Aspergillus fumigatus is a major fungal pathogen of humans but its two closest relatives, Aspergillus fischeri and Aspergillus oerlinghausenensis, are not. Steenwyk et al. examined whether.....
Keywords: secondary metabolites, specialized metabolism, gliotoxin, chemodiversity, pathogenicity
Abstract
Aspergillus fumigatus is a major human pathogen. In contrast, Aspergillus fischeri and the recently described Aspergillus oerlinghausenensis, the two species most closely related to A. fumigatus, are not known to be pathogenic. Some of the genetic determinants of virulence (or “cards of virulence”) that A. fumigatus possesses are secondary metabolites that impair the host immune system, protect from host immune cell attacks, or acquire key nutrients. To examine whether secondary metabolism-associated cards of virulence vary between these species, we conducted extensive genomic and secondary metabolite profiling analyses of multiple A. fumigatus, one A. oerlinghausenensis, and multiple A. fischeri strains. We identified two cards of virulence (gliotoxin and fumitremorgin) shared by all three species and three cards of virulence (trypacidin, pseurotin, and fumagillin) that are variable. For example, we found that all species and strains examined biosynthesized gliotoxin, which is known to contribute to virulence, consistent with the conservation of the gliotoxin biosynthetic gene cluster (BGC) across genomes. For other secondary metabolites, such as fumitremorgin, a modulator of host biology, we found that all species produced the metabolite but that there was strain heterogeneity in its production within species. Finally, species differed in their biosynthesis of fumagillin and pseurotin, both contributors to host tissue damage during invasive aspergillosis. A. fumigatus biosynthesized fumagillin and pseurotin, while A. oerlinghausenensis biosynthesized fumagillin and A. fischeri biosynthesized neither. These biochemical differences were reflected in sequence divergence of the intertwined fumagillin/pseurotin BGCs across genomes. These results delineate the similarities and differences in secondary metabolism-associated cards of virulence between a major fungal pathogen and its nonpathogenic closest relatives, shedding light onto the genetic and phenotypic changes associated with the evolution of fungal pathogenicity.
FUNGAL diseases impose a clinical, economic, and social burden on humans (Drgona et al. 2014; Vallabhaneni et al. 2016; Benedict et al. 2019). Fungi from the genus Aspergillus are responsible for a considerable fraction of this burden, accounting for >250,000 infections annually, with high mortality rates (Bongomin et al. 2017). Aspergillus infections often result in pulmonary and invasive diseases that are collectively termed aspergillosis. Among Aspergillus species, Aspergillus fumigatus is the primary etiological agent of aspergillosis (Latgé and Chamilos 2019).
Even though A. fumigatus is a major pathogen, its closest relatives are not considered pathogenic (Mead et al. 2019a; Steenwyk et al. 2019; Rokas et al. 2020b). Numerous studies have identified genetic determinants that contribute to A. fumigatus pathogenicity, such as the organism’s ability to grow well at higher temperatures and in hypoxic conditions (Kamei and Watanabe 2005; Tekaia and Latgé 2005; Abad et al. 2010; Grahl et al. 2012). Genetic determinants that contribute to pathogenicity could be conceived as analogous to individual “cards” of a “hand” (set of cards) in a card game—that is, individual determinants are typically insufficient to cause disease but can collectively do so (Casadevall 2007).
A. fumigatus biosynthesizes a cadre of secondary metabolites and several metabolites could be conceived as “cards” of virulence because of their involvement in impairing the host immune system, protecting the fungus from host immune cell attacks, or acquiring key nutrients (Shwab et al. 2007; Losada et al. 2009; Yin et al. 2013; Wiemann et al. 2014; Bignell et al. 2016; Knox et al. 2016; Raffa and Keller 2019; Blachowicz et al. 2020). For example, the secondary metabolite gliotoxin has been shown in A. fumigatus to inhibit the host immune response (Sugui et al. 2007; Spikes et al. 2008). Other secondary metabolites implicated in virulence include fumitremorgin, which inhibits the activity of the breast cancer resistance protein (González-Lobato et al. 2010); verruculogen, which modulates the electrophysical properties of human nasal epithelial cells (Khoufache et al. 2007); trypacidin, which is cytotoxic to lung cells and inhibits phagocytosis (Gauthier et al. 2012; Mattern et al. 2015); pseurotin, which inhibits immunoglobulin E (Ishikawa et al. 2009); and fumagillin, which causes epithelial cell damage (Guruceaga et al. 2018) and impairs the function of neutrophils (Fallon et al. 2010, 2011).
By extension, the metabolic pathways responsible for the biosynthesis of secondary metabolites could also be conceived as components of these secondary metabolism-associated “cards” of virulence. Genes in these pathways are typically organized in contiguous sets termed biosynthetic gene clusters (BGCs) (Keller 2019). BGCs are known to evolve rapidly, and their composition can differ substantially across species and strains (Lind et al. 2015, 2017; de Vries et al. 2017; Kjærbølling et al. 2018, 2020; Rokas et al. 2018, 2020a; Vesth et al. 2018). For example, even though A. fumigatus contains 33 BGCs and Aspergillus fischeri contains 48 BGCs, only 10 of those BGCs appear to be shared between the two species (Mead et al. 2019a). Interestingly, one of the BGCs that is conserved between A. fumigatus and A. fischeri is the gliotoxin BGC and both species have been shown to biosynthesize the secondary metabolite, albeit at different amounts (Knowles et al. 2020). These results suggest that the gliotoxin “card” is part of a winning “hand” that facilitates virulence only in the background of the major pathogen A. fumigatus and not in that of the nonpathogen A. fischeri (Knowles et al. 2020).
To date, such comparisons of BGCs and secondary metabolite profiles among A. fumigatus and closely related nonpathogenic species have been few and restricted to single strains (Mead et al. 2019a; Knowles et al. 2020). However, genetic and phenotypic heterogeneity among strains of a single species is an important consideration when studying Aspergillus pathogenicity (Kowalski et al. 2016, 2019; Keller 2017; Ries et al. 2019; Blachowicz et al. 2020; Bastos et al. 2020; Drott et al. 2020; dos Santos et al. 2020; Steenwyk et al. 2020). Examination of multiple strains of A. fumigatus and close relatives—including the recently described closest known relative of A. fumigatus, Aspergillus oerlinghausenensis, whose virulence has yet to be examined but which is not thought to be a human pathogen (Houbraken et al. 2016) and has never been associated with human infections—will increase our understanding of the A. fumigatus secondary metabolism-associated “cards” of virulence.
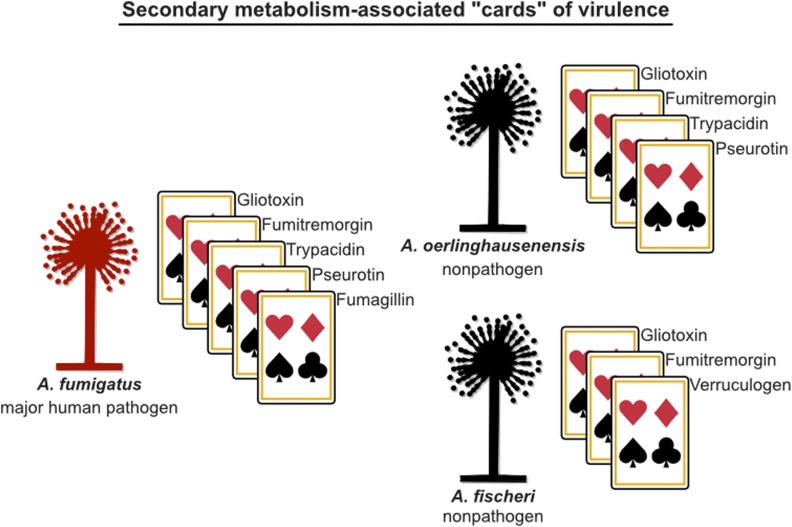
To gain insight into the genomic and chemical similarities and differences in secondary metabolism among A. fumigatus and nonpathogenic close relatives, we characterized variation in BGCs and secondary metabolites produced by A. fumigatus and nonpathogenic close relatives. To do so, we first sequenced and assembled A. oerlinghausenensis CBS 139183T as well as A. fischeri strains NRRL 4585 and NRRL 4161 and analyzed them together with four A. fumigatus and three additional A. fischeri publicly available genomes. We also characterized the secondary metabolite profiles of three A. fumigatus, one A. oerlinghausenensis, and three A. fischeri strains. We observed both variation and conservation among species- and strain-level BGCs and secondary metabolites. We found that the biosynthesis of the secondary metabolites gliotoxin and fumitremorgin, which are both known to interact with mammalian cells (Yamada et al. 2000; González-Lobato et al. 2010; Li et al. 2012; Raffa and Keller 2019), as well as their BGCs, were conserved among pathogenic and nonpathogenic strains. Interestingly, we found only A. fischeri strains, but not A. fumigatus strains, biosynthesized verruculogen, which changes the electrophysical properties of human nasal epithelial cells (Khoufache et al. 2007). Similarly, we found that both A. fumigatus and A. oerlinghausenensis biosynthesized fumagillin and trypacidin, whose effects include broad suppression of the immune response system and lung cell damage (Ishikawa et al. 2009; Fallon et al. 2010, 2011; Gauthier et al. 2012), but A. fischeri did not. Taken together, these results reveal that nonpathogenic close relatives of A. fumigatus also produce some, but not all, of the secondary metabolism-associated cards of virulence known in A. fumigatus. Further investigation of the similarities and differences among A. fumigatus and close nonpathogenic relatives may provide additional insight into the “hand of cards” that enabled A. fumigatus to evolve into a deadly pathogen.
Materials and Methods
Strain acquisition, DNA extraction, and sequencing
Two strains of A. fischeri (NRRL 4161 and NRRL 4585) were acquired from the Northern Regional Research Laboratory (NRRL) at the National Center for Agricultural Utilization Research in Peoria, Illinois, while one strain of A. oerlinghausenensis (CBS 139183T) was acquired from the Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands. These strains were grown in 50 ml of liquid yeast extract soy peptone dextrose (YESD) medium. After ∼7 days of growth on an orbital shaker (100 rpm) at room temperature, the mycelium was harvested by filtering the liquid media through a Corning, 150 ml bottle top, 0.22 µm sterile filter and washed with autoclaved distilled water. All subsequent steps of DNA extraction from the mycelium were performed following protocols outlined previously (Mead et al. 2019b). The genomic DNA from these three strains was sequenced using a NovaSeq S4 at the Vanderbilt Technologies for Advanced Genomes facility (Nashville, Tennessee) using paired-end sequencing (150 bp) strategy with the Illumina TruSeq library kit.
Genome assembly, quality assessment, and annotation
To assemble and annotate the three newly sequenced genomes, we first quality-trimmed raw sequence reads using Trimmomatic, v0.36 (Bolger et al. 2014) using parameters described elsewhere (ILLUMINACLIP:TruSeq3-PE.fa:2:30:10, leading:10, trailing:10, slidingwindow:4:20, minlen:50; Steenwyk and Rokas 2017). The resulting paired and unpaired quality-trimmed reads were used as input to the SPAdes, v3.11.1 (Bankevich et al. 2012), genome assembly algorithm with the “careful” parameter and the “cov-cutoff” set to “auto”.
We evaluated the quality of our newly assembled genomes, using metrics based on continuity of assembly and gene-content completeness. To evaluate genome assemblies by scaffold size, we calculated the N50 of each assembly (or the shortest contig among the longest contigs that account for 50% of the genome assembly’s length) (Yandell and Ence 2012). To determine gene-content completeness, we implemented the BUSCO, v2.0.1 (Waterhouse et al. 2018), pipeline using the “genome” mode. In this mode, the BUSCO pipeline examines assembly contigs for the presence of near-universally single copy orthologous genes (hereafter referred to as BUSCO genes) using a predetermined database of orthologous genes from the OrthoDB, v9 (Waterhouse et al. 2013). We used the OrthoDB database for Pezizomycotina (3156 BUSCO genes). Each BUSCO gene is determined to be present in a single copy, as duplicate sequences, fragmented, or missing. Our analyses indicate the newly sequenced and assembled genomes have high gene-content completeness and assembly continuity (average percent presence of BUSCO genes: 98.80 ± 0.10%; average N50: 451,294.67 ± 9,696.11; Supplemental Material, Figure S1). These metrics suggest these genomes are suitable for comparative genomic analyses.
To predict gene boundaries in the three newly sequenced genomes, we used the MAKER, v2.31.10, pipeline (Holt and Yandell 2011), which creates consensus predictions from the collective evidence of multiple ab initio gene prediction software. Specifically, we created consensus predictions from SNAP, v2006-07-28 (Korf 2004), and AUGUSTUS, v3.3.2 (Stanke and Waack 2003), after training each algorithm individually on each genome. To do so, we first ran MAKER using protein evidence clues from five different publicly available annotations of Aspergillus fungi from section Fumigati. Specifically, we used protein homology clues from A. fischeri NRRL 181 (GenBank accession: GCA_000149645.2), A. fumigatus Af293 (GenBank accession: GCA_000002655.1), Aspergillus lentulus IFM 54703 (GenBank accession: GCA_001445615.1), Aspergillus novofumigatus IBT 16806 (GenBank accession: GCA_002847465.1), and Aspergillus udagawae IFM 46973 (GenBank accession: GCA_001078395.1). The resulting gene predictions were used to train SNAP. MAKER was then rerun using the resulting training results. Using the SNAP trained gene predictions, we trained AUGUSTUS. A final set of gene boundary predictions were obtained by rerunning MAKER with the training results from both SNAP and AUGUSTUS.
To supplement our data set of newly sequenced genomes, we obtained publicly available ones. Specifically, we obtained genomes and annotations for A. fumigatus Af293 (GenBank accession: GCA_000002655.1), A. fumigatus CEA10 (strain synonym: CBS 144.89/ FGSC A1163; GenBank accession: GCA_000150145.1), A. fumigatus HMR AF 270 GenBank accession: GCA_002234955.1), A. fumigatus Z5 (GenBank accession: GCA_001029325.1), A. fischeri NRRL 181 (GenBank accession: GCA_000149645.2). We also obtained assemblies of the recently published A. fischeri genomes for strains IBT 3003 and IBT 3007 (Zhao et al. 2019), which lacked annotations. We annotated the genome of each strain individually using MAKER with the SNAP and AUGUSTUS training results from a close relative of both strains: A. fischeri NRRL 4161. Altogether, our final data set contained a total of 10 genomes from three species: four A. fumigatus strains, one A. oerlinghausenensis strain, and five A. fischeri strains (Table 1).
Table 1. Species and strains used in the present study.
| Genus and species | Strain | Environmental/Clinical | Genomic analysis | Secondary metabolite profiling | Reference |
|---|---|---|---|---|---|
| Aspergillus oerlinghausenensis | CBS 139183T | Environmental | + | + | This study |
| Aspergillus fischeri | NRRL 4585 | Environmental | + | + | This study |
| A. fischeri | NRRL 4161 | Unknown | + | + | This study |
| A. fischeri | NRRL 181 | Environmental | + | + | Fedorova et al. (2008) |
| A. fischeri | IBT 3007 | Environmental | + | − | Zhao et al. (2019) |
| A. fischeri | IBT 3003 | Environmental | + | − | Zhao et al. (2019) |
| Aspergillus fumigatus | Af293 | Clinical | + | + | Nierman et al. (2005) |
| A. fumigatus | CEA10/ CEA17 | Clinical | + | + | Fedorova et al. (2008) |
| A. fumigatus | HMR AF 270 | Clinical | + | − | BioSample: SAMN07177964 |
| A. fumigatus | Z5 | Environmental | + | − | Miao et al. (2015) |
“+” and “−” indicate if BGCs and secondary metabolite profiling was conducted on a particular strain. More specifically “+” indicates the strain was analyzed whereas “−” indicates that the strain was not analyzed.
Maximum likelihood phylogenetics and Bayesian estimation of divergence times
To reconstruct the evolutionary history among the 10 Aspergillus genomes, we implemented a recently developed pipeline (Steenwyk et al. 2019) that relies on the concatenation-approach to phylogenomics (Rokas et al. 2003) and has been successfully used in reconstructing species-level relationships among Aspergillus and Penicillium fungi (Bodinaku et al. 2019; Steenwyk et al. 2019). The first step in the pipeline is to identify single-copy orthologous genes in the genomes of interest, which are ultimately concatenated into a larger phylogenomic data matrix. To identify single copy BUSCO genes across all 10 Aspergillus genomes, we used the BUSCO pipeline with the Pezizomycotina database as described above. We identified 3041 BUSCO genes present at a single copy in all 10 Aspergillus genomes, and created multi-FASTA files for each BUSCO gene that contained the protein sequences for all 10 taxa. The protein sequences of each BUSCO gene were individually aligned using Mafft, v7.4.02 (Katoh and Standley 2013), with the same parameters as described elsewhere (Steenwyk et al. 2019). Nucleotide sequences were then mapped onto the protein sequence alignments using a custom Python, v3.5.2 (https://www.python.org/) script with BioPython v1.7 (Cock et al. 2009). The resulting codon-based alignments were trimmed using trimAl, v1.2.rev59 (Capella-Gutierrez et al. 2009), with the “gappyout” parameter. The resulting trimmed nucleotide alignments were concatenated into a single matrix of 5,602,272 sites and was used as input into IQ-TREE, v1.6.11 (Nguyen et al. 2015). The best-fitting model of substitutions for the entire matrix was determined using Bayesian information criterion values (Kalyaanamoorthy et al. 2017). The best-fitting model was a general time-reversible model with empirical base frequencies that allowed for a proportion of invariable sites and a discrete Gamma model with four rate categories (GTR+I+F+G4) (Tavaré 1986; Yang 1994, 1996; Vinet and Zhedanov 2011). To evaluate bipartition support, we used 5000 ultrafast bootstrap approximations (Hoang et al. 2018).
To estimate divergence times among the 10 Aspergillus genomes, we used the concatenated data matrix and the resulting maximum likelihood phylogeny from the previous steps as input to Bayesian approach implemented in MCMCTree from the PAML package, v4.9d (Yang 2007). First, we estimated the substitution rate across the data matrix using a “GTR+G” model of substitutions (model = 7), a strict clock model, and the maximum likelihood phylogeny rooted on the clade of A. fischeri strains. We imposed a root age of 3.69 million years ago (MYA) according to results from recent divergence time estimates of the split between A. fischeri and A. fumigatus (Steenwyk et al. 2019). We estimated the substitution rate to be 0.005 substitutions per 1 million years. Next, the likelihood of the alignment was approximated using a gradient and Hessian matrix. To do so, we used previously established time constraints for the split between A. fischeri and A. fumigatus (1.85–6.74 MYA) (Steenwyk et al. 2019). Lastly, we used the resulting gradient and Hessian matrix, the rooted maximum likelihood phylogeny, and the concatenated data matrix to estimate divergence times using a relaxed molecular clock (model = 2). We specified the substitution rate prior based on the estimated substitution rate (rgene_gamma = 1 186.63). The “sigma2_gamma” and “finetune” parameters were set to ‘1 4.5′ and “1”, respectively. To collect a high-quality posterior probability distribution, we ran a total of 5.1 million iterations during MCMC analysis, which is 510 times greater than the minimum recommendations (Raftery and Lewis 1995). Our sampling strategy across the 5.1 million iterations was to discard the first 100,000 results followed by collecting a sample every 500th iteration until a total of 10,000 samples were collected.
Identification of gene families and analyses of putative biosynthetic gene clusters
To identify gene families across the 10 Aspergillus genomes, we used a Markov clustering approach. Specifically, we used OrthoFinder, v2.3.8 (Emms and Kelly 2019). OrthoFinder first conducts a blast all-vs-all using the protein sequences of all 10 Aspergillus genomes and NCBI’s Blast+, v2.3.0 (Camacho et al. 2009), software. After normalizing blast bit scores, genes are clustered into discrete orthogroups using a Markov clustering approach (van Dongen 2000). We clustered genes using an inflation parameter of 1.5. The resulting orthogroups were used proxies for gene families.
To identify putative BGCs, we used the gene boundaries predictions from the MAKER software as input into antiSMASH, v4.1.0 (Weber et al. 2015). To identify homologous BGCs across the 10 Aspergillus genomes, we used the software BiG-SCAPE, v20181005 (Navarro-Muñoz et al. 2020). Based on the Jaccard Index of domain types, sequence similarity among domains, and domain adjacency, BiG-SCAPE calculates a similarity metric between pairwise combinations of clusters where smaller values indicate greater BGC similarity. BiG-SCAPE’s similarity metric can then be used as an edge-length in network analyses of cluster similarity. We evaluated networks using an edge-length cutoff from 0.1 to 0.9 with a step of 0.1 (Figure S3). We found networks with an edge-length cutoff of 0.4–0.6 to be similar and based further analyses on a cutoff of 0.5. Because BiG-SCAPE inexplicably split the gliotoxin BGC of the A. fumigatus Af293 strain into two cluster families even though the BGC was highly similar to the gliotoxin BGCs of all other strains, we supplemented BiG-SCAPE’s approach to identifying homologous BGCs with visualize inspection of microsyteny and blast-based analyses using NCBI’s BLAST+, v2.3.0 (Camacho et al. 2009) for BGCs of interest. Similar sequences in microsynteny analyses were defined as at least 100 bp in length, at least 30% similarity, and an expectation value threshold of 0.01. Lastly, to determine if any BGCs have been previously linked to secondary metabolites, we cross referenced BGCs and BGC families with those found in the MIBiG database (Kautsar et al. 2020) as well as previously published A. fumigatus BGCs (Table S2). BGCs not associated with secondary metabolites were considered to likely encode for unknown compounds.
Identification and characterization of secondary metabolite production
General experimental procedures:
The 1H NMR data were collected using a JOEL ECS-400 spectrometer, which was equipped with a JOEL normal geometry broadband Royal probe, and a 24-slot autosampler, and operated at 400 MHz. HRESIMS experiments utilized either a Thermo LTQ Orbitrap XL mass spectrometer or a Thermo Q Exactive Plus (Thermo Fisher Scientific); both were equipped with an electrospray ionization source. A Waters Acquity ultraperformance liquid chromatography (UPLC; Waters Corp.) was utilized for both mass spectrometers, using a BEH C18 column (1.7 μm; 50 mm × 2.1 mm) set to a temperature of 40° and a flow rate of 0.3 ml/min. The mobile phase consisted of a linear gradient of CH3CN-H2O (both acidified with 0.1% formic acid), starting at 15% CH3CN and increasing linearly to 100% CH3CN over 8 min, with a 1.5-min hold before returning to the starting condition. The HPLC separations were performed with Atlantis T3 C18 semipreparative (5 μm; 10 × 250 mm) and preparative (5 μm; 19 × 250 mm) columns, at a flow rate of 4.6 ml/min and 16.9 ml/min, respectively, with a Varian Prostar HPLC system equipped with a Prostar 210 pumps and a Prostar 335 photodiode array (PDA) detector, with the collection and analysis of data using Galaxie Chromatography Workstation software. Flash chromatography was performed on a Teledyne ISCO Combiflash Rf 200 and monitored by both evaporative light scattering detector (ELSD) and PDA detectors.
Chemical characterization:
To identify the secondary metabolites that were biosynthesized by A. fumigatus, A. oerlinghausenensis, and A. fischeri, these strains were grown as large-scale fermentations to isolate and characterize the secondary metabolites. To inoculate oatmeal cereal media (Old Fashioned Breakfast Quaker oats), agar plugs from fungal stains grown on potato dextrose agar; Difco were excised from the edge of the Petri dish culture and transferred to separate liquid seed medium that contained 10 ml YESD broth (2% soy peptone, 2% dextrose, and 1% yeast extract; 5 g of yeast extract, 10 g of soy peptone, and 10 g of d-glucose in 500 ml of deionized H2O) and allowed to grow at 23° with agitation at 100 rpm for 3 days. The YESD seed cultures of the fungi were subsequently used to inoculate solid-state oatmeal fermentation cultures, which were either grown at room temperature (∼23° under 12 hr light/dark cycles for 14 days), 30°, or 37°; all growth at the latter two temperatures was carried out in an incubator (VWR International) in the dark over 4 days. The oatmeal cultures were prepared in 250 ml Erlenmeyer flasks that contained 10 g of autoclaved oatmeal (10 g of oatmeal with 17 ml of deionized H2O and sterilized for 15–20 min at 121°). For all fungal strains, three flasks of oatmeal cultures were grown at all three temperatures, except for A. oerlinghausenensis (CBS 139183T) at room temperature and A. fumigatus (Af293) at 37°. For CBS 139183T, the fungal cultures were grown in four flasks, while for Af293 eight flasks were grown in total. The growths of these two strains were performed differently from the rest because larger amounts of extract were required in order to perform detailed chemical characterization.
The cultures were extracted by adding 60 ml of (1:1) MeOH-CHCl3 to each 250 ml flask, chopping thoroughly with a spatula, and shaking overnight (∼16 hr) at ∼100 rpm at room temperature. The culture was filtered in vacuo, and 90 ml CHCl3 and 150 ml H2O were added to the filtrate. The mixture was stirred for 30 min and then transferred to a separatory funnel. The organic layer (CHCl3) was drawn off and evaporated to dryness in vacuo. The dried organic layer was reconstituted in 100 ml of (1:1) MeOH–CH3CN and 100 ml of hexanes, transferred to a separatory funnel, and shaken vigorously. The defatted organic layer (MeOH–CH3CN) was evaporated to dryness in vacuo.
To isolate compounds, the defatted extract was dissolved in CHCl3, absorbed onto Celite 545 (Acros Organics), and fractioned by normal phase flash chromatography using a gradient of hexane-CHCl3-MeOH. A. fischeri strain NRRL 181 was chemically characterized previously (Mead et al. 2019a; Knowles et al. 2019). A. fumigatus strain Af293, grown at 37°, was subjected to a 12 g column at a flow rate of 30 ml/min and 61.0 column volumes, which yielded four fractions. Fraction 2 was further purified via preparative HPLC using a gradient system of 30:70 to 100:0 of CH3CN-H2O with 0.1% formic acid over 40 min at a flow rate of 16.9 ml/min to yield six subfractions. Subfractions 1, 2 and 5, yielded cyclo(l-Pro-l-Leu) (Li et al. 2008) (0.89 mg), cyclo(l-Pro- l-Phe) (Campbell et al. 2009) (0.71 mg), and monomethylsulochrin (Ma et al. 2004) (2.04 mg), which eluted at ∼5.7, 6.3, and 10.7 min, respectively. Fraction 3 was further purified via preparative HPLC using a gradient system of 40:60 to 65:35 of CH3CN-H2O with 0.1% formic acid over 30 min at a flow rate of 16.9 ml/min to yield four subfractions. Subfractions 1 and 2 yielded pseurotin A (Wang et al. 2011) (12.50 mg) and bisdethiobis (methylthio) gliotoxin (Afiyatullov et al. 2005) (13.99 mg), which eluted at ∼7.5 and 8.0 min, respectively.
A. fumigatus strain CEA10, grown at 37°, was subjected to a 4 g column at a flow rate of 18 ml/min and 90.0 column volumes, which yielded five fractions. Fraction 1 was purified via preparative HPLC using a gradient system of 50:50 to 100:0 of CH3CN-H2O with 0.1% formic acid over 45 min at a flow rate of 16.9 ml/min to yield eight subfractions. Subfraction 1, yielded fumagillin (Halász et al. 2000) (1.69 mg), which eluted at ∼18.5 min. Fraction 2 was purified via semipreparative HPLC using a gradient system of 35:65 to 80:20 of CH3CN-H2O with 0.1% formic acid over 30 min at a flow rate of 4.6 ml/min to yield 10 subfractions. Subfraction 5 yielded fumitremorgin C (Kato et al. 2009) (0.25 mg), which eluted at ∼15.5 min. Fraction 3 was purified via preparative HPLC using a gradient system of 40:60 to 100:0 of CH3CN-H2O with 0.1% formic acid over 30 min at a flow rate of 16.9 ml/min to yield nine subfractions. Subfraction 2 yielded pseurotin A (1.64 mg), which eluted at ∼7.3 min.
A. oerlinghausenensis strain CBS 139183T, grown at RT, was subjected to a 4 g column at a flow rate of 18 ml/min and 90 column volumes, which yielded four fractions. Fraction 3 was further purified via preparative HPLC using a gradient system of 35:65 to 70:30 of CH3CN-H2O with 0.1% formic acid over 40 min at a flow rate of 16.9 ml/min to yield 11 subfractions. Subfractions 3 and 10 yielded spiro [5H,10H-dipyrrolo[1,2-a:1′,2′-d]pyrazine-2-(3H),2′-[2H]indole]-3′,5,10(1′H)-trione (Wang et al. 2008) (0.64 mg) and helvolic acid (Zhao et al. 2010) (1.03 mg), which eluted at ∼11.5 and 39.3 min, respectively (see NMR supporting information; figshare: https://doi.org/10.6084/m9.figshare.12055503).
Metabolite profiling by mass spectrometry:
The metabolite profiling by mass spectrometry, also known as dereplication, was performed as stated previously (El-Elimat et al. 2013). Briefly, UPLC-PDA-electrospray ionization high resolution tandem mass spectrometry (UPLC-PDA-HRMS-MS/MS) was utilized to monitor for secondary metabolites across all strains (Af293, CEA10, CEA17, CBS 139183T, NRRL 181, NRRL 4161, and NRRL 4585). Utilizing positive-ionization mode, ACD MS Manager with add-in software IntelliXtract (Advanced Chemistry Development, Inc.; Toronto, Canada) was used for the primary analysis of the UPLC-MS chromatograms. The data from 19 secondary metabolites are provided in the Supporting Information (see Dereplication table; figshare: https://doi.org/10.6084/m9.figshare.12055503), which for each secondary metabolite lists: molecular formula, retention time (RT), UV-absorption maxima, high-resolution full-scan mass spectra, and MS-MS data (top 10 most intense peaks).
Metabolomics analyses:
Principal component analysis (PCA) analysis was performed on the UPLC-MS data. Untargeted UPLC-MS datasets for each sample were individually aligned, filtered, and analyzed using MZmine 2.20 software (https://sourceforge.net/projects/mzmine/) (Pluskal et al. 2010). Peak detection was achieved using the following parameters, A. fumigatus at (Af293, CEA10, and CEA17): noise level (absolute value), 1×106; minimum peak duration, 0.05 min; m/z variation tolerance, 0.05; and m/z intensity variation, 20%; A. fischeri (NRRL 181, NRRL 4161, and NRRL 4585): noise level (absolute value), 1 × 106; minimum peak duration, 0.05 min; m/z variation tolerance, 0.05; and m/z intensity variation, 20%; and all strains (Af293, CEA10, CEA17, CBS 139183T, NRRL 181, NRRL 4161, and NRRL 4585): noise level (absolute value), 7 × 105; minimum peak duration, 0.05 min; m/z variation tolerance, 0.05; and m/z intensity variation, 20%. Peak list filtering and retention time alignment algorithms were used to refine peak detection. The join algorithm integrated all sample profiles into a data matrix using the following parameters: m/z and RT balance set at 10.0 each, m/z tolerance set at 0.001, and RT tolerance set at 0.5 min The resulting data matrix was exported to Excel (Microsoft) for analysis as a set of m/z–RT pairs with individual peak areas detected in triplicate analyses. Samples that did not possess detectable quantities of a given marker ion were assigned a peak area of zero to maintain the same number of variables for all sample sets. Ions that did not elute between 2 and 8 min and/or had an m/z ratio <200 or >800 Da were removed from analysis. Relative SD was used to understand the quantity of variance between the technical replicate injections, which may differ slightly based on instrument variance. A cutoff of 1.0 was used at any given m/z–RT pair across the technical replicate injections of one biological replicate, and if the variance was greater than the cutoff, it was assigned a peak area of zero. Final chemometric analysis, data filtering (Caesar et al. 2018), and PCA was conducted using Sirius, v10.0 (Pattern Recognition Systems AS; Kvalheim et al. 2011), and dendrograms were created with Python. The PCA scores plots were generated using data from either the three individual biological replicates or the averaged biological replicates of the fermentations. Each biological replicate was plotted using averaged peak areas obtained across four replicate injections (technical replicates).
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Sequence reads and associated genome assemblies generated in this project are available in NCBI’s GenBank database under the BioProject PRJNA577646. Additional descriptions of the genomes including predicted gene boundaries are available through figshare (https://doi.org/10.6084/m9.figshare.12055503). The figshare repository is also populated with other data generated from genomic and natural products analysis. Among genomic analyses, we provide information about predicted BGCs, results associated with network-based clustering of BGCs into cluster families, phylogenomic data matrices, and trees. Among natural products analysis, we provide information that supports methods and results, including NMR spectra.
Results
Conservation and diversity of biosynthetic gene clusters within and between species
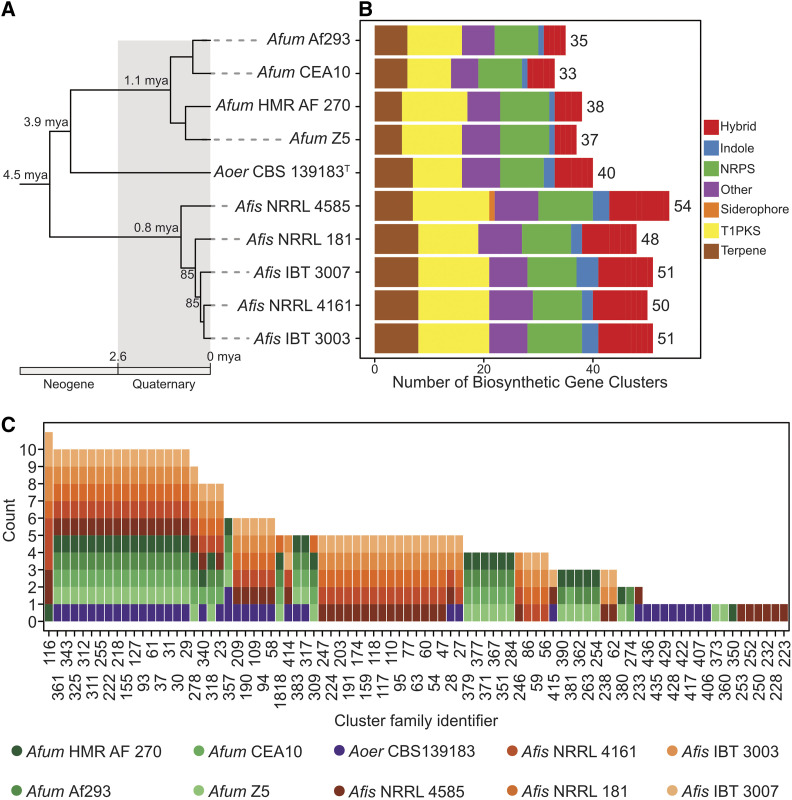
We sequenced and assembled A. oerlinghausenensis CBS 139183T and A. fischeri strains NRRL 4585 and NRRL 4161. Together with publicly available genomes, we analyzed 10 Aspergillus genomes (five A. fischeri strains; four A. fumigatus strains; one A. oerlinghausenensis strain; see Materials and Methods). We found that the newly added genomes were of similar quality to other publicly available draft genomes (average percent presence of BUSCO genes: 98.80 ± 0.10%; average N50: 451,294.67 ± 9696.11; Figure S1). We predicted that A. oerlinghausenensis CBS 139183T, A. fischeri NRRL 4585, and A. fischeri NRRL 4161 have 10,044, 11,152 and 10,940 genes, respectively, numbers similar to publicly available genomes. Lastly, we inferred the evolutionary history of the 10 Aspergillus genomes using a concatenated matrix of 3041 genes (5,602,272 sites) and recapitulated species-level relationships as previously reported (Houbraken et al. 2016). Relaxed molecular clock analyses suggested that A. oerlinghausenensis CBS 139183T diverged from A. fumigatus ∼3.9 (6.4–1.3) MYA and that A. oerlinghausenensis and A. fumigatus split from A. fischeri ∼4.5 (6.8–1.7) MYA (Figure 1A and Figure S2).
Figure 1.
Diverse genetic repertoire of biosynthetic gene clusters and extensive presence and absence polymorphisms between and within species. (A) Genome-scale phylogenomic analysis confirms A. oerlinghausenensis is the closest relative to A. fumigatus. Relaxed molecular clock analyses suggest A. fumigatus, A. oerlinghausenensis, and A. fischeri diverged from one another during the Neogene geologic period. Bipartition support is depicted for internodes that did not have full support. (B) A. fumigatus harbors the lowest number of BGCs compared to its two closest relatives. (C) Network-based clustering of BGCs into cluster families reveal extensive cluster presence and absence polymorphisms between species and strains. Cluster family identifiers are depicted on the x-axis; the number of strains represented in a cluster family are shown on the y-axis; the colors refer to a single strain from each species. Genus and species names are written using the following abbreviations: Afum, A. fumigatus; Aoer, A. oerlinghausenensis; Afis, A. fischeri. Classes of BGCs are written using the following abbreviations: NRPS, nonribosomal peptide synthetase; T1PKS, type I polyketide synthase; Hybrid, a combination of multiple BGC classes.
Examination of the total number of predicted BGCs revealed that A. fischeri has the largest BGC count. Among A. fumigatus, A. oerlinghausenensis, and A. fischeri, we predicted an average of 35.75 ± 2.22, 40, 50.80 ± 2.17 BGCs, respectively, and found they spanned diverse biosynthetic classes (e.g., polyketides, nonribosomal peptides, terpenes, etc.) (Figure 1B). Network-based clustering of BGCs into cluster families (or groups of homologous BGCs) resulted in qualitatively similar networks when we used moderate similarity thresholds (or edge cut-off values; Figure S3A). Using a (moderate) similarity threshold of 0.5, we inferred 88 cluster families of putatively homologous BGCs (Figure 1C).
Examination of BGCs revealed extensive presence and absence polymorphisms within and between species. We identified 17 BGCs that were present in all 10 Aspergillus genomes including the hexadehydroastechrome (HAS) BGC (cluster family 311 or CF311), the neosartoricin BGC (CF61), and other putative BGCs likely encoding unknown products (Figure S3B and Table S1; data available from figshare, https://doi.org/10.6084/m9.figshare.12055503). In contrast, we identified 18 BGCs found in single strains, which likely encode unknown products. Between species, similar patterns of broadly present and species-specific BGCs were observed. For example, we identified 18 BGCs that were present in at least one strain across all species; in contrast, A. fumigatus, A. oerlinghausenensis, and A. fischeri had 16, 8, and 27 BGCs present in at least one strain but absent from the other species, respectively. These results suggest each species has a largely distinct repertoire of BGCs.
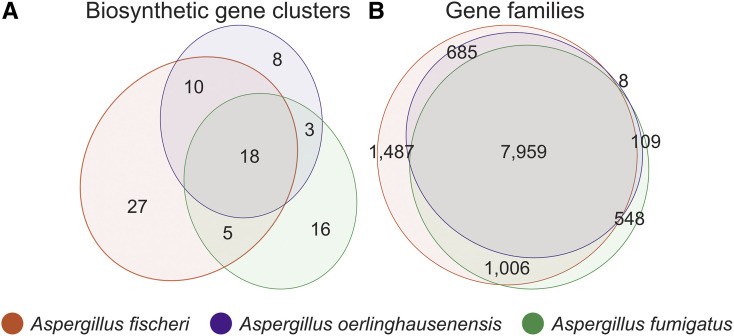
Examination of shared BGCs across species revealed A. oerlinghausenensis CBS139183T and A. fischeri shared more BGCs with each other than either did with A. fumigatus. Surprisingly, we found 10 homologous BGCs between A. oerlinghausenensis CBS 139183T and A. fischeri but only three homologous BGCs shared between A. fumigatus and A. oerlinghausenensis CBS 139183T (Figure 2A and Figure S3C) even though A. oerlinghausenensis is more closely related to A. fumigatus than to A. fischeri (Figure 1A). BGCs shared by A. oerlinghausenensis CBS 139183T and A. fischeri were uncharacterized while BGCs present in both A. fumigatus and A. oerlinghausenensis CBS 139183T included those that encode fumigaclavine and fumagillin/pseurotin. Lastly, to associate each BGC with a secondary metabolite in A. fumigatus Af293, we cross-referenced our list with a publicly available one (Table S2) (Lind et al. 2017). Importantly, all known A. fumigatus Af293 BGCs were represented in our analyses.
Figure 2.
Aspergillus oerlinghausenensis shares more gene families and BGCs with A. fischeri than A. fumigatus. (A) Euler diagram showing species-level shared BGCs. (B) Euler diagram showing species-level shared gene families. In both diagrams, A. oerlinghausenensis shares more gene families or BGCs with A. fischeri than A. fumigatus despite a closer evolutionary relationship. The Euler diagrams show the results for the species-level comparisons, which may be influenced by the unequal numbers of strains used for the three species; strain-level comparisons of BGCs and gene families can be found in Figures 1C and Figure S4, respectively.
At the level of gene families, there were few species-specific gene families in A. oerlinghausenensis (Figure 2B). A. oerlinghausenensis CBS 139183T has only eight species-specific gene families, whereas A. fischeri and A. fumigatus have 1487 and 548 species-specific gene families, respectively. Examination of the best BLAST hits of the eight species-specific gene families suggest that most are hypothetical or uncharacterized fungal genes. To determine if the eight A. oerlinghausenensis CBS 139183T specific gene families were an artifact of using a single representative strain, we conducted and additional ortholog clustering analysis using a single strain of A. fischeri (NRRL 181), a single strain of A. fumigatus (Af293), or a single strain of each species (CBS 139183, NRRL 181, Af293). When using a single strain of A. fischeri or A. fumigatus, there were 23 or 6 gene families unique to each species, respectively. Therefore, the low number of A. oerlinghausenensis-specific gene families likely stems from our use of the genome of a single strain.
Despite a closer evolutionary relationship between A. oerlinghausenensis and A. fumigatus, we found A. oerlinghausenensis shares more gene families with A. fischeri than with A. fumigatus (685 and 109, respectively) suggestive of extensive gene loss in the A. fumigatus stem lineage. Lastly, we observed strain heterogeneity in gene family presence and absence within both A. fumigatus and A. fischeri (Figure S4). For example, the largest intersection that does not include all A. fischeri strains is 493 gene families, which were found in all but one strain, NRRL 181. For A. fumigatus, the largest intersection that does not include all strains is 233 gene families, which were shared by strains Af293 and CEA10.
Within- and between-species variation in secondary metabolite profiles of A. fumigatus and its closest relatives
To gain insight into variation in secondary metabolite profiles within and between species, we profiled A. fumigatus strains Af293, CEA10, and CEA17 (a pyrG1/URA3 derivative of CEA10), A. fischeri strains NRRL 181, NRRL 4585, and NRRL 4161, and A. oerlinghausenensis CBS 139183T for secondary metabolites. Specifically, we used three different procedures, including the isolation and structure elucidation of metabolites, where possible, followed by two different metabolite profiling procedures that use mass spectrometry techniques. Altogether, we isolated and characterized 19 secondary metabolites; 7 from A. fumigatus, 2 from A. oerlinghausenensis, and 10 from A. fischeri (Figure S5). These products encompassed a wide diversity of secondary metabolite classes, such as those derived from polyketide synthases, nonribosomal peptide-synthetases, terpene synthases, and mixed biosynthesis enzymes.
To characterize the secondary metabolites biosynthesized that were not produced in high enough quantity for structural identification through traditional isolation methods, we employed “dereplication” mass spectrometry protocols specific to natural products research on all tested strains at both 30° and 37° (see supporting information, dereplication example; figshare: https://doi.org/10.6084/m9.figshare.12055503) (El-Elimat et al. 2013; Ito and Masubuchi 2014; Gaudêncio and Pereira 2015; Hubert et al. 2017). We found that most secondary metabolites were present across strains of the same species (Table S3); for example, monomethylsulochrin was isolated from A. fumigatus Af293, but, through metabolite profiling, its spectral features were noted also in A. fumigatus strains CEA10 and CEA17. We identified metabolites that were biosynthesized by only one species; for example, pseurotin A was present solely in A. fumigatus strains. Finally, we found several secondary metabolites that were biosynthesized across species, such as fumagillin, which was biosynthesized by A. fumigatus and A. oerlinghausenensis, and fumitremorgin B, which was biosynthesized by strains of both A. oerlinghausenensis and A. fischeri. Together, these analyses suggest that closely related Aspergillus species and strains exhibit variation both within as well as between species in the secondary metabolites produced.
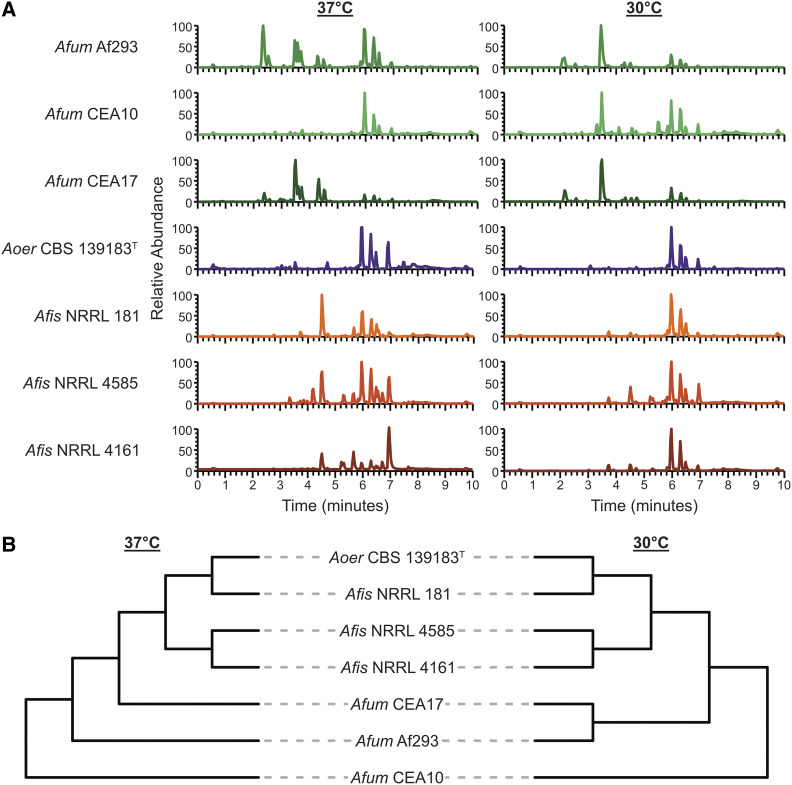
To further facilitate comparisons of secondary metabolite profiles within and between species, we used the 1920 features (i.e., unique m/z – retention time pairs) that were identified from all strains at all temperatures (Figure 3A), to perform hierarchical clustering (Figure 3B) and PCA (Figure S6). Hierarchical clustering at 37° and 30° indicated the chromatogram of A. oerlinghausenensis CBS 139183T is more similar to the chromatogram of A. fischeri than to that of A. fumigatus. PCA results were broadly consistent with the clustering results, but suggested that A. oerlinghausenensis was just as similar to A. fischeri strains as it was to A. fumigatus strains. This difference likely stems from the fact that hierarchical clustering is a total-evidence approach whereas PCA captures most but not all variance in the data (e.g., the two principal components in Figure S6B and S6C capture 84.6% of the total variance). PCA analysis revealed greater variation in secondary metabolite production at 30° Compared to 37° (Figure S6), suggesting there is a more varied response in how BGCs are being utilized at 30°. PCA at both 37° and 30° showed that variation between A. oerlinghausenensis CBS 139183T and A. fischeri strains was largely captured along the second principal component; in contrast, the differences between A. oerlinghausenensis CBS 139183T and A. fumigatus strains are captured along the first principal component (Figure S6, D and E). Taken together, these results suggest that the three A. fischeri strains and A. oerlinghausenensis were the most chemically similar to each other.
Figure 3.
A. oerlinghausenensis and A. fischeri have more similar secondary metabolite profiles than A. fumigatus. (A) UPLC-MS chromatograms of secondary metabolite profiles of A. fumigatus and its closest relatives, A. oerlinghausenensis and A. fischeri at 37° and 30° (left and right, respectively). (B) Hierarchical clustering of chromatograms (1920 total features) reveals A. oerlinghausenensis clusters with A. fischeri and not its closest relative, A. fumigatus at 37° and 30° (left and right, respectively).
In summary, even though A. oerlinghausenensis is phylogenetically more closely related to A. fumigatus than to A. fischeri (Figure 1A), our chemical analyses suggest that the secondary metabolite profile of A. oerlinghausenensis is more similar to the profile of A. fischeri than it is to the profile of A. fumigatus (Figure 3B and Figure S6, B–E). The similarity of secondary metabolite profiles of A. oerlinghausenensis and A. fischeri is consistent with our finding that the genome of A. oerlinghausenensis shares higher numbers of BGCs and gene families with A. fischeri than with A. fumigatus (Figure 2). The broad clustering patterns in secondary metabolite-based plots (Figure S6, B–E) are less robust than, but consistent with, those of BGC-based plots (Figure S6A), suggesting that the observed similarities in the secondary metabolism-associated genotypes of A. oerlinghausenensis and A. fischeri are likely reflected in their chemotypes.
Conservation and divergence among biosynthetic gene clusters implicated in A. fumigatus pathogenicity
Secondary metabolites are known to play a role in A. fumigatus virulence (Raffa and Keller 2019). We therefore conducted a focused examination of specific A. fumigatus BGCs and secondary metabolites that have been previously implicated in the organism’s ability to cause human disease (Table 2). We found varying degrees of conservation and divergence that were associated with the absence or presence of a secondary metabolite. Among conserved BGCs that were also associated with conserved secondary metabolite production, we highlight the mycotoxins gliotoxin and fumitremorgin. Interestingly, we note that only A. fischeri strains synthesized verruculogen—a secondary metabolite that is implicated in human disease and is encoded by the fumitremorgin BGC (Khoufache et al. 2007; Kautsar et al. 2020). Among BGCs that exhibited varying degrees of sequence divergence and divergence in their production of the corresponding secondary metabolites, we highlight those associated with the production of the trypacidin and fumagillin/pseurotin secondary metabolites. We found that nonpathogenic close relatives of A. fumigatus produced some but not all mycotoxins, which provides novel insight into the unique cocktail of secondary metabolites biosynthesized by A. fumigatus.
Table 2. Select A. fumigatus secondary metabolites implicated in modulating host biology.
| Function | Reference(s) | Evidence of biosynthetic gene cluster/ secondary metabolite | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A. fumigatus | A. oerlinghausenensis | A. fischeri | |||||||
| Af293 | CEA10 | CEA17 | CBS 139183T | NRRL 181 | NRRL 4585 | NRRL 4161 | |||
| Gliotoxin | Inhibits host immune response | Sugui et al. (2007) | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| Fumitremorgin | Inhibits the breast cancer resistance protein | González-Lobato et al. (2010) | +/− | +/+ | +/− | +/+ | +/+ | +/+ | +/+ |
| Verruculogen | Changes electrophysical properties of human nasal epithelial cells | Khoufache et al. (2007) | +/− | +/+ | +/− | +/+ | +/+ | +/+ | +/+ |
| Trypacidin | Damages lung cell tissues | Gauthier et al. (2012) | +/+ | +/+ | +/− | +/+ | +/− | −/− | −/− |
| Pseurotin | Inhibits immunoglobulin E | Ishikawa et al. (2009) | +/+ | +/+ | +/+ | +/+ | −/− | −/− | −/− |
| Fumagillin | Inhibits neutrophil function | Fallon et al. (2010, 2011) | +/+ | +/+ | +/+ | +/+ | −/− | −/− | −/− |
A list of select secondary metabolites implicated in human disease and their functional role are described here. All secondary metabolites listed or analogs thereof were identified during secondary metabolite profiling. Plus (+) and minus (−) signs indicate the presence or absence of the BGC and secondary metabolite, respectively. For example, +/+ indicates both BGC presence and evidence of secondary metabolite production, whereas +/− indicates BGC presence but no evidence of secondary metabolite production.
Gliotoxin:
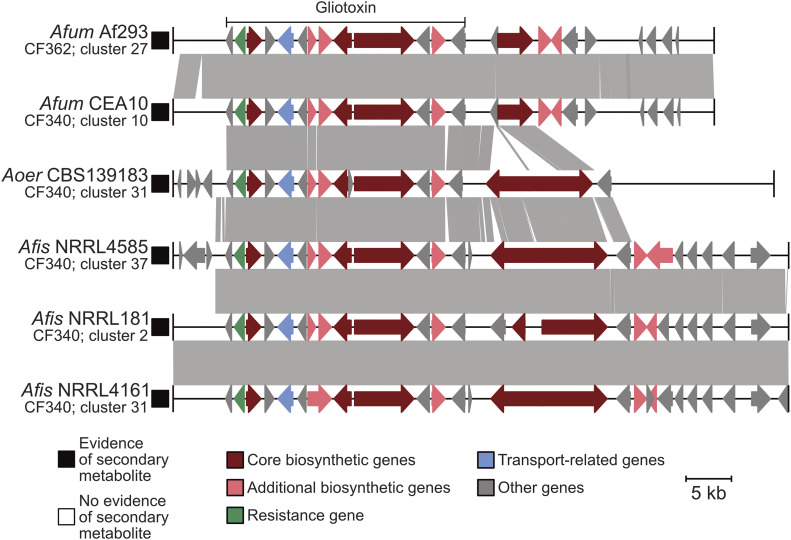
Gliotoxin is a highly toxic compound and known virulence factor in A. fumigatus (Sugui et al. 2007). Nearly identical BGCs encoding gliotoxin are present in all pathogenic (A. fumigatus) and nonpathogenic (A. oerlinghausenensis and A. fischeri) strains examined (Figure 4). Additionally, we found that all examined strains synthesized bisdethiobis(methylthio)gliotoxin a derivative from dithiogliotoxin, involved in the downregulation of gliotoxin biosynthesis (Dolan et al. 2014)—one of the main mechanisms of gliotoxin resistance in A. fumigatus (Kautsar et al. 2020).
Figure 4.
Conservation in the gliotoxin BGC correlates with conserved production of gliotoxin analogs in A. fumigatus and nonpathogenic close relatives. Microsynteny analysis reveals a high degree of conservation in the BGC encoding gliotoxin across all isolates. The known gliotoxin gene cluster boundary is indicated above the A. fumigatus Af293 BGC. Black and white squares correspond to evidence or absence of evidence of secondary metabolite production, respectively. Genes are drawn as arrows with orientation indicated by the direction of the arrow. Gene function is indicated by gene color. Gray boxes between gene clusters indicate BLAST-based similarity of nucleotide sequences defined as being at least 100 bp in length, share at least 30% sequence similarity, and have an expectation value threshold of 0.01. Genus and species names are written using the following abbreviations: Afum, A. fumigatus; Aoer, A. oerlinghausenensis; Afis, A. fischeri. Below each genus and species abbreviation is the cluster family each BGC belongs to and their cluster number.
Fumitremorgin and verruculogen:
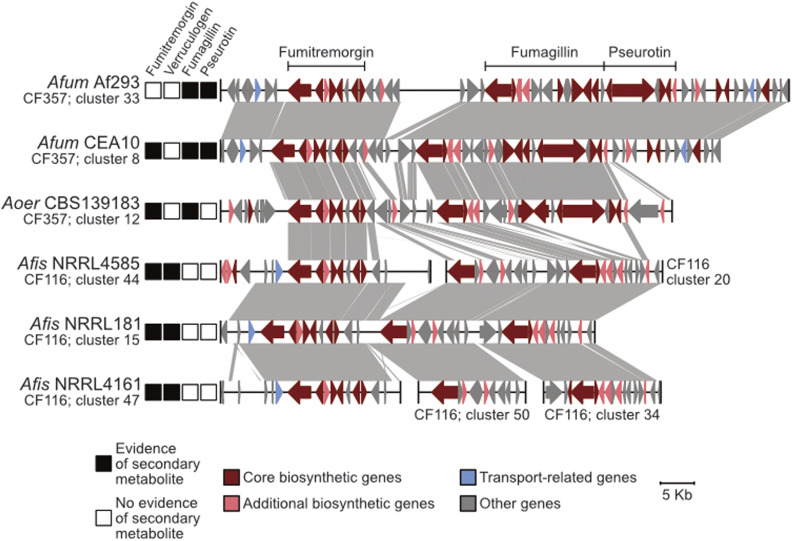
Similarly, there is a high degree of conservation in the BGC that encodes fumitremorgin across all strains (Figure 5). Fumitremorgins have known antifungal activity, are lethal to brine shrimp, and are implicated in inhibiting mammalian proteins responsible for resistance to anticancer drugs in mammalian cells (Raffa and Keller 2019). We found that conservation in the fumitremorgin BGC is associated with the production of fumitremorgins in all isolates examined. The fumitremorgin BGC is also responsible for the production of verruculogen, which is implicated to aid in A. fumigatus pathogenicity by changing the electrophysical properties of human nasal epithelial cells (Khoufache et al. 2007). Interestingly, we found that only A. fischeri strains produced verruculogen under the conditions we analyzed.
Figure 5.
Conservation and divergence in the locus encoding the fumitremorgin and intertwined fumagillin/pseurotin BGCs. Microsynteny analysis reveals conservation in the fumitremorgin BGC across all isolates. Interestingly, only A. fischeri strains synthesize verruculogen, a secondary metabolite also biosynthesized by the fumitremorgin BGC. In contrast, the intertwined fumagillin/pseurotin BGCs are conserved between A. fumigatus and A. oerlinghausenensis but divergent in A. fischeri. BGC conservation and divergence is associated with the presence and absence of a secondary metabolite, respectively. The same convention used in Figure 4 is used to depict evidence of a secondary metabolite, represent genes and broad gene function, BGC sequence similarity, genus and species abbreviations, and BGC cluster families and cluster numbers.
Trypacidin:
Examination of the trypacidin BGC, which encodes a spore-borne and cytotoxic secondary metabolite, revealed a conserved cluster found in four pathogenic and nonpathogenic strains: A. fumigatus Af293, A. fumigatus CEA10, A. oerlinghausenensis CBS 139183T, and A. fischeri NRRL 181 (Figure S7). Furthermore, we found that three of these four isolates (except A. fischeri NRRL 181) biosynthesized a trypacidin analog, monomethylsulochrin. Examination of the microsynteny of the trypacidin BGC revealed that it was conserved across all four genomes with the exception of A. fischeri NRRL l81, which lacked a RING (Really Interesting New Gene) finger gene. Interestingly, RING finger proteins can mediate gene transcription (Poukka et al. 2000). We confirmed the absence of the RING finger protein by performing a sequence similarity search with the A. fumigatus Af293 RING finger protein (AFUA_4G14620; EAL89333.1) against the A. fischeri NRRL 181 genome. In the homologous locus in A. fischeri, we found no significant BLAST hit for the first 23 nucleotides of the RING finger gene suggestive of pseudogenization. Taken together, we hypothesize that presence/absence polymorphisms or a small degree of sequence divergence between otherwise homologous BGCs may be responsible for the presence or absence of a toxic secondary metabolite in A. fischeri NRRL 181. Furthermore, inter- and intra-species patterns of trypacidin presence and absence highlight the importance of strain heterogeneity when examining BGCs.
Fumagillin/pseurotin:
Examination of the intertwined fumagillin/pseurotin BGCs revealed that fumagillin has undergone substantial sequence divergence, and that pseurotin is absent from strains of A. fischeri. The fumagillin/pseurotin BGCs are under the same regulatory control (Wiemann et al. 2013) and biosynthesize secondary metabolites that cause cellular damage during host infection (fumagillin; Guruceaga et al. 2019) and inhibit immunoglobulin E production (pseurotin; Ishikawa et al. 2009). Microsynteny of the fumagillin BGC reveals high sequence conservation between A. fumigatus and A. oerlinghausenensis; however, sequence divergence was observed between A. oerlinghausenensis and A. fischeri (Figure 5). Accordingly, fumagillin production was only observed in A. fumigatus and A. oerlinghausenensis and not in A. fischeri. Similarly, the pseurotin BGC is conserved between A. fumigatus and A. oerlinghausenensis. Rather than sequence divergence, no sequence similarity was observed in the region of the pseurotin cluster in A. fischeri, which may be due to an indel event. Accordingly, no pseurotin production was observed among A. fischeri strains. Despite sequence conservation between A. fumigatus and A. oerlinghausenensis, no evidence of pseurotin biosynthesis was observed in A. oerlinghausenensis, which suggests regulatory decoupling of the intertwined fumagillin/pseurotin BGC. Alternatively, the genes downstream of the A. fumigatus pseurotin BGC, which are absent from the A. oerlinghausenensis locus, may contribute to BGC production and could explain the lack of pseurotin production in A. oerlinghausenensis. Altogether, these results show a striking correlation between sequence divergence and the production (or absence) of secondary metabolites implicated in human disease among A. fumigatus and nonpathogenic closest relatives.
Discussion
Aspergillus fumigatus is a major fungal pathogen nested within a clade (known as section Fumigati) of at least 60 other species, the vast majority of which are nonpathogenic (Steenwyk et al. 2019; Rokas et al. 2020b). Currently, it is thought that the ability to cause human disease evolved multiple times among species in section Fumigati (Rokas et al. 2020b). Secondary metabolites contribute to the success of the major human pathogen A. fumigatus in the host environment (Raffa and Keller 2019) and can therefore be thought of as “cards” of virulence (Casadevall 2007; Knowles et al. 2020). However, whether the closest relatives of A. fumigatus, A. oerlinghausenensis, and A. fischeri, both of which are nonpathogenic, biosynthesize secondary metabolites implicated in the ability of A. fumigatus to cause human disease remained largely unknown. By examining genomic and chemical variation between and within A. fumigatus and its closest nonpathogenic relatives, we identified both conservation and divergence (including within species heterogeneity) in BGCs and secondary metabolite profiles (Figures 1–5, Figures S3 and S5–S8, Table 2, and Table S1 and S3). Examples of conserved BGCs and secondary metabolites include the major virulence factor, gliotoxin (Figure 4), as well as several others (Figure 5 and Figure S7; Table 2 and Table S1 and S3); examples of BGC and secondary metabolite heterogeneity or divergence include pseurotin, fumagillin, and several others (Figure 5, Table 2, and Table S1 and S3). Lastly, we found that the fumitremorgin BGC, which biosynthesizes fumitremorgin in all three species, is also associated with verruculogen biosynthesis in A. fischeri strains (Figure 5).
One of the surprising findings of our study was that although A. oerlinghausenensis and A. fumigatus are evolutionarily more closely related to each other than to A. fischeri (Figure 1), A. oerlinghausenensis and A. fischeri appear to be more similar to each other than to A. fumigatus in BGC composition, gene family content, and secondary metabolite profiles. The power of pathogen–nonpathogen comparative genomics is best utilized when examining closely related species (Fedorova et al. 2008; Jackson et al. 2011; Moran et al. 2011; Mead et al. 2019a; Rokas et al. 2020b). Genomes from additional strains from the closest known nonpathogenic relatives of A. fumigatus, including from the closest species relative A. oerlinghausenensis, A. fischeri, and other nonpathogenic species in section Fumigati will be key for understanding the evolution of A. fumigatus pathogenicity.
Our finding that A. oerlinghausenensis and A. fischeri shares more gene families and BGCs with each other than they do with A. fumigatus (Figures 1C and 2 and Figures S3, S4, and S8) suggests that the evolutionary trajectory of the A. fumigatus ancestor was marked by gene loss. We hypothesize that there were two rounds of gene family and BGC loss in the A. fumigatus stem lineage: (1) gene families and BGCs were lost in the common ancestor of A. fumigatus and A. oerlinghausenensis, and (2) additional losses occurred in the A. fumigatus ancestor. In addition to losses, we note that 548 gene families and 16 BGCs are unique to A. fumigatus, which may have resulted from genetic innovation (e.g., de novo gene formation) or unique gene family and BGC retention (Figure 2 and Figure S8). In line with the larger number of shared BGCs between A. oerlinghausenensis and A. fischeri, we found their secondary metabolite profiles were also more similar (Figure 3 and Figure S6). Notably, the evolutionary rate of the internal branch leading to the A. fumigatus common ancestor is much higher than those in the rest of the branches in our genome-scale phylogeny (Figure S2B), suggesting that the observed gene loss and gene gain/ retention events specific to A. fumigatus may be part of a wider set of evolutionary changes in the A. fumigatus genome. Analyses with a greater number of strains and species will help further test the validity of this hypothesis. More broadly, these results suggest that comparisons of the pathogen A. fumigatus against either the nonpathogen A. oerlinghausenensis (this manuscript) or the nonpathogen A. fischeri (Mead et al. 2019a; Knowles et al. 2020; and this manuscript) will both be instructive in understanding the evolution of A. fumigatus pathogenicity.
When studying Aspergillus pathogenicity, it is important to consider any genetic and phenotypic heterogeneity between strains of a single species (Knox et al. 2016; Kowalski et al. 2016, 2019; Keller 2017; Ries et al. 2019; Blachowicz et al. 2020; Bastos et al. 2020; Drott et al. 2020; dos Santos et al. 2020; Steenwyk et al. 2020). Our finding of strain heterogeneity among gene families, BGCs, and secondary metabolites in A. fumigatus and A. fischeri (Figures 1–3 and Figures S3, S4, S6, and S8) suggests considerable strain-level diversity in each species. For example, we found secondary metabolite profile strain heterogeneity was greater in A. fumigatus than A. fischeri (Figure S6, B–E). These results suggest that strain-specific secondary metabolite profiles may play a role in variation of pathogenicity among A. fumigatus strains. In support of this hypothesis, differential secondary metabolite production has been associated with differences in virulence among isolates of A. fumigatus (Blachowicz et al. 2020). More broadly, our finding supports the hypothesis that strain-level diversity is an important parameter when studying pathogenicity (Kowalski et al. 2016, 2019; Keller 2017; Ries et al. 2019; Blachowicz et al. 2020; Bastos et al. 2020; Drott et al. 2020; dos Santos et al. 2020; Steenwyk et al. 2020).
Secondary metabolites contribute to A. fumigatus virulence through diverse processes including suppressing the human immune system and damaging tissues (Table 2). Interestingly, we found that the nonpathogens A. oerlinghausenensis and A. fischeri produced several secondary metabolites implicated in the ability of A. fumigatus human disease, such gliotoxin, trypacidin, verruculogen, and others (Figures 4 and 5, Figure S7, Table 2 and Table S3). Importantly, our work positively identified secondary metabolites for many structural classes implicated in a previous taxonomic study (Samson et al. 2007). These results suggest that several of the secondary metabolism-associated cards of virulence present in A. fumigatus are conserved in closely related nonpathogens (summarized in Figure 6) as well as in closely related pathogenic species, such as A. novofumigatus (Kjærbølling et al. 2018). Interestingly, disrupting the ability of A. fumigatus to biosynthesize gliotoxin attenuates but does not abolish virulence (Sugui et al. 2007; Dagenais and Keller 2009; Keller 2017), whereas disruption of the ability of A. fischeri NRRL 181 to biosynthesize secondary metabolites, including gliotoxin, does not appear to influence virulence (Knowles et al. 2020). Our findings, together with previous studies, support the hypothesis that individual secondary metabolites are “cards” of virulence in a larger “hand” that A. fumigatus possesses.
Figure 6.
Secondary metabolism-associated “cards” of virulence among A. fumigatus and close relatives. Secondary metabolites contribute to the “hand of cards”’ that enable A. fumigatus to cause disease. Here, we show that the nonpathogenic closest relatives of A. fumigatus possess a subset of the A. fumigatus secondary metabolism-associated cards of virulence. We hypothesize that the unique combination of cards of A. fumigatus contributes to its pathogenicity and that the cards in A. oerlinghausenensis and A. fischeri (perhaps in combination with other nonsecondary-metabolism-associated cards, such as thermotolerance) are insufficient to cause disease. Pathogenic and nonpathogenic species are shown in red and black, respectively. Cartoons of Aspergillus species were obtained from WikiMedia Commons (source: M. Piepenbring) and modified in accordance with the Creative Commons Attribution-Share Alike 3.0 Unported license (https://creativecommons.org/licenses/by-sa/3.0/deed.en).
Acknowledgments
We thank the Rokas, Oberlies, and Goldman laboratories for helpful discussion and support of this work. J.L.S. and A.R. are supported by the Howard Hughes Medical Institute through the James H. Gilliam Fellowships for Advanced Study program. A.R. has additional support from a Discovery grant from Vanderbilt University and the National Science Foundation (DEB-1442113). G.H.G. is supported by the Brazilian funding agencies Fundacão de Amparo a Pesquisa do Estado de São Paulo (FAPESP 2016/07870-9) and Conselho Nacional de Desenvolvimento Cientıfico e Tecnologico (CNPq). N.H.O. is supported by the National Cancer Institute (P01 CA125066). S.L.K. and C.D.R. were supported in part by the National Institutes of Health via the National Center for Complementary and Integrative Health (F31 AT010558) and the National Institute of General Medical Sciences (T34 GM113860), respectively.
Footnotes
Supplemental material available at figshare: https://doi.org/10.6084/m9.figshare.12055503.
These authors contributed equally to this work.
Communicating editor: J. Stajich
Literature Cited
- Abad A., Victoria Fernández-Molina J., Bikandi J., Ramírez A., Margareto J. et al. , 2010. What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 27: 155–182. 10.1016/j.riam.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Afiyatullov S. S., Kalinovskii A. I., Pivkin M. V., Dmitrenok P. S., and Kuznetsova T. A., 2005. Alkaloids from the marine isolate of the fungus Aspergillus fumigatus. Chem. Nat. Compd. 41: 236–238. 10.1007/s10600-005-0122-y [DOI] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M. et al. , 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19: 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos R. W., Valero C., Silva L. P., Schoen T., Drott M. et al. , 2020. Functional Characterization of Clinical Isolates of the Opportunistic Fungal Pathogen Aspergillus nidulans. mSphere 5: e00153-20 10.1128/mSphere.00153-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict K., Jackson B. R., Chiller T., and Beer K. D., 2019. Estimation of direct healthcare costs of fungal diseases in the United States. Clin. Infect. Dis. 68: 1791–1797. 10.1093/cid/ciy776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell E., Cairns T. C., Throckmorton K., Nierman W. C., and Keller N. P., 2016. Secondary metabolite arsenal of an opportunistic pathogenic fungus. Philos. Trans. R. Soc. B Biol. Sci. 371: 20160023 10.1098/rstb.2016.0023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachowicz A., Raffa N., Bok J. W., Choera T., Knox B. et al. , 2020. Contributions of spore secondary metabolites to UV-C protection and virulence vary in different Aspergillus fumigatus strains. MBio 11: e03415-19. 10.1128/mBio.03415-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodinaku, I., J. Shaffer, A. B. Connors, J. L. Steenwyk, M. N. Biango-Daniels et al., 2019 Rapid Phenotypic and Metabolomic Domestication of Wild Penicillium Molds on Cheese. MBio. 10: e02445-19. 10.1128/mBio.02445-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., and Usadel B., 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongomin F., Gago S., Oladele R., and Denning D., 2017. Global and multi-national prevalence of fungal diseases—estimate precision. J. Fungi (Basel) 3: 57 10.3390/jof3040057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caesar L. K., Kvalheim O. M., and Cech N. B., 2018. Hierarchical cluster analysis of technical replicates to identify interferents in untargeted mass spectrometry metabolomics. Anal. Chim. Acta 1021: 69–77. 10.1016/j.aca.2018.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J. et al. , 2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J., Lin Q., Geske G. D., and Blackwell H. E., 2009. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 4: 1051–1059. 10.1021/cb900165y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capella-Gutierrez S., Silla-Martinez J. M., and Gabaldon T., 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A., 2007. Determinants of virulence in the pathogenic fungi. Fungal Biol. Rev. 21: 130–132. 10.1016/j.fbr.2007.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock P. J. A., Antao T., Chang J. T., Chapman B. A., Cox C. J. et al. , 2009. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25: 1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagenais T. R. T., and Keller N. P., 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22: 447–465. 10.1128/CMR.00055-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. P., Riley R., Wiebenga A., Aguilar-Osorio G., Amillis S. et al. , 2017. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biol. 18: 28 10.1186/s13059-017-1151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan S. K., Owens R. A., O’Keeffe G., Hammel S., Fitzpatrick D. A. et al. , 2014. Regulation of nonribosomal peptide synthesis: bis-thiomethylation attenuates gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 21: 999–1012. 10.1016/j.chembiol.2014.07.006 [DOI] [PubMed] [Google Scholar]
- dos Santos R. A. C., Steenwyk J. L., Rivero-Menendez O., Mead M. E., Silva L. P. et al. , 2020. Genomic and phenotypic heterogeneity of clinical isolates of the human pathogens Aspergillus fumigatus, Aspergillus lentulus, and Aspergillus fumigatiaffinis. Front. Genet. 11: 459 10.3389/fgene.2020.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgona L., Khachatryan A., Stephens J., Charbonneau C., Kantecki M. et al. , 2014. Clinical and economic burden of invasive fungal diseases in Europe: focus on pre-emptive and empirical treatment of Aspergillus and Candida species. Eur. J. Clin. Microbiol. Infect. Dis. 33: 7–21. 10.1007/s10096-013-1944-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drott M. T., Bastos R. W., Rokas A., Ries L. N. A., Gabaldón T. et al. , 2020. Diversity of Secondary Metabolism in Aspergillus nidulans Clinical Isolates. mSphere 5: e00156-20 10.1128/mSphere.00156-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Elimat T., Figueroa M., Ehrmann B. M., Cech N. B., Pearce C. J. et al. , 2013. High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products. J. Nat. Prod. 76: 1709–1716. 10.1021/np4004307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D. M., and Kelly S., 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20: 238 10.1186/s13059-019-1832-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallon J. P., Reeves E. P., and Kavanagh K., 2010. Inhibition of neutrophil function following exposure to the Aspergillus fumigatus toxin fumagillin. J. Med. Microbiol. 59: 625–633. 10.1099/jmm.0.018192-0 [DOI] [PubMed] [Google Scholar]
- Fallon J. P., Reeves E. P., and Kavanagh K., 2011. The Aspergillus fumigatus toxin fumagillin suppresses the immune response of Galleria mellonella larvae by inhibiting the action of haemocytes. Microbiology 157: 1481–1488. 10.1099/mic.0.043786-0 [DOI] [PubMed] [Google Scholar]
- Fedorova N. D., Khaldi N., Joardar V. S., Maiti R., Amedeo P. et al. , 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 4: e1000046 10.1371/journal.pgen.1000046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudêncio S. P., and Pereira F., 2015. Dereplication: racing to speed up the natural products discovery process. Nat. Prod. Rep. 32: 779–810. 10.1039/C4NP00134F [DOI] [PubMed] [Google Scholar]
- Gauthier T., Wang X., Sifuentes Dos Santos J., Fysikopoulos A., Tadrist S. et al. , 2012. Trypacidin, a spore-borne toxin from Aspergillus fumigatus, is cytotoxic to lung cells. PLoS One 7: e29906 10.1371/journal.pone.0029906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lobato L., Real R., Prieto J. G., Álvarez A. I., and Merino G., 2010. Differential inhibition of murine Bcrp1/Abcg2 and human BCRP/ABCG2 by the mycotoxin fumitremorgin C. Eur. J. Pharmacol. 644: 41–48. 10.1016/j.ejphar.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Grahl N., Shepardson K. M., Chung D., and Cramer R. A., 2012. Hypoxia and fungal pathogenesis: to air or not to air? Eukaryot. Cell 11: 560–570. 10.1128/EC.00031-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruceaga X., Ezpeleta G., Mayayo E., Sueiro-Olivares M., Abad-Diaz-De-Cerio A. et al. , 2018. A possible role for fumagillin in cellular damage during host infection by Aspergillus fumigatus. Virulence 9: 1548–1561. 10.1080/21505594.2018.1526528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruceaga X., Perez-Cuesta U., Abad-Diaz de Cerio A., Gonzalez O., Alonso R. M. et al. , 2019. Fumagillin, a mycotoxin of Aspergillus fumigatus: biosynthesis, biological activities, detection, and applications. Toxins (Basel) 12: 7 10.3390/toxins12010007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halász J., Podányi B., Vasvári-Debreczy L., Szabó A., Hajdú F. et al. , 2000. Structure elucidation of fumagillin-related natural products. Tetrahedron 56: 10081–10085. 10.1016/S0040-4020(00)00979-0 [DOI] [Google Scholar]
- Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., and Vinh L. S., 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35: 518–522. 10.1093/molbev/msx281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt C., and Yandell M., 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12: 491 10.1186/1471-2105-12-491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbraken J., Weig M., Groß U., Meijer M., and Bader O., 2016. Aspergillus oerlinghausenensis, a new mould species closely related to A. fumigatus. FEMS Microbiol. Lett. 363: fnv236 10.1093/femsle/fnv236 [DOI] [PubMed] [Google Scholar]
- Hubert J., Nuzillard J.-M., and Renault J.-H., 2017. Dereplication strategies in natural product research: how many tools and methodologies behind the same concept? Phytochem. Rev. 16: 55–95. 10.1007/s11101-015-9448-7 [DOI] [Google Scholar]
- Ishikawa M., Ninomiya T., Akabane H., Kushida N., Tsujiuchi G. et al. , 2009. Pseurotin A and its analogues as inhibitors of immunoglobuline E production. Bioorg. Med. Chem. Lett. 19: 1457–1460. 10.1016/j.bmcl.2009.01.029 [DOI] [PubMed] [Google Scholar]
- Ito T., and Masubuchi M., 2014. Dereplication of microbial extracts and related analytical technologies. J. Antibiot. (Tokyo) 67: 353–360. 10.1038/ja.2014.12 [DOI] [PubMed] [Google Scholar]
- Jackson R. W., Johnson L. J., Clarke S. R., and Arnold D. L., 2011. Bacterial pathogen evolution: breaking news. Trends Genet. 27: 32–40. 10.1016/j.tig.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B. Q., Wong T. K. F., von Haeseler A., and Jermiin L. S., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14: 587–589. 10.1038/nmeth.4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei K., and Watanabe A., 2005. Aspergillus mycotoxins and their effect on the host. Med. Mycol. 43: 95–99. 10.1080/13693780500051547 [DOI] [PubMed] [Google Scholar]
- Katoh K., and Standley D. M., 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Suzuki H., Takagi H., Asami Y., Kakeya H. et al. , 2009. Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. ChemBioChem 10: 920–928. 10.1002/cbic.200800787 [DOI] [PubMed] [Google Scholar]
- Kautsar S. A., Blin K., Shaw S., Navarro-Muñoz J. C., Terlouw B. R. et al. , 2020. MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acids Res. 8: D454–D458. 10.1093/nar/gkz882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P., 2017. Heterogeneity confounds establishment of “a” model microbial strain. MBio 8: e00135-17 10.1128/mBio.00135-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller N. P., 2019. Fungal secondary metabolism: regulation, function and drug discovery. Nat. Rev. Microbiol. 17: 167–180. 10.1038/s41579-018-0121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoufache K., Puel O., Loiseau N., Delaforge M., Rivollet D. et al. , 2007. Verruculogen associated with Aspergillus fumigatus hyphae and conidia modifies the electrophysiological properties of human nasal epithelial cells. BMC Microbiol. 7: 5 10.1186/1471-2180-7-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærbølling I., Vesth T. C., Frisvad J. C., Nybo J. L., Theobald S. et al. , 2018. Linking secondary metabolites to gene clusters through genome sequencing of six diverse Aspergillus species. Proc. Natl. Acad. Sci. USA 115: E753–E761. 10.1073/pnas.1715954115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjærbølling I., Vesth T., Frisvad J. C., Nybo J. L., Theobald S. et al. , 2020. A comparative genomics study of 23 Aspergillus species from section Flavi. Nat. Commun. 11: 1106 10.1038/s41467-019-14051-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles S. L., Vu N., Todd D. A., Raja H. A., Rokas A. et al. , 2019. Orthogonal method for double-bond placement via ozone-induced dissociation mass spectrometry (OzID-MS). J. Nat. Prod. 82: 3421–3431. 10.1021/acs.jnatprod.9b00787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, S. L., M. E. Mead, L. P. Silva, H. A. Raja, J. L. Steenwyk et al., 2020 Gliotoxin, a known virulence factor in the major human pathogen Aspergillus fumigatus, is also biosynthesized by its nonpathogenic relative Aspergillus fischeri. MBio. 11: e03361-19. 10.1128/mBio.03361-19 10.1128/mBio.03361-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox B. P., Blachowicz A., Palmer J. M., Romsdahl J., Huttenlocher A. et al. , 2016. Characterization of Aspergillus fumigatus Isolates from Air and Surfaces of the International Space Station. mSphere 1: e00227-16 10.1128/mSphere.00227-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I., 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski C. H., Beattie S. R., Fuller K. K., McGurk E. A., Tang Y.-W. et al. , 2016. Heterogeneity among isolates reveals that fitness in low oxygen correlates with Aspergillus fumigatus virulence. MBio 7: e01515-16 10.1128/mBio.01515-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski C. H., Kerkaert J. D., Liu K.-W., Bond M. C., Hartmann R. et al. , 2019. Fungal biofilm morphology impacts hypoxia fitness and disease progression. Nat. Microbiol. 4: 2430–2441. 10.1038/s41564-019-0558-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvalheim O. M., Chan H., Benzie I. F. F., Szeto Y., Tzang A. H. et al. , 2011. Chromatographic profiling and multivariate analysis for screening and quantifying the contributions from individual components to the bioactive signature in natural products. Chemom. Intell. Lab. Syst. 107: 98–105. 10.1016/j.chemolab.2011.02.002 [DOI] [Google Scholar]
- Latgé J.-P., and Chamilos G., 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin. Microbiol. Rev. 33: e00140-18 10.1128/CMR.00140-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind A. L., Wisecaver J. H., Smith T. D., Feng X., Calvo A. M. et al. , 2015. Examining the evolution of the regulatory circuit controlling secondary metabolism and development in the fungal genus Aspergillus. PLoS Genet. 11: e1005096 10.1371/journal.pgen.1005096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind A. L., Wisecaver J. H., Lameiras C., Wiemann P., Palmer J. M. et al. , 2017. Drivers of genetic diversity in secondary metabolic gene clusters within a fungal species. PLoS Biol. 15: e2003583 10.1371/journal.pbio.2003583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Peng C., Shen Y., Miao X., Zhang H. et al. , 2008. l-Diketopiperazines from Alcaligenes faecalis A72 associated with South China Sea sponge Stelletta tenuis. Biochem. Syst. Ecol. 36: 230–234. 10.1016/j.bse.2007.08.007 [DOI] [Google Scholar]
- Li X.-J., Zhang Q., Zhang A.-L., and Gao J.-M., 2012. Metabolites from Aspergillus fumigatus, an endophytic fungus associated with Melia azedarach, and their antifungal, antifeedant, and toxic activities. J. Agric. Food Chem. 60: 3424–3431. 10.1021/jf300146n [DOI] [PubMed] [Google Scholar]
- Losada L., Ajayi O., Frisvad J. C., Yu J., and Nierman W. C., 2009. Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med. Mycol. 47: S88–S96. 10.1080/13693780802409542 [DOI] [PubMed] [Google Scholar]
- Ma Y., Li Y., Liu J., Song Y., and Tan R., 2004. Anti-Helicobacter pylori metabolites from Rhizoctonia sp. Cy064, an endophytic fungus in Cynodon dactylon. Fitoterapia 75: 451–456. 10.1016/j.fitote.2004.03.007 [DOI] [PubMed] [Google Scholar]
- Mattern D. J., Schoeler H., Weber J., Novohradská S., Kraibooj K. et al. , 2015. Identification of the antiphagocytic trypacidin gene cluster in the human-pathogenic fungus Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 99: 10151–10161. 10.1007/s00253-015-6898-1 [DOI] [PubMed] [Google Scholar]
- Mead, M. E., S. L. Knowles, H. A. Raja, S. R. Beattie, C. H. Kowalski et al., 2019a Characterizing the Pathogenic, Genomic, and Chemical Traits of Aspergillus fischeri, a Close Relative of the Major Human Fungal Pathogen Aspergillus fumigatus. mSphere. 4: e00018-19. 10.1128/mSphere.00018-19 10.1128/mSphere.00018-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mead M. E., Raja H. A., Steenwyk J. L., Knowles S. L., Oberlies N. H. et al. , 2019b Draft genome sequence of the griseofulvin-producing fungus Xylaria flabelliformis strain G536. Microbiol. Resour. Announc. 8: e00890-19 10.1128/MRA.00890-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Liu D., Li G., Li P., Xu Y. et al. , 2015. Genome-wide transcriptomic analysis of a superior biomass-degrading strain of A. fumigatus revealed active lignocellulose-degrading genes. BMC Genomics 16: 459 10.1186/s12864-015-1658-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran G. P., Coleman D. C., and Sullivan D. J., 2011. Comparative genomics and the evolution of pathogenicity in human pathogenic fungi. Eukaryot. Cell 10: 34–42. 10.1128/EC.00242-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Muñoz J. C., Selem-Mojica N., Mullowney M. W., Kautsar S. A., Tryon J. H. et al. , 2020. A computational framework to explore large-scale biosynthetic diversity. Nat. Chem. Biol. 16: 60–68. 10.1038/s41589-019-0400-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L.-T., Schmidt H. A., von Haeseler A., and Minh B. Q., 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32: 268–274. 10.1093/molbev/msu300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman W. C., Pain A., Anderson M. J., Wortman J. R., Kim H. S. et al. , 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438: 1151–1156 [Corrigenda: Nature 439: 502 (2006)]. 10.1038/nature04332 [DOI] [PubMed] [Google Scholar]
- Pluskal T., Castillo S., Villar-Briones A., and Orešič M., 2010. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11: 395 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H., Aarnisalo P., Santti H., Jänne O. A., and Palvimo J. J., 2000. Coregulator small nuclear RING finger protein (SNURF) enhances Sp1- and steroid receptor-mediated transcription by different mechanisms. J. Biol. Chem. 275: 571–579. 10.1074/jbc.275.1.571 [DOI] [PubMed] [Google Scholar]
- Raffa N., and Keller N. P., 2019. A call to arms: Mustering secondary metabolites for success and survival of an opportunistic pathogen. PLOS Pathog. 15: e1007606 10.1371/journal.ppat.1007606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery A. E., and Lewis S. M., 1995. The number of iterations, convergence diagnostics and generic Metropolis algorithms. Pract. Markov Chain Monte Carlo 7: 763–773. 10.1.1.41.6352 [Google Scholar]
- Ries L. N. A., Steenwyk J. L., de Castro P. A., de Lima P. B. A., Almeida F. et al. , 2019. Nutritional heterogeneity among Aspergillus fumigatus strains has consequences for virulence in a strain- and host-dependent manner. Front. Microbiol. 10: 854 10.3389/fmicb.2019.00854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A., Williams B. L., King N., and Carroll S. B., 2003. Genome-scale approaches to resolving incongruence in molecular phylogenies. Nature 425: 798–804. 10.1038/nature02053 [DOI] [PubMed] [Google Scholar]
- Rokas A., Wisecaver J. H., and Lind A. L., 2018. The birth, evolution and death of metabolic gene clusters in fungi. Nat. Rev. Microbiol. 16: 731–744. 10.1038/s41579-018-0075-3 [DOI] [PubMed] [Google Scholar]
- Rokas A., Mead M. E., Steenwyk J. L., Raja H. A., and Oberlies N. H., 2020a Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 37: 868–878. 10.1039/C9NP00045C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokas A., Mead M. E., Steenwyk J. L., Oberlies N. H., and Goldman G. H., 2020b Evolving moldy murderers: Aspergillus section Fumigati as a model for studying the repeated evolution of fungal pathogenicity. PLOS Pathog. 16: e1008315 10.1371/journal.ppat.1008315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R. A., Hong S., Peterson S. W., Frisvad J. C., and Varga J., 2007. Polyphasic taxonomy of Aspergillus section Fumigati and its teleomorph Neosartorya. Stud. Mycol. 59: 147–203. 10.3114/sim.2007.59.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shwab E. K., Bok J. W., Tribus M., Galehr J., Graessle S. et al. , 2007. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot. Cell 6: 1656–1664. 10.1128/EC.00186-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spikes S., Xu R., Nguyen C. K., Chamilos G., Kontoyiannis D. P. et al. , 2008. Gliotoxin production in Aspergillus fumigatus contributes to host‐specific differences in virulence. J. Infect. Dis. 197: 479–486. 10.1086/525044 [DOI] [PubMed] [Google Scholar]
- Stanke M., and Waack S., 2003. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19: ii215–ii225. 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- Steenwyk J., and Rokas A., 2017. Extensive copy number variation in fermentation-related genes among Saccharomyces cerevisiae wine strains. G3 (Bethesda) 7: 1475–1485. 10.1534/g3.117.040105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk J. L., Shen X.-X., Lind A. L., Goldman G. H., and Rokas A., 2019. A Robust phylogenomic time tree for biotechnologically and medically important fungi in the genera Aspergillus and Penicillium. MBio 10: e00925-19 10.1128/mBio.00925-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwyk J. L., Lind A. L., Ries L. N. A., dos Reis T. F., Silva L. P. et al. , 2020. Pathogenic allodiploid hybrids of Aspergillus fungi. Curr. Biol. 30: 2495–2507.e7. 10.1016/j.cub.2020.04.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui J. A., Pardo J., Chang Y. C., Zarember K. A., Nardone G. et al. , 2007. Gliotoxin is a virulence factor of Aspergillus fumigatus : gliP deletion attenuates virulence in mice immunosuppressed with hydrocortisone. Eukaryot. Cell 6: 1562–1569. 10.1128/EC.00141-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S., 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect. Math Life Sci. 17: 57–86. [Google Scholar]
- Tekaia F., and Latgé J.-P., 2005. Aspergillus fumigatus: saprophyte or pathogen? Curr. Opin. Microbiol. 8: 385–392. 10.1016/j.mib.2005.06.017 [DOI] [PubMed] [Google Scholar]
- Vallabhaneni S., Mody R. K., Walker T., and Chiller T., 2016. The global burden of fungal diseases. Infect. Dis. Clin. North Am. 30: 1–11. 10.1016/j.idc.2015.10.004 [DOI] [PubMed] [Google Scholar]
- van Dongen, S., 2000 Graph clustering by flow simulation. Graph Stimul. by flow Clust. PhD thesis: University of Utrecht, Netherlands. 10.1016/j.cosrev.2007.05.001 10.1016/j.cosrev.2007.05.001 [DOI] [Google Scholar]
- Vesth T. C., Nybo J. L., Theobald S., Frisvad J. C., Larsen T. O. et al. , 2018. Investigation of inter- and intraspecies variation through genome sequencing of Aspergillus section Nigri. Nat. Genet. 50: 1688–1695. 10.1038/s41588-018-0246-1 [DOI] [PubMed] [Google Scholar]
- Vinet L., and Zhedanov A., 2011. A ‘missing’ family of classical orthogonal polynomials. J. Phys. A Math. Theor. 44: 085201 10.1088/1751-8113/44/8/085201 [DOI] [Google Scholar]
- Wang F., Fang Y., Zhu T., Zhang M., Lin A. et al. , 2008. Seven new prenylated indole diketopiperazine alkaloids from holothurian-derived fungus Aspergillus fumigatus. Tetrahedron 64: 7986–7991. 10.1016/j.tet.2008.06.013 [DOI] [Google Scholar]
- Wang F.-Z., Li D.-H., Zhu T.-J., Zhang M., and Gu Q.-Q., 2011. Pseurotin A 1 and A 2, two new 1-oxa-7-azaspiro[4.4]non-2-ene-4,6-diones from the holothurian-derived fungus Aspergillus fumigatus WFZ-25. Can. J. Chem. 89: 72–76. 10.1139/V10-157 [DOI] [Google Scholar]
- Waterhouse R. M., Tegenfeldt F., Li J., Zdobnov E. M., and Kriventseva E. V., 2013. OrthoDB: a hierarchical catalog of animal, fungal and bacterial orthologs. Nucleic Acids Res. 41: D358–D365. 10.1093/nar/gks1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse R. M., Seppey M., Simão F. A., Manni M., Ioannidis P. et al. , 2018. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35: 543–548. 10.1093/molbev/msx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Blin K., Duddela S., Krug D., Kim H. U. et al. , 2015. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43: W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P., Guo C.-J., Palmer J. M., Sekonyela R., Wang C. C. C. et al. , 2013. Prototype of an intertwined secondary-metabolite supercluster. Proc. Natl. Acad. Sci. USA 110: 17065–17070. 10.1073/pnas.1313258110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemann P., Lechner B. E., Baccile J. A., Velk T. A., Yin W.-B. et al. , 2014. Perturbations in small molecule synthesis uncovers an iron-responsive secondary metabolite network in Aspergillus fumigatus. Front. Microbiol. 5: 530 10.3389/fmicb.2014.00530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada A., Kataoka T., and Nagai K., 2000. The fungal metabolite gliotoxin: immunosuppressive activity on CTL-mediated cytotoxicity. Immunol. Lett. 71: 27–32. 10.1016/S0165-2478(99)00155-8 [DOI] [PubMed] [Google Scholar]
- Yandell M., and Ence D., 2012. A beginner’s guide to eukaryotic genome annotation. Nat. Rev. Genet. 13: 329–342. 10.1038/nrg3174 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J. Mol. Evol. 39: 306–314. 10.1007/BF00160154 [DOI] [PubMed] [Google Scholar]
- Yang Z., 1996. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol. Evol. 11: 367–372. 10.1016/0169-5347(96)10041-0 [DOI] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Yin W.-B., Baccile J. A., Bok J. W., Chen Y., Keller N. P. et al. , 2013. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J. Am. Chem. Soc. 135: 2064–2067. 10.1021/ja311145n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Latgé J.-P., and Gibbons J. G., 2019. Genome sequences of two strains of the food spoilage mold Aspergillus fischeri. Microbiol. Resour. Announc. 8: e01328-19. 10.1128/MRA.01328-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Mou Y., Shan T., Li Y., Zhou L. et al. , 2010. Antimicrobial metabolites from the endophytic fungus pichia guilliermondii isolated from Paris polyphylla var. yunnanensis. Molecules 15: 7961–7970. 10.3390/molecules15117961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article. Sequence reads and associated genome assemblies generated in this project are available in NCBI’s GenBank database under the BioProject PRJNA577646. Additional descriptions of the genomes including predicted gene boundaries are available through figshare (https://doi.org/10.6084/m9.figshare.12055503). The figshare repository is also populated with other data generated from genomic and natural products analysis. Among genomic analyses, we provide information about predicted BGCs, results associated with network-based clustering of BGCs into cluster families, phylogenomic data matrices, and trees. Among natural products analysis, we provide information that supports methods and results, including NMR spectra.