Realization of the PeachRefPop, the international multisite reference collection in peach, provides an invaluable tool for scientific studies in perennial species.
Abstract
Plants have evolved a range of adaptive mechanisms that adjust their development and physiology to variable external conditions, particularly in perennial species subjected to long-term interplay with the environment. Exploiting the allelic diversity within available germplasm and leveraging the knowledge of the mechanisms regulating genotype interaction with the environment are crucial to address climatic challenges and assist the breeding of novel cultivars with improved resilience. The development of multisite collections is of utmost importance for the conservation and utilization of genetic materials and will greatly facilitate the dissection of genotype-by-environment interaction. Such resources are still lacking for perennial trees, especially with the intrinsic difficulties of successful propagation, material exchange, and living collection maintenance. This work describes the concept, design, and realization of the first multisite peach (Prunus persica) reference collection (PeachRefPop) located across different European countries and sharing the same experimental design. Other than an invaluable tool for scientific studies in perennial species, PeachRefPop provides a milestone in an international collaborative project for the conservation and exploitation of European peach germplasm resources and, ultimately, as a true heritage for future generations.
Since the Roman garden hortus, fruit tree orchards have represented distinctive features of the Mediterranean rural landscape, a synthesis of the interaction among genotype, environment, and human customs (Biasi et al., 2012). The diversity of pedo-climatic conditions and production systems, along with the plasticity of the genotype and human traditions, has shaped the selection of a multitude of local cultivars. These materials represent a cultural and genetic heritage of generations of farmers and a common good to preserve for present and future generations.
Plants have evolved a range of adaptive mechanisms that adjust their development and physiology to variable external conditions, particularly in perennial species subjected to a long-term environmental exposure and interaction. Climate changes are impacting cultivation environments, raising the need for more resilient cultivars able to maintain performance across variable (and often unpredictable) weather conditions (Varshney et al., 2011; Luedeling, 2012; Ramírez and Kallarackal, 2015). Also, increasing the sustainability of fruit production (particularly in terms of resource demands and disease management) requires leveraging knowledge of the interactions between plants, soil, and environmental factors and how they affect productivity and end-product quality (Coakley et al., 1999; Singh et al., 2013; Parajuli et al., 2019).
Peach (Prunus persica) originated in China (Li et al., 2019), later reaching Persia, the Mediterranean Basin, Europe, and the Americas; it is now the third most cultivated fruit tree species in temperate regions. Beside its importance as a crop, peach is a recognized model for genetic and genomic studies in fruit trees, representing the ideal system for addressing two main challenges in fruit tree breeding. (1) Understanding and harnessing the allelic diversity within available genepools. Noteworthy for peach, the intercompatibility with related species of the Amygdalus subgenus (almond [Prunus dulcis], Prunus davidiana, Prunus kansuensis, etc.) has long been considered a source of natural variability, particularly for the introgression of disease resistance (Gradziel, 2002; Foulongne et al., 2003). However, interspecific hybrids have had poor applicability in current breeding programs (Cirilli et al., 2017), although new genomics-based strategies could change this trend (Serra et al., 2016). Conversely, landraces and local ecotypes could be a source of resilience traits more straightforward to introgress, making their preservation and exploitation a suitable strategy for dealing with the changing climatic conditions. (2) Systematic dissection of genotype-by-environment and/or genotype-by-management interactions as primary sources of variability for several important quantitative traits (Myles, 2013; Chagné et al., 2014). This is a critical point for genetic analyses of complex traits, such as genome-wide association studies (GWAS), where germplasm collections are characterized to identify quantitative trait loci (QTLs) across different environments, or genome-wide selection (GS), used to predict genomic estimated breeding values.
The comprehension of genetic, epigenetic, and physiological mechanisms as well as the estimation of genotype-by-environment and/or genotype-by-management effects requires the development of multisite replicated collections and ad hoc experimental designs. The availability of such types of resources is rapidly growing in annual species, while it has not yet been implemented in perennial fruit trees.
During the past century, peach orchard systems have changed dramatically following innovations in orchard design, training systems, and agronomic management (Corelli-Grappadelli and Marini, 2008), other than cultivar evolution. Noteworthy, the first reported modern orchard was a peach plantation established in Massa Lombarda (Ravenna, Italy) at the end of the 19th century using the white-fleshed local cultivar population cv Buco Incavato (Bellucci, 1908). In the last decades, considerable breeding efforts have assisted the intensification of cultivation techniques and the development of horticultural quality concepts with the introduction of novel, fit-for-purpose cultivars (Byrne et al., 2012). In Europe, peach has a long cultivation history, tracing back to the Ancient and Middle Ages and characterized by the isolation and propagation of chance seedlings operated by farmers and amateurs, through which each country set its own pool of locally adapted cultivars (Bassi and Monet, 2008). The paradigm shift to the modern controlled-crosses approach in early United States breeding programs has been the foundation of the dramatic varietal improvement of the last century, beginning with the introduction of seedling materials from China in the mid-19th century (e.g. cv Chinese Cling, progenitor of most modern cultivars; Faust and Timon, 1995; Byrne et al., 2012). The worldwide spread of improved United States materials, favored also by the limited activities in other countries, has resulted in a rapid replacement of landraces and local accessions, particularly in Europe. From the second half of the 20th century, however, novel programs started in several European countries, although they were mostly based on United States breeding stocks with a marginal role for local cultivated germplasm. This led to a consequent loss of many local cultivars, in parallel with a progressive narrowing of the genetic bases in modern cultivars (Aranzana et al., 2010; Verde et al., 2013).
As awareness of genetic erosion in modern plant breeding increased (Fu and Dong, 2015), the conservation and exploitation of genetic resources has become a fundamental aspect in crop breeding (Ford-Lloyd and Jackson, 1986). Considerable efforts have been made in the collection and characterization of many plant germplasms (including fruit tree species), along with the development of approaches for their effective management and utilization (Gepts, 2006). The concept of core collection, a subset of a germplasm collection of a species that captures most of the genetic diversity while reducing redundancy, has represented an ideal solution for reducing costs and increasing the efficiency of conservation programs (Frankel and Brown, 1984). Several allocation methods have been developed for selecting core collections, attempting to maximize allelic richness or allele coverage (MSTRAT, PowerCore, and GenoCore), minimize or maximize genetic distance (GDOpt and SimEli), or simultaneously accommodating for multiple criteria (Core Hunter; Gouesnard et al., 2001; Kim et al., 2007; Thachuk et al., 2009; Odong et al., 2011; Krishnan et al., 2014). However, the effectiveness of the sampling strategies varied depending on the objective of the core collection, the statistical approach for its definition, and the measures for evaluating its quality (Odong et al., 2013). Furthermore, beyond statistical considerations, other aspects are often considered by the institutions hosting the collection, such as historical and socioeconomic importance, relevance for breeding activities, popularity among growers and consumers, and distinctive phenotypic characteristics.
In peach, the absence of wild or feral populations makes ex situ collections the main valuable reservoirs of allelic variability for many traits not yet exploited in current breeding programs. Remarkable progress has been achieved in the phenotypic and genotypic characterization of peach genetic resources (Badenes et al., 2015; Cirilli et al., 2018; Yu et al., 2018), taking advantage of genome sequencing and the development of cutting-edge genotyping tools (Verde et al., 2012, 2017; Aranzana et al., 2019). In the framework of the European collaborative project FruitBreedomics (Laurens et al., 2018), a coordinated characterization of peach collections has been accomplished across relevant European repositories (Micheletti et al., 2015; Hernández Mora et al., 2017), promoting increased utilization of resources and encouraging the sharing of conservation responsibilities. For example, the Prunus Working Group within the Fruit Network in the European Cooperative Program on Plant Genetic Resources is dealing with Prunus spp., including peach (Benediková and Giovannini, 2013). Nevertheless, long-term maintenance of collections remains particularly challenging due to intrinsic vulnerabilities (e.g. direct exposure to environmental variables and pathogens) and costs for in vivo maintenance through vegetative propagation to preserve the original genotypes. Moreover, compliance to phytosanitary requirements hampers the sharing of resources among institutions, each having its own stock of materials, resulting in redundancies or risk of loss for unique accessions.
This article describes the concept, design, and realization of the first multisite peach reference collection, PeachRefPop (PRP), across five locations in three European countries (Italy, Spain, and Greece). Other than an invaluable tool for scientific studies, the PRP provides a milestone of an international collaborative project for the conservation and exploitation of European peach germplasm resources and, ultimately, as a true heritage for future generations.
RESULTS
Criteria for the Construction of a Reference Panel of Peach Accessions and Seedlings
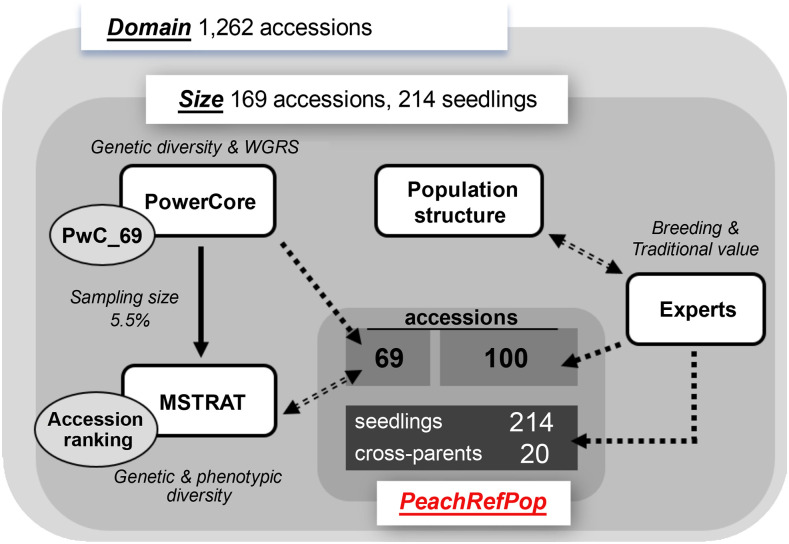
The PRP collection was built with the aim of selecting a reduced germplasm pool, reflecting the original genetic and phenotypic diversity (Fig. 1) and the cultural and socioeconomic value of peach cultivation, for its exploitation in future breeding programs. A four-step procedure was followed (exemplified in Fig. 2): (1) definition of the PRP domain; (2) establishment of PRP size; (3) identification of the selection criteria; and (4) choice and allocation of the entries.
Figure 1.
Overview of the range of phenotypic diversity in the PRP. Images are as follows: columnar and standard tree growth habit (top left and right images, respectively); heart-shaped, round, and flat fruit (top and third rows); range of fruit flesh, skin color, and overcolor (second and bottom rows); variation in flower morphology and color (third and fourth rows); and fruit size variation (fourth row, first and last images).
Figure 2.
Graphical summary of the overall scheme followed for selecting the PRP collection. From the starting panel of 1,262 accessions, 169 accessions were selected combining two sets: 69 accessions extracted from genetic and phenotypic diversity analyses and taking into account the availability of whole-genome resequencing (WGRS) data; and 100 accessions selected by an empirical strategy from an experts panel considering breeding and traditional value along with genetic structure. These were supplemented with 214 seedlings from crossing populations of scientific importance and their respective 20 parents. The total number of entries in the PRP amounts to 403.
Definition of the PRP Domain
To build a resource representing peach diversity and breeding history, the starting point was the genetic material characterized in the framework of the FP7 European project FruitBreedomics in a coordinated effort involving different universities and research institutions across Europe and China. A total of 1,580 Prunus spp. accessions (comprising P. persica and its hybrids with P. davidiana and almond) were phenotyped and genotyped with the 9K single-nucleotide polymorphism (SNP) array, as previously described (Micheletti et al., 2015). The inclusion of only peach (including P. ferganensis; Verde et al., 2012) among all the available Prunus spp. accessions was the leading concept behind the definition of the PRP reference collection. Indeed, as a consequence of many factors (genetic diversity, evolution history, mating system, geographical distribution, etc.), sampling strategies for the inclusion of wild relatives (e.g. species of the Amygdalus subgenus) may substantially differ from those for a cultivated species (e.g. peach; Brown and Marshall, 1995). Moreover, to avoid limitations on the exchange of plant material, the domain was restricted to European repositories. Based on these criteria, the starting panel for building the PRP amounted to a total of 1,262 P. persica accessions (FB_1262). Besides accessions, seedlings from controlled crosses also represent a valuable source of informative materials for both genetic analysis and breeding (or prebreeding) activities. For this reason, 1,467 individuals from 18 progeny and their parents (including an interspecific cross with a P. davidiana accession), also analyzed during the FruitBreedomics project (Hernández Mora et al., 2017), were considered in the construction process.
Establishment of the PRP Size
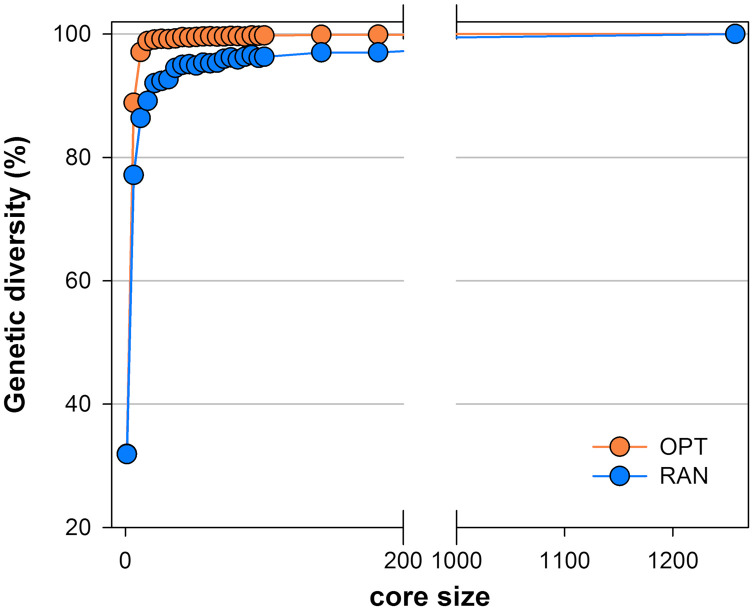
The definition of the size is one of the most critical decisions for the establishment of a reference population. For fruit tree crops, the costs of in vivo maintenance are particularly onerous and, together with long-term space availability in the field, the main limiting factor of running a germplasm collection. In the perspective of analyzing the interactions between genotype and environment and/or management practices, or performing genetic studies such as GWAS and GS, an adequate panel size and experimental design are key factors for the power and reliability of statistical analyses. On the other hand, for agrobiodiversity conservation purposes, the least number of accessions to include in a core set depends on the level of genetic repetitiveness present in the original germplasm pool. The first step toward the establishment of the PRP size was the assessment of the allelic richness and redundancy observed at marker loci. Two series of core collections of incremental size were generated, one based on the genetic diversity (maximization method) the other through random sampling. The maximization procedure (M strategy by Schoen and Brown [1995]) is based on the sampling of the total allelic diversity observed at marker loci in the least number of entries. By plotting the genetic diversity measured over the core size, a convex curve was obtained, indicating the presence of redundancy across the European peach germplasm collection. The inflection point, corresponding to a plateau in the increase of diversity, was observed at the level of core 26. At this core size, 99.9% of the total genetic diversity was captured in the core obtained with the M method in comparison with 93.5% with random sampling (Fig. 3). The outperformance of the optimized versus the random selection was observed across all the core sizes, indicating that the maximization strategy was more efficient and was preferred for conservation purposes in our germplasm.
Figure 3.
Assessment of allelic redundancy observed at marker loci in the starting panel (FB_1262). Core collections of incremental size were generated, based on the maximization (OPT) and random sampling (RAN) methods in MSTRAT software, using a set of 445 SNPs. Data points represent averaged values over five independent repetitions for each size.
According to some recent works in peach (for review, see Aranzana et al., 2019), about 100 to 150 unrelated accessions usually provide an adequate resolution for identifying major loci or developing prediction models. In light of all the above premises, an ideal target number of 400 entries was deemed adequate for allocating a minimum of 150 accessions and a maximum of 250 seedlings from progeny (including the parents) based on the outputs of the selection criteria.
Identification of the Selection Criteria
In spite of the genetic redundancy observed and excluding the rare cases of synonymy, the vast majority of the accessions are not overlapped across the various collections, being conserved for a multitude of reasons and purposes, including scientific research, agrobiodiversity preservation, or support to breeding activities. To reconcile these reasons with the aim of creating a feasible, usable, and multipurpose reference collection to be shared among European institutions, a mixed approach was considered for selecting the accessions. A subset of entries was sampled using an analytical strategy, based on the criteria of maximizing genetic (and phenotypic) diversity, also taking into account the availability of whole-genome resequencing data; the remaining entries were selected using an empirical strategy, leveraging the knowledge of an experts panel (e.g. breeders, experienced scientists, and curators of each repository) and considering the traditional and historical value at national and/or regional levels, the relevance for breeders, growers, and consumers, taking into account agronomical or pomological characteristics. Moreover, to maintain a balanced representation of the genetic structure of the whole collection, the empirical selection of accessions was partially supported by information on population structure (structure and principal component analysis [PCA] data are available from Micheletti et al. [2015]). Complementing the choice of accessions, seedlings were selected based on the availability of detailed genotypic and/or phenotypic information, genetic background, scientific relevance, and, above all, priority traits for breeding.
Choice, Evaluation, and Description of the PRP Accessions
Capturing the maximum amount of genetic diversity present in the entire collection while reducing redundancy was the primary driver for sampling the first PRP subset (the core set). For this purpose, the advanced M method, implemented in the software PowerCore (Kim et al., 2007) through a modified heuristic algorithm, was used to select a core from the initial panel of accessions, based on a set of 3,894 filtered SNPs previously described by Micheletti et al. (2015). After superimposing 17 accessions with available whole-genome resequencing data, an ideal core of 69 accessions (PwC_69) was extracted, representing a sampling size of 5.5% (Supplemental Table S1). Considering the many variables that could affect the actual availability of materials for grafting, a flexible approach was further developed to rank each accession of the whole panel based on genotypic and phenotypic diversity. Four different sets made up of 100 cores of 70 entries each were constructed with MSTRAT by setting different combinations of genotypic data (nine subsets of SNPs extracted approximately every 1.8 Mb to avoid linkage between them) and phenotypic data (seven qualitative and 10 quantitative traits, following transformation of the latter into categories; Supplemental Table S2). Accessions were ranked in groups according to the average frequency of inclusion across the four sets (Supplemental Table S3). Combining the core population extracted by PowerCore with the MSTRAT ranking list resulted in a shortlist of 69 accessions (41 and 28, respectively, indicated as Core_69), ensuring the inclusion of the maximum possible level of genetic diversity. For the completion of the final PRP_X panel, the remaining 100 accessions (Priority_100) were empirically selected by experts, following the above-specified criteria.
Estimates of genetic diversity were used to compare the starting panel FB_1262, the core collection obtained by PowerCore (PwC_69), and the final set of PRP accessions (PRP_X), composed by joining Priority_100 and Core_69 subsets. In addition, Core Hunter software was used to create additional core sets, either of 69 or 169 entries, based on the optimization of various criteria, including allelic coverage (CV_169) and three distance-based algorithms A-NE (AN_69 and AN_169), E-NE (EN_69 and EN_169), and E-E (EE_69 and EE_169). Concerning parameters accounting for allelic diversity, all sets showed high and similar values for the allelic coverage, while the number of effective alleles and expected heterozygosity were slightly lower for the Priority_100 subset (Table 1). The Shannon-Weaver diversity index (SH index) was comparable among the different subsets, ranging between 0.595 in EE_169 and 0.534 in Priority_100. The SH index generally displays higher values in the presence of a reduced redundancy (Peet, 1975). In contrast, values of observed heterozygosity tended to be more variable, ranging from a minimum of 0.202 in PwC_69 to a maximum of 0.318 in AN_69. According to (Odong et al., 2013), distance-based criteria were used for further evaluations, such as the minimization of A-NE distance, particularly indicated for generalist collections (as the PRP), and maximization of either E-E or E-NE, both suitable for core collection representing the extremes of the entire collection. A-NE distance generally tends to decrease along with the increase of core size, being minimized in the AN_169 and AN_69 core sets (0.137 and 0.172, respectively), a priori optimized using this selection criterion. Despite the relative low performance of both Priority_100 and Core_69 (0.188 and 0.195, respectively), the PRP_X set showed low values for this index (0.165), most probably as a consequence of the increased size. Regarding E-E and E-NE, PRP_X (as well as Priority_100) showed lower values, particularly for E-NE distance, indicating the presence of a certain redundancy within the panel.
Table 1. Genetic analysis and parameters for the different core subsets.
A-NE, Average distance between each genotype of the collection and the nearest entry; CV, percentage allelic coverage; E-E, average entry-to-entry distance; E-NE, average distance between each entry and the nearest entry; He, expected heterozygosity; Ho, observed heterozygosity; MR distance, average Modified Rogers genetic distance; Ne, number of effective alleles.
| Set Name | Ne | SH | Ho | He | CV | MR Distance | |||
|---|---|---|---|---|---|---|---|---|---|
| EE | A-NE | E-NE | |||||||
| FB_1262 | 1.621 | 0.547 | 0.292 | 0.367 | 0.995 | 0.285 | 0 | 0.131 | |
| PwC_69 | 1.675 | 0.574 | 0.202 | 0.39 | 0.987 | 0.318 | 0.203 | 0.237 | |
| EE_69 | 1.705 | 0.587 | 0.234 | 0.401 | 0.992 | 0.347 | 0.209 | 0.229 | |
| AN_69 | 1.645 | 0.560 | 0.318 | 0.378 | 0.977 | 0.286 | 0.172 | 0.210 | |
| EN_69 | 1.704 | 0.587 | 0.269 | 0.402 | 0.991 | 0.334 | 0.207 | 0.275 | |
| CV_169 | 1.638 | 0.556 | 0.285 | 0.375 | 0.995 | 0.302 | 0.163 | 0.212 | |
| EE_169 | 1.721 | 0.595 | 0.224 | 0.408 | 0.994 | 0.330 | 0.183 | 0.191 | |
| AN_169 | 1.643 | 0.559 | 0.300 | 0.377 | 0.987 | 0.290 | 0.137 | 0.203 | |
| EN_169 | 1.683 | 0.578 | 0.277 | 0.394 | 0.993 | 0.315 | 0.175 | 0.256 | |
| Core_69 | 1.713 | 0.593 | 0.247 | 0.406 | 0.988 | 0.303 | 0.195 | 0.212 | |
| Priority_100 | 1.597 | 0.534 | 0.283 | 0.356 | 0.979 | 0.277 | 0.188 | 0.179 | |
| PRP_X | 1.647 | 0.563 | 0.270 | 0.379 | 0.988 | 0.290 | 0.165 | 0.180 | |
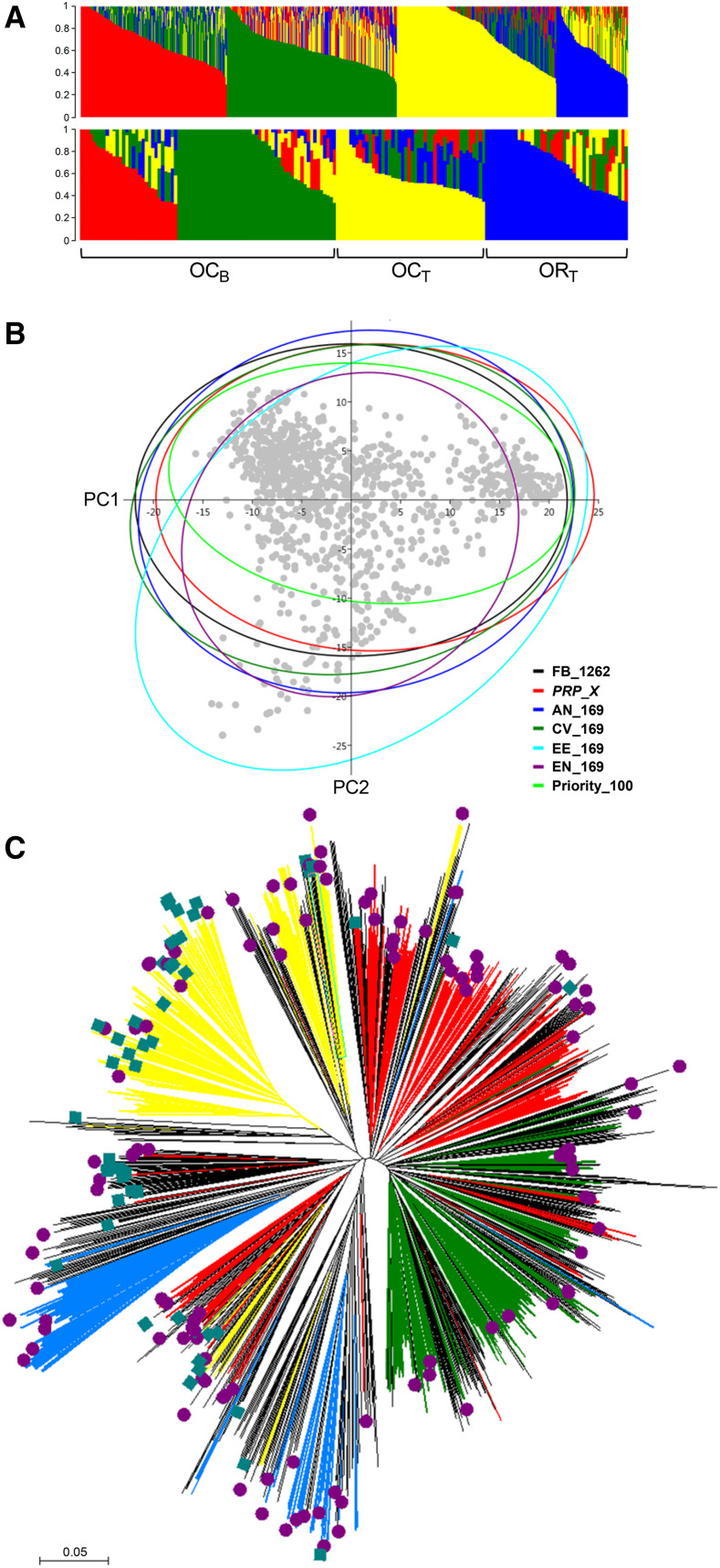
The population structure of peach germplasm was well represented in the PRP_X, in agreement with the presence of clusters of breeding-derived accessions (further separated in peach- and nectarine-type groups), Occidental traditional and admixed entries with prevalent Oriental origins (Fig. 4A). Structure was also preserved in the other core sets, except for that selected through the E-E distance algorithm, tending to oversample the admixed group (Supplemental Fig. S1). PCA was also run to check the distribution of the PRP_X with respect to the other sets, and the first two components explained 15.9% and 8.4%, respectively, of the total variance detected. In the scatterplot, 95% confidence ellipses show almost overlapping areas (except for EE_169), confirming that the PRP_X panel was well distributed to represent the structure of the starting germplasm (Fig. 4B). Finally, a neighbor-joining (NJ) tree, based on the dissimilarity matrix between the whole FB_1262 panel, was also built to assess the distribution of PRP accessions (Fig. 4C).
Figure 4.
Genetic structure and phylogenetic analysis of PRP accessions. A, Population structure estimated in the whole panel (FB_1262) and PRP accessions (PRP_X), as estimated for K (number of a priori cluster) equal to 4. OCB, Occidental breeding; OCT, occidental traditional; ORT, oriental traditional, respectively. B, PCA analysis of the subsets with a core size of 169 entries. Scores for each accession were obtained from the work of Micheletti et al. (2015). The 95% confidence ellipses in the scatterplot were estimated using PAST software. C, NJ phylogenetic tree. Blue squares indicate accessions with traditional and historical value, violet circles indicate the other PRP accessions, and colors reflect the population structure.
A number of accessions of historical and regional importance, mostly belonging to the Occidental traditional cluster, were included. For example, French cultivars dating from the late Middle Ages (cv Grosse Mignonne, cv Millecoton de Septembre, cv Reine des Verges, and cv Brugnon Violet; Okie et al., 2008), traditional nonmelting Spanish cultivars (cv Amarillo de Agosto 1, cv Calante, cv Campiel, cv Jesca, cv Groc Abel, and cv Groc Alto; Badenes et al., 1998; Wünsch et al., 2006), and the Italian cv Crasiommolo Rosso (a white-fleshed nectarine belonging to the ‘Sbergie’ type) and cv Poppa di Venere, first reported at the end of the 18th century (Majoli, 1790). The richness of the Italian peach germplasm was also widely represented by materials from several regions, including Sicily (cv Imera, cv Tardiva di Ficarazzi, cv Settembrina di Bivona, and cv Gialla di Moavero; Marchese et al., 2005), Campania (cv Zingara Nera), Apulia (cv Percoco di Turi), Liguria (cv Michelini), Emilia-Romagna (cv Buco Incavato, cv San Varano 2 and cv San Varano 3, and cv Rosa del West, this last used for the preparation of the famous cocktail Bellini), and Tuscany (cv Regina di Londa; Gallesio, 2003; Monte et al., 2006; Liverani and Giovannini, 2016). Early breeding materials, mainly from United States programs and founders of most of the currently cultivated materials, are also included, along with worldwide commercial cultivars (Supplemental Table S4).
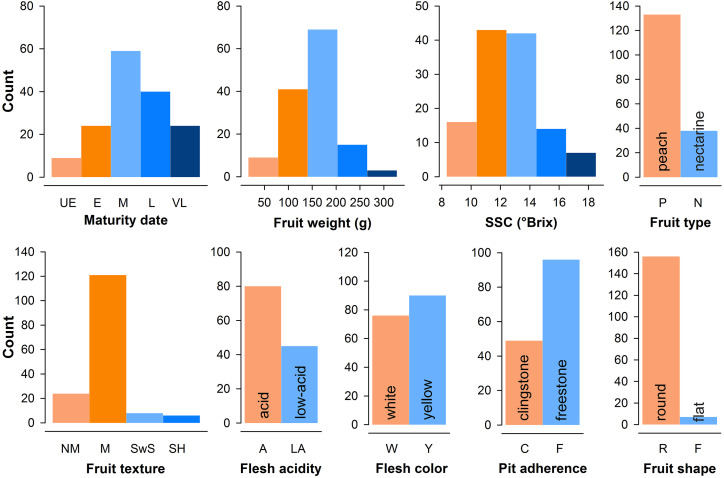
Finally, PRP accessions encompassed a wide range of phenotypic variability for traits related to fruit quality, resistance or tolerance against major diseases (brown rot, powdery mildew, leaf curl, aphids, and Sharka disease), tree growth habit, and phenology (Fig. 5; Supplemental Table S4).
Figure 5.
Distribution of main phenotypic traits in the PRP accessions. In the maturity date plot, UE, E, M, L, and VL indicate ultra-early, early, medium, late, and very late ripening accessions, respectively. SSC, Soluble solids content. In the fruit texture plot, four major texture groups are shown: nonmelting (NM), melting (M), slow softening (SwS), and stony hard (SH).
Choice and Description of the PRP Progeny
Seedlings from 15 cross populations from the research and breeding activities of some European universities and institutions were also added. Most of these materials were already described in depth (Hernández Mora et al., 2017). The leading criterion for the choice of seedlings was the effective segregation of priority traits in peach, mainly related to phenology (fruit developmental period and maturity date), fruit quality (fresh weight, soluble solid content, titratable acidity, texture, and aroma), and disease resistance (brown rot, powdery mildew, green peach aphids, and PPV; Table 2). A range of breeding materials was considered, such as F1, F2, and BC1 populations as well as hybrids with P. davidiana, particularly interesting as a source of PPV resistance (Decroocq et al., 2005).
Table 2. Description of the progeny used for establishing the PRP collection.
Trait abbreviations are as follows: BR, brown rot; FD, flowering date; FW, fruit weight; GPA, green peach aphid; MD, maturity date; PM, powdery mildew; PPV, Plum pox virus; SH, stony hard texture; SSC, soluble solid content; SwS, slow-softening texture; TA, titratable acidity.
| Cross (Parents) | Acronym | Institution | Type of Progeny | Seedling No. | Trait(s) |
|---|---|---|---|---|---|
| ‘Bolero’ × ‘Oro A’ | B × O | UMIL, Milan | F1 | 9 | MD, SSC, FW, skin overcolor, aroma |
| ‘Contender’ × ‘Elegant Lady’ | C × EL | UMIL, Milan | F1 | 14 | BR, MD |
| ‘Max 10’ × ‘Rebus 028’ | M × R | UMIL, Milan | F1 | 9 | MD, TA, SSC, FW, SwS |
| ‘Sweetfire’ × ‘Garcica’ | Sf × G | UMIL, Milan | F1 | 15 | MD, TA, SSC, FW, SwS |
| ‘Belbinette’ × ‘Nectalady’ | Bb × Nl | IRTA, Lleida | F1 | 20 | FD, MD, TA, SSC, FW |
| ‘Big Top’ × ‘Nectaross’ | Bt × Nr | IRTA, Lleida | F1 | 19 | FD, MD, TA, SSC, FW |
| ‘Big Top’ × ‘Armking’ | Bt × Ak | IRTA, Lleida | F1 | 18 | FD, MD, TA, SSC, FW |
| ‘Subirana’ × ‘Feraude’ | PN643 | IRTA, Lleida | F1 | 7 | Fruit shape |
| ‘Summergrand’ × P. davidiana P1908 | SD | INRA, Avignon | F1 | 6 | PM, PPV |
| ‘Zephyr’ × [((‘Summergrand’ (S) × P. davidiana P1908)) × S] | BC2 | INRA, Avignon | BC2 | 13 | FD, PM, PPV, TA, SSC, FW |
| ‘Pamirskij 5’ × ‘Rubira’ | P × R | INRA, Avignon | F2 | 13 | PM, GPA, foliage color |
| FRF 1495 × FRF 1148 (Ma 16-03-059) | POP1376 | CREA, Forli | F1 | 17 | PM, fruit pubescence |
| IFF 983 × Ma 25-01-042 | POP1115 | CREA, Forli | F1 | 17 | TA, SwS, aroma |
| FRF 1695 × FRF 1681 | POP1095 | CREA, Forli | F1 | 19 | SH |
| FRF 813 × FRF 691 | POP1039 | CREA, Forli | F1 | 18 | Skin overcolor |
Experimental Design and Orchard Sites Description
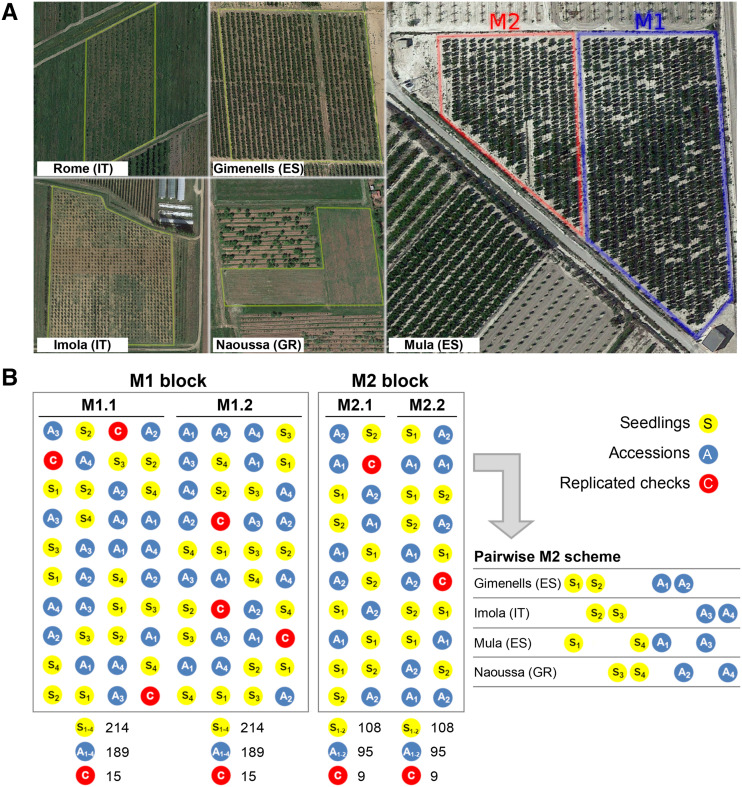
The PRP was established in five institutions from three countries (Greece, Italy, and Spain; Fig. 6A): (1) Institute of Agrifood Research and Technology (IRTA) in Gimenells, Catalonia region, Spain (ES); (2) Murcia Institute of Agri-Food Research and Development (IMIDA) in Mula, Murcia region, Spain (ES); (3) Centro di Ricerca per le Produzioni Vegetali (CRPV) in Imola, Emilia-Romagna region, Italy (IT); (4) Institute of Plant Breeding and Genetic Resources (IPB&GR) in Naoussa, Imathia region, Greece (GR); and (5) Research Centre for Olive, Fruit, and Citrus Crops (CREA) in Rome, Italy (IT).
Figure 6.
Experimental design and PRP orchards layout. A, Google Maps satellite images of the established PRP orchards across the different European sites. B, Experimental design of multisite PRP. A schematic example is provided for the Gimenells location. Accessions (A) and seedlings (S) in each block and subblock were completely randomized. The M1.1 and M1.2 subblocks each include a full copy of the collection (189 accessions, 214 seedlings) plus replicate checks (C) of the accessions cv Big Top, cv Nectaross, and cv Springcrest (five additional trees for each subblock). The M2.1 and M2.2 subblocks include half of the PRP collection and each site has a different half, chosen according to a pairwise design scheme. To this end, accessions (excluding control checks) and seedlings were randomly assigned to eight disjoint subgroups (A1–A4 and S1–S4) of approximately equal size and four of them assigned so that each site shares at least one A or one S group with the other sites. In the example, each M2 subblock at Gimenells is composed of A1 and A2 (46 and 46 accessions, respectively, plus the three checks for a total of 95), S1 and S2 (54 and 54 seedlings, respectively, for a total of 108), and three other additional replicates for each check (nine trees). Gimenells shares the A1 and S1 groups with Mula, S2 with Imola, and A2 with Naoussa.
For each accession and seedling, a single mother plant was propagated through grafting on a common cv GF677 rootstock by the same nursery. Plants were grafted in the same year (2015) to obtain trees of the same age. To ensure an adequate compromise between the number of replicate trees and sustainable costs of maintenance, an augmented design with replicated control checks was adopted in all sites except Rome, hosting a partial copy of the PRP (accessions only, without randomization).
Accessions and seedlings were arranged in two blocks (M1 and M2) according to the following design (Fig. 6B). The M1 block, composed of two subblocks (M1.1 and M1.2) each including the entire PRP collection of 169 accessions plus 20 cross parents (A group) and 214 seedlings (S group), for a total of 403 genotypes in each subblock. Taking into account the physical layout of the design for each location/field combination (i.e. the number of rows and the number of positions per row), each accession and seedling was randomly assigned to a position within a subblock (as illustrated for the Gimenells location in Supplemental Table S5). To assess and correct for spatial variation within and between experimental sites, the three accessions cv Big Top, cv Springcrest, and cv Nectaross from the A group were included with a higher replication and randomly distributed over the M1.1 and M1.2 subblocks (at least five additional trees of each genotype for each subblock). The M2 block, composed of two subblocks (M2.1 and M2.2) each including half of the PRP collection (85 accessions plus 10 cross-parents and 112 seedlings). In each site, the M2.1 and M2.2 subblocks include the same set of entries (i.e. the same half of the collection), randomly assigned in each subblock, plus the replicated control checks previously described (at least three additional trees of each entry for each subblock). The composition of the M2 block is not the same across sites (i.e. each site has a different half of the collection), chosen according to a pairwise design scheme (Fig. 6B). First, excluding control checks, accessions and seedlings of the PRP collection were randomly divided into eight disjoint subgroups, A1 to A4 (of 48, 47, 47, and 48 accessions, respectively) and S1 to S4 (of 54, 53, 54, and 53 seedlings, respectively), then four subgroups were assigned so that each site shares at least one A or one S with the other sites. For example, the Imola location shares A4 and S3 with Naoussa, A3 with Mula, and S2 with Gimenells. This partial replication design is such that, within the full design, all subgroups were well connected.
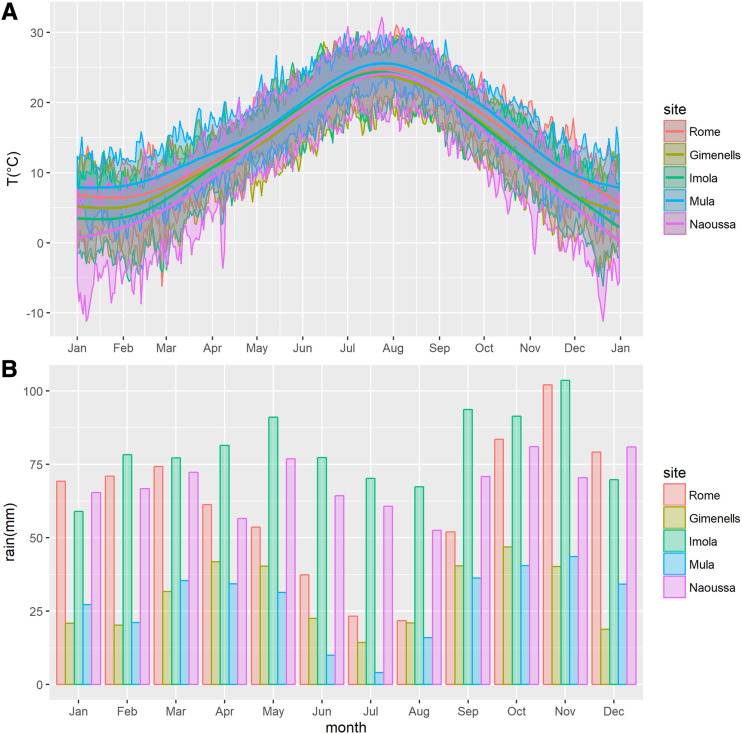
The geographic location of each site as well as the basic climate and soil parameters are shown in Table 3. The sites covered a range of latitude from about 38°N in Mula (southeastern Spain) to 44°N in Imola (northern Italy), while altitude spanned from near sea level in Imola and Rome (53 and 73 m, respectively) to 278 m at the Mula site. Although all sites are included in the Mediterranean zone, climates widely range from semiarid in Mula (warm winter and hot summer) to subcontinental in Imola and Naoussa (with moderately cold winter). Average monthly temperatures (1999–2018 time series) varied from the colder regimes of Naoussa (1.7°C ± 3.9°C and 23.5°C ± 2.9°C in the coldest and hottest months, January and August, respectively) to the warmer conditions of Mula (7.9°C ± 3.7°C and 27.1°C ± 2.6°C, respectively; Fig. 7A). The fulfillment of the chilling requirement (i.e. the period of cold temperatures needed for overcoming endodormancy) is a parameter of utmost relevance for peach reproductive phenology. According to the chilling hours model (Weinberger, 1950), assigning 1 h for each hourly temperature between 0°C and 7.2°C, accumulation patterns widely ranged from 1,762 ± 124 chilling hours at Naoussa to 693 ± 159 chilling hours at Mula. Also, precipitation differently affected the selected sites, with Imola having the wettest conditions (964 ± 218 mm per year) and both Spanish locations having the driest (Gimenells 361 ± 95 and Mula 336 ± 75; Fig. 7B).
Table 3. Basic pedo-climatic features of the five PRP locations.
Features include geographic coordinates, altitude, average annual minimum and maximum temperatures, and cumulative annual precipitation (data series 1999–2018). Chilling accumulation was calculated according to the Chilling Hours (CH) model as the sum of hourly temperatures between 0°C and 7.2°C during the dormant season. S.O.M., Soil organic matter content.
| Site | Geographic Coordinates | Altitude | Climate | Soil | ||||
|---|---|---|---|---|---|---|---|---|
| Average Annual Temperature (Minimum/Maximum) | Cumulative Precipitation | Chilling Accumulation | Texture | pH | S.O.M. | |||
| m | °C | mm | CH | % | ||||
| CREA, Rome (Italy) | 41°47′ N/12°33′ E | 79 | 11.2/18.9 | 731 ± 155 | 1,171 ± 224 | Sandy, loam | 7.7 | 1.9 |
| CRPV, Imola (Italy) | 44°20′ N/11°45′ E | 53 | 9.5/17.9 | 964 ± 218 | 1,753 ± 195 | Silty, loam | 7.2 | 1.5 |
| IMIDA, Mula (Spain) | 38°3 N/1°25′ O | 278 | 12.0/24.5 | 336 ± 75 | 693 ± 159 | Clay | 7.8 | 2.6 |
| IPB&GR, Naoussa (Greece) | 40°37′ N/22°06′ E | 119 | 8.1/17.2 | 818 ± 160 | 1,762 ± 124 | Sandy, loam | 6.8 | 2.5 |
| IRTA, Gimenells (Spain) | 41°39′ N/0°23′ E | 259 | 9.1/18.9 | 361 ± 95 | 1,637 ± 133 | Sandy, loam | 7.7 | 2.6 |
Figure 7.
Climatic profiles of PRP sites. A, Trend of minimum and maximum daily air temperatures at the five PRP locations (averaged from the 1999–2018 time series). Thick lines show smoothed mean temperatures. B, Average monthly precipitation (in millimeters).
DISCUSSION
The concept of the PRP arises from the growing awareness about current and common issues regarding ex situ peach conservation across European institutions. Fluctuations in funding availability and intrinsic constraints of living orchard collections threaten the long-term preservation of diversity resources, causing a progressive loss of valuable materials. Reference or core collections have been designed for several fruit tree species, such as olive (Olea europaea; Khadari et al., 2003; El Bakkali et al., 2013; Belaj et al., 2012), grape (Vitis spp.; Laucou et al., 2011), cherry (Prunus avium; Campoy et al., 2016), apple (Malus domestica; Gross et al., 2013), and apricot (Prunus armeniaca; Krichen et al., 2012). Nevertheless, they have mainly been created for improving resource allocation in the context of a single institution or repository. The development of a transnational and shared strategy provides the most promising opportunity in the conservation approach. Actual establishment of the PRP has required huge coordination efforts and faced the effective availability of materials, the difficulties of their exchange, and the success of clonal propagations (particularly for old, often unique, accessions). The sampling strategy for the PRP has been defined to accommodate multiple purposes while maintaining the maximum possible diversity compared with the starting panel. The final panel of accessions was assembled by the combination of two different subsets: the first (Core_69), ensuring the preservation of the total allele number with the minimum number of accessions, was extracted by widely adopted maximization strategies, either using a class coverage criterion (in PowerCore) or SH index (in MSTRAT), with the latter penalizing redundancy. The second subset, accommodating for other scopes (Priority_100), was chosen by experts with a robust knowledge of the genetic structure in peach, providing a reliable criterion for assisting selection.
As a whole, genetic analysis supports that the PRP composition is highly representative of the diversity of peach germplasms present in European collections, as it retains all the allelic variability present within the starting panel, specifically targets defined genetic clusters according to the genetic structure, and includes most relevant phenotypic traits. Indeed, differences among the various sampling strategies were negligible for allelic coverage, expected heterozygosity, and SH index, revealing a buffer effect toward optimization criteria. Such an effect could be expected, since peach has experienced a severe domestication bottleneck with a reduction of genetic diversity, followed by a strong artificial selection during domestication and modern improvement (Verde et al., 2013; Yu et al., 2018; Li et al., 2019). This is also reflected in the narrow genetic bases of peach germplasm available across the main European repositories. Thus, the high level of allelic redundancy allows selecting many different subpopulations able to retain the same amount of genetic variation. In spite of this, a preliminary validation using a distance-based criterion not used in the selection stage showed a minimized A-NE index, the most indicative for evaluating the quality of multipurpose collections (Odong et al., 2013). Conversely, E-E and, particularly, E-NE indices were less optimized, due to a certain redundancy in the Priority_100 subset (i.e. a higher number of genotypes providing unique alleles). This was mainly due to the inclusion of accessions of traditional and breeding values, respectively belonging to the Occidental traditional and Occidental breeding clusters, characterized by a very narrow genetic background. Clearly, the inclusion of these materials is crucial in the overall perspective of balancing diversity and usefulness, as they integrated various fundamental qualities, such as popularity, prestige, tradition, and breeding. A similar mixed strategy was also recently optimized for creating a core collection for Swiss pear (Pyrus communis) germplasm (Urrestarazu et al., 2019).
Climate challenges in peach-growing areas increase the need for resilient cultivars able to maintain productivity while showing an enhanced capacity for adaptation to suboptimal conditions. Nevertheless, resilience and adaptive traits often have a complex inheritance and a strong interaction with the environment or cultivation practice (Kissoudis et al., 2016). The partitioning of phenotypic variation into genotypic, environmental, and their interaction components involves ad hoc experimental designs and integration of field data on a common set of genetic materials under a range of different environmental/management conditions. Multienvironment trials have been extensively used to study genotype-by-management interactions, carry out GWAS, and develop GS models for complex traits in annual crops (Malosetti et al., 2013; Gutiérrez et al., 2015; Zhu et al., 2018; Bustos-Korts et al., 2019) or study genotype-by-environment interactions in forest trees (Li et al., 2017). In contrast, such experimental designs are lagging in fruit trees, largely because of the need for large and diverse germplasm sets for quantitative genetics analyses and the above-mentioned difficulties in material propagation and exchange. The PRP aims to fill this gap, as the replicated design and the different pedo-climatic conditions across sites are particularly indicated for the dissection of interactions between genotype and environment and/or management practice. The PRP locations cover major climatic zones of the Mediterranean area, from semiarid conditions of southern Spain to subcontinental conditions of northern Italy and Greece, determining a broad range of temperatures and precipitation patterns. In particular, sites are characterized by different rates and amounts of chilling and heat accumulation, which will be particularly useful for the dissection of traits associated with reproductive phenology, such as blooming and fruit ripening time. The proximity of experimental sites to major production areas provides an added value for the translation of scientific outcomes. The inclusion of both accessions and seedlings from various crosses allows the development and testing of statistical approaches for genomics-assisted breeding, such as joint linkage-association analysis (Yu et al., 2008; Lu et al., 2010) and genome-wide selection (Resende et al., 2012; van Nocker and Gardiner, 2014), or systematic QTL validation (Peace et al., 2014). Also, the integration of omics (including epigenomics) data may improve our understanding of physiological changes in response to environmental stimuli and constraints.
The PRP multisite experimental design was established with a complete randomization of genotypes (accessions and seedlings) within each subblock and replicate checks to account for spatial variability. The rationale behind the choice of such a design mainly derived from the possibility of a direct comparison with standard reference varieties. A drawback of this approach is the relatively few degrees of freedom for experimental errors, lowering the power to detect differences among genotypes. The use of α designs (Patterson and Williams, 1976) and derived row-column designs (John and Eccleston, 1986) might be statistically more powerful, especially to estimate contrasts between genotypes and improve the estimation of spatial variation (e.g. due to different soil composition within the orchard). The identification of optimal designs for a large number of genotypes is still challenging (Cullis et al., 1998). The PRP will allow validation of the performance of this experimental design on fruit trees and provide a foundation for future planning of multisite collections.
The PRP has been grafted on a single ‘GF677’ rootstock, a P. amygdalus × P. persica hybrid. ‘GF677’ is the most widespread rootstock, mainly for its growth vigor, excellent affinity, adaptation to limestone soils, and tolerance to drought and replanting (Reighard and Loreti, 2008). While the choice of a single rootstock is justified by the need of simplifying the experimental design, this precludes the assessment of scion-by-rootstock interaction. A number of Prunus spp. rootstocks are currently available for peach, some of them harboring interesting traits for resistance to soil pathogens or abiotic stress conditions. Their integration into feasible experimental designs will be the next challenge.
In perspective, the PRP should fulfill several purposes, from research to education and traineeship of young breeders. A better understanding of diversity is expected to encourage the use of broad-ranging germplasm (maybe also in other existing ex situ collections) in breeding programs. In the last decades, the mission of many agriculture-oriented institutions has shifted from the traditional focus of establishing horticultural collections to a wider target of preserving germplasm resources and agricultural heritage (Hammer et al., 2003; Havens et al., 2006). This objective is of utmost importance for fruit tree species of ancient cultivation history, such as peach. For these reasons, a number of traditional and local cultivars (either old or relatively modern) have been included in the PRP as a safeguard of an integral part of the rural landscape and collective memory. Since information and descriptions about local germplasms are scarce and often restricted to cultivation areas, their choice has been directly handled by the curators of each repository, with the aid of experienced breeders.
MATERIALS AND METHODS
Data Sets
A set of 1,262 accessions was selected as representative of the peach (Prunus persica) germplasm maintained in collections of four different European countries (Supplemental Table S3). The complete list of institutions providing plant materials, SNP genotyping, and phenotypic data for seven monogenic traits has been previously described (Micheletti et al., 2015). SNP genotyping data were obtained from the Genome Database for Rosaceae (https://www.rosaceae.org/publication_datasets; accession no. tfGDR1013). Phenotypic data reported in Supplemental Table S4 were obtained from Micheletti et al. (2015) and the FruitBreedomics database (http://bioinformatics.tecnoparco.org/fruitbreedomics/).
Construction of Core Subsets
The advanced M strategy implemented in PowerCore v1.0 (Kim et al., 2007) using 3,894 SNP markers was carried out to extract a core subset able to capture all the alleles observed in the entire collection. The size of the final core collection depends on the level of variability and redundancy present in the whole panel and cannot be set a priori. Seventeen kernel accessions with available whole-genome resequencing data were superimposed through the preferential selection tool, which retains the accessions defined by the user without validation. The standard M strategy implemented in MSTRAT (Gouesnard et al., 2001) was also applied. The MSTRAT algorithm selects a subset of n accessions from the N accessions of the entire collection by maximizing the number of alleles (and/or trait classes) at each locus. The sampling size estimated with PowerCore was set as the default parameter, and four sets of 100 core collections were constructed by using different combinations of genotypic and phenotypic data. Due to the restraints in the number of variables MSTRAT is able to manage, different subsets of approximately 100 SNPs each were obtained through an ad hoc-developed Perl script program, by extracting one SNP every 1,800 kb, corresponding to the maximum boundary for linkage disequilibrium found in some subpopulations of the original plant material (Micheletti et al., 2015). Seven qualitative and 10 quantitative traits (these last transformed into qualitative categories) were used as phenotypic data. For each run, the core size was set to 70, and 100 independent replicates with 100 iterations were generated. The SH index was used as a second criterion to classify core subsets. Redundancy was assayed through the Redundancy tool implemented in MSTRAT, which samples two different sets of core collections of increasing size, as defined by the user, through the application of the maximization strategy or random sampling. For this analysis, a subset of 445 SNP markers was pruned from the whole set of 4,271 using Plink v1.07 with a window size of 50, a shift of 7, and a variance inflation factor of 2. Redundancy was assayed in the whole panel of accessions with a step of five in the first 100, five repetitions, and 50 iterations. The Mixed Replica search algorithm implemented in the Core Hunter II software (De Beukelaer et al., 2012) was used to generate a core collection of fixed size (either 69 and 169 entries) based on the optimization of the Modified Rogers distance measure, with a weight of 1. For the evaluation of the quality of the different core subsets, genetic distance-based criteria were considered: the average genetic distance between all the entries of each core collection (E-E); the average distance between each entry and the nearest neighboring entry for each core collection (N-E); and the average distance between each genotype of the entire collection and the nearest entry in each core collection (A-NE). The quality of each collection increased for lower values of A-NE (the maximum representation is obtained for AN = 0, when each accession is represented by itself or by an identical duplicate) and higher values both for E-NE (maximizes the average distance between each selected individual and the closest other selected item in the core) and E-E (maximizes the average distance between each pair of selected individuals in the core.).
Genetic Diversity and Population Analyses
Genetic diversity measures were determined using GenAlex 6.41 (Peakall and Smouse, 2006) and include number of effective alleles (the number of equally frequent alleles required to give the observed level of heterozygosity), levels of observed and expected heterozygosity, and the SH index. Allelic coverage was calculated by the function allelic coverage implemented in Core Hunter II software. Population structure was inferred using the model-based clustering algorithm ADMIXTURE v1.22 (Alexander et al., 2009). From SNP data, the software identifies K a priori genetic clusters provided by the user, and for each individual, it estimates the probability of membership to each cluster. A preliminary analysis was performed by inputting successive values of K from 2 to 6. The value of K that maximized the predictive accuracy was chosen based on a 10-fold cross-validation procedure with 10 different fixed initial seeds (Supplemental Fig. S2). PCA data were retrieved from a previous work (Micheletti et al., 2015). The 95% confidence ellipses in the scatterplot were estimated using PAST software (Hammer et al., 2001). The phylogenetic tree was built from a pairwise genetic distance matrix between individuals clustered with the NJ method in TASSEL (Bradbury et al., 2007). Bootstrap replicates and tree reconstruction were performed in MEGA7 software (Kumar et al., 2016).
Experimental Design and Pedo-Climatic Analyses
In the experimental design, randomization was performed with the Genstat software (https://genstat.kb.vsni.co.uk/knowledge-base/hcitegen/). Meteorological time series from 1999 to 2018 were obtained from the European Centre for Medium-Range Weather Forecasts, except for Mula (Murcia, Spain), for which data were available from a nearby weather station. Hourly temperature series were obtained by linear interpolation of available trihourly data and expressed in degrees Celsius. Cumulative precipitations were averaged and expressed in mm per month or year. Chill accumulation was calculated according to the chilling hours model (Weinberger, 1950) as the sum of hourly temperatures between 0°C and 7.2°C during the dormant season (November 15 to March 31). Soil texture was expressed according to U.S. Department of Agriculture classification. Mineral composition, pH, and organic matter content were determined according to standard procedures for soil analysis.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Population structure estimated in the core sets AN_169, EE_169, EN_169, and CV_169.
Supplemental Figure S2. Predictive accuracy (cross-validation error) of population stratification in both PRP_X and FB_1262 as determined by ADMIXTURE software.
Supplemental Table S1. PowerCore output.
Supplemental Table S2. MSTRAT outputs for the four settings.
Supplemental Table S3. Accession ranking by MSTRAT frequencies.
Supplemental Table S4. PRP accession descriptions.
Supplemental Table S5. Layout of subblock M1.1 as illustrated for the Gimenells location.
Acknowledgments
We thank Claudio Buscaroli and Martina Lama (Centro Ricerche Produzioni Vegetali) for field assistance, Remo Chiozzotto (University of Milan) for lab assistance, and Michela Troggio (Fondazione Edmund Mach) for genotypic analyses. We are grateful to Fosco Vesely (University of Milan) for extracting climate series. We thank the Institut National de Recherche pour l’Agriculture’s Prunus Biological Resources Center for preserving and managing the peach collections and the Fruit Tree Experimental Unit (UEA) of Institut National de Recherche pour l’Agriculture-Nouvelle Aquitaine-Bordeaux for growing the trees. We also thank the Italian National Centre of Fruit Germplasm at Consiglio per la Ricerca in Agricoltura e L’Analisi Del’Economia Agraria (Rome) for preserving and maintaining peach collections and the Italian Ministry of Agriculture.
Footnotes
This work was supported by PRIMA (grant no. FREECLIMB to D.B., I.E., P.D., and B.Q.T.), the European Commission (project FruitBreedomics; grant no. FP7–265582 to D.B., L.R., P.A., I.V., T.B., B.Q.T., and W.G.), MAS.PES, an Italian project aimed at apricot and peach breeding (to D.B. and S.F.), the Spanish Ministry of Economy and Competitiveness (MINECO/FEDER grant nos. AGL2012–40228–C02–01 and RTA2015–00050–00–00), and the CERCA Programme-Generalitat of Catalonia.
This work is dedicated to the memory of our colleague Chiara Ferrandi, who recently passed away.
References
- Alexander DH, Novembre J, Lange K(2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana MJ, Abbassi E, Howard W, Arús P (2010) Genetic variation, population structure and linkage disequilibrium in peach commercial varieties. BMC Genet 11: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranzana MJ, Decroocq V, Dirlewanger E, Eduardo I, Gao ZS, Gasic K, Iezzoni A, Jung S, Peace C, Prieto H, et al. (2019) Prunus genetics and applications after de novo genome sequencing: Achievements and prospects. Hortic Res 6: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenes ML, Cambra M, López MM, Batlle I, Iglesias I, Aranzana MJ, López E E, Vives C C, Garcia Brunton J, et al. (2015) A peach germplasm collection for increasing the genetic diversity in European breeding programs. Acta Hortic 1084: 125–129 [Google Scholar]
- Badenes ML, Martínez-Calvo J, Llácer G(1998) Analysis of peach germplasm from Spain. Acta Hortic 465: 243–250 [Google Scholar]
- Bassi D, Monet R(2008) Botany and taxonomy In Layne D, and Bassi D, eds, The Peach: Botany, Production and Uses. CABI, Wallingford, United Kingdom, pp 1–36 [Google Scholar]
- Belaj A, del Carmen Dominguez-García M, Atienza SG, Urdíroz NM, De la Rosa R, Satovic Z, Martín A, Kilian A, Trujillo I, Valpuesta V, et al. (2012) Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits. Tree Genet Genomes 8: 365–378 [Google Scholar]
- Bellucci A.(1908) La Coltivazione del Pesco a Massalombarda. Tip Soc. Mazzini, Ravenna, Italy [Google Scholar]
- Benediková D, Giovannini D(2013) Review on genetic resources in the ECPGR Prunus Working Group. Acta Hortic 981: 43–51 [Google Scholar]
- Biasi R, Botti F, Cullotta S, Barbera G(2012) The role of Mediterranean fruit tree orchards and vineyards in maintaining the traditional agricultural landscapes. Acta Hortic 940: 79–88 [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES(2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Brown AHD, Marshall DR(1995) A Basic Sampling Strategy: Theory And Practice In Collecting Plant Genetic Diversity Technical Guideline. CABI Publishing, Wallingford, United Kingdom, pp 75–91 [Google Scholar]
- Bustos-Korts D, Dawson IK, Russell J, Tondelli A, Guerra D, Ferrandi C, Strozzi F, Nicolazzi EL, Molnar-Lang M, Ozkan H, et al. (2019) Exome sequences and multi-environment field trials elucidate the genetic basis of adaptation in barley. Plant J 99: 1172–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne DH, Raseira MCB, Bassi D, Piagnani MC, Gasic K, Reighard GL, Moreno MA, Perez-Gonzalez S(2012) Peach In Badenes ML, and Byrne DH, eds, Fruit Breeding. Springer, New York, pp 505–570 [Google Scholar]
- Campoy JA, Lerigoleur-Balsemin E, Christmann H, Beauvieux R, Girollet N, Quero-García J, Dirlewanger E, Barreneche T(2016) Genetic diversity, linkage disequilibrium, population structure and construction of a core collection of Prunus avium L. landraces and bred cultivars. BMC Plant Biol 16: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné D, Dayatilake D, Diack R, Oliver M, Ireland H, Watson A, Gardiner SE, Johnston JW, Schaffer RJ, Tustin S(2014) Genetic and environmental control of fruit maturation, dry matter and firmness in apple (Malus × domestica Borkh.). Hortic Res 1: 14046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirilli M, Flati T, Gioiosa S, Tagliaferri I, Ciacciulli A, Gao Z, Gattolin S, Geuna F, Maggi F, Bottoni P, et al. (2018) PeachVar-DB: A curated collection of genetic variations for the interactive analysis of peach genome data. Plant Cell Physiol 59: e2. [DOI] [PubMed] [Google Scholar]
- Cirilli M, Rossini L, Geuna F, Palmisano F, Minafra A, Castrignanò T, Gattolin S, Ciacciulli A, Babini AR, Liverani A, et al. (2017) Genetic dissection of Sharka disease tolerance in peach (P. persica L. Batsch). BMC Plant Biol 17: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coakley SM, Scherm H, Chakraborty S(1999) Climate change and plant disease management. Annu Rev Phytopathol 37: 399–426 [DOI] [PubMed] [Google Scholar]
- Corelli-Grappadelli L, Marini RP(2008) Orchard planting systems In Layne DR, and Bassi D, eds, The Peach: Botany, Production and Uses. CABI, Wallingford, United Kingdom, pp 264–288 [Google Scholar]
- Cullis B, Gogel B, Verbyla A, Thompson R(1998) Spatial analysis of multi‐environment early generation variety trials. Biometrics 54: 1–8 [Google Scholar]
- De Beukelaer H, Smýkal P, Davenport GF, Fack V(2012) Core Hunter II: Fast core subset selection based on multiple genetic diversity measures using Mixed Replica search. BMC Bioinformatics 13: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroocq V, Foulongne M, Lambert P, Gall OL, Mantin C, Pascal T, Schurdi-Levraud V, Kervella J(2005) Analogues of virus resistance genes map to QTLs for resistance to sharka disease in Prunus davidiana. Mol Genet Genomics 272: 680–689 [DOI] [PubMed] [Google Scholar]
- El Bakkali A, Haouane H, Moukhli A, Costes E, Van Damme P, Khadari B(2013) Construction of core collections suitable for association mapping to optimize use of Mediterranean olive (Olea europaea L.) genetic resources. PLoS ONE 8: e61265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust M, Timon B(1995) Origin and dissemination of peach. Hortic Rev (Am Soc Hortic Sci) 17: 331–379 [Google Scholar]
- Ford-Lloyd B, Jackson M(1986) Plant Genetic Resources: An Introduction to Their Conservation and Use. Edward Arnold, London [Google Scholar]
- Foulongne M, Pascal T, Arús P, Kervella J(2003) The potential of Prunus davidiana for introgression into peach [Prunus persica (L.) Batsch] assessed by comparative mapping. Theor Appl Genet 107: 227–238 [DOI] [PubMed] [Google Scholar]
- Frankel OH, Brown AHD(1984) Current plant genetic resources: A critical appraisal In Genetics: New Frontiers, Vol IV Oxford and IBH Publishing, New Delhi, India, pp 1–11 [Google Scholar]
- Fu YB, Dong YB(2015) Genetic erosion under modern plant breeding: Case studies in Canadian crop gene pools In Ahuja MR, and Jain SM, eds, Genetic Diversity and Erosion in Plants. Springer International Publishing, Cham, Switzerland, pp 89–104 [Google Scholar]
- Gallesio G.(2003) Il trattato del pesco di Giorgio Gallesio In Baldini E, ed, Gli Inediti Trattati del Pesco e del Ciliegio: Complementi Scientifici Della “Pomona Italiana” di Giorgio Gallesio. Accademia dei Georgofili, Florence, Italy, pp 9–146 [Google Scholar]
- Gepts P.(2006) Plant genetic resources conservation and utilization. Crop Sci 46: 2278–2292 [Google Scholar]
- Gouesnard B, Bataillon TM, Decoux G, Rozale C, Schoen DJ, David JL(2001) MSTRAT: An algorithm for building germ plasm core collections by maximizing allelic or phenotypic richness. J Hered 92: 93–94 [DOI] [PubMed] [Google Scholar]
- Gradziel TM.(2002) Almond species as sources of new germplasm for peach improvement. Acta Hortic 592: 81–88 [Google Scholar]
- Gross BL, Volk GM, Richards CM, Reeves PA, Henk AD, Forsline PL, Szewc-McFadden A, Fazio G, Chao CT(2013) Diversity captured in the USDA-ARS national plant germplasm system apple core collection. J Am Soc Hortic Sci 138: 375–381 [Google Scholar]
- Gutiérrez L, Germán S, Pereyra S, Hayes PM, Pérez CA, Capettini F, Locatelli A, Berberian NM, Falconi EE, Estrada R, et al. (2015) Multi-environment multi-QTL association mapping identifies disease resistance QTL in barley germplasm from Latin America. Theor Appl Genet 128: 501–516 [DOI] [PubMed] [Google Scholar]
- Hammer K, Arrowsmith N, Gladis T(2003) Agrobiodiversity with emphasis on plant genetic resources. Naturwissenschaften 90: 241–250 [DOI] [PubMed] [Google Scholar]
- Hammer Ø, Harper DA, Ryan PD(2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electronica 4: 9 [Google Scholar]
- Havens K, Vitt P, Maunder M, Guerrant EO, Dixon K(2006) Ex situ plant conservation and beyond. Bioscience 56: 525–531 [Google Scholar]
- Hernández Mora JR, Micheletti D, Bink M, Van de Weg E, Cantín C, Nazzicari N, Caprera A, Dettori MT, Micali S, Banchi E, et al. (2017) Integrated QTL detection for key breeding traits in multiple peach progenies. BMC Genomics 18: 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John JA, Eccleston JA(1986) Row‐column a‐designs. Biometrika 73: 301–306 [Google Scholar]
- Khadari B, Breton C, Moutier N, Roger JP, Besnard G, Bervillé A, Dosba F(2003) The use of molecular markers for germplasm management in a French olive collection. Theor Appl Genet 106: 521–529 [DOI] [PubMed] [Google Scholar]
- Kim KW, Chung HK, Cho GT, Ma KH, Chandrabalan D, Gwag JG, Kim TS, Cho EG, Park YJ(2007) PowerCore: A program applying the advanced M strategy with a heuristic search for establishing core sets. Bioinformatics 23: 2155–2162 [DOI] [PubMed] [Google Scholar]
- Kissoudis C, van de Wiel C, Visser RG, van der Linden G(2016) Future-proof crops: Challenges and strategies for climate resilience improvement. Curr Opin Plant Biol 30: 47–56 [DOI] [PubMed] [Google Scholar]
- Krichen L, Audergon JM, Trifi-Farah N(2012) Relative efficiency of morphological characters and molecular markers in the establishment of an apricot core collection. Hereditas 149: 163–172 [DOI] [PubMed] [Google Scholar]
- Krishnan RR, Sumathy R, Ramesh S, Bindroo B, Naik GV(2014) SimEli: Similarity elimination method for sampling distant entries in development of core collections. Crop Sci 54: 1070–1078 [Google Scholar]
- Kumar S, Stecher G, Tamura K(2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucou V, Lacombe T, Dechesne F, Siret R, Bruno JP, Dessup M, Dessup T, Ortigosa P, Parra P, Roux C, et al. (2011) High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor Appl Genet 122: 1233–1245 [DOI] [PubMed] [Google Scholar]
- Laurens F, Aranzana MJ, Arus P, Bassi D, Bink M, Bonany J, Caprera A, Corelli-Grappadelli L, Costes E, Durel CE, et al. (2018) An integrated approach for increasing breeding efficiency in apple and peach in Europe. Hortic Res 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Cao K, Zhu G, Fang W, Chen C, Wang X, Zhao P, Guo J, Ding T, Guan L, et al. (2019) Genomic analyses of an extensive collection of wild and cultivated accessions provide new insights into peach breeding history. Genome Biol 20: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Suontama M, Burdon RD, Dungey HS(2017) Genotype by environment interactions in forest tree breeding: Review of methodology and perspectives on research and application. Tree Genet Genomes 13: 60–78 [Google Scholar]
- Liverani A, Giovannini D(2016) Pesco In Fideghelli C, ed, Atlante dei Fruttiferi Autoctoni Italiani, Vol III SOI Società di ortoflorofrutticoltura Italiana, Firenze, Italy, pp 1433–1559 [Google Scholar]
- Lu Y, Zhang S, Shah T, Xie C, Hao Z, Li X, Farkhari M, Ribaut J-M, Cao M, Rong T, et al. (2010) Joint linkage-linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. Proc Natl Acad Sci USA 107: 19585–19590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedeling E.(2012) Climate change impacts on winter chill for temperate fruit and nut production: A review. Sci Hortic (Amsterdam) 144: 218–229 [Google Scholar]
- Majoli C.(1790–1810) Plantarum Collectio Juxta Linnaeanum Systema a Lectore Caesare Majolio Hyeronimino Digesta et Depicta. City Library of Forlì, Forlì-Cesena, Italy [Google Scholar]
- Malosetti M, Ribaut JM, van Eeuwijk FA(2013) The statistical analysis of multi-environment data: Modeling genotype-by-environment interaction and its genetic basis. Front Physiol 4: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchese A, Tobutt KR, Caruso T(2005) Molecular characterisation of Sicilian Prunus persica cultivars using microsatellites. J Hortic Sci Biotechnol 80: 121–129 [Google Scholar]
- Micheletti D, Dettori MT, Micali S, Aramini V, Pacheco I, Da Silva Linge C, Foschi S, Banchi E, Barreneche T, Quilot-Turion B, et al. (2015) Whole-genome analysis of diversity and SNP-major gene association in peach germplasm. PLoS ONE 10: e0136803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte M, Sottile F, Barone E, Caruso T, Bazzoni A(2006) The Sicilian peach (Prunus persica L. Batsch) germplasm: Horticultural characteristics and sanitary status. Acta Hortic 713: 57–60 [Google Scholar]
- Myles S.(2013) Improving fruit and wine: What does genomics have to offer? Trends Genet 29: 190–196 [DOI] [PubMed] [Google Scholar]
- van Nocker S, Gardiner SE(2014) Breeding better cultivars, faster: Applications of new technologies for the rapid deployment of superior horticultural tree crops. Hortic Res 1: 14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odong T, van Heerwaarden J, Jansen J, van Hintum TJ, van Eeuwijk F(2011) Statistical techniques for defining reference sets of accessions and microsatellite markers. Crop Sci 51: 2401–2411 [Google Scholar]
- Odong TL, Jansen J, van Eeuwijk FA, van Hintum TJL (2013) Quality of core collections for effective utilisation of genetic resources review, discussion and interpretation. Theor Appl Genet 126: 289–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okie WR, Bacon T, Bassi D(2008) Fresh market cultivar development In Layne DR, and Bassi D, eds, The Peach: Botany, Production and Uses. CABI, Wallingford, United Kingdom, pp 264–288 [Google Scholar]
- Parajuli R, Thoma G, Matlock MD(2019) Environmental sustainability of fruit and vegetable production supply chains in the face of climate change: A review. Sci Total Environ 650: 2863–2879 [DOI] [PubMed] [Google Scholar]
- Patterson HD, Williams ER(1976) A new class of resolvable incomplete block designs. Biometrika 63: 83–92 [Google Scholar]
- Peace CP, Luby JJ, van de Weg WE, Bink MCAM, Iezzoni AF(2014) A strategy for developing representative germplasm sets for systematic QTL validation, demonstrated for apple, peach, and sweet cherry. Tree Genet Genomes 10: 1679–1694 [Google Scholar]
- Peakall ROD, Smouse PE(2006) GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet RK.(1975) Relative diversity indices. Ecology 56: 496–498 [Google Scholar]
- Ramírez F, Kallarackal J(2015) Responses of Fruit Trees to Global Climate Change. Springer International Publishing, Cham, Switzerland [Google Scholar]
- Reighard G, Loreti F (2008) Rootstock development In Layne D, and Bassi D, eds, The Peach: Botany, Production and Uses. CABI, Wallingford, United Kingdom, pp 193–220 [Google Scholar]
- Resende MFR Jr., Muñoz P, Resende MDV, Garrick DJ, Fernando RL, Davis JM, Jokela EJ, Martin TA, Peter GF, Kirst M(2012) Accuracy of genomic selection methods in a standard data set of loblolly pine (Pinus taeda L.). Genetics 190: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen DJ, Brown AHD(1995) Maximising genetic diversity in core collections of wild relatives of crop species In Hodgkin T, Brown AHD, van Hintum TJL, and Morales EAV, eds, Core Collections of Plant Genetic Resources. John Wiley & Sons, Chichester, United Kingdom, pp 55–77 [Google Scholar]
- Serra O, Donoso JM, Picañol R, Batlle I, Howad W, Eduardo I, Arús P(2016) Marker-assisted introgression (MAI) of almond genes into the peach background: A fast method to mine and integrate novel variation from exotic sources in long intergeneration species. Tree Genet Genomes 12: 96 [Google Scholar]
- Singh HCP, Rao NKS, and Shivashankar KS, eds; (2013) Climate-Resilient Horticulture: Adaptation and Mitigation Strategies. Springer India, Bangalore, India, pp 81–88 [Google Scholar]
- Thachuk C, Crossa J, Franco J, Dreisigacker S, Warburton M, Davenport GF(2009) Core Hunter: An algorithm for sampling genetic resources based on multiple genetic measures. BMC Bioinformatics 10: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrestarazu J, Kägi C, Bühlmann A, Gassmann J, Santesteban LG, Frey JE, Kellerhals M, Miranda C(2019) Integration of expert knowledge in the definition of Swiss pear core collection. Sci Rep 9: 8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, Bansal KC, Aggarwal PK, Datta SK, Craufurd PQ(2011) Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant Sci 16: 363–371 [DOI] [PubMed] [Google Scholar]
- Verde I, Abbott AG, Scalabrin S, Jung S, Shu S, Marroni F, Zhebentyayeva T, Dettori MT, Grimwood J, Cattonaro F, et al. (2013) The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet 45: 487–494 [DOI] [PubMed] [Google Scholar]
- Verde I, Bassil N, Scalabrin S, Gilmore B, Lawley CT, Gasic K, Micheletti D, Rosyara UR, Cattonaro F, Vendramin E, et al. (2012) Development and evaluation of a 9K SNP array for peach by internationally coordinated SNP detection and validation in breeding germplasm. PLoS ONE 7: e35668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde I, Jenkins J, Dondini L, Micali S, Pagliarani G, Vendramin E, Paris R, Aramini V, Gazza L, Rossini L, et al. (2017) The Peach v2.0 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 18: 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger JH.(1950) Chilling requirements of peach varieties. Proc Am Soc Hortic Sci 56: 122–128 [Google Scholar]
- Wünsch A, Carrera M, Hormaza JI (2006) Molecular characterization of local Spanish peach [Prunus persica (L.) Batsch] germplasm. Genet Resour Crop Ev 53: 925–932 [Google Scholar]
- Yu J, Holland JB, McMullen MD, Buckler ES(2008) Genetic design and statistical power of nested association mapping in maize. Genetics 178: 539–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Fu J, Xu Y, Zhang J, Ren F, Zhao H, Tian S, Guo W, Tu X, Zhao J, et al. (2018) Genome re-sequencing reveals the evolutionary history of peach fruit edibility. Nat Commun 9: 5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XM, Shao XY, Pei YH, Guo XM, Li J, Song XY, Zhao MA(2018) Genetic diversity and genome-wide association study of major ear quantitative traits using high-density SNPs in maize. Front Plant Sci 9: 966. [DOI] [PMC free article] [PubMed] [Google Scholar]