 Mono- (vaborbactam) and bi-cyclic (taniborbactam) boronate inhibitors bind KPC-2 similarly, but the latter is essentially irreversible.
Mono- (vaborbactam) and bi-cyclic (taniborbactam) boronate inhibitors bind KPC-2 similarly, but the latter is essentially irreversible.
Abstract
Klebsiella pneumoniae carbapenemase-2 (KPC-2) is a serine-β-lactamase (SBL) capable of hydrolysing almost all β-lactam antibiotics. We compare KPC-2 inhibition by vaborbactam, a clinically-approved monocyclic boronate, and VNRX-5133 (taniborbactam), a bicyclic boronate in late-stage clinical development. Vaborbactam inhibition is slowly reversible, whereas taniborbactam has an off-rate indicating essentially irreversible complex formation and a 15-fold higher on-rate, although both potentiate β-lactam activity against KPC-2-expressing K. pneumoniae. High resolution X-ray crystal structures reveal closely related binding modes for both inhibitors to KPC-2, with differences apparent only in positioning of the endocyclic boronate ester oxygen. The results indicate the bicyclic boronate scaffold as both an efficient, long-lasting, KPC-2 inhibitor and capable of supporting further iterations that may improve potency against specific enzyme targets and pre-empt the emergence of inhibitor resistant KPC-2 variants.
Introduction
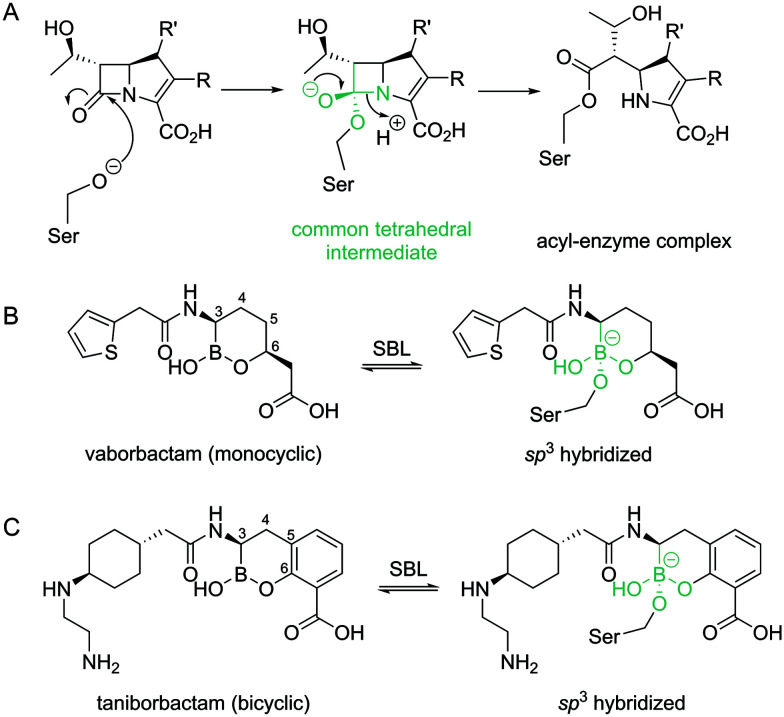
β-Lactamase production by Gram-negative bacteria (GNB) such as Klebsiella pneumoniae, Pseudomonas aeruginosa and Escherichia coli (organisms that are important causes of healthcare-associated infections)1 is a major antibiotic resistance mechanism. β-Lactamases are divided into four classes; classes A, C and D use a nucleophilic serine to hydrolyse β-lactam antibiotics, while class B enzymes employ zinc ions in their active site.2 Of particular clinical importance is the widely disseminated, plasmid-encoded Klebsiella pneumoniae carbapenemase-2 (KPC-2), a class A serine-β-lactamase (SBL).3 The wide spectrum of KPC-2 activity extends to late-generation cephalosporins as well as the last-resort carbapenems such as meropenem.4 Antibiotic hydrolysis by SBLs, like KPC-2, occurs through attack on the β-lactam ring by the nucleophilic serine to form an acyl–enzyme complex via a tetrahedral (sp3 hybridized) intermediate (Fig. 1A), followed by subsequent deacylation to release the inactive hydrolysed product.
Fig. 1. β-Lactam hydrolysis by serine-β-lactamases and cyclic boronate inhibitor structures. (A) Outline mechanism for acyl–enzyme formation during β-lactam hydrolysis by SBLs. The common tetrahedral ‘core’ is in green. (B) Vaborbactam forms a covalent bond to the nucleophilic serine in SBLs, with the sp3 formed boronate mimicking the common tetrahedral intermediate in A. The carbon atoms of the cyclic boronate core are numbered. (C) Formation of the tetrahedral form of taniborbactam (sp3 hybridized boron) on interaction with SBLs.
The combination of a β-lactam antibiotic with a β-lactamase inhibitor is a clinically validated route to overcoming resistance.5 For example, avibactam, a recent clinical introduction, is a non-β-lactam-based diazabicyclooctane (DBO) inhibitor used in combination with the third-generation cephalosporin ceftazidime (Avycaz®) for treatment of complicated urinary tract infections (UTIs) and intra-abdominal infections (IAIs).6 Mechanistically, avibactam forms a reversible, covalent, carbamoyl ester linkage to the nucleophilic serine of SBLs after ring opening.7 Further iterations of the avibactam core are in development, with relebactam recently approved for the treatment of complicated UTIs and IAIs.8 However, DBOs have variable activity, are not active against MBLs,9 and there is evidence now emerging that SBLs (particularly KPC-2) can evolve to resist the action of the DBO combination Avycaz®.10 In addition, some MBLs are known to hydrolyse avibactam, thus highlighting the possibility that MBL production by GNB could contribute to DBO resistance.9 There is therefore continued interest in developing additional classes of non-β-lactam based SBL and MBL inhibitors.11 Boronate-based compounds have long been known as β-lactamase inhibitors.11–16 In 2017 the combination of vaborbactam (a monocyclic boronate, Fig. 1B) with meropenem (Vabomere®) was clinically approved to treat complicated UTIs.5,17 Vaborbactam is particularly potent against the SBL KPC-2, but is not active against class D SBLs,18 and only moderately inhibits some MBLs.18 Further development of cyclic boronates has led to compounds containing bicyclic, rather than monocyclic cores.11,19,20 Bicyclic boronates were shown to inhibit the majority of tested SBLs from all classes, including KPC-2, and some MBLs.19,20 Taniborbactam (Fig. 1C, originally named VNRX-5133), is one such iteration, now in phase 3 clinical trials in combination with cefepime.21 Both mono- and bi-cyclic boronates act as covalent inhibitors, with the mechanism of inhibition involving formation of a sp3 hybridized boron covalently bound to the nucleophilic serine, mimicking the tetrahedral intermediate formed during SBL/MBL catalysis19–21 (Fig. 1). The sp2 hybridised form of boron-based inhibitors has been proposed to mimic the intact β-lactam, facilitating rapid binding to the active site.19–21 Moreover, in crystallographic analyses of the subclass B1 MBL New Delhi β-lactamase-1 (NDM-1) in complex with taniborbactam, an unexpected cyclization of the acylamino oxygen onto the boron of the bicyclic core to give a tricyclic form21 highlighted the ability of boron-based inhibitors to interchange between different forms.
We compare KPC-2 inhibition by vaborbactam and taniborbactam, representatives of clinically relevant mono- and bicyclic boronates, respectively. The kinetic, microbiological and structural data provided describe the mechanism of KPC inhibition by both compounds and inform on future iterations of cyclic boronates which may retain or enhance activity against this clinically important SBL and its growing array of variants.
Results and discussion
Kinetics and microbiology
Cyclic boronates (including vaborbactam and bicyclic analogues of taniborbactam) have previously been shown to exhibit nanomolar IC50 values against some class A β-lactamases, including KPC-2.18 However, to elucidate the inhibition profiles of these two related compounds, detailed kinetic comparisons are required. Accordingly, we directly compared the kinetics of in vitro KPC-2 inhibition by the clinically relevant vaborbactam and taniborbactam (Table 1, Fig. S1†). While both inhibitors exhibit similar IC50s (35/37 nM after 10 min pre-incubation), they differ substantially under more detailed kinetic analysis. Compared to vaborbactam, taniborbactam exhibits a lower Kiapp (calculated with no pre-incubation, 1.3 vs. 8.5 μM) and an increased (15-fold higher) on-rate (k2/K, Fig. S1†).
Table 1. Kinetic parameters for KPC-2 inhibition.
| Inhibitor | IC50 (nM) | K iapp (μM) | k 2/K (M–1 s–1) | k off (s–1) | t 1/2 (min) |
| VAB | 35 (0.025) a | 8.5 (1.1) | 6.7 × 102 (0.2 × 102) | 0.00016 (1.5 × 10–5) | 72 |
| TAN | 37 (0.031) a | 1.3 (0.36) | 1.0 × 104 (0.09 × 104) | 3.4 × 10–10 (5.5 × 10–11) | 3.4 × 107 |
aStandard error of log IC50. VAB: vaborbactam; TAN: taniborbactam.
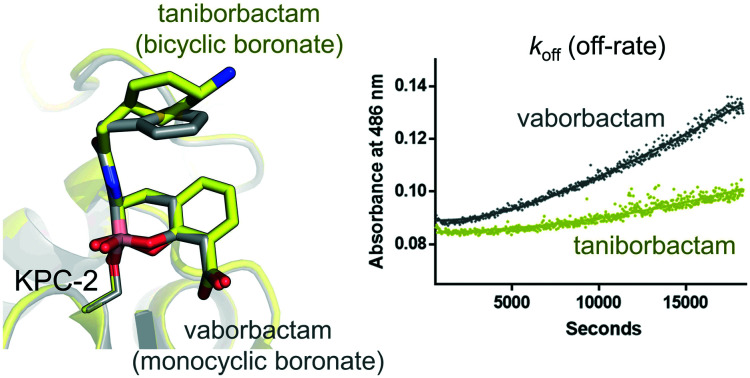
Importantly, the taniborbactam off-rate was extremely slow and difficult to measure, with a half-life (t1/2) indicative of near irreversible covalent complex formation (Fig. S1†). By contrast, vaborbactam (Fig. S1†) exhibited a slow but measurable off rate (0.00016 s–1) and a t1/2 of 72 minutes. These data indicate that taniborbactam likely forms a more stable complex with KPC-2, and is a more effective inhibitor than vaborbactam, at least against purified, recombinant enzyme. Values obtained for vaborbactam are comparable to those we recently reported for the DBO relebactam (calculated in the same way) which exhibits similar Kiapp (1.2 μM) and koff values (0.00087 s–1).22 We also note that a slow but measurable off-rate has recently been observed for inhibition of KPC-2 by non-cyclic phenylboronic acid derivatives, with kinetics for these compounds appearing to more closely resemble those of vaborbactam than taniborbactam.12
Although both vaborbactam and bicyclic boronates potentiate β-lactam activity against KPC-2 producing K. pneumoniae,21,23 direct comparison of their activity is not possible with available data as the two have not to date been tested against the same strain. Accordingly, their combinations with partner β-lactams cefepime or meropenem (β-lactams used clinically, or in trials, in combination with taniborbactam or vaborbactam, respectively) were evaluated against K. pneumoniae Ecl8 expressing KPC-2. Despite the clear improvement in potency of taniborbactam in kinetic experiments, both inhibitors have comparable effects upon this KPC-2 producing strain (Table S1†). The addition of 4 μg ml–1 inhibitor (the same concentration as used for previous taniborbactam MIC determinations24,25) significantly reduced MICs from ≥256 μg ml–1 (cefepime alone) or ≥16 μg ml–1 (meropenem alone) to ≤0.125 μg ml–1, a value well below the clinical breakpoints for susceptibility to these agents (≤2 μg ml–1 and ≤1 μg ml–1 for cefepime and meropenem respectively). Both inhibitors therefore remain viable and potent options for potentiating either cefepime or meropenem activity against KPC-2 producing K. pneumoniae.
Crystal structures
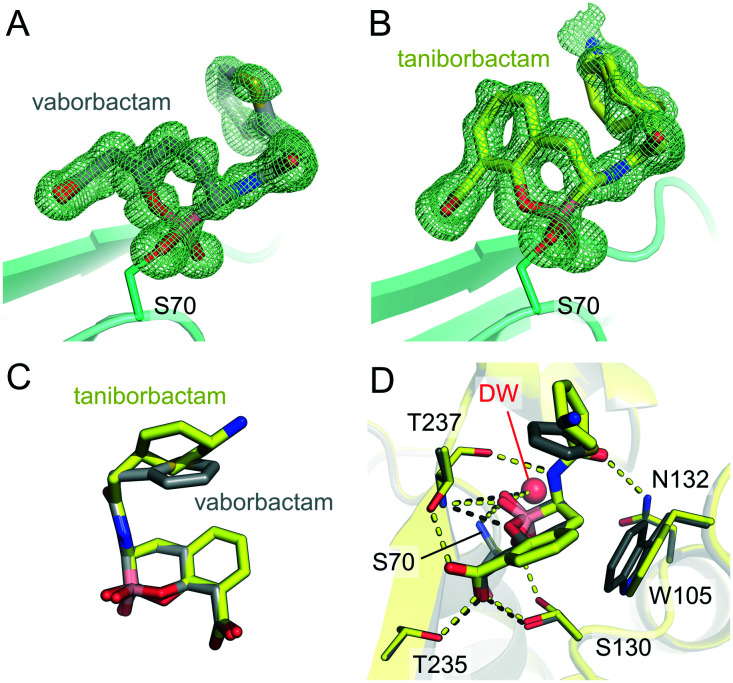
To further understand their mechanisms of inhibition we obtained X-ray crystal structures of KPC-2 complexes with both vaborbactam and taniborbactam, at resolutions of 1.2 Å and 0.99 Å, respectively (Table S2†). In both structures, clear Fo – Fc density in the active site reveals that the cyclic boronates inhibit KPC-2 through formation of a covalent attachment to the catalytic Ser70 (Fig. 2A and B). As previously identified by crystallography of cyclic boronate complexes with SBLs and MBLs,19–21,23,26,27 the boron atom is sp3 hybridized, i.e. in a tetrahedral geometry, thereby mimicking the tetrahedral intermediate formed during β-lactam acylation of SBLs.
Fig. 2. Views from crystal structures of cyclic boronate binding to KPC-2. Fo – Fc density calculated in the absence of ligand is shown as a green mesh, contoured to 3σ, for (A) vaborbactam (grey sticks) and (B) taniborbactam (yellow sticks). (C) Comparison of taniborbactam and vaborbactam in the KPC-2 active site. (D) An overlay of vaborbactam and taniborbactam KPC-2 structures (interactions shown as black and yellow dashes for vaborbactam and taniborbactam, respectively).
Both inhibitors were modelled as dual occupancy due to the presence of more than one conformation of their respective C-3 substituents (Fig. S2†). With vaborbactam, this is evident as a 180° rotation of the thiophene moiety, as was also observed on binding of vaborbactam to CTX-M-15 (a class A SBL) and AmpC (a class C SBL).23 With taniborbactam, the cyclohexane ring adopts two orientations, rotated by approximately 65°, with this flexibility also highlighted by the fact that the ethylamino ring substituent could not be modelled in the final structure due to poorly defined electron density. The same atoms could not be modelled in our previously reported crystal structure of taniborbactam bound to NDM-1,21 an MBL that, like KPC-2, also hydrolyses carbapenems. This contrasts with our previously determined X-ray crystal structure of taniborbactam bound to OXA-10, an enzyme which is unable to hydrolyse carbapenems, in which these atoms were well defined by the electron density.21 Flexibility in the taniborbactam C-3 substituent may therefore solely present when the inhibitor is bound to carbapenemases such as NDM-1 and KPC-2.
The binding modes of vaborbactam and taniborbactam to KPC-2 are similar; both adopt comparable geometries (Fig. 2C) and form almost identical interactions with the protein main chain (Fig. 2D, S3† and Table S3†). The inhibitor C6 carboxylates interact with the side chains of Thr235, Thr237 and Ser130; their acetamido oxygen with the N atom of the Asn132 side chain; and the amino N atoms with the backbone oxygen of Thr237. The boron-bound OH groups interact with the backbone amides of Thr237 and Ser70 that form the oxyanion hole, as well as with a water molecule in the deacylating position (DW), albeit with a (0.23 Å) decrease and (0.35 Å) increase in the distances to Thr237 and DW, respectively, for taniborbactam compared to vaborbactam (Table S3†). A slight movement (1.1 Å) of the indole side chain of Trp105 between the vaborbactam and taniborbactam structures reflects the additional hydrophobic interactions this residue makes with the bicyclic, and not monocyclic, core. Notably, Trp105 is modelled in a single conformation in both the vaborbactam and taniborbactam structures (Fig. 2D). Interactions of KPC-2 Trp105 with β-lactams are suggested to be essential for hydrolysis,28 and KPC-2 Trp105 is indeed stabilised by binding of the β-lactam substrates cefotaxime and faropenem (as observed crystallographically29). Conversely, however, we have previously observed flexibility of Trp105 (as modelled in multiple conformations) in complexes with the DBO inhibitor relebactam.22 There is also a difference in the positioning of the endocyclic boronate ester oxygen for the two cyclic boronates, which may reflect the hybridisation of the adjacent carbon as either sp3 (vaborbactam) or sp2 (taniborbactam). This results in this oxygen being positioned to either hydrogen bond with Thr237 (vaborbactam) or one of the two conformations of Ser130 (taniborbactam) (Fig. 2D and S3†).
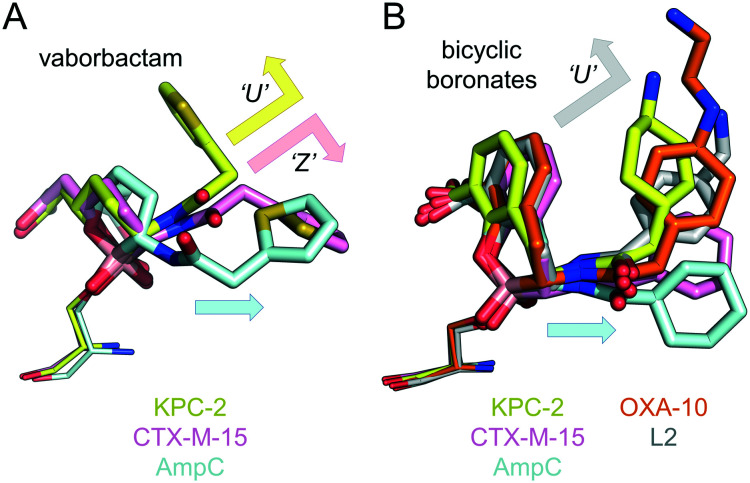
The structures reported here represent the first for any cyclic boronates bound to carbapenem-hydrolysing SBLs. In Fig. 3 we therefore compared them with previously determined crystal structures for cyclic boronates bound to the extended spectrum (class A) SBLs CTX-M-15 (ref. 20 and 23) and S. maltophilia L2,26 the class C SBL AmpC23,27 and the class D SBL OXA-10.21 The vaborbactam core binds KPC-2 almost identically to CTX-M-15,23 both differing from AmpC binding where, unusually, the six-membered cyclic boronic acid ester ring is inverted (i.e. the axial/equatorial conformations of the C-6 carboxymethyl and C-3 amide substituent are switched), resulting in a significantly different binding mode (Fig. 3A).23 Surprisingly, however, the vaborbactam C-3 substituent adopts a different conformation in KPC-2 compared to both CTX-M-15 and AmpC. In KPC-2, the vaborbactam side-chain is ‘U-shaped’, with the C-3 group folded over in a conformation we noted previously as a specific feature of bicyclic boronate binding to SBLs.21
Fig. 3. Cyclic boronate binding across serine-β-lactamases. (A) Overlays of vaborbactam binding to KPC-2 (yellow), CTX-M-15 (PDB 4XUZ, pink) and AmpC (PDB ; 4XUX, blue). Arrows indicate the ‘U’ and ‘Z’-shaped conformations vaborbactam adopts in KPC-2 and CTX-M-15, respectively. The ‘straighter’ conformation in AmpC is represented by a blue arrow. (B) Overlays of bicyclic boronate binding to KPC-2, CTX-M-15 (PDB ; 5T66), AmpC (PDB ; 6I30), OXA-10 (PDB ; 6RTN, orange) and L2 (PDB ; 5NE1, grey).
This contrasts with vaborbactam binding to CTX-M-15, where an alternative ‘Z-shaped’ conformation was observed (Fig. 3A) in a geometry we have previously seen adopted by bicyclic boronates when bound to MBLs.20,21 Indeed, taniborbactam also adopts the ‘U-shaped’ conformation (Fig. 3B), with its overall geometry closely reflecting that of other bicyclic boronates bound to CTX-M-15 (bicyclic boronate 1), L2 (bicyclic boronate 2), OXA-10 (taniborbactam) and AmpC (bicyclic boronate 1). Importantly, the currently available crystal structures indicate none of the bicyclic boronates to adopt the ‘Z-shaped’ conformation in SBLs (Fig. 3B).
In contrast to bicyclic boronates, which have a conserved binding mode across different SBLs, vaborbactam adopts a greater range of conformations in reported SBL complexes. This may be reflected in its substantially poorer spectrum of SBL inhibition when compared to taniborbactam. Against β-lactamases tested to date, vaborbactam has significantly higher IC50s than taniborbactam against TEM-116, AmpC and OXA-10/48 (ref. 21) enzymes.
These crystal structures also provide insights into the significant off-rate differences we observe between vaborbactam and taniborbactam. Release of intact cyclic boronate from KPC-2 would be expected to involve Ser70 protonation and cleavage of the O–B bond, accompanied by the boron atom switching from tetrahedral (sp3) to trigonal (sp2) geometry. Importantly, the positions of Ser70, and its interactions with putative proton donors (Lys73/‘deacylating’ water) are near-identical in the two complexes. Therefore, other factors, such as the environment and positioning of the endocyclic oxygen, which differs in our vaborbactam and taniborbactam structures, may affect inhibitor release. The presence of the adjacent aromatic ring in taniborbactam may also reduce negative charge around the boron atom, disfavouring O–B cleavage. Alternatively, sp2 hybridisation of the adjacent (aromatic) carbon in taniborbactam, compared to the sp3 C6 atom of vaborbactam, may constrain this oxygen and prevent any repositioning required as the geometry about the boron atom rearranges. In addition, the ability of vaborbactam to switch between axial/equatorial conformations,23 when compared with the rigid bicyclic rings of taniborbactam, may contribute to a faster off-rate. Further study will be required to establish the relative importance of these, and other, possibilities.
Experimental
Enzyme kinetics
All enzyme assays were performed at 25 °C in 10 mM HEPES (pH 7.5) and 150 mM NaCl with nitrocefin as a reporter substrate30 (Δε 486 nm = 20 500 M–1 cm–1) and absorbances read within Greiner half area 96-well plates in a POLARstar Omega (BMG LabTech) plate reader. Vaborbactam (MedChemExpress) and taniborbactam (synthesised as previously described21) were dissolved to 100 mM in dimethyl sulfoxide (DMSO) and diluted to the desired concentrations in 10 mM HEPES (pH 7.5) and 150 mM NaCl.
IC50, k2/K, Kiapp and Koff values were calculated using methods and equations as previously described.22 Briefly, IC50 values were determined by following the initial rates of nitrocefin hydrolysis (50 μM) measured after 10-minute preincubation of cyclic boronate and KPC-2. A 10 min preincubation time was chosen based on our previous data that indicates there is little effect on the inhibition of class A β-lactamases by bicylic boronates at longer incubation periods.20 Direct competition assays with a range of concentrations of cyclic boronate and 50 μM nitrocefin were performed to determine both k2/K and Kiapp, reactions were initiated by addition of 1 nM KPC-2. For koff calculations, 1 μM KPC-2 was pre-incubated with 8 μM cyclic boronate for 30 minutes at room temperature. Mixtures were subsequently serially diluted using the jump-dilution method31 and reaction monitored following addition of 50 μM nitrocefin, the final concentration of KPC-2 was 500 pM.
Minimal inhibitory concentration determination
The pUBYT vector containing blaKPC-2 under the ISKpn7 promotor was constructed as previously described.22 MIC values were determined using broth microdilution, in triplicate, in cation-adjusted Mueller–Hinton broth (Sigma) according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. Experiments were performed in microtiter plates (Corning) containing medium with cefepime or meropenem with 4 mg per liter inhibitor (vaborbactam or taniborbactam diluted from 100 mM stock dissolved in DMSO). Plates were incubated overnight at 37 °C for 18 to 24 hours, and the absorbance at 600 nm was read using a POLARstar Omega (BMG LabTech) plate reader.
Crystallisation and structure determination
Recombinant KPC-2 was produced, purified and crystallised as previously described.22 To obtain ligand bound structures, crystals were soaked in mother liquor supplemented with 1 mM vaborbactam or 1 mM taniborbactam for 3 or 16 hours, respectively. Crystals were flash-frozen in liquid nitrogen after brief exposure to mother liquor containing 30% glycerol. Diffraction data were collected at the European Synchrotron Radiation Facility (Grenoble, France) on beamline ID23-1 (vaborbactam soak) or ALBA (Barcelona, Spain) on beamline BL13-XALOC (taniborbactam soak). Data were integrated with DIALS32 (vaborbactam) or XDS33 (taniborbactam) and scaled in Aimless.34 Phases were calculated by molecular replacement in Phaser35 using PDB ID ; 6QW9 (ref. 22) (with ligands removed) as the starting model. Structures were completed with iterative rounds of manual model building in WinCoot36 and refinement in Phenix.37 The final models contained residues 23–294, with their overall structures closely resembling each other (root mean square deviation [RMSD] = 0.16 Å, over 270Cα) and native KPC-2 (PDB ID ; 5UL8,29 RMSD = 0.14 Å/0.17 Å over 270Cα). Geometry restraints for ligands were calculated using eLBOW in Phenix.37 Figures were generated in PyMOL.38
Conclusions
The results show both cyclic boronates to act as KPC-2 inhibitors, potentiating antibiotic activity against laboratory producer strains. However, taniborbactam appears more potent against isolated KPC-2, exhibiting significantly faster inactivation rates, and off-rates suggesting near irreversible inhibition. This finding arises from direct comparison of cyclic boronate inhibition kinetics under identical conditions and highlights the importance of using consistent methods for detailed comparisons across multiple β-lactamases and classes. In addition to the apparent capability of bicyclic boronates to act as more versatile cross-class SBL and MBL inhibitors, this greater potency will be an important consideration in the clinic, particularly against the most difficult to treat Gram-negative pathogens. Given the growing incidence of K. pneumoniae (and other Gram-negative) strains that co-produce KPC-2 alongside MBLs such as NDM-1,39 a taniborbactam combination would be expected to represent a more effective treatment option in these cases.
Our crystal structures reveal the boron-containing cyclic cores of the two inhibitors, together with the associated carboxylate groups, to bind almost identically to KPC-2. The ability of the enzyme to accommodate the substantially different substituent groups, along with structural comparisons of SBL binding modes for mono- and bicyclic compounds, indicates that there may be significant scope for development of further iterations of the cyclic boronate scaffold that retain the ability to act as potent β-lactamase inhibitors. This approach is already bearing fruit for DBOs, with iterations now in development that expand the activity profile and potency.5,22 The need to improve inhibitor activity is becoming increasingly important as enzymes such as KPC-2 accumulate mutations that provide resistance to clinical inhibitors such as avibactam.10 Cyclic boronates represent a potent and versatile β-lactamase inhibitor scaffold to counter the clinical threat and evolution of β-lactamase mediated antibiotic resistance.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
KPC-2:vaborbactam diffraction data were collected at beamline ID23-1 at the European Synchrotron Radiation Facility (ESRF), Grenoble, France. We are grateful to Montserrat Soler Lopez at the ESRF for providing assistance in using beamline ID23-1. KPC-2:taniborbactam diffraction data were collected at BL13-XALOC beamline at ALBA Synchrotron with the collaboration of ALBA staff. Research was supported by BBSRC (SWbioDTP (BB/J014400/1), studentship to C. L. T.) and (BB/S50676X/1), the Wellcome Trust (106244/Z/14/Z), MRC (MR/N002679/1), EPSRC (EP/M022609/1) and the Innovative Medicines Initiative (European Lead factory and ENABLE components).
Footnotes
†Electronic supplementary information (ESI) available: Supplemental figures and tables. See DOI: 10.1039/c9md00557a
References
- Haque M., Sartelli M., McKimm J., Abu Bakar M. Infect. Drug Resist. 2018;11:2321–2333. doi: 10.2147/IDR.S177247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke C. L., Hinchliffe P., Bragginton E. C., Colenso C. K., Hirvonen V. H. A., Takebayashi Y., Spencer J. J. Mol. Biol. 2019;431:3472–3500. doi: 10.1016/j.jmb.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Naas T., Poirel L. Emerging Infect. Dis. 2011;17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit H., Queenan A. M., Anderson G. J., Domenech-Sanchez A., Biddle J. W., Steward C. D., Alberti S., Bush K., Tenover F. C. Antimicrob. Agents Chemother. 2001;45:1151–1161. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush K., Bradford P. A. Nat. Rev. Microbiol. 2019;17:295–306. doi: 10.1038/s41579-019-0159-8. [DOI] [PubMed] [Google Scholar]
- Zhanel G. G., Lawson C. D., Adam H., Schweizer F., Zelenitsky S., Lagace-Wiens P. R., Denisuik A., Rubinstein E., Gin A. S., Hoban D. J., Lynch, 3rd J. P., Karlowsky J. A. Drugs. 2013;73:159–177. doi: 10.1007/s40265-013-0013-7. [DOI] [PubMed] [Google Scholar]
- Ehmann D. E., Jahić H., Ross P. L., Gu R. F., Hu J., Kern G., Walkup G. K., Fisher S. L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R. JAMA, J. Am. Med. Assoc. 2019;322:807–807. [Google Scholar]
- Abboud M. I., Damblon C., Brem J., Smargiasso N., Mercuri P., Gilbert B., Rydzik A. M., Claridge T. D. W., Schofield C. J., Frère J.-M. Antimicrob. Agents Chemother. 2016;60:5655–5662. doi: 10.1128/AAC.00897-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddins M. J., Macesic N., Annavajhala M. K., Stump S., Khan S., McConville T. H., Mehta M., Gomez-Simmonds A., Uhlemann A.-C. Antimicrob. Agents Chemother. 2018;62:e02101–e02117. doi: 10.1128/AAC.02101-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajnc A., Lang P. A., Panduwawala T. D., Brem J., Schofield C. J. Curr. Opin. Chem. Biol. 2019;50:101–110. doi: 10.1016/j.cbpa.2019.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza G., Vicario M., Bellio P., Linciano P., Perilli M., Oliver A., Blazquez J., Cendron L., Tondi D. ChemMedChem. 2018;13:713–724. doi: 10.1002/cmdc.201700788. [DOI] [PubMed] [Google Scholar]
- Werner J. P., Mitchell J. M., Taracila M. A., Bonomo R. A., Powers R. A. Protein Sci. 2017;26:515–526. doi: 10.1002/pro.3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandi F., Caselli E., Morandi S., Focia P. J., Blázquez J., Shoichet B. K., Prati F. J. Am. Chem. Soc. 2003;125:685–695. doi: 10.1021/ja0288338. [DOI] [PubMed] [Google Scholar]
- Strynadka N. C. J., Martin R., Jensen S. E., Gold M., Jones J. B. Nat. Struct. Biol. 1996;3:688–695. doi: 10.1038/nsb0896-688. [DOI] [PubMed] [Google Scholar]
- Rojas L. J., Taracila M. A., Papp-Wallace K. M., Bethel C. R., Caselli E., Romagnoli C., Winkler M. L., Spellberg B., Prati F., Bonomo R. A. Antimicrob. Agents Chemother. 2016;60:1751–1759. doi: 10.1128/AAC.02641-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- US Food & Drug Administration, Highlights of prescribing information. Vabomere™ (meropenem and vaborbactam) for injection, for intravenous use).
- Langley G. W., Cain R., Tyrrell J. M., Hinchliffe P., Calvopiña K., Tooke C. L., Widlake E., Dowson C. G., Spencer J., Walsh T. R., Schofield C. J., Brem J. Bioorg. Med. Chem. Lett. 2019;29:1981–1984. doi: 10.1016/j.bmcl.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem J., Cain R., Cahill S., McDonough M. A., Clifton I. J., Jiménez-Castellanos J.-C., Avison M. B., Spencer J., Fishwick C. W. G., Schofield C. J. Nat. Commun. 2016;7:12406. doi: 10.1038/ncomms12406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill S. T., Cain R., Wang D. Y., Lohans C. T., Wareham D. W., Oswin H. P., Mohammed J., Spencer J., Fishwick C. W., McDonough M. A., Schofield C. J., Brem J. Antimicrob. Agents Chemother. 2017;61:e02260-16. doi: 10.1128/AAC.02260-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajnc A., Brem J., Hinchliffe P., Calvopiña K., Panduwawala T. D., Lang P. A., Kamps J. J. A. G., Tyrrell J. M., Widlake E., Saward B. G., Walsh T. R., Spencer J., Schofield C. J. J. Med. Chem. 2019;62:8544–8556. doi: 10.1021/acs.jmedchem.9b00911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke C. L., Hinchliffe P., Lang P. A., Mulholland A. J., Brem J., Schofield C. J., Spencer J. Antimicrob. Agents Chemother. 2019;63:e00564-19. doi: 10.1128/AAC.00564-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker S. J., Reddy K. R., Totrov M., Hirst G. C., Lomovskaya O., Griffith D. C., King P., Tsivkovski R., Sun D., Sabet M., Tarazi Z., Clifton M. C., Atkins K., Raymond A., Potts K. T., Abendroth J., Boyer S. H., Loutit J. S., Morgan E. E., Durso S., Dudley M. N. J. Med. Chem. 2015;58:3682–3692. doi: 10.1021/acs.jmedchem.5b00127. [DOI] [PubMed] [Google Scholar]
- Daigle D., Hamrick J., Chatwin C., Kurepina N., Kreiswirth B. N., Shields R. K., Oliver A., Clancy C. J., Nguyen M.-H., Pevear D., Xerri L. Open Forum Infect. Dis. 2018;5:S419–S420. [Google Scholar]
- Hackel M., Sahm D. Open Forum Infect. Dis. 2018;5:S416–S417. [Google Scholar]
- Calvopiña K., Hinchliffe P., Brem J., Heesom K. J., Johnson S., Cain R., Lohans C. T., Fishwick C. W. G., Schofield C. J., Spencer J., Avison M. B. Mol. Microbiol. 2017;106:492–504. doi: 10.1111/mmi.13831. [DOI] [PubMed] [Google Scholar]
- Cahill S. T., Tyrrell J. M., Navratilova I. H., Calvopiña K., Robinson S. W., Lohans C. T., McDonough M. A., Cain R., Fishwick C. W. G., Avison M. B., Walsh T. R., Schofield C. J., Brem J. Biochim. Biophys. Acta, Gen. Subj. 2019;1863:742–748. doi: 10.1016/j.bbagen.2019.02.004. [DOI] [PubMed] [Google Scholar]
- Papp-Wallace K. M., Taracila M., Wallace C. J., Hujer K. M., Bethel C. R., Hornick J. M., Bonomo R. A. Protein Sci. 2010;19:1714–1727. doi: 10.1002/pro.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton O. A., Zhang X., Chen Y. J. Med. Chem. 2017;60:3525–3530. doi: 10.1021/acs.jmedchem.7b00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Antimicrob. Agents Chemother. 1972;1:283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland R. A., Basavapathruni A., Moyer M., Scott M. P. Anal. Biochem. 2011;416:206–210. doi: 10.1016/j.ab.2011.05.029. [DOI] [PubMed] [Google Scholar]
- Winter G., Waterman D. G., Parkhurst J. M., Brewster A. S., Gildea R. J., Gerstel M., Fuentes-Montero L., Vollmar M., Michels-Clark T., Young I. D., Sauter N. K., Evans G. Acta Crystallogr., Sect. D: Struct. Biol. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4 Acta Crystallogr., Sect. D: Biol. Crystallogr. 1994;50:760–763. [Google Scholar]
- McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano W. L., The PyMOL user's manual, p452, DeLano Scientific, San Carlos, California, USA, 2002.
- Wei D.-D., Wan L.-G., Liu Y. Genome Announc. 2018;6:e00192-18. doi: 10.1128/genomeA.00192-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.