré, and Loren Rieseberg for developing the sunflower SNPs used in this study, as well as their realignment to the HA412-HOv2 genome assembly.
Footnotes
This work was supported by the National Science Foundation Plant Genome Research Program (grant no. IOS–1444522 to J.M.B. and L.A.D).
Articles can be viewed without a subscription.
References
- Agrawal AA.(2020) A scale-dependent framework for trade-offs, syndromes, and specialization in organismal biology. Ecology 101: e02924. [DOI] [PubMed] [Google Scholar]
- Arnold PA, Nicotra AB, Kruuk LEB(2019) Sparse evidence for selection on phenotypic plasticity in response to temperature. Philos Trans R Soc Lond B Biol Sci 374: 20180185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsova B, Foster KJ, Shelden MC, Bramley H, Watt M(2020) Dynamics in plant roots and shoots minimise stress, save energy and maintain water and nutrient uptake. New Phytol 225: 1111–1119 [DOI] [PubMed] [Google Scholar]
- Auge GA, Penfield S, Donohue K(2019) Pleiotropy in developmental regulation by flowering-pathway genes: is it an evolutionary constraint? New Phytol 224: 55–70 [DOI] [PubMed] [Google Scholar]
- Badouin H, Gouzy J, Grassa CJ, Murat F, Staton SE, Cottret L, Lelandais-Brière C, Owens GL, Carrère S, Mayjonade B, et al. (2017) The sunflower genome provides insights into oil metabolism, flowering and Asterid evolution. Nature 546: 148–152 [DOI] [PubMed] [Google Scholar]
- Bassil E, Zhang S, Gong H, Tajima H, Blumwald E(2019) Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant Physiol 179: 616–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S(2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1–48 [Google Scholar]
- Broadley M, Brown P, Cakmak I, Ma JF, Rengel Z, Zhao F(2012) Beneficial Elements In Marschner P, ed, Marschner’s Mineral Nutrition of Higher Plants, Ed 3 Academic Press, San Diego, CA, pp 249–269 [Google Scholar]
- Browning BL, Browning SR(2016) Genotype imputation with millions of reference samples. Am J Hum Genet 98: 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty K, Sairam RK, Bhaduri D(2016) Effects of different levels of soil salinity on yield attributes, accumulation of nitrogen, and micronutrients in Brassica spp. J Plant Nutr 39: 1026–1037 [Google Scholar]
- Cheeseman JM.(2015) The evolution of halophytes, glycophytes and crops, and its implications for food security under saline conditions. New Phytol 206: 557–570 [DOI] [PubMed] [Google Scholar]
- Cuin TA, Bose J, Stefano G, Jha D, Tester M, Mancuso S, Shabala S(2011) Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant Cell Environ 34: 947–961 [DOI] [PubMed] [Google Scholar]
- Des Marais DL, Hernandez KM, Juenger TE(2013) Genotype-by-environment interaction and plasticity: Exploring genomic responses of plants to the abiotic environment. Annu Rev Ecol Evol Syst 44: 5–29 [Google Scholar]
- Erdei L, Stuiver B (CEE), Kuiper PJC(1980) The effect of salinity on lipid composition and on activity of Ca2+- and Mg2+-stimulated ATPases in salt-sensitive and salt-tolerant Plantago species. Physiol Plant 49: 315–319 [Google Scholar]
- FAO (2005) Global network on integrated soil management for sustainable use of salt-affected soils. FAO Land and Plant Nutrition Management Service, Rome, Italy. http://www.fao.org/ag/agl/agll/spush
- Flexas J, Gago J(2018) A role for ecophysiology in the ‘omics’ era. Plant J 96: 251–259 [DOI] [PubMed] [Google Scholar]
- Flowers TJ.(2004) Improving crop salt tolerance. J Exp Bot 55: 307–319 [DOI] [PubMed] [Google Scholar]
- Fox J, Weisberg S(2011) An R Companion to Applied Regression, Ed 2 Sage, Thousand Oaks, CA [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, et al. (2002) The structure of haplotype blocks in the human genome. Science 296: 2225–2229 [DOI] [PubMed] [Google Scholar]
- Gaxiola RA, Regmi K, Paez-Valencia J, Pizzio G, Zhang S(2016) Plant H(+)-PPases: Reversible enzymes with contrasting functions dependent on membrane environment. Mol Plant 9: 317–319 [DOI] [PubMed] [Google Scholar]
- Guan R, Qu Y, Guo Y, Yu L, Liu Y, Jiang J, Chen J, Ren Y, Liu G, Tian L, et al. (2014) Salinity tolerance in soybean is modulated by natural variation in GmSALT3. Plant J 80: 937–950 [DOI] [PubMed] [Google Scholar]
- Hasegawa PM.(2013) Sodium (Na+) homeostasis and salt tolerance of plants. Environ Exp Bot 92: 19–31 [Google Scholar]
- Hübner S, Bercovich N, Todesco M, Mandel JR, Odenheimer J, Ziegler E, Lee JS, Baute GJ, Owens GL, Grassa CJ, et al. (2019) Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat Plants 5: 54–62 [DOI] [PubMed] [Google Scholar]
- Jung HW, Hwang BK(2007) The leucine-rich repeat (LRR) protein, CaLRR1, interacts with the hypersensitive induced reaction (HIR) protein, CaHIR1, and suppresses cell death induced by the CaHIR1 protein. Mol Plant Pathol 8: 503–514 [DOI] [PubMed] [Google Scholar]
- Katerji N, van Hoorn JW, Hamdy A, Mastrorilli M(2000) Salt tolerance classification of crops according to soil salinity and to water stress day index. Agric Water Manage 43: 99–109 [Google Scholar]
- Koziol L, Rieseberg LH, Kane N, Bever JD(2012) Reduced drought tolerance during domestication and the evolution of weediness results from tolerance-growth trade-offs. Evolution 66: 3803–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W, Boer MP, Malosetti M, Flood PJ, Engel B, Kooke R, Keurentjes JJB, van Eeuwijk FA(2015) Marker-based estimation of heritability in immortal populations. Genetics 199: 379–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusmec A, Srinivasan S, Nettleton D, Schnable PS(2017) Distinct genetic architectures for phenotype means and plasticities in Zea mays. Nat Plants 3: 715–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen RAE, Nikoloski Z(2019) Genetic basis of plasticity in plants. J Exp Bot 70: 739–745 [DOI] [PubMed] [Google Scholar]
- Lenth RV. (2018) emmeans: Estimated marginal means, aka least-squares means. R package version 1.1. https://CRAN.R-project.org/package=emmeans (August 27, 2020)
- Liu C, Niu G, Zhang H, Sun Y, Sun S, Yu F, Lu S, Yang Y, Li J, Hong Z(2018) Trimming of N-glycans by the Golgi-localized α-1,2-mannosidases, MNS1 and MNS2, is crucial for maintaining RSW2 protein abundance during salt stress in Arabidopsis. Mol Plant 11: 678–690 [DOI] [PubMed] [Google Scholar]
- Mandel JR, Dechaine JM, Marek LF, Burke JM(2011) Genetic diversity and population structure in cultivated sunflower and a comparison to its wild progenitor, Helianthus annuus L. Theor Appl Genet 123: 693–704 [DOI] [PubMed] [Google Scholar]
- Mandel JR, Nambeesan S, Bowers JE, Marek LF, Ebert D, Rieseberg LH, Knapp SJ, Burke JM(2013) Association mapping and the genomic consequences of selection in sunflower. PLoS Genet 9: e1003378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangin B, Casadebaig P, Cadic E, Blanchet N, Boniface M-C, Carrère S, Gouzy J, Legrand L, Mayjonade B, Pouilly N, et al. (2017) Genetic control of plasticity of oil yield for combined abiotic stresses using a joint approach of crop modelling and genome-wide association. Plant Cell Environ 40: 2276–2291 [DOI] [PubMed] [Google Scholar]
- Mansour MMF, Salama KHA, Al-Mutawa MM(2003) Transport proteins and salt tolerance in plants. Plant Sci 164: 891–900 [Google Scholar]
- Masalia RR, Temme AA, Torralba NL, Burke JM(2018) Multiple genomic regions influence root morphology and seedling growth in cultivated sunflower (Helianthus annuus L.) under well-watered and water-limited conditions. PLoS One 13: e0204279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser P, Gierth M, Schroeder JI(2002) Molecular mechanisms of potassium and sodium uptake in plants. Plant Soil 247: 43–54 [Google Scholar]
- Mayrose M, Kane NC, Mayrose I, Dlugosch KM, Rieseberg LH(2011) Increased growth in sunflower correlates with reduced defences and altered gene expression in response to biotic and abiotic stress. Mol Ecol 20: 4683–4694 [DOI] [PubMed] [Google Scholar]
- Morton MJL, Awlia M, Al-Tamimi N, Saade S, Pailles Y, Negrão S, Tester M(2019) Salt stress under the scalpel: Dissecting the genetics of salt tolerance. Plant J 97: 148–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R.(2002) Comparative physiology of salt and water stress. Plant Cell Environ 25: 239–250 [DOI] [PubMed] [Google Scholar]
- Munns R.(2005) Genes and salt tolerance: Bringing them together. New Phytol 167: 645–663 [DOI] [PubMed] [Google Scholar]
- Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, Bose J, Byrt CS, Chen Z-H, Foster KJ, et al. (2020a) Energy costs of salt tolerance in crop plants. New Phytol 225: 1072–1090 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Gilliham M, Flowers TJ, Colmer TD(2016) Tissue tolerance: An essential but elusive trait for salt-tolerant crops. Funct Plant Biol 43: 1103–1113 [DOI] [PubMed] [Google Scholar]
- Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, et al. (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30: 360–364 [DOI] [PubMed] [Google Scholar]
- Munns R, Passioura JB, Colmer TD, Byrt CS(2020b) Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol 225: 1091–1096 [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M(2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59: 651–681 [DOI] [PubMed] [Google Scholar]
- Negrão S, Schmöckel SM, Tester M(2017) Evaluating physiological responses of plants to salinity stress. Ann Bot 119: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicotra AB, Atkin OK, Bonser SP, Davidson AM, Finnegan EJ, Mathesius U, Poot P, Purugganan MD, Richards CL, Valladares F, et al. (2010) Plant phenotypic plasticity in a changing climate. Trends Plant Sci 15: 684–692 [DOI] [PubMed] [Google Scholar]
- Pailles Y, Awlia M, Julkowska M, Passone L, Zemmouri K, Negrão S, Schmöckel SM, Tester M (2019) A diversity of traits contributes to salinity tolerance of wild Galapagos tomatoes seedlings. bioRxiv 642876, doi:10.1101/642876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R, Ruano-Rubio V, DePristo MA, Fennell TJ, Carneiro MO, Van der Auwera GA, Kling DE, Gauthier LD, Levy-Moonshine A, Roazen D, et al. (2017) Scaling accurate genetic variant discovery to tens of thousands of samples. bioRxiv 201178, doi:10.1101/01178 [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2019) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
- Ramankutty N, Mehrabi Z, Waha K, Jarvis L, Kremen C, Herrero M, Rieseberg LH(2018) Trends in global agricultural land use: Implications for environmental health and food security. Annu Rev Plant Biol 69: 789–815 [DOI] [PubMed] [Google Scholar]
- Rawson HM, Munns R(1984) Leaf expansion in sunflower as influenced by salinity and short-term changes in carbon fixation. Plant Cell Environ 7: 207–213 [Google Scholar]
- Richards CL, Walls RL, Bailey JP, Parameswaran R, George T, Pigliucci M(2008) Plasticity in salt tolerance traits allows for invasion of novel habitat by Japanese knotweed s. l. (Fallopia japonica and F.xbohemica, Polygonaceae). Am J Bot 95: 931–942 [DOI] [PubMed] [Google Scholar]
- Salt DE, Baxter I, Lahner B(2008) Ionomics and the study of the plant ionome. Annu Rev Plant Biol 59: 709–733 [DOI] [PubMed] [Google Scholar]
- Schilling RK, Tester M, Marschner P, Plett DC, Roy SJ(2017) AVP1: One Protein, Many Roles. Trends Plant Sci 22: 154–162 [DOI] [PubMed] [Google Scholar]
- Schultz E, DeSutter T, Sharma L, Endres G, Ashley R, Honggang B, Markell S, Kraklau A, Franzen D(2018) Response of sunflower to nitrogen and phosphorus in North Dakota. Agron J 110: 685–695 [Google Scholar]
- Sedlazeck FJ, Rescheneder P, von Haeseler A(2013) NextGenMap: Fast and accurate read mapping in highly polymorphic genomes. Bioinformatics 29: 2790–2791 [DOI] [PubMed] [Google Scholar]
- Sella G, Barton NH(2019) Thinking about the evolution of complex traits in the era of genome-wide association studies. Annu Rev Genomics Hum Genet 20: 461–493 [DOI] [PubMed] [Google Scholar]
- Shabala S, Chen G, Chen Z-H, Pottosin I(2019) The energy cost of the tonoplast futile sodium leak. New Phytol 225: 1105–1110 [DOI] [PubMed] [Google Scholar]
- Shabala S, Cuin TA(2008) Potassium transport and plant salt tolerance. Physiol Plant 133: 651–669 [DOI] [PubMed] [Google Scholar]
- Shi D, Sheng Y(2005) Effect of various salt–alkaline mixed stress conditions on sunflower seedlings and analysis of their stress factors. Environ Exp Bot 54: 8–21 [Google Scholar]
- Temme AA, Burns VA, Donovan LA (2019a) Element content and distribution has limited, tolerance metric dependent, impact on salinity tolerance in cultivated sunflower (Helianthus annuus). bioRxiv 872929, doi:10.1101/2019.12.11.872929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temme AA, Kerr KL, Donovan LA(2019b) Vigour/tolerance trade‐off in cultivated sunflower (Helianthus annuus) response to salinity stress is linked to leaf elemental composition. J Agron Crop Sci 205: 508–518 [Google Scholar]
- Todesco M, Owens GL, Bercovich N, Légaré J-S, Soudi S, Burge DO, Huang K, Ostevik KL, Drummond EBM, Imerovski I, et al. (2019) Massive haplotypes underlie ecotypic differentiation in sunflowers. bioRxiv 790279, doi:10.1101/790279 [DOI] [PubMed] [Google Scholar]
- Todesco M, Owens GL, Bercovich M, Légaré J-S, Soudi S, Burge DO, Huang L, Ostevik KL, Drummond EBM, Imerovski I, et al. (2020) Massive haplotypes underlie ecotypic differentiation in sunflowers. Nature 584: 602–607 [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. (2013) From FastQ data to high confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11.10.1-11.10.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit C, König J, Altmann F, Strasser R(2018) Processing of the terminal alpha-1,2-linked mannose residues from oligomannosidic N-glycans is critical for proper root growth. Front Plant Sci 9: 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM(2007) The road to modularity. Nat Rev Genet 8: 921–931 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J(2011) The pleiotropic structure of the genotype-phenotype map: The evolvability of complex organisms. Nat Rev Genet 12: 204–213 [DOI] [PubMed] [Google Scholar]
- Wang Y, Donovan LA, Temme AA (2019) Allometric scaling impacts plasticity in biomass allocation, morphology and anatomical traits due to above and belowground resource limitation in cultivated sunflower (Helianthus annuus L.). bioRxiv 504316, doi:10.1101/504316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YC, Qu GZ, Li HY, Wu YJ, Wang C, Liu GF, Yang CP(2010) Enhanced salt tolerance of transgenic poplar plants expressing a manganese superoxide dismutase from Tamarix androssowii. Mol Biol Rep 37: 1119–1124 [DOI] [PubMed] [Google Scholar]
- Wickham H.(2009) ggplot2: Elegant Graphics for Data Analysis, Ed 1 Springer-Verlag, New York [Google Scholar]
- Wu H, Zhang X, Giraldo JP, Shabala S(2018) It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 431: 1–17 [Google Scholar]
- York LM.(2019) Functional phenomics: An emerging field integrating high-throughput phenotyping, physiology, and bioinformatics. J Exp Bot 70: 379–386 [DOI] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS(2012) A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M(2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44: 821–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Shabala S, Shabala L, Fan Y, Zhou MX(2016) Evaluating predictive values of various physiological indices for salinity stress tolerance in wheat. J Agron Crop Sci 202: 115–124 [Google Scholar]
- Zolla G, Heimer YM, Barak S(2010) Mild salinity stimulates a stress-induced morphogenic response in Arabidopsis thaliana roots. J Exp Bot 61: 211–224 [DOI] [PMC free article] [PubMed] [Google Scholar]