Abstract
Objective
As a brain-specific microRNA, the mechanism of miR-124 in depression has not been clarified so far. The present study aimed to explore the role of miR-124 in depression and its potential targets.
Methods
The depression model was first replicated by the chronic unpredictable mild stress (CUMS) method. miR-124 antagomir was injected into the hippocampus of CUMS rats. Sucrose preference test (SPT), open-field test (OFT), elevated-plus maze (EPM), and forced swimming test (FST) were used to analyze the depression-like behavior. The content of norepinephrine (NE), dopamine (DA) and 5-hydroxytryptamine (5-HT) in the hypothalamus was analyzed by ELISA. qRT-PCR and western blot assay were used for functional analysis.
Results
miR-124 expression was up-regulated in the hippocampus of CUMS -induced depression model rats, while CREB1 and BDNF were down-regulated. Administration of miR-124 antagomir in the hippocampus inhibited miR-124 expression in the hippocampus of CUMS rats. Additionally, SPT, OFT, EPM, and FST also showed that miR-124 antagomir can reduce the depression-like behavior of CUMS rats. Furthermore, miR-124 antagomir injection increased the levels of NE, DA and 5-HT in the hypothalamus of CUMS rats. Moreover, miR-124 antagomir injection increased the expression of cyclic AMP-responsive element-binding protein1 (CREB1) and brain-derived neurotrophic factor (BDNF) in the hippocampus. Using the dual-luciferase reporter assay, it was confirmed that miR-124 directly targets 3'UTR of CREB1 and BDNF genes.
Conclusion
Knockdown of miR-124 can improve depression-like behavior in CUMS-induced depressive rats, which may be related at least in part to the up-regulation of CREB1 and BDNF expression in the hippocampus.
Keywords: miR-124, chronic unpredictable mild stress, depression, cyclic AMP-responsive element binding protein1, brain-derived neurotrophic factor
1. Introduction
Depression is one of the most common mental illnesses, and symptoms include depression, decreased interest, and severe cases with suicidal tendencies [1]. The incidence of depression is increasing year by year. However, current traditional antidepressants may take weeks to months to produce an effective response, and about one-third of patients do not respond to existing treatment strategies [2]. Due to a lack of understanding of the underlying exact molecular mechanisms of depression, antidepressant treatments still
have some limitations. Therefore, the exploration of new targets for the treatment of depression has become one of the important research directions at present.
Epigenetic mechanisms may provide a link between environmental stress factors and gene expression [3]. MicroRNAs (miRNAs) are non-coding RNAs consisting of 22 to 25 nucleotide sequences that have been identified as regulators of biological processes. miRNAs can specifically bind to the 3'untranslated region (UTR) of their target mRNAs to promote the degradation of mRNAs or inhibit their trans-lation [4]. Bioinformatics research suggests that miRNAs may regulate one-third of the transcriptome, which proves that miRNAs play an important role in regulating gene expression. MiRNAs are abundant in brain tissue and play many key roles in regulating brain function, especially in terms of neural regeneration, neural plasticity, and neuron function [5]. Research studies on the regulation of multiple miRNAs in depression have increased recently [6, 7] such as miR-451a, miR-34a-5p, miR-221-3p, and miR-132.
As one of the most abundant miRNAs in the nervous system, miR-124 has been found to be widely expressed in neurons in the brain, retina and spinal cord [8, 9]. According to reports, miR-124 can regulate the regeneration of mature neural cells, thereby playing an important role in promoting neuronal differentiation and regulating synaptic plasticity [10, 11]. Recently, miR-124 has drawn considerable attention due to its role in depression. Cao et al. [12] found that increased expression of miR-124 induces depression in rats under stress. Bahi et al. [13] confirmed that selective inhibition of miR-124a expression in the mouse hippo-campus can produce antidepressant-like effects. He et al. [14] proved that the pathophysiology of miR-124 makes it a potential target for the treatment of depression. However, the role of miR-124 in regulating depression has not yet been clarified.
By establishing the correlation between miRNAs and their target genes, we can better understand the molecular mechanism of depression and provide potential therapeutic targets for clinical treatment of depression [15]. In the present study, we established a chronic unpredictable mild stress (CUMS)-induced depression rat model to study the role of miR-124 in depression and its potential targets. We hope to provide some reference value for the treatment of depression.
2. Materials and Methods
2.1. Laboratory Animals and Groups
Male Sprague-Dawley rats (200-220 g, 8 weeks) were provided by the Laboratory Animal Center of Jiading District Central Hospital affiliated to Shanghai University of Medicine&Health Sciences. The experimental animals were kept in clean animal rooms and were given free food and water, 12h day/night alternating light, temperature (23 ± 1)°C, and humidity (55 ± 2)%. Experiments were performed one week after adaptive feeding. The present study was performed following the Guide for Care and Use of Laboratory Animals and was approved by the Laboratory Animal Center. All experiments were conducted in accordance with the guidelines of Jiading District Central Hospital Affiliated to Shanghai University of Medicine & Health Sciences.
2.2. CUMS Procedure and Intracerebroventricular Injection
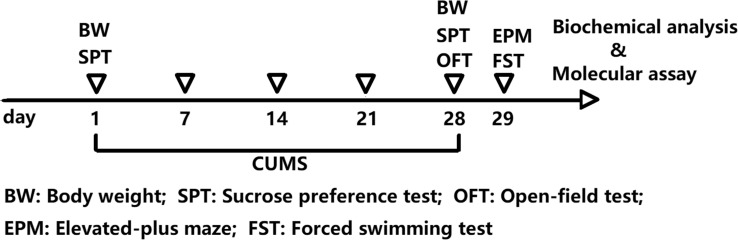
Forty rats were selected and a CUMS model was used to induce depression [16]. Stress methods include restraint, swimming in ice water, fasting, water fasting, day and night reversal, noise stimulation, and tail clamping. Rats received at least one stress stimulus daily. Consecutive appearance of the same stimulus for 4 weeks was avoided. Rats in the control group (n=20) did not receive any stimulation. The changes in bodyweight of the rats were observed and recorded. After the 4 week CUMS model was prepared, ten rats were randomly selected and the depression-like behavior of the rats was detected using the sucrose preference test (SPT), open-field test (OFT), elevated-plus maze (EPM), and forced swimming test (FST). Rats were decapitated and their brains were collected and their hypothalamus and hippocampus tissues were dissected and isolated for biochemical analysis. The experimental design is shown in Fig. (1).
Fig. (1).
Experimental design. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
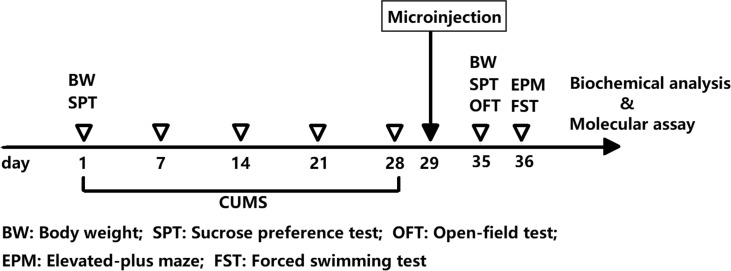
To study the effects of miR-124 on depressive rats, CUMS rats were randomly divided into 3 groups of 10 rats each: CUMS group, CUMS + antago - NC group, and CUMS+antago-miR-124 group. After the last CUMS exposure, intracerebroventricular injection was performed as previously described [17]. In brief, rats were initially anesthetized with sodium pentobarbital [40 mg/kg) and then mounted on a stereotaxic apparatus. A cut was made in the skin on the skull and microdrill was used to make two small holes in the skull above the target position (bilateral hippocampus).
The coordinates are as follows: AP= -4.52 mm; ML= ± 3.20 mm; DV= -3.16 mm below the surface of dura. 20 μL of miR-124 antagomir solution (containing 10 nmol antagomir) or antagomir control (containing 10 nmol negative control) (RiboBio Co., Ltd., Guangzhou, China) was injected bidirectionally into the hippocampus using a microsyringe [18]. The injection was performed for more than 5 min, allowing for slow diffusion. Rats in the control group (n=8) were injected with the same amount of saline in the same manner. Behavioral tests and biochemical analysis were performed one week after the injection was administered. The experimental design is shown in Fig. (2).
Fig. (2).
Experimental design. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.3. SPT
The SPT is a widely-accepted measure of depressant-like behavior [19]. Before the test, rats were kept fasted and water-free, and the rats were adapted to the environment with two bottles of 1% sucrose solution. Within the next 24 h, one of the bottles of 1% sucrose solution was replaced with tap water. On the day of the test, rats were given 200 mL of tap water and 200 mL of a 1% sucrose solution. After 12 h, the bottle was swapped once. After 24 h, tap and sugar bottles were collected. Sucrose consumption/total water consumption was expressed as a sucrose preference.
2.4. OFT
OFT was used to evaluate the movement of animals. The device was a 120 cm x 90 cm x 35 cm opaque open box with a black surrounding. The bottom of the box was divided into 25 squares by white lines. The 16 squares on the periphery were considered as the peripheral site, and the 9 squares on the middle were considered as the central area. In a quiet environment, rats were individually placed in the corners of the device and allowed to explore freely for 5 min. Thirty minutes before the test, the animals were placed in the laboratory to adapt to the environment. After each test, the device was thoroughly cleaned with 70% ethanol. Rat activity data was collected and analyzed by the VideoTrack Animal Behavior Analysis System (VideoTrack 3.0, Viewpoint, French).
2.5. EPM
The anxiety-like behavior of rats was detected using EPM as previously described [20]. In brief, the rats were placed in the middle of the EPM device, and their behavior was recorded by a camera for 5 minutes. The percentage of time the rats spent in the open arms was used as an indicator of anxiety-like behavior. The maze was cleaned after each test.
2.6. FST
FST is a classic method to evaluate the desperate behavior of animals and is considered to show the degree of depression in rodents. As described before [19], rats were placed in a vertical plexiglass cylinder (45 cm × 20 cm] with water temperature (25 ± 1) °C, and depth 30cm. The immobility time of the rat was recorded within 5 min.
2.7. ELISA
Quantitative ELISA kits (ZK-R3347, ZK-R3052 and ZK-R3397; ZIKER, Shenzhen, China) were used to determine the content of norepinephrine (NE), dopamine (DA), and 5-hydroxytryptamine (5-HT) in the hypothalamus. All operations were performed in strict accordance with the kit instructions.
2.8. qRT-PCR
The rat hippocampus tissue was ground and added in cell lysate. The total RNA was extracted from the cells using the RrimeScript® RTreagen Kit (TaKaRa, Dalian, China). An ultraviolet spectrophotometer was used to detect the total RNA purity, and A260/A280 ≥1.80 was used as the qualification standard. One microgram of RNA from each sample was reverse transcribed into cDNA according to the instructions of the DBI-2220 BestarTM qPCR RT Kit (TaKaRa). Real-time PCR was performed using a SYBR® Premix Ex Taq™ Kit (TaKaRa) and the iQ5 Real-Time PCR detection system(Bio-Rad Laboratories, California, USA). PCR reaction conditions were: 95°C 3 min, 95°C 30 s, 52°C 30 s, 73°C 30 s, and 38 consecutive cycles. U6 and GAPDH were used as internal references. The 2-ΔΔCt method was used.
2.9. Western Blot Assay
The total protein extraction kit (Beyotime, Beijing, China) was used to extract total protein from hippocampal tissue. Protein concentration was measured using the Enhanced BCA Protein Assay Kit (Beyotime). A total of 50μg of proteins was loaded and separated by SDS-PAGE. Then, the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon, Millipore, MA). The membranes were blocked to TBST containing 5% non-fat milk for 2 h at room temperature. Then, the membranes were incubated with the primary antibody overnight at 4°C. After having been washed with TBST, the membrane was incubated with HRP-conjugated goat anti-rabbit secondary antibody for 2 h at room temperature. Then, the membrane was developed using enhanced chemiluminescence (ECL) reagent. The greyscale value of each band was analyzed by NIH Image J 7.0. Each experiment was done 3 times.
2.10. In Vitro Luciferase Assay
To elucidate the target role of miR-124, in vitro luciferase experiments were performed to test the interaction between miR-124 and BDNF 3'UTR and CREB1 3'UTR. Genome sequencing of the inserted miR-124 is shown in Figs. (4A and B). Bioinformatics analysis showed that BDNF and CREB1 are potential targets of miR-124. 3'UTR fragments of BDNF and CREB1 were amplified from the human genome by PCR. Corresponding mutant fragments were amplified by overlap extension PCR. Four different recombinant vectors were constructed and synthesized by RiboBio Corporation (China): pmirGLO-CREB1-Wt, pmirGLO-CREB1-Mut, pmirGLO-BDNF-Wt, pmirGLO-BDNF-Mut. The reporter vectors were co-transfected with miR-124 mimic (or miR-NC) into HEK293T cells (Cell Bank of Chinese Academy of Science, Shanghai, China) using Lipofectamine ™ 2000 (Invitrogen).
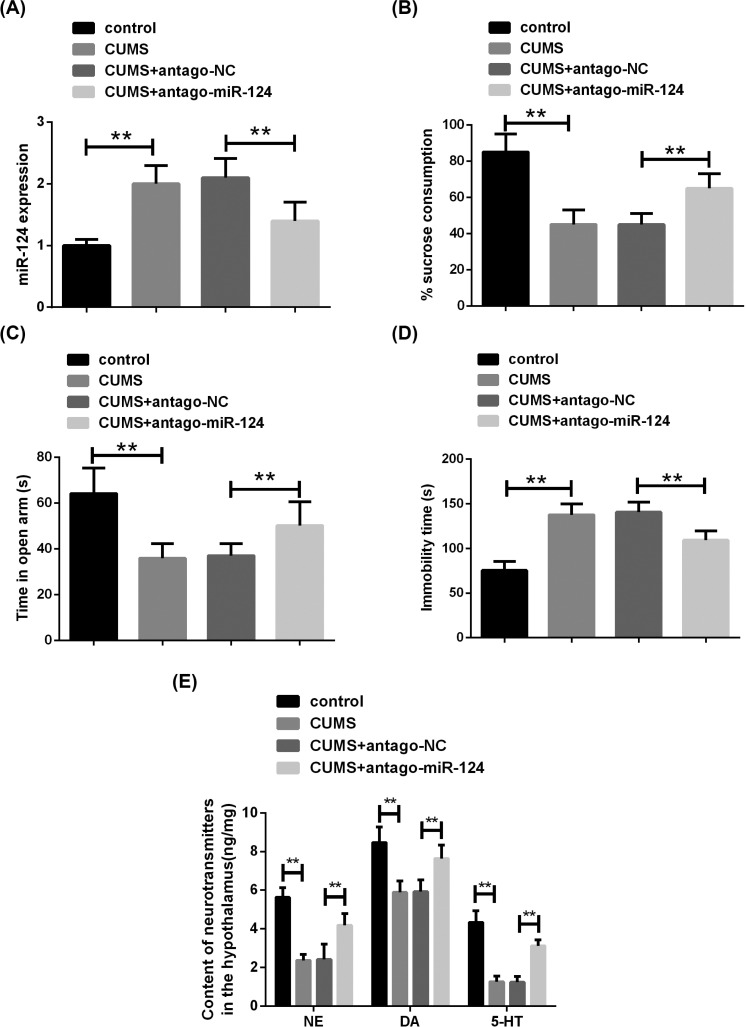
Fig. (4).
The knockdown of miR-124 reduces CUMS-induced depression-like behavior.(A) The expression of miR-124 was detected by qRT-PCR. (B) The sucrose consumption of rats was analyzed by SPT. (C) The time spent in the open arm of rats was evaluated by EPM. (D) The immobility time spent in cold water was assessed by FST. (E) The contents of NE, DA and 5-HT in the hypothalamus of rats were measured by ELISA. n=10. **P<0.01. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.11. Statistical Analysis
All data were presented as the mean ± standard derivation (SD). Data analysis and histograms were done with Graph Pad Prism 5. Differences among experimental groups were determined by one-way analysis of variance followed by Dunn's post-hoc multiple comparisons test. Statistical significance was considered at P<0.05.
3. Results
3.1. Chronic Unpredictable Mild Stress (CUMS) Causes Depression-like Behavior in Rats
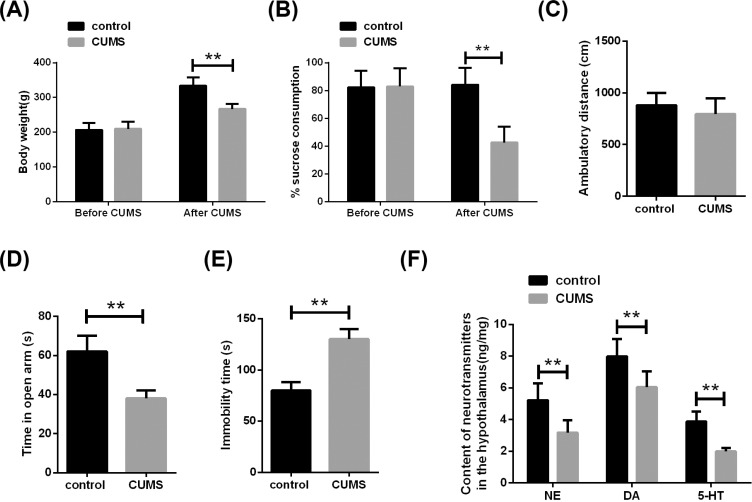
Chronic chronic low-intensity stress is one of the main causes of depression. CUMS induces depression by giving different stimuli to model animals daily, which is close to the cause of depression in humans [21]. In the present study, the depression model was first replicated by the CUMS method (Fig. 1). Weight gain is used to assess the ability of depressed rats to drink freely and to eat food [22]. We first analyzed weight changes in rats after 28 days exposure to CUMS. It was found that after exposure to CUMS, the weight of the rats was significantly lower than before, while the weight of the control group rats had no significant change (Fig. 3A).
Fig. (3).
Chronic unpredictable mild stress (CUMS) causes depression-like behavior in rats.(A) After 28 days of exposure to CUMS, the weight of rats in the CUMS group was significantly lower than control. (B) The sucrose preference test (SPT) showed that the sucrose consumption of rats in the CUMS group was significantly lower than control. (C) Open-field test (OPT) showed that there was no difference in the ambulatory distance between the two groups in the open field. (D) Elevated-plus maze (EPM) showed that the CUMS group spent less time in the open arms than control. (E) The forced swimming test (FST) showed that the time taken by the CUMS group to stand still was significantly higher than the control. (F) The contents of NE, DA and 5-HT in the hypothalamus of rats were measured by ELISA. n=10. **P<0.01. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Next, we test for anxiety-like behavior. SPT showed that exposure to CUMS resulted in a significant reduction in sucrose consumption in rats (Fig. 3B). Additionally, there was no significant difference in the ambulatory distance between the control group and CUMS rats in the open field (Fig. 3C). EPM showed that the CUMS group spent less time in the open arms than control (Fig. 3D). Moreover, FST results showed that the time spent by the CUMS group in resting-state was significantly higher than that of control (Fig. 3E).
Based on the neuromonoamine hypothesis, depression is associated with activities of DA and/or 5-HT metabolism regulation [23]. We further analyzed the content of neurotransmitters in the hypothalamus. It was found that NE, DA, and 5-HT contents in the hypothalamus tissue of rats were significantly reduced when exposed to CUMS (Fig. 3F). These results confirmed that CUMS can induce depression-like behavior in rats.
3.2. Knockdown of miR-124 Reduces CUMS-induced Depression-like Behavior
qRT-PCR showed a significant increase in miR-124 expression in rat hippocampal tissue after exposure to CUMS (Fig. 4A). To study the effect of miR-124 on CUMS rats, miR-124 antagomir was injected into the hippocampus on both sides (Fig. 2). Consistent with expectations, miR-124 antagomir injection significantly reduced miR-124 expression in the hippocampus of CUMS rats (Fig. 2A). Behavioral tests showed that compared to the CUMS group, the sucrose consumption (Fig. 4B) and the time spent in the open arms (Fig. 4C) of rats in the miR-124 antagomir group were markedly increased, while time spent in a resting state (Fig. 4D) of rats was significantly decreased. Simultaneously, the neurotransmitter content in the hypothalamus was detected, and the levels of NE, DA and 5-HT in the miR-124 antagomir group were significantly higher than those in the CUMS group (Fig. 4E). These results suggested that knockdown of miR-124 expression in hippocampal tissue can reduce CUMS-induced depression-like behavior.
3.3. Knockdown of miR-124 Promotes CREB1 and BDNF Expression in the Hippocampus of CUMS Rats
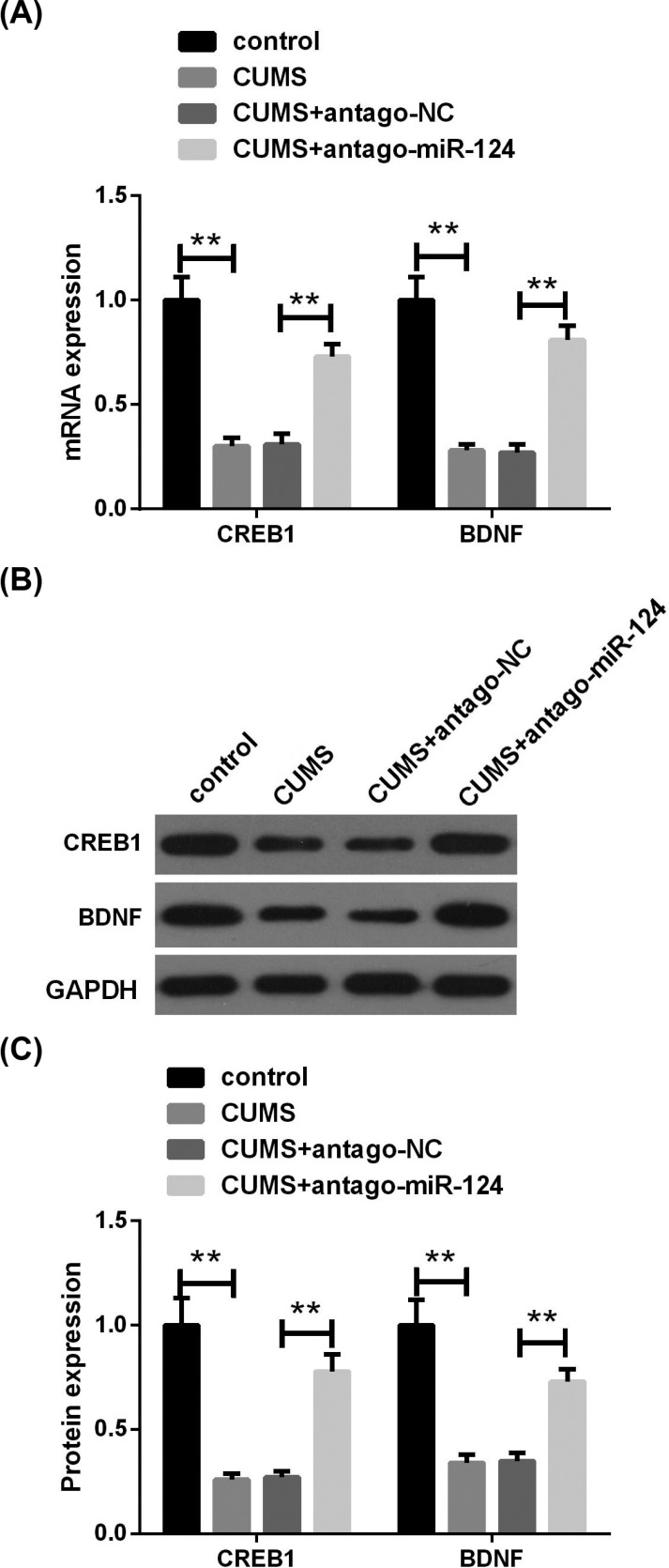
Next, we analyzed the expression of CREB1 and BDNF in the hippocampus. The results of qRT-PCR and western blot assay showed that exposure to CUMS significantly reduced the expression of CREB1 and BDNF mRNA and protein in rat hippocampus (Fig. 5A and B). Moreover, the introduction of miR-124 antagomir in the hippocampus was able to eliminate CUMS-induced reduction of CREB1 and BDNF expression.
Fig. (5).
The knockdown of miR-124 promotes CREB1 and BDNF expression in the hippocampus of CUMS rats. (A) The expressions of CREB1 and BDNF mRNA in the hippocampus tissues were analyzed by qRT-PCR. (B) The expressions of CREB1 and BDNF proteins in hippocampal tissue were assessed by western blot assay. n=10. **P<0.01. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.4. CREB1 and BDNF are Targets of miR-124
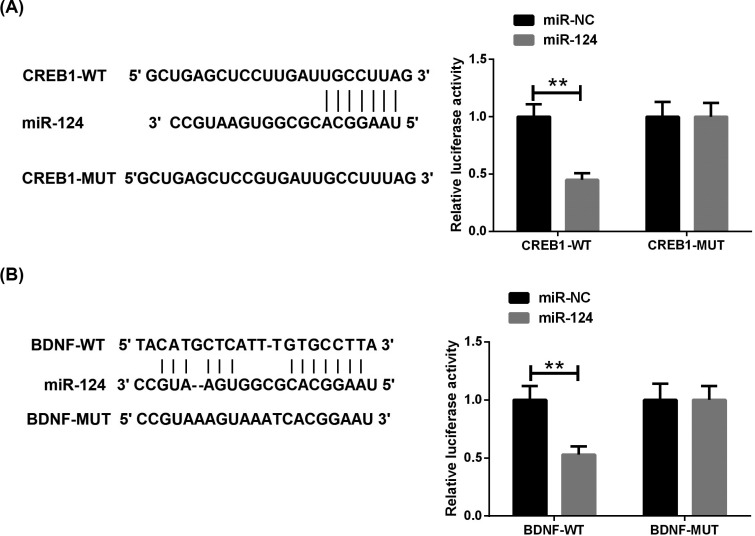
The bioinformatics prediction website shows that CREB1 and BDNF are potential targets for miR-124. Sequences matching miR-124 were found at both CREB1 mRNA 3'UTR and BDNF mRNA 3'UTR (Fig. 6A and B). To confirm that miR-124 targets the 3'UTR of CREB1, we cloned the 3'UTR containing wild- and mutant-CREB1 into a luciferase reporter gene vector, and co-existed the vectors with miR-124 mimic or its negative control into HEK293T cells. The luciferase reporting system showed that the co-transfection of miR-124 mimic with CREB1-Wt resulted in a significant reduction in luciferase activity compared to the control group (Fig. 6A). However, the co-transfection of miR-124 mimic with CREB1-Mut did not affect luciferase activity. Similarly, we observed similar results in the luciferase reporter system co-transfected with miR-124 mimic and BDNF-Wt (Fig. 6B). These results revealed that miR-124 was able to specifically target the 3'UTR regions of CREB1 and BDNF.
Fig. (6).
CREB1 and BDNF are targets of miR-124. (A, B) Relative luciferase activity of reporters, including wild or mutant CREB1 and BDNF1 3′-UTR, co-transfected with miR-NC or miR-124 mimics. **P<0.01. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
4. Discussion
Studies have shown that abnormal expression of miR-124 may be involved in the pathogenesis of depression [24, 25]. However, its related mechanism has not been fully clarified. In this study, by establishing a CUMS depression rat model, we found that the expression of miR-124 in the hippocampus of depression rats was significantly up-regulated. Through SPT, OFT, EPM, and FST measurements, we found that knockdown of miR-124 can improve depression-like behaviors in depressed rats. Simultaneously, the decrease of miR-124 expression in the hippocampus can increase the content of NE, DA and 5-HT in the hypothalamus. Additionally, this study found that knockdown of miR-124 can significantly increase the expression of CREB1 and BDNF in hippocampal tissues. Bioinformatics and luciferase reporter gene analysis also confirmed that CREB1 and BDNF are targets of miR-124. Therefore, we concluded that targeted silencing of miR-124 can exert antidepressant effects by targeting increase in the expression of CREB1 and BDNF in hippocampal tissue.
The “neuronutrition hypothesis” of depression is attracting increasing attention, and the core of this hypothesis is the pathway composed of CREB-BDNF- TrkB. CREB, as a junction in intracellular depression-related signal transduction pathways, can regulate a variety of nervous system functions [26]. Many signaling molecules (such as neurotransmitters, growth factors, etc.) cause phosphorylation of the transcription factor CREB by acting on cAMP- or Ca2+ -activated kinases or directly activating tyrosine kinases. Phosphorylated CREB further enhances the transcription of the downstream target gene BDNF mediated by the cAMP response element (CRE). BDNF then exerts a physiological role by activating a specific receptor, TrkB. Studies have shown that stress and subsequent increase in plasma corticosteroid levels can cause CREB-BDNF-TrkB nutrition pathway activity to decrease, and animal experiments have also found that long-term treatment of multiple antidepressants can not only prevent the weakening of the signal system induced by stress, but also increase its activity [27-29]. Targeting the CREB-BDNF axis may become a therapeutic approach for depression.
It is worth noting that miRNAs have played a role in many of the more widely accepted hypothesis mechanisms of depression, including the neurotrophic hypothesis [30]. Some miRNAs (such as miR-30a-5p and miR-195) in the prefrontal cortex of the human brain have been reported to specifically act on the 3’ UTR of BDNF to inhibit BDNF expression [31]. Lian et al. found that miR-221 promotes the development of depression by regulating the CREB / BDNF axis [32]. We showed that miR-124 knockdown can increase the expression of CREB1 and BDNF in the hippocampus of CUMS rats. Furthermore, bioinformatics and luciferase reporter gene analysis also confirmed the presence of miR-124 binding sites in 3'UTR of CREB1 and BDNF mRNA. Wang et al. [33] showed that miR-124 exerts anti-epileptic functions through targeted regulation of CREB1 expression and activity. Li et al. [34] proved that miR-613 can participate in the occurrence of Alzheimer's disease by regulating the expression of BDNF in the hippocampus of mice. Ma et al. [35] demonstrated that knockdown of miR-1 prevents the decrease of mouse cognitive function by up-regulating the expression of BDNF and CREB in hippocampal tissue. Based on these studies, we speculated that silencing miR-124 can exert antidepressant effects by targeting the promotion of CREB1 and BDNF expression in hippocampal tissue. However, the interaction between CREB1 and BDNF remains to be explored.
conclusion
In summary, miR-124 was up-regulated in the hippocampus of CUMS rats, and miR-124 knockdown improved depression-like behavior in depressive rats, which may be related at least in part to the up-regulation of CREB1 and BDNF expression in the hippocampus. This study may help understand the relationship between miR-124 and depression.
Acknowledgements
Declared none.
Author Contributions
WY and ML conducted most of the experiments and wrote the manuscript. QWZ, JHZ, JC and QYC conducted the experiments and analyzed the data. LXS designed the study and revised the manuscript.
All authors have read and approved the manuscript.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
The study was approved by the ethical committed of the Jiading District Central Hospital Affiliated to Shanghai University of Medicine & Health Sciences, China (Approval no. 2020-3-2).
HUMAN AND ANIMAL RIGHTS
No humans were involved in this study. All the animals procedures were followed in accordance with the Guide for Care and Use of Laboratory Animals.
CONSENT FOR PUBLICATION
Not applicable.
FUNDING
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Zamani M., Alizadeh-Tabari S., Zamani V. Systematic review with meta-analysis: The prevalence of anxiety and depression in patients with irritable bowel syndrome. Aliment. Pharmacol. Ther. 2019;50(2):132–143. doi: 10.1111/apt.15325. [DOI] [PubMed] [Google Scholar]
- 2.Souery D., Serretti A., Calati R., et al. Switching antidepressant class does not improve response or remission in treatment-resistant depression. J. Clin. Psychopharmacol. 2011;31(4):512–516. doi: 10.1097/JCP.0b013e3182228619. [DOI] [PubMed] [Google Scholar]
- 3.Kim Y.K., Ham B.J., Han K.M. Interactive effects of genetic polymorphisms and childhood adversity on brain morphologic changes in depression. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2019;91:4–13. doi: 10.1016/j.pnpbp.2018.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Kosik K.S., Nowakowski T. Evolution of new miRNAs and cerebro-cortical development. Annu. Rev. Neurosci. 2018;41:119–137. doi: 10.1146/annurev-neuro-080317-061822. [DOI] [PubMed] [Google Scholar]
- 5.Gruzdev S.K., Yakovlev A.A., Druzhkova T.A., Guekht A.B., Gulyaeva N.V. The missing link: How exosomes and miRNAs can help in bridging psychiatry and molecular biology in the context of depression, bipolar disorder and schizophrenia. Cell. Mol. Neurobiol. 2019;39(6):729–750. doi: 10.1007/s10571-019-00684-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuang W.H., Dong Z.Q., Tian L.T., et al. MicroRNA-451a, microRNA-34a-5p, and microRNA-221-3p as predictors of response to antidepressant treatment. Brazilian J Med Biol Res Rev. 2018;51:e7212. doi: 10.1590/1414-431X20187212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi S., Yang X., Zhao L., et al. MicroRNA132 associated multimodal neuroimaging patterns in unmedicated major depressive disorder. Brain. 2018;141(3):916–926. doi: 10.1093/brain/awx366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gandy K.A.O., Zhang J., Nagarkatti P., et al. Resveratrol (3, 5, 4′-Trihydroxy-trans-Stilbene) attenuates a mouse model of multiple sclerosis by altering the mir-124/sphingosine kinase 1 axis in encephalitogenic T cells in the brain. J. Neuroimmune Pharmacol. 2019;14:462–477. doi: 10.1007/s11481-019-09842-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu J.Y., Chung K.H., Deo M., Thompson R.C., Turner D.L. MicroRNA miR-124 regulates neurite outgrowth during neuronal differentiation. Exp. Cell Res. 2008;314(14):2618–2633. doi: 10.1016/j.yexcr.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su X., Ye Y., Yang Y., et al. The effect of SPTLC2 on promoting neuronal apoptosis is alleviated by miR-124-3p through TLR4 signalling pathway. Neurochem. Res. 2019;44(9):2113–2122. doi: 10.1007/s11064-019-02849-7. [DOI] [PubMed] [Google Scholar]
- 11.Cheng L.C., Pastrana E., Tavazoie M., Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 2009;12(4):399–408. doi: 10.1038/nn.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao M.Q., Chen D.H., Zhang C.H., Wu Z.Z. Screening of specific microRNA in hippocampus of depression model rats and intervention effect of Chaihu Shugan San. Zhongguo Zhongyao Zazhi. 2013;38(10):1585–1589. [PubMed] [Google Scholar]
- 13.Bahi A., Chandrasekar V., Dreyer J.L. Selective lentiviral-mediated suppression of microRNA124a in the hippocampus evokes antidepressants-like effects in rats. Psychoneuroendocrinology. 2014;46:78–87. doi: 10.1016/j.psyneuen.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 14.He S., Liu X., Jiang K., et al. Alterations of microRNA-124 expression in peripheral blood mononuclear cells in pre- and post-treatment patients with major depressive disorder. J. Psychiatr. Res. 2016;78:65–71. doi: 10.1016/j.jpsychires.2016.03.015. [DOI] [PubMed] [Google Scholar]
- 15.Muñoz-Llanos M., García-Pérez M.A., Xu X., et al. MicroRNA profiling and bioinformatics target analysis in dorsal hippocampus of chronically stressed rats: Relevance to depression pathophysiology. Front. Mol. Neurosci. 2018;11:251. doi: 10.3389/fnmol.2018.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.GL 2019 https://pubmed.ncbi.nlm.nih.gov/31820273/
- 17.Tian F., Yuan C., Yue H. MiR-138/SIRT1 axis is implicated in impaired learning and memory abilities of cerebral ischemia/reperfusion injured rats. Exp. Cell Res. 2018;367(2):232–240. doi: 10.1016/j.yexcr.2018.03.042. [DOI] [PubMed] [Google Scholar]
- 18.Song M.F., Dong J.Z., Wang Y.W., et al. CSF miR-16 is decreased in major depression patients and its neutralization in rats induces depression-like behaviors via a serotonin transmitter system. J. Affect. Disord. 2015;178:25–31. doi: 10.1016/j.jad.2015.02.022. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H., Mohamed N.E., Chan S.J., et al. Absence of stress response in dorsal raphe nucleus in modulator of apoptosis 1-deficient mice. Mol. Neurobiol. 2019;56(3):2185–2201. doi: 10.1007/s12035-018-1205-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fan Y., Tian C., Liu Q., et al. Preconception paternal bisphenol A exposure induces sex-specific anxiety and depression behaviors in adult rats. PLoS One. 2018;13(2):e0192434. doi: 10.1371/journal.pone.0192434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.2019.
- 22.Zhai X.J., Chen F., Chen C., Zhu C.R., Lu Y.N. LC-MS/MS based studies on the anti-depressant effect of hypericin in the chronic unpredictable mild stress rat model. J. Ethnopharmacol. 2015;169:363–369. doi: 10.1016/j.jep.2015.04.053. [DOI] [PubMed] [Google Scholar]
- 23.MM 2019 https://pubmed.ncbi.nlm.nih.gov/31833616/
- 24.Fang Y., Qiu Q., Zhang S., et al. Changes in miRNA-132 and miR-124 levels in non-treated and citalopram-treated patients with depression. J. Affect. Disord. 2018;227:745–751. doi: 10.1016/j.jad.2017.11.090. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q., Sun N.N., Wu Z.Z., Fan D.H., Cao M.Q. Chaihu-Shugan-San exerts an antidepressive effect by downregulating miR-124 and releasing inhibition of the MAPK14 and Gria3 signaling pathways. Neural Regen. Res. 2018;13(5):837–845. doi: 10.4103/1673-5374.232478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma J., Wang L., Yang Y., et al. GNB3 and CREB1 gene polymorphisms combined with negative life events increase susceptibility to major depression in a Chinese Han population. PLoS One. 2017;12(2):e0170994. doi: 10.1371/journal.pone.0170994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razzoli M., Domenici E., Carboni L., et al. A role for BDNF/TrkB signaling in behavioral and physiological consequences of social defeat stress. Genes Brain Behav. 2011;10(4):424–433. doi: 10.1111/j.1601-183X.2011.00681.x. [DOI] [PubMed] [Google Scholar]
- 28.Advani T., Koek W., Hensler J.G. Gender differences in the enhanced vulnerability of BDNF+/- mice to mild stress. Int. J. Neuropsychopharmacol. 2009;12(5):583–588. doi: 10.1017/S1461145709000248. [DOI] [PubMed] [Google Scholar]
- 29.Hoshaw B.A., Malberg J.E., Lucki I. Central administration of IGF-I and BDNF leads to long-lasting antidepressant-like effects. Brain Res. 2005;1037(1-2):204–208. doi: 10.1016/j.brainres.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Li Y.J., Xu M., Gao Z.H., et al. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS One. 2013;8(5):e63648. doi: 10.1371/journal.pone.0063648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mellios N., Huang H.S., Grigorenko A., Rogaev E., Akbarian S. A set of differentially expressed miRNAs, including miR-30a-5p, act as post-transcriptional inhibitors of BDNF in prefrontal cortex. Hum. Mol. Genet. 2008;17(19):3030–3042. doi: 10.1093/hmg/ddn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lian N., Niu Q., Lei Y., Li X., Li Y., Song X. MiR-221 is involved in depression by regulating Wnt2/CREB/BDNF axis in hippocampal neurons. Cell Cycle. 2018;17(24):2745–2755. doi: 10.1080/15384101.2018.1556060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang W, Wang X, Chen L, et al. The microRNA miR-124 suppresses seizure activity and regulates CREB1 activity. 2016. [DOI] [PMC free article] [PubMed]
- 34.Li W., Li X., Xin X., Kan P.C., Yan Y. MicroRNA-613 regulates the expression of brain-derived neurotrophic factor in Alzheimer’s disease. Biosci. Trends. 2016;10(5):372–377. doi: 10.5582/bst.2016.01127. [DOI] [PubMed] [Google Scholar]
- 35.Ma J.C., Duan M.J., Sun L.L., et al. Cardiac over-expression of microRNA-1 induces impairment of cognition in mice. Neuroscience. 2015;299:66–78. doi: 10.1016/j.neuroscience.2015.04.061. [DOI] [PubMed] [Google Scholar]