Arabidopsis OXIDATION RESISTANCE2 levels influence salicylic acid-mediated defense, increasing plant resistance to pathogen infection.
Abstract
Arabidopsis (Arabidopsis thaliana) OXIDATION RESISTANCE2 (AtOXR2) is a mitochondrial protein belonging to the Oxidation Resistance (OXR) protein family, recently described in plants. We analyzed the impact of AtOXR2 in Arabidopsis defense mechanisms against the hemibiotrophic bacterial pathogen Pseudomonas syringae. oxr2 mutant plants are more susceptible to infection by the pathogen and, conversely, plants overexpressing AtOXR2 (oeOXR2 plants) show enhanced disease resistance. Resistance in these plants is accompanied by higher expression of WRKY transcription factors, induction of genes involved in salicylic acid (SA) synthesis, accumulation of free SA, and overall activation of the SA signaling pathway. Accordingly, defense phenotypes are dependent on SA synthesis and SA perception pathways, since they are lost in isochorismate synthase1/salicylic acid induction deficient2 and nonexpressor of pathogenesis-related genes1 (npr1) mutant backgrounds. Overexpression of AtOXR2 leads to faster and stronger oxidative burst in response to the bacterial flagellin peptide flg22. Moreover, AtOXR2 affects the nuclear localization of the transcriptional coactivator NPR1, a master regulator of SA signaling. oeOXR2 plants have increased levels of total glutathione and a more oxidized cytosolic redox cellular environment under normal growth conditions. Therefore, AtOXR2 contributes to establishing plant protection against infection by P. syringae acting on the activity of the SA pathway.
Plants inhabit environments with plenty of challenges like climate changes, variations in soil conditions, and the constant attack of insects and pathogens that use plants as their food source. To deal with pathogens, plants have evolved multiple structural barriers and a sophisticated inducible immune system for pathogen recognition and the activation of downstream defense responses (Jones and Dangl, 2006; Vlot et al., 2009). Briefly, the first defense barrier is raised when plant cell membrane receptors recognize pathogen-associated molecular patterns and generate rapid transcriptional changes inside the plant cell inducing the basal defense or pattern-triggered immunity. Some pathogens release effector molecules into the plant that can interfere with pattern-triggered immunity and are specifically recognized by Resistance proteins of the plant cell, leading to the second induced defense barrier, effector-triggered immunity. Upon pathogen recognition, an oxidative burst characterized by a rapid and transient increase in apoplastic reactive oxygen species (aROS) takes place. This is produced by NADPH oxidases, also called respiratory burst oxidase homologs, within the plasma membrane (Torres and Dangl, 2005; Dubiella et al., 2013). Apoplastic and cytoplasmic ROS are important for defense signaling and can lead to a hypersensitive response and programmed cell death (PCD) at the site of infection (del Río et al., 2006; del Río and López-Huertas, 2016). This is an important feature of the defense response against (hemi)biotrophic pathogens.
ROS play a dual role as prooxidants and antioxidants, acting also as signaling molecules responsible to orchestrate the acclimatization or the defense responses like systemic acquired resistance (Czarnocka and Karpiński, 2018). ROS can perform cellular activities related to gene expression, posttranslational protein modifications, and hormonal regulation (Foyer et al., 2017; Mittler, 2017; Mullineaux et al., 2018; Farooq et al., 2019). Among the chemical and hormonal barriers elicited by plants against pathogens, different antioxidant enzymatic systems and scavengers like ascorbate and glutathione (GSH) have been extensively studied (Foyer and Noctor, 2011; Noctor et al., 2012; Plumb et al., 2018). An increment in ROS production induces increased levels of salicylic acid (SA), and due to the existence of a feedback loop between these two defense players, higher SA levels elicit ROS production in the plant cell. Higher levels of ROS induce a redox imbalance in the cell, promoting NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 (NPR1) monomerization and its migration to the nuclei to induce PATHOGENESIS-RELATED (PR) gene expression in cooperation with TGACG-binding TGA transcription factors (TFs; Kinkema et al., 2000; Lindermayr et al., 2010; Herrera-Vásquez et al., 2015). The increment in the SA levels also exerts a control in the redox-regulated NPR1 nuclear translocation (Wu et al., 2012) by binding to NPR3 and NPR4, sequestrating these components, thereby allowing NPR1 accumulation into the nucleus and thus conferring basal defense levels (Pajerowska-Mukhtar et al., 2013; Ding et al., 2018).
As one of the major sites of ROS production inside the cell, mitochondria play an important role in the perception and transduction of several signals elicited by the cell upon pathogen recognition. Thus, mitochondria amplify the first alarm signal in order to trigger the defense at the cellular level, which may lead to activation of PCD (Lam et al., 2001; Colombatti et al., 2014; Huang et al., 2016; Belt et al., 2017; Van Aken, 2017). Due to their evolutionary origin, plant mitochondria have acquired antioxidant pathways represented by thioredoxins, glutaredoxins, peroxiredoxins, and superoxide dismutase activities (Navrot et al., 2007; Cvetkovska et al., 2013; Dietz et al., 2016; Foyer and Noctor, 2016; Huang et al., 2016; Liebthal et al., 2018).
We recently described the existence of the new OXR (Oxidation Resistance) protein family in Arabidopsis (Arabidopsis thaliana; Colombatti et al., 2019), with six members containing the highly conserved C-terminal TLDc (TBC [Tre2/Bub2/Cdc16], LysM [Lys motif], domain catalytic) domain (Volkert et al., 2000; Doerks et al., 2002; Blaise et al., 2012). One member, AtOXR2, is a mitochondrial protein induced by oxidative stress. Increased expression of AtOXR2 confers plant tolerance to stress conditions generated by plant exposure to methyl viologen, antimycin A, and high light intensity. We postulated that AtOXR2 improves the photosynthetic machinery and elicits basal plant tolerance against situations that produce oxidative imbalances generated by abiotic stress, providing beneficial characteristics for plant growth (Colombatti et al., 2019).
OXR family members have been studied in several eukaryotic organisms from yeast to humans (Homo sapiens; Elliott and Volkert, 2004; Jaramillo-Gutierrez et al., 2010; Oliver et al., 2011; Wang et al., 2012; Kobayashi et al., 2014; Sanada et al., 2014; Yang et al., 2014; Liu et al., 2015; Finelli et al., 2016; Wu et al., 2016; Su et al., 2017; Pomatto et al., 2019; Zhang et al., 2019). Although the mechanism by which OXR proteins exert their action is still unknown, several reports have associated TLDc-containing proteins to the prevention of oxidative damage. Even though they do not possess ROS scavenging activity per se (Oliver et al., 2011; Finelli and Oliver, 2017), TLDc-containing proteins are able to regulate the expression (Yang et al., 2014, 2015; Pomatto et al., 2019) and the activity of some proteins of the antioxidant machinery in mammals, by controlling their oligomerization state (Svistunova et al., 2019). Regarding immune responses, OXR proteins from animals have been implicated in conferring protection against bacterial infection (Finelli and Oliver, 2017). The AtOXR2 homolog in Anopheles gambiae regulates the expression of ROS-detoxifying enzymes and is implicated in eliciting the innate immune responses during Plasmodium spp. infection (Brandt et al., 2008; Jaramillo-Gutierrez et al., 2010). In addition, a gain-of-function mutant in the AtOXR2-homolog gene mustard increases tolerance of Drosophila spp. to Vibrio cholerae infection (Wang et al., 2013).
Due to the relevance of some TLDc-containing proteins from animals in the prevention of infection and the establishment of immune resistance (Yu et al., 2015; Finelli and Oliver, 2017), we decided to analyze the role of AtOXR2 in biotic stress situations in Arabidopsis plants exposed to the hemibiotrophic bacterial pathogen Pseudomonas syringae. Our results show that the SA-mediated defense pathway is constitutively modified in plants with altered AtOXR2, positively impacting Arabidopsis resistance to P. syringae infection.
RESULTS
AtOXR2 Overexpression Mimics the Transcriptional Profile of Plants Responding to Stress
We recently performed a transcriptomic analysis of Arabidopsis plants constitutively overexpressing AtOXR2 (oeOXR2 plants) grown under control conditions. The categories of biological processes statistically overrepresented were those related to response to stimulus (GO:0050896), stress responses (GO:0006950), and PCD (GO:0008219; Colombatti et al., 2019). By using the Signature tool in Genevestigator (Hruz et al., 2008), we compared a list containing the ∼200 most upregulated and downregulated genes from the total of differentially expressed transcripts in oeOXR2 plants (Supplemental Table S1, A and B) against the complete data set of Affymetrix Arabidopsis ATH1 genome arrays (Hruz et al., 2008). Our intention was to find those experimental conditions or particular plant genotypes with a transcriptional profile comparable to one of the oeOXR2 plants. As a result, conditions connected to biotic stress and plant-pathogen interaction from different experiments were overrepresented and highlighted (Supplemental Fig. S1). Thus, at the molecular level, oeOXR2 plants share gene expression signatures with plants infected with the obligate biotrophic pathogen Hyaloperonospora arabidopsidis (Wang et al., 2011) and with the wild type and the enhanced disease resistance1 mutant inoculated with the biotrophic powdery mildew Golovinomyces cichoracearum (Christiansen et al., 2011). The oeOXR2 expression profile is also comparable to those of della mutants challenged with P. syringae pv tomato DC3000 (Pst DC3000; GSE17464), npr1 mutants in comparison with the wild type under control conditions (GSE5745), the npr1/suppressor of npr1-1 inducible1/breast cancer2 triple mutant (GSE23617; Wang et al., 2010), and several recombinant inbred lines (Bayreuth-0 × Shahdara) generated to find natural genetic variation for plant disease resistance responses, after treatment with SA (E-TABM-518; for experimental details, see Supplemental Table S2A). These results allowed us to hypothesize that constitutively increased expression of AtOXR2 generates a transcriptional profile similar to that of the plants that have been challenged with different pathogens or with alterations in hormonal pathways connected with defense against biotic stress.
AtOXR2 Is Induced by SA and Biotic Stress
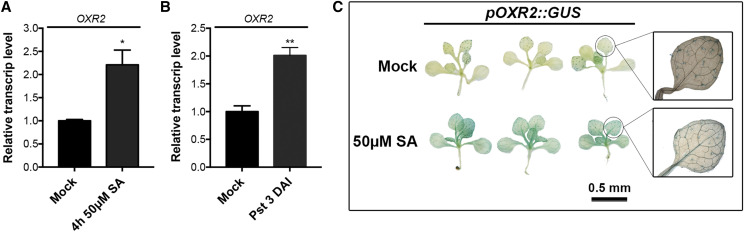
It is well known that TLDc-containing proteins from animals are induced by situations that promote oxidative stress (Oliver et al., 2011; Yang et al., 2014; Yu et al., 2015; Finelli et al., 2016). We recently demonstrated that transcript levels of AtOXR2 are regulated by conditions that generate abiotic stress and oxidative damage, such as exposure to high light, antimycin A, and methyl viologen (Colombatti et al., 2019). We then evaluated the impact of biotic stress on AtOXR2 expression by challenging fully expanded rosette leaves of wild-type plants with the hemibiotrophic pathogen Pst DC3000. We observed an increase in AtOXR2 transcript levels 3 d after infection (DAI; Fig. 1A). We also exposed 15-d-old wild-type plants to a solution containing 50 μm SA and observed an increased expression of AtOXR2 after 4 h of treatment (Fig. 1B). We applied the same conditions to Arabidopsis seedlings expressing the GUS reporter gene under the control of the AtOXR2 promoter region (pOXR2::GUS; Colombatti et al., 2019), which resulted in an increased GUS expression in comparison with mock-treated seedlings (Fig. 1C). This result agrees with publicly available microarray data (Hruz et al., 2008), showing that AtOXR2 is up-regulated during the infection with P. syringae. In contrast, we observed that the expression of AtOXR2 is down-regulated after inoculation with the hemibiotroph oomycete Phytophthora parasitica and the necrotrophic fungus Sclerotinia sclerotiorum (Supplemental Fig. S2; Supplemental Table S2B).
Figure 1.
AtOXR2 is induced both by SA treatment and by Pst DC3000 infection. A and B, AtOXR2 transcript levels were measured in Arabidopsis 10-d-old wild-type seedlings grown in one-half strength Murashige and Skoog (MS) medium and then flooded in mock solution or in a solution containing 50 μm SA plus 0.025% (v/v) Silwet L-77 for 5 min. Samples were taken after 4 h of treatment. Alternatively, 10-d after sowing wild-type seedlings were flood inoculated for 5 min with Pst DC3000 at a concentration of 5 × 108 colony-forming units (CFU) mL−1 plus 0.025% (v/v) Silwet L-77. Mock-inoculated plants were flooded with sterile distilled water containing 0.025% (v/v) Silwet L-77. Samples were taken at 3 DAI. Transcript levels were measured by reverse transcription quantitative PCR (RT-qPCR) and referred to those of wild-type plants in mock treatment. Results are expressed as means ± sd of three independent experiments. Asterisks represent significantly different values by ANOVA, Tukey’s test (*P < 0.05 and **P < 0.01). C, Arabidopsis plants carrying a construct expressing the GUS reporter gene driven by the AtOXR2 promoter (Colombatti et al., 2019) were exposed to one-half-strength Murashige and Skoog (Mock) or to the same solution with 50 μm SA for 1 h. Plants were incubated in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid (X-gluc) staining solution for 3 h and then discolored in a 70% (v/v) ethanol solution to reveal GUS staining.
In parallel, we used the Coexpression Analysis tool available in the Plant Promoter Analysis database (http://plantpan2.itps.ncku.edu.tw/; Chow et al., 2019) to reconstruct a possible regulatory network by identifying TFs coexpressed with AtOXR2 under environmental stresses (threshold > 0.8, P < 0.01; Supplemental Table S3). Among the coexpressed genes, we found several WRKY proteins involved in SA-mediated pathogen defense responses (Eulgem et al., 2000; Pandey and Somssich, 2009; Chen et al., 2019). Therefore, all data obtained about AtOXR2 expression suggest that it can be involved in the defense responses of Arabidopsis against (hemi)biotrophic pathogens.
AtOXR2 Levels Impact Plant Resistance to Pathogen Infection
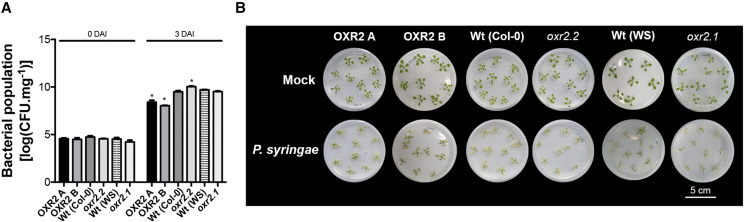
In light of the changes observed in the gene expression, we evaluated the response against Pst DC3000 in plants with altered expression of AtOXR2 previously described by Colombatti et al. (2019). Resistance was tested in 2-week-old overexpressor (oeOXR2) and mutant (oxr2.1 and oxr2.2) Arabidopsis seedlings grown on Murashige and Skoog plates according to Ishiga et al. (2011). Plant phenotypes and bacterial counts were monitored over the course of infection from 1 to 3 DAI. oeOXR2 and oxr2.2 lines showed significantly less and more CFU than wild-type plants at 3 DAI, respectively (Fig. 2A). The growth of oeOXR2 plants, which showed a clear reduction in the signs of infection, was also less affected by the pathogen, while oxr2.2 plants showed higher symptoms of infection (Fig. 2B). The infection assay was also performed using rosette leaves of 4-week-old plants of the three genotypes by infiltrating the bacteria with a syringe, for which comparable results were obtained in oeOXR2 plants at 1 and 3 DAI (Supplemental Fig. S3). These results indicate that AtOXR2 confers increased resistance to P. syringae either when the pathogen is applied on the surface (Fig. 2) or inoculated into the apoplast (Supplemental Fig. S3) of the plants.
Figure 2.
AtOXR2 modifies plant resistance to Pst DC3000. A, A bacterial population of Pst DC3000 was quantified at the beginning of the treatment (0 DAI) and 3 DAI. B, Disease phenotypes of Arabidopsis seedlings flood inoculated with Pst DC 3000 at a concentration of 5 × 108 CFU mL−1. Mock-inoculated plants were flooded with sterile distilled water containing 0.025% (v/v) Silwet L-77. Photographs were taken 3 DAI. Values represent means ± sd from five biological replicates. Asterisks indicate significant differences by ANOVA, lsd Fisher’s test (*P < 0.05) of the different genotypes in comparison with their respective wild type (Columbia-0 [Col-0] or Wassilewskija [WS]).
Flagellin-Triggered aROS Is Modified in Plants with Altered AtOXR2 Expression
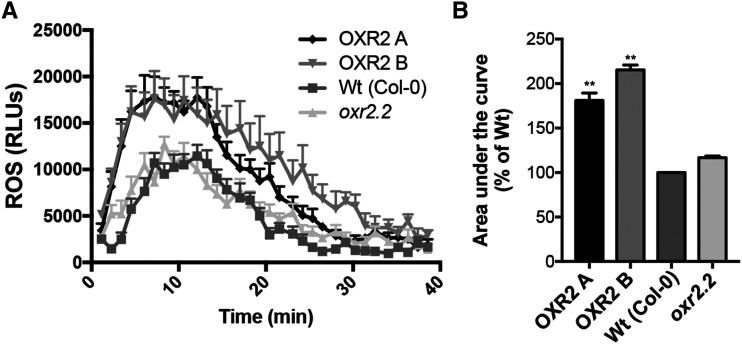
Upon pathogen recognition, an oxidative burst characterized by a rapid and transient aROS production takes place at the site of infection. The sources for this apoplastic oxidative burst are NADPH oxidases of the plasma membrane, also called respiratory burst oxidase homologs (Foreman et al., 2003; Torres and Dangl, 2005). We previously observed that ROS content and lipid peroxidation are increased in oeOXR2 plants under basal growth conditions (Colombatti et al., 2019). We evaluated the impact of modifying AtOXR2 expression on the plant response to pathogen-associated molecular patterns. This was assayed by challenging plants with the bacterially derived peptide elicitor flg22 according to Fabro et al. (2016). We observed that oeOXR2 activated aROS slightly faster than wild-type plants, while oxr2.2 mutants seem to mimic the wild-type behavior (Fig. 3A). In addition, overall aROS levels were higher in oeOXR2 than in control plants (Fig. 3B).
Figure 3.
flg22-induced aROS is increased by AtOXR2 overexpression. A, Time-course kinetics of flg22-triggered aROS burst in wild-type (Wt) Col-0 plants, AtOXR2-overexpressing lines A and B, and the mutant oxr2.2. The aROS burst profiles of 24 leaf discs per genotype were averaged. Results are expressed as means ± sd. B, Quantitation of the area under the curve of flg22-induced ROS performed in the previously mentioned genotypes. Percentages were calculated relative to the wild-type levels. Values represent averages of independent experiments expressed as means ± sd. Asterisks indicate significant differences with regard to the wild type by lsd Fisher’s test (*P < 0.01). RLU, Relative light units.
The SA-Mediated Defense Pathway Is Basally Modified in Plants with Altered AtOXR2 Expression
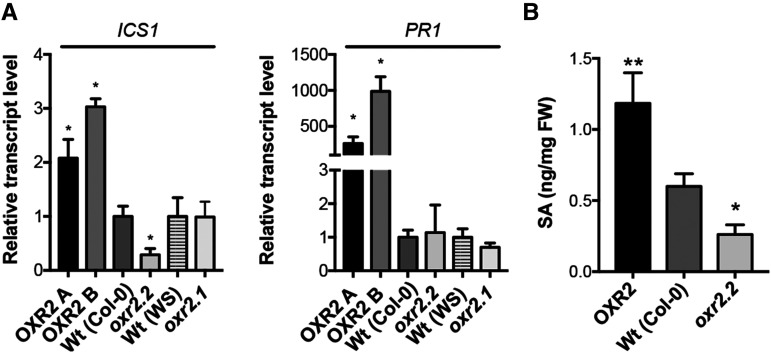
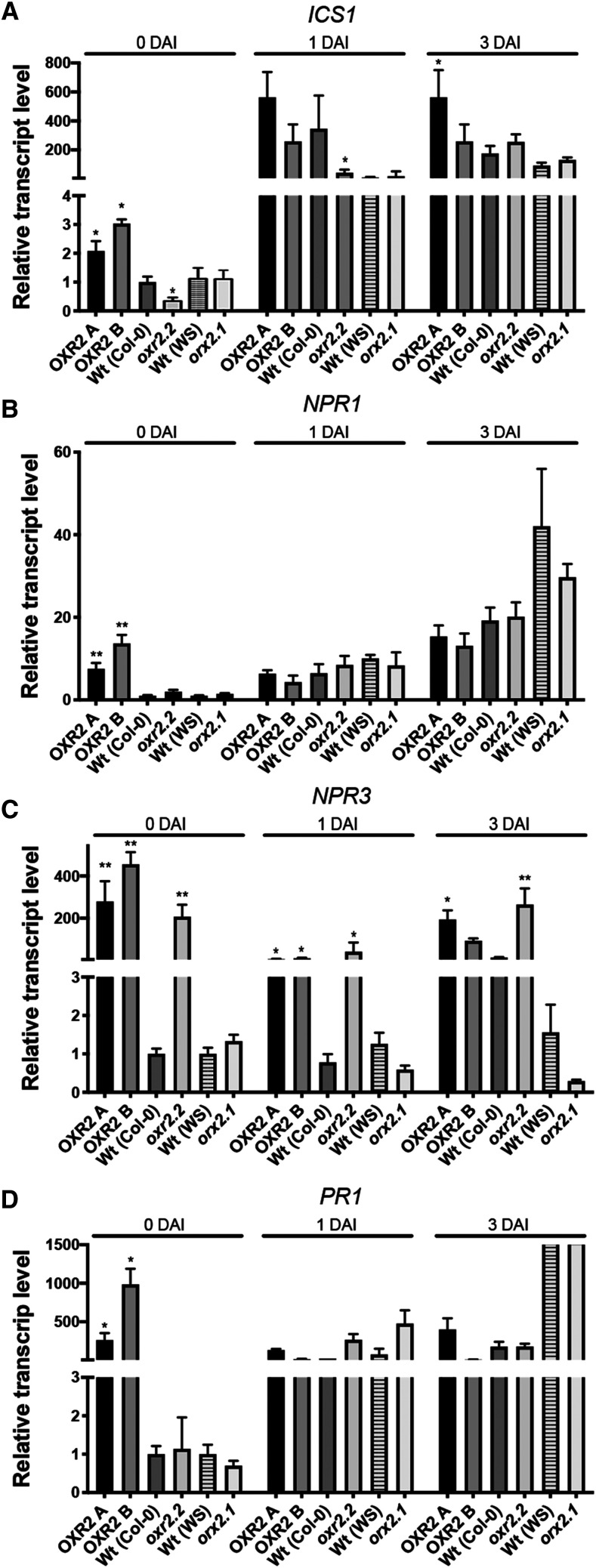
To further investigate the role of AtOXR2 in the defense against biotrophic pathogens, we measured the expression of genes involved in this response in plants with altered expression of AtOXR2 under control conditions. We determined the expression levels of marker genes for SA synthesis and response, like ISOCHORISMATE SYNTHASE1/SALICYLIC ACID INDUCTION DEFICIENT2 (ICS1/SID2) and PATHOGENESISRELATED GENE1 (PR1; Zhang and Li, 2019). The expression levels of ICS1/SID2 and PR1 were increased in oeOXR2 plants grown under normal growth conditions (Fig. 4A). On the other hand, ICS1/SID2 was down-regulated in oxr2.2 plants (Fig. 4A). The increased basal expression of transcripts encoding proteins involved in SA synthesis and pathogen responses in oeOXR2 plants suggested that either SA signaling or SA levels could be affected in oeOXR2 plants. To analyze possible alterations in SA levels, we determined the free SA content by gas chromatography-mass spectrometry (GC-MS) in 3‐week‐old rosette leaves of wild-type, oeOXR2, and oxr2 mutant plants. According to the results, oeOXR2 plants possess higher levels of SA while the oxr2.2 mutant exhibits a reduced SA content (Fig. 4B), suggesting that AtOXR2 impacts the SA content of plants under normal growth conditions.
Figure 4.
AtOXR2 modifies basal SA levels and the expression of genes related to SA synthesis and response. A, Transcript levels for ICS1 and PR1, involved in SA synthesis and response. Expression levels were measured by RT-qPCR in 2-week-old plants grown under normal conditions. Transcript levels are referred to those of wild-type (Wt) plants of the same ecotype (OXR2-A and OXR2-B were referred to wild-type Col-0, and oxr2.1 was referred to wild-type WS). Results are expressed as means ± se. Asterisks indicate significant differences of three biological replicates by ANOVA, lsd Fisher’s test (*P < 0.05). B, Free SA content in plants with altered AtOXR2 expression. SA was measured in rosette leaves from 2-week-old plants by GC-MS. Results are expressed as means ± sd. Asterisks indicate significant differences of five biological replicates of each genotype by ANOVA, lsd Fisher’s test (*P < 0.05 and **P < 0.01). FW, Fresh weight.
oeOXR2 Plants Induce Early Defense Marker Genes
We analyzed the expression of SA pathway signaling and response genes during the course of Pst DC3000 infection in plants with altered levels of AtOXR2 (Fig. 5). We quantified transcripts for ICS1, NPR1, NPR3, and PR1 in the rosette leaves of bacteria-infected plants before (0 DAI) and after 1 or 3 DAI (Fig. 5). ICS1 expression was considerably induced in oeOXR2 plants at 0 DAI, whereas lower expression was observed in the oxr2.2 mutant (Fig. 5A). In the case of the oxr2.1 mutant, perhaps due to the WS ecotype, whose kinetics of gene response may be different, expression levels did not increase significantly throughout the infection (Fig. 5A). Curiously, NPR1 transcript levels remained similar to those of the wild type, except for oeOXR2 lines that exhibited higher expression at basal conditions (Fig. 5B). It is well known that the strongest regulation of NPR1 in response to pathogens occurs at the posttranslational level (Kinkema et al., 2000). Conversely, expression levels of the gene encoding the NPR3 coreceptor and negative regulator were already increased before the infection in oeOXR2 and oxr2.2 mutant plants and remained high throughout the course of infection (Fig. 5C). However, the oxr2 mutant plants were able to raise PR1 expression levels to levels comparable to their respective wild-type plants (Fig. 5D). The overall dynamics of the marker genes’ expression was in agreement with the differential SA content (Fig. 4) and the phenotypes observed in plants with altered expression of AtOXR2 (Fig. 2). Thus, oeOXR2 plants appear to be better prepared for pathogen attack and are able to respond faster, increasing whole-plant resistance, while oxr2 mutants exhibit a delayed response, also at the molecular level (Figs. 3 and 5).
Figure 5.
Molecular analysis of plants with altered AtOXR2 expression during the course of infection with Pst DC3000. Transcript levels of ICS1 (A), NPR1 (B), NPR3 (C), and PR1 (D) were measured by RT-qPCR at different times during the infection. Expression levels are referred to those of wild-type plants of the same ecotype (Col-0 or WS) before the infection (0 DAI). Results are expressed as means ± se. Asterisks indicate significant differences compared with wild-type plants of the same ecotype at the same infection time by ANOVA, lsd Fisher’s test (*P < 0.05 and **P < 0.01).
Figure 6.
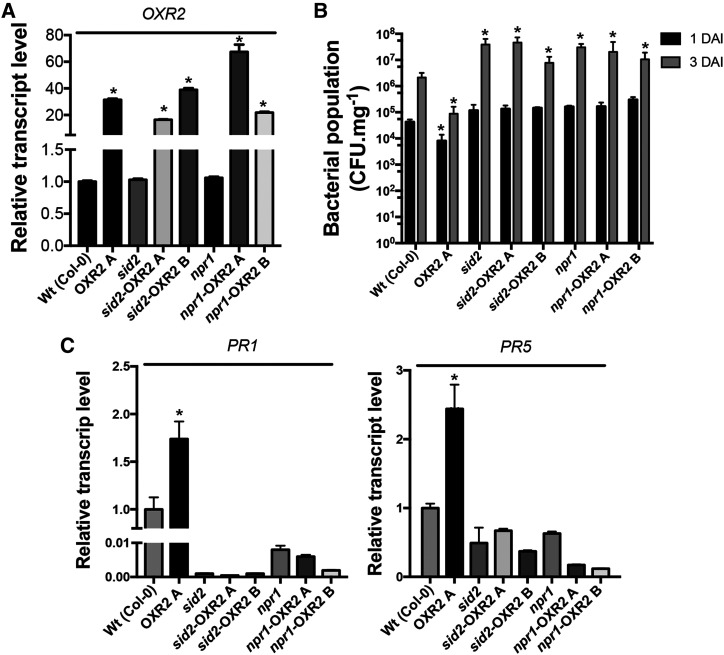
AtOXR2 acts upstream of SID2 and NPR1. A, AtOXR2 expression levels measured by RT-qPCR in rosette leaves of wild-type (Wt), sid2, and npr1 plants, and the respective transformants obtained in the mutant backgrounds (sid2-OXR2A, sid2-OXR2B, npr1-OXR2A, and npr1-OXR2B). Results are expressed as means ± sd of five independent plants. Asterisks indicate significant differences and are referred to the levels of wild-type plants by ANOVA, lsd Fisher’s test (*P < 0.05). B, The enhanced resistance to Pst DC3000 due to AtOXR2 overexpression is lost in the sid2 and npr1 mutant backgrounds. Two-week-old plants were infected with Pst DC3000, and bacterial populations were quantified at 1 and 3 DAI. Results are expressed as means ± sd of five independent biological replicates. Asterisks indicate significant differences and are referred to the levels of wild-type plants at the correspondent DAI by ANOVA, lsd Fisher’s test (*P < 0.05). C, Analysis of transcript levels of pathogen response genes PR1 and PR5. Expression levels were measured by RT-qPCR in the cDNA samples used in A, prepared from plants before the pathogen infection (basal levels, 0 DAI). Results are expressed as means ± sd of five independent plants. Asterisks indicate significant differences referred to the levels of wild-type plants by ANOVA, lsd Fisher’s test (*P < 0.05).
AtOXR2 Acts Upstream of SID2 and NPR1 in Order to Basally Increase SA-Mediated Defense in Arabidopsis
To establish a proper response against pathogens and activate the systemic acquired resistance, plants need (1) to elevate the levels of SA both locally and in the noninfected tissues and (2) to transduce this information for the activation of PR gene expression through the interaction of the NPR1 master regulator with TGA TFs in the nucleus (Lindermayr et al., 2010). In order to understand at which level of the SA-mediated defense pathway AtOXR2 is exerting its action, we overexpressed AtOXR2 in the background of the sid2 (Nawrath and Métraux, 1999) and npr1 (Cao et al., 1994) mutants. Plants with comparable AtOXR2 expression levels (Fig. 6A) were isolated and used to evaluate their behavior after infection with Pst DC3000 in comparison with sid2, npr1, and wild-type plants. The enhanced basal resistance due to AtOXR2 overexpression previously observed in wild-type plants (Fig. 2; Supplemental Fig. S3) was markedly lost in both sid2 and npr1 mutant backgrounds. Quantification of pathogen growth after the infection showed values comparable to those obtained in the sid2 and npr1 mutant plants (Fig. 6B). This remarkable loss of the basal defense phenotype of oeOXR2 plants was also confirmed at the molecular level. The increase of transcript levels for the PR proteins PR1 and PR5 observed in oeOXR2 plants was completely abolished in the background of the sid2 and npr1 mutants (Fig. 6C). These results suggest that AtOXR2 needs intact SA synthesis and SA-mediated defense pathways to increase basal immunity.
AtOXR2 Has an Impact on the Expression of WRKY TFs
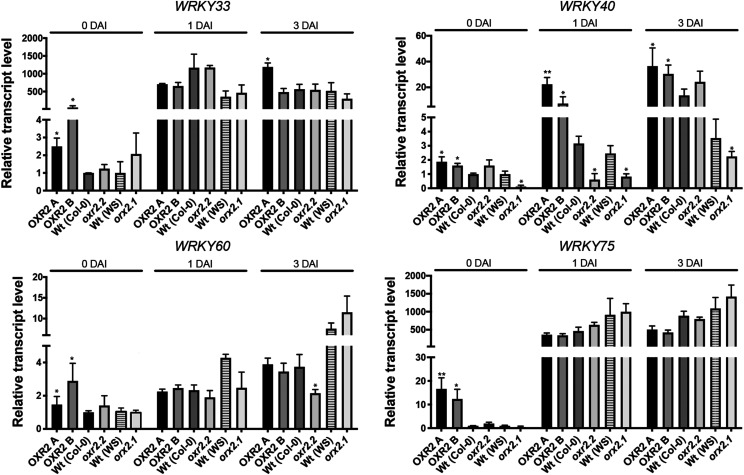
As we mentioned above, the AtOXR2 coexpression network involves several TFs related to plant defense against pathogens, like WRKY TFs (Supplemental Table S3). WRKYs participate in a regulatory network that integrates internal and environmental factors to modulate various aspects of defense responses (Vlot et al., 2009; van Verk et al., 2011; Chen et al., 2019). Recently, a tripartite amplification loop was established between SA, ROS, and WRKY75 during the progression of senescence (Guo et al., 2017). In agreement, we also observed a regulatory positive loop between SA, ROS, and AtOXR2. Like ROS generated by oxidative stress, the presence of SA and pathogen challenge induces AtOXR2 expression (Fig. 1; Colombatti et al., 2019). All these factors, including defense against pathogens, are increased in oeOXR2 plants and decreased or absent in oxr2 mutants (Figs. 2–5). We measured the transcript levels of different members of the WRKY family of TFs in plants with altered AtOXR2 expression under normal growth conditions and during the course of the infection with Pst DC3000 to investigate a possible connection between the elements of the positive loop for the establishment of the basal defense and WRKY expression. In basal conditions, we observed an increased expression of WRKY33, WRKY60, and WRKY75 in oeOXR2 plants but wild-type levels in the mutant, while WRKY40 expression was increased in oeOXR2 plants and decreased in oxr2 mutants (Fig. 7). In the case of oxr2 mutants, there was no significant alteration in WRKY transcript levels after inoculation compared with wild-type plants (Fig. 7). WRKY40 showed a sustained higher level of expression in oeOXR2 plants with regard to the wild type and mutants infected with Pst DC3000.
Figure 7.
AtOXR2 modifies the expression of several WRKY TFs related to the stress response. Transcript levels were measured by RT-qPCR at different times during the infection in 4-week-old plants from each genotype. Expression levels are referred to those of wild-type (Wt) plants of the same ecotype (Col-0 or WS) at basal conditions or before the infection (0 DAI). Asterisks indicate significant differences compared with wild-type plants of the same ecotype at the same infection time by ANOVA, lsd Fisher’s test (*P < 0.05 and **P < 0.01).
NPR1 Shows Nuclear Localization in Untreated oeOXR2 Plants
NPR1 is an essential regulator of SA-mediated pathogen defense pathways. It constitutes an SA receptor, specifically binding the hormone via its Cu-activated Cys521/529 (Wu et al., 2012). NPR1 is regulated posttranslationally, existing as cytosolic oligomers formed through intermolecular disulfide bonds, which are reduced to a monomeric NPR1 isoform due to SA-induced cytoplasmic redox changes upon pathogen infection (Kinkema et al., 2000; Mou et al., 2003). Once in its monomeric form, NPR1 translocates to the nucleus and activates the expression of SA-induced PR and disease resistance genes through the interaction with TGA TFs (Lindermayr et al., 2010; Withers and Dong, 2016; Sun et al., 2018).
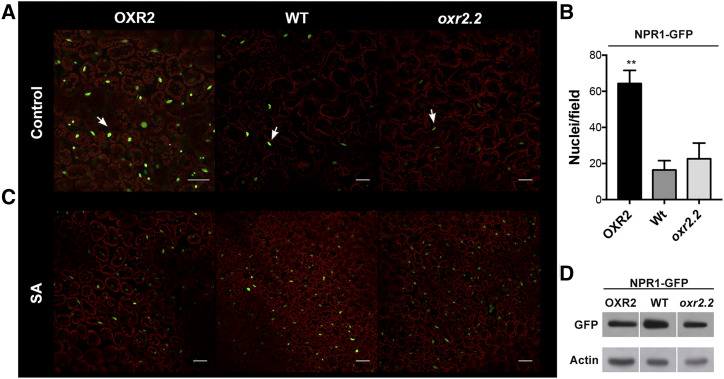
In order to study if AtOXR2 modifies the basal activation state of NPR1, we obtained plants with altered expression of AtOXR2 (oeOXR2 and oxr2.2) in a line constitutively expressing NPR1 fused to GFP (35S::NPR1-GFP; Kinkema et al., 2000). We grew plants in Murashige and Skoog medium and in Murashige and Skoog medium supplemented with SA and evaluated the fluorescence of GFP by confocal laser scanning microscopy (CLSM; Kinkema et al., 2000; Mou et al., 2003; Tada et al., 2008) in different AtOXR2 backgrounds. Wild-type and oxr2.2 seedlings grown under control conditions showed a few nuclei with NPR1-GFP fluorescence. In contrast, an increased number of nuclei of oeOXR2 plants displayed NPR1-GFP fluorescence (Fig. 8A). The quantitation of fluorescent nuclei at the control stage is shown in Figure 8B. To evaluate whether the increased nuclear NPR1-GFP fluorescence observed in the oeOXR2 background under basal conditions may be due to the higher SA levels, we exposed all plants to 24 h of SA treatment. While nuclear NPR1-GFP levels were comparable to those of control plants in SA-treated oeOXR2 plants, the number of fluorescent nuclei and the intensity of the signal became much more intense in wild-type and oxr2.2 plants after SA treatment (Fig. 8C). We also performed a western-blot analysis on these plants to ensure comparable basal levels of NPR1-GFP protein expression in the different AtOXR2 backgrounds evaluated (Fig. 8D). This allowed us to conclude that the increased nuclear fluorescence observed in oeOXR2 plants under control conditions may be related to the higher basal SA levels observed in these plants, which causes a redistribution of NPR1-GFP to the nucleus. We postulate that altered AtOXR2 expression modifies the intracellular environment, inducing a constitutive localization of NPR1 into the nucleus, thus preparing the plants against a potential pathogen attack.
Figure 8.
oeOXR2 plants show NPR1 nuclear accumulation when grown under control conditions. In A and C, oeOXR2, wild-type (WT), and oxr2.2 leaves expressing NPR1-GFP were imaged by CLSM. NPR1-GFP signal in green appears in the nucleus (marked with arrows), and chlorophyll fluorescence of chloroplasts appears in red. A, oeOXR2 plants showed stronger GFP signal than wild-type or oxr2.2 plants under basal conditions. Bars = 50 µm. B, Quantification of the number of nuclei with NPR1:GFP signal per field in different genotypes grown under control conditions in A. Values represent means ± sd (n = 5–10), and asterisks indicate significant differences by ANOVA, lsd Fisher’s test (**P < 0.05). C, After treatment with 0.5 mm SA, NPR1-GFP signals in all genotypes appeared equal. Bars = 50 µm. D, Western blot analysis of NPR1-GFP protein expression in plants with altered levels of AtOXR2. Detection of Actin was used as a loading control.
AtOXR2 Activates the SA Response by Changing the Cellular Redox Environment
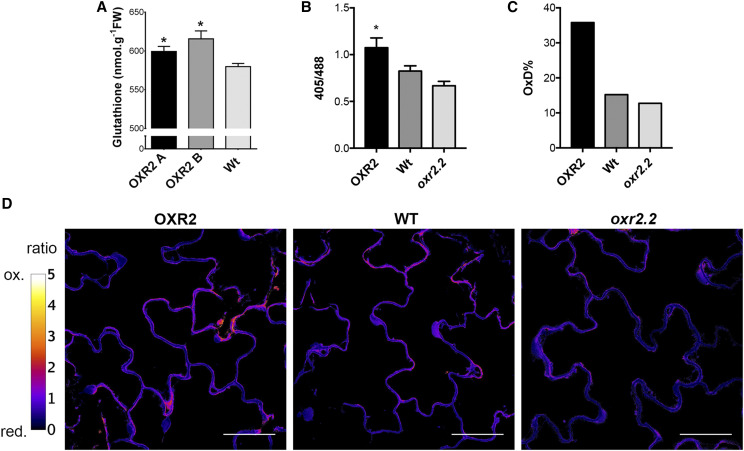
Marked changes in the cellular redox state happen when SA is increased in plants after pathogen recognition. NPR1 activation is accomplished due to cellular redox changes, including (1) an early burst of ROS and (2) a decrease in the cellular reduction potential due to an accumulation of antioxidants such as GSH (Mou et al., 2003). The essential role of GSH in the activation of ROS-dependent SA pathogenesis responses, acting as a signal-transmitter molecule independently of its function as a natural antioxidant, was demonstrated and discussed (Herrera-Vásquez et al., 2015). We previously showed that oeOXR2 plants grown under control conditions possess higher ROS content, measured by using the cell-permeant dye 2′,7′-dichlorodihydrofluorescein diacetate and the increased levels of lipid peroxidation (Colombatti et al., 2019). To explore if the mechanism proposed by Han et al. (2013) could explain the basal nuclear localization state of NPR1 in oeOXR2 plants, and to analyze the impact of possible changes in cellular GSH redox potential on signaling processes during defense, we characterized the cellular redox status of plants with altered levels of AtOXR2. We introduced the Grx1-roGFP2 reporter sensor (Meyer et al., 2007; Albrecht et al., 2014) in oeOXR2 lines and oxr2.2 mutant plants and measured the in vivo redox status by CLSM (Fig. 9). Using the ratio between the fluorescent signal from both channels at 405 and 488 nm (Fig. 9A) and applying the formula described by Schwarzländer et al. (2008), we calculated the degree of oxidation of the GSH pool (Fig. 9B) in the different genetic backgrounds. By this, we determined that the overexpression of AtOXR2 resulted in an increase in the oxidation of the GSH pool (Fig. 9). This shift in the oxidation of the intracellular redox buffer in oeOXR2 plants is in agreement with the increase in the free SA levels (Fig. 4) and the restricted propagation of Pst DC3000 (Fig. 2).
Figure 9.
OXR2 plants show increased oxidation of cytosolic Grx1-roGFP2. A, Total GSH levels measured by a spectrophotometric assay on 4-week-old rosette leaves of plants with altered AtOXR2 expression. Values are expressed nanomole per gram fresh weight (FW) and represent means ± sd from five biological replicates. Asterisks indicate significant differences by ANOVA, lsd Fisher’s test (*P < 0.05). Wt, Wild type. B, 405/488 quantitation in 10-d after sowing plants with altered AtOXR2 expression grown in control conditions. Values represent means ± sd from 10 individual plants. The asterisk indicates a significant difference by ANOVA, lsd Fisher’s test (*P < 0.05). C, Degree of roGFP2 oxidation. D, Confocal images showing the 405/488 ratio. The color scale represents the redox state of the fluorescent sensor from the oxidized (ox.; in white) to the reduced (red.; in black) state. Bars = 50 µm.
DISCUSSION
In this work, we characterized the role of this TLDc-containing protein belonging to the Arabidopsis OXR family in establishing basal plant protection against biotic stress, specifically during infection with the hemibiotrophic pathogen P. syringae.
AtOXR2 is a mitochondrial protein (Colombatti et al., 2019). Several examples exist connecting mitochondrial proteins with plant-pathogen interactions and biotic stress, either related to the mitochondrial electron transport chain (Gleason et al., 2011; Cvetkovska and Vanlerberghe, 2012; Zhu et al., 2014; Belt et al., 2017), mitochondrial membrane proteins such as VOLTAGE-DEPENDENT ANION CHANNEL, PRESEQUENCE TRANSLOCASE-ASSOCIATED PROTEIN IMPORT MOTOR SUBUNIT 16 of the inner mitochondrial membrane, OUTER MITOCHONDRIAL MEMBRANE PROTEIN OF 66 kD, or PENETRATION2 (Tateda et al., 2009, 2011; Huang et al., 2013; Zhang et al., 2014; Fuchs et al., 2016), or mitochondrial proteins involved in amino acid metabolism (Cecchini et al., 2011; Senthil-Kumar and Mysore, 2012; Fabro et al., 2016). These proteins are connected in different stages with alterations in the plant susceptibility to pathogens. In advanced stages of the infection, mitochondria can also promote PCD in order to avoid pathogen propagation to the whole plant (Colombatti et al., 2014; Van Aken, 2017). In general, all the mentioned proteins act by regulating the ROS content at the cellular level. Although the mechanism has not been fully elucidated, mitochondrial ROS, aROS, and the cellular redox balance seem to be essential in the plant response against pathogens (Nie et al., 2015; Fuchs et al., 2016).
ROS are signal molecules contributing to the plant systemic immunity. A rapid ROS increase followed by cellular redox changes plays important roles during the defense pathways established upon plant-pathogen recognition (Alvarez et al., 1998; Vanacker et al., 2000). In general, lower ROS production is associated with higher susceptibility to several pathogens (Cecchini et al., 2011; Gleason et al., 2011; Tateda et al., 2011). Conversely, increased levels of PR proteins and higher SA-triggered ROS content have been linked to enhanced resistance to virulent pathogen attack (Huang et al., 2013; Zhang et al., 2014; Fuchs et al., 2016) and increased susceptibility to the necrotrophic pathogen Botrytis cinerea (Zhu et al., 2014). The scenario observed when overexpressing AtOXR2 in Arabidopsis agrees with those previously published studies. AtOXR2 is induced by SA and biotic stress (Fig. 1). oeOXR2 plants exhibit higher ROS content (Colombatti et al., 2019) with increased resistance to infection by P. syringae (Fig. 2), faster and stronger response to the elicitor flg22 (Fig. 3), basal induction of genes involved in SA perception, synthesis, and responses (Fig. 4), and higher levels of free SA (Fig. 4). The oxr2.2 mutant plants showed the opposite behavior, with lower expression levels of ICS1, less SA content, and increased susceptibility to P. syringae. We did not observe differential behavior against the treatment with flg22 between wild-type and oxr2.2 mutant plants. This is not surprising, as there are several examples of mutants with low SA levels that do not affect early flg22 responses (Yi et al., 2014), particularly the flg22-induced ROS burst (Hung et al., 2014). The higher levels of SA in oeOXR2 plants may also be connected to the elevated tolerance of oeOXR2 plants to abiotic stress recently reported by us (Colombatti et al., 2019). Apart from its role in biotic stress, SA works as a signal during abiotic stress situations generated by high light, drought (Khan et al., 2015), and UV-C light (Seguel et al., 2018). SA accumulation enhances stomatal closing and drought tolerance (Miura et al., 2013; Zhang et al., 2017b), and we previously demonstrated an anticipated stomatal closure and more efficient use of water in oeOXR2 plants (Colombatti et al., 2019).
SA is a key plant defense hormone for biotrophic interactions (Zhang and Li, 2019). Although the PHENYLALANINE AMMONIA-LYASE (PAL) pathway indeed can contribute to SA biosynthesis (Vlot et al., 2009; Huang et al., 2010), we previously demonstrated that PAL transcript levels were down-regulated in oeOXR2 plants (Colombatti et al., 2019). Therefore, the altered levels of SA in oeOXR2 plants and oxr2.2 mutants appear to be entirely due to a misregulation in the ICS1-mediated biosynthesis pathway (Fig. 4). Loss of ICS1 abolishes pathogen-induced SA accumulation (Wildermuth et al., 2001; Genger et al., 2008), and the phenotype of pathogen resistance observed in oeOXR2 plants (Fig. 2) was completely abolished in the sid2 mutant background, deficient in ICS1 (Fig. 6). This allowed us to conclude that the higher basal pathogen defense observed in oeOXR2 plants is dependent on a functional ICS1-mediated SA synthesis pathway.
oeOXR2 plants exhibit higher basal expression of some WRKY TFs (Fig. 7). Different WRKY TFs have been implicated in plant responses to biotic stress (Xu et al., 2006; Pandey and Somssich, 2009; Hu et al., 2012). oeOXR2 plants basally produce higher ROS levels, and the expression of WRKY genes is increased in response to ROS or imbalances in redox homeostasis. Several WRKY TFs, including WRKY40, WRKY60, and WRKY75, which are basally induced in oeOXR2 plants (Fig. 7), bind to the promoter of ICS1/SID2, stimulating its expression and SA synthesis and the activation of SA-sensitive responses (Zhang et al., 2017a). Furthermore, WRKY40 and WRKY63 regulate other mitochondrial proteins involved in plant defense against pathogens (Van Aken et al., 2013; Zhang et al., 2014). Thus, changes in the expression of WRKY genes due to modified ROS levels may be the cause of the basal increase in SA synthesis and the activation of the SA response pathway in oeOXR2 plants. WRKYs are also downstream targets of SA, but it has not been demonstrated that WKRYs must be down-regulated to render the plants susceptible to pathogen infections.
The downstream scenario involving SA perception and responses is also highly regulated. Several reports that analyzed the pathogenesis-related responses in GSH-deficient plants remarked on the importance of GSH content and the intracellular GSH redox state in Arabidopsis disease resistance (Parisy et al., 2007; Dubreuil-Maurizi et al., 2011; Han et al., 2013). GSH is a multifunctional metabolite with parallel functions as an antioxidant, participating in ROS detoxification (Noctor et al., 2018), and as an essential molecule required for connecting the increment in intracellular ROS production with the activation of ICS1-dependent SA signaling pathways (Han et al., 2013). We observed slightly higher levels of GSH disulfide GSSG in leaf extracts of 4-week-old oeOXR2 unstressed plants (Fig. 9) and a moderate in vivo shift to the oxidative GSH redox state (GSSG) in 15-d-old oeOXR2 unstressed plants. This feature is in agreement with the increase in their free SA levels (Fig. 4) and the restricted propagation of Pst DC3000 on these plants (Fig. 2), in agreement with previous reports (Chaouch et al., 2012; Matern et al., 2015).
NPR1 is posttranslationally affected by changes in the cellular redox state generated by the increment of ROS, GSH, and SA levels once the pathogen is detected (Tada et al., 2008). NPR1 is the best-characterized thiol-dependent protein involved in pathogenesis responses that can be either directly or indirectly triggered by the GSH/GSSG ratio (Mou et al., 2003). The regulation of NPR1 is balanced between the action of S-nitrosoglutathione, which promotes its inactivation by oligomerization, and the opposite activities of thioredoxins, which catalyze the SA-induced NPR1 oligomer-to-monomer conversion (Tada et al., 2008). We propose that AtOXR2 generates a state of increased constitutive defense by altering the total GSH content (Fig. 9A) and slightly shifting the cytosolic redox cellular state toward a more oxidizing environment under normal growth conditions (Fig. 9, B–D). The moderate trend we observed toward a more oxidized GSH in the cytosol, concomitant with more reduced NPR1 in the nucleus, is counterintuitive and deserves more investigation. It remains possible that the increase in total GSH content overcomes a certain threshold of reduced GSH, irrespective of the GSH/GSSG ratio, that induces the activation of thioredoxins, thus promoting the translocation of monomeric NPR1 to the nucleus (Figs. 8 and 9). In this sense, human Oxr1 is able to regulate the oligomerization degree of several proteins by different strategies, either through the regulation of the redox environment or through direct interaction with the protein candidate. Thus, Oxr1 regulates posttranslational modifications of Peroxiredoxin2, modulating its oligomerization degree and its antioxidant functions as an H2O2 scavenger (Svistunova et al., 2019). Oxr1 also reduces the aggregation degree of proteins such as Misfolded TAR DNA Binding Protein43 and Fused-In-Sarcoma via protein-protein interactions (Finelli et al., 2015) and regulates the activity and degree of the Glc-6-P isomerase through protein-protein interactions (Finelli et al., 2019). AtOXR2 could exert its function as was previously described for the TLDc-containing proteins from mammals. This can be achieved by increasing the ROS levels, modifying the GSH redox status, and/or increasing the GSH-dependent reducing power, all conditions that promote NPR1 activation (Fig. 10).
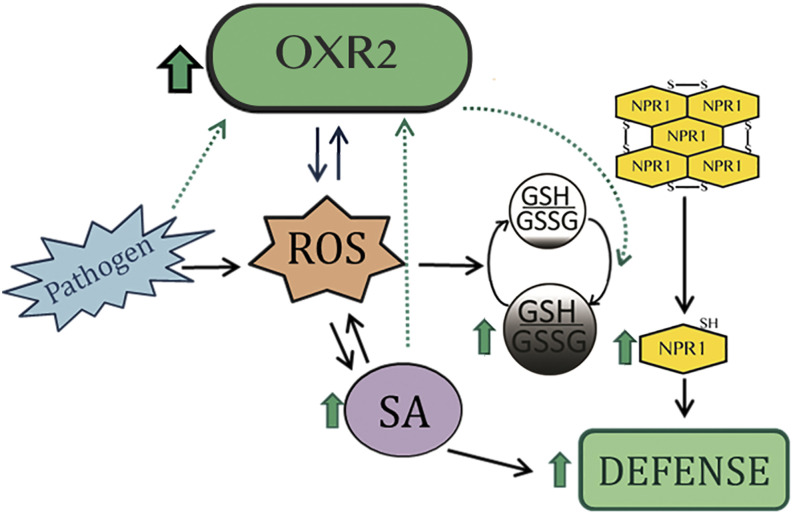
Figure 10.
Model of the role of AtOXR2 in plant defense. AtOXR2 is part of a loop for the activation of the SA-mediated plant defense pathway. By increasing ROS levels, AtOXR2 induces SA synthesis. In turn, SA, pathogens, and oxidative stress (ROS) stimulate the expression of AtOXR2. Increased AtOXR2 expression modifies the intracellular cytosolic redox environment, producing an increment in total GSH content and inducing the translocation of NPR1 to the nucleus, thus increasing defense and preparing plants against a potential pathogen attack. Black arrows correspond to connections known and established by other authors. Blue arrows represent results previously published by our group (Colombatti et al., 2019). Green-dotted arrows indicate original results presented in this work.
There are two parallel signaling pathways downstream of SA represented by NPR3/NPR4 and NPR1. We demonstrated that an intact NPR1-dependent pathway is essential for the plant-pathogen resistance observed in oeOXR2 plants (Fig. 7). NPR1 as well as NPR3/NPR4 are SA receptors, but they play opposite roles in transcriptional regulation of SA-induced defense. While NPR1 is an activator, NPR3/NPR4 functions independently of NPR1 as a transcriptional corepressor of the SA-induced immune responses (Ding et al., 2018). Based on these observations, it is not surprising that NPR3 is also basally up-regulated in oeOXR2 plants (Fig. 5), possibly in an attempt to decrease or slow down the constitutively active NPR1 pathway.
oeOXR2 plants are able to elicit a stronger and faster response to the stress induced by pathogens by modulating the cellular redox environment and interfering with the SA signaling pathway (Boubakri et al., 2016). The mechanism exerted by AtOXR2 to induce basal defense in plants could be comparable with the Oxr1 human homolog. González-Bosch (2018) proposed the same mechanism, induced by fluctuations in the redox balance, for the activation of the master sensors NPR1 and the mammalian Nuclear Factor Erythroid-Derived 2-Related Factor2 (Nrf2). Nrf2 switches between its cytosolic interaction with Kelch ECH Associating Protein1 (Keap1) for proteasomal degradation under nonstressed conditions and its nuclear active localization of a phosphorylated form of Nrf2, due to redox changes in the thiol group within Keap1 during the oxidative stress. Coincidently, human Oxr1 competes with Keap1 for Nrf2 stabilization, increasing its activity and the synthesis of antioxidant enzymes during oxidative stress in mammalian cells (Yang et al., 2014).
In our model (Fig. 10), AtOXR2 would be part of a positive feedback loop generated by increasing ROS levels and inducing the expression of WRKY TFs responsible for regulating the expression of enzymes involved in SA synthesis. Higher SA levels allow plants to be ready for defense while keeping high AtOXR2 levels. Through an increment in ROS and SA levels, and modifying the GSH oxidative pool, AtOXR2 would alter the cellular redox status and therefore NPR1 oligomerization, promoting its nuclear localization. Therefore, oeOXR2 plants are prepared for pathogen attacks without a previous challenge, which is probably the cause of their increased response to flg22 and resistance to bacterial infection. These characteristics of AtOXR2 make it valuable for biotechnological purposes, since its overexpression in plants induces defense mechanisms without penalties in plant fitness and seed and biomass production, as we recently demonstrated (Colombatti et al., 2019).
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants with altered expression of AtOXR2 (oeOXR2 [Col-0] and mutants oxr2.1 [Flag_513D06; WS] and oxr2.2 [SK17762; Col-0]) and plants expressing the promoter region of AtOXR2 followed by GUS were previously described by Colombatti et al. (2019). Arabidopsis sid2 (Nawrath and Métraux, 1999) and npr1 (Cao et al., 1994) mutants were obtained from the Arabidopsis Biological Resource Center. To use GFP as a reporter for NPR1 subcellular localization, we followed the strategy previously established by expressing the NPR1-GFP fusion protein under the control of the constitutive cauliflower mosaic virus 35S promoter (35S::NPR1-GFP; Kinkema et al., 2000; Mou et al., 2003; Tada et al., 2008). To obtain the NPR1-GFP construct, the NPR1 coding sequence was amplified using specific primers (Supplemental Table S4), cloned into the pDONR221 (Invitrogen) vector, and then transferred into the binary destination vector pFK-242 (from the Gateway-compatible pGREEN-IIS vector series) using the Gateway system (Invitrogen). Agrobacterium tumefaciens strain LBA4404 was transformed, and transgenic oeOXR2, wild-type, and oxr2.2 plants expressing the 35S::NPR1-GFP reporter construct were generated by the floral dip procedure (Clough and Bent, 1998). Seeds of Arabidopsis plants carrying the GSH redox couple (GSSG/2GSH) sensor Grx1-roGFP2 (Marty et al., 2009; Aller et al., 2013; Albrecht et al., 2014) were kindly provided by Dr. Markus Schwarzländer. We made reciprocal crosses to obtain several lines expressing the Grx1-roGFP2 in vivo thiol real-time redox reporter in the oeOXR2 and oxr2.2 backgrounds.
Plants were grown on soil at 22°C to 24°C under long-day conditions at a light intensity of 100 µmol m−2 s−1. For SA treatment, 10-d-old seedlings grown on one-half-strength Murashige and Skoog and 0.8% (w/v) agar were flooded with 0.5 mm SA plus 0.025% (v/v) Silwet L-77 for 5 min, and samples were taken at 1, 4, and 6 h after treatment in triplicate pools for GUS detection or RT-qPCR analysis.
Stress and Pathogen Treatments
On Soil
For SA treatment, whole rosettes of 2-week-old plants grown on soil were used. Rosettes were sprayed with a solution containing 0.5 mm SA plus 0.025% (v/v) Silwet L-77 or only 0.025% (v/v) Silwet L-77 (mock treatment). Samples were taken at 4 h after treatment in triplicate pools, immediately frozen in liquid nitrogen, and stored at −80°C for RT-qPCR analysis. For growth curves, fully expanded leaves of 6-week-old plants were syringe inoculated with a suspension of Pseudomonas syringae pv tomato DC3000 at 5 × 105 CFU mL−1 in 10 mm MgCl2. Leaf discs (three per sample with three technical replicates) were collected at 0, 24, and 72 h after inoculation, macerated in 10 mm MgCl2, and 1:10 serial dilutions were plated in Luria-Bertani agar plus rifampicin (100 μg mL−1) and kanamycin (50 μg mL−1). Colonies were counted at 24 h postplating. Statistical analysis was performed with ANOVA.
On Plates
Flood inoculation with Pst DC3000 on petri dishes was performed according to Ishiga et al. (2011), with small variations. Briefly, plants were grown on plates containing one-half-strength Murashige and Skoog and 0.8% (w/v) agar and were grown for 10 d in long-day conditions. A thin layer of water was deposited over 0.8% (w/v) agar to prevent bacterial contamination of the Murashige and Skoogmedium. Infection was carried out by incubating the seedlings for 5 min with a bacterial suspension (5 × 108 CFU mL−1, 0.9% [w/v] NaCl, and 0.025% [v/v] Silwet L-77) or mock solution (0.9% [w/v] NaCl and 0.025% [v/v] Silwet L-77). At 3 DAI, bacterial populations were measured in biological triplicates consisting of five to 10 seedlings per sample and expressed as CFU mg−1 plant tissue.
flg22-Triggered Apoplastic ROS Assay
The flg22 peptide (QRLSTGSRINSAKDDAAGLQIA) was used at 100 nm. Leaf discs (0.38 cm2) were treated with flg22, and ROS levels were determined using the luminol assay as described (Gómez-Gómez et al., 1999) in a microplate reader (Synergy HT; Biotek).
RNA Isolation and Analysis
Total RNA was extracted using Trizol RNA Isolation Reagent (Life Technologies). The RT-qPCR analysis was performed as described by O’Connell (2002) using a cDNA dilution with specific primers (Supplemental Table S4) in a StepOnePlus Real-Time PCR System (Czechowski et al., 2005). The Ct method was used for gene expression calculation with ACTIN2/8 as reference genes (Charrier et al., 2002).
GUS Staining
GUS histochemical assays were performed using the chromogenic substrate X-gluc as described by Hull and Devic (1995). For this, plants were incubated at 37°C with 1 mm X-gluc solution in 100 mm sodium phosphate, pH 7, and 0.1% (v/v) Triton X-100 until satisfactory staining was observed. For tissue clearing, plants were incubated with 70% (v/v) ethanol.
SA Quantification
SA was determined as described by Lovelock et al. (2016). Briefly, 2-week-old full rosettes were homogenized in a mortar, 500 mg was transferred to homogenization buffer (95% [v/v] isopropanol/5% [v/v] glacial acetic acid), and 100 ng of SAD4 (a deuterated compound obtained from Dr. Ehrenstorfer) was added as an internal standard. The homogenate was centrifuged, and the supernatant was dried under a stream of nitrogen gas until an aqueous phase was reached. The sample was then twice extracted with equal volumes of ethyl acetate; pooled ethyl acetate fractions were dried under a nitrogen stream, and SA was derivatized using trimethylsilyldiazomethane (Sigma) before analysis by GC-MS. The GC-MS analysis was carried out on a Varian Saturn 2100 ion-trap mass spectrometer using electron-impact ionization at 70 eV, connected to a Varian CP-3900 gas chromatograph equipped with a CP-8400 autosampler (Varian). SA was identified according to the retention time on GC compared with an authentic methylated standard, and the amount of SA was calculated using the isotope dilution equation with the ions at mass-to-charge ratio 120 (endogenous SA) and 124 (SAD4).
Total GSH Content and GSH Complementation Assay
Total GSH content was estimated according to Queval and Noctor (2007) using 4-week-old Arabidopsis rosette leaves and was expressed as nanomole per gram fresh weight. The results represent means ± se of six independent samples.
CLSM Imaging
To visualize NPR1:GFP fusion reporter expression in wild-type, oeOXR2, and oxr2.2 backgrounds, detached leaves from 4-week-old plants were imaged with a Leica TCS SP8 confocal laser scanning microscope with excitation at 488 nm and detection at 498 to 531 nm for GFP and 600 to 650 nm to proceed by the chlorophyll natural fluorescence subtraction.
In Vivo Imaging of the Cellular Redox State by CLSM and Ratiometric Analysis
CLSM images from 2-week-old wild-type, oeOXR2, and oxr2.2 plants carrying the Grx1-roGFP2 sensor were collected with a Zeiss LSM880 microscope in sequential mode switching between 488 and 405 nm for excitation and 505 to 530 nm for roGFP2 emission detection. For Grx1-roGFP2 sensor calibration, plants were treated with 10 mm dithiothreitol or 10 mm H2O2 to obtain the fully reduced or oxidized form of the roGFP2 protein sensor, respectively, according to Albrecht et al. (2014). Fluorescence quantification and the degree of oxidation of the roGFP2 sensor were carried out as described by Schwarzländer et al. (2008) using Fiji software (Schindelin et al., 2012).
Western Blot
For detection of NPR1-GFP, total protein was extracted from 4-week-old plants of the different genotypes using the protocol described by Martínez-García et al. (1999). Samples were loaded on 8% (w/v) SDS gels and transferred to a polyvinylidene difluoride membrane (GE Healthcare). Afterward, blots were incubated with a 1:5,000 dilution of polyclonal rabbit antibodies against GFP (Agrisera AS15 2987). Actin was used as a loading control. For this, blots were incubated with a 1:10,000 dilution of polyclonal rabbit antibodies against Actin (Agrisera AS132640). Reactions were developed with 1:50,000 goat anti-rabbit IgG (H&L), HRP conjugated (Agrisera, AS09 602) using the Agrisera ECL kit (AS16 ECL-SN).
Statistical Analysis
Data were analyzed by one-way ANOVA, and the means were compared by Tukey’s or Fisher’s (lsd) test. For flg22-triggered apoplastic ROS assay analysis, the Kruskal-Wallis test was performed. Statistical analysis was performed using InfoStat Version 2014 for Windows.
Accession Numbers
Sequence data from the genes mentioned in this article can be found in the Arabidopsis Genome Initiative or the GenBank/EMBL data libraries under the following accession numbers: At1g64280 (NPR1), At2g05590 (OXR2), At1g74710 (ICS1), At3g48090 (EDS1), At2g14610 (PR1), At5g45110 (NPR3), At1g75040 (PR5), At2g23320 (WRKY15), At2g38470 (WRKY33), At1g80840 (WRKY40), At2g25000 (WRKY60), At3g56400 (WRKY70), At5g13080 (WRKY75), and At3g18780 (ACT).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Transcriptional profiles more similar to one of the oeOXR2 plants found using the Signature tool of Genevestigator.
Supplemental Figure S2. Biotic stress conditions that most strongly affect the expression of AtOXR2.
Supplemental Figure S3. AtOXR2 overexpression increases resistance to Pst DC3000.
Supplemental Table S1. Gene list containing the ∼200 most up- and down-regulated genes from the total of misregulated genes in oeOXR2 plants.
Supplemental Table S2. Description of the experiments that yielded a transcriptional profile statistically comparable to that of oeOXR2 plants, according to the Signature tool of Genevestigator.
Supplemental Table S3. TFs statistically coexpressed with AtOXR2 under environmental stresses according to the Coexpression Analysis tool.
Supplemental Table S4. Oligonucleotides used in this study.
Acknowledgments
We thank Dr. Daniel H. Gonzalez (Instituto de Agrobiotecnología del Litoral) for invaluable cooperation on this work and for critical reading of the article. We thank Dr. Markus Schwarzländer (Plant Energy Biology Lab, Institute for Plant Biology and Biotechnology, University of Münster) for providing us the Grx1-roGFP2 sensor plants. We appreciate the help of Rodrigo Vena and the service provided by the Confocal Microscopy Facility of the Instituto de Biología Molecular y Celular de Rosario during the CLSM observations. We thank Centro Universitario Argentino-Alemán-Deutsch-Argentinisches Hochschulzentrum, Facultad de Bioquímica y Ciencias Biológicas (Instituto de Agrobiotecnología del Litoral), and Technische Universität Dresden for supporting R.M. in the frame of the Binational Ph.D. Program.
Footnotes
This work was supported by the Consejo Nacional de Investigaciones Científicas y Técnicas (grant no. PIP2014–2016GI to G.F.), the Agencia Nacional de Promoción Científica y Tecnológica (grant nos. PICT B 2014–0356 and PICT D 2017–0515 to G.F.; PICT 2014–3255 and PICT 2016–2986 to M.E.A.; and PICT 2014–3758 and PICT 2017–2340 to E.W.), and the CAI+D Universidad Nacional del Litoral. G.C., G.F., M.E.A., and E.W. are members of the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET); R.M. and P.T are fellows of the same institution. The stay of R.M. at the Technische Universität Dresden to perform part of the experiments was funded by the Centro Universitario Argentino-Alemán-Deutsch-Argentinisches Hochschulzentrum.
References
- Albrecht SC, Sobotta MC, Bausewein D, Aller I, Hell R, Dick TP, Meyer AJ(2014) Redesign of genetically encoded biosensors for monitoring mitochondrial redox status in a broad range of model eukaryotes. J Biomol Screen 19: 379–386 [DOI] [PubMed] [Google Scholar]
- Aller I, Rouhier N, Meyer AJ(2013) Development of roGFP2-derived redox probes for measurement of the glutathione redox potential in the cytosol of severely glutathione-deficient rml1 seedlings. Front Plant Sci 4: 506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ME, Pennell RI, Meijer PJ, Ishikawa A, Dixon RA, Lamb C(1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- Belt K, Huang S, Thatcher LF, Casarotto H, Singh KB, Van Aken O, Millar AH(2017) Salicylic acid-dependent plant stress signaling via mitochondrial succinate dehydrogenase. Plant Physiol 173: 2029–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaise M, Alsarraf HMAB, Wong JEMM, Midtgaard SR, Laroche F, Schack L, Spaink H, Stougaard J, Thirup S(2012) Crystal structure of the TLDc domain of oxidation resistance protein 2 from zebrafish. Proteins 80: 1694–1698 [DOI] [PubMed] [Google Scholar]
- Boubakri H, Gargouri M, Mliki A, Brini F, Chong J, Jbara M(2016) Vitamins for enhancing plant resistance. Planta 244: 529–543 [DOI] [PubMed] [Google Scholar]
- Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS(2008) Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics 180: 1671–1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X(1994) Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6: 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini NM, Monteoliva MI, Alvarez ME(2011) Proline dehydrogenase contributes to pathogen defense in Arabidopsis. Plant Physiol 155: 1947–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Queval G, Noctor G(2012) AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J 69: 613–627 [DOI] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M(2002) Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol 130: 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li C, Wang H, Guo Z(2019) WRKY transcription factors: Evolution, binding, and action. Phytopathol Res 1: 13 [Google Scholar]
- Chow CN, Lee TY, Hung YC, Li GZ, Tseng KC, Liu YH, Kuo PL, Zheng HQ, Chang WC(2019) PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res 47: D1155–D1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen KM, Gu Y, Rodibaugh N, Innes RW(2011) Negative regulation of defence signalling pathways by the EDR1 protein kinase. Mol Plant Pathol 12: 746–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF(1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colombatti F, Gonzalez DH, Welchen E(2014) Plant mitochondria under pathogen attack: A sigh of relief or a last breath? Mitochondrion 19: 238–244 [DOI] [PubMed] [Google Scholar]
- Colombatti F, Mencia R, Garcia L, Mansilla N, Alemano S, Andrade AM, Gonzalez DH, Welchen E(2019) The mitochondrial oxidation resistance protein AtOXR2 increases plant biomass and tolerance to oxidative stress. J Exp Bot 70: 3177–3195 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Alber NA, Vanlerberghe GC(2013) The signaling role of a mitochondrial superoxide burst during stress. Plant Signal Behav 8: e22749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC(2012) Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol 195: 32–39 [DOI] [PubMed] [Google Scholar]
- Czarnocka W, Karpiński S(2018) Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic Biol Med 122: 4–20 [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR(2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139: 5–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Río LA, López-Huertas E(2016) ROS generation in peroxisomes and its role in cell signaling. Plant Cell Physiol 57: 1364–1376 [DOI] [PubMed] [Google Scholar]
- del Río LA, Sandalio LM, Corpas FJ, Palma JM, Barroso JB(2006) Reactive oxygen species and reactive nitrogen species in peroxisomes: Production, scavenging, and role in cell signaling. Plant Physiol 141: 330–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ, Mittler R, Noctor G(2016) Recent progress in understanding the role of reactive oxygen species in plant cell signaling. Plant Physiol 171: 1535–1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Sun T, Ao K, Peng Y, Zhang Y, Li X, Zhang Y(2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell 173: 1454–1467.e15 [DOI] [PubMed] [Google Scholar]
- Doerks T, Copley RR, Schultz J, Ponting CP, Bork P(2002) Systematic identification of novel protein domain families associated with nuclear functions. Genome Res 12: 47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, Romeis T(2013) Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc Natl Acad Sci USA 110: 8744–8749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil-Maurizi C, Vitecek J, Marty L, Branciard L, Frettinger P, Wendehenne D, Meyer AJ, Mauch F, Poinssot B(2011) Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol 157: 2000–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott NA, Volkert MR(2004) Stress induction and mitochondrial localization of Oxr1 proteins in yeast and humans. Mol Cell Biol 24: 3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T, Rushton PJ, Robatzek S, Somssich IE(2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5: 199–206 [DOI] [PubMed] [Google Scholar]
- Fabro G, Rizzi YS, Alvarez ME(2016) Arabidopsis proline dehydrogenase contributes to flagellin-mediated PAMP-triggered immunity by affecting RBOHD. Mol Plant Microbe Interact 29: 620–628 [DOI] [PubMed] [Google Scholar]
- Farooq MA, Niazi AK, Akhtar J, Saifullah, Farooq M, Souri Z, Karimi N, Rengel Z(2019) Acquiring control: The evolution of ROS-induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol Biochem 141: 353–369 [DOI] [PubMed] [Google Scholar]
- Finelli MJ, Liu KX, Wu Y, Oliver PL, Davies KE(2015) Oxr1 improves pathogenic cellular features of ALS-associated FUS and TDP-43 mutations. Hum Mol Genet 24: 3529–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli MJ, Oliver PL(2017) TLDc proteins: New players in the oxidative stress response and neurological disease. Mamm Genome 28: 395–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli MJ, Paramo T, Pires E, Ryan BJ, Wade-Martins R, Biggin PC, McCullagh J, Oliver PL(2019) Oxidation Resistance 1 modulates glycolytic pathways in the cerebellum via an interaction with glucose-6-phosphate isomerase. Mol Neurobiol 56: 1558–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finelli MJ, Sanchez-Pulido L, Liu KX, Davies KE, Oliver PL(2016) The evolutionarily conserved Tre2/Bub2/Cdc16 (TBC), lysin motif (LysM), domain catalytic (TLDc) domain is neuroprotective against oxidative stress. J Biol Chem 291: 2751–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG, et al. (2003) Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G(2011) Ascorbate and glutathione: The heart of the redox hub. Plant Physiol 155: 2–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G(2016) Stress-triggered redox signalling: What’s in pROSpect? Plant Cell Environ 39: 951–964 [DOI] [PubMed] [Google Scholar]
- Foyer CH, Ruban AV, Noctor G(2017) Viewing oxidative stress through the lens of oxidative signalling rather than damage. Biochem J 474: 877–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R, Kopischke M, Klapprodt C, Hause G, Meyer AJ, Schwarzländer M, Fricker MD, Lipka V(2016) Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in Arabidopsis. Plant Cell 28: 130–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genger RK, Jurkowski GI, McDowell JM, Lu H, Jung HW, Greenberg JT, Bent AF(2008) Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Mol Plant Microbe Interact 21: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB(2011) Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proc Natl Acad Sci USA 108: 10768–10773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Gómez L, Felix G, Boller T(1999) A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J 18: 277–284 [DOI] [PubMed] [Google Scholar]
- González-Bosch C.(2018) Priming plant resistance by activation of redox-sensitive genes. Free Radic Biol Med 122: 171–180 [DOI] [PubMed] [Google Scholar]
- Guo P, Li Z, Huang P, Li B, Fang S, Chu J, Guo H(2017) A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. Plant Cell 29: 2854–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Chaouch S, Mhamdi A, Queval G, Zechmann B, Noctor G(2013) Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid Redox Signal 18: 2106–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera-Vásquez A, Salinas P, Holuigue L(2015) Salicylic acid and reactive oxygen species interplay in the transcriptional control of defense genes expression. Front Plant Sci 6: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P(2008) Genevestigator v3: A reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Dong Q, Yu D(2012) Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 185–186: 288–297 [DOI] [PubMed] [Google Scholar]
- Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou YH, Yu JQ, Chen Z(2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153: 1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Van Aken O, Schwarzländer M, Belt K, Millar AH(2016) The roles of mitochondrial reactive oxygen species in cellular signaling and stress response in plants. Plant Physiol 171: 1551–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Chen X, Liu Y, Roth C, Copeland C, McFarlane HE, Huang S, Lipka V, Wiermer M, Li X(2013) Mitochondrial AtPAM16 is required for plant survival and the negative regulation of plant immunity. Nat Commun 4: 2558. [DOI] [PubMed] [Google Scholar]
- Hull GA, Devic M(1995) The β-glucuronidase (GUS) reporter gene system: Gene fusions; spectrophotometric, fluorometric, and histochemical detection. Methods Mol Biol 49: 125–142 [DOI] [PubMed] [Google Scholar]
- Hung CY, Aspesi P Jr., Hunter MR, Lomax AW, Perera IY(2014) Phosphoinositide-signaling is one component of a robust plant defense response. Front Plant Sci 5: 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS(2011) Arabidopsis seedling flood-inoculation technique: A rapid and reliable assay for studying plant-bacterial interactions. Plant Methods 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Gutierrez G, Molina-Cruz A, Kumar S, Barillas-Mury C(2010) The Anopheles gambiae oxidation resistance 1 (OXR1) gene regulates expression of enzymes that detoxify reactive oxygen species. PLoS ONE 5: e11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL(2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Khan MIR, Fatma M, Per TS, Anjum NA, Khan NA(2015) Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front Plant Sci 6: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X(2000) Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12: 2339–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N, Takahashi M, Kihara S, Niimi T, Yamashita O, Yaginuma T(2014) Cloning of cDNA encoding a Bombyx mori homolog of human oxidation resistance 1 (OXR1) protein from diapause eggs, and analyses of its expression and function. J Insect Physiol 68: 58–68 [DOI] [PubMed] [Google Scholar]
- Lam E, Kato N, Lawton M(2001) Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411: 848–853 [DOI] [PubMed] [Google Scholar]
- Liebthal M, Maynard D, Dietz KJ(2018) Peroxiredoxins and redox signaling in plants. Antioxid Redox Signal 28: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindermayr C, Sell S, Müller B, Leister D, Durner J(2010) Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell 22: 2894–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KX, Edwards B, Lee S, Finelli MJ, Davies B, Davies KE, Oliver PL(2015) Neuron-specific antioxidant OXR1 extends survival of a mouse model of amyotrophic lateral sclerosis. Brain 138: 1167–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovelock DA, Šola I, Marschollek S, Donald CE, Rusak G, van Pée KH, Ludwig-Müller J, Cahill DM(2016) Analysis of salicylic acid-dependent pathways in Arabidopsis thaliana following infection with Plasmodiophora brassicae and the influence of salicylic acid on disease. Mol Plant Pathol 17: 1237–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Monte E, Quail PH(1999) A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J 20: 251–257 [DOI] [PubMed] [Google Scholar]
- Marty L, Siala W, Schwarzländer M, Fricker MD, Wirtz M, Sweetlove LJ, Meyer Y, Meyer AJ, Reichheld J-P, Hell R(2009) The NADPH-dependent thioredoxin system constitutes a functional backup for cytosolic glutathione reductase in Arabidopsis. Proc Natl Acad Sci USA 106: 9109–9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern S, Peskan-Berghoefer T, Gromes R, Kiesel RV, Rausch T(2015) Imposed glutathione-mediated redox switch modulates the tobacco wound-induced protein kinase and salicylic acid-induced protein kinase activation state and impacts on defence against Pseudomonas syringae. J Exp Bot 66: 1935–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, Hell R(2007) Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986 [DOI] [PubMed] [Google Scholar]
- Mittler R.(2017) ROS are good. Trends Plant Sci 22: 11–19 [DOI] [PubMed] [Google Scholar]
- Miura K, Okamoto H, Okuma E, Shiba H, Kamada H, Hasegawa PM, Murata Y(2013) SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J 73: 91–104 [DOI] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X(2003) Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113: 935–944 [DOI] [PubMed] [Google Scholar]
- Mullineaux PM, Exposito-Rodriguez M, Laissue PP, Smirnoff N(2018) ROS-dependent signalling pathways in plants and algae exposed to high light: Comparisons with other eukaryotes. Free Radic Biol Med 122: 52–64 [DOI] [PubMed] [Google Scholar]
- Navrot N, Rouhier N, Gelhaye E, Jacquot JP(2007) Reactive oxygen species generation and antioxidant systems in plant mitochondria. Physiol Plant 129: 185–195 [Google Scholar]
- Nawrath C, Métraux JP(1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11: 1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie S, Yue H, Zhou J, Xing D(2015) Mitochondrial-derived reactive oxygen species play a vital role in the salicylic acid signaling pathway in Arabidopsis thaliana. PLoS ONE 10: e0119853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Chaouch S, Han Y, Neukermans J, Marquez-Garcia B, Queval G, Foyer CH(2012) Glutathione in plants: An integrated overview. Plant Cell Environ 35: 454–484 [DOI] [PubMed] [Google Scholar]
- Noctor G, Reichheld JP, Foyer CH(2018) ROS-related redox regulation and signaling in plants. Semin Cell Dev Biol 80: 3–12 [DOI] [PubMed] [Google Scholar]
- O’Connell J.(2002) The basics of RT-PCR: Some practical considerations. Methods Mol Biol 193: 19–25 [DOI] [PubMed] [Google Scholar]
- Oliver PL, Finelli MJ, Edwards B, Bitoun E, Butts DL, Becker EBE, Cheeseman MT, Davies B, Davies KE(2011) Oxr1 is essential for protection against oxidative stress-induced neurodegeneration. PLoS Genet 7: e1002338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajerowska-Mukhtar KM, Emerine DK, Mukhtar MS(2013) Tell me more: Roles of NPRs in plant immunity. Trends Plant Sci 18: 402–411 [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE(2009) The role of WRKY transcription factors in plant immunity. Plant Physiol 150: 1648–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisy V, Poinssot B, Owsianowski L, Buchala A, Glazebrook J, Mauch F(2007) Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J 49: 159–172 [DOI] [PubMed] [Google Scholar]
- Plumb W, Townsend AJ, Rasool B, Alomrani S, Razak N, Karpinska B, Ruban AV, Foyer CH(2018) Ascorbate-mediated regulation of growth, photoprotection, and photoinhibition in Arabidopsis thaliana. J Exp Bot 69: 2823–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomatto LCD, Sun PY, Yu K, Gullapalli S, Bwiza CP, Sisliyan C, Wong S, Zhang H, Forman HJ, Oliver PL, et al. (2019) Limitations to adaptive homeostasis in an hyperoxia-induced model of accelerated ageing. Redox Biol 24: 101194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Noctor G(2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: Application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Sanada Y, Asai S, Ikemoto A, Moriwaki T, Nakamura N, Miyaji M, Zhang-Akiyama QM(2014) Oxidation resistance 1 is essential for protection against oxidative stress and participates in the regulation of aging in Caenorhabditis elegans. Free Radic Res 48: 919–928 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012) Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, Fricker MD, Müller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, Meyer AJ(2008) Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231: 299–316 [DOI] [PubMed] [Google Scholar]
- Seguel A, Jelenska J, Herrera-Vásquez A, Marr SK, Joyce MB, Gagesch KR, Shakoor N, Jiang SC, Fonseca A, Wildermuth MC, et al. (2018) PROHIBITIN3 forms complexes with ISOCHORISMATE SYNTHASE1 to regulate stress-induced salicylic acid biosynthesis in Arabidopsis. Plant Physiol 176: 2515–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS(2012) Ornithine-delta-aminotransferase and proline dehydrogenase genes play a role in non-host disease resistance by regulating pyrroline-5-carboxylate metabolism-induced hypersensitive response. Plant Cell Environ 35: 1329–1343 [DOI] [PubMed] [Google Scholar]
- Su LD, Zhang QL, Lu Z(2017) Oxidation resistance 1 (OXR1) participates in silkworm defense against bacterial infection through the JNK pathway. Insect Sci 24: 17–26 [DOI] [PubMed] [Google Scholar]
- Sun T, Busta L, Zhang Q, Ding P, Jetter R, Zhang Y(2018) TGACG-BINDING FACTOR 1 (TGA1) and TGA4 regulate salicylic acid and pipecolic acid biosynthesis by modulating the expression of SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1) and CALMODULIN-BINDING PROTEIN 60g (CBP60g). New Phytol 217: 344–354 [DOI] [PubMed] [Google Scholar]
- Svistunova DM, Simon JN, Rembeza E, Crabtree M, Yue WW, Oliver PL, Finelli MJ(2019) Oxidation resistance 1 regulates post-translational modifications of peroxiredoxin 2 in the cerebellum. Free Radic Biol Med 130: 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Spoel SH, Pajerowska-Mukhtar K, Mou Z, Song J, Wang C, Zuo J, Dong X(2008) Plant immunity requires conformational charges of NPR1 via S-nitrosylation and thioredoxins. Science 321: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Watanabe K, Kusano T, Takahashi Y(2011) Molecular and genetic characterization of the gene family encoding the voltage-dependent anion channel in Arabidopsis. J Exp Bot 62: 4773–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Yamashita K, Takahashi F, Kusano T, Takahashi Y(2009) Plant voltage-dependent anion channels are involved in host defense against Pseudomonas cichorii and in Bax-induced cell death. Plant Cell Rep 28: 41–51 [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL(2005) Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol 8: 397–403 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Law S, Narsai R, Whelan J(2013) AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol 162: 254–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H, Carver TLW, Foyer CH(2000) Early H2O2 accumulation in mesophyll cells leads to induction of glutathione during the hyper-sensitive response in the barley-powdery mildew interaction. Plant Physiol 123: 1289–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O.(2017) Mitochondria and cell death. Annu Plant Rev 50: 343–371 [Google Scholar]
- van Verk MC, Bol JF, Linthorst HJ(2011) WRKY transcription factors involved in activation of SA biosynthesis genes. BMC Plant Biol 11: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF(2009) Salicylic acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol 47: 177–206 [DOI] [PubMed] [Google Scholar]
- Volkert MR, Elliott NA, Housman DE(2000) Functional genomics reveals a family of eukaryotic oxidation protection genes. Proc Natl Acad Sci USA 97: 14530–14535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Durrant WE, Song J, Spivey NW, Dong X(2010) Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc Natl Acad Sci USA 107: 22716–22721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Barnaby JY, Tada Y, Li H, Tör M, Caldelari D, Lee DU, Fu XD, Dong X(2011) Timing of plant immune responses by a central circadian regulator. Nature 470: 110–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Berkey CD, Watnick PI(2012) The Drosophila protein mustard tailors the innate immune response activated by the immune deficiency pathway. J Immunol 188: 3993–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hang S, Purdy AE, Watnick PI(2013) Mutations in the IMD pathway and mustard counter Vibrio cholerae suppression of intestinal stem cell division in Drosophila. MBio 4: e00337-e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth MC, Dewdney J, Wu G, Ausubel FM(2001) Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414: 562–565 [DOI] [PubMed] [Google Scholar]
- Withers J, Dong X(2016) Posttranslational modifications of NPR1: A single protein playing multiple roles in plant immunity and physiology. PLoS Pathog 12: e1005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Davies KE, Oliver PL(2016) The antioxidant protein Oxr1 influences aspects of mitochondrial morphology. Free Radic Biol Med 95: 255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Zhang D, Chu JY, Boyle P, Wang Y, Brindle ID, De Luca V, Després C(2012) The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep 1: 639–647 [DOI] [PubMed] [Google Scholar]
- Xu X, Chen C, Fan B, Chen Z(2006) Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18: 1310–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Lin X, Rowe A, Rognes T, Eide L, Bjørås M(2015) Transcriptome analysis of human OXR1 depleted cells reveals its role in regulating the p53 signaling pathway. Sci Rep 5: 17409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Luna L, Sørbø JG, Alseth I, Johansen RF, Backe PH, Danbolt NC, Eide L, Bjørås M(2014) Human OXR1 maintains mitochondrial DNA integrity and counteracts hydrogen peroxide-induced oxidative stress by regulating antioxidant pathways involving p21. Free Radic Biol Med 77: 41–48 [DOI] [PubMed] [Google Scholar]
- Yi SY, Shirasu K, Moon JS, Lee SG, Kwon SY(2014) The activated SA and JA signaling pathways have an influence on flg22-triggered oxidative burst and callose deposition. PLoS ONE 9: e88951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Croze E, Yamaguchi KD, Tran T, Reder AT, Litvak V, Volkert MR(2015) Induction of a unique isoform of the NCOA7 oxidation resistance gene by interferon β-1b. J Interferon Cytokine Res 35: 186–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Van Aken O, Thatcher L, De Clercq I, Duncan O, Law SR, Murcha MW, van der Merwe M, Seifi HS, Carrie C, et al. (2014) The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J 80: 709–727 [DOI] [PubMed] [Google Scholar]
- Zhang R, Miao J, Song Y, Zhang W, Xu L, Chen Y, Zhang L, Gao H, Zhu B, Li J, et al. (2019) Genome-wide association study identifies the PLAG1-OXR1 region on BTA14 for carcass meat yield in cattle. Physiol Genomics 51: 137–144 [DOI] [PubMed] [Google Scholar]
- Zhang S, Li C, Wang R, Chen Y, Shu S, Huang R, Zhang D, Li J, Xiao S, Yao N, et al. (2017a) The Arabidopsis mitochondrial protease FtSH4 is involved in leaf senescence via regulation of WRKY-dependent salicylic acid accumulation and signaling. Plant Physiol 173: 2294–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ivanova A, Vandepoele K, Radomiljac J, Van de Velde J, Berkowitz O, Willems P, Xu Y, Ng S, Van Aken O, et al. (2017b) The transcription factor MYB29 is a regulator of alternative oxidase1a. Plant Physiol 173: 1824–1843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li X(2019) Salicylic acid: Biosynthesis, perception, and contributions to plant immunity. Curr Opin Plant Biol 50: 29–36 [DOI] [PubMed] [Google Scholar]
- Zhu Q, Dugardeyn J, Zhang C, Mühlenbock P, Eastmond PJ, Valcke R, De Coninck B, Öden S, Karampelias M, Cammue BPA, et al. (2014) The Arabidopsis thaliana RNA editing factor SLO2, which affects the mitochondrial electron transport chain, participates in multiple stress and hormone responses. Mol Plant 7: 290–310 [DOI] [PubMed] [Google Scholar]