Four lysin motif receptor kinases controlling rhizobium nodule formation in the nonlegume Parasponia evolved after two ancient duplications.
Abstract
Rhizobium nitrogen-fixing nodule symbiosis occurs in two taxonomic lineages: legumes (Fabaceae) and the genus Parasponia (Cannabaceae). Both symbioses are initiated upon the perception of rhizobium-secreted lipochitooligosaccharides (LCOs), called Nod factors. Studies in the model legumes Lotus japonicus and Medicago truncatula showed that rhizobium LCOs are perceived by a heteromeric receptor complex of distinct Lys motif (LysM)-type transmembrane receptors named NOD FACTOR RECEPTOR1 (LjNFR1) and LjNFR5 (L. japonicus) and LYSM DOMAIN CONTAINING RECEPTOR KINASE3 (MtLYK3)-NOD FACTOR PERCEPTION (MtNFP; M. truncatula). Recent phylogenomic comparative analyses indicated that the nodulation traits of legumes, Parasponia spp., as well as so-called actinorhizal plants that establish a symbiosis with diazotrophic Frankia spp. bacteria share an evolutionary origin about 110 million years ago. However, the evolutionary trajectory of LysM-type LCO receptors remains elusive. By conducting phylogenetic analysis, transcomplementation studies, and CRISPR-Cas9 mutagenesis in Parasponia andersonii, we obtained insight into the origin of LCO receptors essential for nodulation. We identified four LysM-type receptors controlling nodulation in P. andersonii: PanLYK1, PanLYK3, PanNFP1, and PanNFP2. These genes evolved from ancient duplication events predating and coinciding with the origin of nodulation. Phylogenetic and functional analyses associated the occurrence of a functional NFP2-orthologous receptor to LCO-driven nodulation. Legumes and Parasponia spp. use orthologous LysM-type receptors to perceive rhizobium LCOs, suggesting a shared evolutionary origin of LCO-driven nodulation. Furthermore, we found that both PanLYK1 and PanLYK3 are essential for intracellular arbuscule formation of mutualistic endomycorrhizal fungi. PanLYK3 also acts as a chitin oligomer receptor essential for innate immune signaling, demonstrating functional analogy to CHITIN ELECITOR RECEPTOR KINASE-type receptors.
Nitrogen availability is a critical factor for plant growth, but fixed nitrogen in the form of nitrate or ammonia in soils is limited. Plants have acquired different strategies to overcome this limitation. One such strategy is establishing a nodule endosymbiosis with nitrogen-fixing Frankia or rhizobium bacteria. Inside nodules, physiological conditions are created that allow the bacteria to convert atmospheric dinitrogen (N2) into ammonia that can be used by the plant. Carbohydrates of plant origin fuel this energy-demanding process. The unique character of nitrogen-fixing nodule symbiosis has raised the interest of plant researchers for more than a century, ultimately aiming to transfer this trait to nonleguminous crop species (Burrill and Hansen, 1917; Rogers and Oldroyd, 2014; Huisman and Geurts, 2019).
The Frankia spp. and rhizobium nitrogen-fixing nodulation trait occurs in 10 paraphyletic lineages within the orders Fabales, Fagales, Cucurbitales, and Rosales, collectively known as the nitrogen-fixing clade (Soltis et al., 1995). Based on phylogenomic comparisons of nodulating and nonnodulating plant species, it is hypothesized that the nitrogen-fixing nodule symbiosis with rhizobium or Frankia spp. bacteria has a shared evolutionary origin, dating to about 110 million years ago (Griesmann et al., 2018; van Velzen et al., 2018, 2019). Subsequently, the nodulation trait most probably was lost multiple times, which is associated with the pseudogenization of two key genes essential for nodule organogenesis and bacterial infection: the transcription factor NODULE INCEPTION (NIN) and the coiled-coil protein-encoding gene RHIZOBIUM POLAR GROWTH (Griesmann et al., 2018; van Velzen et al., 2018). These two genes likely experienced genetic adaptations, allowing them to function exclusively in nodulation. However, insight into the evolutionary trajectory of signaling receptors involved in the recognition of bacterial signals and subsequent activation of the pathways leading to nodule organogenesis and bacterial infection remains elusive.
The nitrogen-fixing nodulation trait is best studied in the legume models Lotus japonicus and Medicago truncatula (Fabaceae, Fabales). Both these legumes recognize their rhizobium microsymbionts by the structural characteristics of secreted lipochitooligosaccharides (LCOs; also known as Nod factors). Perception of these molecules triggers nodule development (Wang et al., 2012). LCO signaling is also the basis of rhizobium-induced nodulation in the nonlegume genus Parasponia (Cannabaceae, Rosales; Marvel et al., 1987; Op den Camp et al., 2011; van Velzen et al., 2018). Additionally, it was found that diazotrophic Frankia spp. strains of a basal taxonomic lineage (so-called cluster II strains) possess LCO biosynthesis genes, but the nodulating strains of two other taxonomic clusters do not (Pawlowski and Demchenko, 2012; Persson et al., 2015; Nguyen et al., 2016, 2019). LCOs, as well as chitin oligomers (COs), are also used by arbuscular mycorrhiza (AM) fungi to signal their hosts (Maillet et al., 2011; Genre et al., 2013). Perception of these AM signals requires a plant Lys motif (LysM)-type receptor that also is essential for chitin innate immune signaling, such as the CHITIN ELECITOR RECEPTOR KINASE1 (OsCERK1) in rice (Oryza sativa; Miyata et al., 2014; Zhang et al., 2015; He et al., 2019). This suggests that nodulating bacteria coopted LCO signaling from the widespread AM symbiosis and/or innate immune signaling (Parniske, 2008; Gough and Cullimore, 2011; Geurts et al., 2012).
Genetic and biochemical studies in L. japonicus and M. truncatula demonstrated that rhizobium LCOs are perceived specifically by a heteromeric complex containing two distinct LysM-type receptors, named NOD FACTOR RECEPTOR1 (LjNFR1) and LjNFR5 in L. japonicus and LYSM DOMAIN CONTAINING RECEPTOR KINASE3 (MtLYK3) and NOD FACTOR PERCEPTION (MtNFP) in M. truncatula (Limpens et al., 2003; Madsen et al., 2003; Radutoiu et al., 2003, 2007; Arrighi et al., 2006; Broghammer et al., 2012). Other receptors may modulate the LCO response, such as LjNFRe, a homolog of LjNFR1 in L. japonicus (Murakami et al., 2018). The LysM-type receptor family can be divided into two subclasses, named LYK and LYR, characterized by having a functional or dead kinase domain (Arrighi et al., 2006). Together, these make up 11 orthogroups, two of which include legume LCO receptors (Buendia et al., 2018). Within legumes, the orthogroup that includes LjNFR1/MtLYK3 (named LYK-I clade) expanded upon gene duplications, allowing functional separation of rhizobium-induced signaling, AM symbiosis, and chitin-triggered innate immune responses (De Mita et al., 2014; Bozsoki et al., 2017; Buendia et al., 2018; Gibelin-Viala et al., 2019). Likewise, LjNFR5/MtNFP (orthogroup LYR-IA) experienced a gene duplication early in the legume clade (Young et al., 2011; Buendia et al., 2018).
Data on symbiotic LysM-type receptors in nodulating nonlegumes are scarce. Only in Parasponia andersonii has a receptor functioning in nodulation been identified; named PanNFP1, it is a close homolog of LjNFR5/MtNFP (Op den Camp et al., 2011). Besides PanNFP1, Parasponia spp. possess a homologous receptor, named NFP2, that is more closely related to LjNFR5/MtNFP and transcriptionally activated in root nodules. Interestingly, this receptor is pseudogenized in nonnodulating Rosales species (van Velzen et al., 2018). To obtain insight into the evolution of LysM-type LCO receptors that are essential for nodulation, we used P. andersonii as a comparative system to legumes. The genus Parasponia represents five tropical tree species, which form nitrogen-fixing nodules with LCO-producing rhizobium species that also nodulate legumes (van Velzen et al., 2018). Parasponia spp. and legumes diverged at the root of the nitrogen-fixing clade more than 100 million years ago (Li et al., 2015; van Velzen et al., 2019). The microbial symbionts of the ancestral nodulating plants remain elusive, and it is probable that Parasponia spp. and legumes accepted rhizobium as a microbial partner in parallel (van Velzen et al., 2019). In line with this, the genus Parasponia provides a unique comparative system to obtain insight into the evolutionary trajectories of different LCO receptors that are essential for nodulation.
RESULTS
Phylogeny Reconstruction of Orthogroups Representing LysM-Type LCO Receptors
To obtain insight into the LysM-type receptor family of P. andersonii, we analyzed it phylogenetically. We identified 16 P. andersonii genes encoding putative LysM-type receptors that grouped in all except one known orthogroups (Supplemental Fig. S1; Supplemental Table S1). Genetic studies in legumes uncovered only two orthogroups that contain proteins with a known function in rhizobium LCO signaling; these are named LYK-I and LYR-IA (Buendia et al., 2018). P. andersonii has two gene copies in both these orthogroups.
LYK-I is the largest orthogroup, containing the functional legume LCO receptors MtLYK3/LjNFR1 and LjNFRe (Limpens et al., 2003; Radutoiu et al., 2003; Murakami et al., 2018). Besides these, the LYK-I orthogroup also includes chitin innate immune receptors of M. truncatula MtLYK9/MtCERK1, L. japonicus LjCERK6, Arabidopsis (Arabidopsis thaliana) AtCERK1, tomato (Solanum lycopersicum) SlLYK1, and rice OsCERK1 (Limpens et al., 2003; Miya et al., 2007; Wan et al., 2008; Shimizu et al., 2010; Miyata et al., 2014; Zhang et al., 2015; Bozsoki et al., 2017; Carotenuto et al., 2017; Liao et al., 2018; Gibelin-Viala et al., 2019; He et al., 2019). OsCERK1 and MtLYK9/MtCERK1 have also been found to function in AM symbiosis (Miyata et al., 2014; Zhang et al., 2015; Feng et al., 2019; Gibelin-Viala et al., 2019). Two P. andersonii genes are part of this orthogroup, named PanLYK1 and PanLYK3.
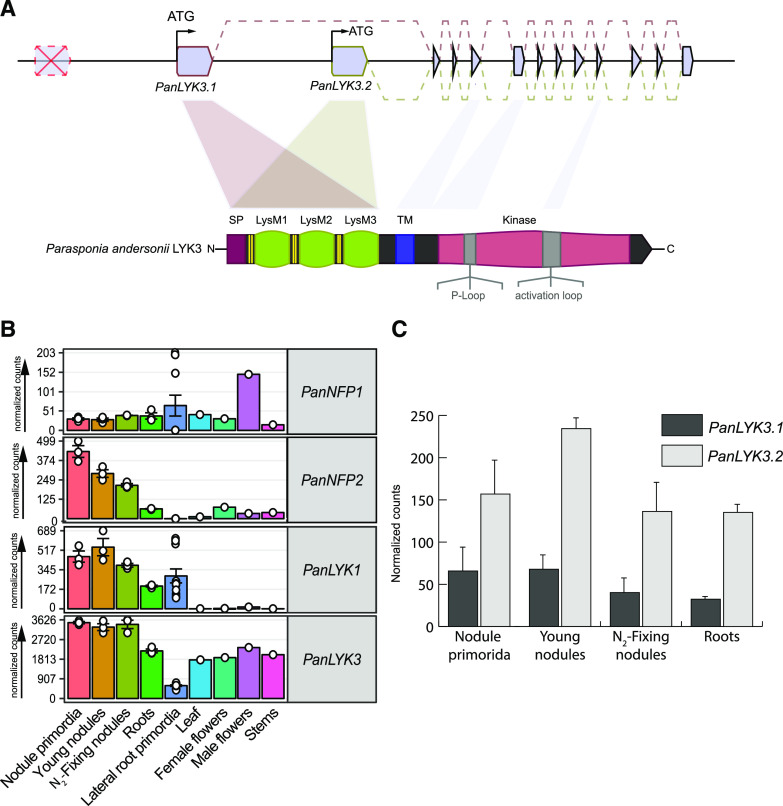
A more exhaustive phylogenetic reconstruction was conducted using gene orthologs of additional species to obtain insight into the evolutionary relationships of these genes when compared with LCO and CO receptors. Notably, LysM-type receptors of the recently sequenced nodulating actinorhizal plants and nonnodulating relatives were included (Griesmann et al., 2018). The resulting phylogeny largely resembled rosid species trees as reconstructed on the basis of plastid-coding genes (Wang et al., 2009; Gonçalves et al., 2019). Our analysis revealed that PanLYK1 and PanLYK3 originated from an ancient duplication, dividing this orthogroup into two subgroups that we named LYK-Ia and LYK-Ib. This duplication does not coincide with the birth of the nitrogen-fixing clade but rather occurred in an ancestral eudicot (Fig. 1; Supplemental Data Set S1). The only studied member in the LYK-Ia orthogroup is tomato SlLYK12, and knockdown of this gene by virus-induced gene silencing substantially reduces mycorrhizal colonization (Liao et al., 2018). The LYK-Ib clade represents several functionally characterized genes, including the chitin innate immune receptors and legume rhizobium LCO receptors. Legumes exhibit an increased number of genes in the LYK-Ib subclade, which are the result of tandem duplications (Limpens et al., 2003; Radutoiu et al., 2003; Zhu et al., 2006). These duplications may have driven neofunctionalization of LCO receptors in legumes (De Mita et al., 2014). In the genus Parasponia, no gene duplications occurred in the LYK-Ib clade (represented by PanLYK3) or in the LYK-Ia clade (represented by PanLYK1). In contrast, P. andersonii PanLYK3 experienced a duplication of exclusively the first exon. To determine whether this duplication is specific for the Parasponia genus, we analyzed the LYK3 genomic region of two additional Parasponia spp. and three nonnodulating species of the closely related genus Trema. This revealed that the duplication of LYK3 exon 1 is present in all species investigated and occurred twice, where the most distal exon 1 copy was lost in P. andersonii (Fig. 2A; Supplemental Fig. S2A). The encoded pre-mRNAs both splice into a shared second exon (Fig. 2). Each exon 1 copy contains a putative transcription and translation start site, which allows for differential expression of the variants (Fig. 2, B and C). Genes of the LYK-I clade have a highly conserved intron-exon structure (Zhang et al., 2009). In most cases, the first exon encodes the extracellular domain comprising the signal peptide and three LysM domains. Therefore, the P. andersonii PanLYK3 gene encodes two protein variants, named PanLYK3.1 and PanLYK3.2, that differ in their extracellular domain (Supplemental Fig. S2B).
Figure 1.
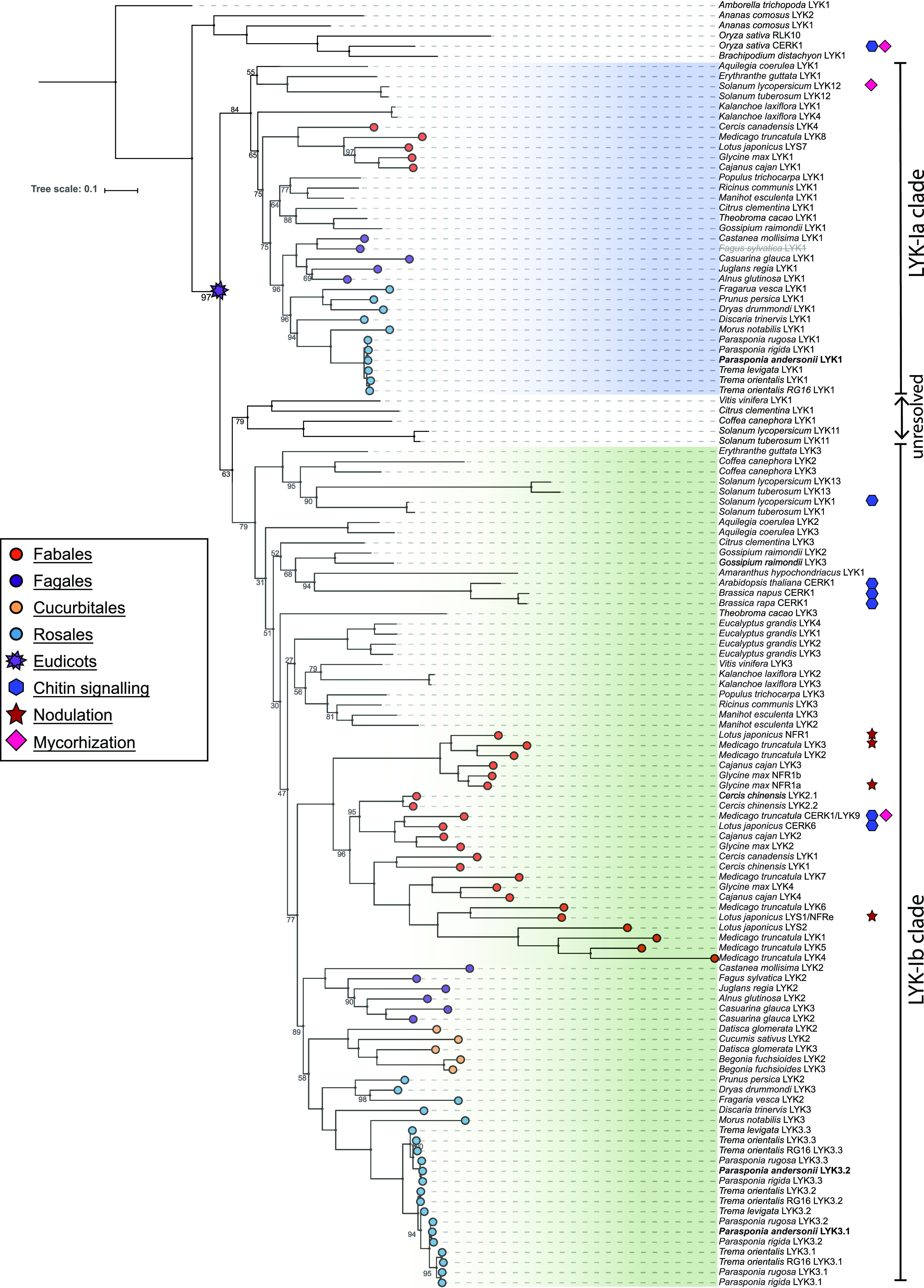
Phylogeny reconstruction of the LYK-I orthogroup, containing known CO and LCO receptors, based on 127 sequences from 47 species. Two main subgroups are recognized in eudicots, LYK-Ia (blue) and LYK-Ib (green). Note the presence of both variants in Aquilegia coeralia, a basal eudicot in the Ranunculales. A subset of proteins is unresolved. P. andersonii proteins are in boldface. Genuses Parasponia and Trema LYK3.1 and LYK3.2 represent protein variants of LYK3. Deduced pseudoproteins are depicted in gray/strikethrough. Proteins with known functions in nodulation, mycorrhization, and/or chitin innate immune signaling are indicated. Bootstrap values indicate IQ-tree UF-bootstrap support%; values >98 are not shown. The scale bar represents substitutions per site. A complete list of species and accession numbers can be found in Supplemental Data Set S1.
Figure 2.
Gene structure and expression of P. andersonii PanLYK3. A, Structure of the PanLYK3 gene model and encoded proteins. PanLYK3 possesses two transcriptional start sites resulting in two protein variants, which differ in the extracellular region containing the LysM domains and are encoded by exon 1. The red cross indicates a third upstream copy of exon 1 lost in P. andersonii but maintained in other Parasponia and Trema species. B, Expression profile of PanNFP1, PanNFP2, PanLYK1, and PanLYK3 in different plant tissues. Expression is given in DESeq2-normalized read counts; error bars represent se of biological replicates. Circles represent individual expression levels. The analysis is based on data presented by van Velzen et al. (2018). C, Relative expression of the PanLYK3.1 and PanLYK3.2 transcriptional variants based on RNA sequencing reads splicing into the second exon. Data are represented as means ± se (n = 3). The analysis is based on data presented by van Velzen et al. (2018).
The LYR-IA orthogroup represents the legume LCO receptors MtNFP, LjNFR5, and pea (Pisum sativum) PsSYM10 (Madsen et al., 2003; Arrighi et al., 2006; Buendia et al., 2016; Miyata et al., 2016). Previously, we have shown that Parasponia spp. harbor two genes in this orthogroup, PanNFP1 and PanNFP2 in P. andersonii, of which the latter is more closely related to MtNFP/LjNFR5 (van Velzen et al., 2018). PanNFP1 and PanNFP2 originated from an ancient duplication. Phylogenetic reconstruction, including additional nodulating and nonnodulating species, supported the occurrence of NFP-I and NFP-II subclades in the LYR-IA orthogroup and showed that this duplication associates with the origin of the nitrogen-fixing clade (Fig. 3; Supplemental Data Set S2). Several actinorhizal species possess gene copies in both NFP subclades, including Datisca glomerata, Dryas drummondii, and Ceanothus thyrsiflorius. All these species nodulate with diazotrophic Frankia spp. of taxonomic cluster II, which possess LCO biosynthesis genes. An NFP-II-type orthologous gene is notably absent in actinorhizal species that are exclusively nodulated by Frankia spp. of cluster I or cluster III that lack LCO biosynthesis genes, such as Alnus glutinosa and Casuarina glauca (Fig. 3; Pawlowski and Demchenko, 2012; Griesmann et al., 2018; Salgado et al., 2018; Nguyen et al., 2019). In line with what was reported for the nonnodulating Rosales species (van Velzen et al., 2018), NFP-II-type pseudogenes can be found in the genomes of the nonnodulating Fagales spp. Chinese chestnut (Castanea mollissima) and European beech (Fagus sylvatica). This shows a strict association of the presence of a functional NFP-II-type gene and LCO-driven nodulation, suggesting that the NFP-II subclade represents LCO receptors that function exclusively in nodulation.
Figure 3.
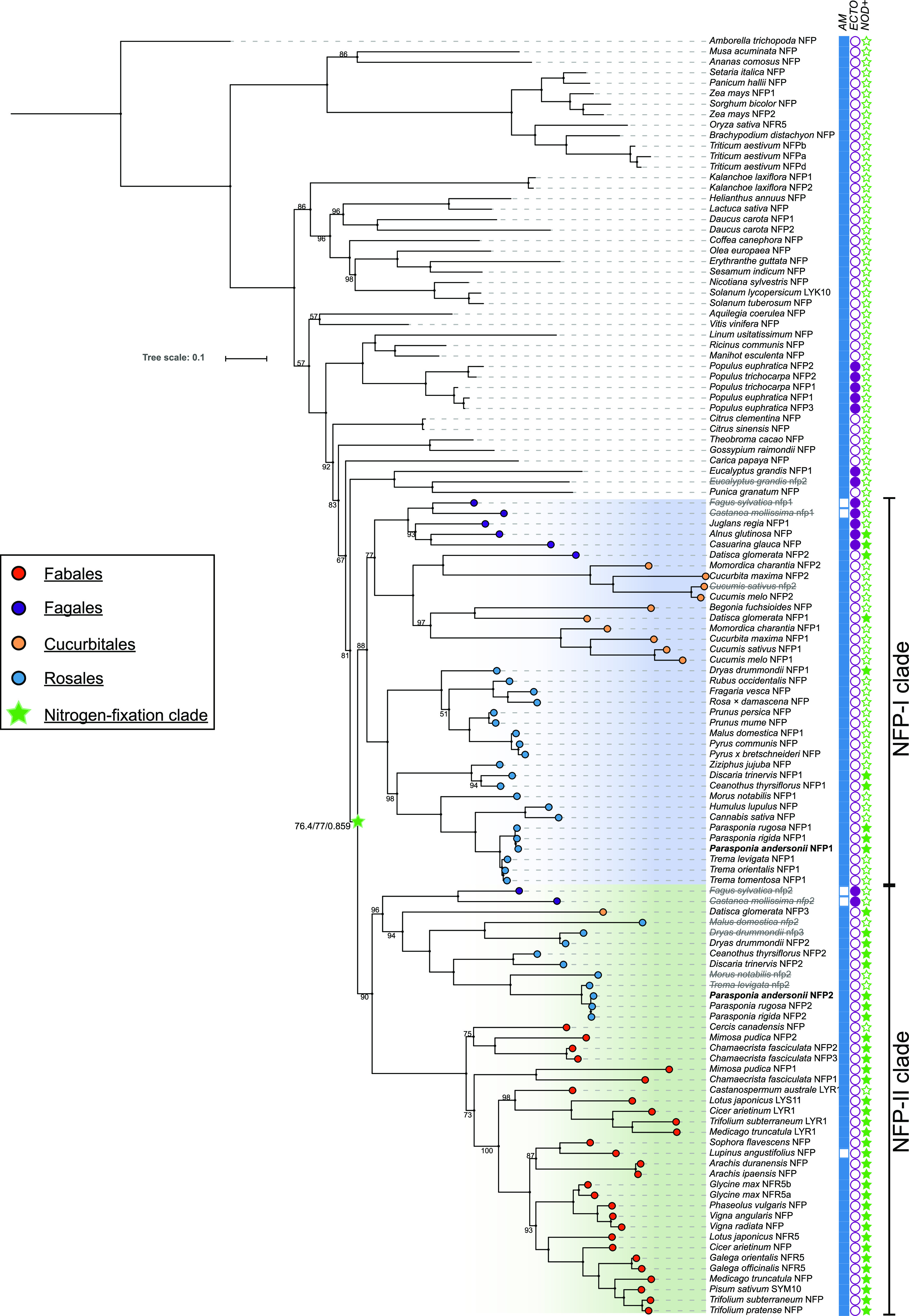
Phylogeny reconstruction of the LYR-IA orthogroup, containing known legume LCO receptors, based on 122 sequences from 87 species. A gene duplication in the root of the nitrogen-fixing clade is recognized, resulting in two subclades named NFP-I (blue) and NFP-II (green). The symbiotic capacities of the species are marked by filled (positive) and unfilled (negative) symbols: AM symbiosis (blue squares), ectomycorrhizal symbiosis (purple circles), and nodulation (green stars). P. andersoniiPanNFP1 and PanNFP2 are in boldface. Deduced pseudoproteins are depicted in gray/strikethrough. Values indicate IQ-tree UF-bootstrap support%; values >98 are not shown. Branch support for the nitrogen-fixing clade indicates aSH-aLRT/UF-Bootstrap/approximate Mr.Bayes support, respectively. The scale bar represents substitutions per site. A list of species and accession numbers can be found in Supplemental Data Set S2.
P. andersonii PanNFP1, PanNFP2, PanLYK1, and PanLYK3 Can Perceive Rhizobium LCOs
Based on the orthologous relation to legume LCO receptors, we considered PanLYK3 (both variants) and PanNFP2 as the most likely candidates to encode rhizobium LCO receptors in P. andersonii. We noted that, in contrast to PanLYK3, PanLYK1 is exclusively expressed in roots and nodule tissue (Fig. 2B), suggesting that this gene may also function in a symbiotic context. Therefore, we decided to include this gene in further studies. Finally, we also included PanNFP1, since an earlier study based on RNA interference (RNAi) in transformed P. andersonii roots showed that this gene functions in nodulation (Op den Camp et al., 2011). To test whether these four P. andersonii genes can function as rhizobium LCO receptors, we conducted two complementary experiments. First, we introduced P. andersonii receptor pairs into a L. japonicus Ljnfr1;Ljnfr5 double mutant, aiming to determine whether these P. andersonii receptors can transcomplement for LCO-induced Ca2+ oscillation. Second, we generated CRISPR-Cas9 knockout mutants in P. andersonii to study their role in nodulation.
We selected L. japonicus for transcomplementation studies because its microbial host Mesorhizobium loti strain R7A can also nodulate P. andersonii (Supplemental Fig. S3, A–C). By using Agrobacterium rhizogenes-mediated root transformation, we tested six combinations of P. andersonii heterodimeric receptor pairs under the control of the promoter and terminator of LjNFR1 and LjNFR5 (Fig. 4). These promoters were shown to be functional in complementation of the L. japonicus Ljnfr1-1;Ljnfr5-2 double mutant (Supplemental Fig. S3, D–H). For the transcomplementation constructs, we included the nucleus-localized calcium sensor R-GECO1.2, allowing visualization of nuclear Ca2+ oscillations (Zhao et al., 2011). In wild-type L. japonicus roots, Ca2+ oscillation was most strong in young root hair cells, whereas this response is not recorded in the Ljnfr1-1;Ljnfr5-2 double mutant (Supplemental Fig. S3, I and J; Supplemental Movie S1; Miwa et al., 2006). Analyzing the transgenic roots expressing P. andersonii receptor combinations revealed that nine out of 11 tested combinations elicit Ca2+ oscillation, although less regular in shape and frequency when compared with the positive control (Fig. 4B; Supplemental Movie S2). Interestingly, the receptor combinations PanLYK1;LjNFR5 and LjNFR1;PanNFP2 did not elicit any Ca2+ oscillation response, whereas both P. andersonii receptors are, at least partially, functional as an M. loti LCO receptor when combined with a P. andersonii counterpart (Fig. 4B). Upon inoculation with M. loti R7A, only nodule-like structures were observed on roots transcomplemented with different P. andersonii receptor combinations (4 weeks postinoculation [wpi]) but not with heterologous receptor pairs (Supplemental Table S2). We sectioned the largest nodule-like structures, which were present on PanLYK3.2;PanNFP2 and PanLYK1;PanNFP1 transformed plants. This showed the absence of intracellular rhizobium infections (Supplemental Fig. S3, K–P). Taken together, the transcomplementation studies of a L. japonicus Ljnfr1;Ljnfr5 mutant indicated that all four P. andersonii receptors, PanLYK1, PanLYK3, PanNFP1, and PanNFP2, have the potential to function as receptors for M. loti LCOs, but none could fully transcomplement a Ljnfr1-1;Ljnfr5-2 double mutant for nodulation.
Figure 4.
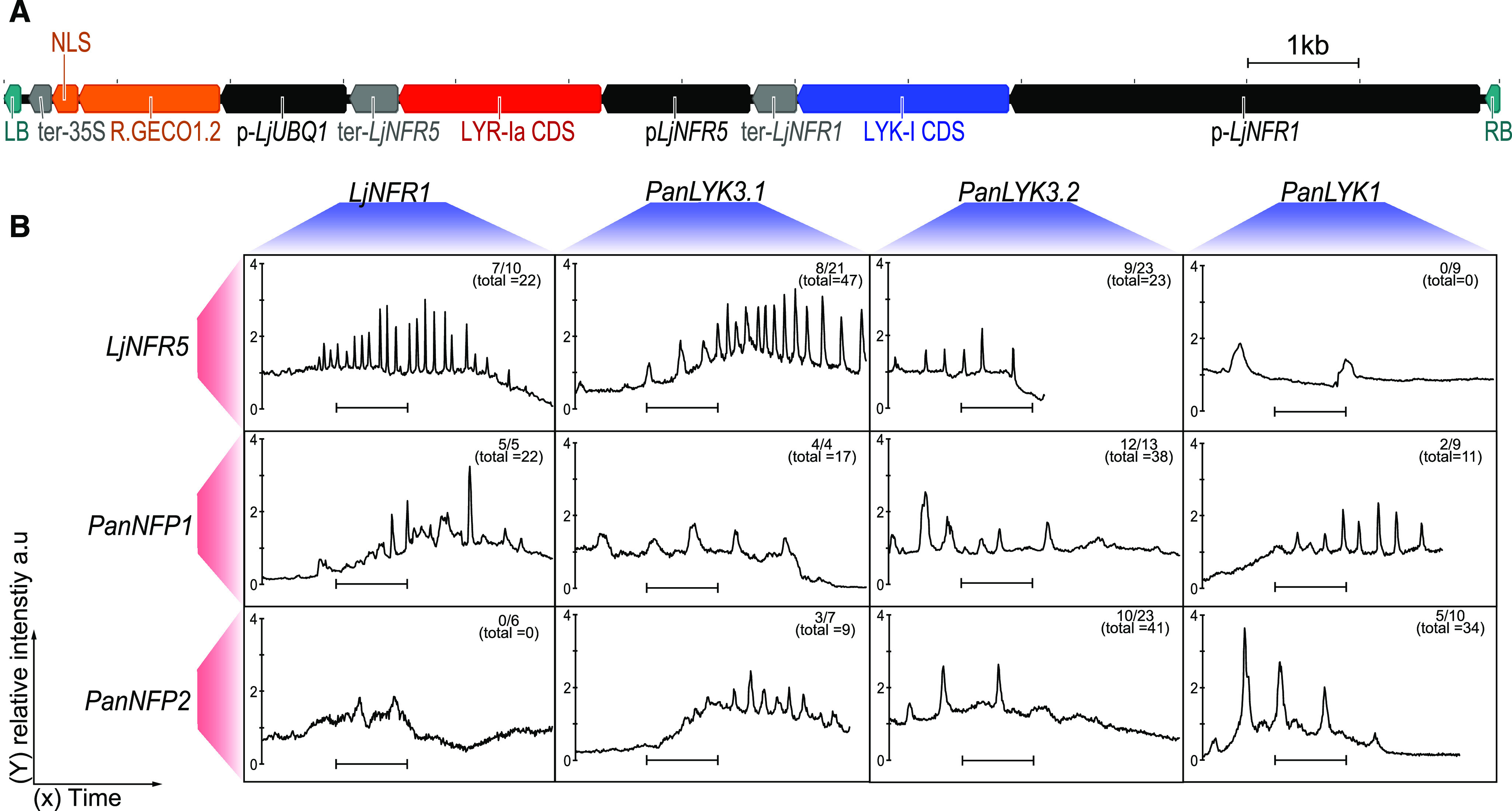
P. andersonii PanNFP1, PanNFP2, PanLYK1, and PanLYK3 complement a L. japonicus Ljnfr1;Ljnfr5 mutant for rhizobium-induced Ca2+ oscillation. A, Schematic representation of the T-DNA region of the binary construct used for A. rhizogenes-based root transformation of a L. japonicus Ljnfr1;Ljnfr5 double mutant. cDNA clones of LYK-I (marked blue) or LYR-Ia (marked red) type genes were cloned in identical fashion. cDNA clones were inserted between native promoter (marked black) pLjNFR1 (4,171 bp) or pLjNFR5 (1,314 bp) and native terminator (marked gray) sequences ter-LjNFR1 (394 bp) or ter-NFR5 (432 bp). pLjUBQ1::R.GECO1.2-nls:CaMV35S-ter (marked orange) was used to visualize nuclear calcium oscillation. The left border (LB) and right border (RB; marked green) flank the T-DNA region. B, Representative traces of nuclear Ca2+ oscillation, as observed in different combinations of LYK-I (red) and LYR-Ia (blue) type receptors introduced in a L. japonicus Ljnfr1;Ljnfr5 double mutant. Note that the receptor combinations PanLYK1;LjNFR5 and LjNFR1;PanNFP2 did not complement for Ca2+ oscillation. Traces were recorded ∼10 min postapplication of LCOs extracted from M. loti R7A (∼10−9 m). Numbers denote spiking roots versus the number of roots analyzed. The numbers in parentheses denote the total numbers of spiking nuclei observed. The y axis is the relative fluorescence intensity compared with the defined baseline in arbitrary units (a.u.). The scale bar = 10 min.
P. andersonii PanNFP1, PanNFP2, PanLYK1, and PanLYK3 Function in Nodulation
We recently established an efficient Agrobacterium tumefaciens-mediated transformation protocol for P. andersonii, which allows the generation of CRISPR-Cas9 mutant plantlets in an ∼3-month time frame (van Zeijl et al., 2018; Wardhani et al., 2019). This enabled us to test by mutagenesis whether PanLYK1, PanLYK3, PanNFP1, and PanNFP2 are essential for rhizobium-induced nodule formation. We aimed to generate small deletions of 100 to 300 bp in the area covering the LysM domains by using two or three single guide RNAs (sgRNAs) that have no potential high-identity off-targets. In the case of PanLYK3, the transmembrane domain was targeted in order to mutate both alternative start variants. Additionally, we targeted specifically PanLYK3.1 and PanLYK3.2 by designing specific guides on the first exon. Selected single guides only had off-targets with at least three mismatches or two insertions/deletions (indels), based on alignments to the P. andersonii reference genome. Shoots regenerated after A. tumefaciens-mediated cocultivation were genotyped using PCR and subsequent sequence analysis to detect potential mutations at the CRISPR target sites. Only T0 shoots with a >75-bp deletion between the two target sites or edits generating a frameshift were considered for propagation and subsequent further evaluation. At least two independent mutant alleles were generated per gene, with the exception of PanLYK3.1, for which only a single suitable allele could be identified (Supplemental Data Set S3). Putative off-target sites that occur in coding sequence regions were amplified by PCR and subsequently sequenced by Sanger sequencing. Subsequently, PanNFP1 was sequenced in PanNFP2 lines and PanNFP2 was sequenced in PanNFP1 lines (Supplemental Data Set S3). No off-target mutations at these locations were identified. The selected tissue culture lines were in vitro propagated and rooted, so they could be used for experimentation.
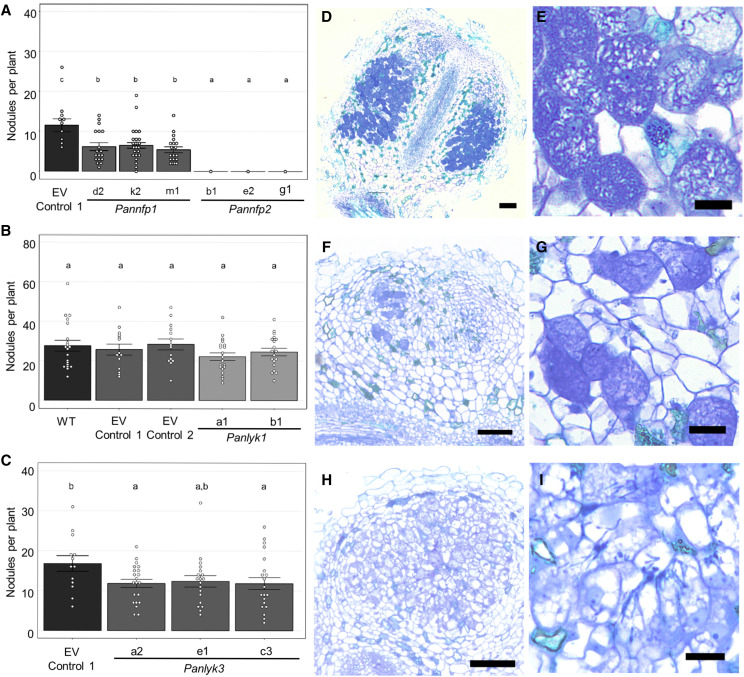
We compared the nodulation phenotypes of Panlyk1, Panlyk3, Pannfp1, and Pannfp2 knockout mutants in independent experiments using empty vector (EV) transformed lines as controls (Fig. 5; Supplemental Fig. S4). All three independent Pannfp2 mutant lines showed to be unable to form nodules or nodule-like structures (5 wpi) with strain Mesorhizobium plurifarium BOR2, demonstrating the requirement for this gene in the nodulation trait (Fig. 5A). Additionally, we noted a reduced nodulation efficiency of all three independent Pannfp1 mutant lines. This is in line with earlier findings using RNAi to target PanNFP1 in A. rhizogenes-transformed P. andersonii roots (Op den Camp et al., 2011), demonstrating that Pannfp1 controls nodulation efficiency but is not essential for rhizobium intracellular infection. Previously, we reported that PanNFP1 RNAi nodules have a strong infection phenotype when inoculated with the Sinorhizobium fredii strain NGR234 (Op den Camp et al., 2011). We did not observe such an infection phenotype in nodules induced by M. plurifarium BOR2 on Pannfp1 knockout mutant plants (Supplemental Fig. S4). In order to determine whether the Pannfp1 infection phenotype is strain dependent, we nodulated plants also with S. fredii NGR234. This strain was shown to be less optimal under the chosen conditions (Agroperlite supplemented with EKM medium [see “Materials and Methods”)] and S. fredii NGR234.pHC60 at OD = 0.05). In an effort to optimize nodulation efficiency with this strain, we used river sand and scored nodulation 8 wpi. Under these conditions, no difference between Pannfp1 and the EV control was observed. Nodules formed on Pannfp1 mutant plants were infected normally (Supplemental Fig. S4).
Figure 5.
P. andersonii Pannfp1, Pannfp2, and Panlyk3 mutants are affected in nodulation. Data are represented as means ± se, and circles represent individual data points. Lowercase letters denote statistical significance based on one-way ANOVA and Tukey’s posthoc contrast (P > 0.05). A, Nodule numbers in P. andersonii CRISPR-Cas9 mutant lines Pannfp1 d2 (n = 18), k2 (n = 31), and m1 (n = 19) and Pannfp2 b1 (n = 19), e2 (n = 10), and g1 (n = 9) at 5 wpi with M. plurifarium BOR2. EV Control 1 (n = 12) represents a positive control line transformed with a binary vector not containing sgRNAs. B, Nodule numbers in P. andersonii CRISPR-Cas9 mutant lines Panlyk1 a1 (n = 19) and b1 (n = 20) at 5 wpi with M. plurifarium BOR2. EV Control 1 (n = 14) and EV Control 2 (n = 14) represent two independent positive control lines transformed with a binary vector not containing sgRNAs. WT (n = 20) represents untransformed plantlets. C, Nodule numbers in P. andersonii CRISPR-Cas9 mutant lines Panlyk3 a2 (n = 21), c3 (n = 21), and e1 (n = 19) at 5 wpi with M. plurifarium BOR2. EV Control 1 (n = 14). EV Control 1 represents a positive control line transformed with a binary vector not containing sgRNAs. D to I, Toluidine Blue-stained sections of representative nodules grown with M. plurifarium BOR2. D, Wild-type P. andersonii transformed with an EV-1 construct expressing Cas9. E, Infected nodule cells containing fixation threads formed on EV-1 plants. F, Infected nodule of Panlyk3 line e2. Note patches of infected cells. G, Infected nodule cells of Panlyk3 line e2 containing fixation threads. H, Empty nodule of Panlyk3 line e2. Note the absence of fully infected cells. I, Nodule cells of Panlyk3 line e2 containing infection threads but no fixation threads. Bars = 100 μm (D, F, and H) and 20 μm (E, G, and I).
Similarly to Pannfp1 mutant plants inoculated with M. plurifarium BOR2, we found a reduced nodulation efficiency in P. andersonii Panlyk3 knockout mutants but not in Panlyk3.1 and Panlyk3.2 variant-specific mutant alleles, nor in Panlyk1 mutants (Fig. 5; Supplemental Fig. S4). To determine whether nodules formed on Panlyk1 and Panlyk3 mutants have an infection phenotype, we analyzed thin sections. In contrast to legumes, P. andersonii does not guide rhizobia in infection threads toward the nodule primordia. Instead, rhizobia enter via apoplastic cracks in epidermis and cortex and only form infection threads to penetrate nodule cells. Once inside, infection threads develop into fixation threads, which are wider, having two phyla of bacteria aligned compared with one in infection threads, and possess a thinner cell wall (Lancelle and Torrey, 1984, 1985). Panlyk1 mutant nodules showed no defects in infection thread structure or the transition from infection threads to fixation threads. In the case of Panlyk3, nodules were relatively small and had diverse phenotypes. Out of 45 sectioned nodules of the line Panlyk3-e2, 22 were infected like the wild type, 15 contained only infection threads but no fixation threads, and eight showed an intermediate phenotype with few infected cells (Fig. 5, F–I; Supplemental Fig. S4). To confirm that the infection phenotype is a result of a full Panlyk3 knockout mutation, we sectioned 28 nodules of the independent knockout line Panlyk3-c3. This revealed similar results: 11 nodules normally infected, 11 contained only infection threads, and six nodules with an intermediate phenotype. Next, we determined whether this infection phenotype is controlled specifically by either PanLYK3.1 or PanLYK3.2, which was shown not to be the case (Supplemental Fig. S4). As ∼50% of the nodules formed on the P. andersonii Panlyk3 mutant plants displayed a wild-type phenotype, this suggests redundancy in gene functioning. Interestingly, S. fredii NGR234 could not nodulate Panlyk3 mutants, which suggests that this strain is fully dependent on PanLYK3-controlled signal transduction (Supplemental Fig. S4).
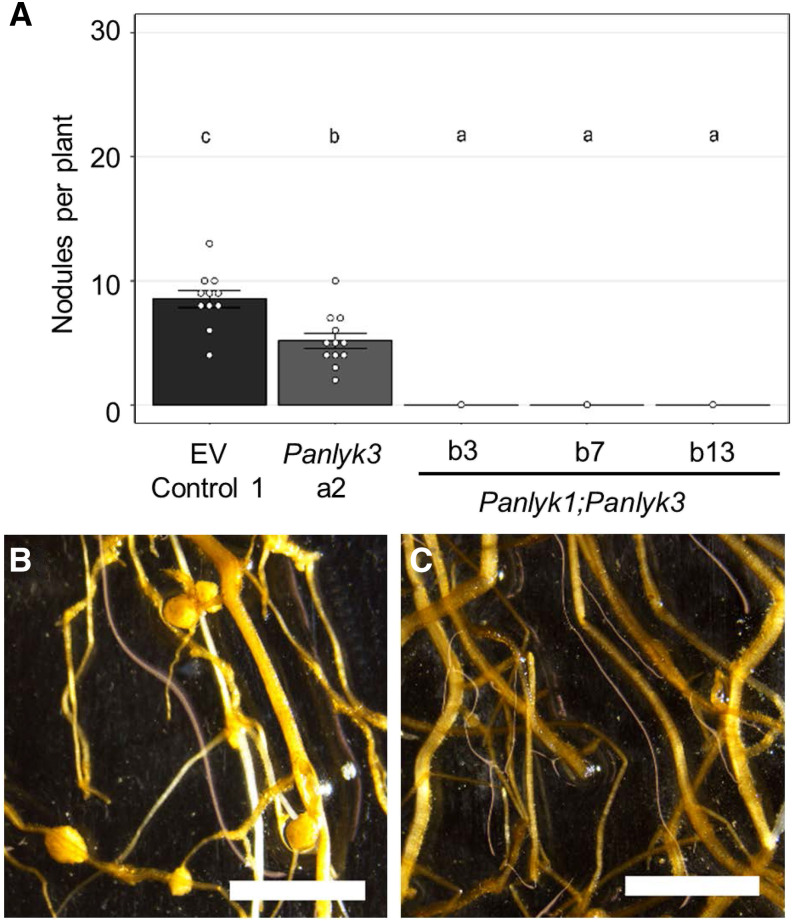
As P. andersonii did not experience any gene duplication events in the LYK-Ib clade, PanLYK1 in the LYK-Ia clade is the closest homolog of PanLYK3. In order to investigate whether the PanLYK1 gene is functionally redundant with PanLYK3 in cases of M. plurifarium BOR2 inoculation, we generated a Panlyk1;Panlyk3 double mutant. To do so, a binary construct with the two sgRNAs targeting PanLYK1 was used for retransformation of the Panlyk3 mutant (line a2). We obtained three independent Panlyk1;Panlyk3 mutants (Supplemental Data Set S3). M. plurifarium BOR2 inoculation experiments revealed that all Panlyk1;Panlyk3 double mutant lines were unable to form any nodule or nodule-like structure (Fig. 6). To confirm that the nodulation-minus phenotype in the Panlyk1;Panlyk3 lines is not due to any off-target mutation, we conducted complementation studies using A. rhizogenes-mediated root transformation. As the putative promoter of PanLYK3 is rather complex due to the occurrence of alternative transcriptional start sites (Fig. 2), we used the LjNFR1 promoter, as well as the constitutive AtUBQ10 and CaMV35S promoters, to drive a CRISPR-resistant allele of PanLYK3.1 (PanLYK3cr). Compound plants carrying transgenic roots expressing PanLYK3cr could be nodulated by M. plurifarium BOR2 (Supplemental Fig. S5). Together, this showed that in P. andersonii, PanLYK1 and PanLYK3 act redundantly in root nodule formation. (For complementation studies of Pannf2, see below.)
Figure 6.
P. andersonii PanLYK1 and PanLYK3 act redundantly in nodulation. A, Average nodule numbers per plant in EV Control 1 (n = 11) and retransformed Panlyk3 line a2 (n = 12) and Panlyk1;Panlyk3 double mutant lines b3 (n = 10), b7 (n = 5), and b13 (n = 10) at 5 wpi with M. plurifarium BOR2. Data are represented as means ± se, and circles represent individual data points. Lowercase letters denote statistical significance based on one-way ANOVA and Tukey’s posthoc contrast (P > 0.05). B, Roots with nodules of EV Control 1 at 5 wpi with M. plurifarium BOR2. C, Roots without nodules of the Panlyk1;Panlyk3 double mutant (line b3) at 5 wpi with M. plurifarium BOR2. Bars = 5 mm.
The results demonstrate that P. andersonii PanLYK1, PanLYK3, PanNFP1, and PanNFP2 function in rhizobium LCO-driven nodulation. PanLYK3 and PanNFP2 are orthologous to legume LjNFR1/MtLYK3 and LjNFR5/MtNFP, indicating a shared evolutionary origin of LCO-driven nodulation in both taxonomic lineages. As PanLYK1 and PanLYK3 evolved from a duplication predating the emergence of the nitrogen-fixing clade, this suggests that LCO signaling is an ancestral function of these LYK-I receptors.
A Repaired Trema levigata NFP2 Pseudogene, But Not PanNFP1, Can Functionally Complement a P. andersonii nfp2 Mutant
PanNFP1 and PanNFP2 differ in expression pattern. Whereas both genes are expressed in root tissue, only PanNFP2 is up-regulated in nodules (Fig. 2B; van Velzen et al., 2018). We questioned whether the difference in symbiotic functioning between both genes is the result of regulatory evolution. To test this, we first identified a functional promoter region of PanNFP2. A. tumefaciens-mediated transformation showed that a 2.75-kb PanNFP2 upstream region can be used to functionally complement the P. andersonii Pannfp2 mutant when using a PanNFP2 CRISPR-resistant allele (PanNFP2cr). Two independent lines formed 7 ± 7 and 4 ± 1 nodules 5 wpi with M. plurifarium BOR2 (Fig. 7). However, when we used PanNFP1 driven by the PanNFP2 promoter, no transcomplementation of the P. andersonii nfp2 mutant phenotype was observed. This suggests that there is a functional difference in the encoded PanNFP1 and PanNFP2 receptors.
Figure 7.
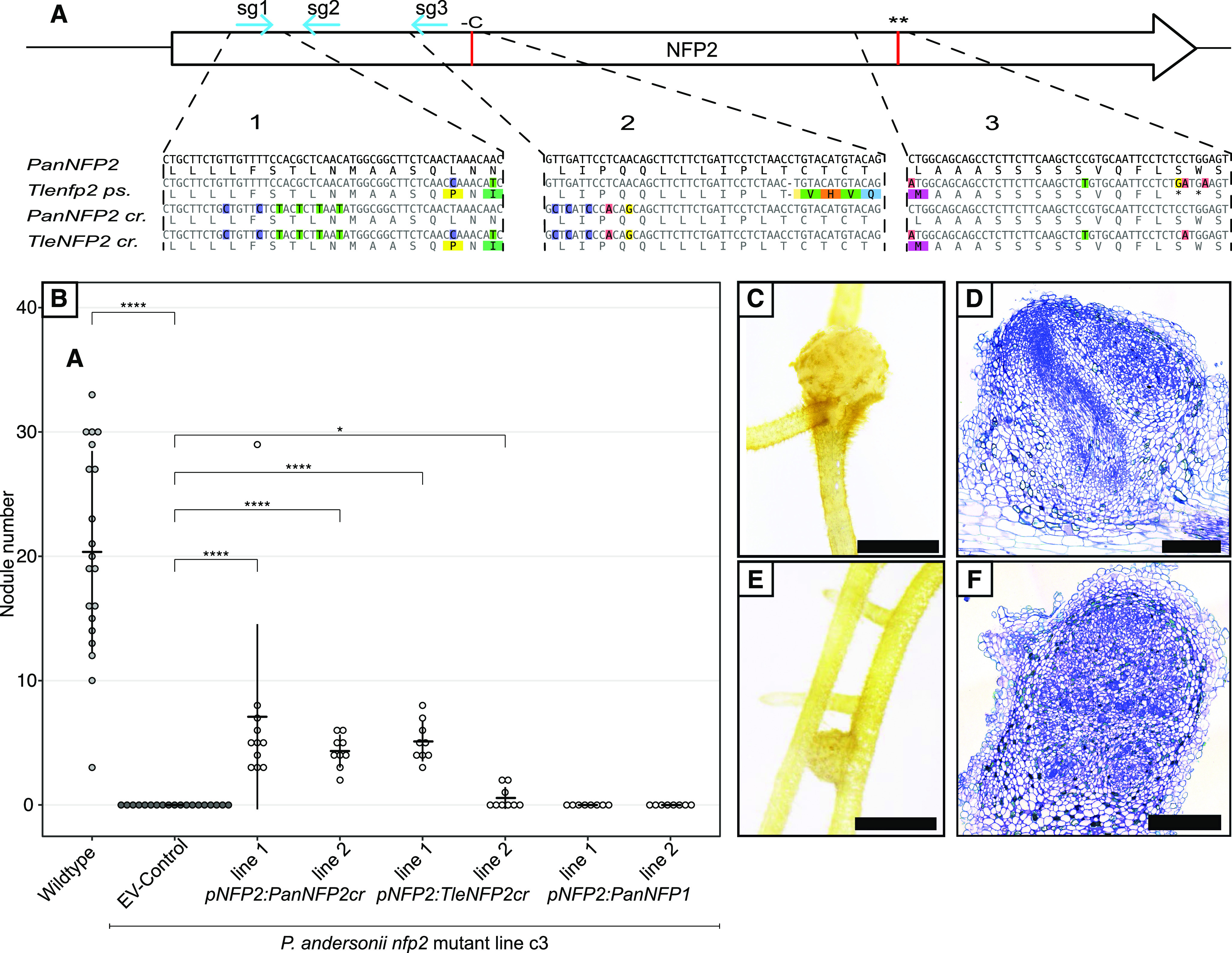
A repaired T. levigata nfp2 pseudogene can replace PanNFP2 for nodule formation. A, Schematic representation of the NFP2 coding region with indicated replacements to avoid CRISPR targeting of inserted NFP2 genes of P. andersonii (PanNFP2cr) and a repaired T. levigata (TleNFP2cr). Blue arrows indicate guide RNA target sites, and red lines indicate T. levigata mutations. Region 1, Replacement of six codons at the sg1 site; region 2, replacement of five codons at the sg3 site plus repair of the T. levigata indel (red line); region 3, repair of the double stop codon in T. levigata (red line, black asterisks). The replacement of five codons at the sg2 site is not shown. B, PanNFP2cr and repaired TleNFP2cr can restore nodulation in the Pannfp2 mutant line C3 when driven by the PanNFP2 promoter, whereas PanNFP1 cannot. Nodulation was scored at 5 wpi with M. plurifarium BOR2. Error bars represent the sd. Asteriks indicate statistical significance by Mann-Whitney-Wilcoxon test (*P < 0.05 and ****P < 0.00001). C and D, Nodule and section of pNFP2:PanNFP2cr line 1. E and F, Nodule and section of pNFP2:TleNFP2cr line 1. Bars = 2 mm (C and E) and 100 μm (D and F).
Next, we questioned whether the nfp2 pseudogene as present in several nonnodulating Rosales species may have encoded a functional symbiosis receptor. To test this, we focused on the nfp2 pseudogene of T. levigata, as it has only three mutations that cause a disturbance of the open reading frame (Fig. 7). We repaired these three mutations, using PanNFP2 as a template, resulting in an engineered CRISPR-resistant TleNFP2cr that encodes for a LysM-type receptor protein of 582 amino acids, similar to PanNFP2 of P. andersonii. We tested whether TleNFP2cr driven by the PanNFP2 promoter can transcomplement the P. andersonii Pannfp2 mutant. A. tumefaciens transformation resulted in two lines that can form functional root nodules 5 wpi with M. plurifarium BOR2. This supports the hypothesis that T. levigata nfp2 encoded a functional symbiosis receptor prior to the pseudogenization of this gene.
P. andersonii PanLYK3 Is Essential for Chitin-Triggered Immune Responses and Controls AM Symbiosis in Coherence with PanLYK1
Next, we aimed to determine whether the P. andersonii LysM-type receptors that control nodulation are also involved in other processes, as this may provide insights into ancestral functions of these genes. Some LysM-type receptors of the LYK-I clade are known to function in chitin-triggered immunity and/or the AM symbiosis: L. japonicus LjCERK6, M. truncatula MtLYK9/MtCERK1, Arabidopsis AtCERK1, tomato SlLYK1, and rice OsCERK1 (Fig. 1; Miya et al., 2007; Wan et al., 2008; Shimizu et al., 2010; Bozsoki et al., 2017; Liao et al., 2018; Feng et al., 2019; Gibelin-Viala et al., 2019; He et al., 2019). Similarly, some experimental evidence using transient silencing assays indicated that LysM-type receptors of the LYR-IA clade function in mycorrhization, including P. andersonii PanNFP1 (Op den Camp et al., 2011). In line with this, we aimed to confirm this phenotype in stable Pannfp1 knockout mutants and determine whether other P. andersonii symbiotic LysM-type receptors may also function in AM symbiosis and/or chitin-induced innate immunity signaling.
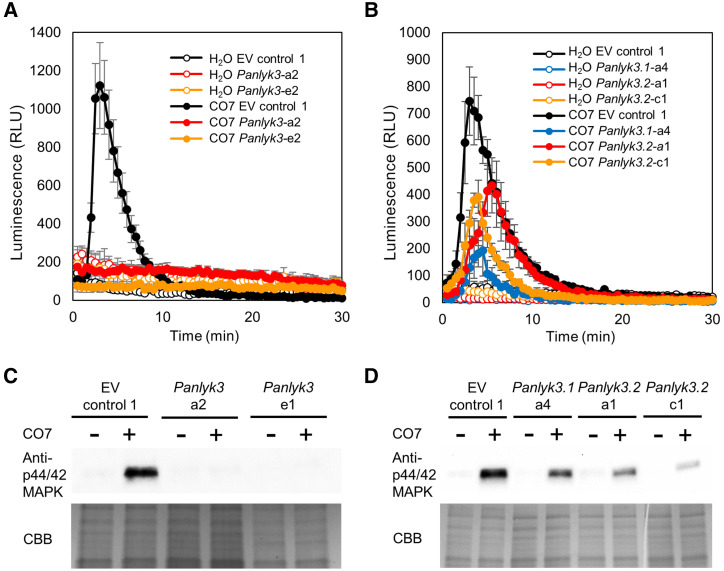
First, we investigated whether the P. andersonii LysM-type receptor mutants are affected in chitin-triggered immunity responses. To do so, two complementary assays were used: a chitin-induced reactive oxygen species (ROS)-burst production and a MITOGEN-ACTIVATED PROTEIN KINASE3 (MAPK3)/MAPK6 phosphorylation assay. Chitin heptamers (CO7) effectively induced a ROS burst in P. andersonii root segments at concentrations of 1 μm or greater when incubated at 28°C, the regular growth temperature of Parasponia spp. (Fig. 8A; Supplemental Fig. S6B). To test whether ROS bursts can also be triggered by rhizobium LCOs, we used the extracts of M. loti R7A and Rhizobium tropici CIAT899. These two strains can nodulate P. andersonii but produce structurally different LCOs (López-Lara et al., 1995; Folch-Mallol et al., 1996). However, neither triggered a ROS burst in P. andersonii roots (Supplemental Fig. S6A). To determine whether CO7-induced ROS bursts were associated with phosphorylation of P. andersonii MAPK3 and MAPK6 homologs, we used an anti-phospho-p44/42 HsMAPK antibody, which detects phosphorylated MAPK3 and MAPK6 of different plant species (Yamaguchi et al., 2013; Bozsoki et al., 2017). P. andersonii possesses a single PanMAPK3 gene and a single PanMAPK6 gene, each of which encodes a protein with a conserved Thr-202/Tyr-204 phosphorylation site (Supplemental Fig. S6C). Upon CO7 application (100 μm, 10 min), a MAPK3/6 phosphorylation pattern can be detected, which is not observed upon application of M. loti or R. tropici LCO extracts (Fig. 8C; Supplemental Fig. S6D). Next, we determined whether P. andersonii LysM-type receptor mutants are affected in response to chitin CO7 oligomers. Pannfp1, Pannfp2, and also a newly created Pannfp1;Pannfp2 double mutant showed a wild-type ROS-burst and MAPK3/6 phosphorylation profile (Supplemental Fig. S6; Supplemental Data Set S3). Similarly, the Panlyk1 mutant showed a ROS-burst and MAPK3/6 phosphorylation profile, as did wild-type root segments (Supplemental Fig. S6, E and F). In contrast, P. andersonii Panlyk3 mutant lines lacked a chitin-triggered ROS burst and showed no p44/42 MAPK phosphorylation (Fig. 8). Individual exon-knockout Panlyk3.1 or Panlyk3.2 mutants both showed ROS production and MAPK3/6 phosphorylation upon application of 100 μm CO7, although at reduced levels (Fig. 8, B and D). Taken together, these data show that PanLYK3, which is the only P. andersonii gene in the LYK-Ib clade, is essential for chitin innate immune signaling in roots.
Figure 8.
P. andersonii PanLYK3 is essential for chitin-triggered immunity responses in roots. A and B, Production of ROS measured upon treatment with 100 μm CO7 (filled circles) or water (open circles) with EV Control 1 plants (black), Panlyk3 line a2 (red), and Panlyk3 line e1 (orange; A) and EV Control 1 plants (black), Panlyk3.1 line a4 (blue), Panlyk3.2 line a1 (red), and Panlyk3.2 line c1 (orange; B). For A and B, data are averages of at least three independent biological replicates ± se. Luminescence is measured in relative light units (RLU). C and D, Phosphorylation of MAPK analyzed by immunoblot using an anti-p44/42 MAPK antibody upon treatment with 100 μm CO7 (top). Equal loading was confirmed by Coomassie Brilliant Blue (CBB) staining (bottom). Results shown are representative of three independent experiments. C, MAPK phosphorylation in root pieces of EV Control 1, Panlyk3 line a2, and Panlyk3 line e1. D, MAPK phosphorylation in root pieces of EV Control 1, Panlyk3.1 line a4, Panlyk3.2 line a1, and Panlyk3.2 line c1.
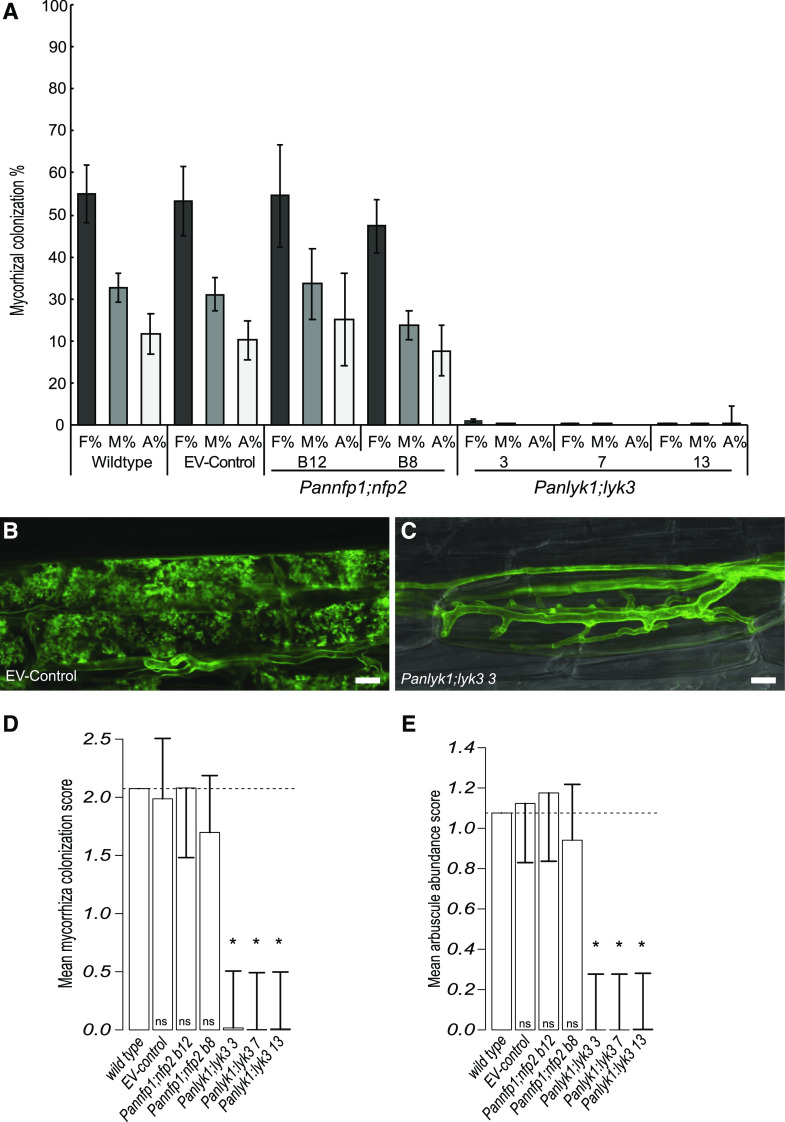
Studies in P. andersonii, tomato, M. truncatula, and rice revealed that LYR-IA and LYK-I putative orthologous genes have functions in AM symbiosis (Miyata et al., 2014, 2016; Zhang et al., 2015; Buendia et al., 2016; Carotenuto et al., 2017; Liao et al., 2018; Feng et al., 2019; Gibelin-Viala et al., 2019; He et al., 2019). Interestingly, we noted that the NFP-I-type gene is pseudogenized in European beech and Chinese chestnut. Both species have lost AM symbiosis in favor of an ectomycorrhizal symbiosis (Fig. 3; Werner et al., 2018). We conducted an RNA sequencing experiment on P. andersonii roots mycorrhized by Rhizophagus irregularis strain DOAM197198. Several marker genes for mycorrhization were shown to be enhanced in expression in mycorrhized P. andersonii root samples, including PanSTR1, PanSTR2, PanPT4, PanVPY, PanD27, PanRAD1, and PanRAM1 (Supplemental Fig. S7). Also, this suggested that PanNFP1 is expressed at a higher level than PanNFP2 under these conditions (Supplemental Fig. S7). However, no significant differential regulation of any of the studied LysM-type receptor-encoding genes was detected between phosphate-starved control roots and mycorrhized root samples (Supplemental Fig. S7). To determine whether P. andersonii symbiotic LysM-type receptors also function in AM symbiosis, we conducted three independent experiments using in vitro-propagated mutant plantlets inoculated with 250 spores of R. irregularis DOAM197198. The average colonization and arbuscule formation frequency were scored 6 wpi. These experiments revealed substantial variation in mycorrhization efficiency between replicates, although no clear impaired AM symbiosis phenotype could be observed in any of the single mutants, including Pannfp1. Strikingly, Panlyk1 showed a significant increase in colonization and arbuscule frequency (Supplemental Fig. S8, A–C). Analyzing both double mutants, Pannfp1;Pannfp2 and Panlyk1;Panlyk3, revealed a strong AM symbiosis phenotype only in the latter (Fig. 9; Supplemental Fig. S8). The fungal colonization of the Panlyk1;Panlyk3 mutant was severely affected, with only a few infections observed. Confocal imaging of wheat germ agglutinin conjugated to Alexa Fluor 488 (WGA-Alexa488)-stained roots showed that besides the level of colonization, also the morphology of the few arbuscules that were formed was affected in Panlyk1;Panlyk3 plants. In wild-type plants, many cortical cells were filled with arbuscules that were finely branched and occupied most of the cell. In contrast, the few hyphae that enter cortical cells in the Panlyk1;Panlyk3 mutant were unable to form mature arbuscules, either because the fungus fails to switch to fine branching or because a limited number of fine branches is made (Fig. 9). As both Panlyk1 and Panlyk3 single mutant plants do not show this impaired mycorrhizal phenotype, we conclude that both genes function in conjunction to control mycorrhizal infection.
Figure 9.
P. andersonii PanLYK1 and PanLYK3 act redundantly in arbuscular mycorrhization. A, The P. andersonii Panlyk1;Panlyk3 double mutant shows a strongly reduced colonization compared with wild-type and control P. andersonii roots. P. andersonii Pannfp1;Pannfp2 mutants are not significantly affected. Frequency and arbuscule abundance classes are according to Trouvelot et al. (1986). F%, Colonization frequency in the root system; M%, intensity of mycorrhizal colonization; A%, arbuscule abundance in the root system. Error bars represent the se of 10 biological replicates scored at 6 wpi using 250 spores of R. irregularis strain DOAM197198 (Trouvelot et al., 1986). B, Highly branched arbuscules formed in EV control plants at 6 wpi stained with WGA-Alexa488. C, Phenotype of stunted arbuscules formed in the Panlyk1;Panlyk3 double mutant stained with WGA-Alexa488. Bars = 10 μm. D and E, Statistical analysis of raw (observed) data: mean colonization frequency score (classes 0–5; D) and mean arbuscule score (classes 0–3; E). Classes are presented according to Trouvelot et al. (1986). Reduced mycorrhizal colonization and arbuscule formation in Panlyk1;Panlyk3 mutants are considered significant compared with the wild type. Error bars represent the Bonferroni-corrected lsd. Error bars nonoverlapping with the mean wild-type value are considered significant. Dashed lines indicate the mean wild-type score. Asterisks mark samples that are significantly different for wild type.
Taken together, these experiments revealed that PanLYK1 and PanLYK3 can function in multiple processes, including rhizobium nodulation (PanLYK1 and PanLYK3), AM symbiosis (PanLYK1 and PanLYK3), and chitin innate immune signaling (PanLYK3). This suggests that no subfunctionalization of these receptors is required to allow functioning in the rhizobium nitrogen-fixing nodulation trait.
DISCUSSION
We used P. andersonii as a comparative system to legumes to obtain insight into the evolutionary trajectory of LysM-type rhizobium LCO receptors. By conducting phylogenetic analysis, transcomplementation studies in a L. japonicus LCO receptor double mutant, and CRISPR-Cas9 mutagenesis in P. andersonii, we identified four LysM-type receptors that function in LCO-driven nodulation in a nonlegume. Two of these, PanLYK3 and PanNFP2, are putative orthologs to known legume rhizobium LCO receptors LjNFR1/MtLYK3 and LjNFR5/MtNFP, respectively. As the Parasponia spp. and legume lineages diverged early in the nitrogen-fixing clade (more than 100 million years ago), the use of orthologous genes for rhizobium LCO perception supports the hypothesis of a shared evolutionary origin of LCO-driven nodulation. In contrast to legumes, symbiotic LysM-type receptors in the genus Parasponia did not experience recent duplication events. Instead, the Parasponia spp. symbiotic LysM-type LCO receptors evolved following two ancient duplications. We hypothesize that the PanNFP1-PanNFP2 duplication associates with the origin of the nitrogen-fixing clade, whereas in the case of PanLYK1 and PanLYK3, the duplication occurred prior to the birth of the nitrogen-fixing clade. This makes it most probable that the capability of these receptors to perceive LCOs predates the evolution of the nitrogen-fixing nodulation trait.
Presently, the NFP1-NFP2 duplication cannot be precisely dated because legumes do not possess an NFP-I-type gene. This can be explained in two scenarios. (1) The NFP1-NFP2 duplication occurred in the root of the nitrogen-fixing clade, and subsequently, the NFP-I-type gene got lost in the Fabales lineage. (2) The NFP1-NFP2 duplication occurred in an ancestor of the Fagales-Cucurbitales-Rosales lineages after the divergence of the Fabales order. The recent finding that ectopic expression of the NFP-type gene of two species outside of the nitrogen-fixing clade (Petunia hybrida PhLYK10 and tomato SlLYK10) can, at least partially, transcomplement the M. truncatula and L. japonicus Mtnfp and Ljnfr5 mutant phenotypes demonstrates that LCO receptor functionality is ancestral to the NFP1-NFP2 duplication (Girardin et al., 2019). The putative promoters of PhLYK10 and SlLYK10 show a nodule-enhanced expression profile similar to that reported for PanNFP2 (Girardin et al., 2019), which may support the second scenario, where the duplication of NFP1-NFP2 occurred only after the divergence of the Fabales clade. However, for such a scenario, it is essential that Fabales represents the most basal lineage in the nitrogen-fixing clade. To date, this remains unknown. For example, a recent phylogenetic study suggests, although with limited statistical support, that Fabales is sister to Fagales (Koenen et al., 2020). The phylogenetic analysis presented here (Fig. 3) suggests that the first scenario is most probable (aSH-aLRT/UF-Bootstrap/approximation with Mr.Bayes support 76.4/77/0.859). Additionally, we searched for amino acid motifs in NFP-I- and NFP-II-type proteins and found an indel region in legume and nonlegume NFP-II-type proteins that is distinct from NFP-I (Supplemental Fig. S9). This also supports the hypothesis that NFP1-NFP2 duplicated at the root of the nitrogen-fixing clade. However, additional experiments are needed to definitively reject either scenario.
Transcomplementation studies in a L. japonicus Ljnfr1;Ljnfr5 double mutant showed that P. andersonii LCO receptors can only partially restore LCO signaling. This only partial complementation we did not anticipate, because of the shared microsymbiont M. loti that can nodulate P. andersonii well as L. japonicus. One explanation for this limited functionality may be that such receptors function in larger multiprotein membrane domain complexes. In such a case, the P. andersonii LCO receptors are not adapted to interact with associated L. japonicus proteins. Additionally, legumes and Parasponia spp. have diverged in the mode of rhizobium infection. Whereas rhizobium penetrates Parasponia spp. roots apoplasticly by crack entry, legumes are generally infected intracellularly via curled root hair cells. Phenotypic analysis of rhizobium infection in legumes suggests that a specific LCO receptor is involved in this process, the so-called entry receptor (Ardourel et al., 1994). Such entry receptors have not yet been fully characterized, but MtLYK3 may carry out such functions, as they control rhizobium infection (Limpens et al., 2003; Smit et al., 2007). It remains elusive whether such entry receptor functioning requires specific adaptations that did not occur in the genus Parasponia LYK3 ortholog.
We showed that an engineered TleNFP2 receptor can functionally complement the P. andersonii Pannfp2 mutant, whereas PanNFP1 cannot. This suggests that the NFP1 and NFP2 receptor proteins have functionally diverged. Based on the finding that NFP-orthologous proteins of P. hybrida (PhLYK10) and tomato (SlLYK10) can complement L. japonicus Ljnfr5 and M. truncatula Mtnfp mutants, it can be hypothesized that, in P. andersonii especially, PanNFP1 has experienced protein adaptations. However, it should be noted that the transcomplementation studies presented here were conducted using the native PanNFP2 promoter, whereas studies conducted with PhLYK10 and SlLYK10 were conducted with CAMV35S (Girardin et al., 2019). Such overexpression may mask differences in substrate affinity and/or specificity, under which native transcriptional regulation is biologically relevant. Our data demonstrate that the ancestor of T. levigata possessed an NFP2 receptor that can function in nodulation.
Mutant analysis in legumes demonstrated that rhizobium nodulation coopted elements of an AM signaling pathway, including the Leucine-rich repeat-type transmembrane receptor kinase L. japonicus SYMBIOTIC RECEPTOR KINASE/ M. truncatula DOES NOT MAKE INFECTIONS2 (MtDMI2), the nuclear envelope-located cation ion channels LjCASTOR, LjPOLLUX/MtDMI1, the nucleus-localized CALCIUM CALMODULIN KINASE LjCCaMK/MtDMI3, and the transcription factor LjCYCLOPS/M. truncatula INTERACTING PROTEIN OF DMI3 (Geurts et al., 2012). However, in legumes, rhizobium and AM fungi were shown to have independent perception mechanisms to activate this common symbiosis signaling pathway. In L. japonicus and M. truncatula, these consist of LjNFR1-LjNFR5/MtLYK3-MtNFP for rhizobium LCOs and MtLYK9/MtCERK1 for AM signals (Geurts et al., 2012; Feng et al., 2019; Gibelin-Viala et al., 2019). MtLYK3 and MtLYK9/MtCERK1 both belong to the LYK-Ib clade and evolved following legume-specific duplication events (Fig. 1; De Mita et al., 2014). The strong phenotype in AM and nodule symbioses of the P. andersonii Panlyk1;Panlyk3 knockout mutant demonstrates that such subfunctionalization is not causal for the evolution of rhizobium LCO receptors. In P. andersonii, both receptors function in conjunction to control AM and rhizobium nodulation. Additionally, PanLYK3 acts as a chitin innate immune receptor. Such multifunctionality has also been reported for MtLYK9/MtCERK1 in M. truncatula and OsCERK1 in rice, which function both in AM symbiosis and chitin innate immune signaling (Miyata et al., 2014; Carotenuto et al., 2017; Feng et al., 2019; Gibelin-Viala et al., 2019). As monocots did not experience the LYK-Ia/LYK-Ib duplication, this demonstrates that performing multiple functions in symbioses and innate immunity was ancestral to species of the nitrogen-fixing clade but functionally diverted in the legume lineage.
The presence of NFP-type genes (LYR-IA orthogroup) in species outside of the nitrogen-fixing clade is associated with the ability to establish an AM symbiosis (Fig. 3; Delaux et al., 2014; Gough et al., 2018). However, corresponding mutants have only a relatively weak phenotype in AM symbiosis (Buendia et al., 2016; Miyata et al., 2016; Girardin et al., 2019). Upon duplication of this gene, the NFP-I and NFP-II subclades may have inherited the ancestral function. As both the P. andersonii PanNFP1 and PanNFP2 receptors can partially complement LCO-induced Ca2+ oscillation in the L. japonicus Ljnfr1;Ljnfr5 double mutant, this supports that receptors of the NFP-I and NFP-II clades can act as an LCO receptor, which may reflect the ancestral function. Our observation that the presence of a functional gene in the NFP-II clade strictly associates with LCO-based nodulation suggests that this gene was coopted to function in this trait. The importance of this LysM-type LCO receptor in the nitrogen-fixing nodulation trait is underlined by the complete block of nodulation in knockout mutants in legumes (e.g. L. japonicus Ljnfr5, M. truncatula Mtnfp, and pea Pssym10) and the genus Parasponia (P. andersonii Pannfp2; Madsen et al., 2003; Arrighi et al., 2006). As the genus Parasponia and legumes diverged at the root of the nitrogen-fixing clade, this suggests that the adaptations in the NFP-II clade are ancient and may have coincided with the birth of the nodulation trait.
The NFP-I-type gene retained, at least in part, its ancestral function, indicated by its presence in nonnodulating species in the nitrogen-fixing clade that can establish an AM symbiosis. In cases where AM symbiosis is replaced by an ectomycorrhizal symbiosis, such as in European beech or Chinese chestnut, the NFP-I-type gene pseudogenized. However, phenotypic studies in stable P. andersonii mutants could not support the functioning of PanNFP1 in AM symbiosis. These findings contradict our earlier observation that this gene functions in arbuscule formation (Op den Camp et al., 2011). The reason for this discrepancy may be due to the RNAi construct used, which may have off-target effects (van Velzen et al., 2018). To determine whether this is the case, we have studied the expression of LysM-type RLK genes in two independent PanNFP1 RNAi experiments. This revealed significant knockdown not only of PanNFP1 but also PanNFP2, which can explain the strong rhizobium nodulation and infection phenotype as reported by Op den Camp et al. (2011). We also found variable expression levels of other LysM-type RLKs, including PanLYK1 and PanLYK3, which may explain the reported mycorrhization phenotype on PanNFP1 RNAi roots (Supplemental Fig. S10). The studies presented here using CRISPR-Cas9 knockout mutant lines revealed substantial biological variation in mycorrhization efficiency of P. andersonii roots, which may have hindered the observation of minor quantitative AM symbiosis phenotypes. To rule out that PanNFP1 and PanNFP2 may function redundantly to control AM symbiosis, we analyzed a Pannfp1;Pannfp2 double mutant. Also, these lines were shown to be effectively mycorrhized. Therefore, we conclude that our current mutant phenotype analysis does not find support for essential functioning of P. andersonii PanNFP1 and PanNFP2 in AM symbiosis.
The study presented here provides insight into the evolutionary trajectory of symbiotic LCO LysM-type receptors. By using P. andersonii as a comparative system to legumes, we revealed two ancestral duplications of LysM-type LCO receptors that predate, and coincide with, the evolution of nitrogen-fixing nodules. The strict association of genes in the NFP-II clade with LCO-driven nodulation strongly suggests that this gene was coopted to function specifically in this symbiosis, making NFP2 a target in approaches to engineer LCO-driven nodulation in nonleguminous plants.
MATERIALS AND METHODS
LysM-Type Receptor Phylogeny Reconstructions
Orthogroups containing LysM-type receptor kinases of Parasponia andersonii, generated in a previous study (van Velzen et al., 2018), were combined and realigned into a single alignment using MAFFTV7.017 (Kazutaka and Schandley, 2013). MrBayes3.2.6 (Ronquist and Huelsenbeck, 2003; Ronquist et al., 2012) was used to calculate phylogenetic relations under default parameters in Geneious R8.1.9 (Biomatters ). Clades were named as published previously (Buendia et al., 2018). For clades LYK-I and LYR-IA, additional putative orthologs were collected from the Phytozome and National Center for Biotechnology Information databases using BLAST with AtCERK and MtNFP protein sequences as queries (Supplemental Table S1; Altschul et al., 1997). Available genomes from Fabales, Fagales, Cucurbitales, and Rosales species were downloaded, and local BLAST analysis was conducted using Geneious R8.1.9 (Biomatters) to search for additional unannotated LYK-I and LYR-IA protein sequences. Pseudogenes were annotated manually based on the closest functional ortholog so that a protein sequence could be deduced. Correct protein sequences were aligned using MAFFTV7.017 and subsequently manually curated. The deduced amino acid sequence was subsequently added to the alignment if the alignment length was at least 70% of the P. andersonii protein. Phylogenetic analysis was performed using IQ-tree (Nguyen et al., 2015; Trifinopoulos et al., 2016), running the modelfinder extension to find the best substitution models (Kalyaanamoorthy et al., 2017). Branch support analysis was done using Sh.aLRT 1,000 replicates, UF-BOOTSTRAP support 1,000 iterations (Kalyaanamoorthy et al., 2017; Hoang et al., 2018), and approximate Bayes support. Branch supports shown are UF-Bootstrap support%. The best fit model for the LYK-I clade was JTT+I+G4, and the best fit model for the LYR-IA clade was JTT+I+G4. Resulting tree files were loaded into Interactive Tree Of Life v3 for editing (Letunic and Bork, 2016). The analysis was run at least three times. Trees were rooted to the outgroup angiosperm species Amborella trichopoda. UF Bootstrap Branch supports >98 were omitted for visual clarity. Gene names, accession numbers, and alignment files of identified homologs can be found in Supplemental Data Set S1 for LYK-I, Supplemental Data Set S2 for LYR-IA, and Supplemental Table S1 for P. andersonii.
LYK3 Alignment and Variant Detection
Genomic LYK3 regions of P. andersonii, Parasponia rigida, Parasponia rugosa, Trema orientalis RG16, T. orientalis RG33, and Trema levigata were extracted from the respective assemblies (van Velzen et al., 2018) and aligned using MAFFTV7.0.17 implemented in Geneious R8.1. Coding sequences of P. andersonii, P. rigida, and P. rugosa LYK3 protein variants were translated and aligned using MAFFTV7.0.17 implemented in Geneious R8.1 (Supplemental Data Set S1).
Vector Constructs
All vectors generated for this study were created using golden gate cloning (Engler et al., 2009). Backbones and binary vectors were derived from the golden gate molecular toolbox (Engler et al., 2014). P. andersonii LysM-type receptor cDNA clones were sequence synthesized as level 0 modules, including silent mutations in golden gate BsaI or BpiI restriction sites. Golden gate-compatible clones of LjNFR1 and LjNFR5 promoters, coding sequences, and terminators were obtained from Aarhus University. The calcium signaling reporter pLjUBQ1:R-GECO1.2 was published previously (Kelner et al., 2018). The generation and assembly of P. andersonii CRISPR constructs were done as published previously (van Zeijl et al., 2018). For hairy root transformation, a modified level 2 standard vector carrying spectinomycin instead of kanamycin resistance was created. All sgRNAs were expressed using the AtU6 promoter. All golden gate binary vectors were verified by restriction digestion and DNA sequencing before transformation. Lists of primers and constructs can be found in Supplemental Tables S3 and S4.
Genotyping and Off-Target Analysis
All sgRNA targets were designed using the Geneious R10 CRISPR design tool, which picks targets on the principles described by Doench et al. (2014). To be selected, guide RNAs must have no potential target sites in the genome with (1) less than three mismatches or (2) less than two indels. Known off-target locations in coding sequence regions were PCR amplified and sequenced. No off-target mutations at these sites were detected. Genotypes and known off-target locations of CRISPR mutants used in this study can be found in Supplemental Data Set S3. Primers used for the creation of sgRNAs and subsequent sequencing of mutants and off-targets are listed in Supplemental Table S4.
Bacterial Strains
We used Mesorhizobium plurifarium BOR2 (van Velzen et al., 2018) and Sinorhizobium fredii NGR234.pHC60 expressing GFP (Trinick and Galbraith, 1980; Cheng and Walker, 1998; Op den Camp et al., 2011) for P. andersonii inoculation experiments. Mesorhizobium loti R7A.pHC60 (Cheng and Walker, 1998; Sullivan et al., 2002) was used for Lotus japonicus inoculations. M. loti R7A and Rhizobium tropici CIAT899 (Martínez-Romero et al., 1991) containing plasmid pMP604 (Spaink et al., 1989) were used for LCO extraction. Agrobacterium rhizogenes strain AR10 (Hansen et al., 1989; Martínez-Romero et al., 1991) was used for L. japonicus root transformation. Agrobacterium tumefaciens strain AGL-1 (Lazo et al., 1991) was used in P. andersonii transformation. Agrobacterium sp. MSU440 was used for P. andersonii hairy root transformation (Cao et al., 2012). The Escherichia coli strain DH5α was used to propagate plasmids and in all subsequent cloning steps.
Rhizobium LCO Isolation
To isolate rhizobium LCOs, the plasmid pMP604 containing an autoactive NodD protein was introduced in M. loti R7A and R. tropici CIAT899 (Spaink et al., 1989; López-Lara et al., 1995). LCOs were extracted from a 750-mL liquid culture, OD600 = 0.5, grown at 28°C in minimal medium (5.75 mm K2HPO4, 7.35 mm KH2PO4, 5.9 mm KNO3, 460 nm CaCl2, 37.5 µm FeCl3, 2.07 mm MgSO4, 20.5 nm biotin, 2.9 nm thiamine-HCl, 8.1 nm nicotinic acid, 4.8 nm pyridoxine-HCl, 2.8 nm myoinositol, 4.6 nm panthotenate, and 1% [w/v] Suc) by the addition of 150 mL of 1-butanol and 1 h of shaking. The butanol phase was transferred and subsequently evaporated (water bath at 40°C). The pellet was dissolved in 75 mL of methanol, tested for Nod factor activity, and stored at −20°C for later use. The concentration of active LCOs was estimated by using LjNIN induction in L. japonicus wild-type cv Gifu roots 3 h postapplication. The lowest active dilution was estimated to be ∼10−10 m.
L. japonicus A. rhizogenes Root Transformation
L. japonicus Ljnfr1-1;Ljnfr5-2 double mutants (Madsen et al., 2003; Radutoiu et al., 2003) were used for LysM complementation assays and cv Gifu wild type as control. Seedlings for A. rhizogenes root transformation were moved to fresh one-half-strength B5 medium and cocultivated for 1 week as described previously using A. rhizogenes strain AR10 (Stougaard et al., 1987; Hansen et al., 1989; Stougaard, 1995). During root emergence, plants were grown on 1% (w/v) agar plates containing one-half-strength B5 medium containing 0.03% (w/v) cefotaxime and 1% (w/v) Suc. Plants were screened for transformed roots using nucleus-localized R.GECO1.2 fluorescence. Shoots with transformed roots were grown in Agroperlite (Maasmond-Westland) supplemented with modified one-half-strength Hoagland medium (Hoagland and Arnon, 1950) containing 0.56 mm NH4NO3 and inoculated with M. loti R7A.pHC60 (expressing GFP) at OD600 = 0.05. Plants were grown at 21°C under a 16-h-light/8-h-dark regime. For calcium oscillation analysis, transformed plants were grown on one-half-strength Hoagland plates with 1% agar containing 0.56 mm NH4NO3 for 1 week. Plants were moved to N-free one-half-strength Hoagland medium 1 week prior to imaging.
Calcium Oscillation Quantification
Calcium spiking experiments were performed on a Leica TCS SP8 HyD confocal microscope equipped with a water lens HC plan-Apochromat CS2 40×/1.0. Transformed root segments expressing R-GECO1.2 were selected and incubated with 500× diluted LCO extract (estimated to represent ∼10−9 m) in nitrate-free one-half-strength Hoagland medium (Hoagland and Arnon, 1950) on a glass slide with coverslip. Images were taken at 5-s intervals for a minimum of 20 min per sample using an excitation wavelength of 552 nm and emission spectrum of 585 to 620 nm. It is possible to monitor a large number of nuclei per root sample. However, only epidermal and especially root hairs were shown to be responsive. Therefore, total nuclei numbers vary largely between samples. Video recordings of imaged root samples were exported to ImageJ1.50i (Collins, 2007). The Geciquant ImageJ plugin was used for background subtraction and region of interest selection (Srinivasan et al., 2015). The average pixel intensities of regions of interest (individual nuclei) were measured. Average pixel values (0–255) per nucleus were plotted, and a background R-GECO1.2 fluorescence baseline of 2 × 1 min (two regions of 12 frames) was selected manually in a region of the trace where no spikes were occurring. Only nuclei with a minimum of three spikes with an amplitude of over 1.5 times background were considered as positive.
P. andersonii Growth Conditions for Propagation, Transformation, Mycorrhization, and Nodulation
Sequenced P. andersonii WU1 trees or their direct descendants were used in all experiments (Op den Camp et al., 2011; van Velzen et al., 2018). Prior to transformation or transfer to tissue culture, P. andersonii trees were grown in a conditioned greenhouse at 28°C, 85% humidity, and a 16/8-h day/night regime. P. andersonii in vitro propagation, transformation, CRISPR-Cas9 mutagenesis, and nodulation assays were done according to van Zeijl et al. (2018). P. andersonii hairy root transformations were performed according to Cao et al. (2012).
P. andersonii Nodulation Assay and Analysis
Rooted tissue culture plantlets for phenotyping assays were grown in crystal-clear polypropylene containers (1 L) with a gas-exchange filter (OS140BOX; Duchefa Biochemie). Pots were half-filled with Agroperlite (Maasmond-Westland) and watered with modified EKM medium (3 mm MES [C6H13NO4], pH 6.6, 2.08 mm MgSO4, 0.88 mm KH2PO4, 2.07 mm K2HPO4, 1.45 mm CaCl2, 0.7 mm Na2SO4, 0.375 mm NH4NO3, 15 μm iron citrate, 6.6 μm MnSO4, 1.5 μm ZnSO4, 1.6 μm CuSO4, 4 μm H3BO3, and 4.1 μm Na2MoO4; Becking, 1983). For nodulation assays, modified EKM medium (Becking, 1983) was inoculated with rhizobia (OD600 = 0.025) prior to planting the shoots. For inoculation with strain S. fredii NGR234.pHC60, containers were half-filled with sterilized river sand and watered with modified EKM medium containing the bacteria at OD600 = 0.05.
All nodules were fixed in buffer containing 4% (w/v) paraformaldehyde mixed with 3% (v/v) glutaraldehyde in 50 mm phosphate (pH 7.4). A vacuum was applied for 2 h during a total 48-h incubation. Fixed nodules were embedded in plastic (Technovit 7100; Heraeus-Kulzer) according to the manufacturer’s recommendations. Sections (5 µm) were made using an RJ2035 microtome (Leica Microsystems). Sections were stained using 0.05% (w/v) Toluidine Blue O. Images were taken with a DM5500B microscope equipped with a DFC425c camera (Leica Microsystems).
P. andersonii Mycorrhization Assay
For mycorrhization experiments, pots were half-filled with sterilized river sand and watered with modified one-half-strength Hoagland medium containing 20 µm potassium phosphate. Pots were inoculated with 250 spores of Rhizopagus irregularis (Agronutrion-DAOM197198). In all experiments, plantlets in pots with closed lids were placed in a climate room at 28°C, 16/8 h day/night. Plants were watered with sterilized, demineralized water. Plants were harvested 6 wpi with R. irregularis (Agronutrion-DAOM197198). Root segments were treated with 10% (w/v) KOH and incubated at 90°C for 20 min. The root samples were then rinsed six times with water and stained with Trypan Blue at 90°C for 5 min. For each mutant, 10 plants were assessed, and from each plant, 30 root segments (each segment approximately 1 cm long) were examined, and mycorrhizal structures (hyphae, vesicles, and arbuscules) were determined using the magnified line intersect method (Trouvelot et al., 1986) using a Leica CTR6000 microscope. For staining with WGA-Alexa488 (Molecular Probes, Thermo Fisher Scientific), roots were incubated in 10% (w/v) KOH at 60°C for 3 h. Then, roots were washed three times in phosphate-buffered saline (150 mm NaCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4, pH 7.4) and incubated in 0.2 μg mL−1 WGA-Alexa488 in phosphate-buffered saline at room temperature for 16 h. For RNA isolation, P. andersonii wild-type plants were grown according to the conditions described above. RNA was isolated according to protocols published previously (Op den Camp et al., 2011; van Velzen et al., 2018). Mock-inoculated plants were harvested as controls. Three independent biological replicates were taken per sample. Expression was determined using RNA sequencing. Reads were mapped using kallisto (Bray et al., 2016). Expression values and differential expression were determined using sleuth (Pimentel et al., 2017). Differentially expressed genes were identified using Benjamini-Hochberg multiple testing correction (q ≤ 0.05).
Quantitative PCR Analysis of PanNFPi cDNA Samples
PanNFPi cDNA samples were generated previously (Op den Camp et al., 2011). Quantitative PCR was performed in 10-μL reactions using 2× iQ SYBR Green Supermix (Bio-Rad). PCR was executed on a CFX Connect optical cycler according to the manufacturer’s protocol (Bio-Rad). Three technical replicates per cDNA sample were used. Data analysis and statistical analysis of biological replicates were performed using CFX Manager 3.0 software (Bio-Rad). Gene expression was normalized against reference genes PanACTIN and PanEF1α. Primers can be found in Supplemental Table S4.
ROS Assay
P. andersonii plantlets were grown on rooting medium (van Zeijl et al., 2018) for 4 weeks at 28°C before the treatment. Roots, submerged in water, were cut into approximately 1-cm pieces. Each well of a black 96-well flat-bottom polystyrene plate (Nunc) was filled with 10 root pieces. Ten replicates per line were analyzed. After filling the wells, the plate was kept 5 h in 28°C. After incubation, the water was replaced with 100 µL of assay solution containing 0.5 µm L-012 (FUJIFILM Wako Chemicals), 10 µg mL−1 horseradish peroxidase (Sigma), and respective elicitors (CO7; ELICITYL) or LCOs extracted from M. loti or R. tropici at the described concentrations. As a mock treatment, 100 µL of water was added. The light emission was immediately measured at 30-s intervals for 30 min using a Clariostar multi-well plate reader. All data are averages of at least three independent biological replicates.
Protein Extraction from P. andersonii and Western Blotting
P. andersonii plantlets were grown on rooting medium (van Zeijl et al., 2018) for 4 weeks at 28°C before the treatment. About 200 mg of roots was cut while submerged in water and collected in a PCR tube. Root segments were incubated for 5 h at 28°C before treatment. Root pieces were treated with water containing 100 μm CO7 (ELICITYL) for 10 min. After incubation, roots were immediately frozen in liquid nitrogen. Samples were homogenized using metal beads. Total root protein was extracted in a buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm KCl, 1 mm EDTA (pH 7.5), 0.1% (w/v) Triton X-100, 1 mm DTT, complete protease inhibitors (Roche), and phosstop (Roche). Amounts of extracted protein were measured with Qubit (Thermo Fisher Scientific), and equal amounts of protein (∼20 μg) were electrophoresed on Mini-PROTEAN TGX stain-free gels (Bio-Rad). A Trans-Blot Turbo Transfer system was used for blotting. To visualize phosphorylated MPK3/MPK6, the antibody for anti-phospho-p44/42 MAPK was used (no. 4370; Cell Signaling Technology). Anti-rabbit antibody (no. 7054; Cell Signaling Technology) was used as a secondary antibody. Equal loading was confirmed by Coomassie Brilliant Blue staining.
Quantification and Statistical Analysis
Nodule number was quantified as mean nodule number ± se for all experiments. Replicate number is denoted in figures or figure legends. Additionally, all individual data points were plotted for graphical visualization of variation. Graphs and statistical analysis were performed using R studio 1.1.456 for nodulation experiments. Statistical tests on nodule numbers was done using one-way ANOVA and Tukey’s posthoc test for multiple comparisons. Statistical significance was defined as P < 0.05. Levene’s test for homogeneity of variance was used prior to running one-way ANOVA. In cases where the normality assumption was violated, alternative tests such as the Mann-Whitney-Wilcoxon test were used as denoted in the figure legends. For the mycorrhization experiment, a standard linear model was used to estimate the difference, and the corresponding lsd, of the knockout mutants with the wild-type control. The lsd with respect to the control was Bonferroni adjusted to correct for multiple testing.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers mentioned in Supplemental Table S1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogeny reconstruction of orthogroups representing LysM-type receptors.
Supplemental Figure S2. Duplication of the LYK3 first exon is conserved among Parasponia spp. and Trema subspecies.
Supplemental Figure S3. Complementation of a L. japonicus Ljnfr1;Ljnfr5 mutant for LCO-induced calcium oscillation.
Supplemental Figure S4. Nodulation is affected in P. andersonii Pannfp1, Pannfp2, and Panlyk3 CRISPR-Cas9 mutants.
Supplemental Figure S5. Complementation of the P. andersonii Panlyk1;Panlyk3 double mutant.
Supplemental Figure S6. CO7 triggered ROS production and MPK phosphorylation in P. andersonii mutant lines.
Supplemental Figure S7. Expression of P. andersonii LysM-type receptors during mycorrhization.
Supplemental Figure S8. P. andersonii LysM-type receptor mutants can establish AM symbiosis.
Supplemental Figure S9. Conserved indels in NFP-II-type receptor proteins.
Supplemental Figure S10. The PanNFPi RNAi construct has off-target activity on PanNFP2 and other LysM-type receptor kinases.
Supplemental Table S1. P. andersonii LysM-type receptors.
Supplemental Table S2. Transcomplementation of L. japonicus Ljnfr1;Ljnfr5 for nodulation.
Supplemental Table S3. Constructs generated in this study.
Supplemental Table S4. Primers used in this study.
Supplemental Data Set S1. Sequence alignment of LYK-I-type receptors in fasta format.
Supplemental Data Set S2. Sequence alignment of LYR-Ia-type receptors in fasta format.
Supplemental Data Set S3. Genotyping and off-target analysis of P. andersonii CRISPR-Cas9 mutants generated in this study.
Supplemental Movie S1. Calcium spiking in root hairs of the L. japonicus Ljnfr1-1;Ljnfr5-2 double mutant complemented with LjNFR1;LjNFR5.
Supplemental Movie S2. Calcium spiking in root hairs of the L. japonicus Ljnfr1-1;Ljnfr5-2 double mutant transcomplemented with PanLYK3.1;LjNFR5.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Jens Stougaard (Aarhus University) and Simona Radutoiu (Aarhus University) for sharing the L. japonicus Ljnfr1-1;Ljnfr5-2 double mutant and the LjNFR1 and LjNFR5 constructs, Maryam Charpentier (John Innes Centre) for sharing R-GECO1.2, Jinling Li (Wageningen University) and Bart Thomma (Wageningen University) for the use of the Clariostar multi-well reader, Joost van Heerwaarden (Wageningen University) for statistical support, and Rens Holmer (Wageningen University) for contributing to Figure 2B. Furthermore, we thank Arianne van Lierop, Jens van Kempen, Stan van der Wal, and Vince Brouwer, who all contributed as Wageningen University undergraduate students to this research.
Footnotes
This work was supported by the Netherlands Organisation for Scientific Research (grant no. 865.13.001 to R.G. and grant no. 863.15.010 to W.K.), the ENSA project funded by the Bill & Melinda Gates Foundation to the University of Cambridge (to R.G.), the Japan Society for the Promotion of Science (Overseas Research Fellowship to K.M.), and the Ministry of Research, Technology, and Higher Education of the Republic of Indonesia (grant no. 8245–ID to Y.P.R.).
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ(1997) Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardourel M, Demont N, Debellé F, Maillet F, de Billy F, Promé JC, Dénarié J, Truchet G(1994) Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6: 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrighi JF, Barre A, Ben Amor B, Bersoult A, Soriano LC, Mirabella R, de Carvalho-Niebel F, Journet EP, Ghérardi M, Huguet T, et al. (2006) The Medicago truncatula lysin motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol 142: 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becking JH(1983) The Parasponia parviflora-Rhizobium symbiosis: Host specificity, growth and nitrogen fixation under various conditions. Plant Soil 75: 309–342 [Google Scholar]
- Bozsoki Z, Cheng J, Feng F, Gysel K, Vinther M, Andersen KR, Oldroyd G, Blaise M, Radutoiu S, Stougaard J(2017) Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception. Proc Natl Acad Sci USA 114: E8118–E8127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L(2016) Erratum: Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 888. [DOI] [PubMed] [Google Scholar]
- Broghammer A, Krusell L, Blaise M, Sauer J, Sullivan JT, Maolanon N, Vinther M, Lorentzen A, Madsen EB, Jensen KJ, et al. (2012) Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA 109: 13859–13864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia L, Girardin A, Wang T, Cottret L, Lefebvre B(2018) LysM receptor-like kinase and LysM receptor-like protein families: An update on phylogeny and functional characterization. Front Plant Sci 9: 1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buendia L, Wang T, Girardin A, Lefebvre B(2016) The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol 210: 184–195 [DOI] [PubMed] [Google Scholar]
- Burrill TJ, Hansen R (1917) Is Symbiosis Possible between Legume Bacteria and Non-Legume Plants? Bulletin University of Illinois Agricultural Experiment Station No. 202.
- Cao Q, den Camp RO, Kalhor MS, Bisseling T, Geurts R(2012) Efficiency of Agrobacterium rhizogenes-mediated root transformation of Parasponia and Trema is temperature dependent. Plant Growth Regul 68: 459–465 [Google Scholar]
- Carotenuto G, Chabaud M, Miyata K, Capozzi M, Takeda N, Kaku H, Shibuya N, Nakagawa T, Barker DG, Genre A(2017) The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol 214: 1440–1446 [DOI] [PubMed] [Google Scholar]
- Cheng HP, Walker GC(1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180: 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ(2007) Introduction to ImageJ for light microscopy. Microsc Microanal 13: 1674–1675 [Google Scholar]
- Delaux PM, Varala K, Edger PP, Coruzzi GM, Pires JC, Ané JM(2014) Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet 10: e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mita S, Streng A, Bisseling T, Geurts R(2014) Evolution of a symbiotic receptor through gene duplications in the legume-rhizobium mutualism. New Phytol 201: 961–972 [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE(2014) Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol 32: 1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S(2009) Golden gate shuffling: A one-pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE 4: e5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Youles M, Gruetzner R, Ehnert TM, Werner S, Jones JDG, Patron NJ, Marillonnet S(2014) A golden gate modular cloning toolbox for plants. ACS Synth Biol 3: 839–843 [DOI] [PubMed] [Google Scholar]
- Feng F, Sun J, Radhakrishnan GV, Lee T, Bozsóki Z, Fort S, Gavrin A, Gysel K, Thygesen MB, Andersen KR, et al. (2019) A combination of chitooligosaccharide and lipochitooligosaccharide recognition promotes arbuscular mycorrhizal associations in Medicago truncatula. Nat Commun 10: 5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch-Mallol JL, Marroqui S, Sousa C, Manyani H, López-Lara IM, van der Drift KM, Haverkamp J, Quinto C, Gil-Serrano A, Thomas-Oates J, et al. (1996) Characterization of Rhizobium tropici CIAT899 nodulation factors: The role of nodH and nodPQ genes in their sulfation. Mol Plant Microbe Interact 9: 151–163 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Balzergue C, Puech-Pagès V, Novero M, Rey T, Fournier J, Rochange S, Bécard G, Bonfante P, et al. (2013) Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol 198: 190–202 [DOI] [PubMed] [Google Scholar]
- Geurts R, Lillo A, Bisseling T(2012) Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis. Curr Opin Plant Biol 15: 438–443 [DOI] [PubMed] [Google Scholar]
- Gibelin-Viala C, Amblard E, Puech-Pages V, Bonhomme M, Garcia M, Bascaules-Bedin A, Fliegmann J, Wen J, Mysore KS, le Signor C, et al. (2019) The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol 223: 1516–1529 [DOI] [PubMed] [Google Scholar]
- Girardin A, Wang T, Ding Y, Keller J, Buendia L, Gaston M, Ribeyre C, Gasciolli V, Auriac MC, Vernié T, et al. (2019) LCO receptors involved in arbuscular mycorrhiza are functional for rhizobia perception in legumes. Curr Biol 29: 4249–4259.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves DJP, Simpson BB, Ortiz EM, Shimizu GH, Jansen RK(2019) Incongruence between gene trees and species trees and phylogenetic signal variation in plastid genes. Mol Phylogenet Evol 138: 219–232 [DOI] [PubMed] [Google Scholar]
- Gough C, Cottret L, Lefebvre B, Bono JJ(2018) Evolutionary history of plant LysM receptor proteins related to root endosymbiosis. Front Plant Sci 9: 923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough C, Cullimore J(2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol Plant Microbe Interact 24: 867–878 [DOI] [PubMed] [Google Scholar]
- Griesmann M, Chang Y, Liu X, Song Y, Haberer G, Crook MB, Billault-Penneteau B, Lauressergues D, Keller J, Imanishi L, et al. (2018) Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science 361: eaat1743. [DOI] [PubMed] [Google Scholar]
- Hansen J, Jørgensen JE, Stougaard J, Marcker KA(1989) Hairy roots: A short cut to transgenic root nodules. Plant Cell Rep 8: 12–15 [DOI] [PubMed] [Google Scholar]
- He J, Zhang C, Dai H, Liu H, Zhang X, Yang J, Chen X, Zhu Y, Wang D, Qi X, et al. (2019) A LysM receptor heteromer mediates perception of arbuscular mycorrhizal symbiotic signal in rice. Mol Plant 12: 1561–1576 [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The Water-Culture Method for Growing Plants without Soil. Circular California Agricultural Experiment Station No. 347.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS(2018) UFBoot2: Improving the ultrafast bootstrap approximation. Mol Biol Evol 35: 518–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman R, Geurts R(2019) A roadmap towards engineered nitrogen-fixing nodule symbiosis. Plant Commun 1: 100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS(2017) ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods 14: 587–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazutaka K, Schandley D(2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30: 772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelner A, Leitão N, Chabaud M, Charpentier M, de Carvalho-Niebel F(2018) Dual color sensors for simultaneous analysis of calcium signal dynamics in the nuclear and cytoplasmic compartments of plant cells. Front Plant Sci 9: 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenen EJM, Ojeda DI, Steeves R, Migliore J, Bakker FT, Wieringa JJ, Kidner C, Hardy OJ, Pennington RT, Bruneau A, et al. (2020) Large-scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near-simultaneous evolutionary origin of all six subfamilies. New Phytol 225: 1355–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancelle SA, Torrey JG(1984) Early development of Rhizobium-induced root nodules of Parasponia rigida. I. Infection and early nodule initiation. Protoplasma 123: 26–37 [Google Scholar]
- Lancelle SA, Torrey JG(1985) Early development of Rhizobium-induced root nodules of Parasponia rigida. II. Nodule morphogenesis and symbiotic development. Can J Bot 63: 25–35 [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA(1991) A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Biotechnology (N Y) 9: 963–967 [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P(2016) Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44: W242–W245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HL, Wang W, Mortimer PE, Li RQ, Li DZ, Hyde KD, Xu JC, Soltis DE, Chen ZD(2015) Large-scale phylogenetic analyses reveal multiple gains of actinorhizal nitrogen-fixing symbioses in angiosperms associated with climate change. Sci Rep 5: 14023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao D, Sun X, Wang N, Song F, Liang Y(2018) Tomato LysM receptor-like kinase SlLYK12 is involved in arbuscular mycorrhizal symbiosis. Front Plant Sci 9: 1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R(2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302: 630–633 [DOI] [PubMed] [Google Scholar]
- López-Lara IM, van den Berg JD, Thomas-Oates JE, Glushka J, Lugtenberg BJJ, Spaink HP(1995) Structural identification of the lipo-chitin oligosaccharide nodulation signals of Rhizobium loti. Mol Microbiol 15: 627–638 [DOI] [PubMed] [Google Scholar]
- Madsen EB, Madsen LH, Radutoiu S, Olbryt M, Rakwalska M, Szczyglowski K, Sato S, Kaneko T, Tabata S, Sandal N, et al. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425: 637–640 [DOI] [PubMed] [Google Scholar]
- Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, et al. (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469: 58–63 [DOI] [PubMed] [Google Scholar]
- Martínez-Romero E, Segovia L, Mercante FM, Franco AA, Graham P, Pardo MA(1991) Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int J Syst Bacteriol 41: 417–426 [DOI] [PubMed] [Google Scholar]
- Marvel DJ, Torrey JG, Ausubel FM(1987) Rhizobium symbiotic genes required for nodulation of legume and nonlegume hosts. Proc Natl Acad Sci USA 84: 1319–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa H, Sun J, Oldroyd GED, Downie JA(2006) Analysis of Nod-factor-induced calcium signaling in root hairs of symbiotically defective mutants of Lotus japonicus. Mol Plant Microbe Interact 19: 914–923 [DOI] [PubMed] [Google Scholar]
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N(2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA 104: 19613–19618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata K, Hayafune M, Kobae Y, Kaku H, Nishizawa Y, Masuda Y, Shibuya N, Nakagawa T(2016) Evaluation of the role of the LysM receptor-like kinase, OsNFR5/OsRLK2 for AM symbiosis in rice. Plant Cell Physiol 57: 2283–2290 [DOI] [PubMed] [Google Scholar]
- Miyata K, Kozaki T, Kouzai Y, Ozawa K, Ishii K, Asamizu E, Okabe Y, Umehara Y, Miyamoto A, Kobae Y, et al. (2014) The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol 55: 1864–1872 [DOI] [PubMed] [Google Scholar]
- Murakami E, Cheng J, Gysel K, Bozsoki Z, Kawaharada Y, Hjuler CT, Sørensen KK, Tao K, Kelly S, Venice F, et al. (2018) Epidermal LysM receptor ensures robust symbiotic signalling in Lotus japonicus. eLife 7: e33506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ(2015) IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32: 268–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Wibberg D, Battenberg K, Blom J, Vanden Heuvel B, Berry AM, Kalinowski J, Pawlowski K(2016) An assemblage of Frankia cluster II strains from California contains the canonical nod genes and also the sulfotransferase gene nodH. BMC Genomics 17: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TV, Wibberg D, Vigil-Stenman T, Berckx F, Battenberg K, Demchenko KN, Blom J, Fernandez MP, Yamanaka T, Berry AM, et al. (2019) Frankia-enriched metagenomes from the earliest diverging symbiotic Frankia cluster: They come in teams. Genome Biol Evol 11: 2273–2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Op den Camp R, Streng A, De Mita S, Cao Q, Polone E, Liu W, Ammiraju JSS, Kudrna D, Wing R, Untergasser A, et al. (2011) LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science 331: 909–912 [DOI] [PubMed] [Google Scholar]
- Parniske M(2008) Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol 6: 763–775 [DOI] [PubMed] [Google Scholar]
- Pawlowski K, Demchenko KN(2012) The diversity of actinorhizal symbiosis. Protoplasma 249: 967–979 [DOI] [PubMed] [Google Scholar]
- Persson T, Battenberg K, Demina IV, Vigil-Stenman T, Vanden Heuvel B, Pujic P, Facciotti MT, Wilbanks EG, O’Brien A, Fournier P, et al. (2015) Candidatus Frankia Datiscae Dg1, the actinobacterial microsymbiont of Datisca glomerata, expresses the canonical nod genes nodABC in symbiosis with its host plant. PLoS ONE 10: e0127630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L(2017) Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425: 585–592 [DOI] [PubMed] [Google Scholar]
- Radutoiu S, Madsen LH, Madsen EB, Jurkiewicz A, Fukai E, Quistgaard EMH, Albrektsen AS, James EK, Thirup S, Stougaard J(2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J 26: 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers C, Oldroyd GED(2014) Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot 65: 1939–1946 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP(2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayers DA, Darling A, Höhna S, Liu L, Suchard MA, Huelsenbeck JP(2012) MrBayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61: 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado MG, van Velzen R, Nguyen TV, Battenberg K, Berry AM, Lundin D, Pawlowski K(2018) Comparative analysis of the nodule transcriptomes of Ceanothus thyrsiflorus (Rhamnaceae, Rosales) and Datisca glomerata (Datiscaceae, Cucurbitales). Front Plant Sci 9: 1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H, Kaku H, et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J 64: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T(2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145: 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Morgan DR, Swensen SM, Mullin BC, Dowd JM, Martin PG(1995) Chloroplast gene sequence data suggest a single origin of the predisposition for symbiotic nitrogen fixation in angiosperms. Proc Natl Acad Sci USA 92: 2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaink HP, Wijffelman CA, Okker RJ, Lugtenberg BE(1989) Localization of functional regions of the Rhizobium nodD product using hybrid nodD genes. Plant Mol Biol 12: 59–73 [DOI] [PubMed] [Google Scholar]
- Srinivasan R, Huang BS, Venugopal S, Johnston AD, Chai H, Zeng H, Golshani P, Khakh BS(2015) Ca2+ signaling in astrocytes from Ip3r2−/− mice in brain slices and during startle responses in vivo. Nat Neurosci 18: 708–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stougaard J(1995) Agobacterium rhizogenes as a vector for transforming higher plants: Application in Lotus corniculatus transformation. Methods Mol Biol 49: 49–61 [DOI] [PubMed] [Google Scholar]
- Stougaard J, Abildsten D, Marcker KA(1987) The Agrobacterium rhizogenes pRi TL-DNA segment as a gene vector system for transformation of plants. Mol Gen Genet 207: 251–255 [Google Scholar]
- Sullivan JT, Trzebiatowski JR, Cruickshank RW, Gouzy J, Brown SD, Elliot RM, Fleetwood DJ, McCallum NG, Rossbach U, Stuart GS, et al. (2002) Comparative sequence analysis of the symbiosis island of Mesorhizobium loti strain R7A. J Bacteriol 184: 3086–3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ(2016) W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44: W232–W235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinick MJ, Galbraith J(1980) The rhizobium requirements of the non-legume parasponia in relationship to the cross-inoculation group concept of legumes. New Phytol 86: 17–26 [Google Scholar]
- Trouvelot A, Kough JL, Gianinazzi-Pearson V(1986) Mesure du taux de mycorhization VA d’un systeme radiculaire: Recherche de methodes d’estimation ayant une signification fonctionnelle. In Gianinazzi-Pearson V, ed, Physiological and Genetic Aspects of Mycorrhizae. INRA Press, Paris, pp 217–221 [Google Scholar]
- van Velzen R, Doyle JJ, Geurts R(2019) A resurrected scenario: Single gain and massive loss of nitrogen-fixing nodulation. Trends Plant Sci 24: 49–57 [DOI] [PubMed] [Google Scholar]
- van Velzen R, Holmer R, Bu F, Rutten L, van Zeijl A, Liu W, Santuari L, Cao Q, Sharma T, Shen D, et al. (2018) Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc Natl Acad Sci USA 115: E4700–E4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zeijl A, Wardhani TAK, Seifi Kalhor M, Rutten L, Bu F, Hartog M, Linders S, Fedorova EE, Bisseling T, Kohlen W, et al. (2018) CRISPR/Cas9-mediated mutagenesis of four putative symbiosis genes of the tropical tree Parasponia andersonii reveals novel phenotypes. Front Plant Sci 9: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G(2008) A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell 20: 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yang S, Tang F, Zhu H(2012) Symbiosis specificity in the legume: Rhizobial mutualism. Cell Microbiol 14: 334–342 [DOI] [PubMed] [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE(2009) Rosid radiation and the rapid rise of angiosperm-dominated forests. Proc Natl Acad Sci USA 106: 3853–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardhani TAK, Roswanjaya YP, Dupin S, Li H, Linders S, Hartog M, Geurts R, van Zeijl A (2019) Transforming, genome editing and phenotyping the nitrogen-fixing tropical Cannabaceae tree Parasponia andersonii. J Vis Exp 150: e59971. [DOI] [PubMed] [Google Scholar]
- Werner GDA, Cornelissen JHC, Cornwell WK, Soudzilovskaia NA, Kattge J, West SA, Kiers ET(2018) Symbiont switching and alternative resource acquisition strategies drive mutualism breakdown. Proc Natl Acad Sci USA 115: 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S, et al. (2013) A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe 13: 347–357 [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480: 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Dong W, Sun J, Feng F, Deng Y, He Z, Oldroyd GED, Wang E(2015) The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J 81: 258–267 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Cannon SB, Stacey G(2009) Evolutionary genomics of LysM genes in land plants. BMC Evol Biol 9: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Araki S, Wu J, Teramoto T, Chang YF, Nakano M, Abdelfattah AS, Fujiwara M, Ishihara T, Nagai T, et al. (2011) An expanded palette of genetically encoded Ca2+ indicators. Science 333: 1888–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Riely BK, Burns NJ, Ané JM(2006) Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172: 2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]