Sperm and oocyte production are differentially regulated to ensure genetic information is accurately passed down from one generation to the next.....
Keywords: BRC-1-BRD-1, crossovers, meiosis, recombination, sex, Genetics of Sex
Abstract
Meiosis is regulated in a sex-specific manner to produce two distinct gametes, sperm and oocytes, for sexual reproduction. To determine how meiotic recombination is regulated in spermatogenesis, we analyzed the meiotic phenotypes of mutants in the tumor suppressor E3 ubiquitin ligase BRC-1-BRD-1 complex in Caenorhabditis elegans male meiosis. Unlike in mammals, this complex is not required for meiotic sex chromosome inactivation, the process whereby hemizygous sex chromosomes are transcriptionally silenced. Interestingly, brc-1 and brd-1 mutants show meiotic recombination phenotypes that are largely opposing to those previously reported for female meiosis. Fewer meiotic recombination intermediates marked by the recombinase RAD-51 were observed in brc-1 and brd-1 mutants, and the reduction in RAD-51 foci could be suppressed by mutation of nonhomologous-end-joining proteins. Analysis of GFP::RPA-1 revealed fewer foci in the brc-1 brd-1 mutant and concentration of BRC-1-BRD-1 to sites of meiotic recombination was dependent on DNA end resection, suggesting that the complex regulates the processing of meiotic double-strand breaks to promote repair by homologous recombination. Further, BRC-1-BRD-1 is important to promote progeny viability when male meiosis is perturbed by mutations that block the pairing and synapsis of different chromosome pairs, although the complex is not required to stabilize the RAD-51 filament as in female meiosis under the same conditions. Analyses of crossover designation and formation revealed that BRC-1-BRD-1 inhibits supernumerary COs when meiosis is perturbed. Together, our findings suggest that BRC-1-BRD-1 regulates different aspects of meiotic recombination in male and female meiosis.
MEIOSIS is essential for sexual reproduction and results in the precise halving of the genome for packaging into gametes. Chromosomes must be accurately segregated during meiosis to ensure that the next generation has the correct genomic complement. In metazoans with defined sexes, the products of meiosis—sperm and oocytes—contribute not only haploid genomes but also unique cellular components to support embryonic development. In addition to the striking morphological differences between sperm and oocytes, the process of meiosis itself exhibits extensive sexual dimorphism with respect to the temporal program of events, the extent and placement of recombination, checkpoint signaling, chromosome segregation, and sex chromosome behavior (Morelli and Cohen 2005; Turner 2007; Nagaoka et al. 2012; Bury et al. 2016; Cahoon and Libuda 2019). However, the underlying mechanisms governing these differences are not well understood.
Meiotic chromosome segregation relies on establishing connections between homologous chromosomes. In most organisms, this is accomplished by the intentional induction of hundreds of double-strand breaks (DSBs) by the conserved topoisomerase Spo11 (Keeney et al. 1997; Dernburg et al. 1998). A subset of meiotic DSBs use a nonsister chromatid as template for repair by homologous recombination (HR) to generate crossovers (COs) that ensure disjunction and promote genetic variation. In almost all animals and plants where it has been examined, COs differ in number, placement, and spacing in the sexes (Lenormand and Dutheil 2005; Gruhn et al. 2013; Stapley et al. 2017; Kianian et al. 2018; Lloyd and Jenczewski 2019).
Knowledge is lacking with respect to the contributions of different pathways to repair of DSBs not destined to form COs and whether their use differs in the sexes. During Caenorhabditis elegans and Drosophila oogenesis, the nonhomologous end joining (NHEJ) pathway for DSB repair is actively inhibited early in meiosis (Joyce et al. 2012; Lemmens et al. 2013; Yin and Smolikove 2013; Lawrence et al. 2016; Girard et al. 2018) but NHEJ and other pathways, including theta-mediated end-joining and single-strand annealing, serve as backups to ensure that all DSBs are repaired in late pachytene before the meiotic divisions (Smolikov et al. 2007; Macaisne et al. 2018). A recent study examining the repair of DNA breaks induced by radiation suggests that mouse spermatocytes switch to a somatic-like repair mode at pachytene, temporarily engaging NHEJ and then HR to repair the damage (Enguita-Marruedo et al. 2019). Interestingly, studies in juvenile male mice suggest that structure-specific nucleases may resolve processed DSBs at the expense of the canonical CO pathway, leading to higher levels of meiotic chromosome mis-segregation (Zelazowski et al. 2017).
Male meiosis in many species has the added challenge of the presence of heteromorphic sex chromosomes. Meiotic DSBs are induced on hemizygous regions of sex chromosomes (Ashley et al. 1995; Moens et al. 1997; Sciurano et al. 2006; Jaramillo-Lambert and Engebrecht 2010), yet they are unable to participate in CO formation due to a lack of a homolog. In C. elegans and the related nematode, Caenorhabditis briggsae, HR using the sister chromatid as repair template, and alternative repair pathways are engaged to repair meiotic DSBs induced on the completely hemizygous X chromosome of males (Checchi et al. 2014; Van et al. 2016). The presence of hemizygous sex chromosomes also complicates analyses of meiotic recombination in mammals as inactivation of many recombination genes impairs meiotic sex chromosome inactivation (MSCI). MSCI is the process whereby hemizygous regions of sex chromosomes acquire heterochromatin marks and are transcriptionally silenced (Turner 2007). MSCI is required for efficient meiotic progression in males, as failure to inactivate sex chromosomes results in elevated apoptosis and elimination of germ cells (Mahadevaiah et al. 2008; Royo et al. 2010).
C. elegans has emerged as an excellent model for meiotic studies, including investigations into the sex-specific regulation of meiotic events. Both the C. elegans hermaphrodite and male germ lines are arranged in a spatiotemporal gradient that in combination with available molecular markers enables recombination progression to be monitored through all stages of meiotic prophase (Shakes et al. 2009; Lui and Colaiacovo 2013; Hillers et al. 2015) (Figure 2A). Additionally, the lack of absolute interdependence of recombination initiation and chromosome synapsis also facilitates analyses of meiotic mutants. C. elegans exists predominantly as a self-fertilizing hermaphrodite (XX); during development, hermaphrodites initially produce sperm and then switch to oocyte production, and thus as adults are functionally female. Males (X0) arise spontaneously due to X chromosome nondisjunction.
Figure 2.
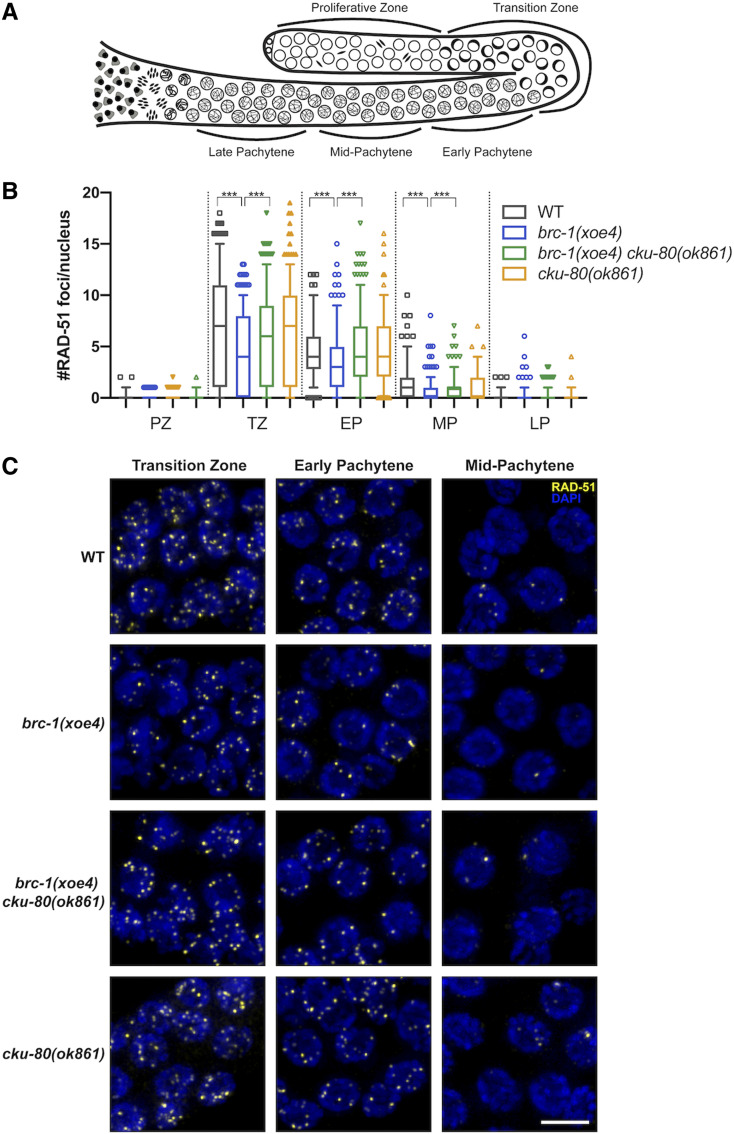
BRC-1-BRD-1 promotes HR at the expense of NHEJ in the male germ line. (A) Cartoon of the spatiotemporal organization of the C. elegans male germ line, modified from Van et al. (2016). (B) Quantification of RAD-51 in indicated regions of the germ line. Box whisker plots show number of RAD-51 foci per nucleus in the different regions. Horizontal line of each box represents the median, top and bottom of each box represents medians of upper and lower quartiles, lines extending above and below boxes indicate SD, and individual data points are outliers from 5 to 95%. Statistical comparisons by Mann–Whitney of WT vs. brc-1(xoe4) and brc-1(xoe4) vs. brc-1(xoe4) cku-80(ok861) in the different regions of the germ line; ***P < 0.0001. All statistical comparisons are shown in Table S3. PZ, proliferative zone; TZ, transition zone; EP, early pachytene; MP, mid-pachytene; LP, late pachytene. Number of germ lines and nuclei scored in each region: WT = 6, PZ = 958; TZ = 413; EP = 266; MP = 252; LP = 219; brc-1(xoe4) = 6, PZ = 848; TZ = 343; EP = 320; MP = 330; LP = 287; brc-1(xoe4) cku-80(ok861) = 6, PZ = 905; TZ = 316; EP = 296; MP = 329; LP = 289; cku-80(ok861) = 4, PZ = 814; TZ = 287; EP = 202; MP = 230; LP = 217. (C) Representative images of nuclei from indicated genotypes and regions of the germ line stained with antibodies against RAD-51 (yellow) and counterstained with DAPI (blue). Images are projections through half of the gonad. Bar, 5 μm.
The hemizygous X chromosome of C. elegans male germ cells undergoes modifications similar to the hemizygous regions of the X and Y of mammalian spermatocytes, including accumulation of repressive chromatin marks resulting in transcriptional silencing (Kelly et al. 2002; Reuben and Lin 2002; Bean et al. 2004; Maine 2010). A C. elegans SETBD1 histone methyltransferase—an ortholog of which has been shown to mediate MSCI in mammals (Hirota et al. 2018)—and a small RNA pathway are important for silencing the X chromosome of male germ cells (She et al. 2009; Bessler et al. 2010; Checchi and Engebrecht 2011). However, the role of many components required for MSCI in mammals, including the tumor suppressor E3 ubiquitin ligase BRCA1 and master checkpoint kinase ATR (Turner et al. 2004; Royo et al. 2013; Broering et al. 2014), have not been analyzed in C. elegans. Here, we examined the requirement for BRCA1-BARD1 (BRC-1-BRD-1) and ATR (ATL-1) in meiotic silencing in C. elegans. Surprisingly our studies revealed that in contrast to mammals, C. elegans BRC-1-BRD-1 is not essential for MSCI. However, X chromosome transcriptional silencing is impaired in the absence of ATL-1, suggesting that while meiotic silencing is conserved, the pathways mediating MSCI have evolved independently. We also found that the meiotic phenotypes of male brc-1 and brd-1 mutants are different than those previously reported in female meiosis (Boulton et al. 2004; Adamo et al. 2008; Janisiw et al. 2018; Li et al. 2018), providing further evidence that recombination is regulated differently in spermatogenic vs. oogenic germ cells (Jaramillo-Lambert and Engebrecht 2010; Checchi et al. 2014). We propose that BRC-1-BRD-1 functions at an early step of meiotic DSB repair in male meiosis, which is similar to one of its established somatic roles in promoting HR at the expense of NHEJ. Additionally, this complex alters the CO landscape when meiosis is perturbed by inhibiting supernumerary COs, rather than promoting extra COs as in female meiosis. Together, our findings indicate that the processing of meiotic DSBs and the regulation of CO patterning are regulated in a sex-specific manner in C. elegans.
Materials and Methods
Genetics
C. elegans var. Bristol (N2), was used as the wild-type strain. Other strains used in this study are listed in Supplemental Materials, Table S1. Some nematode strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources (NIH NCRR). Strains were maintained at 20°.
CRISPR-mediated generation of alleles
zim-3(xoe15) was generated in the Bristol background using guides tacgcctgagaacatgtttt and aaaagatcgtgtgatggtcc with repair template: gtaaataacggttgtcgatacgcctgagaacatgtttttggacatttatcttttctagtaggtttttccatatactttattttattctgaagtttag to delete most of the coding sequence except for exon 7 and 8. External primers cacgacgacaccctcatgta and ttgtgcagagtcgtagcgaa and internal primers cacgacgacaccctcatgta and gctcgtgtacattgagccct were used to genotype for zim-3(xoe15). brc-1(xoe4) was introduced into the Hawaiian background (CB4856) using primers, guides and repair template as described (Li et al. 2018). zim-1(xoe6) was generated in the Bristol and Hawaiian background using guides tccaatcatcacaagtcatc and attcgatgagcttcgtcgtc with repair template tttaaaaatgcagttttaaaagtgtttcattgtcattttatattttccaggcttcgtcgtcgggccgtctgctttttgtaaattgtgtctcatgtgttat to delete the entire coding sequence. External primers cacacatttggctggggtct and atgggcagcagcaagaaagt, and internal primers gctccgtctgcacaaatcct and gttgaaaagcggggaacacc were used to identify zim-1(xoe6). Worms were outcrossed a minimum of two times and analyzed phenotypically by examining progeny viability to confirm correct editing.
Embryonic lethality of male-sired progeny
A single fog-2(q71) female was mated with three males of indicated genotypes on small Escherichia coli OP-50 spots. The mated female was transferred to new plates every 24 hr. Embryonic lethality was determined over 3 days by counting eggs and hatched larvae 24 hr after removing the female and calculating percent as eggs/(eggs + larvae). The progeny of a minimum of 10 mated females were scored.
Cytological analyses
Immunostaining of germ lines was performed as described (Jaramillo-Lambert et al. 2007) except that slides were incubated in 100% ethanol instead of 100% methanol for direct green fluorescent protein (GFP) fluorescence of GFP::COSA-1. The following primary antibodies were used at the indicated dilutions: rabbit anti-Pol2-S2P (1:500; cat #ab5059; RRID: AB_304749; Abcam, Cambridge, MA), rabbit anti-HIM-8 (1:500; cat #4198.00.02; SDIX; Newark, DE; RRID: AB_2616418), rabbit anti-histone H3K4me2 (1:500; cat# 9725; Cell Signaling Technology; Danvers, MA; RRID: AB_10205451), mouse anti-histone H3K9me2 (1:500; Cat# 9753; RRID: AB_659848; AbCam), mouse anti-Pol2-S5P H14 (1:500; cat# MMS-134R; RRID: AB_10119940; Covance, Princeton, NJ), rabbit anti-RAD-51 (1:10,000; cat #2948.00.02; SDIX; RRID: AB_2616441), mouse anti-GFP (1:500; cat #632375; BD Biosciences; San Jose, CA). Secondary antibodies Alexa Fluor 594 donkey anti-rabbit IgG, Alexa Fluor 594 goat anti-mouse IgG, Alexa Fluor 488 goat anti-rabbit IgG, and Alexa Fluor 488 goat anti-mouse IgG from Life Technologies were used at 1:500 dilutions. DAPI (4′,6-diamidino-2-phenylindole; 2 μg/ml; Sigma-Aldrich) was used to counterstain DNA.
Collection of fixed images was performed using an API Delta Vision or an API Delta Vision Ultra deconvolution microscope equipped with an 60×, NA 1.49 objective lens, and appropriate filters for epifluorescence. Z stacks (0.2 μm) were collected from the entire gonad. A minimum of three germ lines was examined for each condition. Images were deconvolved using Applied Precision SoftWoRx batch deconvolution software and subsequently processed and analyzed using Fiji (ImageJ) (Wayne Rasband, NIH).
Quantification of H3K9me2 enrichment on the X chromosome was performed by examining deconvolved three-dimensional (3D) data stacks and binning mid- to late-pachytene nuclei into three categories: enrichment = single strong track of H3K9me2 associated with HIM-8; partial enrichment = diffuse H3K9me2 signal associated with HIM-8; no enrichment = multiple H3K9me2 signals with no HIM-8 association. To quantitate the transcriptional status of the X chromosome in wild type (three germ lines) and the atl-1 mutant (six germ lines), mid- to late pachytene nuclei with a single HIM-8-marked chromosome were examined in deconvolved 3D data stacks for the presence of Pol2-S5P labeling.
RAD-51 foci were quantified in a minimum of three germ lines of age-matched males (18–24 hr post-L4). We divided germ lines into the transition zone (leptotene/zygotene), as counted from the first and last row with two or more crescent-shaped nuclei, and then divided pachytene into three equal parts: early, mid and late (Figure 2A). RAD-51 were quantified from half projections of the germ lines. The number of foci per nucleus was scored for each region.
To assess formation of RAD-51 foci following ionizing radiation (IR) treatment, 18–24 hr post-L4 male worms were exposed to 10 Grays (Gys) of IR; 1 hr post-IR, worms were dissected and gonads fixed for immunofluorescence as above.
GFP::COSA-1 foci were quantified from deconvolved 3D data stacks; late pachytene nuclei were scored individually through z-stacks to ensure that all foci within each individual nucleus were counted.
For live cell imaging (Figure 3, A and C), 18–24 hr post L4 males were anesthetized in 1 mM tetramisole (Sigma-Aldrich) and immobilized between a coverslip and a 2.5% agarose pad on a glass slide. Z-stacks (0.33 μm) were captured on a spinning-disk module of an inverted objective fluorescence microscope [Marianas spinning-disk confocal (SDC) real-time 3D Confocal-TIRF (total internal reflection) microscope; Intelligent Imaging Innovations] with a 100×, 1.46 numerical aperture objective, and a Photometrics QuantiEM electron multiplying charge-coupled device (EMCCD) camera. Z-projections of ∼20–30 z-slices were generated, cropped, and adjusted for brightness in Fiji. GFP::RPA-1 fluorescence was quantified by measuring the mean fluorescence intensity and SD in Fiji for individual nuclei [region of interest (ROI)] in transition zone to mid-pachytene. Coefficient of variation (CV) is defined as SD of intensity divided by mean intensity (Bishop et al. 2015). The CV describes the dispersion of pixel intensity values from a 2D ROI around the mean pixel intensity such that nuclei with more distinct foci will have high CV values, whereas nuclei with more uniform fluorescence will have low CV values.
Figure 3.
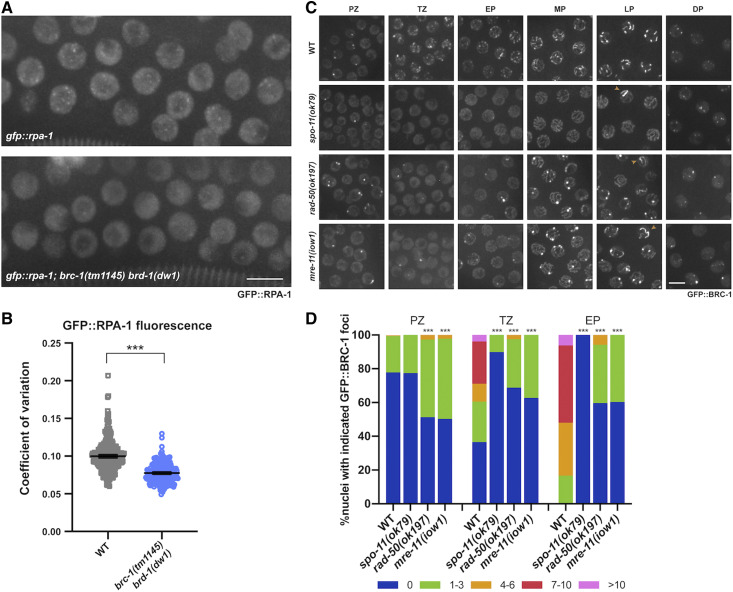
GFP::RPA-1 foci are reduced in the brc-1 brd-1 mutant and GFP::BRC-1 concentration at foci in early meiotic prophase is dependent on meiotic DSB resection. (A) High-magnification images of wild-type and brc-1(tm1145) brd-1(dw1) transition zone/early pachytene nuclei in live worms expressing GFP::RPA-1. Images are projections through half of the gonad. Bar, 5 μm. (B) Coefficient of variation (SD/mean fluorescent intensity) of GFP::RPA-1 fluorescence is shown; six germ lines were analyzed for each genotype. Statistical comparisons between WT and brc-1(tm1145) brd-1(dw1) by Mann–Whitney: ***P < 0.0001. (C) Images of germ cells from live worms expressing GFP::BRC-1 from the indicated genetic backgrounds and gonad regions (PZ, proliferative zone; TZ, transition zone; EP, early pachytene; MP, mid-pachytene, LP, late pachytene, DP, diplotene). Images are projections through half of the gonad. Bar, 5 μm. (D) Number of GFP::BRC-1 foci in PZ, TZ, and EP in wild type and mutants. Numbers were binned as 0, 1–3, 4–6, 7–10, >10. A minimum of three germ lines were quantified for each genotype. Statistical comparisons between WT and mutants by Mann–Whitney: ***P < 0.0001. spo-11(ok79) is statistically different than either rad-50(ok197) or mre-11(iow1): PZ: P < 0.0001; TZ: spo-11(ok79) vs. rad-50(ok197) P = 0.0002; spo-11(ok79) vs. mre-11(iow1) P < 0.0001; EP: spo-11(ok79) vs. rad-50(ok197) P < 0.0001; spo-11(ok79) vs. mre-11(iow1) P = 0.0004.
Meiotic mapping
Meiotic CO frequencies and distribution were assayed using single-nucleotide polymorphism (SNP) markers as in Nabeshima et al. (2004). The SNP markers located at the boundaries of the chromosome domains were chosen based on data from WormBase (WS231), Bazan and Hillers (2011) and Saito et al. (2013). Markers and primers used are listed in Table S2. Hawaiian strain CB4856 males carrying each mutation were crossed to the same mutant strain in the Bristol background. Among the progeny of this cross, male worms were plated individually and crossed to two fog-2(q71) female worms in the Bristol background. Upon successful mating, embryos (Smolikov et al. 2008) together with larva up to L4 stage were collected individually and stored at −80°. Since all three mutant (brc-1, zim-1, brc-1;zim-1) hermaphrodites produce self-fertilized male progeny, the identity of the hybrid Bristol/Hawaiian male was confirmed by PCR, and restriction digest before the collected samples were used for further analysis: individuals were lysed in 5 μl of lysis buffer (50 mM KCl, 10 mM Tris pH 8.2, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween20, 0.01% gelatin; 60 μg of proteinase K/ml was added before use) and diluted to 50 μl volume with molecular biology grade water. PCR was performed using 3–5 μl diluted lysate with Phusion or Taq polymerase in a 15 μl reaction. Half volume of the PCR products was digested overnight with appropriate restriction enzyme and analyzed on 1–2.5% agarose gels. Double crossovers (DCOs) were confirmed either with additional SNPs by a distinctive restriction enzyme digest or by repeating PCR and digestion if no additional SNPs were available for the marker as described in Saito et al. (2013) (Table S2).
Statistical analyses
Statistical analyses and figures were prepared using GraphPad Prism version 8.0 (GraphPad Software). Statistical comparisons of H3K9me2 association with HIM-8 (Figure 1C), absence of Pol2-S5P on HIM-8-marked chromosomes (Figure 1E), RAD-51 (Figure 2B and Figure 5B, and Figure S1A), GFP::RPA-1 fluorescence (Figure 3B), GFP::BRC-1 (Figure 3D), and GFP::COSA-1 foci numbers (Figure 6A) were analyzed by Mann-Whitney. Embryonic lethality (Figure 5A) was analyzed by one-way ANOVA. Fisher exact test on a 2×2 contingency table was used for statistical analyses on genetic map distance, distribution and % multi-COs (Figure 7, B–D). For statistical analyses of interference, χ2 tests on 2×2 contingency tables of observed and expected DCOs were performed (Brady et al. 2018). Detailed descriptions of statistical analyses are indicated in figure legends.
Figure 1.
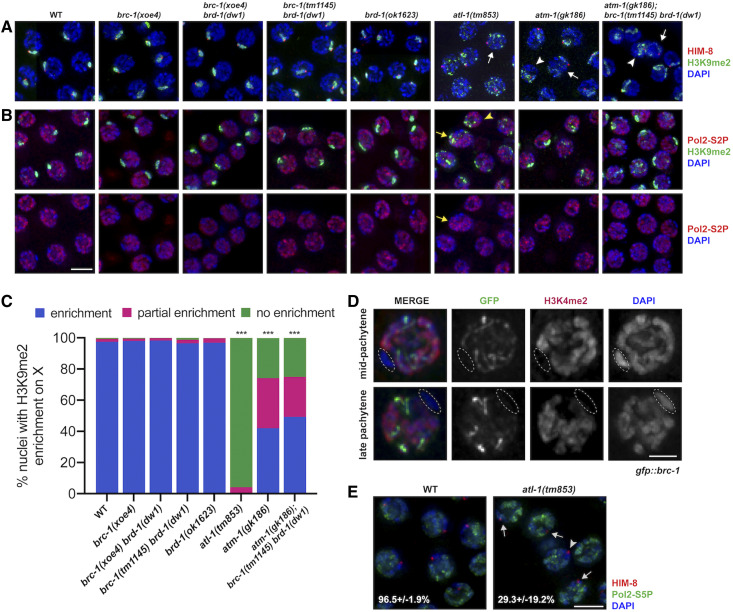
BRC-1-BRD-1 is not required for MSCI. Pachytene nuclei from C. elegans wild-type and indicated mutant male germ lines labeled with (A) anti-H3K9me2 (green; repressive chromatin), anti-HIM-8 (red; X chromosome marker), and counterstained with DAPI (blue); white arrows mark HIM-8 chromosomes largely lacking H3K9me2 while white arrowheads mark HIM-8 chromosomes with diffuse H3K9me2 labeling, or (B) anti-H3K9me2 (green), anti-Pol2-S2P (red; actively transcribing RNA polymerase II), and counterstained with DAPI (blue); lower panel shows anti-Pol2-S2P and DAPI; yellow arrows mark chromatin with both H3K9me2 and Pol2-S2P labeling while yellow arrowheads mark chromatin with neither H3K9me2 nor Pol2-S2p labeling. Images are projections through half of the gonad. Bar, 5 μm. (C) Quantification of enrichment of H3K9me2 on the X chromosome; enrichment = single strong track of H3K9me2 associated with HIM-8 (blue); partial enrichment = diffuse H3K9me2 signal associated with HIM-8 [arrowhead in (A)] (red); no enrichment = multiple H3K9me2 signals with no clear HIM-8 association [arrow in (A)] (green). Statistical comparisons between WT and mutants by Mann-Whitney: ***P < 0.0001. atm-1(gk186) and atm-1(gk186); brc-1(tm1145) brd-1(dw1) were also statistically different from atl-1(tm853) (P < 0.0001). Number of germ lines, nuclei scored: WT = 3, 433; brc-1(xoe4) = 5, 398; brc-1(xoe4) brd-1(dw1) = 6, 654; brc-1(tm1145) brd-1(dw1) = 3, 257; brd-1(ok1623) = 6, 816; atl-1(tm853) = 4, 341; atm-1(gk186) = 3, 333; atm-1(gk186); brc-1(tm1145) brd-1(dw1) = 7, 613. (D) GFP::BRC-1 (green) only localizes to synapsed chromosomes and does not localize to the single X chromosome in male meiotic nuclei. X chromosome (circled) identified by chromosome morphology and lack of anti-H3K4me2 staining (red); nuclei counterstained with DAPI (blue). Bar, 2 μm. (E) Pachytene nuclei labeled with anti-HIM-8 (red; X chromosome marker), anti-Pol2-S5P (green; marking transcriptionally competent chromatin) and counterstained with DAPI (blue); %± SD nuclei containing a X chromosome lacking Pol2-S5P labeling is indicated [arrowhead denotes nucleus without Pol2-S5P on X chromosome; arrows denote nuclei with X chromosome containing Pol2-S5P labeling in atl-1(tm853)]. Bar, 5 μm. Number of germ lines, nuclei scored: WT = 3, 162; atl-1(tm853) = 6, 182. Statistical comparisons between WT and atl-1(tm853) by Mann-Whitney, P = 0.0121.
Figure 5.
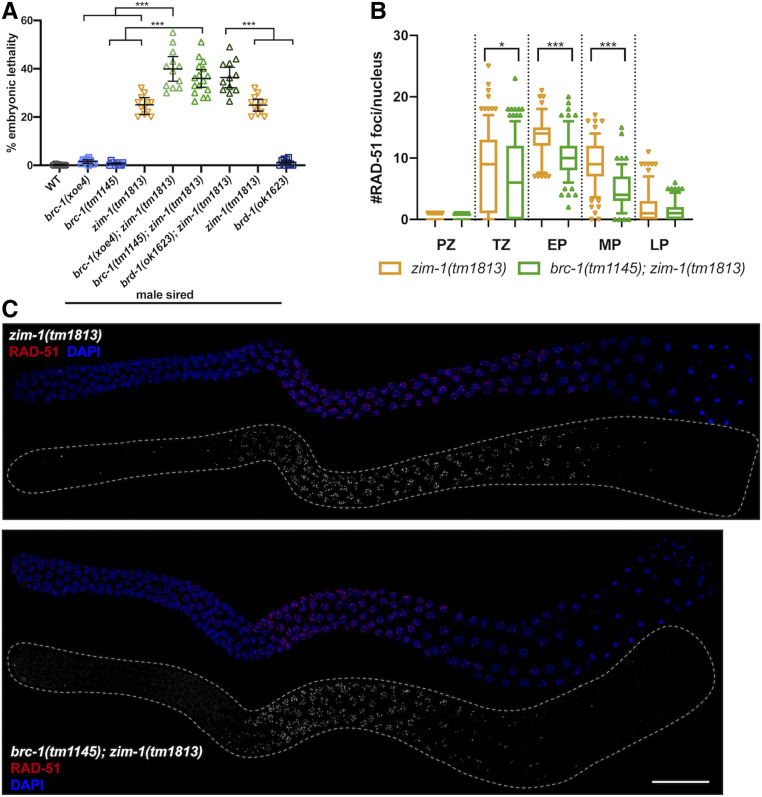
Progeny embryonic lethality is enhanced when sired by brc-1; zim-1 or brd-1; zim-1 double mutant males but RAD-51 stability in not impaired. (A) Embryonic lethality of fog-2(q71) progeny sired by brc-1(xoe4), brc-1(tm1145), zim-1(tm1813), brc-1(xoe4); zim-1(tm1813), brc-1(tm1145); zim-1(tm1813), brd-1(ok1623); zim-1(tm1813), brd-1(ok1623) males. Mean and 95% confidence intervals are shown. The genetic interaction between brc-1 or brd-1 and zim-1 is significant by a one-way ANOVA (***P < 0.0001). A minimum of 10 worms were scored for each genotype. (B) Box whisker plots show average number of RAD-51 foci per nucleus in the different zones. Horizontal line of each box indicates the median, the top and bottom of the box indicates medians of upper and lower quartiles, lines extending above and below boxes indicate SD and individual data points are outliers from 5 to 95%. Statistical comparisons by Mann–Whitney of zim-1(tm1813) vs. brc-1(tm1145); zim-1(tm1813) in the different regions of the germ line: *P < 0.05; ***P < 0.0001. PZ, proliferative zone; TZ, transition zone; EP, early pachytene; MP, mid-pachytene; LP, late pachytene. Numbers of nuclei scored from four germ lines in each zone for zim-1: PZ = 668; TZ = 237; EP = 111; MP = 151; LP = 167 and brc-1; zim-1: PZ = 545; TZ = 318; EP = 155; MP = 137; LP = 149. (C) zim-1(tm1813) and brc-1(tm1145); zim-1(tm1813) mutant germ lines stained with anti-RAD-51 antibody (red) and counterstained with DAPI (blue). Images are projections through half of the gonad. A minimum of four germ lines were imaged. Bar, 20 μm.
Figure 6.
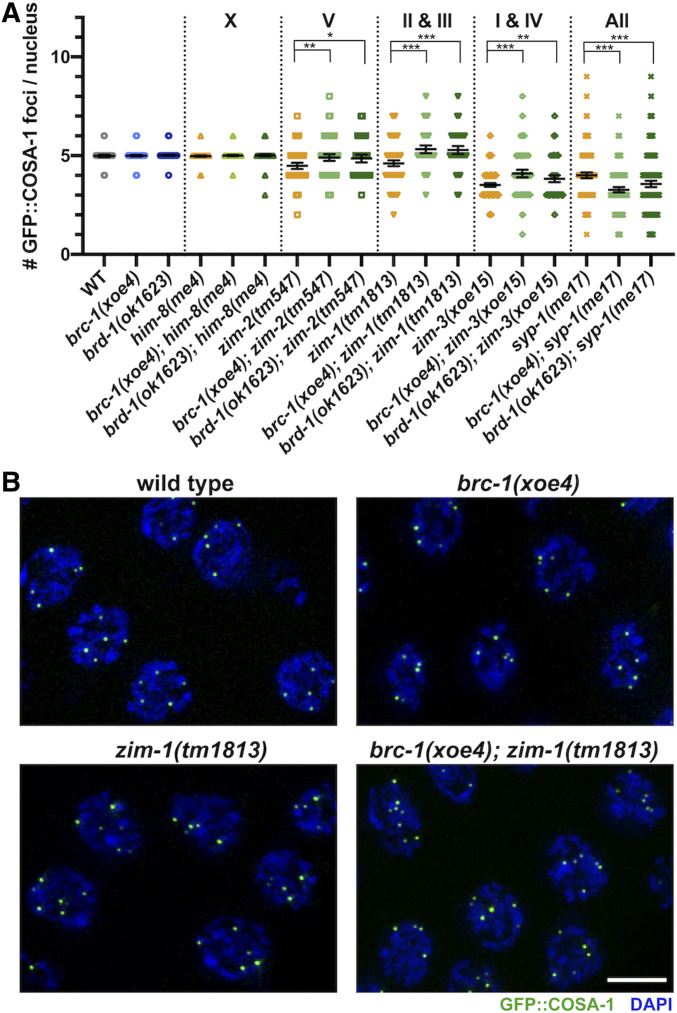
BRC-1-BRD-1 inhibits GFP::COSA-1 marked crossover (CO) precursors when a subset of chromosomes fails to form COs. (A) Number of COSA-1 foci in mid- to late-pachytene in indicated mutants; mean and 95% confidence intervals are shown. Letters/numbers above graph indicate which chromosomes are asynapsed in the different mutants. Statistical comparisons by Mann–Whitney *P < 0.05; **P < 0.001; ***P < 0.0001. Number of nuclei scored: gfp::cosa-1 = 97, gfp::cosa-1; brc-1(xoe4) = 194, gfp::cosa-1; brd-1(ok1623) = 103, gfp::cosa-1; him-8(me4) = 151, gfp::cosa-1; brc-1(xoe4); him-8(me4) = 183; gfp::cosa-1; brd-1(ok1623); him-8(me4) = 172, gfp::cosa-1; zim-2(tm547) = 125, gfp::cosa-1; brc-1(xoe4); zim-2(tm547) = 128; gfp::cosa-1; brd-1(ok1623); zim-2 = 84, gfp::cosa-1; zim-1(tm1813) = 120, gfp::cosa-1; brc-1(xoe4); zim-1(tm1813) = 100, gfp::cosa-1; brd-1(ok1623); zim-1(tm1813) = 97, gfp::cosa-1; zim-3(xoe15) = 308, gfp::cosa-1; brc-1(xoe4); zim-3(xoe15) = 133, gfp::cosa-1; brd-1(ok1623); zim-3(xoe15) = 145, gfp::cosa-1; syp-1(me17) = 271, gfp::cosa-1; brc-1(xoe4); syp-1(me17) = 281, gfp::cosa-1; brd-1(ok1623); syp-1(me17) = 344. (B) Half projections of late pachytene region showing GFP::COSA-1 (green) and DAPI (blue) in wild type, brc-1(xoe4), zim-1(tm1813) and brc-1(xoe4); zim-1(tm1813). Bar, 5 μm.
Figure 7.
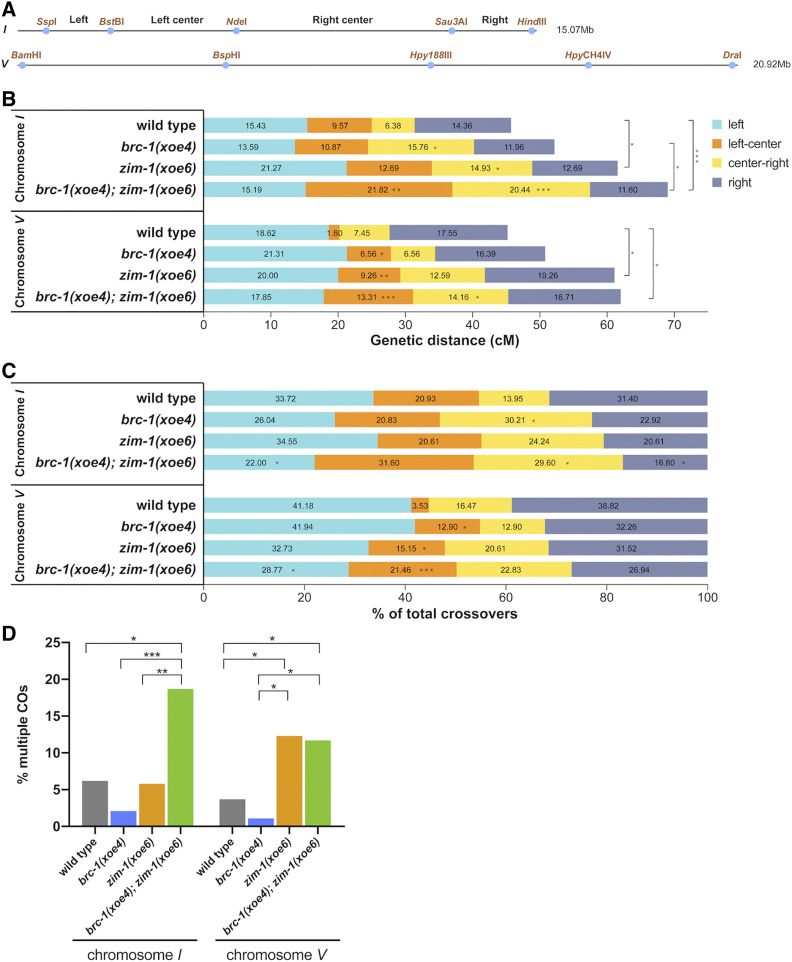
BRC-1 alters the CO landscape in the zim-1 mutant during male meiosis. (A) SNP markers on chromosome I and V used for genotyping; primers and additional information are included in Table S2. (B) CO frequency on chromosome I in wild type (n = 188), brc-1(xoe4) (n = 184), zim-1(xoe6) (n = 268) and brc-1(xoe4); zim-1(xoe6) (n = 362) mutants and on chromosome V in wild type (n = 188), brc-1(xoe4) (n = 183), zim-1(xoe6) (n = 270) and brc-1(xoe4); zim-1(xoe6) (n = 353) mutants. n = number of individuals analyzed per genotype. (C) CO distribution among recombinants on chromosome I and V in wild type, brc-1(xoe4), zim-1(xoe6), and brc-1(xoe4); zim-1(xoe6) mutants. (D) Percent of recombinant chromosomes containing multiple COs calculated as 100 × (DCO + TCOs)/(SCO + DCOs + TCOs). Statistical analyses were conducted using Fisher exact test on 2 × 2 contingency tables, *P < 0.05; **P < 0.001; ***P < 0.0001.
Data availability
Strains and reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12730904.
Results
C. elegans BRC-1-BRD-1 is not required for MSCI
During C. elegans meiosis, the X chromosome accumulates the repressive chromatin mark histone H3 lysine nine dimethylation (H3K9me2) and is transcriptionally silenced similar to MSCI in mammals (Kelly et al. 2002; Reuben and Lin 2002; Bean et al. 2004; Checchi and Engebrecht 2011). In mice, the E3 ubiquitin ligase BRCA1, critical for DNA damage response, is essential for MSCI. As a result, brca1−/− mutant male germ cells inappropriately express X-linked genes leading to pachytene arrest, apoptosis of spermatocytes and infertility (Xu et al. 2003; Turner et al. 2004; Broering et al. 2014). To determine whether C. elegans BRC-1 or its binding partner BRD-1 (Boulton et al. 2004) plays a role in MSCI, we labeled male brc-1, brd-1, and brc-1 brd-1 double mutant germ lines [brc-1(xoe4), brd-1(ok1623), brc-1(xoe4) brd-1(dw1) and brc-1(tm1145) brd-1(dw1) (Polanowska et al. 2006; Janisiw et al. 2018; Li et al. 2018)] with antibodies against H3K9me2 and the X-specific pairing center binding protein HIM-8 (Phillips and Dernburg 2006). The X chromosome, marked by HIM-8, was highly enriched for H3K9me2 in all of the brc-1 and brd-1 mutant combinations, as in wild type, suggesting that enrichment of this repressive chromatin mark on the X chromosome occurs in the absence of BRC-1 and/or BRD-1 (Figure 1, A and C). To examine the transcriptional status of the X chromosome, we colabeled germ lines with antibodies that recognize H3K9me2 and RNA polymerase II phosphorylated on serine 2 (Pol2-S2P), which is associated with transcriptional elongation (Hsin and Manley 2012), and for which we and others previously showed is excluded from the single X chromosome in male germ cells (Kelly et al. 2002; Larson et al. 2016). Pol2-S2P was present throughout the nucleus except for a single track, marked by H3K9me2, in all brc-1 and brd-1 mutants (Figure 1B), suggesting that the X chromosome is transcriptionally silenced in the absence of BRC-1-BRD-1.
In mammals, BRCA1 is observed on asynapsed axes and is enriched on the X–Y sex body (Turner et al. 2004). In C. elegans hermaphrodites, BRC-1 and BRD-1 become associated with fully synapsed chromosomes in pachytene (Polanowska et al. 2006; Janisiw et al. 2018; Li et al. 2018). We examined the localization of BRC-1 in male germ lines expressing an endogenously tagged and fully functional GFP fusion (GFP::BRC-1; Li et al. 2018) and found that it was also associated with tracks corresponding to synapsed chromosomes at pachytene. However, in contrast to the six tracks observed in oocytes, only five tracks were present in spermatocytes, suggesting that BRC-1-BRD-1 does not localize to the asynapsed X chromosome. To verify this, we colabeled male germ lines with antibodies against GFP, to detect GFP::BRC-1, and the activating chromatin mark, H3K4me2, which is enriched on all chromosomes except the X (Reuben and Lin 2002; Bean et al. 2004; Jaramillo-Lambert and Engebrecht 2010; Checchi and Engebrecht 2011), and found that the chromosome lacking H3K4me2 also lacked GFP::BRC-1 (Figure 1D). Thus, contrary to mammals, C. elegans BRC-1-BRD-1 is not enriched on asynapsed sex chromosomes in male germ cells.
During mammalian MSCI, BRCA1 facilitates the recruitment of the phosphoinositide 3-kinase ataxia telangiectasia and RAD3-related (ATR) kinase to sex chromosomes; ATR in turn phosphorylates the histone variant H2AX (γ-H2AX) to facilitate chromosome compaction. Consequently, inactivation of either ATR or H2AX also results in MSCI failure (Fernandez-Capetillo et al. 2003; Turner et al. 2004; Royo et al. 2013). Given that BRC-1-BRD-1 is not essential for MSCI, and no H2AX variant has been identified in the C. elegans genome (Boulton 2006), we next addressed whether the ATR ortholog, ATL-1, is required for enrichment of repressive chromatin on the X chromosome. To that end, we monitored the localization of H3K9me2 and HIM-8 in atl-1(tm853) deletion mutant male germ lines. In contrast to brc-1 or brd-1 mutants, mutation of atl-1 resulted in altered distribution of H3K9me2. In most nuclei (95.9%), there was no clear association between HIM-8 and H3K9me2 (white arrow; Figure 1, A and C), indicating that the X chromosome was not specifically enriched for H3K9me2, and in the remaining nuclei (4.1%), H3K9me2 was associated with HIM-8 but had a much less compact signal (white arrowhead; Figure 1, A and C). Colabeling for Pol2-S2P and H3K9me2 revealed regions of the genome that were enriched for both repressive chromatin and Pol2-S2P (yellow arrow; Figure 1B), as well as regions that were enriched for neither Pol2-S2P nor H3K9me2 (yellow arrowhead; Figure 1B), suggesting that the absence of ATL-1 disrupts the association between repressive chromatin and transcriptional silencing. As H3K9me2 is not a reliable marker of the X chromosome in the atl-1 mutant, we next colabeled wild type and atl-1 mutants with antibodies against HIM-8 and RNA Pol II phosphorylated on serine 5 (Pol2-S5P), which marks transcriptionally competent chromatin (Hsin and Manley 2012), to specifically examine the transcriptional status of the X chromosome. As previously reported (Checchi and Engebrecht 2011), Pol2-S5P is enriched on all chromosomes but the X in wild-type male germ lines. However, in only 29.3 ± 19.2% of atl-1 nuclei Pol2-S5P was not observed on the X chromosome (vs. 96.5 ± 1.9% in wild type; P = 0.0121; arrowhead; Figure 1E). Thus, although BRC-1-BRD-1 does not appear to play a role in MSCI, ATL-1 is important for the correct targeting of H3K9me2 and transcriptional silencing of the X chromosome during C. elegans male meiosis.
ATR participates with the related and partially redundant kinase, ataxia-telangiectasia mutated (ATM) during DNA damage signaling (Abraham 2001). In mice, ATM does not play a role in MSCI (Royo et al. 2013). To determine whether ATM functions in targeting repressive chromatin to the X chromosome in C. elegans, we monitored H3K9me2 and HIM-8 in germ lines of the atm-1(gk186) deletion mutant. While 42.1% of nuclei were wild type with respect to association between HIM-8 and H3K9me2, 32.1% of nuclei showed association between the signals but much more diffuse H3K9me2 labeling, and 25.8% showed no association between HIM-8 and H3K9me2 (Figure 1, A and C). Similarly, Pol2-S2P showed a variable staining pattern with some nuclei containing a single track lacking Pol2-S2P and enriched for H3K9me2, which presumably corresponds to the X chromosome, while in other nuclei no clear chromosome lacking Pol2-S2P was detected (Figure 1B). Thus, in C. elegans, ATL-1, and to a lesser extent ATM-1, are important for accumulation of repressive chromatin and transcriptional silencing of the X chromosome.
To determine whether a function for BRC-1-BRD-1 in the correct targeting of repressive chromatin and transcriptional silencing of the X chromosome can be uncovered in the sensitized atm-1 mutant background, we examined H3K9me2 and HIM-8 as well as H3K9me2 and Pol2-S2P in the atm-1(gk186); brc-1(tm1145) brd-1(dw1) triple mutant (Figure 1, A–C). We found no difference in either H3K9me2 or Pol2-S2P localization between atm-1(gk186) and atm-1(gk186); brc-1(tm1145) brd-1(dw1), consistent with BRC-1-BRD-1 being dispensable for transcriptional silencing of the X chromosome in C. elegans male germ cells.
A subset of meiotic DSBs is repaired by NHEJ in the absence of BRC-1-BRD-1 in male germ cells
BRCA1-BARD1 has been implicated in promoting HR repair in somatic cells; however, its role in meiotic recombination has been controversial and is complicated by the pachytene arrest and apoptotic removal of brca1 mutant spermatocytes due to MSCI failure (Xu et al. 2003; Broering et al. 2014). The finding that neither brc-1 nor brd-1 mutants impair X chromosome transcriptional silencing in C. elegans prompted us to examine the role of BRC-1-BRD-1 in meiotic recombination in the absence of the complications associated with MSCI failure. To that end, we monitored meiotic DSB repair by examining the assembly and disassembly of the recombinase RAD-51 (Rinaldo et al. 2002) in the spatiotemporal organization of the C. elegans male germ line using antibodies against RAD-51 (Colaiácovo et al. 2003; Checchi et al. 2014) (Figure 2A).
brc-1 and brd-1 mutant hermaphrodites exhibit a slight increase in embryonic lethality and male progeny (a readout of X chromosome nondisjunction), and some RAD-51 foci perdure in late meiotic prophase, suggesting that repair of a subset of meiotic DSBs is delayed in the absence of BRC-1-BRD-1 (Boulton et al. 2004; Adamo et al. 2008; Janisiw et al. 2018; Li et al. 2018). In contrast to the appearance of more RAD-51 foci in mid and late pachytene in female germ cells, fewer RAD-51 foci were observed in brc-1, brd-1 or brc-1 brd-1 male germ cells compared to wild type in early meiotic prophase (transition zone) through mid-pachytene (Figure 2, B and C and Figure S1). These results suggest that, in the absence of BRC-1-BRD-1, fewer DSBs are induced, a subset of DSBs is repaired without loading RAD-51, RAD-51 loading is impaired, and/or repair occurs with faster kinetics than wild type. Given a role of BRCA1 in promoting HR at the expense of NHEJ in somatic cells (Daley and Sung 2014), we tested the hypothesis that some meiotic DSBs are repaired by NHEJ in the absence of BRC-1-BRD-1 in male germ cells. To that end, we simultaneously inactivated BRC-1 or BRD-1 and CKU-80 or CKU-70, the C. elegans KU80/KU70 orthologs that mediate NHEJ, and monitored RAD-51 foci throughout the germ line (Figure 2, B and C and Figure S1). When NHEJ was inactivated in the brc-1 or brd-1 mutants, RAD-51 foci were restored to wild-type levels in the transition zone through mid-pachytene in male germ cells. We also observed a small, but statistically significant elevation of RAD-51 foci in late pachytene when both BRC-1-BRD-1 and NHEJ were mutated, suggesting that both of these complexes contribute to repair of lesions at late pachytene (Figure S1B and Table S3), similar to what has been observed in oogenesis (Smolikov et al. 2007; Adamo et al. 2008). Together, these results suggest that BRC-1-BRD-1 functions at, or prior to, RAD-51 assembly to facilitate repair by HR in male germ cells, similar to its proposed role in somatic cells, and, in its absence, some breaks are channeled through NHEJ in early meiotic prophase.
BRC-1-BRD-1 promotes the early processing of meiotic DSBs in male germ cells
Following DSB formation, DNA end resection reveals 3′ single-stranded tails that promote homology search and strand invasion (Ranjha et al. 2018). To examine a potential role of BRC-1-BRD-1 in DNA end resection, we analyzed the localization pattern of RPA-1 (GFP::RPA-1; Sonneville et al. 2012) by live cell imaging. RPA-1 binds single-stranded DNA ends, and its recruitment to DSBs is dependent on resection (Garcia-Muse and Boulton 2005; Sartori et al. 2007; Koury et al. 2018). RPA-1 also associates with post-strand-exchange intermediates (Woglar and Villeneuve 2018). In transition zone to mid-pachytene, where DSBs are formed and processed, we observed abundant foci in addition to strong nucleoplasmic fluorescence in wild-type male germ lines. In brc-1(tm1145) brd-1(dw1) male germ lines, we observed fewer and less intense foci above the nucleoplasmic signal (Figure 3A). To quantify this, we calculated the CV (CV = SD/mean fluorescence intensity), which provides a measure of the extent of foci above the nucleoplasmic signal. Wild type had a significantly higher CV compared to the brc-1 brd-1 mutant (P < 0.0001; Figure 3B), suggesting that fewer RPA-1 molecules accumulated at processed DSBs in the mutant. Taken together, the alteration in both RAD-51 and RPA-1 suggests that BRC-1-BRD-1 facilitates the repair of DSBs by HR most likely through promoting DNA end resection.
To determine whether BRC-1-BRD-1 localizes to DSBs, we examined the localization of GFP::BRC-1 by live cell imaging. In wild-type male germ lines, GFP::BRC-1 was nucleoplasmic and formed a small number of bright foci in proliferating germ cells (Figure 3, C and D). As cells progressed into meiosis, GFP::BRC-1 was observed in multiple foci at transition zone and early pachytene; tracks of fluorescence were also beginning to form at early pachytene (Figure 3, C and D). At mid-pachytene, GFP::BRC-1 was predominantly in tracks, which had begun to concentrate on a chromosomal subdomain. Further concentration into five stretches and then puncta were observed in late pachytene through diplotene. The dynamic localization of GFP::BRC-1 in the male germ line is similar to the hermaphroditic germ line: GFP::BRC-1 foci partially overlap with RAD-51 (Figure S2A), suggesting they mark sites of ongoing meiotic recombination, and the GFP::BRC-1 tracks in pachytene colocalize with the synaptonemal complex (SC) that become concentrated on the short arm, dependent on CO formation (Li et al. 2018).
To test the dependencies of BRC-1 localization on DSB formation and processing, we examined GFP::BRC-1 in spo-11, rad-50 and mre-11 mutants. spo-11 mutants are unable to form meiotic DSBs (Dernburg et al. 1998), and very few GFP::BRC-1 foci were present in transition zone and early pachytene compared to wild type (Figure 3, C and D). At early to mid-pachytene GFP::BRC-1 was observed in tracks in the spo-11 mutant similar to wild type (Figure 3C), as synapsis occurs in the absence of recombination in C. elegans (Dernburg et al. 1998). In late pachytene, GFP::BRC-1 fluorescence did not concentrate on a portion of each chromosome pair as in wild type, consistent with these events being dependent on CO formation. However, in 10.7 ± 3.2% of pachytene nuclei there was enrichment of GFP::BRC-1 on a chromosome track (Figure 3C, arrowhead) with weak fluorescence on the other synapsed chromosomes. This has been observed for GFP::BRC-1 and other synapsis markers in oogenesis and likely represents spo-11-independent lesions capable of recruiting meiotic DNA repair components and altering SC properties (Machovina et al. 2016; Nadarajan et al. 2017; Pattabiraman et al. 2017; Li et al. 2018).
We next examined the requirement for RAD-50 and MRE-11 in recruitment of GFP::BRC-1 to early meiotic foci. RAD-50 and MRE-11 form a complex with NBS-1 (MRX/N complex) and are required for both DSB formation and processing for repair through HR in meiotic cells, in addition to playing a role in repair of lesions generated during DNA replication (Chin and Villeneuve 2001; Hayashi et al. 2007; Girard et al. 2018). In rad-50(ok197) and mre-11(ok179) null mutants, GFP::BRC-1 was observed in fewer foci compared to wild type in transition zone and early pachytene (Figure 3, C and D and Figure S2B). However, in contrast to spo-11, an increased number of nuclei with 1–3 GFP::BRC-1 foci were present in proliferating germ cells and throughout meiotic prophase (Figure 3, C and D), suggesting GFP::BRC-1 is enriched at lesions generated during S phase in these mutant backgrounds. We also observed an earlier appearance and higher percentage of nuclei showing concentrated signal on a subset of chromosomes (rad-50(ok197), 21.17 ± 4.6%), consistent with recruitment of recombination proteins and alteration of the SC properties at mitotic lesions as they progress through meiosis. Together, these results suggest that the enrichment of GFP::BRC-1 to abundant foci in early meiotic prophase is dependent on meiotic DSB formation.
To determine the requirement for DSB end processing in recruiting GFP::BRC-1 to sites of meiotic recombination, we took advantage of a separation-of-function allele, mre-11(iow1); worms harboring this allele are competent for meiotic DSB formation but defective in resection (Yin and Smolikove 2013). As with rad-50(ok197) and mre-11(ok179) null mutants, there was a reduction in meiotic GFP::BRC-1 foci in mre-11(iow1) mutant germ lines (Figure 3, C and D) and a similar number of pachytene nuclei showing concentration of GFP::BRC-1 on a subset of chromosomes [mre-11(iow1), 19.23 ± 2.8%]. These results suggest that accumulation of GFP::BRC-1 into foci in early meiotic prophase requires DSB resection, consistent with BRC-1-BRD-1 functioning at an early step of meiotic DSB processing to promote HR.
RAD-51 loading is dependent on RAD-50 in male meiotic germ cells
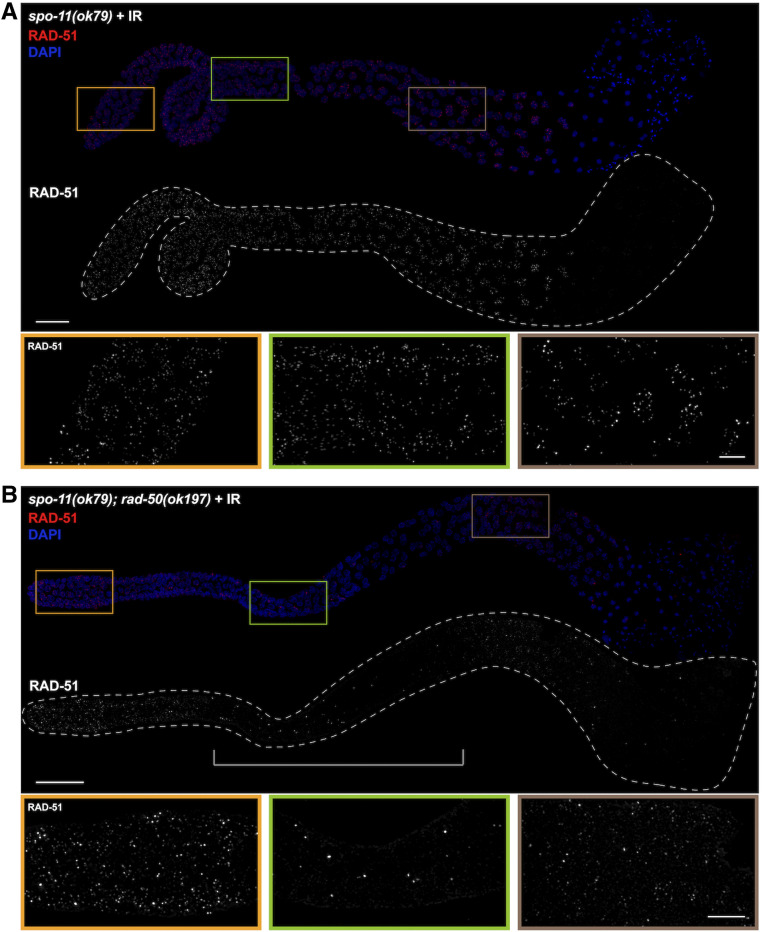
Meiotic recombination occurs in the context of specialized chromosome structure, the chromosomal axes, and fully formed SC, to promote interhomolog COs. Previous analyses in oogenic germ lines revealed a requirement for RAD-50 in loading RAD-51 at DSBs in meiotic prophase (Hayashi et al. 2007). Given the somatic-like role of BRC-1-BRD-1 in promoting HR at the expense of NHEJ in meiotic male germ cells, and the dependency of BRC-1 localization at meiotic DSBs on RAD-50, we next addressed whether male meiosis also requires RAD-50 for loading RAD-51 in the context of synapsed chromosomes. To that end, we analyzed RAD-51 localization in spo-11(ok79) and spo-11(ok79); rad-50(ok197) male germ cells. DNA breaks were induced by exposing worms to 10 Gys of IR; 1 hr post-IR, gonads were dissected and labeled with antibodies against RAD-51 (Hayashi et al. 2007). Abundant RAD-51 foci were observed throughout the germ line in the spo-11 worms, indicating proficient loading of RAD-51 on IR-induced DSBs (Figure 4A). Abundant RAD-51 foci were also observed in irradiated spo-11; rad-50 double mutant germ lines in proliferating germ cells and in mid- to late pachytene/diplotene spermatocytes (Figure 4B). However, in a region extending from the transition zone to mid- to late pachytene very few foci were observed in the irradiated spo-11; rad-50 double-mutant germ lines. Thus, similar to oogenesis, RAD-51 loading is dependent on RAD-50 during meiotic prophase in spermatogenic germ lines. Together, our genetic and cell biological analyses of BRC-1-BRD-1 and DSB processing factors suggest that properties of both somatic and meiotic repair modes exist in male germ cells.
Figure 4.
RAD-51 loading is dependent on RAD-50 in male meiotic germ cells. (A) spo-11(ok79) and (B) spo-11(ok79); rad-50(ok197) male gonads fixed and dissected 1 hr after exposure to 10 Gys IR, stained with RAD-51 antibody (red), and counterstained with DAPI (blue). In the spo-11; rad-50 mutant RAD-51 foci are largely absent in most nuclei in the central portion of the gonad, indicated by the bracket, from the onset of meiotic prophase to mid-pachytene. Images are projections through the entire gonad. Four germ lines were examined. Bar, 20 μm. Insets show selected nuclei from different regions of the germ line; Bar, 5 μm.
BRC-1-BRD-1 is important when CO formation is blocked on a subset of chromosomes during spermatogenesis
In somatic cells, BRCA1 plays a critical role when errors in the cell cycle occur (Takaoka and Miki 2018) and we previously found that removal of BRC-1-BRD-1 during oogenesis impairs progeny viability and RAD-51 stabilization when CO formation is blocked on a subset of chromosomes (Li et al. 2018). To examine the consequence of inactivating BRC-1-BRD-1 under similar conditions during male meiosis, we monitored the viability of progeny sired by mutant zim-1(tm1813) [chromosomes II and III fail to pair and synapse (Phillips and Dernburg 2006)], brc-1(xoe4); zim-1(tm1813), brc-1(tm1145); zim-1(tm1813) and brd-1(ok1623); zim-1(tm1813) males. brc-1(tm1145) is a hypomorphic allele that we previously showed impairs recombination under meiotic checkpoint activating conditions in oogenesis (Li et al. 2018). We used worms carrying the fog-2(q71) mutation for these experiments to eliminate hermaphrodite spermatogenesis, rendering XX animals self-sterile (Schedl and Kimble 1988), so that the contribution of the male parent to embryonic lethality could be assessed unambiguously. Similar to our findings in hermaphrodites (Li et al. 2018), removal of BRC-1 or BRD-1 enhanced the embryonic lethality of zim-1 mutants when mutant sperm were used to fertilize fog-2 ova (Figure 5A; P < 0.0001 by one-way ANOVA). These results suggest that BRC-1-BRD-1 plays important roles to enhance the quality of male germ cells under meiotic checkpoint activating conditions.
Previous analyses in the hermaphrodite germ line revealed that RAD-51 levels are elevated genome wide when the obligate CO is not established on any or all chromosome pairs (Colaiácovo et al. 2003; Carlton et al. 2006; Mets and Meyer 2009). Removal of BRC-1-BRD-1 under these conditions resulted in a “dark zone” of RAD-51 in mid- to late pachytene, which is likely a consequence of premature RAD-51 disassembly (Li et al. 2018). To determine whether BRC-1-BRD-1 promotes RAD-51 filament stability in male germ lines when not all chromosomes are connected by a CO, we monitored RAD-51 levels in zim-1 mutants in the presence and absence of BRC-1-BRD-1. Similar to oogenic germ lines, blocking CO formation on a subset of chromosomes resulted in elevated levels of RAD-51 foci throughout meiotic prophase in male germ lines (Figure 5, B and C). However, in the absence of BRC-1, we did not observe a RAD-51 “dark zone,” suggesting that BRC-1-BRD-1 does not play a role in stabilizing the RAD-51 filament under checkpoint activating conditions in male germ cells (Figure 5C). Quantification of foci revealed reduced RAD-51 levels in brc-1; zim-1 compared to zim-1 (Figure 5B), similar to the reduction in RAD-51 foci observed in brc-1 or brd-1 mutants alone compared to wild-type males (Figure 2B). However, the RAD-51 levels in brc-1; zim-1 were still higher throughout pachytene than in wild-type male germ lines (compare Figure 2B and Figure 5B). These results suggest that BRC-1-BRD-1 promotes meiotic recombination in spermatogenesis using different mechanisms than in oogenesis under meiotic checkpoint activation.
BRC-1-BRD-1 inhibits COSA-1-marked CO designation sites when meiosis is perturbed in male germ cells
In addition to stabilizing the RAD-51 filament, BRC-1-BRD-1 promotes formation of CO precursors marked by the cyclin related COSA-1 (Yokoo et al. 2012) in the zim-1 mutant background in hermaphrodites (Li et al. 2018). To determine whether BRC-1-BRD-1 influences CO designation in male germ cells, we monitored GFP::COSA-1 (Yokoo et al. 2012) in brc-1, brd-1, zim-1, brc-1; zim-1 and brd-1; zim-1 mutant germ lines. Wild-type males mostly exhibit five COSA-1 foci, one on each of the five pairs of autosomes but not on the single X chromosome (Checchi et al. 2014). This pattern was unaltered by removal of either BRC-1 or BRD-1 [WT = 4.99 ± 0.30; brc-1(xoe4) = 4.99 ± 0.30; brd-1(ok1623) = 5.02 ± 0.28; Figure 6A]. As zim-1 mutants have two asynapsed chromosome pairs, we expected to observe three COSA-1 foci; however, we observed an average of 4.61 ± 1.12 COSA-1 foci (Figure 6A). Further, removing BRC-1 or BRD-1 in zim-1 males resulted in significantly more COSA-1 foci (brc-1(xoe4); zim-1(tm1813) = 5.32 ± 0.97; brd-1(ok1623); zim-1(tm1813) = 5.29 ± 0.99) (Figure 6, A and B). This is opposite to what we observed in hermaphrodites, where reduced levels of GFP::COSA-1 was observed in the absence of BRC-1 or BRD-1 in zim-1 mutants (Li et al. 2018).
To examine this further, we monitored GFP::COSA-1 foci in additional mutants that lead to asynapsis of different chromosome pairs. Pairing and synapsis of the X chromosome is impaired in him-8 mutants, zim-2 mutants have asynapsed chromosome Vs and two chromosome pairs, I and IV, fail to pair and synapse in zim-3 mutants (Phillips et al. 2005; Phillips and Dernburg 2006). As expected, mutation of him-8 had no effect on GFP::COSA-1 levels either in the presence or absence of BRC-1-BRD-1, presumably due to the presence of the single X chromosome in male germ cells (Figure 6A). zim-2 and zim-3 mutants showed higher than expected numbers of COSA-1 foci (zim-2(tm547)= 4.48 ± 0.85 observed vs. four expected, zim-3(xoe15)= 3.52 ± 0.80 observed vs. three expected), similar to what we observed in the zim-1 mutant and the number was further increased upon removal of BRC-1-BRD-1 [brc-1(xoe4); zim-2(tm574)= 4.91 ± 0.96, brd-1(ok1623); zim-2(tm574)=4.86 ± 0.93, brc-1(xoe4); zim-3(xoe15)= 4.09 ± 1.12, brd-1(ok1623); zim-3(xoe15)= 3.83 ± 1.10] (Figure 6A). Thus, BRC-1-BRD-1 limits the number of CO precursors in spermatogenesis under circumstances where asynapsed chromosomes are present.
Previous analyses in oogenesis had indicated that when CO formation is completely blocked by mutation of central components of the SC, COSA-1 accumulates at foci that represent aberrant recombination sites (Li et al. 2018; Woglar and Villeneuve 2018; Cahoon et al. 2019; Hurlock et al. 2020). We next examined GFP::COSA-1 in syp-1 mutant males, in which germ cells fail to undergo chromosome synapsis and therefore do not form any interhomolog COs (MacQueen et al. 2002). As observed in hermaphrodites, syp-1 mutant males exhibited a significant number of COSA-1 foci (4.0 ± 1.20) (Figure 6A). However, in the absence of BRC-1 or BRD-1, fewer GFP::COSA-1 foci were observed [brc-1(xoe4); syp-1(me17)=3.27 ± 1.15, brd-1(ok1623); syp-1(me17)=3.56 ± 1.51]. This suggests that unlike the situation where CO formation is inhibited on only a subset of chromosomes, BRC-1-BRD-1 promotes the localization of COSA-1 at recombination sites when no interhomolog COs can form.
BRC-1 influences the CO landscape
Given the effect of BRC-1-BRD-1 on COSA-1 foci in the different mutants, we monitored genetic linkage between SNP markers on chromosomes I and V in male Bristol/Hawaiian hybrid strains to assess whether BRC-1-BRD-1 alters the formation of bona fide COs (Figure 7A). Inactivation of BRC-1 had little effect on the genetic map length of either chromosome I or V [I: WT = 45.74 cM; brc-1(xoe4)= 52.17 cM; V: WT = 45.21 cM; brc-1(xoe4)= 50.82 cM; Figure 7B and Table S4]. In C. elegans, COs are not evenly distributed along the length of the chromosomes but are enriched on the gene-poor arms (Barnes et al. 1995; Lim et al. 2008; Rockman and Kruglyak 2009). Similar to what we reported for oocytes (Li et al. 2018), there is a statistically significant alteration in the distribution of COs in the brc-1 mutant on both chromosomes I and V compared to wild-type males (Figure 7C and Table S4). In the brc-1 mutant, we observed an expansion in the center of the chromosome, with more COs in the center-right interval on chromosome I (30.21% vs. 13.95%; P = 0.0123) and the left-center interval on chromosome V compared to wild type (12.9% vs. 3.53%; P = 0.0304) (Figure 7C and Table S4).
We next monitored linkage between SNP markers in the zim-1 and brc-1; zim-1 mutant males. We observed a significant increase in the genetic map length on both chromosomes I and V in zim-1 and brc-1; zim-1 compared to wild-type males [I: zim-1(xoe6)=61.57 cM P = 0.0014, brc-1(xoe4); zim-1(xoe6)=69.06 cM P = 0.0001; V: zim-1(xoe6)=61.11 cM P = 0.0089, brc-1(xoe4); zim-1(xoe6)=62.04 cM P = 0.0024; Figure 7B and Table S4]. In addition to the expanded genetic maps, CO distributions were also altered. The percentage of COs on the left and right arms of chromosome I were reduced in brc-1; zim-1 compared to wild type (left: 22% vs. 33.7% P = 0.0426; right: 16.8% vs. 31.4% P = 0.0053), while the right-center interval was expanded in brc-1; zim-1 compared to wild-type males (29.6% vs. 13.9% P = 0.004; Figure 7C and Table S4). On chromosome V there was an increased percentage of COs in the left-center interval in zim-1 compared to wild-type males (15.2% vs. 3.5% P = 0.0053), and it was further expanded in brc-1; zim-1 (21.5% P = 0.0001), while the right-center interval had significantly more COs in brc-1; zim-1 compared to brc-1 males (22.83% vs. 12.9% P = 0.045; Figure 7C and Table S4).
A unique feature of C. elegans oogenic meiosis is that, on average, there is a single CO per chromosome pair per meiosis (Albertson et al. 1997; Hillers and Villeneuve 2003; Hammarlund et al. 2005). This is attributed to very strong interference, which is the phenomenon that the presence of one CO at one position decreases the probability of formation of another CO nearby. Analyses in spermatocytes also suggested that there is usually a single CO per chromosome pair (Meneely et al. 2002; Kaur and Rockman 2014); however, Lim et al. (2008) reported that interference was not as strong in male meiosis due to the appearance of closely spaced DCOs. We detected five DCOs on chromosome I and three DCOs on chromosome V in a total of 188 wild-type spermatocytes, which corresponds to 6.2% and 3.7% of total CO events (Figure 7D and Table S4). Fewer DCOs were detected in the brc-1 mutant males, although this was not statistically different (chromosome I: 2 DCO/184, 2.1%; chromosome V: 1 DCO/183, 1.1%; Figure 7D and Table S4). In contrast, we previously detected no DCOs in 187 oocytes in either wild type or brc-1 oocytes (Li et al. 2018). In the zim-1 mutant, we detected nine DCOs in 268 spermatocytes on chromosome I, which corresponds to 5.8% of total CO events and is not significantly different compared to wild type; however, in the brc-1; zim-1 double mutant, a significantly higher percentage of COs were DCOs and triple crossovers (TCOs): 37 DCOs and two TCOs were detected in 362 spermatocytes, which collectively is 18.7% of total CO events (Figure 7D and Table S4). On chromosome V, zim-1 had elevated levels of DCOs and TCOs (18/270, 12.3%) compared to wild type and brc-1 spermatocytes, but this was not further increased in the brc-1; zim-1 double mutant (23/353, 11.7%; Figure 7D and Table S4).
Given the increased frequency of DCOs, we calculated interference. While most intervals had absolute interference of 1 in wild type and brc-1, the detection of DCOs resulted in decreased interference in two intervals on both chromosome I and chromosome V (Table 1). zim-1 mutant males displayed reduced interference in all intervals except the left to left center and left center to right center intervals on chromosome I. Inactivation of BRC-1 in the zim-1 mutant further impaired interference in all intervals on chromosome I, but had a variable effect on chromosome V, although they did not reach statistical significance (Table 1). Taken together, the elevated number of COSA-1 foci and increased numbers of DCOs and TCOs in the brc-1; zim-1 mutant on chromosome I suggest that BRC-1-BRD-1 inhibits supernumerary COs under checkpoint activating conditions.
Table 1. Crossover (CO) interference on chromosome I and V.
| WT (I) | exp DCO freq | obs DCO freq | c.o.c | interference |
|---|---|---|---|---|
| L - LC | 0.0148 | 0.0160 | 1.0805 | −0.0805 |
| L - CR | 0.0098 | 0.0000 | 0.0000 | 1.0000 |
| L - R | 0.0222 | 0.0106 | 0.4802 | 0.5198 |
| LC - R | 0.0138 | 0.0000 | 0.0000 | 1.0000 |
| CR - R | 0.0092 | 0.0000 | 0.0000 | 1.0000 |
| LC - CR | 0.0061 | 0.0000 | 0.0000 | 1.0000 |
| WT (V) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0030 | 0.0000 | 0.0000 | 1.0000 |
| L - CR | 0.0139 | 0.0000 | 0.0000 | 1.0000 |
| L- R | 0.0327 | 0.0106 | 0.3255 | 0.6745 |
| LC - R | 0.0028 | 0.0000 | 0.0000 | 1.0000 |
| CR - R | 0.0131 | 0.0053 | 0.4069 | 0.5931 |
| LC- CR | 0.0012 | 0.0000 | 0.0000 | 1.0000 |
| brc-1 (I) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0148 | 0.0000 | 0.0000 | 1.0000 |
| L - CR | 0.0214 | 0.0054 | 0.2538 | 0.7462 |
| L - R | 0.0162 | 0.0054 | 0.3345 | 0.6655 |
| LC - R | 0.0130 | 0.0000 | 0.0000 | 1.0000 |
| CR - R | 0.0188 | 0.0000 | 0.0000 | 1.0000 |
| LC - CR | 0.0171 | 0.0000 | 0.0000 | 1.0000 |
| brc-1 (V) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0140 | 0.0000 | 0.0000 | 1.0000 |
| L - CR | 0.0140 | 0.0000 | 0.0000 | 1.0000 |
| L - R | 0.0349 | 0.0055 | 0.1564 | 0.8436 |
| LC - R | 0.0107 | 0.0000 | 0.0000 | 1.0000 |
| CR - R | 0.0107 | 0.0000 | 0.0000 | 1.0000 |
| LC - CR | 0.0043 | 0.0000 | 0.0000 | 1.0000 |
| zim-1 (I) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0270 | 0.0000 | 0.0000 | 1.0000 |
| L - CR | 0.0317 | 0.0149 | 0.4702 | 0.5298 |
| L - R | 0.0270 | 0.0112 | 0.4149 | 0.5851 |
| LC - R | 0.0161 | 0.0037 | 0.2318 | 0.7682 |
| CR - R | 0.0189 | 0.0037 | 0.1971 | 0.8029 |
| LC - CR | 0.0189 | 0.0000 | 0.0000 | 1.0000 |
| zim-1 (V) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0185 | 0.0037 | 0.2000 | 0.8000 |
| L - CR | 0.0252 | 0.0148 | 0.5882 | 0.4118 |
| L - R | 0.0385 | 0.0333 | 0.8654 | 0.1346 |
| LC - R | 0.0178 | 0.0111 | 0.6231 | 0.3769 |
| CR - R | 0.0243 | 0.0037 | 0.1527 | 0.8473 |
| LC - CR | 0.0117 | 0.0074 | 0.6353 | 0.3647 |
| brc-1; zim-1 (I) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0332 | 0.0138 | 0.4166 | 0.5834 |
| L - CR | 0.0311 | 0.0304 | 0.9784 | 0.0216 |
| L - R | 0.0176 | 0.0110 | 0.6268 | 0.3732 |
| LC - R | 0.0253 | 0.0193 | 0.7637 | 0.2363 |
| CR - R | 0.0237 | 0.0110 | 0.4659 | 0.5341 |
| LC - CR | 0.0446 | 0.0331 | 0.7431 | 0.2569 |
| brc-1; zim-1 (V) | exp DCO freq | obs DCO freq | c.o.c | interference |
| L - LC | 0.0238 | 0.0170 | 0.7153 | 0.2847 |
| L - CR | 0.0253 | 0.0085 | 0.3362 | 0.6638 |
| L - R | 0.0298 | 0.0227 | 0.7598 | 0.2402 |
| LC - R | 0.0223 | 0.0057 | 0.2546 | 0.7454 |
| CR - R | 0.0237 | 0.0085 | 0.3590 | 0.6410 |
| LC - CR | 0.0189 | 0.0028 | 0.1502 | 0.8498 |
L, left interval; LC, left-center interval; CR, center-right interval; R, right interval; DCO, double crossover; expected DCO: (crossover frequency at interval “A”) × (crossover frequency at interval “B”). c.o.c. (coefficient of coincidence) = actual DCO frequency/expected DCO frequency; Interference = 1- c.o.c. See Table S4 for data used for calculations.
Discussion
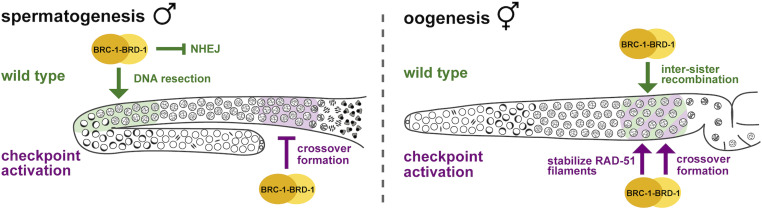
We show here that the BRC-1-BRD-1 complex functions in early processing of meiotic DSBs to promote HR and also inhibits supernumerary COs when some chromosomes are unable to form COs in male meiosis. These functions are distinct from previous analyses in oogenesis and suggests that this complex is differently regulated during male and female meiosis to optimize sperm vs. oocyte production (Figure 8).
Figure 8.
BRC-1-BRD-1 function in spermatogenesis and oogenesis. Model of proposed function of BRC-1-BRD-1 in male and female (hermaphrodite) germ lines. Wild type (green) and checkpoint activation conditions (e.g., zim-1; purple) are shown. During spermatogenesis BRC-1-BRD-1 promotes HR at the expense of NHEJ presumably through regulating DNA end resection in early meiotic prophase, while the complex promotes intersister recombination in late meiotic prophase during oogenesis. Under checkpoint activation, BRC-1-BRD-1 inhibits COSA-1-marked COs in male meiosis, either directly or as a consequence of a role for BRC-1-BRD-1 in promoting intersister repair. In female meiosis, BRC-1-BRD-1 mediates the stability of the RAD-51 filament and promotes COSA-1-marked COs. The different phenotypes observed in brc-1 and brd-1 mutants are likely a consequence of the complex ubiquitinating different substrates dependent on the distinctive temporal regulation of spermatogenesis vs. oogenesis.
Overlapping but distinct meiotic silencing pathways in C. elegans and mammals
Mouse BRCA1 is essential for MSCI, recruiting ATR for H2AX phosphorylation and chromosome compaction (Turner et al. 2004). ATR, in turn, promotes the accumulation of additional BRCA1 and other DNA damage signaling proteins to hemizygous regions of sex chromosomes, perhaps in response to unrepaired meiotic DSBs (Royo et al. 2013; Lu and Yu 2015). Accumulation of DNA damage response components are linked to the recruitment of SETDB1 methyltransferase for H3K9me3 enrichment and gene silencing (Hirota et al. 2018). While C. elegans ATR ortholog and, to a lesser extent, the related ATM checkpoint kinases are critical for targeting H3K9me2 to the hemizygous X chromosome in male germ cells, removal of BRC-1-BRD-1 had no effect on either the deposition of H3K9me2 or lack of transcription on the X chromosome (Figure 1), suggesting that BRC-1-BRD-1 does not mediate MSCI in C. elegans male meiosis.
As master regulators, ATR and ATM phosphorylate a large number of substrates (Matsuoka et al. 2007; Mu et al. 2007); consequently, the observed effect on meiotic silencing is likely to be indirect. Indeed, a recent study revealed that these kinases function in multiple aspects of meiotic recombination during C. elegans oogenesis (Li and Yanowitz 2019). We have shown that the X chromosome in males is refractory to ATM-dependent meiotic DSB formation feedback mechanisms (Checchi et al. 2014), suggesting that the defect in accumulation of H3K9me2 may not be through unrepaired DSBs, as is proposed in mammals. In addition, ATR is normally present at very low levels in the male germ line and accumulates genome-wide in response to exogenous DNA damage or in mutants impaired for recombination or synapsis but is not enriched on the X chromosome (Jaramillo-Lambert et al. 2010), implying an indirect role for this kinase in MSCI. Further, a C. elegans H2AX ortholog has not been identified that can be phosphorylated by ATR/ATM (Boulton 2006). On the other hand, the SETDB1 methyltransferase, MET-2, mediates H3K9me2 deposition and gene silencing of the X chromosome in male germ cells (Bessler et al. 2010; Checchi and Engebrecht 2011), analogous to SETDB1 function in mammals (Hirota et al. 2018). However, in contrast to mice, MET-2 does not accumulate on the X chromosome of male germ cells (Yang et al. 2019). Thus, the mechanisms whereby ATL-1/ATM-1 promote accumulation of H3K9me2 via MET-2 on the X chromosome of males remains to be elucidated but perhaps is linked to a small RNA pathway that is required for meiotic silencing (She et al. 2009). Nonetheless, the overlapping but distinct requirements for components that mediate MSCI in worms and mammals suggest that meiotic silencing is a conserved feature of meiosis in metazoans; however, the pathways used to target repressive chromatin marks have evolved independently.
BRC-1-BRD-1 regulates DSB processing to promote HR in male germ cells
In somatic cells, BRCA1-BARD1 functions in DNA damage signaling and repair to promote genome integrity (Kouznetsova et al. 2009; Li and Greenberg 2012; Savage and Harkin 2015; Takaoka and Miki 2018). Critical to the maintenance of the genome is the choice of pathways for repair of DSBs: HR, NHEJ and other error-prone pathways including microhomology mediated end joining. Whether HR or error-prone pathways are used is largely driven by DNA end resection. Several studies support the hypothesis that BRCA1-BARD1 regulates the choice between repair by HR and NHEJ. Initial evidence for this was based on the observation that brca1−/− embryonic lethality can be rescued by removal of 53BP1, a DNA damage response protein that promotes NHEJ (Cao et al. 2009; Bouwman et al. 2010; Bunting et al. 2010). More recent work has suggested that BRCA1-BARD1 promotes DNA end resection by removing a chromatin barrier through ubiquitination of histone H2A (Densham et al. 2016) and/or through speeding up resection by interaction with CtIP, a protein that promotes end resection (Cruz-García et al. 2014). Studies by other groups also showed that BRCA1 and CtIP work together with the MRX/N complex to mediate resection of complex breaks, and may be important at Spo11-dependent meiotic DSBs (Hartsuiker et al. 2009; Aparicio et al. 2016).
Our analysis of male meiosis reveals that similar to the role of BRCA1-BARD1 in somatic cells, this complex regulates the processing of meiotic DSB to promote repair by HR (Figure 8). First, in the absence of BRC-1-BRD-1, fewer RPA-1 and RAD-51 foci were observed in meiotic prophase (Figure 2 and Figure 3), suggesting BRC-1-BRD-1 functions at or prior to RPA-1/RAD-51 loading onto resected ends. We show that the reduction in RAD-51 foci can be suppressed by mutation of NHEJ proteins, consistent with a role of BRC-1-BRD-1 in regulating the choice between HR and NHEJ. However, a recent study provides evidence that accumulation of deletions in C. elegans brc-1 and brd-1 mutants is a consequence of theta-mediated end joining (Kamp et al. 2020), suggesting that additional error-prone pathways are activated in the absence of BRC-1-BRD-1. Additionally, the localization of BRC-1 to foci in early meiotic prophase, which presumably represent sites of ongoing recombination, is dependent on DNA resection (Figure 3). These findings point to a role for BRC-1-BRD-1 in promoting repair by HR, likely by regulating resection (Figure 8). In mouse spermatocytes, no defect in end resection was detected in a brca1 hypomorphic allele also mutant for p53 (Paiano et al. 2020); thus, it is not clear whether BRCA1-BARD1 function in end resection is a conserved feature of male meiosis. It is also important to note that brc-1 and brd-1 mutants exhibit only subtle meiotic phenotypes, in contrast to the phenotypic consequence of removing components of the resection machinery. Mutation of CtIP (C. elegans COM-1) or components of the MRX/N complex leads to high levels of embryonic lethality and almost a complete absence of RAD-51 loading (Chin and Villeneuve 2001; Hayashi et al. 2007; Lemmens et al. 2013; Girard et al. 2018). Thus, while BRC-1-BRD-1 is not essential for resection, our data are consistent with this complex regulating resection speed or extent, as in somatic cells.
In addition to promoting the processing of DSBs for homologous recombination, BRC-1 also plays a role in CO distribution. Analysis of genetic COs on chromosome I and V revealed that more COs occurred at the chromosome center, and fewer on the arms, as was previously observed in oogenesis (Li et al. 2018). Alteration in CO distribution in the brc-1 mutant may result from changes in the chromatin landscape, which has been linked to BRCA1 function in mammals (Broering et al. 2014; Densham et al. 2016), and has been shown to alter CO patterning (Mézard et al. 2015; Yu et al. 2016). A surprising number of C. elegans meiotic mutants display altered CO distribution (Zetka and Rose 1995; Wagner et al. 2010; Meneely et al. 2012; Saito et al. 2012, 2013; Chung et al. 2015; Hong et al. 2016; Jagut et al. 2016; Janisiw et al. 2020). While the underlying mechanisms are not clear, one possibility is that CO vs. non-CO outcomes are driven by a particular chromatin environment as suggested by Saito and Colaiacovo (2017).
BRC-1-BRD-1 function when male meiosis is perturbed
We show that BRC-1-BRD-1 functions to promote progeny viability when male meiosis is perturbed under conditions when some chromosome pairs fail to pair, synapse, and form a CO (Figure 5). While this is also true for female meiosis, the phenotypic consequences of mutating BRC-1 or BRD-1 when meiosis is perturbed are distinct in the sexes (Figure 8). During female meiosis, removal of BRC-1 or BRD-1 under checkpoint activating conditions leads to premature disassembly of the RAD-51 filament resulting in a “dark zone” of RAD-51 (Li et al. 2018); however, no “dark zone” was observed during male meiosis (Figure 5). While fewer RAD-51 foci were observed in the absence of BRC-1-BRD-1 when meiosis was impaired, this is likely a consequence of the role of BRC-1-BRD-1 in DSB end processing and not in promoting RAD-51 stability, although a subtle role in RAD-51 stability cannot be ruled out. Additionally, while the CO landscape is altered in both male and female meiosis, opposite effects of removing BRC-1-BRD-1 in the zim-1 mutant were observed. In female meiosis, mutation of brc-1 or brd-1 in the zim-1 background led to fewer COSA-1-marked CO designation events, while during male meiosis the numbers increased. One possibility to explain this observation is that destabilization of the RAD-51 filament in the absence of BRC-1-BRD-1 in mid- to late pachytene in female meiosis leads to fewer meiotic recombination intermediates that can be processed into COSA-1-marked CO precursors. On the other hand, the RAD-51 filament remains stable during male meiosis under these conditions such that more recombination intermediates can be processed into COSA-1 marked COs.
In the zim-1 and brc-1; zim-1 mutants, we observed an increase in both the number of CO designation sites (COSA-1 foci) as well as bona fide COs; however, there is no direct correlation between COSA-1 foci and genetic COs. We expected to see three GFP::COSA-1 foci in zim-1 if each chromosome received a single CO as in wild type; we observed an average of 4.6 (note the wide distribution from 3 to 8). This is a 53% increase in COSA-1-marked events genome-wide. If those events were evenly distributed between the three paired chromosomes, we would expect a 17% increase/chromosome. The genetic map distance for both chromosomes I and V was 61 cM in zim-1, compared to 45 cM for wild type, which represents a 35% increase on both chromosomes I and V. Assuming the CO landscape of chromosome IV is similarly altered as chromosomes I and V in the zim-1 mutant, and each CO site is marked by COSA-1, we would expect ∼100% increase in COSA-1 foci. Alternatively, if the chromosome IV CO landscape was unaltered, we would still expect an increase in COSA-1 foci of ∼70%. In either situation, we observed fewer COSA-1 foci than genetic COs, suggesting that not all of the extra COs are marked by COSA-1. In brc-1; zim-1 we observed a 15% increase in COSA-1 foci but only a subtle increase in the genetic map distance compared to zim-1, suggesting that more COs are marked by COSA-1 in the absence of BRC-1. Thus, we propose that BRC-1 alters the type of CO events when some chromosomes cannot achieve a CO. Perhaps under checkpoint-signaling conditions, BRC-1-BRD-1 promotes intersister repair in male meiosis, and, in its absence, more intermediates are channeled into interhomolog COs, similar to the role of BRC-1-BRD-1 in intersister recombination in female meiosis (Adamo et al. 2008; Garcia-Muse et al. 2019). Alternatively, or in addition, BRC-1-BRD-1 may play a direct role in inhibiting interhomolog COs under checkpoint activating conditions.
The alteration in the CO landscape is also reflected in the levels of SCOs and DCOs. On chromosome I, the zim-1 mutant had elevated SCOs, but not DCOs compared to wild type, while removal of BRC-1 in the zim-1 mutant resulted in elevated levels of DCOs at the expense of SCOs. We propose that this reflects a shift from three- and four-strand DCOs, which are included in the SCO class and are presumably not marked by COSA-1, in zim-1, to two-strand DCOs marked by COSA-1 in brc-1; zim-1. In contrast, on chromosome V, the zim-1 mutant showed significantly higher levels of DCOs compared to wild type, but removing BRC-1 had little effect. During female meiosis, inactivation of BRC-1 in the zim-1 mutant background had the opposite effect, i.e., decreasing numbers of DCOs and elevated numbers of SCOs were observed on chromosome V, presumably due to a shift from two-strand DCOs to three- and four-strand DCOs (Li et al. 2018). Thus, there are both chromosome-specific and sex-specific effects on CO patterning when BRC-1 is inactivated. The sex-specific effect is likely due to RAD-51 stability and CO pathway usage. The chromosome-specific effect may be a consequence of size; chromosome I is one of the smallest chromosomes, while chromosome V is the largest chromosome. Recent work in yeast suggests that small chromosomes use multiple mechanisms to ensure the formation of the obligate CO (Murakami et al. 2020). Therefore, the differential impact on chromosome I vs. V may be due to the mechanisms in place to promote CO formation on small chromosomes. Alternatively, other chromosome-specific features may influence which DSBs are converted into COs when BRC-1-BRD-1 is not present to constrain extra CO formation during male meiosis.
Why does removal of BRC-1-BRD-1 enhance embryonic lethality when a subset of chromosomes fails to form a CO? Due to feedback mechanisms, more DSBs are induced when not all homologs are connected by COs (Rosu et al. 2013; Stamper et al. 2013), and, in the absence of BRC-1-BRD-1, more breaks may be repaired through error-prone pathways, potentially leading to an increase in mutations. Additionally, mutation of brc-1 enhanced CO distribution defects as well as the number of DCOs on some chromosomes in the zim-1 mutant background (Figure 7). Alteration in CO position (Altendorfer et al. 2020) as well as elevated CO numbers (Hollis et al. 2020) are deleterious during C. elegans meiosis. This is likely a consequence of the holocentric nature of C. elegans chromosomes and the requirement to establish asymmetric domains as defined by the single CO site for accurate cohesion release and chromosome segregation (de Carvalho et al. 2008; Ferrandiz et al. 2018). Additionally, DSBs on chromosomes that cannot undergo CO formation during male meiosis may fail to be repaired prior to the meiotic divisions due to defects in BRC-1-BRD-1-dependent intersister repair, leading to chromosome fragmentation, loss of genetic material and aneuploid gametes.
Sex-specific regulation of meiosis
Our analyses of BRC-1-BRD-1 reveals several differences between male and female meiosis. First, while there is currently no direct measure of DSB formation in C. elegans, we detected more RAD-51 foci in male vs. female germ cells, suggesting that more DSBs are induced in spermatocytes (Figure 2) (Checchi et al. 2014). Usage of DSBs hotspots in mice has also revealed sex-specific differences (Brick et al. 2018). Second, BRC-1-BRD-1 functions at different steps of meiotic recombination in the sexes in wild-type worms (Figure 8). In males, BRC-1-BRD-1 influences the early processing of DSBs to promote HR, while in females, BRC-1-BRD-1 is engaged in mid- to late pachytene to promote repair of breaks processed and assembled with RAD-51 by intersister recombination (Adamo et al. 2008). How BRC-1-BRD-1 is differentially regulated in the sexes is not known, but the spatiotemporal pattern of BRC-1-BRD-1 function mirrors MAP kinase activation in the male (transition zone/early pachytene) and female (mid- to late-pachytene) germ lines (Lee et al. 2007). Thus, MAP kinase and/or other signaling pathways could regulate the complex in a sex-specific manner to drive ubiquitination of different substrates in spermatogenesis vs. oogenesis.
Overall, C. elegans male meiosis appears to be less tightly regulated compared to female meiosis. For example, we detected DCOs in wild-type male meiosis (Figure 7), but none in oocytes (Li et al. 2018). Further, previous analyses have shown that males undergo meiosis faster and lack germ line apoptosis, one mechanism to enhance gamete quality by removing defective or damaged germ cells (Gartner et al. 2000; Jaramillo-Lambert et al. 2007, 2010). Despite faster kinetics and lack of germline apoptosis, male meiosis has a higher fidelity compared to female meiosis (Jaramillo-Lambert et al. 2010). Why male meiosis appears to lack some regulatory mechanisms yet has a reduced frequency of meiotic errors compared to oogenesis is currently unknown. Future analyses of C. elegans male meiosis may provide insight into the mechanisms that contribute to the fidelity of male gametes.
Acknowledgments
We are grateful to Ben Mallory for generating zim-3(xoe15), Jonathan Amezquita for generating zim-1(xoe6) and Lauren Ahmann, Arshdeep Kaur, and Tara Shahrvini for help with construction of strains. We thank Takamune Saito and Marina Martinez-Garcia for advice on meiotic mapping experiments and the Engebrecht laboratory for thoughtful discussions. We also thank Nicholas Vera for help on quantifying GFP::RPA fluorescence. We thank the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440) for providing strains. This work was supported by NIH GM103860 and GM103860S1 to J.E.
Footnotes
Supplemental material available at figshare: https://doi.org/10.25386/genetics.12730904.
Communicating editor: A. MacQueen
Literature Cited
- Abraham R. T., 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15: 2177–2196. 10.1101/gad.914401 [DOI] [PubMed] [Google Scholar]
- Adamo A., Montemauri P., Silva N., Ward J. D., Boulton S. J. et al. , 2008. BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair. EMBO Rep. 9: 287–292. 10.1038/sj.embor.7401167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson D. G., Rose A. M., and Villeneuve A. M., 1997. Chromosome organization, mitosis, and meiosis, in C. elegans II, edited by Riddle D. L., Blumenthal T., Meyer B. J., and Priess J. R., Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- Altendorfer E., Lascarez-Lagunas L. I., Nadarajan S., Mathieson I. and Colaiacovo M. P., 2020. Crossover position drives chromosome remodeling for accurate meiotic chromosome segregation. Curr. Biol. 30: 1329–1338.e7. 10.1016/j.cub.2020.01.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio T., Baer R., Gottesman M., and Gautier J., 2016. MRN, CtIP, and BRCA1 mediate repair of topoisomerase II-DNA adducts. J. Cell Biol. 212: 399–408. 10.1083/jcb.201504005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T., Plug A. W., Xu J., Solari A. J., Reddy G. et al. , 1995. Dynamic changes in Rad51 distribution on chromatin during meiosis in male and female vertebrates. Chromosoma 104: 19–28. 10.1007/BF00352222 [DOI] [PubMed] [Google Scholar]
- Barnes T. M., Kohara Y., Coulson A., and Hekimi S., 1995. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 141: 159–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan G. C., and Hillers K. J., 2011. SNP-based mapping of crossover recombination in Caenorhabditis elegans. Methods Mol. Biol. 745: 207–222. 10.1007/978-1-61779-129-1_13 [DOI] [PubMed] [Google Scholar]
- Bean C. J., Schaner C. E., and Kelly W. G., 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105. 10.1038/ng1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler J. B., Andersen E. C., and Villeneuve A. M., 2010. Differential localization and independent acquisition of the H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 6: e1000830 10.1371/journal.pgen.1000830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop H. I., Guan D., Bocksteins E., Parajuli L. K., Murray K. D. et al. , 2015. Distinct cell- and layer-specific expressionpatterns and independent regulation of Kv2 channel subtypes in cortical pyramidal neurons. J. Neurosci. 35: 14922–14942. 10.1523/JNEUROSCI.1897-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., 2006. BRCA1-mediated ubiquitylation. Cell Cycle 5: 1481–1486. 10.4161/cc.5.14.2930 [DOI] [PubMed] [Google Scholar]
- Boulton S. J., Martin J. S., Polanowska J., Hill D. E., Gartner A. et al. , 2004. BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr. Biol. 14: 33–39. 10.1016/j.cub.2003.11.029 [DOI] [PubMed] [Google Scholar]
- Bouwman P., Aly A., Escandell J. M., Pieterse M., Bartkova J. et al. , 2010. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 17: 688–695. 10.1038/nsmb.1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady M. M., McMahan S., and Sekelsky J., 2018. Loss of Drosophila Mei-41/ATR alters meiotic crossover patterning. Genetics 208: 579–588. 10.1534/genetics.117.300634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick K., Thibault-Sennett S., Smagulova F., Lam K. G., Pu Y. et al. , 2018. Extensive sex differences at the initiation of genetic recombination. Nature 561: 338–342. 10.1038/s41586-018-0492-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering T. J., Alavattam K. G., Sadreyev R. I., Ichijima Y., Kato Y. et al. , 2014. BRCA1 establishes DNA damage signaling and pericentric heterochromatin of the X chromosome in male meiosis. J. Cell Biol. 205: 663–675. 10.1083/jcb.201311050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting S. F., Callen E., Wong N., Chen H. T., Polato F. et al. , 2010. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141: 243–254. 10.1016/j.cell.2010.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury L., Coelho P. A., and Glover D. M., 2016. From meiosis to mitosis: the astonishing flexibility of cell division mechanisms in early mammalian development. Curr. Top. Dev. Biol. 120: 125–171. 10.1016/bs.ctdb.2016.04.011 [DOI] [PubMed] [Google Scholar]
- Cahoon C. K., and Libuda D. E., 2019. Leagues of their own: sexually dimorphic features of meiotic prophase I. Chromosoma 128: 199–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon C. K., Helm J. M., and Libuda D. E., 2019. Synaptonemal complex central region proteins promote localization of pro-crossover factors to recombination events during Caenorhabditis elegans meiosis. Genetics 213: 395–409. 10.1534/genetics.119.302625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Xu X., Bunting S. F., Liu J., Wang R. H. et al. , 2009. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol. Cell 35: 534–541. 10.1016/j.molcel.2009.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton P. M., Farruggio A. P., and Dernburg A. F., 2006. A link between meiotic prophase progression and crossover control. PLoS Genet. 2: e12 10.1371/journal.pgen.0020012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checchi P. M., and Engebrecht J., 2011. Caenorhabditis elegans histone methyltransferase MET-2 shields the male X chromosome from checkpoint machinery and mediates meiotic sex chromosome inactivation. PLoS Genet. 7: e1002267 10.1371/journal.pgen.1002267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checchi P. M., Lawrence K. S., Van M. V., Larson B. J., and Engebrecht J., 2014. Pseudosynapsis and decreased stringency of meiotic repair pathway choice on the hemizygous sex chromosome of Caenorhabditis elegans males. Genetics 197: 543–560. 10.1534/genetics.114.164152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin G. M., and Villeneuve A. M., 2001. C. elegans mre-11 is required for meiotic recombination and DNA repair but is dispensable for the meiotic G(2) DNA damage checkpoint. Genes Dev. 15: 522–534. 10.1101/gad.864101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung G., Rose A. M., Petalcorin M. I., Martin J. S., Kessler Z. et al. , 2015. REC-1 and HIM-5 distribute meiotic crossovers and function redundantly in meiotic double-strand break formation in Caenorhabditis elegans. Genes Dev. 29: 1969–1979. 10.1101/gad.266056.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A. et al. , 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell 5: 463–474. 10.1016/S1534-5807(03)00232-6 [DOI] [PubMed] [Google Scholar]
- Cruz-García A., Lopez-Saavedra A., and Huertas P., 2014. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 9: 451–459. 10.1016/j.celrep.2014.08.076 [DOI] [PubMed] [Google Scholar]
- Daley J. M., and Sung P., 2014. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 34: 1380–1388. 10.1128/MCB.01639-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Carvalho C. E., Zaaijer S., Smolikov S., Gu Y., Schumacher J. M. et al. , 2008. LAB-1 antagonizes the Aurora B kinase in C. elegans. Genes Dev. 22: 2869–2885. 10.1101/gad.1691208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Densham R. M., Garvin A. J., Stone H. R., Strachan J., Baldock R. A. et al. , 2016. Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23: 647–655. 10.1038/nsmb.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M. et al. , 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. 10.1016/S0092-8674(00)81481-6 [DOI] [PubMed] [Google Scholar]
- Enguita-Marruedo A., Martin-Ruiz M., Garcia E., Gil-Fernandez A., Parra M. T. et al. , 2019. Transition from a meiotic to a somatic-like DNA damage response during the pachytene stage in mouse meiosis. PLoS Genet. 15: e1007439 10.1371/journal.pgen.1007439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O., Mahadevaiah S. K., Celeste A., Romanienko P. J., Camerini-Otero R. D. et al. , 2003. H2AX is required for chromatin remodeling and inactivation of sex chromosomes in male mouse meiosis. Dev. Cell 4: 497–508. 10.1016/S1534-5807(03)00093-5 [DOI] [PubMed] [Google Scholar]
- Ferrandiz N., Barroso C., Telecan O., Shao N., Kim H. M. et al. , 2018. Spatiotemporal regulation of Aurora B recruitment ensures release of cohesion during C. elegans oocyte meiosis. Nat. Commun. 9: 834 (erratum: Nat. Commun. 9: 3558). 10.1038/s41467-018-03229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T., and Boulton S. J., 2005. Distinct modes of ATR activation after replication stress and DNA double-strand breaks in Caenorhabditis elegans. EMBO J. 24: 4345–4355. 10.1038/sj.emboj.7600896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Muse T., Galindo-Diaz U., Garcia-Rubio M., Martin J. S., Polanowska J. et al. , 2019. A meiotic checkpoint alters repair partner bias to permit Inter-sister repair of persistent DSBs. Cell Rep. 26: 775–787.e5. 10.1016/j.celrep.2018.12.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., and Hengartner M. O., 2000. A conserved checkpoint pathway mediates DNA damage–induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. 10.1016/S1097-2765(00)80438-4 [DOI] [PubMed] [Google Scholar]
- Girard C., Roelens B., Zawadzki K. A., and Villeneuve A. M., 2018. Interdependent and separable functions of Caenorhabditis elegans MRN-C complex members couple formation and repair of meiotic DSBs. Proc. Natl. Acad. Sci. USA 115: E4443–E4452. 10.1073/pnas.1719029115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn J. R., Rubio C., Broman K. W., Hunt P. A., and Hassold T., 2013. Cytological studies of human meiosis: sex-specific differences in recombination originate at, or prior to, establishment of double-strand breaks. PLoS One 8: e85075 10.1371/journal.pone.0085075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund M., Davis M. W., Nguyen H., Dayton D., and Jorgensen E. M., 2005. Heterozygous insertions alter crossover distribution but allow crossover interference in Caenorhabditis elegans. Genetics 171: 1047–1056. 10.1534/genetics.105.044834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E., Mizuno K., Molnar M., Kohli J., Ohta K. et al. , 2009. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol. Cell. Biol. 29: 1671–1681. 10.1128/MCB.01182-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M., Chin G. M., and Villeneuve A. M., 2007. C. elegans germ cells switch between distinct modes of double-strand break repair during meiotic prophase progression. PLoS Genet. 3: e191 10.1371/journal.pgen.0030191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers K. J., and Villeneuve A. M., 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13: 1641–1647. 10.1016/j.cub.2003.08.026 [DOI] [PubMed] [Google Scholar]
- Hillers, K. J., V. Jantsch, E. Martinez-Perez and J. L. Yanowitz, 2015 Meiosis. (May 4, 2017), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.178.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed]
- Hirota T., Blakeley P., Sangrithi M. N., Mahadevaiah S. K., Encheva V. et al. , 2018. SETDB1 links the meiotic DNA damage response to sex chromosome silencing in mice. Dev. Cell 47: 645–659.e6. 10.1016/j.devcel.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis, J. A., M. L. Glover, A. Schlientz, C. K. Cahoon, B. Bowerman et al., 2020 Excess crossovers impede faithful meiotic chromosome segregation in C. elegans. bioRxiv doi: 10.1101/2020.01.30.927640 (Preprint posted January 31, 2020). [DOI] [PMC free article] [PubMed]
- Hong Y., Sonneville R., Agostinho A., Meier B., Wang B. et al. , 2016. The SMC-5/6 complex and the HIM-6 (BLM) helicase synergistically promote meiotic recombination intermediate processing and chromosome maturation during Caenorhabditis elegans meiosis. PLoS Genet. 12: e1005872 10.1371/journal.pgen.1005872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J. P., and Manley J. L., 2012. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 26: 2119–2137. 10.1101/gad.200303.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlock M. E., Cavka I., Kursel L. E., Haversat J., Wooten M. et al. , 2020. Identification of novel synaptonemal complex components in C. elegans. J. Cell Biol. 219: e201910043 10.1083/jcb.201910043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagut M., Hamminger P., Woglar A., Millonigg S., Paulin L. et al. , 2016. Separable roles for a Caenorhabditis elegans RMI1 homolog in promoting and antagonizing meiotic crossovers ensure faithful fhromosome inheritance. PLoS Biol. 14: e1002412 10.1371/journal.pbio.1002412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisiw E., Dello Stritto M. R., Jantsch V., and Silva N., 2018. BRCA1-BARD1 associate with the synaptonemal complex and pro-crossover factors and influence RAD-51 dynamics during Caenorhabditis elegans meiosis. PLoS Genet. 14: e1007653 10.1371/journal.pgen.1007653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janisiw, E., M. Raices, F. Balmir, L. P. Paz, A. Baudrimont et al., 2020 Poly(ADP-ribose) glycohydrolase promotes formation and homology-directed repair of meiotic DNA double-strand breaks independent of its catalytic activity. bioRxiv doi: 10.1101/2020.03.12.988840 (Preprint posted March 13, 2020)
- Jaramillo-Lambert A., and Engebrecht J., 2010. A single unpaired and transcriptionally silenced X chromosome locally precludes checkpoint signaling in the Caenorhabditis elegans germ line. Genetics 184: 613–628. 10.1534/genetics.109.110338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Ellefson M., Villeneuve A. M., and Engebrecht J., 2007. Differential timing of S phases, X chromosome replication, and meiotic prophase in the C. elegans germ line. Dev. Biol. 308: 206–221. 10.1016/j.ydbio.2007.05.019 [DOI] [PubMed] [Google Scholar]
- Jaramillo-Lambert A., Harigaya Y., Vitt J., Villeneuve A., and Engebrecht J., 2010. Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr. Biol. 20: 2078–2089. 10.1016/j.cub.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce E. F., Paul A., Chen K. E., Tanneti N., and McKim K. S., 2012. Multiple barriers to nonhomologous DNA end joining during meiosis in Drosophila. Genetics 191: 739–746. 10.1534/genetics.112.140996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp J. A., van Schendel R., Dilweg I. W., and Tijsterman M., 2020. BRCA1-associated structural variations are a consequence of polymerase theta-mediated end-joining. Nat. Commun. 11: 3615 10.1038/s41467-020-17455-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur T., and Rockman M. V., 2014. Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics 196: 137–148. 10.1534/genetics.113.158857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney S., Giroux C. N., and Kleckner N., 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88: 375–384. 10.1016/S0092-8674(00)81876-0 [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M. H., Kim S. K. et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kianian P. M. A., Wang M., Simons K., Ghavami F., He Y. et al. , 2018. High-resolution crossover mapping reveals similarities and differences of male and female recombination in maize. Nat. Commun. 9: 2370 10.1038/s41467-018-04562-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koury E., Harrell K., and Smolikove S., 2018. Differential RPA-1 and RAD-51 recruitment in vivo throughout the C. elegans germline, as revealed by laser microirradiation. Nucleic Acids Res. 46: 748–764. 10.1093/nar/gkx1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouznetsova A., Wang H., Bellani M., Camerini-Otero R. D., Jessberger R. et al. , 2009. BRCA1-mediated chromatin silencing is limited to oocytes with a small number of asynapsed chromosomes. J. Cell Sci. 122: 2446–2452. 10.1242/jcs.049353 [DOI] [PubMed] [Google Scholar]
- Larson B. J., Van M. V., Nakayama T., and Engebrecht J., 2016. Plasticity in the meiotic epigenetic landscape of sex chromosomes in Caenorhabditis species. Genetics 203: 1641–1658. 10.1534/genetics.116.191130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence K. S., Tapley E. C., Cruz V. E., Li Q., Aung K. et al. , 2016. LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. J. Cell Biol. 215: 801–821. 10.1083/jcb.201604112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Ohmachi M., Arur S., Nayak S., Francis R. et al. , 2007. Multiple functions and dynamic activation of MPK-1 extracellular signal-regulated kinase signaling in Caenorhabditis elegans germline development. Genetics 177: 2039–2062. 10.1534/genetics.107.081356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmens B. B., Johnson N. M., and Tijsterman M., 2013. COM-1 promotes homologous recombination during Caenorhabditis elegans meiosis by antagonizing Ku-mediated non-homologous end joining. PLoS Genet. 9: e1003276 10.1371/journal.pgen.1003276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenormand T., and Dutheil J., 2005. Recombination difference between sexes: a role for haploid selection. PLoS Biol. 3: e63 10.1371/journal.pbio.0030063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M. L., and Greenberg R. A., 2012. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem. Sci. 37: 418–424. 10.1016/j.tibs.2012.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Saito T. T., Martinez-Garcia M., Deshong A. J., Nadarajan S. et al. , 2018. The tumor suppressor BRCA1-BARD1 complex localizes to the synaptonemal complex and regulates recombination under meiotic dysfunction in Caenorhabditis elegans. PLoS Genet. 14: e1007701 10.1371/journal.pgen.1007701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., and Yanowitz J. L., 2019. ATM and ATR influence meiotic crossover formation through antagonistic and overlapping functions in Caenorhabditis elegans. Genetics 212: 431–443. 10.1534/genetics.119.302193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J. G., Stine R. R., and Yanowitz J. L., 2008. Domain-specific regulation of recombination in Caenorhabditis elegans in response to temperature, age and sex. Genetics 180: 715–726. 10.1534/genetics.108.090142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A., and Jenczewski E., 2019. Modelling sex-specific crossover patterning in Arabidopsis. Genetics 211: 847–859. 10.1534/genetics.118.301838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L. Y., and Yu X., 2015. Double-strand break repair on sex chromosomes: challenges during male meiotic prophase. Cell Cycle 14: 516–525. 10.1080/15384101.2014.998070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui D. Y., and Colaiacovo M. P., 2013. Meiotic development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757: 133–170. 10.1007/978-1-4614-4015-4_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaisne N., Kessler Z., and Yanowitz J. L., 2018. Meiotic double-strand break proteins influence repair pathway utilization. Genetics 210: 843–856. 10.1534/genetics.118.301402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machovina T. S., Mainpal R., Daryabeigi A., McGovern O., Paouneskou D. et al. , 2016. A surveillance system ensures crossover formation in C. elegans. Curr. Biol. 26: 2873–2884. 10.1016/j.cub.2016.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Colaiacovo M. P., McDonald K., and Villeneuve A. M., 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16: 2428–2442. 10.1101/gad.1011602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah S. K., Bourc’his D., de Rooij D. G., Bestor T. H., Turner J. M. et al. , 2008. Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation. J. Cell Biol. 182: 263–276. 10.1083/jcb.200710195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine E. M., 2010. Meiotic silencing in Caenorhabditis elegans. Int. Rev. Cell Mol. Biol. 282: 91–134. 10.1016/S1937-6448(10)82002-7 [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R. 3rd, Hurov K. E. et al. , 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166. 10.1126/science.1140321 [DOI] [PubMed] [Google Scholar]
- Meneely P. M., Farago A. F., and Kauffman T. M., 2002. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics 162: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneely P. M., McGovern O. L., Heinis F. I., and Yanowitz J. L., 2012. Crossover distribution and frequency are regulated by him-5 in Caenorhabditis elegans. Genetics 190: 1251–1266. 10.1534/genetics.111.137463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mets D. G., and Meyer B. J., 2009. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell 139: 73–86. 10.1016/j.cell.2009.07.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mézard C., Jahns M. T., and Grelon M., 2015. Where to cross? New insights into the location of meiotic crossovers. Trends Genet. 31: 393–401. 10.1016/j.tig.2015.03.008 [DOI] [PubMed] [Google Scholar]
- Moens P. B., Chen D. J., Shen Z., Kolas N., Tarsounas M. et al. , 1997. Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106: 207–215. 10.1007/s004120050241 [DOI] [PubMed] [Google Scholar]
- Morelli M. A., and Cohen P. E., 2005. Not all germ cells are created equal: aspects of sexual dimorphism in mammalian meiosis. Reproduction 130: 761–781. 10.1530/rep.1.00865 [DOI] [PubMed] [Google Scholar]
- Mu J. J., Wang Y., Luo H., Leng M., Zhang J., et al. , 2007. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J. Biol. Chem. 282: 17330–17334. 10.1074/jbc.C700079200 [DOI] [PubMed] [Google Scholar]
- Murakami H., Lam I., Huang P. C., Song J., van Overbeek M., et al. , 2020. Multilayered mechanisms ensure that short chromosomes recombine in meiosis. Nature 582: 124–128. 10.1038/s41586-020-2248-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., and Hillers K. J., 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292. 10.1534/genetics.104.030700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajan S., Lambert T. J., Altendorfer E., Gao J., Blower M. D. et al. , 2017. Polo-like kinase-dependent phosphorylation of the synaptonemal complex protein SYP-4 regulates double-strand break formation through a negative feedback loop. eLife 6: e23437 10.7554/eLife.23437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S. I., Hassold T. J., and Hunt P. A., 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13: 493–504. 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiano J., Wu W., Yamada S., Sciascia N., Callen E. et al. , 2020. ATM and PRDM9 regulate SPO11-bound recombination intermediates during meiosis. Nat. Commun. 11: 857 10.1038/s41467-020-14654-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D., Roelens B., Woglar A., and Villeneuve A. M., 2017. Meiotic recombination modulates the structure and dynamics of the synaptonemal complex during C. elegans meiosis. PLoS Genet. 13: e1006670 10.1371/journal.pgen.1006670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., and Dernburg A. F., 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11: 817–829. 10.1016/j.devcel.2006.09.020 [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P. et al. , 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063. 10.1016/j.cell.2005.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polanowska J., Martin J. S., Garcia-Muse T., Petalcorin M. I., and Boulton S. J., 2006. A conserved pathway to activate BRCA1-dependent ubiquitylation at DNA damage sites. EMBO J. 25: 2178–2188. 10.1038/sj.emboj.7601102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjha L., Howard S. M., and Cejka P., 2018. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma 127: 187–214. 10.1007/s00412-017-0658-1 [DOI] [PubMed] [Google Scholar]
- Reuben M., and Lin R., 2002. Germline X chromosomes exhibit contrasting patterns of histone H3 methylation in Caenorhabditis elegans. Dev. Biol. 245: 71–82. 10.1006/dbio.2002.0634 [DOI] [PubMed] [Google Scholar]
- Rinaldo C., Bazzicalupo P., Ederle S., Hilliard M., and La Volpe A., 2002. Roles for Caenorhabditis elegans rad-51 in meiosis and in resistance to ionizing radiation during development. Genetics 160: 471–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockman M. V., and Kruglyak L., 2009. Recombinational landscape and population genomics of Caenorhabditis elegans. PLoS Genet. 5: e1000419 10.1371/journal.pgen.1000419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosu S., Zawadzki K. A., Stamper E. L., Libuda D. E., Reese A. L. et al. , 2013. The C. elegans DSB-2 protein reveals a regulatory network that controls competence for meiotic DSB formation and promotes crossover assurance. PLoS Genet. 9: e1003674 10.1371/journal.pgen.1003674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H., Polikiewicz G., Mahadevaiah S. K., Prosser H., Mitchell M. et al. , 2010. Evidence that meiotic sex chromosome inactivation is essential for male fertility. Curr. Biol. 20: 2117–2123. 10.1016/j.cub.2010.11.010 [DOI] [PubMed] [Google Scholar]
- Royo H., Prosser H., Ruzankina Y., Mahadevaiah S. K., Cloutier J. M. et al. , 2013. ATR acts stage specifically to regulate multiple aspects of mammalian meiotic silencing. Genes Dev. 27: 1484–1494. 10.1101/gad.219477.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., and Colaiacovo M. P., 2017. Regulation of crossover frequency and distribution during meiotic recombination. Cold Spring Harb. Symp. Quant. Biol. 82: 223–234. 10.1101/sqb.2017.82.034132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Mohideen F., Meyer K., Harper J. W., and Colaiacovo M. P., 2012. SLX-1 is required for maintaining genomic integrity and promoting meiotic noncrossovers in the Caenorhabditis elegans germline. PLoS Genet. 8: e1002888 10.1371/journal.pgen.1002888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T. T., Lui D. Y., Kim H. M., Meyer K., and Colaiacovo M. P., 2013. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLoS Genet. 9: e1003586 10.1371/journal.pgen.1003586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori A. A., Lukas C., Coates J., Mistrik M., Fu S. et al. , 2007. Human CtIP promotes DNA end resection. Nature 450: 509–514. 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage K. I., and Harkin D. P., 2015. BRCA1, a ‘complex’ protein involved in the maintenance of genomic stability. FEBS J. 282: 630–646. 10.1111/febs.13150 [DOI] [PubMed] [Google Scholar]
- Schedl T., and Kimble J., 1988. fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119: 43–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciurano R. B., Rahn M. I., Pigozzi M. I., Olmedo S. B., and Solari A. J., 2006. An azoospermic man with a double-strand DNA break-processing deficiency in the spermatocyte nuclei: case report. Hum. Reprod. 21: 1194–1203. 10.1093/humrep/dei479 [DOI] [PubMed] [Google Scholar]
- Shakes D. C., Wu J. C., Sadler P. L., Laprade K., Moore L. L. et al. , 2009. Spermatogenesis-specific features of the meiotic program in Caenorhabditis elegans. PLoS Genet. 5: e1000611 10.1371/journal.pgen.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She X., Xu X., Fedotov A., Kelly W. G., and Maine E. M., 2009. Regulation of heterochromatin assembly on unpaired chromosomes during Caenorhabditis elegans meiosis by components of a small RNA-mediated pathway. PLoS Genet. 5: e1000624 10.1371/journal.pgen.1000624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Eizinger A., Hurlburt A., Rogers E., Villeneuve A. M. et al. , 2007. Synapsis-defective mutants reveal a correlation between chromosome conformation and the mode of double-strand break repair during Caenorhabditis elegans meiosis. Genetics 176: 2027–2033. 10.1534/genetics.107.076968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolikov S., Schild-Prufert K., and Colaiacovo M. P., 2008. CRA-1 uncovers a double-strand break-dependent pathway promoting the assembly of central region proteins on chromosome axes during C. elegans meiosis. PLoS Genet. 4: e1000088 10.1371/journal.pgen.1000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R., Querenet M., Craig A., Gartner A., and Blow J. J., 2012. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J. Cell Biol. 196: 233–246. 10.1083/jcb.201110080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper E. L., Rodenbusch S. E., Rosu S., Ahringer J., Villeneuve A. M. et al. , 2013. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLoS Genet. 9: e1003679 10.1371/journal.pgen.1003679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapley J., Feulner P. G. D., Johnston S. E., Santure A. W., and Smadja C. M., 2017. Variation in recombination frequency and distribution across eukaryotes: patterns and processes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372: 20160455 [corrigenda: Philos. Trans. R. Soc. Lond. B Biol. Sci. 373: 20170360 (2018)]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka M., and Miki Y., 2018. BRCA1 gene: function and deficiency. Int. J. Clin. Oncol. 23: 36–44. 10.1007/s10147-017-1182-2 [DOI] [PubMed] [Google Scholar]
- Turner J. M., 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. 10.1242/dev.000018 [DOI] [PubMed] [Google Scholar]
- Turner J. M., Aprelikova O., Xu X., Wang R., Kim S. et al. , 2004. BRCA1, histone H2AX phosphorylation, and male meiotic sex chromosome inactivation. Curr. Biol. 14: 2135–2142. 10.1016/j.cub.2004.11.032 [DOI] [PubMed] [Google Scholar]
- Van M. V., Larson B. J., and Engebrecht J., 2016. To break or not to break: sex chromosome hemizygosity during meiosis in Caenorhabditis. Genetics 204: 999–1013. 10.1534/genetics.116.194308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C. R., Kuervers L., Baillie D. L. and Yanowitz J. L., 2010. xnd-1 regulates the global recombination landscape in Caenorhabditis elegans. Nature 467: 839–843. 10.1038/nature09429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., and Villeneuve A. M., 2018. Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell 173: 1678–1691.e16. 10.1016/j.cell.2018.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Aprelikova O., Moens P., Deng C. X., and Furth P. A., 2003. Impaired meiotic DNA-damage repair and lack of crossing-over during spermatogenesis in BRCA1 full-length isoform deficient mice. Development 130: 2001–2012. 10.1242/dev.00410 [DOI] [PubMed] [Google Scholar]
- Yang B., Xu X., Russell L., Sullenberger M. T., Yanowitz J. L. et al. , 2019. A DNA repair protein and histone methyltransferase interact to promote genome stability in the Caenorhabditis elegans germ line. PLoS Genet. 15: e1007992 10.1371/journal.pgen.1007992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y., and Smolikove S., 2013. Impaired resection of meiotic double-strand breaks channels repair to nonhomologous end joining in Caenorhabditis elegans. Mol. Cell. Biol. 33: 2732–2747. 10.1128/MCB.00055-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoo R., Zawadzki K. A., Nabeshima K., Drake M., Arur S. et al. , 2012. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149: 75–87. 10.1016/j.cell.2012.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Kim Y., and Dernburg A. F., 2016. Meiotic recombination and the crossover assurance checkpoint in Caenorhabditis elegans. Semin. Cell Dev. Biol. 54: 106–116. 10.1016/j.semcdb.2016.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazowski M. J., Sandoval M., Paniker L., Hamilton H. M., Han J. et al. , 2017. Age-dependent alterations in meiotic recombination cause chromosome segregation errors in spermatocytes. Cell 171: 601–614.e13. 10.1016/j.cell.2017.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M. C., and Rose A. M., 1995. Mutant rec-1 eliminates the meiotic pattern of crossing over in Caenorhabditis elegans. Genetics 141: 1339–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and reagents are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are represented fully within the article and its tables and figures. Supplemental material available at figshare: https://doi.org/10.25386/genetics.12730904.