Abstract
The gene based therapeutics and drug targets have shown incredible and appreciable advances in alleviating human sufferings and complexities. Epigenetics simply means above genetics or which controls the organism beyond genetics. At present it is very clear that all characteristics of an individual are not determined by DNA alone, rather the environment, stress, life style and nutrition play a vital part in determining the response of an organism. Thus, nature (genetic makeup) and nurture (exposure) play equally important roles in the responses observed, both at the cellular and organism levels. Epigenetics influence plethora of complications at cellular and molecular levels that includes cancer, metabolic and cardiovascular complications including neurological (psychosis) and neurodegenerative disorders (Alzheimer’s disease, Parkinson disease etc.). The epigenetic mechanisms include DNA methylation, histone modification and non coding RNA which have substantial impact on progression and pathways linked to Alzheimer’s disease. The epigenetic mechanism gets deregulated in Alzheimer’s disease and is characterized by DNA hyper methylation, deacetylation of histones and general repressed chromatin state which alter gene expression at the transcription level by upregulation, downregulation or silencing of genes. Thus, the processes or modulators of these epigenetic processes have shown vast potential as a therapeutic target in Alzheimer’s disease.
Keywords: Alzheimer’s disease, epigenetics, amyloid β, DNA methylation, histone, non coding RNA
1. Introduction
Alzheimer’s disorder (AD) is the progressive neurodegenerative disorder affecting primarily elderly population and is the most common form of dementia globally [1]. AD is characterized by progressive and continuous neurodegeneration in the brain, especially in the hippocampus, temporal lobe, frontal lobes and frontal cortex regions of the brain. Neurodegeneration in these regions inflicts the clinical manifestations of the AD, which includes dementia, learning impairment and progressive cognitive dysfunction [1]. Pathologically, AD is characterized by the development and accumulation of extracellular aggregates of amyloid β (Aβ) peptide in the brain accompanied by intracellular neurofibrillary tangles, which are generated by the hyperphosphorylated tau proteins within the neurons [2]. Clinically, AD is the 6th leading cause of mortality worldwide [3] and short-term memory loss is the most frequently reported clinical symptom in patients suffering from AD [4]. As the disease progresses, symptoms like mood disorder, unpredicted mood swings, reduced self-care, linguistic difficulties, social isolation, depression, etc. become evident [4]. Life expectancy of the patients diagnosed with AD cannot be predicted perfectly; however, on an average such patients may have 3 to 9 years before AD is diagnosed and eventually the patient dies [5, 6].
The factors which lead to the initiation and uncontrolled progression of AD are not well understood till date. However, the involvement of genetic factors in the pathogenesis of AD has been established in approximately 70% of cases and involves multiple genes [7]. Mutation in the genes regulating the production of Aβ peptide (PSEN1, APP and PSEN2 genes) has been recognized as the cause for the development of early-onset AD, which affects individuals having age less than 65 years. However, these mutations provide justification for just 1% of the cases and the etiology of the rest of the cases remains unexplained [8]. Late onset AD (LOAD) affects people above 65 years of age and accounts for approximately 95% clinical cases [8]. The etiology of LOAD is also unknown and the involvement of genetic factors in it is highly suspected. Researchers have reported that the mutations in the APOE gene are one of the major risk factors for the development and progression of LOAD, however, mutations alone in the APOE gene cannot explain completely the pathogenesis of LOAD [9].
Continuous research exploring the genome-wise association of AD has identified more than 20 susceptibility LOAD loci which explain its pathogenesis to some extent. The majority of these genes encodes for the proteins which are involved in the inflammatory pathway, cholesterol metabolism pathway and endosomal vesicle recycling pathways, however, individually neither of these pathways are considered a major risk factor for the development and progression of AD [1]. This along with the several studies conducted on the monozygotic twins suggest that several other risk factors besides genetic factors are involved in the pathogenesis of AD [8]. Further, approximately one-third cases of AD indicate low educational status, diabetes mellitus, depression, hypertension, obesity, smoking and lack of cognitive activity as the major contributing risk factor for AD [10]. Besides, the involvement of environmental factors, like the use of pesticides, emotional trauma, and head injuries, drastic changes in the lifestyle and dietary habits, in the pathogenesis of AD has been reported [11].
Although the pathogenesis and symptoms related to AD have been established, the pathways that lead to the development of these symptoms and pathogenesis of AD are poorly known. Besides, there is no cure for AD in current time and the available therapeutics provide only symptomatic relief without affecting the underlying pathogenesis of AD [12-14]. AD is a result of a complex interplay between genetic and environmental factors. Therefore, the pathogenesis of AD could be efficiently explained by the epigenetic mechanisms, which mediates the interaction between environmental factors and the genome response. In this review, authors have reviewed and discussed the major studies which provide a comprehensive detail towards the association of epigenetics in the pathogenesis of AD.
2. Epigenetic mechanisms involved in AD
Epigenetics is referred to any heritable changes altering the expression of gene/s without the alterations in the actual DNA sequence. Therefore, unlike genetic code, epigenetic code is specific to a particular cell or a tissue that can show changes with respect to different environmental factors, aging or a disease condition [15]. Epigenetic involves the response generated to environmental factors and this is the major difference between the epigenetics and genetic variations, which generally are independent of the environmental stimuli [16]. There are three major epigenetic mechanisms that regulate the interplay between genes and environmental factors, which include DNA methylation, histone modification and noncoding RNA molecules, mediated regulation of gene expression (Fig. 1).
Fig. (1).
Epigenetic is the core area of variety of disorders including AD. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
2.1. DNA methylation
Epigenetics is all about the lessons of structural changes of the chromatin which modify the phenotype without changing the genotype. DNA methylation is the most studied epigenetic biological phenomenon that controls the genetic expression and involves the addition of a methyl group to DNA. The process does not alter the DNA sequence yet changes the activity of DNA, gene expression and function of genes. DNA methylation is the most widely investigated mechanism of epigenetics for its involvement with several disorders like AD, Parkinsonism, depression etc [17-20]. DNA methylation involves the addition of a methyl residue to the DNA chain with the help of an enzyme known as DNA methyltransferases. The most common and frequent site of methylation by this enzyme is the cytosine residue leading to the formation of the 5-methylcytosine (5-mC) in a CpG dinucleotide clusters [18, 21]. Sites of the CpG clusters are referred as CpG islands, and when any such island located on the promoter region of a gene undergoes methylation, it results in the repression of the expression of that gene [22]. Further, CpG dinucleotides are located on the centromeric sequences, also known as repetitive sequences, and methylation of these CpG dinucleotidesis responsible for stabilizing the chromosome and for preventing translocation events [23]. Another epigenetic modification of the cytosine residue, mediated by its oxidation, is known as hydroxyl methylcytosine (5-hmC). 5-hmC is broadly distributed in the CNS and is known to be associated with several neurodegenerative disorders and neuro developmental abnormalities [24].
The first conclusive evidence of the involvement of the altered DNA methylation during AD was provided by West et al. in 1995. They reported that during AD, methylation levels of APP promoter region in the temporal lobe are significantly lowered when compared to a healthy individual and a patient suffering from Pick’s disease [25]. This was followed by a series of in-vitro and in-vivo studies involving cell lines and animals. These studies suggested that the alterations in the various environmental factors, such as vitamin B-group deficiency and exposure to heavy metals such as lead, lead to the methylation in the genes which are associated with the synthesis of Aβ peptide [26-28]. These findings were followed by the investigation of the DNA methylation in AD samples at either global or gene specific level. The association of global DNA methylation with the pathogenesis of AD is a debatable question. Investigators studied the content of 5-mC or 5-hmC as a measure of global DNA methylation and these reports from the past investigations have generated conflicting conclusions regarding the involvement of global DNA methylation in AD [29-37] and these reports confirm that the levels of both 5-mC and 5-hmC are significantly altered in different brain regions during the early stages of AD. These results suggest that global DNA methylation may be associated with the early onset of AD or during initial phases of AD or dementia [37].
The search for epigenetic involvement in the pathogenesis of AD was investigated directly on the postmortem AD brain samples, to achieve a better understanding. This search was initially focused on the genes which were related to dementia, LOAD or to the pathogenic markers of AD. These genes include those involved in the production of Aβ peptide (BACE1, APP, PSEN2 and PSEN1), genes involved in the formation of neurofibrillary tangle(GSK3B and MAPT) and gene involved in the pathogenesis of LOAD (APOE) [25, 38-41]. However, not much success was achieved through these studies as no clear cut and conclusive evidence of epigenetic alterations in the expression of these gene during AD was reported. Further, similar findings were reported from the blood samples of the living AD patients, suggesting the limited epigenetic involvement of these genes in the pathogenesis of AD [42, 43]. Further, several studies focused on the genes involved in the synaptic plasticity, inflammation, and other pathogenic pathways of AD to identify an epigenetic marker of AD. These studies were conducted on both blood samples of AD patients and neuronal DNA samples [42, 44-46]. However, these studies did not reveal any conclusive epigenetic biomarker due to having limited population size and lack of reproducibility of results, resulting in conflicting and contradictive outcomes [47, 48]. Some success was achieved with the TREM2 and BDNF genes studied in the blood and neuronal DNA of AD patients. Epigenetic changes in these genes can be considered as the potential biomarkers of AD in terms of DNA methylation [49-55]. These observations were reinforced with the observations in which it was suggested that methylation of BDNF in the promoter region resulted in the conversion of amnestic mild cognitive impairment condition (a stage prior to AD) to confirmed AD development [54, 55].
Neurodegeneration is a continuous process and it increases with age and a broad examination of more than 5000 genes demonstrated that changes in 5hmC with age can be linked directly to the development and progression of AD and specific loci have been identified on chromosomes which are primarily affected in AD patients [56, 57].
A search for an epigenetic biomarker for AD has been extensively studied through epigenome wide methylation and hydroxymethylation studies (EWAS). These studies have revealed a large number of differentially methylated (DMRs) or hydroxyl methylated (DhMRs) regions in AD brain when compared with control brains [58, 59]. These studies associated the involvement of gene PPT2-EGFL8, LOC100507547, PPT2, PRDM16, PRRT1, CDH23, RNF39, and C10orf105 in the pathogenesis of AD. All these genes encode for various proteins that are involved in the regulation of various cellular pathways, such as synaptic plasticity, intracellular communication etc. [60-62]. For a better understanding of the epigenetic association with the AD, researchers have used the samples from monozygotic and dizygotic twin pairs. These studies identified the crucial association of epigenetic modification of ADARB2 gene in the hippocampus with AD pathogenesis [63]. Further, impaired synaptic plasticity, learning dysfunction and memory impairment are also reported in ADARB2 mutant models [64]. Further, DNA hydroxyl methylation investigation of the postmortem AD dorso-lateral prefrontal cortex revealed several DhMRs associated with AD neuropathogenesis. These included a signification association of 517 DhMRs with neuritic plaques and significant involvement of 60DhMRs with the development of neurofibrillary tangles [65]. Further, Bernstein et al. (2016) conducted a similar study on DNA extracted from the prefrontal cortex of postmortem AD patients and demonstrated significant association of 325 genes containing DhMRs [66].
Recent literature has depicted importance of mitochondrial DNA methylation in the pathogenesis of AD and these inputs reaffirm the fact that mitochondrial DNA can be influenced by the epigenetic mechanisms and these changes are crucial for AD progression [67]. It is evident that mitochondrial dysfunction occurs during AD and this may be attributed to some extent due to the disruption of regulatory epigenetic mechanisms [68]. Bradley et al. (2013) compared the global 5-hmC levels in the mitochondrial DNA in samples of temporal gyrus taken from the healthy individuals and AD patients, and reported an increase in global 5-hmC levels in AD patients [29]. Further, investigation of the DNA samples from the living patients of LOAD revealed a significant interaction of epigenetic modification of mitochondrial DNA in the pathogenesis of AD as indicated by the significant reduction of the methylation in the D-loop of mitochondrial DNA when compared to the matched control subjects [69].
Although the association of differential methylation with the pathogenesis and progression of AD is now evident, there are some conflicting reports which open the conflicting debate regarding the involvement of DNA methylation in the pathogenesis of AD. A study conducted by Nagata et al. (2018) demonstrated no conclusive evidence of differential methylation at the NEP promoter of post-mortem AD brain samples [70]. NEP is a metallo-protease enzyme which is found to be deficit in AD patients and is involved in the degradation of β-amyloid proteins. Consequently, NEP deficiency leads to the accumulation of amyloid plaques leading to the AD pathogenesis [71]. Although, the debate persists in the association of NEP with AD, overall, the association of methylation with the pathogenesis of AD cannot be ruled out and is backed up by conclusive evidence which clearly suggests the strong epigenetics-AD association and need to extensively explore epigenetic interventions for AD.
2.2. Histone Modification and AD
Histone proteins are associated closely with the DNA and are the most abundant protein within the nucleus. Histone proteins wrap the double stranded DNA around them and provide stability to DNA along with the site for gene transcription. Histone aggregates to form an octamer and DNA is wrapped around this octamer to form the nucleosome. Histone proteins are prone to several post-translational modifications on its N-terminal, which include ubiquitination, acetylation, ADP ribosylation phosphorylation and methylation [72-76]. Histone proteins get acetylated by histone deacetylases (HDACs) and histone acetyltransferases (HATs). Likewise, methylation of the histone proteins is achieved with the help of histone demethylases (HDMs) and histone methyltransferases (HMTs). These enzymes are important for various modifications on the histone proteins, which in turn is responsible for the activation and deactivation of various genes and therefore, is an important mechanism, which regulates the gene expression [77]. Histone modification can occur on amino acids like lysine, where its acetylation generates a relaxed chromatin structure. This relaxed structure allows greater access to the activators of the transcription for particular gene expression, and likewise, deacetylation may hinder gene expression [78]. Several studies conducted on the transgenic mouse models of AD had revealed prominent evidence of the association of histone modifications with the AD. In these studies, enhanced acetylation of the histone H4 occurs at the lysine 12 (H4K12ac) residue during the early stages when amyloid protein starts to show aggregation in the brain. Further, acetylation of the histone H4 at the lysine 12 (H4K12ac) was also observed during amnestic mild cognitive impairment, suggesting it as an early stage AD biomarker [79]. Besides, histone modifications at H3K27me3 and H3K4me3 have also been demonstrated to be crucial in the pathogenesis of AD through a genome-wide methylation study conducted in the brain of AD patients [62]. These findings are strongly supported by the in-vivo findings, where, treating animals with HDAC inhibitors resulted in improvement in learning and memory functions [74, 80] and decreased the levels of Aβ in the brain [81].
2.3. Non-coding RNA and AD
Along with the histone modification and DNA methylation, short and long non-protein coding RNAs (ncRNA) have also been demonstrated as the potential regulator of the epigenetic pathogenesis of AD [82, 83]. BACE1 is responsible for the generation of Aβ peptide in the brain. BACE1 is characterized by the miRNAs and it has been reported that these RNAs are markedly deregulated in the brain and blood of AD patients [84-86]. It was also demonstrated that miR-29c improves memory and learning physiology in the neurons of the hippocampus region [86], and therefore may be considered as a therapeutic target for the management of AD. Further, miRNAs involved in the lipid metabolism (miR-33) and neuroinflammation (miR-34a and miR-155) have been identified to be involved in the epigenetic pathogenesis of AD [87-89]. Further, many studies demonstrate various ncRNAs involved in apoptosis, synaptic plasticity and Aβ production when evaluated in the brains of the AD patients, suggesting a prominent role in the pathogenesis of AD [90-96]. All these observations suggest that ncRNA could be a potential target for the early diagnosis and management of AD, however, extensive research is required.
2.4. Tau Protein and Epigenetics
The development of neurofibrillary tangles is another hallmark of the AD pathogenesis. Neurofibrillary tangles are produced due to the hyperphosphorylation of tau proteins leading to their aggregations to produce tangles which lead to the development of several taupathies such as AD, dementia and frontotemporal dementia [97, 98]. Interestingly, hyperphosphorylated tau proteins and oligomeric tau proteins are found in close association with PTK2B. These observations are made in the brain sample of AD patients, and also in the brain samples of transgenic mice suggesting a strong genetic insight to the tau pathogenesis of AD. Therefore, PTK2B can be considered as a biomarker for the early pathogenesis of tau hyperphosphorylation etiology of AD [99]. Three transcriptional activator sites, namely AP2, SP1 and GCF, are known to play a vital role in the epigenetics of tau hyperphosphorylation and have been investigated to a greater extent. Although no difference in the methylation status of the AP2 site was observed with age, the other two sites, SP1 and GCF, decrease significantly with the increasing age and thereby downregulate tau expression [100]. Further, it is now established that the methylation reactions occurring in the cytoplasm can directly affect the phosphorylation process and therefore the formation of the neurofibrillary tangles is also affected by these methylation reactions. Homocysteine is known to inhibit enzyme methyltransferase at high concentrations and it also inhibits hypo-methylation of protein phosphare 2A (PP2A), which is necessary for inhibition of the formation of neurofibrillary tangles as these pathways have the potential to dephosphorylate the phosphorylated tau protein [101, 102]. Glycogen synthase kinase 3β (GSK3β) belongs to a family of kinases and is known to phosphorylate tau in the brain. Moreover, increased expression of its regulating genes has been observed in the brain of AD subjects [103]. Various studies conducted on the cell lines, embryonic stem cells and transgenic models of AD revealed that a decrease in the methylation in the promoter region of GSK3β results in the enhanced expression of GSK3β [103, 104].
3. Epigenetic Insights to Aging
Aging is an evolutionary conserved biological phenomenon affecting all living organisms and may be affected by alterations in the epigenome. Aging is a continuous phenomenon and one of the leading causes of dementia, neurodegeneration and related disorders such as AD and Parkinsonism [105]. Although, exact mechanism how dementia, memory loss and AD are triggered with advanced age yet valuable research inputs suggest that epigenetic changes over time may progress to AD. It has been reported that the aging process is accompanied by loss of heterochromatin (H3K9me3) and aging leads to a reduction in DNA methylation [106-108]. Further, aging is known to induce epigenetic modifications in euchromatic regions which primarily include significant changes in the transcription repressive mark (H3K27me3), transcription elongation mark (H3K36me3) and the active mark (H3K4me3). Changes in these marks results in the altered expression of genes and therefore leads to the development of progressive functional loss with aging [109-111]. There are some other studies which suggest that methylation status of the brain changes drastically with age [112], which may be due to the interplay of epigenetic and environmental factors. Although it is now evident that age has a major impact on the epigenetic expression on various locations in the DNA, the exact etiology still remains unclear and needs to be explored further [113].
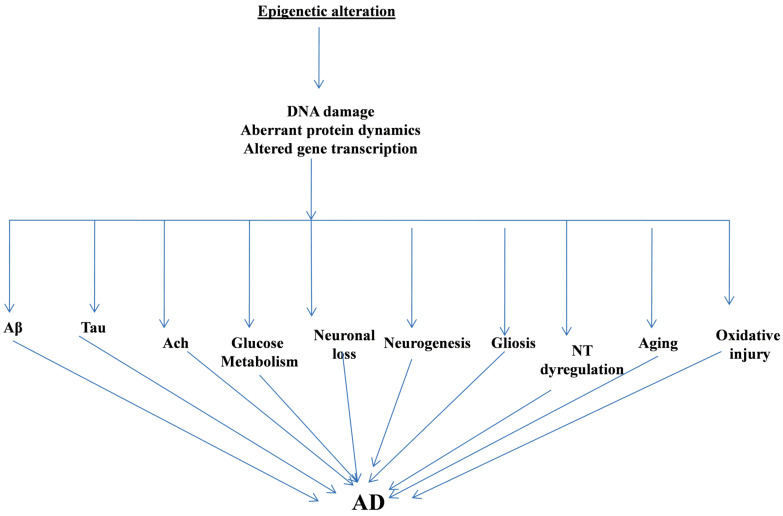
Along with the genetic risk factors, the involvement of environmental factors in the pathogenesis of AD cannot be ruled out [114, 115]. McCartney et al. (2018) performed an extensive study of DNA methylation-based aging which involved epigenetic comparison of biological epigenetic profile age versus actual chronological age. For this, they utilized more than 5000 individuals to evaluate genetic and environmental risk factors for AD and revealed a significant involvement of lifestyle risk factors in the pathogenesis of AD. They reported that lifestyle factors such as increased body mass index, hypertension, socioeconomic status, smoking habits etc. demonstrated a significant association with AD and these factors adversely influenced age altered epigenetic profile and therefore contributed to the pathogenesis and progression of AD [115]. Fig. 2 gives a graphic presentation of the interaction of epigenetical markers at various levels in AD.
Fig. (2).
The Epigenetic Alterations influence the core complexities of Alzheimer’s Disease. Aβ: Amyloid Beta; Ach: Acetylcholine; NT: Neurotransmitter; AD: Alzheimer's disease. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
4. Epigenetics and AD Therapy
As evident from the above discussion, epigenetics influence the pathogenesis and progression of AD to a significant extent. Currently, the exact pathogenesis for the development and progression of AD is not known completely and the currently available therapeutics only provide symptomatic relief from the disorder rather than curing it. Looking into the significant association between epigenetics and AD pathogenesis, modulation of epigenetic pathways through epigenetic therapy may result in a better understanding of AD pathogenesis and better management of AD. Although not much success has been achieved so far in the management of AD, epigenetic therapy has successfully and efficiently utilized for the management of cancer in past years [116]. In this context, many inhibitors of histone deacetylation and DNA methylation have been approved by the US FDA for the treatment of blood tumors [117].
4.1. DNA Methylation and AD Therapy
DNA methylation has been associated with neuronal phenomenon like learning and memory functions and AD. Many genes have been identified which are hypo-methylated during AD. As a therapy for the management of AD, some success has been achieved by supplementing the diet of AD patients with DNA methyl donors. In one such study, scientists have reported an improvement in the cognitive functions with growing age, which generally show a marked decline, when supplementing diet with folate for 3 years [118]. Likewise, increasing the levels of vitamin B12 in the plasma has been linked to lowering the risk for the development of cognitive dysfunction, dementia and homocysteine-associated memory loss [119]. S-adenosyl-L-methionine and L-methyl-folate have shown some success during idiopathic dementia when supplemented through diet. S-adenosyl-L-methionine therapy improved patients who were suffering from major depression [120] and also showed beneficial effects in improving learning, long term potentiation and neurological complications associated with exposure of lead to the population [121]. Tet1 maintains the methylation status of the brain by controlling 5hmC production. It is now considered as a promising therapeutic target for AD as its inhibition leads to cognitive enhancement and deletion of Tet1 is associated with improved memory consolidation and retrieval [122]. During AD, neurons express both hyper and hypo-methylation at different loci which may be region-specific in the brain. Due to this, drugs for epigenetic modification may exert opposite effects. Therefore, further research for the gene specific epigenetic modulators is needed which may significantly improve the AD therapeutics.
Recently, Smith et al. (2018) demonstrated for the first time that differential methylation of HOX gene is significantly involved in the pathogenesis of AD. Their study involved three independent cohorts and demonstrated extensive hyper-methylation across HOXA cluster in the samples collected from the superior temporal gyrus and prefrontal cortex [123]. Moreover, hypo methylation of APP is also an important implication in the AD pathogenesis which results in the enhanced accumulation of amyloid plaques [124]. Methylation in the numerous other genes has now been identified to be associated with the pathogenesis and progression of AD, which includes ANK1, MCF2L, S100B, LRRC8B, MAP2 and STK32C [124].
4.2. Histone Modifications and AD Therapy
HDAC inhibitors are the most widely studied for AD therapy and have shown some promising results as well. Extensively studied inhibitors of the HDAC include sodium 4-phenylbutyrate, vorinostat and valproic acid. These drugs are known to affect tau hyperphosphorylation and Aβ plaque deposition. Valproic acid has demonstrated the potential to reduce Aβ plaque deposition in both cell line studies and on transgenic animal model of AD [125]. Further, valproic acid was demonstrated to inhibit GSK-3β-mediated γ-secretase which cleaves the amyloid precursor protein, thereby improving the behavioral dysfunction in transgenic mouse model of AD [126]. Sodium 4-phenylbutyrate improved memory and learning function in a transgenic mouse model of AD by lowering the level of Aβ, clearing deposited Aβ and by decreasing tau hyperphosphorylation. The beneficial effects of sodium 4-phenylbutyrate were attributed to its potential to increase histone acetylation in the brain, increase dendritic spine density in the hippocampus, alleviating endoplasmic reticulum stress and improving synaptic plasticity in the brain [127,128]. Several studied conducted on vorinostat demonstrated its potential to increase H4K12 acetylation and restore genes induced during learning and memory formation [129]. Vorinostat, along with sodium valproate and sodium butyrate, is a class I HDAC inhibitor and all three of them restored contextual memory in the transgenic AD model by increasing the histone H4 acetylation levels [130]. Besides, there are several studies conducted on a transgenic mouse model of AD which demonstrate that the inhibition of HDAC improves cognition, restore cognitive deficits and memory function [131]. Despite several conclusive evidence of the involvement of histone modifications in the AD etiology, the clinical application of this has gained a limited success due to several issues, amongst which toxicity is the major concern. HDAC inhibitors are known to induce cell death, cell cycle arrest and apoptosis in several cancer studies and have similar effects on neurons as well [132-134]. Further, another limiting factor for the use of histone modifying agents in AD is the untoward effect on the immune system and lower blood brain barrier permeability of HDAC inhibitors [135-137].
Epigenetic regulation of PU.1 transcription factor has been reported to have a significant potential for the therapeutics of AD. PU.1 is essential for the microglia gene expression and for the development of myeloid cells and it has been demonstrated that reduction in the levels of PU.1 corresponds to the delaying the AD onset [138, 139]. Further FDA approved HDAC inhibitors such as Vorinostat, have been reported to reduce PU.1 expression in microglia of humans. Vorinostat, along with other HDAC inhibitors, which are capable of knocking down PU.1 expression, can reduce microglia mediated responses, which primarily include excessive neuronal inflammation, and therefore could prove to be a highly useful therapeutic strategy [139, 140]. Likewise, promising results have been observed for the use of HDAC inhibitors for the management of AD in mouse models, which has been extrapolated to clinical settings as well. RGFP-966 is an HDAC inhibitor and it has been demonstrated to reverse tau hyperphosphorylation, lower the β-amyloid protein levels and improve spatial learning and memory in triple transgenic AD mice. RGFP-966 increased BDNF expression reduced tau phosphorylation and reduced tau acetylation which may contribute to its beneficial effects during AD. Further, in an attempt to extrapolate these findings to the clinical settings, RGFP-966 was tested in the stem cells derived from two AD subjects and compared to two healthy individuals. Although the sample size in this study is low, it revealed some important findings where the accumulation of amyloid β and tau hyperphosphorylation demonstrated a significant reduction in primary neurons [141]. Therefore, to utilize the full therapeutic potential of the histone-modifying agents, extensive research is still needed.
4.3. Non-coding RNAs for AD Therapy
ncRNAs are widely expressed in the brain and are the key regulators of neuronal functions, neuronal developmental processes. During AD, abnormal expression of these ncRNAs occurs in the brain and other regions such as blood and CSF. Therefore, these ncRNAs could be a crucial tool for the early diagnosis of AD and for AD therapeutics. Recent studies demonstrated that enhanced expression of miR-124 and mir-195 lower the levels of Aβ via BACE1 and therefore can be a crucial therapeutic target for the management of AD [142, 143]. Another such therapy for the AD based on ncRNAs is through the utilization of anti-miRNAs, miRNA mimics and miRNA precursor analogs. miRNA mimics have been reported to down-regulate the expression of the target protein and gene, which includes BACE gene and BACE1 protein. Further, it is very difficult to target these therapeutic molecules to the desired cell, which is challenging and limits the use for AD [144, 145]. Although, ncRNAs seems to be potential therapeutic agents, their use is limited by several factors which include targeting to a specific cell, blood brain barrier etc. therefore extensive research is still required for utilizing the complete therapeutic potential of this therapeutic strategy.
5. Epigenetic therapeutics beyond the focus of AD
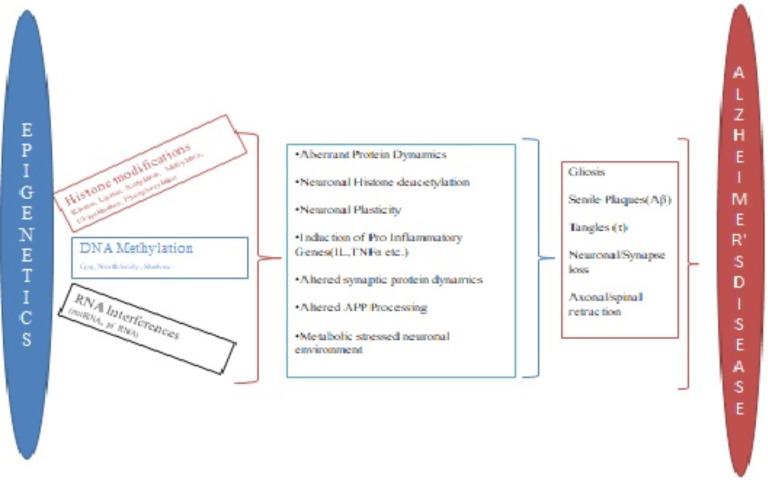
Epigenetic therapies focused on methylation has not only opened a promising new therapeutic strategy for AD, but also has emerged as a therapeutic alternative for the management of other neurodegenerative disorders as well. Evaluation of the differential methylation in the samples form AD subjects has revealed striking similarities with other neurodegenerative disorders such as vascular dementia, Huntington’s disorder, bipolar disorder, Parkinson’s disorder, Lewy-bodies dementia, etc. [146-148]. Many epigenetic similarities were observed in the post-mortem frontal cortex region of the brains of patients suffering from AD and bipolar disorders when evaluated for the genes associated with CpG methylation, DNA methylation and histone modifications. Both these disorders demonstrate increased global DNA methylation, increased histone H3 phosphorylation and increased methylation at BDNF promoter, along with reduced methylation of COX-2 promoter gene. Further, CpG methylation of synaptic markers is also associated with both conditions with the slight difference such as methylation of the synaptophysin promoter occurs only in AD and methylation of drebin promoter occurs only in the case of bipolar disorder. In both these disorders, the epigenetic factors have been associated with the increase in the transcription and translation of the neuro-inflammatory markers such as microglial activation markers, TNF-α, astrocytic and IL-1β [52], along with the increased levels of other markers of neuro-inflammation (IL1β, CD11b, GFAP, and COX2) and reduced levels of neurotrophic BDNF, presynaptic and postsynaptic synaptophysin and drebin [148]. Further, ANK1 hypermethylation is observed during AD, Parkinson’s disorder and Huntington’s disorder. Vascular Dementia or Lewy bodies Dementia also show enhanced ANK1 hypermethylation, however, this is only observed when there is coexisting AD pathogenesis [147]. These observations demonstrate that the epigenetics could emerge as a promising therapeutic strategy for the management of neurodegenerative conditions, beyond the focus of AD. Fig. 3 gives a simple schematic presentation of the influence of major epigenetic alterations on the core pathological steps in the progression of Alzheimer’s disease
Fig. (3).
The schematic presentation of altered epigenetical pathways on alzheimer’s disease. (A higher resolution / colour version of this figure is available in the electronic copy of the article).
CONCLUSION AND FUTURE PERSPECTIVE
In this review, we have focused on providing a concise overview of the progress made so far in the epigenetic implications during the pathogenesis and therapeutics of AD. Research work on identifying epigenetic insights into the pathogenesis and progression of AD is still in its initial phase, but still, has revealed some commendable advancement which is capable of changing the therapeutics of AD tremendously. Epigenetic modifications have been extensively documented in the peripheral blood samples and post-mortem brain samples from AD patients which suggest the significance of epigenetic modifications in the development and progression of AD pathogenesis. From this review, the existence of a very strong and direct association between AD and epigenetic dysregulation can be concluded, especially that of DNA methylation and histone modifications which include acetylation and deacetylation. This review also concludes that targeting epigenetic markers can be a crucial therapeutic strategy for the management and early diagnosis of neurodegenerative disorders like AD. However, certain conflicting findings which debate the association of epigenetic modifications and AD cannot be ruled out, though very few such findings are available, which may be due to lesser sample size, different experimental techniques employed or due variations in samples used for testing [48].
One of the prime reasons for some conflicting reports may be the fact that different cells or neuron types in different regions of the brain demonstrate different methylation levels. Such a difference has been reported in the same regions of the brain as well [148]. Conclusive evidence for epigenetic association with AD has been developed from the peripheral blood samples and post-mortem brain samples of the AD patients. This has provided us with the potential peripheral biomarkers for AD progression and can have marked therapeutic potential in early diagnosis and treatment of AD [47]. Although, some correlation between peripheral epigenetic modifications and central epigenetic modification has been established from the samples collected from AD patients, still, it is unclear to what extent the peripheral changes can be correlated correctly to the changes in CNS and to the AD progression. However, these findings are still in their initial stages and need extensive research before confirming them as a potential biomarker to check the progression of AD. A recent study suggests that just 8% of the peripheral epigenetic changes can be correlated to the epigenetic changes observed in the brain, suggesting that only a fraction of peripheral epigenetic changes can be correlated to the CNS [149]. Although this study was conducted on patients with epilepsy rather than AD, the same trend can be expected for AD which may critically limit the use of peripheral markers for the therapeutics of AD.
The therapeutic strategy for the management of AD is not fully effective to date. No doubt currently available therapeutic can slow down the progression of AD, but they only provide symptomatic relief from AD and it continues to progress. Epigenetic modulators, such as HDAC inhibitors, have demonstrated therapeutic potential and have been approved for the management of AD. However, these are also in their initial stages and extensive research needs to be done to derive conclusive evidence for the benefit of these strategies for AD management. Despite all these possible debates, extensive research targeting epigenetics has revealed very interesting findings and their modulations with the help of some herbal or synthetic intervention can provide us with the solution to prevent the progression of AD, besides reversing the damage already inflicted by AD progression. Taking into consideration that epigenetic changes are reversible, whether they are due to environmental factors, age or due to other risk factors, targeting these changes may be a promising strategy for the management of AD and other neurodegenerative conditions [80, 150].
Overall, it is clear from the literature and extensive research so far that epigenetic modulations can be a crucial therapeutic strategy for the management of AD yet the in-depth analysis of mechanisms involved is highly desired. This may be achieved by taking a larger sample size, with necessary age, environmental and disease controls. It will also be interesting to identify some peripheral biomarkers which can show a significant correlation with the CNS aspects of AD and therefore can be a vital tool for the prevention and early diagnosis of AD.
Acknowledgements
The authors are grateful to the Chitkara College of Pharmacy, Chitkara University, Rajpura, Patiala, Punjab, India for providing the necessary facilities to carry out the research work.
Consent for Publication
Not applicable.
Funding
None.
Conflict of Interest
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Lane C.A.H.J., Hardy J., Schott J.M. Alzheimer’s disease. Eur. J. Neurol. 2018;25(1):59–70. doi: 10.1111/ene.13439. [DOI] [PubMed] [Google Scholar]
- 2.Vlassenko A.G., Benzinger T.L., Morris J.C. PET amyloid-beta imaging in preclinical Alzheimer’s disease. Biochim. Biophys. Acta. 2012;1822(3):370–379. doi: 10.1016/j.bbadis.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.2019.
- 4.Burns A., Iliffe S. Alzheimer’s disease. BMJ. 2009;338:b158. doi: 10.1136/bmj.b158. [DOI] [PubMed] [Google Scholar]
- 5.Querfurth H.W., LaFerla F.M. Alzheimer’s disease. N. Engl. J. Med. 2010;362(4):329–344. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 6.Todd S., Barr S., Roberts M., Passmore A.P. Survival in dementia and predictors of mortality: a review. Int. J. Geriatr. Psychiatry. 2013;28(11):1109–1124. doi: 10.1002/gps.3946. [DOI] [PubMed] [Google Scholar]
- 7.Ballard C., Gauthier S., Corbett A., Brayne C., Aarsland D., Jones E. Alzheimer’s disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- 8.Pimenova A.A., Raj T., Goate A.M. Untangling genetic risk for Alzheimer’s Disease. Biol. Psychiatry. 2018;83(4):300–310. doi: 10.1016/j.biopsych.2017.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanzi R.E. The genetics of Alzheimer disease. Cold Spring Harb. Perspect. Med. 2012;2(10):2. doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Killin L.O., Starr J.M., Shiue I.J., Russ T.C. Environmental risk factors for dementia: a systematic review. BMC Geriatr. 2016;16(1):175. doi: 10.1186/s12877-016-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Migliore L., Coppedè F. Genetics, environmental factors and the emerging role of epigenetics in neurodegenerative diseases. Mutat. Res. 2009;667(1-2):82–97. doi: 10.1016/j.mrfmmm.2008.10.011. [DOI] [PubMed] [Google Scholar]
- 12.Van Bulck M., Sierra-Magro A., Alarcon-Gil J., Perez-Castillo A., Morales-Garcia J.A. Novel Approaches for the treatment of Alzheimer’s and Parkinson’s Disease. Int. J. Mol. Sci. 2019;20(3):719. doi: 10.3390/ijms20030719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindsley C.W. Alzheimer’s disease: development of disease-modifying treatments is the challenge for our generation. ACS Chem. Neurosci. 2012;3(11):804–805. doi: 10.1021/cn300190f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitra S., Behbahani H., Eriksdotter M. Innovative Therapy for Alzheimer’s Disease-With Focus on Biodelivery of NGF. Front. Neurosci. 2019;13:38. doi: 10.3389/fnins.2019.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feil R., Fraga M.F. Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 2012;13(2):97–109. doi: 10.1038/nrg3142. [DOI] [PubMed] [Google Scholar]
- 16.Mirbahai L., Chipman J.K. Epigenetic memory of environmental organisms: a reflection of lifetime stressor exposures. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014;764-765:10–17. doi: 10.1016/j.mrgentox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Fuke C., Shimabukuro M., Petronis A., Sugimoto J., Oda T., Miura K., Miyazaki T., Ogura C., Okazaki Y., Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann. Hum. Genet. 2004;68(Pt 3):196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- 18.Mastroeni D., Grover A., Delvaux E., Whiteside C., Coleman P.D., Rogers J. Epigenetic changes in Alzheimer’s disease: decrements in DNA methylation. Neurobiol. Aging. 2010;31(12):2025–2037. doi: 10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vanyushin B.F., Nemirovsky L.E., Klimenko V.V., Vasiliev V.K., Belozersky A.N. The 5-methylcytosine in DNA of rats. Tissue and age specificity and the changes induced by hydrocortisone and other agents. Gerontologia. 1973;19(3):138–152. doi: 10.1159/000211967. [DOI] [PubMed] [Google Scholar]
- 20.Wilson V.L., Smith R.A., Ma S., Cutler R.G. Genomic 5-methyldeoxycytidine decreases with age. J. Biol. Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]
- 21.Moore L.D., Le T., Fan G. methylation and its basic function. Neuropsychopharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christopher M.A., Kyle S.M., Katz D.J. Neuroepigenetic mechanisms in disease. Epigenetics Chromatin. 2017;10(1):47. doi: 10.1186/s13072-017-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zampieri M., Ciccarone F., Calabrese R., Franceschi C., Burkle A., Caiafa P. Reconfiguration of DNA methylation in aging. Mech. Ageing Dev. 2015;151:60–70. doi: 10.1016/j.mad.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 24.Cheng Y., Bernstein A., Chen D., Jin P. 5-Hydroxymethylcytosine: A new player in brain disorders? Exp. Neurol. 2015;268:3–9. doi: 10.1016/j.expneurol.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.West R.L., Lee J.M., Maroun L.E. Hypomethylation of the amyloid precursor protein gene in the brain of an Alzheimer’s disease patient. J. Mol. Neurosci. 1995;6(2):141–146. doi: 10.1007/BF02736773. [DOI] [PubMed] [Google Scholar]
- 26.Eid A., Bihaqi S.W., Renehan W.E., Zawia N.H. Developmental lead exposure and lifespan alterations in epigenetic regulators and their correspondence to biomarkers of Alzheimer’s disease. Alzheimers Dement. (Amst.) 2016;2:123–131. doi: 10.1016/j.dadm.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuso A., Seminara L., Cavallaro R.A., D’Anselmi F., Scarpa S. S-adenosylmethionine/homocysteine cycle alterations modify DNA methylation status with consequent deregulation of PS1 and BACE and beta-amyloid production. Mol. Cell. Neurosci. 2005;28(1):195–204. doi: 10.1016/j.mcn.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Wu J., Basha M.R., Brock B., Cox D.P., Cardozo-Pelaez F., McPherson C.A., Harry J., Rice D.C., Maloney B., Chen D., Lahiri D.K., Zawia N.H. Alzheimer’s disease (AD)-like pathology in aged monkeys after infantile exposure to environmental metal lead (Pb): evidence for a developmental origin and environmental link for AD. J. Neurosci. 2008;28(1):3–9. doi: 10.1523/JNEUROSCI.4405-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bradley-Whitman M.A., Lovell M.A. Epigenetic changes in the progression of Alzheimer’s disease. Mech. Ageing Dev. 2013;134(10):486–495. doi: 10.1016/j.mad.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chouliaras L., Mastroeni D., Delvaux E., Grover A., Kenis G., Hof P.R., Steinbusch H.W., Coleman P.D., Rutten B.P., van den Hove D.L. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging. 2013;34(9):2091–2099. doi: 10.1016/j.neurobiolaging.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Condliffe D., Wong A., Troakes C., Proitsi P., Patel Y., Chouliaras L., Fernandes C., Cooper J., Lovestone S., Schalkwyk L., Mill J., Lunnon K. Cross-region reduction in 5-hydroxymethylcytosine in Alzheimer’s disease brain. Neurobiol. Aging. 2014;35(8):1850–1854. doi: 10.1016/j.neurobiolaging.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coppieters N., Dieriks B.V., Lill C., Faull R.L., Curtis M.A., Dragunow M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol. Aging. 2014;35(6):1334–1344. doi: 10.1016/j.neurobiolaging.2013.11.031. [DOI] [PubMed] [Google Scholar]
- 33.Di Francesco A., Arosio B., Falconi A., Micioni Di Bonaventura M.V., Karimi M., Mari D., Casati M., Maccarrone M., D’Addario C. Global changes in DNA methylation in Alzheimer’s disease peripheral blood mononuclear cells. Brain Behav. Immun. 2015;45:139–144. doi: 10.1016/j.bbi.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Hernández H.G., Mahecha M.F., Mejía A., Arboleda H., Forero D.A. Global long interspersed nuclear element 1 DNA methylation in a Colombian sample of patients with late-onset Alzheimer’s disease. Am. J. Alzheimers Dis. Other Demen. 2014;29(1):50–53. doi: 10.1177/1533317513505132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lashley T., Gami P., Valizadeh N., Li A., Revesz T., Balazs R. Alterations in global DNA methylation and hydroxymethylation are not detected in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2015;41(4):497–506. doi: 10.1111/nan.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastroeni D., McKee A., Grover A., Rogers J., Coleman P.D. Epigenetic differences in cortical neurons from a pair of monozygotic twins discordant for Alzheimer’s disease. PLoS One. 2009;4(8):e6617. doi: 10.1371/journal.pone.0006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ellison E.M., Abner E.L., Lovell M.A. Multiregional analysis of global 5-methylcytosine and 5-hydroxymethylcytosine throughout the progression of Alzheimer’s disease. J. Neurochem. 2017;140(3):383–394. doi: 10.1111/jnc.13912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrachina M., Ferrer I. DNA methylation of Alzheimer disease and tauopathy-related genes in postmortem brain. J. Neuropathol. Exp. Neurol. 2009;68(8):880–891. doi: 10.1097/NEN.0b013e3181af2e46. [DOI] [PubMed] [Google Scholar]
- 39.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 40.Iwata A., Nagata K., Hatsuta H., Takuma H., Bundo M., Iwamoto K., Tamaoka A., Murayama S., Saido T., Tsuji S. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum. Mol. Genet. 2014;23(3):648–656. doi: 10.1093/hmg/ddt451. [DOI] [PubMed] [Google Scholar]
- 41.Wang S.C., Oelze B., Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3(7):e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Piaceri I., Raspanti B., Tedde A., Bagnoli S., Sorbi S., Nacmias B. Epigenetic modifications in Alzheimer’s disease: cause or effect? J. Alzheimers Dis. 2015;43(4):1169–1173. doi: 10.3233/JAD-141452. [DOI] [PubMed] [Google Scholar]
- 43.Tannorella P., Stoccoro A., Tognoni G., Petrozzi L., Salluzzo M.G., Ragalmuto A., Siciliano G., Haslberger A., Bosco P., Bonuccelli U., Migliore L., Coppedè F. Methylation analysis of multiple genes in blood DNA of Alzheimer’s disease and healthy individuals. Neurosci. Lett. 2015;600:143–147. doi: 10.1016/j.neulet.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 44.Humphries C., Kohli M.A., Whitehead P., Mash D.C., Pericak-Vance M.A., Gilbert J. Alzheimer disease (AD) specific transcription, DNA methylation and splicing in twenty AD associated loci. Mol. Cell. Neurosci. 2015;67(67):37–45. doi: 10.1016/j.mcn.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sanchez-Mut J.V., Aso E., Panayotis N., Lott I., Dierssen M., Rabano A., Urdinguio R.G., Fernandez A.F., Astudillo A., Martin-Subero J.I., Balint B., Fraga M.F., Gomez A., Gurnot C., Roux J.C., Avila J., Hensch I., Ferrer T.K., Esteller M. DNA methylation map of mouse and human brain identifies target genes in Alzheimer’s disease. Brain. 2013;136:3018–3027. doi: 10.1093/brain/awt237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu L., Chibnik L.B., Srivastava G.P., Pochet N., Yang J., Xu J., Kozubek J., Obholzer N., Leurgans S.E., Schneider J.A., Meissner A., De Jager P.L., Bennett D.A. Association of Brain DNA methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 with pathological diagnosis of Alzheimer disease. JAMA Neurol. 2015;72(1):15–24. doi: 10.1001/jamaneurol.2014.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fransquet P.D., Lacaze P., Saffery R., McNeil J., Woods R., Ryan J. Blood DNA methylation as a potential biomarker of dementia: A systematic review. Alzheimers Dement. 2018;14(1):81–103. doi: 10.1016/j.jalz.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wen K.X., Miliç J., El-Khodor B., Dhana K., Nano J., Pulido T., Kraja B., Zaciragic A., Bramer W.M., Troup J., Chowdhury R., Ikram M.A., Dehghan A., Muka T., Franco O.H. The role of DNA methylation and histone modifications in neurodegenerative diseases: A systematic review. PLoS One. 2016;11(12):e0167201. doi: 10.1371/journal.pone.0167201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Celarain N., Sánchez-Ruiz de Gordoa J., Zelaya M.V., Roldán M., Larumbe R., Pulido L., Echavarri C., Mendioroz M. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer’s disease hippocampus. Clin. Epigenetics. 2016;8:37. doi: 10.1186/s13148-016-0202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nagata T., Kobayashi N., Ishii J., Shinagawa S., Nakayama R., Shibata N., Kuerban B., Ohnuma T., Kondo K., Arai H., Yamada H., Nakayama K. Association between DNA methylation of the BDNF promoter region and clinical presentation in Alzheimer’s Disease. Dement. Geriatr. Cogn. Disord. Extra. 2015;5(1):64–73. doi: 10.1159/000375367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozaki Y., Yoshino Y., Yamazaki K., Sao T., Mori Y., Ochi S., Yoshida T., Mori T., Iga J.I., Ueno S.I. DNA methylation changes at TREM2 intron 1 and TREM2 mRNA expression in patients with Alzheimer’s disease. J. Psychiatr. Res. 2017;92:74–80. doi: 10.1016/j.jpsychires.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 52.Rao J.S., Keleshian V.L., Klein S., Rapoport S.I. Epigenetic modifications in frontal cortex from Alzheimer’s disease and bipolar disorder patients. Transl. Psychiatry. 2012;2:e132. doi: 10.1038/tp.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith.; A.R.; Smith, R.G.; Condliffe, D.; Hannon, E.; Schalkwyk, L.; Mill, J.; Lunnon, K. Increased DNA methylation near TREM2 is consistently seen in the superior temporal gyrus in Alzheimer’s disease brain. Neurobiol. Aging. 2016;47:35–40. doi: 10.1016/j.neurobiolaging.2016.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xie B., Liu Z., Liu W., Jiang L., Zhang R., Cui D., Zhang Q., Xu S. DNA Methylation and Tag SNPs of the BDNF gene in conversion of amnestic mild cognitive impairment into Alzheimer’s disease: A Cross-Sectional Cohort Study. J. Alzheimers Dis. 2017;58(1):263–274. doi: 10.3233/JAD-170007. a. [DOI] [PubMed] [Google Scholar]
- 55.Xie B., Xu Y., Liu Z., Liu W., Jiang L., Zhang R., Cui D., Zhang Q., Xu S. Elevation of peripheral BDNF promoter methylation predicts conversion from amnestic mild cognitive impairment to Alzheimer’s Disease: A 5-Year longitudinal study. J. Alzheimers Dis. 2017;56(1):391–401. doi: 10.3233/JAD-160954. b. [DOI] [PubMed] [Google Scholar]
- 56.Irier H.A., Jin P. Dynamics of DNA methylation in aging and Alzheimer’s disease. DNA Cell Biol. 2012;31(1) Suppl. 1:S42–S48. doi: 10.1089/dna.2011.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song C.X., Szulwach K.E., Fu Y., Dai Q., Yi C., Li X., Li Y., Chen C.H., Zhang W., Jian X., Wang J., Zhang L., Looney T.J., Zhang B., Godley L.A., Hicks L.M., Lahn B.T., Jin P., He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat. Biotechnol. 2011;29(1):68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bakulski K.M., Dolinoy D.C., Sartor M.A., Paulson H.L., Konen J.R., Lieberman A.P., Albin R.L., Hu H., Rozek L.S. Genome-wide DNA methylation differences between late-onset Alzheimer’s disease and cognitively normal controls in human frontal cortex. J. Alzheimers Dis. 2012;29(3):571–588. doi: 10.3233/JAD-2012-111223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lunnon K., Smith R., Hannon E., De Jager P.L., Srivastava G., Volta M., Troakes C., Al-Sarraj S., Burrage J., Macdonald R., Condliffe D., Harries L.W., Katsel P., Haroutunian V., Kaminsky Z., Joachim C., Powell J., Lovestone S., Bennett D.A., Schalkwyk L.C., Mill J. Methylomic profiling implicates cortical deregulation of ANK1 in Alzheimer’s disease. Nat. Neurosci. 2014;17(9):1164–1170. doi: 10.1038/nn.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Jager P.L., Srivastava G., Lunnon K., Burgess J., Schalkwyk L.C., Yu L., Eaton M.L., Keenan B.T., Ernst J., McCabe C., Tang A., Raj T., Replogle J., Brodeur W., Gabriel S., Chai H.S., Younkin C., Younkin S.G., Zou F., Szyf M., Epstein C.B., Schneider J.A., Bernstein B.E., Meissner A., Ertekin-Taner N., Chibnik L.B., Kellis M., Mill J., Bennett D.A. Alzheimer’s disease: early alterations in brain DNA methylation at ANK1, BIN1, RHBDF2 and other loci. Nat. Neurosci. 2014;17(9):1156–1163. doi: 10.1038/nn.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Roubroeks J.A.Y., Smith R.G., van den Hove D.L.A., Lunnon K. Epigenetics and DNA methylomic profiling in Alzheimer’s disease and other neurodegenerative diseases. J. Neurochem. 2017;143(2):158–170. doi: 10.1111/jnc.14148. [DOI] [PubMed] [Google Scholar]
- 62.Watson C.T., Roussos P., Garg P., Ho D.J., Azam N., Katsel P.L., Haroutunian V., Sharp A.J. Genome-wide DNA methylation profiling in the superior temporal gyrus reveals epigenetic signatures associated with Alzheimer’s disease. Genome Med. 2016;8(1):5. doi: 10.1186/s13073-015-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Konki M., Malonzo M., Karlsson I., Lindgren N., Ghimire B., Smolander J., Scheinin N., Ollikainen M., Laiho A., Elo L.L. Peripheral blood DNA methylation differences in twin pairs discordant for Alzheimer’s disease. Clin. Epigenetics. 2019;11(1):130. doi: 10.1186/s13148-019-0729-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mladenova D., Barry G., Konen L.M., Pineda S.S., Guennewig B., Avesson L., Zinn R., Schonrock N., Bitar M., Jonkhout N., Crumlish L., Kaczorowski D.C., Gong A., Pinese M., Franco G.R., Walkley C.R., Vissel B., Mattick J.S. Adar3 is involved in learning and memory in mice. Front. Neurosci. 2018;12:243. doi: 10.3389/fnins.2018.00243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao J., Zhu Y., Yang J., Li L., Wu H., De Jager P.L., Jin P., Bennett D.A. A genome-wide profiling of brain DNA hydroxymethylation in Alzheimer’s disease. Alzheimers Dement. 2017;13(6):674–688. doi: 10.1016/j.jalz.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bernstein A.I., Lin Y., Street R.C., Lin L., Dai Q., Yu L., Bao H., Gearing M., Lah J.J., Nelson P.T., He C., Levey A.I., Mullé J.G., Duan R., Jin P. 5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer’s disease modulate Tau-induced neurotoxicity. Hum. Mol. Genet. 2016;25(12):2437–2450. doi: 10.1093/hmg/ddw109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mposhi A., Van der Wijst M.G., Faber K.N., Rots M.G. Regulation of mitochondrial gene expression, the epigenetic enigma. Front. Biosci. 2017;22:1099–1113. doi: 10.2741/4535. [DOI] [PubMed] [Google Scholar]
- 68.Hroudová J., Singh N., Fišar Z. Mitochondrial dysfunctions in neurodegenerative diseases: relevance to Alzheimer’s disease. BioMed Res. Int. 2014;2014:175062. doi: 10.1155/2014/175062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stoccoro A., Siciliano G., Migliore L., Coppedè F. Decreased methylation of the mitochondrial d-loop region in late-onset Alzheimer’s Disease. J. Alzheimers Dis. 2017;59(2):559–564. doi: 10.3233/JAD-170139. [DOI] [PubMed] [Google Scholar]
- 70.Nagata K., Mano T., Murayama S., Saido T.C., Iwata A. DNA methylation level of the neprilysin promoter in Alzheimer’s disease brains. Neurosci. Lett. 2018;670:8–13. doi: 10.1016/j.neulet.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 71.Turner A.J., Fisk L., Nalivaeva N.N. Targeting amyloid-degrading enzymes as therapeutic strategies in neurodegeneration. Ann. N. Y. Acad. Sci. 2004;1035:1–20. doi: 10.1196/annals.1332.001. [DOI] [PubMed] [Google Scholar]
- 72.Anderson K.W., Turko I.V. Histone post-translational modifications in frontal cortex from human donors with Alzheimer’s disease. Clin. Proteomics. 2015;12:26. doi: 10.1186/s12014-015-9098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer A., Sananbenesi F., Mungenast A., Tsai L.H. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol. Sci. 2010;31(12):605–617. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 74.Gräff J., Rei D., Guan J.S., Wang W.Y., Seo J., Hennig K.M., Nieland T.J., Fass D.M., Kao P.F., Kahn M., Su S.C., Samiei A., Joseph N., Haggarty S.J., Delalle I., Tsai L.H. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483(7388):222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narayan P.J., Lill C., Faull R., Curtis M.A., Dragunow M. Increased acetyl and total histone levels in post-mortem Alzheimer’s disease brain. Neurobiol. Dis. 2015;74:281–294. doi: 10.1016/j.nbd.2014.11.023. [DOI] [PubMed] [Google Scholar]
- 76.Zhang K., Schrag M., Crofton A., Trivedi R., Vinters H., Kirsch W. Targeted proteomics for quantification of histone acetylation in Alzheimer’s disease. Proteomics. 2012;12(8):1261–1268. doi: 10.1002/pmic.201200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kouskouti A., Talianidis I. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 2005;24(2):347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bannister A.J., Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21(3):381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plagg B., Ehrlich D., Kniewallner K.M., Marksteiner J., Humpel C. Increased acetylation of histone H4 at Lysine 12 (H4K12) in monocytes of transgenic alzheimer’s mice and in human patients. Curr. Alzheimer Res. 2015;12(8):752–760. doi: 10.2174/1567205012666150710114256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coppedè F. The potential of epigenetic therapies in neurodegenerative diseases. Front. Genet. 2014;5:220. doi: 10.3389/fgene.2014.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sung Y.M., Lee T., Yoon H., DiBattista A.M., Song J.M., Sohn Y., Moffat E.I., Turner R.S., Jung M., Kim J., Hoe H.S. Mercaptoacetamide-based class II HDAC inhibitor lowers Aβ levels and improves learning and memory in a mouse model of Alzheimer’s disease. Exp. Neurol. 2013;239:192–201. doi: 10.1016/j.expneurol.2012.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maoz R., Garfinkel B.P., Soreq H. Alzheimer’s Disease and ncRNAs. Adv. Exp. Med. Biol. 2017;978:337–361. doi: 10.1007/978-3-319-53889-1_18. [DOI] [PubMed] [Google Scholar]
- 83.Peschansky V.J., Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. doi: 10.4161/epi.27473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hébert S.S., Horré K., Nicolaï L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA. 2008;105(17):6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lei X., Lei L., Zhang Z., Zhang Z., Cheng Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2015;8(2):1565–1574. [PMC free article] [PubMed] [Google Scholar]
- 86.Yang G., Song Y., Zhou X., Deng Y., Liu T., Weng G., Yu D., Pan S. MicroRNA-29c targets β-site amyloid precursor protein-cleaving enzyme 1 and has a neuroprotective role in vitro and in vivo. Mol. Med. Rep. 2015;12(2):3081–3088. doi: 10.3892/mmr.2015.3728. [DOI] [PubMed] [Google Scholar]
- 87.Guedes J.R., Custódia C.M., Silva R.J., de Almeida L.P., Pedroso de Lima M.C., Cardoso A.L. Early miR-155 upregulation contributes to neuroinflammation in Alzheimer’s disease triple transgenic mouse model. Hum. Mol. Genet. 2014;23(23):6286–6301. doi: 10.1093/hmg/ddu348. [DOI] [PubMed] [Google Scholar]
- 88.Kim J., Yoon H., Horie T., Burchett J.M., Restivo J.L., Rotllan N., Ramírez C.M., Verghese P.B., Ihara M., Hoe H.S., Esau C., Fernández-Hernando C., Holtzman D.M., Cirrito J.R., Ono K., Kim J. MicroRNA-33 Regulates ApoE Lipidation and Amyloid-β metabolism in the brain. J. Neurosci. 2015;35(44):14717–14726. doi: 10.1523/JNEUROSCI.2053-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schipper H.M., Maes O.C., Chertkow H.M., Wang E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Bio. 2007;1:263–274. doi: 10.4137/GRSB.S361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ciarlo E., Massone S., Penna I., Nizzari M., Gigoni A., Dieci G., Russo C., Florio T., Cancedda R., Pagano A. An intronic ncRNA-dependent regulation of SORL1 expression affecting Aβ formation is upregulated in post-mortem Alzheimer’s disease brain samples. Dis. Model. Mech. 2013;6(2):424–433. doi: 10.1242/dmm.009761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G., Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14(7):723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Magistri M., Velmeshev D., Makhmutova M., Faghihi M.A. Transcriptomics Profiling of Alzheimer’s Disease reveal neurovascular defects, altered amyloid-β homeostasis, and deregulated expression of long noncoding RNAs. J. Alzheimers Dis. 2015;48(3):647–665. doi: 10.3233/JAD-150398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Massone S., Vassallo I., Fiorino G., Castelnuovo M., Barbieri F., Borghi R., Tabaton M., Robello M., Gatta E., Russo C., Florio T., Dieci G., Cancedda R., Pagano A. 17A, a novel non-coding RNA, regulates GABA B alternative splicing and signaling in response to inflammatory stimuli and in Alzheimer disease. Neurobiol. Dis. 2011;41(2):308–317. doi: 10.1016/j.nbd.2010.09.019. [DOI] [PubMed] [Google Scholar]
- 94.Mus E., Hof P.R., Tiedge H. Dendritic BC200 RNA in aging and in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 2007;104(25):10679–10684. doi: 10.1073/pnas.0701532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scheckel C., Drapeau E., Frias M.A., Park C.Y., Fak J., Zucker-Scharff I., Kou Y., Haroutunian V., Ma’ayan A., Buxbaum J.D., Darnell R.B. Regulatory consequences of neuronal ELAV-like protein binding to coding and non-coding RNAs in human brain. eLife. 2016;5:5. doi: 10.7554/eLife.10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou X., Xu J. Identification of Alzheimer’s disease-associated long noncoding RNAs. Neurobiol. Aging. 2015;36(11):2925–2931. doi: 10.1016/j.neurobiolaging.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 97.Alonso A., Zaidi T., Novak M., Grundke-Iqbal I., Iqbal K. Hyperphosphorylation induces self-assembly of tau into tangles of paired helical filaments/straight filaments. Proc. Natl. Acad. Sci. USA. 2001;98(12):6923–6928. doi: 10.1073/pnas.121119298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lovestone S., Reynolds C.H. The phosphorylation of tau: a critical stage in neurodevelopment and neurodegenerative processes. Neuroscience. 1997;78(2):309–324. doi: 10.1016/s0306-4522(96)00577-5. [DOI] [PubMed] [Google Scholar]
- 99.Dourlen P., Fernandez-Gomez F.J., Dupont C., Grenier-Boley B., Bellenguez C., Obriot H., Caillierez R., Sottejeau Y., Chapuis J., Bretteville A., Abdelfettah F., Delay C., Malmanche N., Soininen H., Hiltunen M., Galas M.C., Amouyel P., Sergeant N., Buée L., Lambert J.C., Dermaut B. Functional screening of Alzheimer risk loci identifies PTK2B as an in vivo modulator and early marker of Tau pathology. Mol. Psychiatry. 2017;22(6):874–883. doi: 10.1038/mp.2016.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tohgi H., Utsugisawa K., Nagane Y., Yoshimura M., Ukitsu M., Genda Y. The methylation status of cytosines in a tau gene promoter region alters with age to downregulate transcriptional activity in human cerebral cortex. Neurosci. Lett. 1999;275(2):89–92. doi: 10.1016/S0304-3940(99)00731-4. [DOI] [PubMed] [Google Scholar]
- 101.Sontag E., Nunbhakdi-Craig V., Sontag J.M., Diaz-Arrastia R., Ogris E., Dayal S., Lentz S.R., Arning E., Bottiglieri T. Protein phosphatase 2A methyltransferase links homocysteine metabolism with tau and amyloid precursor protein regulation. J. Neurosci. 2007;27(11):2751–2759. doi: 10.1523/JNEUROSCI.3316-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang C.E., Tian Q., Wei W., Peng J.H., Liu G.P., Zhou X.W., Wang Q., Wang D.W., Wang J.Z. Homocysteine induces tau phosphorylation by inactivating protein phosphatase 2A in rat hippocampus. Neurobiol. Aging. 2008;29(11):1654–1665. doi: 10.1016/j.neurobiolaging.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 103.Nicolia V., Fuso A., Cavallaro R.A., Di Luzio A., Scarpa S. B vitamin deficiency promotes tau phosphorylation through regulation of GSK3beta and PP2A. J. Alzheimers Dis. 2010;19(3):895–907. doi: 10.3233/JAD-2010-1284. [DOI] [PubMed] [Google Scholar]
- 104.Popkie A.P., Zeidner L.C., Albrecht A.M., D’Ippolito A., Eckardt S., Newsom D.E., Groden J., Doble B.W., Aronow B., McLaughlin K.J., White P., Phiel C.J. Phosphatidylinositol 3-kinase (PI3K) signaling via glycogen synthase kinase-3 (Gsk-3) regulates DNA methylation of imprinted loci. J. Biol. Chem. 2010;285(53):41337–41347. doi: 10.1074/jbc.M110.170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xie A., Gao J., Xu L., Meng D. 2014.
- 106.Cruickshanks H.A., McBryan T., Nelson D.M., Vanderkraats N.D., Shah P.P., van Tuyn J., Singh Rai T., Brock C., Donahue G., Dunican D.S., Drotar M.E., Meehan R.R., Edwards J.R., Berger S.L., Adams P.D. Senescent cells harbour features of the cancer epigenome. Nat. Cell Biol. 2013;15(12):1495–1506. doi: 10.1038/ncb2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Salta E., Sierksma A., Vanden Eynden E., De Strooper B. miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 2016;8(9):1005–1018. doi: 10.15252/emmm.201606520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhang W., Li J., Suzuki K., Qu J., Wang P., Zhou J., Liu X., Ren R., Xu X., Ocampo A., Yuan T., Yang J., Li Y., Shi L., Guan D., Pan H., Duan S., Ding Z., Li M., Yi F., Bai R., Wang Y., Chen C., Yang F., Li X., Wang Z., Aizawa E., Goebl A., Soligalla R.D., Reddy P., Esteban C.R., Tang F., Liu G.H., Belmonte J.C. Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science. 2015;348(6239):1160–1163. doi: 10.1126/science.aaa1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cheung I., Shulha H.P., Jiang Y., Matevossian A., Wang J., Weng Z., Akbarian S. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc. Natl. Acad. Sci. USA. 2010;107(19):8824–8829. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu L., Cheung T.H., Charville G.W., Hurgo B.M., Leavitt T., Shih J., Brunet A., Rando T.A. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4(1):189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pu M., Ni Z., Wang M., Wang X., Wood J.G., Helfand S.L., Yu H., Lee S.S. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 2015;29(7):718–731. doi: 10.1101/gad.254144.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Numata S., Ye T., Hyde T.M., Guitart-Navarro X., Tao R., Wininger M., Colantuoni C., Weinberger D.R., Kleinman J.E., Lipska B.K. DNA methylation signatures in development and aging of the human prefrontal cortex. Am. J. Hum. Genet. 2012;90(2):260–272. doi: 10.1016/j.ajhg.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Klein H.U., Bennett D.A., De Jager P.L. The epigenome in Alzheimer’s disease: current state and approaches for a new path to gene discovery and understanding disease mechanism. Acta Neuropathol. 2016;132(4):503–514. doi: 10.1007/s00401-016-1612-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eid A., Bihaqi S.W., Hemme C., Gaspar J.M., Hart R.P., Zawia N.H. Histone acetylation maps in aged mice developmentally exposed to lead: epigenetic drift and Alzheimer-related genes. Epigenomics. 2018;10(5):573–583. doi: 10.2217/epi-2017-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCartney D.L., Stevenson A.J., Walker R.M., Gibson J., Morris S.W., Campbell A., Murray A.D., Whalley H.C., Porteous D.J., McIntosh A.M., Evans K.L., Deary I.J., Marioni R.E. Investigating the relationship between DNA methylation age acceleration and risk factors for Alzheimer’s disease. Alzheimers Dement. (Amst.) 2018;10:429–437. doi: 10.1016/j.dadm.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sato T., Cesaroni M., Chung W., Panjarian S., Tran A., Madzo J., Okamoto Y., Zhang H., Chen X., Jelinek J., Issa J.J. Transcriptional selectivity of epigenetic therapy in cancer. Cancer Res. 2017;77:470–481. doi: 10.1158/0008-5472.CAN-16-0834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang J., Yu J.T., Tan M.S., Jiang T., Tan L. Epigenetic mechanisms in Alzheimer’s disease: implications for pathogenesis and therapy. Ageing Res. Rev. 2013;12(4):1024–1041. doi: 10.1016/j.arr.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 118.Durga J., van Boxtel M.P., Schouten E.G., Kok F.J., Jolles J., Katan M.B., Verhoef P. Effect of 3-year folic acid supplementation on cognitive function in older adults in the FACIT trial: a randomised, double blind, controlled trial. Lancet. 2007;369(9557):208–216. doi: 10.1016/S0140-6736(07)60109-3. [DOI] [PubMed] [Google Scholar]
- 119.Haan M.N., Miller J.W., Aiello A.E., Whitmer R.A., Jagust W.J., Mungas D.M., Allen L.H., Green R. Homocysteine, B vitamins, and the incidence of dementia and cognitive impairment: results from the sacramento area latino study on aging. Am. J. Clin. Nutr. 2007;85(2):511–517. doi: 10.1093/ajcn/85.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Werneke U., Turner T., Priebe S. Complementary medicines in psychiatry: review of effectiveness and safety. Br. J. Psychiatry. 2006;188(188):109–121. doi: 10.1192/bjp.188.2.109. [DOI] [PubMed] [Google Scholar]
- 121.Cao X.J., Huang S.H., Wang M., Chen J.T., Ruan D.Y. S-adenosyl-L-methionine improves impaired hippocampal long-term potentiation and water maze performance induced by developmental lead exposure in rats. Eur. J. Pharmacol. 2008;595(1-3):30–34. doi: 10.1016/j.ejphar.2008.07.061. [DOI] [PubMed] [Google Scholar]
- 122.Kumar D., Aggarwal M., Kaas G.A., Lewis J., Wang J., Ross D.L., Zhong C., Kennedy A., Song H., Sweatt J.D. Tet1 oxidase regulates neuronal gene transcription, active dna hydroxy-methylation, object location memory, and threat recognition memory. Neuroepigenetics. 2015;4:12–27. doi: 10.1016/j.nepig.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Smith R.G., Hannon E., De Jager P.L., Chibnik L., Lott S.J., Condliffe D., Smith A.R., Haroutunian V., Troakes C., Al-Sarraj S., Bennett D.A., Powell J., Lovestone S., Schalkwyk L., Mill J., Lunnon K. Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimers Dement. 2018;14(12):1580–1588. doi: 10.1016/j.jalz.2018.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gasparoni G., Bultmann S., Lutsik P., Kraus T.F.J., Sordon S., Vlcek J., Dietinger V., Steinmaurer M., Haider M., Mulholland C.B., Arzberger T., Roeber S., Riemenschneider M., Kretzschmar H.A., Giese A., Leonhardt H., Walter J. DNA methylation analysis on purified neurons and glia dissects age and Alzheimer’s disease-specific changes in the human cortex. Epigenetics Chromatin. 2018;11(1):41. doi: 10.1186/s13072-018-0211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Su Y., Ryder J., Li B., Wu X., Fox N., Solenberg P., Brune K., Paul S., Zhou Y., Liu F., Ni B. Lithium, a common drug for bipolar disorder treatment, regulates amyloid-beta precursor protein processing. Biochemistry. 2004;43(22):6899–6908. doi: 10.1021/bi035627j. [DOI] [PubMed] [Google Scholar]
- 126.Qing H., He G., Ly P.T., Fox C.J., Staufenbiel M., Cai F., Zhang Z., Wei S., Sun X., Chen C.H., Zhou W., Wang K., Song W. Valproic acid inhibits Abeta production, neuritic plaque formation, and behavioral deficits in Alzheimer’s disease mouse models. J. Exp. Med. 2008;205(12):2781–2789. doi: 10.1084/jem.20081588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ricobaraza A., Cuadrado-Tejedor M., Marco S., Pérez-Otaño I., García-Osta A. Phenylbutyrate rescues dendritic spine loss associated with memory deficits in a mouse model of Alzheimer disease. Hippocampus. 2012;22(5):1040–1050. doi: 10.1002/hipo.20883. [DOI] [PubMed] [Google Scholar]
- 128.Ricobaraza A., Cuadrado-Tejedor M., Perez-Mediavilla A., Frechilla D., Del Rio J., Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an Alzheimer’s disease mouse model. Neuropsychopharmacology. 2009;34:1721–1732. doi: 10.1038/npp.2008.229. [DOI] [PubMed] [Google Scholar]
- 129.Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R.C., Cota P., Wittnam J.L., Gogol-Doering A., Opitz L., Salinas-Riester G., Dettenhofer M., Kang H., Farinelli L., Chen W., Fischer A. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328(5979):753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 130.Kilgore M., Miller C.A., Fass D.M., Hennig K.M., Haggarty S.J., Sweatt J.D., Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Green K.N., Steffan J.S., Martinez-Coria H., Sun X., Schreiber S.S., Thompson L.M., LaFerla F.M. Nicotinamide restores cognition in Alzheimer’s disease transgenic mice via a mechanism involving sirtuin inhibition and selective reduction of Thr231-phosphotau. J. Neurosci. 2008;28(45):11500–11510. doi: 10.1523/JNEUROSCI.3203-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brahe C., Vitali T., Tiziano F.D., Angelozzi C., Pinto A.M., Borgo F., Moscato U., Bertini E., Mercuri E., Neri G. Phenylbutyrate increases SMN gene expression in spinal muscular atrophy patients. Eur. J. Hum. Genet. 2005;13(2):256–259. doi: 10.1038/sj.ejhg.5201320. [DOI] [PubMed] [Google Scholar]
- 133.Marks P.A. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin. Investig. Drugs. 2010;19(9):1049–1066. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Marks P.A., Xu W.S. Histone deacetylase inhibitors: Potential in cancer therapy. J. Cell. Biochem. 2009;107(4):600–608. doi: 10.1002/jcb.22185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kelly-Sell M.J., Kim Y.H., Straus S., Benoit B., Harrison C., Sutherland K., Armstrong R., Weng W.K., Showe L.C., Wysocka M., Rook A.H. The histone deacetylase inhibitor, romidepsin, suppresses cellular immune functions of cutaneous T-cell lymphoma patients. Am. J. Hematol. 2012;87(4):354–360. doi: 10.1002/ajh.23112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rossi L.E., Avila D.E., Spallanzani R.G., Ziblat A., Fuertes M.B., Lapyckyj L., Croci D.O., Rabinovich G.A., Domaica C.I., Zwirner N.W. Histone deacetylase inhibitors impair NK cell viability and effector functions through inhibition of activation and receptor expression. J. Leukoc. Biol. 2012;91(2):321–331. doi: 10.1189/jlb.0711339. [DOI] [PubMed] [Google Scholar]
- 137.Salminen A., Tapiola T., Korhonen P., Suuronen T. Neuronal apoptosis induced by histone deacetylase inhibitors. Brain Res. Mol. Brain Res. 1998;61(1-2):203–206. doi: 10.1016/S0169-328X(98)00210-1. [DOI] [PubMed] [Google Scholar]
- 138.Huang K.L., Marcora E., Pimenova A.A., Di Narzo A.F., Kapoor M., Jin S.C., Harari O., Bertelsen S., Fairfax B.P., Czajkowski J., Chouraki V., Grenier-Boley B., Bellenguez C., Deming Y., McKenzie A., Raj T., Renton A.E., Budde J., Smith A., Fitzpatrick A., Bis J.C., DeStefano A., Adams H.H.H., Ikram M.A., van der Lee S., Del-Aguila J.L., Fernandez M.V., Ibañez L., Sims R., Escott-Price V., Mayeux R., Haines J.L., Farrer L.A., Pericak-Vance M.A., Lambert J.C., van Duijn C., Launer L., Seshadri S., Williams J., Amouyel P., Schellenberg G.D., Zhang B., Borecki I., Kauwe J.S.K., Cruchaga C., Hao K., Goate A.M. International Genomics of Alzheimer’s Project; Alzheimer’s Disease Neuroimaging Initiative. A common haplotype lowers PU.1 expression in myeloid cells and delays onset of Alzheimer’s disease. Nat. Neurosci. 2017;20(8):1052–1061. doi: 10.1038/nn.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rustenhoven J., Smith A.M., Smyth L.C., Jansson D., Scotter E.L., Swanson M.E.V., Aalderink M., Coppieters N., Narayan P., Handley R., Overall C., Park T.I.H., Schweder P., Heppner P., Curtis M.A., Faull R.L.M., Dragunow M.P.U. 1 regulates Alzheimer’s disease-associated genes in primary human microglia. Mol. Neurodegener. 2018;13(1):44. doi: 10.1186/s13024-018-0277-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jian W., Yan B., Huang S., Qiu Y. Histone deacetylase 1 activates PU.1 gene transcription through regulating TAF9 deacetylation and transcription factor IID assembly. FASEB J. 2017;31(9):4104–4116. doi: 10.1096/fj.201700022R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Janczura K.J., Volmar C.H., Sartor G.C., Rao S.J., Ricciardi N.R., Lambert G., Brothers S.P., Wahlestedt C. Inhibition of HDAC3 reverses Alzheimer’s disease-related pathologies in vitro and in the 3xTg-AD mouse model. Proc. Natl. Acad. Sci. USA. 2018;115(47):E11148–E11157. doi: 10.1073/pnas.1805436115. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 142.Fang M., Wang J., Zhang X., Geng Y., Hu Z., Rudd J.A., Ling S., Chen W., Han S. The miR-124 regulates the expression of BACE1/β-secretase correlated with cell death in Alzheimer’s disease. Toxicol. Lett. 2012;209(1):94–105. doi: 10.1016/j.toxlet.2011.11.032. [DOI] [PubMed] [Google Scholar]
- 143.Zhu H.C., Wang L.M., Wang M., Song B., Tan S., Teng J.F., Duan D.X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-β production by targeting BACE1. Brain Res. Bull. 2012;88(6):596–601. doi: 10.1016/j.brainresbull.2012.05.018. [DOI] [PubMed] [Google Scholar]
- 144.Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011;29(4):341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 145.Junn E., Mouradian M.M. MicroRNAs in neurodegenerative diseases and their therapeutic potential. Pharmacol. Ther. 2012;133(2):142–150. doi: 10.1016/j.pharmthera.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Rao J.S., Rapoport S.I., Kim H.W. Altered neuroinflammatory, arachidonic acid cascade and synaptic markers in postmortem Alzheimer’s disease brain. Transl. Psychiatry. 2017;7(5):e1127. doi: 10.1038/tp.2017.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Smith A.R., Smith R.G., Burrage J., Troakes C., Al-Sarraj S., Kalaria R.N., Sloan C., Robinson A.C., Mill J., Lunnon K. A cross-brain regions study of ANK1 DNA methylation in different neurodegenerative diseases. Neurobiol. Aging. 2019;74:70–76. doi: 10.1016/j.neurobiolaging.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 148.Phipps A.J., Vickers J.C., Taberlay P.C., Woodhouse A. Neurofilament-labeled pyramidal neurons and astrocytes are deficient in DNA methylation marks in Alzheimer’s disease. Neurobiol. Aging. 2016;45:30–42. doi: 10.1016/j.neurobiolaging.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 149.Walton E., Hass J., Liu J., Roffman J.L., Bernardoni F., Roessner V., Kirsch M., Schackert G., Calhoun V., Ehrlich S. Correspondence of DNA methylation between blood and brain tissue and its application to schizophrenia research. Schizophr. Bull. 2016;42(2):406–414. doi: 10.1093/schbul/sbv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cacabelos R., Torrellas C., Carrera I., Cacabelos P., Corzo L., Fernández-Novoa L., Tellado I., Carril J.C., Aliev G. Novel therapeutic strategies for dementia. CNS Neurol. Disord. Drug Targets. 2016;15(2):141–241. doi: 10.2174/1871527315666160202121548. [DOI] [PubMed] [Google Scholar]