A transcriptomic database of 4498 differentially expressed genes revealed that ABI4 participates in brassinosteroid response by binding to the BAK1 promoter and inhibiting transcription.
Abstract
Brassinosteroids (BRs) are plant growth-promoting steroid hormones. BRs affect plant growth by regulating panels of downstream genes. Much effort has been made to establish BR-regulated gene expression networks, but there is little overlap among published expression networks. In this study, we built an optimal BR-regulated gene expression network using the model plant Arabidopisis (Arabidopisis thaliana). Seven- and 24-d-old seedlings of the constitutive photomorphogenesis and dwarfism mutant and brassinosteroid-insensitive 1-701 (bri1-701) BRI1-like receptor genes1 (brl1) brl3 triple mutant seedlings were treated with brassinolide and RNA sequencing (RNA-seq) was used to detect differentially expressed genes. Using this approach, we generated a transcriptomic database of 4,498 differentially expressed genes and identified 110 transcription factors that specifically respond to BR at different stages. We also found that, among the identified BR-responsive transcription factors, ABSCISIC ACID-INSENSlTIVE4 (ABI4), an ethylene response factor transcription factor, inhibits BR-regulated growth. Compared to wild-type plants, the abi4-102 mutant was less sensitive to brassinazole and more sensitive to BR. Next, we performed a chromatin immunoprecipitation followed by high-throughput sequencing assay and established that ABI4 binds directly to the BRI1-associated receptor kinase1 promoter and inhibits transcription. These results provide insight into BR-responsive gene functions in regulating plant growth at different stages and may serve as a basis for predicting gene function, selecting candidate genes, and improving the understanding of BR regulatory pathways.
Brassinosteroids (BRs) are growth-promoting steroid hormones with important roles in plant development. BR biosynthesis and signal defective mutants typically present round and dark green leaf, short petiole, dwarf, and male sterility phenotypes. Many components of the BR signaling pathway have been described. When BR is perceived by the BRASSINOSTEROID INSENSITIVE1 (BRI1) protein, a transmembrane Ser/Thr kinase, BRI1 is activated and BR signals are transduced through the phosphorylation of downstream proteins (Li and Chory, 1997). BRASSINOSTEROID INSENSITIVE2 (BIN2) is another negative regulator of BR response that has a conserved kinase domain and C-terminal domain (Li et al., 2001; Li and Nam, 2002). In the absence of BR, BIN2 phosphorylates transcription factors BRASSINAZOLE-RESISTANT1 (BZR1) and BRI1-EMS-SUPPRESSOR1 (BES1) to inhibit their activity (He et al., 2002; Wang et al., 2002; Yin et al., 2002).
Plant hormones often act through transcription factors to regulate downstream gene expression. BR regulates plant development through transcription factors that either induce or repress downstream genes. Many transcription factors have been identified as participating in downstream BR signaling pathways. BES1 and BZR1 are two important transcription factors in the BR signaling pathway. BES1 shares 88% identity with BZR1 and has similar protein domains: a nuclear localization signal in the N-terminal, a Ser-rich domain in the central part, and a PEST domain in the C terminus (Yin et al., 2002). BES1/BZR1 also interacts with many transcription factors, such as BIM1, MYB30, MYBL2, and HAT1, to induce or reduce the expression of downstream genes and integrate BR and other signaling pathways (Yin et al., 2005; Li et al., 2009; Ye et al., 2012; Zhang et al., 2014).
To fully understand how BR regulates plant growth and division through downstream genes, multiple research groups have identified the direct target genes of BES1 and BZR1 through microarrays and chromatin immunoprecipitation (ChIP) on Affymetrix tiling arrays (ChIP-chip) assays. Yu et al. (2011) used the bes1-D mutant for a ChIP-chip experiment and identified 1609 putative BES1 target genes. Sun et al. (2010) performed a ChIP-chip analysis using BZR1-CFP transgene plants, and 2260 loci linked to 3410 genes were identified as BZR1 targets. However, fewer than expected BR-regulated genes have been identified, and we believe that there are other, as-yet-undiscovered, transcription factors involved in gene regulation in the BR signaling pathway, and more BR-responsive genes need to be identified.
Microarray studies using BR mutants have identified large numbers of BR-responsive genes, and these data suggest that BR regulates multiple cellular processes and interacts with other pathways (Goda et al., 2002, 2004; Müssig et al., 2002; Nemhauser et al., 2004; Guo et al., 2009; Sun et al., 2010; Yu et al., 2011). Müssig et al. (2002) performed Affymetrix Arabidopsis (Arabidopsis thaliana) genome arrays using wild type, dwf1-6, and CONSTITUTIVE PHOTOMORPHOGENESIS and DWARFISM (CPD)-antisense plants grown under two conditions to identify BR responsive genes, identifying several BR-regulated genes (Goda et al., 2002, 2004; Mussig et al., 2002). Goda et al. (2002, 2004) performed microarray studies where BR-deficient mutants (de-etiolated2 [det2] and bri1-5) were treated with brassinolide (BL) to identify BL-regulated genes. Although many microarrays and ChIP-chip analyses have aimed to identify BR-responsive genes, the overlap among the gene sets has been poor. This lack of overlap among gene lists is likely a result of the different experimental conditions, plant tissues, and developmental stages used, as well as insufficient sensitivity of the tools.
In this study, we have used a comparative RNA sequencing (RNA-seq) approach to identify BR-responsive differentially expressed genes (DEGs) in Arabidopsis. For this, we made use of the cpd mutant, and to avoid falsely identifying non-BR-induced genes, we make use of the bri1-701 brl1 brl3 triple mutant as a negative control. Analyses of RNA-seq data for 7- and 24-d-old seedlings identified 3,002 and 1,496 DEGs, respectively. Among the DEGs, we identified 110 genes encoding transcription factors that specifically respond to BR at different stages. Through phenotype analysis, we found that ABI4, one of the 110 transcription factors, inhibits BR-regulated growth, demonstrating that BR regulates downstream genes through ABI4. By ChIP followed by high-throughput sequencing (ChIP-seq), we show that ABI4 directly binds to the BAK1 promoter and inhibits BAK1 transcription. Our data support the notion that BR affects plant growth by using transcription factors to regulate the expression of downstream genes. These data help integrate the BR transcriptional network and guide future works addressing plant responses to BRs.
RESULTS
Characterization of cpd and bri1-701 brl1 brl3 Triple Mutants
cpd is a severe BR biosynthesis mutant and displays small, dwarf, dark green and rounded leaf, and sterility phenotypes. Here, we use the salk_023532 transfer DNA insertion mutant of CPD (hereafter cpd), which has a BR-sensitive phenotype. When cpd seeds were sown on one-half strength Murashige and Skoog (MS) medium with 1% Suc, 0.8% agar, and 1 μm BL, the seedlings were as sensitive as wild type (Fig. 1A). bri1-701 is the transfer DNA insertion mutant of BR receptor BRI1 and is sterile. BRL1 and BRL3 are BRI1 homologs that can bind BL with high affinity (Caño-Delgado et al., 2004). The bri1-701 brl1 brl3 triple null mutant showed the same phenotype with cpd and is insensitive to BL (Fig. 1A). When cpd and bri1-701 brl1 brl3 were grown in soil for 24 d, they also showed the same retarded phenotype (Fig. 1B).
Figure 1.
cpd and bir1-701 brl1 brl3 showed the same significantly retarded phenotype. A, The phenotype of 7-d-old wild type, bri1-701 brl1 brl3, and cpd grown in one-half strength MS medium and one-half strength MS + 1 μm BL. Bar = 1cm. B, The phenotype of 24-d-old wild type, bri1-701 brl1 brl3, and cpd grown in soil.
BL treatments induce gene expression changes, and this offers a convenient route to detecting DEGs, typically through the RNA-seq. As cpd is a strong BR biosynthesis mutant, the BR-induced genes were inhibited significantly, and the expression of BR-related genes are very low. When treated with BL, the gene expression will change obviously; it is convenient to detect the DEGs through the RNA-seq. These RNA-seq datasets can be used to investigate the molecular responses to short-term BR treatment in Arabidopsis. Because of long-term BR deficiency, there might be secondary effects in BR-deficient mutants, and the expression of stress-related genes and BR-related genes that are not transducted by BR receptor BRI1 might change when treated with BR. Therefore, to avoid such genes in our list of DEGs, here we use the bri1-701 brl1 brl3 triple mutant (that presents the same phenotypes as cpd) as a negative control.
RNA-seq Analysis of BL-Treated and Nontreated cpd and bri1-701 brl1 brl3 Mutants
To identify DEGs during the response to BRs, an RNA-seq study was performed using 7-d-old cpd and bri1-701 brl1 brl3 seedlings. At this developmental stage, cells are rapidly elongating and dividing. Before collecting the plant material, the seedlings were inoculated with 1 μm BL for 2 h. Expression analysis identified 3002 genes that were significantly differentially regulated (Fig. 2, A and B; Supplemental Tables S1 and S2).
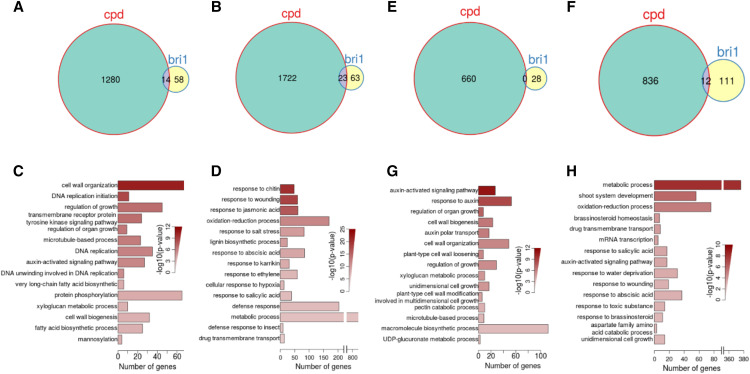
Figure 2.
BR responsiveness in BL-treated 7- and 24-d-old cpd and bri1-701 brl1 brl3 seedlings determined by RNA-seq analysis. A and B, Venn diagrams show the numbers of genes upregulated (A) or downregulated (B) in BL-treated 7-d-old cpd seedlings. E and F, Venn diagrams show the numbers of genes upregulated (E) or downregulated (F) in BL-treated 24-d-old cpd seedlings. C and D, The top 15 enriched GO terms in the category of biological process among genes upregulated in 7-d-old cpd seedlings (C), downregulated in 7-d-old cpd seedlings (D). G and H, The top 15 enriched GO terms in the category of biological process among genes upregulated in 24-d-old cpd seedlings (G) and down-regulated in 24-d-old cpd seedlings (H). In each barplot, the x axis represents the number of regulated genes annotated to each respective GO term, and color shading shows the statistical significance (negative log10-transformed P values) of the enrichment of the respective GO term obtained by performing Fisher’s exact test.
To obtain an overview of the functional pathways in which the BR-responsive genes are involved, Gene Ontology (GO) enrichment analysis for biological processes was performed for the RNA-seq data (Fig. 2, C and D). The most significantly enriched GO terms in the category of biological process (P < 0.05) for upregulated DEGs of the 7-d-old seedlings included cell wall organization, DNA replication initiation, and regulation of growth (Fig. 2C). The highly significant enrichment of genes involved in cell wall organization among upregulated DEGs suggested a regulatory function of BR on cell growth and cell division through regulating cell wall organization and DNA replication initiation. In the 1,722 BR downregulated genes, maximum downregulation was identified for genes responding to chitin and wounding. The enrichment of jasmonic acid, ethylene, and abscisic acid response genes indicates antagonism between these and BR.
To determine whether the developmental stage of the materials contributed to the discrepancies in the detection of the BR-regulated genes, we also performed another RNA-seq assay using 24-d-old seedlings. At this stage, vegetative tissues and cell elongation has slowed, and reproductive tissue cells begin to divide and elongate. A total of 1,496 genes were identified with significantly altered expression in BR-treated cpd plants (Fig. 2, E and F; Supplemental Tables S1 and S3). We identified 660 upregulated genes and 836 downregulated genes. GO analysis of the genes was also performed to facilitate the global analysis of BR-regulated gene expression and evaluate the gene functions at the 24-d-old stage. Upregulated genes were enriched with genes involved in the auxin-activated signaling pathway, regulation of organ growth, and cell wall biogenesis, among others. Among commonly downregulated genes, metabolic process, shoot system development, oxidation-reduction process, and BR homeostasis genes were significantly enriched (Fig. 2, G and H). Taken together, these transcriptome results demonstrate that BR had a significant effect on the transcription of a subset of genes, through multiple mechanisms at both the 7- and 24-d-old stages.
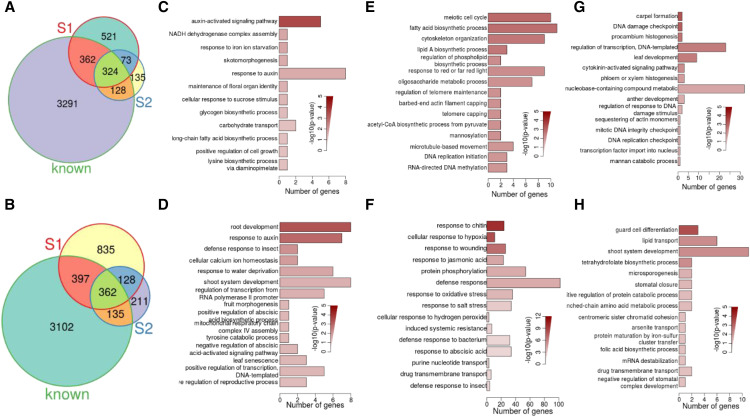
A total of 1,940 BR upregulated genes and 2558 BR downregulated genes were detected in the two examined different stage seedlings. Among these, 20.5% of the genes (397 of 1,940) and 19.2% (490 of 2,558) were commonly up- or down-regulated at both developmental stages (Fig. 3, A and B). When comparing the 3,002 DEGs of 7-d-old seedlings with 1496 DEGs from 24-d-old seedlings, significantly more BR-responsive genes were up- or downregulated at the 7- than 24-d-old stage, indicating that more genes respond to BR at the rapid cell elongation and division stage. We also found that only 397 were upregulated at both developmental stages (representing 31% [397 of 1,280] of the upregulated genes in the 7-d-old seedlings list and 60% [397 of /660] of the upregulated genes in the 24-d-old seedlings list) and that 490 genes were downregulated at both developmental stages (representing 28.5% [490 of 1,722] of the downregulated genes in the 7-d-old seedlings list and 58.6% [490 of 836] of the downregulated genes in the 24-d-old seedlings list; Fig. 3, A and B). From these comparisons, we can see that the common genes regulated both in 7- and 24-d-old seedlings are less than specifically regulated genes in 7- and 24-d-old seedlings. This comparison and the poor overlap between them suggest that BR responses at different stages might have both common and distinct mechanisms.
Figure 3.
Specific DEGs in BR-treated cpd and bri1-701 brl1 brl3. A and B, Comparison of genes upregulated (A) or downregulated (B) in 7-d-old seedlings (S1), 24-d-old seedlings (S2) of cpd, and previously identified BR-responsive genes (known). C to H, Top 15 (or 14 for F) significantly enriched GO terms in the category of biological process for genes upregulated (C) or downregulated (D) by BR in both 7-d-old and 24-d-old seedlings. Specifically upregulated (E) or specifically downregulated (F) by BR in 7-d-old seedlings. Genes specifically upregulated (G) or specifically downregulated (H) by BR in 24-d-old seedlings. For each barplot, the x axis represents the number of BR-regulated genes annotated to each respective GO term, with color shading showing the statistical significance (negative log10-transformed P values) of the enrichment of the respective GO term obtained by performing Fisher’s exact test. DR, Defense response; DSR, double-strand break.
Specific DEGs Identified in 7- and 24-d-Old Seedlings
Many microarrays and ChIP-chip analyses of BR-responsive genes have been performed by multiple research groups using various BR mutants at different development stages, collectively identifying 8,102 BR-regulated genes (4,105 BR-induced genes and 3,997 BR-repressed genes; Goda et al., 2004; Nemhauser et al., 2004; Guo et al., 2009; Yu et al., 2011). Comparing our results with those 8,102 BR-regulated genes, we found that greater overlap for the 24-d-old seedlings list (68.5% upregulated and 59.4% downregulated) than for the 7-d-old seedlings list (53.6% upregulated and 44.1% downregulated; Fig. 3, A and B) revealed that stage specification is a very important factor for the discrepancies of gene expression. To identify stage-specific effects of BR, we compared 729 upregulated and 1,174 downregulated BR-responsive genes newly identified in different stages and looked for genes that were specifically regulated in 7- and 24-d-old seedlings. Among them, 1,356 DEGs (521 upregulated and 835 downregulated) were uniquely identified in 7-d-old seedlings, and 346 DEGs (135 upregulated and 211 downregulated) were uniquely identified in 24-d-old seedlings, and 201 genes were commonly regulated (Supplemental Tables S4 and S5). Examination of the 73 identified upregulated genes shared between the two stages revealed several key functional genes, including genes involved in the auxin-activated signaling pathway, NADH dehydrogenase complex assembly, and response to iron ion starvation (Fig. 3C). Root development, response to auxin, and defense response to insect genes were downregulated both at the 7- and 24-d-old stages (Fig. 3D). The meiotic cell cycle, fatty acid biosynthetic process, and cytoskeleton organization genes were upregulated specifically at the 7-d-old stage (Fig. 3E), suggesting the importance of the meiotic cell cycle in the BR-regulated tissues. Response to chitin, cellular response to hypoxia, and response to wounding genes were downregulated by BR specifically at the 7-d-old stage (Fig. 3F). Among the genes upregulated only in the 24-d-old seedlings, carpel formation, DNA damage checkpoint, and procambium histogenesis genes were significantly enriched (Fig. 3G). Guard cell differentiation, lipid transport, and shoot system development genes were downregulated only at the 24-d-old stage (Fig. 3H). GO enrichments showed it is quite different between specific upregulated or downregulated genes in 7-d-old and 24-d-old seedlings. Our transcriptome results demonstrate that BR-responsive programs in seedlings at different stages have many active processes specifically.
Confirmation of the RNA-seq Results
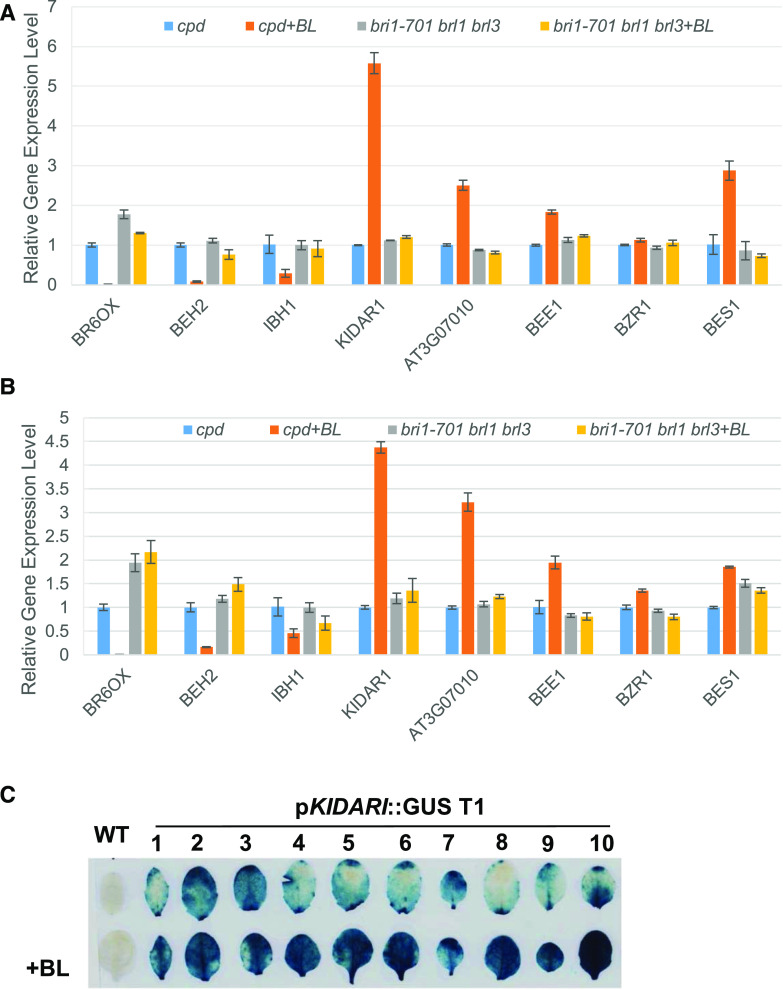
To validate our RNA-seq data, eight genes were randomly selected for reverse transcription quantitative PCR (RT-qPCR) analysis: BR6OX2, BEH2, IBH1, KIDAR1, AT3G07010, BEE1, BZR1, and BES1. From the RT-qPCR results, BL treatment significantly reduced the gene expression of BR6OX2, BEH2, and IBH1 both in 7- and 24-d-old cpd seedlings (Fig. 4, A and B). KIDAR1, BEE1, AT3G07010, BZR1, and BES1 gene expression were higher after BL treatment in cpd seedlings and plants (Fig. 4, A and B). And in BL-treated bri1-701 brl1 brl3 seedlings, the expression of these genes did not change significantly (Fig. 4, A and B). The RT-qPCR results were consistent with those of the RNA-seq. These results indicate that our RNA-seq data are reliable. We also detected the expression of KIDAR1 by fusing its putative promoter region to a GUS gene. In the T1 transgenic plants harboring this fusion, GUS staining signals were detected in rosette leaves, and the GUS signal could be significantly enhanced by BL treatment (Fig. 4C).
Figure 4.
Validation of the RNA-seq results. A, RT-qPCR analysis of BR6OX2, BEH2, IBH1, KIDAR1, AT3G07010, BEE1, BZR1, and BES1 in the 7-d-old cpd and bri1-701 brl1 brl3 seedlings treated with or without 1 µm BL for 2 h. B, RT-qPCR analysis of BR6OX2, BEH2, IBH1, KIDAR1, AT3G07010, BEE1, BZR1, and BES1 in the 24-d-old cpd and bri1-701 brl1 brl3 seedlings treated with or without 1 µm BL for 2 h. For each sample, the RT-qPCR assays were repeated three times, and the error bars denote ± sd. C, Effects of BR on the expression of pKIDAR1::GUS transgene plants. The top row shows the GUS staining of wild-type (WT) and 10 pKIDAR1::GUS T1 plants. The bottom row shows the 1-μm BL treatment increased the GUS signal of pKIDAR1::GUS transgene plants.
Transcription Factors That Respond to BR in Arabidopsis Seedlings
We found that the BR-responsive genes identified in this study have little overlap with the 1,609 BES1 and 3,410 BZR1 targets (Supplemental Fig. S1, A and B; Supplemental Table S6), indicating that other transcription factors also participate in BR-regulating gene expression. Through further investigation of our RNA-seq data, we identified 110 transcription factors among the BR-responsive genes, including transcription factors belonging to the ethylene response factor (ERF; n = 34), basic helix-loop-helix (bHLH; n = 14), HDZIP (n = 9), MYB (n = 26), C2H2 (n = 14), GRF (n = 4), GATA (n = 4), ZFHD (n = 3), and BES1 (n = 2) families. Among them, 28 ERF genes were downregulated at both the 7- and the 24-d-old stages and six ERFs and two BES1 homolog genes were upregulated at the 24-d-old stage. Fourteen bHLH genes and three ZFHD genes were specifically upregulated at the 7-d-old stage. Twenty-six MYB genes and 14 C2H2 genes were specifically downregulated at the 7-d-old stage. Nine HDZIP genes were specifically downregulated at the 24-d-old stage. Four GRF genes and four GATA genes were specifically upregulated at the 24-d-old stage (Table 1; Supplemental Table S7). We also identify several known BR-related transcription factors that are upregulated by BR, but only at defined developmental stages. BR Enhanced Expression1 (BEE1), BEE2, and BEE3, which encode putative bHLH proteins (Friedrichsen et al., 2002), were specifically upregulated in the 7-d-old stage. Another bHLH transcription factor, AT1G26945 (KIDAR1/PRE6), was also specifically upregulated in the 7-d-old stage. We also found that AIF1 was upregulated specifically at the 24-d-old stage and is involved in BR-regulated cell elongation and growth. These results suggest that different tissues recruit different transcriptional factors to respond to and transmit the BR signal at a different stage. Furthermore, transcription factor genes were more likely downregulated by BR treatment than upregulated, and they were more likely to be downregulated in 7- than 24-d-old seedlings. Therefore, future work must now aim to understand the functions of these transcription factors that have been newly implicated in BR signaling.
Table 1. Transcription factors regulated by BR at different stages.
Number in parentheses indicates the number of this kind of gene detected in our RNA-seq result.
| Transcription Factor | 7-d-old Seedlings | 24-d-old Seedlings |
|---|---|---|
| ERF (28) | Down | Down |
| ERF (6) | – | Up |
| bHLH (14) | Up | – |
| HDZIP (9) | – | Down |
| MYB (26) | Down | – |
| C2H2 (14) | Down | – |
| GRF (4) | – | UP |
| GATA (4) | – | UP |
| ZFHD (3) | Up | – |
| BES1 (2) | Down | Down |
abi4 Is Hypersensitive to BR and Insensitive to BRZ
Different ERFs respond to BR specifically at different stage suggested an important role of them and prompted us to investigate the role of ERFs in BR response. For this, we ordered all the mutants of BR-responsive ERFs and found the abi4-102 (cs3837) mutant has a longer petiole than wild type (which is one of the typical phenotypes of plants overproducing BRs; Fig. 5A). Next, we performed BR response assays using the BR biosynthesis inhibitor brassinazole (BRZ). We found that abi4-102 is relatively insensitive to BRZ, both in the dark and light (Fig. 5, B to D). ABI4-overexpressing plants (ABI4-cFLAG) showed a dwarf phenotype and were hypersensitive to BRZ both in the dark and light (Fig. 5, E to H). In a root elongation assay, abi4-102 is more sensitive to BL than wild type, and ABI4-cFLAG is almost as sensitive to BL as wild type (Fig. 5I). To test whether the phenotype of abi4-102 was correlated with the strength of BR signaling, we measured the expression levels of the BR-related marker genes, including the BR-suppressed genes CPD, DWF4, and a BR-induced gene Saur_AC1 by RT-qPCR. Compared to the wild type, in abi4-102, the expression of CPD and DWF4 were decreased a bit, while the expression of Saur_AC1 was much higher, and in ABI4-cFLAG, the expression of CPD and DWF4, Saur-AC1 are recovered (Fig. 5J), indicating that the BR signaling outputs were increased in abi4-102. These results indicate that in abi4-102, the BR signal is amplified, suggesting that BR regulates gene expression through the ABI4 transcription factor.
Figure 5.
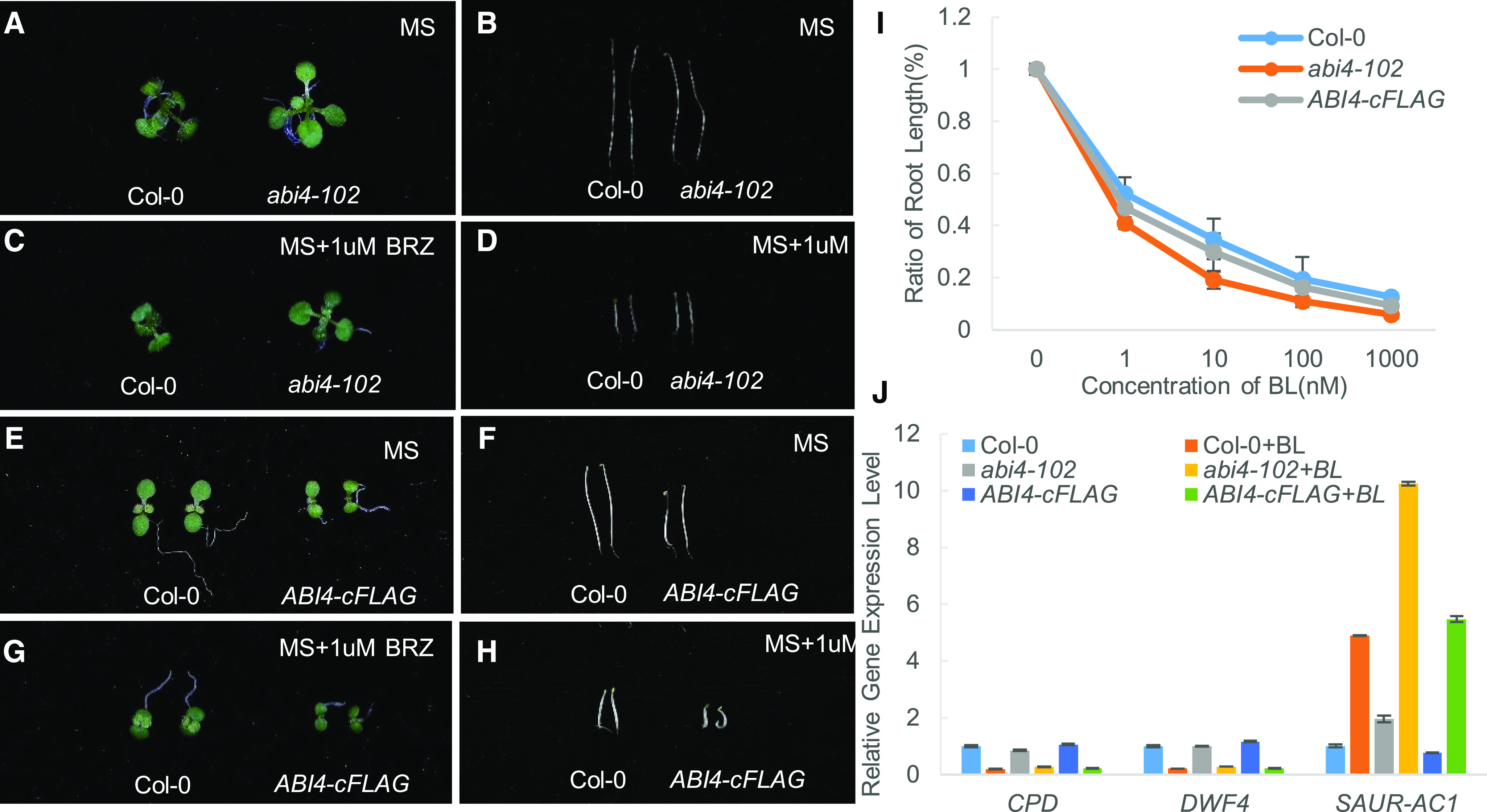
abi4-102 was hypersensitive to BR and hyposensitive to BRZ. A to D, Phenotype of abi4-102 grown in one-half strength MS in light (A), grown in one-half strength MS in dark (B), grown in one-half strength MS + 1 μm BRZ in light (C), and grown in one-half strength MS + 1 μm BRZ in dark (D). E to H, Phenotype of ABI4-cFLAG grown in one-half strength MS in light (E), grown in one-half strength MS in dark (F), grown in one-half strength MS + 1 μm BRZ in light (G), and grown in one-half strength MS + 1 μm BRZ in dark (H). I, Quantification of root length of Col-0, abi4-102, and ABI4-cFLAG grown in one-half strength MS containing different concentration of BL. Each data point represents the average of 25 seedlings of duplicated experiments, and the error bars denote ± sd. J, Gene expression level of CPD, DWF4, and SAUR-AC1 in treated with or without 1 µm BL of Col-0, abi4-102, and ABI4-cFLAG. For each sample, the RT-qPCR assays were repeated three times, and the error bars denote ± sd.
ABI4 Inhibits BR-Regulated Plant Growth by Binding and Inhibiting the Expression of BAK1
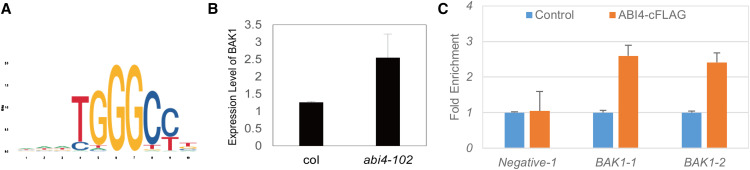
To test whether ABI4 represses downstream BR-related genes and to identify candidate target genes, we performed a ChIP-seq assay for ABI4. For the ChIP-seq assay, a FLAG antibody was used to pull down the putative DNA sequences from transgene ABI4-FLAG seedlings. ABI4-FLAG was enriched in the 2-kb upstream regions of a total of 2343 genes in Arabidopsis seedlings (Supplemental Fig. S2; Supplemental Table S8). From the ChIP-seq analysis, we found that ABI4 binds TGGGCC motif (Fig. 6A), and ABI4 directly binds BRI1 Associated receptor Kinase1 (BAK1) promoter and the homologs of BAK1, SERK1, and SERK2, while ABI5 and RD26, known targets of ABI4, were used as positive controls. The binding peaks and fold enrichment values for these genes are given in Table 2 and Supplemental Figure S3. We used RT-qPCR to test whether the binding of ABI4 to BAK1 affects BAK1 transcript levels. In the abi4-102 mutant, BAK1 transcript levels were increased relative to wild type (Fig. 6B). ChIP-qPCR assay showed that ABI4 bound to the promoter regions of BAK1 (Fig. 6C). Taken together, our data indicate that BR regulates plant growth through the ABI4 transcription factor, which binds to the BAK1 promoter and inhibits BAK1 gene expression.
Figure 6.
ABI4 binds BAK1 promoter and inhibit transcription. A, Sequence logo for ABI4 binding motif in the promoters of ABI4-targeting genes. The height of each letter represents the frequency of the base at that position. B, Gene expression level of BAK1 in abi4-102. For each sample, the RT-qPCR assays were repeated three times, and the error bars denote ± sd. C, ChIP-qPCR results indicating that the promoter fragments of BAK1 can be amplified from the immunoprecipitates pulled down by the anti-FLAG antibody. For each sample, the ChIP-qPCR assays were repeated three times, and the error bars denote ± sd.
Table 2. Direct binding peaks of ABI4.
Chr, Chromosome.
| Gene | Peak Localization | Fold Change | P Value |
|---|---|---|---|
| BAK1 | Chr4: 16090492–16091002 | 2.47047 | 2.17E−06 |
| SERK1 | Chr1: 27017836–27018111 | 2.08315 | 3.29E−05 |
| SERK2 | Chr1: 12458754–12459029 | 1.99309 | 0.00039852 |
| ABI5 | Chr2: 15207369–15207633 | 2.74754 | 4.57E−08 |
| RD26 | Chr4: 13707566–13707853 | 2.73776 | 7.32E-−08 |
DISCUSSION
In this study, we aimed to investigate the transcriptomic response of Arabidopsis to BR and hoped to identify new genes and proteins involved in BR-mediated regulation of plant growth. Many microarray and ChIP-immunoprecipitation (IP) studies by multiple groups have investigated BR regulation, but RNA-seq analysis has been relatively underutilized in the search for BR-responsive genes. Also, to our knowledge, very few studies searching for BR-responsive genes have used strong BR-defective mutants, such as cpd and bri1-701 brl1 brl3, as genetic tools. In this work, instead of analyzing the long-term BL treatment response, we focused on identifying genes differentially expressed soon after treatment and compared lists of BR-regulated DEGs at two developmental stages (7- and 24-d-old seedlings). We found that BR positively regulates cell tip growth, cell proliferation, cell wall polysaccharide biosynthesis, DNA methylation and auxin responses, and BR biosynthesis. BR also repressed the ethylene biosynthesis and signaling pathway, ABA signaling pathway, jasmonic acid biosynthesis and signaling pathway, and fungus defense responses. Among the DEGs responsive to short-term BR treatment, 15,57 of 3,002 are BR-responsive genes found in 7-d-old seedlings, and 547 of 1,496 are found in 24-d-old seedlings. We also found that the transcript levels of an AP2/ERF transcription-factor-encoding gene, ABI4, were down-regulated and that ABI4 can bind to the promoter of BAK1, thereby inhibiting its expression and plant growth. Taken together, our results contribute to a more comprehensive understanding of the BR regulatory network.
From these results, we can see that the number of DEGs identified for the 24-d-old plants was less than that for the 7-d-old seedlings, which might be a consequence of the developmental stage of these mutants, in which all organs are stuck in the maturation phase and the change of vegetative stage to reproductive stage of plants. Differences in gene regulation patterns across reproductive stages have been reported (Zhang et al., 2017). The 24-d-old cpd is transforming from the vegetative stage to the reproductive stage, and regulation of some genes within this context might have a stronger negative impact on gene expression, so there are fewer DEGs than at the 7-d-old stage. In addition, it is possible that many genes related to plant development have already achieved maximal expression levels by 24 d, and thus the BL treatment does not result in any further increase in expression.
BES1 is the key transcription factor of the BR signaling pathway and has six homologs: BES1, BZR1, BEH1, BEH2, BEH3, and BEH4. We found that only BEH1 and BEH2 were downregulated by BR treatment (Supplemental Table S7). The bes1-D and bzr1-1D mutant plants display different phenotypes when grown in light because of their different biological functions in plant development (Wang et al., 2002; Yin et al., 2002). The distinct functions of BES1 and BZR1 indicate the different functions of these gene families. While BZR1 has dual roles in regulating BR biosynthesis and growth responses (He et al., 2005), BEH1 to BEH4 might also function differently. BES1 and BZR1 have been well studied, but the functions of BEH1 to BEH4 are unclear. Our results verify the differences between these families and help to understand the biological functions of BEH1 to BEH4. In future work, it would be interesting to compare the affinities of BEH1, BEH2, BEH3, and BEH4 for gene promoters.
ERF is one of the largest transcription factor families in Arabidopsis. The ERF transcription factors have an AP2/ERF-type DNA-binding domain, and diverse functions have been described for ERFs at various developmental stages (Chandler 2018). ERF can be regulated by many hormones such as ethylene, auxin, and ABA (Fujimoto et al., 2000). The significantly changed expression of ERF indicated the cross talk between these hormones. Recently, a stress-inducible AP2/ERF transcription factor TINY was proved to inhibit BR-regulated growth and positively regulates drought responses (Xie et al., 2019), indicating the role of ERF in the interaction between BR and ABA. Now, we also found 34 ERFs (including TINY) can be regulated by BR. Twenty-eight ERFs were downregulated at the 7-d-old seedling stage and the 24-d-old stage specifically, and six ERFs were upregulated in 24-d-old seedlings, indicating important and specific functions for ERF in BR-regulating plant growth. Here, we found that one of the AP2/ERF transcription factors, ABI4, also inhibits BR-regulated growth by inhibiting BAK1 expression. BAK1 was the coreceptor of the BR-signaling pathway; the binding and inhibiting of BAK1 by ABI4 might represent a feedback mechanism for BR signaling. These results indicate that the BR might recruit different ERFs to bind and regulate different developmental response genes at different stages and interacted with different hormones through different ERFs.
BR and ABA function antagonistically through multiple signaling components during different development processes such as seed germination and root length. Many papers have been published revealing the interaction and cross talk between BR and ABA. BR-deficient and perception mutants showed hypersensitivity to ABA in seed germination (Steber and McCourt 2001; Zhang et al., 2009). BIN2, a negative regulator of BR signaling, phosphorylates and stabilizes ABI5 to regulate ABA response during seed germination (Hu and Yu, 2014). BIN2 can also phosphorylate SnRK2.3, a positive regulator of ABA signaling, on T180 to promote its kinase activity to promote ABA signaling (Cai et al., 2014). Wang et al. (2018) found that ABA inhibits BR signaling by ABI1 and ABI2, which dephosphorylate BIN2 to regulate its activity. Here, we find another transcription factor of the ABA signaling pathway, ABI4, is also involved in BR response through binding and inhibiting the expression of BAK1. ABI4 was an ABA-responsive gene, the BR responsiveness of ABI4 and the BR-related phenotypes indicated ABI4 might also participate in the cross talk between BR and ABA. ABA could probably regulate BR signaling through ABI4 regulating the gene expression of BAK1. While few studies have detected the cross talk between BAK1 and other proteins, it should be a new start of future work.
We also identified that genes involved in DNA replication initiation, cutin biosynthetic process, and chloride transmembrane transport were significantly upregulated, whereas genes involved in response to wounding, response to chitin, response to jasmonic acid, defense response to insect, response to karrikin, and ethylene-activated signaling were significantly downregulated. BR regulates cell elongation and cell division synergistically with another plant hormone. Here, we report that in BR-treated cpd seedlings, there are significant changes in the expression of 35 genes involved in DNA replication, 60 wounding response genes, 48 chitin response genes, and 49 jasmonic acid response genes. Although we do not know how BR regulates DNA replication and response to wounding, chitin, jasmonic acid, and karrikin, we believe the data presented here will contribute to future work aiming to describe the mechanisms underlying these regulatory pathways.
Thus, we identified many specific genes’ response to BR at different stages and integrated the BR transcriptional network. From the RNA-seq data, we identified that 110 transcriptional factors specifically respond to BR at different stages, and moreover, we identified that ABI4 participates in the BR response and showed a BR-hypersensitive and BRZ-insensitive phenotype. ChIP-seq assay showed that ABI4 directly binds BAK1 promoter and inhibits its expression. Our work has emphasized the importance of investigating the functional roles of BR-responsive genes and the feasibility of RNA-seq to identify genes and proteins involved in BR-mediated regulation of plant growth.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) plants cpd (salk_023532) and bri1-701 brl1 brl3, abi4-102 (cs3738) were used in this study. Seeds were grown on one-half strength MS medium (pH 5.8) with 0.8% (w/v) agar and 1% (w/v) Suc for 14 d at 22°C. The plants were grown at 22°C under a 16 h light/8 h dark photoperiod.
BRZ and BL Treatment
For the BRZ (Cayman Chemical) and BL (Wako Chemicals) treatment assays, the plants were treated with either 1 μm BL or mock control for 2 h. Wild-type seeds were grown on one-half strength MS medium with 1 μm BRZ, 1 μm BL for 7 d in light and 5 d in dark, followed by observation of the seedlings.
RNA Extraction and RNA-seq
Total RNA was extracted from 7- and 24-d-old seedlings using the QIAGEN RNAprep plant kit. Oligo (dT) magnetic beads were used to enrich the mRNA. mRNA was fragmented using a fragment buffer treatment. The first-strand complementary DNA (cDNA) was synthesized by random hexamer primers using the mRNA fragments the template. Buffer, deoxy-ribonucleoside triphosphate, RNase H, and DNA polymerase I were used to synthesize the second strand. The double-strand cDNAs, purified with QiaQuick PCR extraction kit, were used for end repair and base A addition. Finally, sequencing adaptors were ligated to the fragments. All low-quality reads (FASTq value < 13) were removed, and 3p and 5p adapter sequences were trimmed using genome analyzer pipeline (Fasteris). The remaining low-quality reads with “n” were removed using a Python script. The fragments were purified by agarose gel electrophoresis and PCR amplified to produce the sequencing library. All reads were pair-end sequenced with an average insert size of 160 bp and typical read-length of 90 bp. The RNA-seq was performed using an Illumina HiSeq 2000 platform.
Differential Expression Analysis
The read count data for all genes and samples were imported to edgeR once to perform global normalization, calculation of the common dispersion factor, and then the estimation of gene-specific dispersion parameters (i.e. tag-wise dispersion). Differential gene expression between each pair of BR-treated plants and controls was evaluated by calling the exact test. Raw P values were adjusted to form multiple comparison effects using the q value (false discovery rate) method. The cutoff for significant differential expression was set as > 1.5 absolute fold change and q < 0.01. We used the software package topology-based GO scoring (topGO, v2.26.0) of the R package to conduct GO enrichment analysis, with the gene-to-GO association data obtained from the GO database (submitted by TAIR at April 1, 2016).
Gene Expression Analysis by RT-qPCR
Total RNA was extracted from seedlings using the QIAGEN RNAprep plant kit. First-strand cDNA was synthesized using the iScript gDNA clear cDNA synthesis kit (Bio-Rad). PCR primers were designed using Perlprimer soft. PCRs were performed with a Bio-Rad CFX96 real-time system. PCR reactions were performed in a total volume of 20 μL, with 1 μL of first-strand cDNAs and 0.4 μL of each primer. The amplification program was as follows: 95°C for 2 min, and 39 cycles of 95°C for 15 s, 55°C for the 30 s. The UBOX gene was used as a control to normalize the level of total RNA. Primers for real-time PCR are listed in Supplemental Table S9. The 2−ΔΔCT values from three technical replicates from each biological replicate were used for the statistical analysis (Schefe et al., 2006).
ChIP and ChIP-seq Libraries
For ChIP-seq, 12-d-old seedlings of p35S:ABI4-cFLAG plants were selected for the ChIP experiment. Anti-FLAG (Abmart) was used to precipitate the DNA. ChIP assays were performed as described (Hansen et al., 2017), with minor modifications.
ChIP-Seq Assays and ChIP-qPCR
ChIP sequencing library preparation and data analysis were conducted by LC-Bio. FastQC (v0.11.5; https://www.softpedia.com/get/Science-CAD/FastQC.shtml) was used for quality control analysis of the sequencing reads to generate an next generation sequencing quality control report. Trimmomatic (v0.36; http://www.mybiosoftware.com/trimmomatic-0-30-flexible-read-trimming-tool-illumina-ngs-data.html) was used to clean the raw reads and filter out the adaptor and low-quality reads and alignment of the reads to the genome. The reference genome for Arabidopsis (TAIR10) was downloaded from ftp://ftp.arabidopsis.org/home/tair/Sequences/whole_chromosomes/ (reference genome). For each sample, we mapped the clean sequence reads to the reference genome using the STAR (v2.5.3a) program (https://github.com/alexdobin/STAR/releases). RSeQC (v2.6; https://www.ibp.ucla.edu/research/xiao/RASER.html) was used to evaluate mapped reads distribution, coverage uniformity, and strand specificity. MACS2 (v2.1.1; https://pypi.org/project/MACS2/) was used to call peaks, giving robust and high-resolution ChIP-seq peak predictions. Peaks were annotated as related genes using Homer (v4.10; http://homer.ucsd.edu/homer/). deepTools (v2.4.1) was used to plot gene coverage of the reads near transcription start site and transcription end site. ChIPseeker (v1.5.1) was used to depict the reads distribution on chromosomes. Motif and analyze transcription factors were searched by Homer (v4.10).
Three to five microliter immunoprecipitated products were used for ChIP-qPCR. Each immunoprecipitation was performed three times independently, with the input being used as the control. The primers for ChIP-qPCR are listed in Supplemental Table S9.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: BRI1 (AAC49810.1); CPD (NM120651); DWF4 (AF044216); SAUR-AC1 (S70188.1); and ABI4 (AF040959.1).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Comparison among BR-regulated genes with BES1 target genes, and BZR1 target genes.
Supplemental Figure S2. Genome-wide binding profiles from ChIP-seq analysis.
Supplemental Figure S3. Direct targets of ABI4.
Supplemental Table S1. Genes differentially expressed in cpd versus BL treated cpd.
Supplemental Table S2. Genes differentially expressed in 7-d-old cpd versus BL-treated 7-d-old cpd.
Supplemental Table S3. Genes differentially expressed in 24-d-old cpd versus BL-treated 24-d-old cpd.
Supplemental Table S4. Genes specifically up-regulated by BR at different stage.
Supplemental Table S5. Genes specifically down-regulated by BR at different stage.
Supplemental Table S6. Comparison of BES1 and BZR1 target genes with BR-responsive genes.
Supplemental Table S7. Transcription factors responsive to BR.
Supplemental Table S8. Putative target genes of ABI4 in Arabidopsis seedlings.
Supplemental Table S9. Primers used in this study.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
The authors thank Dr. Jia Li for seeds of Arabidopsis bri1-701 brl1 brl3 mutants, the Arabidopsis Biological Resource Center for seeds of cpd and abi4-102, and the PSC sequence core facility for the RNAseq. They also thank additional members of Dr. Li’s lab for stimulatory discussion throughout this study.
Footnotes
This work was supported by the National Natural Science Foundation of China (grant nos. 31870253 to J.L. and 31600219 to X.L.) and the Shanghai Center for Plant Stress Biology to J.L.
Articles can be viewed without a subscription.
References
- Cai Z, Liu J, Wang H, Yang C, Chen Y, Li Y, Pan S, Dong R, Tang G, de Dios Barajas-Lopez J, et al. (2014) GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc Natl Acad Sci USA 111: 9651–9656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-García S, Cheng JC, Nam KH, Li J, Chory J(2004) BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 131: 5341–5351 [DOI] [PubMed] [Google Scholar]
- Chandler JW.(2018) Class VIIIb APETALA2 ethylene response factors in plant development. Trends Plant Sci 23: 151–162 [DOI] [PubMed] [Google Scholar]
- Friedrichsen DM, Nemhauser J, Muramitsu T, Maloof JN, Alonso J, Ecker JR, Furuya M, Chory J(2002) Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 162: 1445–1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M(2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S(2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S(2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 130: 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Li L, Ye H, Yu X, Algreen A, Yin Y(2009) Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc Natl Acad Sci USA 106: 7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X(2017) CTCF and cohesin regulate chromatin loop stability with distinct dynamics. eLife 6: 25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY(2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY(2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Yu D(2014) BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26: 4394–4408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chory J(1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 90: 929–938 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH(2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J(2001) BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol 127: 14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y(2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J 58: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T(2002) Brassinosteroid-regulated gene expression. Plant Physiol 129: 1241–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J(2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H(2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression’s CT difference” formula. J Mol Med (Berl) 84: 901–910 [DOI] [PubMed] [Google Scholar]
- Steber CM, McCourt P(2001) A role for brassinosteroids in germination in Arabidopsis. Plant Physiol 125: 763–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan XY, Cao DM, Tang W, He K, Zhu JY, He JX, Bai MY, Zhu S, Oh E, et al. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Tang J, Liu J, Hu J, Liu J, Chen Y, Cai Z, Wang X(2018) Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol Plant 11: 315–325 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, et al. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Xie Z, Nolan T, Jiang H, Tang B, Zhang M, Li Z, Yin Y(2019) The AP2/ERF transcription factor TINY modulates brassinosteroid-regulated plant growth and drought responses in Arabidopsis. Plant Cell 31: 1788–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Li L, Guo H, Yin Y(2012) MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc Natl Acad Sci USA 109: 20142–20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J(2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J(2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, et al. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang D, Ye H, Guo H, Johnson A, Zhang M, Lin H, Yin Y(2014) Transcription factor HAT1 is phosphorylated by BIN2 kinase and mediates brassinosteroid repressed gene expression in Arabidopsis. Plant J 77: 59–70 [DOI] [PubMed] [Google Scholar]
- Zhang S, Cai Z, Wang X(2009) The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc Natl Acad Sci USA 106: 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SS, Yang H, Ding L, Song ZT, Ma H, Chang F, Liu JX(2017) Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 29: 1007–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]