Lignification of Arabidopsis stems is accompanied by precise localization of laccases and peroxidases to particular regions of the cell wall and differential production of apoplastic H2O2.
Abstract
Lignin, a critical phenolic polymer in secondary cell walls of plant cells, enables strength in fibers and water transportation in xylem vessel elements. Secreted enzymes, namely laccases (LACs) and peroxidases (PRXs), facilitate lignin polymerization by oxidizing lignin monomers (monolignols). Previous work in Arabidopsis (Arabidopsis thaliana) demonstrated that AtLAC4 and AtPRX64 localized to discrete lignified cell wall domains in fibers, although the spatial distributions of other enzymes in these large gene families are unknown. Here, we show that characteristic sets of putative lignin-associated LACs and PRXs localize to precise regions during stem development, with LACs and PRXs co-occurring in cell wall domains. AtLAC4, AtLAC17, and AtPRX72 localized to the thick secondary cell wall of xylem vessel elements and fibers, whereas AtLAC4, AtPRX64, and AtPRX71 localized to fiber cell corners. Interestingly, AtLAC4 had a transient cell corner localization early in fiber development that disappeared in the mature stem. In contrast with these secondary cell wall localizations, AtLAC10, AtPRX42, AtPRX52, and AtPRX71 were found in nonlignified tissues. Despite ubiquitous PRX occurrence in cell walls, PRX oxidative activity was restricted to lignifying regions during development, which suggested regulated production of apoplastic hydrogen peroxide. Relative amounts of apoplastic reactive oxygen species differed between lignified cell types, which could modulate PRX activity. Together, these results indicate that precise localization of oxidative enzymes and differential distribution of oxidative substrates, such as hydrogen peroxide, provide mechanisms to control spatiotemporal deposition of lignin during development.
The evolution of secondary cell walls (SCWs), rich in cellulose, hemicellulose, and lignin, was essential for plant colonization of land (Weng and Chapple, 2010). Lignin constitutes up to 18% to 35% of the overall plant biomass (Sarkanen and Ludwig, 1971) and provides structural strength to the cell wall, allowing efficient water transportation in xylem vessel elements and mechanical support in fiber cells for upright growth. Lignin is composed of the monolignols p-coumaryl, coniferyl, and sinapyl alcohols, which are produced from phenylalanine in the cytoplasm and exported to the cell wall (Boerjan et al., 2003). The final stage of lignin polymerization requires oxidation of monolignols by secreted enzymes, namely laccases (LACs) and peroxidases (PRXs), which initiates random cross-linking to form the lignin polymer. LACs are glycosylated multi-copper oxidoreductases that oxidize phenolic compounds through the reduction of molecular oxygen to water (Berthet et al., 2012), whereas class-III PRXs are glycosylated heme-containing oxidoreductases that oxidize phenolic compounds using hydrogen peroxide (H2O2; Shigeto and Tsutsumi, 2016).
Despite acting in a fundamental step in lignin polymerization, identifying and functionally characterizing the specific LACs and PRXs involved in lignification has been hindered by the large gene families of these enzymes, overlapping expression profiles, broad substrate specificities, and functional redundancy (Turlapati et al., 2011; Shigeto and Tsutsumi, 2016). In Arabidopsis (Arabidopsis thaliana), there are 17 LACs and 73 PRXs, and the isoenzymes involved in lignification and their spatial distributions in plant tissues are poorly characterized.
Previous mutant analyses by Berthet et al. (2011) demonstrated that simultaneous deletion of Arabidopsis LACCASE4 (AtLAC4) and AtLAC17 decreased lignin content by 40% and led to an irregular xylem phenotype (Berthet et al., 2011). Furthermore, Arabidopsis triple lac4-2/lac11/lac17 mutants exhibit dwarfing and have reduced stem and root lignin, providing strong genetic evidence for AtLAC4, AtLAC11, and AtLAC17 in lignification (Zhao et al., 2013). In addition, AtPRX64 is required for lignification of the root Casparian strip (Lee et al., 2013). Several other PRXs are implicated in lignification, including AtPRX2, AtPRX4, AtPRX17, AtPRX25, AtPRX52, AtPRX71, and AtPRX72, due to altered lignin content or composition in mutant plant lines (Herrero et al., 2013; Fernández-Pérez et al., 2015, 2015a, 2015b; Shigeto et al., 2015; Cosio et al., 2017).
The root Casparian strip is still lignified in triple lac4-2/lac11/lac17 mutants, indicating that LACs and PRXs are nonredundant in lignified tissues (Zhao et al., 2013). Fluorescent tagging of Arabidopsis AtLAC4 and AtPRX64 provided further evidence for the nonoverlapping role of these enzymes during lignification, as AtLAC4-mCherry localizes to the thick lignified SCW of fiber cells in stems, whereas AtPRX64-mCherry localizes to the lignified cell corner and middle lamella between adjacent fiber cells (Yi Chou et al., 2018). The spatial separation to different regions of the cell wall suggests separate roles of these enzymes during lignification, although it is unknown whether other LACs and PRXs show similar discrete localizations in lignified cell walls. Another possibility for the nonredundancy of LACs and PRXs is different substrate specificities, although most LACs and PRXs show promiscuous activity in vitro (Sterjiades et al., 1993; Shigeto et al., 2014). Alternatively, LACs and PRXs could act during different temporal stages due to differential availability of H2O2 for PRX activity (Sterjiades et al., 1993; Pandey et al., 2016; Laitinen et al., 2017). However, there are few studies that have tested these hypotheses directly.
SCW lignification is controlled in space and time throughout growth, with only specific cell types becoming lignified while neighboring cells remain unlignified (Ehlting et al., 2005). The high spatial and temporal regulation over lignification has been demonstrated using fluorescent monolignol analogs in Arabidopsis stems, where different monolignols accumulate in specific tissue types and cell wall layers during development (Pandey et al., 2016). The cellular mechanisms directing lignin polymerization are currently unknown, but plant cells may control where lignin is deposited by specifying where lignin-associated LACs and PRXs are found (Donaldson, 2001; Schuetz et al., 2014). Due to the large number of LAC and PRX enzymes, different oxidative enzymes could be coordinating lignification in different cell types, regions of the cell wall, or stages of growth.
In this study, bioinformatics analyses were performed to choose eight highly expressed LACs and PRXs putatively involved in lignification. LACs and PRXs were expressed under their native promoters, tagged with the red fluorescent protein mCherry, and localized during different stages of Arabidopsis stem development. The localization analyses indicated that specific LACs and PRXs colocalize within subdomains of lignified cell walls, depending on cell type and developmental stage. A combination of PRX and reactive oxygen species (ROS) histochemistry suggested that regulated production of ROS in lignifying cell walls could control PRX activity spatially and temporally throughout stem growth. Thus, lignin polymerization in a particular region is coordinated by not only distinctive localizations of LACs and PRXs to cell wall domains but also monolignol and/or H2O2 substrate availability.
RESULTS
Identification of Putative Lignin-Associated LACs and PRXs
Previously, AtLAC4-mCherry was localized to the thick SCW, whereas AtPRX64-mCherry was only found in the cell corners/middle lamella between adjacent fiber cells in Arabidopsis stems (Yi Chou et al., 2018). To test whether the other LACs and PRXs followed this pattern, candidate LAC or PRX genes that are expressed in the stem and likely to be implicated in developmental lignification were identified with bioinformatics. Three criteria were used: (1) reported lignin deficient phenotypes in single-, double-, and triple-lac or -prx mutant lines; (2) high expression in the lignifying stem; and (3) coexpression with other SCW-associated genes. Initial candidates of LACs and PRXs putatively involved in lignification were identified from previous mutant analyses, including AtLAC4, AtLAC11, AtLAC12, AtLAC17, AtPRX2, AtPRX4, AtPRX25, AtPRX52, AtPRX64, AtPRX71, and AtPRX72 (Supplemental Tables S1–S3 and references there-in). This list was further refined using publicly available bioinformatics tools, incorporating gene expression data from two independent microarray datasets using Arabidopsis stem tissue (Supplemental Fig. S1; Schmid et al., 2005; Hall and Ellis, 2013) and coexpression networks of selected genes using Genevestigator (Supplemental Figs. S2 and S3; Hruz et al., 2008; Waese et al., 2017). Coexpression gene lists were annotated using The Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org) and were assessed for involvement in SCW biosynthesis. The combination of mutant phenotypes, transcript abundance in the stem, and coexpression network analyses therefore provided multiple lines of evidence for the selected LACs and PRXs to be involved in lignification (Table 1).
Table 1. Putative lignin-associated LACs and PRXs identified using a combination of expression, co-expression, and mutant phenotyping data.
Expression and co-expression data were generated from Genevestigator and the eFP browser (Hruz et al., 2008; Waese et al., 2017). n.d., No data.
| Gene | Loci | Tissue Expression | Co-expression | Stem Mutant Analysis | Reference |
|---|---|---|---|---|---|
| AtLAC4 | At2g38080 | Stem, root, leaf, hypocotyl, flower, silique | AtLAC11; AtLAC17; SCW cellulose and xylan biosynthetic genes | Decreased lignin; irregular xylem phenotype | Brown et al., 2005; Berthet et al., 2011 |
| AtLAC10 | At5g01190 | Stem, root, leaf, flower | SCW cellulose and xylan biosynthetic genes | No observable phenotype | Berthet et al., 2012 |
| AtLAC11 | At5g03260 | Stem, root, leaf, hypocotyl, flower | AtLAC4; AtLAC17; SCW cellulose biosynthetic genes | No observable phenotype | Berthet et al., 2011; Zhao et al., 2013 |
| AtLAC12 | At5g05390 | Stem, root, pollen, silique | AtLAC2; SCW cellulose and xylan biosynthetic genes | Decreased lignin | Berthet et al., 2012 |
| AtLAC17 | At5g60020 | Stem, root, leaf, hypocotyl, flower | AtLAC4; AtLAC11; SCW cellulose and xylan biosynthetic genes | Decreased lignin | Berthet et al., 2011 |
| AtPRX42 | At4g21960 | Stem, leaf, hypocotyl, root, silique, flower | Primary cell wall cellulose and pectin biosynthesis or modification genes | n.d. | n.d. |
| AtPRX52 | At5g05340 | Stem, leaf, root, flower | AtCCoAOMT; cell wall signaling and defense genes | Decreased lignin | Fernández-Pérez et al., 2015b |
| AtPRX64 | At5g42180 | Stem, root, silique, flower | AtMYB63; root endodermis and Casparian strip-related genes | n.d. | Lee et al., 2013 |
| AtPRX71 | At5g64120 | Stem, leaf, hypocotyl, root, flower | Response to biotic and abiotic stress; nutrient transport and metabolism | Altered lignin composition | Shigeto et al., 2013 |
| AtPRX72 | At5g66390 | Stem, root, leaf, hypocotyl, flower | Root endodermis and Casparian strip-related genes | Decreased lignin; irregular xylem phenotype | Herrero et al., 2013; Fernández-Pérez et al., 2015a |
For the LAC genes, transcriptomic data showed moderate to high expression of AtLAC4, AtLAC10, AtLAC11, AtLAC12, and AtLAC17 in the inflorescence stem (Supplemental Fig. S1) and coexpression with other LACs and SCW cellulose and hemicellulose biosynthetic genes (Supplemental Fig. S2), providing evidence for involvement in lignification and SCW formation. Additionally, amino acid phylogenetic analyses showed that all identified putative lignin-associated LACs, other than AtLAC12, cluster together (Supplemental Fig. S2).
In contrast with the LAC genes, bioinformatics analysis of PRXs in Arabidopsis stem tissues revealed highly variable expression among genes, with AtPRX42 and AtPRX64 comparable with the most highly expressed LAC genes and other PRXs expressed at very low levels (Supplemental Fig. S1). Due to low expression of AtPRX2, AtPRX4, AtPRX17, and AtPRX25 in stem tissues, these genes were not included in further analyses. AtPRX42, AtPRX52, AtPRX64, AtPRX71, and AtPRX72 were consistently detected in Arabidopsis stems and therefore selected as putative lignin-associated PRXs for further study (Table 1). The selected PRXs did not provide as clear coexpression and evolutionary relationships as the LAC genes, with broad expression in various plant tissues, diverse coexpression networks, and low amino acid similarity between candidates (Supplemental Fig. S3). Despite differences among the PRXs, each selected gene showed potential involvement in lignification due to expression in the stem, coexpression networks, or previous mutant phenotyping (Table 1).
Characteristic Sets of LACs and PRXs Are Secreted to Specific Cell Types and Regions of the Cell Wall during Stem Development
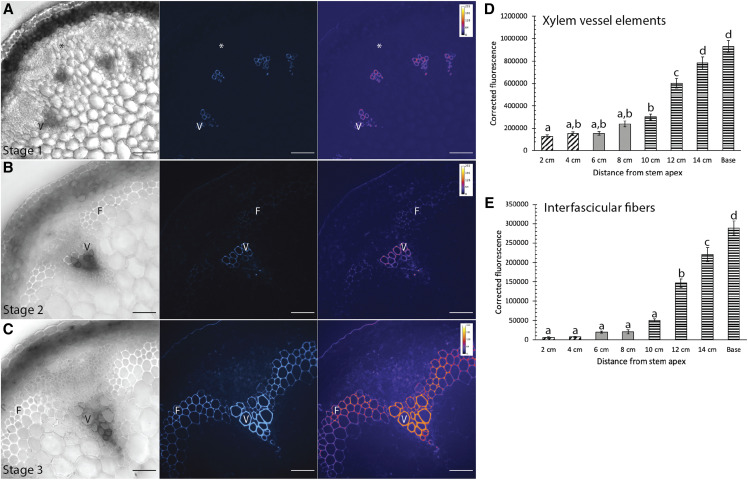
Despite earlier qualitative observations of lignin deposition in growing Arabidopsis stems (Ehlting et al., 2005), quantitative measurement of lignin deposition in different cell types during stem development has not been performed. To provide context to the localizations of the oxidative enzymes, it was first necessary to document lignin deposition to cell types and regions of the cell wall during stem development. The location and relative amount of lignin was assessed using ultraviolet (UV) lignin autofluorescence, detected using spinning disk confocal microscopy, from eight developmental timepoints of 17-cm-tall, 5-week-old Columbia-0 (Col-0) stems (Supplemental Fig. S4). Three discrete developmental stages were selected for further analysis, based on the location of UV lignin autofluorescence and presence of a thick SCW. Stage 1 was defined as the youngest tissue at the apical tip of the stem (2–4 cm), where there was low UV lignin autofluorescence in the SCW of xylem vessel elements and fibers were unlignified (Fig. 1A). Stage 2 was defined as the middle of the stem (6–8 cm), where lignin levels increased in the SCW of xylem vessel elements and UV lignin autofluorescence was first detected in the cell corners and middle lamella of interfascicular fibers (Fig. 1B). At this stage, fibers showed a slightly thickened SCW relative to neighboring nonlignified cells. Finally, stage 3 was defined as older, mature tissues (10 cm to the base of the stem), where bulk lignin deposition was observed in the thick SCW of both xylem vessel elements and fibers (Fig. 1C). Quantification of UV lignin autofluorescence intensity showed that lignin autofluorescence in xylem vessel elements was very low during Stage 1 but significantly increased to greater intensities in stages 2 and 3 (Fig. 1D), whereas UV lignin autofluorescence for fibers was first detected during stage 2 and significantly increased during stage 3 (Fig. 1E). These stages consequently represent three major developmental timepoints for lignin deposition in the stem, both for different cell types and regions of the SCW.
Figure 1.
Lignin location and amount differs during three stages of Arabidopsis stem development. A to C, Representative brightfield images, corresponding UV lignin autofluorescence (blue), and falsely colored heat map in stem cross-sections from stage 1 (A), stage 2 (B), and stage 3 (C). Scale bars = 50 µm. D and E, Quantification of corrected lignin UV autofluorescence from stem cross-sections for eight developmental timepoints. Cross-hatched bars indicate stage 1, solid gray bars indicate stage 2, and horizontal bars indicate stage 3. D, Corrected fluorescence for xylem vessel elements increases throughout development. E, Corrected lignin UV autofluorescence for fibers increases throughout development. A one-way ANOVA was used to compare developmental stages; n > 30 cells for each developmental stage from three biological replicates; lowercase letters indicate significantly different values according to Tukey-Kramer post-hoc tests (P < 0.0001). Bars ± se.
The three developmental stages were used as the framework to test if the localization of candidate LACs and PRXs identified in Table 1 generally followed the pattern established in mature tissues, where tagged AtPRX64 localized to lignified cell corners and AtLAC4 localized to the thick SCW (Yi Chou et al., 2018). The localization of fluorescently tagged AtLAC10, AtLAC17, AtPRX42, AtPRX52, AtPRX71, and AtPRX72 under their respective native promoters was examined in stem cross-sections from each of the developmental stages. Several attempts were made to transform prLAC11::LAC11-mCherry and prLAC12::LAC12-mCherry into Arabidopsis, but unfortunately there was strong silencing of the transgene in all lines examined (data not shown). As a positive control, the localization of tagged candidate LACs and PRXs was compared with previously characterized prLAC4::LAC4-mCherry (AtLAC4-mCherry) and prPRX64::PRX64-mCherry (AtPRX64-mCherry) lines (Lee et al., 2013; Schuetz et al., 2014; Yi Chou et al., 2018).
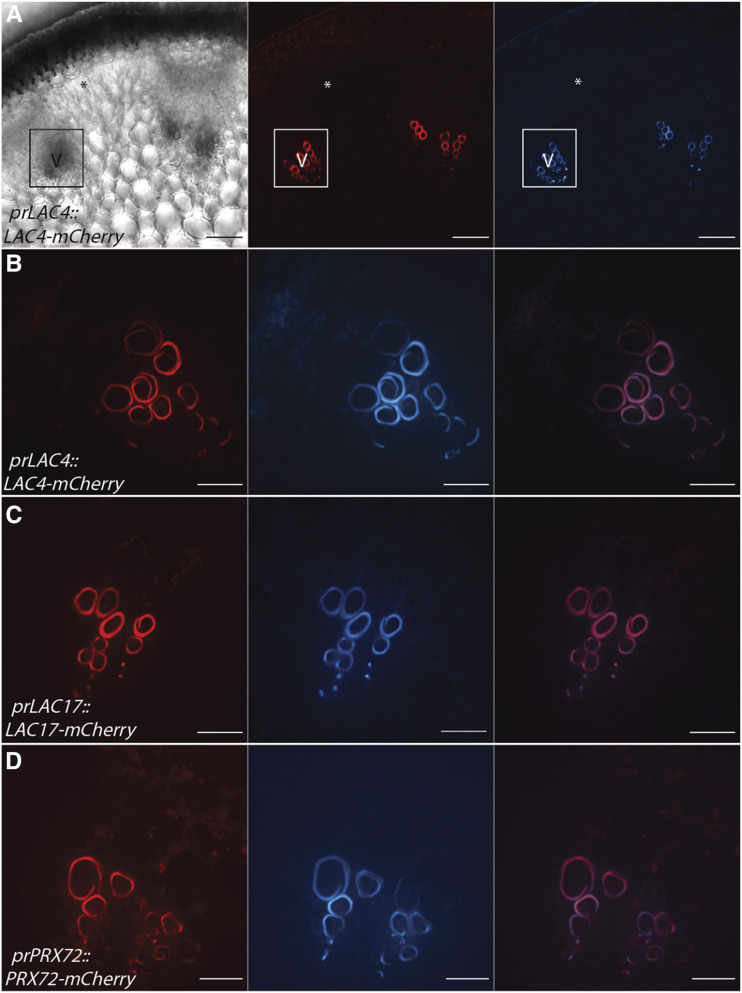
The only lignified cells in the stem in stage 1 were the xylem vessel elements (Fig. 1A), and three oxidative enzymes, AtLAC4, AtLAC17, and AtPRX72, localized to the SCW (Fig. 2). The localization of AtLAC4-mCherry and AtLAC17-mCherry to the thick SCW of protoxylem vessel elements (Fig. 2, A–C) is consistent with previous localization studies in experimentally induced protoxylem (Schuetz et al., 2014). In addition to the LACs, AtPRX72-mCherry also localized to the SCW of lignified protoxylem (Fig. 2D). Of the eight oxidative enzymes assessed, only this set of AtLAC4, AtLAC17, and AtPRX72 were detected in stem cross-sections during this early developmental stage. None of the tagged candidate enzymes localized to the unlignified primary cell walls in the interfascicular region (Fig. 2).
Figure 2.
AtLAC4-mCherry, AtLAC17-mCherry, and AtPRX72-mCherry localize to the SCW of xylem vessel elements during stage 1. Representative images depicting brightfield, the tagged LAC or PRX under its respective native promoter (red), UV lignin autofluorescence (blue), and merged image are shown. V, Xylem vessel element. Asterisks indicate cells that will differentiate into interfascicular fibers. A, Low magnification of AtLAC4-mCherry provides context for localization. Outlined region is vascular bundle with early lignifying xylem vessel elements. B, High magnification of AtLAC4-mCherry in the SCW of xylem vessel elements. C, High magnification of AtLAC17-mCherry in the SCW of xylem vessel elements. D, High magnification of AtPRX72-mCherry in the SCW of xylem vessel elements. Scale bars = 50 µm (A) and 20 µm (B–D).
During stage 2, lignin was detected in the middle lamella/cell corners of fibers and thus presented the opportunity to study initiation of lignin deposition in a particular region of the cell wall (Fig. 3A). As AtPRX64-mCherry was previously found in lignified cell corners of fibers in mature stems, we hypothesized that AtPRX64 may be co-secreted with primary cell wall and middle lamella components such as pectin early in development. Unexpectedly, AtPRX64 only localized to the cell corners of fibers at the onset of cell corner lignification and SCW thickening (Fig. 3B). At this stage, AtPRX71-mCherry and AtLAC4-mCherry were also specifically localized to the lignifying cell corners of fibers (Fig. 3, C and D). The cell-corner localization of AtLAC4-mCherry in fiber cell corners contrasts with previous analysis in mature tissues, where it was only observed in the fiber thick SCW (Yi Chou et al., 2018). To ensure that the localization of AtLAC4-mCherry in fiber cell corners represents dynamic cell wall protein changes and not simply the loss of cell corner signal due to SCW fluorescence, all imaging was done with identical microscope settings. Similar to stage 1, AtLAC4-mCherry, AtLAC17-mCherry, and AtPRX72-mCherry were collectively localized to the thick SCW of xylem vessel elements in vascular bundles (Supplemental Fig. S5). Unlike AtLAC4-mCherry, AtLAC17-mCherry and AtPRX72-mCherry were not observed in interfascicular fibers at this developmental stage. These data therefore identify another set of oxidative enzymes (AtPRX64, AtPRX71, and AtLAC4) that collectively occupy the cell corners, representing the first lignifying cell wall domain of fibers.
Figure 3.
AtLAC4-mCherry, AtPRX64-mCherry, and AtPRX71-mCherry are secreted to early lignifying cell corners of fibers at stage 2. A, Overview of a Col-0 stem cross-section at stage 2, with brightfield image, UV lignin autofluorescence (blue), and falsely colored heat map. Low magnification view of stem cross-section with higher magnification inset of outlined box is shown. B to D, Representative images depicting the tagged LAC or PRX under its respective native promoter (red), UV lignin autofluorescence (blue), and merged image. Inset shows higher magnification region of outlined box. B, AtPRX64-mCherry localizes to the cell corners of fibers. C, AtPRX71-mCherry localizes to the cell corners of fibers. D, AtLAC4-mCherry localizes to the SCW of xylem vessel elements and the cell corners of fibers. F, Interfascicular fibers; V, xylem vessel elements. Scale bars = 50 µm and 15 µm (insets).
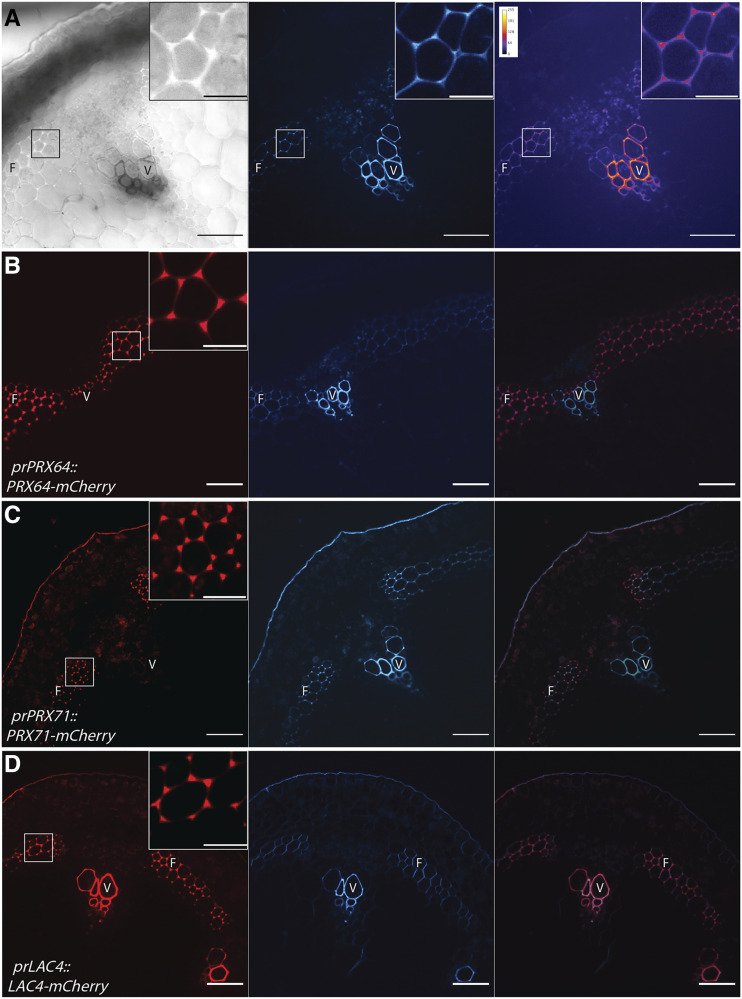
During stage 3 at the mature base of the stem, SCWs of both xylem vessel elements and fibers showed high UV lignin autofluorescence (Fig. 1C). Five tagged LACs and PRXs were observed in lignifying xylem vessel elements and fibers at maturity (Fig. 4; Supplemental Fig. S6). Consistent with earlier developmental stages, AtLAC4-mCherry, AtLAC17-mCherry, and AtPRX72-mCherry were observed in xylem vessel SCWs (Fig. 4, A–F). Interestingly, this same set of enzymes were also observed in the thickened SCW of interfascicular and xylary fibers, which were prominent at this stage of development (Fig. 4, A–F). The cell corner and middle lamella signal of AtLAC4-mCherry from stage 2 was absent (Fig. 4B). Similar to stage 2, AtPRX64-mCherry and AtPRX71-mCherry localized to the cell corners of fibers (Fig. 4, G–I). Unlike the homogenous localization of AtPRX64-mCherry to every fiber cell corner, AtPRX71-mCherry showed highest concentration in the outer fiber cell layer near the endodermis, which is the cell layer bordering photosynthetic cortex cells (Fig. 4J). These data demonstrated that the set of oxidative enzymes found in the thick SCW of Arabidopsis stems, in both vessel and fiber cell types, are AtLAC4, AtLAC17, and AtPRX72. In contrast with stage 2, mature stems contained a different set of oxidative enzymes (AtPRX64 and AtPRX71) in the cell corners/middle lamella, with the outer-most fiber cell layer showing an accumulation of oxidative enzymes at the cell corner junctions.
Figure 4.
LACs and PRXs are found in both the thick SCW and cell corners of lignified xylem vessel elements and fibers during Stage 3. Representative images depicting the tagged LAC or PRX under its native promoter (red), UV lignin autofluorescence (blue), and merged image are shown. F, Fiber; P, pith; V, xylem vessel element. Arrowheads indicate cell corner localization near endodermal layer. A and B, AtLAC4-mCherry localizes to the SCW of xylem vessel elements (A) and fibers (B). C and D, AtLAC17-mCherry localizes to the SCW of xylem vessel elements (C) and fibers (D). E and F, AtPRX72-mCherry localizes to the SCW of xylem vessel elements (E) and fibers (F). G and H, AtPRX64-mCherry is absent from xylem vessel elements (G) but localizes to the cell corners of fibers (H). I and J, AtPRX-71-mCherry localizes to the cell corners of xylem vessel elements (I) and fibers (J). Scale bars = 20 µm.
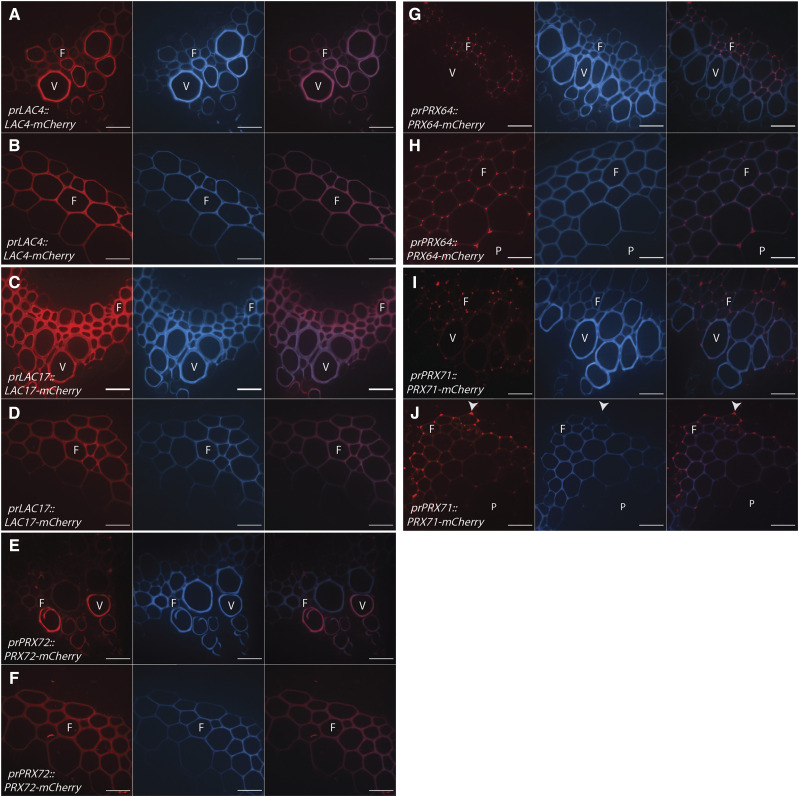
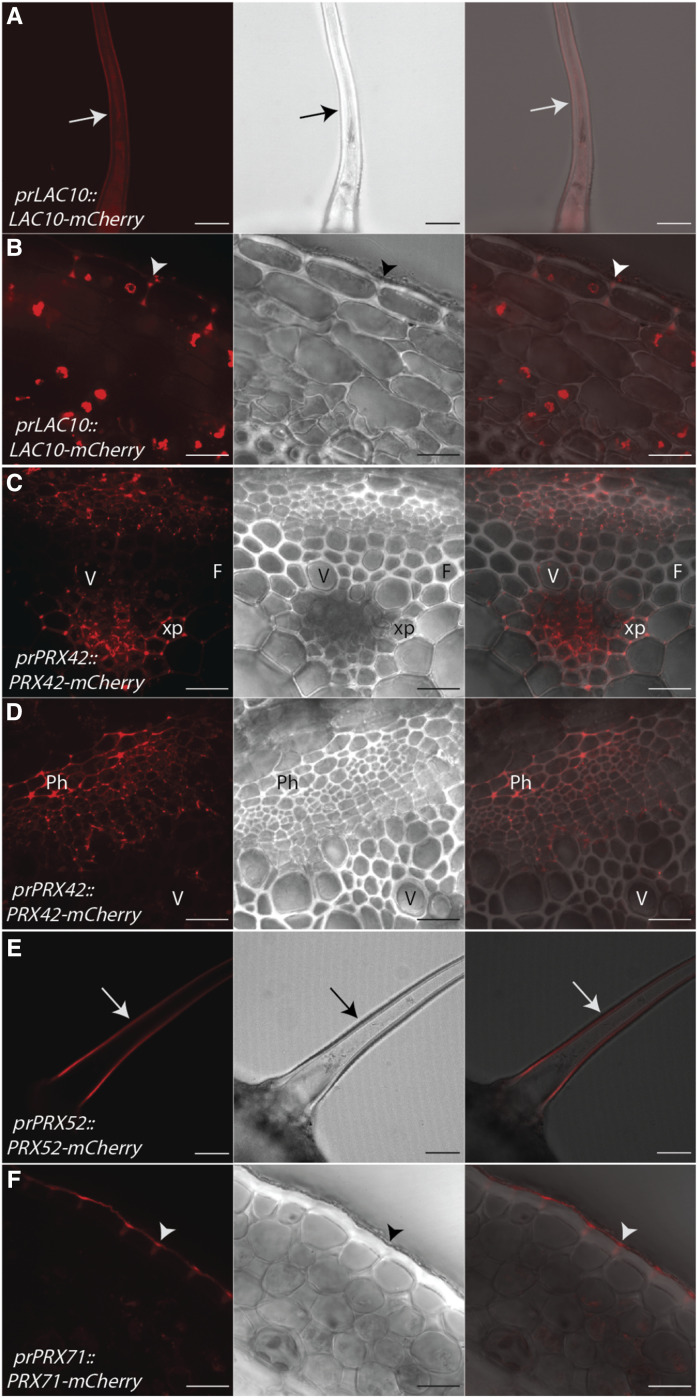
Among the candidate LACs and PRXs, several also localized to nonlignifying cell types during stage 3. AtLAC10-mCherry localized to both the primary cell wall of trichomes and epidermal cells (Fig. 5, A and B). AtPRX42-mCherry localized to the cell corners of nonlignifying xylem parenchyma and phloem cells (Fig. 5, C and D). AtPRX52-mCherry exclusively localized to the primary cell wall of trichomes (Fig. 5E) and was not observed in any other cell type. AtPRX71-mCherry also localized to the primary cell wall of epidermal cells (Fig. 5F; Supplemental Fig. S6). Therefore, certain enzymes can localize to both lignified and unlignified cell wall regions (e.g. AtPRX71), whereas other enzymes exclusively localize to lignified cell walls (e.g. AtLAC4, AtLAC17, AtPRX64, and AtPRX72) or primary cell walls (e.g. AtLAC10, AtPRX42, and AtPRX52) in stem tissues.
Figure 5.
LACs and PRXs localize to nonlignifying cell types in the inflorescence stem of Arabidopsis. Representative images depicting the tagged LAC or PRX under its native promoter (red), brightfield, and merged image are shown. Arrows indicate trichomes and arrowheads indicate epidermal cells. F, Fiber; Ph, phloem; V, xylem vessel element; xp, xylem parenchyma. A and B, AtLAC10-mCherry localizes to trichomes (A) and cell corners of epidermal cells (B). C and D, AtPRX42-mCherry localizes to the cell corners of nonlignifying xylem parenchyma cells (C) and the primary cell wall of phloem cells (D). E, AtPRX52-mCherry localizes to the primary cell wall of trichomes. F, AtPRX71-mCherry localizes to the cell corners of epidermal cells. Scale bars = 50 µm (A and E) and 20 µm (B–D and F).
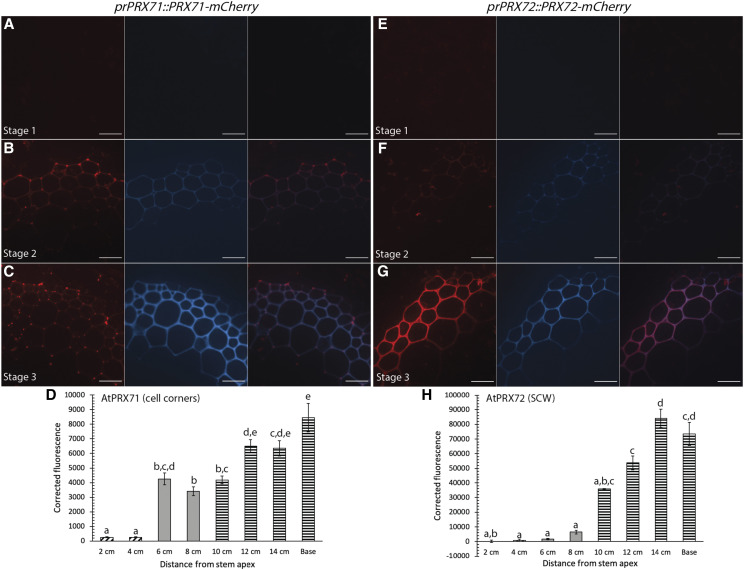
The observation that AtPRX71 and AtPRX72 localized to cell corners and the thick SCW, respectively, allowed investigation into how two members of the same gene family accumulate in fibers during development. AtPRX71-mCherry first appeared at very low fluorescence intensity in the lignified cell corners during stage 2 and the fluorescence signal significantly increased in Stage 3 (Fig. 6, A–D), suggesting that AtPRX71-mCherry was continually secreted to cell corners during progressive lignification of the SCW. AtPRX72-mCherry was first detected in the SCW of fibers during stage 3, and the fluorescence intensity of the tagged enzyme in the SCW significantly increased from 10 cm to the mature base of the stem (Fig. 6, E–H). Therefore, the increasing fluorescence intensity of AtPRX71-mCherry in cell corners and AtPRX72-mCherry in the SCW indicated that different proteins could simultaneously localize to distinct regions of the cell wall, even during lignification of the SCW.
Figure 6.
AtPRX71-mCherry and AtPRX72-mCherry accumulate in different regions of fiber cell walls throughout development of the stem. Representative images depicting tagged PRXs under the respective native promoter (red), UV lignin autofluorescence (blue), and merged image. A to C, AtPRX71-mCherry increases in fluorescence intensity in the cell corners of fibers in stage 1 (A), stage 2 (B), and stage 3 (C) during stem development. D, Quantification of AtPRX71-mCherry cell-corner fluorescence intensity. E to G, AtPRX72-mCherry increases in fluorescence intensity in the SCW of fibers in stage 1 (E), stage 2 (F), and stage 3 (G) during stem development. H, Quantification of AtPRX72-mCherry SCW fluorescence intensity. Corrected fluorescence was measured for n > 30 cells for each developmental timepoint from three biological replicates and compared using a one-way ANOVA; lowercase letters indicate significantly different values according to Tukey-Kramer post-hoc tests (P < 0.05). Bars ± se. Cross-hatched bars indicate stage 1, solid gray bars indicate stage 2, and horizontal bars indicate stage 3. Scale bars = 20 µm.
The presence of the mCherry tags on these cell wall enzymes might introduce artifacts that would alter their localization. We had previously shown that AtLAC4-mCherry rescues the irregular xylem phenotype of lac4-2/lac17 double mutants (Schuetz et al., 2014). Here, we demonstrate that AtLAC17-mCherry also rescues the irregular xylem phenotype of lac4-2/lac17 double mutants (Supplemental Fig. S7). If the mCherry tag was determining cell wall localization the enzymes, then we would predict that the localization of the secreted mCherry protein would give the same localization as the tagged proteins. To test where the fluorescent tag alone was localized, mCherry was targeted to the cell wall using an N-terminal signal sequence directing it through the default secretion pathway (prUBQ10::sec-mCherry; Yi Chou et al., 2018). It appeared in cell corners of cells with thickened SCW as well as nonlignified primary cell walls (Supplemental Fig. S8). This localization is distinct from the SCW localization patterns found with AtLAC4-mCherry, AtLAC17-mCherry, or AtPRX72-mCherry, implying that mCherry did not direct their localization. Similarly, the observation that PRXs have contrasting localizations in the cell corners and SCWs (Fig. 6) is not consistent with the mCherry tag driving localization.
PRX Enzymatic Activity and Production of Apoplastic Reactive Oxygen Species Is Limited to Lignifying Regions in Developing Arabidopsis Stems
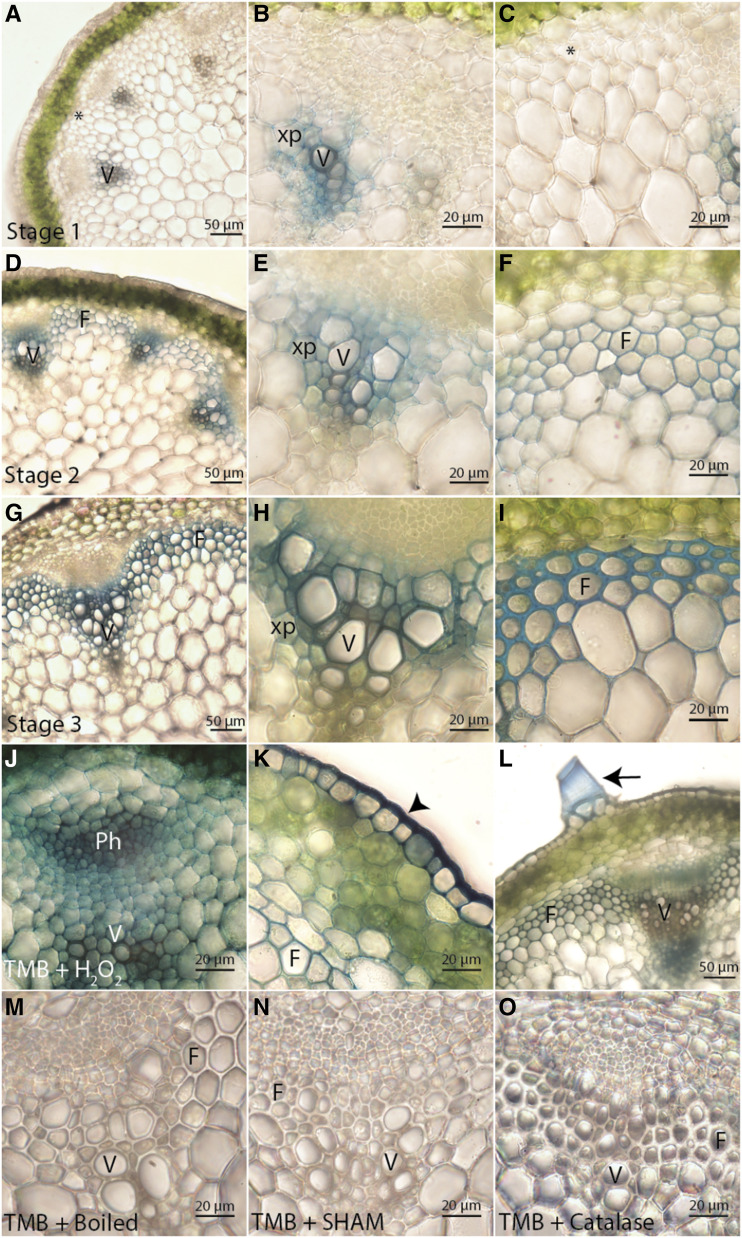
From the localization analyses, it was evident that the appearance of multiple tagged oxidative enzymes correlated with lignification in distinct cell types and cell wall regions during development. However, it was unclear whether these enzymes were active. To visualize where PRX oxidative enzymatic activity was occurring during development, cross-sections of 17-cm-tall Col-0 stems were incubated with an artificial PRX substrate (3,3’,5,5’-tetramethylbenzidine [TMB]), which is oxidized by PRXs in the presence of H2O2 to form a blue precipitate (Ros-Barceló, 1998). For all developmental stages, a blue precipitate formed specifically in regions of lignification (Fig. 7, A–I). At stage 1, a blue precipitate was apparent in the SCW of xylem vessel elements and in the primary cell walls of neighboring xylem parenchyma cells, but was absent in the primary cell walls of unlignified fibers (Fig. 7, A–C). During stage 2, a blue precipitate formed in the SCWs of xylem vessel elements and in the early lignifying cell corners of interfascicular fibers (Fig. 7, D–F). During stage 3, a blue precipitate formed in both the middle lamella and SCWs of xylem vessel elements and fibers (Fig. 7, G–I). These results provided direct evidence for PRX oxidative activity corresponding to the regions where AtPRX64-mCherry, AtPRX71-mCherry, and AtPRX72-mCherry were localized.
Figure 7.
PRXs are ubiquitous in cell walls but only show oxidative activity in lignifying regions during stem development. PRX oxidation of TMB results in a blue precipitate in Col-0 stems. F, Fibers; Ph, phloem; V, xylem vessel elements; xp, xylem parenchyma. Asterisks indicate cells that will differentiate into interfascicular fibers, arrowheads indicate epidermis, and arrows indicate trichome. A to I, PRX enzymatic activity localizes specifically to regions of lignin deposition in stage 1 (A–C), stage 2 (D–F), and stage 3 (G–I) of stem development. J to L, Addition of H2O2 causes TMB oxidation in cell walls of both lignifying and non-lignifying cell types, including xylem and phloem cells (J); fibers, cortex, and epidermal cells (K); and trichomes (L). M to O, Control experiments demonstrate TMB oxidation can be inhibited by boiling (M), treating with the PRX inhibitor SHAM (N), and scavenging H2O2 with catalase (O).
To test for the presence and location of other PRXs in stem tissues, Col-0 stem cross-sections were incubated with TMB and exogenous H2O2, which allowed visualization of any PRX capable of oxidizing TMB. Following incubation in H2O2, a blue precipitate was detected in all regions of the cell wall for xylem, phloem, cortex, epidermal, pith, and trichome cells at every developmental stage (Fig. 7, J–L). TMB and H2O2 histochemistry therefore suggested that secreted PRXs are found in the cell wall of all cell types, but the amount of apoplastic H2O2 is limiting the oxidative activity of these enzymes under normal growth conditions. There was no formation of blue precipitate in control experiments using boiled stem cross-sections, the PRX inhibitor salicylhydroxamic acid (SHAM), or the H2O2 scavenger catalase, confirming that formation of a blue TMB precipitate is specific for PRX oxidative activity and H2O2 (Fig. 7, M–O).
The results using the artificial substrate TMB indicated that PRX activity is restricted to regions with production of ROS, such as H2O2, in lignifying cell walls. Extracellular H2O2 is produced through the activity of plasma membrane-bound NADPH oxidases (Respiratory burst oxidase homologs [Rboh] in plants), which produce superoxide anions (O2−; Torres and Dangl, 2005). O2− can be converted into H2O2 spontaneously or through the enzyme superoxide dismutase. To visualize production of total ROS in stems, stem cross-sections from each of the eight developmental stages were incubated in the fluorescent dye 2’,7’-dichlorodihydrofluorescein diacetate (H2DCF), which detects the presence of ROS (Kalyanaraman et al., 2012). The nonfluorescent H2DCF is permeable through the plasma membrane, where it is cleaved by esterases to a carboxylate anion. Oxidation of this product produces the fluorescent molecule dichlorofluorescein (DCF) that can be visualized using confocal microscopy. Initial tests using H2DCF on stem cross-sections immediately after sectioning resulted in a super-saturated, nonspecific fluorescent signal in every cell type (Supplemental Fig. S9), likely due to ROS production following wounding. To minimize the wounding response, cross-sections were incubated in H2O for 24 h before staining and imaging. After 24 h, overall H2DCF fluorescence intensity was highly reduced and showed specific production in lignifying cell walls (Fig. 8A; Supplemental Fig. S9). Control experiments using the NADPH oxidase inhibitor diphenyleneiodonium chloride (DPI), the PRX inhibitor SHAM, and the H2O2 scavenger catalase demonstrated that H2DCF fluorescence is dependent on NADPH oxidases, PRX activity, and ROS such as H2O2 (Fig. 8B; Supplemental Fig. S9).
Figure 8.
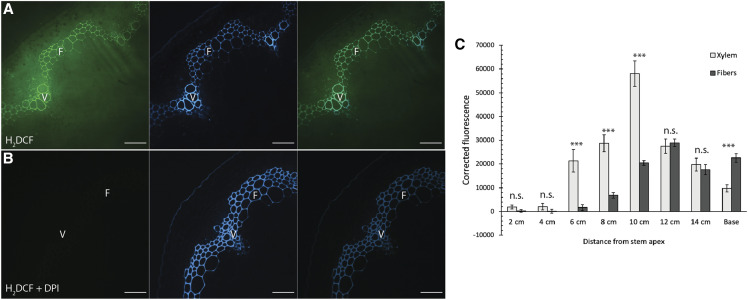
Xylem vessel elements and fibers have differential ROS accumulation during development. A and B, Representative images of cross-sections from the base of 17-cm-tall Col-0 stems incubated in H2DCF. Oxidation of H2DCF from ROS results in production of a green fluorescent product, which is compared with the location of UV lignin autofluorescence (blue). F, Interfascicular fibers; V, xylem vessel elements. A, Col-0 stems incubated in H2DCF show accumulation of ROS in SCWs of xylem vessel elements and interfascicular fibers. B, Treatment with the NADPH oxidase inhibitor DPI abolishes H2DCF signal. Scale bars = 50 µm. C, Quantification of H2DCF fluorescence intensity in the cell wall of xylem vessel elements and fibers at different timepoints of stem development. A two-way t-test was used to compare xylem vessel elements and fibers for a given developmental timepoint (***P < 0.001). n.s., Not significant. n > 20 cells for four biological replicates. Bars ± se.
To quantify ROS production or accumulation during stem growth, cell wall H2DCF fluorescence intensity was measured in xylem vessel elements and interfascicular fibers for the eight developmental timepoints of Col-0 stems (Fig. 8C). Fluorescence intensity of xylem vessel elements was low during stage 1 (2–4 cm), but substantially increased to a maximum at 10 cm before decreasing to lower levels in more mature tissues. Interfascicular fibers had negligible fluorescence intensity in cell walls until stage 2 (8 cm), and this signal increased during stage 3. The H2DCF fluorescence intensity in the SCW of xylem vessel elements was significantly higher than fibers from 6 to 10 cm, but at the base of the stem the fluorescence intensity of fibers was significantly higher than that of xylem vessel elements. Therefore, H2DCF staining showed that xylem vessel elements had highest accumulation of ROS early in development and fibers showed higher accumulation of ROS later in development following SCW deposition.
DISCUSSION
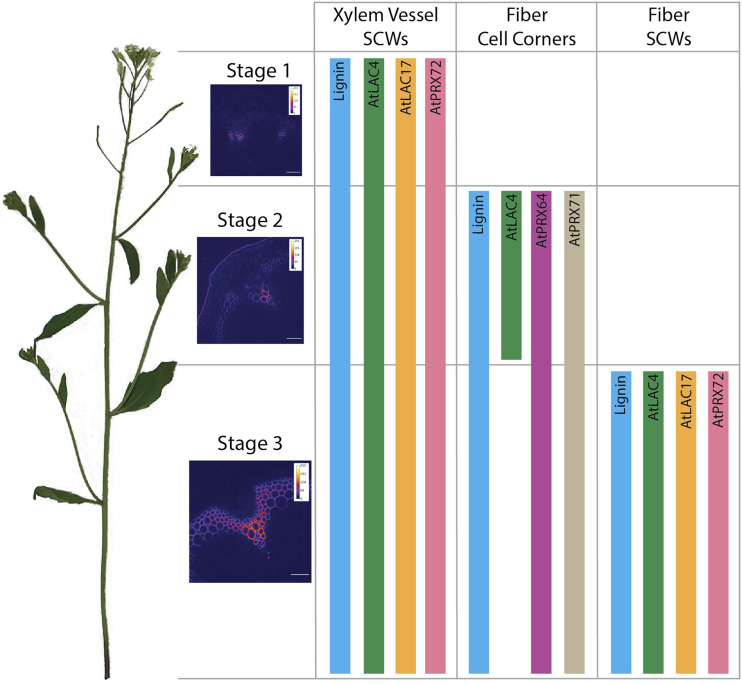
Before this work, the localization of a limited number of Arabidopsis oxidative enzymes involved in lignification was known (Lee et al., 2013; Schuetz et al., 2014; Yi Chou et al., 2018; Khandal et al., 2020). The goal of this work was twofold: first, to identify the localization of other putative lignin-associated LACs and PRXs, and second, to examine how the localization of these enzymes change throughout developmental lignification. Our results demonstrate that characteristic sets of LACs and PRXs colocalize within particular cell types and regions of the cell wall and that the set of LACs or PRXs in a cell wall region can be either stable or dynamic during development (Fig. 9). Overall, these data contribute to a refined model of SCW lignification, where individual LACs or PRXs contribute to lignification as part of a mixed set of oxidative enzymes embedded in specific polysaccharide environments.
Figure 9.
Summary of the endogenous localization patterns of tagged LACs and PRXs in the inflorescence stem of Arabidopsis. Stem development is divided into three stages, which show progressive lignification of SCWs of xylem vessel elements, cell corners of fibers, and SCWs of fibers. Representative heat maps of UV lignin autofluorescence depict the three stages. Scale bars = 50 µm. AtLAC4-mCherry, AtLAC17-mCherry, and AtPRX72-mCherry localize to SCWs of both xylem vessel elements and fibers. AtLAC4-mCherry transiently localizes to the cell corners of fibers during stage 2. AtPRX64-mCherry and AtPRX71-mCherry localize to the cell corners of fibers in stages 2 and 3.
Distinct Subsets of LACs and PRXs Localize to Specific Tissue Types and Regions of the Cell Wall during Development
The SCW set of AtLAC4, AtLAC17, and AtPRX72 were consistently observed in the thick SCW of both xylem vessel elements and interfascicular fibers (Figs. 2 and 4; Supplemental Fig. S5). Interestingly, AtPRX72-mCherry is the first and only identified PRX to localize to the thick SCW in Arabidopsis. This SCW localization is supported by mutant phenotyping, as prx72 mutants show a significant reduction in stem lignin and a slight irregular xylem phenotype (Herrero et al., 2013). In addition, AtPRX72 expression is increased in the lac4-2/lac11/lac17 triple mutants, implying a possible functional linkage to AtLAC4, AtLAC11, and AtLAC17 (Zhao et al., 2013). However, increased AtPRX72 expression in the lac4-2/lac11/lac17 triple mutants is not sufficient to rescue the lignin defect, indicating that LACs and PRXs can still have distinct roles in lignification.
The cell corner/middle lamella set of tagged AtPRX64 and AtPRX71 exclusively localized to cell corners of interfascicular fibers and appeared upon initial cell corner lignification (Fig. 3). Consistent with these results, Wi et al. (2005) documented a substantial increase in PRX immunolabeling in the middle lamella of alfalfa (Medicago sativa) fiber cells following initiation of lignification. The observation that tagged AtLAC4 was found transiently in the early cell corners of fibers and later exclusively in the SCW of mature tissues suggests a role in lignification of both regions of the cell wall during development. The short-lived nature of AtLAC4-mCherry cell corner localization compared with the stable localization of AtLAC4-mCherry in SCWs of mature tissues also highlights the importance of studying oxidative enzyme localization at different developmental stages.
Other LACs and PRXs, AtLAC10, AtPRX42, AtPRX52, and AtPRX71, localized to nonlignifying tissues, indicating their involvement in different biological processes (Fig. 5). The absence of AtPRX52-mCherry from lignifying cell types was unexpected, as prx52 mutants were reported to have a reduction in total stem lignin and a change in lignin composition (Fernández-Pérez et al., 2015b). AtPRX52 expression was also highly up-regulated in cells at regions of organ abscission, denoting that AtPRX52 may play a specialized role in creating the lignin honeycomb pattern observed during separation of plant organs (Lee et al., 2018). AtPRX42 is one of the most highly expressed PRXs in the Arabidopsis stem and localized to the primary cell walls of nonlignifying xylem parenchyma and phloem cells (Fig. 5; Supplemental Fig. S1). PRX activity in phloem tissues is well-documented in numerous species and is thought to be involved in response to biotic and abiotic stresses (Walz et al., 2002). Understanding where and when these oxidative enzymes are localized therefore provides new approaches for exploring their mutant phenotypes and functions.
Targeted Localization of Oxidative Enzymes in Cell Wall Domains
The localization patterns of specific LACs and PRXs in different tissue types and regions of the cell wall throughout stem development indicates that gene expression and secretion of these lignin-associated oxidative enzymes is highly regulated. Several mechanisms have been proposed for the localization of cell wall proteins, including direct binding to cell wall polysaccharides or proteins. For example, a protein scaffold containing AtPRX64, AtCASP1, and the NADPH oxidase AtRbohF is required for localized lignin polymerization in the Casparian strip of Arabidopsis roots (Lee et al., 2013). Another PRX, AtPRX36, was also shown to bind to specific motifs of demethylesterified pectin in primary cell walls of seed coats, providing a platform for restricted PRX activity in a given location (Francoz et al., 2019). It is therefore possible that different lignin-associated LACs and PRXs have specific binding domains within SCWs or cell corners, although this has not been studied in stem tissues.
Alternatively, mobility in the cell wall may impact localization, as proteins could diffuse to different regions of the cell wall following secretion. The mobility of enzymes can differ based on the structure of the protein as well as the composition and organization of the surrounding polysaccharide matrix. Fluorescence recovery after photobleaching experiments provide evidence that the SCW can impede protein movement, that is, AtLAC4-mCherry was mobile when ectopically expressed in the primary cell wall but was immobile in highly lignified SCWs (Yi Chou et al., 2018). The observation that tagged AtPRX71 and AtPRX72 both accumulated in their respective regions of the cell wall throughout progressive fiber lignification (Fig. 6) suggests that tagged AtPRX71 could diffuse through the lignified SCW to the cell corners, whereas AtPRX72 is restricted to the SCW.
Balancing Monolignol Availability and Oxidative Enzyme Properties to Facilitate Cell Wall Lignification
It is interesting to view the localization of lignin-associated PRXs and LACs through the lens of classic and recent studies utilizing tagged monolignols to track the stepwise deposition of lignin. Classic autoradiography of radioactive monolignols (Terashima and Fukushima, 1988), fluorescently tagged monolignol studies (Tobimatsu et al., 2013), click chemistry of monolignols (Pandey et al., 2016), and a new bioorthogonal labeling imaging sequential strategy (Lion et al., 2017) have demonstrated early and strong lignin deposition in the cell corners and middle lamella. This is consistent with the appearance of tagged AtLAC4, AtPRX64, and AtPRX71 in the cell corners during initial fiber lignification (Fig. 3), as well as the persistent localization of AtPRX64 and AtPRX71 in cell corners and middle lamella (Fig. 4). As lignification proceeds, the thick SCW is lignified, and this study identifies the set of oxidative enzymes (AtLAC4, AtLAC17, and AtPRX72) as the key players during this process.
How specific subdomains of lignin are laid down in different parts of the SCW, for example, enrichment of p-coumaryl monolignols to form H-lignin in the cell corners (Donaldson, 2001; Gierlinger, 2014), could be influenced by a combination of monolignol availability and/or the substrate preferences of the oxidative enzymes present. When different ratios of fluorescent monolignol analogs are applied to flax stems, the total incorporation reflects the monolignols available, although nonoverlapping cell wall domains preferentially incorporate one subunit (Lion et al., 2017). In Cleome hassleriama, seed coat lignin switches from coniferyl alcohol-rich G-lignin to a homopolymer of caffeyl alcohol, which is associated with predominant expression of genes involved in producing one type of monolignol over the other (Zhuo et al., 2019). Monolignol availability is clearly an important factor in determining lignin composition, but it is harder to assess the roles of the oxidative enzymes. There is limited information about Arabidopsis LAC substrate specificity, mainly conjectures from mutant analyses (Supplemental Tables S1–S3). Relatively few PRXs from Arabidopsis have been characterized, but a detailed study of substrate specificity of recombinant AtPRX2, AtPRX25, and AtPRX71 showed that they could oxidize coniferyl alcohol, sinapyl alcohol, and potentially the lignin polymer (Shigeto et al., 2014). However, the substrate preference from in vitro experiments do not necessarily inform on enzyme function within the complex cell wall matrix. Overall, these studies suggest that oxidative enzymes are relatively promiscuous, with the ability to oxidize the diverse phenolic substrates that they encounter. The precise lignin composition in cell wall microdomains is therefore likely determined by the combination of the properties of the set of oxidative enzymes anchored in that cell wall matrix and monolignol availability at critical developmental stages.
ROS Production and PRX Activity Correlates with Cell Wall Lignification during Stem Development
Despite the observed localization of tagged PRXs to lignifying cell types, the presence of a PRX does not necessarily reflect oxidative activity unless H2O2 is present. Both TMB histochemistry (H2O2-dependent enzymatic activity; Fig. 7) and the ROS-sensitive dye H2DCF (Fig. 8) demonstrated that production of apoplastic H2O2 correlates with tagged PRX localizations and lignin deposition. Remarkably, the addition of exogenous H2O2 demonstrated that PRXs are ubiquitous in all plant cell walls, but only PRXs in lignifying cell walls normally have access to H2O2. The universal presence of PRXs in cell walls was not unexpected, as PRXs have been implicated in response to abiotic and biotic stresses, cell elongation, cell stiffening, auxin metabolism, and lignification (Shigeto and Tsutsumi, 2016).
Analysis of H2DCF along stem development showed that xylem vessel elements have highest apoplastic ROS in the middle of the stem but lower amounts in mature tissues, which could correspond to programmed cell death of these cell types (Karlsson et al., 2005). Fibers, which are long-lived relative to xylem vessel elements, could take over the production and accumulation of ROS in later stages and act in combination with xylem parenchyma to produce H2O2 for post-mortem lignification of xylem vessel elements (Pesquet et al., 2013; Smith et al., 2013). These results suggest that lignifying cell types have different amounts of ROS, which may influence PRX oxidative activity. In cell walls with low apoplastic H2O2 concentrations, LAC activity could predominate over PRX activity as both LACs and PRXs are found within the same regions of lignified cell walls.
Conclusion
Altogether, these results indicate that both LACs and PRXs are involved in Arabidopsis stem lignification and that the location of oxidative enzyme and availability of substrates, such as H2O2, dictate lignin deposition spatially and temporally throughout growth. Restricted production of apoplastic ROS in lignifying tissues could control PRX activity throughout development, whereas regulation over LAC oxidative activity may be more dependent on targeted secretion to certain cell types and regions of the cell wall. In addition, these results support the previously detailed view (Zhao et al., 2013) that LACs and PRXs are nonredundant due to differential and specific localization patterns.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) seeds were surface-sterilized using chlorine gas and plated on germination media (1× Murashige-Skoog, 1% [w/v] Suc, 1X Gamborg’s Vitamin mix, 0.05% [w/v] MES, at pH 5.8) and 0.75% (w/v) agar. Seeds were kept for 3 to 4 d at 4°C in the dark, then grown at 21°C under 24-h light for 7 d before being transferred to soil (Sungro Sunshine Mix 4). Seedlings on soil were grown at 21°C under 18-h light cycles. Previously characterized prLAC4::LAC4-mCherry, prLAC17::LAC17-mCherry, and prUBQ10::sec-mCherry seeds were evaluated (Schuetz et al., 2014; Yi Chou et al., 2018). prPRX64::PRX64-mCherry seeds (Lee et al., 2013) were provided by Dr. Niko Geldner (University of Lausanne, Switzerland).
Molecular Biology
The open reading frame and ∼2 kb of the upstream putative promoter sequence for AtLAC10 (At5g01190), AtPRX42 (At4g21960), AtPRX52 (At5g05340), AtPRX71 (At5g64120), and AtPRX72 (At5g66390) was amplified from genomic DNA using KAPA HiFi Hotstart ReadyMix (Kapa Biosystem). Invitrogen Gateway attB adaptor sequences with a modified stop codon were added to gene products for addition of a C-terminal fusion protein. Promoter and gene fragments were cloned into a modified pMDC111-mCherry vector (Schuetz et al., 2014) and transformed into Agrobacterium tumefaciens strain GV3101. Arabidopsis Columbia-0 (Col-0) plants were transformed using the floral dip method. Results were consistent for at least three independent lines per construct. T2-generation stems were analyzed for LAC/PRX localizations.
Stem Sampling Protocol
Arabidopsis inflorescence stems were sampled at a height of 16 to 18 cm, approximately 5 weeks after germination. Stems were divided into 2-cm pieces (starting measurement from the apical tip of the stem) for a total of eight developmental timepoints. Stem hand cross-sections were cut using a double-edged razor. For mCherry imaging, stem cross-sections were fixed in a solution of 4% (w/v) paraformaldehyde buffered in 1× PME (0.5 m PIPES pH 7.4, 0.05 m MgSO4, 0.05 m EGTA at pH 7.5) for 1 h at room temperature, rinsed three times in 1× TBST (10 mm Tris pH 7.0, 0.25 m NaCl, 0.1% [w/v] Tween20), and stored in 1× TBST at 4°C for up to a month.
To demonstrate functionality of prLAC17::LAC17-mCherry, basal cross-sections from mature stems were stained with 0.01% (w/v) toluidine blue solution (Ted Pella Inc) for 2 to 3 minutes, rinsed with water, and imaged with the Leica DMR microscope. For phloroglucinol staining, stem cross-sections were treated with 10% (w/v) phloroglucinol (Sigma Aldrich) in ethanol and two drops of concentrated hydrochloric acid. Samples were covered with a coverslip and allowed to react for an additional 5 minutes before being imaged.
TMB Histochemistry
The 16- to 18-cm tall Col-0 stems were sampled as per developmental analyses and incubated in 100 μm TMB (Acros Organics) for 1 h in the dark, with gentle shaking. For TMB + H2O2 analyses, cross-sections were incubated in 100 μm TMB and 100 μm H2O2 for 1 h in the dark, with gentle shaking. PRX enzymatic activity could be visualized by formation of a blue precipitate. Stem sections were rinsed twice in water, imaged on the Leica DMR compound microscope, and repeated for five biological replicates.
To test for protein function, stems were boiled in a 100°C water bath for 10 minutes, then incubated in TMB, as above. To inhibit PRXs, stems were pretreated in 100 mm SHAM (Sigma-Aldrich) for 1 h with shaking, then incubated in TMB. To remove endogenous H2O2, stems were incubated in 300 U mL−1 bovine liver catalase (Sigma) for 2 h with shaking, then incubated with TMB. Control experiments were performed with at least three biological replicates with consistent results.
H2DCF Staining
The 16- to 18-cm tall Col-0 stems were sectioned as per developmental analysis and incubated in water for ∼24 h at room temperature to reduce the wounding response. A 10-mm stock solution of H2DCF (Thermo Fisher Scientific) was prepared in dimethyl sulfoxide and stored at −20°C in the dark. Stem sections were incubated in a working solution of 50 μm H2DCF in 50 mm NaH2PO4 (pH 6.0) for 15 minutes in the dark. Staining solution was made fresh the day of use.
Controls for H2DCF staining were performed on stem cross-sections from the base of 17-cm-tall Col-0 plants. To test for ROS-induced wounding response, stem cross-sections were immediately placed in H2DCF solution after sectioning and imaged. All other control experiments were first incubated in water for 24 h to reduce the wounding response. To inhibit PRXs, cross-sections were pretreated in 100 mm SHAM for 1 h while shaking, then incubated in H2DCF as above. To inhibit NADPH oxidases, cross-sections were pretreated in 1 mm DPI (Sigma-Aldrich) for 2 h while shaking, then incubated in H2DCF. To remove endogenous H2O2, cross-sections were incubated in 300 U mL−1 bovine liver catalase for 2 h while shaking, then incubated in H2DCF. Data is representative of three biological replicates for four independent experiments.
Microscopy
Histochemistry treatments of Arabidopsis stem cross-sections were imaged using a Leica DMR compound microscope. Images were taken using a Canon EOS Rebel T5 camera and EOSUtility software. All images were processed using FIJI.
Fluorescence confocal microscopy was conducted on a Perkin-Elmer UltraView VoX spinning disk confocal mounted on a Leica DM16000 inverted microscope with a Hamamatsu 9100-02 CCD camera. The 405/UV filter (excitation 405 nm, emission 440–510 nm) was used to image lignin autofluorescence, 488/GFP filter (excitation 488 nm, emission 525 nm) to image H2DCF, and 561/red fluorescent protein filter (excitation 561 nm, emission 595–625 nm) to image mCherry-tagged enzymes. Images were processed using Volocity image analysis software (Improvision) and FIJI. Heatmaps to visualize fluorescence intensity were generated on FIJI by applying a look up table filter to a 32-bit image.
Corrected Fluorescence
Images were taken on the Perkin-Elmer UltraView VoX spinning disk confocal mounted on a Leica DM16000 inverted microscope with a Hamamatsu 9100-02 charge-coupled device camera. Consistent microscope conditions (exposure time, sensitivity, and beam intensity) were used to image all developmental stages and biological replicates in the same plant line and were optimized to minimize pixilated regions. Corrected fluorescence minus background signal was calculated from images using FIJI according to the methodology of Yi Chou et al. (2018). Areas of interest (SCWs versus cell corners) were drawn by hand. To test for statistically significant differences between developmental stages or genotypes, a Student’s t-test or ANOVA followed by a Tukey-Kramer post hoc test was performed.
Bioinformatics and Phylogenetic Reconstruction
Tissue expression data for LACs and PRXs was collected from the BAR ePlant browser (bar.utoronto.ca/eplant; Waese et al., 2017) and Genevestigator (www.genevestigator.ethz.ch; Hruz et al., 2008). Lists of significantly coexpressed genes from Genevestigator were annotated using TAIR. Amino-acid sequences for 17 LACs (Pourcel et al., 2005; Cai et al., 2006) and 73 Class-III PRXs from Arabidopsis were downloaded from TAIR and aligned using MUSCLE. Maximum likelihood phylogenetic trees were reconstructed on MEGAX, with 1,000 bootstrap replicates. The LAC phylogenetic tree used a Trametes versicolor LAC as an outgroup (TvLAC; GenBank: AAL07440.1), whereas the PRX phylogenetic tree is unrooted.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this article are as follows: At2g38080 for AtLAC4, At5g01190 for AtLAC10, At5g03260 for AtLAC11, At5g05390 for AtLAC12, At5g60020 for AtLAC17, At4g21960 for AtPRX42, At5g05340 for AtPRX52, At5g64120 for AtPRX71, and At5g66390 for AtPRX72.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. LACs and PRXs are differentially expressed in the inflorescence stem of Arabidopsis.
Supplemental Figure S2. Putative lignin-associated LACs are identified through bioinformatics approaches.
Supplemental Figure S3. Putative lignin-associated PRXs are identified through bioinformatics approaches.
Supplemental Figure S4. Lignin is deposited to different cell types and regions of the cell wall during development of the Arabidopsis stem.
Supplemental Figure S5. AtLAC4-mCherry, AtLAC17-mCherry, and AtPRX72-mCherry localize to the SCW of xylem vessel elements during Stage 2.
Supplemental Figure S6. LACs and PRXs localize to both the SCW and cell corners of xylem vessel elements and fibers during Stage 3.
Supplemental Figure S7. AtLAC17-mCherry complements the irregular xylem phenotype of lac4-2/lac17 mutants.
Supplemental Figure S8. prUBQ10::sec-mCherry localizes to the cell corners and primary cell walls of all cell types at the base of 17 cm tall Arabidopsis stems.
Supplemental Figure S9. H2DCF fluorescence signal in Arabidopsis stems is dependent on wounding, PRXs, NADPH oxidases, and H2O2.
Supplemental Table S1. Arabidopsis LACCASE single T-DNA insertion line mutants and associated lignin-related phenotypes.
Supplemental Table S2. Arabidopsis PEROXIDASE single mutants and associated lignin-related phenotypes.
Supplemental Table S3. Arabidopsis LACCASE and PEROXIDASE double and triple mutants and associated lignin-related phenotypes.
Acknowledgments
We thank Niko Geldner for providing the prPRX64::PRX64-mCherry plant line. The technical support of the UBC Bioimaging Facility, especially Kevin Hodgson, is also gratefully acknowledged.
Footnotes
This work was supported by a Canadian Natural Sciences and Engineering Research Council Discovery Grant (to A.L.S.) and CGS-M postgraduate scholarship (to N.H.).
Articles can be viewed without a subscription.
References
- Berthet S, Demont-Caulet N, Pollet B, Bidzinski P, Cézard L, Le Bris P, Borrega N, Hervé J, Blondet E, Balzergue S, et al. (2011) Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell 23: 1124–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet S, Thevenin J, Baratiny D, et al. (2012) Role of plant laccases in lignin polymerization In Jouanin L, and Lapierre C, eds, Lignins: Biosynthesis, Biodegradation and Bioengineering. Academic Press, London, pp 145–172 [Google Scholar]
- Boerjan W, Ralph J, Baucher M(2003) Lignin biosynthesis. Annu Rev Plant Biol 54: 519–546 [DOI] [PubMed] [Google Scholar]
- Brown DM, Zeef LAH, Ellis J, Goodacre R, Turner SR(2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Davis EJ, Ballif J, Liang M, Bushman E, Haroldsen V, Torabinejad J, Wu Y(2006) Mutant identification and characterization of the laccase gene family in Arabidopsis. J Exp Bot 57: 2563–2569 [DOI] [PubMed] [Google Scholar]
- Cosio C, Ranocha P, Francoz E, Burlat V, Zheng Y, Perry SE, Ripoll JJ, Yanofsky M, Dunand C(2017) The class III peroxidase PRX17 is a direct target of the MADS-box transcription factor AGAMOUS-LIKE15 (AGL15) and participates in lignified tissue formation. New Phytol 213: 250–263 [DOI] [PubMed] [Google Scholar]
- Donaldson LA.(2001) Lignification and lignin topochemistry - an ultrastructural view. Phytochemistry 57: 859–873 [DOI] [PubMed] [Google Scholar]
- Ehlting J, Mattheus N, Aeschliman DS, Li E, Hamberger B, Cullis IF, Zhuang J, Kaneda M, Mansfield SD, Samuels L, et al. (2005) Global transcript profiling of primary stems from Arabidopsis thaliana identifies candidate genes for missing links in lignin biosynthesis and transcriptional regulators of fiber differentiation. Plant J 42: 618–640 [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez F, Pomar F, Pedreño MA, Novo-Uzal E(2015a) Suppression of Arabidopsis peroxidase 72 alters cell wall and phenylpropanoid metabolism. Plant Sci 239: 192–199 [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez F, Pomar F, Pedreño MA, Novo-Uzal E(2015b) The suppression of AtPrx52 affects fibers but not xylem lignification in Arabidopsis by altering the proportion of syringyl units. Physiol Plant 154: 395–406 [DOI] [PubMed] [Google Scholar]
- Fernández-Pérez F, Vivar T, Pomar F, Pedreño MA, Novo-Uzal E(2015) Peroxidase 4 is involved in syringyl lignin formation in Arabidopsis thaliana. J Plant Physiol 175: 86–94 [DOI] [PubMed] [Google Scholar]
- Francoz E, Ranocha P, Le Ru A, Martinez Y, Fourquaux I, Jauneau A, Dunand C, Burlat V(2019) Pectin demethylesterification generates platform that anchors peroxidases to remodel plant cell wall domains. Dev Cell 48: 261–276.e8 [DOI] [PubMed] [Google Scholar]
- Gierlinger N.(2014) Revealing changes in molecular composition of plant cell walls on the micron-level by Raman mapping and vertex component analysis (VCA). Front Plant Sci 5: 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall H, Ellis B(2013) Transcriptional programming during cell wall maturation in the expanding Arabidopsis stem. BMC Plant Biol 13: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero J, Fernández-Pérez F, Yebra T, Novo-Uzal E, Pomar F, Pedreño MÁ, Cuello J, Guéra A, Esteban-Carrasco A, et al. (2013) Bioinformatic and functional characterization of the basic peroxidase 72 from Arabidopsis thaliana involved in lignin biosynthesis. Planta 237: 1599–1612 [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P(2008) Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinforma 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman B, Darley-Usmar V, Davies KJA, Dennery PA, Forman HJ, Grisham MB, Mann GE, Moore K, Roberts LJ II, Ischiropoulos H(2012) Measuring reactive oxygen and nitrogen species with fluorescent probes: challenges and limitations. Free Radic Biol Med 52: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson M, Melzer M, Prokhorenko I, Johansson T, Wingsle G(2005) Hydrogen peroxide and expression of hipI-superoxide dismutase are associated with the development of secondary cell walls in Zinnia elegans. J Exp Bot 56: 2085–2093 [DOI] [PubMed] [Google Scholar]
- Khandal H, Singh AP, Chattopadhyay D(2020) The MicroRNA397b-LACCASE2 module regulates root lignification under water and phosphate deficiency. Plant Physiol 182: 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitinen T, Morreel K, Delhomme N, Gauthier A, Schiffthaler B, Nickolov K, Brader G, Lim KJ, Teeri TH, Street NR, et al. (2017) A key role for apoplastic H2O2 in Norway spruce phenolic metabolism. Plant Physiol 174: 1449–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Rubio MC, Alassimone J, Geldner N(2013) A mechanism for localized lignin deposition in the endodermis. Cell 153: 402–412 [DOI] [PubMed] [Google Scholar]
- Lee Y, Yoon TH, Lee J, Jeon SY, Lee JH, Lee MK, Chen H, Yun J, Oh SY, Wen X, et al. (2018) A lignin molecular brace controls precision processing of cell walls critical for surface integrity in Arabidopsis. Cell 173: 1468–1480.e9 [DOI] [PubMed] [Google Scholar]
- Lion C, Simon C, Huss B, Blervacq AS, Tirot L, Toybou D, Spriet C, Slomianny C, Guerardel Y, Hawkins S, et al. (2017) BLISS: A bioorthogonal dual-labeling strategy unravel lignification dynamics in plants. Cell Chem Biol 24: 326–338 [DOI] [PubMed] [Google Scholar]
- Pandey JL, Kiemle SN, Richard TL, Zhu Y, Cosgrove DJ, Anderson CT(2016) Investigating biochemical and developmental dependencies of lignification with a click-compatible monolignol analog in Arabidopsis thaliana stems. Front Plant Sci 7: 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquet E, Zhang B, Gorzsás A, Puhakainen T, Serk H, Escamez S, Barbier O, Gerber L, Courtois-Moreau C, Alatalo E, et al. (2013) Non-cell-autonomous postmortem lignification of tracheary elements in Zinnia elegans. Plant Cell 25: 1314–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcel L, Routaboul JM, Kerhoas L, Caboche M, Lepiniec L, Debeaujon I(2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17: 2966–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ros-Barceló A.(1998) The generation of H2O2 in the xylem of Zinnia elegans is mediated by an NADPH-oxidase-like enzyme. Planta 207: 207–216 [Google Scholar]
- Sarkanen K, Ludwig C(1971) Lignins: Occurrence, Formation, Structure and Reactions. Wiley-Interscience, New York [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU(2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Schuetz M, Benske A, Smith RA, Watanabe Y, Tobimatsu Y, Ralph J, Demura T, Ellis B, Samuels AL(2014) Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiol 166: 798–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeto J, Itoh Y, Hirao S, Ohira K, Fujita K, Tsutsumi Y(2015) Simultaneously disrupting AtPrx2, AtPrx25 and AtPrx71 alters lignin content and structure in Arabidopsis stem. J Integr Plant Biol 57: 349–356 [DOI] [PubMed] [Google Scholar]
- Shigeto J, Nagano M, Fujita K, Tsutsumi Y(2014) Catalytic profile of Arabidopsis peroxidases, AtPrx-2, 25 and 71, contributing to stem lignification. PLoS One 9: e105332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeto J, Tsutsumi Y(2016) Diverse functions and reactions of class III peroxidases. New Phytol 209: 1395–1402 [DOI] [PubMed] [Google Scholar]
- Shigeto J, Kiyonaga Y, Fujita K, Kondo R, Tsutsumi Y(2013) Putative cationic cell-wall-bound peroxidase homologues in Arabidopsis, AtPrx2, AtPrx25, and AtPrx71, are involved in lignification. J Agric Food Chem 61: 3781–3788 [DOI] [PubMed] [Google Scholar]
- Smith RA, Schuetz M, Roach M, Mansfield SD, Ellis B, Samuels L(2013) Neighboring parenchyma cells contribute to Arabidopsis xylem lignification, while lignification of interfascicular fibers is cell autonomous. Plant Cell 25: 3988–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterjiades R, Dean JFD, Gamble G, Himmelsbach DS, Eriksson KEL(1993) Extracellular laccases and peroxidases from sycamore maple (Acer pseudoplatanus) cell-suspension cultures: Reactions with monolignols and lignin model compounds. Planta 190: 75–87 [Google Scholar]
- Terashima N, Fukushima K(1988) Heterogeneity in formation of lignin-XI: An autoradiographic study of the heterogeneous formation and structure of pine lignin. Wood Sci Technol 22: 259–270 [Google Scholar]
- Tobimatsu Y, Wagner A, Donaldson L, Mitra P, Niculaes C, Dima O, Kim JI, Anderson N, Loque D, Boerjan W, et al. (2013) Visualization of plant cell wall lignification using fluorescence-tagged monolignols. Plant J 76: 357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL(2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Turlapati PV, Kim K-W, Davin LB, Lewis NG(2011) The laccase multigene family in Arabidopsis thaliana: Towards addressing the mystery of their gene function(s). Planta 233: 439–470 [DOI] [PubMed] [Google Scholar]
- Waese J, Fan J, Pasha A, Yu H, Fucile G, Shi R, Cumming M, Kelley LA, Sternberg MJ, Krishnakumar V, et al. (2017) ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 29: 1806–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J(2002) Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J 31: 189–197 [DOI] [PubMed] [Google Scholar]
- Weng JK, Chapple C(2010) The origin and evolution of lignin biosynthesis. New Phytol 187: 273–285 [DOI] [PubMed] [Google Scholar]
- Wi SG, Singh AP, Lee KH, Kim YS(2005) The pattern of distribution of pectin, peroxidase and lignin in the middle lamella of secondary xylem fibres in alfalfa (Medicago sativa). Ann Bot 95: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Chou E, Schuetz M, Hoffmann N, Watanabe Y, Sibout R, Samuels AL(2018) Distribution, mobility, and anchoring of lignin-related oxidative enzymes in Arabidopsis secondary cell walls. J Exp Bot 69: 1849–1859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Nakashima J, Chen F, Yin Y, Fu C, Yun J, Shao H, Wang X, Wang ZY, Dixon RA(2013) Laccase is necessary and nonredundant with peroxidase for lignin polymerization during vascular development in Arabidopsis. Plant Cell 25: 3976–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo C, Rao X, Azad R, Pandey R, Xiao X, Harkelroad A, Wang X, Chen F, Dixon RA(2019) Enzymatic basis for C-lignin monomer biosynthesis in the seed coat of Cleome hassleriana. Plant J 99: 506–520 [DOI] [PubMed] [Google Scholar]