In barley, ROP INTERACTING PARTNER b scaffold protein acts downstream of the signaling ROP GTPase and powdery mildew susceptibility factor RACB and influences susceptibility to this pathogen.
Abstract
Rho of Plants (ROP) G-proteins are key components of cell polarization processes in plant development. The barley (Hordeum vulgare) ROP protein RACB is a susceptibility factor in the interaction of barley with the barley powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh). RACB also drives polar cell development, and this function might be coopted during the formation of fungal haustoria in barley epidermal cells. To understand RACB signaling during the interaction of barley with Bgh, we searched for potential downstream interactors of RACB. Here, we show that ROP INTERACTIVE PARTNER b (RIPb; synonym: INTERACTOR OF CONSTITUTIVE ACTIVE ROP b) directly interacts with RACB in yeast and in planta. Overexpression of RIPb supports the susceptibility of barley to Bgh. RIPb further interacts with itself at microtubules. However, the interaction with activated RACB largely takes place at the plasma membrane. Both RIPb and RACB are recruited to the site of fungal attack around the neck of developing haustoria, suggesting locally enhanced ROP activity. We further assigned different functions to different domains of the RIPb protein. The N-terminal coiled-coil CC1 domain is required for microtubule localization, while the C-terminal coiled-coil CC2 domain is sufficient to interact with RACB and to fulfill a function in susceptibility at the plasma membrane. Hence, RIPb appears to be localized at microtubules and is then recruited by activated RACB for a function at the plasma membrane during formation of the haustorial complex.
The interaction of plants with powdery mildew fungi is a model for the biology of cell-autonomous responses to fungal parasites (Dörmann et al., 2014). The powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh) is a biotrophic ascomycete specifically adapted to barley (Hordeum vulgare) and grows on the plant’s surface. In the beginning of its life cycle, Bgh has to penetrate an epidermal cell in order to establish a haustorium for nutrient uptake (Hahn et al., 1997; Voegele et al., 2001) and to provide a surface for the translocation of virulence effector proteins into the host cell (Catanzariti et al., 2007). During all stages of fungal invasion, the epidermal host cell stays intact. Host cytosol and fungal haustorium are separated by the extrahaustorial matrix and the plant-derived extrahaustorial membrane.
Plant host cells polarize in very early phases of the interaction with fungi. A reorganization of the cytoskeleton was shown in different pathosystems as well as the accumulation of peroxisomes, mitochondria, Golgi bodies, and the endoplasmic reticulum at the site of pathogen attack (Kobayashi et al., 1997; Takemoto et al., 2003, 2006; Koh et al., 2005; Fuchs et al., 2016). This is accompanied by relocation of the nucleus to the site of attack (Gross et al., 1993; Scheler et al., 2016). Polarization is considered important for effective defense, in particular for the focal formation of papilla or cell wall appositions, which requires localized deposition of callose, other cell wall glucans, and phenolic compounds at the attempted penetration site (McLusky et al., 1999; Hückelhoven, 2007; Chowdhury et al., 2014). However, it is reasonable to assume that host cell polarization is also important for successful pathogen establishment, for instance for the generation of the extrahaustorial membrane (Scheler et al., 2016; Kwaaitaal et al., 2017).
ROP GTPases (Rho of Plants; also called RAC for rat sarcoma-related C3 botulinum toxin substrate) are small monomeric G-proteins that form a plant-specific RHO subfamily. ROPs can cycle between an actively signaling GTP-bound state and an inactive GDP-bound state and are crucial for the polarity of diverse types of plant cells (Feiguelman et al., 2018). While activation is mediated by Guanosine Nucleotide Exchange Factors (GEFs), enabling the exchange of GDP to GTP, inactivation is facilitated by GTPase Activating Proteins (GAPs), which activate the intrinsic GTPase function of the G-protein, leading to GTP hydrolysis. ROPs seem to fulfill different functions depending on particular downstream factors called ROP effectors. For instance, Arabidopsis (Arabidopsis thaliana) ROP2 suppresses light-induced stomata opening by interacting with ROP Interactive CRIB Motif Containing Protein7 (RIC7), which in turn interacts with and inhibits the exocyst vesicle-tethering complex subunit Exo70B1 (Hong et al., 2016). ROP2 is additionally involved in pavement cell lobe interdigitation by interacting with RIC4 for actin assembly in lobes and at the same time inhibiting RIC1, which organizes microtubules together with the katanin KTN1 downstream of ROP6 (Fu et al., 2005; Lin et al., 2013). In these pathways, RIC proteins are considered scaffolds for connecting activated ROPs with downstream effector proteins in G-protein signaling.
Another class of downstream interactors are ROP Interactive Partners (RIPs), first called Interactors of Constitutive Active ROPs (ICRs; Lavy et al., 2007; Li et al., 2008). ICRs/RIPs represent a second class of plant-specific proteins connecting ROP signaling to downstream effectors. So far, little is known about these proteins. Arabidopsis knockout plants of ICR1/RIP1 have defects in pavement cell development, root hair development, as well as root meristem maintenance, showing the involvement of ICR1/RIP1 in different developmental processes. ICR1/RIP1 seems to be able to interact with different ROP proteins and was found to interact downstream with SEC3a of the exocyst complex and thereby possibly controlling the localization of the auxin transporter PIN1 (Lavy et al., 2007; Hazak et al., 2010, 2014). Additionally, it was reported that ICR1/RIP1 acts in pollen tube formation, where it interacts with ROP1 at the plasma membrane of the pollen tube tip (Li et al., 2008). RIP3 (also called ICR5 or MIDD1 for Microtubule Depletion Domain1) plays a key role in xylem cell development in Arabidopsis. During the formation of the secondary cell wall in progenitor cells, RIP3 interacts with ROP11 and the kinesin KIN13A, which leads to local microtubule depletion and the formation of secondary wall pits (Mucha et al., 2010; Oda et al., 2010; Oda and Fukuda, 2012, 2013).
ROP GTPases also play a role as signaling components in plant defense (Ono et al., 2001; Chen et al., 2010). For instance, upon chitin perception, the receptor kinase CERK1 phosphorylates RacGEF1. RacGEF1 in turn activates RAC1, which supports immunity to Magnaporthe oryzae (Akamatsu et al., 2013).
The barley ROP protein RACB is involved in root hair outgrowth and controls asymmetric cell division of subsidiary cells in stomata development (Scheler et al., 2016). RACB and RACB-associated proteins influence arrays and the stability of F-actin and the microtubule cytoskeleton (Opalski et al., 2005; Hoefle et al., 2011; Huesmann et al., 2012). In addition to its function in polar cell development, RACB is also a susceptibility factor in the interaction with the powdery mildew fungus Bgh. Overexpression of constitutively activated RACB (CA RACB) enhances the penetration success of Bgh into barley epidermal cells, and silencing of RACB leads to a decreased penetration rate (Schultheiss et al., 2002, 2003; Hoefle et al., 2011). RACB’s function in susceptibility seems not to be dependent on defense suppression but rather on the exploitation of developmental signaling mechanisms of the host (Scheler et al., 2016). A Bgh effector candidate, ROP-Interactive Peptide1 (ROPIP1), binds directly to activated RACB. Expression of ROPIP1 in barley cells negatively influences microtubule stability and leads to an increased penetration rate of Bgh into barley epidermal cells (Nottensteiner et al., 2018). RACB further interacts with the class VI receptor-like cytoplasmic kinase ROP-Binding Kinase1 (RBK1). Activated RACB supports the in vitro kinase activity of RBK1, but RBK1 acts in resistance rather than susceptibility. This seems to be explained by the interaction of RBK1 with S-Phase Kinase1-Associated-Like Protein (SKP1-like), which is part of an E3-ubiquitin ligase complex, and both RBK1 and SKP1-like can limit the abundance of the RACB protein (Huesmann et al., 2012; Reiner et al., 2016). Another interactor of RACB is the Microtubule-Associated ROP GTPase Activating Protein1 (MAGAP1), a CRIB-motif containing ROP-GAP. MAGAP1 and RACB recruit each other to the cell periphery and to the microtubule cytoskeleton, and MAGAP1 apparently counters the susceptibility effect of RACB, while silencing of MAGAP1 leads to increased susceptibility to Bgh (Hoefle et al., 2011).
In this study, we identified barley RIPb as another downstream interactor of RACB. We investigated the effect of RIPb on susceptibility by transient overexpression and RNA interference (RNAi) knockdown of RIPb in single epidermal cells and the interaction between RIPb and RACB by yeast two-hybrid assays and ratiometric bimolecular fluorescence complementation (BiFC). RIPb and RACB colocalize and presumably interact at the plasma membrane, at the microtubule cytoskeleton, and at the site of fungal invasion. To further investigate the structure-function relationship of RIPb, we tested a series of RIPb truncations regarding their function in the interaction of barley with Bgh and their role in protein-protein interaction.
RESULTS
Identification of ICR/RIP Proteins in Barley
Previous studies have shown that ICR/RIP proteins are a class of proteins with little sequence similarity (Li et al., 2008). All ICR/RIP proteins identified so far in Arabidopsis contain an N-terminal QEEL motif and a C-terminal QWRKAA motif. These motifs are present in respective N- and C-terminal coiled-coil (CC) domains. Based on this, we performed bioinformatic analyses and identified three high-confidence genes coding for ICR/RIP proteins in barley (Supplemental Fig. S1). It appears that in several grasses the first Glu in the QEEL motif is exchanged to Asp (QDEL). Because we did not observe a clear orthology to individually numbered Arabidopsis ICR/RIP proteins and phylogenetic analysis was ambiguous as well, we named the barley proteins RIPa/ICRa (HORVU3Hr1G087430), RIPb/ICRb (HORVU1Hr1G012460), and RIPc/ICRc (HORVU3Hr1G072880; Supplemental Fig. S2). We also identified three ICR/RIP proteins in rice (Oryza sativa) containing the QDEL motif as well as the QWRKAA motif (Os01g61760, Os05g03120, and OsJ_03509; Yu et al., 2005). Alignments of the barley ICRs/RIPs with the ICR/RIP proteins from rice and the five ICR/RIP proteins previously identified in Arabidopsis (ICR1/RIP1 [At1g17140], ICR2/RIP2 [At2g37080], ICR3/RIP5 [At5g60210], ICR4/RIP4 [At1g78430], and ICR5/RIP3/MIDD1 [At3g53350]) show little overall amino acid sequence conservation between the grasses and Arabidopsis, except for the conserved QD/EEL motif at the more N-terminal part and the QWRKAA motif at the more C-terminal part of the protein. The latter was shown to be necessary for ROP interaction (Lavy et al., 2007). The alignment also shows conservation of several Lys residues at the very C terminus, which were shown before to be important for membrane localization of other ICR/RIP proteins (Supplemental Fig. S1; Li et al., 2008).
Phylogenetic analysis shows that HvICRa/HvRIPa and HvICRb/HvRIPb are more closely related to each other than to HvICRc/ HvRIPc, which is located on an independent branch of the tree (Supplemental Fig. S2). Each ICR/RIP from rice (O. sativa ssp. japonica) and Brachypodium distachyon appear to be orthologous to HvICRa/HvRIPa, HvICRb/HvRIPb, and HvICRc/HvRIPc, respectively (Supplemental Fig. S2).
RIPb Influences the Susceptibility of Barley to Bgh
Semiquantitative reverse transcription PCR showed that all three barley ICRs/RIPs are transcribed in whole leaves and the epidermis, with RIPb showing the highest RNA levels and RIPa being only weakly expressed. Samples from inoculated leaves showed that Bgh infection does not alter the expression of any of the three barley ICRs/RIPs (Supplemental Fig. S3).
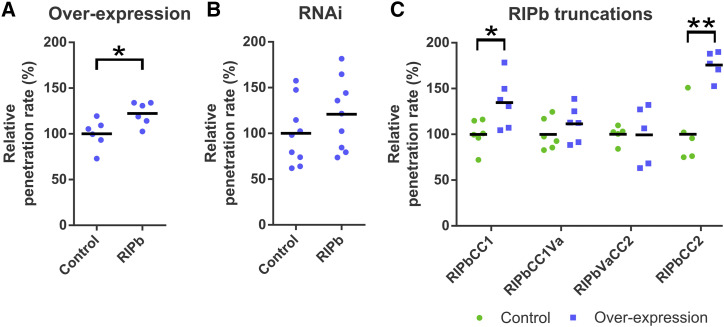
To investigate if one of the ICRs/RIPs influences the susceptibility of barley to Bgh, we tested the penetration efficiency of Bgh into transiently transformed epidermal cells. We introduced either an overexpressing construct under the control of the cauliflower mosaic virus (CaMV) 35S promoter or a posttranscriptional gene-silencing construct into these cells. Overexpression of RIPa or RIPc had no significant effect on susceptibility (Supplemental Fig. S4A; barley ICR/RIP proteins are called RIPs from now on for reasons of simplicity). Overexpression of RIPb, however, significantly and consistently increased the penetration rate of Bgh into transformed cells by 22% compared with cells transformed with the empty vector control (Fig. 1A). RNAi-mediated silencing of RIPb did not significantly change the penetration rate of Bgh into the transformed cells (Fig. 1B).
Figure 1.
The effect of RIPb on the interaction of barley and Bgh was tested by biolistic transformation of epidermal cells of 7-d-old barley leaves and determination of the penetration rate of Bgh into the transformed cells 24 h after inoculation. Overexpression constructs for RIPb (A) as well as an RNAi silencing construct for RIPb (B) and overexpression constructs for RIPb truncations (C) were introduced. As a control, the respective empty vectors were used. Values represent means of results of individual independent experiments (n ≥ 5) relative to the mean of each respective control set as 100%. *, P < 0.05 and **, P < 0.01, Student’s t test.
RIPb Interacts with RACB
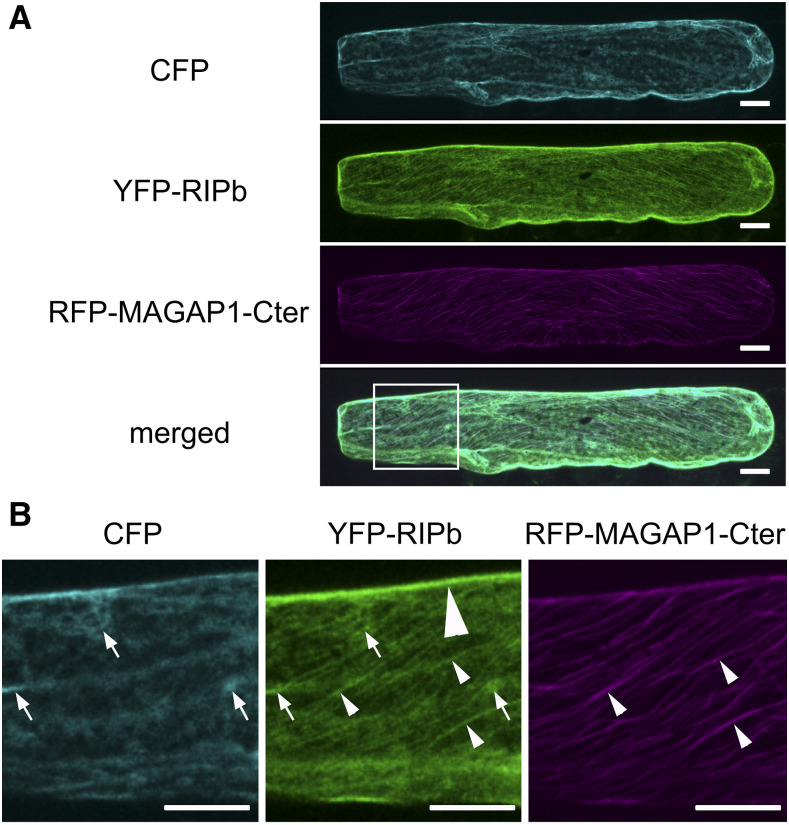
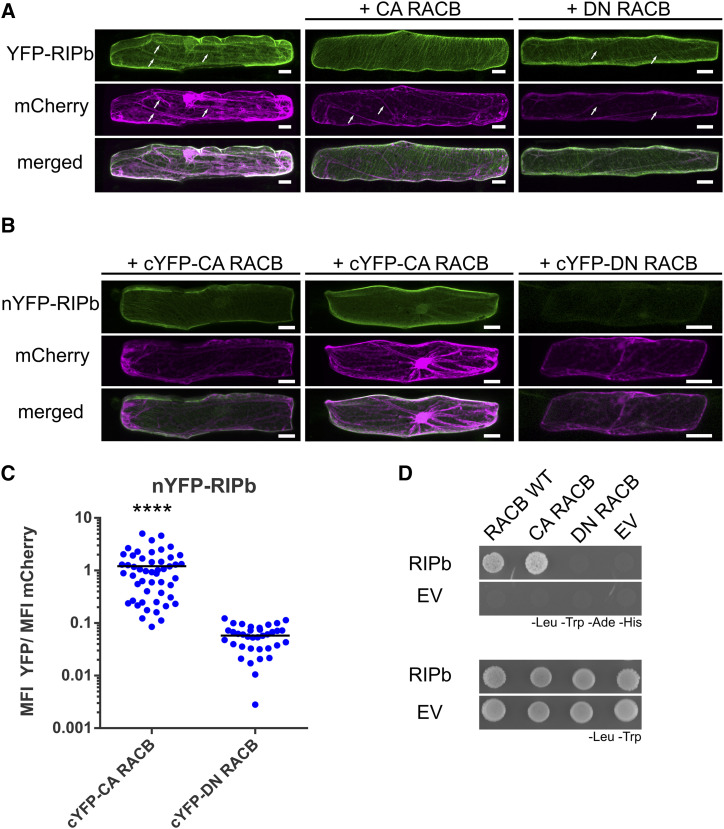
In order to determine the subcellular localization of RIPb, we transiently expressed a YFP-tagged fusion protein of RIPb in single epidermal cells via biolistic transformation. YFP-RIPb was detected in the cytoplasm, at cytoskeleton structures, and at the cell periphery. Coexpression experiments showed partial colocalization of YFP-RIPb and the barley microtubule marker MAGAP1-Cter at cortical microtubules (Fig. 2). The RFP-MAGAP1-Cter marker does not interact with ROPs because it lacks the ROP-interacting CRIB and the GAP domains but still contains the microtubule-interacting C-terminal part of MAGAP1 (Hoefle et al., 2011). Recorded images further confirmed that YFP-RIPb is also present in the cytosol, where it colocalized with additionally coexpressed soluble CFP, and at the cell periphery or plasma membrane (Fig. 2). Coexpression with CA RACB-G15V resulted in depleted cytosolic localization of YFP-RIPb when compared with soluble red-fluorescing mCherry. At the same time, microtubule localization and cell peripheral localization were still clearly detectable. Cytoplasmic depletion of YFP-RIPb was not observed when we coexpressed dominant negative RACB-T20N (DN RACB; Fig. 3A). This change in RIPb localization might be best explained if RACB recruits RIPb to the cell periphery/plasma membrane. To test this, we coexpressed YFP-RIPb with the plasma membrane marker pm-rk (Nelson et al., 2007; Weis et al., 2013) either alone or in the presence of CA RACB (Supplemental Fig. S5). YFP-RIPb alone showed some overlapping signal with pm-rk, but the peak in the signal profile was slightly displaced due to additional cytosolic signal (Supplemental Fig. S5A). However, in the presence of CA RACB, we recorded a shift of the YFP-RIPb peak toward the peak of the plasma membrane marker (Supplemental Fig. S5B). This supports that RIPb gets recruited to the plasma membrane by activated RACB but also shows that RIPb itself localized close to or attached to the plasma membrane. Ratiometric BiFC experiments further supported the interaction of RIPb with RACB. YFP fluorescence was reconstituted when nYFP-RIPb and cYFP-CA RACB were coexpressed in leaf epidermal cells (Fig. 3, B and C). By contrast, coexpression of nYFP-RIPb and cYFP-DN RACB did not result in clear BiFC, and the strength of the signals was on average less than 10% of the signals recorded for the interaction with CA RACB (Fig. 3, B and C). We observed that the complemented CA RACB-RIPb YFP complex signals either at the plasma membrane or at cortical microtubules and the plasma membrane but hardly in the cytosol when compared with mCherry (Fig. 3B). We further confirmed a direct interaction between both wild-type RACB and CA RACB with RIPb (Fig. 3D) in yeast. These experiments together suggest a direct interaction between RIPb and RACB in planta.
Figure 2.
Subcellular localization of RIPb in planta. A, A barley epidermal cell was transiently cotransformed with CFP as a cytosolic marker, YFP-tagged RIPb (YFP-RIPb), and RFP-MAGAP1-Cter as a microtubule marker. Images show z-stacks of optical sections visualized by the confocal laser scanning microscope of the upper half of the cell. Bars = 20 µm. B, Magnification of the part of the cell highlighted in A (merged channel). Please note the YFP-RIPb signal in the cytoplasm (arrows; compare with CFP), at the upper cell periphery (big arrowhead; highlighted only in the YFP-RIPb channel), and at microtubules (long narrow arrowheads; compare with RFP-MAGAP1-Cter). The brightness of the images was equally increased for display purposes. Images show z-stacks of 10 confocal sections of each 2-µm increment. Images represent typical cell recordings of 10 cells per experiment and from three independent transformation experiments with similar results. Bars = 20 µm.
Figure 3.
RACB and RIPb interact in yeast and in planta. A, Single epidermal cells were transiently transformed by particle bombardment. YFP-RIPb and the cytosolic transformation marker mCherry were expressed alone or coexpressed with CA RACB or DN RACB. Images were taken 24 h after bombardment and show representative z-stacks of xy optical sections of the upper half of the cells. White arrows show cytosolic strands. Images represent typical cell recordings of 20 cells per experiment and from three independent transformation experiments with similar results. Bars = 20 µm. B, For BiFC experiments, protein fusions of RIPb, CA RACB, and DN RACB with split-YFP tags were coexpressed. Images were taken 24 h after bombardment. Images show z-stacks of 10 optical sections of the upper half of the cells. Images represent typical cell recordings (n > 30) from two independent transformation experiments with similar results. Bars = 20 µm. C, For the quantification of BiFC experiments, images were taken with constant settings and signal intensity (mean fluorescence intensity [MFI]) was measured over a region of interest at the cell periphery. The ratio between YFP and mCherry fluorescence signals was calculated. The findings shown are from one out of two replicates with similar results. For each replicate, more than 30 cells were measured. ****, P < 0.0001, Student’s t test. D, RIPb was tested in a yeast two-hybrid assay for its interaction with barley wild-type RACB (RACB WT), CA RACB, and DN RACB. As a control, the interaction with the respective empty vectors (EV) was tested. For the identification of interactions, synthetic defined medium lacking Leu (-Leu), Trp (-Trp), adenine (-Ade), and His (-His) was used. For the identification of transformed cells, synthetic dextrose medium lacking Leu and Trp was used.
RIPb Truncations Show Distinct Subcellular Localization and Function
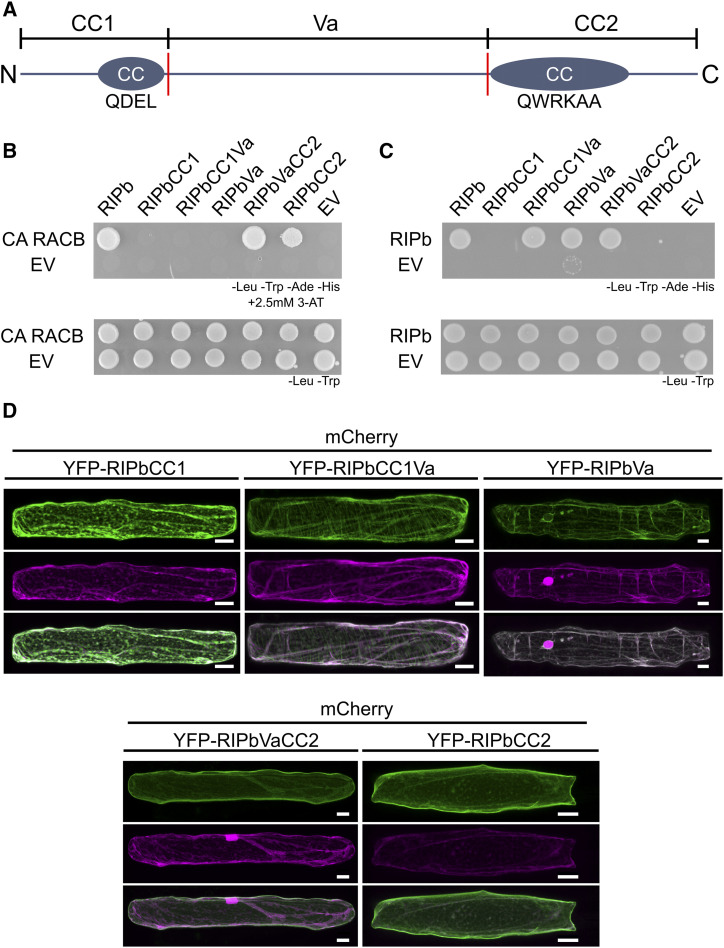
All predicted ICR/RIP proteins from barley, rice, Arabidopsis, and B. distachyon contain an N-terminal CC domain with the QD/EEL motif as well as a C-terminal CC domain with the QWRKAA motif (Fig. 4A; Supplemental Fig. S1). Based on this and with regard to previous studies (Mucha et al., 2010), we created RIPb truncations based on the first CC domain (CC1), the central variable region (Va), and the second CC domain (CC2). In yeast, only constructs containing the CC2 domain and hence the QWRKAA motif interacted with CA RACB, as was shown before for the interaction of Arabidopsis ROPs and ICRs/RIPs (Fig. 4B; Lavy et al., 2007; Mucha et al., 2010). RIPbCC2 was also able to interact with CA RACB but not with DN RACB in BiFC assays, and this interaction took place at the plasma membrane (Supplemental Fig. S6). To confirm this, we again coexpressed a YFP fusion of RIPbCC2 with the plasma membrane marker pm-rk either alone or in the presence of CA RACB (Supplemental Fig. S5). Similar to full-length RIPb, YFP-RIPbCC2 showed colocalization with pm-rk but the peak was slightly shifted in the signal profile, probably due to additional cytosolic signal (Supplemental Fig. S5C). Coexpression with CA RACB again shifted the signal toward the plasma membrane, resulting in an overlay of the two peaks in the signal profile (Supplemental Fig. S5D). RIPb was also able to interact with itself in yeast. The Va region may be important for this, since only-full length RIPb and truncations containing this region were able to interact in yeast (Fig. 4C).
Figure 4.
Structure-function relationship of RIPb. A, Domain structure and truncations of RIPb. The CC1-domain stretches from amino acids (aa) 1 to 132 and contains the N-terminal CC domain with the QDEL motif (circles). The Va region starts at amino acid 133 at ends at amino acid 420. The CC2 domain stretches from amino acid 421 to the end at amino acid 612. The CC2 domain also represents a CC structure and contains the QWRKAA motif. Red lines mark borders of the Va domain that was or was not included in the truncations as indicated. B, RIPb truncations were tested in yeast two-hybrid assays for their interaction with CA RACB or RIPb (shown in C). As controls, the interaction with the respective empty vector (EV) was tested. For the identification of interactions, synthetic defined medium without Leu (-Leu), Trp (-Trp), adenine (-Ade), and His (-His) was used, together with 2,5 mm 3-aminotriazole (3-AT) to reduce background growth in the combinations containing the RIPbVa truncation. For the identification of transformed cells, synthetic dextrose medium without Leu and Trp was used. D, Single epidermal cells were transiently transformed with different RIPb truncations tagged to YFP. Images show z-stacks of 15 optical sections of the upper half of cells. Images represent typical cell recordings of four to 18 cells per experiment and from two independent transformation experiments with similar results. Bars = 20 µm.
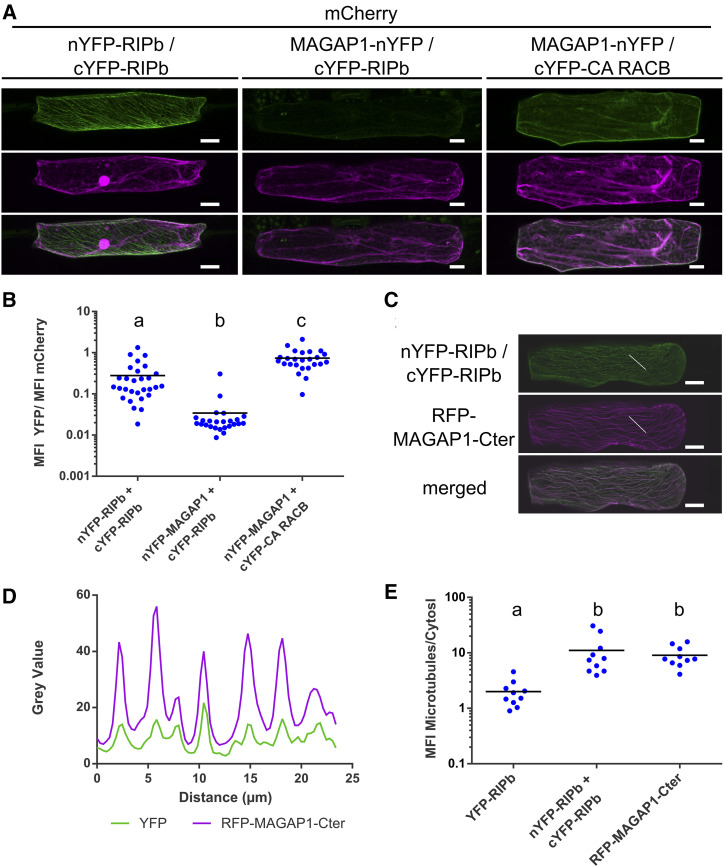
In order to look for specific subcellular localizations in planta, we created YFP-tagged protein fusions of these truncations. Like YFP-RIPbCC2, YFP-RIPbVaCC2 localized strongly to the cell periphery, presumably the plasma membrane, with weak cytosolic background (Fig. 4D). YFP-RIPbCC1Va was located in the cytosol and at the microtubules. Additionally, coexpression experiments with YFP-RIPbCC1Va and CA RACB show that YFP-RIPbCC1Va was not positioned at the cell periphery despite the presence of CA RACB (Supplemental Fig. S7), suggesting that the CC2 domain is necessary for the recruitment by CA RACB. However, YFP-RIPbCC1 and YFP-RIPbVa were detected in the cytosol (Fig. 4D). Hence, both the CC1 domain and the Va domain appear to be required but not sufficient for microtubule association. Double mutation of Asp-85 and Glu-86 of the QDEL motif did not lead to a loss of microtubule localization (Supplemental Fig. S7B). The QDEL motif itself might therefore not be necessary for microtubule localization. Since the Va domain was required for dimerization and microtubule association, RIPb might localize to the microtubules as a dimer or oligomer. This was further supported because BiFC signals recorded after coexpression of nYFP-RIPb with cYFP-RIPb occur mainly at the microtubules and show less cytosolic background (Fig. 5A) when compared with YFP-RIPb alone, which may be detected both in its monomeric and its dimeric/oligomeric forms (Fig. 2). Quantification of reconstituted YFP fluorescence showed significantly stronger signal intensities for the nYFP-RIPb/cYFP-RIPb homodimer compared with the coexpression of MAGAP1-nYFP with cYFP-RIPb, which are not supposed to interact and were used as a negative control. As a positive control for the negative control, we coexpressed MAGAP1-nYFP with cYFP-CA RACB, which again showed high YFP complementation signals (Fig. 5, A and B). Signal quantification showed high signal overlap between the complemented YFP fluorescence signal and the microtubule marker RFP-MAGAP1-Cter over a linear region of interest (Fig. 5, C and D). Since there appeared to be little cytosolic background in the nYFP-RIPb/cYFP-RIPb BiFC images, we measured the ratio between microtubule and cytosolic signal within each cell. We then compared signal ratios within YFP-RIPb-expressing cells and those expressing the nYFP-RIPb/cYFP-RIPb BiFC pair. As a positive control for microtubule localization, we also measured the signal ratio for the microtubule marker RFP-MAGAP1-Cter. The data show that the microtubule/cytosol ratio for nYFP-RIPb/cYFP-RIPb BiFC was far higher than that of YFP-RIPb, indicating a more exclusive microtubule localization of the dimer/oligomer (Fig. 5E). In fact, the signal ratio for nYFP-RIPb/cYFP-RIPb was similar to that of the microtubule marker RFP-MAGAP1-Cter.
Figure 5.
RIPb-RIPb interaction at microtubules. A, Single epidermal cells were transiently transformed by particle bombardment with split-YFP constructs in the combinations nYFP-RIPb and cYFP-RIPb, MAGAP1-nYFP and cYFP-RIPb, as well as MAGAP1-nYFP and cYFP-RIPb. Images represent typical cell recordings of 10 cells per experiment and from three independent transformation experiments with similar results. Images represent z-stacks of 10 confocal sections of each 2-µm increment. B, Quantification of BiFC signals from images were taken with constant settings. Signal intensity (mean fluorescence intensity [MFI]) was measured over a region of interest at the cell periphery. The ratio between split-YFP and free mCherry signal was calculated. Signals were measured in 10 cells for each construct. Letters indicate significance by one-way ANOVA (Tukey’s multiple comparison test; P < 0.05). C and D, Coexpression of nYFP-RIPb and cYFP-RIPb with RFP-MAGAP1-Cter. Image brightness was equally increased for display purposes (C), but signal intensities over a region of interest (white lines) were measured using original data (D). Bars = 20 µm. E, Ratio between the microtubule signal and the cytosolic signal was measured for YFP-RIPb alone, BiFC signal for RIPb-RIPb interaction, and the microtubule marker RFP-MAGAP1-Cter. Letters indicate significance by one-way ANOVA (Tukey’s multiple comparison test; P < 0.05). Microtubule and cytosolic signals were measured at three different regions of interest in the cell. Measurements were made in single optical sections of the z-stack. Note the logarithmic scales.
Results from Lavy et al. (2007) and Mucha et al. (2010) suggest that ICRs/RIPs lacking a functional QWRKAA motif lose the ability to interact with ROPs and that either the CC1 or CC2 domain can bind to further downstream signaling components. This indicates that RIPb might be able to fulfill a ROP signaling function through one of these domains. To test the functionality of RIPb truncations, we tested their effect on the penetration success of Bgh on barley. Interestingly, overexpression of RIPbCC2 strongly increased susceptibility by about 75% (Fig. 1C). In contrast, overexpression of the CC2 domain of RIPa did not lead to a significant increase in susceptibility (Supplemental Fig. S4B). The effect of RIPbCC2 completely disappeared when we expressed RIPbVaCC2, containing additionally the Va domain. The CC1 domain alone also increased susceptibility by about 35%, and this effect was also reduced when we expressed the longer RIPbCC1Va truncation (Fig. 1C). In order to investigate the possible influence of protein levels on the influence on susceptibility, we measured the fluorescence intensity of YFP-tagged fusion proteins relative to an internal mCherry control in single transformed cells. We found that the fluorescence intensities of full-length RIPb and YFP-RIPbVaCC2 were lower than those of RIPbCC1 and RIPbCC2 (Supplemental Fig. S8). Hence, protein expression levels might have influenced the strength of induced susceptibility, but CC1 and particularly CC2 alone were sufficient to support fungal penetration success.
Oda et al. (2010) showed that in Arabidopsis RIP3/ICR5/MIDD1 is involved in local microtubule depolymerization during xylem cell development. Microtubule depletion might also influence the outcome of the interaction of barley and Bgh. Indeed, we recently showed that barley RIPa influences microtubule organization when coexpressed with barley RAC1 and MAGAP1 (Hoefle et al., 2020). To see if this could be the case for RIPb in barley, we coexpressed RIPb and RIPbCC2, individually, with the microtubule marker RFP-MAGAP1-Cter and evaluated microtubule organization as described by Nottensteiner et al. (2018). In the empty vector control, we found 68% of microtubules in a well-organized parallel state, while about 17% of cells showed disordered but intact microtubules and the rest of the cells showed fragmented microtubules (Supplemental Fig. S9). In cells expressing RIPb or RIPbCC2, we observed a similar pattern. The amount of fragmented microtubules was a little lower in cells expressing RIPb and a little higher in cells expressing RIPbCC2, but no statistically significant changes were observed.
RACB and RIPb Colocalize at the Site of Fungal Attack
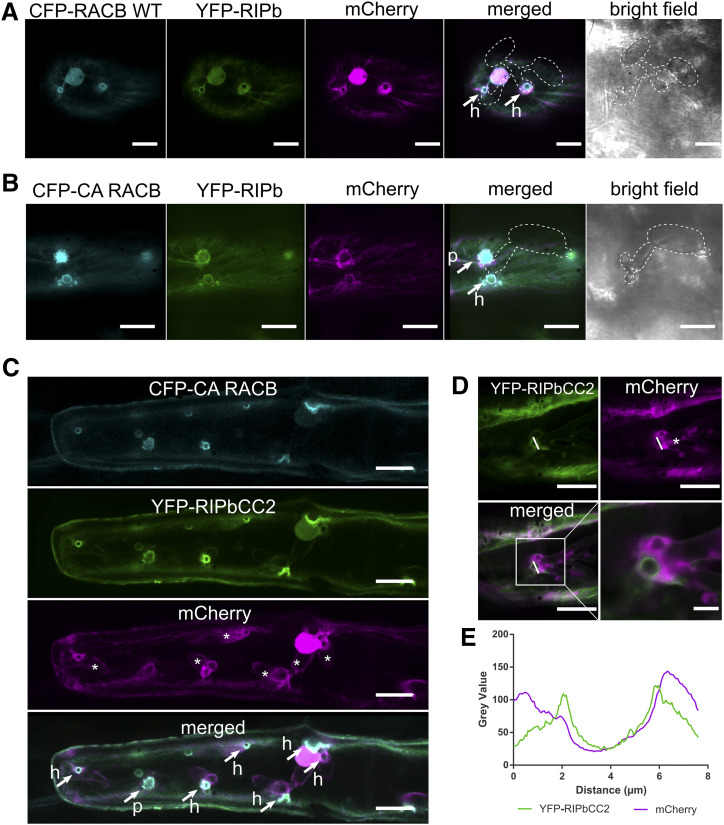
Since RIPb and RACB can interact in planta and both proteins can influence barley susceptibility to Bgh, we wanted to know whether RIPb and RACB would localize to the sites of fungal penetration. Therefore, we transiently coexpressed YFP-RIPb and CFP-RACB in single epidermal cells and inoculated the leaves with conidia of Bgh. At 24 h after inoculation, we observed ring-like accumulations of both YFP-RIPb and CFP-RACB at the site of fungal penetration around the haustorial neck. Cytosolic mCherry appeared also in regions at the site of attack but was less spatially confined than YFP-RIPb and CFP-RACB (Fig. 6A). We observed even more pronounced fluorescence at infection sites when YFP-tagged RIPb was coexpressed with CFP-CA RACB. In this context, we detected clear accumulation of RIPb and CA RACB at the site of fungal penetration, although independent of the outcome of the penetration attempt. If the penetration was successful, a clear ring-like localization pattern around the haustorial neck could be observed. However, if the fungal penetration was not successful, we detected a more fringed accumulation of both proteins, possibly representing membrane domains around papilla protrusions (Fig. 6B). Since RIPbCC2 had a stronger influence on fungal penetration success than full-length RIPb, we also imaged YFP-RIPbCC2 when coexpressed with CFP-CA RACB. Interestingly, there was a very strong localization of both proteins around the haustorial neck region in penetrated cells but also in some instances at sites of repelled fungal attempts (Fig. 6C). The ring-like accumulation of RIPbCC2 around the haustorial neck was also visible later at 48 h after the inoculation (Fig. 6D). There was also constantly local aggregation of cytoplasm at the sites of attack, but measurements of the ring-like YFP-RIPbCC2 fluorescence showed that signal intensities were clearly more confined to the cell periphery compared with cytosolic mCherry fluorescence (Fig. 6E).
Figure 6.
RIPb and RACB colocalize at sites of fungal attack. A and B, Transiently transformed epidermis cells were inoculated with Bgh. YFP-RIPb colocalizes with wild-type RACB (CFP-RACB WT; A) as well CFP-CA RACB (B) at the site of fungal attack at 24 h after inoculation. mCherry was used as a cytosolic marker. Confocal laser scanning micrographs show z-stacks of the upper part of the cells (five confocal sections of each 2-µm increment). Transmission channel images show a single optical section. C, YFP-RIPCC2 colocalizes with CFP-CA RACB 24 h after inoculation at the site of fungal attack. mCherry was used as a cytosolic marker. Arrows mark sites of fungal penetration attempts that either succeeded with the formation of a haustorium (h) or failed in a nonpenetrated papilla (p). Asterisks indicate haustorial bodies. D, Single epidermal cells were transiently transformed with YFP-RIPbCC2 and mCherry. Images were taken 48 h after inoculation with Bgh. The asterisk indicates a haustorial body. E, Signal intensities at the haustorial neck over the region of interest (white lines in D). Images in A to D show z-stacks of 15 confocal sections of each 2-µm increment. Images represent typical cell recordings of at least five cells per experiment and from more than 10 independent transformation experiments with similar results. Bars = 20 µm in A to C and to 5 µm in D.
DISCUSSION
ICR/RIP proteins are considered scaffold proteins in ROP signaling. Like RICs, ICRs/RIPs might be key factors in the branching of ROP signaling in plants. It appears that so far most described downstream functions of ROPs are mediated through either RIC or ICR/RIP proteins. ICRs/RIPs contain a characteristic QWRKAA motif in the CC2 domain, which was previously described as the motif responsible for ROP interaction (Lavy et al., 2007). Our results support this, since only full-length RIPb and truncations containing this motif interacted with RACB and were subcellularly recruited by CA RACB (Figs. 3 and 4; Supplemental Figs. S5–S7). The CC2 domain is part of all predicted ICRs/RIPs form Arabidopsis, rice, B. distachyon, and barley. All identified ICRs/RIPs from these four species also contain a conserved QD/EEL motif located in an N-terminal CC1 domain (Supplemental Fig. S1). The function of this motif, however, remains more elusive. Although the CC1 domain is important for the microtubule localization of RIPb (Fig. 4), amino acid exchanges in the QDEL motif did not result in a loss of microtubule association (Supplemental Fig. S7).
Phylogenetic analyses show that both rice and B. distachyon possess putative orthologs of each of the three barley ICRs/RIPs, implying possible conserved function of the ICRs/RIPs in grasses (Supplemental Figs. S1 and S2). However, the five ICR/RIP proteins of Arabidopsis show no clear phylogenetic relation to the grass ICRs/RIPs. It would be interesting to see whether Arabidopsis and grass ICRs/RIPs have similar functions or may have evolved in different directions, as the little sequence conservation suggests. In this context, it is noteworthy that barley RIPa has functions reminiscent of Arabidopsis RIP3/ICR5/MIDD1 in microtubule organization, as recently shown (Hoefle et al., 2020). Hence, although these proteins are little conserved when compared between monocots and dicots, they may share at least some conserved functions in ROP signaling. By contrast, when comparing barley RIPa and RIPb, only RIPb appears to influence the interaction outcome, whereas RIPa might have a greater influence on microtubule patterns (Hoefle et al., 2020). This suggests that ICR/RIP proteins have functionally diversified in barley.
For this study, we focused on a possible RACB signaling mechanism via ICR/RIP proteins during the interaction of barley and Bgh. Barley RIPb interacts with CA and wild-type RACB in yeast, supporting that it is a potential downstream interactor of RACB. Overexpression of RIPb but not of RIPa or RIPc increased the penetration rate of Bgh into transformed epidermal barley cells (Fig. 1; Supplemental Fig. S4A). RIPb silencing had no significant effect on the interaction between epidermal cells and Bgh (Fig. 1B). This might be due to residual transcript or protein amounts of RIPb after transient knockdown or due to a convergence in RACB downstream signaling that could compensate for the lack of RIPb during the interaction. For instance, RIC171 might act as an alternative downstream interactor of RACB (Schultheiss et al., 2008), and it is possible that even more interactors of RACB are involved, because ROP proteins are considered signaling hubs (Nibau et al., 2006). Hence, silencing of only one signaling branch might not have a significant effect on the interaction, whereas overexpression could support a certain RACB downstream branch and therefore has an effect. Additionally, RACB is not the only barley ROP that can support fungal penetration success (Schultheiss et al., 2003), and hence, even RACB-independent ROP signaling could compensate for RACB-RIPb functions in RIPb-silenced cells.
RIPb shows diverse subcellular localizations. In addition to cytosolic localization, we observed localization at the plasma membrane and at the microtubule cytoskeleton (Fig. 2). The N-terminal CC1 domain is necessary but not sufficient for microtubule localization, since the RIPbVaCC2 truncation lacking the CC1 domain did not localize to microtubules. The CC1 domain alone also did not show microtubule localization. The central Va domain alone was insufficient for microtubule association but it appeared to be required for both microtubule association and RIPb-RIPb interaction (Fig. 4). BiFC experiments further suggested that the RIPb-RIPb interaction takes mainly place at microtubules (Fig. 5). Interestingly, truncated versions of RIPb, which contain the Va domain, did not induce susceptibility when overexpressed, whereas RIPbCC1 and particularly RIPbCC2 induced susceptibility, similar to or much stronger than the full-length protein. We therefore speculate that dimerization or oligomerization of RIPb at microtubules might have a regulatory purpose, potentially by sequestration of inactive RIPb. However, interpretation of these results is complicated because the amount of expressed protein truncations also differed and could partially explain the differences in efficacy. Nevertheless, overexpression of the RIPbCC2 domain resulted in a very strong increase in the susceptibility of barley epidermal cells to Bgh. Lavy et al. (2007) showed that the QWRKAA motif in the CC2 domain of Arabidopsis AtICR1/AtRIP1 is not only necessary for ROP interaction but also for the interaction with the downstream interactor AtSEC3, indicating that the CC2 domain might be able to fulfill the signaling function of AtICR1/AtRIP1. In a follow-up publication by the same group, they also show that the last 40 amino acids of AtRIP1 are also required for interaction with CMI1 (Ca2+-dependent modulator of ICR1; Hazak et al., 2019). This might be comparable to RIPbCC2, if RIPbCC2 would bind in addition to RACB a yet to be identified downstream protein. By contrast, overexpression of the CC2 domain of RIPa did not result in a significant increase in susceptibility (Supplemental Fig. S4B), and therefore this effect appears specific for RIPb. RIPbCC2 was able to interact with RACB in yeast and in planta (Fig. 4; Supplemental Fig. S6). Furthermore, RIPb did not localize to the cell periphery anymore without the CC2 domain (RIPbCC1Va) even in the presence of CA RACB (Supplemental Fig. S7). These findings together suggest that the CC2 domain of RIPb is responsible both for ROP interaction and for a function that may take place at the plasma membrane.
The N-terminal CC1 domain of RIPb is required for microtubule association but might interact with signaling components as well. This could explain the susceptibility effect of the overexpression of RIPbCC1, although the CC1 domain itself does not interact with RACB (Figs. 1C and 4C). Interestingly, the CC1 domain of Arabidopsis AtRIP3/ICR5/MIDD1 is required for interaction with KINESIN13A (Mucha et al., 2010). It could hence be that RIPb also fulfills a dual function via different domains of the protein.
BiFC experiments showed interaction between RACB and RIPb at the microtubules and at the plasma membrane. Since RACB alone does not localize to microtubules (Schultheiss et al., 2003), it seems that RIPb is able to recruit RACB to microtubules when overexpressed. The interaction between the susceptibility-inducing CC2 domain and RACB, on the other hand, takes place at the plasma membrane (Supplemental Figs. S5 and S6). These results suggest that RACB also recruits RIPb to the plasma membrane during susceptibility signaling and that recruitment of RACB to microtubules perhaps limits this effect. We speculate that in this experimental setup, recruitment of RACB to microtubules brings RACB into proximity of microtubule-located MAGAP1, which presumably inactivates RACB (Hoefle et al., 2011). This might explain why full-length RIPb has a less strong effect on susceptibility when compared with RIPbCC2, which cannot recruit RACB to the microtubules. We found that protein levels of YFP-RIPbCC2 are higher than the levels of full-length YFP-RIPb when transiently expressed in epidermal cells (Supplemental Fig. S6). Since both constructs are driven by a CaMV35S promoter, a different posttranscriptional regulation or protein turnover might be the most plausible explanation for this. The difference in protein levels can influence the effect that both proteins have, which would also confirm our notion that RIPbCC2 might be a less regulated functional version of RIPb. However, since RIPbVaCC2 showed similar protein levels to RIPb but had no influence on the outcome of the interaction between barley and Bgh, it is unlikely that protein levels alone explain the different efficacies of overexpression constructs.
We observed colocalization of RIPb and RACB and of RIPbCC2 and RACB at the site of fungal attack. In interactions where the fungus was able to penetrate the host cell, a ring of RIPb and RACB or CA RACB around the haustorial neck at the plasma membrane was observed. However, we also observed signals at repelled penetration attempts around the formed papilla, indicating that the accumulation of these two proteins alone is not sufficient to render all cells susceptible. RACB possesses a C-terminal CSIL motif, which is predicted to mediate protein prenylation at the Cys residue and is necessary for plasma membrane association and function in susceptibility (Schultheiss et al., 2003). Additionally, RACB has a polybasic stretch close to the C terminus (Schultheiss et al., 2003) shown for other ROPs to be involved in lipid interaction (Platre et al., 2019) and a conserved Cys at position Cys-158, which is S-acylated in activated Arabidopsis AtROP6 (Sorek et al., 2017). Hence, lipid modification and interaction with negatively charged phospholipids together may bring activated RACB to specific membrane domains, to which it then recruits proteins that execute ROP signaling function. Phosphatidylserine and phosphoinositides are often involved in defining areas of cell polarization in membranes, such as during root hair and pollen tube tip growth (Helling et al., 2006; Kusano et al., 2008; Platre et al., 2019), and ROPs are known to moderate the phosphorylation pattern of phosphoinositides during polarization (Kost et al., 1999). We hence speculate that localization of ROP signaling components at the site of interaction reflects domains of enriched negatively charged phospholipids.
The exact effect of RACB-RIPb signaling on the interaction remains unknown so far. However, the finding that Arabidopsis ICRs/RIPs interact with proteins of the exocyst complex and KINESIN13A opens the possibility that barley ICRs/RIPs also modify the cytoskeleton or membrane trafficking, both being key to resistance and susceptibility in powdery mildew interactions (Hückelhoven and Panstruga, 2011; Dörmann et al., 2014), although we found no strong evidence that the microtubule cytoskeleton might be affected by RIPb (Supplemental Fig. S9). We cannot finally exclude that RIPb would influence microtubule organization in attacked cells, but this is difficult to assess because RIPb supports the fungus and Bgh on its own has the potential to influence microtubule structure (Hoefle et al., 2011; Nottensteiner et al., 2018). Together, our data support a hypothesis according to which RIPb is localized at microtubules, from which it is recruited to RACB signaling hotspots at the plasma membrane by activated RACB. There it might interact with further proteins of the RACB signaling pathway but also with RACB-independent factors to facilitate fungal entry into barley epidermal cells. The fact that the putative fungal effector ROPIP1 binds RACB and destabilizes barley microtubules (Nottensteiner et al., 2018) adds another level of complexity, on which ROPIP1 may facilitate the release of RIPb from microtubules for its function in susceptibility.
CONCLUSION
Over the last years, the impact of susceptibility factors for plant-pathogen interactions has become more and more obvious. The barley susceptibility factor RACB might be a key player in cellular polarization during fungal invasion. Here, we identified RIPb as a potential downstream interactor of activated RACB in susceptibility. RACB and RIPb might be involved in fine-tuning of cell polarization to the advantage of the fungus. It will be important to identify further interactors of RIPb and of its strongly susceptibility-supporting CC2 domain. This may establish a deep understanding of the components and mechanisms of subcellular reorganization in the cell cortex, which support the biotrophic parasite Bgh in accommodation of its haustorium in an intact epidermal cell.
MATERIALS AND METHODS
Biological Material
Barley (Hordeum vulgare) ‘Golden Promise’ was used in all experiments. Plants were grown under long-day conditions with 16 h of light and 8 h of dark with a relative humidity of 65% and light intensity of 150 µmol s−1 m−2 at a temperature of 18°C.
The powdery mildew fungus Blumeria graminis f. sp. hordei race A6 was cultivated on wild-type cv Golden Promise plants under the conditions described above and inoculated by blowing spores into a plastic tent that was positioned over healthy plants or transformed leaf segments.
Cloning Procedures
HvRIPb (HORVU1Hr1G012460) was amplified from cDNA using primers Ripb-EcoRI_fwd and Ripb-BamHI_rev (Supplemental Table S1), introducing EcoRI and BamHI restriction sites, respectively. HvRIPa (HORVU3Hr1G087430) was amplified from cDNA using primers RipaXbaI_fwd and RipaXbaI_rev, introducing the XbaI restriction site at the 5′ and 3′ ends. HvRIPc (HORVU3Hr1G072880) was amplified from cDNA using primers RipcXbaI_fwd and RipcPstI_rev, introducing restriction sites for XbaI at the 5′ end and for SalI at the 3′ end. The amplified products were ligated into the pGEM-T easy vector (Promega) by blunt-end cloning according to the manufacturer’s instructions and sequenced. HvRIPb truncations spanned the following amino acids: HvRIPbCC1 from amino acids 1 to 132, HvRIPbVa from amino acids 133 to 420, and HvRIPbCC2 from amino acids 420 to 612. HvRIPb truncations for yeast two-hybrid analysis were amplified from pGEM-T easy containing full-length RIPb using primers with EcoRI and BamHI restriction sites. RIPbCC1 was amplified using primers Ripb-EcoRI_fwd and RipbCC1BamHI_rev, RIPbCC1Va with primers Ripb-EcoRI_fwd and RipbVaBamHI_rev, RIPbVa with primers RipbVaEcoRI_fwd and RipbVaBamHI_rev, RIPbVaCC2 with primers RipbVaEcoRI_fwd and Ripb-BamHI_rev, and RIPbCC2 with primer RipbC2EcoRI_fwd. Each reverse primer introduced a stop codon. For yeast two-hybrid assays, HvRIPb and HvRIPb truncations were subcloned from the pGEM-T easy vector into pGADT7 and pGBKT7 plasmids (Clontech Laboratories) using the EcoRI and BamHI restriction sites. For overexpression constructs and constructs for protein localization, the pUC18-based vector pGY1, containing a CaMV35S promoter, was used (Schweizer et al., 1999). From the pGEM-T easy vector, HvRIPb was further amplified with primers Ripb-XbaI_fwd and Ripb-SalI_rev, containing XbaI and SalI restriction sites, respectively. Using those restriction sites, HvRIPb was then ligated into the pGY1 plasmid and the pGY1-YFP plasmid for N-terminal YFP fusion. HvRIPa and HvRIPc were subcloned form pGEM-T easy into pGY1 using the XbaI restriction site for HvRIPa and the XbaI and PstI restriction sites for HvRIPc. An overexpression construct for HvRIPaCC2 was produced by introducing attB attachment sites for Gateway cloning. For this, a first PCR was performed with primers GW1-RipaCC2_fwd and GW1-Ripa_rev using the pGEM-T easy construct as template. A subsequent second PCR was performed using primers Gate2_F and Gate2_R to introduce attB attachment sites for Gateway cloning. The construct was then cloned by BP clonase reaction using Gateway BP Clonase II (Invitrogen) into the pDONR223 entry vector (Invitrogen). From there, HvRIPaCC2 was cloned by LR clonase reaction with Gateway LR Clonase II (Invitrogen) into pGY1-GW, a modified pGY1 vector containing the Gateway cassette. The pGY1-GW plasmid was constructed using the Gateway Vector Conversion System (Invitrogen) according to the manufacturer’s instructions.
For BiFC, HvRIPb was amplified from the pGEM-T easy vector using the primers Ripb-SpeI_fwd and Ripb-SalI_rev with restriction sites for SpeI and SalI, respectively. The construct was then digested with SpeI and SalI and ligated into pUC-SPYNE(R)173 and pUC-SPYCE(MR) plasmids (Waadt et al., 2008) using these restriction sites.
A 538-bp-long RNAi sequence for HvRIPb was amplified, using primers RipbRNAi_fwd and RipbRNAi_rev, and introduced into the pIPKTA38 vector by blunt-end cloning using the SmaI restriction site (Douchkov et al., 2005). This plasmid was used as an entry vector to clone the RNAi sequence into the pIPKTA30N vector for double-strand RNA formation via Gateway LR Clonase II (Invitrogen) reaction according to the manufacturer’s instruction.
All HvRIPb truncations were introduced into the pGY1-YFP plasmid for N-terminal YFP fusion using the following primers: for HvRIPbCC1, primers Ripb-XbaI_fwd and RipbC1-SalI_rev; for HvRIPbCC1Va, primers Ripb-XbaI_fwd and RipbVa-SalI_rev; for HvRIPbVa, primers RipbVa-XbaI_fwd and RipbVa-SalI_rev; for HvRIPbVaCC2, primers RipbVa-XbaI_fwd and Ripb-SalI_rev; and for HvRIPbCC2, primers RipbC2-XbaI_fwd and Ripb-SalI_rev. All forward primers introduce an XbaI restriction site and all reverse primers contain a SalI restriction site, which were used for the ligation into pGY1-YFP. The same products and restriction sites were used for ligation into the pGY1 vector except for HvRIPbCC1Va. For HvRIPbCC1Va, primers GW-Ripb_fwd and GW1-RipbC1Va_rev were used for amplification followed by a second PCR with primers Gate2_F and Gate2_R to introduce attB attachment sites for Gateway cloning. The construct was then cloned by BP clonase reaction using Gateway BP Clonase II (Invitrogen) into the pDONR223 entry vector (Invitrogen). From there, HvRIPbCC1Va was cloned by LR clonase reaction with Gateway LR Clonase II (Invitrogen) into pGY1-GW.
Transient Transformation of Barley Cells
Barley epidermal cells were transiently transformed by biolistic particle bombardment using PDS-1000/HE (Bio-Rad). For this, 7-d-old primary leaves of barley were cut and placed on 0.8% (w/v) water agar. Per shot, 302.5 µg of 1-µm gold particles (Bio-Rad) was coated with 1 µg of plasmid; 0.5 µg of plasmid per shot was used for cytosolic transformation markers. After the addition of plasmids to the gold particles, CaCl2 was added to a final concentration of 0.5 m. Finally, 3 µL of 2 mg mL−1 protamine (Sigma) was added to the mixture per shot. After incubation for half an hour at room temperature, gold particles were washed twice with 500 µL of ethanol, in the first step with 70% (v/v) ethanol and in the second step with 100% (v/v) ethanol. After washing, the gold particles were resuspended in 6 µL of 100% (v/v) ethanol per shot and placed on the macrocarrier for bombardment.
Alignments and Phylogenetic Analysis
Sequences of Arabidopsis (Arabidopsis thaliana) ICR/RIP proteins were used to identify barley ICRs/RIPs using the IPK Barley BLAST Server (https://webblast.ipk-gatersleben.de/barley_ibsc/viroblast.php). ICRs/RIPs from rice (Oryza sativa ssp. japonica) were identified using the BLAST tool on the Rice Genome Annotation Project (http://rice.plantbiology.msu.edu/home_faq.shtml; Kawahara et al., 2013). ICRs/RIPs from Brachypodium distachyon were identified by BLAST search on EnsemblPLANTs (https://plants.ensembl.org/index.html). The alignment of ICR/RIP protein sequences was done with ClustalO (https://www.ebi.ac.uk/Tools/msa/clustalo/) and displayed with Jalview (jalview 2.10.5). A phylogenetic maximum likelihood tree was generated using the PhyML tool in the program seaview (v4.7).
Determination of Susceptibility
Transiently transformed barley leaves were inoculated with Bgh 24 h after bombardment for overexpression constructs and 48 h after bombardment for gene-silencing constructs. At 24 h after inoculation, the penetration rate into the transformed cells was determined by fluorescence microscopy as described before (Hückelhoven et al., 2003). For independent experiments, 50 to 230 cells were judged depending on the transformation efficacy for each control and test construct. Efficacy of the RIPb gene-silencing construct was checked before use in the susceptibility assays. Therefore, we checked their ability to significantly reduce the fluorescence signal intensity of coexpressed YFP-RIPb versus mCherry (both under the control of a CaMV35S promoter) proteins by ratiometric measurement of both proteins when constructs were codelivered into barley epidermal cells.
Protein Localization and Protein-Protein Interaction in Planta
Localization of HvRIPb and colocalization of HvRIPb and HvRACB were determined by transiently transforming barley epidermal cells with plasmids encoding fluorophore fusion proteins. Imaging was done with a Leica TCS SP5 microscope equipped with hybrid HyD detectors. CFP was excited at 458 nm and detected between 465 and 500 nm. YFP was excited at 514 nm and detected between 525 and 570 nm. Excitation of mCherry and RFP was done at 561 nm and detection between 570 and 610 nm.
For ratiometric quantification of BiFC experiments, mean fluorescence intensity was measured over a region of interest at the cell periphery. Background signal was subtracted and the ratio between YFP and mCherry signals was calculated. At least 25 cells were analyzed per construct for each experiment. Images were taken 24 to 48 h after transformation by particle bombardment.
To evaluate the microtubule-to-cytosol signal ratio, 10 cells per construct were measured. Mean fluorescence intensity in each cell was measured either on cytosolic strands or along microtubules on three different regions of interest each in single imaging plains. Average mean fluorescence intensity was calculated for cytosol and microtubules individually. Afterward, the ratio between average microtubule signal and average cytosolic signal was calculated.
Yeast Two-Hybrid Assays
For targeted yeast two-hybrid assays, HvRIPb and its truncations were introduced into pGADT7. Introduction of HvRACB into pGBKT7 was as described by Schultheiss et al. (2008). Constructs were transformed into yeast strain AH109 following the small-scale LiAc yeast transformation procedure from the Yeast Protocol Handbook (Clontech).
RNA Extraction and Semiquantitative PCR
RNA was extracted from barley tissue using the TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. One microgram of RNA was reverse transcribed with the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions.
For semiquantitative PCR, 2 µL of cDNA transcribed from RNA of pealed epidermis from barley leaves was used. Samples were taken from leaves 24 h after inoculation with Bgh or from uninoculated leaves of the same age. A 209-bp fragment of RIPa was amplified with an annealing temperature (Ta) of 58°C with primers Ripa_sqPCR4_fwd and Ripa_sqPCR5_rev (Supplemental Table S1). For RIPb, a 181-bp fragment was amplified at a Ta of 56°C using primers Ripb_sqPCR9_fwd and RIPb_sqPCR10_rev. For RIPc, a 168-bp fragment was amplified at a Ta of 58°C using primers Ripc_sqPCR4_fwd and Ripc_sqPCR5_rev. As a control, HvUbc was amplified at a Ta of 61°C using primers HvUBC2_fwd and HvUBC2_rev.
Statistics
For bilateral comparisons, unpaired two-sided Student’s t test was performed. For multiple comparisons, a one-way ANOVA followed by Tukey’s test was performed.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: RACB (CAC83043), MAGAP1 (CCE60914), RIPa/ICRa_HORVU3Hr1G087430 (BAJ94758), RIPb/ICRb_HORVU1Hr1G012460 (BAJ91555), and RIPc/ICRc_HORVU3Hr1G072880 (BAJ96551).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Alignment of amino acid sequences of barley ICR/RIP proteins with ICR/RIP proteins of Arabidopsis and rice.
Supplemental Figure S2. Phylogenetic relationship of ICR/RIP proteins.
Supplemental Figure S3. Semiquantitative PCR shows transcription levels of HvRIPa, HvRIPb, and HvRIPc in barley epidermis.
Supplemental Figure S4. Effect of RIPa and RIPc on the interaction of barley and Bgh.
Supplemental Figure S5. RIPb gets recruited to the plasma membrane by RACB.
Supplemental Figure S6. The CC2 domain of RIPb interacts with RACB in planta.
Supplemental Figure S7. RIPbCC1Va cannot be recruited to the cell periphery by RACB.
Supplemental Figure S8. Stability of YFP-RIPb fusion proteins.
Supplemental Figure S9. RIPb influence on cortical microtubules.
Supplemental Table S1. Primer list.
Acknowledgments
We thank Johanna Hofer for technical assistance and Dr. Caroline Höfle, Dr. Michaela Stegmann, and Clara Igisch for their support of this work (all Technical University of Munich, Phytopathology).
Footnotes
This work was supported by the Deutsche Forschungsgemeinschaft (grant nos. HU886/8 and SFB924 to R.H.).
References
- Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K, Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y, et al. (2013) An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe 13: 465–476 [DOI] [PubMed] [Google Scholar]
- Catanzariti AM, Dodds PN, Ellis JG(2007) Avirulence proteins from haustoria-forming pathogens. FEMS Microbiol Lett 269: 181–188 [DOI] [PubMed] [Google Scholar]
- Chen L, Shiotani K, Togashi T, Miki D, Aoyama M, Wong HL, Kawasaki T, Shimamoto K(2010) Analysis of the Rac/Rop small GTPase family in rice: Expression, subcellular localization and role in disease resistance. Plant Cell Physiol 51: 585–595 [DOI] [PubMed] [Google Scholar]
- Chowdhury J, Henderson M, Schweizer P, Burton RA, Fincher GB, Little A(2014) Differential accumulation of callose, arabinoxylan and cellulose in nonpenetrated versus penetrated papillae on leaves of barley infected with Blumeria graminis f. sp. hordei. New Phytol 204: 650–660 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Kim H, Ott T, Schulze-Lefert P, Trujillo M, Wewer V, Hückelhoven R(2014) Cell-autonomous defense, re-organization and trafficking of membranes in plant-microbe interactions. New Phytol 204: 815–822 [DOI] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P(2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18: 755–761 [DOI] [PubMed] [Google Scholar]
- Feiguelman G, Fu Y, Yalovsky S(2018) ROP GTPases structure-function and signaling pathways. Plant Physiol 176: 57–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Gu Y, Zheng Z, Wasteneys G, Yang Z(2005) Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700 [DOI] [PubMed] [Google Scholar]
- Fuchs R, Kopischke M, Klapprodt C, Hause G, Meyer AJ, Schwarzländer M, Fricker MD, Lipka V(2016) Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28: 130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross P, Julius C, Schmelzer E, Hahlbrock K(1993) Translocation of cytoplasm and nucleus to fungal penetration sites is associated with depolymerization of microtubules and defence gene activation in infected, cultured parsley cells. EMBO J 12: 1735–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M, Neef U, Struck C, Göttfert M, Mendgen K(1997) A putative amino acid transporter is specifically expressed in haustoria of the rust fungus Uromyces fabae. Mol Plant Microbe Interact 10: 438–445 [DOI] [PubMed] [Google Scholar]
- Hazak O, Bloch D, Poraty L, Sternberg H, Zhang J, Friml J, Yalovsky S(2010) A rho scaffold integrates the secretory system with feedback mechanisms in regulation of auxin distribution. PLoS Biol 8: e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Mamon E, Lavy M, Sternberg H, Behera S, Schmitz-Thom I, Bloch D, Dementiev O, Gutman I, Danziger T, et al. (2019) A novel Ca2+-binding protein that can rapidly transduce auxin responses during root growth. PLoS Biol 17: e3000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazak O, Obolski U, Prat T, Friml J, Hadany L, Yalovsky S(2014) Bimodal regulation of ICR1 levels generates self-organizing auxin distribution. Proc Natl Acad Sci USA 111: E5471–E5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling D, Possart A, Cottier S, Klahre U, Kost B(2006) Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. Plant Cell 18: 3519–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Börnke F, Hensel G, Kumlehn J, Hückelhoven R(2011) A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell 23: 2422–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, McCollum C, Hückelhoven R(2020) Barley ROP-Interactive Partner-a organizes into RAC1- and MICROTUBULE-ASSOCIATED ROP-GTPASE ACTIVATING PROTEIN 1-dependent membrane domains. BMC Plant Biol 20: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Jeon BW, Kim SY, Hwang JU, Lee Y(2016) The ROP2-RIC7 pathway negatively regulates light-induced stomatal opening by inhibiting exocyst subunit Exo70B1 in Arabidopsis. New Phytol 209: 624–635 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R.(2007) Cell wall-associated mechanisms of disease resistance and susceptibility. Annu Rev Phytopathol 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Dechert C, Kogel KH(2003) Overexpression of barley BAX inhibitor 1 induces breakdown of mlo-mediated penetration resistance to Blumeria graminis. Proc Natl Acad Sci USA 100: 5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R, Panstruga R(2011) Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol 14: 738–746 [DOI] [PubMed] [Google Scholar]
- Huesmann C, Reiner T, Hoefle C, Preuss J, Jurca ME, Domoki M, Fehér A, Hückelhoven R(2012) Barley ROP binding kinase1 is involved in microtubule organization and in basal penetration resistance to the barley powdery mildew fungus. Plant Physiol 159: 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Kobayashi I, Funaki Y, Fujimoto S, Takemoto T, Kunoh H(1997) Dynamic reorganization of microfilaments and microtubules is necessary for the expression of non-host resistance in barley coleoptile cells. Plant J 11: 525–537 [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S(2005) Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J 44: 516–529 [DOI] [PubMed] [Google Scholar]
- Kost B, Lemichez E, Spielhofer P, Hong Y, Tolias K, Carpenter C, Chua NH(1999) Rac homologues and compartmentalized phosphatidylinositol 4,5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J Cell Biol 145: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano H, Testerink C, Vermeer JE, Tsuge T, Shimada H, Oka A, Munnik T, Aoyama T(2008) The Arabidopsis Phosphatidylinositol Phosphate 5-Kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 20: 367–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Nielsen ME, Böhlenius H, Thordal-Christensen H(2017) The plant membrane surrounding powdery mildew haustoria shares properties with the endoplasmic reticulum membrane. J Exp Bot 68: 5731–5743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M, Bloch D, Hazak O, Gutman I, Poraty L, Sorek N, Sternberg H, Yalovsky S(2007) A novel ROP/RAC effector links cell polarity, root-meristem maintenance, and vesicle trafficking. Curr Biol 17: 947–952 [DOI] [PubMed] [Google Scholar]
- Li S, Gu Y, Yan A, Lord E, Yang ZB(2008) RIP1 (ROP Interactive Partner 1)/ICR1 marks pollen germination sites and may act in the ROP1 pathway in the control of polarized pollen growth. Mol Plant 1: 1021–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Cao L, Zhou Z, Zhu L, Ehrhardt D, Yang Z, Fu Y(2013) Rho GTPase signaling activates microtubule severing to promote microtubule ordering in Arabidopsis. Curr Biol 23: 290–297 [DOI] [PubMed] [Google Scholar]
- McLusky SR, Bennett MH, Beale MH, Lewis MJ, Gaskin P, Mansfield JW(1999) Cell wall alterations and localized accumulation of feruloyl-3′-methoxytyramine in onion epidermis at sites of attempted penetration by Botrytis allii are associated with actin polarisation, peroxidase activity and suppression of flavonoid biosynthesis. Plant J 17: 523–534 [Google Scholar]
- Mucha E, Hoefle C, Hückelhoven R, Berken A(2010) RIP3 and AtKinesin-13A: A novel interaction linking Rho proteins of plants to microtubules. Eur J Cell Biol 89: 906–916 [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A(2007) A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J 51: 1126–1136 [DOI] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY(2006) RAC/ROP GTPases: ‘Hubs’ for signal integration and diversification in plants. Trends Plant Sci 11: 309–315 [DOI] [PubMed] [Google Scholar]
- Nottensteiner M, Zechmann B, McCollum C, Hückelhoven R(2018) A barley powdery mildew fungus non-autonomous retrotransposon encodes a peptide that supports penetration success on barley. J Exp Bot 69: 3745–3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Fukuda H(2012) Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336 [DOI] [PubMed] [Google Scholar]
- Oda Y, Fukuda H(2013) Rho of plant GTPase signaling regulates the behavior of Arabidopsis kinesin-13A to establish secondary cell wall patterns. Plant Cell 25: 4439–4450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y, Iida Y, Kondo Y, Fukuda H(2010) Wood cell-wall structure requires local 2D-microtubule disassembly by a novel plasma membrane-anchored protein. Curr Biol 20: 1197–1202 [DOI] [PubMed] [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K(2001) Essential role of the small GTPase Rac in disease resistance of rice. Proc Natl Acad Sci USA 98: 759–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalski KS, Schultheiss H, Kogel KH, Hückelhoven R(2005) The receptor-like MLO protein and the RAC/ROP family G-protein RACB modulate actin reorganization in barley attacked by the biotrophic powdery mildew fungus Blumeria graminis f.sp. hordei. Plant J 41: 291–303 [DOI] [PubMed] [Google Scholar]
- Platre MP, Bayle V, Armengot L, Bareille J, Marquès-Bueno MDM, Creff A, Maneta-Peyret L, Fiche JB, Nollmann M, Miège C, et al. (2019) Developmental control of plant Rho GTPase nano-organization by the lipid phosphatidylserine. Science 364: 57–62 [DOI] [PubMed] [Google Scholar]
- Reiner T, Hoefle C, Hückelhoven R(2016) A barley SKP1-like protein controls abundance of the susceptibility factor RACB and influences the interaction of barley with the barley powdery mildew fungus. Mol Plant Pathol 17: 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheler B, Schnepf V, Galgenmüller C, Ranf S, Hückelhoven R(2016) Barley disease susceptibility factor RACB acts in epidermal cell polarity and positioning of the nucleus. J Exp Bot 67: 3263–3275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R(2002) A small GTP-binding host protein is required for entry of powdery mildew fungus into epidermal cells of barley. Plant Physiol 128: 1447–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss H, Dechert C, Kogel KH, Hückelhoven R(2003) Functional analysis of barley RAC/ROP G-protein family members in susceptibility to the powdery mildew fungus. Plant J 36: 589–601 [DOI] [PubMed] [Google Scholar]
- Schultheiss H, Preuss J, Pircher T, Eichmann R, Hückelhoven R(2008) Barley RIC171 interacts with RACB in planta and supports entry of the powdery mildew fungus. Cell Microbiol 10: 1815–1826 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Christoffel A, Dudler R(1999) Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J 20: 541–552 [DOI] [PubMed] [Google Scholar]
- Sorek N, Poraty L, Sternberg H, Buriakovsky E, Bar E, Lewinsohn E, Yalovsky S(2017) Corrected and republished from: Activation status-coupled transient S-acylation determines membrane partitioning of a plant Rho-related GTPase. Mol Cell Biol 37: e00333-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR(2003) GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J 33: 775–792 [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR(2006) Re-organization of the cytoskeleton and endoplasmic reticulum in the Arabidopsis pen1-1 mutant inoculated with the non-adapted powdery mildew pathogen, Blumeria graminis f. sp. hordei. Mol Plant Pathol 7: 553–563 [DOI] [PubMed] [Google Scholar]
- Voegele RT, Struck C, Hahn M, Mendgen K(2001) The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc Natl Acad Sci USA 98: 8133–8138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Schmidt LK, Lohse M, Hashimoto K, Bock R, Kudla J(2008) Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J 56: 505–516 [DOI] [PubMed] [Google Scholar]
- Weis C, Hückelhoven R, Eichmann R(2013) LIFEGUARD proteins support plant colonization by biotrophic powdery mildew fungi. J Exp Bot 64: 3855–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Wang J, Lin W, Li S, Li H, Zhou J, Ni P, Dong W, Hu S, Zeng C, et al. (2005) The genomes of Oryza sativa: A history of duplications. PLoS Biol 3: e38. [DOI] [PMC free article] [PubMed] [Google Scholar]