A European sequence-indexed library of Mutator insertions provides an important resource for functional genetics studies of maize.
Abstract
Sequence-indexed insertional libraries in maize (Zea mays) are fundamental resources for functional genetics studies. Here, we constructed a Mutator (Mu) insertional library in the B73 inbred background designated BonnMu. A total of 1,152 Mu-tagged F2-families were sequenced using the Mu-seq approach. We detected 225,936 genomic Mu insertion sites and 41,086 high quality germinal Mu insertions covering 16,392 of the annotated maize genes (37% of the B73v4 genome). On average, each F2-family of the BonnMu libraries captured 37 germinal Mu insertions in genes of the Filtered Gene Set (FGS). All BonnMu insertions and phenotypic seedling photographs of Mu-tagged F2-families can be accessed via MaizeGDB.org. Downstream examination of 137,410 somatic and germinal insertion sites revealed that 50% of the tagged genes have a single hotspot, targeted by Mu. By comparing our BonnMu (B73) data to the UniformMu (W22) library, we identified conserved insertion hotspots between different genetic backgrounds. Finally, the vast majority of BonnMu and UniformMu transposons was inserted near the transcription start site of genes. Remarkably, 75% of all BonnMu insertions were in closer proximity to the transcription start site (distance: 542 bp) than to the start codon (distance: 704 bp), which corresponds to open chromatin, especially in the 5′ region of genes. Our European sequence-indexed library of Mu insertions provides an important resource for functional genetics studies of maize.
The genome of the maize (Zea mays) inbred line B73v4 is predicted to contain 44,117 genes, including 39,179 high confidence protein encoding gene models and 4,938 genes encoding for large intergenic noncoding RNAs, transfer RNAs, and microRNAs (Jiao et al., 2017). To date only a few hundred of these genes have been functionally characterized (Schnable and Freeling, 2011). The identification of specific mutants in forward genetic screens and the subsequent cloning of the underlying genes has been the method of choice to determine gene functions for several decades (Candela and Hake, 2008). Until now, reverse genetics using specific gene sequences as a starting point for the identification of the corresponding mutants (Candela and Hake, 2008) has mainly been used to independently validate candidate genes identified by forward genetics experiments (e.g. Hochholdinger et al., 2008; Nestler et al., 2014). Nevertheless, reverse genetics has also been successfully applied to identify developmental mutants of genes without prior knowledge of their phenotype (Xu et al., 2015). Genome-wide insertional mutagenesis is a tool to create loss-of-function mutations for virtually all genes in a genome by the insertion of DNA into the gene of interest. Sequence-indexed collections of such mutants have been generated for Arabidopsis (Arabidopsis thaliana; Alonso et al., 2003), rice (Oryza sativa; Hirochika et al., 2004), and maize (Fernandes et al., 2004; McCarty et al., 2005; Liang et al., 2019).
Endogenous transposons or transposable elements (TEs) are mobile DNA sequences initially discovered in maize by McClintock (1951). Once activated, a transposon can move from one location in the genome to another, thereby disrupting genes. The two major classes of transposons are class I retrotransposons and class II DNA transposons. The main difference between these classes is their mode of transposition. Class I retrotransposons first require transcription of their DNA to an RNA intermediate, which is then reversely transcribed into DNA, before it is reinserted into a new genomic location (for review, see Feschotte et al., 2002). This mechanism is catalyzed by a reverse transcriptase. In contrast, the TE itself is the transposition intermediate for class II DNA transposons. Both classes have in common that they consist of autonomous and nonautonomous TEs. Autonomous TEs can transpose by themselves, because they encode a transposase, whereas nonautonomous TEs require another autonomous transposon to move. Class I transposons are predominantly intergenic, contributing to plant genome size (Bennetzen, 2000). In contrast, class II TEs insert preferentially in and around genes (Dietrich et al., 2002; Fernandes et al., 2004; Settles et al., 2004), which is a prerequisite for genome-wide insertional mutagenesis screens.
The most active class II transposon family in maize is the family of Mutator (Mu) elements (Lisch, 2002). Discovered by Robertson (1978), MuDR, an autonomous TE, controls the transposition of itself and 16 classes of nonautonomous Mu elements (Lisch, 2015). All Mu transposons share highly conserved 215-bp terminal inverted repeats (TIRs) at both ends of the element and upon insertion they generate 9-bp target site duplications directly flanking the Mu transposon sequences. For untargeted insertional mutagenesis, Mu exhibits a number of advantages over other maize class II transposon families, such as Activator/Dissociation (Ac/Ds) or Enhancer/Suppressor-mutator (En/Spm), including its high transposition rate and high-copy number (Chandler and Hardeman, 1992). While Ac/Ds transposons exhibit a preference for regional mutagenesis (i.e. transposition into closely linked genes; Brutnell, 2002), Mu elements randomly target genes throughout the maize genome (Lisch, 2002). As such, Mu insertion site frequencies strongly correlate with gene density (Liu et al., 2009; Schnable et al., 2009; Springer et al., 2018).
In the past, adapter-mediated (Frey et al., 1998) and thermal asymmetric interlaced-PCR (Liu and Whittier, 1995) have been used to amplify Mu-flanking fragments for sequencing. The presence of highly conserved TIR sequences at both ends of the transposons is ideally suited to design Mu-specific primers used in thermal asymmetric interlaced-PCR. This PCR technique combines nested TIR-specific primers and degenerative arbitrary primers to amplify sequences neighboring known sequences by using high and low annealing temperature cycles (Settles et al., 2004). More recently, high-throughput next generation sequencing enables efficient, reproducible, and sequence-based identification and mapping of transposon insertion sites in maize (McCarty et al., 2013; Liu et al., 2016; Liang et al., 2019).
Thus far, several transposon-tagged maize populations have been generated using the Mu transposon system (Fernandes et al., 2004; Stern et al., 2004; McCarty et al., 2005; McCarty et al., 2013; Liang et al., 2019). Nevertheless, to date only about 52% of the annotated maize gene models (Liang et al., 2019) have mapped Mu insertions. Among these collections, the insertion-tagged UniformMu population (McCarty et al., 2005) has been developed by backcrossing an active MuDR element eight times into the genetically uniform inbred line W22. A genetically uniform genetic inbred background is one key feature of the UniformMu resource, because it allows to distinguish between parental and newly created mutations. Therefore, a uniform background aids to identification of insertions related to new phenotypes (McCarty et al., 2005). A large set of more than 14,000 UniformMu lines are publicly available (Liu et al., 2016). Another Mu insertional library, called ChinaMu, has been generated by crossing a Mu-starter line to the inbred line B73 (Liang et al., 2019). The Mu-tagged sequences of 2,581 F2-Mu lines were isolated by a Mu tag enrichment approach and sequenced by high-throughput sequencing.
Here we used the Mutator-seq approach (Mu-seq; McCarty et al., 2013; Liu et al., 2016) to construct multiplexed sequencing libraries in the inbred line B73 that guarantees unbiased coverage of mutations in the maize genome. The Mu-seq reverse genetics approach enables the identification of F2-families carrying transposon insertions in genes of interest in a sequence-indexed transposon tagged population. These newly identified transposon induced mutations can be used for subsequent molecular and genetic analyses. Furthermore, this European database (BonnMu) of sequence-indexed Mu transposon insertion sites in B73 complements the UniformMu and ChinaMu resources and allows for forward and reverse genetics experiments. Besides the sequence-indexed Mu insertional libraries from North America (UniformMu) and Asia (ChinaMu), our resource of transposon induced mutations will offer easier access for European researchers but also for maize geneticists around the globe.
RESULTS
Mu-seq Library Alignment Statistics of BonnMu
To generate a sequence-indexed collection of transposon tagged maize mutants, the Mu-seq protocol (McCarty et al., 2013; Liu et al., 2016) was applied to generate two libraries each containing 576 Mu active B73 F2-families (BonnMu) of maize. These 1,152 mutagenized F2-families were pooled according to a 24 × 24 grid system per library (see “Materials and Methods”). Sequencing of the two libraries yielded 55 and 90 million raw read pairs (Table 1; Supplemental Dataset S1). Each Mu-seq library was parsed according to the individual 6-bp barcodes of the 48 multiplexed pools (Supplemental Table S1), resulting in an averaged output of 1,151,003 and 1,871,525 read pairs per pool (Supplemental Dataset S1). Downstream statistical analysis of the two Mu-seq libraries are summarized in Table 1. After U-adapter and TIR sequences were trimmed, 83% and 89% (Table 1) of the remaining reads (43 and 65 million) were aligned to the B73 Filtered Gene Set (FGS) AGPv4.36, containing 44,117 high confidence gene models. After duplicate read removal, 18 and 23 million read pairs remained and were used to identify genomic insertion sites, in (1) any of the 48 pools (somatic and germinal insertions, named union in Table 1) and (2) at intersections of one row and one column pool (germinal insertions, named intersection in Table 1). For insertion site identification, reads were counted in each of the 48 pools. Only insertion sites supported by at least four reads were used for further analyses. In total, 225,936 genomic insertion sites were detected in the two Mu-seq libraries (Table 1; Supplemental Dataset S2). Among those insertion sites, 61% (137,410 of 225,936 insertions) mapped to FGS genes, tagging in total 22,362 genes (Table 1). Thus, there were on average six somatic or germinal insertions per tagged FGS gene. Further parsing of insertion sites identified 110,730 insertion sites in exons of FGS genes, 40,140 of such sites in the coding sequence, 75,201 sites in 5′ untranslated region (UTR), 7,125 insertion sites in 3′ UTR, and 25,203 insertion sites in introns of FGS genes. In addition, 38,609 insertion sites ≤500 bp upstream of FGS genes and 88,562 intergenic insertions were detected (Table 1). Based on the genome annotation of AGPv4.36, insertion sites can be assigned to more than one feature (e.g. both the 5′ UTR and exonic region), within a gene. To calculate the number of insertion sites in introns, the union of insertion sites located in exons/coding sequence (cds) and UTRs was subtracted from the number of insertion sites in FGS genes. Therefore, the number of insertion sites located in introns cannot be recalculated from Table 1.
Table 1. Mu-seq library alignment statistics.
| Statistical Analysis | Mu-seq 1 | Mu-seq 2 | Combined |
|---|---|---|---|
| Raw read pairs | 89,833,184 | 55,248,154 | – |
| Read pairs after trimminga | 65,773,604 | 43,651,352 | – |
| Average alignment rate | 89% | 83% | – |
| Remaining read pairs after removing duplicate readsb | 23,251,540 | 18,785,728 | – |
| Union (somatic and germinal insertions)c | |||
| Insertion sites | 135,108 | 112,205 | 225,936 |
| Insertion sites in FGS genes | 81,698 | 68,413 | 137,410 |
| Insertion sites in exons of FGS genes | 66,270 | 55,221 | 110,730 |
| Insertion sites in cds of FGS genes | 22,743 | 19,743 | 40,140 |
| Insertion sites in 5‘ UTR of FGS genes | 46,362 | 37,945 | 75,201 |
| Insertion sites in 3‘ UTR of FGS genes | 4,085 | 3,361 | 7,125 |
| Insertion sites in introns of FGS genes | 14,586 | 12,404 | 25,203 |
| Insertion sites upstream of FGS genes (≤ 500 bp) | 23,268 | 19,411 | 38,609 |
| Intergenic insertion sites | 53,410 | 43,792 | 88,562 |
| Mu-tagged FGS genes | 19,162 | 17,923 | 22,362 |
| Intersection (germinal insertions)d | |||
| Insertion sites | 43,225 | 24,234 | 66,750 |
| Insertion sites in noncoding DNA | 16,462 | 9,323 | 25,664 |
| Insertions sites in FGS genes | 26,762 | 14,912 | 41,086 |
| Mu-tagged FGS genes | 13,590 | 9,538 | 16,392 |
| Mu-tagged FGS genes, affected in their cds | 5,605 | 3,487 | 7,582 |
Mu-seq reads were trimmed and aligned to the B73v4 genome as described in the “Materials and Methods” section.
Duplicate reads were removed as described in the “Materials and Methods” section.
All insertion sites identified in any of the samples. Insertion sites were identified as described in the “Materials and Methods” section.
Insertion sites identified in only one row and one column sample, respectively.
Finally, to only consider heritable (germinal) insertions the information from the different pools was combined to track down Mu-tagged genes in specific F2-families. Each F2-family is represented by the intersection of one column and one row pool (according to the 24 × 24 grid system; Supplemental Table S1). By that criterion, we identified 66,750 germinal insertion sites in the two Mu-seq libraries, of which 38% (25,664 of 66,750) mapped to noncoding DNA (Table 1). The remaining 62% (41,086 of 66,750) of the insertion sites were located in genes of the FGS (Table 1; Supplemental Dataset S3). On average, each F2-family harbored 37 germinal insertions in FGS genes (Supplemental Tables S2 and S3; Supplemental Dataset S3). Furthermore, 46% of mutated FGS genes (7,582 of 16,392 FGS genes) contained insertions in coding sequences.
European-Based BonnMu Complements the North American UniformMu and the Asian ChinaMu Resource
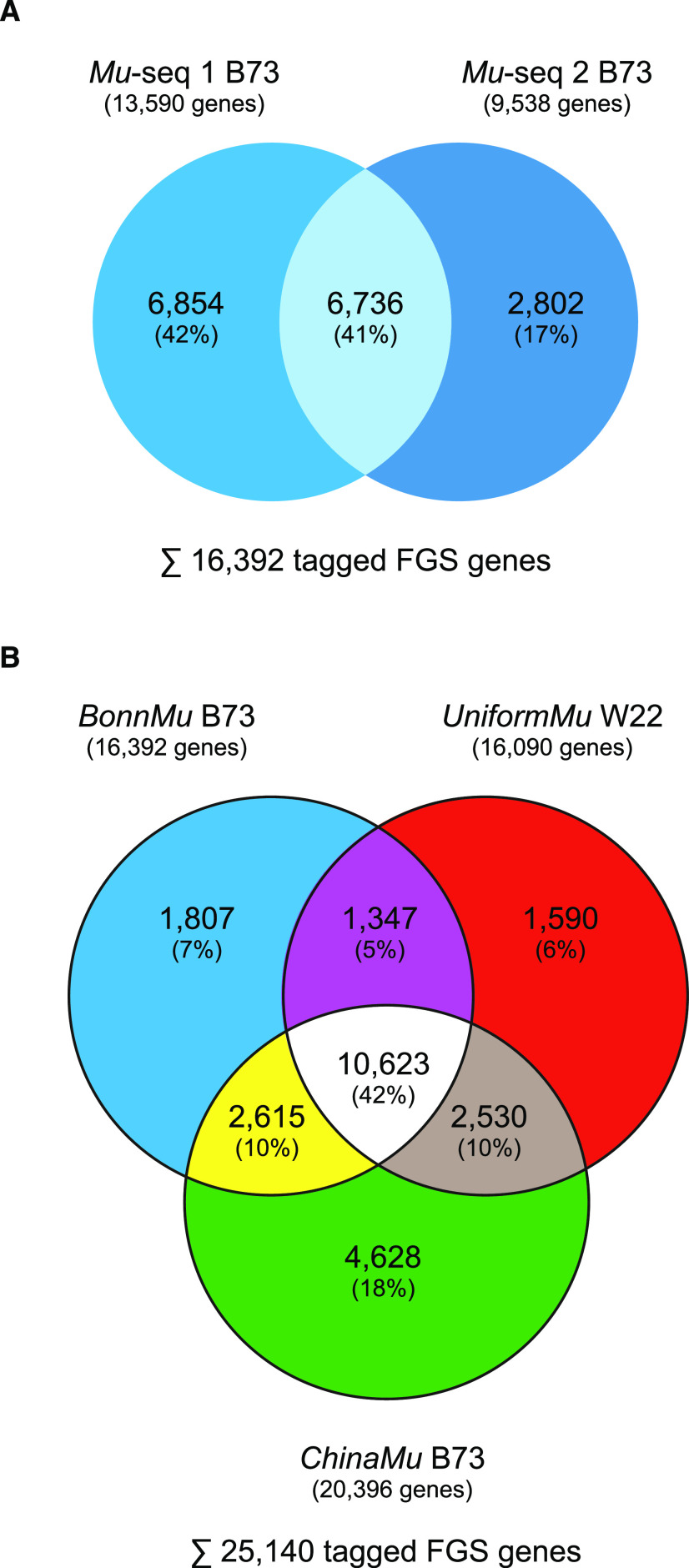
Based on the analysis of two Mu-seq libraries in B73 (BonnMu), Mu insertions affected 16,392 of 44,117 (37%; Fig. 1A; Table 1) high-confidence gene models of maize. Of the 16,392 genes, 41% (6,763) were identified in F2-families of both Mu-seq libraries, whereas 59% (9,656) of the genes were uniquely detected in one of the Mu-seq libraries (Fig. 1A). We observed a significantly higher overlap of Mu-tagged genes in both Mu-seq libraries than expected (2,938; Supplemental Table S4). This finding supports the notion that genes are not randomly targeted by Mu transposons, but that there is a preference for Mu tagging of specific genes.
Figure 1.
Overlap of FGS genes tagged by Mu transposon insertions. A, Tagged FGS genes among the two Mu-seq libraries in the B73 background. B, FGS genes with Mu insertions in the three available Mu-tagged resources in the B73 (blue, BonnMu; green, ChinaMu) and W22 inbred backgrounds (red, UniformMu).
The 16,392 genes of the BonnMu collection in the B73 background was compared with 16,090 Mu-tagged genes of the UniformMu resource in the W22 background (McCarty et al., 2013) and to 20,396 genes tagged in B73 in the ChinaMu dataset (Liang et al., 2019) and personal communication with R. Song). The three collections cover 57% (25,140 of 44,117) of the genes present in the B73 reference genome AGPv4.36 (Fig. 1B). A considerable proportion of Mu-tagged genes, that is, 42% (10,623 of 25,140) were hit in all three collections. This number of overlapping genes was significantly higher than expected by pure chance (2,724; Supplemental Table S4), suggesting that at least some genes are preferentially targeted by Mu insertions.
BonnMu insertions can be browsed at the MaizeGDB Web site (https://www.maizegdb.org; Portwood et al., 2019) as previously described for the UniformMu database (Liu et al., 2016). A unique feature of BonnMu is that photos of seedling phenotypes of segregating F2-families are linked to the database entries of each transposon insertion. To access and visualize B73 insertions in maizegdb.org, a gene model identifier is needed for the genome browser view. When “select track” and “BonnMu” are clicked, insertion sites become visible in the locus. Clicking on the insertion site links to the Mu insertion identifiers (e.g. BonnMu0000812) and the respective F2-Mu-seq family (e.g. BonnMu-2-A-0596) and its respective phenotype 10 d after germination. Among the analyzed F2-families various leaf color mutants were identified such as albino (e.g. BonnMu-2-A-0784) or pale green (e.g. BonnMu-2-A-0784). The mutation rate, detected on the basis of albino and pale green leaf phenotype, was 13% (149 of 1,152 F2-families). Furthermore, shoot gravitropism mutants (e.g. BonnMu-2-A-0596, BonnMu-2-A-0598) were identified.
The Number of Mu Insertions Correlates with Gene Length in a Size-Dependent Manner
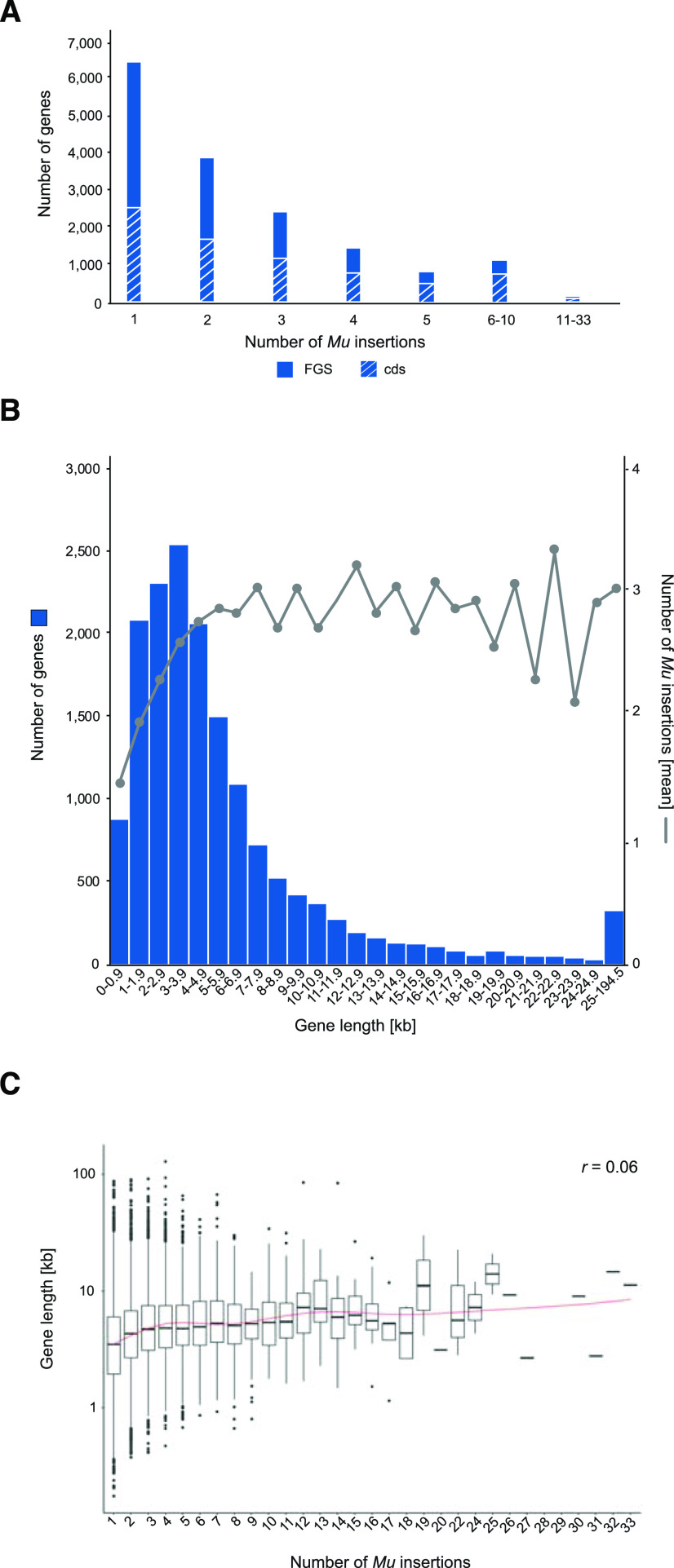
The present dataset contains 41,086 Mu insertions in 16,392 B73 high confidence gene models (Table 1; Supplemental Dataset S3). More specifically, 39% (6,455 of 16,392) of the high-confidence gene models were tagged by a unique insertion (Fig. 2A), whereas 61% (9,937 of 16,392) contained at least two Mu insertions. A similar distribution was observed when the 7,582 FGS genes harboring insertions in their cds (Table 1; Supplemental Dataset S3) were analyzed. Among those, 33% (2,535 of 7,582) had a unique insertion, whereas 67% (5,047 of 7,582) were tagged by at least two Mu insertions (Fig. 2A). An extreme example is a gene encoding an adaptor protein 4 (AP-4) complex subunit µ (Zm00001d002925), which harbored 33 independent insertions in its cds (Supplemental Dataset S3).
Figure 2.
Number of Mu insertions and its correlation to the length of affected FGS genes of B73. A, Distribution and number of Mu insertions among 16,392 affected FGS genes and the subset of 7,582 genes harboring insertions in the cds of FGS genes. B, Number of tagged genes and associated mean number of Mu insertions plotted against the corresponding gene size. C, Length distribution of affected genes plotted against the individual number of Mu insertions (Pearson correlation coefficient, r = 0.06).
The length of the affected genes ranged between 72 bp and 195 kb (Fig. 2B; Supplemental Dataset S3). The three shortest genes (72 bp) encode transfer RNAs (Zm00001d006818, Zm00001d007104, and Zm00001d034123), whereas the largest gene of 195 kb (Zm00001d001010) represents a large intergenic noncoding RNA. We observed 882 tagged genes (5% of all 16,392 Mu-tagged FGS genes) in a size range of 72 to 999 bp (Fig. 2B). Then, the size and the number of tagged genes increased, with a peak of 2,548 affected genes (16%) having a size between 3 and 3.999 kb. After this peak the number of affected genes decreased, while their length increased. Overall, 12,510 tagged genes (76%) had a size between 72 bp and 6.999 kb, whereas only 3,882 genes (24%) were >7 kb. In contrast with the observed distribution of the 16,392 tagged genes according to their gene length, the size distribution of all 44,117 B73v4 high-confidence gene models differed. Although 47% of all B73v4 gene models had a size of <2 kb, only 18% of all Mu-tagged genes of the present dataset ranged in that size (Supplemental Fig. S1). In contrast, 53% of all B73v4 gene models had a length between 2 and 194.5 kb, whereas among all Mu-tagged genes 82% ranged between these lengths.
Next, we tested the hypothesis that the number of Mu insertions increased with the length of the affected genes. Although a strong linear correlation of insertion number with gene size was detected in the gene size range of 72 bp to 3.999 kb, there was no correlation with gene size for genes >4 kb (Fig. 2B). Hence, the calculated Pearson correlation over all groups of gene sizes revealed no correlation (r = 0.06; Fig. 2C). This abrupt transition in gene-size dependence at 4 kb was also detected in the W22 dataset (Supplemental Fig. S2; Supplemental Dataset S4). Due to the fact that a large subset of 48% affected genes (7,838 B73 and 7,790 W22 genes) have a size of <4 kb, the gene-size-dependent correlation is important to consider.
Mu Insertion Hotspots Are Conserved Among Affected Genes of B73 and W22
Next, we investigated if Mu transposons show preferences regarding their insertion sites in the genes. Therefore, we examined all insertion sites in any of the samples, including somatic and germinal insertions of the present BonnMu datasest (Table 1; Supplemental Dataset S2). The majority of these insertion sites (61%; 137,410 of 225,936) was located in FGS genes. Each of the 22,362 Mu-tagged genes was divided into bins of 200 bp in size. Then, all insertion sites were sorted into the corresponding bins. Each bin with at least one insertion site was counted as a hotspot. In total, 38% (8,502 of 22,362 genes) of the Mu-tagged genes had one hotspot, which was predominantly targeted by the transposons (Supplemental Fig. S3A). The number of hotspots per affected gene ranged from 1 to 15, with a median number of two hotspots, independent of gene length (Supplemental Fig. S3B).
To investigate if insertion hotspots are conserved among affected genes of our Mu-seq repository in B73 and the UniformMu repository in the W22 genetic background, one exemplary W22 Mu-seq dataset (SRA accession SRP028545) was reanalyzed. A total of 558 identified insertion sites in W22 supported by at least two reads could be assigned to 423 FGS genes.
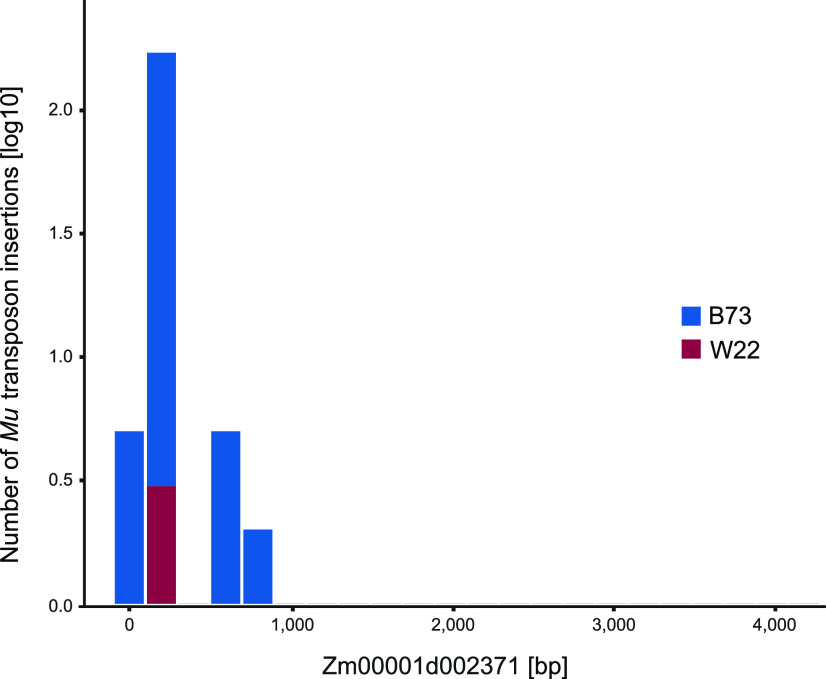
Among these 423 Mu-tagged genes in W22, 413 (98%) were also affected in B73. For the overlapping 413 genes a hotspot analysis was performed. Because we were interested in only conserved hotspots of new Mu insertion events, we first corrected the affected genes for endogenous Mu insertions that are present in the genomes of B73 and W22. About 40 canonical Mu elements exist in the two genomes (Springer et al., 2018; D. R. McCarty, personal communication; Supplemental Dataset S5). Due to the fact that such parental Mu elements are fixed in every B73/W22 plant, such elements account for a large proportion of reads. Hence, the endogenous elements could contribute to an apparent hotspot overlap between B73 and W22. After correcting for endogenous Mu elements, 22,348 B73 genes and 420 W22 genes remained. Among those, 412 genes overlapped between the two datasets and were divided into bins of 200 bp in length. Then, Mu transposon insertion sites identified in B73 and W22 were sorted into the corresponding bin. Out of 14,736 bins, 293 were targeted in both B73 and W22, 1,406 only in B73, 138 only in W22, and 12,899 in neither B73 nor W22 (Table 2). A χ2-test was performed to test the hypothesis that there is an association between insertion sites in B73 and W22. The observed value of overlapping bins in both B73 and W22 (293) significantly exceeded the expected value of 50 bins, demonstrating that there are conserved insertion site hotspots in both genomes. This observation is exemplarily shown in Figure 3 for gene Zm00001d002371. Here, Mu transposon insertions were concentrated around two hotspots within that gene in B73 and one overlapping hotspot in W22.
Table 2. Number of expected and observed unique and overlapping bins (i.e. 200-bp sized regions of genes) targeted by Mu transposon insertions in B73 and W22.
| Genotype | B73 YES | B73 NO | Total | Observed/Expected |
|---|---|---|---|---|
| W22 YES | 293a | 138 | 431 | Observed |
| 50 | 381 | – | Expectedb | |
| W22 NO | 1,406 | 12,899 | 14,305 | Observed |
| 1,649 | 12,656 | – | Expectedb | |
| Total | 1,699 | 13,037 | 14,736 | – |
P < 2.2×e−16 (obtained from χ2-testing).
Expected value of bins being hit in B73 and W22 was calculated as total number of bins identified in W22 (431)* total number of bins identified in B73 (1,699) / total number of bins (14,736).
Figure 3.
A Mu insertion hotspot in the representative gene Zm00001d002371 affected in the B73 and W22 genomes. The number of Mu insertions is plotted against location in Zm00001d002371.
Mu Transposons Preferentially Insert near the Transcription Start Site of Genes in B73 and W22
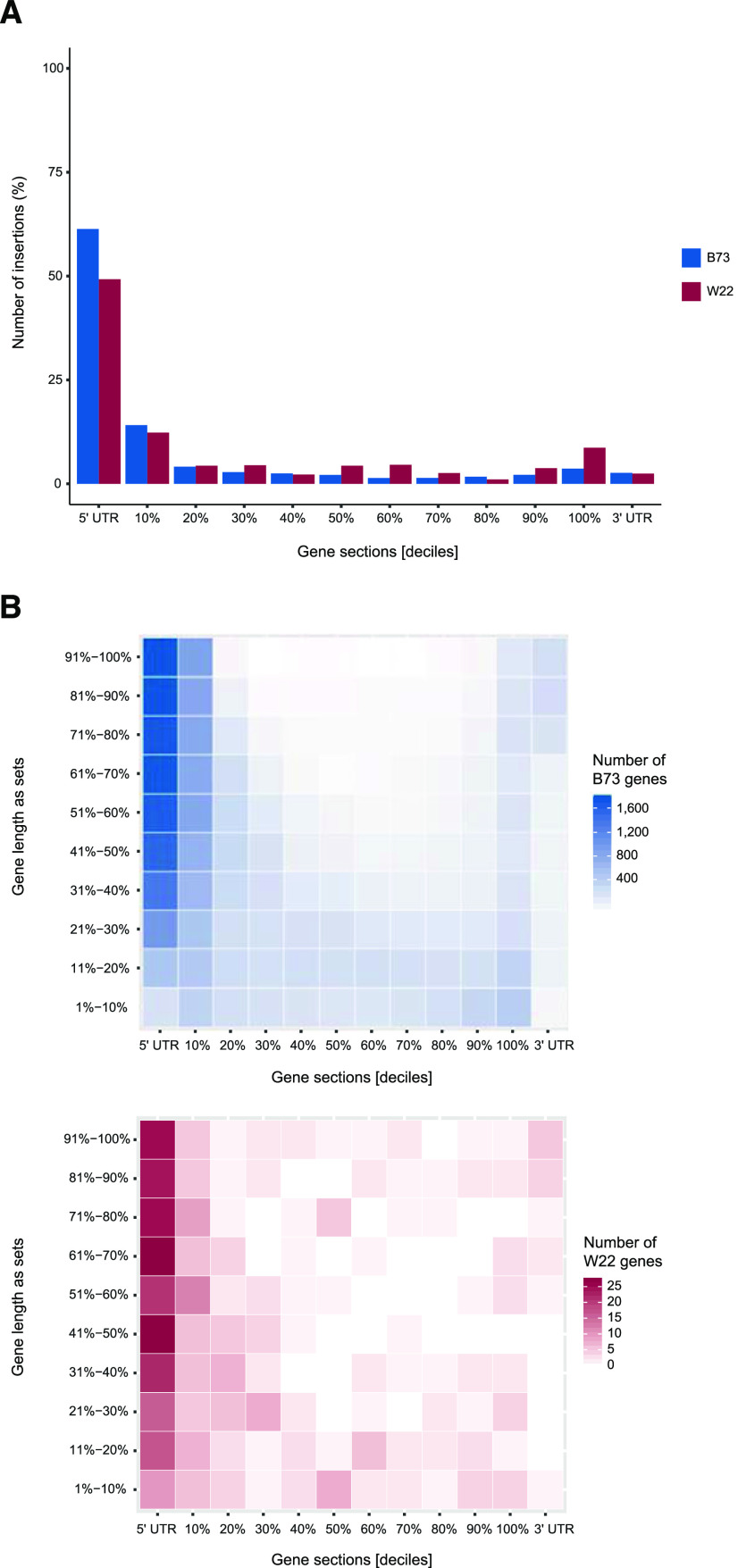
To investigate if Mu transposons show preferences regarding their insertion sites in genes, the union of all B73 Mu transposon insertion sites located in FGS genes was analyzed. A total of 110,730 of the insertion sites were located in exons of FGS genes (Table 1). Whereas 40,140 of the insertion sites were assigned to the cds of FGS genes, the majority of insertion sites (75,201) were located in the 5′ UTR of FGS genes.
To examine Mu transposon insertion site preferences in B73 compared with W22, for each affected FGS gene with annotated cds, the cds was divided into deciles. Subsequently, all insertion sites located in the cds of FGS genes were sorted into the corresponding cds decile. Insertion sites located up- or downstream of the cds were designated as insertions in the 5′ or 3′ UTR, respectively. The majority of Mu insertion sites was detected in the 5′ UTR and the first decile of the cds in both B73 (76%) and W22 (62%), with another smaller peak at the end of the cds (Fig. 4A). To account for the length of the FGS genes, all affected genes were sorted into gene sets based on their length (Supplemental Fig. S4A). Each of the sets (deciles) contained 2,152 affected B73 and either 42 or 43 affected W22 genes. Whereas in shorter genes of B73 (deciles 1% to 10%, and 11% to 20%) Mu insertions were evenly distributed among the gene, longer genes showed an insertion peak at the start region (i.e. the 5′ UTR and first 10% of the genes’ cds; Fig. 4B). Another small peak of insertions was detected at the end of the cds (100%) of B73 genes.
Figure 4.
Distribution of Mu transposon insertion sites in FGS genes. A, Proportion of Mu insertions located in each decile of the cds, 5′ UTR, and 3′ UTR (B73, blue; W22, red). B, All FGS genes targeted by Mu transposons in B73 and W22 (SRP028545) were divided into gene sets based on their length (1% to 10% represent the shortest set of genes and 91% to 100% represent the longest genes) and the number of genes with insertions in each cds decile, 5′ UTR and 3′ UTR (B73, blue; W22, red).
Although the W22 dataset (SRP028545) only contained 423 genes, a similar preference of Mu insertions to target the 5′ UTR of the genes was observed (Fig. 4B). The observation that Mu insertions were evenly distributed among the gene of shorter genes, especially in B73, can be explained by the very short 5′ UTR of those genes. By comparing the length of the 5′ UTRs of each of the ten gene sets it became clear that the first two gene sets (1% to 10%, 135–1214 bp; 11% to 20%, 1215–1872 bp) had very short or even no 5′ UTRs (Supplemental Fig. S4B). Therefore, the absolute distance of insertions to the 5′ UTRs in those genes was small, even if the insertion was close to the 3′ end of the gene.
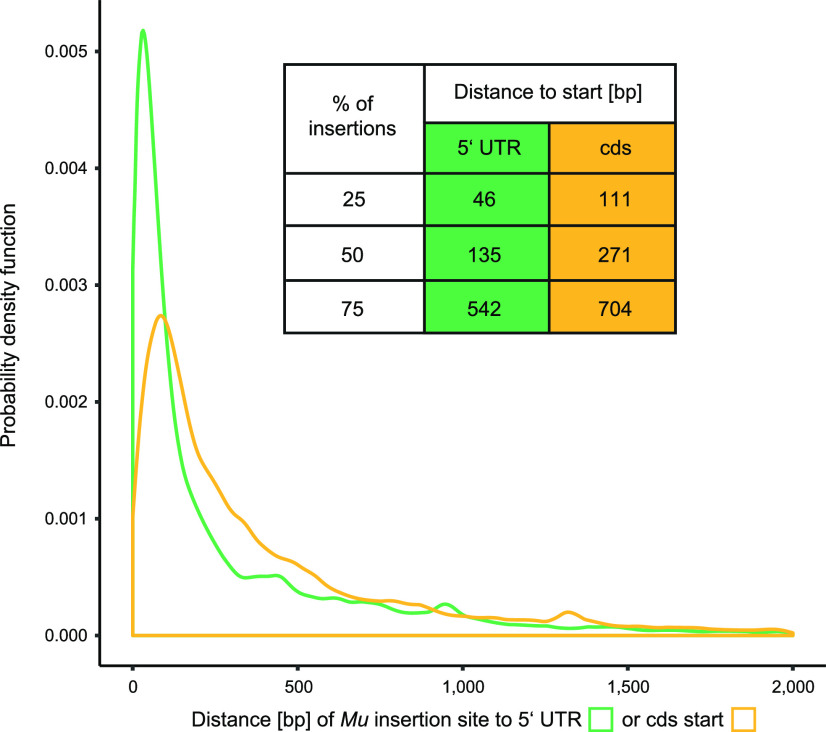
To verify if the insertion sites were located closer to the 5′ UTR start or closer to the start codon (ATG) of the affected genes, the distance (bp) from insertions inside or upstream of FGS genes to both the 5′ UTR start and cds start codon was calculated for the B73 dataset. The density plot (Fig. 5) represents only insertion sites located 500 bp upstream of the 5′ UTR to 2,000 bp downstream of the genes’ start codon. Irrespective of this cutoff, the vast majority of 93% of all insertion sites are in the range of the above-mentioned distance to the 5′ UTR and 90% of all insertion sites are in such a distance to the cds. This result suggested that the majority of Mu insertion sites are close to the 5′ UTR and the cds start of the affected genes. This is also visible in the density plot (Fig. 5) where the majority of Mu insertion sites peaked in close proximity to the 5′ UTR and cds start. The slope of the curve revealed that the Mu transposon insertions are closer to the 5′ UTR than to the genes’ start codon. Finally, 25% of all Mu insertion sites were located in close proximity to the 5′ UTR (46 bp) and to the cds start (111 bp; Fig. 5). When 50% of all insertion sites were considered, the distance was on average 135 bp to the 5′ UTR and 271 bp to the cds. Remarkably, 75% of all Mu insertion sites were located near the 5′ UTR (542 bp) and close to the start codon (704 bp) of the affected genes. In summary, this data revealed that the majority of Mu insertion sites are located closer to the 5′ UTR TSS than to the cds start of affected genes.
Figure 5.
Distance of Mu insertion sites to the start of the 5′ UTR (transcription start site [TSS]) and the cds of affected B73 genes. Density plot illustrating the distances of the Mu insertion sites to the 5′ UTR start (green) and the start of the cds (yellow). Only insertion sites located between 500 bp upstream of the 5′ UTR and 2,000 bp downstream of the ATG were considered. Calculated distances of Mu insertion sites to the 5′ UTR and the start of affected genes are given in the table.
DISCUSSION
Construction of the European sequence-indexed insertional library (BonnMu) in maize discovered 41,086 high quality germinal Mu insertions in 16,392 high confidence gene models (Fig. 1), tagging 37% of the B73 genome. This number is comparable with the North American UniformMu resource (McCarty et al., 2013; Liu et al., 2016), covering 16,090 genes, which were tagged by 43,943 germinal Mu insertions. Recently, an Asian insertional library (ChinaMu; Liang et al., 2019) identified 66,565 high-quality germinal insertions representing 20,396 annotated maize genes. In comparison with UniformMu and ChinaMu, 1,807 genes were newly tagged by BonnMu. Hence, with our new European sequence-indexed resource, the percentage of Mu-tagged maize genes in public repositories increased from 52% to 57%. Although, this number is still less than that of Arabidopsis (74% of the genes covered; Alonso et al., 2003), it is comparable with rice (about 60% tagged genes; Wang et al., 2013). However it is worth noting that different techniques, such as transfer-DNA insertional mutagenesis (Alonso et al., 2003; Toki et al., 2006), two-component transposon system Ac/Ds based mutagenesis (van Enckevort et al., 2005), or Tos17 retrotransposon mutagenesis (Miyao et al., 2003) were applied to construct insertional libraries in Arabidopsis and rice. Nevertheless, because adequate Mu-tagged F2-families are available in the three global resources (Liang et al., 2019) expanding the datasets is already being implemented to increase the coverage of tagged genes. As described by Liang et al. (2019) we also detected an increase in the number of insertional alleles per gene reducing the efficiency of tagging new genes. A reduction of efficient new gene tagging was also observed in insertional libraries of rice, where more than 246,000 insertions only covered 60% of the genes (Wang et al., 2013). Therefore, our data and previous studies (Kolesnik et al., 2004; Hsing et al., 2007; Zhang, 2007; Liang et al., 2019) suggest that it is difficult, if not impossible, to achieve whole-genome saturation using insertional mutagenesis.
Maize is an excellent model for transposon genetics and mutagenesis studies (Settles et al., 2004). Several endogenous DNA transposon systems, such as Ac/Ds (McClintock, 1950) and Mu transposons (Robertson, 1978), are highly active in maize and can be easily adopted for large-scale mutagenesis. Genic transposon insertions are usually large enough to cause substantial disruptions of gene function, which are genetically stable (McCarty and Meeley, 2009). Moreover, transposon insertions are relatively easy to identify using molecular genetics and high-throughput sequencing. Nevertheless, early constructed Mu-based mutation libraries in maize (Bensen et al., 1995; May et al., 2003; Fernandes et al., 2004) had to deal with (1) inefficient recovery and identification of newly transposed insertions and (2) the challenge to discriminate between germinal and the high background of somatic insertions. Recent studies have overcome both of these challenges. The Mu-seq method (McCarty et al., 2013; this study) applies high-throughput NextGen sequencing of Mu-tag enriched replicated samples to identify germinal Mu insertions. More recently, Liang et al. (2019) achieved Mu-tag enrichment utilizing probe hybridization, coupled with high-throughput sequencing to efficiently recover Mu insertions in F2-families of maize. To distinguish germinal from somatic insertions Liang et al. (2019) used a practical criterion (i.e. normalized Mu-flanking sequence tag read counts), in addition to the replicated samples as were used for the UniformMu analysis (McCarty et al., 2013).
Since the present European BonnMu and the North American UniformMu insertional library was constructed using the Mu-seq approach, Mu insertion sites of BonnMu were analyzed and compared in more detail with the UniformMu library. Due to multiple-round backcrossing between the mutagenic parents and the uniform W22 inbred line, the UniformMu lines captured on average five new germinal insertions (McCarty et al., 2005), already excluding about 40 ancestral insertions in the W22 genome (D. R. McCarty, personal communication; Supplemental Dataset S5). However, the BonnMu libraries presented here captured 37 germinal insertions in genes of the FGS per F2-family on average (Supplemental Tables S2 and S3; Supplemental Dataset S3).
For about 50% of the BonnMu and UniformMu-tagged genes, we observed a gene-size– dependent number of insertions (Fig. 2B; Supplemental Fig. S2). On average, 2.5 insertions were detected per Mu-tagged gene, and we observed an association between insertion sites in B73 and W22 (i.e. hotspots of insertions), which are conserved among tagged genes of both genomes (Fig. 3). The availability of the W22v2 assembly (Springer et al., 2018) allows for comparisons with the B73v4 reference genome at multiple scales, such as Mu transposon composition. In total, 75% to 80% of the genes are present at syntenic locations in both maize inbred lines (Springer et al., 2018), making a comparison suitable. To provide additional support for the notion that insertion hotspots are conserved among tagged genes in B73 and W22, it will be essential to analyze and compare a larger W22-Mu-seq dataset.
Previous studies revealed that Mu insertions concentrate in genomic regions with epigenetic marks of open chromatin near the TSS (Dietrich et al., 2002; Liu et al., 2009; Springer et al., 2018). Indeed, Mu elements do exhibit a strong preference for 5′ UTRs of genes (Fig. 4; Dietrich et al., 2002; Liang et al., 2019). Even more precisely, we demonstrated that 50% of all BonnMu insertions are located within 135 bp of the TSS (Fig. 5). Pericentromeric regions, which are rich in heterochromatin, display low frequencies of Mu insertions and meiotic recombinations (Liu et al., 2009), suggesting an association of site selection for Mu insertions with chromatin structure. Indeed, locations for novel transposon insertions can be sensitive to chromatin structure (Naito et al., 2009; Sultana et al., 2017; Springer et al., 2018). For instance, DNA methylation patterns were analyzed by whole-genome bisulfite sequencing and chromatin accessibility throughout the W22 genome using a Micrococcal Nuclease assay (Springer et al., 2018). The enrichment of open chromatin, based on epigenetic marks, such as Cytosin methylation (CHH, where H = A, T, or C) and Micrococcal Nuclease treatment at the TSS and the 3′ UTR of genes (Regulski et al., 2013; Springer et al., 2018) corresponds with the majority of BonnMu insertions detected near the 5′ UTR region of genes (Figs. 4 and 5) and a smaller peak near the 3′ UTR (Fig. 4A). Hence, it is likely that chromatin structure plays a key role in site selection for Mu insertions. Further indirect support of the hypothesis that open chromatin drives Mu insertion sites preferences is shown in Figure 1. The high number of observed Mu-tagged genes overlapping among the libraries suggests that the genes are not randomly selected by Mu. To further test the hypothesis that open chromatic marks drives Mu insertion site preferences, chemical treatment to promote chromatin opening (Baubec et al., 2009) could be applied, thereby facilitating Mu insertions in heterochromatic (i.e. inaccessible regions). Another indirect strategy to support this hypothesis is to test if the subset of genes that is not yet Mu tagged in any of the published libraries belongs to transcriptionally silenced genes with densely methylated chromatin.
CONCLUSION
In summary, our publicly available European BonnMu insertional library provides a starting point for forward and reverse genetics studies in maize. To facilitate functional genomics study in maize, specific mutants and respective phenotypes of segregating F2-families can be identified based on gene sequences of interest through MaizeGDB.org as described in Liu et al. (2016). Detected on the basis of albino and pale green leaf phenotypes, the mutation rate was ∼13%, which is consistent with previously published mutation rates (Robertson, 1983), suggesting a high transposon activity in these stocks. The European BonnMu resource is still expanding to archive additional genes tagged by germinal Mu insertions. To accelerate the identification of Mu insertion sites of future Mu-seq libraries, we created a Mu-seq workflow utility (MuWU), which is publicly available at https://github.com/Crop-Bioinformatics-Bonn/MuWU. Our MuWU makes use of bioconda (Grüning et al., 2018) and snakemake (Köster and Rahmann, 2012) to provide an easy to install and completely automated framework for the detection and annotation of Mu insertion sites.
MATERIAL AND METHODS
Plant Material
The Mu-tagged maize (Zea mays) F1-population was generated in the summer season of 2014 by crossing an active Mu line (Mu4 per se; Robertson, 1983) into B73 at the experimental field station Campus Klein Altendorf (University of Bonn). The F1-generation, which is heterozygous for the new Mu insertions, was self-pollinated during winter nursery 2014 to 2015 in Chile and in the summer season 2015 at Campus Klein Altendorf to produce segregating F2-families.
Mu-seq Library Production
For construction of two multiplexed Mu-seq libraries we used 1,152 (2 × 576) F2-families in the B73 background and followed the previously described Mu-seq approach (McCarty et al., 2013). Briefly, each of the two Mu-seq libraries contained 576 F2-families, which were pooled according to a two-dimensional 24 × 24 grid design (Supplemental Table S1). Hence, the two Mu-seq libraries consisted of 1,152 (2 × 24 × 24) F2-families. Construction of the Mu-seq libraries started with germination of 2 × 576 maize F2-families (6 seeds per F2-family) in paper rolls (Hetz et al., 1996). Sampling of leaf-tissue was carried out 10 to 12 d after germination, according to the 24 × 24 grid design, where each family contributed to one distinct column and one distinct row pool. In total, there were 24 row and 24 column pools, which created 48 multiplexed leaf samples. Depending on the germination rate, 3 to 6 seedlings were sampled per F2-family. If less than 3 seedlings germinated, seeds of these F2-families were regerminated to complement the number of seedlings. A probability was calculated of 99% (Supplemental Table S5) to obtain at least one mutant allele per tagged gene among the 3 to 6 germinated plants per F2-family. To ensure the specific identification of heritable germinal insertions at intersections of rows and columns in the grid, leaf samples of row and column pools were taken from independent somatic cell lineages, that is, from alternate leaves of each seedling. By utilizing this approach, nonheritable, somatic insertions appeared only in a single axis of the grid and they were excluded from further analyses. Subsequently, gDNA was isolated from these 48 pools (Nalini et al., 2003) and randomly fragmented to a size of about 1 kb using 1 to 3 cycles of 3 min sonication using an Ultrasonication unit followed by 3 min rigorous vortexing. The single stranded overhang of the randomly sheared gDNA fragments was filled in by an enzyme mix to create blunt ends that enabled ligation of a double-stranded universal (U) adapter. Afterward, Mu-flanking amplicons were enriched by ligation-mediated PCR using a Mu-TIR-specific and a specific primer for the ligated U adapter (PCR I). Two subsequent rounds of PCR (PCR II and III) used nested TIR primers in combination with Illumina sequencing adapter primers to successively incorporate the adapters, necessary for sequencing of the fragments. To reduce the number of very short Mu-flanking fragments, PCR II products were purified to a size range of 300 to 800 bp on 1.2% (w/v) agarose gels. To avoid sample contamination, each of the 48 PCR II products was loaded on a separate gel. The final PCR III completed the required sequencing adapters and incorporated a 6-bp barcode allowing multiplexing of the 48 pools. Finally, each Mu-seq library was quantified using a Bioanalyzer, DNA 7500 chip (Agilent Technologies). The final concentrations of the two Mu-seq libraries were 73 and 102 nm. Paired-end sequencing (100 bp) of the multiplexed Mu-seq libraries was performed in two separate lanes of a HiSeq 2500 (Illumina) sequencer.
Identification of Mu Insertion Sites in B73
Adapters and TIR sequences were cut from both ends of the raw Mu-seq reads with cutadapt v2.3 (-e 0.2; Martin, 2011). Low-quality bases were filtered with Trimmomatic v0.36 (SLIDINGWINDOW:4:15; MINLEN:12; Bolger et al., 2014) and mapped to the maize reference genome AGPv4.36 (Jiao et al., 2017) with Bowtie2 v2.5.0 (Langmead and Salzberg, 2012). Duplicate reads were removed with the Picard MarkDuplicates tool v2.5.0 (Picard Toolkit, 2019: https://github.com/broadinstitute/picard). Mu insertion sites were identified using a customized Python script based on the characteristic 9 bp overlap of Mu insertion flanking sequences (McCarty et al., 2013). An insertion site was selected if the 9-bp overlap was supported by at least two reads on each side. To detect insertions that can be assigned to a specific maize F2-family, all insertion sites with matching genomic coordinates in only one row and one column sample were selected. To identify Mu insertion sites inside or upstream of genes of the B73 Filtered Gene Set v4.36 (FGS; Jiao et al., 2017), the Bioconductor R packages IRanges v2.16 and GenomicRanges v1.36 (Lawrence et al., 2013), as well as ChIPpeakAnno v3.16.1 (Zhu et al., 2010) were subsequently used. Only Mu insertion sites located inside or upstream (≤500 bp) of FGS genes were considered. The Pearson correlation coefficient was calculated in R v3.5.2 to test the hypothesis that the number of Mu insertions increased with the length of the affected genes.
To determine the expected number of FGS genes being Mu tagged among BonnMu libraries and global sequence-indexed libraries (Fig. 1; Supplemental Table S4), a generalized linear model with a log-link and Poisson distribution was fitted under the assumption of independence between the three available Mu-seq resources using proc genmod in SAS 9.4 (SAS Institute). For each comparison, the model under independence comprised main effects for each of the compared resources. The expected and observed numbers of Mu-tagged FGS genes were compared using a χ2-test for the two- or three-way interaction effect of the compared resources, respectively. Significant differences were determined at a significance level of α = 0.05. Because it was not known which genome versions were used to align the reads of the UniformMu or the ChinaMu datasets, the reference genome AGPv4.36, used in this study, was applied. Hence, only 16,066 UniformMu genes and 20,133 ChinaMu genes (Fig. 1B) were considered for the calculation of expected values.
Identification of Mu Insertion Sites in W22
Single-end raw Mu-seq reads in W22 background (SRA accession SRP028545) were trimmed with cutadapt v2.3 (-e 0.2; Martin, 2011). The barcode and the TIR sequence were cut from the beginning of each read. Reads without TIR sequence were discarded and bases were cut from the end of the remaining reads with Trimmomatic v0.36 (Bolger et al., 2014) if they fell below a quality score of 20 (TRAILING:20; MINLEN:12). Reads were mapped to the maize. reference genome W22 v2.0 (Springer et al., 2018) with Bowtie2 v2.5.0 (Langmead and Salzberg, 2012), and duplicate reads were subsequently removed using the Picard MarkDuplicates tool v2.5.0 (Picard Toolkit, 2019: https://github.com/broadinstitute/picard). Reads that mapped to more than one position were excluded from the deduplicated SAM files and Mu insertion sites were identified using a customized Python script. Genomic coordinates were tagged as Mu transposon insertion sites if the alignment start of at least two reads matched at the same position. Insertion sites located inside W22 v2.0 genes (Springer et al., 2018) were identified with the R Bioconductor packages IRanges v2.16, GenomicRanges v1.36 (Lawrence et al., 2013), and ChIPpeakAnno v3.16.1 (Zhu et al., 2010), and all insertion sites located inside W22 genes were selected.
Analysis of Mu Insertion Sites in B73 and W22
To analyze hotspots of Mu insertion sites in B73 and W22, all insertion sites were sorted into corresponding bins. To this end, the length of each gene was divided into bins of 200 bp in size and the number of bins (i.e. hotspots) containing Mu transposon insertions was counted for each gene. To compare hotspots between B73 and W22, Mu flanking sequences in W22 were extracted (50 bp upstream – 50 bp downstream of the insertion site) using BEDTools v2.25.0 (Quinlan and Hall, 2010) and aligned to the maize. reference genome AGPv4.36 (Jiao et al., 2017) with Bowtie2 v2.5.0 (Langmead and Salzberg, 2012). Multireads (i.e. reads aligning to multiple genomic locations) were excluded and B73 coordinates of W22 reads were further analyzed to identify W22 Mu insertion sites within genes of the B73 FGS (Jiao et al., 2017) as described above. Identified insertion sites in W22 and B73 were filtered for endogenous Mu elements. To this end, matches between sequences of endogenous Mu elements (Springer et al., 2018) and Mu insertion flanking sequences (200 bp upstream – 200 bp downstream of the insertion site) were identified with BLAST v2.3.0 (Camacho et al., 2009). Matches that exceeded the defined threshold (pident ≥ 95; bitscore ≥ 50) were excluded from further analyses. Remaining Mu-tagged genes that were hit in both B73 and W22 were divided into 200-bp bins. The filtered Mu insertions were sorted into the corresponding bins. A χ2-test (α ≤ 0.01) was performed on the confusion matrix given in Table 2 in R v3.5.2 to test the hypothesis that there was an association between hotspots in B73 and W22.
To analyze insertion site preferences of Mu transposons, the longest transcript for each affected gene in either B73 or W22 was selected and the cds was divided into deciles based on the B73 FGS (Jiao et al., 2017). Genes without annotated cds were excluded from further analyses. All identified insertion sites located in the cds, including endogenous Mu elements, were sorted into the corresponding cds decile. Insertion sites located within FGS genes, but up- or downstream of the cds, were counted as an insertion site in the 5′ UTR and 3′ UTR, respectively. To determine the distance from each Mu insertion site to the TSS, the longest transcript of each FGS gene was selected. For each Mu insertion site identified in B73 inside or upstream (≤500 bp) of B73 FGS genes, the absolute distance to both the cds start and, if available, the 5′ UTR start of the associated gene was calculated.
Accession Numbers
Raw sequencing data are stored at the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) with the accession number PRJNA608624.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Size distribution of 16,392 affected FGS genes in comparison to the size distribution of all 44,117 B73v4 genes.
Supplemental Figure S2. Number of affected W22 genes and associated number of Mu insertions plotted against the corresponding gene size.
Supplemental Figure S3. Number of regions (200 bp bins) in FGS genes preferentially targeted by Mu transposons.
Supplemental Figure S4. Affected genes sorted into gene sets according to their length (deciles).
Supplemental Table S1. Grid design for 24 x 24 F2-families, i.e. 576 F2-families per Mu-seq library.
Supplemental Table S2. Number of germinal Mu insertions in FGS genes per 576 F2-families (Mu-seq 1).
Supplemental Table S3. Number of germinal Mu insertions in FGS genes per 576 F2-families (Mu-seq 2).
Supplemental Table S4. Number of observed and expected Mu-tagged genes among sequence-indexed Mu insertional libraries (related to Fig. 1).
Supplemental Table S5. Calculated probability to obtain at least one mutant allele per tagged gene among the 3-6 germinated plants per F2-family.
Supplemental Dataset S1. Number of raw read pairs per row and column sample sequenced per Mu-seq library with HiSeq2500 (Illumina) sequencer.
Supplemental Dataset S2. A total of 225,936 genomic insertion sites detected in the two Mu-seq libraries.
Supplemental Dataset S3. A total of 41,086 Mu insertion sites detected in 16,392 genes of the FGS.
Supplemental Dataset S4. List of 43,943 UniformMu insertions within 16,090 genes in B73.
Supplemental Dataset S5. Canonical Mu elements in W22 and B73.
Acknowledgments
We thank Hans-Peter Piepho (University of Hohenheim) for statistical assistance and Patrick S. Schnable (Iowa State University) for providing seeds of Mu4 per se. We also appreciate the help of Peng Yu (University of Bonn) in this project. We thank Karl-Josef Wiesel (Campus Klein Altendorf, University of Bonn), Christa Schulz, and Helmut Rehkopf (University of Bonn) for technical assistance.
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant no. MA 8427/1–1 to C.M.), the Bundesministerium für Bildung und Forschung (grant no. 031 B195C to F.H.), and the United States Department Agriculture, Agricultural Research Service to J.L.P. and C.T.H.
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Baubec T, Pecinka A, Rozhon W, Mittelsten Scheid O(2009) Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J 57: 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen JL.(2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42: 251–269 [PubMed] [Google Scholar]
- Bensen RJ, Johal GS, Crane VC, Tossberg JT, Schnable PS, Meeley RB, Briggs SP(1995) Cloning and characterization of the maize An1 gene. Plant Cell 7: 75–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B(2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutnell TP.(2002) Transposon tagging in maize. Funct Integr Genomics 2: 4–12 [DOI] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL(2009) BLAST+: Architecture and applications. BMC Bioinformatics 10: 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candela H, Hake S(2008) The art and design of genetic screens: maize. Nat Rev Genet 9: 192–203 [DOI] [PubMed] [Google Scholar]
- Chandler VL, Hardeman KJ(1992) The Mu elements of Zea mays. Adv Genet 30: 77–122 [DOI] [PubMed] [Google Scholar]
- Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, Nikolau BJ, Schnable PS(2002) Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes J, Dong Q, Schneider B, Morrow DJ, Nan GL, Brendel V, Walbot V(2004) Genome-wide mutagenesis of Zea mays L. using RescueMu transposons. Genome Biol 5: R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Jiang N, Wessler SR(2002) Plant transposable elements: Where genetics meets genomics. Nat Rev Genet 3: 329–341 [DOI] [PubMed] [Google Scholar]
- Frey M, Stettner C, Gierl A(1998) A general method for gene isolation in tagging approaches: Amplification of insertion mutagenised sites (AIMS). Plant J 13: 717–721 [Google Scholar]
- Grüning B, Dale R, Sjödin A, Chapman BA, Rowe J, Tomkins-Tinch CH, Valieris R, Köster J; Bioconda Team (2018) Bioconda: Sustainable and comprehensive software distribution for the life sciences. Nat Methods 15: 475–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz W, Hochholdinger F, Schwall M, Feix G(1996) Isolation and characterization of rtcs, a maize mutant deficient in the formation of nodal roots. Plant J 10: 845–857 [Google Scholar]
- Hirochika H, Guiderdoni E, An G, Hsing YI, Eun MY, Han CD, Upadhyaya N, Ramachandran S, Zhang Q, Pereira A, et al. (2004) Rice mutant resources for gene discovery. Plant Mol Biol 54: 325–334 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Wen TJ, Zimmermann R, Chimot-Marolle P, da Costa e Silva O, Bruce W, Lamkey KR, Wienand U, Schnable PS(2008) The maize (Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J 54: 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsing YI, Chern CG, Fan MJ, Lu PC, Chen KT, Lo SF, Sun PK, Ho SL, Lee KW, Wang YC, et al. (2007) A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol Biol 63: 351–364 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Peluso P, Shi J, Liang T, Stitzer MC, Wang B, Campbell MS, Stein JC, Wei X, Chin CS, et al. (2017) Improved maize reference genome with single-molecule technologies. Nature 546: 524–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnik T, Szeverenyi I, Bachmann D, Kumar CS, Jiang S, Ramamoorthy R, Cai M, Ma ZG, Sundaresan V, Ramachandran S(2004) Establishing an efficient Ac/Ds tagging system in rice: Large-scale analysis of Ds flanking sequences. Plant J 37: 301–314 [DOI] [PubMed] [Google Scholar]
- Köster J, Rahmann S(2012) Snakemake--a scalable bioinformatics workflow engine. Bioinformatics 28: 2520–2522 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL(2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, Carey VJ(2013) Software for computing and annotating genomic ranges. PLOS Comput Biol 9: e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Zhou L, Tang Y, Li N, Song T, Shao W, Zhang Z, Cai P, Feng F, Ma Y, et al. (2019) A sequence-indexed Mutator insertional library for maize functional genomics study. Plant Physiol 181: 1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D.(2002) Mutator transposons. Trends Plant Sci 7: 498–504 [DOI] [PubMed] [Google Scholar]
- Lisch D.(2015) Mutator and MULE transposons. Microbiol Spectr 3: MDNA3-0032-2014 [DOI] [PubMed] [Google Scholar]
- Liu P, McCarty DR, Koch KE(2016) Transposon mutagenesis and analysis of mutants in UniformMu maize (Zea mays). Curr Protoc Plant Biol 1: 451–465 [DOI] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM, Fu Y, Nettleton D, Schnable PS(2009) Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet 5: e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Whittier RF(1995) Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25: 674–681 [DOI] [PubMed] [Google Scholar]
- Martin M.(2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12 [Google Scholar]
- May BP, Liu H, Vollbrecht E, Senior L, Rabinowicz PD, Roh D, Pan X, Stein L, Freeling M, Alexander D, et al. (2003) Maize-targeted mutagenesis: A knockout resource for maize. Proc Natl Acad Sci USA 100: 11541–11546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Latshaw S, Wu S, Suzuki M, Hunter CT, Avigne WT, Koch KE(2013) Mu-seq: Sequence-based mapping and identification of transposon induced mutations. PLoS One 8: e77172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty DR, Meeley RB(2009) Transposon resources for forward and reverse genetics in maize In Bennetzen J.L., and Hake S., eds, Handbook of Maize. Springer, New York, pp 561–584 [Google Scholar]
- McCarty DR, Settles AM, Suzuki M, Tan BC, Latshaw S, Porch T, Robin K, Baier J, Avigne W, Lai J, et al. (2005) Steady-state transposon mutagenesis in inbred maize. Plant J 44: 52–61 [DOI] [PubMed] [Google Scholar]
- McClintock B.(1950) The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA 36: 344–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B.(1951) Mutable loci in maize. Carnegie Institution of Washington Yearbook 50: 174–181 [Google Scholar]
- Miyao A, Tanaka K, Murata K, Sawaki H, Takeda S, Abe K, Shinozuka Y, Onosato K, Hirochika H(2003) Target site specificity of the Tos17 retrotransposon shows a preference for insertion within genes and against insertion in retrotransposon-rich regions of the genome. Plant Cell 15: 1771–1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, Richardson AO, Okumoto Y, Tanisaka T, Wessler SR(2009) Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461: 1130–1134 [DOI] [PubMed] [Google Scholar]
- Nalini E, Bhagwat SG, Jawali N(2003) A simple method for isolation of DNA from plants suitable for long term storage and DNA marker analysis. BARC Newsletter 249: 208–214 [Google Scholar]
- Nestler J, Liu S, Wen TJ, Paschold A, Marcon C, Tang HM, Li D, Li L, Meeley RB, Sakai H, et al. (2014) Roothairless5, which functions in maize (Zea mays L.) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. Plant J 79: 729–740 [DOI] [PubMed] [Google Scholar]
- Portwood JL II, Woodhouse MR, Cannon EK, Gardiner JM, Harper LC, Schaeffer ML, Walsh JR, Sen TZ, Cho KT, Schott DA, et al. (2019) MaizeGDB 2018: The maize multi-genome genetics and genomics database. Nucleic Acids Res 47(D1): D1146–D1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM(2010) BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regulski M, Lu Z, Kendall J, Donoghue MTA, Reinders J, Llaca V, Deschamps S, Smith A, Levy D, McCombie WR, et al. (2013) The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res 23: 1651–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson DS.(1978) Characterization of a mutator system in maize. Mutat Res 51: 21–28 [Google Scholar]
- Robertson DS.(1983) A possible dose-dependent inactivation of Mutator (Mu) in maize. Mol Gen Genet 191: 86–90 [Google Scholar]
- Schnable JC, Freeling M(2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Settles AM, Latshaw S, McCarty DR(2004) Molecular analysis of high-copy insertion sites in maize. Nucleic Acids Res 32: e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Anderson SN, Andorf CM, Ahern KR, Bai F, Barad O, Barbazuk WB, Bass HW, Baruch K, Ben-Zvi G, et al. (2018) The maize W22 genome provides a foundation for functional genomics and transposon biology. Nat Genet 50: 1282–1288 [DOI] [PubMed] [Google Scholar]
- Stern DB, Hanson MR, Barkan A(2004) Genetics and genomics of chloroplast biogenesis: Maize as a model system. Trends Plant Sci 9: 293–301 [DOI] [PubMed] [Google Scholar]
- Sultana T, Zamborlini A, Cristofari G, Lesage P(2017) Integration site selection by retroviruses and transposable elements in eukaryotes. Nat Rev Genet 18: 292–308 [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H(2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- van Enckevort LJ, Droc G, Piffanelli P, Greco R, Gagneur C, Weber C, González VM, Cabot P, Fornara F, Berri S, et al. (2005) EU-OSTID: A collection of transposon insertional mutants for functional genomics in rice. Plant Mol Biol 59: 99–110 [DOI] [PubMed] [Google Scholar]
- Wang N, Long T, Yao W, Xiong L, Zhang Q, Wu C(2013) Mutant resources for the functional analysis of the rice genome. Mol Plant 6: 596–604 [DOI] [PubMed] [Google Scholar]
- Xu C, Tai H, Saleem M, Ludwig Y, Majer C, Berendzen KW, Nagel KA, Wojciechowski T, Meeley RB, Taramino G, et al. (2015) Cooperative action of the paralogous maize lateral organ boundaries (LOB) domain proteins RTCS and RTCL in shoot-borne root formation. New Phytol 207: 1123–1133 [DOI] [PubMed] [Google Scholar]
- Zhang Q.(2007) Strategies for developing green super rice. Proc Natl Acad Sci USA 104: 16402–16409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR(2010) ChIPpeakAnno: A bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]