Abstract
The control of body and organ growth is essential for the development of adults with proper size and proportions, which is important for survival and reproduction. In animals, adult body size is determined by the rate and duration of juvenile growth, which are influenced by the environment. In nutrient-scarce environments in which more time is needed for growth, the juvenile growth period can be extended by delaying maturation, whereas juvenile development is rapidly completed in nutrient-rich conditions. This flexibility requires the integration of environmental cues with developmental signals that govern internal checkpoints to ensure that maturation does not begin until sufficient tissue growth has occurred to reach a proper adult size. The Target of Rapamycin (TOR) pathway is the primary cell-autonomous nutrient sensor, while circulating hormones such as steroids and insulin-like growth factors are the main systemic regulators of growth and maturation in animals. We discuss recent findings in Drosophila melanogaster showing that cell-autonomous environment and growth-sensing mechanisms, involving TOR and other growth-regulatory pathways, that converge on insulin and steroid relay centers are responsible for adjusting systemic growth, and development, in response to external and internal conditions. In addition to this, proper organ growth is also monitored and coordinated with whole-body growth and the timing of maturation through modulation of steroid signaling. This coordination involves interorgan communication mediated by Drosophila insulin-like peptide 8 in response to tissue growth status. Together, these multiple nutritional and developmental cues feed into neuroendocrine hubs controlling insulin and steroid signaling, serving as checkpoints at which developmental progression toward maturation can be delayed. This review focuses on these mechanisms by which external and internal conditions can modulate developmental growth and ensure proper adult body size, and highlights the conserved architecture of this system, which has made Drosophila a prime model for understanding the coordination of growth and maturation in animals.
Keywords: checkpoint, critical weight, DILP8, Drosophila, ecdysone, insulin, metamorphosis, prothoracic gland, PTTH, timing, FlyBook
THE nature of the mechanisms by which animals control the growth of their bodies and their different parts to produce adults of correct size and proportions is a fundamental question. Studies in Drosophila have provided insight into these questions through the identification of systems that link body and organ growth to environmental and developmental cues. This research illustrates how organs exchange external- and internal-status information via circulating hormones, and how this information is integrated by the neuroendocrine circuitry regulating insulin-like growth factor and steroid hormone signaling, the two main factors that underlie developmental growth regulation and coordination.
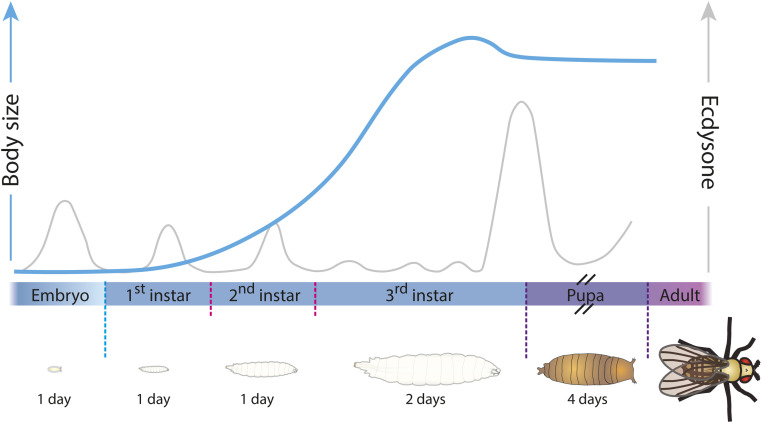
In many animals, growth is largely restricted to the juvenile stage, and adult body size is therefore determined by the size at which the juvenile undergoes maturation (Tennessen and Thummel 2011). Intrinsic developmental programs that determine species-specific size are modulated by environmental cues to produce adults with proper size and proportions in changing environments. These environmental factors affect the rate of growth as well as the timing of maturation, which ends the juvenile growth period. In Drosophila, almost all growth occurs in the larval stage, which is terminated by pupariation, which marks the onset of metamorphosis, the transition to adulthood comparable with mammalian puberty (Figure 1) (Yamanaka et al. 2013a; Boulan et al. 2015; Juarez-Carreño et al. 2018). In Drosophila, nutritional status is linked to a checkpoint called critical weight (CW) that occurs early in the final larval instar, which is important for determining final body size (Mirth and Riddiford 2007). Insulin regulates CW and is the primary hormone mediating systemic growth control in response to nutrient sensing, while cellular nutrient sensing is mediated by the Target of Rapamycin (TOR) pathway. The main nutrient-sensing tissue is the fat body, which receives information from cellular levels of amino acids through TOR as well as other environmental conditions including oxygen levels (Colombani et al. 2003; Texada et al. 2019a). In response to these cues, the fat body secretes adipokines that mediate systemic growth responses through their regulatory effects on insulin signaling (Rajan and Perrimon 2012; Sano et al. 2015; Agrawal et al. 2016; Delanoue et al. 2016; Koyama and Mirth 2016; Texada et al. 2019a).
Figure 1.
The development of D. melanogaster. A fertilized Drosophila embryo spends roughly 1 day developing into a mobile, feeding larva (under normal conditions). After hatching, the larva feeds for the next 4 days, growing to 200 times its initial size; to accommodate this dramatic growth, the larva sheds its cuticle twice during this time in molts that separate the first, second, and third larval “instars.” After larval growth is complete, the animal wanders away from its food source to find a location suitable for the 4-day metamorphosis period, during which time the animal survives on stored material while its larval tissues are degraded and adult structures finish their development. The adult emerges (“ecloses”) once this process is complete. Pulses of the insect steroid hormone ecdysone regulate the animal’s progression through these developmental stages.
The steroid ecdysone is the key factor regulating developmental transitions in Drosophila. Pulses of ecdysone control molting and metamorphosis (Figure 1), while between pulses, the lower, basal level of ecdysone negatively regulates the growth of larval tissues by antagonizing insulin signaling (Colombani et al. 2005; Yamanaka et al. 2013a; Moeller et al. 2017). Thus, the interaction between insulin and ecdysone controls final body size. In addition to the nutritional checkpoint at CW, the larval growth period is determined by a checkpoint that assesses the growth status of imaginal tissues, primordia that give rise during metamorphosis to adult body structures (Rewitz et al. 2013). Imaginal disc damage or growth retardation inhibits ecdysone production, and thus induces a delay in pupariation to allow regeneration and compensatory growth, thereby maintaining proper organ proportions. Recently, DILP8 was identified as the hormone released by discs that delays pupariation in response to tissue damage (Colombani et al. 2012; Garelli et al. 2012). As with nutrition, the main focal points of the developmental checkpoint activated by disc-derived DILP8 are the regulation of insulin and ecdysone signaling. Thus, multiple developmental and nutritional signals converge on neuroendocrine hubs, regulating insulin and ecdysone, to couple environment and growth to maturation. Recent studies of Drosophila have provided new perspectives and uncovered remarkable conservation of these pathways, providing the framework for understanding how animals coordinate organ and body growth with developmental transitions. Here, we review recent findings that link environmental factors, organ growth, maturation timing, and body size in Drosophila, along with the cellular and systemic signals that regulate body and organ growth in the fly.
Regulation of Cell Size and Number
Achieving an appropriate size is a critical aspect of development for individual cells, tissues, organs, and whole animals. Body and tissue size can be thought of as the product of growth rate and growth duration; it can also be thought of as the product of cell number and cell size. The processes that mediate systemic growth and proliferation control, including nutrition-linked hormones that modulate insulin production and release, or developmental assessments that time developmental transitions, are discussed further below. These systemic factors act through their effects within individual cells, where the information they convey is integrated with intracellular pathways that reflect each cell’s tissue context and its internal metabolic state. Through the combined effects of these layers of control, cells regulate their own size, through modulating the uptake of raw materials and the synthesis of new cellular components, and their number, by controlling cell proliferation and apoptosis.
At the finest level of growth and proliferation control, each cell must sense its own metabolite levels and use these data to evaluate whether it possesses the necessary raw ingredients for the production of more proteins, membrane lipids, and genomic DNA before inducing cell growth or mitosis. The main intracellular sensory apparatus underlying this control is the TOR pathway, an evolutionarily ancient system predating the divergence of fungi and animals, that integrates a wide variety of intracellular growth-governing inputs. In metazoans, the pathway is termed the “mammalian” or “mechanistic” TOR (mTOR) pathway, and it also incorporates extracellular growth-factor signals into its operation. At the next organizational level, of cells within an organized epithelium, each cell must coordinate its own growth and division with that of its local neighbors. Cells perceive their local tissue context through the intermediation of intercellular junctions and cytoskeletal strain induced by tissue movement and growth, and this information is transduced into regulatory activity through the conserved Hippo/Warts/Yorkie pathway. This pathway governs the expression of genes controlling growth, proliferation, and apoptosis in response to cell-to-cell contact and tissue organization. Locally acting growth factors and morphogens such as Wingless (Wg) and Decapentaplegic (Dpp) sculpt tissue growth at this level of organization as well. The broadest level of growth control, that of the entire organism, relies on systemic hormonal growth factors such as the Drosophila insulin-like peptides (DILPs) and the insect steroid hormone ecdysone, acting through their respective intracellular pathways to modulate cellular activity. These signaling systems interact mechanistically with one another and across organizational levels; for example, TOR activity in the cells of the fat body leads to modulation of DILP release to regulate systemic growth, and Hippo signaling in imaginal tissues indirectly regulates the production of ecdysone. The first section of this review summarizes the cellular mechanics of major growth-regulatory pathways such as the TOR, Hippo, insulin, and ecdysone. In the second section, these pathways will be put into an organismal context, describing how they are coordinated throughout the organism to regulate body size in response to environmental conditions.
Although this review is focused on developmental growth, it is important to mention that cell growth and proliferation are not restricted to the larval stages, but also occur in adults to maintain tissue homeostasis and to support reproduction. Like juvenile growth, adult growth is influenced by physiological needs and environmental cues. For example, mating induces growth in the reproductive systems of both males and females, and the adult gut undergoes remodeling in response to environmental conditions, mating, and infection to maintain tissue homeostasis (Leiblich et al. 2012, 2019; Ameku and Niwa 2016; Ameku et al. 2018; Colombani and Andersen 2020). Adult tissue growth and oogenesis are governed by cell-intrinsic and systemic mechanisms similar to those of juveniles, including TOR, insulin and ecdysone, juvenile hormone (JH), cytokines, TNF-α, and transforming growth factor-β (TGF-β) (Petryk et al. 2003; Ono et al. 2006; Knapp and Sun 2017; Colombani and Andersen 2020). The mechanisms that govern growth patterning within imaginal discs are also not covered here.
The intracellular TOR pathway
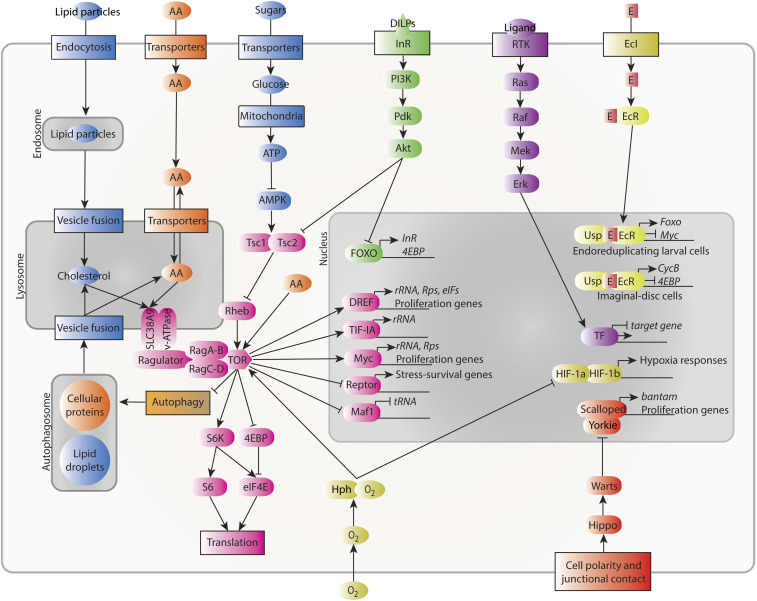
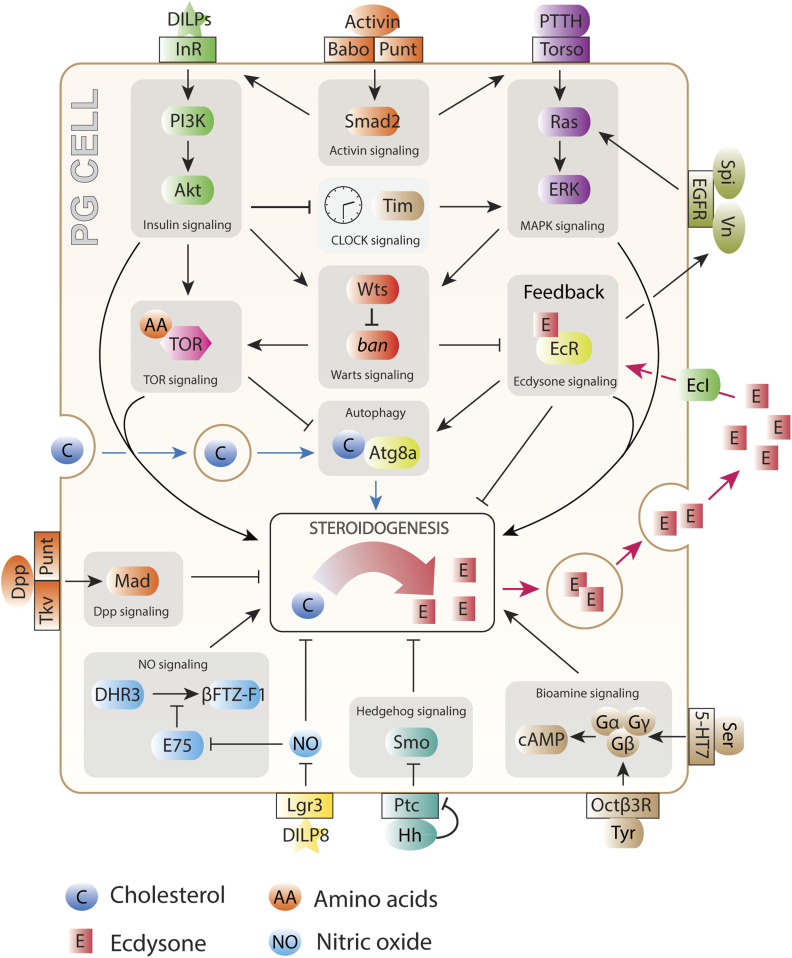
A cell requires raw materials such as amino acids, sugars, and oxygen to survive, grow, and proliferate. These metabolic inputs do not merely allow cell activity by their presence or block it through their deficiency however; their levels are sensed by intracellular mechanisms that accordingly promote or inhibit the processes that require them. The TOR pathway is the primary hub through which intracellular nutritional levels influence cell-autonomous growth, regulating diverse processes including gene expression, protein synthesis, and nutrient metabolism (Figure 2). The central player of this pathway, the kinase TOR itself, acts as a member of two protein complexes differentiated by their accessory proteins: mTOR Complex 1 (mTORC1), which mediates cell growth, and mTORC2, which largely regulates the cytoskeleton and is not discussed here, although it does have effects on growth in the fly as well [e.g., Wang et al. (2012) and Kuo et al. (2015)]. mTORC1 comprises TOR and the accessory proteins Raptor (Hara et al. 2002; Kim et al. 2002) and Lst8 (Kim et al. 2003), which regulate the interaction of the complex with target proteins as well as the kinase activity of TOR itself (for simplicity, we will use “TOR” to refer to mTORC1 from now on). TOR activation primarily takes place on the outer membrane of lysosomes and requires simultaneous activating input through two independent pathways. One of these is primarily thought of as responding to external growth-factor stimulation, and the other as generally mediating nutrient-sufficiency signals, but both nutritional and growth-factor inputs impinge upon both forks. Thus, TOR acts as a cellular coincidence detector integrating nutritional sufficiency and growth-factor stimulation to promote cellular growth and proliferation.
Figure 2.
Intracellular signaling pathways govern cell growth and proliferation. Cholesterol (blue), amino acids (orange), sugars (blue), and oxygen (olive) feed into growth regulation through cell-autonomous regulation of TOR signaling (pink; some aspects of the TOR pathway in this diagram are mammal-specific, such as SLC38A9-mediated regulation; there is no close Drosophila ortholog of this protein). Local signaling via the Hippo/Warts pathway (reddish orange) responds to cell–cell junctions and epithelial organization, and receptor tyrosine kinase signaling (purple) responds to systemic or local signals. Systemic signaling through insulin-like factors (green) and ecdysone (E, yellow) also governs cell activity. Pathways are shown terminating in the nucleus with transcription factor activity. Not all pathway components are shown, and most links between pathways are not shown. eIFs, eukaryotic initiation factors; Rps, ribosomal proteins; rRNA, ribosomal RNA; tRNA, transfer RNA.
The hormone-sensitive fork: the tuberous sclerosis complex proteins and Rheb:
One branch of the TOR activation pathway came to light through its human medical importance. Human genetic association studies of the tuberous sclerosis complex (TSC) of diseases, which produce benign tumors in diverse tissues, identified two underlying loci, Tsc1 and Tsc2 (European Chromosome 16 Tuberous Sclerosis Consortium 1993; Povey et al. 1994; van Slegtenhorst et al. 1997). Tsc1 and Tsc2 bind one another in the TSC complex (TSCC) (van Slegtenhorst et al. 1998) and in mammals can bind a third protein, TBC1d7 (Dibble et al. 2012). However, this protein has not been associated with human TSC disease and, in the fly, TBC1d7 does not seem to regulate TOR, instead affecting growth through insulin-related means (Ren et al. 2018).
Drosophila mosaic genetic screens for loss-of-function overgrowth phenotypes led to the identification of mutations in Tsc1 (Ito and Rubin 1999) and Tsc2 (Gao and Pan 2001; Potter et al. 2001; Tapon et al. 2001) as driving aberrations in cell size and cell cycle control. These reports positioned the TSCC epistatic to insulin signaling downstream of that pathway’s intermediating kinase, Akt, and later observations in the fly (Gao et al. 2002) and human cell culture (Tee et al. 2002) further positioned the TSCC upstream of TOR activity. Tsc2 was noted to exhibit similarity to GTPase-activating proteins (GAPs), which increase the rate of GTP hydrolysis by their target GTPases, and four contemporaneous reports in Drosophila identified the small GTPase Rheb (Ras homolog enriched in brain; Yamagata et al. 1994) as the target of Tsc2’s GAP activity: an RNA interference (RNAi)-based screen of potential Tsc2-target GTPases for loss of S6K phosphorylation in Drosophila S2 cells identified Rheb as a driver of TOR activity (Zhang et al. 2003); genome-wide overexpression screens in the midgut (Patel et al. 2003) and the eye disc (Saucedo et al. 2003) identified Rheb as a growth promoter; and both loss-of-function and overexpression screens for growth phenotypes in the eye identified Rheb (Stocker et al. 2003). Rheb was also identified in a human-cell-culture screen of GTPases for those whose activity is elevated in Tsc2 nulls (Garami et al. 2003).
Rheb is localized to the external lysosomal surface via an attached lipid group (Tee et al. 2003; Buerger et al. 2006). As a small GTPase, Rheb binds GTP, undergoing a conformational change and becoming active in the process; in this case, becoming competent to activate TOR. Rheb:GTP remains in this competent state until its endogenous GTPase activity, accelerated by TSCC’s Rheb-GAP functionality, hydrolyzes the bound GTP to GDP, switching Rheb back to its noncompetent state. At some point, the spent GDP is replaced with a fresh GTP molecule, restarting the activity cycle. In mammalian cell culture, under conditions unfavorable to growth including amino acid starvation, growth-factor deprivation, and energetic and hypoxic stress, the TSC complex is recruited to the lysosomal surface by the Rag GTPases (see below) (Demetriades et al. 2014; Menon et al. 2014; Demetriades et al. 2016). There it inhibits the TOR-activating ability of Rheb by promoting GTP hydrolysis as well as by blocking its reactivation through GDP exchange, remaining bound as a guanine-dissociation inhibitor (GDI) (Garami et al. 2003; Inoki et al. 2003a; Tee et al. 2003; Zhang et al. 2003; Marshall et al. 2009; Demetriades et al. 2014; Menon et al. 2014).
Unlike Rheb inhibition mediated by the TSCC’s Rheb-GAP and -GDI functionality, the reactivation step by which Rheb-bound GDP is replaced by GTP is not well understood. Guanine exchange may occur in an unassisted manner because of the higher ratio of GTP to GDP in cells (Im et al. 2002), once TSCC’s GDI activity is relieved. The protein Tctp has been reported to act as a growth promoter and guanine-exchange factor for Rheb (that is, as a Rheb-GEF) in the fly (Hsu et al. 2007; Le et al. 2016) and in human cells (Dong et al. 2009), but other reports are in tension with these results (Rehmann et al. 2008; Wang et al. 2008b). The mechanism(s) by which Rheb:GTP activates TOR on the lysosome are also not precisely clear, and this may involve several routes, including (1) induction of a conformational change in TOR that promotes its activity (Yang et al. 2017); (2) displacement of endogenous TOR-binding inhibitory proteins (Bai et al. 2007); and (3) the local generation of the charged membrane lipid phosphatidic acid, which promotes TOR lysosomal recruitment and activity (Fang et al. 2001, 2003; Sun et al. 2008; Veverka et al. 2008; Toschi et al. 2009).
Growth-factor signaling appears to impinge on the TOR pathway in part through actions on the TSC complex. The kinase Akt, a downstream effector of signaling induced by insulin and other growth factors, phosphorylates Tsc2 in mammalian cell culture, and prevention of this phosphorylation blocks the activation of S6K downstream of mTORC1; Akt and S6K are discussed below (Inoki et al. 2002; Manning et al. 2002). In the fly as well, Akt appears to phosphorylate Tsc2 (Potter et al. 2002; Dong and Pan 2004), but this does not appear to alter levels of S6K phosphorylation (Dong and Pan 2004). Overexpression of nonphosphorylable and pseudophosphorylated Tsc2 proteins (in addition to endogenous Tsc2) in the eye disc leads to Akt-dependent defects in cell growth and proliferation (Potter et al. 2002), but in another report, expression of similar constructs at roughly wild-type levels in a Tsc2-null background caused no effects on cell growth or animal survival (Dong and Pan 2004). Tsc1 is also phosphorylated by Akt, but blocking the phosphorylation sites on both Tsc1 and Tsc2 has no effect on fly growth or survival, although it does lead to a reduction in body lipid levels (Schleich and Teleman 2009). These data suggest that, at least under rich laboratory conditions, the biological impact of Akt-mediated Tsc1/2 phosphorylation is minor in Drosophila, acting to fine-tune metabolism, or is obscured by redundant mechanisms.
The nutrient-sensitive fork: the Rag GTPases:
Since Rheb is associated with the lysosomal membrane, Rheb:GTP can only activate TOR when TOR is also localized to the lysosome. The recruitment of TOR is controlled by a parallel nutrient-sensitive pathway associated with the lysosomal membrane. This branch of the TOR-activation system, like the TSCC/Rheb branch, is centered on small GTPases, and GAP and GEF proteins that govern their activity state. Compared to the Rheb fork, this half of the TOR-activation system has been explored relatively sparsely in Drosophila.
The central GTPases of the Saccharomyces cerevisiae system are Gtr1 and Gtr2; mammals possess two paralogs of each of these, RagA and RagB (Gtr1-type) plus RagC and RagD (Gtr2-type), and the Drosophila genome encodes one of each (RagA-B and RagC-D). In mammals and the fly, two Rag proteins—one Gtr1-like and one Gtr2-like—are bound to and regulated by the “Ragulator” complex, which is associated with the lysosomal membrane via contacts with the membrane-integral vesicular H+-ATPase; in Saccharomyces, the unrelated EGO complex performs this role. Myriad metabolic and physiological inputs regulate the Rags via the GAP/GEF activity of the Ragulator complex and other proteins. Conditions favorable for growth promote a configuration of RagA-B:GTP + RagC-D:GDP, which recruits cytoplasmic TOR to the surface of the lysosome (Sancak et al. 2008, 2010), where it may be activated by Rheb:GTP.
Amino acid levels regulate TOR activity via an array of influences on the Rag/Ragulator complex. The Ragulator complex itself acts as a RagA/B-GEF in response to amino acids, promoting part of the TOR-recruiting guanine configuration (Bar-Peled et al. 2012). The GATOR1 complex is a RagA/B-GAP, tending to inhibit TOR, and the related GATOR2 complex inhibits GATOR1, thus disinhibiting TOR recruitment (Bar-Peled et al. 2013). Individual amino acids affect TOR recruitment through dedicated channels; the branched-chain amino acid leucine appears to be especially important, activating TOR through several mechanisms. The stress-responsive Sestrin proteins inhibit GATOR2, thus disinhibiting GATOR1 and blocking TOR recruitment; leucine relieves the Sestrin-mediated inhibition of GATOR2, thus promoting TOR recruitment (Chantranupong et al. 2014; Parmigiani et al. 2014; Kim et al. 2015; Kimball et al. 2016). Mammalian Folliculin and FNIP1/2 also promote TOR recruitment in the presence of leucine (Petit et al. 2013; Tsun et al. 2013; Wu et al. 2016). Deletion of the Drosophila folliculin ortholog Bhd leads to slow growth and developmental arrest that can be rescued by expression of human Folliculin or through leucine supplementation; leucine rescue can be blocked by rapamycin, consistent with a role for BHD in regulating TOR activity in response to amino acids (Wu et al. 2016). LeuRS, the leucyl-transfer RNA (tRNA) synthetase, also acts as a leucine sensor, localizing to the lysosomal membrane in the presence of leucine and altering the Rag configuration in both mammals and yeast, albeit through different mechanisms in these species (Bonfils et al. 2012; Han et al. 2012; Choi et al. 2017; Kim et al. 2017); although the LeuRS protein exists in the fly, no reports concerning its effects on TOR activity have been published.
The arginine-sensing CASTOR proteins function analogously to Sestrin (Chantranupong et al. 2016), whereas the methionine sensor SAMTOR binds to and activates GATOR1 under conditions of low S-adenosyl-methionine concentration (Gu et al. 2017). The lysosomal amino acid transporter SLC38A9 interacts with Ragulator and is required for arginine sufficiency to activate TOR (Jung et al. 2015; Rebsamen et al. 2015; Wang et al. 2015; Wyant et al. 2017; Shen and Sabatini 2018). SLC38A9 also underlies cholesterol-mediated TOR regulation (Castellano et al. 2017). The presence of these sensors, like that of Rheb and Tsc1/2 (not TOR-related and not present in S. cerevisiae, respectively) and Ragulator, is varied across taxa (Tatebe and Shiozaki 2017; Wolfson and Sabatini 2017). For example, no close Drosophila orthologs of SLC38A9 or the CASTOR proteins are apparent. Whether these proteins’ functionalities are absent as well, or if their roles are played by nonhomologous systems, will be an interesting subject of future research.
TOR receives many additional inputs reflecting diverse metabolic variables. Properly formed initiator tRNAMet and successful translation initiation appear to promote TOR activity, and growth in flies and yeast (Rojas-Benitez et al. 2015). High abundance of uncharged tRNAs (that is, those carrying no amino acid), suggesting low amino acid abundance and sensed by the kinase GCN2, leads to TOR inhibition (Ye et al. 2015; Averous et al. 2016). Intracellular energy levels are sensed by AMPK, which is inhibited by a high ATP:ADP ratio; AMPK phosphorylates and activates Tsc2’s Rheb-GAP activity in human cells (Inoki et al. 2003b) and flies (Kim and Lee 2015). Low ATP also inhibits the formation of TTT-Pontin/Reptin protein assemblies that are required for the formation of TOR complexes in mouse embryonic fibroblasts (Kim et al. 2013) and flies (David-Morrison et al. 2016). Oxygen promotes TOR activity and cell and organismal growth [reviewed in Ellisen (2005) and Magdalena Romero et al. (2007)], and mechanical stimulation promotes TOR activity as well, via a phosphatidic acid-mediated mechanism (Hornberger et al. 2006; O’Neil et al. 2009; You et al. 2014; Lin and Liu 2019).
The effects of TOR activity:
Once both branches of the activation pathway are engaged, TOR becomes activated on the lysosome surface. Activated TOR acts to increase cellular growth and proliferation by indirectly increasing the expression of ribosomal components such as ribosomal RNA (rRNA) and ribosomal proteins; enhancing messenger RNA (mRNA) translation initiation; and promoting translation efficiency by upregulating tRNA expression (Figure 2). By regulating the activity of transcription factors, TOR also promotes the expression of proliferation-inducing genes, such as those involved in the cell cycle and the replication of DNA, and, in the fly, it also downregulates the Reptor-mediated expression of genes required for survival under stressful conditions (Tiebe et al. 2015). Furthermore, TOR-mediated phosphorylation of autophagy-inducing proteins inhibits this intracellular recycling process (Ganley et al. 2009; Hosokawa et al. 2009; Jung et al. 2009).
Activation of TOR promotes the synthesis of ribosomal components through several routes. It promotes the expression of the transcription factor DREF, which upregulates many genes required for tasks related to cell growth and proliferation, such as cell cycle progression, DNA replication, and gene expression (Hyun et al. 2005; Thao et al. 2008; Killip and Grewal 2012). DREF-binding sites are recognizable in the promoters of 18 of 25 Drosophila rRNA-processing genes and 31 of 77 ribosomal-protein genes, and loss of Dref function reduces the expression of these factors and blocks TOR-mediated growth (Killip and Grewal 2012). TOR activity also promotes the expression and activity of the RNA polymerase (Pol) I transcription factor TIF-IA (Grewal et al. 2007; Ghosh et al. 2014), leading to increased expression of rRNA. Thus, TOR promotes the biosynthesis of ribosomes to support increased protein production. Moreover, TOR promotes tRNA expression, and thus increases translation efficiency, through inhibiting Maf1, a suppressor of RNA Pol III (Murawski et al. 1994; Pluta et al. 2001; Cieśla et al. 2007; Marshall et al. 2012). DREF sites are present near 26 of 50 genes encoding translation-initiation factors (Killip and Grewal 2012).
Furthermore, TOR directly phosphorylates the ribosomal protein S6 kinase (S6K), which then phosphorylates ribosomal protein S6, leading to increased translation (Brown et al. 1995; Watson et al. 1996; Montagne et al. 1999; Zhang et al. 2000). S6K also phosphorylates and activates eukaryotic Initiation Factor 4E (eIF4E), which binds to the mRNA 5′ cap structure, promoting mRNA ribosomal recruitment and translation initiation (Raught et al. 2004). In parallel, TOR also directly phosphorylates and inactivates the translation inhibitor eIF4E-Binding Protein (4E-BP, encoded in Drosophila by Thor) (Heesom and Denton 1999). By promoting translation initiation and increasing translation efficiency, TOR thereby induces the cell to put its newly synthesized ribosomes to use, leading to increased synthesis of protein.
TOR-mediated translation control also has regulatory effects beyond the bulk production of cellular content. For example, increased translation of the transcription factor E2F1 promotes rhythmic oscillations in its abundance and underlies nutrition-dependent endocycling in the larval salivary gland (Zielke et al. 2011). Somewhat surprisingly, given their seeming centrality, neither S6K (Montagne et al. 1999) nor 4E-BP (Bernal et al. 2004; Teleman et al. 2005) is required for viability in the fly under normal conditions. Although S6K-null mutants are slow to develop and rarely survive to adulthood, as small and short-lived animals, Thor/4EBP-null animals exhibit no growth-rate or size defect, instead showing only adipose defects. Likewise, mice null for 4E-BP1, 4E-BP2, or both are viable with only behavioral or metabolic defects (Tsukiyama-Kohara et al. 2001; Banko et al. 2005; Le Bacquer et al. 2007), and mice lacking either one of two S6K paralogs, but not both, are viable (Shima et al. 1998; Pende et al. 2000, 2004).
The proto-oncogenic transcription factor Myc
The growth-promoting transcription factor Myc was identified as an ortholog to a sequence within the avian myelocytoma virus (Colby et al. 1983; Schweinfest et al. 1988; Gallant et al. 1996; Johnston et al. 1999). Myc—generally but not always in conjunction with its cofactor Max (Steiger et al. 2008)—promotes cell growth in a variety of ways [reviewed in Gallant (2013)]. One of them is through the upregulation of genes encoding rRNA, ribosomal proteins, and other ribosome-biosynthesis genes. Overexpression of Myc leads to upregulation of many genes, including 70 related to ribosome biogenesis, in larval tissues and in wing-disc cells (Grewal et al. 2005). TOR indirectly promotes Myc stability, increasing the expression of growth-promoting genes, and activates cell cycle-control proteins, allowing proliferation (Diehl et al. 1998; Alt et al. 2000; Armstrong et al. 2001; Welcker et al. 2004; Parisi et al. 2011; Stein et al. 2011). Indeed, much of the growth-promoting activity of TOR appears to be funneled through Myc. More than 90% of genes found to be regulated downstream of TOR in the fly have a nearby Myc-binding E-box (Guertin et al. 2006; Teleman et al. 2008; Parisi et al. 2011). Thus, the TOR complex integrates information about levels of amino acids, energy, oxygen, and cholesterol—inputs required for the generation of more cellular material—with signals conveyed via insulin and growth-factor pathways, and promotes gene expression, ribosomal biogenesis, and protein synthesis to drive cell growth and proliferation.
The Hippo local signaling system
Within many tissues, such as developing imaginal discs, cells lie in an epithelial plane, in contact with a basement substrate and with their neighbors through various types of junctional complexes, which serve both to provide orientation axes to individual cells as well as to transmit mechanical forces between them. These axes direct the growth and division axes of epithelial cells, and the physical tension generated by cell growth and movement is transduced back into regulatory activity (Bosveld et al. 2012; Pan et al. 2016, 2018). In general, cell contacts inhibit cell proliferation, and loss of these contacts, such as through wounding, promotes cellular growth and division. The signaling system underlying this phenomenon is the conserved Hippo pathway. The central nodes of this system are the kinase Hippo; the Hippo target Warts, also a kinase; and the Warts target Yorkie, a transcriptional coactivator required for expression of many growth-, proliferation-, and survival-promoting genes. Mechanical and environmental stimuli consistent with proper tissue embedding, such as cytoskeletal tension, proper planar cell polarity, and maintenance of cell-to-cell junctions, lead to the activation of Warts, which phosphorylates and deactivates Yorkie, thus preventing the expression of genes promoting growth and proliferation. Loss of a cell’s tissue context thus leads to inhibition of Warts, activation of Yorkie, and induction of target-gene expression. The details of the mechanisms leading to Warts activation and Yorkie inhibition are reviewed elsewhere (Fulford et al. 2018; Misra and Irvine 2018; Ma et al. 2019).
Phosphorylation of Yorkie leads to its exclusion from the nucleus into the cytoplasm, where it can be sequestered by interactions with other members of the Hippo–Warts pathway. Deactivation of Warts thus promotes nuclear Yorkie localization (Dong et al. 2007; Oh and Irvine 2008; Badouel et al. 2009; Oh et al. 2009; Ren et al. 2010; Manning et al. 2018). However, Yorkie has no DNA-binding domain of its own and regulates gene expression through its association with tissue-specific transcription factors including Scalloped (Sd), Homothorax (Hth), Teashirt (Tsh), and the Dpp mediator Mad (Goulev et al. 2008; Wu et al. 2008; Zhang et al. 2008; Peng et al. 2009; Oh and Irvine 2011). In the absence of nuclear Yorkie, the protein Tgi acts as an inhibitory cofactor of Sd, leading to repression of Yorkie target genes (Guo et al. 2013; Koontz et al. 2013). Interestingly, in the developing wing disc, TOR gates Yorkie-mediated gene expression, only releasing Yorkie from “seclusion” at chromatin sites distant from its target-gene promoters when nutritional levels are adequate (Parker and Struhl 2015).
Yorkie promotes the expression of genes required for cell growth and proliferation, including cyclins and inhibitors of apoptosis (Tapon et al. 2002; Huang et al. 2005a; Shimizu et al. 2008; Wu et al. 2008; Zhang et al. 2008; Verghese et al. 2012; Zhang and Cohen 2013). Yorkie promotes the expression of Myc in conjunction with its tissue-specific binding partners Sd in the wing and Hth in the notum, leading to a growth and cell-competitive phenotype. In a negative-feedback loop, Myc inhibits Yorkie expression (Neto-Silva et al. 2010; Ziosi et al. 2010). Yorkie also promotes the expression of Hippo-pathway proteins that inhibit its own function, thus forming a second negative-feedback loop (Cho et al. 2006; Hamaratoglu et al. 2006; Genevet et al. 2010).
A major Yorkie target is the microRNA bantam, which is required for Yorkie-driven growth in Drosophila (Hipfner et al. 2002; Brennecke et al. 2003; Nolo et al. 2006; Thompson and Cohen 2006; Peng et al. 2009). As a microRNA, bantam induces the degradation of complementary target transcripts, and known bantam targets include those encoding Mad (Robins et al. 2005; Kane et al. 2018), Tgi (Shen et al. 2015), the apoptosis promoter Hid (Brennecke et al. 2003), the transcriptional repressor Capicua (Herranz et al. 2012), and the cell cycle inhibitor Tribbles, which blocks the G2/M transition (Gerlach et al. 2019). Through its downregulation of these and other targets, bantam promotes cell survival and proliferation. Through this system, tissue type (via the availability of Yorkie cofactors), multicellular context (via junctional components of the Hippo pathway), and intracellular nutrition (via TOR signaling) are funneled through the activity of a single growth-promoting transcriptional effector, Yorkie.
Local growth-factor signaling
In addition to local signaling mediated by junctional contacts and the Hippo pathway, cell growth and proliferation are modulated by short- and long-range signaling factors including Dpp (Hamaratoglu et al. 2014; Restrepo et al. 2014); Hedgehog (Hh; Robbins et al. 2012; Briscoe and Therond 2013); Wingless (Wg; Swarup and Verheyen 2012; Bejsovec 2018); the TNF-α ortholog Eiger [reviewed by La Marca and Richardson (2020)]; and many ligands for receptor tyrosine kinases (RTKs) [reviewed in Shilo (2014)]. Space does not allow a full account of these pathways and their interactions with one another, but the RTKs are of special interest here, as they mediate several signals driving ecdysone production (see below). Ligand binding leads to RTK dimerization, which induces Ras-GEF activity in receptor accessory proteins, promoting GTP loading of Ras. Ras:GTP then activates a cascade of mitogen-activated protein kinases (MAPKs)—Pole hole/Raf, Dsor1/Mek, and Rolled/Erk—leading to phosphorylation of various targets, including transcription factors and RSK/S6KII. For example, the transcriptional repressor Capicua is inhibited downstream of signaling through Epidermal Growth Factor Receptor (EGFR) (Roch et al. 2002; Tseng et al. 2007) and the receptor Torso (Ajuria et al. 2011), leading to derepression of target genes and inducing either differentiation or proliferation. This decision is influenced by Hippo signaling: when Hippo is active—when cells are properly embedded in tissue—Ras activity leads to cell differentiation, whereas if Hippo is inactive, Ras induces proliferation (Pascual et al. 2017). This “reprogramming” of Ras effects is mediated by interactions between Ras/MAPK and Hippo-pathway components including Capicua, Yorkie, and bantam (Herranz et al. 2012; Pascual et al. 2017; Simón-Carrasco et al. 2018).
The intracellular insulin-signaling pathway
In addition to cell-autonomous and local growth control, organisms require systemic regulation and coordination of growth and development. This is mediated by circulating factors including insulin-like proteins, which are the major growth- and metabolism-regulating hormones in flies and mammals, and steroid hormones, which determine developmental progression in addition to affecting growth. The organismal effects of these hormones, whose production and release are governed by numerous internal and environmental cues, are brought about through their intracellular signaling actions.
Whereas mammals express both an insulin receptor and several receptors for insulin-like growth factors (IGFs), allowing metabolism- and growth-governing signals to be interpreted separately, Drosophila cells express a single insulin receptor (InR), an RTK that transduces signals carried via multiple DILPs (Fernandez et al. 1995; Chen et al. 1996; Scanga et al. 2000; Brogiolo et al. 2001; Britton et al. 2002; Ikeya et al. 2002). DILP binding induces InR dimerization and cross-phosphorylation, which leads to the activation of phosphatidylinositol 3-kinase (PI3K) and the generation of second-messenger membrane lipids (Yenush et al. 1996; Goberdhan et al. 1999; Verdu et al. 1999). PI3K’s effects are antagonized by PTEN, which dephosphorylates these lipids and reduces signaling flux (Goberdhan et al. 1999; Gao et al. 2000). Membrane phosphoinositides recruit and activate protein kinase B (PKB or Akt) and phosphoinositide-dependent kinase (Pdk), which phosphorylate and further activates Akt at the membrane (Verdu et al. 1999; Cho et al. 2001; Rintelen et al. 2001; Radimerski et al. 2002; Lizcano et al. 2003). Active Akt then disassociates from the membrane and phosphorylates a range of target proteins, altering their activity.
One of the primary Akt targets is the transcription factor Forkhead Box O (FOXO), which promotes the expression of genes required for adaptation to low-nutrition conditions. When insulin signaling is active, Akt phosphorylates FOXO, leading to its exclusion from the nucleus (Junger et al. 2003; Kramer et al. 2003; Puig et al. 2003). One of the primary growth-related FOXO targets downregulated by insulin signaling is the translational inhibitor 4E-BP (encoded in the fly by Thor), a negative regulator of growth (Junger et al. 2003). FOXO also upregulates InR expression, establishing a feedback loop to sensitize cellular responses to insulin in nutrient-scarce conditions with low signaling through this pathway (Puig and Tjian 2005). Insulin signaling also promotes growth via lift of FOXO-mediated repression of Myc and though Akt-mediated promotion of Myc stability (Welcker et al. 2004; Teleman et al. 2008). Furthermore, as discussed above, Akt-mediated phosphorylation of Tsc1 and Tsc2 may have TOR-activation effects in the fly. In other systems, Akt also phosphorylates the endogenous TOR substrate-like inhibitor PRAS40, leading to its dissociation from TOR (Sancak et al. 2007; Haar et al. 2007; Wang et al. 2007, 2008a; Yang et al. 2017), although in flies this appears to be relevant only in the ovary (Pallares-Cartes et al. 2012); Akt also inhibits GATOR1 in mammalian cell culture (Padi et al. 2019), promoting TOR recruitment to the lysosome. Thus, insulin signaling promotes cell growth and proliferation via control of gene expression and protein synthesis, in large part via Akt, which regulates FOXO, Myc, and the TOR pathway.
Intracellular signaling downstream of ecdysone
In developing insects, cell proliferation and differentiation must be tightly orchestrated to achieve proper development before metamorphosis. During this period, extensive changes take place in the regulation of these processes. The molting-inducing steroid hormone ecdysone is therefore also a key regulator of cell proliferation. Ecdysone regulates gene expression through a heterodimeric receptor complex comprising the nuclear ecdysone receptor (EcR) and its partner Ultraspiracle (Usp), which together bind to ecdysone-response elements in the promoters of target genes (Riddiford et al. 2000; King-Jones and Thummel 2005). Usp is an ortholog of the vertebrate retinoid X receptor (RXR) (Oro et al. 1990; Yao et al. 1992), and the retinoic-acid signaling pathway is a key regulator of cell differentiation in vertebrate cells (Breitman et al. 1980). Ecdysone inhibits growth in larval cells (Colombani et al. 2005; Delanoue et al. 2010) while stimulating the growth of imaginal disc cells (Mirth et al. 2009; Oliveira et al. 2014; Herboso et al. 2015), at least partially via interactions with DILP and TOR signaling, including EcR-mediated repression of Myc (Delanoue et al. 2010). Ecdysone also promotes the expression of FOXO (Colombani et al. 2005), perhaps via dDOR, whose expression in the fat body is upregulated by ecdysone but negatively regulated by insulin signaling (Francis et al. 2010). This intracellular cross talk between ecdysone and insulin signaling partially explains their antagonistic effects on growth; these two axes interact at a systemic level as well, discussed below.
During the final feeding stage of larval development, ecdysone induces the growth and proliferation of imaginal disc cells, partially through repression of 4EBP (Herboso et al. 2015). In the eye discs of feeding larvae, reduced ecdysone signaling inhibits cell proliferation due to dramatically decreased expression of the mitotic inducer cyclin B (Zelhof et al. 1997; Brennan et al. 1998). Ecdysone also acts through Wg and the zinc-finger transcription factor Crooked legs (Crol) to control wing-disc cell proliferation by indirectly regulating cyclin B (Mitchell et al. 2008, 2013). Furthermore, the EcR coactivator Taiman (Tai) appears to interact with Hippo signaling: Tai binds to the Hippo effector Yorkie and upregulates both Hippo target genes as well as genes specifically targeted by the Tai:Yki complex to control cell proliferation in the developing wing pouch (Zhang et al. 2015). Taken together, ecdysone is required to stimulate cell proliferation and growth in imaginal disc cells of feeding larvae.
In contrast, after the cessation of feeding at the wandering stage, which is induced by a pulse of ecdysone, the response of imaginal disc cells to ecdysone changes considerably. Imaginal discs show reduced cell proliferation after pupation (Graves and Schubiger 1982; Schubiger and Palka 1987; Sustar and Schubiger 2005); cells of the wing and leg discs temporarily arrest in G2 prior to permanently exiting the cell cycle (Graves and Schubiger 1982; Schubiger and Palka 1987). Cell cycle arrest and exit seem to be related to the expression of the ecdysone-inducible pupal specifier Broad (Br). Br represses string, encoding the Drosophila ortholog of the G2/M cell cycle promoter Cdc25, and the lack of String induces G2 arrest (Guo et al. 2016). Then, as the pulse of ecdysone subsides, string is derepressed, stimulating a final, synchronized cell division (Guo et al. 2016). Thus, ecdysone appears to regulate cell proliferation and growth in a stage- and concentration-dependent manner to coordinate the size of developing imaginal discs.
Body-Size Control
While local growth regulation ensures that individual organs grow to achieve the correct size, organization, and shape, systemic growth control ensures that they grow in correct proportion to each other and to the entire organism. Local growth-controlling mechanisms also provide instructive cues to the systemic regulatory axes. This two-way communication is mediated by circulating signals that act globally and coordinate growth across the entire body.
Duration and rate of growth
Holometabolous insects such as Drosophila develop through an embryonic stage followed by a series of larval stages called instars, which are separated by molts in which the animal replaces its old cuticle with a new, larger one to accommodate further growth (Figure 1). Wild-type Drosophila reared at 25° with a normal oxygen level and adequate nutrition complete embryogenesis and the first two larval instars (L1 and L2) in ∼1 day per stage, and the third and final instar (L3) lasts 2 days. During these four feeding days, the animal can increase in size by ∼200-fold (Robertson 1966) before wandering and pupariation end the juvenile growth period. After another 4 days of metamorphic development, adults emerge (eclose) from the pupal case and do not further increase their body size, although some cell growth and proliferation continues to maintain homeostasis and reproductive capacity, as mentioned above. The final adult size is thus determined by larval growth, which is quite plastic within species-specific limits and is a function of two key parameters, the rate and the duration of growth. These are regulated by environmental and internal cues that converge onto two key systemic axes: the insulin-like signaling system and the steroid ecdysone signaling system.
As in mammals, insulin-like signaling in Drosophila regulates cellular nutrient uptake and storage, metabolism, and cellular and organismal growth at the systemic level, in response to nutritional and environmental cues. The major systemic growth- and metabolism-regulating DILPs are released into the hemolymph by the so-called insulin-producing cells (IPCs) of the brain in response to a variety of inputs (Figure 3 and Table 1). Developmental progression, on the other hand, is largely regulated by steroid signaling in both mammals and insects. In Drosophila, diverse regulatory mechanisms control the production and release of the steroid ecdysone by the cells of the larval prothoracic gland (PG; Figure 4, Figure 5, Table 2, and Table 3). Pulses of ecdysone drive developmental progression through larval molts (ecdyses) and into metamorphosis; lower, basal levels of ecdysone inhibit the growth of larval tissues and promote the growth of the imaginal discs. These systems, and their upstream regulatory mechanisms, are discussed below.
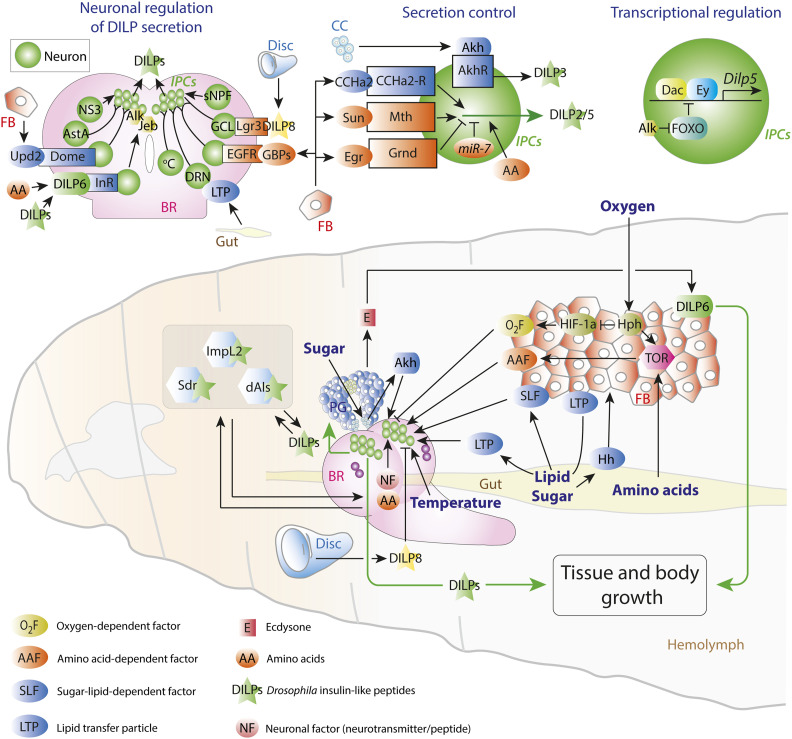
Figure 3.
Regulation of insulin expression, release, and activity in the Drosophila larva. The larval insulin-producing cells (IPCs, small green spheres) of the brain (pink) receive a multitude of regulatory inputs (see also Table 1). Bottom panel: signals released by the fat body (FB), the gut, the developing imaginal tissues, and the prothoracic gland (PG) act on the IPCs to regulate DILP expression and release. Top left panel: input from neurons that sense temperature, disc development, and humoral factors act on the IPCs. BR, brain; DRN, DILP2-recruiting neurons; GCL, growth-coordinating Lgr3+ neurons. Top middle panel: Akh/AkhR signaling in the larval IPCs promotes DILP3 release; DILP2 and DILP5 are regulated by fat-derived activating factors CCHa2 and Stunted, which signal “nutrition,” and the inhibitor Eiger, which conveys “starvation.” Top, right panel: little is known about the cis-regulation of insulin gene expression. Dachshund and Eyeless, like their mammalian homologs Dach1/2 and Pax6, promote insulin expression, specifically of Dilp5. This expression is inhibited by FOXO, and signaling through the receptor Alk derepresses Dilp5 in response to the ligand Jelly Belly released by glia of the blood/brain barrier during starvation.
Table 1. Factors that act upon IPCs in the larva, the adult, or both.
| IPC-influencing factor | Larval data | Adult data |
|---|---|---|
| Adipokinetic hormone (Akh) | Akh from the CC mediates trehalose-induced release of DILP3 but not DILP2; Kim and Neufeld (2015). | No adult data |
| AdipoR ligand (unknown) | Ligand and source unknown; AdipoR in IPCs regulates DILP secretion and metabolism, but has no effect on body size; Kwak et al. (2013). | Ligand and source unknown; IPC AdipoR regulates metabolism, survival, Dilp3 expression, and DILP release; Kwak et al. (2013). |
| Autonomous sugar sensing | No; sensing occurs via Akh relay; Kim and Neufeld (2015). | Yes, through a mechanism involving inhibition of KATP channels and Ca2+ increase; Kréneisz et al. (2010). |
| Autonomous amino acid sensing | Via leucine transporters Minidiscs and JhI-21 and the GDH pathway; Manière et al. (2016); Ziegler et al. (2018). | No adult data. |
| Allatostatin A (AstA) | AstA-R2 regulates both IPCs and APCs; Bowser and Tobe (2005); Hentze et al. (2015). AstA-R1 regulates DILP2/5 release but not expression; Deveci et al. (2019). | AstA-R2 regulates both IPCs and APCs. AstA-R2 RNAi in IPCs downregulates Dilp2 but not Dilp3, in females but not males; Hentze et al. (2015). |
| CCHamide-2 (CCHa2) | From gut and fat; regulated by dietary sugar and TOR; via CCHa2-R, promotes DILP2 and DILP5 release and Dilp5 expression; Ren et al. (2015); Sano et al. (2015). | CCHa2 null affects insulin expression in the pupa via an undetermined route; Ren et al. (2015). |
| Dawdle (Daw) | Daw from undetermined source(s) promotes DILP release, probably indirectly; Ghosh and O’Connor (2014). | Dawdle signaling in muscle remotely promotes insulin release via an unknown route; Bai et al. (2013). |
| DILPs (via InR) | No larval data on DILP-specific feedback. | IPC DILPs and fat-body DILP6 regulate one another; Gronke et al. (2010); Bai et al. (2012). |
| DILP8 | GCL neurons presynaptic to IPCs inhibit Dilp3 and Dilp5 expression; Vallejo et al. (2015). | No adult data. |
| Dopamine | No larval data. | DopR1-RNAi in IPCs prevents dormancy; Andreatta et al. (2018). |
| Ecdysone | Dominant negative EcR in IPCs appears to block DILP release; Buhler et al. (2018). | No adult data. |
| Eiger (Egr) | Released from the fat body under starvation; acts via Grindelwald receptor to inhibit DILP2/5 release; Agrawal et al. (2016). | No adult IPC data. |
| Female-specific independent of Transformer (FIT) | Not expressed in larvae; Sun et al. (2017). | From fat body of head; induced by protein feeding via TOR; affects IPCs through unknown route; Sun et al. (2017). |
| GABA | GABA-B-R2 is present in IPCs, but RNAi does not alter size; Enell et al. (2010). | GABA-B-R2 is present in adult IPCs, and RNAi leads to increased anti-DILP staining, altered metabolism, and increased stress sensitivity; Enell et al. (2010). |
| Growth-blocking peptides (GBPs) | Expressed in fat body in response to amino acids and TOR; act via EGFR-expressing “IPC-connecting neurons”; Koyama and Mirth (2016); Meschi et al. (2019). | GBP receptor Mthl10 is expressed in IPCs; global Mthl10 RNAi blocks DILP2 release from IPCs, at least indirectly, Mthl10 is broadly expressed; Sung et al. (2017). |
| Hugin (Hug) | Subesophageal-zone Hugin neurons synapse on the IPCs, which express the Hugin receptor PK2-R1; Schlegel et al. (2016). | No adult data. |
| Hypoxia (unknown signals) | From fat body, primarily regulating Dilp3 expression and release of all DILPs; Texada et al. (2019a). | No adult data. |
| Jelly Belly (Jeb) | From cholinergic neurons, via Alk; Okamoto and Nishimura (2015). | No adult data. |
| Leucokinin (Lk) | No larval data. | From neuronal source; receptor Lkr is expressed in IPCs and regulates Dilp expression, Zandawala et al. (2018); and sleep, Yurgel et al. (2019). |
| Limostatin (Lst) | No larval data. | From CC in response to carbohydrate restriction; suppresses DILP expression and release via PK1-R (LstR); Alfa et al. (2015). |
| Lipid particles | Lipids from yeast but not plants cause particle accumulation on DILP2-recruiting neurons presynaptic to IPCs, and this increases DILP release; Brankatschk et al. (2014). | No adult data. |
| Octopamine/tyramine | Oamb-RNAi does not alter adult size; Luo et al. (2014). | Receptor OAMB is expressed in IPCs and regulates sleep and metabolism; Crocker et al. (2010); Erion et al. (2012). Oamb-RNAi increases Dilp3 expression; Luo et al. (2014). |
| Pigment-dispersing factor (PDF) | No larval data. | PDF from clock neurons increases cAMP levels via PDFR to block dormancy; Nagy et al. (2019). |
| Serotonin | 5-HT1A-GAL4 is not expressed in feeding third-instar larval IPCs, and 5-HT1A-RNAi animals are of normal size; Luo et al. (2012). | 5-HT1A-GAL4 is expressed in IPCs; 5-HT1-RNAi leads to increased DILP staining in IPCs and reduces starvation survival; Luo et al. (2012); 5-HT1A-RNAi increases expression of Dilp2 and Dilp5; Luo et al. (2014); Andreatta et al. (2018). |
| Short neuropeptide F (sNPF) | sNPF peptides 1 and 2, but not 3 or 4, act on IPCs via sNPF-R (shown via anti-sNPF-R) and govern Dilp expression; Lee et al. (2008); Lee et al. (2009). However, IPCs do not express sNPF-R-GAL4; Kapan et al. (2012); Carlsson et al. (2013) (same line in both). | IPCs express sNPF-R-GAL4; Kapan et al. (2012). sNPF from sugar-sensitive upstream neurons activates the IPCs and inhibits the APCs via sNPF-R; Oh et al. (2019). sNPF from clock neurons increases cAMP and Ca2+ levels, likely directly, to block dormancy; Nagy et al. (2019). Bidirectional sNPF/DILP feedback governs feeding; Sudhakar et al. (2020). See larval papers as well. |
| Stunted (Sun) | Expressed in fat body in response to feeding via Spargel/PGC1, not via TOR. TOR does promote translation or release. Acts via Methuselah receptor to promote DILP release; Delanoue et al. (2016). | No adult data. |
| Tachykinin (Tk) | TkR99D perhaps present in larval IPCs; Birse et al. (2011), but no functional data reported. | Source undefined, but Tk+ neurons terminate near IPC projections; suppresses Dilp2 and promotes Dilp3 in starvation via TkR99D; Birse et al. (2011). |
| Taotie neurons | No larval data. | Activation of peptidergic Taotie neurons (named for a Chinese mythological “gluttonous ogre”) upstream of IPCs inhibits feeding and DILP release; Zhan et al. (2016). |
| Temperature | Cold-activated sensory neurons presynaptic to the IPCs promote DILP expression and release; Li and Gong (2015). | No adult data. |
| Unpaired-2 (Upd2) | Expressed in fat body in response to sugars and lipids; acts via Domeless receptor in presynaptic GABAergic neurons; Rajan and Perrimon (2012). | Expressed in fat body in response to sugars and lipids; acts via Domeless receptor in presynaptic GABAergic neurons; Rajan and Perrimon (2012). |
APC, Akh-producing cell; CC, corpora cardiaca; EcR, ecdysone receptor; InR, insulin receptor; IPC, insulin-producing cell; RNAi, RNA interference.
Figure 4.
Regulation and effects of ecdysone (E) production in Drosophila larvae. A network of signals regulates E production in the prothoracic gland (PG). Nutritional influences (relayed by Hh, AstA, Crz, and amino acid-regulated serotonergic neurons) act on the IPCs, the PTTHn (prothoracicotropic hormone-producing neurons), and the PG; signals from the developing imaginal discs (DILP8 and Dpp) act on these cells as part of the growth-coordination mechanism. Light and internal clocks (not shown) regulate the PG and the PTTHn. E feeds back onto the PG to upregulate and then downregulate its own production, and onto the PTTHn to promote PTTH expression. Peaks of E act to promote developmental transitions, and basal levels block the growth of larval tissues while promoting disc growth. E entry is mediated by the E importer, EcI. See also Table 2 and Table 3.
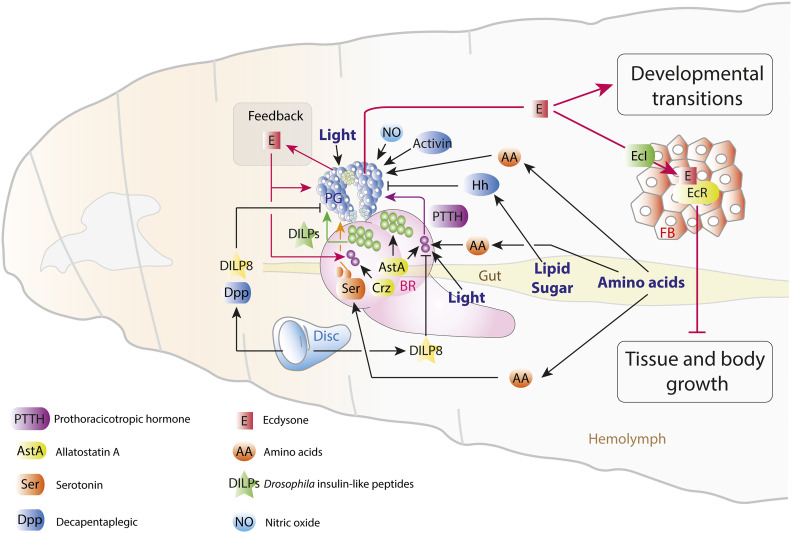
Figure 5.
Pathways affecting ecdysone (E) synthesis and release in the Drosophila prothoracic gland (PG). A broad array of autonomous and external cues govern the production and release of E, both at basal levels that regulate the growth of larval and imaginal tissues as well as in the peaks of synthesis that govern developmental transitions. DILP and PTTH signals carry nutritional and developmental information; the competence of the PG to respond to these signals is regulated by Activin signaling. Nutrition also affects the PG through the TOR and Warts pathways, as well as through inputs from serotonergic neurons and gut-derived Hedgehog (Hh). The metabolic state of the PG regulates cholesterol trafficking for steroidogenesis, and the developmental state of imaginal tissues is conveyed directly to the PG by the disc-derived factors DILP8 and Dpp (as well as by indirect means such as PTTH). PG-autonomous molecular clocks interface with external clock input (not shown) to organize E pulses. Feedback through E (via Ecdysone Importer, EcI) and EGF-like ligands drives and sculpts E peaks. See also Table 3.
Table 2. Factors that regulate PTTH expression or release in Drosophila.
| PTTH-influencing factor | Comments |
|---|---|
| Allatostatin A (AstA) | Released by AstA neurons presynaptic to PTTHn and insulin-producing cells, and promotes PTTH release via AstA-R1; Deveci et al. (2019); commentary in Pan and O’Connor (2019). |
| Amino acids | Glial expression of the amino acid transporter Sobremesa (Sbm; “upon the table,” the Spanish tradition of relaxation after a heavy meal) is required for proper PTTH expression; Galagovsky et al. (2018). |
| Corazonin (Crz) | During mid-L3, nutrition-mediating octopaminergic input to Corazonin-releasing cells presynaptic to the PTTHn induces Crz release, PTTH release, and basal ecdysone synthesis, limiting larval growth but not affecting timing; Imura et al. (2020). |
| DILP8 | Growing discs release DILP8, which acts via growth-coordinating Lgr3-expressing neurons presynaptic to PTTHn; Colombani et al. (2015); Garelli et al. (2015); Vallejo et al. (2015). |
| Ecdysone | Ecdysone promotes Ptth expression in a feedback loop contributing to the metamorphosis-triggering surge of PTTH and ecdysone; Christensen et al. (2020). |
| Juvenile hormone (JH) | Appears to be unimportant for PTTH signaling in the fly; Mirth et al. (2014); although not in Manduca; Nijhout and Williams (1974). However, reporters of JH receptor expression are active in PTTH cells; Baumann et al. (2017). |
| Photoperiod/sNPF | Photoperiod affects PTTH; Truman (1972). Projections of clock neurons expressing PDF and sNPF overlap with those of the PTTHn in larval and pharate-adult brains; McBrayer et al. (2007); Selcho et al. (2017). Clock output sNPF from these neurons acts on PTTHn via sNPF-R; Selcho et al. (2017). |
| Retinoids | Retinoids are released by damaged discs and inhibit Ptth expression Halme et al. (2010). |
Table 3. Factors that regulate the PG in Drosophila.
| PG-influencing factor | Comments |
|---|---|
| Activin/TGF-β | Regulates torso and InR expression via Baboon/Smad2; Gibbens et al. (2011). |
| Decapentaplegic (Dpp)/TGF-β | Dpp released by growing discs represses ecdysone synthesis via Thickveins (Tkv), at least in part through effects on FOXO and ban; Setiawan et al. (2018). |
| DILPs (via InR) | Insulin signaling in the PG promotes ecdysone synthesis through effects on TOR, Warts signaling (via bantam), and cholesterol trafficking; Caldwell et al. (2005); Colombani et al. (2005); Mirth et al. (2005); Boulan et al. (2013); Moeller et al. (2017); Texada et al. (2019b). A small insulin-induced peak appears to be associated with critical weight; Shingleton et al. (2005); Koyama et al. (2014). |
| DILP8 | DILP8 secreted by damaged or unevenly growing discs acts directly on PG via Lgr3 and nitric oxide signaling to repress basal ecdysone synthesis, and thus growth of undamaged imaginal tissues; Caceres et al. (2011); Jaszczak et al. (2015); Jaszczak et al. (2016). |
| Ecdysone | Ecdysone feedback via EcR to Br-Z4 (positive feedback) and Br-Z1 (negative feedback) drives and terminates the metamorphic ecdysone pulse; Moeller et al. (2013). EcR promotes autophagic mobilization of cholesterol for ecdysone synthesis; Texada et al. (2019b). EcR promotes expression of EGF-like ligands Vein and Spitz; Cruz et al. (2020). |
| EGF-like signals | Autocrine signaling via ligands Spitz and Vein, induced by ecdysone feedback, drives the MAPK pathway and promotes the metamorphic ecdysone peak; Cruz et al. (2020). |
| Hedgehog (lipid-associated) | Released from enterocytes under starvation conditions and inhibits expression of phantom and spookier; Rodenfels et al. (2014). |
| Juvenile hormone (JH) | Inhibits basal ecdysone synthesis via Kr-h1 (Zhang et al. 2018) but does not appear to affect timing in Drosophila; Mirth et al. (2014). |
| PG-endogenous clock | A PG-autonomous clock, interacting with central circadian rhythms and insulin signaling, is required for steroidogenesis; commentary in Danielsen and Rewitz (2016); Di Cara and King-Jones (2016). |
| Prothoracicotropic hormone (PTTH) | Promotes PG-cell growth, endoreduplication, and Halloween-gene expression via receptor tyrosine kinase Torso; King-Jones et al. (2005, 2016); McBrayer et al. (2007); Rewitz et al. (2009b); Ghosh et al. (2010); Ou et al. (2011, 2016); Rewitz and O’Connor (2011); Ohhara et al. (2017); Shimell et al. (2018). |
| Serotonin | Serotonergic neurons receive input from SEZ/SOG and arborize more densely on the PG under well-fed conditions. Serotonin acts via receptor 5-HT7 (Shimada-Niwa and Niwa 2014) to promote ecdysone synthesis. |
| TOR signaling | Loss blocks pupariation; activation rescues nutritional delay; Layalle et al. (2008). |
| Drives endoreduplication via Snail; Ohhara et al. (2017); Zeng et al. (2020). | |
| Regulates autophagic cholesterol trafficking; Pan et al. (2019); Texada et al. (2019b). | |
| Tyramine | Autocrine tyramine signaling through Octβ3R is required for intracellular DILP and PTTH transduction and ecdysone synthesis; Ohhara et al. (2015). |
EcR, ecdysone receptor; InR, insulin receptor; PG, prothoracic gland.
Tissue and body growth must be tightly linked to developmental progression, to ensure that sufficient growth has occurred before irreversible developmental transitions are initiated. Numerous intersections between the insulin and ecdysone systems underlie some aspects of this coordination, which in Drosophila involves at least two checkpoint mechanisms: (1) a nutritional checkpoint called CW, which ensures that the feeding larva has accumulated enough reserves to survive the nonfeeding metamorphosis stage, and (2) a developmental checkpoint that assesses the growth status of the imaginal discs within the larva to ensure that maturation does not begin until damaged or slow-growing discs have regenerated and are sufficiently developed in proportion to one another. Later sections of this review build on the descriptions below of the insulin and ecdysone systems to examine the mechanisms (Figure 6) by which these larval checkpoints allow the organism to assess its size and proportions.
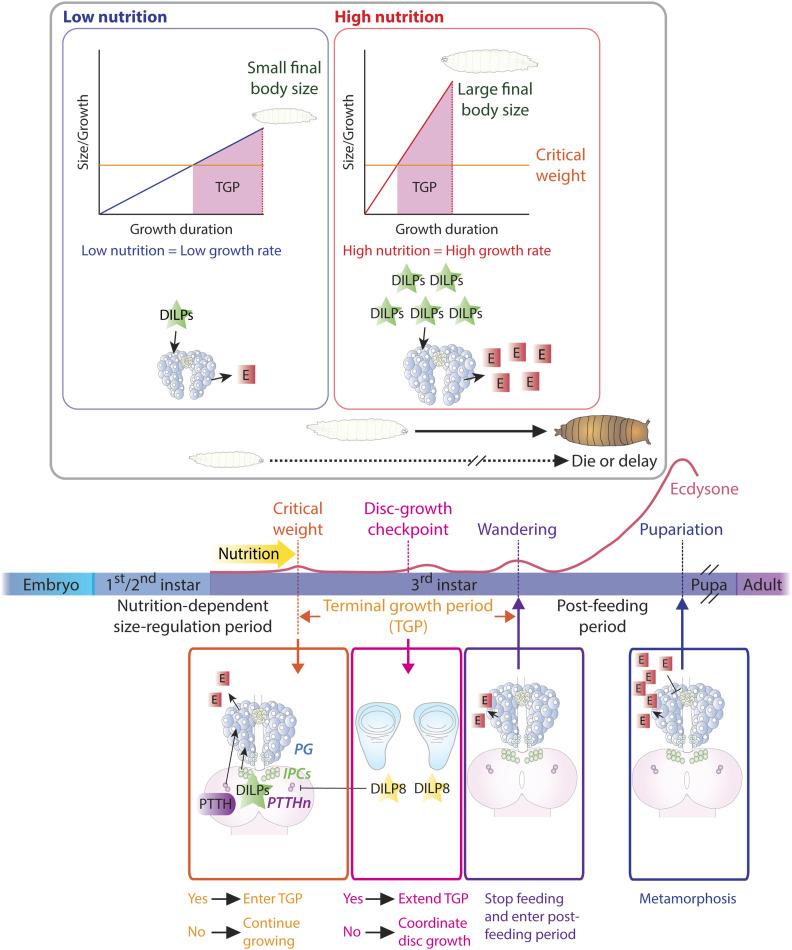
Figure 6.
Developmental checkpoints determine developmental timing in Drosophila. Top: at the onset of the final (third) instar, the resumption of larval feeding after the previous molt stimulates a small rise in ecdysone (E) production via insulin together with PTTH. This small ecdysone pulse results in attainment of the nutrition-sensitive developmental checkpoint critical weight (CW), which begins the subsequent nonnutrition-sensitive feeding period called the terminal growth period (TGP). In post-CW larvae, further nutrition intake is not necessary to undergo metamorphosis. Larvae will not proceed into metamorphosis until they reach CW, which reflects their ability to survive through pupal life on stored nutrients alone. After CW, the larva continues to grow during TGP if food is present. Under poor conditions (left), the larva grows slowly, reaching CW later; slow growth continues during TGP, leading to small adults. This slow growth and delayed maturation results from low production of insulin and ecdysone. Right: under rich conditions, the larva feeds well and produces more insulin, which induces fast growth. The animal soon reaches CW and continues to grow quickly during TGP. Insulin also promotes earlier peaks of ecdysone, which accelerate developmental timing. As a result, adults emerge more quickly, with larger bodies. Bottom: organ growth also affects developmental timing. Developing discs secrete DILP8, which affects the timing of PTTH secretion and indirectly controls the timing of growth cessation by regulating the timing of the second small ecdysone pulse. DILP8 also regulates the growth of all discs simultaneously by acting directly on the prothoracic gland (PG) to regulate basal ecdysone levels.
The insulin system: coupling of growth to nutritional and environmental inputs
Many environmental factors modulate growth and development, including nutrition, temperature, and oxygen level (Beadle et al. 1938; Partridge et al. 1994; Nunney and Cheung 1997; French et al. 1998; Peck and Maddrell 2005; Callier and Nijhout 2011; Harrison et al. 2015; Texada et al. 2019a). Larvae raised under low-oxygen or nutritionally poor conditions grow slowly and give rise to smaller adults, despite a prolonged growth period (Callier et al. 2013; Texada et al. 2019a). At lower temperatures, Drosophila larvae also extend their developmental time but produce larger adults (Li and Gong 2015), indicating that temperature and nutrient/oxygen levels affect growth through different means. Variation in growth conditions also leads to adults with altered body proportions—allometry—indicating that organs respond tissue-specifically to growth-affecting environmental cues (Shingleton et al. 2009, 2017); this phenomenon is discussed at the end of this review.
Diet has a huge influence on growth, and Drosophila can be raised under a range of nutritional conditions that produce adults of different sizes and proportions. Dietary amino acids are indispensable for growth and development, and the amount of protein in the diet is inversely related to developmental time. Essential amino acids are usually obtained mostly from dietary yeast, and the amount of dietary protein also influences vitamin requirements (Sang 1962). Newly hatched larvae fed a protein-free, sugar-only diet cannot grow, whereas larvae reared on diets containing amino acids but lacking nucleotide precursors, lipids, or vitamins can grow and develop to the late-L2 stage (Britton and Edgar 1998). Dietary carbohydrates and lipids also influence larval growth and development. Carbohydrate-rich diets negatively affect growth and delay pupariation in Drosophila, and this dietary condition has been used to model aspects of type 2 diabetes and obesity, as well as to understand the connections between diet, metabolic disorders, and cancer development (Musselman et al. 2011; Pasco and Leopold 2012; Hirabayashi et al. 2013; Barry and Thummel 2016). The effects of high-sugar diets on development are mainly mediated by the insulin pathway and include increased lipid storage and insulin resistance. While the effect of high sugar on developmental timing may not be relevant for normal ecological and physiological conditions, it may be important for understanding how human disorders such as diabetes and obesity can affect the timing of puberty. Like amino acids, the neutral lipid cholesterol is also essential for development in Drosophila. Although cholesterol is a biochemical precursor to ecdysone, which generally slows larval growth, increased dietary cholesterol promotes body growth (Carvalho et al. 2010; Lee et al. 2010), suggesting that it has a systemic growth-promoting effect independent of its ecdysone-related role.
All of these growth-governing environmental factors converge on the insulin and TOR pathways described above. Many of their effects arise from the modulation of DILP secretion, which is regulated cell-autonomously by nutrients, by central mechanisms such cold and nutrient sensing within the nervous system, and by humoral factors released by peripheral organs such as the fat body, which functions as a sensor of nutrient and oxygen levels. Thus, this coordination of growth depends on the exchange of information between cells and organs sensing external and internal conditions, and target cells such as the IPCs that integrate these messages to exert systemic control over growth (Figure 3 and Table 1). The growth of tissues such as the muscles, which is driven by nutritional inputs via insulin and TOR, then feeds back to affect systemic body growth. Body growth is systemically slowed by muscle-growth inhibition (Demontis and Perrimon 2009), and DILP release is inhibited by physiological perturbation of adult muscle (Demontis and Perrimon 2010; Bai et al. 2013), suggesting that complex interplay and feedback between organ growth and growth-regulatory mechanisms ensures coordinated responses across the entire body.
Control of systemic growth through DILP signaling:
Eight genes encoding insulin-like proteins—Dilp1 through Dilp8—have been identified in Drosophila based on their characteristic six-cysteine insulin/relaxin-like motif (Brogiolo et al. 2001; Ikeya et al. 2002; Colombani et al. 2012; Garelli et al. 2012; Liu et al. 2016). All eight DILPs are thought to be synthesized as preprohormones containing an N-terminal signal sequence and a prohormone comprising two peptide segments, the A and B chains, flanking an intervening “C peptide.” Within each molecule, the six conserved cysteines link the A and B chains through disulfide bonds. Proteolytic processing removes the C peptide of insulin- and relaxin-family proteins, but this peptide remains intact in mature IGF-like hormones. DILP1 through DILP5 are most closely related to vertebrate insulin, whereas DILP6 is the only IGF-like peptide in Drosophila (Okamoto et al. 2009). These six DILPs are believed to act through the single insulin RTK InR (Fernandez et al. 1995; Chen et al. 1996; Brogiolo et al. 2001), although only DILP2 and DILP5 have been assayed biochemically for InR activity (Sajid et al. 2011; Lin et al. 2017; Post et al. 2018a). DILP7 and DILP8 appear to be more closely related to human relaxin-family molecules than to insulin/IGF. DILP8 does not act through InR but rather through the G protein-coupled receptor (GPCR) Lgr3 (Colombani et al. 2015; Garelli et al. 2015; Vallejo et al. 2015; Jaszczak et al. 2016), a relaxin-receptor-like protein containing an extracellular ligand-binding leucine-rich-repeat domain (Van Hiel et al. 2015). The receptor for DILP7 has not been identified, but evolutionary genomics suggests it may act through another leucine-rich-repeat-containing GPCR family member, Lgr4 (Veenstra et al. 2012), while genetic evidence is also consistent with a role for InR here (Ikeya et al. 2002; Linneweber et al. 2014).
The DILPs exhibit diverse spatiotemporal patterns of expression and are regulated by different developmental and nutritional cues (Brogiolo et al. 2001; Ikeya et al. 2002; Colombani et al. 2012; Garelli et al. 2012; Liu et al. 2016). The main systemically acting growth-regulating DILPs—2, 3, and 5—are primarily produced by the IPCs, a bilateral cluster of neurosecretory cells in the larval and adult brain (Brogiolo et al. 2001; Ikeya et al. 2002). Ablation of these cells in the larva causes growth retardation and developmental delay (Rulifson et al. 2002). These cells also transiently express DILP1 during the nonfeeding pupal-to-adult transition and in diapausing flies (Liu et al. 2016). Other tissues also express DILPs for local or systemic growth control. DILP2 is expressed by imaginal discs, while DILP3 is also expressed by the musculature of the larval midgut (Veenstra et al. 2008; Amcheslavsky et al. 2014). DILP5 is expressed under stress conditions by the principal cells of the renal Malpighian tubules (Söderberg et al. 2011). DILP6 is expressed in a nutrient-dependent manner by glia cells and in the larval fat body in response to ecdysone and starvation through FOXO-dependent regulation to promote growth under nutritionally restricted conditions, including during the nonfeeding metamorphosis process (Okamoto et al. 2009; Slaidina et al. 2009; Bai et al. 2012; Okamoto and Nishimura 2015).
Regulation of IPC activity and functional role of DILPs:
Because Drosophila only express one known receptor (InR) for the growth- and metabolism-regulating DILPs, DILP signaling regulates both cell growth and metabolism during larval development, thus performing the roles of both mammalian insulin and IGFs. Funneling these two functions through one receptor may seem to present a challenge during periods of growth that require sustained insulin signaling along with simultaneous maintenance of hemolymph sugar homeostasis. This challenge seems to be met by selective DILP expression and release, as well as by functional differences between the DILPs, allowing them to mediate responses to distinct nutritional cues (Figure 3 and Table 1). For example, whereas DILP2 loss induces a strong growth defect, the loss of DILP3 does not, only leading to delayed development under conditions with low dietary yeast (Kim and Neufeld 2015), indicating that DILP3 is required for normal growth on amino acid-poor diets.
DILP expression:
The IPC-derived DILPs vary independently in their expression over developmental time. Under constant-feeding laboratory conditions, Dilp2 is highly expressed in the first instar, with levels falling toward wandering, Dilp3 is expressed at low levels until the midthird instar, when it is strongly upregulated, and Dilp5 rises from a low level through the first instar and remains elevated until wandering (Slaidina et al. 2009; Okamoto and Nishimura 2015). Nutritional cues also affect DILP expression and release independently in both the larva and the adult (Ikeya et al. 2002, 2009; Kim and Neufeld 2015; Post and Tatar 2016). Expression of Dilp3 and Dilp5 in the larval IPCs is downregulated by starvation (Ikeya et al. 2002). Although Dilp2 expression is somewhat independent of nutrient availability and appears to be unchanged by starvation in the L3 stage, expression of both Dilp2 and Dilp5 are upregulated by a chronic high-sugar diet in the larval stages (Pasco and Leopold 2012). In adults, Dilp2 expression increases with increased ratios of carbohydrates to protein in the diet, Dilp3 expression peaks in diets with high sugar-to-protein ratios, and Dilp5 appears to increase with caloric value (Post and Tatar 2016). Furthermore, Dilp expression is regulated by multiple hormonal inputs (Figure 3 and Table 1) and by complex feedback regulation (Broughton et al. 2008; Grönke et al. 2010; Bai et al. 2012; Post et al. 2018b).
The DILPs share homology with mammalian insulin at the level of their transcriptional regulation. The transcription factor Eyeless (Ey) and its interaction partner Dachshund (Dac) control IPC differentiation and regulate Dilp5 expression. Their mammalian orthologs Pax6 and Dach1/Dach2 function similarly in pancreatic β-cells (Clements et al. 2008; Okamoto et al. 2012). In the Drosophila larval IPCs, Dilp5 expression is repressed by FOXO, which inhibits Ey:Dac-mediated Dilp5 transcription (Figure 3) (Okamoto and Nishimura 2015). This conservation underscores the homology between the IPCs and mammalian β cells, suggesting that flies can be a useful model for understanding molecular mechanisms of β-cell function and insulin-mediated metabolic and growth control.
DILP release:
In mammals, the release of insulin from β cells is directly influenced by sugars and amino acids. High blood-sugar levels strongly induce insulin secretion via induction of ATP synthesis and the closure of ATP-sensitive K+ channels, leading to voltage-gated calcium influx and vesicle release. A similar mechanism allows adult Drosophila IPCs to respond directly to sugar levels (Kréneisz et al. 2010). However, larval IPCs do not appear to respond autonomously to hemolymph sugars. Instead, release of DILP3, but not DILP2, is induced by Adipokinetic hormone (Akh, the Drosophila functional analog of glucagon) released by the Akh-producing cells (APCs) of the larval corpora cardiaca (CC) (Kim and Neufeld 2015), which autonomously respond to hemolymph sugar levels (Figure 3) (Kim and Rulifson 2004; Braco et al. 2012).
Dietary amino acids, especially branched-chain amino acids (BCAAs) such as leucine, also have strong insulinergic effects and directly stimulate secretion from mammalian β cells. In Drosophila larvae, DILP2 secretion is also coupled to amino acid levels, especially of BCAAs (Géminard et al. 2009), via two mechanisms. As an indirect route of control, the fat body senses amino acids and remotely induces DILP release; this is discussed further below. The larval IPCs also autonomously respond to leucine by secreting DILP2 and DILP5. Leucine is imported into the IPCs via the proteins Minidiscs (Mnd) (Manière et al. 2016) and JH inducible-21 (JhI-21) (Ziegler et al. 2018), homologous with the mammalian L-type amino acid transporter LAT1, which mediates leucine-stimulated insulin secretion from mammalian β cells (Cheng et al. 2016). Imported leucine allosterically activates the glutamate dehydrogenase (GDH) pathway, which is required for leucine-induced DILP2/5 secretion (Manière et al. 2016). In mammals, this GDH-dependent pathway is known to lead to increased production of the Krebs cycle intermediate α-ketoglutarate and thus to increased ATP generation, which induces insulin release (Gao et al. 2003; Fahien and Macdonald 2011). Notably, stimulation of DILP secretion by amino acid sensing in the IPCs appears to be TOR-independent (Manière et al. 2016). In contrast to DILP2 and DILP5, release of DILP3 is not affected by amino acids, but sugars selectively induce the release of DILP3 (Kim and Neufeld 2015). Thus, in addition to their independent transcriptional regulation by nutrient conditions, DILP2 and DILP3 appear to be segregated into different secretory vesicles in the IPCs, possibly providing another mode of selective release of individual DILPs in response to distinct nutritional cues. The exact mechanism by which DILPs are trafficked and sorted into secretory granules is generally not known, but it involves Hobbit, a conserved protein named for its reduced-body-size phenotype, which was recently shown to be required for DILP secretion in Drosophila (Neuman and Bashirullah 2018). Secretion of DILPs is also regulated by the highly conserved microRNA miR-7 in the IPCs, which inhibits the production and secretion of DILPs (Agbu et al. 2020). miR-7 regulates body size at least in part though effects mediated by DILP2. miR-7 does not directly target Dilp transcripts but rather regulates insulin production by affecting the F-actin capping protein α (CPA), a mechanism that is also conserved in mammalian β cells.
Modulation of circulating DILP activity:
Insulin signaling is also regulated after DILP release by several secreted proteins that bind selectively with DILPs and thereby modulate their stability, availability, and activity (Figure 3). Ecdysone-inducible gene L2 (ImpL2), a member of the immunoglobulin family related to mammalian IGF-binding proteins, and the Drosophila acid-labile subunit ortholog dALS/Convoluted can form complexes with circulating DILP2 and DILP5, sequestering them and thereby negatively regulating systemic growth. DILP3 has a greater affinity for Secreted decoy receptor (Sdr), which is structurally similar to the ligand-binding domain of InR, and interacts with several DILPs and antagonizes their action (Arquier et al. 2008; Honegger et al. 2008; Okamoto et al. 2013). Furthermore, the DILPs act with different kinetics on InR and thereby drive different outputs of the downstream effector pathway. DILP2 transiently activates Akt phosphorylation, whereas DILP5 leads to sustained phosphorylation downstream of receptor binding, suggesting that two related DILPs have the capacity to elicit unique downstream signaling outputs. Indeed, DILP2 signaling promotes deactivation of glycogen phosphorylase, the rate-limiting enzyme in glycogen breakdown, whereas DILP5 does not (Post et al. 2018a). Although it has been proposed that some DILPs may work as InR antagonists, DILPs 1–7 promote developmental growth when ubiquitously expressed, suggesting that they possess growth-promoting activity (Ikeya et al. 2002).
Glial and neuronal relays controlling DILP signaling:
In addition to direct nutrient sensing in the IPCs, DILP production and release are also regulated by nonautonomous signals relayed from central and peripheral tissues. Signaling from glial cells of the larval blood/brain barrier (BBB) regulates nutrient-dependent IPC Dilp5 expression, which is required to sustain body growth under restrictive nutrient conditions (Okamoto and Nishimura 2015). These glial cells sense circulating amino acid levels via intracellular TOR signaling and through circulating DILPs at the interface between surface glia and the hemolymph, and they secrete DILP6 in response to sufficient levels. This DILP6 acts on certain cholinergic neurons of the brain, leading to their release of Jelly belly (Jeb) onto the IPCs, which express the Jeb-binding RTK Anaplastic lymphoma kinase (Alk). Activation of Alk in the IPCs induces PI3K signaling, which relieves FOXO-mediated inhibition of Ey- and Dac-dependent Dilp5 expression. Dilp2 expression is not regulated by this signaling, which provides a mechanism by which DILP signaling is differentially coordinated with nutrient conditions.
Furthermore, neuronally derived short Neuropeptide F (sNPF) promotes growth through ERK-mediated regulation of Dilp expression in the larval IPCs (Lee et al. 2008). Although the nutritional cues conveyed by sNPF are not clear, Drosophila sNPF regulates feeding like its mammalian homolog neuropeptide Y (NPY) (Lee et al. 2004; Root et al. 2011; Carlsson et al. 2013; Ko et al. 2015; Selcho et al. 2017), which suggests that this neuromodulatory peptide may link feeding behavior with systemic growth control. In the adult brain, a pair of sugar-sensing neurons sense sugar levels and release sNPF onto both the IPCs and the APCs, activating the former and inhibiting the latter, and thus coordinating the uptake and usage of energetic species with their storage or release (Oh et al. 2019).
Larval IPC activity is also regulated by serotonergic neurons through a process that involves NS3, a nucleostemin-family GTPase. NS3 is required in serotonergic neurons whose axonal projections are closely apposed to the IPC for proper regulation of body growth (Kaplan et al. 2008). Ns3 mutants accumulate DILP2 in the IPCs and exhibit strongly reduced body size, suggesting that this serotonergic circuit controls DILP2 secretion. Consistent with this notion, the serotonin receptor 5-HT1A is expressed in the adult IPCs and modulates DILP signaling, downregulating expression of Dilp2 and Dilp5 with no effect on Dilp3 (Luo et al. 2012, 2014). However, the 5-HT1A reporter is not expressed in the larval IPCs, suggesting that a different receptor may function in the larva (Luo et al. 2012). The larval IPCs also receive synaptic input from neurons expressing the feeding-associated peptide Hugin, and the IPCs express the Hugin receptor PK2-R1, although the functional significance of this link is unknown (Melcher and Pankratz 2005; Bader et al. 2007; Schlegel et al. 2016). DILP8-responsive growth-coordinating Lgr3-positive (GCL) neurons (Vallejo et al. 2015) and Allatostatin-A (AstA)-releasing neurons (Deveci et al. 2019) synapse onto the IPCs as well as the prothoracicotropic hormone (PTTH)-producing neurons (PTTHn), which are discussed in detail below. A second AstA receptor, AstA-R2, is expressed in the larval and adult IPCs and APCs; AstA-R2 RNAi in the adult IPCs alters the expression of Dilp2 but not Dilp3 (Hentze et al. 2015). The larval IPCs also receive input from cold-activated thermosensory neurons (Li and Gong 2015), which promotes DILP production and secretion, enhancing growth at low temperatures. This neuronal mechanism explains, in part, the inverse relationship between temperature and body size in Drosophila.
Peripheral organs relaying nutrient and oxygen status to the IPCs:
Although the larval IPCs sense amino acids autonomously, the effects of nutrient availability on Drosophila larval growth are thought to be mediated primarily by signals relayed to the IPCs from the fat body (Figure 3 and Table 1). Fat-body nutrient sensing relies in many cases on TOR, which regulates the release of several humoral factors that regulate the IPCs, first illustrated in organ co-culture experiments demonstrating that one or more TOR-dependent fat-body-derived humoral factors couple DILP2 and DILP5 secretion with amino acid intake (Géminard et al. 2009). The fat body’s influence over the IPCs has since been shown to be mediated by several humoral factors. Among these factors are Growth-blocking peptides (GBP1 and -2) (Koyama and Mirth 2016), which are structurally similar to epidermal growth factor (EGF)-like ligands and activate EGFR in neurons that synapse upon the IPCs and stimulate DILP secretion in response to nutrient intake (Meschi et al. 2019). Insulin secretion is also stimulated by the protein Stunted (Sun), which acts directly on the IPCs via its receptor Methuselah (Mth) (Delanoue et al. 2016). The Drosophila TNF-α homolog Eiger inhibits DILP secretion via its receptor Grindelwald in the IPCs (Agrawal et al. 2016); under dietary protein restriction, TNF-α-converting enzyme (TACE) is activated in the fat body via the relief of TOR-mediated inhibition, leading to the cleavage and release of Eiger into the hemolymph.
In addition to these amino acid and TOR-dependent humoral signals, the fat body also responds to dietary sugars and lipids by releasing Unpaired-2 (Upd2), a leptin-like factor that acts through regulation of JAK/STAT signaling in GABAergic neurons presynaptic to the IPCs. Upd2, via its receptor Domeless, inhibits these GABAergic neurons, which relieves their inhibition of IPC activity, thereby disinhibiting DILP2 and DILP5 secretion (Rajan and Perrimon 2012). Furthermore, CCHamide-2 (CCHa2) is required for normal levels of transcription of Dilp5 but not Dilp2, and is required for secretion of both DILP2 and DILP5 in response to sugar (Sano et al. 2015). Larvae lacking this hormone grow slowly and are developmentally delayed (Ren et al. 2015).
Oxygen is another factor essential for growth and development. In many organisms, including Drosophila, low oxygen levels slow systemic growth, delay development, and reduce body size (Palos and Blasko 1979; Frazier et al. 2001; Henry and Harrison 2004; Peck and Maddrell 2005; Callier and Nijhout 2011, 2013). The transcription factor HIF-1a is a primary effector of the conserved metazoan oxygen-sensing pathway. Under normoxia, HIF-1a is rapidly degraded by HIF-1a prolyl hydroxylase (Hph) in a process dependent on molecular oxygen. Thus, in hypoxia, HIF-1a is stabilized and induces transcriptional responses that modulate growth and metabolism. The Drosophila larval fat body is also the primary internal sensor of oxygen (Texada et al. 2019a), releasing one or more as-yet unidentified hypoxia-induced HIF-1a-dependent humoral factors that strongly inhibit IPC DILP expression and secretion, leading to a reduction in circulating DILP2 levels more pronounced than that observed with total starvation (Texada et al. 2019a). Hph is also required for nutrient-dependent activation of TOR in the fat body, linking the pathways mediating nutrient and oxygen sensing in this organ. Taken together, these studies show that the fat body coordinates organismal growth with environmental conditions through its ability to sense nutrients and oxygen, and to release systemic endocrine factors.
Other dietary components such as lipids are essential for growth. Besides serving as building blocks for molecular synthesis and as an energy-storage medium, lipids have multiple regulatory functions and serve as important signaling molecules (Horner et al. 2009; Bujold et al. 2010; Senyilmaz et al. 2015). In Drosophila, lipids derived from yeast exert a strong influence over organismal growth (Carvalho et al. 2010) and can act as nutritional signals on specific neurons in the brain to modulate DILP2 secretion (Brankatschk et al. 2014). This involves a fat–gut–brain relay, in which dietary lipids are acquired from the gut and delivered to tissues by circulating apoB-containing lipoproteins, some of which are transported across the glial BBB and accumulate on certain neurons that synapse upon the IPCs. These lipoproteins include Lipophorin (Lpp), the major carrier of circulating lipids in the hemolymph, and Lipid transfer particle (LTP), another apoB-family lipoprotein (Palm et al. 2012). Interestingly, these proteins are produced by the fat body, suggesting that they might also be involved in signaling nutritional status to the neuroendocrine system that initiates metamorphosis, which may provide another mechanism to explain the relationship between timing of maturation and adiposity. Furthermore, dietary cholesterol promotes systemic growth through a mechanism that is likely independent of its role as a substrate for ecdysone biosynthesis (Carvalho et al. 2010). Since insects are cholesterol auxotrophs, they must somehow sense the dietary and cellular availability of this molecule and couple that to growth-regulatory pathways. Cholesterol binds the Drosophila nuclear receptor Hr96, which controls dietary uptake through actions in the midgut and is essential for maintaining cholesterol homeostasis (Horner et al. 2009; Bujold et al. 2010). Furthermore, recent studies have found that cholesterol activates TOR signaling in mammalian cells (Castellano et al. 2017), suggesting that TOR, perhaps in the fat body, may also integrate information about cellular cholesterol levels with amino acid and oxygen levels.
The mammalian intestine senses food-derived nutrients and also comprises the largest endocrine organ. In response to food intake, the enteroendocrine cells (EECs) of the mammalian gut release hormones that modulate insulin secretion from β cells (Paternoster and Falasca 2018). Early studies showed that the gut potentiates glucose-stimulated insulin secretion; glucose ingested and absorbed via the gut stimulates a greater increase in insulin secretion than glucose injected into circulation. This “incretin” effect is largely due to hormones secreted by the gut such as glucagon-like peptide-1 (GLP-1), which modulates β-cell activity. Whether the gut contributes to systemic growth control through the regulation of insulin signaling during Drosophila development is not known, but the hormone Activin, a TGF-β family member, is produced by the EECs of the larval midgut in response to high-sugar diets and acts on the fat body to promote Akh receptor (AkhR) expression, and thus Akh signaling, suggesting that this gut-to-fat relay mechanism controls sugar homeostasis during development (Song et al. 2017). A gut-derived lipid-associated form of the morphogen Hh, released in response to nutrient insufficiency by the absorptive enterocytes, has also been shown to act systemically (Rodenfels et al. 2014). Circulating Hh slows larval growth and acts on the fat body to promote lipolysis (Zhang et al. 2020). At the same time, Hh regulates the timing of pupariation through direct effects on PG ecdysone production (Rodenfels et al. 2014), discussed below (Figures 4 and 5 and Table 3), thereby coordinating growth and maturation according to nutrition.
The ecdysone signaling system: coordinating growth and developmental maturation
Ecdysone is synthesized in the cells of the larval PG, part of a composite organ called the ring gland in Drosophila. This gland is situated anterior to the brain and also comprises the CC (producing Akh) and the corpora allata (CA, the source of sesquiterpenoid JH). Identification of most of the genes mediating ecdysone biosynthesis—first described by Nüsslein-Volhard, Wieschaus, and their colleagues in their Nobel-winning embryonic-patterning work—was based on their characteristic embryonic lethal phenotype, termed the “Halloween” phenotype (Jurgens et al. 1984; Nüsslein-Volhard et al. 1984; Chavez et al. 2000). These genes encode conserved enzymes that convert dietary sterols such as cholesterol into ecdysone. The early steps in the pathway are mediated by Neverland (Nvd), Spook (Spo) or Spookier (Spok) depending on the developmental stage, and Shroud (Sro). These enzymes convert cholesterol into 5-β-ketodiol through a series of steps, some of which are not yet fully understood: the so-called “black box” (Gilbert et al. 2002; Ono et al. 2006; Yoshiyama et al. 2006; Niwa et al. 2010; Yoshiyama-Yanagawa et al. 2011). β-Ketodiol is then converted by Phantom (Phm), Disembodied (Dib), and Shadow (Sad) into ecdysone, which is imported into vesicles and exocytotically released from the PG into the hemolymph (Warren et al. 2002, 2004; Niwa et al. 2004, 2005; Rewitz et al. 2006b; Yamanaka et al. 2015). In peripheral tissues, ecdysone is converted to its more-active form 20-hydroxyecdysone by the action of the 20-monooxygenase Shade (Petryk et al. 2003; Rewitz et al. 2006a) (henceforth, ecdysone will be used to refer to both ecdysone per se and 20-hydroxyecdysone). Although “textbook” nuclear-hormone ligands enter cells via simple diffusion, ecdysone diffusion into receptive cells must be facilitated by the transporter protein Ecdysone importer (EcI) in Drosophila (Okamoto et al. 2018), which is also required for the passage of ecdysone across the BBB into the brain (Okamoto and Yamanaka 2020).
The PG integrates a variety of signals that regulate ecdysone synthesis and release (Figure 5 and Table 3). The neuropeptide PTTH is a primary regulator of ecdysone synthesis in the PG via effects mediated by its RTK, Torso (McBrayer et al. 2007; Rewitz et al. 2009b). The neurons that produce PTTH, the PTTHn, also integrate multiple cues to regulate PTTH expression and release (Figure 4 and Table 2). Systemic DILP signaling to the PG and intracellular TOR signaling within the gland mediate nutritional control of ecdysone production. These pathways regulate PG ecdysone production through effects on cell growth, genome amplification, Halloween-gene expression, and the availability of cholesterol. Other factors regulate the competence of the PG to respond to PTTH and DILP signals, and the regulation of vesicle-mediated ecdysone release is another layer of control.
Control of prothoracicotropic hormone release:
The neuropeptide PTTH is an important factor stimulating ecdysone production in the PG and thus controlling the initiation of metamorphosis. PTTH was the first insect hormone to be identified, based on classical studies by Kopeć (1922). These studies later led to the discovery of neurosecretory cells and suggested that insect molting is controlled by a humoral factor from the brain (Wigglesworth 1940, 1964). Later studies in lepidopterans elucidated the nature and structure of PTTH, leading to the classical dogma of insect endocrinology (Steel and Davey 1985; Kawakami et al. 1990; Kataoka et al. 1991; Rybczynski 2005). According to this model, developmental transitions such as metamorphosis are controlled by the release of PTTH from the brain, under the influence of JH, that stimulates the PG to produce and release ecdysone. In this scheme, the principal event determining the timing of metamorphosis is the release of PTTH, a decision that is controlled by the integration of cues in the brain. These classical lepidopteran studies subsequently facilitated the characterization of Drosophila PTTH, produced by two pairs of PG-innervating neurosecretory cells (the PTTHn) in the larval brain (McBrayer et al. 2007), and its receptor, Torso (Rewitz et al. 2009b).
Photoperiod and inputs that relay organ-growth and nutritional status (see below; Figure 4 and Table 2) are thought to regulate PTTH secretion. Photoperiod affects PTTH secretion in a wide range of insects (Truman 1972), and in Drosophila, circadian influence is believed to be mediated by input from clock neurons producing the neuropeptide sNPF, which synapse with the PTTHn (Siegmund and Korge 2001; McBrayer et al. 2007; Selcho et al. 2017). Clock defects affect the rhythmicity of Ptth-expression oscillations during L3, which is believed to affect the generation of correctly timed ecdysone pulses. PTTH also acts on peripheral light-sensing organs to control larval light-avoidance behavior (Gong et al. 2010, 2019; Yamanaka et al. 2013b), thereby coordinating behavioral and developmental transitions.
DILP8 secreted from the imaginal discs induces pupariation delay by inhibiting PTTH secretion (discussed in more detail below under the Disc checkpoint section). The DILP8-responsive GCL neurons of the larval brain synapse onto the PTTHn and inhibit PTTH release, thereby delaying metamorphosis (Colombani et al. 2015; Garelli et al. 2015; Vallejo et al. 2015). Lgr3 is expressed broadly in the larval and adult nervous systems, beyond the GCLs, indicating that it has other functions within the animal. Indeed, some neuronal Lgr3 expression is female-specific in neurons that govern mating receptivity (Meissner et al. 2016). The GCL neurons also interact with the IPCs to control the secretion of DILPs, suggesting that DILP8 coordinates growth and maturation in Drosophila by relaying disc growth status to both the PTTHn and the IPCs (Vallejo et al. 2015). Larval insulin/PTTH coordination is also mediated by the neuropeptide AstA released by two bilateral neurons that contact the PTTHn and IPCs (Deveci et al. 2019; Pan and O’Connor 2019). AstA signaling, acting through AstA receptor 1 (AstA-R1), is required in the PTTHn to promote PTTH secretion and normal developmental timing, and in the IPCs to promote DILP secretion and systemic growth. This suggests that AstA signaling coordinates juvenile growth and the onset of maturation in Drosophila through its simultaneous activation of two neuroendocrine centers in the brain. AstA and its receptors are orthologous with the mammalian peptide kisspeptin (KISS) and its receptor, GPR54 (Felix et al. 2015); KISS:GPR54 directly regulates the gonadotropin-releasing hormone (GnRH)-expressing neurons of the brain that induce sex-steroid production, and this system is believed to function as a neuroendocrine switch for the initiation of puberty (Sisk and Foster 2004). Furthermore, a recent report shows that neurons producing the neuropeptide Corazonin (Crz) directly contact the PTTHn and regulate the secretion of PTTH via the Corazonin receptor, CrzR (Imura et al. 2020). While loss of AstA signaling in the PTTHn delays pupariation, suggesting that AstA neurons control the maturation-inducing ecdysone peak that determines growth duration, the Crz receptor signaling in the PTTHn controls basal ecdysone production that negatively regulates the larval growth rate. Crz and CrzR are thought to be orthologous with GnRH/GnRHR (Hauser and Grimmelikhuijzen 2014). Thus, functional similarity and sequence conservation suggest that the overall architecture of the neuroendocrine systems that coordinate growth and maturation has been evolutionarily conserved, including roles for the AstA/KISS and Crz/GnRH system. AstA is regulated in response to nutrient supply in adult Drosophila (Hentze et al. 2015), providing a possible mechanistic link between nutrition and PTTH secretion. The Crz neurons are also likely to receive nutrient cues via octopaminergic input from the feeding-control center of the subesophageal zone (SEZ). Another link between PTTH and nutrition is described in a recent report showing that PTTH secretion is controlled by amino acid levels (Galagovsky et al. 2018). When the Solute-carrier-family-7 (SLC7)-type amino acid transporter Sobremesa (Sbm) is impaired in glia, PTTH secretion is attenuated via an unidentified link, thus reducing ecdysone production and delaying pupariation by 1 day, phenocopying the Ptth-null phenotype (Shimell et al. 2018).
Studies in lepidopterans support the existence of feedback regulation within the PTTH–ecdysone axis (Sakurai 2005; Hossain et al. 2006), reminiscent of the mammalian pathway in which steroids feed back to modulate upstream components, including GnRH, of the hypothalamic–pituitary–gonadal (HPG) axis (Acevedo-Rodriguez et al. 2018). Feedback regulation of the PTTH–PG axis was recently confirmed based on findings that ecdysone-mediated feedback via EcR in the PTTHn upregulates PTTH expression toward the end of larval life (Christensen et al. 2020). Activation of this feedback circuitry is required to produce the PTTH surge that times pupariation, further supporting overall conservation of the neuroendocrine system that controls developmental maturation. Data are consistent with a model in which ecdysone slowly accumulates, perhaps initiated by nutritional factors, until its concentration reaches a critical threshold, which induces (via EcR) a surge of PTTH release, triggering generation of the large maturation-inducing ecdysone peak. Future studies should determine whether JH also participates in the regulation of PTTH secretion in Drosophila.
Integration of signals within the PG:
The Drosophila PG has become a prime model for studying the regulation of steroid-hormone production and release in response to developmental and environmental cues. Since the initiation of metamorphosis is irreversible, and potentially lethal if undertaken prematurely, many developmental and environmental cues feed into the PG (Figure 4, Figure 5, and Table 3). These cues govern the ecdysone biosynthetic pathway over long and short time scales through increased transcription of the biosynthetic genes (in part due to endoreduplication), as well as translational and post-translation regulation. They also act through mechanisms that make cholesterol substrate available for ecdysone biosynthesis.
Although it is generally believed that the metamorphosis-triggering ecdysone peak results from PTTH-mediated stimulation of the PG, a growing body of evidence shows that many additional environmental and developmental inputs are integrated within the cells of the PG to determine the pattern of ecdysone synthesis (Figure 4, Figure 5, and Table 3). PTTH stimulates ecdysone production by acting as a trophic factor that promotes PG growth as well as by directly upregulating the genes of the ecdysone-biosynthetic pathway. Ablation of the PTTHn reduces the size of PG cells, and mutations in the gap gene giant can lead to stochastic elimination of PTTH production in one of the PTTHn pairs, which reduces the growth of the PG lobe innervated by that pair, suggesting that PTTH is released from synaptic terminals directly onto the PG and acts in a local manner (Ghosh et al. 2010; Shimell et al. 2018). Loss of PTTH signaling has little effect on the timing of the first two larval molts and mainly affects the duration of the L3 stage, prolonging it by roughly 1 day and leading to larval overgrowth (McBrayer et al. 2007; Rewitz et al. 2009b; Shimell et al. 2018). Interestingly, ablation of the PTTHn altogether induces a much more dramatic 5-day delay in pupariation, suggesting that the PTTHn may produce additional signals that stimulate ecdysone production in the PG (McBrayer et al. 2007).
While factors such as PTTH do promote the growth of the PG through increased polyploidy, they also induce ecdysone biosynthesis within minutes in lepidopterans in a process that depends on rapid translation (Rybczynski 2005) and possibly post-translational modifications, suggesting that cell size and transcriptional upregulation are not the only means by which ecdysone biosynthesis is regulated. In Manduca, PTTH stimulation leads to rapid phosphorylation of Spo, which is believed to catalyze a step in the rate-limiting black-box reaction (Rewitz et al. 2009a). Although great progress has been made in this area, questions remain regarding the exact mechanisms by which ecdysone biosynthesis is regulated at the post-transcriptional level and how biochemical intermediates transit between subcellular organelles such as the endoplasmic reticulum (ER)—where Nvd, Spo/Spok, and Phm reside—and the mitochondria, where the later steps catalyzed by Dib and Sad take place. The regulatory pathways acting on the PG to control ecdysone production include PTTH, insulin and TOR, Warts, TGF-β, EGF, DILP8 and nitric oxide (NO), the circadian clock, EcR/USP, tyramine, serotonin, and Hh, all of which are discussed in detail below.
Developmental cues and size-sensing in the PG:
A number of studies have shown that the PG, like the fat body, is a hub for nutritional signals, integrating them to govern ecdysone production and thus developmental progression. PG-cell-autonomous TOR function directly couples nutrient sensing with ecdysone production. Early studies in Manduca suggested an important role for S6K (Song and Gilbert 1994), supported by observation of S6 phosphorylation in response to PTTH stimulation (Rewitz et al. 2009a). When TOR activity is mildly inhibited in the Drosophila PG, reduced ecdysone production leads to delayed pupariation and thus to larval overgrowth (Layalle et al. 2008). Conversely, activation of TOR signaling in the PG partially suppresses the developmental delay induced by poor nutrition, suggesting that TOR mediates nutrient sensing in the PG. In contrast, mild reduction in insulin signaling in the PG increases the larval growth rate as a consequence of lowering basal ecdysone, but does not affect the ecdysone peaks that govern the timing of pupariation (Colombani et al. 2005). However, when insulin signaling is more strongly inhibited in the PG, low ecdysone production delays pupariation (Caldwell et al. 2005). Activation of insulin signaling via overexpression of InR strongly accelerates pupariation (K. Rewitz, unpublished data), similar to ectopic activation of the PTTH/Torso/MAPK pathway via expression of constitutively active Ras in the PG (Rewitz et al. 2009b).
TOR signaling promotes endoreduplicative genome amplification in the PG during L3 via the transcription factor Snail, which is important for the CW checkpoint, described below (Ohhara et al. 2017; Zeng et al. 2020). Several transcription factors have also been shown to regulate the expression of the ecdysone-biosynthetic genes specifically. Ventral veins lacking (Vvl) and Knirps (Kni) regulate these genes in the PG, and Vvl may function as a master transcriptional regulator required to maintain their expression during larval development (Danielsen et al. 2014). Furthermore, torso and InR are downregulated in the PG when Vvl or Kni are impaired, suggesting that these transcription factors are important for the PG’s competence to respond to PTTH and insulin. The nuclear receptors DHR3, Ftz-f1, E75, EcR, and Usp also regulate ecdysone synthesis (Bialecki et al. 2002; Parvy et al. 2005; Caceres et al. 2011). E75 functions as a sensor of NO, which blocks its ability to repress DHR3. DHR3 then induces the expression of Ftz-f1, which positively regulates expression of the ecdysone-biosynthetic genes and metamorphosis-inducing ecdysone peak. Several transcription factors have been shown to regulate the expression of single or multiple ecdysone-biosynthetic genes in the PG. It is unclear how some of these factors regulate gene expression, but Molting defective (Mld), Séance (Sean), and Ouija board (Ouib) cooperatively regulate Nvd and Spok by binding response elements in the nvd and spok enhancers (Danielsen et al. 2014; Komura-Kawa et al. 2015; Niwa and Niwa 2016; Uryu et al. 2018). Furthermore, Vvl, Kni, Krüppel homolog 1 (Kr-h1), and EcR also appear to bind the promoters of the ecdysone-biosynthetic genes (Moeller et al. 2013; Danielsen et al. 2014; Zhang et al. 2018). While Vvl and Kni seem to be important for spatial regulation to set and maintain expression of the ecdysone-biosynthetic genes, Kr-h1 may mediate the suppressive effects of JH signaling on ecdysone production. Together with EcR, Kr-h1 may be responsible for temporal control of biosynthetic-gene expression during L3 to generate the large metamorphosis-inducing ecdysone pulse. The upregulation of these genes in late L3 involves EcR-mediated ecdysone feedback that activates expression of isoform Z4 of the transcription factor Br (Br-Z4) in the PG, which in turn upregulates expression of the Halloween genes (Moeller et al. 2013). This regulatory feedback circuit may work as a switch by which the nutrient-dependent rise in ecdysone levels in the beginning of L3 leads to the irreversible nutrition-independent activation of the endocrine system at CW. When ecdysone levels reach a threshold, it generates self-sustaining feedback that generates the maturation-inducing ecdysone pulse. The end of the ecdysone pulse is sculpted by negative feedback through EcR and Br, in this case isoform Z1 (Br-Z1), which downregulates Halloween-gene expression and thus terminates the production of ecdysone. Ecdysone also induces expression of the cytochrome P450 enzyme Cyp18a1, whose 26-hydroxylase activity inactivates both ecdysone and 20-hydroxyecdysone in peripheral tissues (Rewitz et al. 2010).
Once CW has been attained (discussed below) and the neuroendocrine cascade has been activated, insulin signaling is no longer required for ecdysone biosynthesis, but nutritional conditions continue to modulate ecdysone production. Indeed, total starvation after CW accelerates pupariation, suggesting that other signals act to regulate ecdysone synthesis post-CW. PTTH signaling and TOR activity in the PG act both before and after CW to regulate ecdysone-pulse timing, suggesting that these signals modulate the duration of the terminal growth period (TGP) to control final body size (Layalle et al. 2008; Shimell et al. 2018). The growth rate during this period is modulated by basal ecdysone synthesis, controlled by the insulin and PTTH pathways (Colombani et al. 2005; Moeller et al. 2017; Imura et al. 2020). Inhibition of insulin or PTTH signaling in the PG reduces basal ecdysone production during the L3 stage, which increases body size by derepressing the growth rate (discussed below under Interactions between ecdysone and insulin signaling). This effect of insulin in the PG is mediated by the Warts pathway, which promotes ecdysone synthesis by inhibiting the expression of the effector microRNA bantam, which itself inhibits ecdysone production (Boulan et al. 2013). The Warts/bantam pathway modulates TOR and EcR signaling, which controls an autophagic process that traffics cholesterol for steroidogenesis (Texada et al. 2019b,c). Interestingly, this nutrient-dependent autophagic cholesterol-trafficking process also seems to play a role in the CW checkpoint (Pan et al. 2019). Suppression of autophagy in the PG causes a shift in this nutritional checkpoint, suggesting that autophagy-dependent regulation of cholesterol availability in the PG is involved in mediating the starvation-response switch that occurs at CW. The availability of cholesterol in the PG also is regulated by uptake, which is promoted by TOR activity and repressed by EcR signaling (Danielsen et al. 2016). In the PG, cholesterol uptake and trafficking are regulated by the Niemann-Pick type C-1a (Npc1a) protein, the fatty-acid elongase Stuck in traffic (Sit), and the glutathione S-transferase Noppera-bo (Nobo) (Huang et al. 2005b; Enya et al. 2014; Danielsen et al. 2016). TOR and EcR regulate Npc1a and Sit, suggesting that TOR signaling and ecdysone feedback coordinate substrate availability and delivery with biosynthetic activity, to couple ecdysone production to nutritional conditions and development. Like the Halloween genes, Npc1a is regulated by Br-Z4 in the PG (Xiang et al. 2010), suggesting that EcR-mediated feedback through Br-Z4 coordinates cholesterol uptake with ecdysone biosynthesis (Moeller et al. 2013). In addition to Npc1a- and Sit-mediated cholesterol uptake via the endosomal/lysosomal pathway, PG cells also appear to obtain cholesterol via Sensory neuron membrane protein 1 (Snmp1), a Scavenger Receptor Class B type I (SR-BI) family member that mediates lipid uptake. Snmp1-mediated lipid uptake is regulated by Ftz-f1 activity, which is modulated by SUMOylation of Ftz-f1 by Smt3 (Talamillo et al. 2008, 2013). In the PG, loss of Smt3 reduces lipid droplets and ecdysone production, leading to larval developmental arrest. This provides potentially yet another layer of regulation of sterol metabolism in the steroidogenic PG, which may be conserved across species.
Other signals regulating ecdysone production:
Superimposed on the central insulin/TOR and PTTH pathways, multiple other signal pathways contribute to the regulation of ecdysone production in the PG. One such pathway involves nutrient-responsive serotonergic neurons that project to the PG and modulate the timing of ecdysone release (Shimada-Niwa and Niwa 2014). These and neurons receive input from the feeding center in the SEZ, and their density of arborization is correlated with nutrition; under limiting conditions, these cells only sparsely innervate the PG, whereas under high-nutrient conditions, they densely arborize on the PG. Serotonin released onto the PG acts through the GPCR 5-HT7 to raise intracellular levels of cAMP, a second messenger that has been suggested to regulate ecdysone production in Manduca (Rybczynski 2005). Nutrient information is also relayed directly to the PG by midgut-derived Hh, which (as mentioned above) signals nutritional deprivation (Rodenfels et al. 2014). Circulating Hh signals to the fat body to mobilize energy and reduce larval growth under starvation. In parallel it signals directly to the PG, inhibiting ecdysone release, which delays pupariation and allows prolonged growth. These findings suggest that in addition to the central nutrient sensor in the fat body, nutrient sensing by another organ, namely the gut, is involved in coupling growth and developmental timing to nutritional conditions.
Ecdysone production in the PG is also regulated by TGF-β signaling mediated by Activin and Dpp. Increased Activin signaling in the PG leads to precocious metamorphosis, whereas inactivation of this pathway via impairment of the downstream effector dSmad2 blocks pupariation, resulting in continued larval growth and formation of giant L3 larvae (Gibbens et al. 2011). Activin promotes ecdysone production via the receptor Baboon (Babo), which controls expression of torso and InR, suggesting that Activin regulates the competence of the PG to receive insulin and PTTH signals. Consistent with this, the giant-larva phenotype caused by impaired dSmad2 signaling can be rescued by activation of either PTTH or insulin signaling in the PG, suggesting that Activin acts a competence factor for the PG to respond to these metamorphosis-inducing signals.
Developing imaginal discs release the TGF-β ligand Dpp, which acts on the PG via the receptor Thickveins (Tkv) and the effector Mad to downregulate Halloween-gene expression (Setiawan et al. 2018). Dpp signaling in the PG falls as the discs grow, suggesting that disc-derived Dpp may function as an additional checkpoint signal that conveys the discs’ growth status to the endocrine system. Consistent with this, inactivation of this pathway in the PG abrogates the CW checkpoint. When Mad is impaired in the PG, larvae starved shortly after the L2–L3 transition, before reaching CW, proceed to undergo pupariation. Increased release of Dpp from the discs delays pupariation and leads to larval overgrowth. Dpp signaling regulates ecdysone production, at least in part, by interacting with FOXO and bantam, with increased Dpp signaling leading to increased nuclear FOXO localization and bantam activity. This suggests that Dpp signaling modulates insulin signaling and is consistent with the role of Dpp in setting the CW checkpoint, which depends on insulin-mediated ecdysone production in the PG.
In parallel with the Dpp-mediated disc signal, disc-derived DILP8 also acts on the PG via Lgr3 (Jaszczak et al. 2015, 2016). DILP8 binding induces the production of the second messenger NO, which blocks basal ecdysone production and therefore reduces disc growth, but does not affect pupariation timing like E75-mediated NO signaling (Caceres et al. 2011). Thus, the imaginal discs signal their developmental status to the PG through several routes: DILP8 and Dpp act directly on the PG, and DILP8 also via a neuronal relay acts through effects on PTTH and DILPs. It is important to note that some imaginal cells in the larva are not organized into disc structures (Zhou and Riddiford 2002; Minakuchi et al. 2008), and that these cells may also contribute signals such as DILP8 and Dpp that coordinate growth across the body.
Furthermore, ecdysone production is regulated by circadian clocks, both directly by a PG-autonomous system and indirectly from the central brain timekeeper via inputs from the PTTHn. PG-specific disruption of the core oscillatory components of the clock, Timeless and Period, inhibits ecdysone production, blocks pupariation, and leads to larval overgrowth (Danielsen and Rewitz 2016; Di Cara and King-Jones 2016; Ou et al. 2016). These clock components are required for upregulation of the ecdysone-biosynthetic genes, and they interact in the PG with insulin and PTTH signaling. This suggests that synchronization of the local PG clock with the hormonal output of central timekeepers is required for the proper timing of ecdysone production.
Other local mechanisms regulating ecdysone production in the PG involve autocrine signaling through the biogenic amine tyramine and through EGF-like ligands. If the G protein-coupled monoamine receptor Octopamine receptor β3 (Octβ3R), which binds tyramine and its derivative octopamine, is disrupted, or if the synthesis of tyramine (but not of octopamine) is blocked in the PG, ecdysone production around CW is impaired due to loss of biosynthetic-gene expression (Ohhara et al. 2015). Furthermore, autocrine EGF signaling induces ecdysone production toward the end of larval development (Cruz et al. 2020). When the EGF-like ligands Spitz and Vein or their receptor Torpedo/EGFR are inhibited in the PG, reduced ecdysone production results in developmental arrest at the L3 stage. This effect seems to result from impaired PG growth and lower expression of the ecdysone-biosynthetic genes. EGFR signals through the MAPK pathway, the same pathway used by PTTH/Torso, supporting the central role of this is pathway in the regulation of ecdysone production. PG expression of Spitz and Vein is induced by ecdysone, indicating that they are upregulated by, and part of, the positive feedback circuit (as also discussed above) that generates the large ecdysone pulse that triggers pupariation. These results suggest that a rise in ecdysone level pre-CW, which is controlled by insulin and PTTH, initiates an ecdysone-mediated feed-forward mechanism in the PG at CW that activates EGF signaling. In turn, EGF/EGFR/MAPK then acts synergistically with PTTH/Torso/MAPK signaling to ensure sustained ecdysone production during the midlate L3 stage, which initiates metamorphosis. EGFR also regulates the localization of ecdysone-containing secretory vesicles in the PG prior to release (Cruz et al. 2020).
Interactions between ecdysone and insulin signaling:
In Drosophila, activation of the neuroendocrine cascade that ultimately generates the pupariation-inducing ecdysone pulse determines the duration of the growth period (Colombani et al. 2005; Mirth et al. 2005; Koyama et al. 2014). Interestingly, however, before this peak, lower, basal levels of ecdysone negatively regulate the growth of larval tissues by modulating peripheral insulin signaling (Colombani et al. 2005; Boulan et al. 2013; Moeller et al. 2017; Texada et al. 2019b). This cross talk appears to arise mainly through EcR-mediated effects in the fat body (Figure 4). An increased circulating ecdysone level, achieved either by direct ecdysone feeding or through activation of the insulin-signaling pathway in the PG, reduces peripheral insulin signaling, which can be rescued by knockdown of EcR in the fat body. This suggests that ecdysone modulates organismal growth rate through a fat-body relay that attenuates systemic insulin signaling.
In the fat tissue, ecdysone acts via EcR to inhibit Myc function. This local action modulates systemic insulin signaling and global growth, suggesting that a humoral message expressed or released downstream of Myc relays information to control insulin signaling and organismal growth rate (Delanoue et al. 2010). The ability of fat-body Myc activity to affect IPC DILP2 secretion depends on stearoyl-CoA desaturase (Desat1) activity (Parisi et al. 2013). Desat1 is involved in fatty-acid production and affects Myc’s ability to promote lipid storage, which suggests that triglyceride synthesis in the fat body may affect the humoral message that controls DILP2 release. These findings are especially interesting in light of the relationship between adiposity and maturation timing discussed below, which suggests that humoral signals reflecting fat storage are linked to the neuroendocrine pathways regulating CW attainment via insulin secretion from the IPCs.
Another mechanism by which ecdysone modulates insulin-dependent systemic growth involves the conserved microRNA miR-8 (Hyun et al. 2009). Fat-body miR-8 is required for normal larval growth, and miR-8 mutants exhibit reduced body size, owing to a decreased growth rate (Hyun et al. 2009). miR-8 cell-autonomously upregulates PI3K signaling in the fat body by inhibiting u-shaped (ush; mammalian FOG2), which encodes a PI3K-signaling inhibitor (Jin et al. 2012). This promotes systemic growth and peripheral insulin signaling through a noncell-autonomous mechanism. In Drosophila, miR-8 is repressed by ecdysone, and deletion of miR-8 abolishes ecdysone-mediated modulation of insulin signaling and systemic growth (Jin et al. 2012). This suggests that ecdysone suppresses body growth toward the end of larval life through regulation of fat-body miR-8, whose expression decreases during the L3 stage as the ecdysone level gradually increases, leading to the upregulation of Ush that inhibits insulin signaling.
While TOR and EcR in the fat body modulate systemic growth through their indirect regulation of insulin signaling, insulin signaling itself in this tissue cell-autonomously regulates the growth of the fat cells but does not seem to influence body growth, at least in normal physiological conditions (Colombani et al. 2003). The same logic appears to apply to another mechanism that regulates body size through modulation of insulin signaling, in this case as a response to low tissue oxygen levels. In this case, growth of the larva beyond the capacity of the tracheal airway system to deliver sufficient oxygen to the fat body leads to inhibition of insulin release and body growth, mediated by one or more hypoxia-induced fat-body factors (Texada et al. 2019a). However, the growth of the tracheal terminal cells themselves seems to be insulin-independent, as they must be to allow their growth to catch up with that of the rest of the (insulin-dependent) body. This suggests that mechanisms through which certain organs remotely regulate insulin-driven body growth are independent of insulin. In these organs, such as the fat body, the function of insulin seems to be limited to the control of tissue growth and energy storage to maintain body-wide metabolic homeostasis.
Cross talk between ecdysone and insulin signaling explains how increasing ecdysone levels inhibit the growth of larval tissues, leading to an overall attenuation of body growth toward the end of larval life. Nutrition sensed through TOR in the fat body leads, via humoral signals, to increased insulin secretion from the IPCs; increased insulin signaling in the PG promotes ecdysone production through effects mediated by the Warts pathway. This leads to increasing ecdysone levels that feed back to the fat body via EcR, and this leads to a relay of information back to the IPCs to suppress insulin secretion, inhibiting body growth at the end of larval development. However, while ecdysone negatively regulates body growth, this hormone promotes the growth of the imaginal discs, which continues after body growth ends.
Developmental and nutritional checkpoints
Mechanisms have evolved to postpone the onset of maturation until the larva is large enough to survive the nonfeeding pupal stage and produce a properly sized adult. This nutritional checkpoint is known as CW. Another developmental checkpoint has evolved to ensure that the imaginal discs, growing within the larva, attain correct proportions and size before the initiation of pupariation. If these structures are damaged, or if they grow out of proportion with one another, a “disc checkpoint” mechanism delays maturation to allow these tissues additional time to grow or heal. Together these two checkpoints, which rely on insulin and ecdysone regulation (Figure 6), ensure that adults emerge with correct body size and proportions.
CW:
CW, first described by Beadle et al. (1938), is a nutrient-dependent body-size checkpoint that animals must pass through before metamorphosis can be initiated (Mirth and Riddiford 2007). Once this checkpoint has been passed, a neuroendocrine cascade is initiated that commits the larva to undergoing metamorphosis irrespective of further nutrition. CW is a mostly fixed, genetically determined species-specific body size, defined as the size after which starvation no longer delays metamorphosis; it is attained roughly 8 hr after the L2–L3 transition in wild-type Drosophila. Prior to this point, a developing larva can compensate for poor nutritional conditions by delaying metamorphosis, but beyond this point, metamorphosis will occur after a fixed time interval—the TGP—regardless of nutritional input (Figure 6). However, nutrition still influences the growth rate during the TGP. Since CW does not vary with nutrition, the variable amount of growth achieved during the TGP largely determines the final adult size. Drosophila larvae can grow quickly during this period, which means that environmental conditions can have a huge influence on final body size. Inactivation of insulin signaling through loss of InR function during the period before CW, but not after, extends the larval growth period without affecting final organ or body size (Shingleton et al. 2005), because terminal growth is not affected. However, after CW, inactivation of insulin signaling reduces adult size without altering developmental timing. This observation suggests that reaching the CW checkpoint depends on insulin signaling, which is required for growth but not developmental progression after CW. Interestingly, starvation after CW attainment has not only been shown not to delay development but indeed to accelerate pupariation, thereby shortening the TGP in Drosophila (Stieper et al. 2008). This allows the animal to speed up development into adulthood under low-nutrition conditions that prevent further growth, once it has passed CW and therefore has sufficient energy to survive through the nonfeeding metamorphic stage. The underlying mechanism is not known, but it may be associated with a switch in energy allocation controlled by the CW checkpoint (Hironaka et al. 2019).
In Drosophila and other holometabolous insects, the growth of imaginal disc tissues is to some extent decoupled from nutrient intake. Although starvation after CW blocks the growth of larval tissues, imaginal discs continue their growth during post-CW starvation. These tissues grow rapidly after CW during the TGP, and indeed they continue to grow after the end of the feeding stage and the cessation of larval growth. The growth period of adult structures is therefore longer than the growth period for larval body growth. The growth strategy of disc tissues is also different from larval tissues, since mitotic tissues such as the brain and imaginal discs increase in size by proliferation, while polyploid larva-specific tissues grow by increasing their ploidy and cell size. In contrast to the growth of larva-specific tissues, which largely depends on systemic insulin signaling, imaginal tissues grow in an ecdysone-dependent manner (Mirth et al. 2009; Herboso et al. 2015; Dye et al. 2017). Although imaginal discs autonomously require insulin/PI3K signaling (Leevers et al. 1996; Britton and Edgar 1998; Brogiolo et al. 2001; Britton et al. 2002), their growth may be regulated by autocrine signaling through DILP2, which is ubiquitously expressed in imaginal discs and may protect their growth from nutrient-dependent variations in systemic insulin signaling. The nutrient-independent growth of disc tissues after CW may also be mediated by their dependence on ecdysone for growth. Attainment of CW may therefore be associated with a switch from energy storage by lipogenesis to utilization by lipolysis in response to starvation, which causes the mobilization of energy from the fat body to be directed toward the growth of adult precursor tissues. Consistent with this, starvation after CW is associated with decreased activity of Sterol regulatory element-binding protein (SREBP), a master regulator of lipogenesis (Xie et al. 2015). This may be related to the antagonistic effects of insulin and ecdysone signaling, discussed above. Starvation after CW leads to a precocious increase in ecdysone signaling (Lee et al. 2018), which explains why starvation after CW accelerates the pupariation of Drosophila larvae.
Almost simultaneously with CW in Drosophila, another size checkpoint called “minimum viable weight” (MVW) occurs, which is the size at which energy stores in the fat body are sufficient to permit survival through metamorphosis (Mirth and Riddiford 2007). MVW occurs shortly before CW and is distinguishable due to extreme developmental delay of metamorphosis in starved conditions, while post-CW larva undergo metamorphosis without any delay in these conditions. An alternative explanation for the MVW phenomenon is that MVW attainment may be the time of PG reactivation rather than acquisition of sufficient energy storage in the fat body, although it is not clear whether this PG inactivation/reactivation happens at the onset of the final larval instar in Drosophila (Xu et al. 2020).
The link between the attainment of CW and the activation of the neuroendocrine cascade which leads to increased circulating ecdysone levels – and ultimately to metamorphosis – was elucidated in the lepidopteran Manduca sexta (Nijhout and Williams 1974). In this insect, a drop in JH during the final larval instar is permissive for PTTH secretion at the following photoperiodic gate. This in turn induces ecdysone secretion, leading to the cessation of growth and the initiation of metamorphosis. In Drosophila, this link between CW and the neuroendocrine cascade has not been fully demonstrated. One difference seems to involve JH, which in Drosophila does not influence CW and has little effect on the timing of metamorphosis (Mirth et al. 2014). Although JH does not determine the growth-period duration in the fly, elimination of JH by ablation of the CA leads to a reduction in the larval growth rate mediated through elevation of FOXO activity, discussed above (Mirth et al. 2014). CA-ablated larvae also exhibit elevated ecdysone levels, which negatively affects the growth rate through the suppression of insulin signaling. Although the mechanism by which JH modulates insulin signaling is unclear, it may involve this increase in ecdysone levels. However, whether JH regulates PTTH in Drosophila as it does in Manduca is an interesting yet unresolved question.
Assessment of CW:
CW is likely based on assessment of nutritional status rather than actual body mass per se, at least in Drosophila. The apt questions here are: (1) in what tissues and by what means do animals sense their nutritional status, and (2) what are the signals from these tissues that initiate the neuroendocrine cascade leading to metamorphosis? Early studies indicated that the primary developmental timer that controls the timing of metamorphosis resides in the (nonimaginal) larval tissues, since removal of the imaginal discs does not affect CW or delay pupariation (Simpson et al. 1980; Poodry and Woods 1990; Stieper et al. 2008). This suggests that sensing CW involves an organ or organs other than the discs that sense nutritional status. As described above, the fat body senses its own nutritional status via TOR and releases a variety of humoral signals that regulate the IPCs, controlling growth and ecdysone production via the DILPs. Because the primary function of this nutrient-sensing fat-resident system is to couple nutrients with DILP signaling, it seems likely to underlie CW sensing, at least in part.
Several mechanisms are involved in CW assessment. A primary one of these is insulin-dependent ecdysone production, which drives a small nutrient-sensitive ecdysone peak early in the L3 stage that is believed to trigger the CW transition (Koyama et al. 2014). Insulin signaling resulting from feeding early in L3 relieves repression of ecdysone production in the PG by alleviating activity of the FOXO-Usp complex, which is activated transiently by the L2–L3 molt, during which feeding is blocked. These observations suggest that the effect of nutritional status on CW is mediated by insulin-regulated ecdysone synthesis in the PG. In this model, fat-body nutrient-sensing mechanisms convert nutritional cues into endocrine signals that activate the core maturation-inducing insulin and ecdysone systems. Similarly to CW and maturation in Drosophila, human weight and body fat mass correlate with the timing of menarche, which led to the use of the term CW for humans (Frisch and Revelle 1970, 1971; Ahmed et al. 2009). Indeed, childhood obesity as associated with early puberty (Kaplowitz 2008), suggesting a relationship between body fat and puberty timing. This implies that the neuroendocrine pathways initiating maturation in humans are linked to signals reflecting adiposity, similar to the TOR-dependent adipokines released from the Drosophila fat body that feed into the neuroendocrine system via insulin-mediated ecdysone synthesis. Consistently with this, the mammalian adipokine leptin, which is correlated with adiposity, is an important factor for the initiation of puberty (Farooqi 2002). One Drosophila analog of leptin is the nutrient-dependent adipokine Upd2, which is released from the fat body and regulates insulin secretion from the IPCs (Rajan and Perrimon 2012). Based on recent insights from Drosophila and the strong link between body fat and early puberty in humans, we speculate that Drosophila CW is related to larval body fat, rather than size or weight per se. In accord with this, a large portion of the body mass accumulated during the larval growth period is attributable to the fat body (Church and Robertson 1966), indicating that accumulation of body fat is an important factor for surviving metamorphosis and producing an adult of proper size and maximized fitness.
In addition to nutrient sensing via the fat body, the PG itself integrates nutritional signals, as described above. Animals with active insulin or TOR signaling in the PG reach CW earlier and at a smaller size (Mirth et al. 2005; Pan et al. 2019), suggesting that these pathways are part of the size-assessment mechanism that determines when CW has been attained. TOR-mediated regulation of endocycling in the PG also plays a role in the CW checkpoint (Ohhara et al. 2017). Activation of genome amplification by endoreduplication via TOR and the transcription factor Snail correlates with the attainment of CW. Since endoreduplication is an irreversible process that increases the ploidy of the PG cells, this mechanism translates nutritional cues into increased transcription that commits the PG to producing ecdysone at the CW transition. The importance of endoreduplication within the cells of the PG to the regulation of ecdysone is further supported by the finding that lysine demethylase 5 (Kdm5) is specifically required for endoreduplication in the PG through its transcriptional upregulation of torso (Drelon et al. 2019). TOR also regulates autophagy-mediated cholesterol trafficking that affects CW (Pan et al. 2019; Texada et al. 2019b). Larvae with genetically activated TOR in the PG pupariate without delay even when starved as early, pre-CW L3 larvae (2–4 hr after the L2–L3 transition), suggesting that activation of TOR in the PG mostly eliminates the CW checkpoint.
Like insulin/TOR-pathway activity, PTTH signaling is also involved in setting the CW checkpoint (McBrayer et al. 2007; Shimell et al. 2018). Loss of Ptth leads to a roughly 12-hr delay in the attainment of this checkpoint (i.e., Ptth mutants reach CW roughly 20 hr after the L2–L3 transition), which increases CW from 0.8 mg/larva in wild-types to 1.9 mg/larva in Ptth mutants. Furthermore, the TGP is prolonged from ∼35 hr in wild-types to ∼47 hr for Ptth mutants, showing that PTTH is also involved in determining the length of the TGP, similarly to TOR (Layalle et al. 2008). The PG also receives nutritional cues mediated by PTTH, which activates the Ras/Raf/MAP kinase (MAPK) pathway (Rewitz et al. 2009a,b; Galagovsky et al. 2018). The nuclear receptor DHR4 is a key target of PTTH/Torso/MAPK signaling in the PG and is believed to drive circadian oscillatory signaling in the PG that generates the temporally defined ecdysone pulses during the L3 stage (Ou et al. 2011, 2016; Rewitz and O’Connor 2011). DHR4 blocks ecdysone biosynthesis in the PG and is negatively regulated by PTTH signaling, which causes the translocation of DHR4 from the nucleus to the cytoplasm. DHR4 undergoes three rounds of nucleocytoplasmic shuttling in the PG during the L3 stage that coincide with the occurrence of three small ecdysone pulses prior the large ecdysone peak that triggers pupariation (Figure 6). Although the existence of these low-level ecdysone peaks has been challenged by findings indicating a stepwise increase in ecdysone during L3 (Lavrynenko et al. 2015), the small peak coinciding with CW has been detected in several independent studies (Warren et al. 2006; Ou et al. 2011; Koyama et al. 2014). Furthermore, the nucleocytoplasmic shuttling of DHR4 indicates oscillations in PTTH signaling coinciding with these low-level pulses, which are also supported indirectly by transcriptional changes in ecdysone-regulated genes during the early L3 stage (Andres et al. 1993). When DHR4 is disrupted in the PG, ecdysone levels rise prematurely after the L2–L3 transition, triggering accelerated development. Consistent with this, DHR4 mutants reach CW early at a smaller size (King-Jones et al. 2005), suggesting that in the PG, DHR4 plays an essential role in mediating PTTH responses to the attainment of CW. Although the exact interactions between the insulin/TOR and PTTH pathways are poorly understood, they all promote endoreduplication in the PG, which is believed to activate a transcriptional program at CW that commits the PG to synthesize ecdysone (Ohhara et al. 2017).
Common to these pathways is their effect on the rate of basal ecdysone synthesis, supporting the idea that CW corresponds to a threshold level of ecdysone that triggers nutrient-independent feedback activation of the metamorphosis-inducing neuroendocrine cascade. Insulin/TOR and PTTH signals convey different informational cues, and while it is clear that the information conveyed by the insulin/TOR pathway is nutritional in nature, it is less apparent what information PTTH might carry in pre-CW early-L3 animals. While PTTH communicates photoperiod information and imaginal disc developmental cues, recent work suggests that it carries nutritional information as well (Galagovsky et al. 2018). Furthermore, the PTTHn have extensive dendritic arbors, which suggests that they receive diverse inputs and therefore may represent an additional processing hub for the integration of extrinsic or intrinsic cues.
Although CW is not altered by nutrition, other environmental factors such as oxygen and temperature do have effects on CW (Callier et al. 2013; Ghosh et al. 2013), implying that they affect ecdysone production. As discussed in detail above, these environmental cues are sensed by central and peripheral mechanisms and integrated via the IPCs. Since insulin is involved in setting CW and also affects ecdysone production, this provides a mechanism by which environmental factors can modulate CW. Information about temperature and oxygen might also be integrated directly by the PG to affect CW. In other insects, reports indicate that ecdysone production is inhibited PG-autonomously and PTTH-independently by hypoxia (DeLalio et al. 2015) and lower temperatures mimicking the overwintering phase (Meola and Adkisson 1977). Another interesting possibility is that these factors may modulate CW through effects on PTTH release. Taken together, the mechanisms of CW assessment are complex, and although they have not been completely defined, they clearly depend on interplay between nutrient-sensing and neuroendocrine pathways.
Disc checkpoint:
Organ growth must be coordinated across the entire body and with developmental transitions to ensure that different organs have each attained an appropriate size before maturation can be initiated. In a broad range of insect species, the growth of juvenile appendages or imaginal discs (in hemi- and holometabolans, respectively) is tightly coupled with the timing of molting, including metamorphic molts (Hackney and Cherbas 2014). This coordination is ensured in Drosophila by a developmental checkpoint that monitors the growth of imaginal disc tissues. Early studies established that growing or damaged imaginal discs produce a factor that can delay pupariation. Growth-altering genetic perturbations to the discs, radiation-induced damage to these tissues, tumor-like abnormal growth, or transplantation of damaged discs delays the metamorphosis of Drosophila larvae, whereas complete X-ray-induced ablation of the discs does not (Russell 1974; Simpson and Scheinderman 1975; Simpson et al. 1980; Poodry and Woods 1990). These studies suggested that proliferating discs emit a humoral signal that inhibits ecdysone production, thus signaling local growth perturbations to the neuroendocrine system that control developmental timing (Bourgin et al. 1956; Stieper et al. 2008). This delay allows damaged or slow-growing discs the time to regenerate or to catch up to their appropriate size. Foundational studies also showed that discs transplanted from a developing larva into the abdomen of an adult would terminate growth at their normal size (Bryant and Levinson 1985), suggesting that their growth is governed in large part by organ-intrinsic mechanisms. However, perturbation of one disc in an animal slows the growth of the undamaged discs, thus maintaining appropriate proportionality between organs. This coordination of growth also occurs between different compartments within the same disc, suggesting that the undamaged compartments slow their growth while the injured part regenerates to retain tissue shape (Repiso et al. 2013). The disc-health checkpoint therefore delays developmental progression to coordinate growth between regenerating and intact tissues to maintain correct final proportions.
Disc damage induced during early larval life does not delay the first two molts, indicating that the developmental checkpoint for disc growth, like the CW checkpoint, only operates during the L3 stage (Halme et al. 2010). The ability of disc damage to delay pupariation depends on the timepoint within the L3 stage at which the injury is sustained. Tissue damage induced after the CW transition is still able to delay pupariation, suggesting that the disc checkpoint is distinct from the CW checkpoint. The disc checkpoint seems to coincide with the midthird-instar transition, a developmental time point after CW (Figure 6) that is associated with widespread gene-expression changes, including the activation of the Salivary gland secretion 3 (Sgs3) “glue” gene (Hackney et al. 2012). After a larva passes through this transition, tissue damage can no longer delay its onset of pupariation. Accordingly, the capacity of tissues to regenerate is correlated with this developmental or regenerative checkpoint and is lost ∼24 hr before pupariation (Halme et al. 2010). Like the nutritional checkpoint for CW, the disc checkpoint involves PTTH, the secretion of which is inhibited by regenerating discs, thus extending the larval growth period (Halme et al. 2010). The two checkpoints (CW and disc growth) therefore ensure the attainment of both sufficient nutrient storage and organ growth, based on assessment of internal and external cues by the neuroendocrine system, before the onset of maturation is permitted.
Coupling developmental timing to imaginal disc growth:
While disruption of retinoid biosynthesis reduces the delay in pupariation after disc damage, indicating that retinoids contribute to this mechanism (Halme et al. 2010), two Drosophila studies identified DILP8 as the signal released by damaged discs that is necessary and sufficient to induce developmental delay and for the growth coordination of distal tissues during regeneration (Colombani et al. 2012; Garelli et al. 2012). DILP8 is cell-autonomously expressed in response to disc growth perturbations, and loss of DILP8 rescues the developmental delay caused by disc overgrowth or damage, whereas overexpression of DILP8 is sufficient to delay pupariation without affecting disc integrity. The mechanism by which disc-derived DILP8 delays pupariation is the indirect blockage of ecdysone biosynthesis in the PG, acting via a neuronal relay through GCL neurons in the larval brain that inhibits PTTH secretion. These neurons express the DILP8 receptor Lgr3 and synapse upon the PTTHn and the IPCs, but not the PG (Colombani et al. 2015; Garelli et al. 2015; Vallejo et al. 2015; Jaszczak et al. 2016). Loss of Lgr3 in the GCLs is sufficient to prevent DILP8-induced developmental delay. Imaginal disc damage extends development by increasing the duration of L3, similar to loss of PTTH (McBrayer et al. 2007; Halme et al. 2010; Parker and Shingleton 2011; Hackney et al. 2012; Shimell et al. 2018), suggesting that PTTH signaling is the key target by which DILP8 acts to delay development. However, DILP8 overexpression has also been reported to reduce the growth rate (Garelli et al. 2012), which cannot be explained by effects of PTTH inhibition; reduced PTTH signaling limits ecdysone production, which, if anything, should increase the larval growth rate. This apparent paradox might be explained by the parallel inhibitory action of the GCL neurons on the IPCs (Vallejo et al. 2015), which may reduce secretion of growth-promoting DILPs from these cells.
In addition to slowing larval growth and prolonging larval development, disc aberrations also inhibit the growth of undamaged compartments within the same disc and of other imaginal tissues to maintain proportionality (Stieper et al. 2008; Parker and Shingleton 2011). DILP8 secretion by slow-growing discs is also necessary for this intra- and interorgan growth coordination, which depends on remote action of DILP8 via neuronal Lgr3 activity and is mediated by the systemic effects of ecdysone (Colombani et al. 2015; Vallejo et al. 2015), which promotes growth of imaginal discs (Colombani et al. 2005; Herboso et al. 2015; Dye et al. 2017; Moeller et al. 2017). Feeding with ecdysone prevents DILP8-mediated growth reduction in intact discs (Boulan et al. 2019), suggesting that DILP8 secreted from abnormally growing discs suppresses the growth of intact imaginal discs by limiting ecdysone signaling. Although the activation of Lgr3-expressing neurons in the brain mediates the growth coordination of undamaged discs, Lgr3 is also required in the PG itself for growth coordination—not for pupariation delay—during regeneration (Jaszczak et al. 2015, 2016).
The DILP8-Lgr3 signaling system was discovered and characterized based on tissue-damage responses and capacity for tissue regeneration, which has led to the general notion that it functions as a regeneration checkpoint. However, DILP8-Lgr3 signaling in normal development seems more likely to function as part of a surveillance system, conceptually analogous to cell cycle checkpoints, that regulates developmental progression. The role of this developmental checkpoint is to enable the neuroendocrine system to assess the growth status of the discs to determine whether the animal is ready to proceed with maturation. This ensures that disc tissues have completed enough development before progression and, at the same time, coordinates the size of all discs to ensure symmetry of the different body parts. Consistent with this view, lack of Dilp8 or Lgr3 leads to acceleration of pupariation and increases the frequency of asymmetric growth in paired organs (Garelli et al. 2012; Colombani et al. 2015; Garelli et al. 2015; Vallejo et al. 2015).
Induction of DILP8 in growing or damaged discs is mediated by several pathways in response to tissue stress and regeneration. Activation of the c-Jun N-terminal kinase (JNK) pathway in the discs is necessary in neoplastic growth conditions for DILP8 induction and pupariation delay, which also depends on cytokine Unpaired-1 (Upd1)-mediated activation of the JAK/STAT pathway in regenerating discs (Colombani et al. 2012; Katsuyama et al. 2015). In slow-growth conditions such as those caused by loss of ribosomal protein genes (Minute mutants), the stress-responsive transcription factor Xrp1 is required for remote nonautonomous growth inhibition of other discs by DILP8 (Boulan et al. 2019). During normal development, Yorkie and Scalloped, the transcriptional effectors of the Hippo pathway (described above), directly regulate DILP8 expression, coupling normal growth and DILP8 expression (Boone et al. 2016). This mechanism may contribute to organ size-sensing via changes in cytoskeletal strain and cell-to-cell contacts that arise because of disc cell growth and proliferation (Bosveld et al. 2012; Pan et al. 2016, 2018). This provides developmental stability by correcting minor stochastic disc growth variations. Thus, different types of tissue perturbation activate distinct pathways that converge on the regulation of DILP8 in the discs and, during normal development, DILP8-Lgr3 signaling fine-tunes developmental timing and adjusts tissue growth, promoting the development of individuals with appropriate body size, symmetry, and proportions. The mechanisms by which DILP8 links organ growth status to systemic growth responses rely on effects mediated by PTTH and ecdysone, the neuroendocrine system controlling developmental timing.
Allometry and scaling of organ growth and body size
Although the mechanisms described above ensure the proper size of adult structures and appropriate proportionality between them, these sizes and proportions also vary with environmental conditions. As overall body size increases, some organs may grow in strict proportion with it (“isometry”), whereas other organs may disproportionally increase or decrease in size (hyper- or hypoallometry). This variable size relationship, morphological allometry, reflects growth-regulation sensitivity that varies from organ to organ. These scaling phenomena are evident throughout the living world. For example, in dung beetles and rhinoceros beetles, nutrition-dependent signaling strongly affects organ scaling, and in these species, male cuticular “horns,” which are used in courtship battles, grow in a hyperallometric relationship with body size (Arrow 1951; Emlen 1994). Above a certain threshold body size, males develop disproportionally larger horns, whereas males below this size have, like females, very small horns (Emlen 1997a,b).
Similarly, some of the organs of Drosophila adults also show different scaling relationships to the overall body size in response to environmental conditions. Changes in nutrition affect the size of some organs like the wings and legs in isometric proportion to body size (Shingleton 2005; Shingleton et al. 2009). This proportional scaling is mainly mediated by nutrition-dependent insulin/TOR signaling. In contrast, other organs such as the central nervous system (CNS) and the genitalia are less sensitive to changes in nutrition and develop to an approximately similar size, irrespective of increased body size (Shingleton 2005; Cheng et al. 2011; Dreyer and Shingleton 2011; Tang et al. 2011). This allows tissues whose function is highly size-dependent to compensate for the effects of nutritional input (Shingleton 2010; Koyama et al. 2013). Interestingly, insulin/TOR signaling activities are modified in both of these organs to render them less sensitive to changes in nutrition, although this occurs through two distinct molecular mechanisms. The genital disc’s reduced nutritional sensitivity arises through modified intracellular insulin signaling, in which expression of the negative effector FOXO is reduced (Tang et al. 2011). This reduction makes the genital disc less sensitive to the low insulin/TOR activity that occurs under poor nutritional conditions. In fact, FOXO overexpression restores nutrition dependence to genital discs, which results in reduced genital size in adult males (Tang et al. 2011). The reduced nutritional plasticity of the genital discs likely ensures consistent, and thus morphologically compatible, genital structures under various environmental conditions, which is crucial for mating success.
The nutritional insensitivity of the CNS, commonly referred to as “brain sparing,” arises through a different insulin-modifying molecular mechanism. Glial cells supporting proliferating neuroblasts in the CNS constitutively secrete Jeb, which binds to its receptor Alk in neuroblasts, leading to activation of the PI3K pathway and inhibition of 4E-BP, thereby bypassing InR and TOR in poorly fed larvae (Cheng et al. 2011). Thus, the alteration of insulin/TOR activity seems to be a common adaptive mechanism to modify scaling relationships between organs. In addition to insulin/TOR, the TGF-β signaling pathway was recently shown to affect organ scaling. Mutations in the Activin gene disproportionally affect the growth of larval muscles compared to other tissues, leading to undersized adults (Moss-Taylor et al. 2019). Activin derived from motor neurons is locally delivered to muscles and is essential for proper tissue scaling and final body proportions.
Each environmental variable (temperature vs. nutrition, for example) can induce independent effects on allometric relationships. Although changes in nutritional conditions proportionally change both adult leg size and wing size, changes in temperature much more strongly affect wing size than leg size (Azevedo et al. 2002; Shingleton et al. 2009; McDonald et al. 2018). The molecular mechanisms underlying differential sensitivity to environmental stimuli are not clear, but temperature-driven plasticity is, at least partially, regulated in an organ-specific manner via the regulation of cell proliferation (Azevedo et al. 2002; Shingleton et al. 2009; McDonald et al. 2018). Because the molecular mechanism underlying temperature-dependent scaling appears to differ from the ones mediating nutrition-dependent proportionality, which include the insulin/TOR pathway, one might speculate that temperature-sensitive scaling relationships could involve distinct molecular pathways, such as the TGF-β signaling pathway.
Concluding Remarks
The ability of multicellular organisms to coordinate the growth of their individual organs and their whole body, and to terminate growth at the appropriate size, is essential for generating adults with body sizes and proportions that maximize fitness under varying environmental conditions. Species- and tissue-specific genetic frameworks specify broad body and organ growth parameters, which are adjusted through the integration of cues from the environment, such as temperature and the availability of oxygen and nutrients. The last decades of research on growth control in Drosophila have shown that two endocrine axes, the insulin and ecdysone signaling systems, determine adult size and body proportions by regulating the rate and duration of growth—that is, the timing of maturation—during the larval stages. Through these systems, growth and maturation are coordinated by key checkpoints that monitor nutritional and tissue-development status signals. These checkpoint mechanisms can extend the larval growth period by delaying metamorphosis until (1) nutritional stores are sufficient to ensure survival through the nonfeeding metamorphosis process, and (2) adult tissue primordia have developed symmetrically and sufficiently to produce animals with correct proportions.
These control mechanisms depend on interorgan communication mediated by signals reflecting external and internal conditions, and the integration of these signals by neuroendocrine hubs that control insulin and ecdysone signaling. The insulin- and steroid-signaling systems themselves are evolutionarily ancient, and recent studies suggest that the higher-order architecture of maturation-inducing signaling that acts via these systems is conserved between mammals and insects as well. This conservation makes Drosophila a prime system for understanding how environmental influences can regulate growth and body size. The physiological responses of Drosophila to nutritional changes mimic those observed in mammals, including humans, suggesting that studies of Drosophila are useful for understanding the mechanism by which nutritional cues systemically affect cell growth and proliferation, developmental progression, and body morphology.
Acknowledgments
Work in the Rewitz laboratory is supported by Danish Council for Independent Research (Det Frie Forskningsråd) Natural Sciences grant 8021-00055B, Lundbeck Foundation grant 2019-772, and Novo Nordisk Foundation grant 0054632 to K.R.
Footnotes
Communicating editor: C. Thummel
Literature Cited
- Acevedo-Rodriguez A., Kauffman A. S., Cherrington B. D., Borges C. S., Roepke T. A. et al. , 2018. Emerging insights into hypothalamic-pituitary-gonadal axis regulation and interaction with stress signalling. J. Neuroendocrinol. 30: e12590 10.1111/jne.12590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agbu P., Cassidy J. J., Braverman J., Jacobson A., and Carthew R. W., 2020. MicroRNA miR-7 regulates secretion of insulin-like peptides. Endocrinology 161: bqz040. 10.1210/endocr/bqz040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal N., Delanoue R., Mauri A., Basco D., Pasco M. et al. , 2016. The Drosophila TNF Eiger is an adipokine that acts on insulin-producing cells to mediate nutrient response. Cell Metab. 23: 675–684. 10.1016/j.cmet.2016.03.003 [DOI] [PubMed] [Google Scholar]
- Ahmed M. L., Ong K. K., and Dunger D. B., 2009. Childhood obesity and the timing of puberty. Trends Endocrinol. Metab. 20: 237–242. 10.1016/j.tem.2009.02.004 [DOI] [PubMed] [Google Scholar]
- Ajuria L., Nieva C., Winkler C., Kuo D., Samper N. et al. , 2011. Capicua DNA-binding sites are general response elements for RTK signaling in Drosophila. Development 138: 915–924. 10.1242/dev.057729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa R. W., Park S., Skelly K. R., Poffenberger G., Jain N. et al. , 2015. Suppression of insulin production and secretion by a decretin hormone. Cell Metab. 21: 323–334 [corrigenda: Cell Metab. 27: 479 (2018)]. 10.1016/j.cmet.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt J. R., Cleveland J. L., Hannink M., and Diehl J. A., 2000. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 14: 3102–3114. 10.1101/gad.854900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amcheslavsky A., Song W., Li Q., Nie Y., Bragatto I. et al. , 2014. Enteroendocrine cells support intestinal stem-cell-mediated homeostasis in Drosophila. Cell Rep. 9: 32–39. 10.1016/j.celrep.2014.08.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameku T., and Niwa R., 2016. Mating-induced increase in germline stem cells via the neuroendocrine system in female Drosophila. PLoS Genet. 12: e1006123 10.1371/journal.pgen.1006123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameku T., Yoshinari Y., Texada M. J., Kondo S., Amezawa K. et al. , 2018. Midgut-derived neuropeptide F controls germline stem cell proliferation in a mating-dependent manner. PLoS Biol. 16: e2005004 10.1371/journal.pbio.2005004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreatta G., Kyriacou C. P., Flatt T., and Costa R., 2018. Aminergic signaling controls ovarian dormancy in Drosophila. Sci. Rep. 8: 2030 10.1038/s41598-018-20407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres A. J., Fletcher J. C., Karim F. D., and Thummel C. S., 1993. Molecular analysis of the initiation of insect metamorphosis: a comparative study of Drosophila ecdysteroid-regulated transcription. Dev. Biol. 160: 388–404. 10.1006/dbio.1993.1315 [DOI] [PubMed] [Google Scholar]
- Armstrong J. L., Bonavaud S. M., Toole B. J., and Yeaman S. J., 2001. Regulation of glycogen synthesis by amino acids in cultured human muscle cells. J. Biol. Chem. 276: 952–956. 10.1074/jbc.M004812200 [DOI] [PubMed] [Google Scholar]
- Arquier N., Geminard C., Bourouis M., Jarretou G., Honegger B. et al. , 2008. Drosophila ALS regulates growth and metabolism through functional interaction with insulin-like peptides. Cell Metab. 7: 333–338. 10.1016/j.cmet.2008.02.003 [DOI] [PubMed] [Google Scholar]
- Arrow G. J., 1951. Horned Beetles, a Study of the Fantastic in Nature, W. Junk, The Hague: 10.1007/978-94-017-6178-9 [DOI] [Google Scholar]
- Averous J., Lambert-Langlais S., Mesclon F., Carraro V., Parry L. et al. , 2016. GCN2 contributes to mTORC1 inhibition by leucine deprivation through an ATF4 independent mechanism. Sci. Rep. 6: 27698 10.1038/srep27698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo R. B., French V., and Partridge L., 2002. Temperature modulates epidermal cell size in Drosophila melanogaster. J. Insect Physiol. 48: 231–237. 10.1016/S0022-1910(01)00168-8 [DOI] [PubMed] [Google Scholar]
- Bader R., Colomb J., Pankratz B., Schrock A., Stocker R. F. et al. , 2007. Genetic dissection of neural circuit anatomy underlying feeding behavior in Drosophila: distinct classes of hugin-expressing neurons. J. Comp. Neurol. 502: 848–856. 10.1002/cne.21342 [DOI] [PubMed] [Google Scholar]
- Badouel C., Gardano L., Amin N., Garg A., Rosenfeld R. et al. , 2009. The FERM-domain protein Expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell 16: 411–420. 10.1016/j.devcel.2009.01.010 [DOI] [PubMed] [Google Scholar]
- Bai X., Ma D., Liu A., Shen X., Wang Q. J. et al. , 2007. Rheb activates mTOR by antagonizing its endogenous inhibitor, FKBP38. Science 318: 977–980. 10.1126/science.1147379 [DOI] [PubMed] [Google Scholar]
- Bai H., Kang P., and Tatar M., 2012. Drosophila insulin-like peptide-6 (dilp6) expression from fat body extends lifespan and represses secretion of Drosophila insulin-like peptide-2 from the brain. Aging Cell 11: 978–985. 10.1111/acel.12000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai H., Kang P., Hernandez A. M., and Tatar M., 2013. Activin signaling targeted by insulin/dFOXO regulates aging and muscle proteostasis in Drosophila. PLoS Genet. 9: e1003941 10.1371/journal.pgen.1003941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banko J. L., Poulin F., Hou L., DeMaria C. T., Sonenberg N. et al. , 2005. The translation repressor 4E–BP2 is critical for eIF4F complex formation, synaptic plasticity, and memory in the hippocampus. J. Neurosci. 25: 9581–9590. 10.1523/JNEUROSCI.2423-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Schweitzer L. D., Zoncu R., and Sabatini D. M., 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150: 1196–1208. 10.1016/j.cell.2012.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A. D., Chen W. W., Ottina K. A. et al. , 2013. A tumor suppressor complex with GAP activity for the rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1106. 10.1126/science.1232044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry W. E., and Thummel C. S., 2016. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. Elife 5: e11183 10.7554/eLife.11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann A. A., Texada M. J., Chen H. M., Etheredge J. N., Miller D. L. et al. , 2017. Genetic tools to study juvenile hormone action in Drosophila. Sci. Rep. 7: 2132 10.1038/s41598-017-02264-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G., Tatum E., and Glancy C., 1938. Food level in relation to rate of development and eye pigmentation in Drosophila melanogaster. Biol. Bull. 75: 447–462. 10.2307/1537573 [DOI] [Google Scholar]
- Bejsovec A., 2018. Wingless signaling: a genetic journey from morphogenesis to metastasis. Genetics 208: 1311–1336. 10.1534/genetics.117.300157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal A., Schoenfeld R., Kleinhesselink K., and Kimbrell D. A., 2004. Loss of Thor, the single 4E-BP gene of Drosophila, does not result in lethality. D. I. S. 87: 81–84. [Google Scholar]
- Bialecki M., Shilton A., Fichtenberg C., Segraves W. A., and Thummel C. S., 2002. Loss of the ecdysteroid-inducible E75A orphan nuclear receptor uncouples molting from metamorphosis in Drosophila. Dev. Cell 3: 209–220. 10.1016/S1534-5807(02)00204-6 [DOI] [PubMed] [Google Scholar]
- Birse R. T., Soderberg J. A., Luo J., Winther A. M., and Nassel D. R., 2011. Regulation of insulin-producing cells in the adult Drosophila brain via the tachykinin peptide receptor DTKR. J. Exp. Biol. 214: 4201–4208. 10.1242/jeb.062091 [DOI] [PubMed] [Google Scholar]
- Bonfils G., Jaquenoud M., Bontron S., Ostrowicz C., Ungermann C. et al. , 2012. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 46: 105–110. 10.1016/j.molcel.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Boone E., Colombani J., Andersen D. S., and Leopold P., 2016. The Hippo signalling pathway coordinates organ growth and limits developmental variability by controlling dilp8 expression. Nat. Commun. 7: 13505 10.1038/ncomms13505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosveld F., Bonnet I., Guirao B., Tlili S., Wang Z. et al. , 2012. Mechanical control of morphogenesis by Fat/Dachsous/Four-jointed planar cell polarity pathway. Science 336: 724–727. 10.1126/science.1221071 [DOI] [PubMed] [Google Scholar]
- Boulan L., Martin D., and Milan M., 2013. Bantam miRNA promotes systemic growth by connecting insulin signaling and ecdysone production. Curr. Biol. 23: 473–478. 10.1016/j.cub.2013.01.072 [DOI] [PubMed] [Google Scholar]
- Boulan L., Milan M., and Leopold P., 2015. The systemic control of growth. Cold Spring Harb. Perspect. Biol. 7: a019117. 10.1101/cshperspect.a019117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulan L., Andersen D., Colombani J., Boone E. and Leopold P., 2019. Inter-organ growth coordination is mediated by the Xrp1-Dilp8 axis in Drosophila. Dev. Cell 49: 811–818.e4. 10.1016/j.devcel.2019.03.016 [DOI] [PubMed] [Google Scholar]
- Bourgin R. C., Krumins R., and Quastler H., 1956. Radiation-induced delay of pupation in Drosophila. Radiat. Res. 5: 657–673. 10.2307/3570585 [DOI] [PubMed] [Google Scholar]
- Bowser P. R., and Tobe S. S., 2005. Immunocytochemical analysis of putative allatostatin receptor (DAR-2) distribution in the CNS of larval Drosophila melanogaster. Peptides 26: 81–87. 10.1016/j.peptides.2004.08.026 [DOI] [PubMed] [Google Scholar]
- Braco J. T., Gillespie E. L., Alberto G. E., Brenman J. E., and Johnson E. C., 2012. Energy-dependent modulation of glucagon-like signaling in Drosophila via the AMP-activated protein kinase. Genetics 192: 457–466. 10.1534/genetics.112.143610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brankatschk M., Dunst S., Nemetschke L., and Eaton S., 2014. Delivery of circulating lipoproteins to specific neurons in the Drosophila brain regulates systemic insulin signaling. Elife 3: e02862 10.7554/eLife.02862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitman T. R., Selonick S. E., and Collins S. J., 1980. Induction of differentiation of the human promyelocytic leukemia cell line (HL-60) by retinoic acid. Proc. Natl. Acad. Sci. USA 77: 2936–2940. 10.1073/pnas.77.5.2936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C. A., Ashburner M., and Moses K., 1998. Ecdysone pathway is required for furrow progression in the developing Drosophila eye. Development 125: 2653–2664. [DOI] [PubMed] [Google Scholar]
- Brennecke J., Hipfner D. R., Stark A., Russell R. B., and Cohen S. M., 2003. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113: 25–36. 10.1016/S0092-8674(03)00231-9 [DOI] [PubMed] [Google Scholar]
- Briscoe J., and Therond P. P., 2013. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol. 14: 416–429. 10.1038/nrm3598 [DOI] [PubMed] [Google Scholar]
- Britton J. S., and Edgar B. A., 1998. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development 125: 2149–2158. [DOI] [PubMed] [Google Scholar]
- Britton J. S., Lockwood W. K., Li L., Cohen S. M., and Edgar B. A., 2002. Drosophila’s insulin/PI3-kinase pathway coordinates cellular metabolism with nutritional conditions. Dev. Cell 2: 239–249. 10.1016/S1534-5807(02)00117-X [DOI] [PubMed] [Google Scholar]
- Brogiolo W., Stocker H., Ikeya T., Rintelen F., Fernandez R. et al. , 2001. An evolutionarily conserved function of the Drosophila insulin receptor and insulin-like peptides in growth control. Curr. Biol. 11: 213–221. 10.1016/S0960-9822(01)00068-9 [DOI] [PubMed] [Google Scholar]
- Broughton S., Alic N., Slack C., Bass T., Ikeya T. et al. , 2008. Reduction of DILP2 in Drosophila triages a metabolic phenotype from lifespan revealing redundancy and compensation among DILPs. PLoS One 3: e3721 10.1371/journal.pone.0003721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown E. J., Beal P. A., Keith C. T., Chen J., Shin T. B. et al. , 1995. Control of p70 s6 kinase by kinase activity of FRAP in vivo. Nature 377: 441–446 (erratum: Nature 378: 644). 10.1038/377441a0 [DOI] [PubMed] [Google Scholar]
- Bryant P. J., and Levinson P., 1985. Intrinsic growth control in the imaginal primordia of Drosophila, and the autonomous action of a lethal mutation causing overgrowth. Dev. Biol. 107: 355–363. 10.1016/0012-1606(85)90317-3 [DOI] [PubMed] [Google Scholar]
- Buerger C., DeVries B., and Stambolic V., 2006. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem. Biophys. Res. Commun. 344: 869–880. 10.1016/j.bbrc.2006.03.220 [DOI] [PubMed] [Google Scholar]
- Buhler K., Clements J., Winant M., Bolckmans L., Vulsteke V. et al. , 2018. Growth control through regulation of insulin signalling by nutrition-activated steroid hormone in Drosophila. Development 145: dev165654. 10.1242/dev.165654 [DOI] [PubMed] [Google Scholar]
- Bujold M., Gopalakrishnan A., Nally E., and King-Jones K., 2010. Nuclear receptor DHR96 acts as a sentinel for low cholesterol concentrations in Drosophila melanogaster. Mol. Cell. Biol. 30: 793–805. 10.1128/MCB.01327-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres L., Necakov A. S., Schwartz C., Kimber S., Roberts I. J. et al. , 2011. Nitric oxide coordinates metabolism, growth, and development via the nuclear receptor E75. Genes Dev. 25: 1476–1485. 10.1101/gad.2064111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell P. E., Walkiewicz M., and Stern M., 2005. Ras activity in the Drosophila prothoracic gland regulates body size and developmental rate via ecdysone release. Curr. Biol. 15: 1785–1795. 10.1016/j.cub.2005.09.011 [DOI] [PubMed] [Google Scholar]
- Callier V., and Nijhout H. F., 2011. Control of body size by oxygen supply reveals size-dependent and size-independent mechanisms of molting and metamorphosis. Proc. Natl. Acad. Sci. USA 108: 14664–14669. 10.1073/pnas.1106556108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callier V., and Nijhout H. F., 2013. Body size determination in insects: a review and synthesis of size- and brain-dependent and independent mechanisms. Biol. Rev. Camb. Philos. Soc. 88: 944–954. 10.1111/brv.12033 [DOI] [PubMed] [Google Scholar]
- Callier V., Shingleton A. W., Brent C. S., Ghosh S. M., Kim J. et al. , 2013. The role of reduced oxygen in the developmental physiology of growth and metamorphosis initiation in Drosophila melanogaster. J. Exp. Biol. 216: 4334–4340. 10.1242/jeb.093120 [DOI] [PubMed] [Google Scholar]
- Carlsson M. A., Enell L. E., and Nassel D. R., 2013. Distribution of short neuropeptide F and its receptor in neuronal circuits related to feeding in larval Drosophila. Cell Tissue Res. 353: 511–523. 10.1007/s00441-013-1660-4 [DOI] [PubMed] [Google Scholar]
- Carvalho M., Schwudke D., Sampaio J. L., Palm W., Riezman I. et al. , 2010. Survival strategies of a sterol auxotroph. Development 137: 3675–3685. 10.1242/dev.044560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano B. M., Thelen A. M., Moldavski O., Feltes M., van der Welle R. E. et al. , 2017. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355: 1306–1311. 10.1126/science.aag1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L., Wolfson R. L., Orozco J. M., Saxton R. A., Scaria S. M. et al. , 2014. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 9: 1–8. 10.1016/j.celrep.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L., Scaria S. M., Saxton R. A., Gygi M. P., Shen K. et al. , 2016. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165: 153–164. 10.1016/j.cell.2016.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez V. M., Marques G., Delbecque J. P., Kobayashi K., Hollingsworth M. et al. , 2000. The Drosophila disembodied gene controls late embryonic morphogenesis and codes for a cytochrome P450 enzyme that regulates embryonic ecdysone levels. Development 127: 4115–4126. [DOI] [PubMed] [Google Scholar]
- Chen C., Jack J., and Garofalo R. S., 1996. The Drosophila insulin receptor is required for normal growth. Endocrinology 137: 846–856. 10.1210/endo.137.3.8603594 [DOI] [PubMed] [Google Scholar]
- Cheng L. Y., Bailey A. P., Leevers S. J., Ragan T. J., Driscoll P. C. et al. , 2011. Anaplastic lymphoma kinase spare organ growth during nutrient restriction in Drosophila. Cell 146: 435–447. 10.1016/j.cell.2011.06.040 [DOI] [PubMed] [Google Scholar]
- Cheng Q., Beltran V. D., Chan S. M., Brown J. R., Bevington A. et al. , 2016. System-L amino acid transporters play a key role in pancreatic beta-cell signalling and function. J. Mol. Endocrinol. 56: 175–187. 10.1530/JME-15-0212 [DOI] [PubMed] [Google Scholar]
- Cho K. S., Lee J. H., Kim S., Kim D., Koh H. et al. , 2001. Drosophila phosphoinositide-dependent kinase-1 regulates apoptosis and growth via the phosphoinositide 3-kinase-dependent signaling pathway. Proc. Natl. Acad. Sci. USA 98: 6144–6149. 10.1073/pnas.101596998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E., Feng Y., Rauskolb C., Maitra S., Fehon R. et al. , 2006. Delineation of a Fat tumor suppressor pathway. Nat. Genet. 38: 1142–1150. 10.1038/ng1887 [DOI] [PubMed] [Google Scholar]
- Choi H., Son J. B., Kang J., Kwon J., Kim J. H. et al. , 2017. Leucine-induced localization of Leucyl-tRNA synthetase in lysosome membrane. Biochem. Biophys. Res. Commun. 493: 1129–1135. 10.1016/j.bbrc.2017.09.008 [DOI] [PubMed] [Google Scholar]
- Christensen, C. F., T. Koyama, S. Nagy, E. T. Danielsen, M. J. Texada et al., Ecdysone-dependent feedback regulation of prothoracicotropic hormone times developmental maturation. Development 2020: dev.188110. DOI: 10.1242/dev.188110. [DOI] [PMC free article] [PubMed]
- Church R. B., and Robertson F. W., 1966. A biochemical study of the growth of Drosophila melanogaster. J. Exp. Zool. 12: 852–858. [Google Scholar]
- Cieśla M., Towpik J., Graczyk D., Oficjalska-Pham D., Harismendy O. et al. , 2007. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol. Cell. Biol. 27: 7693–7702. 10.1128/MCB.01051-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements J., Hens K., Francis C., Schellens A., and Callaerts P., 2008. Conserved role for the Drosophila Pax6 homolog Eyeless in differentiation and function of insulin-producing neurons. Proc. Natl. Acad. Sci. USA 105: 16183–16188. 10.1073/pnas.0708330105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby W. W., Chen E. Y., Smith D. H., and Levinson A. D., 1983. Identification and nucleotide sequence of a human locus homologous to the v-myc oncogene of avian myelocytomatosis virus MC29. Nature 301: 722–725. 10.1038/301722a0 [DOI] [PubMed] [Google Scholar]
- Colombani J., and Andersen D. S., 2020. The Drosophila gut: a gatekeeper and coordinator of organism fitness and physiology. Wiley Interdiscip. Rev. Dev. Biol. DOI: 10.1002/wdev.378. 10.1002/wdev.378 [DOI] [PubMed] [Google Scholar]
- Colombani J., Raisin S., Pantalacci S., Radimerski T., Montagne J. et al. , 2003. A nutrient sensor mechanism controls Drosophila growth. Cell 114: 739–749. 10.1016/S0092-8674(03)00713-X [DOI] [PubMed] [Google Scholar]
- Colombani J., Bianchini L., Layalle S., Pondeville E., Dauphin-Villemant C. et al. , 2005. Antagonistic actions of ecdysone and insulins determine final size in Drosophila. Science 310: 667–670. 10.1126/science.1119432 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., and Leopold P., 2012. Secreted peptide Dilp8 coordinates Drosophila tissue growth with developmental timing. Science 336: 582–585. 10.1126/science.1216689 [DOI] [PubMed] [Google Scholar]
- Colombani J., Andersen D. S., Boulan L., Boone E., Romero N. et al. , 2015. Drosophila Lgr3 couples organ growth with maturation and ensures developmental stability. Curr. Biol. 25: 2723–2729. 10.1016/j.cub.2015.09.020 [DOI] [PubMed] [Google Scholar]
- Crocker A., Shahidullah M., Levitan I. B., and Sehgal A., 2010. Identification of a neural circuit that underlies the effects of octopamine on sleep:wake behavior. Neuron 65: 670–681. 10.1016/j.neuron.2010.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz J., Martin D., and Franch-Marro X., 2020. Egfr signaling is a major regulator of ecdysone biosynthesis in the Drosophila prothoracic gland. Curr. Biol. 30: 1547–1554.e4. 10.1016/j.cub.2020.01.092 [DOI] [PubMed] [Google Scholar]
- Danielsen E. T., and Rewitz K. F., 2016. Developmental biology: when less damage causes more harm. Curr. Biol. 26: R855–R858. 10.1016/j.cub.2016.07.068 [DOI] [PubMed] [Google Scholar]
- Danielsen E. T., Moeller M. E., Dorry E., Komura-Kawa T., Fujimoto Y. et al. , 2014. Transcriptional control of steroid biosynthesis genes in the Drosophila prothoracic gland by ventral veins lacking and knirps. PLoS Genet. 10: e1004343 10.1371/journal.pgen.1004343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielsen E. T., Moeller M. E., Yamanaka N., Ou Q., Laursen J. M. et al. , 2016. A Drosophila genome-wide screen identifies regulators of steroid hormone production and developmental timing. Dev. Cell 37: 558–570. 10.1016/j.devcel.2016.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Morrison G., Xu Z., Rui Y. N., Charng W. L., Jaiswal M. et al. , 2016. WAC regulates mTOR activity by acting as an adaptor for the TTT and pontin/reptin complexes. Dev. Cell 36: 139–151. 10.1016/j.devcel.2015.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLalio L. J., Dion S. M., Bootes A. M., and Smith W. A., 2015. Direct effects of hypoxia and nitric oxide on ecdysone secretion by insect prothoracic glands. J. Insect Physiol. 76: 56–66. 10.1016/j.jinsphys.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Delanoue R., Slaidina M., and Leopold P., 2010. The steroid hormone ecdysone controls systemic growth by repressing dMyc function in Drosophila fat cells. Dev. Cell 18: 1012–1021. 10.1016/j.devcel.2010.05.007 [DOI] [PubMed] [Google Scholar]
- Delanoue R., Meschi E., Agrawal N., Mauri A., Tsatskis Y. et al. , 2016. Drosophila insulin release is triggered by adipose Stunted ligand to brain Methuselah receptor. Science 353: 1553–1556. 10.1126/science.aaf8430 [DOI] [PubMed] [Google Scholar]
- Demetriades C., Doumpas N., and Teleman A. A., 2014. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156: 786–799. 10.1016/j.cell.2014.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demetriades C., Plescher M., and Teleman A. A., 2016. Lysosomal recruitment of TSC2 is a universal response to cellular stress. Nat. Commun. 7: 10662 10.1038/ncomms10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., and Perrimon N., 2009. Integration of insulin receptor/Foxo signaling and dMyc activity during muscle growth regulates body size in Drosophila. Development 136: 983–993. 10.1242/dev.027466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demontis F., and Perrimon N., 2010. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell 143: 813–825. 10.1016/j.cell.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveci D., Martin F. A., Leopold P., and Romero N. M., 2019. AstA signaling functions as an evolutionary conserved mechanism timing juvenile to adult transition. Curr. Biol. 29: 813–822.e4. 10.1016/j.cub.2019.01.053 [DOI] [PubMed] [Google Scholar]
- Dibble C. C., Elis W., Menon S., Qin W., Klekota J. et al. , 2012. TBC1D7 is a third subunit of the TSC1–TSC2 complex upstream of mTORC1. Mol. Cell 47: 535–546. 10.1016/j.molcel.2012.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cara F., and King-Jones K., 2016. The circadian clock is a key driver of steroid hormone production in Drosophila. Curr. Biol. 26: 2469–2477. 10.1016/j.cub.2016.07.004 [DOI] [PubMed] [Google Scholar]
- Diehl J. A., Cheng M., Roussel M. F., and Sherr C. J., 1998. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12: 3499–3511. 10.1101/gad.12.22.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., and Pan D., 2004. Tsc2 is not a critical target of Akt during normal Drosophila development. Genes Dev. 18: 2479–2484. 10.1101/gad.1240504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N. et al. , 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Yang B., Li Y., Zhong C., and Ding J., 2009. Molecular basis of the acceleration of the GDP-GTP exchange of human ras homolog enriched in brain by human translationally controlled tumor protein. J. Biol. Chem. 284: 23754–23764. 10.1074/jbc.M109.012823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drelon C., Rogers M. F., Belalcazar H. M., and Secombe J., 2019. The histone demethylase KDM5 controls developmental timing in Drosophila by promoting prothoracic gland endocycles. Development 146: dev182568. 10.1242/dev.182568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer A. P., and Shingleton A. W., 2011. The effect of genetic and environmental variation on genital size in male Drosophila: canalized but developmentally unstable. PLoS One 6: e28278 10.1371/journal.pone.0028278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye N. A., Popovic M., Spannl S., Etournay R., Kainmuller D. et al. , 2017. Cell dynamics underlying oriented growth of the Drosophila wing imaginal disc. Development 144: 4406–4421. 10.1242/dev.155069 [DOI] [PubMed] [Google Scholar]
- Ellisen L. W., 2005. Growth control under stress: mTOR regulation through the REDD1-TSC pathway. Cell Cycle 4: 1500–1502. 10.4161/cc.4.11.2139 [DOI] [PubMed] [Google Scholar]
- Emlen D. J., 1994. Environmental control of horn length dimorphism in the beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae). Proc. Biol. Sci. 256: 131–136. 10.1098/rspb.1994.0060 [DOI] [Google Scholar]
- Emlen D. J., 1997a Alternative reproductive tactics and male dimorphism in the horned beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae). Behav. Ecol. Sociobiol. 41: 335–341. 10.1007/s002650050393 [DOI] [Google Scholar]
- Emlen D. J., 1997b Diet alters male horn allometry in the beetle Onthophagus acuminatus (Coleoptera: Scarabaeidae). Proc. Biol. Sci. 264: 567–574. 10.1098/rspb.1997.0081 [DOI] [Google Scholar]
- Enell L. E., Kapan N., Soderberg J. A., Kahsai L., and Nassel D. R., 2010. Insulin signaling, lifespan and stress resistance are modulated by metabotropic GABA receptors on insulin producing cells in the brain of Drosophila. PLoS One 5: e15780 10.1371/journal.pone.0015780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enya S., Ameku T., Igarashi F., Iga M., Kataoka H. et al. , 2014. A halloween gene noppera-bo encodes a glutathione S-transferase essential for ecdysteroid biosynthesis via regulating the behaviour of cholesterol in Drosophila. Sci. Rep. 4: 6586 10.1038/srep06586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erion R., DiAngelo J. R., Crocker A., and Sehgal A., 2012. Interaction between sleep and metabolism in Drosophila with altered octopamine signaling. J. Biol. Chem. 287: 32406–32414. 10.1074/jbc.M112.360875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Chromosome 16 Tuberous Sclerosis Consortium , 1993. Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell 75: 1305–1315. 10.1016/0092-8674(93)90618-Z [DOI] [PubMed] [Google Scholar]
- Fahien L. A., and Macdonald M. J., 2011. The complex mechanism of glutamate dehydrogenase in insulin secretion. Diabetes 60: 2450–2454. 10.2337/db10-1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y., Vilella-Bach M., Bachmann R., Flanigan A., and Chen J., 2001. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science 294: 1942–1945. 10.1126/science.1066015 [DOI] [PubMed] [Google Scholar]
- Fang Y., Park I. H., Wu A. L., Du G., Huang P. et al. , 2003. PLD1 regulates mTOR signaling and mediates Cdc42 activation of S6K1. Curr. Biol. 13: 2037–2044. 10.1016/j.cub.2003.11.021 [DOI] [PubMed] [Google Scholar]
- Farooqi I. S., 2002. Leptin and the onset of puberty: insights from rodent and human genetics. Semin. Reprod. Med. 20: 139–144. 10.1055/s-2002-32505 [DOI] [PubMed] [Google Scholar]
- Felix R. C., Trindade M., Pires I. R., Fonseca V. G., Martins R. S. et al. , 2015. Unravelling the evolution of the allatostatin-type A, KISS and galanin peptide-receptor gene families in bilaterians: insights from Anopheles mosquitoes. PLoS One 10: e0130347 10.1371/journal.pone.0130347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez R., Tabarini D., Azpiazu N., Frasch M., and Schlessinger J., 1995. The Drosophila insulin receptor homolog: a gene essential for embryonic development encodes two receptor isoforms with different signaling potential. EMBO J. 14: 3373–3384. 10.1002/j.1460-2075.1995.tb07343.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis V. A., Zorzano A., and Teleman A. A., 2010. dDOR is an EcR coactivator that forms a feed-forward loop connecting insulin and ecdysone signaling. Curr. Biol. 20: 1799–1808. 10.1016/j.cub.2010.08.055 [DOI] [PubMed] [Google Scholar]
- Frazier M. R., Woods H. A., and Harrison J. F., 2001. Interactive effects of rearing temperature and oxygen on the development of Drosophila melanogaster. Physiol. Biochem. Zool. 74: 641–650. 10.1086/322172 [DOI] [PubMed] [Google Scholar]
- French V., Feast M., and Partridge L., 1998. Body size and cell size in Drosophila: the developmental response to temperature. J. Insect Physiol. 44: 1081–1089. 10.1016/S0022-1910(98)00061-4 [DOI] [PubMed] [Google Scholar]
- Frisch R. E., and Revelle R., 1970. Height and weight at menarche and a hypothesis of critical body weights and adolescent events. Science 169: 397–399. 10.1126/science.169.3943.397 [DOI] [PubMed] [Google Scholar]
- Frisch R. E., and Revelle R., 1971. Height and weight at menarche and a hypothesis of menarche. Arch. Dis. Child. 46: 695–701. 10.1136/adc.46.249.695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulford A., Tapon N., and Ribeiro P. S., 2018. Upstairs, downstairs: spatial regulation of Hippo signalling. Curr. Opin. Cell Biol. 51: 22–32. 10.1016/j.ceb.2017.10.006 [DOI] [PubMed] [Google Scholar]
- Galagovsky D., Depetris-Chauvin A., Manière G., Geillon F., Berthelot-Grosjean M. et al. , 2018. Sobremesa L-type amino acid transporter expressed in glia is essential for proper timing of development and brain growth. Cell Rep. 24: 3156–3166.e4. 10.1016/j.celrep.2018.08.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P., 2013. Myc function in Drosophila. Cold Spring Harb. Perspect. Med. 3: a014324 10.1101/cshperspect.a014324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallant P., Shiio Y., Cheng P. F., Parkhurst S. M., and Eisenman R. N., 1996. Myc and Max homologs in Drosophila. Science 274: 1523–1527. 10.1126/science.274.5292.1523 [DOI] [PubMed] [Google Scholar]
- Ganley I. G., Lam D. H., Wang J., Ding X., Chen S., and Jiang X., 2009. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284: 12297–12305. 10.1074/jbc.M900573200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., and Pan D., 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15: 1383–1392. 10.1101/gad.901101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Neufeld T. P., and Pan D., 2000. Drosophila PTEN regulates cell growth and proliferation through PI3K-dependent and -independent pathways. Dev. Biol. 221: 404–418. 10.1006/dbio.2000.9680 [DOI] [PubMed] [Google Scholar]
- Gao X., Zhang Y., Arrazola P., Hino O., Kobayashi T. et al. , 2002. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nat. Cell Biol. 4: 699–704. 10.1038/ncb847 [DOI] [PubMed] [Google Scholar]
- Gao Z., Young R. A., Li G., Najafi H., Buettger C. et al. , 2003. Distinguishing features of leucine and alpha-ketoisocaproate sensing in pancreatic beta-cells. Endocrinology 144: 1949–1957. 10.1210/en.2002-0072 [DOI] [PubMed] [Google Scholar]
- Garami A., Zwartkruis F. J., Nobukuni T., Joaquin M., Roccio M. et al. , 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11: 1457–1466. 10.1016/S1097-2765(03)00220-X [DOI] [PubMed] [Google Scholar]
- Garelli A., Gontijo A. M., Miguela V., Caparros E., and Dominguez M., 2012. Imaginal discs secrete insulin-like peptide 8 to mediate plasticity of growth and maturation. Science 336: 579–582. 10.1126/science.1216735 [DOI] [PubMed] [Google Scholar]
- Garelli A., Heredia F., Casimiro A. P., Macedo A., Nunes C. et al. , 2015. Dilp8 requires the neuronal relaxin receptor Lgr3 to couple growth to developmental timing. Nat. Commun. 6: 8732 10.1038/ncomms9732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géminard C., Rulifson E. J., and Léopold P., 2009. Remote control of insulin secretion by fat cells in Drosophila. Cell Metab. 10: 199–207. 10.1016/j.cmet.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Genevet A., Wehr M. C., Brain R., Thompson B. J., and Tapon N., 2010. Kibra is a regulator of the Salvador/Warts/Hippo signaling network. Dev. Cell 18: 300–308. 10.1016/j.devcel.2009.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach S. U., Sander M., Song S., and Herranz H., 2019. The miRNA bantam regulates growth and tumorigenesis by repressing the cell cycle regulator tribbles. Life Sci. Alliance 2: e201900381. 10.26508/lsa.201900381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A. C., and O’Connor M. B., 2014. Systemic Activin signaling independently regulates sugar homeostasis, cellular metabolism, and pH balance in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 111: 5729–5734. 10.1073/pnas.1319116111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., McBrayer Z., and O’Connor M. B., 2010. The Drosophila gap gene giant regulates ecdysone production through specification of the PTTH-producing neurons. Dev. Biol. 347: 271–278. 10.1016/j.ydbio.2010.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. M., Testa N. D., and Shingleton A. W., 2013. Temperature-size rule is mediated by thermal plasticity of critical size in Drosophila melanogaster. Proc. Biol. Sci. 280: 20130174 10.1098/rspb.2013.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A., Rideout E. J., and Grewal S. S., 2014. TIF-IA-dependent regulation of ribosome synthesis in drosophila muscle is required to maintain systemic insulin signaling and larval growth. PLoS Genet. 10: e1004750 10.1371/journal.pgen.1004750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbens Y. Y., Warren J. T., Gilbert L. I., and O’Connor M. B., 2011. Neuroendocrine regulation of Drosophila metamorphosis requires TGFbeta/Activin signaling. Development 138: 2693–2703. 10.1242/dev.063412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert L. I., Rybczynski R., and Warren J. T., 2002. Control and biochemical nature of the ecdysteroidogenic pathway. Annu. Rev. Entomol. 47: 883–916. 10.1146/annurev.ento.47.091201.145302 [DOI] [PubMed] [Google Scholar]
- Goberdhan D. C., Paricio N., Goodman E. C., Mlodzik M., and Wilson C., 1999. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev. 13: 3244–3258. 10.1101/gad.13.24.3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Liu J., Guo C., Zhou Y., Teng Y. et al. , 2010. Two pairs of neurons in the central brain control Drosophila innate light preference. Science 330: 499–502. 10.1126/science.1195993 [DOI] [PubMed] [Google Scholar]
- Gong C., Ouyang Z., Zhao W., Wang J., Li K. et al. , 2019. A neuronal pathway that commands deceleration in Drosophila larval light-avoidance. Neurosci. Bull. 35: 959–968. 10.1007/s12264-019-00349-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulev Y., Fauny J. D., Gonzalez-Marti B., Flagiello D., Silber J. et al. , 2008. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr. Biol. 18: 435–441. 10.1016/j.cub.2008.02.034 [DOI] [PubMed] [Google Scholar]
- Graves B. J., and Schubiger G., 1982. Cell cycle changes during growth and differentiation of imaginal leg discs in Drosophila melanogaster. Dev. Biol. 93: 104–110. 10.1016/0012-1606(82)90243-3 [DOI] [PubMed] [Google Scholar]
- Grewal S. S., Li L., Orian A., Eisenman R. N., and Edgar B. A., 2005. Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat. Cell Biol. 7: 295–302. 10.1038/ncb1223 [DOI] [PubMed] [Google Scholar]
- Grewal S. S., Evans J. R., and Edgar B. A., 2007. Drosophila TIF-IA is required for ribosome synthesis and cell growth and is regulated by the TOR pathway. J. Cell Biol. 179: 1105–1113. 10.1083/jcb.200709044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönke S., Clarke D.-F., Broughton S., Andrews T. D., and Partridge L., 2010. Molecular evolution and functional characterization of Drosophila insulin-like peptides. PLoS Genet. 6: e1000857 10.1371/journal.pgen.1000857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin D. A., Guntur K. V., Bell G. W., Thoreen C. C., and Sabatini D. M., 2006. Functional genomics identifies TOR-regulated genes that control growth and division. Curr. Biol. 16: 958–970. 10.1016/j.cub.2006.03.084 [DOI] [PubMed] [Google Scholar]
- Guo T., Lu Y., Li P., Yin M. X., Lv D. et al. , 2013. A novel partner of Scalloped regulates Hippo signaling via antagonizing Scalloped-Yorkie activity. Cell Res. 23: 1201–1214. 10.1038/cr.2013.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Flegel K., Kumar J., McKay D. J., and Buttitta L. A., 2016. Ecdysone signaling induces two phases of cell cycle exit in Drosophila cells. Biol. Open 5: 1648–1661. 10.1242/bio.017525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X., Orozco J. M., Saxton R. A., Condon K. J., Liu G. Y. et al. , 2017. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358: 813–818. 10.1126/science.aao3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney J. F., and Cherbas P., 2014. Injury response checkpoint and developmental timing in insects. Fly (Austin) 8: 226–231. 10.1080/19336934.2015.1034913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney J. F., Zolali-Meybodi O., and Cherbas P., 2012. Tissue damage disrupts developmental progression and ecdysteroid biosynthesis in Drosophila. PLoS One 7: e49105 10.1371/journal.pone.0049105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halme A., Cheng M., and Hariharan I. K., 2010. Retinoids regulate a developmental checkpoint for tissue regeneration in Drosophila. Curr. Biol. 20: 458–463. 10.1016/j.cub.2010.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaratoglu F., Willecke M., Kango-Singh M., Nolo R., Hyun E. et al. , 2006. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat. Cell Biol. 8: 27–36 (erratum: Nat. Cell Biol. 8: 100). 10.1038/ncb1339 [DOI] [PubMed] [Google Scholar]
- Hamaratoglu F., Affolter M., and Pyrowolakis G., 2014. Dpp/BMP signaling in flies: from molecules to biology. Semin. Cell Dev. Biol. 32: 128–136. 10.1016/j.semcdb.2014.04.036 [DOI] [PubMed] [Google Scholar]
- Han J. M., Jeong S. J., Park M. C., Kim G., Kwon N. H. et al. , 2012. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 149: 410–424. 10.1016/j.cell.2012.02.044 [DOI] [PubMed] [Google Scholar]
- Hara K., Maruki Y., Long X., Yoshino K., Oshiro N. et al. , 2002. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110: 177–189. 10.1016/S0092-8674(02)00833-4 [DOI] [PubMed] [Google Scholar]
- Harrison J. F., Shingleton A. W., and Callier V., 2015. Stunted by developing in hypoxia: linking comparative and model organism studies. Physiol. Biochem. Zool. 88: 455–470. 10.1086/682216 [DOI] [PubMed] [Google Scholar]
- Hauser F., and Grimmelikhuijzen C. J., 2014. Evolution of the AKH/corazonin/ACP/GnRH receptor superfamily and their ligands in the Protostomia. Gen. Comp. Endocrinol. 209: 35–49. 10.1016/j.ygcen.2014.07.009 [DOI] [PubMed] [Google Scholar]
- Heesom K. J., and Denton R. M., 1999. Dissociation of the eukaryotic initiation factor-4E/4E–BP1 complex involves phosphorylation of 4E–BP1 by an mTOR-associated kinase. FEBS Lett. 457: 489–493. 10.1016/S0014-5793(99)01094-7 [DOI] [PubMed] [Google Scholar]
- Henry J. R., and Harrison J. F., 2004. Plastic and evolved responses of larval tracheae and mass to varying atmospheric oxygen content in Drosophila melanogaster. J. Exp. Biol. 207: 3559–3567. 10.1242/jeb.01189 [DOI] [PubMed] [Google Scholar]
- Hentze J. L., Carlsson M. A., Kondo S., Nassel D. R., and Rewitz K. F., 2015. The Neuropeptide Allatostatin A Regulates Metabolism and Feeding Decisions in Drosophila. Sci. Rep. 5: 11680 10.1038/srep11680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herboso L., Oliveira M. M., Talamillo A., Perez C., Gonzalez M. et al. , 2015. Ecdysone promotes growth of imaginal discs through the regulation of Thor in D. melanogaster. Sci. Rep. 5: 12383 10.1038/srep12383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz H., Hong X., and Cohen S. M., 2012. Mutual repression by bantam miRNA and Capicua links the EGFR/MAPK and Hippo pathways in growth control. Curr. Biol. 22: 651–657. 10.1016/j.cub.2012.02.050 [DOI] [PubMed] [Google Scholar]
- Hipfner D. R., Weigmann K., and Cohen S. M., 2002. The bantam gene regulates Drosophila growth. Genetics 161: 1527–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirabayashi S., Baranski T. J., and Cagan R. L., 2013. Transformed Drosophila cells evade diet-mediated insulin resistance through wingless signaling. Cell 154: 664–675. 10.1016/j.cell.2013.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hironaka K. I., Fujimoto K., and Nishimura T., 2019. Optimal scaling of critical size for metamorphosis in the genus Drosophila. iScience 20: 348–358. 10.1016/j.isci.2019.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger B., Galic M., Kohler K., Wittwer F., Brogiolo W. et al. , 2008. Imp-L2, a putative homolog of vertebrate IGF-binding protein 7, counteracts insulin signaling in Drosophila and is essential for starvation resistance. J. Biol. 7: 10 10.1186/jbiol72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornberger T. A., Chu W. K., Mak Y. W., Hsiung J. W., Huang S. A. et al. , 2006. The role of phospholipase D and phosphatidic acid in the mechanical activation of mTOR signaling in skeletal muscle. Proc. Natl. Acad. Sci. USA 103: 4741–4746. 10.1073/pnas.0600678103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner M. A., Pardee K., Liu S., King-Jones K., Lajoie G. et al. , 2009. The Drosophila DHR96 nuclear receptor binds cholesterol and regulates cholesterol homeostasis. Genes Dev. 23: 2711–2716. 10.1101/gad.1833609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A. et al. , 2009. Nutrient-dependent mTORC1 association with the ULK1-Atg13–FIP200 complex required for autophagy. Mol. Biol. Cell 20: 1981–1991. 10.1091/mbc.e08-12-1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M., Shimizu S., Fujiwara H., Sakurai S., and Iwami M., 2006. EcR expression in the prothoracicotropic hormone-producing neurosecretory cells of the Bombyx mori brain. FEBS J. 273: 3861–3868. 10.1111/j.1742-4658.2006.05398.x [DOI] [PubMed] [Google Scholar]
- Hsu Y. C., Chern J. J., Cai Y., Liu M., and Choi K. W., 2007. Drosophila TCTP is essential for growth and proliferation through regulation of dRheb GTPase. Nature 445: 785–788. 10.1038/nature05528 [DOI] [PubMed] [Google Scholar]
- Huang J., Wu S., Barrera J., Matthews K., and Pan D., 2005a The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122: 421–434. 10.1016/j.cell.2005.06.007 [DOI] [PubMed] [Google Scholar]
- Huang X., Suyama K., Buchanan J., Zhu A. J., and Scott M. P., 2005b A Drosophila model of the Niemann-Pick type C lysosome storage disease: dnpc1a is required for molting and sterol homeostasis. Development 132: 5115–5124. 10.1242/dev.02079 [DOI] [PubMed] [Google Scholar]
- Hyun J., Jasper H., and Bohmann D., 2005. DREF is required for efficient growth and cell cycle progression in Drosophila imaginal discs. Mol. Cell. Biol. 25: 5590–5598. 10.1128/MCB.25.13.5590-5598.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S., Lee J. H., Jin H., Nam J., Namkoong B. et al. , 2009. Conserved microRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139: 1096–1108. 10.1016/j.cell.2009.11.020 [DOI] [PubMed] [Google Scholar]
- Ikeya T., Galic M., Belawat P., Nairz K., and Hafen E., 2002. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr. Biol. 12: 1293–1300. 10.1016/S0960-9822(02)01043-6 [DOI] [PubMed] [Google Scholar]
- Ikeya T., Broughton S., Alic N., Grandison R., and Partridge L., 2009. The endosymbiont Wolbachia increases insulin/IGF-like signalling in Drosophila. Proc. Biol. Sci. 276: 3799–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im E., von Lintig F. C., Chen J., Zhuang S. H., Qui W. S. et al. , 2002. Rheb is in a high activation state and inhibits B-Raf kinase in mammalian cells. Oncogene 21: 6356–6365. 10.1038/sj.onc.1205792 [DOI] [PubMed] [Google Scholar]
- Imura E., Shimada-Niwa Y., Nishimura T., Hückesfeld S., Schlegel P. et al. , 2020. The Corazonin-PTTH neuronal axis controls systemic body growth by regulating basal ecdysteroid biosynthesis in Drosophila melanogaster. Curr. Biol. 30: 2156–2165.e5. 10.1016/j.cub.2020.03.050 [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Zhu T., Wu J., and Guan K. L., 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4: 648–657. 10.1038/ncb839 [DOI] [PubMed] [Google Scholar]
- Inoki K., Li Y., Xu T., and Guan K. L., 2003a Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17: 1829–1834. 10.1101/gad.1110003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Zhu T., and Guan K. L., 2003b TSC2 mediates cellular energy response to control cell growth and survival. Cell 115: 577–590. 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- Ito N., and Rubin G. M., 1999. Gigas, a Drosophila homolog of tuberous sclerosis gene product-2, regulates the cell cycle. Cell 96: 529–539. 10.1016/S0092-8674(00)80657-1 [DOI] [PubMed] [Google Scholar]
- Jaszczak J. S., Wolpe J. B., Dao A. Q., and Halme A., 2015. Nitric oxide synthase regulates growth coordination during Drosophila melanogaster imaginal disc regeneration. Genetics 200: 1219–1228. 10.1534/genetics.115.178053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczak J. S., Wolpe J. B., Bhandari R., Jaszczak R. G., and Halme A., 2016. Growth coordination during Drosophila melanogaster imaginal disc regeneration is mediated by signaling through the relaxin receptor Lgr3 in the prothoracic gland. Genetics 204: 703–709. 10.1534/genetics.116.193706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H., Kim V. N., and Hyun S., 2012. Conserved microRNA miR-8 controls body size in response to steroid signaling in Drosophila. Genes Dev. 26: 1427–1432. 10.1101/gad.192872.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston L. A., Prober D. A., Edgar B. A., Eisenman R. N., and Gallant P., 1999. Drosophila myc regulates cellular growth during development. Cell 98: 779–790. 10.1016/S0092-8674(00)81512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez-Carreño S., Morante J., and Dominguez M., 2018. Systemic signalling and local effectors in developmental stability, body symmetry, and size. Cell Stress 2: 340–361. 10.15698/cst2018.12.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M. et al. , 2009. ULK-Atg13–FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20: 1992–2003. 10.1091/mbc.e08-12-1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J., Genau H. M., and Behrends C., 2015. Amino acid-dependent mTORC1 regulation by the lysosomal Membrane Protein SLC38A9. Mol. Cell. Biol. 35: 2479–2494. 10.1128/MCB.00125-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junger M. A., Rintelen F., Stocker H., Wasserman J. D., Vegh M. et al. , 2003. The Drosophila forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2: 20 10.1186/1475-4924-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens G., Wieschaus E., Nüsslein-Volhard C., and Kluding H., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: II. Zygotic loci on the third chromosome. Wilehm Roux Arch. Dev. Biol. 193: 283–295. 10.1007/BF00848157 [DOI] [PubMed] [Google Scholar]
- Kane N. S., Vora M., Padgett R. W., and Li Y., 2018. bantam microRNA is a negative regulator of the Drosophila decapentaplegic pathway. Fly (Austin) 12: 105–117. 10.1080/19336934.2018.1499370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapan N., Lushchak O. V., Luo J., and Nassel D. R., 2012. Identified peptidergic neurons in the Drosophila brain regulate insulin-producing cells, stress responses and metabolism by coexpressed short neuropeptide F and corazonin. Cell. Mol. Life Sci. 69: 4051–4066. 10.1007/s00018-012-1097-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. D., Zimmermann G., Suyama K., Meyer T., and Scott M. P., 2008. A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size. Genes Dev. 22: 1877–1893. 10.1101/gad.1670508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplowitz P. B., 2008. Link between body fat and the timing of puberty. Pediatrics 121: S208–S217. 10.1542/peds.2007-1813F [DOI] [PubMed] [Google Scholar]
- Kataoka H., Nagasawa H., Isogai A., Ishizaki H., and Suzuki A., 1991. Prothoracicotropic hormone of the silkworm, Bombyx mori: amino acid sequence and dimeric structure. Agric. Biol. Chem. 55: 73–86. [PubMed] [Google Scholar]
- Katsuyama T., Comoglio F., Seimiya M., Cabuy E., and Paro R., 2015. During Drosophila disc regeneration, JAK/STAT coordinates cell proliferation with Dilp8-mediated developmental delay. Proc. Natl. Acad. Sci. USA 112: E2327–E2336. 10.1073/pnas.1423074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A., Kataoka H., Oka T., Mizoguchi A., Kimura-Kawakami M. et al. , 1990. Molecular cloning of the Bombyx mori prothoracicotropic hormone. Science 247: 1333–1335. 10.1126/science.2315701 [DOI] [PubMed] [Google Scholar]
- Killip L. E., and Grewal S. S., 2012. DREF is required for cell and organismal growth in Drosophila and functions downstream of the nutrition/TOR pathway. Dev. Biol. 371: 191–202. 10.1016/j.ydbio.2012.08.020 [DOI] [PubMed] [Google Scholar]
- Kimball S. R., Gordon B. S., Moyer J. E., Dennis M. D., and Jefferson L. S., 2016. Leucine induced dephosphorylation of Sestrin2 promotes mTORC1 activation. Cell. Signal. 28: 896–906. 10.1016/j.cellsig.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., and Neufeld T. P., 2015. Dietary sugar promotes systemic TOR activation in Drosophila through AKH-dependent selective secretion of Dilp3. Nat. Commun. 6: 6846 10.1038/ncomms7846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., and Lee J. H., 2015. Identification of an AMPK phosphorylation site in Drosophila TSC2 (gigas) that regulate cell growth. Int. J. Mol. Sci. 16: 7015–7026. 10.3390/ijms16047015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. K., and Rulifson E. J., 2004. Conserved mechanisms of glucose sensing and regulation by Drosophila corpora cardiaca cells. Nature 431: 316–320. 10.1038/nature02897 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R. et al. , 2002. MTOR interacts with Raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110: 163–175. 10.1016/S0092-8674(02)00808-5 [DOI] [PubMed] [Google Scholar]
- Kim D. H., Sarbassov D. D., Ali S. M., Latek R. R., Guntur K. V. P. et al. , 2003. G beta L, a positive regulator of the rapamycin-sensitive pathway required for the nutrient-sensitive interaction between raptor and mTOR. Mol. Cell 11: 895–904. 10.1016/S1097-2765(03)00114-X [DOI] [PubMed] [Google Scholar]
- Kim J. H., Lee C., Lee M., Wang H., Kim K. et al. , 2017. Control of leucine-dependent mTORC1 pathway through chemical intervention of leucyl-tRNA synthetase and RagD interaction. Nat. Commun. 8: 732 10.1038/s41467-017-00785-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. S., Ro S. H., Kim M., Park H. W., Semple I. A. et al. , 2015. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 5: 9502 (erratum: Sci. Rep. 5: 14029) 10.1038/srep09502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. G., Hoffman G. R., Poulogiannis G., Buel G. R., Jang Y. J. et al. , 2013. Metabolic stress controls mTORC1 lysosomal localization and dimerization by regulating the TTT-RUVBL1/2 complex. Mol. Cell 49: 172–185. 10.1016/j.molcel.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K., and Thummel C. S., 2005. Nuclear receptors–a perspective from Drosophila. Nat. Rev. Genet. 6: 311–323. 10.1038/nrg1581 [DOI] [PubMed] [Google Scholar]
- King-Jones K., Charles J. P., Lam G., and Thummel C. S., 2005. The ecdysone-induced DHR4 orphan nuclear receptor coordinates growth and maturation in Drosophila. Cell 121: 773–784. 10.1016/j.cell.2005.03.030 [DOI] [PubMed] [Google Scholar]
- Knapp E., and Sun J., 2017. Steroid signaling in mature follicles is important for Drosophila ovulation. Proc. Natl. Acad. Sci. USA 114: 699–704. 10.1073/pnas.1614383114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komura-Kawa T., Hirota K., Shimada-Niwa Y., Yamauchi R., Shimell M. et al. , 2015. The Drosophila zinc finger transcription factor Ouija board controls ecdysteroid biosynthesis through specific regulation of spookier. PLoS Genet. 11: e1005712 10.1371/journal.pgen.1005712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koontz L. M., Liu-Chittenden Y., Yin F., Zheng Y., Yu J. et al. , 2013. The Hippo effector Yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev. Cell 25: 388–401. 10.1016/j.devcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopeć S., 1922. Studies on the necessity of the brain for the inception of insect metamorphosis. Biol. Bull. 42: 323–342. 10.2307/1536759 [DOI] [Google Scholar]
- Ko K. I., Root C. M., Lindsay S. A., Zaninovich O. A., Shepherd A. K. et al. , 2015. Starvation promotes concerted modulation of appetitive olfactory behavior via parallel neuromodulatory circuits. Elife 4: e08298 10.7554/eLife.08298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., and Mirth C. K., 2016. Growth-blocking peptides as nutrition-sensitive signals for insulin secretion and body size regulation. PLoS Biol. 14: e1002392 10.1371/journal.pbio.1002392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Mendes C. C., and Mirth C. K., 2013. Mechanisms regulating nutrition-dependent developmental plasticity through organ-specific effects in insects. Front. Physiol. 4: 263 10.3389/fphys.2013.00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Rodrigues M. A., Athanasiadis A., Shingleton A. W., and Mirth C. K., 2014. Nutritional control of body size through FoxO-Ultraspiracle mediated ecdysone biosynthesis. Elife 3: e03091 10.7554/eLife.03091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M., Davidge J. T., Lockyer J. M., and Staveley B. E., 2003. Expression of Drosophila FOXO regulates growth and can phenocopy starvation. BMC Dev. Biol. 3: 5 10.1186/1471-213X-3-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kréneisz O., Chen X., Fridell Y.-W., and Mulkey D. K., 2010. Glucose increases activity and Ca2+ in insulin-producing cells of adult Drosophila. Neuroreport 21: 1116–1120. 10.1097/WNR.0b013e3283409200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo Y., Huang H., Cai T., and Wang T., 2015. Target of Rapamycin Complex 2 regulates cell growth via Myc in Drosophila. Sci. Rep. 5: 10339 10.1038/srep10339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak S. J., Hong S. H., Bajracharya R., Yang S. Y., Lee K. S. et al. , 2013. Drosophila adiponectin receptor in insulin producing cells regulates glucose and lipid metabolism by controlling insulin secretion. PLoS One 8: e68641 10.1371/journal.pone.0068641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Marca J. E., and Richardson H. E., 2020. Two-faced: roles of JNK signalling during tumourigenesis in the Drosophila model. Front. Cell Dev. Biol. 8: 42 10.3389/fcell.2020.00042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavrynenko O., Rodenfels J., Carvalho M., Dye N. A., Lafont R. et al. , 2015. The ecdysteroidome of Drosophila: influence of diet and development. Development 142: 3758–3768. 10.1242/dev.124982 [DOI] [PubMed] [Google Scholar]
- Layalle S., Arquier N., and Leopold P., 2008. The TOR pathway couples nutrition and developmental timing in Drosophila. Dev. Cell 15: 568–577. 10.1016/j.devcel.2008.08.003 [DOI] [PubMed] [Google Scholar]
- Le T. P., Vuong L. T., Kim A. R., Hsu Y. C., and Choi K. W., 2016. 14–3-3 proteins regulate Tctp-Rheb interaction for organ growth in Drosophila. Nat. Commun. 7: 11501 10.1038/ncomms11501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bacquer O., Petroulakis E., Paglialunga S., Poulin F., Richard D. et al. , 2007. Elevated sensitivity to diet-induced obesity and insulin resistance in mice lacking 4E–BP1 and 4E–BP2. J. Clin. Invest. 117: 387–396. 10.1172/JCI29528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. S., You K. H., Choo J. K., Han Y. M., and Yu K., 2004. Drosophila short neuropeptide F regulates food intake and body size. J. Biol. Chem. 279: 50781–50789. 10.1074/jbc.M407842200 [DOI] [PubMed] [Google Scholar]
- Lee K. S., Kwon O. Y., Lee J. H., Kwon K., Min K. J. et al. , 2008. Drosophila short neuropeptide F signalling regulates growth by ERK-mediated insulin signalling. Nat. Cell Biol. 10: 468–475. 10.1038/ncb1710 [DOI] [PubMed] [Google Scholar]
- Lee K. S., Hong S. H., Kim A. K., Ju S. K., Kwon O. Y. et al. , 2009. Processed short neuropeptide F peptides regulate growth through the ERK-insulin pathway in Drosophila melanogaster. FEBS Lett. 583: 2573–2577. 10.1016/j.febslet.2009.07.024 [DOI] [PubMed] [Google Scholar]
- Lee M. J., Park M. S., Hwang S., Hong Y. K., Choi G. et al. , 2010. Dietary hempseed meal intake increases body growth and shortens the larval stage via the upregulation of cell growth and sterol levels in Drosophila melanogaster. Mol. Cells 30: 29–36. 10.1007/s10059-010-0085-0 [DOI] [PubMed] [Google Scholar]
- Lee G. J., Han G., Yun H. M., Lim J. J., Noh S. et al. , 2018. Steroid signaling mediates nutritional regulation of juvenile body growth via IGF-binding protein in Drosophila. Proc. Natl. Acad. Sci. USA 115: 5992–5997. 10.1073/pnas.1718834115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leevers S. J., Weinkove D., MacDougall L. K., Hafen E., and Waterfield M. D., 1996. The Drosophila phosphoinositide 3-kinase Dp110 promotes cell growth. EMBO J. 15: 6584–6594. 10.1002/j.1460-2075.1996.tb01049.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiblich A., Marsden L., Gandy C., Corrigan L., Jenkins R. et al. , 2012. Bone morphogenetic protein- and mating-dependent secretory cell growth and migration in the Drosophila accessory gland. Proc. Natl. Acad. Sci. USA 109: 19292–19297. 10.1073/pnas.1214517109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiblich A., Hellberg J., Sekar A., Gandy C., Mendes C. C. et al. , 2019. Mating induces switch from hormone-dependent to hormone-independent steroid receptor-mediated growth in Drosophila secondary cells. PLoS Biol. 17: e3000145 10.1371/journal.pbio.3000145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., and Gong Z., 2015. Cold-sensing regulates Drosophila growth through insulin-producing cells. Nat. Commun. 6: 10083 10.1038/ncomms10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin F., Hossain M. A., Post S., Karashchuk G., Tatar M. et al. , 2017. Total solid-phase synthesis of biologically active Drosophila insulin-like peptide 2 (DILP2). Aust. J. Chem. 70: 208–212. 10.1071/CH16626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S. S., and Liu Y. W., 2019. Mechanical stretch induces mTOR recruitment and activation at the phosphatidic acid-enriched macropinosome in muscle cell. Front. Cell Dev. Biol. 7: 78 10.3389/fcell.2019.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneweber G. A., Jacobson J., Busch K. E., Hudry B., Christov C. P. et al. , 2014. Neuronal control of metabolism through nutrient-dependent modulation of tracheal branching. Cell 156: 69–83. 10.1016/j.cell.2013.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liao S., Veenstra J. A., and Nassel D. R., 2016. Drosophila insulin-like peptide 1 (DILP1) is transiently expressed during non-feeding stages and reproductive dormancy. Sci. Rep. 6: 26620 10.1038/srep26620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizcano J. M., Alrubaie S., Kieloch A., Deak M., Leevers S. J. et al. , 2003. Insulin-induced Drosophila S6 kinase activation requires phosphoinositide 3-kinase and protein kinase B. Biochem. J. 374: 297–306. 10.1042/bj20030577 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Luo J., Becnel J., Nichols C. D., and Nassel D. R., 2012. Insulin-producing cells in the brain of adult Drosophila are regulated by the serotonin 5–HT1A receptor. Cell. Mol. Life Sci. 69: 471–484. 10.1007/s00018-011-0789-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J., Lushchak O. V., Goergen P., Williams M. J., and Nassel D. R., 2014. Drosophila insulin-producing cells are differentially modulated by serotonin and octopamine receptors and affect social behavior. PLoS One 9: e99732 10.1371/journal.pone.0099732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S., Meng Z., Chen R., and Guan K. L., 2019. The Hippo pathway: biology and pathophysiology. Annu. Rev. Biochem. 88: 577–604. 10.1146/annurev-biochem-013118-111829 [DOI] [PubMed] [Google Scholar]
- Manière G., Ziegler A. B., Geillon F., Featherstone D. E., and Grosjean Y., 2016. Direct sensing of nutrients via a LAT1-like transporter in Drosophila insulin-producing cells. Cell Rep. 17: 137–148. 10.1016/j.celrep.2016.08.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning B. D., Tee A. R., Logsdon M. N., Blenis J., and Cantley L. C., 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-Kinase/Akt pathway. Mol. Cell 10: 151–162. 10.1016/S1097-2765(02)00568-3 [DOI] [PubMed] [Google Scholar]
- Manning S. A., Dent L. G., Kondo S., Zhao Z. W., Plachta N. et al. , 2018. Dynamic fluctuations in subcellular localization of the Hippo pathway effector Yorkie in vivo. Curr. Biol. 28: 1651–1660.e4. 10.1016/j.cub.2018.04.018 [DOI] [PubMed] [Google Scholar]
- Marshall C. B., Ho J., Buerger C., Plevin M. J., Li G. Y. et al. , 2009. Characterization of the intrinsic and TSC2-GAP-regulated GTPase activity of Rheb by real-time NMR. Sci. Signal. 2: ra3 10.1126/scisignal.2000029 [DOI] [PubMed] [Google Scholar]
- Marshall L., Rideout E. J., and Grewal S. S., 2012. Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J. 31: 1916–1930. 10.1038/emboj.2012.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrayer Z., Ono H., Shimell M., Parvy J. P., Beckstead R. B. et al. , 2007. Prothoracicotropic hormone regulates developmental timing and body size in Drosophila. Dev. Cell 13: 857–871. 10.1016/j.devcel.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald J. M. C., Ghosh S. M., Gascoigne S. J. L., and Shingleton A. W., 2018. Plasticity through canalization: the contrasting effect of temperature on trait size and growth in Drosophila. Front. Cell Dev. Biol. 6: 156 10.3389/fcell.2018.00156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner G. W., Luo S. D., Dias B. G., Texada M. J., and Baker B. S., 2016. Sex-specific regulation of Lgr3 in Drosophila neurons. Proc. Natl. Acad. Sci. USA 113: E1256–E1265. 10.1073/pnas.1600241113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher C., and Pankratz M. J., 2005. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3: e305 10.1371/journal.pbio.0030305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S., Dibble C. C., Talbott G., Hoxhaj G., Valvezan A. J. et al. , 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156: 771–785. 10.1016/j.cell.2013.11.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meola R. W., and Adkisson P. L., 1977. Release of prothoracicotropic hormone and potentiation of developmental ability during diapause in bollworm, Heliothis-Zea. J. Insect Physiol. 23: 683–688. 10.1016/0022-1910(77)90084-1 [DOI] [Google Scholar]
- Meschi E., Leopold P., and Delanoue R., 2019. An EGF-responsive neural circuit couples insulin secretion with nutrition in Drosophila. Dev. Cell 48: 76–86.e5. 10.1016/j.devcel.2018.11.029 [DOI] [PubMed] [Google Scholar]
- Minakuchi C., Zhou X., and Riddiford L. M., 2008. Kruppel homolog 1 (Kr-h1) mediates juvenile hormone action during metamorphosis of Drosophila melanogaster. Mech. Dev. 125: 91–105. 10.1016/j.mod.2007.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K., and Riddiford L. M., 2007. Size assessment and growth control: how adult size is determined in insects. Bioessays 29: 344–355. 10.1002/bies.20552 [DOI] [PubMed] [Google Scholar]
- Mirth C., Truman J. W., and Riddiford L. M., 2005. The role of the prothoracic gland in determining critical weight for metamorphosis in Drosophila melanogaster. Curr. Biol. 15: 1796–1807. 10.1016/j.cub.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Mirth C. K., Truman J. W., and Riddiford L. M., 2009. The ecdysone receptor controls the post-critical weight switch to nutrition-independent differentiation in Drosophila wing imaginal discs. Development 136: 2345–2353. 10.1242/dev.032672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirth C. K., Tang H. Y., Makohon-Moore S. C., Salhadar S., Gokhale R. H. et al. , 2014. Juvenile hormone regulates body size and perturbs insulin signaling in Drosophila. Proc. Natl. Acad. Sci. USA 111: 7018–7023. 10.1073/pnas.1313058111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra J. R., and Irvine K. D., 2018. The Hippo signaling network and its biological functions. Annu. Rev. Genet. 52: 65–87. 10.1146/annurev-genet-120417-031621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell N., Cranna N., Richardson H., and Quinn L., 2008. The Ecdysone-inducible zinc-finger transcription factor Crol regulates Wg transcription and cell cycle progression in Drosophila. Development 135: 2707–2716. 10.1242/dev.021766 [DOI] [PubMed] [Google Scholar]
- Mitchell N. C., Lin J. I., Zaytseva O., Cranna N., Lee A. et al. , 2013. The Ecdysone receptor constrains wingless expression to pattern cell cycle across the Drosophila wing margin in a cyclin B-dependent manner. BMC Dev. Biol. 13: 28 10.1186/1471-213X-13-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller M. E., Danielsen E. T., Herder R., O’Connor M. B., and Rewitz K. F., 2013. Dynamic feedback circuits function as a switch for shaping a maturation-inducing steroid pulse in Drosophila. Development 140: 4730–4739. 10.1242/dev.099739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller M. E., Nagy S., Gerlach S. U., Soegaard K. C., Danielsen E. T. et al. , 2017. Warts signaling controls organ and body growth through regulation of ecdysone. Curr. Biol. 27: 1652–1659.e4. 10.1016/j.cub.2017.04.048 [DOI] [PubMed] [Google Scholar]
- Montagne J., Stewart M. J., Stocker H., Hafen E., Kozma S. C. et al. , 1999. Drosophila S6 kinase: a regulator of cell size. Science 285: 2126–2129. 10.1126/science.285.5436.2126 [DOI] [PubMed] [Google Scholar]
- Moss-Taylor L., Upadhyay A., Pan X., Kim M.-J., and O’Connor M. B., 2019. Body size and tissue-scaling is regulated by motoneuron-derived Activinβ in Drosophila melanogaster. Genetics 213: 1447–1464. 10.1534/genetics.119.302394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murawski M., Szczesniak B., Zoladek T., Hopper A. K., Martin N. C. et al. , 1994. maf1 mutation alters the subcellular localization of the Mod5 protein in yeast. Acta Biochim. Pol. 41: 441–448. 10.18388/abp.1994_4691 [DOI] [PubMed] [Google Scholar]
- Musselman L. P., Fink J. L., Narzinski K., Ramachandran P. V., Hathiramani S. S. et al. , 2011. A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis. Model. Mech. 4: 842–849. 10.1242/dmm.007948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy D., Cusumano P., Andreatta G., Anduaga A. M., Hermann-Luibl C. et al. , 2019. Peptidergic signaling from clock neurons regulates reproductive dormancy in Drosophila melanogaster. PLoS Genet. 15: e1008158 10.1371/journal.pgen.1008158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neto-Silva R. M., de Beco S., and Johnston L. A., 2010. Evidence for a growth-stabilizing regulatory feedback mechanism between Myc and Yorkie, the Drosophila homolog of Yap. Dev. Cell 19: 507–520. 10.1016/j.devcel.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman S. D., and Bashirullah A., 2018. Hobbit regulates intracellular trafficking to drive insulin-dependent growth during Drosophila development. Development 145: dev161356. 10.1242/dev.161356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijhout H. F., and Williams C. M., 1974. Control of moulting and metamorphosis in the tobacco hornworm, Manduca sexta (L.): cessation of juvenile hormone secretion as a trigger for pupation. J. Exp. Biol. 61: 493–501. [DOI] [PubMed] [Google Scholar]
- Niwa Y. S., and Niwa R., 2016. Ouija board: a transcription factor evolved for only one target in steroid hormone biosynthesis in the fruit fly Drosophila melanogaster. Transcription 7: 196–202. 10.1080/21541264.2016.1210370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa R., Matsuda T., Yoshiyama T., Namiki T., Mita K. et al. , 2004. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J. Biol. Chem. 279: 35942–35949. 10.1074/jbc.M404514200 [DOI] [PubMed] [Google Scholar]
- Niwa R., Sakudoh T., Namiki T., Saida K., Fujimoto Y. et al. , 2005. The ecdysteroidogenic P450 Cyp302a1/disembodied from the silkworm, Bombyx mori, is transcriptionally regulated by prothoracicotropic hormone. Insect Mol. Biol. 14: 563–571. 10.1111/j.1365-2583.2005.00587.x [DOI] [PubMed] [Google Scholar]
- Niwa R., Namiki T., Ito K., Shimada-Niwa Y., Kiuchi M. et al. , 2010. Non-molting glossy/shroud encodes a short-chain dehydrogenase/reductase that functions in the ‘Black Box’ of the ecdysteroid biosynthesis pathway. Development 137: 1991–1999. 10.1242/dev.045641 [DOI] [PubMed] [Google Scholar]
- Nolo R., Morrison C. M., Tao C., Zhang X., and Halder G., 2006. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr. Biol. 16: 1895–1904. 10.1016/j.cub.2006.08.057 [DOI] [PubMed] [Google Scholar]
- Nunney L., and Cheung W., 1997. The effect of temperature on body size and fecundity in female Drosophila melanogaster: evidence for adaptive plasticity. Evolution 51: 1529–1535. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Wieschaus E., and Kluding H., 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster: I. Zygotic loci on the second chromosome. Wilehm Roux Arch. Dev. Biol. 193: 267–282. 10.1007/BF00848156 [DOI] [PubMed] [Google Scholar]
- Oh H., and Irvine K. D., 2008. In vivo regulation of Yorkie phosphorylation and localization. Development 135: 1081–1088. 10.1242/dev.015255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., and Irvine K. D., 2011. Cooperative regulation of growth by Yorkie and Mad through bantam. Dev. Cell 20: 109–122. 10.1016/j.devcel.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H., Reddy B. V., and Irvine K. D., 2009. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev. Biol. 335: 188–197. 10.1016/j.ydbio.2009.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Lai J. S., Mills H. J., Erdjument-Bromage H., Giammarinaro B. et al. , 2019. A glucose-sensing neuron pair regulates insulin and glucagon in Drosophila. Nature 574: 559–564. 10.1038/s41586-019-1675-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhara Y., Shimada-Niwa Y., Niwa R., Kayashima Y., Hayashi Y. et al. , 2015. Autocrine regulation of ecdysone synthesis by β3-octopamine receptor in the prothoracic gland is essential for Drosophila metamorphosis. Proc. Natl. Acad. Sci. USA 112: 1452–1457. 10.1073/pnas.1414966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohhara Y., Kobayashi S., and Yamanaka N., 2017. Nutrient-dependent endocycling in steroidogenic tissue dictates timing of metamorphosis in Drosophila melanogaster. PLoS Genet. 13: e1006583 10.1371/journal.pgen.1006583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., and Nishimura T., 2015. Signaling from glia and cholinergic neurons controls nutrient-dependent production of an insulin-like peptide for Drosophila body growth. Dev. Cell 35: 295–310. 10.1016/j.devcel.2015.10.003 [DOI] [PubMed] [Google Scholar]
- Okamoto N., and Yamanaka N., 2020. Steroid hormone entry into the brain requires a membrane transporter in Drosophila. Curr. Biol. 30: 359–366.e3. 10.1016/j.cub.2019.11.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Yamanaka N., Yagi Y., Nishida Y., Kataoka H. et al. , 2009. A fat body-derived IGF-like peptide regulates postfeeding growth in Drosophila. Dev. Cell 17: 885–891. 10.1016/j.devcel.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Nishimori Y., and Nishimura T., 2012. Conserved role for the Dachshund protein with Drosophila Pax6 homolog Eyeless in insulin expression. Proc. Natl. Acad. Sci. USA 109: 2406–2411. 10.1073/pnas.1116050109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Nakamori R., Murai T., Yamauchi Y., Masuda A. et al. , 2013. A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila. Genes Dev. 27: 87–97. 10.1101/gad.204479.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N., Viswanatha R., Bittar R., Li Z., Haga-Yamanaka S. et al. , 2018. A membrane transporter is required for steroid hormone uptake in Drosophila. Dev. Cell 47: 294–305.e7. 10.1016/j.devcel.2018.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M. M., Shingleton A. W., and Mirth C. K., 2014. Coordination of wing and whole-body development at developmental milestones ensures robustness against environmental and physiological perturbations. PLoS Genet. 10: e1004408 10.1371/journal.pgen.1004408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil T. K., Duffy L. R., Frey J. W., and Hornberger T. A., 2009. The role of phosphoinositide 3-kinase and phosphatidic acid in the regulation of mammalian target of rapamycin following eccentric contractions. J. Physiol. 587: 3691–3701. 10.1113/jphysiol.2009.173609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono H., Rewitz K. F., Shinoda T., Itoyama K., Petryk A. et al. , 2006. Spook and Spookier code for stage-specific components of the ecdysone biosynthetic pathway in Diptera. Dev. Biol. 298: 555–570. 10.1016/j.ydbio.2006.07.023 [DOI] [PubMed] [Google Scholar]
- Oro A. E., McKeown M., and Evans R. M., 1990. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature 347: 298–301. 10.1038/347298a0 [DOI] [PubMed] [Google Scholar]
- Ou Q., Magico A., and King-Jones K., 2011. Nuclear receptor DHR4 controls the timing of steroid hormone pulses during Drosophila development. PLoS Biol. 9: e1001160 10.1371/journal.pbio.1001160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou Q., Zeng J., Yamanaka N., Brakken-Thal C., O’Connor M. B. et al. , 2016. The insect prothoracic gland as a model for steroid hormone biosynthesis and regulation. Cell Rep. 16: 247–262. 10.1016/j.celrep.2016.05.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padi S. K. R., Singh N., Bearss J. J., Olive V., Song J. H. et al. , 2019. Phosphorylation of DEPDC5, a component of the GATOR1 complex, releases inhibition of mTORC1 and promotes tumor growth. Proc. Natl. Acad. Sci. USA 116: 20505–20510. 10.1073/pnas.1904774116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallares-Cartes C., Cakan-Akdogan G., and Teleman A. A., 2012. Tissue-specific coupling between insulin/IGF and TORC1 signaling via PRAS40 in Drosophila. Dev. Cell 22: 172–182. 10.1016/j.devcel.2011.10.029 [DOI] [PubMed] [Google Scholar]
- Palm W., Sampaio J. L., Brankatschk M., Carvalho M., Mahmoud A. et al. , 2012. Lipoproteins in Drosophila melanogaster–assembly, function, and influence on tissue lipid composition. PLoS Genet. 8: e1002828 10.1371/journal.pgen.1002828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palos L. A., and Blasko G., 1979. Effect of hypoxia on the development of Drosophila melanogaster (Meigen). Aviat. Space Environ. Med. 50: 411–412. [PubMed] [Google Scholar]
- Pan X., and O’Connor M. B., 2019. Developmental maturation: Drosophila AstA signaling provides a kiss to grow up. Curr. Biol. 29: R161–R164. 10.1016/j.cub.2019.01.040 [DOI] [PubMed] [Google Scholar]
- Pan X., Neufeld T. P., and O’Connor M. B., 2019. A tissue- and temporal-specific autophagic switch controls drosophila pre-metamorphic nutritional checkpoints. Curr. Biol 29: 2840–2851.e4. 10.1016/j.cub.2019.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Heemskerk I., Ibar C., Shraiman B. I., and Irvine K. D., 2016. Differential growth triggers mechanical feedback that elevates Hippo signaling. Proc. Natl. Acad. Sci. USA 113: E6974–E6983. 10.1073/pnas.1615012113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Alégot H., Rauskolb C., and Irvine K. D., 2018. The dynamics of Hippo signaling during Drosophila wing development. Development 145: dev165712. 10.1242/dev.165712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F., Riccardo S., Daniel M., Saqcena M., Kundu N. et al. , 2011. Drosophila insulin and target of rapamycin (TOR) pathways regulate GSK3 beta activity to control Myc stability and determine Myc expression in vivo. BMC Biol. 9: 65 10.1186/1741-7007-9-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F., Riccardo S., Zola S., Lora C., Grifoni D. et al. , 2013. dMyc expression in the fat body affects DILP2 release and increases the expression of the fat desaturase Desat1 resulting in organismal growth. Dev. Biol. 379: 64–75. 10.1016/j.ydbio.2013.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker N. F., and Shingleton A. W., 2011. The coordination of growth among Drosophila organs in response to localized growth-perturbation. Dev. Biol. 357: 318–325. 10.1016/j.ydbio.2011.07.002 [DOI] [PubMed] [Google Scholar]
- Parker J., and Struhl G., 2015. Scaling the Drosophila wing: TOR-dependent target gene access by the Hippo pathway transducer Yorkie. PLoS Biol. 13: e1002274 10.1371/journal.pbio.1002274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parmigiani A., Nourbakhsh A., Ding B., Wang W., Kim Y. C. et al. , 2014. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 9: 1281–1291. 10.1016/j.celrep.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L., Barrie B., Fowler K., and French V., 1994. Evolution and development of body size and cell size in Drosophila melanogaster in response to temperature. Evolution 48: 1269–1276. 10.1111/j.1558-5646.1994.tb05311.x [DOI] [PubMed] [Google Scholar]
- Parvy J. P., Blais C., Bernard F., Warren J. T., Petryk A. et al. , 2005. A role for betaFTZ-F1 in regulating ecdysteroid titers during post-embryonic development in Drosophila melanogaster. Dev. Biol. 282: 84–94. 10.1016/j.ydbio.2005.02.028 [DOI] [PubMed] [Google Scholar]
- Pasco M. Y., and Leopold P., 2012. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PLoS One 7: e36583 10.1371/journal.pone.0036583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual J., Jacobs J., Sansores-Garcia L., Natarajan M., Zeitlinger J. et al. , 2017. Hippo reprograms the transcriptional response to Ras signaling. Dev. Cell 42: 667–680.e4. 10.1016/j.devcel.2017.08.013 [DOI] [PubMed] [Google Scholar]
- Patel P. H., Thapar N., Guo L., Martinez M., Maris J. et al. , 2003. Drosophila Rheb GTPase is required for cell cycle progression and cell growth. J. Cell Sci. 116: 3601–3610. 10.1242/jcs.00661 [DOI] [PubMed] [Google Scholar]
- Paternoster S., and Falasca M., 2018. Dissecting the physiology and pathophysiology of glucagon-like peptide-1. Front. Endocrinol. (Lausanne) 9: 584 10.3389/fendo.2018.00584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck L. S., and Maddrell S. H., 2005. Limitation of size by hypoxia in the fruit fly Drosophila melanogaster. J. Exp. Zool. A Comp. Exp. Biol. 303: 968–975. [DOI] [PubMed] [Google Scholar]
- Pende M., Kozma S. C., Jaquet M., Oorschot V., Burcelin R. et al. , 2000. Hypoinsulinaemia, glucose intolerance and diminished beta-cell size in S6K1-deficient mice. Nature 408: 994–997. 10.1038/35050135 [DOI] [PubMed] [Google Scholar]
- Pende M., Um S. H., Mieulet V., Sticker M., Goss V. L. et al. , 2004. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24: 3112–3124. 10.1128/MCB.24.8.3112-3124.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H. W., Slattery M., and Mann R. S., 2009. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 23: 2307–2319. 10.1101/gad.1820009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C. S., Roczniak-Ferguson A., and Ferguson S. M., 2013. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J. Cell Biol. 202: 1107–1122. 10.1083/jcb.201307084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk A., Warren J. T., Marques G., Jarcho M. P., Gilbert L. I. et al. , 2003. Shade is the Drosophila P450 enzyme that mediates the hydroxylation of ecdysone to the steroid insect molting hormone 20-hydroxyecdysone. Proc. Natl. Acad. Sci. USA 100: 13773–13778. 10.1073/pnas.2336088100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta K., Lefebvre O., Martin N. C., Smagowicz W. J., Stanford D. R. et al. , 2001. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 5031–5040. 10.1128/MCB.21.15.5031-5040.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poodry C. A., and Woods D. F., 1990. Control of the developmental timer for Drosophila pupariation. Roux Arch. Dev. Biol. 199: 219–227. 10.1007/BF01682081 [DOI] [PubMed] [Google Scholar]
- Post S., and Tatar M., 2016. Nutritional geometric profiles of insulin/IGF expression in Drosophila melanogaster. PLoS One 11: e0155628 10.1371/journal.pone.0155628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S., Karashchuk G., Wade J. D., Sajid W., De Meyts P. et al. , 2018a Drosophila insulin-like peptides DILP2 and DILP5 differentially stimulate cell signaling and glycogen phosphorylase to regulate longevity. Front. Endocrinol. 9: 245 10.3389/fendo.2018.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post S., Liao S., Yamamoto R., Veenstra J. A., Nassel D. R. et al. , 2018b Drosophila insulin-like peptide dilp1 increases lifespan and glucagon-like Akh expression epistatic to dilp2. Aging Cell 18: e12863 10.1111/acel.12863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter C. J., Huang H., and Xu T., 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105: 357–368. 10.1016/S0092-8674(01)00333-6 [DOI] [PubMed] [Google Scholar]
- Potter C. J., Pedraza L. G., and Xu T., 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4: 658–665. 10.1038/ncb840 [DOI] [PubMed] [Google Scholar]
- Povey S., Burley M. W., Attwood J., Benham F., Hunt D. et al. , 1994. Two loci for tuberous sclerosis: one on 9q34 and one on 16p13. Ann. Hum. Genet. 58: 107–127. 10.1111/j.1469-1809.1994.tb01881.x [DOI] [PubMed] [Google Scholar]
- Puig O., and Tjian R., 2005. Transcriptional feedback control of insulin receptor by dFOXO/FOXO1. Genes Dev. 19: 2435–2446. 10.1101/gad.1340505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Marr M. T., Ruhf M. L., and Tjian R., 2003. Control of cell number by Drosophila FOXO: downstream and feedback regulation of the insulin receptor pathway. Genes Dev. 17: 2006–2020. 10.1101/gad.1098703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radimerski T., Montagne J., Rintelen F., Stocker H., van der Kaay J. et al. , 2002. dS6K-regulated cell growth is dPKB/dPI(3)K-independent, but requires dPDK1. Nat. Cell Biol. 4: 251–255. 10.1038/ncb763 [DOI] [PubMed] [Google Scholar]
- Rajan A., and Perrimon N., 2012. Drosophila cytokine unpaired 2 regulates physiological homeostasis by remotely controlling insulin secretion. Cell 151: 123–137 [corrigenda: Cell 152: 1197 (2013)]. 10.1016/j.cell.2012.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raught B., Peiretti F., Gingras A. C., Livingstone M., Shahbazian D. et al. , 2004. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 23: 1761–1769. 10.1038/sj.emboj.7600193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebsamen M., Pochini L., Stasyk T., de Araujo M. E., Galluccio M. et al. , 2015. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 519: 477–481. 10.1038/nature14107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann H., Bruning M., Berghaus C., Schwarten M., Kohler K. et al. , 2008. Biochemical characterisation of TCTP questions its function as a guanine nucleotide exchange factor for Rheb. FEBS Lett. 582: 3005–3010. 10.1016/j.febslet.2008.07.057 [DOI] [PubMed] [Google Scholar]
- Ren F., Zhang L., and Jiang J., 2010. Hippo signaling regulates Yorkie nuclear localization and activity through 14–3-3 dependent and independent mechanisms. Dev. Biol. 337: 303–312. 10.1016/j.ydbio.2009.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G. R., Hauser F., Rewitz K. F., Kondo S., Engelbrecht A. F. et al. , 2015. CCHamide-2 is an orexigenic brain-gut peptide in Drosophila. PLoS One 10: e0133017 10.1371/journal.pone.0133017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren S., Huang Z., Jiang Y., and Wang T., 2018. dTBC1D7 regulates systemic growth independently of TSC through insulin signaling. J. Cell Biol. 217: 517–526. 10.1083/jcb.201706027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repiso A., Bergantinos C., and Serras F., 2013. Cell fate respecification and cell division orientation drive intercalary regeneration in Drosophila wing discs. Development 140: 3541–3551. 10.1242/dev.095760 [DOI] [PubMed] [Google Scholar]
- Restrepo S., Zartman J. J., and Basler K., 2014. Coordination of patterning and growth by the morphogen DPP. Curr. Biol. 24: R245–R255. 10.1016/j.cub.2014.01.055 [DOI] [PubMed] [Google Scholar]
- Rewitz K., and O’Connor M. B., 2011. Timing is everything: PTTH mediated DHR4 nucleocytoplasmic trafficking sets the tempo of Drosophila steroid production. front. Endocrinology 2: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz K. F., Rybczynski R., Warren J. T., and Gilbert L. I., 2006a Developmental expression of Manduca shade, the P450 mediating the final step in molting hormone synthesis. Mol. Cell. Endocrinol. 247: 166–174. 10.1016/j.mce.2005.12.053 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Rybczynski R., Warren J. T., and Gilbert L. I., 2006b Identification, characterization and developmental expression of Halloween genes encoding P450 enzymes mediating ecdysone biosynthesis in the tobacco hornworm, Manduca sexta. Insect Biochem. Mol. Biol. 36: 188–199. 10.1016/j.ibmb.2005.12.002 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Larsen M. R., Lobner-Olesen A., Rybczynski R., O’Connor M. B. et al. , 2009a A phosphoproteomics approach to elucidate neuropeptide signal transduction controlling insect metamorphosis. Insect Biochem. Mol. Biol. 39: 475–483. 10.1016/j.ibmb.2009.04.005 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., Gilbert L. I., and O’Connor M. B., 2009b The insect neuropeptide PTTH activates receptor tyrosine kinase torso to initiate metamorphosis. Science 326: 1403–1405. 10.1126/science.1176450 [DOI] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., and O’Connor M. B., 2010. Steroid hormone inactivation is required during the juvenile-adult transition in Drosophila. Dev. Cell 19: 895–902. 10.1016/j.devcel.2010.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rewitz K. F., Yamanaka N., and O’Connor M. B., 2013. Developmental checkpoints and feedback circuits time insect maturation. Curr. Top. Dev. Biol. 103: 1–33. 10.1016/B978-0-12-385979-2.00001-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford L. M., Cherbas P., and Truman J. W., 2000. Ecdysone receptors and their biological actions. Vitam. Horm. 60: 1–73. 10.1016/S0083-6729(00)60016-X [DOI] [PubMed] [Google Scholar]
- Rintelen F., Stocker H., Thomas G., and Hafen E., 2001. PDK1 regulates growth through Akt and S6K in Drosophila. Proc. Natl. Acad. Sci. USA 98: 15020–15025. 10.1073/pnas.011318098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins H., Li Y., and Padgett R. W., 2005. Incorporating structure to predict microRNA targets. Proc. Natl. Acad. Sci. USA 102: 4006–4009. 10.1073/pnas.0500775102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins D. J., Fei D. L., and Riobo N. A., 2012. The Hedgehog signal transduction network. Sci. Signal. 5: re6 10.1126/scisignal.2002906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson F. W., 1966. The ecological genetics of growth in Drosophila. 8. Adaptation to a new diet. Genet. Res. 8: 165–179. 10.1017/S0016672300010028 [DOI] [PubMed] [Google Scholar]
- Roch F., Jimenez G., and Casanova J., 2002. EGFR signalling inhibits Capicua-dependent repression during specification of Drosophila wing veins. Development 129: 993–1002. [DOI] [PubMed] [Google Scholar]
- Rodenfels J., Lavrynenko O., Ayciriex S., Sampaio J. L., Carvalho M. et al. , 2014. Production of systemically circulating Hedgehog by the intestine couples nutrition to growth and development. Genes Dev. 28: 2636–2651. 10.1101/gad.249763.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Benitez D., Thiaville P. C., de Crecy-Lagard V., and Glavic A., 2015. The levels of a universally conserved tRNA modification regulate cell growth. J. Biol. Chem. 290: 18699–18707. 10.1074/jbc.M115.665406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero N. M., Dekanty A., and Wappner P., 2007. Cellular and developmental adaptations to hypoxia: a Drosophila perspective. Methods Enzymol. 435: 123–144. 10.1016/S0076-6879(07)35007-6 [DOI] [PubMed] [Google Scholar]
- Root C. M., Ko K. I., Jafari A., and Wang J. W., 2011. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 145: 133–144. 10.1016/j.cell.2011.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulifson E. J., Kim S. K., and Nusse R., 2002. Ablation of insulin-producing neurons in flies: growth and diabetic phenotypes. Science 296: 1118–1120. 10.1126/science.1070058 [DOI] [PubMed] [Google Scholar]
- Russell M. A., 1974. Pattern formation in the imaginal discs of a temperature-sensitive cell-lethal mutant of Drosophila melanogaster. Dev. Biol. 40: 24–39. 10.1016/0012-1606(74)90104-3 [DOI] [PubMed] [Google Scholar]
- Rybczynski R., 2005. The prothoracicotropic hormone, pp. 61–123 in Comprehensive Molecular Insect Science, edited by Gilbert L. I., Iatrou K., and Gill S.. Pergamon Press, Oxford: 10.1016/B0-44-451924-6/00033-8 [DOI] [Google Scholar]
- Sajid W., Kulahin N., Schluckebier G., Ribel U., Henderson H. R. et al. , 2011. Structural and biological properties of the Drosophila insulin-like peptide 5 show evolutionary conservation. J. Biol. Chem. 286: 661–673. 10.1074/jbc.M110.156018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai S., 2005. Feedback regulation of prothoracic gland activity, pp. 409–431 in Comprehensive Molecular Insect Science, edited by Gilbert L. I., Iatrou K., and Gill S. S.. Pergamon Press, Oxford: 10.1016/B0-44-451924-6/00041-7 [DOI] [Google Scholar]
- Sancak Y., Thoreen C. C., Peterson T. R., Lindquist R. A., Kang S. A. et al. , 2007. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 25: 903–915. 10.1016/j.molcel.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C. et al. , 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501. 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S. et al. , 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303. 10.1016/j.cell.2010.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang J. H., 1962. Relationships between protein supplies and B-vitamin requirements, in axenically cultured Drosophila. J. Nutr. 77: 355–368. 10.1093/jn/77.3.355 [DOI] [PubMed] [Google Scholar]
- Sano H., Nakamura A., Texada M. J., Truman J. W., Ishimoto H. et al. , 2015. The nutrient-responsive hormone CCHamide-2 controls growth by regulating insulin-like peptides in the brain of Drosophila melanogaster. PLoS Genet. 11: e1005209 (erratum: PLoS Genet 11: e1005481). 10.1371/journal.pgen.1005209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo L. J., Gao X., Chiarelli D. A., Li L., Pan D. et al. , 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5: 566–571 (erratum: Nat Cell Biol 5: 680). 10.1038/ncb996 [DOI] [PubMed] [Google Scholar]
- Scanga S. E., Ruel L., Binari R. C., Snow B., Stambolic V. et al. , 2000. The conserved PI3′K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 19: 3971–3977. 10.1038/sj.onc.1203739 [DOI] [PubMed] [Google Scholar]
- Schlegel P., Texada M. J., Miroschnikow A., Schoofs A., Huckesfeld S. et al. , 2016. Synaptic transmission parallels neuromodulation in a central food-intake circuit. Elife 5: e16799 10.7554/eLife.16799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleich S., and Teleman A. A., 2009. Akt phosphorylates both Tsc1 and Tsc2 in Drosophila, but neither phosphorylation is required for normal animal growth. PLoS One 4: e6305 10.1371/journal.pone.0006305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M., and Palka J., 1987. Changing spatial patterns of DNA replication in the developing wing of Drosophila. Dev. Biol. 123: 145–153. 10.1016/0012-1606(87)90436-2 [DOI] [PubMed] [Google Scholar]
- Schweinfest C. W., Fujiwara S., Lau L. F., and Papas T. S., 1988. c-myc can induce expression of G0/G1 transition genes. Mol. Cell. Biol. 8: 3080–3087. 10.1128/MCB.8.8.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selcho M., Millan C., Palacios-Munoz A., Ruf F., Ubillo L. et al. , 2017. Central and peripheral clocks are coupled by a neuropeptide pathway in Drosophila. Nat. Commun. 8: 15563 10.1038/ncomms15563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyilmaz D., Virtue S., Xu X., Tan C. Y., Griffin J. L. et al. , 2015. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 525: 124–128. 10.1038/nature14601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan L., Pan X., Woods A. L., O’Connor M. B., and Hariharan I. K., 2018. The BMP2/4 ortholog Dpp can function as an inter-organ signal that regulates developmental timing. Life Sci. Alliance 1: e201800216 10.26508/lsa.201800216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K., and Sabatini D. M., 2018. Ragulator and SLC38A9 activate the Rag GTPases through noncanonical GEF mechanisms. Proc. Natl. Acad. Sci. USA 115: 9545–9550. 10.1073/pnas.1811727115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Guo X., Yan H., Lu Y., Ji X. et al. , 2015. A miR-130a-YAP positive feedback loop promotes organ size and tumorigenesis. Cell Res. 25: 997–1012. 10.1038/cr.2015.98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilo B. Z., 2014. The regulation and functions of MAPK pathways in Drosophila. Methods 68: 151–159. 10.1016/j.ymeth.2014.01.020 [DOI] [PubMed] [Google Scholar]
- Shima H., Pende M., Chen Y., Fumagalli S., Thomas G. et al. , 1998. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17: 6649–6659. 10.1093/emboj/17.22.6649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada-Niwa Y., and Niwa R., 2014. Serotonergic neurons respond to nutrients and regulate the timing of steroid hormone biosynthesis in Drosophila. Nat. Commun. 5: 5778 10.1038/ncomms6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimell M., Pan X., Martin F. A., Ghosh A. C., Leopold P. et al. , 2018. Prothoracicotropic hormone modulates environmental adaptive plasticity through the control of developmental timing. Development 145: dev159699. 10.1242/dev.159699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Ho L. L., and Lai Z. C., 2008. The mob as tumor suppressor gene is essential for early development and regulates tissue growth in Drosophila. Genetics 178: 957–965. 10.1534/genetics.107.081570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton A. W., 2005. Body-size regulation: combining genetics and physiology. Curr. Biol. 15: R825–R827. 10.1016/j.cub.2005.10.006 [DOI] [PubMed] [Google Scholar]
- Shingleton A. W., 2010. The regulation of organ size in Drosophila: physiology, plasticity, patterning and physical force. Organogenesis 6: 76–87. 10.4161/org.6.2.10375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton A. W., Das J., Vinicius L., and Stern D. L., 2005. The temporal requirements for insulin signaling during development in Drosophila. PLoS Biol. 3: e289 10.1371/journal.pbio.0030289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton A. W., Estep C. M., Driscoll M. V., and Dworkin I., 2009. Many ways to be small: different environmental regulators of size generate distinct scaling relationships in Drosophila melanogaster. Proc. Biol. Sci. 276: 2625–2633. 10.1098/rspb.2008.1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingleton A. W., Masandika J. R., Thorsen L. S., Zhu Y., and Mirth C. K., 2017. The sex-specific effects of diet quality vs. quantity on morphology in Drosophila melanogaster. R. Soc. Open Sci. 4: 170375 10.1098/rsos.170375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegmund T., and Korge G., 2001. Innervation of the ring gland of Drosophila melanogaster. J. Comp. Neurol. 431: 481–491. [DOI] [PubMed] [Google Scholar]
- Simón-Carrasco L., Jiménez G., Barbacid M., and Drosten M., 2018. The Capicua tumor suppressor: a gatekeeper of Ras signaling in development and cancer. Cell Cycle 17: 702–711. 10.1080/15384101.2018.1450029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson P., and Scheinderman H. A., 1975. Isolation of temperature sensitive mutations blocking clone development in Drosophila melanogaster, and effects of a temperature sensitive cell lethal mutation on pattern formation in imaginal disks. Wihelm Roux Arch. Dev. Biol. 178: 247–275. 10.1007/BF00848432 [DOI] [PubMed] [Google Scholar]
- Simpson P., Berreur P., and Berreur-Bonnenfant J., 1980. The initiation of pupariation in Drosophila: dependence on growth of the imaginal discs. J. Embryol. Exp. Morphol. 57: 155–165. [PubMed] [Google Scholar]
- Sisk C. L., and Foster D. L., 2004. The neural basis of puberty and adolescence. Nat. Neurosci. 7: 1040–1047. 10.1038/nn1326 [DOI] [PubMed] [Google Scholar]
- Slaidina M., Delanoue R., Gronke S., Partridge L., and Leopold P., 2009. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev. Cell 17: 874–884. 10.1016/j.devcel.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg J. A. E., Birse R. T., and Nässel D. R., 2011. Insulin production and signaling in renal tubules of Drosophila is under control of tachykinin-related peptide and regulates stress resistance. PLoS One 6: e19866 10.1371/journal.pone.0019866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q., and Gilbert L. I., 1994. S6 phosphorylation results from prothoracicotropic hormone stimulation of insect prothoracic glands: a role for S6 kinase. Dev. Genet. 15: 332–338. 10.1002/dvg.1020150404 [DOI] [PubMed] [Google Scholar]
- Song W., Cheng D., Hong S., Sappe B., Hu Y. et al. , 2017. Midgut-derived activin regulates glucagon-like action in the fat body and glycemic control. Cell Metab. 25: 386–399. 10.1016/j.cmet.2017.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel C. G. H., and Davey K. G., 1985. Integration in the insect endocrine system, pp. 1–36 in Comprehensive Insect Physiology, Biochemistry and Pharmacology, edited by Kerkut G. A., and Gilbert L. I.. Pergamon Press, Oxford. [Google Scholar]
- Steiger D., Furrer M., Schwinkendorf D., and Gallant P., 2008. Max-independent functions of Myc in Drosophila melanogaster. Nat. Genet. 40: 1084–1091. 10.1038/ng.178 [DOI] [PubMed] [Google Scholar]
- Stein J., Milewski W. M., Hara M., Steiner D. F., and Dey A., 2011. GSK-3 inactivation or depletion promotes beta-cell replication via down regulation of the CDK inhibitor, p27 (Kip1). Islets 3: 21–34. 10.4161/isl.3.1.14435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieper B. C., Kupershtok M., Driscoll M. V., and Shingleton A. W., 2008. Imaginal discs regulate developmental timing in Drosophila melanogaster. Dev. Biol. 321: 18–26. 10.1016/j.ydbio.2008.05.556 [DOI] [PubMed] [Google Scholar]
- Stocker H., Radimerski T., Schindelholz B., Wittwer F., Belawat P. et al. , 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5: 559–565. 10.1038/ncb995 [DOI] [PubMed] [Google Scholar]
- Sudhakar S. R., Pathak H., Rehman N., Fernandes J., Vishnu S. et al. , 2020. Insulin signalling elicits hunger-induced feeding in Drosophila. Dev. Biol. 459: 87–99. 10.1016/j.ydbio.2019.11.013 [DOI] [PubMed] [Google Scholar]
- Sun J., Liu C., Bai X., Li X., Li J. et al. , 2017. Drosophila FIT is a protein-specific satiety hormone essential for feeding control. Nat. Commun. 8: 14161 10.1038/ncomms14161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Fang Y., Yoon M. S., Zhang C., Roccio M. et al. , 2008. Phospholipase D1 is an effector of Rheb in the mTOR pathway. Proc. Natl. Acad. Sci. USA 105: 8286–8291. 10.1073/pnas.0712268105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung E. J., Ryuda M., Matsumoto H., Uryu O., Ochiai M. et al. , 2017. Cytokine signaling through Drosophila Mthl10 ties lifespan to environmental stress. Proc. Natl. Acad. Sci. USA 114: 13786–13791. 10.1073/pnas.1712453115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sustar A., and Schubiger G., 2005. A transient cell cycle shift in Drosophila imaginal disc cells precedes multipotency. Cell 120: 383–393. 10.1016/j.cell.2004.12.008 [DOI] [PubMed] [Google Scholar]
- Swarup S., and Verheyen E. M., 2012. Wnt/Wingless signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 4: a007930. 10.1101/cshperspect.a007930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamillo A., Sanchez J., Cantera R., Perez C., Martin D. et al. , 2008. Smt3 is required for Drosophila melanogaster metamorphosis. Development 135: 1659–1668. 10.1242/dev.020685 [DOI] [PubMed] [Google Scholar]
- Talamillo A., Herboso L., Pirone L., Perez C., Gonzalez M. et al. , 2013. Scavenger receptors mediate the role of SUMO and Ftz-f1 in Drosophila steroidogenesis. PLoS Genet. 9: e1003473 10.1371/journal.pgen.1003473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H. Y., Smith-Caldas M. S., Driscoll M. V., Salhadar S., and Shingleton A. W., 2011. FOXO regulates organ-specific phenotypic plasticity in Drosophila. PLoS Genet. 7: e1002373 10.1371/journal.pgen.1002373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N., Ito N., Dickson B. J., Treisman J. E., and Hariharan I. K., 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105: 345–355. 10.1016/S0092-8674(01)00332-4 [DOI] [PubMed] [Google Scholar]
- Tapon N., Harvey K. F., Bell D. W., Wahrer D. C. R., Schiripo T. A. et al. , 2002. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell 110: 467–478. 10.1016/S0092-8674(02)00824-3 [DOI] [PubMed] [Google Scholar]
- Tatebe H., and Shiozaki K., 2017. Evolutionary conservation of the components in the TOR signaling pathways. Biomolecules 7: 77 10.3390/biom7040077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A. R., Fingar D. C., Manning B. D., Kwiatkowski D. J., Cantley L. C. et al. , 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 99: 13571–13576. 10.1073/pnas.202476899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A. R., Manning B. D., Roux P. P., Cantley L. C., and Blenis J., 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13: 1259–1268. 10.1016/S0960-9822(03)00506-2 [DOI] [PubMed] [Google Scholar]
- Teleman A. A., Chen Y. W., and Cohen S. M., 2005. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 19: 1844–1848. 10.1101/gad.341505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teleman A. A., Hietakangas V., Sayadian A. C., and Cohen S. M., 2008. Nutritional control of protein biosynthetic capacity by insulin via Myc in Drosophila. Cell Metab. 7: 21–32. 10.1016/j.cmet.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Tennessen J. M., and Thummel C. S., 2011. Coordinating growth and maturation - insights from Drosophila. Curr. Biol. 21: R750–R757. 10.1016/j.cub.2011.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texada M. J., Jorgensen A. F., Christensen C. F., Koyama T., Malita A. et al. , 2019a A fat-tissue sensor couples growth to oxygen availability by remotely controlling insulin secretion. Nat. Commun. 10: 1955 10.1038/s41467-019-09943-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texada M. J., Malita A., Christensen C. F., Dall K. B., Faergeman N. J. et al. , 2019b Autophagy-mediated cholesterol trafficking controls steroid production. Dev. Cell 48: 659–671.e4. 10.1016/j.devcel.2019.01.007 [DOI] [PubMed] [Google Scholar]
- Texada M. J., Malita A., and Rewitz K., 2019c Autophagy regulates steroid production by mediating cholesterol trafficking in endocrine cells. Autophagy 15: 1478–1480. 10.1080/15548627.2019.1617608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao D. T., Seto H., and Yamaguchi M., 2008. Drosophila Myc is required for normal DREF gene expression. Exp. Cell Res. 314: 184–192. 10.1016/j.yexcr.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Thompson B. J., and Cohen S. M., 2006. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell 126: 767–774. 10.1016/j.cell.2006.07.013 [DOI] [PubMed] [Google Scholar]
- Tiebe M., Lutz M., De La Garza A., Buechling T., Boutros M. et al. , 2015. REPTOR and REPTOR-BP regulate organismal metabolism and transcription downstream of TORC1. Dev. Cell 33: 272–284. 10.1016/j.devcel.2015.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toschi A., Lee E., Xu L., Garcia A., Gadir N. et al. , 2009. Regulation of mTORC1 and mTORC2 complex assembly by phosphatidic acid: competition with rapamycin. Mol. Cell. Biol. 29: 1411–1420. 10.1128/MCB.00782-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truman J. W., 1972. Physiology of insect rhythms. 1. Circadian organization of endocrine events underlying molting cycle of larval tobacco hornworms. J. Exp. Biol. 57: 805–820. [DOI] [PubMed] [Google Scholar]
- Tseng A. S., Tapon N., Kanda H., Cigizoglu S., Edelmann L. et al. , 2007. Capicua regulates cell proliferation downstream of the receptor tyrosine kinase/ras signaling pathway. Curr. Biol. 17: 728–733. 10.1016/j.cub.2007.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K., Poulin F., Kohara M., DeMaria C. T., Cheng A. et al. , 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E–BP1. Nat. Med. 7: 1128–1132. 10.1038/nm1001-1128 [DOI] [PubMed] [Google Scholar]
- Tsun Z. Y., Bar-Peled L., Chantranupong L., Zoncu R., Wang T. et al. , 2013. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 52: 495–505. 10.1016/j.molcel.2013.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uryu O., Ou Q., Komura-Kawa T., Kamiyama T., Iga M. et al. , 2018. Cooperative control of ecdysone biosynthesis in Drosophila by transcription factors Séance, Ouija Board, and Molting Defective. Genetics 208: 605–622. 10.1534/genetics.117.300268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallejo D. M., Juarez-Carreno S., Bolivar J., Morante J., and Dominguez M., 2015. A brain circuit that synchronizes growth and maturation revealed through Dilp8 binding to Lgr3. Science 350: aac6767 10.1126/science.aac6767 [DOI] [PubMed] [Google Scholar]
- Vander Haar E., Lee S.-I., Bandhakavi S., Griffin T. J., and Kim D.-H., 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat. Cell Biol. 9: 316–323. 10.1038/ncb1547 [DOI] [PubMed] [Google Scholar]
- Van Hiel M. B., Vandersmissen H. P., Proost P., and Vanden Broeck J., 2015. Cloning, constitutive activity and expression profiling of two receptors related to relaxin receptors in Drosophila melanogaster. Peptides 68: 83–90. 10.1016/j.peptides.2014.07.014 [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M., de Hoogt R., Hermans C., Nellist M., Janssen B. et al. , 1997. Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science 277: 805–808. 10.1126/science.277.5327.805 [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M., Nellist M., Nagelkerken B., Cheadle J., Snell R. et al. , 1998. Interaction between hamartin and tuberin, the TSC1 and TSC2 gene products. Hum. Mol. Genet. 7: 1053–1057. 10.1093/hmg/7.6.1053 [DOI] [PubMed] [Google Scholar]
- Veenstra J. A., Agricola H. J., and Sellami A., 2008. Regulatory peptides in fruit fly midgut. Cell Tissue Res. 334: 499–516. 10.1007/s00441-008-0708-3 [DOI] [PubMed] [Google Scholar]
- Veenstra J. A., Rombauts S., and Grbic M., 2012. In silico cloning of genes encoding neuropeptides, neurohormones and their putative G-protein coupled receptors in a spider mite. Insect Biochem. Mol. Biol. 42: 277–295. 10.1016/j.ibmb.2011.12.009 [DOI] [PubMed] [Google Scholar]
- Verdu J., Buratovich M. A., Wilder E. L., and Birnbaum M. J., 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat. Cell Biol. 1: 500–506. 10.1038/70293 [DOI] [PubMed] [Google Scholar]
- Verghese S., Bedi S., and Kango-Singh M., 2012. Hippo signalling controls Dronc activity to regulate organ size in Drosophila. Cell Death Differ. 19: 1664–1676. 10.1038/cdd.2012.48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veverka V., Crabbe T., Bird I., Lennie G., Muskett F. W. et al. , 2008. Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR. Oncogene 27: 585–595. 10.1038/sj.onc.1210693 [DOI] [PubMed] [Google Scholar]
- Wang L., Harris T. E., Roth R. A., and Lawrence J. C. Jr, 2007. PRAS40 regulates mTORC1 kinase activity by functioning as a direct inhibitor of substrate binding. J. Biol. Chem. 282: 20036–20044. 10.1074/jbc.M702376200 [DOI] [PubMed] [Google Scholar]
- Wang L., Harris T. E., and Lawrence J. C. Jr, 2008a Regulation of proline-rich Akt substrate of 40 kDa (PRAS40) function by mammalian target of rapamycin complex 1 (mTORC1)-mediated phosphorylation. J. Biol. Chem. 283: 15619–15627. 10.1074/jbc.M800723200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tsun Z. Y., Wolfson R. L., Shen K., Wyant G. A. et al. , 2015. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 347: 188–194. 10.1126/science.1257132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Blumhagen R., Lao U., Kuo Y., and Edgar B. A., 2012. LST8 regulates cell growth via target-of-rapamycin complex 2 (TORC2). Mol. Cell. Biol. 32: 2203–2213. 10.1128/MCB.06474-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Fonseca B. D., Tang H., Liu R., Elia A. et al. , 2008b Re-evaluating the roles of proposed modulators of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 283: 30482–30492. 10.1074/jbc.M803348200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. T., Petryk A., Marques G., Jarcho M., Parvy J. P. et al. , 2002. Molecular and biochemical characterization of two P450 enzymes in the ecdysteroidogenic pathway of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 99: 11043–11048. 10.1073/pnas.162375799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren J. T., Petryk A., Marques G., Parvy J. P., Shinoda T. et al. , 2004. Phantom encodes the 25-hydroxylase of Drosophila melanogaster and Bombyx mori: a P450 enzyme critical in ecdysone biosynthesis. Insect Biochem. Mol. Biol. 34: 991–1010. 10.1016/j.ibmb.2004.06.009 [DOI] [PubMed] [Google Scholar]
- Warren J. T., Yerushalmi Y., Shimell M. J., O’Connor M. B., Restifo L. L. et al. , 2006. Discrete pulses of molting hormone, 20-hydroxyecdysone, during late larval development of Drosophila melanogaster: correlations with changes in gene activity. Dev. Dyn. 235: 315–326. 10.1002/dvdy.20626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson K. L., Chou M. M., Blenis J., Gelbart W. M., and Erikson R. L., 1996. A Drosophila gene structurally and functionally homologous to the mammalian 70-kDa s6 kinase gene. Proc. Natl. Acad. Sci. USA 93: 13694–13698. 10.1073/pnas.93.24.13694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J., Grim J. E., Harper J. W. et al. , 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. USA 101: 9085–9090 [corrigenda: Proc. Natl. Acad. Sci. USA 103: 504 (2006)]. 10.1073/pnas.0402770101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigglesworth V. B., 1940. The determination of characters at metamorphosis in Rhodnius prolixus. J. Exp. Biol. 17: 201–222. [Google Scholar]
- Wigglesworth V. B., 1964. The hormonal regulation of growth and reproduction in insects, pp. 247–336 in Advances in Insect Physiology, edited by Beament J. W., Academic Press, New York. [Google Scholar]
- Wolfson R. L., and Sabatini D. M., 2017. The dawn of the age of amino acid sensors for the mTORC1 pathway. Cell Metab. 26: 301–309. 10.1016/j.cmet.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Liu Y., Zheng Y., Dong J., and Pan D., 2008. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev. Cell 14: 388–398. 10.1016/j.devcel.2008.01.007 [DOI] [PubMed] [Google Scholar]
- Wu X., Zhao L., Chen Z., Ji X., Qiao X. et al. , 2016. FLCN maintains the leucine level in lysosome to stimulate mTORC1. PLoS One 11: e0157100 10.1371/journal.pone.0157100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyant G. A., Abu-Remaileh M., Wolfson R. L., Chen W. W., Freinkman E. et al. , 2017. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 171: 642–654.e12. 10.1016/j.cell.2017.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y., Liu Z., and Huang X., 2010. br regulates the expression of the ecdysone biosynthesis gene npc1. Dev. Biol. 344: 800–808. 10.1016/j.ydbio.2010.05.510 [DOI] [PubMed] [Google Scholar]
- Xie X. J., Hsu F. N., Gao X., Xu W., Ni J. Q. et al. , 2015. CDK8-cyclin C mediates nutritional regulation of developmental transitions through the ecdysone receptor in Drosophila. PLoS Biol. 13: e1002207 (erratum PLoS Biol. 13: e1002250). 10.1371/journal.pbio.1002207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L. C., Nunes C., Wang V. R., Saito A., Chen T. et al. , 2020. Distinct nutritional and endocrine regulation of prothoracic gland activities underlies divergent life history strategies in Manduca sexta and Drosophila melanogaster. Insect Biochem. Mol. Biol. 119: 103335 10.1016/j.ibmb.2020.103335 [DOI] [PubMed] [Google Scholar]
- Yamagata K., Sanders L. K., Kaufmann W. E., Yee W., Barnes C. A. et al. , 1994. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J. Biol. Chem. 269: 16333–16339. [PubMed] [Google Scholar]
- Yamanaka N., Rewitz K. F., and O’Connor M. B., 2013a Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58: 497–516. 10.1146/annurev-ento-120811-153608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Romero N. M., Martin F. A., Rewitz K. F., Sun M. et al. , 2013b Neuroendocrine control of Drosophila larval light preference. Science 341: 1113–1116. 10.1126/science.1241210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Marques G., and O’Connor M. B., 2015. Vesicle-mediated steroid hormone secretion in Drosophila melanogaster. Cell 163: 907–919. 10.1016/j.cell.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Jiang X., Li B., Yang H. J., Miller M. et al. , 2017. Mechanisms of mTORC1 activation by RHEB and inhibition by PRAS40. Nature 552: 368–373. 10.1038/nature25023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T. P., Segraves W. A., Oro A. E., McKeown M., and Evans R. M., 1992. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71: 63–72. 10.1016/0092-8674(92)90266-F [DOI] [PubMed] [Google Scholar]
- Ye J., Palm W., Peng M., King B., Lindsten T. et al. , 2015. GCN2 sustains mTORC1 suppression upon amino acid deprivation by inducing Sestrin2. Genes Dev. 29: 2331–2336. 10.1101/gad.269324.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yenush L., Fernandez R., Myers M. G. Jr., Grammer T. C., Sun X. J. et al. , 1996. The Drosophila insulin receptor activates multiple signaling pathways but requires insulin receptor substrate proteins for DNA synthesis. Mol. Cell. Biol. 16: 2509–2517. 10.1128/MCB.16.5.2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiyama T., Namiki T., Mita K., Kataoka H., and Niwa R., 2006. Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133: 2565–2574. 10.1242/dev.02428 [DOI] [PubMed] [Google Scholar]
- Yoshiyama-Yanagawa T., Enya S., Shimada-Niwa Y., Yaguchi S., Haramoto Y. et al. , 2011. The conserved Rieske oxygenase DAF-36/Neverland is a novel cholesterol-metabolizing enzyme. J. Biol. Chem. 286: 25756–25762. 10.1074/jbc.M111.244384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J. S., Lincoln H. C., Kim C. R., Frey J. W., Goodman C. A. et al. , 2014. The role of diacylglycerol kinase zeta and phosphatidic acid in the mechanical activation of mammalian target of rapamycin (mTOR) signaling and skeletal muscle hypertrophy. J. Biol. Chem. 289: 1551–1563. 10.1074/jbc.M113.531392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurgel M. E., Kakad P., Zandawala M., Nassel D. R., Godenschwege T. A. et al. , 2019. A single pair of leucokinin neurons are modulated by feeding state and regulate sleep-metabolism interactions. PLoS Biol. 17: e2006409 10.1371/journal.pbio.2006409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandawala M., Yurgel M. E., Texada M. J., Liao S., Rewitz K. F. et al. , 2018. Modulation of Drosophila post-feeding physiology and behavior by the neuropeptide leucokinin. PLoS Genet. 14: e1007767 10.1371/journal.pgen.1007767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelhof A. C., Ghbeish N., Tsai C., Evans R. M., and McKeown M., 1997. A role for ultraspiracle, the Drosophila RXR, in morphogenetic furrow movement and photoreceptor cluster formation. Development 124: 2499–2506. [DOI] [PubMed] [Google Scholar]
- Zeng J., Huynh N., Phelps B., and King-Jones K., 2020. Snail synchronizes endocycling in a TOR-dependent manner to coordinate entry and escape from endoreplication pausing during the Drosophila critical weight checkpoint. PLoS Biol. 18: e3000609 10.1371/journal.pbio.3000609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y. P., Liu L., and Zhu Y., 2016. Taotie neurons regulate appetite in Drosophila. Nat. Commun. 7: 13633 10.1038/ncomms13633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Robinson B. S., Xu W., Yang L., Yao B. et al. , 2015. The ecdysone receptor coactivator Taiman links Yorkie to transcriptional control of germline stem cell factors in somatic tissue. Dev. Cell 34: 168–180. 10.1016/j.devcel.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. B., Stallock J. P., Ng J. C., Reinhard C., and Neufeld T. P., 2000. Regulation of cellular growth by the Drosophila target of rapamycin dTOR. Genes Dev. 14: 2712–2724. 10.1101/gad.835000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liu Y., Jiang K., and Jia J., 2020. Hedgehog signaling promotes lipolysis in adipose tissue through directly regulating Bmm/ATGL lipase. Dev. Biol. 457: 128–139. 10.1016/j.ydbio.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Ren F., Zhang Q., Chen Y., Wang B. et al. , 2008. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev. Cell 14: 377–387. 10.1016/j.devcel.2008.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Song W., Li Z., Qian W., Wei L. et al. , 2018. Kruppel homolog 1 represses insect ecdysone biosynthesis by directly inhibiting the transcription of steroidogenic enzymes. Proc. Natl. Acad. Sci. USA 115: 3960–3965. 10.1073/pnas.1800435115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., and Cohen S. M., 2013. The Hippo pathway acts via p53 and microRNAs to control proliferation and proapoptotic gene expression during tissue growth. Biol. Open 2: 822–828. 10.1242/bio.20134317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gao X., Saucedo L. J., Ru B., Edgar B. A. et al. , 2003. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat. Cell Biol. 5: 578–581. 10.1038/ncb999 [DOI] [PubMed] [Google Scholar]
- Zhou X., and Riddiford L. M., 2002. Broad specifies pupal development and mediates the ‘status quo’ action of juvenile hormone on the pupal-adult transformation in Drosophila and Manduca. Development 129: 2259–2269. [DOI] [PubMed] [Google Scholar]
- Ziegler A. B., Maniere G., and Grosjean Y., 2018. JhI-21 plays a role in Drosophila insulin-like peptide release from larval IPCs via leucine transport. Sci. Rep. 8: 1908 10.1038/s41598-018-20394-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Kim K. J., Tran V., Shibutani S. T., Bravo M. J. et al. , 2011. Control of Drosophila endocycles by E2F and CRL4(CDT2). Nature 480: 123–127. 10.1038/nature10579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziosi M., Baena-Lopez L. A., Grifoni D., Froldi F., Pession A. et al. , 2010. dMyc functions downstream of Yorkie to promote the supercompetitive behavior of hippo pathway mutant cells. PLoS Genet. 6: e1001140 10.1371/journal.pgen.1001140 [DOI] [PMC free article] [PubMed] [Google Scholar]