Abstract
Introduction
To halt the spread of coronary artery disease (CAD), the number one killer in the world, requires primary prevention. Fifty percent of all Americans are expected to experience a cardiac event; the challenge is identifying those at risk. 40 to 60% of predisposition to CAD is genetic. The first genetic risk variant, 9p21, was discovered in 2007. Genome-Wide Association Studies has since discovered hundreds of genetic risk variants. The genetic burden for CAD can be expressed as a single number, Genetic Risk Score (GRS). Assessment of GRS to risk stratify for CAD was superior to conventional risk factors in several large clinical trials assessing statin therapy, and more recently in a population of nearly 500,000 (UK Biobank). Studies were performed based on prospective genetic risk stratification for CAD. These studies showed that a favorable lifestyle was associated with a 46% reduction in cardiac events and programmed exercise, a 50% reduction in cardiac events. Genetic risk score is superior to conventional risk factors, and is markedly attenuated by lifestyle changes and drug therapy. Genetic risk can be determined at birth or any time thereafter.
Conclusion
Utilizing the GRS to risk stratify young, asymptomatic individuals could provide a paradigm shift in the primary prevention of CAD and significantly halt its spread.
Keywords: Genome-wide association studies, coronary artery disease, genetic risk stratification, genetics, primary, prevention, genetic risk variants, genomic
1. INTRODUCTION
The sequencing of the human genome in 2001 [1], with further completion in 2004 [2], has significantly enhanced our opportunities to prevent and manage human diseases. Epidemiologists have documented that predisposition to Coronary Artery Disease (CAD) is due to both acquired (lifestyle and environmental) [3], and genetic factors [4-7]. It has been estimated for some time that 40% to 60% of predisposition for CAD is due to inherited factors [7]. It was also hypothesized that CAD, like most common diseases, would be due to multiple genetic risk variants that occur commonly, with each contributing only minimal risk. To pursue the genetic structure of polygenic common diseases, such as CAD, would require methods different from the genetic linkage analysis of related pedigrees, utilized in the pursuit of genes responsible for rare Mendelian disorders. This review will summarize the developments that led to the application of Genome-Wide case-control association studies based on the indirect method utilizing DNA markers spanning the human genome. This directed to the discovery of hundreds of genetic risk variants predisposing to CAD, as well as the elucidation of many of the genetic variants regulating plasma lipids. The availability of genetic variants enabled Mendelian Randomization [8] studies to help in the future discovery of new drugs. For CAD, it has enabled the development of algorithms to predict and risk-stratify in young asymptomatic individuals. Risk stratification for CAD based on genetic risk factors could provide a paradigm shift in primary prevention of CAD and significantly attenuate its spread.
1.1. Rare Genetic Disorders and Linkage Analysis
In the latter part of the 20th century, genetic research was dominated by rare inherited monogenetic disorders. These disorders by definition occur in less than one percent of the population [9]. The mutations are highly penetrant and generally occur in protein-coding regions of the genome. The responsible mutation was shown to induce the phenotype, which led to these disorders referred as Mendelian single-gene disorders. This does not exclude the possibility that its expression and phenotypic characteristics can be influenced by other genes [10]. Proof that the single mutation can induce the disorder was amply demonstrated by the offspring of transgenic animals expressing the expected phenotype. The human mutated gene responsible for the disease is inserted into the egg of the mouse and the wild type human gene is inserted into a control group. The first gene [11] responsible for Familiar Hypertrophic Cardiomyopathy (FHCM) was mapped to its chromosomal location, 14q1, [11, 12] and utilizing positional cloning the mutant gene was cloned and sequenced to identify the mutation [13]. Subsequently, the mutant gene was expressed in a mouse. The hearts of the offspring from the mouse exhibited the sarcomere disarray and fibrosis similar to that observed in human FHCM [14]. We developed a transgenic rabbit [15] using human FHCM mutation in the myosin heavy chain gene and the offspring exhibited cardiac sarcomere disarray, fibrosis, along with cardiac hypertrophy and premature sudden cardiac death. Using genetic linkage analysis, we discovered a gene responsible for Wolff-Parkinson-White (WPW) syndrome [16] and expressed it in a transgenic mouse, which exhibited a phenotype of prolonged PR interval and significant glycogen infiltration of the myocardium similar to that observed in the humans [17]. Similar approaches were taken for other inherited cardiomyopathies, such as right ventricular dysrhythmia cardiomyopathy [18]. Thus, the manifestation of the phenotype in the offspring of genetically engineered animals proved the mutation was both necessary and sufficient to induce the phenotype.
Chromosomal mapping of genes responsible for these rare diseases, utilized genetic linkage analysis [19], a method designed for Mendelian disorders. This consisted of collecting pedigrees affected with the disease of at least two generations, and preferably three generations. The pedigree of affected and unaffected individuals was genotyped using 50 to 100 DNA markers. Those markers that segregated with affected individuals, more than by chance, indicated the marker was in close physical proximity to the gene responsible for the phenotype. Subsequent positional cloning and sequencing of the chromosomal region identified the causal mutation by showing it was present only in those affected. This technique is dependent on obtaining a two to three generation pedigree and requires relatively few DNA markers since it is performed in individuals all of which are closely or distantly related. It is claimed there are over 8,000 rare Mendelian disorders with the chromosomal location and/or mutation discovered in over 4,000 [20].
1.2. Candidate Gene Approach is Inappropriate for Common Polygenic Disorders
It was realized in the 90s that common disorders such as CAD, diabetes, and hypertension are due to a combination of environmental and genetic factors. In addition, these disorders would most likely be due to multiple common genetic variants, each contributing only minimal risk. This is in sharp contrast to rare Mendelian inherited disorders, in which a single mutation can induce the phenotype. Consideration of these observations, and the epidemiology of chronic, common polygenic disorders, it became apparent that genetic linkage analysis would not be the most appropriate technique to pursue their genetic architecture. Linkage analysis has low power except when a single locus explains a substantial fraction of the disease. A more appropriate technique would be that of the Direct or Indirect case-control association study. An unbiased case-control association study, to have adequate power, would require massive sample sizes together with hundreds of thousands of DNA markers distributed throughout the human genome.
Such diverse DNA markers were not available, and even if they were, platforms did not exist to analyze such massive data. So, attempts were made using the candidate gene approach. This consisted of choosing a protein with a function that influenced coronary atherosclerosis or myocardial infarction. Thus, one or more forms (alleles) of the gene encoding this protein would be expected to predispose to CAD. Association studies were performed in selected populations with the disease to determine if one or more alleles of a particular selected gene were associated with the disease. The sample size was usually small, and the p-value selected was that of 0.05, with no attempts to replicate in an independent population. The candidate gene association approach of one or a few candidate genes examined only a small fraction of the extensive sequence variation in the genome of each patient. This was recognized at the time as a biased approach, and significant concerns were expressed as to whether this would provide reproducible trustworthy data. We now know that none of the candidate genes claimed to predispose to CAD was confirmed by the unbiased approach of GWAS [21, 22].
1.3. Case-Control Association Studies Appropriate for Polygenic Disorders
A comprehensive search for genetic variants predisposing to a polygenic disease such as CAD ideally requires exploring all genetic variation in a large number of affected and unaffected individuals. Investigators in the field of genomics and genetics emphasized the indirect case-control association approach based on polymorphic DNA markers selected to span the whole of the human genome [23-26]. The case-control association study is simplistic in concept. One genotypes DNA markers in a control population without the disease and a population selected for having the disease of interest, which for this review is CAD. The frequency of each marker in the CAD group versus the control group is determined. Markers occurring more frequently in the CAD group than the control are interpreted to be a risk predisposing to CAD. It is preferable to refer to such sequence as a genetic risk variant rather than a gene, since the sequence may be in non-protein or protein coding regions. The DNA marker itself will most likely not be the causal mutation, but rather in close physical proximity to the causal mutation. Since the mutation is detected indirectly, it is referred to as an Indirect case-control association study
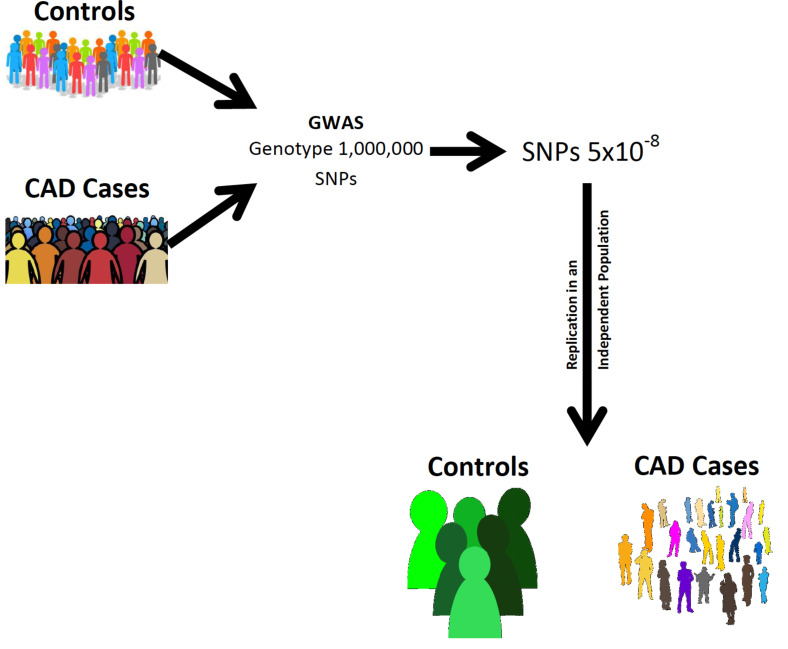
While the case-control association study approach concept is simplistic, to provide meaningful, reliable, and interpretable results, it must be properly designed. A symposium [27], held at the University of Southern California in April 2005, was dedicated to the case-control association study and the markers required to span the genome, referred to as Genome-Wide Association Studies (GWAS). To have adequate power would require a marker evenly distributed throughout the genome at intervals of at least 3,000 bases. This would require at least a million markers, but selecting a p-value of 0.05 would inherently give you 50,000 false positives. This led to the recommendation of a Bonferroni correction whereby one million markers divided into 0.05 would give a much smaller p-value of 0.00000005 (5x10-8). The other less stringent approach would be to use a false discovery rate of less than five percent. The Bonferroni correction was accepted as the preferred stringency and became known as genome-wide significance. To satisfy such stringent p-values would require sample sizes in the thousands, and even then would only detect SNPs that occur in the population with a frequency of ≥ 5%. There was much discussion regarding it being performed in phases rather than in a single study. This would decrease the cost and the sample size, making it more practical. The ultimate decision was to perform it in two stages. The first stage would be to genotype the majority of the sample size using the Bonferroni corrected p-value. Those SNPs reaching statistical significance with a p-value of 5x10-8 would be genotyped for replication in a smaller, similar, but independent population (Fig. 1). Only those SNPs reaching statistical significance in the replication population would be considered as genetic risk variants for CAD. In summary, an indirect, two-stage GWAS appeared to be the most appropriate for the pursuit of genetic variants predisposing to CAD. The stage was set to entice the development of informative markers, distributed throughout the genome, and technological innovations such that high throughput genotyping of microarrays encrypted with hundreds of thousands of SNPs would lend itself to algorithms for rapid analysis [28-30].
Fig. (1).
Genome-Wide Association Study for CAD. A sample size in the thousands for cases and controls is required. Ideally, one requires a marker about every 3,000 base pairs, which is in the form of SNPs selected to cover the genome as much as possible. Those SNPs occurring with greater frequency in the CAD cases, having a p-value of ≤ 5x10-8, are genotyped again to confirm that the marker reflects at a sequence that is a genetic risk variant for CAD. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
1.4. Selection of Informative DNA Markers for GWAS
It was evident that the DNA markers should be single nucleotide polymorphisms since they are the most common form of DNA sequence variation in the human genome [24], and are somewhat evenly distributed throughout the genome. The number of SNPs per human genome is fairly constant, at about 5,000,000 SNPs [31-33]. If one used all of the 5,000,000 SNPs identified in a single individual as DNA markers to span the genome, it should provide excellent coverage for that individual, but may not be adequate coverage for the remainder of the population. This is because there are billions of SNPs circulating in the general population from which is selected the 5,000,000 for each individual.
Will it be feasible using 500,000 or millions of SNPs to have adequate genome coverage for screening of risk variants of CAD in a large unrelated population? Certain observations may make it more plausible. The hypothesis that predisposition to common diseases is due to SNPs that occur commonly. Secondly, the genome consists of independent blocks of DNA, referred to as haplotypes [31-33]. Selecting a single SNP as a representative of the haplotype, rather than all of the SNPs, would not only increase the power but reduce cost and be time-efficient.
1.5. Hypothesis: Common DNA Variants Predispose to Common Polygenic Diseases-CAD
It is hypothesized that DNA variants predisposing to CAD, or any common polygenic disease are common, meaning they occur frequently in the population. Common variants are defined as those with a frequency in the population of ≥ 0.5%. There is a strong rationale to support the “common variant theory”. CAD is a common disease, being the most common cause of death throughout the world, whether it is in low, middle, or high-income countries. The acquired and environmental risk factors contributing to CAD, as described by the Framingham [34] study include, hypertension, hypercholesterolemia, obesity, smoking, diabetes, age, and a sedentary way of life. These risk factors predispose to CAD regardless of race, ethnicity, or geographical location, as shown by the INTERHEART trial involving over 52 countries [35].
The reduction of these risk factors is associated with decreased cardiac events and mortality also regardless of ethnicity, race, or geographical location. The HOPE trial showed lowering plasma cholesterol in populations from 21 countries consistently reduced cardiac events [36].
Coronary atherosclerosis is the underlying pathology for CAD. Cholesterol is a major culprit, which once engulfed and oxidized by macrophages, induces inflammation and plaque formation. Cardiac events such as myocardial infarction, or sudden cardiac death, are usually precipitated by plaque rupture and thrombosis. CAD is not sex-dependent in that it is the most common cause of death in both males and females throughout the world. The onset of CAD in females is delayed by about a decade due to the protective effect of hormones in the premenopausal phase. In the U.S, and probably in most countries, if one lives a normal lifespan, 50% [37] are expected to experience a cardiac event. CAD is due to a combination of risk factors resulting from lifestyle, environmental, and genetic factors. Epidemiologists for decades have claimed genetic factors are responsible for about 50% of predisposition to CAD [7]. In light of this extensive data obtained from epidemiological, pathophysiological, and clinical trials, CAD has a common underlying pathology, with similar risk factors throughout the world. CAD is universally observed as an insidious disease, which progresses with age. The central culprit is plasma LDL-C, which doubles the risk of CAD with each additional decade of exposure [38]. It is reasonable to hypothesize that genetic predisposition is transmitted by variants that are common in the population. GWASs were designed with the power to pursue genetic risk variants for CAD occurring with a frequency of ≥ 5%.
1.6. Selection of SNPs Representative of Haplotypes Distributed Throughout the Genome
The DNA sequence of any two human genomes of Homo sapiens is 99% identical [39-42]. The remaining one percent is responsible for the transmission of the unique features of each individual. This consists of about 30 million base pairs, which is classified as Structural Variants (SVs) or SNPs. The SVs consists of large chunks of DNA ≥ 50 kbp and accounts for most of the DNA sequence variation in the one percent. This is because of the size of the SVs, inversions and the variation in the number of repeat copies. The contribution of SV variants to disease or other unique attributes appears to be very little, however, our knowledge of the functional roles of SV remains meager. This leaves the five million SNPs as the major component responsible for the unique features of humans, including predisposition to disease. Current knowledge suggests the SNPs transmit 80 to 90 percent of the unique attributes of humans [32]. This is supported by the observation that evolution, induced by new mutations, is transmitted to each generation primarily by SNPs. The source of these mutations, which are responsible for evolution, is due to DNA copying [43, 44] errors made in the process of the normal turnover and replacement of germ-line DNA. DNA turns over every few days and is synthesized by adding one nucleotide at a time. When an error occurs that leads to a new mutation, they are primarily substitution of a single nucleotide, resulting in 96% of the mutations being a single nucleotide polymorphism (SNP). It is estimated that there is only one mistake per one billion nucleotides added. These errors do add up over time, such that each individual of a new generation inherits 40 to 60 new mutations, which are in the forms of SNPs [23, 45].
The transmission of the genome to offspring, as demonstrated by Mendel, is such that all DNA sequences, including genes, are inherited independently and randomly during mitosis. This normal transmission is referred to as being in genetic linkage equilibrium, but there are exceptions. DNA sequences that are in close physical proximately tend to be co-inherited, rather than by independent randomized assortment, referred to as linkage disequilibrium (LD). The architecture of the genome is such that stretches of DNA sequence, which may vary in length from 10,000 base pairs to more than 100,000 base pairs, referred to as haplotypes, are co-inherited [32, 33]. All of the sequences, including the SNPs, on a particular haplotype, travel together during genetic transmission, and all are in LD with each other. Determining and identifying SNPs representative of haplotypes is very important to the design of GWAS to identify disease-related variants. Let us assume a 20,000 haplotype has an SNP every 500 base pairs, for a total of 40 SNPs. All of the SNPs on any particular haplotype will be co-inherited as a block and are in LD with any and all of the SNPs on that particular haplotype. Selection of any one of the 40 SNPs present on a 20,000 base pair haplotype will provide the same information as selecting all 40. Selecting SNPs as tags for haplotypes reduces several-fold the number of SNPs required to span the genome without loss of power to detect disease-related variants.
1.7. The Japanese, Single Nucleotide Polymorphism Project and the HapMap Project
To develop a genomic map of non-redundant SNPs that is representative of haplotypes distributed throughout the genome would require a dedicated group with appropriate multidisciplinary expertise, infrastructure, and funding. The first to initiate this effort was the Japanese single nucleotide polymorphisms (JSNP) in April 2000. This project was initiated in collaboration with the Human Genome Center (HGC), Institute of Medical Science (IMS), University of Tokyo, and the Japan Science and Technology Agency (JST) [46]. Its mission was to identify 150,000 SNP distributed throughout the human genome within two years and make them available to the public. A total of 190,562 SNPs in 24 subjects were discovered and made available through http://snp.ims.u-tokyo.ac.jp/ [47] in 2002. Around the same time, shortly after sequencing the human genome, the international HapMap project was launched in the U.S. on October 2003 [32]. The HapMap project was a natural extension of the Human Genome Project. It would focus on DNA sequence differences amongst individuals. It was formed to create a public, genome-wide database of common human variations. This would be primarily a catalog of SNPs spanning the whole genome. One obvious need was to utilize the SNPs as DNA markers in genetic studies pursing the genotypes associated with known clinical phenotypes. The HapMap project was formed as an international consortium involving several countries, including Japan. The HapMap project included the infrastructure and expertise of the major genome researchers in the world to provide a database of 1,000,000 genetic variations for representative populations: Africans, European Caucasians, Chinese, and Japanese. The African Yoruba people were in Ibadan, Nigeria. The European Caucasians were Utah, USA populations, collected in the 1980s by the centred’Etude de Polymorphism Humain (CEPH). The Chinese consisted of unrelated Han Chinse in Beijing, China. The Japanese were unrelated, self-identified Japanese population living in Tokyo, Japan.
The HapMap project made available over one million SNPs in 2004 [31], and over three million SNPs in 2007 [48], carefully selected to be representative of haplotypes without redundancy and relatively evenly distributed throughout the genome. This was followed by the 1,000 genome project consortium [33], which released their results in 2015. Whole-genome sequencing and exon targeted deep sequencing, resulted in a genome map that would serve as a global reference for human genetic variation. The results were obtained from 26 populations originating from Africa, East Asia, Europe, South Asia, and the Americas. They characterized a total of 88 million variants, comprising 84.7 million SNPs, 3.6 million short insertions and deletions (INDELs), and 60,000 structural variants [33] (Table 1).
Table 1.
1000 genome project consortium.
| 88 Million Variants |
| 84.7 Million SNPs |
| 3.6 Million Short Insertions/Deletions (INDELs) |
| 60,000 Structural Variants |
The investigators observed that a typical genome differs from the referenced genome at four million to five million sites and the variants at these sites are 99% due to SNPs and short INDELs [33]. The majority of the 84 million SNPs are rare, with 64 million having a frequency less than 0.5%, 12 million with a frequency between 0.5% and 5%, and 8 million occur commonly, namely with a frequency of greater than 5%. Important to note for GWAS in pursuit of disease-related genetic variants, the majority of variants observed in a single genome are common with less than 4% of the variants having a frequency in the population of less than 0.5% [33]. If indeed common variants are primarily responsible for predisposition to common diseases, sequencing or genotyping rare variants might not improve significantly the power to identify disease-related variants. The results of these studies suggest that common variants are shared across the world, and rare variants are typically restricted to closely related populations. They predicted the average genome to have about 2,000 variants associated with complex traits [33].
1.8. Development of Microarrays with Millions of SNPs to Genotype for GWAS
Development of microarrays encrypted with millions of SNPs as DNA markers made it possible to span the human genome at intervals of 3,000 base pairs. Several commercial microarrays became available with hundreds of thousands to millions of SNPs, enabling high throughput genotyping [30]. The availability of millions of SNPs also made it possible to customize microarrays in the pursuit of disease-related variants. Secondly, it became possible to impute additional SNPs onto the array and further increase the density of SNPs for GWAS. Just as the last 2 decades of the 20th century was a golden era for monogenic disorders, the initial decade of the 21st century will be the golden era for polygenic disorders.
1.9. Discovery of 9p21, the First Genetic Risk Variant for CAD
The first genetic risk variant, 9p21, for CAD was discovered in 2007, simultaneously by two independent groups [49 50], followed very shortly by a third group [51]. Multiple investigators confirmed 9p21 as a CAD risk factor in Caucasians [51-53]. Internationally, 9p21 as a risk factor for CAD was confirmed by the Chinese [54, 55], Koreans [56, 57], Italians [58], Japanese [57], and South East Asians including Pakistan and India [59, 60]. Limited studies in African Americans indicated 9p21 is not a risk factor for CAD [59]. The 9p21 risk variant for CAD was associated with a 25% increased relative risk per copy and found to be extremely common, estimated to occur in about 75% of the world’s population. The risk transmitted by 9p21 is independent of known conventional risk factors, such as cholesterol and diabetes. The common occurrence of 9p21 was in-keeping with the hypothesis that genetic risk variants predisposing to CAD occur commonly. It also emphasized the risk imparted by a single genetic risk variant is minimal. Analysis of the 9p21 region indicated it was in a non-protein coding region. These observations were pertinent to the design of subsequent GWAS studies in pursuing other CAD genetic risk variants, which strongly indicated the genetic risk would involve multiple variants with minimal effect size and could be distributed amongst protein-coding and non-protein coding regions. An international consortium was formed including investigators from Canada, U.S, UK, Iceland, and Germany [61].
This consortium was referred to as Coronary ARtery DIsease Genome-wide Replication and Meta-analysis (CARDIoGRAM) [61]. This consortium would subsequently be joined by others designated as CARDIoGRAMPlusC4D [60]. The sample size was initially 88,000 cases and controls and would increase rapidly. The CARDIoGRAMPlusC4D was the largest collaboration ever in cardiology and together with independent investigators, have discovered hundreds of genetic risk variants predisposing to CAD and are detailed in several reviews [60, 62-71]. These genetic risk variants predisposing to CAD provided interesting insights into the genetic architecture of CAD predisposition and were similar to the architectural predisposition for many other common polygenic diseases such as hypertension. The CAD risk variants exhibit features similar to risk variants for other common, chronic polygenic diseases. (1) Genetic risk variants for CAD occurs common, being present in more than 50% of the population. (2) The risk per variant was minimal, averaging less than a 10% increase in relative risk for CAD. (3) Interestingly, over two-thirds of the risk variants mediate the risk for CAD independently of known risk factors, and the mechanism of risk mediation remains unknown. (4) Over 80% of the genetic risk variants for CAD are located in DNA regions that do not code for proteins. The implication being these variants mediate their risk through a regulatory influence on protein-coding genes, located upstream or downstream (cis-acting), or even on other chromosomes (trans-acting). Studies clearly indicate that the burden of risk for CAD, and for other common chronic diseases, is determined by the total number of risk variants inherited, rather than anyone specific variant.
2. Genetic Variant of Unknown Function – Potential for New Drug Targets
Genetic discoveries responsible for rare disorders have already had a significant impact on the treatment of CAD. Cholesterol has long been suspected to be a major culprit in the underlying pathology responsible for CAD. Brown and Goldstein in 1970 discovered a mutation in the gene encoding the receptor that removes low-density lipoprotein cholesterol from the plasma (LDL-R). The mutation was associated with hypercholesterolemia and premature cardiac events in individuals in their second or third decade of life [72]. This significantly supported the cholesterol hypothesis for CAD and enhanced the development of statins. Statin, the first drug to inhibit cholesterol synthesis, was developed and it became the main drug for primary and secondary prevention of CAD, with a world budget of > 70 billion dollars. Recently another genetic disorder has led to the development of new therapy for reducing LDL-C and preventing CAD [73]. In 2003, an enzyme was discovered, PCSK9, which increases the degradation of LDL-R, leading to increased plasma levels of LDL-C and an increased incidence of CAD [74]. Shortly after Abifadel et al. [75] discovered a gain of function mutation in PCSK9, which was associated with hypercholesterolemia and increased CAD. This was followed by a discovery of a loss of function mutation in an African population [76] with a frequency of 2.6%. The loss of function was associated with a 28% reduction in the mean plasma LDL-C and an 88% reduction of CAD. A new drug was developed utilizing a monoclonal antibody [77] that inhibits PCSK9, and within 10 years clinical studies were completed [77, 78] showing up to 50 percent reduction in plasma LDL-C. A large placebo-controlled clinical trial was performed, referred to as the FOURIER (Further Cardiovascular Outcomes Research With PCSK9 Inhibition in Subjects With Elevated Risk) trial, which evaluated the PCSK9 drug Evolocumab [79]. The trial was ended earlier than planned and observed a significant reduction in cardiovascular events, but no significant decline in mortality. One reason for not observing a mortality benefit was stopping the trial earlier than planned, and it lacked power due to an inadequate sample size. The ODYSSEY OUTCOMES (Evaluation of Cardiovascular Outcomes After an Acute Coronary Syndrome During Treatment With Alirocumab) trial with the PCSK9 drug Alirocumab had a longer follow up period showing remarkably similar results, except there was also a significant reduction in mortality [80]. The combined data from these two large trials led to FDA approval for routine clinical use of these two drugs.
We previously indicated that over 200 genetic risk variants have been discovered that predispose to CAD. Only about 1/3 of these risk variants mediate their effect through the traditional risk factors [71]. This observation has important implications for the pathophysiology of CAD and its prevention and treatment. These genetic risk variants acting through unknown mechanisms to increase CAD, will ultimately lead to discovery of new pathways with multiple targets for the development of novel drugs to specifically mitigate the risk. This is ample proof that while cholesterol is the major culprit, other factors are also contributing to the pathogenesis of CAD, which has yet to be elucidated. Considerable evidence suggests that inflammation is part of the underlying pathology [81]. Several of the genetic risk variants, such as the interleukins (IL6R), and others such as CXCL12, SH2B3, MRAS, and PLG, participate in various inflammatory pathways [69, 82]. This is confirmed by the large clinical trial CANTOS [83].
2.1. Mendelian Randomizations Could Shorten the Journey to Drug Discovery and Approval
A major component of FDA’s approach in determining approval of a drug for clinical application is the result of randomized, placebo-controlled clinical trials. These trials frequently require 3 to 5 years to perform and coupled with the time for pre-clinical development, could take up to 10-12 years for clinical approval [8]. Only about five percent of compounds selected are approved by the FDA. The availability of genetic risk variants for CAD enables Mendelian Randomization which could be complementary to the RCT and significantly decrease the time required from discovery to clinical application of a new drug [84]. Essentially all genes, or genetic variants, have multiple forms referred to as alleles. The function of these alleles, with respect to each other, may have gain-of-function, loss-of-function, or be neutral. Genetic variants are randomized at conception and remain fixed throughout life with no confounding factors. If one has a genetic variant known to increase plasma HDL-C, at age 50 they will have had 50 years of exposure to increased plasma HDL-C. If HDL-C is a causative protective agent against CAD, then this individual should have fewer cardiac events. Such a study, referred to as a Mendelian Randomization, was performed by us in 2012 [85]. GWAS have made available more than 160 genetic variants that regulate plasma lipid levels [86, 87]. It has been dogma since the 1960s [88] that HDL-C is protective of coronary artery disease. Several interventions that increase HDL-C, including statins, niacin, exercise, and alcohol, also decrease plasma levels of LDL-C [89, 90]. Interpretation of these data is confounded as to whether the effect is increased plasma HDL-C, or decreased plasma LDL-C. We utilized a genetic variant in LIPG p.ASN396SER associated solely with increased plasma concentration of HDL-C [85] in a large sample size with replication in an independent population.
The genetic variant increased plasma HDL-C but had no effect on other plasma lipids. There was no increase or decrease in cardiac events which indicated HDL-C is not associated with protection or risk of CAD [85]. Several recent studies, including RCT, suggested that plasma HDL-C levels are not associated with MI or CAD [91]. While it is possible that other proteins involved in the HDL-C complex play a role, this study clearly indicated the assumption of plasma HDL-C as a protective factor must be reassessed. Several subsequent RCTs have confirmed that increased plasma HDL-C is not protective of CAD [92-97]. A recent meta-analysis of 39 randomized trials involving 117,411 patients was assessed to determine the effect of niacin, fibrates, and cholesterol ester transfer protein (CETP) inhibitors on cardiovascular events [98]. All interventions increased plasma HDL-C, but neither niacin, fibrates, nor CETP inhibitors exhibited any effects on cardiac events. The investigators concluded that the minimal benefit of niacin and fibrates is due to decreasing plasma LDL-C concentrations. In the past decade, plasma HDL-C has been the target of several clinical trials, despite increasing plasma levels of HDL, all have had a negative effect on cardiac events [92, 95, 96]. It is of note that MR studies all consistently showed plasma LDL-C and triglycerides are significant risk factors for CAD [91]. If someone discovered a molecule that has the potential to decrease coronary artery disease, Mendelian randomization can be used to ascertain whether the target of this drug is causally related to CAD [84]. If the target is not casual the effort is not likely to be worthwhile. MR studies performed on targets traditionally associated with CAD, such as folic acid [99], uric acid [100], and fibrinogen, showed these compounds are not causative and should not be used as drug targets to prevent or manage CAD.
2.2. Genetic Risk for CAD is Proportional to the Number of Variants Inherited and can be Expressed in a Single Number
The risk associated with each genetic variant for CAD is minimal, averaging less than 10% increased relative risk. The total genetic risk burden is proportional to the number of risk variants inherited [71, 101]. Utilizing all of the genetic risk variants for CAD, one can derive a single number that summarizes the total genetic risk burden for CAD. To determine this number, one must first determine the number of genetic risk variants inherited by each individual. The range in the number of copies inherited for each variant per individual is from 0 to 2. It is 0 if neither parent transmits a copy of the risk variant, 1 if a single parent transmits a copy of the risk variant, and 2 if both parents transmit a copy of the risk variant. The risk of each variant is the odds ratio previously determined by GWAS and weighted by the natural log of that odds ratio [102]. The genetic risk score is the summation of all these products (number of copies times the log of the odds ratio).
2.3. The Need to Risk Stratify for Primary Prevention of CAD
CAD has been shown to be preventable in multiple clinical trials. Interventions, such as changes in lifestyle and drug therapy, to decrease plasma cholesterol have consistently shown a 40-60% reduction in cardiac events [3, 36]. In the US, CAD and its sequelae have decreased by 50% in the past 30 years [37]. Secondary prevention after a cardiac event has been very successful by reducing the traditional risk factors. Primary prevention must be based on risk stratification to determine who would benefit most from preventive measures. 50% of the population [37] will experience a cardiac event in their lifetime, however, nearly everyone in their 40s, males and females have a significantly increased plasma LDL-C [103-105]. They are asymptomatic, and most have no risk factors other than increased plasma LDL-C. To significantly reduce the spread of CAD, one must decrease the prevalence, which requires primary prevention. Our traditional risk factors and methods used to predict 10-year risk for CAD, including the Framingham Risk Score, Reynolds Score, and Pooled Cohort Equations of ACC and AHA are all age-dependent and become more accurate with age. Blood pressure is age-dependent, as is cholesterol and diabetes. The genetic risk score (GRS) is independent of age and can be determined at any time at birth or any time thereafter. One’s DNA does not change in one’s lifetime. One of the objectives of discovering genetic risk variants for CAD was to develop a genetic risk score to risk stratify CAD that is independent of age, and would determine who would benefit most from primary preventative measures.
2.4. Retrospective Assessment of a Genetic Risk Score in Clinical Trials for Statins
The development and assessment of the genetic risk score have paralleled the effort to discover genetic risk variants predisposing to CAD. Attempts to predict using 9p21 when it was first discovered were disappointing and abandoned [106]. As more genetic risk variants became available, other attempts were made to risk stratify [107-109]. Studies appeared on how to weigh the risk of variants and incorporate genetic and acquired risk prediction of CAD [102]. Prediction based on just 12 genetic risk variants (all of genome significance) were encouraging, but the additional benefit over that of traditional risk factors was small [110]. The increase in the number of genetic variants enabled the effort to continue. It was appreciated that the total risk burden of CAD, like most polygenic disorders, was related to the total number of variants inherited, rather than any single variant. Inherent, in this statement, was the desire for a greater number of risk variants in the hopes of improving prediction.
In 2015, Mega et al. [111] utilized 27 genetic risk variants for CAD (all of genome significance) and genotyped a population of 48,421 individuals who had enrolled in various clinical trials to assess efficacy and safety of statins to reduce cardiac events. There were four clinical trials, two of which involved primary prevention, and the other two involved secondary prevention. Following genotyping with the genetic risk variants, individuals were classified into low, intermediate, and high genetic risks. The individuals with the highest GRS were in the high risk group and had the most benefit from statin therapy. The GRS was found to be equally effective in stratifying for risk of CAD in both primary and secondary prevention. The discriminatory power of GRS, over that of traditional risk factors, is indicated by the observation that the number to be treated with a statin was only 25 to prevent a cardiac event. Similarly, individuals enrolled in the West of Scotland Coronary Prevention Study (WOSCOP) were genotyped and stratified by GRS into low, intermediate, and high risk. The group with the high GRS score had a relative risk reduction of 44%, compared to a relative risk reduction of 24% in others [112]. Based on the results of WOSCOP, one would need to treat only 13 individuals with a statin versus 38 individuals in the low risk group. Risk stratification for CAD based on traditional risk factors would require statin treatment of 100 individuals to prevent two cardiac events [113]. The investigators concluded that the GRS increased discriminatory power for risk stratification of CAD over that of traditional risk factors, and was relatively independent of traditional risk factors. Stratification with GRS was more effective in identifying those individuals in which statin therapy would be most effective. Abraham et al. [114], utilizing a microarray of 49,310 SNPs based on the CARDIoGRAMPlusC4D consortium, assessed its predictive power in five prospective population cohorts. The microarray included SNPs of genome-wide significance, but also included thousands of SNPs of less than genome-wide significance shown to be in linkage disequilibrium. Individuals with the higher GRS were at higher risk for coronary artery disease compared to those with low GRS scores. Furthermore, they confirmed the previous observations that the GRS is relatively independent of traditional risk factors.
2.5. Prospective Assessment of a Genetic Risk Score for CAD
The evolution of larger sample sizes, together with increasing numbers of SNPs as markers, led to discovery of over 200 genetic risk variants for CAD. These variants have all reached genome-wide significance, and have been replicated in an independent population. There are hundreds more found by GWAS to be statistically significant with less than 5% false discovery rate. If one combines both groups of genetic risk variants, it would still only account for about 38 percent of inheritability [101]. To increase the number of genetic risk variants for CAD, two approaches were taken. Inouye et al. [115] reduced statistical stringency and included those with a false discovery rate of only 5%, which resulted in a microarray containing 1.7 million risk variants for CAD. The group at Boston utilized a computerized algorithm, LDpred [116], to predict genetic variants that associate with predisposition for CAD. Further pruning was performed to ensure all SNPs were in linkage equilibrium to avoid redundancy of markers [117]. These investigators put together a microarray with 6.6 million genetic risk variants predisposing to CAD. It is realized by both groups that many of these variants have minimal effect and maybe redundant. It is highly likely that the number of causal genetic risk variants is more likely to be a few thousand, rather than a few million. However, the statistical basis for predictive algorithms indicates that adding SNPs with no risk do not dilute the power to predict risk. The addition of SNPs with even minimally associated increased risk that is non-redundant should increase predictive power [118].
The era of large databases has ushered in the development of biobanks which collect phenotypic and genotypic data. A well advanced biobank is that of the UK Biobank. They have collected data on over 500,000 individuals in the UK, and the data is available through its public website. Khera et al., utilizing a sample size of 288,978 and the 6.6 million microarrays, showed that 8% of the population inherited a threefold increase risk for CAD, and 0.5% inherited a fivefold risk for CAD [117]. The group with the highest genetic risk, and the high risk for CAD, would not have been identified using traditional risk factors for CAD. Only 20% of the individuals with increased risk had hypercholesterolemia, and only 28% had hypertension. A family history of CAD was observed in only 35% of the individuals with the high GRS. The 1.7 million microarray by Inouye et al. [115] utilized a sample size of nearly 500,000, again selected from the UK Biobank. They observed that in the top 20% risk group of the GRS, there was a fourfold increased risk for CAD. Genetic risk stratification using the GRS, based on either 1.7 million genetic risk variants or 6.6 million, confirmed increased predictive power over that of previous microarrays, which have used either 27, 50, or 49,000 [115, 117] genetic risk variants.
2.6. Lifestyle Changes Reduce Genetic Risk for CAD
There has long been a myth regarding genetic factors. The myth being that once it is in your genes, it cannot be treated. This of course is not true, and genetic predisposition has long been treated with the same therapies that we use to treat acquired and environmental factors. Genes themselves live a very provincial life, being restricted to the nucleus. The plebiscites that carry out the functions of genes are mediated by the protein derived from the mRNA template that leaves the nucleus and attaches to the ribosomes in the cytoplasm. Statin therapy, which inhibits the activity of the rate-limiting enzyme 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA), inhibits synthesis of cholesterol, and indirectly blocks the function of the gene encoding for this enzyme.
The development of a sensitive prediction method for the genetic predisposition of CAD has major implications for the prevention of CAD, the world’s number one killer. This is based on the assumption that we can reverse the genetic risk in those individuals at high risk for CAD. We previously presented the data on several large clinical trials in which their genome was genotyped retrospectively for genetic risk variants predisposing to CAD. Results of these studies consistently showed the genetic risk score selected those individuals at greatest risk for CAD and would benefit most from statin therapy.
A major intervention in the prevention of CAD (primary and secondary) is that of lifestyle changes. These behavioral changes are often difficult to initiate and even more difficult to sustain. A randomized clinical trial performed by Khera et al. [119] consisted of 55,685 participants and a microarray with 50 genetic risk variants. The participants were prospectively genotyped and a genetic risk score was determined and used to risk stratify into low, intermediate, and high risk. The end point was comparing a favorable lifestyle with that of an unfavorable lifestyle. A favorable lifestyle consisted of no obesity, a healthy diet, frequent exercise, and no current smoking. An unfavorable lifestyle had at least two of these unfavorable components. Results showed the top 20%, with a high GRS, had a 91% higher risk of cardiac events than those with a low GRS. Individuals with a favorable lifestyle, and a high GRS, had a 40% lower risk for cardiac events than an unfavorable lifestyle.
Tikkanen et al. [120] performed genetic risk stratification to assess the effect of physical activity on the genetic risk for CAD. The UK Biobank provided 468,095 individuals to be tested. The individuals performed handgrip for three seconds and a cardiorespiratory test of exercise on a stationary bicycle, during which oxygen consumption was monitored. Genetic risk was determined and categorized into low, intermediate, or high. The individuals with the highest GRS had the most benefit from exercise, with a 49% lower risk for CAD.
Two additional studies completed recently confirmed the discriminatory power of the GRS to stratify for risk of CAD. The 14,298 patients enrolled in the FOURIER trial [121] were genotyped with a microarray having 27 risk variants for 6 million SNPs predisposing to CAD. Individuals with intermediate and high genetic risk for CAD had 1.23 and 1.65 fold increased hazard for major coronary events respectively. In patients receiving Evolocumab, there was a 13% relative risk reduction in the group with traditional risk factors, but without high genetic risk, and 31% relative risk reduction in patients with high genetic risk regardless of traditional clinical risk factors. The patients with the highest genetic risk score had the greatest risk and the greater benefit from lowering of plasma cholesterol with Evolocumab. The GRS was found to be independent of, and superior to, traditional risk factors for risk stratification of CAD. It is of interest that utilizing a microarray of 27 risk SNPs were just as effective as the 6 million microarrays. A similar study was performed in the ODYSSEY trial [122] using a microarray with over 6 million SNPs in a sample size of 11,953. The group with the highest GRS had the highest risk for CAD and the relative reduction of cardiac events by Alirocumab was 37% in the high GRS, versus a 13% reduction in the low GRS group.
Multiple studies has shown consistent positive results utilizing the genetic risk score to risk stratify for CAD. The GRS has been shown to be superior to prediction programs (Framingham Risk Score, Pooled Cohort Equations, and Reynolds Score) based on traditional risk factors and relatively independent of traditional risk factors. However, two recent studies showed less favorable results, one genotyping the UK Biobank population [123] of 352,660, and the other a U.S. population [124] of 7,237. Both populations were genotyped with an array containing over 6 million genetic risk variants for CAD. The studies concluded the GRS for risk stratifying for CAD was statistically better than traditional risk factors, but the difference was small. They recognized that the GRS has advantages in the young over traditional methods, but recommended it not be used for routine clinical applications at this time. An accompanying editorial [125] also was in agreement with this recommendation. It is difficult to reconcile these results with previous studies. The investigators emphasized that the pretest sample was more appropriately characterized, which may account for some of the differences. Nevertheless, even these two studies show the GRS is equal or slightly better than the traditional risk methods.
The GRS has significantly improved over the past 5 years and will continue to do so. The GRS provides an unparalleled opportunity to assess risk in younger individuals at a time when traditional risk factors are mostly lacking. An excellent example of this is the premenopausal female in her 40s with a plasma LDL-C of 160 mg/dL, and no other risk factors. This is a missed opportunity for primary prevention, which could be remedied in those individuals with increase LDL-C and a high GRS [126].
2.7. Limitations to the Current GRS
The GRS for CAD has been evaluated as a risk stratifying tool in over one million individuals, and with minor exceptions [125], has been shown to be successful in selecting individuals at high risk for CAD, and those who would benefit most from lifestyle changes [119] and cholesterol lowering agents [121, 122]. Furthermore, genetic risk for CAD, as determined by the GRS, is markedly reduced by lifestyle changes [119, 120] and cholesterol lowering agents [121, 122]. The GRS for CAD is superior to those risk methods based on traditional risk factors, and is also relatively independent of traditional risk factors. There are limitations to routine applications of the GRS. (1) It has been assessed primarily in individuals of European descent. (2) The test and validation population in most of these studies have been the same. (3) While more and more genetic risk variants are included to derive the GRS, it is likely that more are yet to be discovered. (4) The genetic risk variants are primarily tags, rather than the casual mutations. (5) Many of the studies assessing the GRS have been in clinical trials in which everyone is known to have CAD since a diagnosis of CAD is required for enrollment.
2.8. The Future, A Paradigm Shift in Primary Prevention of CAD
To reduce the spread of CAD will be necessary to implement primary prevention to decrease the prevalence. Lifestyle changes and cholesterol lowering agents are relatively inexpensive, and proven to be effective for primary prevention throughout the world. The barrier is selecting those who would benefit from primary prevention. Plasma cholesterol, a major culprit of CAD, is increased significantly in the general adult population, with the average plasma LDL-C being 147 mg/dL in males, and 121 mg/dL in females in their 40s [105]. The clinical practice guidelines for cardiology recommends the plasma LDL-C should be ≤ 70 mg/dL and multiple clinical trials show cardiac events and mortality are further reduced at plasma LDL-C levels of 30-60 mg/dL [127, 128]. Epidemiologists indicate during your lifetime it is expected that 50% will experience a cardiac event [37].
conclusion
The GRS is a simple test, it is inexpensive, can be tested in blood or saliva, and be applied throughout the world. Secondly, we have simple and inexpensive preventive measures proven to be effective in reducing genetic risk for CAD. It is reasonable to assume the GRS, as a risk stratifying tool, will detect a significant proportion of the 50% who will benefit most from primary prevention. Current predictive programs, such as the Pooled Cohort Equation, are age-dependent because they are based on traditional risk factors, many of which are not present in the young when most needed for primary prevention. The GRS is independent of age and relatively independent of traditional risk factors. All of these features give the GRS an advantage over traditional risk factors for primary prevention, but most importantly, it enables a prediction of CAD risk to be determined at birth or anytime thereafter. One’s DNA does not change in a lifetime, neither does your genetic predisposition. The potential of the GRS to induce a paradigm shift in primary prevention could markedly halt the spread of CAD.
Acknowledgements
Declared none.
Consent for Publication
Not applicable.
Funding
The author has received research funding from the Canadian Institutes of Health Research, CIHR no. MOP P82810, the Canada Foundation for Innovation, the CFI, no. 11966, and the Dignity Health Foundation.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J.P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann Y., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J.C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R.H., Wilson R.K., Hillier L.W., McPherson J.D., Marra M.A., Mardis E.R., Fulton L.A., Chinwalla A.T., Pepin K.H., Gish W.R., Chissoe S.L., Wendl M.C., Delehaunty K.D., Miner T.L., Delehaunty A., Kramer J.B., Cook L.L., Fulton R.S., Johnson D.L., Minx P.J., Clifton S.W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J.F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R.A., Muzny D.M., Scherer S.E., Bouck J.B., Sodergren E.J., Worley K.C., Rives C.M., Gorrell J.H., Metzker M.L., Naylor S.L., Kucherlapati R.S., Nelson D.L., Weinstock G.M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D.R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H.M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R.W., Federspiel N.A., Abola A.P., Proctor M.J., Myers R.M., Schmutz J., Dickson M., Grimwood J., Cox D.R., Olson M.V., Kaul R., Raymond C., Shimizu N., Kawasaki K., Minoshima S., Evans G.A., Athanasiou M., Schultz R., Roe B.A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W.R., de la Bastide M., Dedhia N., Blöcker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J.A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D.G., Burge C.B., Cerutti L., Chen H.C., Church D., Clamp M., Copley R.R., Doerks T., Eddy S.R., Eichler E.E., Furey T.S., Galagan J., Gilbert J.G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L.S., Jones T.A., Kasif S., Kaspryzk A., Kennedy S., Kent W.J., Kitts P., Koonin E.V., Korf I., Kulp D., Lancet D., Lowe T.M., McLysaght A., Mikkelsen T., Moran J.V., Mulder N., Pollara V.J., Ponting C.P., Schuler G., Schultz J., Slater G., Smit A.F., Stupka E., Szustakowki J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y.I., Wolfe K.H., Yang S.P., Yeh R.F., Collins F., Guyer M.S., Peterson J., Felsenfeld A., Wetterstrand K.A., Patrinos A., Morgan M.J., de Jong P., Catanese J.J., Osoegawa K., Shizuya H., Choi S., Chen Y.J., Szustakowki J., International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431(7011):931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 3.Wald N.J., Law M.R. 2003.
- 4.Zdravkovic S., Wienke A., Pedersen N.L., Marenberg M.E., Yashin A.I., De Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J. Intern. Med. 2002;252(3):247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 5.Wienke A., Holm N.V., Skytthe A., Yashin A.I. The heritability of mortality due to heart diseases: a correlated frailty model applied to Danish twins. Twin Res. 2001;4(4):266–274. doi: 10.1375/twin.4.4.266. [DOI] [PubMed] [Google Scholar]
- 6.Marenberg M.E., Risch N., Berkman L.F., Floderus B., de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994;330(15):1041–1046. doi: 10.1056/NEJM199404143301503. [DOI] [PubMed] [Google Scholar]
- 7.Chan L., Boerwinkle E. Gene-environment interactions and gene therapy in atherosclerosis. Cardiology. 1994;2:130–137. [Google Scholar]
- 8.DiMasi J.A., Grabowski H.G., Hansen R.W. The cost of drug development. N. Engl. J. Med. 2015;372(20):1972. doi: 10.1056/NEJMc1504317. [DOI] [PubMed] [Google Scholar]
- 9.Roberts R. Genetics of coronary artery disease. Circ. Res. 2014;114(12):1890–1903. doi: 10.1161/CIRCRESAHA.114.302692. [DOI] [PubMed] [Google Scholar]
- 10.Daw E.W., Chen S.N., Czernuszewicz G., Lombardi R., Lu Y., Ma J., Roberts R., Shete S., Marian A.J. Genome-wide mapping of modifier chromosomal loci for human hypertrophic cardiomyopathy. Hum. Mol. Genet. 2007;16(20):2463–2471. doi: 10.1093/hmg/ddm202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hejtmancik J.F., Brink P.A., Towbin J., Hill R., Brink L., Tapscott T., Trakhtenbroit A., Roberts R. Localization of gene for familial hypertrophic cardiomyopathy to chromosome 14q1 in a diverse US population. Circulation. 1991;83(5):1592–1597. doi: 10.1161/01.CIR.83.5.1592. [DOI] [PubMed] [Google Scholar]
- 12.Jarcho J.A., McKenna W., Pare J.A., Solomon S.D., Holcombe R.F., Dickie S., Levi T., Donis-Keller H., Seidman J.G., Seidman C.E. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. N. Engl. J. Med. 1989;321(20):1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 13.Geisterfer-Lowrance A.A., Kass S., Tanigawa G., Vosberg H.P., McKenna W., Seidman C.E., Seidman J.G. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990;62(5):999–1006. doi: 10.1016/0092-8674(90)90274-I. [DOI] [PubMed] [Google Scholar]
- 14.Geisterfer-Lowrance A.A.T., Christe M., Conner D.A., Ingwall J.S., Schoen F.J., Seidman C.E., Seidman J.G. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272(5262):731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- 15.Marian A.J., Wu Y., Lim D-S., McCluggage M., Youker K., Yu Q-T., Brugada R., DeMayo F., Quinones M., Roberts R. A transgenic rabbit model for human hypertrophic cardiomyopathy. J. Clin. Invest. 1999;104(12):1683–1692. doi: 10.1172/JCI7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gollob M.H., Roberts R. AMP-activated protein kinase and familial Wolff-Parkinson-White syndrome: new perspectives on heart development and arrhythmogenesis. Eur. Heart J. 2002;23(9):679–681. doi: 10.1053/euhj.2001.2954. [DOI] [PubMed] [Google Scholar]
- 17.Sidhu J.S., Rajawat Y.S., Rami T.G., Gollob M.H., Wang Z., Yuan R., Marian A.J., DeMayo F.J., Weilbacher D., Taffet G.E., Davies J.K., Carling D., Khoury D.S., Roberts R. Transgenic mouse model of ventricular preexcitation and atrioventricular reentrant tachycardia induced by an AMP-activated protein kinase loss-of-function mutation responsible for Wolff-Parkinson-White syndrome. Circulation. 2005;111(1):21–29. doi: 10.1161/01.CIR.0000151291.32974.D5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad F., Li D., Karibe A., Gonzalez O., Tapscott T., Hill R., Weilbaecher D., Blackie P., Furey M., Gardner M., Bachinski L.L., Roberts R. Localization of a gene responsible for arrhythmogenic right ventricular dysplasia to chromosome 3p23. Circulation. 1998;98(25):2791–2795. doi: 10.1161/01.CIR.98.25.2791. [DOI] [PubMed] [Google Scholar]
- 19.Marian A.J., Brugada R., Roberts R. Chapter 55: Mendelian basis of congenital and other cardiovascular disease. Hurst’s. Heart. 2015;•••:14e. [Google Scholar]
- 20.Ku C.S., Naidoo N., Pawitan Y. Revisiting Mendelian disorders through exome sequencing. Hum. Genet. 2011;129(4):351–370. doi: 10.1007/s00439-011-0964-2. [DOI] [PubMed] [Google Scholar]
- 21.Morgan T.M., Krumholz H.M., Lifton R.P., Spertus J.A. Nonvalidation of reported genetic risk factors for acute coronary syndrome in a large-scale replication study. JAMA. 2007;297(14):1551–1561. doi: 10.1001/jama.297.14.1551. [DOI] [PubMed] [Google Scholar]
- 22.Anand S.S., Xie C., Paré G., Montpetit A., Rangarajan S., McQueen M.J., Cordell H.J., Keavney B., Yusuf S., Hudson T.J., Engert J.C., INTERHEART Investigators Genetic variants associated with myocardial infarction risk factors in over 8000 individuals from five ethnic groups: The INTERHEART Genetics Study. Circ Cardiovasc Genet. 2009;2(1):16–25. doi: 10.1161/CIRCGENETICS.108.813709. [DOI] [PubMed] [Google Scholar]
- 23.Kruglyak L., Nickerson D.A. Variation is the spice of life. Nat. Genet. 2001;27(3):234–236. doi: 10.1038/85776. [DOI] [PubMed] [Google Scholar]
- 24.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet. 1999;22(2):139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 25.Hirschhorn J.N., Daly M.J. Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 2005;6(2):95–108. doi: 10.1038/nrg1521. [DOI] [PubMed] [Google Scholar]
- 26.Wang W.Y., Barratt B.J., Clayton D.G., Todd J.A. Genome-wide association studies: theoretical and practical concerns. Nat. Rev. Genet. 2005;6(2):109–118. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 27.Thomas D.C., Haile R.W., Duggan D. Recent developments in genomewide association scans: a workshop summary and review. Am. J. Hum. Genet. 2005;77(3):337–345. doi: 10.1086/432962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dandona S., Stewart A.F.R., Roberts R. Genomics in coronary artery disease: past, present and future. Can. J. Cardiol. 2010;26(Suppl. A):56A–59A. doi: 10.1016/S0828-282X(10)71064-3. [DOI] [PubMed] [Google Scholar]
- 29.Roberts R. A customized genetic approach to the number one killer: coronary artery disease. Curr. Opin. Cardiol. 2008;23(6):629–633. doi: 10.1097/HCO.0b013e32830e6b4e. [DOI] [PubMed] [Google Scholar]
- 30.Roberts R. New gains in understanding coronary artery disease, Interview with Dr Robert Roberts. Affymetrix Microarray Bull. 2007;3(2):1–4. [Google Scholar]
- 31.Altshuler D., Donnelly P. International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibbs R., Belmont J., Hardenbol P. International HapMap Consortium. The International HapMap Project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 33.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R. 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kannel W.B., Dawber T.R., Kagan A., Revotskie N., Stokes J., III Factors of risk in the development of coronary heart disease--six year follow-up experience. The Framingham Study. Ann. Intern. Med. 1961;55(1):33–50. doi: 10.7326/0003-4819-55-1-33. [DOI] [PubMed] [Google Scholar]
- 35.Yusuf S., Hawken S., Ounpuu S. Dans, T.; Avezum, A.; Lanas, F.; McQueen, M.; Budaj, A.; Pais, P.; Varigos, J.; Lisheng, L. INTERHEART Study Investigators. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004;364(9438):937–952. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 36.Yusuf S., Bosch J., Dagenais G., Zhu J., Xavier D., Liu L., Pais P., López-Jaramillo P., Leiter L.A. Dans, A.; Avezum, A.; Piegas, L.S.; Parkhomenko, A.; Keltai, K.; Keltai, M.; Sliwa, K.; Peters, R.J.; Held, C.; Chazova, I.; Yusoff, K.; Lewis, B.S.; Jansky, P.; Khunti, K.; Toff, W.D.; Reid, C.M.; Varigos, J.; Sanchez-Vallejo, G.; McKelvie, R.; Pogue, J.; Jung, H.; Gao, P.; Diaz, R.; Lonn, E. HOPE-3 Investigators. Cholesterol lowering in intermediate-risk persons without cardiovascular disease. N. Engl. J. Med. 2016;374(21):2021–2031. doi: 10.1056/NEJMoa1600176. [DOI] [PubMed] [Google Scholar]
- 37.Murray C.J., Lopez A.D. Measuring the global burden of disease. N. Engl. J. Med. 2013;369(5):448–457. doi: 10.1056/NEJMra1201534. [DOI] [PubMed] [Google Scholar]
- 38.Navar-Boggan A.M., Peterson E.D., D’Agostino R.B.S., Sr, Neely B., Sniderman A.D., Pencina M.J. Hyperlipidemia in early adulthood increases long-term risk of coronary heart disease. Circulation. 2015;131(5):451–458. doi: 10.1161/CIRCULATIONAHA.114.012477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaisson M.J.P., Sanders A.D., Zhao X., Malhotra A., Porubsky D., Rausch T., Gardner E.J., Rodriguez O.L., Guo L., Collins R.L., Fan X., Wen J., Handsaker R.E., Fairley S., Kronenberg Z.N., Kong X., Hormozdiari F., Lee D., Wenger A.M., Hastie A.R., Antaki D., Anantharaman T., Audano P.A., Brand H., Cantsilieris S., Cao H., Cerveira E., Chen C., Chen X., Chin C.S., Chong Z., Chuang N.T., Lambert C.C., Church D.M., Clarke L., Farrell A., Flores J., Galeev T., Gorkin D.U., Gujral M., Guryev V., Heaton W.H., Korlach J., Kumar S., Kwon J.Y., Lam E.T., Lee J.E., Lee J., Lee W.P., Lee S.P., Li S., Marks P., Viaud-Martinez K., Meiers S., Munson K.M., Navarro F.C.P., Nelson B.J., Nodzak C., Noor A., Kyriazopoulou-Panagiotopoulou S., Pang A.W.C., Qiu Y., Rosanio G., Ryan M., Stütz A., Spierings D.C.J., Ward A., Welch A.E., Xiao M., Xu W., Zhang C., Zhu Q., Zheng-Bradley X., Lowy E., Yakneen S., McCarroll S., Jun G., Ding L., Koh C.L., Ren B., Flicek P., Chen K., Gerstein M.B., Kwok P.Y., Lansdorp P.M., Marth G.T., Sebat J., Shi X., Bashir A., Ye K., Devine S.E., Talkowski M.E., Mills R.E., Marschall T., Korbel J.O., Eichler E.E., Lee C. Multi-platform discovery of haplotype-resolved structural variation in human genomes. Nat. Commun. 2019;10(1):1784. doi: 10.1038/s41467-018-08148-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sudmant P.H., Rausch T., Gardner E.J., Handsaker R.E., Abyzov A., Huddleston J., Zhang Y., Ye K., Jun G., Fritz M.H., Konkel M.K., Malhotra A., Stütz A.M., Shi X., Casale F.P., Chen J., Hormozdiari F., Dayama G., Chen K., Malig M., Chaisson M.J.P., Walter K., Meiers S., Kashin S., Garrison E., Auton A., Lam H.Y.K., Mu X.J., Alkan C., Antaki D., Bae T., Cerveira E., Chines P., Chong Z., Clarke L., Dal E., Ding L., Emery S., Fan X., Gujral M., Kahveci F., Kidd J.M., Kong Y., Lameijer E.W., McCarthy S., Flicek P., Gibbs R.A., Marth G., Mason C.E., Menelaou A., Muzny D.M., Nelson B.J., Noor A., Parrish N.F., Pendleton M., Quitadamo A., Raeder B., Schadt E.E., Romanovitch M., Schlattl A., Sebra R., Shabalin A.A., Untergasser A., Walker J.A., Wang M., Yu F., Zhang C., Zhang J., Zheng-Bradley X., Zhou W., Zichner T., Sebat J., Batzer M.A., McCarroll S.A., Mills R.E., Gerstein M.B., Bashir A., Stegle O., Devine S.E., Lee C., Eichler E.E., Korbel J.O. 1000 Genomes Project Consortium. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaisson M.J.P., Wilson R.K., Eichler E.E. Genetic variation and the de novo assembly of human genomes. Nat. Rev. Genet. 2015;16(11):627–640. doi: 10.1038/nrg3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eichler E.E. Genetic variation, comparative genomics, and the diagnosis of disease. N. Engl. J. Med. 2019;381(1):64–74. doi: 10.1056/NEJMra1809315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhangale T.R., Rieder M.J., Livingston R.J., Nickerson D.A. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 2005;14(1):59–69. doi: 10.1093/hmg/ddi006. [DOI] [PubMed] [Google Scholar]
- 44.Carlson C. Considerations for SNP selection. In: Winer M.P., editor. Genetic Variation: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 263–281. [Google Scholar]
- 45.Sun J.X., Helgason A., Masson G., Ebenesersdóttir S.S., Li H., Mallick S., Gnerre S., Patterson N., Kong A., Reich D., Stefansson K. A direct characterization of human mutation based on microsatellites. Nat. Genet. 2012;44(10):1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hirakawa M., Tanaka T., Hashimoto Y., Kuroda M., Takagi T., Nakamura Y. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 2002;30(1):158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haga H., Yamada R., Ohnishi Y., Nakamura Y., Tanaka T. Gene-based SNP discovery as part of the Japanese Millennium Genome Project: identification of 190,562 genetic variations in the human genome. Single-nucleotide polymorphism. J. Hum. Genet. 2002;47(11):605–610. doi: 10.1007/s100380200092. [DOI] [PubMed] [Google Scholar]
- 48.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A., Belmont J.W., Boudreau A., Hardenbol P., Leal S.M., Pasternak S., Wheeler D.A., Willis T.D., Yu F., Yang H., Zeng C., Gao Y., Hu H., Hu W., Li C., Lin W., Liu S., Pan H., Tang X., Wang J., Wang W., Yu J., Zhang B., Zhang Q., Zhao H., Zhao H., Zhou J., Gabriel S.B., Barry R., Blumenstiel B., Camargo A., Defelice M., Faggart M., Goyette M., Gupta S., Moore J., Nguyen H., Onofrio R.C., Parkin M., Roy J., Stahl E., Winchester E., Ziaugra L., Altshuler D., Shen Y., Yao Z., Huang W., Chu X., He Y., Jin L., Liu Y., Shen Y., Sun W., Wang H., Wang Y., Wang Y., Xiong X., Xu L., Waye M.M., Tsui S.K., Xue H., Wong J.T., Galver L.M., Fan J.B., Gunderson K., Murray S.S., Oliphant A.R., Chee M.S., Montpetit A., Chagnon F., Ferretti V., Leboeuf M., Olivier J.F., Phillips M.S., Roumy S., Sallée C., Verner A., Hudson T.J., Kwok P.Y., Cai D., Koboldt D.C., Miller R.D., Pawlikowska L., Taillon-Miller P., Xiao M., Tsui L.C., Mak W., Song Y.Q., Tam P.K., Nakamura Y., Kawaguchi T., Kitamoto T., Morizono T., Nagashima A., Ohnishi Y., Sekine A., Tanaka T., Tsunoda T., Deloukas P., Bird C.P., Delgado M., Dermitzakis E.T., Gwilliam R., Hunt S., Morrison J., Powell D., Stranger B.E., Whittaker P., Bentley D.R., Daly M.J., de Bakker P.I., Barrett J., Chretien Y.R., Maller J., McCarroll S., Patterson N., Pe’er I., Price A., Purcell S., Richter D.J., Sabeti P., Saxena R., Schaffner S.F., Sham P.C., Varilly P., Altshuler D., Stein L.D., Krishnan L., Smith A.V., Tello-Ruiz M.K., Thorisson G.A., Chakravarti A., Chen P.E., Cutler D.J., Kashuk C.S., Lin S., Abecasis G.R., Guan W., Li Y., Munro H.M., Qin Z.S., Thomas D.J., McVean G., Auton A., Bottolo L., Cardin N., Eyheramendy S., Freeman C., Marchini J., Myers S., Spencer C., Stephens M., Donnelly P., Cardon L.R., Clarke G., Evans D.M., Morris A.P., Weir B.S., Tsunoda T., Mullikin J.C., Sherry S.T., Feolo M., Skol A., Zhang H., Zeng C., Zhao H., Matsuda I., Fukushima Y., Macer D.R., Suda E., Rotimi C.N., Adebamowo C.A., Ajayi I., Aniagwu T., Marshall P.A., Nkwodimmah C., Royal C.D., Leppert M.F., Dixon M., Peiffer A., Qiu R., Kent A., Kato K., Niikawa N., Adewole I.F., Knoppers B.M., Foster M.W., Clayton E.W., Watkin J., Gibbs R.A., Belmont J.W., Muzny D., Nazareth L., Sodergren E., Weinstock G.M., Wheeler D.A., Yakub I., Gabriel S.B., Onofrio R.C., Richter D.J., Ziaugra L., Birren B.W., Daly M.J., Altshuler D., Wilson R.K., Fulton L.L., Rogers J., Burton J., Carter N.P., Clee C.M., Griffiths M., Jones M.C., McLay K., Plumb R.W., Ross M.T., Sims S.K., Willey D.L., Chen Z., Han H., Kang L., Godbout M., Wallenburg J.C., L’Archevêque P., Bellemare G., Saeki K., Wang H., An D., Fu H., Li Q., Wang Z., Wang R., Holden A.L., Brooks L.D., McEwen J.E., Guyer M.S., Wang V.O., Peterson J.L., Shi M., Spiegel J., Sung L.M., Zacharia L.F., Collins F.S., Kennedy K., Jamieson R., Stewart J. International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McPherson R., Pertsemlidis A., Kavaslar N., Stewart A., Roberts R., Cox D.R., Hinds D.A., Pennacchio L.A., Tybjaerg-Hansen A., Folsom A.R., Boerwinkle E., Hobbs H.H., Cohen J.C. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316(5830):1488–1491. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helgadottir A., Thorleifsson G., Manolescu A., Gretarsdottir S., Blondal T., Jonasdottir A., Jonasdottir A., Sigurdsson A., Baker A., Palsson A., Masson G., Gudbjartsson D.F., Magnusson K.P., Andersen K., Levey A.I., Backman V.M., Matthiasdottir S., Jonsdottir T., Palsson S., Einarsdottir H., Gunnarsdottir S., Gylfason A., Vaccarino V., Hooper W.C., Reilly M.P., Granger C.B., Austin H., Rader D.J., Shah S.H., Quyyumi A.A., Gulcher J.R., Thorgeirsson G., Thorsteinsdottir U., Kong A., Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 51.Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saxena R., Voight B.F., Lyssenko V., Burtt N.P., de Bakker P.I., Chen H., Roix J.J., Kathiresan S., Hirschhorn J.N., Daly M.J., Hughes T.E., Groop L., Altshuler D., Almgren P., Florez J.C., Meyer J., Ardlie K., Bengtsson Boström K., Isomaa B., Lettre G., Lindblad U., Lyon H.N., Melander O., Newton-Cheh C., Nilsson P., Orho-Melander M., Råstam L., Speliotes E.K., Taskinen M.R., Tuomi T., Guiducci C., Berglund A., Carlson J., Gianniny L., Hackett R., Hall L., Holmkvist J., Laurila E., Sjögren M., Sterner M., Surti A., Svensson M., Svensson M., Tewhey R., Blumenstiel B., Parkin M., Defelice M., Barry R., Brodeur W., Camarata J., Chia N., Fava M., Gibbons J., Handsaker B., Healy C., Nguyen K., Gates C., Sougnez C., Gage D., Nizzari M., Gabriel S.B., Chirn G.W., Ma Q., Parikh H., Richardson D., Ricke D., Purcell S. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 53.Broadbent H.M., Peden J.F., Lorkowski S., Goel A., Ongen H., Green F., Clarke R., Collins R., Franzosi M.G., Tognoni G., Seedorf U., Rust S., Eriksson P., Hamsten A., Farrall M., Watkins H., PROCARDIS consortium Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet. 2008;17(6):806–814. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 54.Ding H., Xu Y., Wang X., Wang Q., Zhang L., Tu Y., Yan J., Wang W., Hui R., Wang C.Y., Wang D.W. 9p21 is a shared susceptibility locus strongly for coronary artery disease and weakly for ischemic stroke in Chinese Han population. Circ Cardiovasc Genet. 2009;2(4):338–346. doi: 10.1161/CIRCGENETICS.108.810226. [DOI] [PubMed] [Google Scholar]
- 55.Zhou L., Zhang X., He M., Cheng L., Chen Y., Hu F.B., Wu T. Associations between single nucleotide polymorphisms on chromosome 9p21 and risk of coronary heart disease in Chinese Han population. Arterioscler. Thromb. Vasc. Biol. 2008;28(11):2085–2089. doi: 10.1161/ATVBAHA.108.176065. [DOI] [PubMed] [Google Scholar]
- 56.Shen G.Q., Li L., Rao S., Abdullah K.G., Ban J.M., Lee B.S. Four SNPs on chromosome 9p21 in a South Korean population implicate a genetic locus that confers high cross-race risk for development of coronary artery disease. Arterioscler. Thromb. Vasc. Biol. 2008;28(2):360–365. doi: 10.1161/ATVBAHA.107.157248. [DOI] [PubMed] [Google Scholar]
- 57.Hinohara K., Nakajima T., Takahashi M., Hohda S., Sasaoka T., Nakahara K.I., Chida K., Sawabe M., Arimura T., Sato A., Lee B.S., Ban J.M., Yasunami M., Park J.E., Izumi T., Kimura A. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J. Hum. Genet. 2008;53(4):357–359. doi: 10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 58.Shen G.Q., Rao S., Martinelli N., Li L., Olivieri O., Corrocher R., Abdullah K.G., Hazen S.L., Smith J., Barnard J., Plow E.F., Girelli D., Wang Q.K. Association between four SNPs on chromosome 9p21 and myocardial infarction is replicated in an Italian population. J. Hum. Genet. 2008;53(2):144–150. doi: 10.1007/s10038-007-0230-6. [DOI] [PubMed] [Google Scholar]
- 59.Assimes T.L., Knowles J.W., Basu A., Iribarren C., Southwick A., Tang H., Absher D., Li J., Fair J.M., Rubin G.D., Sidney S., Fortmann S.P., Go A.S., Hlatky M.A., Myers R.M., Risch N., Quertermous T. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE study. Hum. Mol. Genet. 2008;17(15):2320–2328. doi: 10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coronary Artery Disease (C4D) Genetics Consortium. A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43(4):339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- 61.Preuss M., König I.R., Thompson J.R., Erdmann J., Absher D., Assimes T.L., Blankenberg S., Boerwinkle E., Chen L., Cupples L.A., Hall A.S., Halperin E., Hengstenberg C., Holm H., Laaksonen R., Li M., März W., McPherson R., Musunuru K., Nelson C.P., Burnett M.S., Epstein S.E., O’Donnell C.J., Quertermous T., Rader D.J., Roberts R., Schillert A., Stefansson K., Stewart A.F., Thorleifsson G., Voight B.F., Wells G.A., Ziegler A., Kathiresan S., Reilly M.P., Samani N.J., Schunkert H., CARDIoGRAM Consortium CARDIoGRAM Consortium. Design of the Coronary ARtery DIsease Genome-Wide Replication And Meta-Analysis (CARDIoGRAM) Study: a genome-wide association meta-analysis involving more than 22 000 cases and 60 000 controls. Circ Cardiovasc Genet. 2010;3(5):475–483. doi: 10.1161/CIRCGENETICS.109.899443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., Barrett J.H., König I.R., Stevens S.E., Szymczak S., Tregouet D.A., Iles M.M., Pahlke F., Pollard H., Lieb W., Cambien F., Fischer M., Ouwehand W., Blankenberg S., Balmforth A.J., Baessler A., Ball S.G., Strom T.M., Braenne I., Gieger C., Deloukas P., Tobin M.D., Ziegler A., Thompson J.R., Schunkert H., WTCCC and the Cardiogenics Consortium Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Howson J.M.M., Zhao W., Barnes D.R., Ho W.K., Young R., Paul D.S., Waite L.L., Freitag D.F., Fauman E.B., Salfati E.L., Sun B.B., Eicher J.D., Johnson A.D., Sheu W.H.H., Nielsen S.F., Lin W.Y., Surendran P., Malarstig A., Wilk J.B., Tybjærg-Hansen A., Rasmussen K.L., Kamstrup P.R., Deloukas P., Erdmann J., Kathiresan S., Samani N.J., Schunkert H., Watkins H., Do R., Rader D.J., Johnson J.A., Hazen S.L., Quyyumi A.A., Spertus J.A., Pepine C.J., Franceschini N., Justice A., Reiner A.P., Buyske S., Hindorff L.A., Carty C.L., North K.E., Kooperberg C., Boerwinkle E., Young K., Graff M., Peters U., Absher D., Hsiung C.A., Lee W.J., Taylor K.D., Chen Y.H., Lee I.T., Guo X., Chung R.H., Hung Y.J., Rotter J.I., Juang J.J., Quertermous T., Wang T.D., Rasheed A., Frossard P., Alam D.S., Majumder A.A.S., Di Angelantonio E., Chowdhury R., Chen Y.I., Nordestgaard B.G., Assimes T.L., Danesh J., Butterworth A.S., Saleheen D. CARDIoGRAMplusC4D; EPIC-CVD. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017;49(7):1113–1119. doi: 10.1038/ng.3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schunkert H., König I.R., Kathiresan S. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43(4):333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Harst P., Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ. Res. 2018;122(3):433–443. doi: 10.1161/CIRCRESAHA.117.312086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nelson C.P., Goel A., Butterworth A.S., Kanoni S., Webb T.R., Marouli E., Zeng L., Ntalla I., Lai F.Y., Hopewell J.C., Giannakopoulou O., Jiang T., Hamby S.E., Di Angelantonio E., Assimes T.L., Bottinger E.P., Chambers J.C., Clarke R., Palmer C.N.A., Cubbon R.M., Ellinor P., Ermel R., Evangelou E., Franks P.W., Grace C., Gu D., Hingorani A.D., Howson J.M.M., Ingelsson E., Kastrati A., Kessler T., Kyriakou T., Lehtimäki T., Lu X., Lu Y., März W., McPherson R., Metspalu A., Pujades-Rodriguez M., Ruusalepp A., Schadt E.E., Schmidt A.F., Sweeting M.J., Zalloua P.A., AlGhalayini K., Keavney B.D., Kooner J.S., Loos R.J.F., Patel R.S., Rutter M.K., Tomaszewski M., Tzoulaki I., Zeggini E., Erdmann J., Dedoussis G., Björkegren J.L.M., Schunkert H., Farrall M., Danesh J., Samani N.J., Watkins H., Deloukas P., EPIC-CVD Consortium CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat. Genet. 2017;49(9):1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 67.Nikpay M., Goel A., Won H.H., Hall L.M., Willenborg C., Kanoni S., Saleheen D., Kyriakou T., Nelson C.P., Hopewell J.C., Webb T.R., Zeng L., Dehghan A., Alver M., Armasu S.M., Auro K., Bjonnes A., Chasman D.I., Chen S., Ford I., Franceschini N., Gieger C., Grace C., Gustafsson S., Huang J., Hwang S.J., Kim Y.K., Kleber M.E., Lau K.W., Lu X., Lu Y., Lyytikäinen L.P., Mihailov E., Morrison A.C., Pervjakova N., Qu L., Rose L.M., Salfati E., Saxena R., Scholz M., Smith A.V., Tikkanen E., Uitterlinden A., Yang X., Zhang W., Zhao W., de Andrade M., de Vries P.S., van Zuydam N.R., Anand S.S., Bertram L., Beutner F., Dedoussis G., Frossard P., Gauguier D., Goodall A.H., Gottesman O., Haber M., Han B.G., Huang J., Jalilzadeh S., Kessler T., König I.R., Lannfelt L., Lieb W., Lind L., Lindgren C.M., Lokki M.L., Magnusson P.K., Mallick N.H., Mehra N., Meitinger T., Memon F.U., Morris A.P., Nieminen M.S., Pedersen N.L., Peters A., Rallidis L.S., Rasheed A., Samuel M., Shah S.H., Sinisalo J., Stirrups K.E., Trompet S., Wang L., Zaman K.S., Ardissino D., Boerwinkle E., Borecki I.B., Bottinger E.P., Buring J.E., Chambers J.C., Collins R., Cupples L.A., Danesh J., Demuth I., Elosua R., Epstein S.E., Esko T., Feitosa M.F., Franco O.H., Franzosi M.G., Granger C.B., Gu D., Gudnason V., Hall A.S., Hamsten A., Harris T.B., Hazen S.L., Hengstenberg C., Hofman A., Ingelsson E., Iribarren C., Jukema J.W., Karhunen P.J., Kim B.J., Kooner J.S., Kullo I.J., Lehtimäki T., Loos R.J.F., Melander O., Metspalu A., März W., Palmer C.N., Perola M., Quertermous T., Rader D.J., Ridker P.M., Ripatti S., Roberts R., Salomaa V., Sanghera D.K., Schwartz S.M., Seedorf U., Stewart A.F., Stott D.J., Thiery J., Zalloua P.A., O’Donnell C.J., Reilly M.P., Assimes T.L., Thompson J.R., Erdmann J., Clarke R., Watkins H., Kathiresan S., McPherson R., Deloukas P., Schunkert H., Samani N.J., Farrall M. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015;47(10):1121–1130. doi: 10.1038/ng.3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Khera A.V., Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat. Rev. Genet. 2017;18(6):331–344. doi: 10.1038/nrg.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kessler T., Vilne B., Schunkert H. The impact of genome-wide association studies on the pathophysiology and therapy of cardiovascular disease. EMBO Mol. Med. 2016;8(7):688–701. doi: 10.15252/emmm.201506174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.CARDIoGRAMplusC4D Consortium. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 2013;45(1):25–33. doi: 10.1038/ng.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Assimes T.L., Roberts R. Genetics: implications for prevention and management of coronary artery disease. J. Am. Coll. Cardiol. 2016;68(25):2797–2818. doi: 10.1016/j.jacc.2016.10.039. [DOI] [PubMed] [Google Scholar]
- 72.Brown M.S., Goldstein J.L. Familial hypercholesterolemia: A genetic defect in the low-density lipoprotein receptor. N. Engl. J. Med. 1976;294(25):1386–1390. doi: 10.1056/NEJM197606172942509. [DOI] [PubMed] [Google Scholar]
- 73.Roberts R. PCSK9 inhibition--a new thrust in the prevention of heart disease: genetics does it again. Can. J. Cardiol. 2013;29(8):899–901. doi: 10.1016/j.cjca.2013.04.014. [DOI] [PubMed] [Google Scholar]
- 74.Seidah N.G., Benjannet S., Wickham L., Marcinkiewicz J., Jasmin S.B., Stifani S., Basak A., Prat A., Chretien M. The secretory proprotein convertase neural apoptosis-regulated convertase 1 (NARC-1): liver regeneration and neuronal differentiation. Proc. Natl. Acad. Sci. USA. 2003;100(3):928–933. doi: 10.1073/pnas.0335507100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Abifadel M., Varret M., Rabès J.P., Allard D., Ouguerram K., Devillers M., Cruaud C., Benjannet S., Wickham L., Erlich D., Derré A., Villéger L., Farnier M., Beucler I., Bruckert E., Chambaz J., Chanu B., Lecerf J.M., Luc G., Moulin P., Weissenbach J., Prat A., Krempf M., Junien C., Seidah N.G., Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat. Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 76.Cohen J.C., Boerwinkle E., Mosley T.H., Jr, Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 77.Ni Y.G., Di Marco S., Condra J.H., Peterson L.B., Wang W., Wang F., Pandit S., Hammond H.A., Rosa R., Cummings R.T., Wood D.D., Liu X., Bottomley M.J., Shen X., Cubbon R.M., Wang S.P., Johns D.G., Volpari C., Hamuro L., Chin J., Huang L., Zhao J.Z., Vitelli S., Haytko P., Wisniewski D., Mitnaul L.J., Sparrow C.P., Hubbard B., Carfí A., Sitlani A.A. PCSK9-binding antibody that structurally mimics the EGF(A) domain of LDL-receptor reduces LDL cholesterol in vivo. J. Lipid Res. 2011;52(1):78–86. doi: 10.1194/jlr.M011445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stein E.A., Mellis S., Yancopoulos G.D., Stahl N., Logan D., Smith W.B., Lisbon E., Gutierrez M., Webb C., Wu R., Du Y., Kranz T., Gasparino E., Swergold G.D. Effect of a monoclonal antibody to PCSK9 on LDL cholesterol. N. Engl. J. Med. 2012;366(12):1108–1118. doi: 10.1056/NEJMoa1105803. [DOI] [PubMed] [Google Scholar]
- 79.Sabatine M.S., Giugliano R.P., Keech A.C., Honarpour N., Wiviott S.D., Murphy S.A., Kuder J.F., Wang H., Liu T., Wasserman S.M., Sever P.S., Pedersen T.R., FOURIER Steering Committee and Investigators Evolocumab and clinical outcomes in patients with cardiovascular disease. N. Engl. J. Med. 2017;376(18):1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 80.Schwartz G.G., Szarek M., Bhatt D.L., Bittner V., Diaz R., Edelberg J., Goodman S.G. 2018. [Google Scholar]
- 81.Libby P. Interleukin-1 beta as a target for atherosclerosis therapy: biological basis of CANTOS and beyond. J. Am. Coll. Cardiol. 2017;70(18):2278–2289. doi: 10.1016/j.jacc.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hansson G.K. Inflammation and atherosclerosis: the end of a controversy. Circulation. 2017;136(20):1875–1877. doi: 10.1161/CIRCULATIONAHA.117.030484. [DOI] [PubMed] [Google Scholar]
- 83.Ridker P.M., Everett B.M., Thuren T., MacFadyen J.G., Chang W.H., Ballantyne C., Fonseca F., Nicolau J., Koenig W., Anker S.D., Kastelein J.J.P., Cornel J.H., Pais P., Pella D., Genest J., Cifkova R., Lorenzatti A., Forster T., Kobalava Z., Vida-Simiti L., Flather M., Shimokawa H., Ogawa H., Dellborg M., Rossi P.R.F., Troquay R.P.T., Libby P., Glynn R.J., CANTOS Trial Group Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 2017;377(12):1119–1131. doi: 10.1056/NEJMoa1707914. [DOI] [PubMed] [Google Scholar]
- 84.Roberts R. Mendelian randomization studies promise to shorten the journey to FDA approval. JACC Basic Transl. Sci. 2018;3(5):690–703. doi: 10.1016/j.jacbts.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Voight B.F., Peloso G.M., Orho-Melander M., Frikke-Schmidt R., Barbalic M., Jensen M.K., Hindy G., Hólm H., Ding E.L., Johnson T., Schunkert H., Samani N.J., Clarke R., Hopewell J.C., Thompson J.F., Li M., Thorleifsson G., Newton-Cheh C., Musunuru K., Pirruccello J.P., Saleheen D., Chen L., Stewart A., Schillert A., Thorsteinsdottir U., Thorgeirsson G., Anand S., Engert J.C., Morgan T., Spertus J., Stoll M., Berger K., Martinelli N., Girelli D., McKeown P.P., Patterson C.C., Epstein S.E., Devaney J., Burnett M.S., Mooser V., Ripatti S., Surakka I., Nieminen M.S., Sinisalo J., Lokki M.L., Perola M., Havulinna A., de Faire U., Gigante B., Ingelsson E., Zeller T., Wild P., de Bakker P.I., Klungel O.H., Maitland-van der Zee A.H., Peters B.J., de Boer A., Grobbee D.E., Kamphuisen P.W., Deneer V.H., Elbers C.C., Onland-Moret N.C., Hofker M.H., Wijmenga C., Verschuren W.M., Boer J.M., van der Schouw Y.T., Rasheed A., Frossard P., Demissie S., Willer C., Do R., Ordovas J.M., Abecasis G.R., Boehnke M., Mohlke K.L., Daly M.J., Guiducci C., Burtt N.P., Surti A., Gonzalez E., Purcell S., Gabriel S., Marrugat J., Peden J., Erdmann J., Diemert P., Willenborg C., König I.R., Fischer M., Hengstenberg C., Ziegler A., Buysschaert I., Lambrechts D., Van de Werf F., Fox K.A., El Mokhtari N.E., Rubin D., Schrezenmeir J., Schreiber S., Schäfer A., Danesh J., Blankenberg S., Roberts R., McPherson R., Watkins H., Hall A.S., Overvad K., Rimm E., Boerwinkle E., Tybjaerg-Hansen A., Cupples L.A., Reilly M.P., Melander O., Mannucci P.M., Ardissino D., Siscovick D., Elosua R., Stefansson K., O’Donnell C.J., Salomaa V., Rader D.J., Peltonen L., Schwartz S.M., Altshuler D., Kathiresan S. Plasma HDL cholesterol and risk of myocardial infarction: a mendelian randomisation study. Lancet. 2012;380(9841):572–580. doi: 10.1016/S0140-6736(12)60312-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Teslovich T.M., Musunuru K., Smith A.V., Edmondson A.C., Stylianou I.M., Koseki M., Pirruccello J.P., Ripatti S., Chasman D.I., Willer C.J., Johansen C.T., Fouchier S.W., Isaacs A., Peloso G.M., Barbalic M., Ricketts S.L., Bis J.C., Aulchenko Y.S., Thorleifsson G., Feitosa M.F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J.Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D.C., Lange L.A., Smith J.D., Song K., Zhao H. J.; Yuan, X.; Luan, J.; Lamina, C.; Ziegler, A.; Zhang, W.; Zee, R.Y.; Wright, A.F.; Witteman, J.C.; Wilson, J.F.; Willemsen, G.; Wichmann, H.E.; Whitfield, J.B.; Waterworth, D.M.; Wareham, N.J.; Waeber, G.; Vollenweider, P.; Voight, B.F.; Vitart, V.; Uitterlinden, A.G.; Uda, M.; Tuomilehto, J.; Thompson, J.R.; Tanaka, T.; Surakka, I.; Stringham, H.M.; Spector, T.D.; Soranzo, N.; Smit, J.H.; Sinisalo, J.; Silander, K.; Sijbrands, E.J.; Scuteri, A.; Scott, J.; Schlessinger, D.; Sanna, S.; Salomaa, V.; Saharinen, J.; Sabatti, C.; Ruokonen, A.; Rudan, I.; Rose, L.M.; Roberts, R.; Rieder, M.; Psaty, B.M.; Pramstaller, P.P.; Pichler, I.; Perola, M.; Penninx, B.W.; Pedersen, N.L.; Pattaro, C.; Parker, A.N.; Pare, G.; Oostra, B.A.; O’Donnell, C.J.; Nieminen, M.S.; Nickerson, D.A.; Montgomery, G.W.; Meitinger, T.; McPherson, R.; McCarthy, M.I.; McArdle, W.; Masson, D.; Martin, N.G.; Marroni, F.; Mangino, M.; Magnusson, P.K.; Lucas, G.; Luben, R.; Loos, R.J.; Lokki, M.L.; Lettre, G.; Langenberg, C.; Launer, L.J.; Lakatta, E.G.; Laaksonen, R.; Kyvik, K.O.; Kronenberg, F.; König, I.R.; Khaw, K.T.; Kaprio, J.; Kaplan, L.M.; Johansson, A.; Jarvelin, M.R.; Janssens, A.C.; Ingelsson, E.; Igl, W.; Kees Hovingh, G.; Hottenga, J.J.; Hofman, A.; Hicks, A.A.; Hengstenberg, C.; Heid, I.M.; Hayward, C.; Havulinna, A.S.; Hastie, N.D.; Harris, T.B.; Haritunians, T.; Hall, A.S.; Gyllensten, U.; Guiducci, C.; Groop, L.C.; Gonzalez, E.; Gieger, C.; Freimer, N.B.; Ferrucci, L.; Erdmann, J.; Elliott, P.; Ejebe, K.G.; Döring, A.; Dominiczak, A.F.; Demissie, S.; Deloukas, P.; de Geus, E.J.; de Faire, U.; Crawford, G.; Collins, F.S.; Chen, Y.D.; Caulfield, M.J.; Campbell, H.; Burtt, N.P.; Bonnycastle, L.L.; Boomsma, D.I.; Boekholdt, S.M.; Bergman, R.N.; Barroso, I.; Bandinelli, S.; Ballantyne, C.M.; Assimes, T.L.; Quertermous, T.; Altshuler, D.; Seielstad, M.; Wong, T.Y.; Tai, E.S.; Feranil, A.B.; Kuzawa, C.W.; Adair, L.S.; Taylor, H.A., Jr; Borecki, I.B.; Gabriel, S.B.; Wilson, J.G.; Holm, H.; Thorsteinsdottir, U.; Gudnason, V.; Krauss, R.M.; Mohlke, K.L.; Ordovas, J.M.; Munroe, P.B.; Kooner, J.S.; Tall, A.R.; Hegele, R.A.; Kastelein, J.J.; Schadt, E.E.; Rotter, J.I.; Boerwinkle, E.; Strachan, D.P.; Mooser, V.; Stefansson, K.; Reilly, M.P.; Samani, N.J.; Schunkert, H.; Cupples, L.A.; Sandhu, M.S.; Ridker, P.M.; Rader, D.J.; van Duijn, C.M.; Peltonen, L.; Abecasis, G.R.; Boehnke, M.; Kathiresan, S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Willer C.J., Schmidt E.M., Sengupta S., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., Buchkovich M.L., Mora S., Beckmann J.S., Bragg-Gresham J.L., Chang H.Y., Demirkan A., Den Hertog H.M., Do R., Donnelly L.A., Ehret G.B., Esko T., Feitosa M.F., Ferreira T., Fischer K., Fontanillas P., Fraser R.M., Freitag D.F., Gurdasani D., Heikkilä K., Hyppönen E., Isaacs A., Jackson A.U., Johansson Å., Johnson T., Kaakinen M., Kettunen J., Kleber M.E., Li X., Luan J., Lyytikäinen L.P., Magnusson P.K.E., Mangino M., Mihailov E., Montasser M.E., Müller-Nurasyid M., Nolte I.M., O’Connell J.R., Palmer C.D., Perola M., Petersen A.K., Sanna S., Saxena R., Service S.K., Shah S., Shungin D., Sidore C., Song C., Strawbridge R.J., Surakka I., Tanaka T., Teslovich T.M., Thorleifsson G., Van den Herik E.G., Voight B.F., Volcik K.A., Waite L.L., Wong A., Wu Y., Zhang W., Absher D., Asiki G., Barroso I., Been L.F., Bolton J.L., Bonnycastle L.L., Brambilla P., Burnett M.S., Cesana G., Dimitriou M., Doney A.S.F., Döring A., Elliott P., Epstein S.E., Ingi Eyjolfsson G., Gigante B., Goodarzi M.O., Grallert H., Gravito M.L., Groves C.J., Hallmans G., Hartikainen A.L., Hayward C., Hernandez D., Hicks A.A., Holm H., Hung Y.J., Illig T., Jones M.R., Kaleebu P., Kastelein J.J.P., Khaw K.T., Kim E., Klopp N., Komulainen P., Kumari M., Langenberg C., Lehtimäki T., Lin S.Y., Lindström J., Loos R.J.F., Mach F., McArdle W.L., Meisinger C., Mitchell B.D., Müller G., Nagaraja R., Narisu N., Nieminen T.V.M., Nsubuga R.N., Olafsson I., Ong K.K., Palotie A., Papamarkou T., Pomilla C., Pouta A., Rader D.J., Reilly M.P., Ridker P.M., Rivadeneira F., Rudan I., Ruokonen A., Samani N., Scharnagl H., Seeley J., Silander K., Stančáková A., Stirrups K., Swift A.J., Tiret L., Uitterlinden A.G., van Pelt L.J., Vedantam S., Wainwright N., Wijmenga C., Wild S.H., Willemsen G., Wilsgaard T., Wilson J.F., Young E.H., Zhao J.H., Adair L.S., Arveiler D., Assimes T.L., Bandinelli S., Bennett F., Bochud M., Boehm B.O., Boomsma D.I., Borecki I.B., Bornstein S.R., Bovet P., Burnier M., Campbell H., Chakravarti A., Chambers J.C., Chen Y.I., Collins F.S., Cooper R.S., Danesh J., Dedoussis G., de Faire U., Feranil A.B., Ferrières J., Ferrucci L., Freimer N.B., Gieger C., Groop L.C., Gudnason V., Gyllensten U., Hamsten A., Harris T.B., Hingorani A., Hirschhorn J.N., Hofman A., Hovingh G.K., Hsiung C.A., Humphries S.E., Hunt S.C., Hveem K., Iribarren C., Järvelin M.R., Jula A., Kähönen M., Kaprio J., Kesäniemi A., Kivimaki M., Kooner J.S., Koudstaal P.J., Krauss R.M., Kuh D., Kuusisto J., Kyvik K.O., Laakso M., Lakka T.A., Lind L., Lindgren C.M., Martin N.G., März W., McCarthy M.I., McKenzie C.A., Meneton P., Metspalu A., Moilanen L., Morris A.D., Munroe P.B., Njølstad I., Pedersen N.L., Power C., Pramstaller P.P., Price J.F., Psaty B.M., Quertermous T., Rauramaa R., Saleheen D., Salomaa V., Sanghera D.K., Saramies J., Schwarz P.E.H., Sheu W.H., Shuldiner A.R., Siegbahn A., Spector T.D., Stefansson K., Strachan D.P., Tayo B.O., Tremoli E., Tuomilehto J., Uusitupa M., van Duijn C.M., Vollenweider P., Wallentin L., Wareham N.J., Whitfield J.B., Wolffenbuttel B.H.R., Ordovas J.M., Boerwinkle E., Palmer C.N.A., Thorsteinsdottir U., Chasman D.I., Rotter J.I., Franks P.W., Ripatti S., Cupples L.A., Sandhu M.S., Rich S.S., Boehnke M., Deloukas P., Kathiresan S., Mohlke K.L., Ingelsson E., Abecasis G.R., Global Lipids Genetics Consortium Discovery and refinement of loci associated with lipid levels. Nat. Genet. 2013;45(11):1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gofman J.W., Young W., Tandy R. Ischemic heart disease, atherosclerosis, and longevity. Circulation. 1966;34(4):679–697. doi: 10.1161/01.CIR.34.4.679. [DOI] [PubMed] [Google Scholar]
- 89.Gadi R., Amanullah A., Figueredo V.M. HDL-C: does it matter? an update on novel HDL-directed pharmaco-therapeutic strategies. Int. J. Cardiol. 2013;167(3):646–655. doi: 10.1016/j.ijcard.2012.05.052. [DOI] [PubMed] [Google Scholar]
- 90.Boden W.E., Probstfield J.L., Anderson T., Chaitman B.R., Desvignes-Nickens P., Koprowicz K., McBride R., Teo K., Weintraub W., AIM-HIGH Investigators Niacin in patients with low HDL cholesterol levels receiving intensive statin therapy. N. Engl. J. Med. 2011;365(24):2255–2267. doi: 10.1056/NEJMoa1107579. [DOI] [PubMed] [Google Scholar]
- 91.Wright R.S. Recent clinical trials evaluating benefit of drug therapy for modification of HDL cholesterol. Curr. Opin. Cardiol. 2013;28(4):389–398. doi: 10.1097/HCO.0b013e328362059d. [DOI] [PubMed] [Google Scholar]
- 92.Lincoff A.M., Nicholls S.J., Riesmeyer J.S., Barter P.J., Brewer H.B., Fox K.A.A., Gibson C.M., Granger C., Menon V., Montalescot G., Rader D., Tall A.R., McErlean E., Wolski K., Ruotolo G., Vangerow B., Weerakkody G., Goodman S.G., Conde D., McGuire D.K., Nicolau J.C., Leiva-Pons J.L., Pesant Y., Li W., Kandath D., Kouz S., Tahirkheli N., Mason D., Nissen S.E., ACCELERATE Investigators Evacetrapib and cardiovascular outcomes in highrisk vascular disease. N. Engl. J. Med. 2017;376(20):1933–1942. doi: 10.1056/NEJMoa1609581. [DOI] [PubMed] [Google Scholar]
- 93.Kawashiri M.A., Tada H., Nomura A., Yamagishi M. Mendelian randomization: its impact on cardiovascular disease. J. Cardiol. 2018;72(4):307–313. doi: 10.1016/j.jjcc.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 94.Barter P.J., Caulfield M., Eriksson M., Grundy S.M., Kastelein J.J., Komajda M., Lopez-Sendon J., Mosca L., Tardif J.C., Waters D.D., Shear C.L., Revkin J.H., Buhr K.A., Fisher M.R., Tall A.R., Brewer B., ILLUMINATE Investigators Effects of torcetrapib in patients at high risk for coronary events. N. Engl. J. Med. 2007;357(21):2109–2122. doi: 10.1056/NEJMoa0706628. [DOI] [PubMed] [Google Scholar]
- 95.Schwartz G.G., Olsson A.G., Abt M., Ballantyne C.M., Barter P.J., Brumm J., Chaitman B.R., Holme I.M., Kallend D., Leiter L.A., Leitersdorf E., McMurray J.J., Mundl H., Nicholls S.J., Shah P.K., Tardif J.C., Wright R.S. Effects of dalcetrapib in patients with a recent acute coronary syndrome. N. Engl. J. Med. 2012;367(22):2089–2099. doi: 10.1056/NEJMoa1206797. [DOI] [PubMed] [Google Scholar]
- 96.Bowman L., Hopewell J.C., Chen F., Wallendszus K., Stevens W., Collins R., Wiviott S.D., Cannon C.P., Braunwald E., Sammons E., Landray M.J., HPS3/TIMI55–REVEAL Collaborative Group Effects of anacetrapib in patients with atherosclerotic vascular disease. N. Engl. J. Med. 2017;377(13):1217–1227. doi: 10.1056/NEJMoa1706444. [DOI] [PubMed] [Google Scholar]
- 97.Nomura A., Won H.H., Khera A.V., Takeuchi F., Ito K., McCarthy S., Emdin C.A., Klarin D., Natarajan P., Zekavat S.M., Gupta N., Peloso G.M., Borecki I.B., Teslovich T.M., Asselta R., Duga S., Merlini P.A., Correa A., Kessler T., Wilson J.G., Bown M.J., Hall A.S., Braund P.S., Carey D.J., Murray M.F., Kirchner H.L., Leader J.B., Lavage D.R., Manus J.N., Hartze D.N., Samani N.J., Schunkert H., Marrugat J., Elosua R., McPherson R., Farrall M., Watkins H., Juang J.J., Hsiung C.A., Lin S.Y., Wang J.S., Tada H., Kawashiri M.A., Inazu A., Yamagishi M., Katsuya T., Nakashima E., Nakatochi M., Yamamoto K., Yokota M., Momozawa Y., Rotter J.I., Lander E.S., Rader D.J., Danesh J., Ardissino D., Gabriel S., Willer C.J., Abecasis G.R., Saleheen D., Kubo M., Kato N., Ida Chen Y.D., Dewey F.E., Kathiresan S. Proteintruncating variants at the cholesteryl ester transfer protein gene and risk for coronary heart disease. Circ. Res. 2017;121(1):81–88. doi: 10.1161/CIRCRESAHA.117.311145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keene D., Price C., Shun-Shin M.J., Francis D.P. Effect on cardiovascular risk of high density lipoprotein targeted drug treatments niacin, fibrates, and CETP inhibitors: meta-analysis of randomised controlled trials including 117,411 patients. BMJ. 2014;349:g4379. doi: 10.1136/bmj.g4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang H.T., Lee M., Hong K.S., Ovbiagele B., Saver J.L. Efficacy of folic acid supplementation in cardiovascular disease prevention: an updated meta-analysis of randomized controlled trials. Eur. J. Intern. Med. 2012;23(8):745–754. doi: 10.1016/j.ejim.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 100.Keenan T., Zhao W., Rasheed A., Ho W.K., Malik R., Felix J.F., Young R., Shah N., Samuel M., Sheikh N., Mucksavage M.L., Shah O., Li J., Morley M., Laser A., Mallick N.H., Zaman K.S., Ishaq M., Rasheed S.Z., Memon F.U., Ahmed F., Hanif B., Lakhani M.S., Fahim M., Ishaq M., Shardha N.K., Ahmed N., Mahmood K., Iqbal W., Akhtar S., Raheel R., O’Donnell C.J., Hengstenberg C., März W., Kathiresan S., Samani N., Goel A., Hopewell J.C., Chambers J., Cheng Y.C., Sharma P., Yang Q., Rosand J., Boncoraglio G.B., Kazmi S.U., Hakonarson H., Köttgen A., Kalogeropoulos A., Frossard P., Kamal A., Dichgans M., Cappola T., Reilly M.P., Danesh J., Rader D.J., Voight B.F., Saleheen D. Causal assessment of serum urate levels in cardiometabolic diseases through a Mendelian randomization study. J. Am. Coll. Cardiol. 2016;67(4):407–416. doi: 10.1016/j.jacc.2015.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Erdmann J., Kessler T., Munoz Venegas L., Schunkert H. A decade of genome-wide association studies for coronary artery disease: the challenges ahead. Cardiovasc. Res. 2018;114(9):1241–1257. doi: 10.1093/cvr/cvy084. [DOI] [PubMed] [Google Scholar]
- 102.Goldstein B.A., Knowles J.W., Salfati E., Ioannidis J.P., Assimes T.L. Simple, standardized incorporation of genetic risk into non-genetic risk prediction tools for complex traits: coronary heart disease as an example. Front. Genet. 2014;5:254. doi: 10.3389/fgene.2014.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fujita H., Okada T., Inami I., Makimoto M., Hosono S., Minato M., Takahashi S., Mugishima H., Yamamoto T. Low-density lipoprotein profile changes during the neonatal period. J. Perinatol. 2008;28(5):335–340. doi: 10.1038/jp.2008.8. [DOI] [PubMed] [Google Scholar]
- 104.Pac-Kozuchowska E. Evaluation of lipids, lipoproteins and apolipoproteins concentrations in cord blood serum of newborns from rural and urban environments. Ann. Agric. Environ. Med. 2007;14(1):25–29. [PubMed] [Google Scholar]
- 105.Swiger K.J., Martin S.S., Blaha M.J. Narrowing sex differences in lipoprotein cholesterol subclasses following mid-life: the very large database of lipids (VLDL-10B). J. Am. Heart Assoc. 2014;3(2):e000851. doi: 10.1161/JAHA.114.000851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Paynter N.P., Chasman D.I., Buring J.E., Shiffman D., Cook N.R., Ridker P.M. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann. Intern. Med. 2009;150(2):65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ripatti S., Tikkanen E., Orho-Melander M., Havulinna A.S., Silander K., Sharma A., Guiducci C., Perola M., Jula A., Sinisalo J., Lokki M.L., Nieminen M.S., Melander O., Salomaa V., Peltonen L., Kathiresan S. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376(9750):1393–1400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tikkanen E., Havulinna A.S., Palotie A., Salomaa V., Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler. Thromb. Vasc. Biol. 2013;33(9):2261–2266. doi: 10.1161/ATVBAHA.112.301120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Thanassoulis G., Peloso G.M., Pencina M.J., Hoffmann U., Fox C.S., Cupples L.A., Levy D., D’Agostino R.B., Hwang S.J., O’Donnell C.J. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet. 2012;5(1):113–121. doi: 10.1161/CIRCGENETICS.111.961342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davies R.W., Dandona S., Stewart A.F., Chen L., Ellis S.G., Tang W.H., Hazen S.L., Roberts R., McPherson R., Wells G.A. Improved prediction of cardiovascular disease based on a panel of single nucleotide polymorphisms identified through genome-wide association studies. Circ Cardiovasc Genet. 2010;3(5):468–474. doi: 10.1161/CIRCGENETICS.110.946269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mega J.L., Stitziel N.O., Smith J.G., Chasman D.I., Caulfield M., Devlin J.J., Nordio F., Hyde C., Cannon C.P., Sacks F., Poulter N., Sever P., Ridker P.M., Braunwald E., Melander O., Kathiresan S., Sabatine M.S. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385(9984):2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Natarajan P., Young R., Stitziel N.O., Padmanabhan S., Baber U., Mehran R., Sartori S., Fuster V., Reilly D.F., Butterworth A., Rader D.J., Ford I., Sattar N., Kathiresan S. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135(22):2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Torkamani A., Wineinger N.E., Topol E.J. The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 2018;19(9):581–590. doi: 10.1038/s41576-018-0018-x. [DOI] [PubMed] [Google Scholar]
- 114.Abraham G., Havulinna A.S., Bhalala O.G., Byars S.G., De Livera A.M., Yetukuri L., Tikkanen E., Perola M., Schunkert H., Sijbrands E.J., Palotie A., Samani N.J., Salomaa V., Ripatti S., Inouye M. Genomic prediction of coronary heart disease. Eur. Heart J. 2016;37(43):3267–3278. doi: 10.1093/eurheartj/ehw450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Inouye M., Abraham G., Nelson C.P., Wood A.M., Sweeting M.J., Dudbridge F., Lai F.Y., Kaptoge S., Brozynska M., Wang T., Ye S., Webb T.R., Rutter M.K., Tzoulaki I., Patel R.S., Loos R.J.F., Keavney B., Hemingway H., Thompson J., Watkins H., Deloukas P., Di Angelantonio E., Butterworth A.S., Danesh J., Samani N.J. UK Biobank CardioMetabolic Consortium CHD Working Group. Genomic risk prediction of coronary artery disease in 480,000 adults. J. Am. Coll. Cardiol. 2018;72(16):1883–1893. doi: 10.1016/j.jacc.2018.07.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vilhjálmsson B.J., Yang J., Finucane H.K., Gusev A., Lindström S., Ripke S., Genovese G., Loh P.R., Bhatia G., Do R., Hayeck T., Won H.H., Kathiresan S., Pato M., Pato C., Tamimi R., Stahl E., Zaitlen N., Pasaniuc B., Belbin G., Kenny E.E., Schierup M.H., De Jager P., Patsopoulos N.A., McCarroll S., Daly M., Purcell S., Chasman D., Neale B., Goddard M., Visscher P.M., Kraft P., Patterson N., Price A.L., Schizophrenia Working Group of the Psychiatric Genomics Consortium Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) study. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97(4):576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Khera A.V., Chaffin M., Aragam K.G., Haas M.E., Roselli C., Choi S.H., Natarajan P., Lander E.S., Lubitz S.A., Ellinor P.T., Kathiresan S. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018;50(9):1219–1224. doi: 10.1038/s41588-018-0183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang Y., Qi G., Park J-H., Chatterjee N. Estimation of complex effect-size distributions using summary-level statistics from genome-wide association studies across 32 complex traits. Nat. Genet. 2018;50(9):1318–1326. doi: 10.1038/s41588-018-0193-x. [DOI] [PubMed] [Google Scholar]
- 119.Khera A.V., Emdin C.A., Drake I., Natarajan P., Bick A.G., Cook N.R., Chasman D.I., Baber U., Mehran R., Rader D.J., Fuster V., Boerwinkle E., Melander O., Orho-Melander M., Ridker P.M., Kathiresan S. Genetic risk, adherence to a healthy lifestyle, and coronary disease. N. Engl. J. Med. 2016;375(24):2349–2358. doi: 10.1056/NEJMoa1605086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tikkanen E., Gustafsson S., Ingelsson E. Associations of fitness, physical activity, strength, and genetic risk with cardiovascular disease: longitudinal analyses in the UK Biobank study. Circulation. 2018;137(24):2583–2591. doi: 10.1161/CIRCULATIONAHA.117.032432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marston N.A., Kamanu F.K., Nordio F., Gurmu Y., Roselli C., Sever P.S., Pedersen T.R., Keech A.C., Wang H., Lira Pineda A., Giugliano R.P., Lubitz S.A., Ellinor P.T., Sabatine M.S., Ruff C.T. Predicting benefit from evolocumab therapy in patients with atherosclerotic disease using a genetic risk score: Results From the FOURIER Trial. Circulation. 2020;141(8):616–623. doi: 10.1161/CIRCULATIONAHA.119.043805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Damask A., Steg P.G., Schwartz G.G., Szarek M., Hagström E., Badimon L., Chapman M.J., Boileau C., Tsimikas S., Ginsberg H.N., Banerjee P., Manvelian G., Pordy R., Hess S., Overton J.D., Lotta L.A., Yancopoulos G.D., Abecasis G.R., Baras A., Paulding C. Regeneron genetics center and the ODYSSEY OUTCOMES investigators. Patients with high genome-wide polygenic risk scores for coronary artery disease may receive greater clinical benefit from alirocumab treatment in the ODYSSEY OUTCOMES trial. Circulation. 2020;141(8):624–636. doi: 10.1161/CIRCULATIONAHA.119.044434. [DOI] [PubMed] [Google Scholar]
- 123.Elliott J., Bodinier B., Bond T.A., Chadeau-Hyam M., Evangelou E., Moons K.G.M., Dehghan A., Muller D.C., Elliott P., Tzoulaki I. Predictive accuracy of a polygenic risk score-enhanced prediction model vs a clinical risk score for coronary artery disease. JAMA. 2020;323(7):636–645. doi: 10.1001/jama.2019.22241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mosley J.D., Gupta D.K., Tan J., Yao J., Wells Q.S., Shaffer C.M., Kundu S., Robinson-Cohen C., Psaty B.M., Rich S.S., Post W.S., Guo X., Rotter J.I., Roden D.M., Gerszten R.E., Wang T.J. Predictive accuracy of a polygenic risk score compared with a clinical risk score for incident coronary heart disease. JAMA. 2020;323(7):627–635. doi: 10.1001/jama.2019.21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Khan S.S., Cooper R., Greenland P. Do polygenic risk scores improve patient selection for prevention of coronary artery disease? JAMA. 2020;323(7):614–615. doi: 10.1001/jama.2019.21667. [DOI] [PubMed] [Google Scholar]
- 126.Roberts R. Genetic risk stratification: tipping point for global primary prevention of coronary artery disease. Circulation. 2018;137(24):2554–2556. doi: 10.1161/CIRCULATIONAHA.118.034732. [DOI] [PubMed] [Google Scholar]
- 127.Manolio T.A., Collins F.S. The HapMap and genome-wide association studies in diagnosis and therapy. Annu. Rev. Med. 2009;60(1):443–456. doi: 10.1146/annurev.med.60.061907.093117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Baigent C., Blackwell L., Emberson J., Holland L.E., Reith C., Bhala N., Peto R., Barnes E.H., Keech A., Simes J., Collins R., Cholesterol Treatment Trialists’ (CTT) Collaboration Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 2010;376(9753):1670–1681. doi: 10.1016/S0140-6736(10)61350-5. [DOI] [PMC free article] [PubMed] [Google Scholar]