Abstract
We investigate the evolutionary rescue of a microbial population in a gradually deteriorating environment, through a combination of analytical calculations and stochastic simulations. We consider a population destined for extinction in the absence of mutants, which can survive only if mutants sufficiently adapted to the new environment arise and fix. We show that mutants that appear later during the environment deterioration have a higher probability to fix. The rescue probability of the population increases with a sigmoidal shape when the product of the carrying capacity and of the mutation probability increases. Furthermore, we find that rescue becomes more likely for smaller population sizes and/or mutation probabilities if the environment degradation is slower, which illustrates the key impact of the rapidity of environment degradation on the fate of a population. We also show that our main conclusions are robust across various types of adaptive mutants, including specialist and generalist ones, as well as mutants modeling antimicrobial resistance evolution. We further express the average time of appearance of the mutants that do rescue the population and the average extinction time of those that do not. Our methods can be applied to other situations with continuously variable fitnesses and population sizes, and our analytical predictions are valid in the weak-to-moderate mutation regime.
Keywords: adaptation, deteriorating environment, evolutionary rescue, variable fitness, variable population size
UNDERSTANDING how a population of living organisms can survive in a gradually deteriorating environment is a fundamental question in evolution (Uecker and Hermisson 2011; Waxman 2011; Peischl and Kirkpatrick 2012), which is particularly relevant in the pressing context of climate change (Bell and Gonzalez 2009; Chevin et al. 2010; Pauls et al. 2013; Botero et al. 2015; Nadeau et al. 2017). Addressing this question is also important in order to understand antimicrobial resistance evolution, which often occurs in a variable environment, as when antimicrobials are added to a medium or given to a patient (Lin and Kussell 2016; Levin-Reisman et al. 2017). In fact, even when an antimicrobial is added instantaneously, yielding an abrupt environment switch, the resulting fitness decrease is gradual (Lin and Kussell 2016). In a deteriorating environment, the fitness of wild-type organisms decreases with time. In the simple case of asexual microorganisms, their division rate can then become smaller than their death rate, which yields a decrease of population size, eventually leading to extinction (Coates et al. 2018). However, the population can be rescued by a mutation that is better adapted to the new environment, and restores positive population growth (or several such mutations): this phenomenon is called evolutionary rescue (Martin et al. 2013; Gonzalez et al. 2013; Alexander et al. 2014; Carlson et al. 2014; Barton and Etheridge 2018).
A gradually deteriorating environment impacts the population size and the fitness of the wild-type organism, which can both strongly impact the fate of a mutation (Uecker and Hermisson 2011). The decay of the wild-type population simultaneously entails a decreased frequency of mutant appearance, which can hinder rescue, and a decreased competition for existing mutants, known as competitive release (Wargo et al. 2007; Kouyos et al. 2014), which can facilitate rescue. Studying the evolutionary rescue of a population in a gradually deteriorating environment requires accounting for simultaneous continuous time variations of fitness, population size and population composition, which makes it complex. Varying patterns of selection have recently been the focus of significant interest, mainly in the case of switches between different environment states, highlighting their strong effect on evolution (Kussell et al. 2006; Mustonen and Lässig 2008; Rivoire and Leibler 2011; Cvijović et al. 2015; Melbinger and Vergassola 2015; Hufton et al. 2016; Skanata and Kussell 2016; Mayer et al. 2017; Wienand et al. 2017; Danino et al. 2018; Marrec and Bitbol 2018, 2020; Meyer and Shnerb 2018; Trubenová et al. 2019). Despite its practical relevance, the case of a fitness varying continuously over time for a given genotype has been comparatively less studied, with a focus on stabilizing selection (Burger and Lynch 1995; Kopp and Hermisson 2007, 2009a,b; Gomulkiewicz and Houle 2009; Alexander et al. 2014; Matuszewski et al. 2014) or on the fate of a single beneficial mutation (Uecker and Hermisson 2011; Waxman 2011; Peischl and Kirkpatrick 2012). Furthermore, most theoretical works on evolutionary rescue consider an abrupt environment change (Orr and Unckless 2008; Bell and Gonzalez 2009; Martin et al. 2013; Anciaux et al. 2018). Here, we address evolutionary rescue in a gradually changing environment, which deteriorates from the point of view of wild-type organisms.
Adaptation to a new environment can occur in multiple ways. A specialist mutant that is particularly well-adapted to this new environment can emerge, e.g., a thermophilic mutant in the case of a temperature rise. Another possibility is the appearance of a generalist mutant, which is able to grow in both the initial and the final environments, while being less fit than specialists in their respective favorite environments (Donaldson-Matasci et al. 2008; Mayer et al. 2017; Sachdeva et al. 2020; Wang and Dai 2019). Yet another one regards mutants that are less fit in the final environment than in the initial one, but still sufficiently fit to be able to grow in the final environment. The latter case can model the evolution of antimicrobial resistance as drug concentration is increased from zero to a value that is above the minimum inhibitory concentration of the sensitive microbes, but below that of the resistant microbes (Gullberg et al. 2011; Yu et al. 2018).
In the present work, we consider a microbial population subjected to a gradual environment deterioration, such that the fitness and the size of the wild-type population are gradually decaying, and that extinction would be certain in the absence of adaptation. We study the fixation probability of generalist and specialist adaptive mutants as a function of the time when they appear during the environment deterioration, and we also consider a model of antimicrobial resistance evolution. We obtain an expression for the overall probability that the population is rescued by an adaptive mutation, thereby avoiding extinction. We investigate the dependence of the rescue probability on the rapidity of the environment deterioration, as well as on population size and mutation probability. We also compare different types of mutants. We further express the average time of appearance of the mutants that do rescue the population and the average extinction time of those that do not.
Model and Methods
Population model
We consider a population of asexual microorganisms with carrying capacity K, corresponding to the maximum population size that the environment can sustain, given, e.g., the nutrients available. We assume that two types of microorganisms can exist in this population: wild-type (W) and mutant (M). The division rate of each organism is assumed to be logistic (Verhulst 1838), and reads , where N represents the total population size, while the time-dependent fitness fi (t) with i = W or i = M represents the maximal possible division rate of the (wild-type or mutant) organism at time t, which would be reached if . The death rates of W and M organisms are denoted by gW and gM, respectively. Note that (Malthusian) fitness is usually measured as the exponential growth rate at the population scale, and that fitness often refers to the overall outcome of both survival and selection: under such definitions, fitness would in fact correspond to fi(t) − gi here. However, here we will not consider any variability of death rates, and, thus, for the sake of simplicity, fitness will refer to fi(t) throughout. While we assume that the variability of the environment impacts fitnesses and not death rates, our approach can be easily extended to variable death rates. We further assume that W microorganisms can mutate into M microorganisms, with the mutation probability μ upon each division. We do not consider back mutations. Note that, because mutations occur upon division, the number of mutants appearing per unit time depends both on the population size and on the fitness of W microorganisms. Importantly, our model incorporates both variations of population size (population dynamics) and of composition (population genetics) (Melbinger et al. 2010; Melbinger and Vergassola 2015; Huang et al. 2015). Throughout, we consider the fitness of W microorganisms in the initial environment as the reference fitness and set it to 1. Therefore, our time unit corresponds to the inverse of this fitness (which is the maximum division rate we consider).
We start from a microbial population composed of wild-type microorganisms and no mutants. Specifically, our simulations include a phase of initial growth, which can model, e.g., the development of an infection starting from the bottleneck at transmission (Abel et al. 2015). In practice, we will start our simulations with Supplemental Material, Figure S5 demonstrates that our results do not depend on this particular choice, since starting with gives the same results as starting with which corresponds to the stationary population size in the initial environment within a deterministic description. Note, however, that if we started with a very small number of W microorganisms (i.e., 1 or 2), we would need to take into account rapid stochastic extinctions of the population (Ovaskainen and Meerson 2010): we will not consider this regime.
Fitnesses in a deteriorating environment
To model the impact of a continuously deteriorating environment on the fitness of W microorganisms, we choose the Hill function:
| (1) |
where n is the Hill coefficient and θ the inflection point, such that fW(θ) = 0.5. This sigmoidal function represents a transition between two different environments, by decreasing from the reference fitness value fW(0) = 1 toward 0 as t increases, with a steepness that is tunable via n. Specifically, the decay is more abrupt manner for larger values of n (see Figure 1A). The Hill function is quite generic in biological contexts, e.g., it is a good model for cooperative reactions, and for the pharmacodynamics of antimicrobials (Regoes et al. 2004). Moreover, Equation 1 allows us to recover the case of an abrupt environment change as a limiting case when n → ∞. Because it is n that sets the timescale of the environmental change occurring around θ, we will vary n at a fixed (and large) value of θ. Note that employing Equation 1 implies environment changes with rates symmetric with respect to θ. But, crucially, the methods presented here do not depend on the exact function chosen, and can be applied to other forms of environment degradation beyond Equation 1.
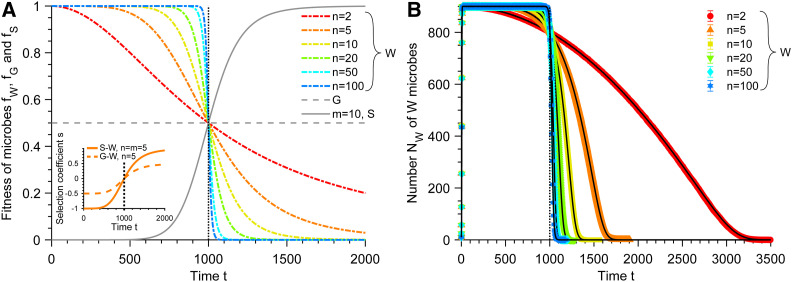
Figure 1.
Fitnesses and wild-type population in a deteriorating environment. (A) Fitnesses fW, fG, and fS of the wild-type organisms (W), generalist (G), and specialist (S) mutants vs. time t (see Equations 1 and 2). Several values of the Hill coefficient n are shown for W. Inset: selection coefficient for both types of mutants M = G or S vs. time t, shown with n = 5 (and m = 5 for S mutants). (B) Number NW of W microbes vs. time t for different values of n (same colors as in A). Data points correspond to averages over 103 replicate stochastic simulations, and error bars (smaller than markers) represent 95% confidence intervals. Black solid curves correspond to numerical integrations of Equation 3. Parameter values: K = 103, and θ = 103. Vertical dotted line in both panels: t = θ.
We will mainly consider two types of adaptive mutants. First, generalist mutants, denoted by G, are not impacted by gradual changes of the environment and have a constant fitness fG. We choose fG = 0.5 so that G mutants and W organisms have the same time-averaged fitness. Second, specialist mutants, denoted by S, have a fitness described by an increasing Hill function, so that they are better adapted to the final environment, in contrast to W organisms:
| (2) |
We take the same point of inflection θ for W and S, as it marks the midst of the environmental transition. Conversely, we allow different Hill coefficients n and m, reflecting a different sensitivity of W and S individuals to environmental change (see Figure 1A). Note that S mutants, G mutants, and W organisms have the same time-averaged fitness over a time window that is symmetric around θ, and that G mutants are in fact S mutants with m = 0. The selection coefficient, defined as the fitness difference between mutant and wild type (see Figure 1A, inset), switches from negative to positive at the inflection point, more steeply when n and m are large, and with a wider range for S mutants than for G mutants.
In section 2.1 of the Supporting Information, we also consider another type of mutant in order to model antimicrobial resistance evolution. We focus on the case where drug concentration is increased from zero to a value that is above the minimum inhibitory concentration of the sensitive microbes but below that of the resistant microbes (Gullberg et al. 2011; Yu et al. 2018). Then, resistant mutants are able to grow in the final environment and rescue the population.
Methods
We present both analytical and numerical results. Our analytical results are obtained using methods from stochastic processes, especially from birth–death processes with time varying rates (Nissen-Meyer 1966; Bailey 1964; Uecker and Hermisson 2011; Alexander and Bonhoeffer 2012; Parzen 1999). Importantly, our predictions make quite minimal assumptions and hold in the weak-to-moderate mutation regime where Our simulations employ a Gillespie algorithm (Gillespie 1976, 1977), and incorporate all individual stochastic division, mutation, and death events with the associated rates. In principle, the time variability of the division rates imposes a difficulty (Thanh and Priami 2015), but the short duration of time intervals between individual events allows us to neglect rate variations between events (see Supporting Information, section 10 for details). Our model allows us to fully account for the stochasticity of mutation occurrence and establishment (Ewens 1979; Rouzine et al. 2001; Fisher 2007; Patwa and Wahl 2008; Weissman et al. 2009), as well as that of population extinction (Coates et al. 2018; Teimouri and Kolomeisky 2019; Alexander and MacLean 2013.
In our analytical calculations, we will often make a deterministic approximation for the evolution of the number NW of W individuals, while the evolution of the mutant population will be described in a fully stochastic manner. Indeed, mutants are in small numbers when they appear, while they generally arise in a large population of W organisms. In the deterministic limit, NW satisfies the following ordinary differential equation:
| (3) |
This description is appropriate for very large NW, and Equation 3 can be derived from the complete stochastic model in this limit (see Supporting Information, section 8 and Van Kampen 1981; Gardiner 1985).
Figure 1B compares the predictions from Equations 1 and 3 to the results of stochastic simulations (see Supporting Information Section 10.1), and demonstrates the validity of the deterministic approximation in this regime. Figure 1B also illustrates that in the absence of mutants, the population of W individuals always goes extinct, due to the fact that fitness fW tends to 0 while death rate is nonzero (gW > 0). Moreover, the bigger the Hill coefficient n, the faster the W population goes extinct.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article or Supplemental Material. Matlab implementations of numerical simulations available at https://doi.org/10.5281/zenodo.3993272. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12860975.
Results
Fixation probability of mutants: on the importance of good timing
In a deteriorating environment, mutants will have different fates depending on when they appear. Therefore, before investigating overall rescue probabilities, we address the fixation probability pfix(t0) of a mutant as a function of the time t0 when it appears during the environment deterioration. Competition with wild-type organisms is felt by mutants through their division rate At the early stages when competition matters, i.e., when the logistic term is important, the number of mutants is small with respect to the number of wild-type microorganisms, and thus the division rate of mutants can be approximated by Furthermore, at these early stages, the number of wild-type microorganisms NW is large enough to be described in a deterministic framework (see Models and Methods, Equation 3 and Figure 1). We retain a full stochastic description for mutants, which are in small numbers just after the mutation arises (Parzen 1999; Uecker and Hermisson 2011; Alexander and Bonhoeffer 2012), and we introduce the probability of having i mutants at time t knowing that there is 1 mutant at time t0. The fixation probability of the mutants can then be obtained from the probability generating function which satisfies Solving the partial differential equation governing the evolution of (see Supporting Information, section 1) yields (Parzen 1999; Uecker and Hermisson 2011; Alexander and Bonhoeffer 2012)
| (4) |
where
| (5) |
Numerical integration of Equation 4 is discussed in section 9 of the Supporting Information.
Figure 2 shows the fixation probability pfix of a mutant vs. the time t0 at which it appears during the deterioration of the environment. A very good agreement is obtained between the results of our stochastic simulations and the analytical prediction of Equation 4. This holds both when t0 < θ, while mutants are less fit than W organisms, and when t0 > θ, where the opposite is true. In Figure S4, we provide additional results for the fixation probability of generalist mutants with different fitness values fG, which thus become effectively beneficial sooner or later during the environment deterioration, illustrating that Equation 4 holds in these various cases.
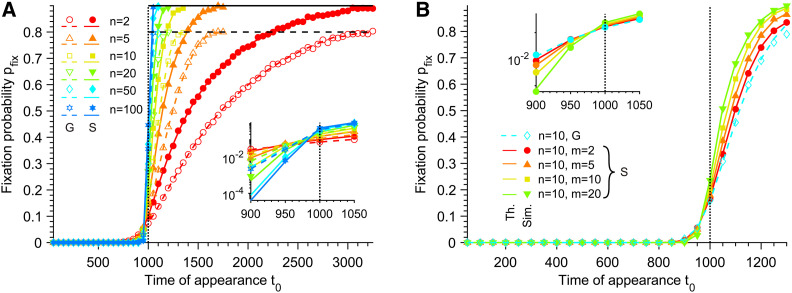
Figure 2.
Fixation probability of mutants. (A) Fixation probability pfix of G and S mutants vs. their time of appearance t0 in the deteriorating environment, for different Hill coefficients n characterizing the steepness of the environment deterioration (see Equation 1). Here, S mutants satisfy m = n, i.e., they have the same sensitivity to the environment as W organisms (see Equation 2). Horizontal dashed line: Horizontal solid line: Data are shown for t0 < τW, where τW is the average extinction time of the W population in the absence of mutation. (B) Fixation probability pfix of different types of mutants vs. their time of appearance t0 in the deteriorating environment, for a fixed Hill coefficient n = 10 characterizing the decay of fW (see Equation 1). G mutants and S mutants with different Hill coefficients m (see Equation 2), corresponding to different sensitivities to the changing environment, are considered. In both panels, markers correspond to averages over 104 replicate stochastic simulations (“Sim.”). Dashed and solid lines correspond to numerical integrations of Equation 4 (“Th.”) for G and S mutants, respectively. Parameter values: K = 103, and θ = 103. Vertical dotted lines: t0 = θ. Main panels: linear scale; insets: semilogarithmic scale.
Figure 2 shows that pfix strongly increases with t0: mutants appearing later in the environmental degradation are much more likely to fix. This reflects both the increasing intrinsic fitness advantage of mutants due to the environment transition, and the decreasing competition with the W population that decays as the environment deteriorates for W organisms. Note that variations of selection coefficients only, or of competition pressure only, were previously addressed (Uecker and Hermisson 2011), and that an increase in fixation probability with mutant appearance time was described under decreasing competition (Alexander and Bonhoeffer 2012). Figure 2A shows that the increase of pfix is strong around the inflection point θ, and is steeper for larger Hill coefficients n characterizing the fitness decay of the wild-type organisms (see Equation 1). Furthermore, for each value of n, sufficiently before θ, generalist (G) mutants are more likely to fix than specialist (S) mutants with m = n (see Models and Methods, Equation 2), because then fG > fS. Conversely, S mutants are more likely to fix than G mutants sufficiently after θ because fG < fS. Note that in section seven of the Supporting Information, we provide analytical approximations for the fixation probability with large Hill coefficients n,m → ∞. Finally, Figure 2B shows that for t0 > θ, pfix increases with the Hill coefficient m characterizing the steepness of the fitness transition for S mutants, and all S mutants are more likely to fix than G mutants, consistently with the fact that G mutants correspond to S mutants with m = 0 (see Equation 2).
For large t0, if the W population is not yet extinct, the fixation probability pfix in Equation 4 converges to (resp. 1 − gS) for G (resp. S) mutants, which is corroborated by our simulation results (see Figure 2A and Figure S4A). This simple limit can be interpreted as follows: mutants appearing just before the extinction of the W population face negligible competition, and thus they survive and fix unless they undergo rapid stochastic extinction (Ovaskainen and Meerson 2010; Coates et al. 2018; Marrec and Bitbol 2020). Note that pfix is constructed so that mutant lineages that undergo rapid stochastic extinctions are counted as not fixing in the population. Importantly, even though the fixation probability pfix at a given t0 becomes larger as n is increased, mutants appearing just before the extinction of the W population (which occurs faster as n is increased, see Figure 1B) have a fixation probability independent of n (see Figure 2A and Figure S1).
Rescue probability
So far, we investigated the fate of a given mutant lineage as a function of its appearance time during the environment degradation. Let us now address whether mutants can rescue the population or not. For a mutation probability μ at division, both the occurrence of a new mutation and its subsequent fixation probability depend on the number and division rate of W organisms. We thus consider the probability paf (t) that a mutant appears between 0 and t and fixes. The rescue probability pr corresponds to the probability that a mutant appears and fixes before the microbial population goes extinct, and is thus given by . Using Bayes’ rule, the probability that a mutant appears between t and t + dt and fixes (which is equal to the probability that no mutant destined for fixation appeared before, and that a mutant destined for fixation then appears), denoted by can be written as:
| (6) |
where is the probability that no mutant destined for fixation appeared before, while dpnaf (t) is the probability that a mutant appears between t and t + dt and fixes, provided that no mutant destined for fixation appeared before. The latter can be calculated by considering that the population is fully or mostly wild type at time t, i.e., which is expected to be valid in most cases, except in the strong-mutation regime where multiple mutant lineages arise almost simultaneously. Then, where is the number of mutants that appear between t and t + dt in a fully wild-type population (see Figure S6). Thus,
| (7) |
We again take a deterministic description for NW(t) (see Equation 3), and the fitness fW(t) of W organisms is given by Equation 1. Then, integrating Equation 7 with paf(0) = 0 yields with
| (8) |
Taking the limit t → ∞ then gives the rescue probability
| (9) |
where
| (10) |
Note that if , Equation 9 reduces to , which would be obtained by neglecting possible earlier fixations. Note also that, since mutant lineages undergoing rapid stochastic extinction are counted as not fixing in pfix, they are correctly counted as not able to rescue the population. Numerical integration of Equations 9–10 is discussed in section 9 of the Supporting Information.
Figure 3 shows the rescue probability pr vs. the mutation probability μ at each division. It demonstrates a very good agreement between our analytical prediction in Equation 9 and results from our stochastic simulations (see Supporting Information, section 10.3). We observe a sigmoidal increase of pr as μ increases, with a transition between a small-μ regime, where the population almost certainly goes extinct, and a large-μ regime, where it is almost certainly rescued by adaptive mutants. Figure 3A further shows that this transition is strongly impacted by the rapidity of the environment degradation, which is modeled via the Hill coefficient n (see Equation 1). Specifically, the faster the environment degradation, the bleaker the prospect is for the population, and the larger μ becomes necessary to allow its rescue. This is related to the rapidity of extinction of the W population in the absence of mutations: for small n, the population decay is slower, allowing a larger window of opportunity for mutants to appear and to be selected (see Figure 1). Increasing n does not substantially affect the steepness of pr, but rather shifts the transition between small and large pr toward larger μ, because the associated faster decay of the W population mainly decreases the total number of mutants that appear (see Figure S6), with little impact on their fixation probabilities at the end of the process (see Figure 2A and Figure S1). Note that our prediction in Equation 9 is valid far beyond the weak-mutation regime While our assumption that when the rescuing mutant arises can fail for rescue is almost certain as this regime is reached. In the limit n → ∞ of an instantaneous environment degradation, discussed in detail in section seven of the Supporting Information, the transition from large to small pr occurs for (see Figure 3A and Figure S11A). Indeed, preexisting mutations then become necessary to population rescue, as no division occurs after the abrupt environment transition. In section 7.2 of the Supporting Information, we further show that Equation 9 generalizes the predictions in our previous work (Marrec and Bitbol 2020) regarding the probability of extinction of a microbial population subjected to abrupt additions of antimicrobial, beyond the weak-mutation regime (see Figure S11B).
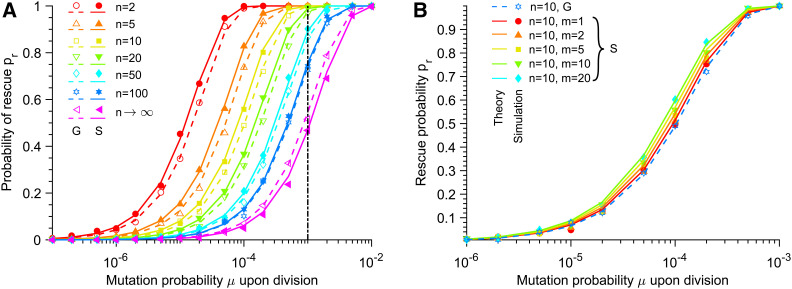
Figure 3.
Rescue probability. (A) Rescue probability pr of a W population in a deteriorating environment by G or S mutants, vs. mutation probability μ upon division. Different Hill coefficients n characterizing the steepness of the environment deterioration (see Equation 1) are considered. Here, S mutants satisfy m = n, i.e., they have the same sensitivity to the environment as W organisms (see Equation 2). Vertical dash-dotted line: Kμ = 1. (B) Rescue probability pr by different types of mutants vs. mutation probability μ upon division. A fixed Hill coefficient n = 10 characterizing the decay of fW (see Equation 1) is chosen, but G mutants and S mutants with different Hill coefficients m (see Equation 2) are considered. In both panels, markers correspond to averages over 104 replicate stochastic simulations (“Simulation”). Dashed and solid lines correspond to numerical integrations of Equation 9 (“Theory”) for G and S mutants, respectively. Parameter values: K = 103, and θ = 103.
In Figure 3A, we also compare G mutants and S mutants satisfying m = n (see Equation 2) for each n, and we find that S mutants are slightly more successful at rescuing the population than G mutants, unless n is very large. This is because S mutants that occur for t > θ have a larger selective advantage than G mutants, and, thus, a larger fixation probability (see Figure 2A). Note that for very steep environment changes, the situation reverses (see Figure 3A and Figure S10), because the decay of the W population is so fast that mutants occurring for t < θ are more likely to be the ones that rescue the population. Consistently, Figure 3B further shows that specialists with a larger Hill coefficient m, such that fitness increases more steeply during the environment transition (see Equation 2), are slightly more efficient at rescuing the population. The impact of n on the rescue probability is stronger than that of m, because n controls the rapidity of the decay of the wild-type population, which directly impacts the number of mutants that appear during this decay (see Figure S6).
Apart from the detailed differences we just described, Figure 3 demonstrates that the mutant type affects rescue probability quite little. In section 2.1 of the Supporting Information, we consider yet another mutant type, aiming to model antimicrobial resistance evolution, and we find that our results are also qualitatively robust to this variant. Overall, the key ingredients are that wild-type organisms are doomed to extinction in the absence of mutants, while mutants are fit enough in the final environment to be able to grow and rescue the population. If this holds, the detailed time evolution of mutant fitness matters little.
Time of appearance of the mutants that fix
The fixation probability of a mutant depends strongly on the time at which it appears during the environment degradation (see Figure 2). But when do the mutants that fix and rescue the population appear? The probability density function of the time of appearance of a mutant that fixes can be obtained from paf (see Equation 7 and below) through where normalization is ensured by 1/pr (we focus on cases where rescue occurs). Indeed, is the cumulative distribution function of Thus,
| (11) |
where
| (12) |
is shown in Figure S7 for different Hill coefficients n characterizing the steepness of the environment deterioration. It illustrates that rescuing mutants tend to appear later as n is decreased, because the decay of the W population is slower in these cases.
Equation 11 allows to express the average time of appearance of the mutants that fix:
| (13) |
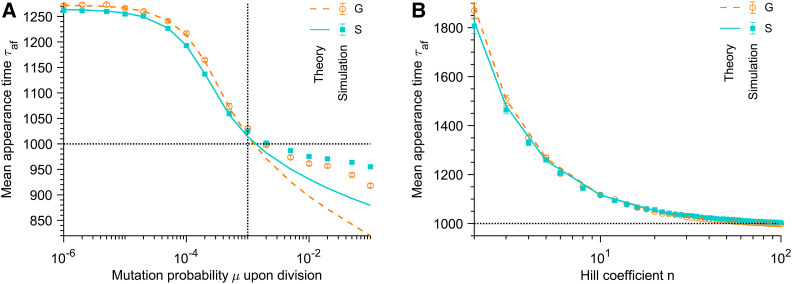
Figure 4 shows the average time τaf of appearance of the mutants that fix, and demonstrates a very good agreement between our analytical prediction in Equation 13 and the results of our stochastic simulations in the weak-to-moderate mutation regime (Recall that our calculations assume that when the rescuing mutant appears, which can fail when Kμ is large.) Figure 4A shows that τaf decreases as the mutation probability μ upon division is increased: this is because more mutants appear for larger μ. In addition, τaf is larger than the inflection time θ for which confirms that the mutants that fix tend to be beneficial ones (see Figure 2), and is consistent with the fact that S mutants, which are more beneficial than G mutants for t > θ, are more efficient at rescuing the population (see Figure 3). Besides, when τaf > θ, S mutants that fix appear earlier than G mutants that fix: this is also due to their larger selective advantage, and, consistently, the opposite holds for τaf < θ, when G mutants are fitter than S mutants (see Equation 1). In addition, Figure 4B shows that τaf decreases as the Hill coefficient n, which characterizes the steepness of the environment degradation (see Equation 1) is increased. Indeed, for large n, the population gets extinct quickly and rescue needs to occur fast if it occurs at all.
Figure 4.
Time of appearance of the mutants that fix. (A) Average time τaf of appearance of a G or S mutant that fixes vs. mutation probability μ upon division. The Hill coefficient characterizing the steepness of the environment deterioration (see Equation 1) is n = 5. Vertical dotted line: Kμ = 1. (B) Average time τaf of appearance of a G or S mutant that fixes vs. Hill coefficient n. The mutation probability upon division is μ = 10−5. In both panels, markers correspond to averages over 103–104 replicate stochastic simulations (“Simulation”). Dashed and solid lines correspond to numerical integrations of Equation 13 (“Theory”) for G and S mutants, respectively. Parameter values: K = 103, and θ = 103. Horizontal dotted lines: τaf = θ.
While we have focused mainly on mutants that fix and rescue the population, in section 6 of the Supporting Information, we also investigate the mean time to extinction of the lineages of mutants that do not fix. This time is longest for mutants appearing close to the inflection point θ of the environment transition, which corresponds to the time when the fitness difference between W organisms and mutants is smallest. Intuitively, mutants that are strongly deleterious or beneficial have their fates sealed faster than neutral ones. Furthermore, in the framework of the Moran process (with constant population size and fitnesses), extinction times are longest for neutral mutants (Ewens 1979; Teimouri and Kolomeisky 2019; Teimouri et al. 2019). While the time to extinction is not crucial to our study of rescue by a single mutation, it can become relevant to more complex processes involving several mutations, e.g., to the crossing of fitness valleys or plateaus (Weissman et al. 2009; Bitbol and Schwab 2014).
Impact of population size on rescue
So far, we have discussed population rescue at a given carrying capacity K. What is the impact of K on rescue?
First, our analytical expression of the fixation probability pfix of mutants in Equation 4 depends on K only via the function ρ introduced in Equation 5. But ρ depends on the number of wild-type microbes NW (t) and on the carrying capacity K only through the ratio , whose dynamics is independent from K (see Equation 3). Therefore, pfix is expected to be independent from K. Figure S8A confirms that it is the case: the simulation results obtained for different values of K collapse on the same curves. In addition, they are in very good agreement with the predictions from Equation 4. Note that Eq. S13 shows that the mean extinction time of the lineages of mutants that do not fix is also independent from population size, which is confirmed by Figure S9B.
Let us now turn to the rescue probability pr. Equations 9 and 10 demonstrate that pr depends on population size only via the product Therefore, the relevant parameter is Kμ. Figure S8B confirms that pr only depends on K via Kμ: the simulation results obtained for different values of K collapse on the same curves when they are plotted as a function of Kμ, and feature a good agreement with Equation 9. For larger K, smaller mutation probabilities per division suffice to ensure larger rescue probabilities, because more mutants appear in larger populations, but, more precisely, what really matters for rescue is the value of Kμ. This finding extends previous results regarding abrupt environment change (Martin et al. 2013).
Finally, Equations 12 and 13 show that, for the mean time τaf of appearance of a mutant that fixes, the relevant parameter is also Kμ. Figure S8C confirms this: the simulation results obtained by varying μ at constant K or by varying K at constant μ collapse when they are plotted as a function of Kμ, in good agreement with Equation 13.
Overall, the main quantities that characterize population rescue, namely the rescue probability pr and the mean time τaf of appearance of a mutant that fixes, are governed by Kμ. Hence, the impact of population size and mutation probability is mainly felt through this parameter.
Discussion
In this paper, we investigated the evolutionary rescue of a microbial population in a gradually deteriorating environment, characterized by a sigmoidal decay down to zero of the fitness of wild-type organisms, with a tunable steepness. The population is thus destined for extinction in the absence of adaptive mutants. We showed that mutants that appear later during the environment deterioration have a higher probability to fix, due to an increase of their intrinsic fitness advantage and to competitive release (Wargo et al. 2007; Kouyos et al. 2014). However, the decay of the wild-type population also entails that mutants are less likely to appear at such late stages. We demonstrated that the overall rescue probability of the population increases with a sigmoidal shape as the product Kμ of the carrying capacity K and of the mutation probability μ is increased, which extends previous results regarding abrupt environment change (Martin et al. 2013). In the limit of an instantaneous environment degradation, the increase of rescue probability occurs for Kμ ≈ 1, as preexisting mutations become necessary for rescue. Importantly, much smaller values of Kμ suffice for rescue if the environment degradation, and thus the population decay, are slower, consistently with previous studies on the rate of fitness decay in the regime of stabilizing selection (Burger and Lynch 1995; Gomulkiewicz and Houle 2009). We also found that our main conclusions are robust to the exact type of mutant considered (generalist, specialist, or modeling antimicrobial resistance evolution), provided that mutants are fit enough in the final environment to be able to rescue the microbial population, which is doomed to extinction in the absence of mutants. We further characterized the rescue process by investigating the average time of appearance of the mutants that do rescue the population, which also depends on the parameter Kμ, and the average extinction time of those that do not, which is longest when mutants are almost neutral.
In all cases, we provided both analytical expressions and stochastic simulation results, and obtained a very good agreement between them. Our analytical expressions were obtained with assumptions that hold in the weak-to-moderate mutation regime as we only required the wild-type population to be much larger than the mutant one upon the appearance of the successful mutant lineage. Our methods can be applied to other situations with continuously variable fitnesses and population sizes. Our predictions could be tested in controlled evolution experiments, e.g., in the context of antimicrobial resistance evolution, especially by varying population size and/or by studying strains with different mutation rates.
Overall, our study quantitatively confirms the key impact of the rapidity of environment degradation on the fate of a population, with fast degradation bringing the harshest prospects for population survival. This point confirms and extends previous theoretical results regarding a trait under stabilizing selection with a gradually moving optimum (Burger and Lynch 1995), as well as experimental (Lindsey et al. 2013) and numerical (Wu et al. 2014) results in the context of antibiotic resistance. Very large populations can almost always escape extinction because they have a wide range of existing mutants, while smaller ones (or rarely mutating ones, since what matters is Kμ) can be rescued by adaptive mutations only if the environment changes slowly enough. The case of not-too-large populations is practically very important because real populations tend to have complex structures (van Marle et al. 2007), and competition is local, which decreases their effective size, at least on timescales shorter than those of large-scale migrations and/or mixing. Accordingly, an exciting extension would be to consider the impact of spatial structure (Bitbol and Schwab 2014; Cooper et al. 2015; Nahum et al. 2015) on evolutionary rescue (Uecker et al. 2014; Czuppon et al. 2019) in a gradually deteriorating environment. In cases where one aims to avoid rescue, our results entail that environment changes should be made as fast as possible. For instance, in order to avoid antimicrobial resistance evolution, gradually increasing doses of antimicrobial should be avoided. In addition, our results on the fixation probability of mutants and on the mean time of appearance of mutants that fix could be exploited in evolution experiments, e.g., to guide the timing of mutagen use to potentially favor the appearance of rescue mutants. The average time to extinction of mutants that do not fix can also matter in practice, as another environment change occurring within this time after their appearance might rescue them. Importantly, here, we have considered rescue by a single mutation. However, more adaptations can be accessible in several mutation steps, and, thus, considering rescue in a gradually deteriorating environment in the presence of fitness valleys (Weinreich and Chao 2005; Weissman et al. 2009) or on more complete fitness landscapes (Poelwijk et al. 2007; Szendro et al. 2013) would also be very interesting from a theoretical point of view. Studying the interplay between time variability of the environment and spatial heterogeneities would also be interesting in this context, given that static antimicrobial gradients can favor resistance evolution (Zhang et al. 2011; Greulich et al. 2012; Hermsen et al. 2012; Baym et al. 2016), in particular by stepwise accumulation of several mutations.
Acknowledgments
LM acknowledges funding by a graduate fellowship from EDPIF (École Doctorale Physique en Île-de-France).
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12860975.
Communicating editor: L. Wahl
Literature Cited
- Abel S., Abel zur Wiesch P., Davis B. M., and Waldor M. K.. 2015. Analysis of Bottlenecks in Experimental Models of Infection. PLoS Pathog. 11: e1004823 10.1371/journal.ppat.1004823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander H. K. and Bonhoeffer S., 2012. Pre-existence and emergence of drug resistance in a generalized model of intra-host viral dynamics. Epidemics 4: 187–202. 10.1016/j.epidem.2012.10.001 [DOI] [PubMed] [Google Scholar]
- Alexander H. K. and MacLean R. C.. 2013. Stochastic bacterial population dynamics prevent the emergence of antibiotic resistance. bioRxiv. . (Preprint posted January 11, 2019) 10.1101/458547 [DOI] [Google Scholar]
- Alexander H. K., Martin G., Martin O. Y., and Bonhoeffer S.. 2014. Evolutionary rescue: linking theory for conservation and medicine. Evol. Appl. 7: 1161–1179. 10.1111/eva.12221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anciaux Y., Chevin L. M., Ronce O., and Martin G., 2018. evolutionary rescue over a fitness landscape. Genetics 209: 265–279. 10.1534/genetics.118.300908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey N. T. J., 1964. The Elements of Stochastic Processes with Applications to the Natural Sciences. John Wiley and Sons, New York. [Google Scholar]
- Barton N. and Etheridge A.. 2018. Establishment in a new habitat by polygenic adaptation. Theor. Popul. Biol. 122: 110–127. [DOI] [PubMed] [Google Scholar]
- Baym M., Lieberman T. D., Kelsic E. D., Chait R., Gross R. et al. , 2016. Spatiotemporal microbial evolution on antibiotic landscapes. Science 353: 1147–1151. 10.1126/science.aag0822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. and Gonzalez A., 2009. Evolutionary rescue can prevent extinction following environmental change. Ecol. Lett. 12: 942–948. [DOI] [PubMed] [Google Scholar]
- Bitbol A. F. and Schwab D. J., 2014. Quantifying the role of population subdivision in evolution on rugged fitness landscapes. PLOS Comput. Biol. 10: e1003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botero C. A., Weissing F. J., Wright J., and Rubenstein D. R., 2015. Evolutionary tipping points in the capacity to adapt to environmental change. Proc. Natl. Acad. Sci. USA 112: 184–189. 10.1073/pnas.1408589111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger R. and Lynch M., 1995. Evolution and extinction in a changing environment. Evolution 49: 151–163. [DOI] [PubMed] [Google Scholar]
- Carlson S. M., Cunningham C. J., and Westley P. A., 2014. Evolutionary rescue in a changing world. Trends Ecol. Evol. 29: 521–530. 10.1016/j.tree.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Chevin L. M., Lande R., and Mace G. M., 2010. Adaptation, plasticity, and extinction in a changing environment: towards a predictive theory. PLoS Biol. 8: e1000357 10.1371/journal.pbio.1000357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates J., Park B. R., Le D., Simsek E., Chaudhry W., and Kim M., 2018. Antibiotic-induced population fluctuations and stochastic clearance of bacteria. eLife 7: e32976 10.7554/eLife.32976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. D., Neuhauser C., Dean A. M., and Kerr B., 2015. Tipping the mutation-selection balance: limited migration increases the frequency of deleterious mutants. J. Theor. Biol. 380: 123–133. 10.1016/j.jtbi.2015.05.003 [DOI] [PubMed] [Google Scholar]
- Cvijović I., Good B. H., Jerison E. R., and Desai M. M., 2015. Fate of a mutation in a fluctuating environment. Proc. Natl. Acad. Sci. USA 112: E5021–E5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuppon P., Blanquart F., Uecker H., and Debarre F.. 2019. The effect of habitat choice on evolutionary rescue in subdivided populations. bioRxiv. . (Preprint posted October 6, 2019) 10.1101/738898 [DOI] [PubMed] [Google Scholar]
- Danino M., Kessler D. A., and Shnerb N. M., 2018. Stability of two-species communities: drift, environmental stochasticity, storage effect and selection. Theor. Popul. Biol. 119: 57–71. 10.1016/j.tpb.2017.11.003 [DOI] [PubMed] [Google Scholar]
- Donaldson-Matasci M., Lachmann M., and Bergstrom C., 2008. Phenotypic diversity as an adaptation to environmental uncertainty. Evol. Ecol. Res. 10: 493–515. [Google Scholar]
- Ewens W. J., 1979. Mathematical Population Genetics. Springer-Verlag, NY. [Google Scholar]
- Fisher, D. S., 2007 Evolutionary Dynamics. In J.-P. Bouchaud, M. Mézard, and J. Dalibard (Eds.), Les Houches, Session LXXXV, Complex Systems. Elsevier, Amsterdam.
- Gardiner C. W., 1985. Handbook of Stochastic Methods for Physics, Chemistry and the Natural Sciences. Springer, NY. [Google Scholar]
- Gillespie D. T., 1976. A general method for numerically simulating the stochastic time evolution of coupled chemical reactions. J. Comput. Phys. 22: 403–434. [Google Scholar]
- Gillespie D. T., 1977. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 81: 2340–2361. [Google Scholar]
- Gomulkiewicz R. and Houle D., 2009. Demographic and genetic constraints on evolution. Am. Nat. 174: E218–E229. [DOI] [PubMed] [Google Scholar]
- Gonzalez A., Ronce O., Ferriere R., and Hochberg M. E., 2013. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120404 10.1098/rstb.2012.0404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greulich P., Waclaw B., and Allen R. J., 2012. Mutational pathway determines whether drug gradients accelerate evolution of drug-resistant cells. Phys. Rev. Lett. 109: 088101 10.1103/PhysRevLett.109.088101 [DOI] [PubMed] [Google Scholar]
- Gullberg E., Cao S., Berg O. G., Ilbäck C., Sandegren L. et al. , 2011. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. 7: e1002158 10.1371/journal.ppat.1002158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsen R., Deris J. B., and Hwa T., 2012. On the rapidity of antibiotic resistance evolution facilitated by a concentration gradient. Proc. Natl. Acad. Sci. USA 109: 10775–10780. 10.1073/pnas.1117716109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Hauert C., and Traulsen A., 2015. Stochastic game dynamics under demographic fluctuations. Proc. Natl. Acad. Sci. USA 112: 9064–9069. 10.1073/pnas.1418745112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufton P. G., Lin Y. T., Galla T., and McKane A. J., 2016. Intrinsic noise in systems with switching environments. Phys. Rev. E 93: 052119 10.1103/PhysRevE.93.052119 [DOI] [PubMed] [Google Scholar]
- Kopp M. and Hermisson J., 2007. Adaptation of a quantitative trait to a moving optimum. Genetics 176: 715–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M. and Hermisson J., 2009a The genetic basis of phenotypic adaptation I: fixation of beneficial mutations in the moving optimum model. Genetics 182: 233–249. 10.1534/genetics.108.099820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp M. and Hermisson J., 2009b The genetic basis of phenotypic adaptation II: the distribution of adaptive substitutions in the moving optimum model. Genetics 183: 1453–1476. 10.1534/genetics.109.106195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyos R. D., Metcalf J. E., Birger R., Klein E. Y., Abel zur P. et al. , 2014. The path of least resistance: aggressive or moderate treatment? Proc. Biol. Sci. 281: 20140566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E., Leibler S., and Grosberg A., 2006. Polymer-population mapping and localization in the space of phenotypes. Phys. Rev. Lett. 97: 068101 10.1103/PhysRevLett.97.068101 [DOI] [PubMed] [Google Scholar]
- Levin-Reisman I., Ronin I., Gefen O., Braniss I., Shoresh N., and Balaban N. Q., 2017. Antibiotic tolerance facilitates the evolution of resistance. Science 355: 826–830. 10.1126/science.aaj2191 [DOI] [PubMed] [Google Scholar]
- Lin W. H. and Kussell E., 2016. Complex interplay of physiology and selection in the emergence of antibiotic resistance. Curr. Biol. 26: 1486–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey H. A., Gallie J., Taylor S., and Kerr B., 2013. Evolutionary rescue from extinction is contingent on a lower rate of environmental change. Nature 494: 463–467. 10.1038/nature11879 [DOI] [PubMed] [Google Scholar]
- Marrec L. and Bitbol A.-F., 2018. Quantifying the impact of a periodic presence of antimicrobial on resistance evolution in a homogeneous microbial population of fixed size. J. Theor. Biol. 457: 190–198. 10.1016/j.jtbi.2018.08.040 [DOI] [PubMed] [Google Scholar]
- Marrec L. and Bitbol A.-F., 2020. Resist or perish: fate of a microbial population subjected to a periodic presence of antimicrobial. PLOS Comput. Biol. 16: e1007798 10.1371/journal.pcbi.1007798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Aguilee R., Ramsayer J., Kaltz O., and Ronce O., 2013. The probability of evolutionary rescue: towards a quantitative comparison between theory and evolution experiments. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368: 20120088 10.1098/rstb.2012.0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszewski S., Hermisson J., and Kopp M., 2014. Fisher’s geometric model with a moving optimum. Evolution 68: 2571–2588. 10.1111/evo.12465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Mora T., Rivoire O., and Walczak A. M, 2017. Transitions in optimal adaptive strategies for populations in fluctuating environments. Phys Rev E 96: 032412. [DOI] [PubMed] [Google Scholar]
- Melbinger A., Cremer J., and Frey E., 2010. Evolutionary game theory in growing populations. Phys. Rev. Lett. 105: 178101 10.1103/PhysRevLett.105.178101 [DOI] [PubMed] [Google Scholar]
- Melbinger A. and Vergassola M., 2015. , The impact of environmental fluctuations on evolutionary fitness functions. Sci. Rep. 5: 15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer I. and Shnerb N. M., 2018. Noise-induced stabilization and fixation in fluctuating environment. Sci. Rep. 8: 9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustonen V. and Lässig M., 2008. Molecular evolution under fitness fluctuations. Phys. Rev. Lett. 100: 108101. [DOI] [PubMed] [Google Scholar]
- Nadeau C. P., Urban M. C., and Bridle J. R., 2017. Climates past, present, and yet-to-come shape climate change vulnerabilities. Trends Ecol. Evol. (Amst.) 32: 786–800. 10.1016/j.tree.2017.07.012 [DOI] [PubMed] [Google Scholar]
- Nahum J. R., Godfrey-Smith P., Harding B. N, Marcus J. H., Carlson-Stevermer J., and Kerr B., 2015. A tortoise-hare pattern seen in adapting structured and unstructured populations suggests a rugged fitness landscape in bacteria. Proc. Natl. Acad. Sci. USA 112: 7530–7535. 10.1073/pnas.1410631112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen-Meyer S, 1966. Analysis of effects of antibiotics on bacteria by means of stochastic models. Biometrics 22: 761–780. [Google Scholar]
- Orr H. A. and Unckless R. L., 2008. Population extinction and the genetics of adaptation. Am. Nat. 172: 160–169. [DOI] [PubMed] [Google Scholar]
- Ovaskainen O. and Meerson B., 2010. Stochastic models of population extinction. Trends Ecol. Evol. 25: 643–652. [DOI] [PubMed] [Google Scholar]
- Parzen E.1999. Stochastic processes. SIAM, Philadelphia. [Google Scholar]
- Patwa Z. and Wahl L. M., 2008. The fixation probability of beneficial mutations. J. R. Soc. Interface 5: 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauls S. U., Nowak C., Bálint M., and Pfenninger M., 2013. The impact of global climate change on genetic diversity within populations and species. Mol. Ecol. 22: 925–946. 10.1111/mec.12152 [DOI] [PubMed] [Google Scholar]
- Peischl S. and Kirkpatrick M., 2012. Establishment of new mutations in changing environments. Genetics 191: 895–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poelwijk F. J., Kiviet D. J., Weinreich D. M., and Tans S. J., 2007. Empirical fitness landscapes reveal accessible evolutionary paths. Nature 445: 383–386. 10.1038/nature05451 [DOI] [PubMed] [Google Scholar]
- Regoes R. R., Wiuff C., Zappala R. M., Garner K. N., Baquero F., and Levin B. R., 2004. Pharmacodynamic functions: a multiparameter approach to the design of antibiotic treatment regimens. Antimicrob. Agents Chemother. 48: 3670–3676. 10.1128/AAC.48.10.3670-3676.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivoire O., ad S., Leibler, 2011. The value of information for populations in varying environments. J. Stat. Phys. 142: 1124–1166. [Google Scholar]
- Rouzine I. M., Rodrigo A., and Coffin J. M., 2001. Transition between stochastic evolution and deterministic evolution in the presence of selection: general theory and application to virology. Microbiol. Mol. Biol. Rev. 65: 151–185. 10.1128/MMBR.65.1.151-185.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva V., Husain K., Sheng J., Wang S., and Murugan A., 2020. Tuning environmental timescales to evolve and maintain generalists. Proc. Natl. Acad. Sci. USA 117: 12693–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skanata A. and Kussell E., 2016. Evolutionary phase transitions in random environments. Phys. Rev. Lett. 117: 038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szendro I. G., Schenk M. F., Franke J., Krug J., and de Visser J. A. G. M., 2013. Quantitative analyses of empirical fitness landscapes. J. Stat. Mech. 2013: P01005. [Google Scholar]
- Teimouri H. and Kolomeisky A. B., 2019. Theoretical investigation of stochastic clearance of bacteria: first-passage analysis. J. R. Soc. Interface 16: 20180765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teimouri H., Kochugaeva M. P., and Kolomeisky A. B., 2019Elucidating the correlations between cancer initiation times and lifetime cancer risks. Sci. Rep. 9: 18940 10.1038/s41598-019-55300-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh V. and Priami C., 2015. Simulation of biochemical reactions with time-dependent rates by the rejection-based algorithm. J. Chem. Phys. 143: 054104. [DOI] [PubMed] [Google Scholar]
- Trubenová B., Krejca M. S., Lehre P. K., and Kötzing T., 2019. Surfing on the seascape: adaptation in a changing environment. Evolution 73: 1356–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uecker H. and Hermisson J., 2011. On the fixation process of a beneficial mutation in a variable environment. Genetics 188: 915–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uecker H., Otto S. P., and Hermisson J., 2014. Evolutionary rescue in structured populations. Am. Nat. 183: 17–35. 10.1086/673914 [DOI] [PubMed] [Google Scholar]
- Van Kampen, N. 1981 Stochastic Processes in Physics and Chemistry, North-Holland, Amsterdam. [Google Scholar]
- van Marle G., Gill M. J., Kolodka D., McManus L., Grant T., and Church D. L., 2007. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology 4: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhulst P.-F., 1838. Notice sur la loi que la population suit dans son accroissement. Curr. Math. Phys 110: 113. [Google Scholar]
- Wang S. and Dai L., 2019. Evolving generalists in switching rugged landscapes. PLOS Comput. Biol. 15: e1007320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo A. R., Huijben S., de Roode J. C., Shepherd J., and Read A. F., 2007. Competitive release and facilitation of drug-resistant parasites after therapeutic chemotherapy in a rodent malaria model. Proc. Natl. Acad. Sci. USA 104: 19914–19919. 10.1073/pnas.0707766104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman D., 2011. A Unified Treatment of the Probability of Fixation when Population Size and the Strength of Selection Change Over Time. Genetics 188: 907–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinreich D. M. and Chao L., 2005. Rapid evolutionary escape in large populations from local peaks on the Wrightian fitness landscape. Evolution 59: 1175–1182. [PubMed] [Google Scholar]
- Weissman D. B., Desai M. M., Fisher D. S., and Feldman M. W., 2009. The rate at which asexual populations cross fitness valleys. Theor. Popul. Biol. 75: 286–300. 10.1016/j.tpb.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienand K., Frey E., and Mobilia M., 2017. Evolution of a fluctuating population in a randomly switching environment. Phys. Rev. Lett. 119: 158301 10.1103/PhysRevLett.119.158301 [DOI] [PubMed] [Google Scholar]
- Wu Y., Saddler C. A., Valckenborgh F., and Tanaka M. M., 2014. Dynamics of evolutionary rescue in changing environments and the emergence of antibiotic resistance. J. Theor. Biol. 340: 222–231. 10.1016/j.jtbi.2013.09.026 [DOI] [PubMed] [Google Scholar]
- Yu G., Baeder D. Y., Regoes R. R., and Rolff J., 2018. Predicting drug resistance evolution: insights from antimicrobial peptides and antibiotics. Proc. Biol. Sci.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Lambert G., Liao D., Kim H., Robin K., Tung C., Pourmand N., and Austin R. H., 2011. Acceleration of emergence of bacterial antibiotic resistance in connected microenvironments. Science 333: 1764–1767. 10.1126/science.1208747 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article or Supplemental Material. Matlab implementations of numerical simulations available at https://doi.org/10.5281/zenodo.3993272. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12860975.