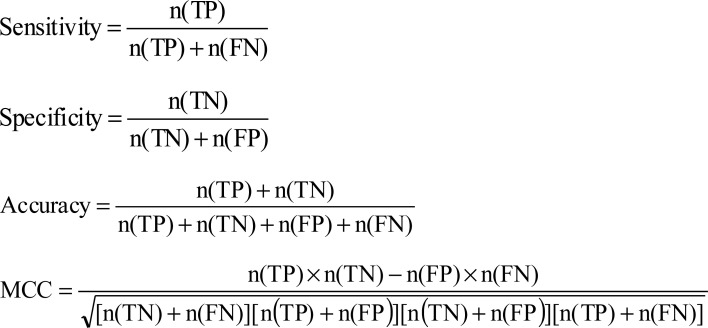Abstract
Protein-protein interactions (PPIs) are the physical connections between two or more proteins via electrostatic forces or hydrophobic effects. Identification of the PPIs is pivotal, which contributes to many biological processes including protein function, disease incidence, and therapy design. The experimental identification of PPIs via high-throughput technology is time-consuming and expensive. Bioinformatics approaches are expected to solve such restrictions. In this review, our main goal is to provide an inclusive view of the existing sequence-based computational prediction of PPIs. Initially, we briefly introduce the currently available PPI databases and then review the state-of-the-art bioinformatics approaches, working principles, and their performances. Finally, we discuss the caveats and future perspective of the next generation algorithms for the prediction of PPIs.
Keywords: Protein-protein interactions, PPIs database, sequence features, feature selection, machine learning, bioinformatics
1. Introduction
Protein-protein interactions (PPIs) are the physical connections between two or more proteins via electrostatic forces or hydrophobic effects [1]. PPIs play a vital role in diverse biological developments, including immune response, DNA transcription and replication, metabolic cycles, and signal transduction pathways [2-4]. To identify the PPIs responsible for such concerted functions is needed [4-6]. Different studies have suggested that PPIs occur between two species such as human-bacteria, human-virus, and plant-pathogen [7-12]. As a result, an understanding of the molecular mechanisms involved in PPIs is very critical for the design of new medicine and therapeutic targets.
Proteins often form complexes with other proteins to perform certain tasks [13-15]. PPIs occur at almost every level of cellular functions and provide a global picture of biological progressions [12, 15, 16]. Particularly, a protein complex with multiple subunits [4, 17-20] assists as an efficient subnetwork inside the whole PPI networks [3, 21]. Due to the development of high-throughput sequencing technologies, the identification of PPIs in specific species (PPIs of intraspecies), confirmed by extensive experiments, has enlarged [3, 22-25]. On the other hand, the identification of PPIs between different species (PPIs of interspecies) is limited. While the identification of PPI in both the intra- and inter-species is required for understanding biological functions, mechanisms by which PPI affects the functions of a cell remains to be revealed [26-32]. Many large-scale experiments have been achieved to identify PPIs based on the molecular signature proteins [18, 32-39]. The experimental investigations are often laborious and time-consuming, making it difficult to perceive all potential PPIs. All these restrictions could be solved by bioinformatics approaches in the era of artificial intelligence.
Traditional computational algorithms of intraspecies PPIs are often used to deduce the possible associations of interrogating protein pairs [40-44]. These approaches are usually denoted as the interlog mapping [43, 45], the DDI-based method [41, 42] and the DMI-based method [40]. Meanwhile, in recent decades, machine learning (ML)-based approaches have been booming [10, 46-50] that use the amino acid sequence [51, 52], evolutionary profiles [53, 54], physicochemical properties [47, 55], and structure information [56] of the protein pairs. The interspecies PPI prediction is a relatively earlier stage research topic and more challenging task than the intraspecies PPI prediction. Recently, some of the interspecies prediction models have been developed with increases in experimentally verified data [10, 21, 57].
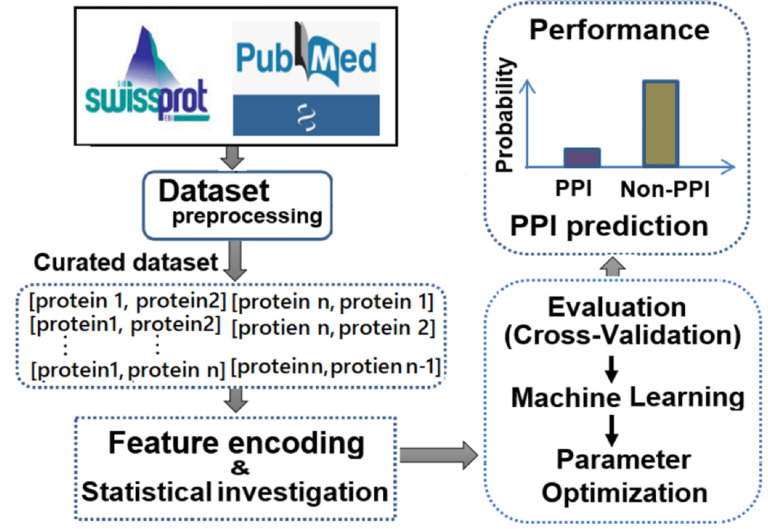
In this review, we provide an inclusive assessment of the state-of-the-art ML approaches for sequence-based PPIs prediction, as shown in Fig. (1), and discuss their benefits and shortcomings to aid readers select the best PPI predictor for their purpose. Moreover, we present the future perspectives of ML-based PPI predictions.
Fig. (1).
A general framework of ML-based PPI prediction. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2. Databases of PPIS
Many PPI databases are available, e.g., APID [58], TAIR [59], HIPPIE [60], PPIM [61], BioGrid [62, 63], and DIP [64] (Table 1). The APID is the most updated, available database, which delivers an inclusive and curated assortment of PPIs for over 1100 organisms. It includes more than 500 experimentally identified PPIs for each of 30 species.
Table 1.
Currently available databases for PPIs.
| Database | Description | Year | Database URL |
|---|---|---|---|
| DIP | Several species PPIs that are manually curated | 2002 | https://dip.doe-mbi.ucla.edu/dip/Main.cgi |
| TAIR | PPI annotations for Arabidopsis thaliana | 2007 | https://www.arabidopsis.org/portals/proteome/proteinInteract.jsp |
| PPIM | PPI database for Maize | 2016 | comp-sysbio.org/ppim/ |
| PPIM | 2,762,560 interactions among 14,000 proteins | 2016 | https://dbaasp.org/home |
| HIPPIE | Human PPI references | 2017 | http://cbdm.uni-mainz.de/hippie/ |
| BioGRID | 400,000 PPIs collected from the experimentations and primary literatures | 2018 | https://openwetware.org/wiki/Protein-protein_interaction_databases#BioGRID |
| APID | Agile protein intercoms database for bacterial PPIs | 2019 | http://compsysbio.org/bacteriome/ |
| APID | It integrates the existing public resources and provides PPI information of more than 1100 organisms | 2019 | http://apid.dep.usal.es |
3. Dataset preprocessing
To build a high-quality dataset is a crucial step for the sequence-based PPI prediction via ML algorithms. The datasets are normally collected from the Swiss-Prot /UniProtKB. In particular, the experimentally identified PPI pairs were considered as positive samples. Sequentially, all the positive pairs of PPIs were randomly crossed to make negative samples, assuming that the randomly shuffled proteins are very unlikely to be positive PPIs. The optimal numbers of negative samples were considered on training data through several statistical investigations [65]. Then the redundancy of the curated sequence datasets was considered. If two sets of PPIs contain similar sequences, either of them is deleted. Recently, Sun et al. used different subcellular locations for generating the negative samples, while considering the experimentally verified PPIs as positive samples [49]. They used the non-interaction pairs as negative samples by pairing proteins in diverse subcellular locations. First, the Swiss-Prot database (version 57.3) was used. Second, the annotated sequences with uncertain or indeterminate subcellular location terms, such as “possible”, “maybe”, “potential”, or “by similarity”, were accessed from the human protein. Finally, two or more locations were excluded from the annotated sequences. Due to the possibility of sequence homolog, 50% homology reduction was performed. This approach had a slight advantage over the random generation of negative samples.
To establish a computational tool for accurately predicting PPIs, one of the major challenges is to handle imbalance positive and negative samples [46]. To solve the potentially imbalanced problem, the negative PPI samples are randomly pooled from the entire negative samples to keep a ratio of positive to negative samples [61]. However, exact solutions of dataset imbalance problems are still indispensable issues.
Overfitting and underfitting problems may exist in the datasets. When the datasets are highly homologous, they can cause overestimation in the prediction model. Generally, scientists cluster the composed protein sequences with an identity threshold of 60%, 50%, 40%, and 30% by using CD-HIT [66] or BlasClust (http://nebc.nox.ac.uk/bioinformatics/docs/blastclust.html) to solve the bias problems. However, the curated datasets may contain some correlated sequences that the CD-HIT and BalstClust miss. Exact sequence homolog reduction methods are still an important issue. Another problem is data underfitting, when the prediction model uses a very small dataset as the input. Therefore, the dataset should not be too small.
4. Sequence encoding methods
Generally, PPI prediction methods created the input data by combining the two feature vectors of protein pairs in a row [67-69]. Feature encoding is one of the major phases for predicting PPIs that encodes protein pairs as numeric feature vectors. Appropriate feature descriptors enable to accurately predict PPIs. Recently, a number of computational approaches have been developed as an alternative to experimental methods for identifying potential PPIs (Table 2). Due to the sequence diversity, some of the PPIs may be feeble [70-77]. To solve this issue, the PSI-BLAST [78, 79] can be used to produce an outline profile by using a position-specific scoring matrix (PSSM). The given profiles reproduce the variation and conservation through the evolutionary information between protein sequences [80, 81]. These appearances may be suitable for a particular PPI classification problem. Khatun et al. have used autocorrelation and amino acid compositional features for analyzing Zea mays PPI sequences [46]. Recently, the DLPred used diverse sequence information including PSSM, Hydropathy index (HI), AAindex, conservation scores and 3D-1D scores. Chou’s pseudo amino acid composition (PseAAC) is used to encode the positional-wide composition of PPIs [82].
Table 2.
Currently available tools for PPI prediction.
| Predictor |
ML
Algorithms |
Encoding Methods | Testing Methods | Accuracy | Year | Predictor URL | References |
|---|---|---|---|---|---|---|---|
| Pred_PPI | SVM | Auto covariance | Jackknife | 90.67% (human), 88.99% (yeast), 90.09% (Drosophila), 92.73% (E. coli), 97.51% (C. eleganse) |
2010 | http://cic.scu.edu.cn/bioinfor-matics/predict_ppi/default.html | [72] |
| Hotpoint | SVM | PseAAC and local alignment kernel | 5-fold CV | 70% | 2010 | http://prism.ccbb.ku.edu.tr/hotpoint/ | [89] |
| PSOPIA | Domain-based | Sequence similarity | 10-fold CV | 70-85% | 2014 | http://mizuguchilab.org/PSOPIA | [80] |
| NIP | SVM | G-gap dipeptide compositions | Jackknife | 92.67% | 2016 | http://mlda.swu.edu.cn/codes.php?name=NIP | [70] |
| SPRINT | SVM | k-mer | 10-fold | N/A | 2017 | https://github.com/lucian-ilie/SPRINT/ | [71] |
| SIPMA | RF | Autocorrelation, AAC, PseAAC |
10-fold CV | 89.9% | 2018 | http://kurata14.bio.kyutech.ac.jp/SIPMA/ | [46] |
| DPPI | Deep learning | Sequence features | 10-fold CV | 96% | 2018 | https://github.com/hashemifar/DPPI/ | [77] |
| PPI-Detect | SVM | BPF and sequence features | 10-fold CV | 91.40% | 2018 | https://ppi-detect.zmb.uni-due.de/ | [47] |
| DLPred | Deep learning | PSSM, HI, AAindex, sequence conservation score, and 3D-1D scores. | 10-fold CV | 73.68% | 2019 | http://qianglab.scst.suda.edu.cn/dlp/ | [75] |
| GWORVMBIG | Optimizer-Based Relevance Vector Machine | PSSM and evolutionary encoding | 5-fold CV | NA | 2019 | http://219.219.62.123:8888/GWORVMBIG | [76] |
| DAMpred | Neural-Network | Protein structure encoding | 10-fold | 86% | 2019 | https://zhanglab.ccmb.med.umich.edu/DAMpred | [73] |
| FCTP-WSRC | SVM and Weighted sparse leraning | Auto covariance and KNN | 5-fold CV | 96.67%, 99.82%, and 98.09% for H. pylori, Human and Yeast | 2020 | https://github.com/wowkiekong/PPI-prediction | [74] |
Several physicochemical features available for PPI prediction are introduced, including hydropathy indexes, physical properties, physicochemical characteristics, pKa, conservation score, and 3D-1D scores. The employed physicochemical features are a pKa value of the amino acid residues, hydrophobicity/hydrophilicity, negatively/positively charged, and uncharged residues, a volume of amino acid side chains, and control of functional groups such as methyl, benzyl, and thioether groups. In the amino acid index (AAindex) database, 544 physicochemical properties are stored as numerical indexes [83, 84]. Khatun et al. proposed a sequence-based algorithm using autocorrelation (AC) for PPIs prediction in Zea Mays [46]. One advantage of this method is that AC considers long-range interaction features of amino acids which are responsible for PPI identification. Recently, several structure-based prediction methods have employed the domain information, secondary structure states, polar surface locations, solvent accessibility and hydrophobicity [25].
Generally, the features are extracted by the two different structure-and sequence-based methods. In most cases of PPIs, the protein sequence data has been used more often than the structure data. There is an alarming limitation because proteins are essentially stated as sequences with unfixed length. In the PPI identification, it is necessary to fix the sequence size s (s is the static number with D dimensions of amino acids information). Thus, every PPI sequence is signified as a feature vector of size Dxs. When the length of a PPI sequence is shorter than s, zero is added to the remaining elements of the feature vector. It requires a long computational time to generate the feature vectors. Furthermore, many ML algorithms classify the high-dimensional dataset very properly. Therefore, precise sequence encoding schemes are necessary for valuable perdition.
Furthermore, several domain-based approaches have been developed [85-90] that use the domain-domain interaction scores evaluated by diverse ML algorithms including a relevance vector machine and SVM. These approaches consider the proportion of an important domain or domain co-occurrence relationships, but they do not employ the entire domain evidence [84], which is crucial to the understanding a global view of the PPI.
5. Machine learning algorithm
To detect the potential PPIs via the sequence-based prediction models, several ML algorithms are employed, such as deep learning (DL), support vector machines (SVM), and random forest (RF). Most of the existing predictors use three types of ML algorithms: DL, SVM, and RF. The description of these algorithms is as follows.
5.1. Deep Learning
Deep learning (DL) consists of several approaches including Recurrent Neural Networks (RNN), Deep Belief Networks (DBNs), and Deep Neural Networks (DNN). Different DL algorithms are suitable for different specific applications. For instance, to the analysis of sequential information, RNNs are appropriate. The DBNs are decent at examining inside associations in high-dimensional data. To predict PPIs, DNN is one of the most suitable ML algorithms [49]. The DNN input should be the vectors with a fixed dimension. The main parts of the DNN component are to remove highly homologous samples and eliminate noise, and to decrease data dimensions. DNN architectures are assembled layer-by-layer with a greedy algorithm. DNN helps to pick out unravel features to improve performance.
5.2. Support Vector Machine
To classify the PPI datasets, SVM or kernel machines are used [89]. The SVM maximizes the margins that are related to the inevitability of its classification. The objective of this classifier is likely to have small margins [90] using a labeled of the training dataset. SVM is very influential and can classify problems with random density information, although it needs large memory requirements and complex format. The SVM is a little bit slow to train and assess the high dimensional features via radial basis function kernel. Another disadvantage is that the parameters significantly alter the results. We refer to more details [90-92].
5.3. Random Forest
The RF algorithm involves numerous ensemble decision trees that categorizes the two-class prediction problem [93-97]. On the training model, each decision tree is built using the casual feature vectors that are sampled from a dataset in every node in a tree independently. Then each classification tree is entirely grown via randomly selected variables. To categorize a new entity, the response vector keeps each of the trees in the forest. Allowing the majority voting, one class is allocated to the entity. The RF is an effective algorithm when there exist a large number of features and
datasets, and can rank important features for accurate classification [98, 99]. The RF is widely used in computational biology research [46, 90, 99-103].
5.4. Combined Model
For a real-world prediction task, the feature sets are combined to enhance the prediction performance [104-110]. The feasibility of different feature sets is evaluated by diverse statistical learning algorithms. Then the evaluation scores are integrated by using various statistical strategies such as logistic regression [111], weight score [112] and multiple linear regression [113]. Moreover, recently the meta-classifiers (e.g. combined different ML algorithms) have widely been used in bioinformatics research to enhance the prediction performance [100, 114].
6. Evaluation
6.1. Measure
To examine the performance of different ML classifiers, many statistical measurements were used, including accuracy, specificity, sensitivity, and Matthew`s correlation coefficient (MCC). These assume a two-class binary classification problem, in which the outputs (PPI or non-PPI) are categorized either as PPI (+) or non-PPI (-). Four consequences will be provided (Table 3). True positive (TP) signifies that the real value is ‘+’ and predicted class is ‘+’; false positive (FP) signifies that the real value is ‘-’ and predicted class is ‘+’. False negative (FN) occurs when the real value is ‘+’ and outcome is ‘-’; true negative (TN) occurs when both the real and prediction results are ‘-’.
Table 3.
Contingency table.
| Confusion Matrix or 2×2 Contingency Table | |||
|---|---|---|---|
| Tested/Estimated/Predicted Results | Total Samples | True Condition | |
| Positive (+) | Negative (-) | ||
| Positive (+) | n(TP) | n(FP) | |
| Negative (-) | n(FN) | n(TN) | |
n(TP) and n(FP) represent the numbers of correctly and incorrectly predicted positive samples, respectively. n(TN) and n(FN) represent the numbers of the correctly and incorrectly predicted negative samples, respectively.
The four measures are defined by:
The values of sensitivity, specificity, and accuracy lie between 0 and 1 and MCC between -1 and 1, a higher value signifies better estimate.
6.2. Parameter Optimization
After applying ML algorithms, threshold value selection is an important step for the precise prediction of PPIs and non-PPIs. The performance of the prediction model by using the training samples was assessed with a stepwise change in specificity [46, 115, 116]. Typically, high specificity decreases sensitivity. Users need to set different threshold values in their algorithms to understand the exact level of performance. However, existing methods did not set different threshold values, but used a fixed threshold value so that the specificity or sensitivity value was within a certain range. In this case, ordinary users cannot understand exact performances. Therefore, developers should control specificity or sensitivity by changing the threshold of the ML scores via a cross-validation test.
6.3. Training and Independent Datasets
Generally, the independent, test dataset used 10-30% samples randomly selected out of the whole PPI samples and the rest of the samples were considered as a training dataset. To evaluate the model performances, initially, a cross-validation test was executed on the training data [117, 118]. In this process, the samples are separated into n sub-groups, and each group is consecutively evaluated n times after training with the other groups. For example, the training dataset is divided into 10 groups. It is an ordinarily accepted number. Among the 10 groups, one group was selected for a test and the other 9 groups were used for training. The predicted PPIs with maximal scores were set to positive samples and the PPIs with low scores were regarded as negative samples. Particularly, a jackknife or a 10-fold CV test was used to predict existing PPI prediction (Table 2) [119, 120].
7. Caveats of the exiting bioinformatics algorithms
Even though much advancement has been done for the expansion of PPI prediction algorithms [121-129], some challenges and limitations need to be addressed. Firstly, the accuracy reported by CV tests is hard to reproduce, unless the source codes and ML parameters regarding sequence encoding methods are provided. However, if developers provide a standalone program or web application, the performances could be evaluated based on independent datasets. Unfortunately, few reported methods provided their source codes or datasets (Table 2). Therefore, it is highly recommended to provide the datasets and source codes while publishing a new methodology [119]. Secondly, most existing algorithms removed identical sequences and considered the remaining proteins as a dataset. A few studies have used the dataset including the proteins showing higher sequence identity (>30%). Using such high sequence similarity dataset might cause overfitting problems and overestimate the prediction accuracy. Hence, to develop a reliable prediction model, it is highly recommended to utilize low sequence identity cut-off (<30%), which has been extensively used in various sequence-based predictions. Thirdly, most of the publicly available methods use their own independent dataset to assess prediction performances. To conduct a fair comparison, it is essential to build unique or independent dataset. It is necessary to check whether the prediction model identifies unseen PPIs. Finally, half of the existing PPI tools are not publicly available. To get reliable performances without any knowledge of mathematics and statistics, online services are particularly valuable. Therefore, state-of art accessible services or software should be freely accessible to the users.
Future perspectives and conclusion
Due to the advancement in sequencing technology, it is essential to develop computational methods to enable fast and precise prediction of unseen PPIs from a large number of candidate proteins. Several ML-based methods have been proposed (Table 2). A future study requires the construction of unbiased datasets with larger size and independent dataset for validating the proposed models, and the development of new encoding schemes. Of note, it is arguable that the addition of structure-based, side-chain orientation of amino acids or evolutionary information can advance the prediction performance. It is also important to integrate different feature encodings [129-133] such as chemical properties, multivariate mutual information, K-nearest neighbors, and pseudo amino acid configuration and to explore ML algorithms [134-138] including light gradient boosting, extreme gradient boosting, and deep learning.
Acknowledgements
We thank the reviewers for their great comments in helping to improve this manuscript.
Consent for Publication
Not applicable.
Funding
This work was supported by the Grant-in-Aid for Scientific Research (B) (19H04208) and partially supported by the developing key technologies for discovering and manufacturing pharmaceuticals used for next-generation treatments and diagnoses both from the Ministry of Economy, Trade and Industry, Japan (METI) and from Japan Agency for Medical Research and Development (AMED).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.De Las Rivas J., Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLOS Comput. Biol. 2010;6(6):e1000807. doi: 10.1371/journal.pcbi.1000807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu X., Yang Z., Sang S., Lin H., Wang J., Xu B. Detection of protein complexes from multiple protein interaction networks using graph embedding. Artif. Intell. Med. 2019;96:107–115. doi: 10.1016/j.artmed.2019.04.001. [DOI] [PubMed] [Google Scholar]
- 3.Dos Santos Vasconcelos C.R., de Lima Campos T., Rezende A.M. Building protein-protein interaction networks for Leishmania species through protein structural information. BMC Bioinformatics. 2018;19(1):85. doi: 10.1186/s12859-018-2105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caterino M., Ruoppolo M., Mandola A., Costanzo M., Orrù S., Imperlini E. Protein-protein interaction networks as a new perspective to evaluate distinct functional roles of voltage-dependent anion channel isoforms. Mol. Biosyst. 2017;13(12):2466–2476. doi: 10.1039/C7MB00434F. [DOI] [PubMed] [Google Scholar]
- 5.Xiao H., Yang L., Liu J., Jiao Y., Lu L., Zhao H. Protein-protein interaction analysis to identify biomarker networks for endometriosis. Exp. Ther. Med. 2017;14(5):4647–4654. doi: 10.3892/etm.2017.5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Planas-Iglesias J., Marin-Lopez M.A., Bonet J., Garcia-Garcia J., Oliva B. iLoops: a protein-protein interaction prediction server based on structural features. Bioinformatics. 2013;29(18):2360–2362. doi: 10.1093/bioinformatics/btt401. [DOI] [PubMed] [Google Scholar]
- 7.Ammari MG, Gresham CR, McCarthy FM, Nanduri B. 2016. [DOI] [PMC free article] [PubMed]
- 8.Ohue M., Matsuzaki Y., Uchikoga N., Ishida T., Akiyama Y. MEGADOCK: an all-to-all protein-protein interaction prediction system using tertiary structure data. Protein Pept. Lett. 2014;21(8):766–778. doi: 10.2174/09298665113209990050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goel R., Harsha H.C., Pandey A., Prasad T.S. Human Protein Reference Database and Human Proteinpedia as resources for phosphoproteome analysis. Mol. Biosyst. 2012;8(2):453–463. doi: 10.1039/C1MB05340J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lian X., Yang S., Li H., Fu C., Zhang Z. Machine-learning-based predictor of human-bacteria protein-protein interactions by incorporating comprehensive host-network properties. J. Proteome Res. 2019;18(5):2195–2205. doi: 10.1021/acs.jproteome.9b00074. [DOI] [PubMed] [Google Scholar]
- 11.Liu C., Liu L., Zhou C., Zhuang J., Wang L., Sun Y., Sun C. Protein-protein interaction networks and different clustering analysis in Burkitt’s lymphoma. Hematology. 2018;23(7):391–398. doi: 10.1080/10245332.2017.1409947. [DOI] [PubMed] [Google Scholar]
- 12.Hanna E.M., Zaki N., Amin A. Detecting protein complexes in protein interaction networks modeled as gene expression biclusters. PLoS One. 2015;10(12):e0144163. doi: 10.1371/journal.pone.0144163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giurgiu M., Reinhard J., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Ruepp A. CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res. 2019;47(D1):D559–D563. doi: 10.1093/nar/gky973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruepp A., Waegele B., Lechner M., Brauner B., Dunger-Kaltenbach I., Fobo G., Frishman G., Montrone C., Mewes H.W. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2010;38(Database issue):D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaake R.M., Wang X., Huang L. Profiling of protein interaction networks of protein complexes using affinity purification and quantitative mass spectrometry. Mol. Cell. Proteomics. 2010;9(8):1650–1665. doi: 10.1074/mcp.R110.000265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochoa D., García-Gutiérrez P., Juan D., Valencia A., Pazos F. Incorporating information on predicted solvent accessibility to the co-evolution-based study of protein interactions. Mol. Biosyst. 2013;9(1):70–76. doi: 10.1039/C2MB25325A. [DOI] [PubMed] [Google Scholar]
- 17.Marsh J.A., Teichmann S.A. Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 2015;84:551–575. doi: 10.1146/annurev-biochem-060614-034142. [DOI] [PubMed] [Google Scholar]
- 18.Yeh F.L., Tung L., Chang T.H. Detection of protein-protein interaction within an RNA-protein complex via unnatural-amino-acid-mediated photochemical crosslinking. Methods Mol. Biol. 2016;1421:175–189. doi: 10.1007/978-1-4939-3591-8_15. [DOI] [PubMed] [Google Scholar]
- 19.Pham C.D. Detection of protein-protein interaction using bimolecular fluorescence complementation assay. Methods Mol. Biol. 2015;1278:483–495. doi: 10.1007/978-1-4939-2425-7_32. [DOI] [PubMed] [Google Scholar]
- 20.Lavallee-Adam M., Coulombe B., Blanchette M. Detection of locally over-represented GO terms in protein-protein interaction networks. J. Comput. Biol. 2010;17(3):443–457. doi: 10.1089/cmb.2009.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S., Fu C., Lian X., Dong X., Zhang Z. Understanding human-virus protein-protein interactions using a human protein complex-based analysis framework. mSystems. 2019;4(2):e00303–e00318. doi: 10.1128/mSystems.00303-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saha S., Prasad A., Chatterjee P., Basu S., Nasipuri M. Protein function prediction from protein-protein interaction network using gene ontology based neighborhood analysis and physico-chemical features. J. Bioinform. Comput. Biol. 2018;16(6):1850025. doi: 10.1142/S0219720018500257. [DOI] [PubMed] [Google Scholar]
- 23.Zhai J.X., Cao T.J., An J.Y., Bian Y.T. Highly accurate prediction of protein self-interactions by incorporating the average block and PSSM information into the general PseAAC. J. Theor. Biol. 2017;432:80–86. doi: 10.1016/j.jtbi.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Teng W.J., Zhou C., Liu L.J., Cao X.J., Zhuang J., Liu G.X., Sun C.G. Construction of a protein-protein interaction network of Wilms’ tumor and pathway prediction of molecular complexes. Genet. Mol. Res. 2016;15(2):1–9. doi: 10.4238/gmr.15028365. [DOI] [PubMed] [Google Scholar]
- 25.Aumentado-Armstrong T.T., Istrate B., Murgita R.A. Algorithmic approaches to protein-protein interaction site prediction. Algorithms Mol. Biol. 2015;10:7. doi: 10.1186/s13015-015-0033-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taghipour S., Zarrineh P., Ganjtabesh M., Nowzari-Dalini A. Improving protein complex prediction by reconstructing a high-confidence protein-protein interaction network of Escherichia coli from different physical interaction data sources. BMC Bioinformatics. 2017;18(1):10. doi: 10.1186/s12859-016-1422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keane H., Ryan B.J., Jackson B., Whitmore A., Wade-Martins R. Protein-protein interaction networks identify targets which rescue the MPP+ cellular model of Parkinson’s disease. Sci. Rep. 2015;5:17004. doi: 10.1038/srep17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji C., Cao X., Yao C., Xue S., Xiu Z. Protein-protein interaction network of the marine microalga Tetraselmis subcordiformis: prediction and application for starch metabolism analysis. J. Ind. Microbiol. Biotechnol. 2014;41(8):1287–1296. doi: 10.1007/s10295-014-1462-z. [DOI] [PubMed] [Google Scholar]
- 29.Wang L., Tam J.P., Liu D.X. Biochemical and functional characterization of Epstein-Barr virus-encoded BARF1 protein: interaction with human hTid1 protein facilitates its maturation and secretion. Oncogene. 2006;25(31):4320–4331. doi: 10.1038/sj.onc.1209458. [DOI] [PubMed] [Google Scholar]
- 30.Amoutzias G.D., Robertson D.L., Bornberg-Bauer E. The evolution of protein interaction networks in regulatory proteins. Comp. Funct. Genomics. 2004;5(1):79–84. doi: 10.1002/cfg.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ivanic J., Yu X., Wallqvist A., Reifman J. Influence of protein abundance on high-throughput protein-protein interaction detection. PLoS One. 2009;4(6):e5815. doi: 10.1371/journal.pone.0005815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurst R., Hook B., Slater M.R., Hartnett J., Storts D.R., Nath N. Protein-protein interaction studies on protein arrays: effect of detection strategies on signal-to-background ratios. Anal. Biochem. 2009;392(1):45–53. doi: 10.1016/j.ab.2009.05.028. [DOI] [PubMed] [Google Scholar]
- 33.Park H., Kang H., Ko W., Lee W., Jo K., Lee H.S. FRET-based analysis of protein-nucleic acid interactions by genetically incorporating a fluorescent amino acid. Amino Acids. 2015;47(4):729–734. doi: 10.1007/s00726-014-1900-2. [DOI] [PubMed] [Google Scholar]
- 34.Xu B., Guan J., Wang Y., Wang Z. Essential protein detection by random walk on weighted protein-protein interaction networks. IEEE/ACM Trans. Comput. Biol. Bioinformatics. 2019;16(2):377–387. doi: 10.1109/TCBB.2017.2701824. [DOI] [PubMed] [Google Scholar]
- 35.Zaki N., Alashwal H. Improving the Detection of Protein Complexes by Predicting Novel Missing Interactome Links in the Protein-Protein Interaction Network.; Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Annual Conference 2018; 2018. pp. 5041–5044. [DOI] [PubMed] [Google Scholar]
- 36.Liu W., Ma L., Jeon B., Chen L., Chen B. A Network Hierarchy-Based method for functional module detection in protein-protein interaction networks. J. Theor. Biol. 2018;455:26–38. doi: 10.1016/j.jtbi.2018.06.026. [DOI] [PubMed] [Google Scholar]
- 37.Liu T.Y., Chou W.C., Chen W.Y., Chu C.Y., Dai C.Y., Wu P.Y. Detection of membrane protein-protein interaction in planta based on dual-intein-coupled tripartite split-GFP association. Plant J. 2018;94(3):426–438. doi: 10.1111/tpj.13874. [DOI] [PubMed] [Google Scholar]
- 38.Song B., Wang F., Guo Y., Sang Q., Liu M., Li D., Fang W., Zhang D. Protein-protein interaction network-based detection of functionally similar proteins within species. Proteins. 2012;80(7):1736–1743. doi: 10.1002/prot.24066. [DOI] [PubMed] [Google Scholar]
- 39.Subramanian C., Xu Y., Johnson C.H., von Arnim A.G. In vivo detection of protein-protein interaction in plant cells using BRET. Methods Mol. Biol. 2004;284:271–286. doi: 10.1385/1-59259-816-1:271. [DOI] [PubMed] [Google Scholar]
- 40.Pang E., Lin K. Yeast protein-protein interaction binding sites: prediction from the motif-motif, motif-domain and domain-domain levels. Mol. Biosyst. 2010;6(11):2164–2173. doi: 10.1039/c0mb00038h. [DOI] [PubMed] [Google Scholar]
- 41.Singhal M., Resat H. A domain-based approach to predict protein-protein interactions. BMC Bioinformatics. 2007;8:199. doi: 10.1186/1471-2105-8-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dyer M.D., Murali T.M., Sobral B.W. Computational prediction of host-pathogen protein-protein interactions. Bioinformatics. 2007;23(13):i159–i166. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- 43.Burgoyne N.J., Jackson R.M. Predicting protein interaction sites: binding hot-spots in protein-protein and protein-ligand interfaces. Bioinformatics. 2006;22(11):1335–1342. doi: 10.1093/bioinformatics/btl079. [DOI] [PubMed] [Google Scholar]
- 44.Tachiki H., Kato R., Kuramitsu S. DNA binding and protein-protein interaction sites in MutS, a mismatched DNA recognition protein from Thermus thermophilus HB8. J. Biol. Chem. 2000;275(52):40703–40709. doi: 10.1074/jbc.M007124200. [DOI] [PubMed] [Google Scholar]
- 45.Yu H., Luscombe N.M., Lu H.X., Zhu X., Xia Y., Han J.D., Bertin N., Chung S., Vidal M., Gerstein M. Annotation transfer between genomes: protein-protein interologs and protein-DNA regulogs. Genome Res. 2004;14(6):1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khatun M.S., Hasan M.M., Mollah M.N.H., Kurata H. 2018. [Google Scholar]
- 47.Romero-Molina S., Ruiz-Blanco Y.B., Harms M., Münch J., Sanchez-Garcia E. PPI-Detect: A support vector machine model for sequence-based prediction of protein-protein interactions. J. Comput. Chem. 2019;40(11):1233–1242. doi: 10.1002/jcc.25780. [DOI] [PubMed] [Google Scholar]
- 48.An J.Y., You Z.H., Zhou Y., Wang D.F. Sequence-based prediction of protein-protein interactions using gray wolf optimizer-based relevance vector machine. Evol. Bioinform. Online. 2019;15:1176934319844522. doi: 10.1177/1176934319844522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun T., Zhou B., Lai L., Pei J. Sequence-based prediction of protein protein interaction using a deep-learning algorithm. BMC Bioinformatics. 2017;18(1):277. doi: 10.1186/s12859-017-1700-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xia J.F., Han K., Huang D.S. Sequence-based prediction of protein-protein interactions by means of rotation forest and autocorrelation descriptor. Protein Pept. Lett. 2010;17(1):137–145. doi: 10.2174/092986610789909403. [DOI] [PubMed] [Google Scholar]
- 51.Huang Y.A., You Z.H., Chen X., Chan K., Luo X. Sequence-based prediction of protein-protein interactions using weighted sparse representation model combined with global encoding. BMC Bioinformatics. 2016;17(1):184. doi: 10.1186/s12859-016-1035-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eid F.E., ElHefnawi M., Heath L.S. DeNovo: virus-host sequence-based protein-protein interaction prediction. Bioinformatics. 2016;32(8):1144–1150. doi: 10.1093/bioinformatics/btv737. [DOI] [PubMed] [Google Scholar]
- 53.Hamp T., Rost B. Evolutionary profiles improve protein-protein interaction prediction from sequence. Bioinformatics. 2015;31(12):1945–1950. doi: 10.1093/bioinformatics/btv077. [DOI] [PubMed] [Google Scholar]
- 54.Zahiri J., Yaghoubi O., Mohammad-Noori M., Ebrahimpour R., Masoudi-Nejad A. PPIevo: protein-protein interaction prediction from PSSM based evolutionary information. Genomics. 2013;102(4):237–242. doi: 10.1016/j.ygeno.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 55.Zahiri J., Mohammad-Noori M., Ebrahimpour R., Saadat S., Bozorgmehr J.H., Goldberg T., Masoudi-Nejad A. LocFuse: human protein-protein interaction prediction via classifier fusion using protein localization information. Genomics. 2014;104(6 Pt B):496–503. doi: 10.1016/j.ygeno.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Neuvirth H., Raz R., Schreiber G. ProMate: a structure based prediction program to identify the location of protein-protein binding sites. J. Mol. Biol. 2004;338(1):181–199. doi: 10.1016/j.jmb.2004.02.040. [DOI] [PubMed] [Google Scholar]
- 57.Yang S., Li H., He H., Zhou Y., Zhang Z. Critical assessment and performance improvement of plant-pathogen protein-protein interaction prediction methods. Brief. Bioinform. 2019;20(1):274–287. doi: 10.1093/bib/bbx123. [DOI] [PubMed] [Google Scholar]
- 58.Alonso-Lopez D., Campos-Laborie F.J., Gutierrez M.A., Lambourne L., Calderwood M.A., Vidal M., De Las Rivas J. APID database: redefining protein-protein interaction experimental evidences and binary interactomes. Database (Oxford) 2019;••• doi: 10.1093/database/baz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poole R.L. The TAIR database. Methods Mol. Biol. 2007;406:179–212. doi: 10.1007/978-1-59745-535-0_8. [DOI] [PubMed] [Google Scholar]
- 60.Alanis-Lobato G., Andrade-Navarro M.A., Schaefer M.H. HIPPIE v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res. 2017;45(D1):D408–D414. doi: 10.1093/nar/gkw985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu G., Wu A., Xu X.J., Xiao P.P., Lu L., Liu J., Cao Y., Chen L., Wu J., Zhao X.M. PPIM: a protein-protein interaction database for maize. Plant Physiol. 2016;170(2):618–626. doi: 10.1104/pp.15.01821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oughtred R., Stark C., Breitkreutz B.J., Rust J., Boucher L., Chang C., Kolas N., O’Donnell L., Leung G., McAdam R., Zhang F., Dolma S., Willems A., Coulombe-Huntington J., Chatr-Aryamontri A., Dolinski K., Tyers M. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47(D1):D529–D541. doi: 10.1093/nar/gky1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chatr-Aryamontri A., Oughtred R., Boucher L., Rust J., Chang C., Kolas N.K., O’Donnell L., Oster S., Theesfeld C., Sellam A., Stark C., Breitkreutz B.J., Dolinski K., Tyers M. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45(D1):D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xenarios I., Salwínski L., Duan X.J., Higney P., Kim S.M., Eisenberg D. DIP, the Database of Interacting Proteins: a research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002;30(1):303–305. doi: 10.1093/nar/30.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hashemifar S., Neyshabur B., Khan A.A., Xu J. Predicting protein-protein interactions through sequence-based deep learning. Bioinformatics. 2018;34(17):i802–i810. doi: 10.1093/bioinformatics/bty573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fu L., Niu B., Zhu Z., Wu S., Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen K.H., Wang T.F., Hu Y.J. Protein-protein interaction prediction using a hybrid feature representation and a stacked generalization scheme. BMC Bioinformatics. 2019;20(1):308. doi: 10.1186/s12859-019-2907-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ray S., Alberuni S., Maulik U. Computational prediction of HCV-human protein-protein interaction via topological analysis of HCV infected PPI modules. IEEE Trans. Nanobioscience. 2018;17(1):55–61. doi: 10.1109/TNB.2018.2797696. [DOI] [PubMed] [Google Scholar]
- 69.Sze-To A., Fung S., Lee E.A., Wong A.K.C. Prediction of protein-protein interaction via co-occurring aligned pattern clusters. Methods. 2016;110:26–34. doi: 10.1016/j.ymeth.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 70.Zhang L., Yu G., Guo M., Wang J. Predicting protein-protein interactions using high-quality non-interacting pairs. BMC Bioinformatics. 2018;19(Suppl. 19):525. doi: 10.1186/s12859-018-2525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y., Ilie L. SPRINT: ultrafast protein-protein interaction prediction of the entire human interactome. BMC Bioinformatics. 2017;18(1):485. doi: 10.1186/s12859-017-1871-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guo Y., Li M., Pu X., Li G., Guang X., Xiong W., Li J. PRED_PPI: a server for predicting protein-protein interactions based on sequence data with probability assignment. BMC Res. Notes. 2010;3:145. doi: 10.1186/1756-0500-3-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Quan L., Wu H., Lyu Q., Zhang Y. DAMpred: recognizing disease-associated nsSNPs through bayes-guided neural-network model built on low-resolution structure prediction of proteins and protein-protein interactions. J. Mol. Biol. 2019;431(13):2449–2459. doi: 10.1016/j.jmb.2019.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kong M., Zhang Y., Xu D., Chen W., Dehmer M. FCTP-WSRC: protein-protein interactions prediction via weighted sparse representation based classification. Front. Genet. 2020;11:18. doi: 10.3389/fgene.2020.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murakami Y., Mizuguchi K. Homology-based prediction of interactions between proteins using Averaged One-Dependence Estimators. BMC Bioinformatics. 2014;15:213. doi: 10.1186/1471-2105-15-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Islam M.M., Alam M.J., Ahmed F.F., Hasan M.M., Mollah M.N.H. Improved prediction of protein-protein interaction mapping on Homo sapiens by using amino acid sequence features in a supervised learning framework. Protein Pept. Lett. 2020;••• doi: 10.2174/0929866527666200610141258. [DOI] [PubMed] [Google Scholar]
- 77.Mosharaf M.P., Hassan M.M., Ahmed F.F., Khatun M.S., Moni M.A., Mollah M.N.H. Computational prediction of protein ubiquitination sites mapping on Arabidopsis thaliana. Comput. Biol. Chem. 2020;85:107238. doi: 10.1016/j.compbiolchem.2020.107238. [DOI] [PubMed] [Google Scholar]
- 78.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hasan M.M., Khatun M.S. Recent progress and challenges for protein pupylation sites prediction. EC Proteomics and Bioinformatics. 2017;2(1):36–45. [Google Scholar]
- 80.Murakami Y., Mizuguchi K. PSOPIA: Toward more reliable protein-protein interaction prediction from sequence information.; 2017 International Conference on Intelligent Informatics and Biomedical Sciences (ICIIBMS); Okinawa. 2017. pp. 255–261. [Google Scholar]
- 81.Li Z.W., You Z.H., Chen X., Gui J., Nie R. Highly accurate prediction of protein-protein interactions via incorporating evolutionary information and physicochemical characteristics. Int. J. Mol. Sci. 2016;17(9):E1396. doi: 10.3390/ijms17091396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.You Z-H. MengChu Zhou; Xin Luo; Shuai, L. Highly efficient framework for predicting interactions between proteins. IEEE Trans. Cybern. 2017;47(3):731–743. doi: 10.1109/TCYB.2016.2524994. [DOI] [PubMed] [Google Scholar]
- 83.Kawashima S., Pokarowski P., Pokarowska M., Kolinski A., Katayama T., Kanehisa M. AAindex: amino acid index database, progress report 2008. Nucleic Acids Res. 2008;36(Database issue):D202–D205. doi: 10.1093/nar/gkm998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawashima S., Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 2000;28(1):374. doi: 10.1093/nar/28.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Narykov O., Bogatov D., Korkin D. DISPOT: a simple knowledge-based protein domain interaction statistical potential. Bioinformatics. 2019;35(24):5374–5378. doi: 10.1093/bioinformatics/btz587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li X., Yang L., Zhang X., Jiao X. Prediction of protein-protein interactions based on domain. Comput. Math. Methods Med. 2019;2019:5238406. doi: 10.1155/2019/5238406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wojcik J., Schächter V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics. 2001;17(Suppl. 1):S296–S305. doi: 10.1093/bioinformatics/17.suppl_1.S296. [DOI] [PubMed] [Google Scholar]
- 88.Kim W.K., Park J., Suh J.K. Large scale statistical prediction of protein-protein interaction by potentially interacting domain (PID) pair. Genome Informatics; 2002. pp. 22–50. [PubMed] [Google Scholar]
- 89.Hayashida M., Kamada M., Song J., Akutsu T. Conditional random field approach to prediction of protein-protein interactions using domain information. BMC Syst. Biol. 2011;5(Suppl. 1):S8. doi: 10.1186/1752-0509-5-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghadie M.A., Lambourne L., Vidal M., Xia Y. Domain-based prediction of the human isoform interactome provides insights into the functional impact of alternative splicing. PLOS Comput. Biol. 2017;13(8):e1005717. doi: 10.1371/journal.pcbi.1005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hasan M.M., Zhou Y., Lu X., Li J., Song J., Zhang Z. Computational identification of protein pupylation sites by using profile-based composition of k-spaced amino acid pairs. PLoS One. 2015;10(6):e0129635. doi: 10.1371/journal.pone.0129635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hasan M.M., Kurata H. 2018. [Google Scholar]
- 93.Liaw A. Wiener: Classification and regression by random forest. R News. 2002;2:18–22. [Google Scholar]
- 94.Su R., Hu J., Zou Q., Manavalan B., Wei L. Empirical comparison and analysis of web-based cell-penetrating peptide prediction tools. Brief. Bioinform. 2019;21(2):408–420. doi: 10.1093/bib/bby124. [DOI] [PubMed] [Google Scholar]
- 95.Shoombuatong W., Schaduangrat N., Pratiwi R., Nantasenamat C. THPep: a machine learning-based approach for predicting tumor homing peptides. Comput. Biol. Chem. 2019;80:441–451. doi: 10.1016/j.compbiolchem.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 96.Schaduangrat N., Nantasenamat C., Prachayasittikul V., Shoombuatong W. Meta-iAVP: a sequence-based meta-predictor for improving the prediction of antiviral peptides using effective feature representation. Int. J. Mol. Sci. 2019;20(22):E5743. doi: 10.3390/ijms20225743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Win T.S., Malik A.A., Prachayasittikul V., Wikberg S. J.E.; Nantasenamat, C.; Shoombuatong, W. HemoPred: a web server for predicting the hemolytic activity of peptides. Future Med. Chem. 2017;9(3):275–291. doi: 10.4155/fmc-2016-0188. [DOI] [PubMed] [Google Scholar]
- 98.Manavalan B., Subramaniyam S., Shin T.H., Kim M.O., Lee G. Machine-learning-based prediction of cell-penetrating peptides and their uptake efficiency with improved accuracy. J. Proteome Res. 2018;17(8):2715–2726. doi: 10.1021/acs.jproteome.8b00148. [DOI] [PubMed] [Google Scholar]
- 99.Manavalan B., Shin T.H., Kim M.O., Lee G. PIP-EL: a new ensemble learning method for improved proinflammatory peptide predictions. Front. Immunol. 2018;9:1783. doi: 10.3389/fimmu.2018.01783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Boopathi V., Subramaniyam S., Malik A., Lee G., Manavalan B., Yang D.C. mACPpred: a support vector machine-based meta-predictor for identification of anticancer peptides. Int. J. Mol. Sci. 2019;20(8):E1964. doi: 10.3390/ijms20081964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics. 2018;35(16):2757–2765. doi: 10.1093/bioinformatics/bty1047. [DOI] [PubMed] [Google Scholar]
- 102.Hasan M.M., Khatun M.S., Mollah M.N.H., Yong C., Guo D. A systematic identification of species-specific protein succinylation sites using joint element features information. Int. J. Nanomedicine. 2017;12:6303–6315. doi: 10.2147/IJN.S140875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hasan M.M., Yang S., Zhou Y., Mollah M.N. SuccinSite: a computational tool for the prediction of protein succinylation sites by exploiting the amino acid patterns and properties. Mol. Biosyst. 2016;12(3):786–795. doi: 10.1039/C5MB00853K. [DOI] [PubMed] [Google Scholar]
- 104.Hasan M.M., Schaduangrat N., Basith S., Lee G., Shoombuatong W., Manavalan B. HLPpred-Fuse: improved and robust prediction of hemolytic peptide and its activity by fusing multiple feature representation. Bioinformatics. 2020;36(11):3350–3356. doi: 10.1093/bioinformatics/btaa160. [DOI] [PubMed] [Google Scholar]
- 105.Hasan M.M., Manavalan B., Shoombuatong W., Khatun M.S., Kurata H. i6mA-Fuse: improved and robust prediction of DNA 6 mA sites in the Rosaceae genome by fusing multiple feature representation. Plant Mol. Biol. 2020;103(1-2):225–234. doi: 10.1007/s11103-020-00988-y. [DOI] [PubMed] [Google Scholar]
- 106.Hasan M.M., Manavalan B., Khatun M.S., Kurata H. i4mC-ROSE, a bioinformatics tool for the identification of DNA N4-methylcytosine sites in the Rosaceae genome. Int. J. Biol. Macromol. 2019;157:752–758. doi: 10.1016/j.ijbiomac.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 107.Khatun S., Hasan M., Kurata H. Efficient computational model for identification of antitubercular peptides by integrating amino acid patterns and properties. FEBS Lett. 2019;593(21):3029–3039. doi: 10.1002/1873-3468.13536. [DOI] [PubMed] [Google Scholar]
- 108.Khatun M.S., Hasan M.M., Kurata H. PreAIP: computational prediction of anti-inflammatory peptides by integrating multiple complementary features. Front. Genet. 2019;10:129. doi: 10.3389/fgene.2019.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hasan M.M., Rashid M.M., Khatun M.S., Kurata H. Computational identification of microbial phosphorylation sites by the enhanced characteristics of sequence information. Sci. Rep. 2019;9(1):8258. doi: 10.1038/s41598-019-44548-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hasan M.M., Khatun M.S., Kurata H. Large-scale assessment of bioinformatics tools for lysine succinylation sites. Cells. 2019;8(2):E95. doi: 10.3390/cells8020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hasan M.M., Khatun M.S., Mollah M.N.H., Yong C., Dianjing G. NTyroSite: computational identification of protein nitrotyrosine sites using sequence evolutionary features. Molecules. 2018;23(7):E1667. doi: 10.3390/molecules23071667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hasan M.M., Guo D., Kurata H. Computational identification of protein S-sulfenylation sites by incorporating the multiple sequence features information. Mol. Biosyst. 2017;13(12):2545–2550. doi: 10.1039/C7MB00491E. [DOI] [PubMed] [Google Scholar]
- 113.Hasan M.M., Khatun M.S., Kurata H. A comprehensive review of in silico analysis for protein S-sulfenylation sites. Protein Pept. Lett. 2018;25(9):815–821. doi: 10.2174/0929866525666180905110619. [DOI] [PubMed] [Google Scholar]
- 114.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. mAHTPred: a sequence-based meta-predictor for improving the prediction of anti-hypertensive peptides using effective feature representation. Bioinformatics. 2019;35(16):2757–2765. doi: 10.1093/bioinformatics/bty1047. [DOI] [PubMed] [Google Scholar]
- 115.Hasan M.M., Manavalan B., Khatun M.S., Kurata H. Prediction of S-nitrosylation sites by integrating support vector machines and random forest. Molecular Omics. 2019;15(6):451–458. doi: 10.1039/c9mo00098d. [DOI] [PubMed] [Google Scholar]
- 116.Hasan M.M., Kurata H. GPSuc: Global Prediction of Generic and Species-specific Succinylation Sites by aggregating multiple sequence features. PLoS One. 2018;13(10):e0200283. doi: 10.1371/journal.pone.0200283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Win T.S., Schaduangrat N., Prachayasittikul V., Nantasenamat C., Shoombuatong W. PAAP: a web server for predicting antihypertensive activity of peptides. Future Med. Chem. 2018;10(15):1749–1767. doi: 10.4155/fmc-2017-0300. [DOI] [PubMed] [Google Scholar]
- 118.Simeon S., Shoombuatong W., Anuwongcharoen N., Preeyanon L., Prachayasittikul V., Wikberg J.E., Nantasenamat C. osFP: a web server for predicting the oligomeric states of fluorescent proteins. J. Cheminform. 2016;8:72. doi: 10.1186/s13321-016-0185-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Shoombuatong W., Prachayasittikul V., Anuwongcharoen N., Songtawee N., Monnor T., Prachayasittikul S., Prachayasittikul V., Nantasenamat C. Navigating the chemical space of dipeptidyl peptidase-4 inhibitors. Drug Des. Devel. Ther. 2015;9:4515–4549. doi: 10.2147/DDDT.S86529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang B., Li J., Quan L., Chen Y., Lü Q. Sequence-based prediction of protein-protein interaction sites by simplified long short-term memory network. Neurocomputing. 2019;86:100. doi: 10.1016/j.neucom.2019.05.013. [DOI] [Google Scholar]
- 121.Tabei Y. Scalable prediction of compound-protein interaction on compressed molecular fingerprints. Mol. Inform. 2020;39(1-2):e1900130. doi: 10.1002/minf.201900130. [DOI] [PubMed] [Google Scholar]
- 122.Ruas F.A.D., Guerra-Sá R. In silico prediction of protein-protein interaction network induced by Manganese II in Meyerozyma guilliermondii. Front. Microbiol. 2020;11:236. doi: 10.3389/fmicb.2020.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Basith Mail S., Manavalan B., Shin T.H., Lee D., Lee G. Evolution of machine learning algorithms in the prediction and design of anticancer peptides. Curr. Protein Pept. Sci. 2020;••• doi: 10.2174/1389203721666200117171403. [DOI] [PubMed] [Google Scholar]
- 124.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. Meta-4mCpred: a sequence-based meta-predictor for accurate DNA 4mC site prediction using effective feature representation. Mol. Ther. Nucleic Acids. 2019;16:733–744. doi: 10.1016/j.omtn.2019.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alkan F., Erten C. SiPAN: simultaneous prediction and alignment of protein-protein interaction networks. Bioinformatics. 2015;31(14):2356–2363. doi: 10.1093/bioinformatics/btv160. [DOI] [PubMed] [Google Scholar]
- 126.Aloy P., Russell R.B. InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19(1):161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 127.Li Z., Nie R., You Z., Cao C., Li J. Using discriminative vector machine model with 2DPCA to predict interactions among proteins. BMC Bioinformatics. 2019;20(S25) Suppl. 25:694. doi: 10.1186/s12859-019-3268-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Göbel U., Sander C., Schneider R., Valencia A. Correlated mutations and residue contacts in proteins. Proteins. 1994;18(4):309–317. doi: 10.1002/prot.340180402. [DOI] [PubMed] [Google Scholar]
- 129.Chen W., Feng P., Song X., Lv H., Lin H. iRNA-m7G: identifying N7-methylguanosine sites by fusing multiple features. Mol. Ther. Nucleic Acids. 2019;18:269–274. doi: 10.1016/j.omtn.2019.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shatabda S., Saha S., Sharma A., Dehzangi A. iPHLoc-ES: identification of bacteriophage protein locations using evolutionary and structural features. J. Theor. Biol. 2017;435:229–237. doi: 10.1016/j.jtbi.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 131.Charoenkwan P., Shoombuatong W., Lee H.C., Chaijaruwanich J., Huang H.L., Ho S.Y. SCMCRYS: predicting protein crystallization using an ensemble scoring card method with estimating propensity scores of P-collocated amino acid pairs. PLoS One. 2013;8(9):e72368. doi: 10.1371/journal.pone.0072368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chowdhury S.Y., Shatabda S., Dehzangi A. iDNAProt-ES: identification of DNA-binding proteins using evolutionary and structural features. Sci. Rep. 2017;7(1):14938. doi: 10.1038/s41598-017-14945-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hasan M.M., Khatun M.S. 2018.
- 134.Chen X., Huang L., Xie D., Zhao Q. EGBMMDA: extreme gradient boosting machine for MiRNA-disease association prediction. Cell Death Dis. 2018;9(1):3. doi: 10.1038/s41419-017-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Li F., Chen J., Leier A., Marquez-Lago T., Liu Q., Wang Y., Revote J., Smith A.I., Akutsu T., Webb G.I. DeepCleave: a deep learning predictor for caspase and matrix metalloprotease substrates and cleavage sites. Bioinformatics. 2019;••• doi: 10.1093/bioinformatics/btz721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Manavalan B., Basith S., Shin T.H., Wei L., Lee G. AtbPpred: A robust sequence-based prediction of anti-tubercular peptides using extremely randomized trees. Comput. Struct. Biotechnol. J. 2019;17:972–981. doi: 10.1016/j.csbj.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Basith S., Manavalan B., Hwan Shin T., Lee G. Machine intelligence in peptide therapeutics: A next-generation tool for rapid disease screening. Med. Res. Rev. 2020;••• doi: 10.1002/med.21658. [DOI] [PubMed] [Google Scholar]
- 138.Manavalan B., Shin T.H., Kim M.O., Lee G. AIPpred: sequence-based prediction of anti-inflammatory peptides using random forest. Front. Pharmacol. 2018;9:276. doi: 10.3389/fphar.2018.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]