Abstract
Introduction
Plants do not grow in isolation, rather they are hosts to a variety of microbes in their natural environments. While, few thrive in the plants for their own benefit, others may have a direct impact on plants in a symbiotic manner. Unraveling plant-microbe interactions is a critical component in recognizing the positive and negative impacts of microbes on plants. Also, by affecting the environment around plants, microbes may indirectly influence plants. The progress in sequencing technologies in the genomics era and several omics tools has accelerated in biological science. Studying the complex nature of plant-microbe interactions can offer several strategies to increase the productivity of plants in an environmentally friendly manner by providing better insights. This review brings forward the recent works performed in building omics strategies that decipher the interactions between plant-microbiome. At the same time, it further explores other associated mutually beneficial aspects of plant-microbe interactions such as plant growth promotion, nitrogen fixation, stress suppressions in crops and bioremediation; as well as provides better insights on metabolic interactions between microbes and plants through omics approaches. It also aims to explore advances in the study of Arabidopsis as an important avenue to serve as a baseline tool to create models that help in scrutinizing various factors that contribute to the elaborate relationship between plants and microbes. Causal relationships between plants and microbes can be established through systematic gnotobiotic experimental studies to test hypotheses on biologically derived interactions.
Conclusion
This review will cover recent advances in the study of plant-microbe interactions keeping in view the advantages of these interactions in improving nutrient uptake and plant health.
Keywords: Plant-microbe interactions, plant stress response, genomics, transcriptomics, proteomics, metabolomics
1. Introduction
Microbial interactions play an important role in sustaining several ecosystems. Description and knowledge about these interactions between microorganisms and other biotic factors is an important step towards understanding the association and functions of different microbial communities. Plant-microbe interaction is of utmost importance in sustaining the balance in an ecosystem compared to the other microbial interactions. Several inorganic and organic compounds are produced by plants, which leads to the development of a nutrient-enriched environment that is beneficial for the profound colonization of a variety of microbes. Plant related microbial communities depend on the microenvironment provided by crops. Classification of plants can be done based on microenvironments like carposphere, spermosphere, phyllosphere and endorhiza. Microenvironments can be preserved by several environmental parameters for specified organisms. The survival mechanism of microbial populations determines their impact on the plant system [1]. This datum was discovered by scientists through dormant pathogenic bacteria that usually get colonized despite their capacity to be a good incubator for seed microbiome. However, plants interact with different microbes existing in water or wind and can colonize the phyllosphere [2, 3]. Microbial communities can have a positive or negative impact on the physiology of plants indirectly or directly through several interactions like amensalism, commensalism, mutualism and pathogenesis. Based on their impact, microbes may be classified as endophytic or epiphytic. Almost all plants on earth house endophytes, which is one instance of plant-microbe interactions. In this scenario, bacteria live in a non-competitive environment of the host plant tissue without causing any damage to the host cells [4]. The microflora of endophytes including bacteria and fungi are present beyond surface sterilization of several plant parts like shoot, root, seed or nodules. These endophytes may originate from the seed, nodules, root or shoot, rhizosphere or aerial parts of the plant [5, 6]. Rhizospheric region is a good source of root endophytes [5-7]. Endophytic microorganisms take entry into the plant tissue by the degradation of cellulose or local fractures in the root system. The endophytes colonize the intracellular spaces and thus get isolated from all the other compartments. Endophytic bacteria may be gram-negative or gram-positive and can be isolated from various tissues of different types of plant species. Several facultative endophytes are found in Arabidopsis, maize, wheat, sorghum, potato and cotton. Various species of microbes have been isolated from each plant. Very few studies have been performed that elucidate the interactions between plants and microbes through computational approaches, molecular variations, avirulence protein (Avr) and virulent gene interactions [7-9]. A number of laboratory-scale studies including pot experiments and growth assays have been performed to study the different interactions between plants and microbes [10-12]. Genomics and Bioinformatics are efficient tools to study and predict the interactions between microbes and plants for experimental validation [13-15]. The predictions will be based on various kinds of informational data, involving measurement of the abundance of species from high throughput sequencing or re-constructed metabolic models for several species’ communities. More reports associated with genome engineering, gene editing, and advanced technologies related to plant-microbe interactions have been discussed [14, 15]. The present review gives insights into the types of plant-microbe interactions in the environment and envisages the concept of plant-microbe interaction with respect to genomics, metatranscriptomics and metabolomics.
1.1. Plant Microbiome
Microbiome refers to the microbial community related to a variety of unique environments [16, 17]. Parallel sequencing emerged as a key enabler to study plant-microbe interactions as it enabled and promoted the research on microbial communities associated with plants. There has been a strong interest to try describing the make-up of microbial communities residing in the plants. Identifying and understanding the plant microbiomes has provided significant value to the advancement of agricultural practices and the creation of new biotechnological techniques for therapeutic and diagnostic purposes. In agricultural sectors, microbes are employed for improving plant health [18]. The plants are holobionts comprising of associated microbes and host. Although research related to microbes impacting plant health is not new, there has been a surge of research into this area to identify new pathways of impact and symbiotic benefits. The development in these technologies has enhanced the interest in plant microbiota as shown by various reviews [6, 14, 19-21]. Amplicon-depended profiling approaches of the community give a better understanding of the structure of the community and phylogenetic variations in plant-related microbes and are appropriate for examining key biological and environmental conditions that shape its configuration [19]. But, detailed information regarding plant microbiome remains to be explored. Further advancements in computational and sequencing techniques will drive functional research on plant-microbiome interactions.
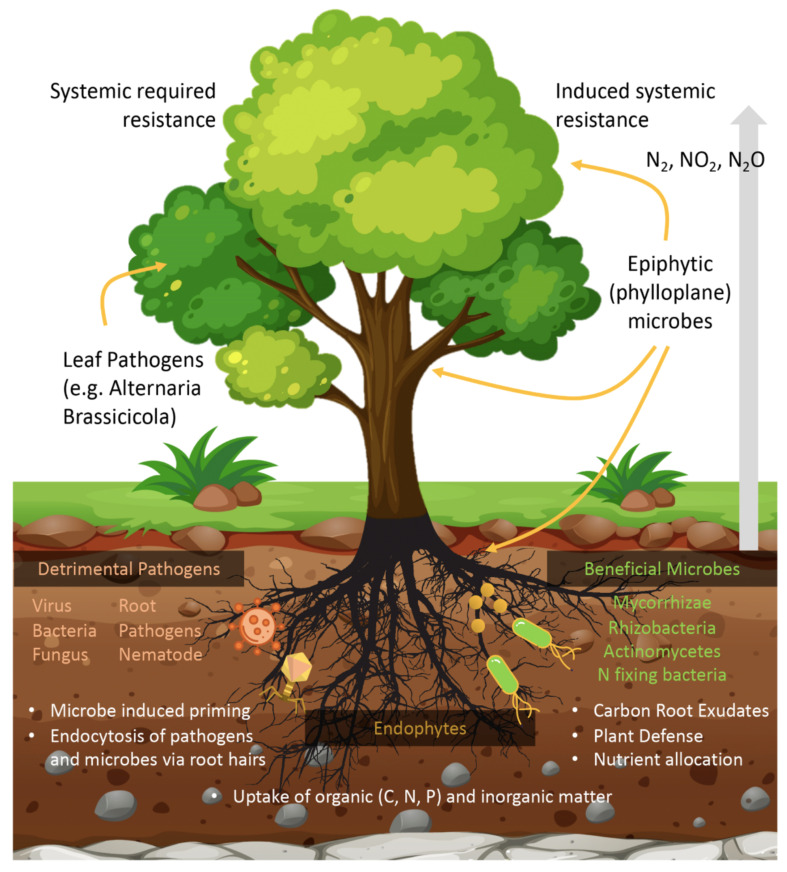
Plant microbiome exists in three diverse habitats when considering plants as the host of the microbial population. These include phyllosphere - above the ground stem and leaf surfaces, endosphere - tissues present within the plant and rhizosphere - root surfaces and marginal growth layer around the roots (Fig. 1). The noticeably different atmosphere generated by the plant anatomy presents a unique microbial constitution within each area [22]. Interactions between plant and host microbiota cover a whole spectrum, which extends from mutualistic to parasitic and commensalism relationships (Fig. 1). Different studies conducted on the microbes from the leaf or root endosphere, phyllosphere and rhizosphere have revealed several traits of microbes from the host point of view [11, 12, 23-25]. Interactions between plant and microbes can be pathogenic (disease-causing) or beneficial such as nitrogen fixation, plant growth promotion, bioremediation or stress combating in plants (Table 1) [26-33].
Fig. (1).
Illustration depicting various kinds of interactions between plant-pathogen and plant and beneficial organisms in the environment. The figure depicts interactions between plants, mycorrhizal fungi and bacteria. There are different types of interactions between plants and microbes ranging from beneficial interactions (nitrogen fixation, allocation of nutrients and plant defense) to plant-pathogen interactions (inducing bacterial, fungal and viral infections in plants). (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Table 1.
Different types of interaction between plant and microbes.
| S. No. | Types of Plant-Microbe Interaction | Description |
|---|---|---|
| i. | Plant growth-promoting microorganisms (PGPMs) | Microbes which benefit plants majorly through improved acquisition of nutrients by nitrogen fixation [26, 27], solubilizing inorganic phosphate [28, 29], organic phosphorous mineralization and production of siderophores for the uptake of iron [30]. Plant growth-promoting bacteria (PGPM) can enhance the accessibility of water and minerals by synthesizing growth regulators such as gibberellins [31] and auxin. |
| ii. | Plant disease suppression by rhizobacteria | Rhizobacteria can prevent plant diseases by producing incompatible compounds against phytopathogens (antibiotics competition, siderophores production) through priming. Priming helps in enhancing the defense system of the plants and inducing the resistance against pathogens. Various bacterial traits were discovered as triggers of induced systemic resistance [32] like cell envelope components, flagellae, phenolic compounds, antibiotics and quorum sensing. |
| iii. | Mycorrhizae | Arbuscular mycorrhizal fungi have the ability to spread the plant root systems and enhance the accessibility of the roots to nutrients with low mobility in plants. The roots of the plant are interconnected by hyphal networks of mycorrhizae. This network assists in resource exchange and also supplies about 90% of nitrogen [33]. |
| iv. | Rhizobia | Nitrogen entry into soil can be identified by interactions between legume and rhizobium. Plants metabolize the ammonium that is produced by the reduction of nitrogenase from atmospheric dinitrogen by nitrogen-fixing bacteria. |
| v. | Plant-Pathogen Interaction | (a) Biotrophic Pathogens: This type of interaction requires living plant tissue. Fungal haustoria are invaginations in the cell membrane of the living host for the extraction of nutrients. |
| (b) Necrotrophic Pathogens: Production of cellulolytic enzymes or toxins causes necrosis in the infected tissues taking up nutrients from the dead spots. | ||
| (c) Hemibiotrophic pathogens: These pathogens include both the biotrophic and necrotrophic pathogens. An early biotrophic infection phase is followed by the necrotrophic spreading phase. |
2. Plant-Microbe Beneficial Interaction
Plants exist with a complex network of interactions with microbes where some are beneficial while others are detrimental. Various sorts of plant-microbe interactions are present such as mutualism, commensalism and parasitism [34, 35]. Interaction between plants and various microbes such as bacteria and fungi is known to be beneficial for both and thus draws the attention of various researchers due to its potential application in agriculture. Several studies have been carried out related to interactions between plants and various other organisms such as arbuscular mycorrhizal fungi (AMF), diazotrophs, bacilli, Pseudomonas, phosphate solubilizing fungi, cellulose-degrading bacteria and plant growth-promoting rhizobacteria (PGPR) [11, 12, 36]. Advantages delivered by microbes vary depending on the plant species. In general, microbes support the growth of plants by fixing nitrogen in legumes thus enhancing the supply of nutrients such as iron, copper, sulphur, phosphorous. They also promote the production of plant hormones for checking bacterial and fungal diseases and assisting in bioremediation of polluted soil. Novel techniques for the protection of crops are dependent on the utilization of advantageous organisms, applied as biocontrol agents and biofertilizers [6, 18]. This is an important technique to control plant diseases and might result in a substantial decrease in the use of chemical fertilisers that contribute to environmental pollution. Microbial inoculants have been widely used in modern agriculture as biocontrol agents and as biofertilizers. Linking plant phenotype to the expression of gene and proteins and for the production of metabolites and accrual is one of the major impediments for enhancing the global agricultural production.
2.1. Nitrogen Fixation
A good example of an interaction between plant and microbes is nitrogen fixation [37-39]. This process is the natural form of fertilization and it promotes the research in the field of sustainable agriculture [39-41]. Transcriptome analysis of Azoarcus sp. strain has been carried out, which is an obligate nitrogen-fixing endophyte nitrogenase (nif) gene inside the roots of rice. Proteomics and metabolomics are ideal methods to observe the symbiotic interaction between the root nodules and the nitrogen-fixing bacteria. These studies provide a wide spectrum of metabolites/ proteins released by the symbionts. During the initial phase, both symbiotic partners release signals into the soil and several secondary
metabolites such as isoprenoids, phenolic acids, alkaloids and flavonoids that are sensed by microbial receptors and roots leading to consequent morphological and physiological changes. The best example of a symbiotic relationship is nitrogen fixation which results in effective communication between both symbionts. Flavonoids serve as primary signals and are released by the host plants that act as chemoattractants [42], activating the expression of rhizobia node genes. The function of flavonoids in the nodulation process has been studied by several researchers [42-44]. Van Noorden et al. [45] observed the interaction of Sinorhizobium meliloti with 131 identified proteins in Medicago truncatula. They further reported the auxin treatment performed by various redox-associated proteins and late embryogenesis proteins during the nodule formation and isoflavone reductase. Lery et al. [46] studied the proteomic profile of interaction between Gluconacetobacter diazotrophicus and sugarcane using mass spectrometry. Enhanced nitrogen fixation can be attributed to the overexpression of signal proteins by SP70-1143 genotype of sugarcane. The presence of G. diazotrophicus along with glutamate ammonia lyase in SP70-1143 plant roots demonstrated the efficiency of nitrogen metabolism and induction of bacterial proteins. The symbiosis between plant and bacteria during nodule formation was brought forward by Salavati et al. [47]. They used MALDI/mass spectrometric imaging (MSI) to study the distribution of root nodules in Medicago trunculata and its role in nitrogen fixation when associated with Sinorhizobium meliloti. Variations in metabolite profiling between nodules and the root along with differences in nitrogen-fixing and non-fixing nodules were also reported. Moreover, metabolite profile of M. truncatula extracts identified in the first hour of the metabolite suppression exhibited resemblance to oxylipin in planta nod factor treatment. Jasmonic acid and oxylipin metabolite inhibited the signaling of nod factor [48]. In Bradyrhizobium japonicum, 2610 metabolites present in the nitrogen-fixing bacterium were inoculated inside soya bean root hairs and were further studied using GC-MS.
2.2. Growth Promotion
The growth of rice is promoted by Pseudomonas fluorescens. MS-analysis revealed that P. fluorescens strain KH-1 induces proteins such as P23 co-chaperone, ribulose bisphosphate carboxylase, thioredoxin H, and glutathione S-transferase [49]. Gel-dependent proteomic approach revealed that inoculation with Sinorhizonium meliloti resulted in a higher level of photosynthetic proteins [50]. Transcriptomic analysis of the interaction between Petunia hybrida and arbuscular mycorrhiza showed a novel function of phosphate solubilization in the host’s repressed essential symbiotic genes. This result brings forth the beneficial impact of symbiosis on plant growth and crop yield through the modification of phosphorus concentrations. Secondary metabolites protect against environmental stressors while enhancing plant development and growth. In M. truncatula, analysis of the metabolites in mycorrhizal and non-mycorrhizal roots depicted a rise in the quantity of certain amino acids (Glutamine, Aspartate and Arginine), isoflavonoids, fatty acids (oleic acids, palmitic acid) and accretion of cell wall-bound tyrosol and apocarotenoids, particularly in the roots. Schliemann et al. [51] observed a contrast between the normal and symbiosis dependent secondary metabolism in M. trunculata. Inoculation by mycorrhiza in Anadenanthera colubrina seedlings enhanced the concentrations of metabolites such as phenols, flavonoids, and total tannins [52]. The roots of Lotus japonicus roots showed synergetic association to Mesorhizobium loti, and inoculation of secondary metabolites in its roots resulted in fourteen different phenolic acids in comparison with non-inoculated plants [53]. Popovici et al. [54] reported the changes in root phenolics when Actinobacterium frankia interacts with the Myriacaceae plant species as observed through HPLC analysis. These studies bring forward strain dependant changes in the secondary metabolism of host plants.
2.3. Bioremediation
Bioremediation using microbes is an important topic in agriculture as it helps reduce the impact on the environment. Trace metal pollution poses a foremost risk to agriculture, animal and human health. Phytoremediation of heavy metals from polluted soil can be attained using plant growth-promoting bacteria [12, 55, 56]. Cheng et al. [57] observed the behaviour of bacteria Pseudomonas putida UW4 towards nickel contamination and reported various mechanisms involving stresses due to the adaptation and efflux of heavy metals proteins. 130 leaf proteins were expressed in the interaction between arsenic-treated AMF such as Gigaspora margarita and Glomus mosseae and Pteris vittata. Bona et al. [58] observed and identified the major functions of glycolytic enzymes and arsenic (As) transporter (e.g. PgPOR29) during As metabolism. Aloui et al. [59] performed 2-DE/MALDI-TOF-dependent proteomic analysis which revealed that mitigation of cadmium toxicity through M. trunculata shoot on exposure to mycorrhiza was supported by a rise in the level of chaperones and photosynthetic proteins. Considerable reduction in concentrations of Zn and Cu in mycorrhizal-inoculated poplar plants was observed by Lingua et al. [60] supporting the phytoremediation characteristics of mycorrhiza. They used mass spectroscopy to find that the growth of mycorrhizal plants on metal polluted soils led to the upregulation of a small Hsp, Hsp70, large subunit of RuBisCO and downregulation of 43 spots mostly associated with oxidative stress and carbohydrate metabolism. Farinati et al. [61] observed a decrease in concentrations of Cd and Zn and modification of shoot proteome of the Arabidopsis helleri plants when cultivated in metal-polluted areas along with the bacterial strains (isolated from the rhizospheric region of polluted soil). These results show that strain screening for various functions can help develop effective usage of biocontrol mediators.
2.4. Stress Suppression in Crops
There are several stress influences such as drought, salinity, deficit of nutrients, pests, diseases and contamination which can bring a change in the interactions of plants and microbes in the rhizosphere. These stress factors trigger the signaling molecules in which phytohormones have a major function. Signal processing is used to trace the signal input that allows plants to retort towards environmental restraints. Plants are subjected to various stresses concurrently, thus a meta-analysis reveals a composite regulation of plant immunity and growth. The way phytohormones interact with the signaling network is essential to understand interactions between plant-microbiome systems during stressed conditions. This interpretation is essential to develop biotechnological approaches for optimizing plant acclimatisation techniques and to enhance the microbial activity of soil for assuagement in crops [62]. Gomes et al. [63] observed a promotion in the growth of the host due to elevated photosynthetic proteins, transport proteins and chaperones such as GroEL and DnaK during temperature stress, drought and metal toxicity. Recent researches have shown that changes in plant morphology, physiology, transporter action, exudation profiles of roots and vicissitudes might trigger the plant to employ microbes with stress-reducing abilities and potentially enhancing the crop under stress conditions. The stress factors lead to negative effects on the productivity and functioning of agricultural systems and rhizospheric microbes are important in leading plants to sustain in hostile conditions [64].
3. Molecular Techniques for Decoding Plant-microbiome interactions
In the past, the absence of proper methods led to limited knowledge on mechanisms behind plant-microbe interactions in the rhizosphere. There were problems associated with the profiling of a wide array of methods in which a large variety of microbial communities involved were unculturable [65]. An abundance of culture-neutral molecular techniques have been popularized over time and are presently being used for deciphering the diversity of microbes and rhizospheric microenvironments and to gather more knowledge related to the molecular basis of the plant-microbe associations (Fig. 2). These modern molecular techniques are important for evaluating the effects of perturbations activated by the biotic and abiotic stress on the diversity of soil microbiome and plant-microbial interfaces in the current worldwide change.
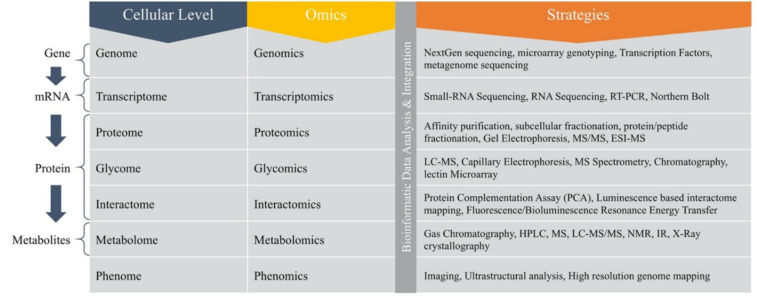
Fig. (2).
Omics approaches and strategies for studying Plant-Microbe Interactions. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
A major advancement related to the molecular ecology technique for analysis of microbial diversity of soil is based on the isolation of microbes. The entire microbiome of soil can be analysed directly for the extraction of DNA/RNA and the biochemical markers. DNA exemplifies the phylogenetic identity and the functional ability of microbes. Hirsch and Mauchline [66] reported about the extraction of phospholipid fatty acids from the cell membrane and stated them to be biochemical markers acting as indicators of the microbial community structure of the soil. Culture-based studies are generally utilized to determine the genetic and functional variations in the microbial communities of the soil or the rhizosphere and were undertaken by a number of researchers [67, 68].
Molecular approach first involves the process of DNA and RNA extraction from the soil. Labeling of nucleic acids can be performed before their abstraction from environmental samples for scrutiny. The collective microbial genome constituting isolation of DNA from microbial communities is referred to as a metagenome. Metagenomics involves isolation and cloning of large fragments of DNA that comprises of various operons and genes. The total DNA extraction from the environmental samples can be analysed by various methods depending on the cloning procedures, amplification of PCR, high-throughput sequencing and microarray hybridization. Closing dependent techniques result in building of metagenomic libraries which can be used for screening of functional and structural genes or for observing phenotypic characters associated with proteins such as enzymes and profiling of secondary metabolites. Bioinformatics software is mostly used in soil metagenomic studies. Microbial diversity can be quantitatively determined using the PCR technique. Small ribosomal subunit sequences act as molecular markers of the target microbial groups. Predominantly, a comparative gene examination of 18S or 16S ribosomal rRNA is the most common molecular technique for recognising microbes. Several microbial genomes have rRNA gene as a general gene marker.
Well protected segments of rRNA sequence in the gene can be utilized to build general primers for amplifying the gene obtained from DNA extraction of environmental samples. Analysis of 18S and 16S rRNA gene sequences is the foundation for comparing richness, evenness, composition and assembly of the microbial groups. PCR products share comparable or indistinguishable variable regions and are known as operational taxonomic units (OTUs). Diversity of the PCR products is defined by distinguished molecular processes that allow a molecular impression for determining the make-up of a specific microbial group. Diversity of the amplicon is determined by sequencing and cloning methods. Sequence evaluation of 16S or 18S rDNA amplicons can be done using high throughput next-generation sequencing. The 3rd generation sequencing technology is dependent on a single DNA molecule. Techniques other than fingerprinting used for monitoring the abundance of specific taxonomic groups in microbial communities are functional gene microarray dependent PhytoChip and Geochip techniques [67]. The functional variations in the microbial community may be analysed by amplification of certain functional genes important for specific metabolic processes. Strategies depending on the abundance of transcripts were employed during studies of functional diversity and were monitored by the qRT-PCR. The functional gene arrays evaluate the expression of the transcripts from various genes and are used to determine the activity of some functional microbial actions. The shotgun sequencing involves microarrays of DNA constituting environmental cDNA and helps differentiate the response of soil microbiome towards external effects at the level of transcription. High-throughput phenotyping of plants helps to analyse plant functions affected by microbial community activities. However, the effects of plant genotype on the functioning and variability of microbial communities can be assessed by molecular ecology techniques. Thus, the important molecular techniques such as next-generation sequencing, transcriptomics, proteomics, metabolomics to study interactions between plant and microbes are discussed in detail (Fig. 2).
3.1. Deciphering the Plant Microbiome through Next-generation Sequencing
Recent developments in the plant-microbiota researches validate that plant PGPM is an essential member of the plant microbiota. Though, information related to PGPM is quite sparse with respect to reports associated with separate isolates under laboratory conditions [19] as well as how microbial communities contribute to the growth of plants. Microbial mechanisms which promote the growth of plants include increase in nutrient requirement from soil, tolerance to abiotic stress, protection from pathogen indirectly and increase in nutrient enhancement from soil. These organisms are referred to as plant growth-promoting microorganisms. It was clearly observed through investigations that PGPM forms an integral part of the plant microbiota. The better knowledge and understanding related to plant-microbe interaction can be established by several approaches discussed in the following sections.
The natural habitat of plants constitutes a variety of microorganisms. A good correlation between the upper and lower layers of the earth constitutes a huge number of microorganisms [69]. Some microbes grow well under stressed conditions and benefit the plants while natural ecosystems are better managed by mosses and PGPM. Thus, via NGS, plants can be studied with respect to taxome, ecological function and interactome involving transcriptomics, genomics, and metabolomics studies of microbes which will eventually lead to the identification of the mechanism for their survival and interactions [22].
Many NGS studies have been conducted to decipher the bacterial communities that support plant growth. Endophyte-promoting activity can be concluded by the presence of compounds such as IAA (indole-3-acetic acid), phosphatases, ACC (1-aminocyclopropane1-carboxylate) deaminase and siderophores. Analysis of tissues from various parts of the plants (flowers, leaves, stem and roots) and rhizospheric soil samples for plant growth-promoting bacteria [70-73] concluded that the microbiota composition varies from tissue to tissue [74, 75]. In general, these studies have displayed the presence of Proteobacteria in larger amounts in plant-related ecosystems, accompanied by significant amounts of Firmicutes, Bacteroidetes, Actinobacteria and Acidobacteria. But, these methods to identify the bacterial communities have limitations that need to be taken into account: (i) The ease of DNA extraction is dependent on spores and membranes of the bacteria present in the community, thus making community profiling a function of the extracted DNA rather than real quantity of various bacteria present in the microbiome [76]. (ii) The 16S rRNA sequencing is a good method to identify phyla but not very suitable to classify species or genus [77]. (iii) Bias could creep in based on the selection of sample, recovery method and treatment [78, 79].
3.2. The Meta-omics Approaches
The meta-omics study not only provides information about expressed gene levels via genomics and transcriptomics but also about post-translational changes through proteomics (Fig. 2; Table 2) [80-84]. Metabolites are a result of cellular processes which can be obtained by applying the metabolomics method. Thus, there exists a combination of omics methods that can connect all the aspects of variations at the cellular level. These studies provide a complete view of the complex dynamics of cellular systems involved in plants and microbe interactions. For instance, the bacteroid, Bradyrhizobium japonicum found in the soya bean root nodules was examined by accumulating datasets of transcriptomics and proteomics [85]. Several proteins were discovered depending on the dataset of various kinds of bacterial metabolism occurring during symbiosis. Ali et al. [86] used a combination of proteomic and transcriptomic methods to analyse the compatible and incompatible associations and interactions of Solanum tuberosum with Phytophthora infestans. Modifications in the range of 1700 transcripts and 1000 expressed proteins were identified through the integration of these techniques.
Table 2.
Molecular biology techniques and their pros and cons for studying plant-microbe interactions.
| NGS Techniques | Softwares | Pros | Cons | Purposes | References |
|---|---|---|---|---|---|
| Genomics and Metagenomics | Assembly: Meta Velvet and Ray Meta, IDBA-UD Profiling: AMPHORA, MetaPhlAn mOTU Function Analysis: IMG/M, MG-RAST And CAMERA |
Profiling is unbiased; Uncultured microbial studies can be performed; Correlation can be observed between genes of diverse organisms from similar environment |
Less information related to sequencing of the marker genes; Less abundance anticipating functions of the gene are not equivalent to protein content expressed |
New species along with their taxonomic profile will be discovered; Potential, metabolic, functional and evolutionary relationships will be derived; Helps in genome reconstruction |
[80-84] |
| Transcriptomics and Metatranscriptomics | Mapping: BWA-SW; Bowtie2 de Novo Assembly: IDBA-MT Function Analysis: CAMERA and MG-RAST |
Novel transcripts determination and sensitive methods for detection; Aids in detecting simultaneous expression of the host and microbial gene; Simple to perform; Metatranscriptomics allows capturing of transcriptomes in case of un-cultured bacteria |
Low concentration of rRNA presence in the samples; Difficult to assign transcripts to specific organisms; High complexity of the community and host transcriptome leads to low sensitivity; Extensive genome is needed for mapping |
Analysis of Pathways and active function study | |
| Proteogenomics | Mascot: Protein Identification; MG-RAST and camera |
Proper estimation of functional activities in comparison to transcriptomics; Results in Semi-quantification of the proteins in the environment; More accurate to perform qualitative protein analysis |
Reference genes needed for identification of proteins; Difficulty in sample preparation process; Quantity of various proteins cannot be compared; Difficult for performing in plants due to host contamination |
Determination of functions and analysis of pathways | |
| Metabolomics | Metabolite study | Metabolites produced due to plant-microbe interactions can be identified; Nominal bias analysis of various compounds |
Size of reference public databases is limited; Different metabolites with their functions give similar signals during Mass spectrometry; Similarity between primary metabolites of microbes and plants results in their difficult determination |
||
| Marker gene | Amplicon Noise, mothur, QIIME | Classification of new and rare species | Problems during amplification in PCR | Novel species with taxonomic profiling can be discovered |
3.2.1. Genomics and Metagenomics
Genomics involves the study of the complete genome of organisms and helps to incorporate elements from genetics. The process utilizes a combination of DNA sequencing techniques, recombinant DNA and bioinformatics approach for sequencing, assembling and analysing the structural and functional characteristics of genomes. Metagenomics helps in real-time characterization of complex DNA mixtures obtained from the habitat of microbes [87]. Metagenomics establishes the base for various other techniques to investigate RNA such as metaproteomics, metatranscriptomics, and
metabolomics, all delivering new insights on the metabolic capacity of certain microbiota. The phyllosphere microbiota composition along with the expression of microbial traits in the habitat was studied by Delmotte et al. [88]. They collectively used metaproteogenomics in which metagenomic information aided the annotation of the metaproteomic data. They further researched the phyllosphere of soya bean, Arabidopsis and clover and found that despite 130 million years of divergence among the families of Brassicaceae and Fabaceae, about 70% of the phyllosphere metaproteome was conserved. Their outcome suggested the presence of a sizeable core microbiome along with slight host-specific roles of the microbiome. The metagenome of root endosphere was studied through the bacterial cells attained from surface-sterilization of rice roots, giving information about the metabolic potential of the microbial community residing inside the plant roots [89]. On coalescing the transcriptional examination of some microbial genes with metagenomics, it was established that bacterial microbiota within the plant roots can perform primary steps of the nitrogen cycle in plants. Knief et al. [90] compared the traits between the rhizosphere environment of rice plants grown in the paddy field and the microbiota using metaproteogenomics. The presence of di-nitrogenase reductase genes and proteins depicted habitat-specific microbial traits caused by nitrogen fixation in the rhizosphere. However, the technique has some limitations due to the presence of rRNA in the samples.
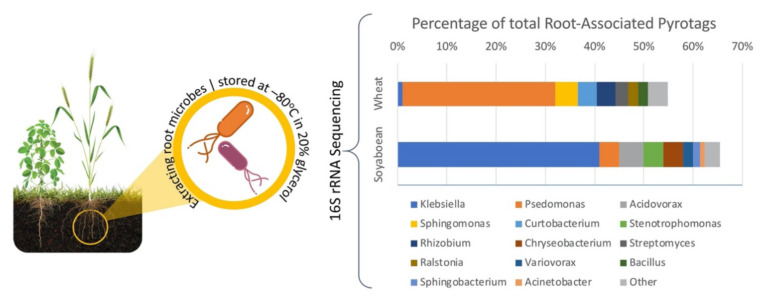
Rascovan et al. [91] studied and explored root microbial communities associated with wheat and soybean in field conditions using 16S rRNA pyrosequencing. Fig. (3) gives a comparative overview of the metagenomic and cultured bacteria datasets. They extracted bacterial isolates from rhizospheres, to test the in-vitro presence of PGP traits. They compared the dominant bacterial communities linked with roots from both crops and found that a high proportion of them (60-70%) exhibit >97% resemblance to bacteria from the rhizospheric isolated samples. Pseudomonas and Bacillus genera represented the highest proportion of the full dataset (~20%). Considering only a Blast best hit of ≥ 90% similarity, they observed a total of 100 different genera of bacterial communities. This study brought forward the fact that microbiome composition is a function of its soil environment. The root-associated microbiome in wheat plants had much more variability when compared with soybean. This may be attributed to the architecture of wheat roots which can grow up to five primary roots in order to capture water and nutrients, as opposed to soybean plant which has a simpler root structure.
Fig. (3).
Metagenomics study to identify microbial community associated with wheat and soybean plant roots. Readings from root-associated (RA) pyrotag were compared with Sanger 16 rRNA sequences from rhizospheric isolates using BlastN. Adopted and Modified from Rascovan et al. [91]. Open Access under a Creative Commons Attribution 4.0 International License. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4911569/pdf/srep28084.pdf. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
Thus, plant microbiome deciphering needs large scale sequencing coverage along with a replication of experiments for robust analysis, and substantial computational and financial support [92, 93]. Moreover, the interpretation of data on RNA, microbial DNA, and protein sequence gets hampered due to reduced rates in the high-confidence annotation. Thomas et al. [74] reported that only a few sequences can be retrieved from a non-human environment and about 50% can be annotated functionally with available data. Despite these limitations, it is important to underline that meta investigations deliver initial inferences and supplementary experiments are required to disclose if the practical diversification of plant microbiota is a reason or an outcome of studied plant phenotype [22].
3.2.2. Transcriptomics to Analyse Plant-microbe Interactions
Various studies related to patterns of gene expression of microbial symbionts have been performed [94-96] with respect to a single partner and only some have discussed the method of transcriptional profiling [97]. It has been hypothesized that beneficial fungi exhibit less impact on expression profiles of host gene in the absence of pathogens (Pseudomonas syringae or Blumeria graminis sp.) [98] in Piriformospora indica-barley-powdery mildew. Breuillin et al. [99] researched the transcriptional profiling of AMF in Petunia hybrida and observed a novel function for phosphate requiring symbiotic genes in the host. The study was important in terms of phosphorous management under field set-up to enhance yield and plant growth by leveraging both phosphorous supply and useful effects of symbiotic interaction. Plants can interact with saprophytic fungi which breaks down the organic material and releases minerals that can be taken up by plants [100]. Fungi further help in organic acids production and extrusion of proton causing a drop in pH of the soil. This enables the phosphate precipitation into soil solution making it available for root uptake [101]. Also, there are many studies related to transcriptional profiling of beneficial microorganisms like Pseudomonas on plants under lab conditions [102, 103]. Substantial variations were observed in the transcript levels of the Arabidopsis shoots when inoculated with pathogen Pseudomonas syringae in the leaves and Pseudomonas fluorescens WCS417r-mediated ISR inoculated in the roots [104]. Studies have reported that variations in the Arabidopsis transcriptome occur as a result of different traits [FPT9601-T5; MLG45] of Pseudomonas fluorescens. Moreover, more studies associated with multiple interactions between plants and microbes are being promoted due to the advancement of these techniques.
3.2.2.1. Transcriptome Analysis of Plant-pathogen Interactions
Microarrays have been used to study the plant-microbe associations and interactions using transcriptome analysis [105, 106]. Xu et al. [107] performed a study using RNA-sequencing to observe plants' ability to produce genes mediating defense reactions against Verticillium dahlia, a root blighting pathogen. They uncovered a network of signal transduction pathways using microarray-based plant transcriptomics, which gets triggered due to elicitors constituting pathogen-associated molecular patterns (PAMPs) and signaling compounds such as SA, ABA, JA, and ET [108, 109]. These studies have resulted in the discovery of key regulatory and resistance genes [110].
Plants have an efficient defense response towards microbes. A multilayer defense response triggers in the microbes and plants on the entry of a microbe into a plant. This response relies on the reserves required to sustain the defense. The induced responses need fewer resources compared to the constitutive response. Mostly the environmental surroundings stimulate plants to deliver the constitutive response. The major consequence of plant-microbe interaction is guided by genotypes of host and microbes and environmental circumstances. Interaction of plants with pathogenic microbes such as bacteria, viruses and fungi result in infectious ailments impacting only the plant kingdom. The benefits related to such interactions aid in understanding a natural phenomenon which affects daily life and could result in their application in sustainable resources, environmental clean-up, the effect on atmospheric gases, and reducing environmental impact. Plant defense mechanisms comprise a mixture of inducible and constitutive responses. Constitutive responses comprise biochemical defenses and barriers, whereas inducible responses are local in nature and advance with systematic action mode initiating from recognizing the pathogen to the defense expression of genes. The prevailing biochemical defenses include pathogen identification by the host plant, signal transduction and gene expression. Plant tissues act against the pathogen by programmed cell death in a localised response. However, in systemic defense, the signal spreads from the interaction site, facilitated by various molecules that work as plant messengers (for example, ethylene and nitric oxide). The messenger molecules are important in increased expression of profiling information for different stresses under in vitro conditions. Various plant-pathogen interactions studied through the omics approach are presented in Table 3.
Table 3.
Study of plant-pathogen interactions by omics techniques.
| Pathogen | Host Plant | References |
|---|---|---|
| Transcriptomics | ||
| Hyaloperonospora arabidopsidis | Arabidopsis thaliana | [111] |
| Sclerotinia sclerotiorum | Pisum sativum | [112] |
| Phytophthora infestans | Solanum tuberosum | [113] |
| Botryosphaeria dothidea | Populus tomentosa | [114] |
| Blumeria graminis f. sp. tritici | Triticum | [115] |
| Botrytis cinerea | Lactuca sativa | [116] |
| Mycosphaerella musicola | Musa acuminata | [117] |
| Phytoplasma | Cocos nucifera | [118] |
| Phytoplasma | Glycine max | [119] |
| Marssonina coronaria | Malus domestica | [120] |
| Marssonina coronaria | Malus baccata | [121] |
| Cladosporium fulvum | Lycopersicon esculentum | [122] |
| Diplocarpon rosae | Rose Plant | [123] |
| Podosphaera pannosa | Rose Plant | [124] |
| Colletotrichum graminicola | Zea mays | [125] |
| Proteomics | ||
| Pseudomonas syringae pv. tomato | Solanum lycopersicum | [126] |
| Puccinia hordei | Hordeum vulgare | [127] |
| Candidatus Phytoplasma vitis | Vitis vinifera | [128] |
| Mung bean yellow mosaic virus | Vigna mungo | [129] |
| Pseudomonas syringae pv. actinidiae | Actinidia chinensis | [130] |
| Erwinia amylovora | Apple rootstocks | [131] |
| Magnaporthe oryzae | Oryza sativa | [132] |
| Botryosphaeria dothidea | Populus | [133] |
| Moniliophthora perniciosa | Theobroma cacao L. | [134] |
| Verticillium dahliae | Tomato Plants | [135] |
| Cladosporium fulvum | Tomato Plants | [136] |
| Metabolomics | ||
| Fusarium tucumaniae | Glycine max | [137] |
| Rhizoctonia solani | Glycine max | [138] |
| Botrytis cinerea | Vitis vinifera | [139] |
| Pseudomonas syringae pv. tomato | Arabidopsis thaliana | [140] |
| Phytophthora infestans | Solanum tuberosum | [141] |
| Fusarium proliferatum | Asparagus officinalis | [142] |
| Fusarium graminearum | Triticum | [143] |
| Plasmopara viticola | Grapevine | [144] |
| Verticillium dahliae | Arabidopsis thaliana | [145] |
| Tuta absoluta | Solanum lycopersicum | [146] |
| Pyricularia oryzae | Rice | [147] |
| Phytophthora sojae | Soybean Hypocotyls | [148] |
Kawahara et al. [147] reported on the transcriptome investigation of blast fungus and rice in infected plant tissue. 240 transcripts of secreted proteins, encoding fungi such as glycosyl hydrolases, LysM domain-constituting protein and cutinases were identified which behave as effector genes in instigating the early infection processes. The pathogenesis associated proteins and phytoalexin biosynthetic genes showed upregulation in rice. Transcriptome characterization of Sclerotina sclerotiorum and pea interaction discovered 142 ESTs with encoded secretory peptides and 93ESTs responsible for virulence, and 277 pea ESTs important for biotic and abiotic stresses [94]. De Cremer et al. [116] observed the downregulation and upregulation of terpenoid and phenylpropanoid pathway genes and photosynthetic genes on incubation of B. cinerea with lettuce for 48 h. Liao et al. [114] studied the involvement of genes in energy metabolism and in the redox reaction transcripts encrypting glutathione S-transferase (GST) which were considerably amassed in Arabidopsis on interacting with Botryosphaeria dothidea. Aragona et al. [148] performed functional characterization of RNA sequence data for analysing effectors and virulent mechanisms of the pathogen. Zhao et al. [121] identified prime regulatory genes associated with resistance response towards Cladosporium fulvum in the tomato plants having Cf-19. Transcriptomics was used to analyse variations between the resistant response of plants constituting Cf-19 gene (CGN18423) and the susceptible plants carrying Cf-0 gene at intervals of 0, 7 and 20 days after the inoculation. Neu et al. [122] observed the interactions between roses and biotrophic and hemibiotrophic leaf pathogen. They observed the transcriptomic changes using next-generation sequencing supported by an extensive cDNA analysis which was further validated by high-throughput qPCR.
Transcriptomics has played a key role in interpreting the concept of fungi related diseases in plants. The studies conducted by Hane et al. [149] helped in identifying genes among the pathogens (such as AG8, AG3 and AG1-1A) with functions that are unique to R. solani. Transcriptomic analysis of plant-related bacteria with the help of gene expression microarray approach using RNA sequencing technology reveals that genes are expressed differently under some conditions. Transcriptomics studies related to plant and bacteria studies were performed on separate bacteria cultures from the host plant. RNA sequencing is used to detect genes responding to the plant extract [150]. However, the plant transcriptomes considerably outnumber the bacterial transcriptomes that are basically house-keeping ribosomal RNAs. Thus, attaining proper strength of bacterial mRNA transcripts for the sequencing and differential gene expression is quite problematic. Various bacterial isolates transcriptomics studies denoting bacterial gene and plant gene expression were performed by Roux et al. [151], Pankievicz et al. [152] and Paungfoo-Lonhienne et al. [153]. Two closely related methods for enriching the P. syringae transcriptome in Arabidopsis leaf infection model were employed by Nobori et al. [154]. They used a novel isolation buffer which facilitates the stabilization of the bacterial RNA during leaf grinding.
The technique involved filtration and centrifugation to separate the bacterial cells from the plant cells before RNA isolation. Other methods involved depleting the plant-derived transcripts by customized probes. RNA sequencing technique helps in detecting transcriptome regulations like antisense RNA, gene operons, riboswitches, small non-coding RNA, and antisense RNA [155, 156].
Metatranscriptomics involves sequencing of whole bacterial community from the environmental samples. It results in an understanding of the transcriptional state of several microbes. In Arabidopsis, metatranscriptomics was used for identifying the bacterial genes in the rhizosphere which gets expressed differently during the development [157, 158]. Metatranscriptomics was performed on willow rhizosphere to discover the influence of microbial communities in the field of phytoremediation, presenting the earliest examples of such a discovery in plant science [159]. The main advantage of using this approach was to recognize the possibility of microbial traits occurring in a plant microbiome without the cultivation of their members. However, there are few limitations related to this approach such as the complexity of plant microbiome and inherent traits [19] due to the contamination of plant-associated microbiota specimens with DNA, RNA or proteins of the host. Around 90% of plant sequences were attained from DNA preparations of root samples from Arabidopsis [19].
With the cost of sequencing declining, the use of transcriptomics and metatranscriptomics has increased to gather knowledge about gene expression of bacteria. Transcriptomic investigation aids in the changing aspects and the regulation of actively transcribed genes for detection, thus posing an advantage over genomic analysis. Metatranscriptomics though has some limitations such as transcripts can seldom be allocated to specific microbes constituting good quality reference genomes. Hybridization based nano string technology is an alternative to sequencing-based transcriptomic approaches which allows improvement in bacterial transcript detection in mixed plant microbiota samples of a transcript. Thus, improvement in the methods for enrichment and bacterial transcripts detection is relevant to a wide range of plant-bacteria systems, and in future these techniques will enhance the knowledge of plant-related bacterial functions.
3.2.3. Proteomics
Proteomics approaches are mostly related to liquid chromatography and tandem mass spectrometry techniques and are used to study plant-pathogen interactions. Proteomics plays an important role in protein identification and their changes upon infection. It reveals a variety of bacterial proteins in the environment using semi-quantitative methodology. These methods include analysis of collected samples, protein isolation, extraction and fractionation through mass spectroscopy and further assessment against a proteome database. Proteomics determines the constituents of functional protein produced by the cell rather than identifying the possibility to make them. It also provides information on the exact number of active pathways in the sample. Some studies related to proteomics have brought forward plant-pathogen interaction. Rph15 gene is an important strain in the development of resistance breeding. It is resistant to about 350 isolates of Puccinia hordei, a pathogen causing leaf rust foliar disease in barley. Bernardo et al. [125] used LC-MS/MS analysis to profile the protein and observe the resistant and susceptible isogenic lines and study the Rph 15-based defense response. Various pathogen-associated proteins were discovered in Rph 15 resistant line at 4 dpi which are associated with carbohydrates metabolism, protein degradation, defense mechanism and photosynthesis.
Margaria et al. [126] performed a proteomic study through 2D gel analysis of Flavescence doree (a grapevine disease occurring due to phytoplasma) infecting the grapevine. This study identified 48 proteins that were expressed differently. Isocitrate dehydrogenase and glutathione S-transferase proteins hold an important antioxidant function in infected plants. The response of plants towards pathogenic fungus was studied by several authors [160, 161]. Kundu et al. [127] observed the proteomic profiling of Vigna mungo, when mung bean interacted with the yellow mosaic virus. They detected the expression of 109 different proteins. They observed that electron transports of the photosystem II were main targets during the pathogenesis process and during the downregulation of photosynthetic proteins in some genotypes. Infection of tomato by Pseudomonas syringae results in bacterial speck disease and was observed by iTRAQ proteomic methods which further identified 2,369 proteins in the leaves of tomato, among which 477 proteins are Pst responsive.
Parker et al. [124] observed the major upregulated proteins such as glutathione S-transferase, thioredoxin and superoxide dismutase. Several literature reviews based on the proteomic applications to study interactions between plant and pathogens were studied by Delaunois et al. [162], Ashwin et al. [163] and Wright [164]. Li et al. [165] observed 38 proteins expressed differently in banana infected with Fusarium oxysporum (Cubense tropical race 4, Foc4). They found that antifungal protein production, PR proteins, cell-wall strengthening proteins were basically concerned with resistant genotypes. Proteins which are involved in scavenging of ROS, PCD and photosynthesis were highly impacted. Broad-spectrum analysis and screening of proteins in various strains and genotypes would result in the selection of resistant plants and beneficial strains for future utilization in agricultural sectors. Li et al. [131] performed proteomic studies to compare the plant-pathogen interactions between susceptible and the resistant ecotypes of poplar infected by Botryosphaeria dothidea. They identified the resistant proteins of B. dothidea by observing the molecular mechanisms of poplar and pathogen interactions. The susceptible and resistant ecotypes of poplar towards B. dothidea were studied by nanoflow liquid chromatography and tandem MS using label-free quantitative method. They identified 588 proteins categorized into 21 biological processes involving 80 metabolic enzymes, 72 types of hydrolytic enzymes and 29 proteins of unknown function. The interactions between Cacao genotypes and pathogen Moniliophthora perniciosa were studied by Santos and his co-workers [132] using proteomics. They compared the proteomic changes between the two genotypes of cacao by observing resistance and susceptibility towards witches’ broom disease on inoculation of M. perniciosa at intervals of 72 hrs (early stages) and 45 days of biotrophic and necrotrophic stages of the plant-pathogen interaction.
Metaproteomics can be used for measuring the metaproteome of phyllosphere in forest trees [166] to observe different proteins produced by PGP bacterial strains as a response to root exudates [167]. It also recognizes the proteins and organisms important for oxidation and nitrogen fixation in paddy fields [168]. Proteomics might be restricted due to low protein value, less concentration and low sensitivity owing to host proteins and complicated microbes. Metaproteogenomics was used in the Vorholt lab where proteins occurring in the microbial communities were determined from the metagenomes generated by plant microbiota. This technique increased the number of proteins which can be identified by publicly available databases [90]. Use of proteomics for describing plant-related communities of bacteria has limitations owing to several factors like comparatively low bacterial protein, lower expression levels in complex plant-related samples and subsequent detection limits and the requirement for a complete reference database. More proteomics studies should be carried out concerning plant-bacteria interactions.
3.2.4. Metabolomics
Metabolomics is also a novel technique for studying plant-microbe interactions. Several bacterial genes like nodulation genes produce Nod factors during nodulation of roots which affect the host plant or the metabolism by microbes. The variations in levels of a metabolite can be observed by specific treatments. This technique was used for demonstrating the chemical exudation process from grassroots during development, thus affecting the assembly of the rhizospheric community. This chemical succession by bacterial enrichment in the substrate preferred the consumption of exuded metabolites, primarily the aromatic organic acids [169]. Negrel et al. [142] identified the lipid markers of Plasmopara viticola infection in grapevines through metabolomics studies (non-targeted). P. viticola results in downy mildew in grapevines causing a lot of damage. They used MS-based metabolomic non-targeted approach for identifying the potential of specific metabolites of Plasmopara. The infection of Phytophthora sojae caused in soybean hypocotyls was studied by Zhu et al. [146] using metabolomics. They examined the metabolic variations between two hypocotyl lines of soybean, i.e. susceptible line (S), 06-070583 and resistant line (R), Nannong 10-1at points of 12 hpi and 36 hpi on inoculation of P. sojae and further observed the metabolic variations between S and R line. They observed 90 different accumulated metabolites and variations between S and R lines. Thus, metabolomics has been used to study a lot of plant-pathogen interactions [135-146].
However, there are many limitations related to metabolomic analysis in plant-microbe systems which have not been widely accepted. Moreover, the cost involved, equipment and technical expertise important for performing metabolic studies make it a less viable approach. Also, there are only a few public metabolomic reference databases which make it difficult to associate a given metabolite to a particular organism. Still, metabolomics is an influential tool for detecting and quantifying small molecules and the molecular variations at the interface of plant-microbe interaction.
3.2.5. Interaction Transcriptomics between Plants and Associated Microorganisms
Reports concerning the transcriptional profiling of plants and microbes are still rare thus, more research should be done to study the close association between plants and microbes. Nodule formation in interactions between legume and rhizobium is well documented by gene expression and developmental changes in the roots [170]. Despite dual genome symbiosis chips development, the analysis of gene expression of the host and symbiotic microbe was mostly concerned with the study of nitrogen-fixing nodules. Brechenmacher et al. [171] reported that the interactions between the root of the plants and AMF are characterized for expression of the gene in the plant roots, however few genes are involved in the mycorrhiza constituents. Thus, DNA microarrays containing both microbial and host genes have been developed. Analysis of microarrays was carried out by an Affymetrix Gene Chip comprising 38000 genes of soybean and 15800 genes of Phytophthora sojae [172]. Analysis of transcriptome was further performed between fungal pathogen of the rice blast, Magnaporthe oryzae and rice which resulted in the determination of biotrophy-related effector proteins which might help the plant during hyphal invasion [173]. Such analysis should be supplemented with a proper bioinformatics approach involving databases generated by plant-related Gene Ontology Consortium [174]. Few examples include genomes of Pseudomonas syringae, i.e., Dickeya dadantii 3937, pv tomato DC3000, Magnaporthe oryzae fungus, few oomycete and Agrobacterium tumefaciens.
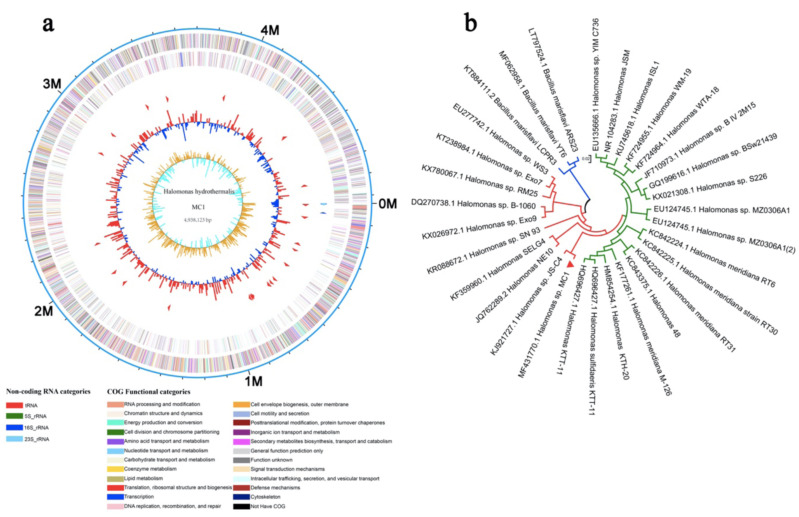
Zhang et al. [175] performed a transcriptome analysis of Halomonas sp. strain MC1(an endophyte) to study its plant growth-promoting potential on Mesembryanthemum crystallinum (Ice Plant) and identify the genes involved in providing salt tolerance to the plant. The BLAST analysis of 16S rDNA sequencing of strain MC1 followed by the construction of its phylogenetic tree revealed its close relationship with genus Halomonas sp. (Fig. 4). They were able to demonstrate that under salt stress, the bacteria strain MC1 itself was able to adapt and resist salt stress, thus enabling Ice Plant to survive under these conditions. This was achieved by MC1’s capability to regulate metabolic and cellular processes as well as regulating carboxylic and organic acid catabolic activities. This indicates a shift from other studies performed [176, 177] that highlight the capability of bacteria to induce salt stress resistance in plants.
Fig. (4).
Genome-wide assessment. (a) Circular plot of reads mapped to the Halomonas sp. MC1 genome. (b) Phylogenetic tree constructed on the basis of 16S rDNA sequences of neighboring species using the neighbor-joining method. The bars represent 0.02 substitutions per nucleotide position [175]. Open Access under a Creative Commons Attribution License. No changes or alterations were made in the figure. https://www.mdpi.com/2076-2607/8/1/88/htm. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
3.2.6. Multispecies Transcriptomics for Deciphering Plant-Microbe Interactions
A better understanding of the relationship between plant and microbial genes observed through multi-species transcriptomics has helped in developing new approaches towards disease resistance. A multidisciplinary technique to get an inclusive knowledge about the approaches of systems biology is clearly required. Datasets from transcriptomics, genomics, proteomics and metabolomics studies lead to identification and integration of prime biological processes and making predictions through modeling. This method helps in developing an understanding of ectomycorrhizal interactions among the roots of Populus tremuloides (Aspen) and Laccaria bicolor. Larsen and his co-workers [178]
reported short read next-generation transcriptomic sequencing data obtained from ectomycorrhiza to identify well expressed genes responsible for fixed pathways of metabolism. Ectomycorrhizal metabolome model was developed for predicting different metabolites (glutamate, allantoin and glycine) synthesized from fungus L. bicolor and may be utilized by aspen. Consequently, aspen provides sugars such as glucose and fructose to the fungus. These results suggest the involvement of transcriptomic data from these complex systems [178]. Thus, this will help in the identification of both RNA molecules and the functions of individual genes and will lead to the proteins and metabolites production during the interactions between plant and microbes. A simplified form of this method can be applied using genome-based models which are applicable for metabolic flux analysis. Here, the microbial communities act as a closely associated superorganism where the genome-based models for the interacting plants integrate compartmentalization levels and differentiate metabolic processes in mitochondria, peroxisomes, cytoplasm, vacuole and chloroplasts. The genome-based models were developed based on the key metabolites of C4 plants, Arabidopsis and 25 species of bacteria [179]. Quantitative data from metabolite profiling and expression of gene (metabolomics) can be involved in both types of models.
4. Information gaps and future perspectives
Plants constitute an enormous treasure of microbiome, which remains to be explored for the identification of associated bacterial communities. This may be interesting for plants that exist under unique ecosystems or with exceptional lifestyles (carnivores, parasites). Though there has been a huge progress in understanding plant microbiome, there is a need to develop further understanding of the processes that surmount to community formation and function in plants. Metagenomic examination and comparison of plant-related communities result in novel functional and phylogenetic views. Proteomes, transcriptomes and initial metagenomes have been extensively studied [90]. A fascinating example of plant-microbe interaction is the coexistence of photosynthesis in plant and microbes on the leaves of Tamarix plant [180]. Benefits of a microbial community can be assessed through functional analysis. Other useful information that can be derived through this analysis unravels induction pathways and activation patterns between strains and ecotypes. Amplicon sequencing of 16SrRNA gene segments reveals important information related to dominant colonizers. The quantities of gene amplicon depend on the primer efficiency, extraction methods and their variation in copy-number.
The study of plant microbiome can be an effective tool to formulate strategies for sustainable agriculture, e.g. biocontrol, developing biofertilizers using microbial inoculants, products for stress protection. The future holds great prospects for plant microbiome in plant biotechnology and breeding. The extension of the plant microbiome as an integrated biomarker should be considered to maximize the benefits of the entire microbiome. Better plant microbiome cataloguing may serve as a key to prevent the outbreak of diseases in plants as well as human pathogens transgression in plants. Recent studies have proven the involvement of the human microbiome in diseases and the association of pathogen outbreak with support from pathogenic species. While human pathogens have been extensively studied, plant pathogens have remained a mystery [2]. Plant microbiome may also hold the key to improving other microbiomes. Comparing microbiome structures may yield phylogenetic diversity identification among similar/related microbiomes e.g. microbiomes between human and plant habitats. Food not only delivers essential nutrients, but also microorganisms into the human microbiome. Studies have revealed the heavy influence of surrounding vegetation and human inhabitants on the domestic microbiome. These complex natured interactions and connections among microbiomes need further exploration.
Conclusion
The methods involving omics have strengthened our knowledge in studying interactions between plant and microbes in the field of bioremediation, stress tolerance and nitrogen fixation. Intensive research to discover the methods and impacts of plant and pathogen interaction to increase defense response of the host have been observed in several crop systems. Changes detected under abiotic and biotic stress levied on the host correspond to the modification of the reactive oxygen species (ROS) sifting molecular constituents. Early research focused on unraveling nitrogen fixation methodology and functions of flavonoids. Observations are underway to identify the most efficient strains and possible influences at play in the process. Omics has proven effective in characterizing known constituents such as nod factors, inositol monophosphatase, nifH, and fixA. Omics studies have also been carried out to identify unknown proteins that help in fixing nitrogen using genistein (a known isoflavone for nodulation regulation). During interactions between plant and pathogen, ROS-scavenging and PR constituents’ synthesis are primarily improved. The profile of biological constituents and their microbial interactions can be studied using the omics technique. These researches should be formulated because of the close linkage between gene expression in parasitism and symbiosis between microbes and plants. These studies help to identify resistant genes and their respective Avr proteins during the interaction. The analysis of end products of toxins produced by the pathogen helps in the evolution of strategies to increase the productivity of plants. Omics technology helps in analysing the complex cellular mechanism during the interactions of plant and microbes in various ways. Genomics helps in comparing different hosts, microbe and interaction between host and microbe. The evolution of new strains and their interactions with plants can be researched by applying comparative and integrated genomics which can make significant progress in developing sustainable agriculture strategies.
Acknowledgements
Authors would like to thank the Department of Chemistry and Biotechnology, Tallinn University of Technology, for providing research facilities. The authors would like to acknowledge the English editing of the manuscript by Dr. Urvashi Kuhad, Department of English, University of Delhi, New Delhi, India. The authors would also like to acknowledge Alex Prigojine, a native English speaker for revising the language content and grammar of the manuscript.
Consent for Publication
Not applicable.
Funding
None.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Olanrewaju O.S., Ayangbenro A.S., Glick B.R., Babalola O.O. Plant health: feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019;103(3):1155–1166. doi: 10.1007/s00253-018-9556-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fürnkranz M., Lukesch B., Müller H., Huss H., Grube M., Berg G. Microbial diversity inside pumpkins: microhabitat-specific communities display a high antagonistic potential against phytopathogens. Microb. Ecol. 2012;63(2):418–428. doi: 10.1007/s00248-011-9942-4. [DOI] [PubMed] [Google Scholar]
- 3.Bragina A., Cardinale M., Berg C., Berg G. Vertical transmission explains the specific Burkholderia pattern in Sphagnum mosses. Front. Microbiol. 2014;18:434–394. doi: 10.3389/fmicb.2013.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manias D., Verma A., Soni D.K. Isolation and characterization of endophytes: Biochemical and molecular approach. Prospects for Sustainable Agriculture, Woodhead Publishing Series in Food Science, Technology and Nutrition; 2020. pp. 1–14. [Google Scholar]
- 5.Kumar V., Baweja M., Singh P.K., Shukla P. Recent developments in system biology and metabolic engineering of plant-microbe interactions. Front. Plant Sci. 2016;7:1421. doi: 10.3389/fpls.2016.01421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishra A., Mishra S.P., Arshi A., Agarwal A., Dwivedi S.K. In: Plant-microbe interactions for bioremediation and phytoremediation of environmental pollutants and agro-ecosystem development. Bioremediation of Industrial Waste for Environmental Safety; Bharagava, R. Saxena G., editor. Singapore: Springer; 2020. [Google Scholar]
- 7.Imam J., Alam S., Mandal N.P., Variar M., Shukla P. Molecular screening for identification of blast resistance genes in North East and Eastern Indian rice germplasm (Oryza sativa L.) with PCR based makers. Euphytica. 2013;196:199–211. doi: 10.1007/s10681-013-1024-x. [DOI] [Google Scholar]
- 8.Imam J., Mahto D., Mandal N.P., Maiti D., Shukla P., Variar M. Molecular analysis of Indian rice germplasm accessions with resistance to blast pathogen pages. J. Crop Improv. 2014;28:729–739. doi: 10.1080/15427528.2014.921261. [DOI] [Google Scholar]
- 9.Imam J., Alam S., Mandal N.P., Maiti D., Variar M., Shukla P. Molecular diversity and mating type distribution of the rice blast pathogen Magnaporthe oryzae in North-East and Eastern India. Indian J. Microbiol. 2015;55:108–113. doi: 10.1007/s12088-014-0504-6. [DOI] [Google Scholar]
- 10.Ren X-M., Guo S.J., Tian W., Chen Y., Han H., Chen E., Li B.L., Li Y.Y., Chen Z-J. Effects of plant growth-promoting bacteria (PGPB) inoculation on the growth, antioxidant activity, Cu uptake, and bacterial community structure of rape (Brassica napus L.) grown in cu-contaminated agricultural soil. Front. Microbiol. 2019;10:1455. doi: 10.3389/fmicb.2019.01455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rani R., Kumar V., Usmani Z., Gupta P., Chandra A. Influence of plant growth promoting rhizobacterial strains Paenibacillus sp. IITISM08, Bacillus sp. PRB77 and Bacillus sp. PRB101 using Helianthus annuus on degradation of endosulfan from contaminated soil. Chemosphere. 2019;225:479–489. doi: 10.1016/j.chemosphere.2019.03.037. [DOI] [PubMed] [Google Scholar]
- 12.Gupta P., Kumar V., Usmani Z., Rani R., Chandra A., Gupta V.K. Implications of plant growth promoting Klebsiella sp. CPSB4 and Enterobacter sp. CPSB49 in luxuriant growth of tomato plants under chromium stress. Chemosphere. 2020;240:124944. doi: 10.1016/j.chemosphere.2019.124944. [DOI] [PubMed] [Google Scholar]
- 13.Lima-Mendez G., Faust K., Henry N., Decelle J., Colin S., Carcillo F., Chaffron S., Ignacio-Espinosa J.C., Roux S., Vincent F., Bittner L., Darzi Y., Wang J., Audic S., Berline L., Bontempi G., Cabello A.M., Coppola L., Cornejo-Castillo F.M., d’Ovidio F., De Meester L., Ferrera I., Garet-Delmas M.J., Guidi L., Lara E., Pesant S., Royo-Llonch M., Salazar G., Sánchez P., Sebastian M., Souffreau C., Dimier C., Picheral M., Searson S., Kandels-Lewis S., Gorsky G., Not F., Ogata H., Speich S., Stemmann L., Weissenbach J., Wincker P., Acinas S.G., Sunagawa S., Bork P., Sullivan M.B., Karsenti E., Bowler C., de Vargas C., Raes J. Tara Oceans coordinators. Ocean plankton. Determinants of community structure in the global plankton interactome. Science. 2015;348(6237):1262073. doi: 10.1126/science.1262073. [DOI] [PubMed] [Google Scholar]
- 14.Frantzeskakis L., Di Pietro A., Rep M., Schirawski J., Wu C-H., Panstruga R. Rapid evolution in plant-microbe interactions - a molecular genomics perspective. New Phytol. 2020;225(3):1134–1142. doi: 10.1111/nph.15966. [DOI] [PubMed] [Google Scholar]
- 15.Terauchi R. KokiFujisaki, K.; Shimizu, M.; Oikawa, K.; Takeda, T.; Takagi, H.; Abe, A.; Okuyama, Y.; Yoshida, K.; Saitoh, H. Using genomics tools to understand plant resistance against pathogens: A case study of Magnaporthe-rice interactions. Applied Plant Biotechnology for Improving Resistance to Biotic Stress; 2020. pp. 181–188. [Google Scholar]
- 16.Gilbert J.A., Jansson J.K., Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cordero J., de Freitas J.R., Germida J.J. Bacterial microbiome associated with the rhizosphere and root interior of crops in Saskatchewan, Canada. Can. J. Microbiol. 2020;66(1):71–85. doi: 10.1139/cjm-2019-0330. [DOI] [PubMed] [Google Scholar]
- 18.Parray J.A., Shameem N. Sustainable agriculture: Advances in Plant Metabolome and Microbiome. United Kingdom: Academic Press, Elsevier; 2020. [Google Scholar]
- 19.Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 20.Berg G., Mahnert A., Moissl-Eichinger C. Beneficial effects of plant-associated microbes on indoor microbiomes and human health? Front. Microbiol. 2014;5:15. doi: 10.3389/fmicb.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wille L., Messmer M.M., Studer B., Hohmann P. Insights to plant-microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ. 2019;42(1):20–40. doi: 10.1111/pce.13214. [DOI] [PubMed] [Google Scholar]
- 22.Schlaeppi K., Bulgarelli D. The plant microbiome at work. Mol. Plant Microbe Interact. 2015;28(3):212–217. doi: 10.1094/MPMI-10-14-0334-FI. [DOI] [PubMed] [Google Scholar]
- 23.Berg G., Rybakova D., Grube M., Köberl M. The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 2016;67(4):995–1002. doi: 10.1093/jxb/erv466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orozco-Mosqueda M.D.C., Rocha-Granados M.D.C., Glick B.R., Santoyo G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 2018;208:25–31. doi: 10.1016/j.micres.2018.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Compant S., Samad A., Faist H., Sessitsch A. A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019;19:29–37. doi: 10.1016/j.jare.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang C., Nasir F., Ma L., Tian C. Legume Nitrogen Fixation in Soils with Low Phosphorus Availability. Cham: Springer; 2017. Molecular communication and nutrient transfer of arbuscular mycorrhizal fungi, symbiotic nitrogen-fixing bacteria, and host plant in tripartite symbiosis. [Google Scholar]
- 27.Taiwo L.B., Oyedele A.O., Ailenokhuoria B.V., Okareh O.T. In: Plant-microbes relationships in soil ecological system and benefits accruable to food health. Field Crops: Sustainable Management by PGPR. Sustainable Development and Biodiversity; Maheshwari, D. Dheeman S., editor. Vol. 23 Cham: Springer; 2019. [Google Scholar]
- 28.Alori E.T., Glick B.R., Babalola O.O. Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 2017;8:971. doi: 10.3389/fmicb.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y., Zhang J., Zhang J., Xu W., Mou Z. Characteristics of inorganic phosphate-solubilizing bacteria from the sediments of a eutrophic lake. Int. J. Environ. Res. Public Health. 2019;16(12):2141. doi: 10.3390/ijerph16122141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chhabra S., Dowling D.N. Endophyte-promoted nutrient acquisition: phosphorus and iron. In: Doty S., editor. Functional Importance of the Plant Microbiome. Cham: Springer; 2017. [Google Scholar]
- 31.Parewa H.P., Meena V.S., Jain L.K., Choudhary A. Sustainable crop production and soil health management through plant growth-promoting rhizobacteria. In: Meena V., editor. Role of Rhizospheric Microbes in Soil. Singapore: Springer; 2018. [Google Scholar]
- 32.Islam S., Akanda A.M., Prova A., Islam Md. T.; Md. Hossain, M. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front. Microbiol. 2016;••• doi: 10.3389/fmicb.2015.01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth R., Paszkowski U. Plant carbon nourishment of arbuscular mycorrhizal fungi. Curr. Opin. Plant Biol. 2017;39:50–56. doi: 10.1016/j.pbi.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 34.Finkel O.M., Castrillo G., Herrera Paredes S., Salas González I., Dangl J.L. Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 2017;38:155–163. doi: 10.1016/j.pbi.2017.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hiruma K., Kobae Y., Toju H. Beneficial associations between Brassicaceae plants and fungal endophytes under nutrient-limiting conditions: evolutionary origins and host-symbiont molecular mechanisms. Curr. Opin. Plant Biol. 2018;44:145–154. doi: 10.1016/j.pbi.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 36.Berruti A., Lumini E., Balestrini R., Bianciotto V. Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes. Front. Microbiol. 2016;6:1559. doi: 10.3389/fmicb.2015.01559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pathan S.I., Ceccherini M.T., Sunseri F., Lupini A. In: Rhizosphere as hotspot for plant-soil-microbe interaction. Carbon and Nitrogen Cycling in Soil; Datta, R.; Meena, R.; Pathan, S. Ceccherini M., editor. Springer; 2020. [Google Scholar]
- 38.Padda K.P., Puri A., Chanway C. Endophytic nitrogen fixation - a possible ‘hidden’ source of nitrogen for lodgepole pine trees growing at unreclaimed gravel mining sites. FEMS Microbiol. Ecol. 2019;95(11):fiz172. doi: 10.1093/femsec/fiz172. [DOI] [PubMed] [Google Scholar]
- 39.Bashandy S.R., Abd‐Alla M.H., Bagy M.M.K. Integrating Green Chemistry and Sustainable Engineering. Scrivener Publishing LLC; 2019. Biological Nitrogen Fixation and Biofertilizers as Ideal Potential Solutions for Sustainable Agriculture. [Google Scholar]
- 40.Pankievicz V.C.S., Irving T.B., Maia L.G.S., Ané J-M. Are we there yet? the long walk towards the development of efficient symbiotic associations between nitrogen-fixing bacteria and non-leguminous crops. BMC Biol. 2019;17(1):99. doi: 10.1186/s12915-019-0710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mahmud K., Makaju S., Ibrahim R., Missaoui A. Current progress in nitrogen fixing plants and microbiome research. Plants (Basel) 2020;9(1):97. doi: 10.3390/plants9010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C.W., Murray J.D. The role of flavonoids in nodulation host-range specificity: an update. Plants (Basel) 2016;5(3):33. doi: 10.3390/plants5030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathesius U. Flavonoid functions in plants and their interactions with other organisms. Plants (Basel) 2018;7(2):30. doi: 10.3390/plants7020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu Y.C., Qin X.M., Xiao J.X., Tang L., Wei C.Z., Wei J.J., Zheng Y. Intercropping influences component and content change of flavonoids in root exudates and nodulation of Faba bean. J. Plant Interact. 2017;12(1):187–192. doi: 10.1080/17429145.2017.1308569. [DOI] [Google Scholar]
- 45.van Noorden G.E., Kerim T., Goffard N., Wiblin R., Pellerone F.I., Rolfe B.G., Mathesius U. Overlap of proteome changes in Medicago truncatula in response to auxin and Sinorhizobium meliloti. Plant Physiol. 2007;144(2):1115–1131. doi: 10.1104/pp.107.099978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lery L.M.S., Hemerly A.S., Nogueira E.M., von Krüger W.M.A., Bisch P.M. Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium Gluconacetobacter diazotrophicus and sugarcane. Mol. Plant Microbe Interact. 2011;24(5):562–576. doi: 10.1094/MPMI-08-10-0178. [DOI] [PubMed] [Google Scholar]
- 47.Salavati A., Shafeinia A., Klubicova K., Bushehri A.A., Komatsu S. Proteomic insights into intra- and intercellular plant-bacteria symbiotic association during root nodule formation. Front. Plant Sci. 2013;4:28. doi: 10.3389/fpls.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Bousquet A., Harris J.M. Abscisic acid and lateral root organ defective/NUMEROUS INFECTIONS AND POLYPHENOLICS modulate root elongation via reactive oxygen species in Medicago truncatula. Plant Physiol. 2014;166(2):644–658. doi: 10.1104/pp.114.248542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kandasamy S., Loganathan K., Muthuraj R., Duraisamy S., Seetharaman S., Thiruvengadam R., Ponnusamy B., Ramasamy S. Understanding the molecular basis of plant growth promotional effect of Pseudomonas fluorescens on rice through protein profiling. Proteome Sci. 2009;7(47):47. doi: 10.1186/1477-5956-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chi F., Yang P., Han F., Jing Y., Shen S. Proteomic analysis of rice seedlings infected by Sinorhizobium meliloti 1021. Proteomics. 2010;10(9):1861–1874. doi: 10.1002/pmic.200900694. [DOI] [PubMed] [Google Scholar]
- 51.Schliemann W., Ammer C., Strack D. Metabolite profiling of mycorrhizal roots of Medicago truncatula. Phytochemistry. 2008;69(1):112–146. doi: 10.1016/j.phytochem.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 52.Pedone-Bonfim M.V., Lins M.A., Coelho I.R., Santana A.S., Silva F.S., Maia L.C. Mycorrhizal technology and phosphorus in the production of primary and secondary metabolites in cebil (Anadenanthera colubrina (Vell.) Brenan) seedlings. J. Sci. Food Agric. 2013;93(6):1479–1484. doi: 10.1002/jsfa.5919. [DOI] [PubMed] [Google Scholar]
- 53.Rispail N., Hauck B., Bartholomew B., Watson A.A., Nash R.J., Webb K.J. Secondary metabolite profiling of the model legume Lotus japonicus during its symbiotic interaction with Mesorhizobium loti. Symbiosis. 2010;50:119–128. doi: 10.1007/s13199-010-0053-7. [DOI] [Google Scholar]
- 54.Popovici J., Walker V., Bertrand C., Bellvert F., Fernandez M.P., Comte G. Strain specificity in the Myricaceae-Frankia symbiosis is correlated to plant root phenolics. Funct. Plant Biol. 2011;38(9):682. doi: 10.1071/FP11144. [DOI] [PubMed] [Google Scholar]
- 55.Hrynkiewicz K., Zloch M., Kowalkowski T., Baum C., Buszewski B. Efficiency of microbially assisted phytoremediation of heavy-metal contaminated soils. Environ. Rev. 2018;26(3):316–332. doi: 10.1139/er-2018-0023. [DOI] [Google Scholar]
- 56.Mendoza-Hernández J.C., Vázquez-Delgado O.R., Castillo-Morales M., Varela-Caselis J.L., Santamaría-Juárez J.D., Olivares-Xometl O., Arriola Morales J., Pérez-Osorio G. Phytoremediation of mine tailings by Brassica juncea inoculated with plant growth-promoting bacteria. Microbiol. Res. 2019;228:126308. doi: 10.1016/j.micres.2019.126308. [DOI] [PubMed] [Google Scholar]
- 57.Cheng Z., Wei Y.Y., Sung W.W., Glick B.R., McConkey B.J. Proteomic analysis of the response of the plant growth-promoting bacterium Pseudomonas putida UW4 to nickel stress. Proteome Sci. 2009;7:18. doi: 10.1186/1477-5956-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bona E., Cattaneo C., Cesaro P., Marsano F., Lingua G., Cavaletto M., Berta G. Proteomic analysis of Pteris vittata fronds: two arbuscular mycorrhizal fungi differentially modulate protein expression under arsenic contamination. Proteomics. 2010;10(21):3811–3834. doi: 10.1002/pmic.200900436. [DOI] [PubMed] [Google Scholar]
- 59.Aloui A., Recorbet G., Robert F., Schoefs B., Bertrand M., Henry C., Gianinazzi-Pearson V., Dumas-Gaudot E., Aschi-Smiti S. Arbuscular mycorrhizal symbiosis elicits shoot proteome changes that are modified during cadmium stress alleviation in Medicago truncatula. BMC Plant Biol. 2011;11:75. doi: 10.1186/1471-2229-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lingua G., Bona E., Todeschini V., Cattaneo C., Marsano F., Berta G., Cavaletto M. Effects of heavy metals and arbuscular mycorrhiza on the leaf proteome of a selected poplar clone: a time course analysis. PLoS One. 2012;7(6):e38662. doi: 10.1371/journal.pone.0038662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Farinati S., DalCrso G., Panigati M., Furini A. Interaction between selected bacterial strains and Arabidopsis halleri modulates shoot proteome and cadmium and zinc accumulation. J. Exp. Bot. 2011;62(10):3433–3447. doi: 10.1093/jxb/err015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pozo M.J., López-Ráez J.A., Azcón-Aguilar C., García-Garrido J.M. Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 2015;205(4):1431–1436. doi: 10.1111/nph.13252. [DOI] [PubMed] [Google Scholar]
- 63.Gomes D.F., da Silva Batista J.S., Rolla A.A., da Silva L.P., Bloch C., Galli-Terasawa L.V., Hungria M. Proteomic analysis of free-living Bradyrhizobium diazoefficiens: highlighting potential determinants of a successful symbiosis. BMC Genomics. 2014;15:643. doi: 10.1186/1471-2164-15-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barea J.M. Future challenges and perspectives for applying microbial biotechnology in sustainable agriculture based on a better understanding of plant microbiome interactions. J. Soil Sci. Plant Nutr. 2015;15(2):261–282. doi: 10.4067/S0718-95162015005000021. [DOI] [Google Scholar]
- 65.Carvalhais L.C., Dennis P.G., Badri D.V., Tyson G.W., Vivanco J.M., Schenk P.M. Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS One. 2013;8(2):e56457. doi: 10.1371/journal.pone.0056457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirsch P.R., Mauchline T.H. Who’s who in the plant root microbiome? Nat. Biotechnol. 2012;30(10):961–962. doi: 10.1038/nbt.2387. [DOI] [PubMed] [Google Scholar]
- 67.Barret M., Tan H., Egan F., Morrissey J.P., Reen J., O’Gara F. F.J, de Bruijn Molecular Microbial Ecology of the Rhizosphere. Hoboken, New Jersey, USA: Wiley Blackwell; 2013. Exploiting new systems-based strategies to elucidate plant-bacterial interactions in the rhizosphere. pp. 57–68. [DOI] [PubMed] [Google Scholar]
- 68.Schreiter S., Eltlbany N., Smalla K. Microbial communities in the rhizosphere analyzed by cultivation independent DNA-based methods. In: Lugtenberg B., editor. Principles of Plant-Microbe Interactions. Heidelberg: Springer International Publishing Switzerland; 2015. pp. 289–298. [Google Scholar]
- 69.Igiehon N.O., Babalola O.O. Below-ground-above-ground Plant-microbial Interactions: focusing on Soybean, Rhizobacteria and Mycorrhizal Fungi. Open Microbiol. J. 2018;12:261–279. doi: 10.2174/1874285801812010261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertani I., Abbruscato P., Piffanelli P., Subramoni S., Venturi V. Rice bacterial endophytes: isolation of a collection, identification of beneficial strains and microbiome analysis. Environ. Microbiol. Rep. 2016;8(3):388–398. doi: 10.1111/1758-2229.12403. [DOI] [PubMed] [Google Scholar]
- 71.Junker R.R., Keller A. Microhabitat heterogeneity across leaves and flower organs promotes bacterial diversity. FEMS Microbiol. Ecol. 2015;91(9):fiv097. doi: 10.1093/femsec/fiv097. [DOI] [PubMed] [Google Scholar]
- 72.Romero F.M., Marina M., Pieckenstain F.L. The communities of tomato (Solanum lycopersicum L.) leaf endophytic bacteria, analyzed by 16S-ribosomal RNA gene pyrosequencing. FEMS Microbiol. Lett. 2014;351(2):187–194. doi: 10.1111/1574-6968.12377. [DOI] [PubMed] [Google Scholar]
- 73.Trujillo M.E., Riesco R., Benito P., Carro L. Endophytic Actinobacteria and the interaction of Micromonospora and nitrogen fixing plants. Front. Microbiol. 2015;6:1341. doi: 10.3389/fmicb.2015.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fonseca-García C., Coleman-Derr D., Garrido E., Visel A., Tringe S.G., Partida-Martínez L.P. The cacti microbiome: interplay between habitat-filtering and host-specificity. Front. Microbiol. 2016;7:150. doi: 10.3389/fmicb.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rosenberg E., Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. MBio. 2016;7(2):e01395. doi: 10.1128/mBio.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hart M.L., Meyer A., Johnson P.J., Ericsson A.C. Comparative evaluation of DNA extraction methods from faeces of multiple host species for downstream next-generation sequencing. PLoS One. 2015;10(11):e0143334. doi: 10.1371/journal.pone.0143334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franzén O., Hu J., Bao X., Itzkowitz S.H., Peter I., Bashir A. Improved OTU-picking using long-read 16S rRNA gene amplicon sequencing and generic hierarchical clustering. Microbiome. 2015;3:43. doi: 10.1186/s40168-015-0105-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Felczykowska A., Krajewska A., Zielińska S., Łoś J.M. Sampling, metadata and DNA extraction - important steps in metagenomic studies. Acta Biochim. Pol. 2015;62(1):151–160. doi: 10.18388/abp.2014_916. [DOI] [PubMed] [Google Scholar]
- 79.Glassing A., Dowd S.E., Galandiuk S., Davis B., Jorden J.R., Chiodini R.J. Changes in 16S RNA Gene microbial community profiling by concentration of prokaryotic DNA. J. Microbiol. Methods. 2015;119:239–242. doi: 10.1016/j.mimet.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 80.Sunagawa S., Mende D.R., Zeller G., Izquierdo-Carrasco F., Berger S.A., Kultima J.R., Coelho L.P., Arumugam M., Tap J., Nielsen H.B., Rasmussen S., Brunak S., Pedersen O., Guarner F., de Vos W.M., Wang J., Li J., Doré J., Ehrlich S.D., Stamatakis A., Bork P. Metagenomic species profiling using universal phylogenetic marker genes. Nat. Methods. 2013;10(12):1196–1199. doi: 10.1038/nmeth.2693. [DOI] [PubMed] [Google Scholar]
- 81.Shade A., McManus P.S., Handelsman J. Unexpected diversity during community succession in the apple flower microbiome. MBio. 2013;4(2):e00602–e00612. doi: 10.1128/mBio.00602-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaul S., Sharma T., Dhar M.K. “Omics” tools for better understanding the plant-endophyte interactions. Front. Plant Sci. 2016;••• doi: 10.3389/fpls.2016.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumari A., Sumer S., Jalan B., Lyngdoh P. In: Nongbri, Laskar, M.A. Impact of Next-Generation Sequencing Technology in Plant-Microbe Interaction Study. Springer International Publishing AG, Kalia. Kumar P., editor. Vol. C Microbial Applications; 2017. [Google Scholar]
- 84.Chaterji S., Koo J., Li N., Meyer F., Grama A., Bagchi S. Federation in genomics pipelines: techniques and challenges. Brief. Bioinform. 2019;20(1):235–244. doi: 10.1093/bib/bbx102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delmotte N., Ahrens C.H., Knief C., Qeli E., Koch M., Fischer H.M., Vorholt J.A., Hennecke H., Pessi G. An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics. 2010;10(7):1391–1400. doi: 10.1002/pmic.200900710. [DOI] [PubMed] [Google Scholar]
- 86.Ali A., Alexandersson E., Sandin M., Resjo S., Lenman M., Hedley P., Levander F., Andreasson E. Quantitative proteomics and transcriptomics of potato in response to Phytophthora infestans in compatible and incompatible interactions. BMC Genomics. 2014;19:15–497. doi: 10.1186/1471-2164-15-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mitra S. 2019. Multiple Data Analyses and Statistical Approaches for analyzing data from metagenomic studies and clinical trials. [DOI] [PubMed] [Google Scholar]
- 88.Delmotte N., Knief C., Chaffron S., Innerebner G., Roschitzki B., Schlapbach R., von Mering C., Vorholt J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA. 2009;106(38):16428–16433. doi: 10.1073/pnas.0905240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sessitsch A., Hardoim P., Döring J., Weilharter A., Krause A., Woyke T., Mitter B., Hauberg-Lotte L., Friedrich F., Rahalkar M., Hurek T., Sarkar A., Bodrossy L., van Overbeek L., Brar D., van Elsas J.D., Reinhold-Hurek B. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol. Plant Microbe Interact. 2012;25(1):28–36. doi: 10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- 90.Knief C., Delmotte N., Chaffron S., Stark M., Innerebner G., Wassmann R., von Mering C., Vorholt J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6(7):1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rascovan N., Carbonetto B., Perrig D., Díaz M., Canciani W., Abalo M., Alloati J., González-Anta G., Vazquez M.P. Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep. 2016;6(1):28084. doi: 10.1038/srep28084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas T., Gilbert J., Meyer F. Metagenomics - a guide from sampling to data analysis. Microb. Inform. Exp. 2012;2(1):3. doi: 10.1186/2042-5783-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lucaciu R., Pelikan C., Gerner S.M., Zioutis C., Köstlbacher S., Marx H., Herbold C.W., Schmidt H., Rattei T. A bioinformatics guide to plant microbiome analysis. Front. Plant Sci. 2019;10:1313. doi: 10.3389/fpls.2019.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liao H.L., Chen Y., Vilgalys R. Metatranscriptomic Study of common and host-specific patterns of gene expression between pines and their symbiotic ectomycorrhizal fungi in the Genus Suillus. PLoS Genet. 2016;12(10):e1006348. doi: 10.1371/journal.pgen.1006348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas J., Kim H.R., Rahmatallah Y., Wiggins G., Yang Q., Singh R., Glazko G., Mukherjee A. 2019. [DOI] [PMC free article] [PubMed]
- 96.Oliveira V.H.D., Ullah I., Dunwell J.M., Tibbett M. Mycorrhizal symbiosis induces divergent patterns of transport and partitioning of Cd and Zn in Populus trichocarpa. Environ. Exp. Bot. 2020;171:103925. doi: 10.1016/j.envexpbot.2019.103925. [DOI] [Google Scholar]
- 97.Ithal N., Recknor J., Nettleton D., Hearne L., Maier T., Baum T.J., Mitchum M.G. Parallel genome-wide expression profiling of host and pathogen during soybean cyst nematode infection of soybean. Mol. Plant Microbe Interact. 2007;20(3):293–305. doi: 10.1094/MPMI-20-3-0293. [DOI] [PubMed] [Google Scholar]
- 98.Brotman Y., Lisec J., Méret M., Chet I., Willmitzer L., Viterbo A. Transcript and metabolite analysis of the Trichoderma-induced systemic resistance response to Pseudomonas syringae in Arabidopsis thaliana. Microbiology. 2012;158(Pt 1):139–146. doi: 10.1099/mic.0.052621-0. [DOI] [PubMed] [Google Scholar]
- 99.Breuillin F., Schramm J., Hajirezaei M., Ahkami A., Favre P., Druege U., Hause B., Bucher M., Kretzschmar T., Bossolini E., Kuhlemeier C., Martinoia E., Franken P., Scholz U., Reinhardt D. Phosphate systemically inhibits development of arbuscular mycorrhiza in Petunia hybrida and represses genes involved in mycorrhizal functioning. Plant J. 2010;64(6):1002–1017. doi: 10.1111/j.1365-313X.2010.04385.x. [DOI] [PubMed] [Google Scholar]
- 100.Medina J., Monreal C.M., Orellana L., Calabi-Floody M., González M.E., Meier S., Borie F., Cornejo P., Cornejo P. Influence of saprophytic fungi and inorganic additives on enzyme activities and chemical properties of the biodegradation process of wheat straw for the production of organo-mineral amendments. J. Environ. Manage. 2020;255:109922. doi: 10.1016/j.jenvman.2019.109922. [DOI] [PubMed] [Google Scholar]
- 101.Scervino J.M., Mesa M.P., Ivana Della Monica I.D., Recchi M., Moreno N.S., Godeas A. Soil fungal isolates produce different organic acid patterns involved in phosphate salts solubilisation. Biol. Fertil. Soils. 2010;46:755–763. doi: 10.1007/s00374-010-0482-8. [DOI] [Google Scholar]
- 102.Planchamp C., Glauser G., Mauch-Mani B. Root inoculation with Pseudomonas putida KT2440 induces transcriptional and metabolic changes and systemic resistance in maize plants. Front. Plant Sci. 2015;5(1):719. doi: 10.3389/fpls.2014.00719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yasmin S., Hafeez F.Y., Mirza M.S., Rasul M., Arshad H.M.I., Zubair M., Iqbal M. Biocontrol of bacterial leaf blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front. Microbiol. 2017;8:1895. doi: 10.3389/fmicb.2017.01895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Verhagen B.W.M., Glazebrook J., Zhu T., Chang H-S., van Loon L.C., Pieterse C.M. The transcriptome of rhizobacteria-induced systemic resistance in arabidopsis. Mol. Plant Microbe Interact. 2004;17(8):895–908. doi: 10.1094/MPMI.2004.17.8.895. [DOI] [PubMed] [Google Scholar]
- 105.Utsumi Y., Tanaka M., Kurotani A., Yoshida T., Mochida K., Matsui A., Ishitani M., Sraphet S., Whankaew S., Asvarak T., Narangajavana J., Triwitayakorn K., Sakurai T., Seki M. Cassava (Manihot esculenta) transcriptome analysis in response to infection by the fungus Colletotrichum gloeosporioides using an oligonucleotide-DNA microarray. J. Plant Res. 2016;129(4):711–726. doi: 10.1007/s10265-016-0828-x. [DOI] [PubMed] [Google Scholar]
- 106.Sham A., Moustafa K., Al-Shamisi S., Alyan S., Iratni R., AbuQamar S. Microarray analysis of Arabidopsis WRKY33 mutants in response to the necrotrophic fungus Botrytis cinerea. PLoS One. 2017;12(2):e0172343. doi: 10.1371/journal.pone.0172343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xu L., Zhu L., Tu L., Liu L., Yuan D., Jin L., Long L., Zhang X. Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 2011;62(15):5607–5621. doi: 10.1093/jxb/err245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Romero F.M., Marina M., Pieckenstain F.L., Rossi F.R., Gonzalez M.R., Vignatti P., Garriz A. In: Gaining insight into plant responses to beneficial and pathogenic microorganisms using metabolomic and transcriptomic approaches. Metabolic Engineering for Bioactive Compounds; Kalia, V. Saini A., editor. Singapore: Springer; 2017. [Google Scholar]
- 109.Farooq M.A., Niazi A.K., Akhtar J. Saifullah; Farooq, M.; Souri, Z.; Karimi, N.; Rengel, Z. Acquiring control: the evolution of ROS-Induced oxidative stress and redox signaling pathways in plant stress responses. Plant Physiol. Biochem. 2019;141:353–369. doi: 10.1016/j.plaphy.2019.04.039. [DOI] [PubMed] [Google Scholar]
- 110.Schwessinger B., Zipfel C. News from the frontline: recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008;11(4):389–395. doi: 10.1016/j.pbi.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 111.Asai S., Rallapalli G., Piquerez S.J., Caillaud M.C., Furzer O.J., Ishaque N., Wirthmueller L., Fabro G., Shirasu K., Jones J.D. Expression profiling during arabidopsis/downy mildew interaction reveals a highly-expressed effector that attenuates responses to salicylic acid. PLoS Pathog. 2014;10(10):e1004443. doi: 10.1371/journal.ppat.1004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhuang X., McPhee K.E., Coram T.E., Peever T.L., Chilvers M.I. Rapid transcriptome characterization and parsing of sequences in a non-model host-pathogen interaction; pea-Sclerotinia sclerotiorum. BMC Genomics. 2012;13(668):668. doi: 10.1186/1471-2164-13-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gyetvai G., Sønderkær M., Göbel U., Basekow R., Ballvora A., Imhoff M., Kersten B., Nielsen K.L., Gebhardt C. The transcriptome of compatible and incompatible interactions of potato (Solanum tuberosum) with Phytophthora infestans revealed by DeepSAGE analysis. PLoS One. 2012;7(2):e31526. doi: 10.1371/journal.pone.0031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liao W., Ji L., Wang J., Chen Z., Ye M., Ma H., An X. Identification of glutathione S-transferase genes responding to pathogen infestation in Populus tomentosa. Funct. Integr. Genomics. 2014;14(3):517–529. doi: 10.1007/s10142-014-0379-y. [DOI] [PubMed] [Google Scholar]
- 115.Xin M., Wang X., Peng H., Yao Y., Xie C., Han Y., Ni Z., Sun Q. Transcriptome comparison of susceptible and resistant wheat in response to powdery mildew infection. Genomics Proteomics Bioinformatics. 2012;10(2):94–106. doi: 10.1016/j.gpb.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.De Cremer K., Mathys J., Vos C., Froenicke L., Michelmore R.W., Cammue B.P., De Coninck B. RNAseq-based transcriptome analysis of Lactuca sativa infected by the fungal necrotroph Botrytis cinerea. Plant Cell Environ. 2013;36(11):1992–2007. doi: 10.1111/pce.12106. [DOI] [PubMed] [Google Scholar]
- 117.Passos M.A., de Cruz V.O., Emediato F.L., de Teixeira C.C., Azevedo V.C., Brasileiro A.C., Amorim E.P., Ferreira C.F., Martins N.F., Togawa R.C., Júnior G.J., da Silva O.B., Jr, Miller R.N. Analysis of the leaf transcriptome of Musa acuminata during interaction with Mycosphaerella musicola: gene assembly, annotation and marker development. BMC Genomics. 2013;14:78. doi: 10.1186/1471-2164-14-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nejat N., Cahill D.M., Vadamalai G., Ziemann M., Rookes J., Naderali N., Ziemann M., Rookes J., Naderali N. Transcriptomics-based analysis using RNA-Seq of the coconut (Cocos nucifera) leaf in response to yellow decline phytoplasma infection. Mol. Genet. Genomics. 2015;290(5):1899–1910. doi: 10.1007/s00438-015-1046-2. [DOI] [PubMed] [Google Scholar]
- 119.Jaiswal S., Jadhav P.V., Jasrotia R.S., Kale P.B., Kad S.K., Moharil M.P., Dudhare M.S., Kheni J., Deshmukh A.G., Mane S.S., Nandanwar R.S., Penna S., Manjaya J.G., Iquebal M.A., Tomar R.S., Kawar P.G., Rai A., Kumar D. Transcriptomic signature reveals mechanism of flower bud distortion in witches’-broom disease of soybean (Glycine max). BMC Plant Biol. 2019;19(1):26. doi: 10.1186/s12870-018-1601-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Feng H., Li H., Zhang M., Song Y., Yuan G., Han Q., Huang L. Responses of Fuji (Malus domestica) and Shandingzi (Malus baccata) apples to Marssonina coronaria infection revealed by comparative transcriptome analysis. Physiol. Mol. Plant Pathol. 2019;106:87–95. doi: 10.1016/j.pmpp.2018.12.007. [DOI] [Google Scholar]
- 121.Zhao T., Liu W., Zhao Z., Yang H., Bao Y., Zhang D., Wang Z., Jiang J., Xu Y., Zhang H., Li J., Chen Q., Xu X. Transcriptome profiling reveals the response process of tomato carrying Cf-19 and Cladosporium fulvum interaction. BMC Plant Biol. 2019;19(1):572. doi: 10.1186/s12870-019-2150-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Neu E., Domes H.S., Menz I., Kaufmann H., Linde M., Debener T. Interaction of roses with a biotrophic and a hemibiotrophic leaf pathogen leads to differences in defense transcriptome activation. Plant Mol. Biol. 2019;99(4-5):299–316. doi: 10.1007/s11103-018-00818-2. [DOI] [PubMed] [Google Scholar]
- 123.Bhat N.N., Bilal Padder B.A., Barthelson R.A., Andrabi K.I. Compendium of Colletotrichum graminicola responsive infection-induced transcriptomic shifts in the maize. Plant Gene. 2019;17:100166. doi: 10.1016/j.plgene.2018.11.001. [DOI] [Google Scholar]
- 124.Parker J., Koh J., Yoo M.J., Zhu N., Feole M., Yi S., Chen S. Quantitative proteomics of tomato defense against Pseudomonas syringae infection. Proteomics. 2013;13(12-13):1934–1946. doi: 10.1002/pmic.201200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bernardo L., Prinsi B., Negri A.S., Cattivelli L., Espen L., Valè G. Proteomic characterization of the Rph15 barley resistance gene-mediated defence responses to leaf rust. BMC Genomics. 2012;13:642. doi: 10.1186/1471-2164-13-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Margaria P., Abbà S., Palmano S. Novel aspects of grapevine response to phytoplasma infection investigated by a proteomic and phospho-proteomic approach with data integration into functional networks. BMC Genomics. 2013;14:38. doi: 10.1186/1471-2164-14-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kundu S., Chakraborty D., Kundu A., Pal A. Proteomics approach combined with biochemical attributes to elucidate compatible and incompatible plant-virus interactions between Vigna mungo and Mungbean Yellow Mosaic India Virus. Proteome Sci. 2013;11(15):15. doi: 10.1186/1477-5956-11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Petriccione M., Di Cecco I., Arena S., Scaloni A., Scortichini M. Proteomic changes in Actinidia chinensis shoot during systemic infection with a pandemic Pseudomonas syringae pv. actinidiae strain. J. Proteomics. 2013;78:461–476. doi: 10.1016/j.jprot.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 129.Holtappels M., Noben J.P., Van Dijck P., Valcke R. Fire blight host-pathogen interaction: proteome profiles of Erwinia amylovora infecting apple rootstocks. Sci. Rep. 2018;8(1):11689. doi: 10.1038/s41598-018-30064-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Meng Q., Gupta R., Min C.W., Kwon S.W., Wang Y., Je B.I., Kim Y.J., Jeon J.S., Agrawal G.K., Rakwal R., Kim S.T. Proteomics of rice-Magnaporthe oryzae interaction: what have we learned so far? Front. Plant Sci. 2019;10:1383. doi: 10.3389/fpls.2019.01383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li Y., Feng Y., Lü Q., Yan D., Liu Z., Zhang X. Comparative proteomic analysis of plant-pathogen interactions in resistant and susceptible poplar ecotypes infected with Botryosphaeria dothidea. Phytopathology. 2019;109(12):2009–2021. doi: 10.1094/PHYTO-12-18-0452-R. [DOI] [PubMed] [Google Scholar]
- 132.Dos Santos E.C., Pirovani C.P., Correa S.C., Micheli F., Gramacho K.P. The pathogen Moniliophthora perniciosa promotes differential proteomic modulation of cacao genotypes with contrasting resistance to witches´ broom disease. BMC Plant Biol. 2020;20(1):1. doi: 10.1186/s12870-019-2170-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hu X., Puri K.D., Gurung S., Klosterman S.J., Wallis C.M., Britton M., Johnson B.D., Phinney B., Salemi M., Short D.P.G., Subbarao K.V. Proteome and metabolome analyses reveal differential responses in tomato-Verticillium dahliae-interactions. J. Proteomics. 2019;207:15–103449. doi: 10.1016/j.jprot.2019.103449. [DOI] [PubMed] [Google Scholar]
- 134.Jashni K.M., Burgt A., Battaglia E., Mehrabi R., Collemare J., Pierre J.G.M. de Wit. Transcriptome and proteome analyses of proteases in biotroph fungal pathogen Cladosporium fulvum. J. Plant Pathol. 2019;102:377–386. doi: 10.1007/s42161-019-00433-0. [DOI] [Google Scholar]
- 135.Scandiani M.M., Luque A.G., Razori M.V., Ciancio Casalini L., Aoki T., O’Donnell K., Cervigni G.D., Spampinato C.P. Metabolic profiles of soybean roots during early stages of Fusarium tucumaniae infection. J. Exp. Bot. 2015;66(1):391–402. doi: 10.1093/jxb/eru432. [DOI] [PubMed] [Google Scholar]
- 136.Aliferis K.A., Faubert D., Jabaji S. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One. 2014;9(11):e111930. doi: 10.1371/journal.pone.0111930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hong Y.S., Martinez A., Liger-Belair G., Jeandet P., Nuzillard J.M., Cilindre C. Metabolomics reveals simultaneous influences of plant defence system and fungal growth in Botrytis cinerea-infected Vitis vinifera cv. Chardonnay berries. J. Exp. Bot. 2012;63(16):5773–5785. doi: 10.1093/jxb/ers228. [DOI] [PubMed] [Google Scholar]
- 138.Allwood J.W., Clarke A., Goodacre R., Mur L.A. Dual metabolomics: a novel approach to understanding plant-pathogen interactions. Phytochemistry. 2010;71(5-6):590–597. doi: 10.1016/j.phytochem.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 139.Yogendra K.N., Ajjamada C.K., Sarmiento F., Rodriguez E.B., Mosquera T. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Funct. Plant Biol. 2014;42(3):284–298. doi: 10.1071/FP14177. [DOI] [PubMed] [Google Scholar]
- 140.Waskiewicz A., Irzykowska L., Drzewiecka K., Bocianowski J., Dobosz B., Weber Z., Karolewski Z., Krzyminiewski R., Golinski P. Plant-pathogen interactions during infection process of Asparagus with Fusarium spp. Cent. Eur. J. Biol. 2013;8(11):1065–1076. [Google Scholar]
- 141.Pasquet J.C., Chaouch S., Macadré C., Balzergue S., Huguet S., Martin-Magniette M.L., Bellvert F., Deguercy X., Thareau V., Heintz D., Saindrenan P., Dufresne M. Differential gene expression and metabolomic analyses of Brachypodium distachyon infected by deoxynivalenol producing and non-producing strains of Fusarium graminearum. BMC Genomics. 2014;15:629. doi: 10.1186/1471-2164-15-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Negrel L., Halter D., Wiedemann-Merdinoglu S., Rustenholz C., Merdinoglu D., Hugueney P., Baltenweck R. Identification of lipid markers of Plasmopara viticola infection in grapevine using a non-targeted metabolomic approach. Front. Plant Sci. 2018;9:360. doi: 10.3389/fpls.2018.00360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Su X., Lu G., Guo H., Zhang K., Li X., Cheng H. The dynamic transcriptome and metabolomics profiling in Verticillium dahliae inoculated Arabidopsis thaliana. Sci. Rep. 2018;8(1):15404. doi: 10.1038/s41598-018-33743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.de Falco B., Manzo D., Incerti G., Garonna A.P., Ercolano M., Lanzotti V. Metabolomics approach based on NMR spectroscopy and multivariate data analysis to explore the interaction between the leafminer Tuta absoluta and tomato (Solanum lycopersicum). Phytochem. Anal. 2019;30(5):556–563. doi: 10.1002/pca.2850. [DOI] [PubMed] [Google Scholar]
- 145.Azizi P., Osman M., Hanafi M.M., Sahebi M., Yusop M.R., Taheri S. Adaptation of the metabolomics profile of rice after Pyricularia oryzae infection. Plant Physiol. Biochem. 2019;144:466–479. doi: 10.1016/j.plaphy.2019.10.014. [DOI] [PubMed] [Google Scholar]
- 146.Zhu L., Zhou Y., Li X., Zhao J., Guo N., Xing H. Metabolomics analysis of soybean hypocotyls in response to Phytophthora sojae infection. Front. Plant Sci. 2018;9:1530. doi: 10.3389/fpls.2018.01530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kawahara Y., Oono Y., Kanamori H., Matsumoto T., Itoh T., Minami E. Simultaneous RNA-seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One. 2012;7(11):e49423. doi: 10.1371/journal.pone.0049423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aragona M., Minio A., Ferrarini A., Valente M.T., Bagnaresi P., Orrù L., Tononi P., Zamperin G., Infantino A., Valè G., Cattivelli L., Delledonne M. De novo genome assembly of the soil-borne fungus and tomato pathogen Pyrenochaeta lycopersici. BMC Genomics. 2014;15:313. doi: 10.1186/1471-2164-15-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hane J.K., Anderson J.P., Williams A.H., Sperschneider J., Singh K.B. Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. 2014;10(5):e1004281. doi: 10.1371/journal.pgen.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Coutinho B.G., Licastro D., Mendonça-Previato L., Cámara M., Venturi V. Plant-influenced gene expression in the rice endophyte Burkholderia kururiensis M130. Mol. Plant Microbe Interact. 2015;28(1):10–21. doi: 10.1094/MPMI-07-14-0225-R. [DOI] [PubMed] [Google Scholar]
- 151.Roux B., Rodde N., Jardinaud M-F., Timmers T., Sauviac L., Cottret L., Carrère S., Sallet E., Courcelle E., Moreau S., Debellé F., Capela D., de Carvalho-Niebel F., Gouzy J., Bruand C., Gamas P. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014;77(6):817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- 152.Pankievicz V.C.S., Camilios-Neto D., Bonato P., Balsanelli E., Tadra-Sfeir M.Z., Faoro H., Chubatsu L.S., Donatti L., Wajnberg G., Passetti F., Monteiro R.A., Pedrosa F.O., Souza E.M. RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol. Biol. 2016;90(6):589–603. doi: 10.1007/s11103-016-0430-6. [DOI] [PubMed] [Google Scholar]
- 153.Paungfoo-Lonhienne C., Lonhienne T.G.A., Yeoh Y.K., Donose B.C., Webb R.I., Parsons J., Liao W., Sagulenko E., Lakshmanan P., Hugenholtz P., Schmidt S., Ragan M.A. Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci. Rep. 2016;6:37389. doi: 10.1038/srep37389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nobori T., Velásquez A.C., Wu J., Kvitko B.H., Kremer J.M., Wang Y., He S.Y., Tsuda K. Transcriptome landscape of a bacterial pathogen under plant immunity. Proc. Natl. Acad. Sci. USA. 2018;115(13):E3055–E3064. doi: 10.1073/pnas.1800529115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Saberi F., Kamali M., Najafi A. Yazdanparast. A.; Moghaddam, M.M. Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications. Cell. Mol. Biol. Lett. 2016;21:6. doi: 10.1186/s11658-016-0007-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hör J., Gorski S.A., Vogel J. Bacterial RNA biology on a genome scale. Mol. Cell. 2018;70(5):785–799. doi: 10.1016/j.molcel.2017.12.023. [DOI] [PubMed] [Google Scholar]
- 157.Chaparro J.M., Badri D.V., Vivanco J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014;8(4):790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Chapelle E., Mendes R., Bakker P.A.H., Raaijmakers J.M. Fungal invasion of the rhizosphere microbiome. ISME J. 2016;10(1):265–268. doi: 10.1038/ismej.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yergeau E., Sanschagrin S., Maynard C., St-Arnaud M., Greer C.W. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2014;8(2):344–358. doi: 10.1038/ismej.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Robison F.M., Turner M.F., Jahn C.E., Schwartz H.F., Prenni J.E., Brick M.A., Heuberger A.L. Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum. Plant Cell Environ. 2018;41(9):2141–2154. doi: 10.1111/pce.13176. [DOI] [PubMed] [Google Scholar]
- 161.Nordzieke D.E., Fernandes T.R., El Ghalid M., Turrà D., Di Pietro A. NADPH oxidase regulates chemotropic growth of the fungal pathogen Fusarium oxysporum towards the host plant. New Phytol. 2019;224(4):1600–1612. doi: 10.1111/nph.16085. [DOI] [PubMed] [Google Scholar]
- 162.Delaunois B., Jeandet P., Clément C., Baillieul F., Dorey S., Cordelier S. Uncovering plant-pathogen crosstalk through apoplastic proteomic studies. Front. Plant Sci. 2014;5:249. doi: 10.3389/fpls.2014.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ashwin N.M.R., Barnabas L., Ramesh Sundar A., Malathi P., Viswanathan R., Masi A., Agrawal G.K., Rakwal R. Advances in proteomic technologies and their scope of application in understanding plant-pathogen interactions. J. Plant Biochem. Biotechnol. 2017;26:371–386. doi: 10.1007/s13562-017-0402-1. [DOI] [Google Scholar]
- 164.Wright M.H. Chemical proteomics of host-microbe interactions. Proteomics. 2018;18(18):e1700333. doi: 10.1002/pmic.201700333. [DOI] [PubMed] [Google Scholar]
- 165.Li X., Bai T., Li Y., Ruan X., Li H. Proteomic analysis of Fusarium oxysporum sp. cubense tropical race 4-inoculated response to Fusarium wilts in the banana root cells. Proteome Sci. 2013;11(1):41. doi: 10.1186/1477-5956-11-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lambais M.R., Barrera S.E., Santos E.C., Crowley D.E., Jumpponen A. Phyllosphere metaproteomes of trees from the Brazilian Atlantic forest show high levels of functional redundancy. Microb. Ecol. 2017;73(1):123–134. doi: 10.1007/s00248-016-0878-6. [DOI] [PubMed] [Google Scholar]
- 167.Kierul K., Voigt B., Albrecht D., Chen X.H., Carvalhais L.C., Borriss R. Influence of root exudates on the extracellular proteome of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Microbiology. 2015;161(Pt 1):131–147. doi: 10.1099/mic.0.083576-0. [DOI] [PubMed] [Google Scholar]
- 168.Bao Z., Okubo T., Kubota K., Kasahara Y., Tsurumaru H., Anda M., Ikeda S., Minamisawa K. Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl. Environ. Microbiol. 2014;80(16):5043–5052. doi: 10.1128/AEM.00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhalnina K., Louie K.B., Hao Z., Mansoori N., da Rocha U.N., Shi S., Cho H., Karaoz U., Loqué D., Bowen B.P., Firestone M.K., Northen T.R., Brodie E.L. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018;3(4):470–480. doi: 10.1038/s41564-018-0129-3. [DOI] [PubMed] [Google Scholar]
- 170.de Bruijn F. 2019.
- 171.Brechenmacher L., Weidmann S., van Tuinen D., Chatagnier O., Gianinazzi S., Franken P., Gianinazzi-Pearson V. Expression profiling of up-regulated plant and fungal genes in early and late stages of Medicago truncatula-Glomus mosseae interactions. Mycorrhiza. 2004;14(4):253–262. doi: 10.1007/s00572-003-0263-4. [DOI] [PubMed] [Google Scholar]
- 172.Moy P., Qutob D., Chapman B.P., Atkinson I., Gijzen M. Patterns of gene expression upon infection of soybean plants by Phytophthora sojae. Mol. Plant Microbe Interact. 2004;17(10):1051–1062. doi: 10.1094/MPMI.2004.17.10.1051. [DOI] [PubMed] [Google Scholar]
- 173.Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as Biotrophy-associated secreted proteins in rice blast disease. Plant Cell. 2009;21(4):1273–1290. doi: 10.1105/tpc.107.055228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Torto-Alalibo T., Collmer C.W., Gwinn-Giglio M., The Plant-Associated Microbe Gene Ontology (PAMGO) Consortium The Plant-Associated Microbe Gene Ontology (PAMGO) Consortium: community development of new Gene Ontology terms describing biological processes involved in microbe-host interactions. BMC Microbiol. 2009;9(Suppl. 1):S1. doi: 10.1186/1471-2180-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang J., Wang P., Tian H., Tao Z., Guo T. Transcriptome analysis of ice plant growth-promoting endophytic Bacterium Halomonas sp. Strain MC1 to identify the genes involved in salt tolerance. Microorganisms. 2020;8(1):88. doi: 10.3390/microorganisms8010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Orozco-Mosqueda M.D.C., Duan J., DiBernardo M., Zetter E., Campos-García J., Glick B.R., Santoyo G. The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 2019;10:1392. doi: 10.3389/fmicb.2019.01392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Niu S-Q., Li H-R., Pare P.W., Aziz M., Wang S-M., Shi H., Li J., Han Q-Q., Guo S-Q., Li J. Induced growth promotion and higher salt tolerance in the halophyte grass Puccinelliatenuiflora by beneficial rhizobacteria. Plant Soil. 2016;407:217–230. doi: 10.1007/s11104-015-2767-z. [DOI] [Google Scholar]
- 178.Larsen P.E., Sreedasyam A., Trivedi G., Podila G.K., Cseke L.J., Collart F.R., Collart F.R. Using next generation transcriptome sequencing to predict an ectomycorrhizal metabolome. BMC Syst. Biol. 2011;5:70. doi: 10.1186/1752-0509-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Dal’Molin C.G.O., Quek L.E., Palfreyman R.W., Brumbley S.M., Nielsen L.K. C4GEM, a genome-scale metabolic model to study C4 plant metabolism. Plant Physiol. 2010;154(4):1871–1885. doi: 10.1104/pp.110.166488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Atamna-Ismaeel N., Finkel O., Glaser F., von Mering C., Vorholt J.A., Koblížek M., Belkin S., Béjà O. Bacterial anoxygenic photosynthesis on plant leaf surfaces. Environ. Microbiol. Rep. 2012;4(2):209–216. doi: 10.1111/j.1758-2229.2011.00323.x. [DOI] [PubMed] [Google Scholar]