One BAHD acyltransferase specifically mediates the acylation of shikonin, whereas a second BAHD acyltransferase acylates only alkannin, an enantiomer of shikonin, in Lithospermum erythrorhizon.
Abstract
Several Boraginaceae plants produce biologically active red naphthoquinone pigments, derivatives of the enantiomers shikonin and alkannin, which vary in acyl groups on their side chains. Compositions of shikonin/alkannin derivatives vary in plant species, but the mechanisms generating the diversity of shikonin/alkannin derivatives are largely unknown. This study describes the identification and characterization of two BAHD acyltransferases, shikonin O-acyltransferase (LeSAT1) and alkannin O-acyltransferase (LeAAT1), from Lithospermum erythrorhizon, a medicinal plant in the family Boraginaceae that primarily produces the shikonin/alkannin derivatives acetylshikonin and β-hydroxyisovalerylshikonin. Enzyme assays using Escherichia coli showed that the acylation activity of LeSAT1 was specific to shikonin, whereas the acylation activity of LeAAT1 was specific to alkannin. Both enzymes recognized acetyl-CoA, isobutyryl-CoA, and isovaleryl-CoA as acyl donors to produce their corresponding shikonin/alkannin derivatives, with both enzymes showing the highest activity for acetyl-CoA. These findings were consistent with the composition of shikonin/alkannin derivatives in intact L. erythrorhizon plants and cell cultures. Genes encoding both enzymes were preferentially expressed in the roots and cell cultures in the dark in pigment production medium M9, conditions associated with shikonin/alkannin production. These results indicated that LeSAT1 and LeAAT1 are enantiomer-specific acyltransferases that generate various shikonin/alkannin derivatives.
Shikonin and alkannin, an enantiomeric pair of red naphthoquinone derivatives, are specialized plant metabolites produced by a limited number of plants of the family Boraginaceae. Because shikonin/alkannin derivatives have various biological activities, including wound healing, antimicrobial, antiadenovirus, and anticancer properties, plants producing these derivatives, including Alkanna tinctoria in Europe and Lithospermum erythrorhizon and Arnebia euchroma in East Asia, have been utilized for centuries as crude sources of traditional medicines (Papageorgiou et al., 2006; Yazaki, 2017; Boulos et al., 2019). Compositions of shikonin/alkannin derivatives vary in plant species. Intact L. erythrorhizon plants mainly produce acetylshikonin and β-hydroxyisovalerylshikonin, whereas A. tinctoria and A. euchroma plants produce high amounts of β,β-dimethylacrylalkannin, and Onosma confertum, a Boraginales plant from China, biosynthesizes mainly β,β-dimethylacrylshikonin and acetylshikonin (Assimopoulou et al., 2006; Zhou et al., 2011). In addition, many other derivatives with a variety of acyl groups with branched chains have been isolated (Papageorgiou et al., 1999; Kumar et al., 2013). Although the biological activities of shikonin/alkannin derivatives from various plants have been reported to differ (Chen et al., 2002; Lee et al., 2008; Hasenoehrl et al., 2017; Mitra and Dash, 2018), the relationships of biological activities with the acylation of shikonin/alkannin and the configurations of these compounds remain unclear.
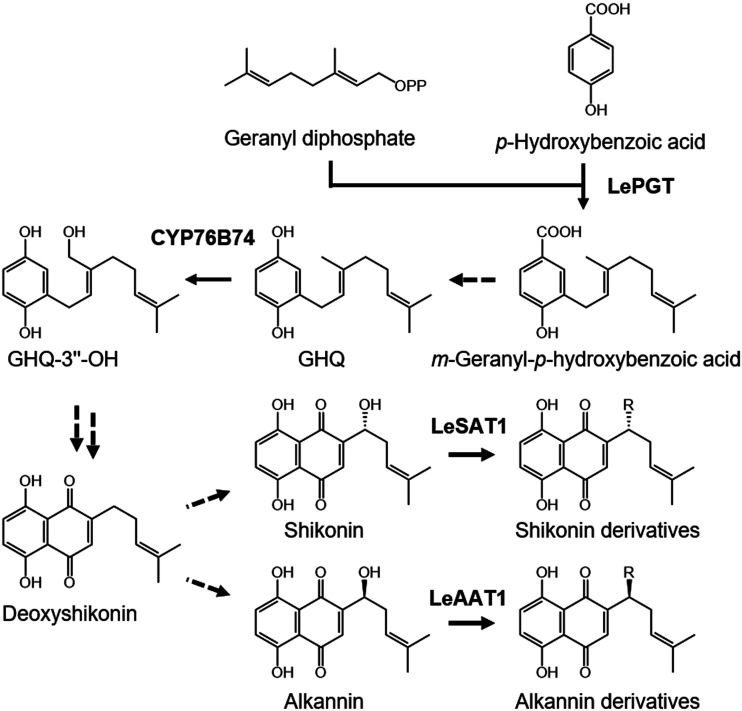
Over the last decades, methods of culturing cells of Boraginales plants that produce large amounts of shikonin/alkannin derivatives have been established (Tabata et al., 1974; Fukui et al., 1983; Wang et al., 2014; Malik et al., 2016), and these cultured cells have been used to study shikonin/alkannin biosynthesis pathways (Fig. 1). To date, two enzymes specifically involved in the shikonin/alkannin biosynthesis pathway have been identified and characterized. p-Hydroxybenzoic acid geranyltransferase in L. erythrorhizon catalyzes the coupling of p-hydroxybenzoic acid and geranyl diphosphate to produce m-geranyl-p-hydroxybenzoic acid (Yazaki et al., 2002), the first step in forming the basic carbon skeleton leading to shikonin/alkannin. This step constitutes the key junction of two representative pathways, the phenylpropanoid and mevalonate pathways. CYP76B74, a recently reported cytochrome P450 in A. euchroma, hydroxylates the 3″ position of geranylhydroquinone (GHQ) to yield 3″-hydroxygeranylhydroquinone (GHQ-3′'-OH; Wang et al., 2019). After the latter step, however, little is known about the enzymes catalyzing the cyclization reaction required for the formation of the naphthoquinone structure or the enzymes catalyzing the acylation reactions of shikonin and alkannin, which generate various shikonin/alkannin derivatives.
Figure 1.
Biosynthesis pathways of shikonin/alkannin derivatives. Dashed arrows indicate the reaction steps for which enzymes have not yet been identified. LeSAT1 and LeAAT1 were identified in this study. LePGT, L. erythrorhizon p-hydroxybenzoic acid geranyltransferase.
To identify and characterize specific genes and proteins involved in shikonin/alkannin biosynthesis, we previously performed a comparative proteome analysis using cultured L. erythrorhizon cells and identified several candidate genes, including acyltransferases (Takanashi et al., 2019). These acyltransferases belong to the BAHD acyltransferase family that transfers acyl moieties from various acyl-activated CoA thioester donors to acceptor molecules, primarily phenolic compounds, contributing to the diversity of plant secondary metabolites (D’Auria, 2006; Bontpart et al., 2015; Kusano et al., 2019).
This study reports the identification and characterization of two BAHD acyltransferases, shikonin O-acyltransferase (LeSAT1) and alkannin O-acyltransferase (LeAAT1), which mediate the enantiomer-specific acylation of shikonin or alkannin in L. erythrorhizon (Fig. 1). Substrate specificity analysis showed that both enzymes could transfer acyl groups from acetyl-CoA, isobutyryl-CoA, and isovaleryl-CoA. Our findings indicate that LeSAT1 and LeAAT1 are key enzymes in the production of diverse shikonin/alkannin derivatives.
RESULTS
Chiral Composition of Shikonin/Alkannin Derivatives and Acylation Activity in L. erythrorhizon-Cultured Cells
Plants belonging to the family Boraginaceae produce a variety of shikonin/alkannin derivatives. Their compositions differ among plant species, and even among individuals within the same species. To determine the composition of shikonin/alkannin derivatives in L. erythrorhizon cells cultured in M9 medium, high-performance liquid chromatography (HPLC) analysis with a chiral column was performed. The most abundant compounds were acetylshikonin (44.6%) and β-hydroxyisovalerylshikonin (26.8%; Table 1), followed by their enantiomers acetylalkannin (15.3%) and β-hydroxyisovalerylalkannin (5.7%). Other shikonin derivatives, including isobutyrylshikonin (3.7%) and isovalerylshikonin (0.8%), were also detected. These results showed that cultured L. erythrorhizon cells mainly produced shikonin rather than alkannin derivatives, consistent with results obtained using intact L. erythrorhizon plants and callus cultures (Fukui et al., 1983; Zhou et al., 2011).
Table 1. Composition of shikonin/alkannin derivatives in L. erythrorhizon-cultured cells.
Data are means ± sd of three independently prepared cultured cells.
| Compounds | Structures of Acyl Groups | Proportion |
|---|---|---|
| % | ||
| Acetylshikonin | OCOCH3 | 44.6 ± 2.7 |
| β-Hydroxyisovalerylshikonin | OCOCH2C(CH3)2OH | 26.8 ± 2.2 |
| Acetylalkannin | OCOCH3 | 15.3 ± 0.4 |
| β-Hydroxyisovalerylalkannin | OCOCH2C(CH3)2OH | 5.7 ± 0.3 |
| Isobutyrylshikonin | OCOCH(CH3)2 | 3.7 ± 0.5 |
| Isovalerylshikonin | OCOCH2CH(CH3)2 | 0.8 ± 0.1 |
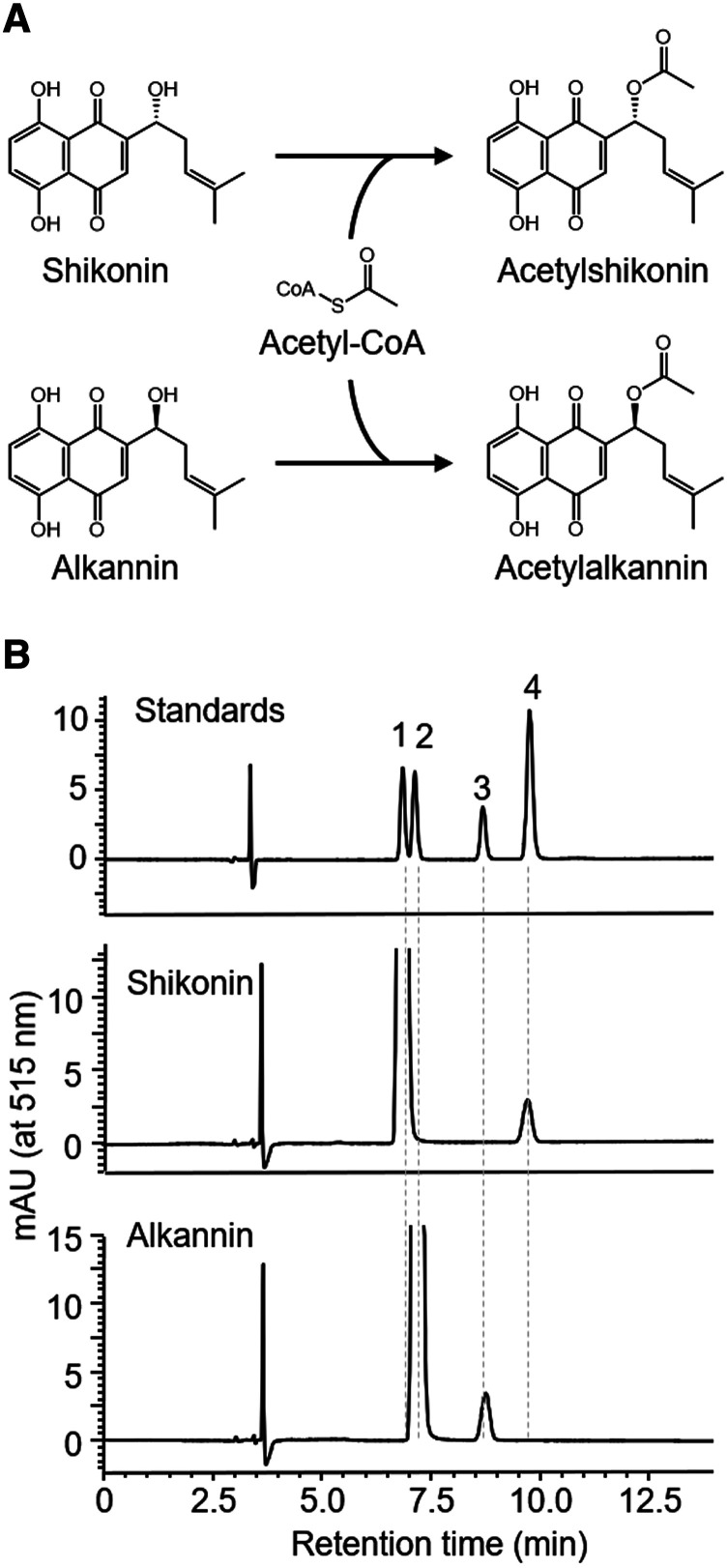
To characterize the shikonin acylation activity in cell-free extracts of cultured L. erythrorhizon cells, enzyme assays were performed, in which extracts were incubated with shikonin or alkannin and the acyl donor acetyl-CoA (Fig. 2A). These extracts acetylated both shikonin and alkannin to a comparable extent (Fig. 2B). Moreover, extracts of cultured L. erythrorhizon cells acylated shikonin and alkannin using isobutyryl-CoA and isovaleryl-CoA as acyl donors (Supplemental Fig. S1), suggesting that cultured L. erythrorhizon cells possess at least one acyltransferase with broad substrate specificity, with various acyl donors and shikonin and alkannin as acyl acceptors.
Figure 2.
Shikonin and alkannin acetylation activities in desalted cell-free extracts of cultured L. erythrorhizon cells, as analyzed by chiral HPLC. A, Reaction scheme for acetylation. B, HPLC chromatograms of the acetylation products. Labeled compounds: 1, shikonin; 2, alkannin; 3, acetylalkannin; and 4, acetylshikonin.
Identification of an Acyltransferase Gene
Our previous comparative transcriptome and proteome analyses of L. erythrorhizon identified four acyltransferase candidates expressed in shikonin-producing cultured cells (Takanashi et al., 2019). The expression of three candidates, comp39163_c0_seq2, comp63127_c0_seq2, and comp89737_c13_seq3, were induced specifically when cells in M9 medium were cultured in the dark, conditions that allowed high levels of shikonin/alkannin derivatives to be produced. The fourth candidate, comp92813_c0_seq1, was detected in cells cultured in M9 medium regardless of light irradiation, a condition under which shikonin/alkannin production is strongly suppressed (Supplemental Fig. S2). These four candidates belong to the BAHD acyltransferase family, enzymes that transfer acyl moieties from a variety of acyl-activated CoA thioester donors to an acceptor molecule (D’Auria, 2006). Of those four genes, two, comp63127_c0_seq2 and comp92813_c0_seq1, had open reading frames of more than 1,200 bp, the average length of BAHD acyltransferase genes, and all five motifs conserved in clade III BAHD acyltransferases (Supplemental Fig. S3; St-Pierre and De Luca, 2000; Tuominen et al., 2011).The other two, comp39163_c0_seq2, and comp89737_c13_seq3, lacked one and two of the five conserved motifs, respectively. The former two genes were therefore cloned and further analyzed.
In Vitro Assay of Two BAHD Acyltransferases Using Escherichia coli
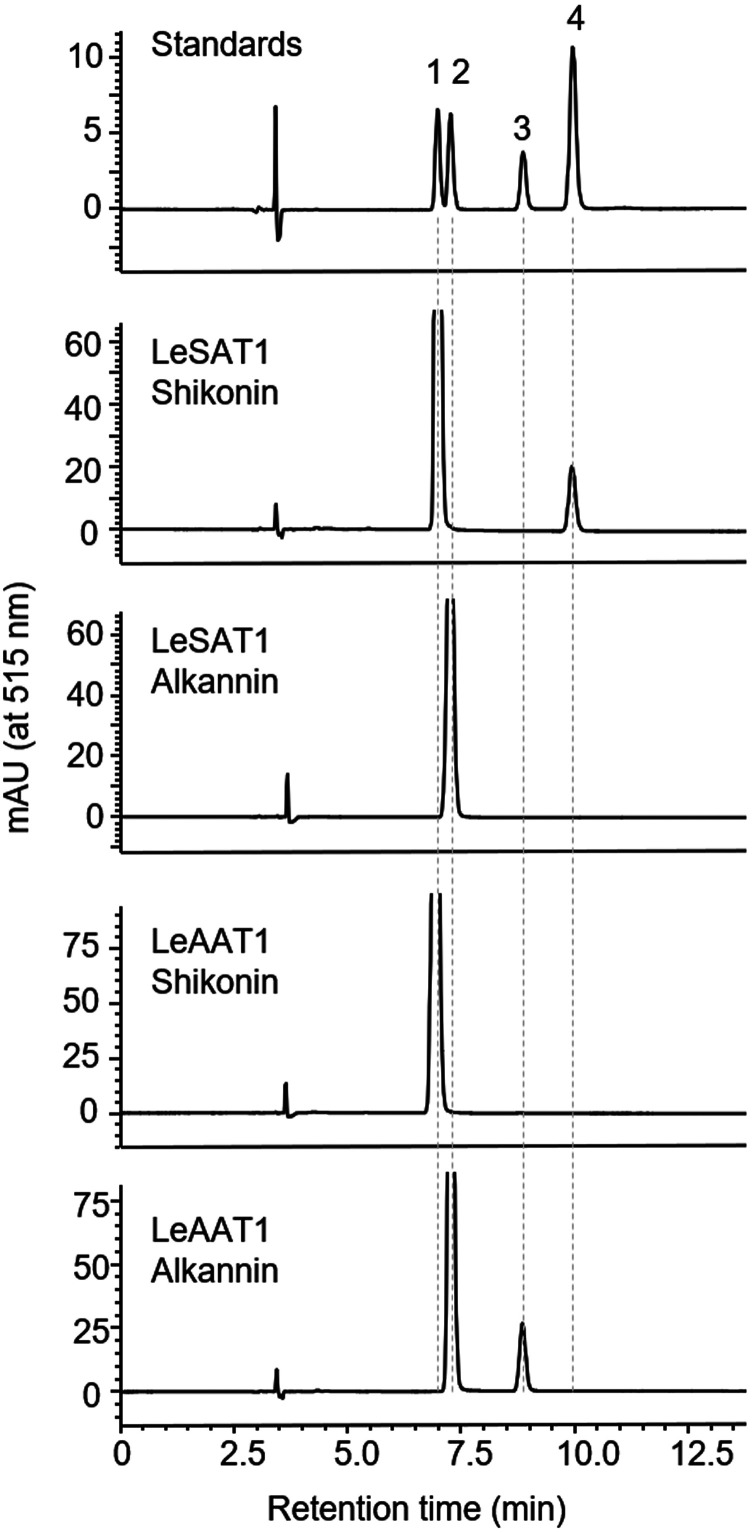
The enzyme activities of the two candidate BAHDs were assessed in a heterologous expression system using E. coli. The coding sequences of both genes were optimized for E. coli expression. Because recombinant protein encoded by the comp63127_c0_seq2 gene was not soluble without molecular chaperons, the chaperon proteins GroEL and GroES were coexpressed. Due to the strong interaction between recombinant protein encoded by the comp63127_c0_seq2 and GroEL, both proteins were eluted together in subsequent purification by nickel affinity chromatography (Supplemental Fig. S4). In contrast, expression of protein encoded by the comp92813_c0_seq1 was independent of GroEL (Supplemental Fig. S4). The enzymatic activities of these proteins were assessed with acetyl-CoA as the acyl donor and shikonin or alkannin as the acyl acceptor. HPLC analysis showed that the acyltransferase encoded by the comp63127_c0_seq2 gene recognized shikonin, but not alkannin, as an acceptor substrate and generated acetylshikonin as the sole product (Fig. 3). By contrast, the BAHD acyltransferase encoded by the comp92813_c0_seq1 gene recognized alkannin, but not shikonin, as an acceptor substrate (Fig. 3). These results indicated that these two BAHD acetyltransferases, designated LeSAT1 and LeAAT1, had distinct functions in the biosynthesis of shikonin/alkannin derivatives.
Figure 3.
Stereospecific acetylation of shikonin and alkannin by LeSAT1 and LeAAT1, as analyzed by chiral HPLC. LeSAT1 mediated the acetylation of shikonin, whereas LeAAT1 mediated the acetylation of alkannin. Labeled compounds: 1, shikonin; 2, alkannin; 3, acetylalkannin; and 4, acetylshikonin.
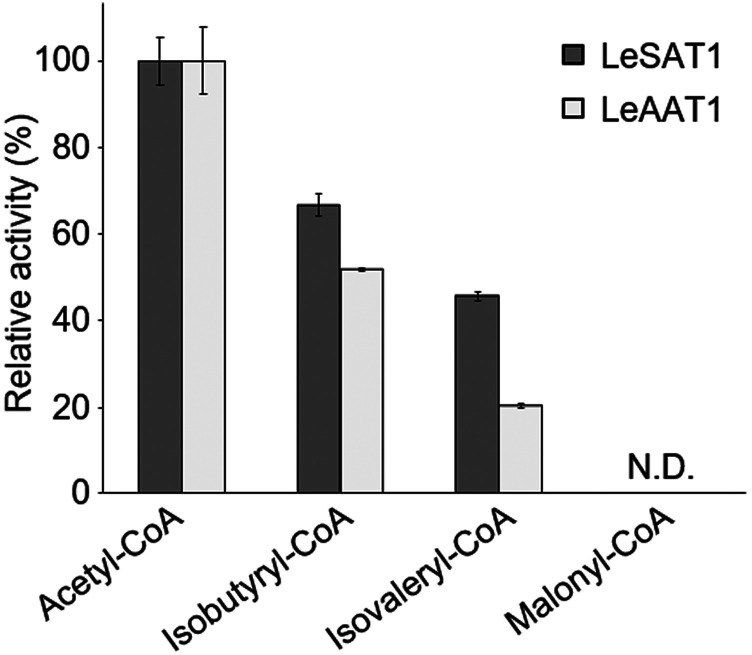
To investigate their substrate specificities, LeSAT1 or LeAAT1 was incubated with acetyl-CoA and three intermediates in the shikonin/alkannin biosynthesis pathway, GHQ, GHQ-3″-OH, and deoxyshikonin (Fig. 1). Neither of these enzymes acetylated any of these intermediates. The acyl donor specificity of LeSAT1 and LeAAT1 was also analyzed. LeSAT1 catalyzed the acylation of shikonin, using isobutyryl-CoA and isovaleryl-CoA, but not malonyl-CoA as acyl donors and producing isobutyrylshikonin and isovalerylshikonin, respectively (Fig. 4). The enzyme activities of LeSAT1 using isobutyryl-CoA and isovaleryl-CoA as acyl donor were approximately two-thirds and one-half of those obtained with acetyl-CoA. Similar results were obtained in analyzing the substrate specificity of LeAAT1 using alkannin as the acyl acceptor (Fig. 4). These results were consistent with the native enzymatic activities detected in the cultured cells (Supplemental Fig. S1), and agreed with the composition of distribution of the most abundant acetylated shikonin/alkannin derivatives in L. erythrorhizon (Table 1).
Figure 4.
Substrate specificities of LeSAT1 and LeAAT1, using shikonin and alkannin, respectively, as acyl acceptors. Both enzymes showed an inverse relationship of acylation activities with the size of donor acyl chains. N.D., Not detected. Values are means ± sd of three technical replicates.
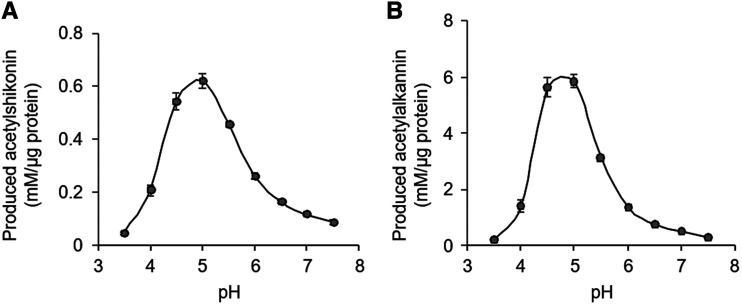
These substrate specificities of LeSAT1 and LeAAT1 were also confirmed by kinetic analysis. The apparent affinities (Km) of LeSAT1 and LeAAT1 for their acceptor substrates were calculated as 159 and 54 μm, respectively. Both enzymes showed higher catalytic rate constant (kcat) values when shorter acyl-CoAs were used as the acyl donors (Table 2). The catalytic efficiency (kcat/Km) of LeAAT1 with respect to alkannin was 12.7 mm−1 s−1, which is similar to those reported for clade III BAHD acytransferases, such as Ss5MaT2 in Salvia splendens, which had a catalytic efficiency of 13.5 mm−1 s−1 in synthesizing monodemalonylsalvianin (Suzuki et al., 2004) and EcCS in Erythroxylum coca, which has a catalytic efficiency of 26.0 mm−1 s−1 in the synthesis of methylecgonine (Schmidt et al., 2015). Interestingly, the catalytic efficiency of LeSAT1 was ∼10% of that of LeAAT1, which may have been due to the contamination by the chaperon protein in enzyme preparation of LeSAT1. This resulted in apparently higher protein concentrations and lower kcat for enzyme activity due to the amount of nonrelevant proteins (Supplemental Fig. S4). Investigation of the pH dependency showed that the acetylation activities of both LeSAT1 and LeAAT1 were maximal at pH 5.0, a moderately acidic condition (Fig. 5).
Table 2. Steady-state kinetic parameters of the recombinant LeSAT1 and LeAAT1.
Data are means ± sd of three independently prepared samples with three technical replicates for each sample. Shikonin and alkannin were incubated with 0.25 mm of acetyl-CoA, and acyl-CoAs were incubated with 0.5 mm of shikonin or alkannin.
| Enzyme | Substrate | Product | Km | kcat | kcat/Km |
|---|---|---|---|---|---|
| µm | s−1 | mm−1 s−1 | |||
| LeSAT1 | Shikonin | Acetylshikonin | 159 ± 25 | 0.185 ± 0.031 | 1.23 ± 0.32 |
| Acetyl-CoA | Acetylshikonin | 53 ± 4 | 0.093 ± 0.017 | 1.77 ± 0.27 | |
| Isobutyryl-CoA | Isobutyrylshikonin | 78 ± 18 | 0.083 ± 0.018 | 1.07 ± 0.11 | |
| Isovaleryl-CoA | Isovalerylshikonin | 39 ± 5 | 0.048 ± 0.007 | 1.24 ± 0.13 | |
| LeAAT1 | Alkannin | Acetylalkannin | 54 ± 13 | 0.68 ± 0.27 | 12.7 ± 4.4 |
| Acetyl-CoA | Acetylalkannin | 92 ± 39 | 0.94 ± 0.30 | 11.9 ± 3.6 | |
| Isobutyryl-CoA | Isobutyrylalkannin | 116 ± 60 | 0.78 ± 0.30 | 7.87 ± 2.5 | |
| Isovaleryl-CoA | Isovalerylalkannin | 20 ± 7 | 0.14 ± 0.04 | 9.72 ± 4.1 |
Figure 5.
pH dependence of acetylation activity. Acylation by LeSAT1 (A) and LeAAT1 (B) was assessed. Both enzymes showed maximal acetylation activity at pH 5.0. Values are means ± sd of three technical replicates.
Organ-Specific Expression of LeSAT1 and LeAAT1
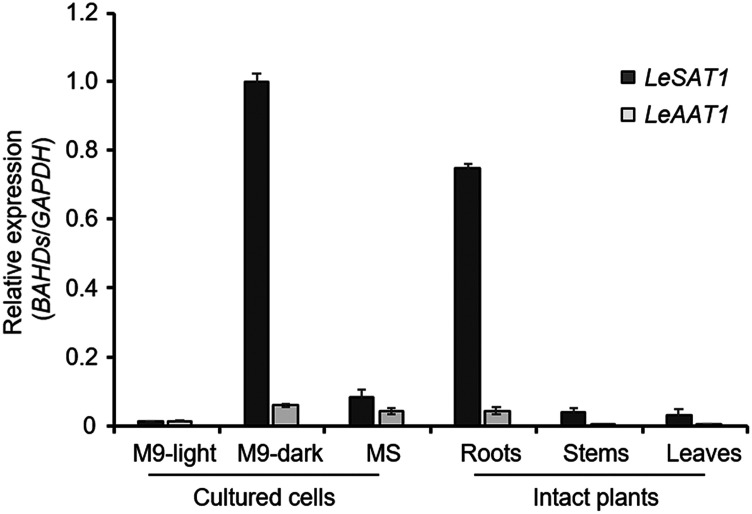
The expression profiles of LeSAT1 and LeAAT1 in intact plants and cultured cells of L. erythrorhizon were investigated by extracting total RNA from the roots, stems, and leaves of intact plants, and from cells cultured under three different growth conditions. Reverse-transcription quantitative PCR (RT-qPCR) analysis showed that both genes were preferentially expressed in roots of intact plants and in cells cultured in M9 medium in the dark, conditions under which shikonin/alkannin derivatives were synthesized and their accumulations were highest (Fig. 6; Wu et al., 2017; Takanashi et al., 2019). To assess the shikonin/alkannin acetylation activity in each organ, cell-free extracts of roots, stems, and leaves were incubated with shikonin or alkannin in the presence of acyl donor acetyl-CoA. The acetylation activity was detected only in roots (Supplemental Fig. S5), which is consistent with the RT-qPCR analysis (Fig. 6).
Figure 6.
Organ-specific expression of LeSAT1 and LeAAT1. The LeSAT1 and LeAAT1 expression was assayed by RT-qPCR in three different tissues and three cell culture conditions. GAPDH expression was used as the control. LeSAT1 and LeAAT1 were specifically expressed in tissues and cells with high levels of production of shikonin/alkannin derivatives. Values are means ± sd of three technical replicates performed individually on two independently prepared samples.
DISCUSSION
This study identified two BAHD acyltransferases, LeSAT1 and LeAAT1, as enzymes that catalyze the last steps of the shikonin/alkannin biosynthesis pathway in L. erythrorhizon. LeSAT1 converts shikonin to acetylshikonin, the most abundant shikonin derivative in these cells, whereas LeAAT1 is involved in the biosynthesis of acetylalkannin, the most abundant alkannin derivative in L. erythrorhizon.
Although the ratio of shikonin to alkannin derivatives in cultured L. erythrorhizon cells was 8:2, our crude enzyme assay showed that L. erythrorhizon cells produced almost equal amounts of shikonin and alkannin derivatives. These results suggested that the ratio of shikonin to alkannin derivatives was not determined by the stereospecific acylation of shikonin or alkannin but by the stereospecific hydroxylation of deoxyshikonin, in which two separate stereoselective hydroxylases may be involved.
Crude enzyme assays showed an inverse relationship between acylation activities and the lengths of acyl-CoAs. These results were similar to the substrate specificities of LeSAT1 and LeAAT1, although the relative activity using isovaleryl-CoA was not, suggesting that other BAHD acyltransferases may be involved in the biosynthesis of shikonin/alkannin derivatives. Involvement of other BAHD acyltransferases was also suggested by the results of RT-qPCR analysis, which showed that the relative expression of LeSAT1 was at least 10-fold higher than that of LeAAT1 in cells grown in M9 medium in the dark, as well as being consistent with the results of crude enzyme assays, which showed acylation of shikonin and alkannin was similar in L. erythrorhizon. Unfortunately, we were unable to compare the kinetics of LeSAT1 and LeAAT1 because LeSAT1 had to be copurified with a chaperon protein, GroEL. All attempts to remove GroEL from LeSAT1, including the addition of ATP to the binding buffer and separation on an ion-exchange resin after chromatography on a nickel column, were unsuccessful.
Shikonin/alkannin derivatives are synthesized inside L. erythrorhizon cells and accumulate in apoplasts. Secretion may be mediated by a vesicle-mediated transport system, as shown by electron microscopy and treatment with inhibitors (Tsukada and Tabata, 1984; Tatsumi et al., 2016). Studies using 14C-labeled deoxyshikonin and shikonin suggest that a hydroxylase and an acetyltransferase directly convert deoxyshikonin to acetylshikonin in a coordinated manner with localization to vesicle membranes (Okamoto et al., 1995). Because BAHD acyltransferases do not have transit peptides or transmembrane domains, these soluble proteins are thought to localize to the cytosol (D’Auria, 2006). In L. erythrorhizon cells, LeSAT1 and LeAAT1 may interact with a membrane-bound hydroxylase, forming a multienzyme complex called a metabolon, which can directly convert deoxyshikonin to acylated shikonin/alkannin derivatives. This is similar to other pathways involved in the biosynthesis of specialized metabolites, in which soluble enzymes are anchored by membrane-bound cytochrome P450 (Mucha et al., 2019; Nakayama et al., 2019).
Cultured L. erythrorhizon cells mainly produce acetylshikonin and β-hydroxyisovalerylshikonin but also biosynthesize isobutyrylshikonin and isovalerylshikonin. Analysis of their substrate specificities showed that LeSAT1 and LeAAT1 had maximum activities when acetyl-CoA was the acyl donor, although isobutyryl-CoA and isovaleryl-CoA were also accepted. These findings suggested that the variety of shikonin/alkannin derivatives in L. erythrorhizon may be determined by the substrate specificities of LeSAT1 and LeAAT1. Compositions of shikonin/alkannin derivatives differ among species of plants in the family Boraginaceae (Fukui et al., 1983; Assimopoulou et al., 2006; Zhou et al., 2011). It would be interesting to investigate whether the substrate specificities of orthologs of LeSAT1 and LeAAT1 correlate with the compositions of shikonin/alkannin derivatives in other plants of this family. Recent omics approaches using several Boraginaceae, such as A. euchroma and Echium plantagineum, may facilitate the identification of orthologs of LeSAT1 and LeAAT1 (Wang et al., 2014; Wu et al., 2017; Tang et al., 2020).
LeSAT1 and LeAAT1 show moderate amino acid identity, 64%, with respect to each other, with both proteins sharing 32% to 36% amino acid identities with BAHDs involved in the biosynthesis of acylsucrose in cultivated tomato (Solanum lycopersicum; SlASAT1, SlASAT2, and SlASAT3; Schilmiller et al., 2015; Fan et al., 2016). Acylsugars in S. lycopersicum are biosynthesized by consecutive acylations by four BAHD acyltransferases. The enzyme activities of these acyltransferases differ in position of acylation on sugar cores and also in preference for acyl donors (Fan et al., 2016). Compared with orthologs in cultivated tomato, these orthologs in wild tomato species (Solanum pennellii, Solanum pimpinellifolium, Solanum galapagense, etc.) differ in their order of acylation and substrate specificities (Fan et al., 2017; Moghe et al., 2017; Nadakuduti et al., 2017), resulting in a large variety of acylsugar structures in plants of the family Solanaceae (Fan et al., 2019). Studies using wild tomato and related species show that BAHD acyltransferases involved in acylsugar biosynthesis originated from alkaloid biosynthesis genes 50 to 80 million years ago and evolved subsequently (Moghe et al., 2017). Similar functional analyses of BAHD acyltransferases in the family Boraginaceae may reveal the origin of family members and the molecular evolutionary process of BAHDs responsible for the diversity of shikonin/alkannin derivatives.
BAHD acyltransferases in Catharanthus roseus also share 31% to 38% amino acid identities with LeSAT1 and LeAAT1. Two BAHD acyltransferases, CrMAT (Laflamme et al., 2001) and CrTAT (Carqueijeiro et al., 2018) involved in monoterpenoid indole alkaloid biosynthesis in C. roseus, have been recharacterized as the enantiomer-specific enzymes (Williams et al., 2019) acting upon the side-chain hydroxyl groups of (+)-vincadifformine and (−)-tabersonine, respectively. Separate enantiospecific cytochrome P450s are also involved in the formation of respective substrates for CrMAT and CrTAT (Williams et al., 2019). This also leads to the possibility that separate hydroxylases may act on deoxyshikonin to form shikonin and alkannin substrates used by LeSAT1 and LeAAT1 reactions, respectively.
MATERIALS AND METHODS
Plant Materials and Chemicals
Lithospermum erythrorhizon plants were germinated and grown in a cultivation chamber at 23°C under a 16-h light/8-h dark photoperiod for ∼4 months. Leaves, stems, and main roots were collected and stored at −80°C. L. erythrorhizon cell cultures were maintained as described in Takanashi et al. (2019). GHQ was synthesized as reported in Baeza et al. (2012), and GHQ-3″-OH was synthesized from homogentisic acid γ-lactone, described in detail in Supplemental Protocol S1. Shikonin, alkannin, and their derivatives were purchased from Nagara Science. Acetyl-CoA was purchased from Fujifilm Wako, and isobutyryl-CoA, isovaleryl-CoA, and malonyl-CoA were from Sigma-Aldrich.
Detection of Shikonin/Alkannin Derivatives in Cultured Media
Cultured cells grown on modified Murashige and Skoog medium were transferred to M9 medium for induction of shikonin/alkannin production. After incubation for 2 weeks on a rotary shaker (80 rpm) at 23°C in the dark, shikonin/alkannin derivatives were extracted from 3 mL of cultured M9 medium using an equal volume of ethyl acetate. This extract was evaporated to dryness, dissolved in 100 μL of methanol, and centrifuged at 14,400g for 5 min. The supernatant was analyzed by HPLC using an SPD-M20A system (Shimadzu) equipped with a CHIRALPAK IH-3 column (4.6 mm internal diameter × 250 mm; Daicel) at a flow rate of 1.0 mL min−1 at 35°C. Shikonin/alkannin derivatives were separated by isocratic elution with 60% (v/v) acetonitrile containing 0.4% (v/v) formic acid.
Extraction of Crude Enzyme from Cultured Cells and Intact Plants
L. erythrorhizon cells were cultured for 2 weeks in M9 medium. Liquid paraffin was overlaid onto the cultured medium for 3 d to remove shikonin/alkannin derivatives. Pigment-producing cells were collected by suction filtration and stored at −80°C. To prepare cell-free extracts, the cells and each organ of intact plants were homogenized in buffer containing 2.5 mm of acetic acid, 50 mm of sodium acetate, 100 mm of NaCl, 10 mm of dithiothreitol, 1 mm of phenylmethylsulfonyl fluoride, 0.5% (w/v) sodium ascorbate, and 1% (w/v) polyvinylpolypyrrolidone (pH 6.0). After centrifugation at 10,000g for 30 min at 4°C, the supernatants were filtered through a cotton-funnel, recentrifuged, and desalted on a PD-10 column (GE Healthcare).
Heterologous Expression of LeSAT1 and LeAAT1 in Escherichia coli
The full-length open reading frames of LeSAT1 and LeAAT1 were codon-optimized for expression in E. coli. Full-length LeSAT1 and LeSAT1 were digested with NdeI and SalI and cloned into the NdeI/SalI sites of pCold I (Takara Bio). These constructs were used to transform E. coli Origami strain. For LeSAT1 expression, the pGro7 plasmid (Takara Bio) expressing the molecular chaperonins GroEL and GroES was cotransformed, and the transformants cultivated in 300 mL of lysogeny broth medium at 37°C. When OD600 reached 0.6 to 0.8, protein expression was induced by cold shock on ice for 30 min and the addition of 1 mm of isopropyl β-d-1-thiogalactopyranoside, and the bacteria were further cultivated at 15°C for 12 to 24 h.
Protein Extraction and Purification of LeSAT1 and LeAAT1 Expressed in E. coli
All of the following steps were performed at ∼4°C. Harvested cells were sonicated in lysis buffer containing 50 mm of Tris-HCl (pH 7.6), 5% (v/v) glycerol, 20 mm of MgCl2, 1% (v/v) TWEEN 20, and the cell debris was removed by centrifugation at 10,000g for 20 min. The supernatant was applied to an affinity column filled with 2 mL of Ni resin (Ni Sepharose 6 Fast Flow; GE Healthcare). The column was washed twice with 4 mL of binding buffer containing 50 mm of Tris-HCl (pH 7.6), 10% (v/v) glycerol, 500 mm of NaCl, and 20 mm of imidazole for the purification of LeSAT1. For LeAAT1, 50 mm of imidazole was used instead of 20 mm of imidazole. His-tagged LeSAT1 or LeAAT1 was eluted four times each with 2 mL of elution buffer containing 50 mm of Tris-HCl at pH 7.6, 10% (v/v) glycerol, 500 mm of NaCl, and 500 mm of imidazole. For LeSAT1 purification, 4 mL of binding buffer supplemented with 10 mm of ATP was applied to the column, and the column was incubated for 1 h at room temperature before elution. Purified enzyme fractions were desalted on a PD-10 column containing 50 mm of MES (pH 6.5) buffer for enzyme assays.
Enzyme Assays
Enzyme assays were performed in 50 mm of MES at pH 5.5 (50 μL) containing 10 mm of sodium ascorbate, 0.3% (v/v) Triton-X, enzyme fraction (25 μL), and substrates (250 μm of acyl-donor and 500 μm of acyl-acceptor). After incubation at 30°C for 5 min, the products were extracted in 50 μL of ethyl acetate and centrifuged at 14,400g for 5 min. Supernatants were analyzed by HPLC using a CHIRALPAK IH-3 column (Daicel), as described above. The optimum pH for acetylation activity was determined using a buffer solution containing 3,3′-dimethylglutaric acid, Tris, and 2-amino-2-methyl-1,3-propanediol. The kinetic parameters of shikonin and alkannin (2.5–250 μm each) and acyl-CoAs (2.5–500 µm) were determined using 250 µm of acetyl-CoA and 500 μm of shikonin or alkannin, respectively.
RNA Extraction and RT-qPCR Analysis
Total RNA was isolated from cultured cells and intact plants using RNeasy Plant Mini Kits (Qiagen) or ISOSPIN Plant RNA (Nippon Gene). After treatment with DNase I, total RNAs were reverse-transcribed using ReverTra Ace qPCR RT Master Mix (Toyobo), and RT-qPCR was performed using a KOD SYBR qPCR Mix (Toyobo) with gene-specific primers (Supplemental Table S1) on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific). Gene expression levels of each sample were normalized relative to GAPDH mRNA as an internal standard and calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001), combined with the standard-curve method (Larionov et al., 2005).
Accession Numbers
Sequence data from this article can be obtained from the DNA Data Bank of Japan (https://www.ddbj.nig.ac.jp/index-e.html) under the accession numbers: LC520137 (LeSAT1) and LC520138 (LeAAT1).
Supplemental Data
The following supplemental information is available.
Supplemental Figure S1. Native acylation activity of shikonin and alkannin in desalted cell-free extracts of cultured L. erythrorhizon cells.
Supplemental Figure S2. Omics analyses identification of four BAHD acyltransferases in L. erythrorhizon cells.
Supplemental Figure S3. Comparison of the structures of four BAHD acyltransferases in L. erythrorhizon cells.
Supplemental Figure S4. Purification of recombinant enzymes of LeSAT1 and LeAAT1.
Supplemental Figure S5. Native acetylation activity of shikonin and alkannin in desalted cell-free extracts of roots, stems, and leaves.
Supplemental Table S1. Oligonucleotide primers used in RT-qPCR analyses.
Supplemental Protocol S1. Synthesis of GHQ-3″-OH and GHQ.
Acknowledgments
The authors thank Dr. Tsuyoshi Nakagawa (Shimane University) for the series of pGWB plasmids, Amato Pharmaceutical Products, for providing the L. erythrorhizon seeds, Dr. Takao Koeduka (Yamaguchi University) for helpful advice on protein expression in E. coli, and Kyoko Ohmine and Akiko Fujihashi (Kyoto University) for instrumental analyses.
Footnotes
This work was supported by the Japan Society for the Promotion of Science (KAKENHI grant no. 19K05825 to K.T.); the Research Institute for Sustainable Humanosphere, Kyoto University (Exploratory Research on Sustainable Humanosphere Science research grant to B.W., K.Y., and K.T.); the Research Institute for Sustainable Humanosphere, Kyoto University (Mission Research on Sustainable Humanosphere research grant to K.T., B.W., and K.Y.); and the Institute for Chemical Research, Kyoto University (Collaborative Research Program grant no. 2018–72 to K.T. and B.W.).
Articles can be viewed without a subscription.
References
- Assimopoulou AN, Karapanagiotis I, Vasiliou A, Kokkini S, Papageorgiou VP(2006) Analysis of alkannin derivatives from Alkanna species by high-performance liquid chromatography/photodiode array/mass spectrometry. Biomed Chromatogr 20: 1359–1374 [DOI] [PubMed] [Google Scholar]
- Baeza E, Catalán K, Peña-Cortés H, Espinoza L, Villena J, Carrasco H(2012) Synthesis of geranylhydroquinone derivatives with potential cytotoxic activity. Quim Nova 35: 523–526 [Google Scholar]
- Bontpart T, Cheynier V, Ageorges A, Terrier N(2015) BAHD or SCPL acyltransferase? What a dilemma for acylation in the world of plant phenolic compounds. New Phytol 208: 695–707 [DOI] [PubMed] [Google Scholar]
- Boulos JC, Rahama M, Hegazy MF, Efferth T(2019) Shikonin derivatives for cancer prevention and therapy. Cancer Lett 459: 248–267 [DOI] [PubMed] [Google Scholar]
- Carqueijeiro I, Dugé de Bernonville T, Lanoue A, Dang TT, Teijaro CN, Paetz C, Billet K, Mosquera A, Oudin A, Besseau S, et al. (2018) A BAHD acyltransferase catalyzing 19-o-acetylation of tabersonine derivatives in roots of Catharanthus roseus enables combinatorial synthesis of monoterpene indole alkaloids. Plant J 94: 469–484 [DOI] [PubMed] [Google Scholar]
- Chen X, Yang L, Oppenheim JJ, Howard MZ(2002) Cellular pharmacology studies of shikonin derivatives. Phytother Res 16: 199–209 [DOI] [PubMed] [Google Scholar]
- D’Auria JC.(2006) Acyltransferases in plants: A good time to be BAHD. Curr Opin Plant Biol 9: 331–340 [DOI] [PubMed] [Google Scholar]
- Fan P, Leong BJ, Last RL(2019) Tip of the trichome: Evolution of acylsugar metabolic diversity in Solanaceae. Curr Opin Plant Biol 49: 8–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P, Miller AM, Liu X, Jones AD, Last RL(2017) Evolution of a flipped pathway creates metabolic innovation in tomato trichomes through BAHD enzyme promiscuity. Nat Commun 8: 2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan P, Miller AM, Schilmiller AL, Liu X, Ofner I, Jones AD, Zamir D, Last RL(2016) In vitro reconstruction and analysis of evolutionary variation of the tomato acylsucrose metabolic network. Proc Natl Acad Sci USA 113: E239–E248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui H, Tsukada M, Mizukami H, Tabata M(1983) Formation of stereoisomeric mixtures of naphthoquinone derivatives in Echium lycopsis callus cultures. Phytochemistry 22: 453–456 [Google Scholar]
- Hasenoehrl C, Schwach G, Ghaffari-Tabrizi-Wizsy N, Fuchs R, Kretschmer N, Bauer R, Pfragner R(2017) Anti-tumor effects of shikonin derivatives on human medullary thyroid carcinoma cells. Endocr Connect 6: 53–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Kumar R, Kishore K(2013) Onosma L.: A review of phytochemistry and ethnopharmacology. Pharmacogn Rev 7: 140–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Li H, Minami H, Kato Y, Tabata H, Yazaki K(2019) Evolutionary developments in plant specialized metabolism, exemplified by two transferase families. Front Plant Sci 10: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme P, St-Pierre B, De Luca V(2001) Molecular and biochemical analysis of a Madagascar periwinkle root-specific minovincinine-19-hydroxy-o-acetyltransferase. Plant Physiol 125: 189–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov A, Krause A, Miller W(2005) A standard curve-based method for relative real time PCR data processing. BMC Bioinformatics 6: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Lee HJ, Magesh V, Nam D, Lee EO, Ahn KS, Jung MH, Ahn KS, Kim DK, Kim JY, et al. (2008) Shikonin, acetylshikonin, and isobutyroylshikonin inhibit VEGF-induced angiogenesis and suppress tumor growth in Lewis lung carcinoma-bearing mice. Yakugaku Zasshi 128: 1681–1688 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD(2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Malik S, Bhushan S, Sharma M, Ahuja PS(2016) Biotechnological approaches to the production of shikonins: A critical review with recent updates. Crit Rev Biotechnol 36: 327–340 [DOI] [PubMed] [Google Scholar]
- Mitra S, Dash R(2018) Structural dynamics and quantum mechanical aspects of shikonin derivatives as CREBBP bromodomain inhibitors. J Mol Graph Model 83: 42–52 [DOI] [PubMed] [Google Scholar]
- Moghe GD, Leong BJ, Hurney SM, Daniel Jones A, Last RL(2017) Evolutionary routes to biochemical innovation revealed by integrative analysis of a plant-defense related specialized metabolic pathway. eLife 6: e28468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucha S, Heinzlmeir S, Kriechbaumer V, Strickland B, Kirchhelle C, Choudhary M, Kowalski N, Eichmann R, Hückelhoven R, Grill E, et al. (2019) The formation of a camalexin biosynthetic metabolon. Plant Cell 31: 2697–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadakuduti SS, Uebler JB, Liu X, Jones AD, Barry CS(2017) Characterization of trichome-expressed BAHD acyltransferases in Petunia axillaris reveals distinct acylsugar assembly mechanisms within the Solanaceae. Plant Physiol 175: 36–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T, Takahashi S, Waki T(2019) Formation of flavonoid metabolons: Functional significance of protein-protein interactions and impact on flavonoid chemodiversity. Front Plant Sci 10: 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto T, Yazaki K, Tabata M(1995) Biosynthesis of shikonin derivatives from L-phenylalanine via deoxyshikonin in Lithospermum cell cultures and cell-free extracts. Phytochemistry 38: 83–88 [Google Scholar]
- Papageorgiou VP, Assimopoulou AN, Couladouros EA, Hepworth D, Nicolaou KC(1999) The chemistry and biology of alkannin, shikonin, and related naphthazarin natural products. Angew Chem Int Ed Engl 38: 270–301 [DOI] [PubMed] [Google Scholar]
- Papageorgiou VP, Assimopoulou AN, Samanidou VF, Papadoyannis IN(2006) Recent advances in chemistry, biology and biotechnology of alkannins and shikonins. Curr Org Chem 10: 2123–2142 [Google Scholar]
- Schilmiller AL, Moghe GD, Fan P, Ghosh B, Ning J, Jones AD, Last RL(2015) Functionally divergent alleles and duplicated Loci encoding an acyltransferase contribute to acylsugar metabolite diversity in Solanum trichomes. Plant Cell 27: 1002–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt GW, Jirschitzka J, Porta T, Reichelt M, Luck K, Torre JCP, Dolke F, Varesio E, Hopfgartner G, Gershenzon J, et al. (2015) The last step in cocaine biosynthesis is catalyzed by a BAHD acyltransferase. Plant Physiol 167: 89–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, De Luca V(2000) Evolution of acyltransferase genes: Origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism In Romeo JT, Ibrahim R, Varin L, and De Luca V, eds, Recent Advances in Phytochemistry. Elsevier Science, Oxford, pp 285–315 [Google Scholar]
- Suzuki H, Sawada S, Watanabe K, Nagae S, Yamaguchi MA, Nakayama T, Nishino T(2004) Identification and characterization of a novel anthocyanin malonyltransferase from scarlet sage (Salvia splendens) flowers: An enzyme that is phylogenetically separated from other anthocyanin acyltransferases. Plant J 38: 994–1003 [DOI] [PubMed] [Google Scholar]
- Tabata M, Mizukami H, Hiraoka N, Konoshima M(1974) Pigment formation in callus cultures of Lithospermum erythrorhizon. Phytochemistry 13: 927–932 [Google Scholar]
- Takanashi K, Nakagawa Y, Aburaya S, Kaminade K, Aoki W, Saida-Munakata Y, Sugiyama A, Ueda M, Yazaki K(2019) Comparative proteomic analysis of Lithospermum erythrorhizon reveals regulation of a variety of metabolic enzymes leading to comprehensive understanding of the shikonin biosynthetic pathway. Plant Cell Physiol 60: 19–28 [DOI] [PubMed] [Google Scholar]
- Tang CY, Li S, Wang YT, Wang X(2020) Comparative genome/transcriptome analysis probes Boraginales’ phylogenetic position, WGDs in Boraginales, and key enzyme genes in the alkannin/shikonin core pathway. Mol Ecol Resour 20: 228–241 [DOI] [PubMed] [Google Scholar]
- Tatsumi K, Yano M, Kaminade K, Sugiyama A, Sato M, Toyooka K, Aoyama T, Sato F, Yazaki K(2016) Characterization of shikonin derivative secretion in Lithospermum erythrorhizon hairy roots as a model of lipid-soluble metabolite secretion from plants. Front Plant Sci 7: 1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M, Tabata M(1984) Intracellular localization and secretion of naphthoquinone pigments in cell cultures of Lithospermum erythrorhizon. Planta Med 50: 338–341 [DOI] [PubMed] [Google Scholar]
- Tuominen LK, Johnson VE, Tsai CJ(2011) Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Guo LP, Xie T, Yang J, Tang JF, Li X, Wang X, Huang LQ(2014) Different secondary metabolic responses to MeJA treatment in shikonin-proficient and shikonin-deficient cell lines from Arnebia euchroma (Royle) Johnst. Plant Cell Tissue Organ Cult 119: 587–598 [Google Scholar]
- Wang S, Wang R, Liu T, Lv C, Liang J, Kang C, Zhou L, Guo J, Cui G, Zhang Y, et al. (2019) CYP76B74 catalyzes the 3″-hydroxylation of geranylhydroquinone in shikonin biosynthesis. Plant Physiol 179: 402–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D, Qu Y, Simionescu R, De Luca V(2019) The assembly of (+)-vincadifformine- and (−)-tabersonine-derived monoterpenoid indole alkaloids in Catharanthus roseus involves separate branch pathways. Plant J 99: 626–636 [DOI] [PubMed] [Google Scholar]
- Wu FY, Tang CY, Guo YM, Bian ZW, Fu JY, Lu GH, Qi JL, Pang YJ, Yang YH(2017) Transcriptome analysis explores genes related to shikonin biosynthesis in Lithospermeae plants and provides insights into Boraginales’ evolutionary history. Sci Rep 7: 4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K.(2017) Lithospermum erythrorhizon cell cultures: Present and future aspects. Plant Biotechnol (Tokyo) 34: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K, Kunihisa M, Fujisaki T, Sato F(2002) Geranyl diphosphate:4-hydroxybenzoate geranyltransferase from Lithospermum erythrorhizon. Cloning and characterization of a key enzyme in shikonin biosynthesis. J Biol Chem 277: 6240–6246 [DOI] [PubMed] [Google Scholar]
- Zhou W, Jiang HdaG, Peng Y, Li SS(2011) Comparative study on enantiomeric excess of main alkannin/shikonin derivatives isolated from the roots of three endemic Boraginaceae plants in China. Biomed Chromatogr 25: 1067–1075 [DOI] [PubMed] [Google Scholar]