For >60 years, percutaneous kidney biopsy specimens have been evaluated by light microscopy, immunofluorescence (IF), and electron microscopy to diagnose medical kidney diseases. Shortly thereafter, immunohistochemistry and in situ hybridization were developed to analyze proteins and nucleic acids, respectively, but only minor advances in diagnostic methods have been made since. Although biopsy specimens provide important information about diagnosis, prognosis, and response to treatment, there are inherent limitations in the current methods of interpretation. Accuracy and reproducibility of findings are suboptimal; for example, interobserver reproducibility is only in the moderate κ coefficient range of agreement for lupus nephritis, despite published definitions (1). Even something as fundamental as counting glomeruli is unreliable in clinical and research settings (2). Providing reproducible findings, such as tubulointerstitial fibrosis, requires careful biopsy specimen assessment, and this can be time consuming. Therefore, more reliable and accurate methods for determining kidney biopsy specimen findings with greater speed and reproducibility are of great interest.
The existing terminology in artificial intelligence (AI) is complex and seemingly overlapping. AI encompasses machine learning and deep learning. Machine learning uses computer algorithms to identify expert-defined and labeled data patterns (“ground-truth” annotations, such as glomeruli in digitized whole-slide images). This training is applied in future decision making (“supervised learning”) and improves with continuous exposure to new annotated data. Deep learning passes data through multiple layers of processing and transformation (neural networks), and can extract pertinent features without human intervention (“self-” or “unsupervised” learning). Due to a more complex algorithmic structure, deep learning is capable of identifying more complicated and subtle data features and relations. Convolutional neural networks are a subclass of deep and machine learning where multiple layers of processing mimic the neural connectivity of the human visual cortex and are better suited to complex tasks, such as image recognition (Figure 1) (3). In fact, convolutional neural networks can identify and analyze not only known histologic features, but also novel and subvisual features not typically considered of diagnostic value or apparent to the human eye.
Figure 1.
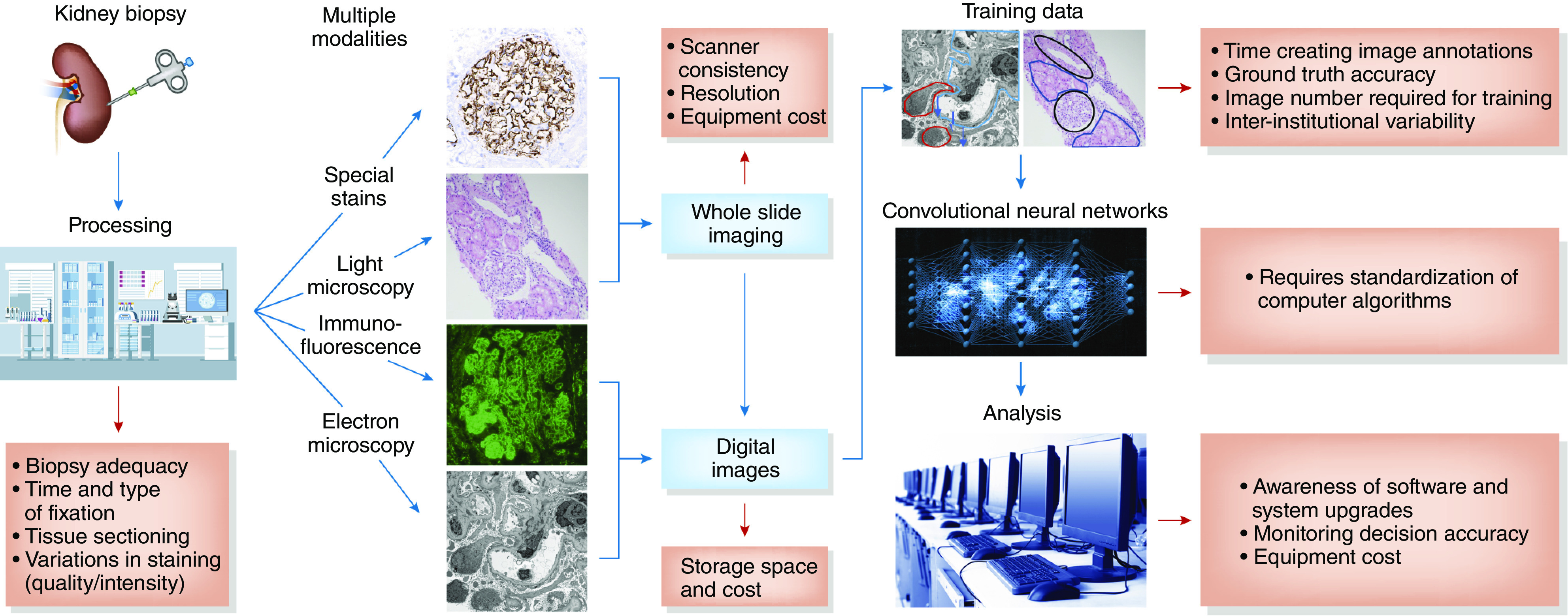
There are correlative obstacles (red arrows) in the proposed workflow (blue arrows and boxes) using convolutional neural networks in the evaluation of kidney biopsy specimens. These obstacles to reproducible and accurate clinical implementation are detailed in the red boxes.
In this issue of CJASN, the study by Ligabue et al. (4) describes one of the first attempts of applying convolutional neural networks to glomerular IF interpretation in kidney biopsy specimens. The ground truth for eight immune reactant locations (mesangial, capillary wall), distributions (segmental, global), appearances (coarse and fine granular), and intensities (0–3+) were taken from 2542 kidney biopsy specimen reports generated over 18 years by two of the study pathologists. Images of glomeruli for all immune reactants from each case, taken at the same exposure time, were used for training (11,059), validation (200), and test (1000) sets. Additionally, 180 images were assessed by three pathologists and convolutional neural networks. Convolutional neural networks correctly identified glomeruli in the images, an important first step. Compared with ground truth, the best performance for convolutional neural networks was location identification as mesangial or continuous regular capillary wall with good to excellent accuracy (0.84, 0.81), area under the curve (AUC; 0.89, 0.87), and fair recall (sensitivity; 0.78, 0.77). Distribution identification as global or segmental had good results for accuracy (0.82, 0.81) and AUC (0.89, 0.81), and fair to good results for recognizing global staining (recall, 0.74; precision [positive predictive value], 0.87; F1 score [overall accuracy measure], 0.79), but was poor for segmental distribution (recall, 0.50; precision, 0.36; F1 score, 0.42). Appearance had good to excellent accuracy (0.84, 0.94) and AUC (0.85, 0.83) for coarse and fine granular staining by convolutional neural networks, although some performance measures were poor (precision, 0.34, 0.43; F1 score, 0.44, 0.47). The authors did not address other IF patterns, including pseudolinear/linear, discontinuous capillary wall, amorphous/globular (i.e., IgM in segmental sclerosis, amyloid, thrombi), and urinary space (i.e., fibrin in crescents) staining. It is possible that training on more images with these findings would allow accurate identification.
The intensity of staining was determined by convolutional neural networks as the highest probability for the ground-truth intensity score. Predictions of correct intensity were overall good, with the best predictions for 0 and 3+, and the furthest from the actual value for 1.5+ and 2.5+. For overall interpretation comparing ground truth, three pathologists, and convolutional neural networks using the Cohen κ coefficient, all comparisons essentially showed moderate agreement (0.39–0.56), with convolutional neural networks equal to pathologist performance. However, analysis by convolutional neural networks was much faster than the pathologists’ analysis (62.5 milliseconds versus 7.3+1.2 seconds per image). A confounding factor for intensity prediction is the accuracy of ground truth given variability in antibody staining and microscope filament intensity, image burnout under viewing, camera exposure consistency, and human interpretation. In this study, two pathologists provided the initial ground-truth reports, yet, on image review, had only moderate κ scores when compared with ground truth. Therefore, it is unknown if the moderate convolutional neural networks scores were due to suboptimal AI performance or due to inconsistencies in the ground-truth intensity scores. The issue of ground-truth accuracy is recognized for supervised learning and needs to be addressed before widespread use of convolutional neural networks can be implemented.
AI has supported prediction, classification, and clinical decision making in several medical fields (5). In oncopathology, for example, convolutional neural networks can simultaneously incorporate a wide range of adjunctive testing, demonstrated in analyses of histology with cancer biomarkers and quantitative immunohistochemical staining to provide diagnostic and prognostic information for a variety of malignancies (6). The early applications of AI in nephropathology used computer algorithms to successfully relate single histologic parameters such as interstitial fibrosis to clinical outcomes in CKD. While this association is not novel, this study demonstrated that convolutional neural networks can outperform pathologists in determining the percentage of estimated interstitial fibrosis (7). Use in glomerular diseases has required more sophisticated approaches. In a study by Naumovic et al. (8) looking at idiopathic membranous nephropathy, multiple semiquantifiable and machine-assessed histologic factors (global glomerulosclerosis, tubulointerstitial scarring) were integrated with demographic, clinical, and biochemical parameters to evaluate different therapies and clinical outcomes. Ginley et al. (9) used convolutional neural networks to classify diabetic glomerulosclerosis through analysis of glomerular color, texture, inter- and intrastructural distances, and feature containment. They found moderate agreement of convolutional neural networks with ground truth and pathologists, and similar moderate agreement among pathologists (9). This is similar to the results of the study by Ligabue et al. (4).
There are several challenges to using AI in assessment of kidney biopsy specimens (Figure 1). These include accounting for preanalytic variables, such as tissue fixation and preparation, stain coloration and intensity, and scanning quality and consistency (image capture) (10). Accurate training of convolutional neural networks requires knowing how much training data are required. There are several well-curated, digitized slide sets that could be used; however, it takes considerable time to perform the reproducible ground-truth annotations necessary for effective pattern recognition. The training dataset also must include reliable data for light, IF, and electron microscopy, and other special stains. Seamless integration of AI into routine pathologic evaluation must be generalizable, requiring standardization of computer algorithms across different institutions and vigilant equipment maintenance, including monitoring of computer operating systems and software upgrades. Additionally, the algorithms deep learning uses are “black boxes” that currently are not truly understood, raising clinical, regulatory, and legal concerns about how decisions are reached.
The application of AI to kidney biopsy specimen interpretation is in its infancy, and there are many obstacles to overcome before wide use in routine clinical practice is possible. However, there are profound potential benefits—including accuracy, reproducibility, incorporating important novel or subvisual features, and speed—which can provide more diagnostic information, treatment targets, and outcome predictions in glomerular and other kidney diseases. In this regard, the paper by Ligabue et al. (4) moves us one step closer to this goal. However, achieving this goal will require continuous oversight for quality control and assurance around the different components of tissue preparation, digital imaging, and computational analysis. In addition, there will be a continued need for nephropathologists to ensure synthesis of computationally generated data with clinical, molecular, and likely other findings to provide optimal, clinically useful information. Thus, AI can be leveraged to enhance research and clinical practice in nephrology. We should welcome these advances while being mindful of the effort necessary for careful monitoring and appropriate use in the care of patients with kidney disease.
Disclosures
All authors have nothing to disclose.
Funding
None.
Acknowledgments
The content of this article reflects the personal experience and views of the author(s) and should not be considered medical advice or recommendation. The content does not reflect the views or opinions of the American Society of Nephrology (ASN) or CJASN. Responsibility for the information and views expressed herein lies entirely with the author(s).
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
See related article, “Evaluation of the Classification Accuracy of the Kidney Biopsy Direct Immunofluorescence through Convolutional Neural Networks,” on pages 1445–1454.
References
- 1.Furness PN, Taub N: Interobserver reproducibility and application of the ISN/RPS classification of lupus nephritis-A UK-wide study. Am J Surg Pathol 30: 1030–1035, 2006. [DOI] [PubMed] [Google Scholar]
- 2.Rosenberg AZ, Palmer M, Merlino L, Troost JP, Gasim A, Bagnasco S, Avila-Casado C, Johnstone D, Hodgin JB, Conway C, Gillespie BW, Nast CC, Barisoni L, Hewitt SM: The application of digital pathology to improve accuracy in glomerular enumeration in renal biopsies. PLoS One 11: e0156441, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serag A, Ion-Margineanu A, Qureshi H, McMillan R, Saint Martin MJ, Diamond J, O’Reilly P, Hamilton P: Translational AI and deep learning in diagnostic pathology. Front Med (Lausanne) 6: 185, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ligabue G, Pollastri F, Fontana F, Leonelli M, Furci L, Giovanella S, Alfano G, Cappelli G, Testa F, Bolelli F, Grana C, Magistroni R: Evaluation of the classification accuracy of the kidney biopsy direct immunofluorescence through convolutional neural networks. Clin J Am Soc Nephrol 15: 1445–1454, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lisboa PJ: A review of evidence of health benefit from artificial neural networks in medical intervention. Neural Netw 15: 11–39, 2002. [DOI] [PubMed] [Google Scholar]
- 6.Khosravi P, Kazemi E, Imielinski M, Elemento O, Hajirasouliha I: Deep convolutional neural networks enable discrimination of heterogeneous digital pathology images. EBioMedicine 27: 317–328, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolachalama VB, Singh P, Lin CQ, Mun D, Belghasem ME, Henderson JM, Francis JM, Salant DJ, Chitalia VC: Association of pathological fibrosis with renal survival using deep neural networks. Kidney Int Rep 3: 464–475, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naumovic R, Furuncic D, Jovanovic D, Stosovic M, Basta-Jovanovic G, Lezaic V: Application of artificial neural networks in estimating predictive factors and therapeutic efficacy in idiopathic membranous nephropathy. Biomed Pharmacother 64: 633–638, 2010. [DOI] [PubMed] [Google Scholar]
- 9.Ginley B, Lutnick B, Jen KY, Fogo AB, Jain S, Rosenberg A, Walavalkar V, Wilding G, Tomaszewski JE, Yacoub R, Rossi GM, Sarder P: Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 30: 1953–1967, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Acs B, Rantalainen M, Hartman J: Artificial intelligence as the next step towards precision pathology. J Intern Med 288: 62–81, 2020. [DOI] [PubMed] [Google Scholar]


