Summary
From 2002 to 2019, three deadly human coronaviruses (hCoVs), severe acute respiratory syndrome coronavirus (SARS‐CoV), Middle Eastern respiratory syndrome coronavirus (MERS‐CoV) and severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) emerged to produce outbreaks of SARS, MERS and coronavirus disease 2019 (Covid‐19), respectively. All three hCoVs are members of the Betacoronavirus genus in the subfamily Orthocoronavirinae and share many similarities in virology and epidemiology. However, the pattern and scale of Covid‐19 global spread is similar to 2009 pandemic H1N1 influenza (H1N1pdm09), rather than SARS or MERS. Covid‐19 exhibits high viral shedding in the upper respiratory tract at an early stage of infection, and has a high proportion of transmission competent individuals that are pre‐symptomatic, asymptomatic and mildly symptomatic, characteristics seen in H1N1pdm09 but not in SARS or MERS. These two traits of Covid‐19 and H1N1pdm09 result in reduced efficiency in identification of transmission sources by symptomatic screening and play important roles in their ability to spread unchecked to cause pandemics. To overcome these attributes of Covid‐19 in community transmission, identifying the transmission source by testing for virus shedding and interrupting chains of transmission by social distancing and public masking are required.
Keywords: Covid‐19, MERS, pandemic influenza, pre‐symptomatic infection, SARS, viral shedding
Abbreviations
- ACE2
angiotensin‐converting enzyme‐2
- CoVs
coronaviruses
- Covid‐19
coronavirus disease‐2019
- GSH
golden Syrian hamster
- H1N1pdm09
2009 pandemic H1N1 influenza A
- MERS
Middle Eastern respiratory syndrome
- RNA
ribonucleic acid
- RT‐PCR
reverse transcriptase polymerase chain reaction
- SARS
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome coronavirus‐2
- URT
upper respiratory tract
- WHO
World Health Organization
1. INTRODUCTION
There are seven different coronaviruses (CoVs) that can infect humans, four of which (OC43, 229E, NL63 and HKU1) cause only mild illness. The other three cause life‐threatening severe acute respiratory syndrome. Severe acute respiratory syndrome (SARS), caused by SARS‐CoV, was first identified in Guangdong province, China in November 2002. By the end of the SARS epidemic in July 2003, a total of 8096 cases and 774 deaths were confirmed in 27 countries. 1 Middle East respiratory syndrome (MERS), caused by the MERS‐CoV, emerged in 2012 in Saudi Arabia and also affected 27 countries. 2 By March 2020, 2521 laboratory‐confirmed MERS cases were reported globally to the World Health Organization (WHO) and 866 associated deaths were recorded with a case‐fatality rate of 34.3%. 3 The most recent emerged human coronavirus (hCoV), severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2), was first identified in Wuhan city, Hubei province, China in December 2019 and has caused the ongoing global pandemic of Coronavirus Disease 2019 (Covid‐19). 4 In the first 4 months outbreak by 1 April 2020, there were 823,626 confirmed cases and 40,598 deaths in 205 countries and territories. 5 By 27 July 2020, 16,114,449 of infections and 646,641 deaths had been recorded. 6
The most recent influenza pandemic, H1N1 influenza (2009 pandemic H1N1 influenza [H1N1pdm09]) was first detected in Mexico in February 2009. The disease spread to 73 countries in the first 4 months. 7 By 31 August 2010, 18,449 laboratory‐confirmed deaths from H1N1pdm09 were reported to WHO, a figure widely believed to be greatly underestimated due to significant numbers of undiagnosed influenza and related deaths. 8 Covid‐19 has been showing a similar transmission speed and extent as H1N1pdm09, rather than SARS or MERS. This review aims to identify the most important factors that contribute to the unprecedented spread of Covid‐19 by comparing the features and parameters that affect the spread of these three deadly hCoVs and H1N1pdm09.
2. GENOMIC AND VIROLOGICAL FEATURES OF SARS‐CoV, MERS‐CoV AND SARS‐CoV‐2
These CoVs belong to the subfamily Orthocoronavirinae in the family Coronaviridae of the order Nidovirales. The subfamily includes four genera: α, β, γ and δ‐CoVs. All three of SARS‐CoV, MERS‐CoV and SARS‐CoV‐2 belong to the β‐CoV genera and are reported to originate from bats. 9 The genetic similarity of SARS‐CoV‐2 to SARS‐CoV and MERS‐CoV is 79% and 50%, respectively. 10
The virus particles of CoVs are approximately 50–200 nm in diameter, enveloped and contain positive‐sense ribonucleic acid (RNA) genomes of 26–32 kb. Virions contain four major viral structural proteins known as the spike (S), envelope (E), membrane (M) and nucleocapsid (N). The N protein enshrouds the RNA genome while the S, E and M proteins together assemble in the viral envelope. 10 , 11 The S protein is a large multifunctional class I viral transmembrane protein that lies on the virion surface imparting a halo‐ or crown‐like appearance when viewed by electron microscopy. 12 The viral S protein mediates attachment and fusion of the viral and cellular plasma membranes, leading to viral entry. Therefore, S protein is a critical determinant of host range and tissue tropism. 12 , 13 The S proteins of SARS‐CoV‐2 and SARS‐CoV are phylogenetically and closely related with an amino acid sequence identity of 77%. Both proteins directly bind to and use angiotensin‐converting enzyme‐2 (ACE2) as a cell receptor and the binding affinity of SARS‐CoV‐2 S protein with ACE2 is 10 to 20‐fold higher than that of SARS‐CoV S protein, indicating that SARS‐CoV‐2 infects the same types of host cells as SARS‐CoV, but with a higher efficiency. 11 , 14 , 15 , 16 MERS‐CoV and H1N1pdm09 employ dipeptidyl peptidase 4 (DPP4) and α2,6 sialic acids as a receptor for cell entry, respectively. 17 , 18 , 19 While these cellular receptors distribute in the respiratory tract to serve the original viral entry, ACE2 and DPP4 are expressed more broadly in various tissue and organs (Table 1), indicating that the three hCoVs can infect cells outside of the respiratory track.
TABLE 1.
Comparisons of virological, epidemiological and clinical features of SARS, MERS, Covid‐19 and H1N1pdm09
| SARS | MERS | Covid‐19 | H1N1pdm09 | |
|---|---|---|---|---|
| Speed and scale of spread | Nov 2002 to Jul 2003; total confirmed cases: 8096; confirmed death: 774; countries and territories spread: 27 1 | Oct 2012 to Mar 2020; total confirmed cases: 2521; confirmed death: 866; countries and territories spread: 27. 3 | Jan 2020 to 27 Jul 2020; total confirmed cases: 16,114,449; confirmed death: 646,641 countries and territories spread: 214 107 | In first 4 months' epidemic since Feb 2009; 73 countries and territories spread confirmed. 7 By 31 Aug 2010; 18,449 confirmed deaths reported to WHO. 8 |
| Virus origin | Bat 9 | Bat 108 | Bat 10 , 109 | Swine 110 |
| Cell receptor | ACE2 17 | DPP4 17 | ACE2 11 , 15 | α2,6‐SA 18 |
| Receptor distribution | Respiratory tract epithelium; arterial and venous endothelium; arterial smooth muscle; small intestine, alveolar monocytes and macrophages 111 | Respiratory tract epithelium; kidney, small intestine; liver and prostate; activated leucocyte 111 | Respiratory tract epithelium; arterial and venous endothelium; arterial smooth muscle; small intestine, alveolar monocytes and macrophages 111 , 16 | Respiratory tract ciliated cells, cuboidal cells and alveolar type II pneumocytes 49 |
| Mean of incubation period (days) and 95% CI | 4.7 (4.3–5.1) 112 | 5.8 (5.0–6.5) 112 | 4.9 (4.4–5.5) 112 | 1.4 (1–1.8) 113 |
| Reproduction number R 0 (days) | 2–4 28 | South Korea: 2–5 114 Saudi Arabia: 0.45 115 | 1.4–6.49 27 | 1.2–3.1 20 |
| Mean serial interval (days) | 8–12 116 | Korean: 12.6, global: 7–12 53 | 3.95∼7.5 4 | 0.8–3.3 20 , 42 |
| Proportion of asymptomatic infection | Serological testing based: 11%–13% 68 , 69 | Virus RNA testing based: 12.5%–25.1% 77 | Virus RNA testing based. Japanese citizens evacuated from Wuhan: 33.3%. 88 A prospective study in Nanjing: 29.7% 89 | Virus RNA testing based Household studies: 10%–45%. 42 , 80 A prospective household study: 45% 42 |
| Proportion of mild symptomatic cases | 4–25% 68 | 21% 117 | China: 81% 50 | 92% were outpatients 118 |
| Proportion of cases who had fever at admission | 99% 111 | 84% 111 | 43.8% at the time of symptomatic onset and 87.9% in hospitalised patients 31 | 94% 119 |
| Proportion of severe cases | 20–30% 1 | 50–89% 1 | China: 19% 50 | 6.5% 7 |
| Case‐fatality rate | Worldwide: 9.6%, mainland China: 6.4%, and Hong Kong: 17% 117 | Worldwide (WHO): 34.5% and South Korea: 20.4% 117 | By 8 Jul 2020 Worldwide: 4.0% USA: 3.5% Brazil: 3.6% China: 5.4% Singapore: <0.1% Italy: 14.3% Germany: 4.4% 6 | 0.2–1.3% 19 |
3. EPIDEMIOLOGICAL AND CLINICAL FEATURES OF SARS, MERS, Covid‐19 AND H1N1pdm09
3.1. Transmission routes and incubation period
Respiratory virus infections, like influenza, SARS, MERS and Covid‐19 predominately transmit by close person‐to‐person contact via respiratory droplets, direct contact and airborne particles. 2 , 20 , 21 Aerosol transmission was reported to have an important role in the spread of SARS‐CoV, MERS‐CoV, SARS‐CoV‐2 and influenza viruses. 21 Aerosols are droplets of less than 5 µm that can remain airborne for a prolonged period of time. 22 In experimental conditions, dynamic aerosol efficiency of SARS‐CoV‐2 surpassed those of SARS‐CoV and MERS‐CoV, and respirable‐sized aerosols of SARS‐CoV‐2 retained infectivity and virion integrity for up to 16 h. 23 Although coughing and sneezing produce more aerosols per breathing manoeuvre than normal breathing, normal breathing can generate aerosols, 24 implying the transmission competency of pre‐symptomatic, asymptomatic and mild infections. The incubation period, defined as the number of days from virus exposure to symptom onset, is similar among these three hCoVs, and shorter for H1N1pdm09 (Table 1).
3.2. Reproduction number R0
Reproduction number R0 is defined as the number of secondary cases resulting from a single initial case and is an index of viral infectiousness. 25 The R0 value of a spreading virus is dynamic during an outbreak and affected by numerous biological, sociobehavioral and environmental factors. 26 R0 value at early stage of an outbreak, prior to implementation of interventions, is an indicator of viral infectiousness, while the R0 value at later stages of outbreak is generally more a reflection of the effectiveness of control measures. R0 values are also greatly affected by the calculation methods 27 and the nature of transmission events, such as an outbreak in a healthcare settings, in an aircraft and by superspreading events. 28 , 29 , 30 The R0 values for SARS‐CoV, MERS‐CoV, SARS‐CoV‐2 and H1N1pdm09 varied considerably between different studies, which complicate comparisons between these viruses. In 12 studies by 7 February 2020, the R0 of SARS‐CoV‐2 ranged from 1.4 to 6.9, with a mean of 3.28 and a median of 2.79 27 , and the R0 values of SARS‐CoV, MERS‐CoV and H1N1pdm09 are in a similar range (Table 1), implying that the infectiousness of these four viruses is not significantly different.
3.3. Serial interval
Serial interval is an epidemiological term used to describe the time between successive cases in a string of transmissions from a primary case symptom onset to a secondary case symptom onset. If the observed mean serial interval is shorter than the observed mean of incubation period, this suggests that the transmission may have been caused by infected persons before symptom onset (pre‐symptomatic transmission). The mean of serial interval of Covid‐19 (3.95–7.5 days) 4 is shorter than SARS (8–12 days) and MERS (7–12 days; Table 1). Given the similar incubation periods of these three hCoVs, the short serial interval of Covid‐19 indicates faster transmission of SARS‐Cov‐2 compared to SARS‐CoV and MERS‐CoV, and suggests that pre‐symptomatic transmission is possible.
3.4. Disease spectrum
Similar to H1N1pdm09, Covid‐19 has higher proportions of asymptomatic and mild infection compared to SARS and MERS (Table 1). In contrast to SARS, MERS and H1N1pdm09, Covid‐19 showed a very low proportion of cases with fever at the early stage of infection, 43.8% at the time of symptom onset and 87.9% at hospital admission. 31 This unique feature of Covid‐19 can result in reduced effectiveness of symptomatic identification of infected individuals, especially when relying on body temperature measurement.
Compared to SARS and MERS, the rates of severe cases and case‐fatality of Covid‐19 are lower, but significantly higher than H1N1pdm09. The case‐fatality rates of Covid‐19 are remarkably divergent in different counties and regions, and the reasons for this remarkable difference warrant further investigations (Table 1).
4. DYNAMICS OF VIRAL SHEDDING OF SARS, MERS, Covid‐19 AND H1N1pdm09
Viral shedding refers to release of virus into the environment from a body where the virus replicates. Viral shedding is essential for the spread of infection between hosts. For respiratory viruses such as hCoVs and influenza, viral shedding from the respiratory tract, especially upper respiratory tract (URT), is one of the factors determining viral infectiousness and transmissibility. For RNA viruses, viral shedding can be determined by detection of viral genomic RNA, viral protein or isolation of infectious virus using cell culture. Reverse transcriptase polymerase chain reaction (RT‐PCR) is the most sensitive and broadly used method to detect viral RNA from URT secretion, which is an indicator of hCoV and influenza virus shedding.
4.1. Viral shedding of SARS and MERS peaked in the second week after symptom onset
While there is no available date of SARS‐CoV detection from pre‐symptomatic SARS patients, viral shedding can be detected from nasopharyngeal aspirates of patients at the first day of onset 32 and the positive rate peaked at 6–11 days of illness. Viral load also peaked during 12–14 days of illness 33 , 34 (Figure 1a).
FIGURE 1.
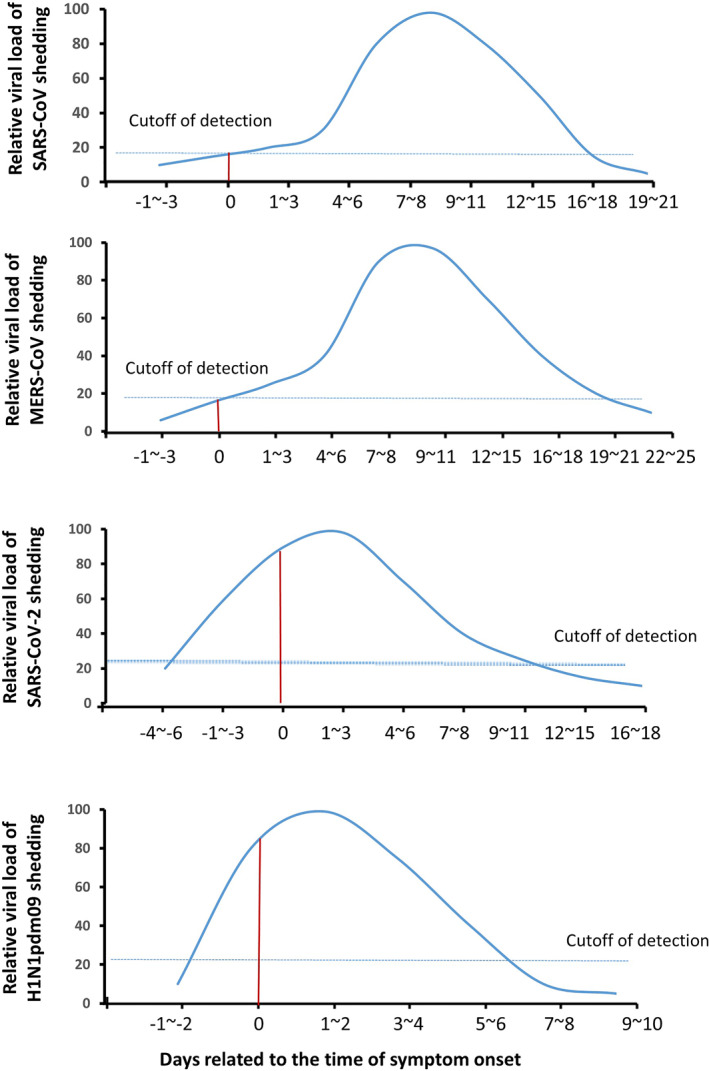
Schematic diagram of viral shedding dynamic in upper respiratory tract of SARS, MERS, Covid‐19 and H1N1pdm09. Y‐axis represents relative viral loads to the peak level and X‐axis represents the days related to the time of symptom onset. Viral shedding of SARS 32 , 33 , 34 and MERS 35 , 36 starts from the time of symptom onset and peaked in the second week of illness and mostly became undetectable after 3 weeks since symptom onset. Viral shedding of Covid‐19 37 , 38 and H1N1pdm09 41 , 42 , 43 starts from the incubation period and peaked around the time of symptom onset. The red line indicates the viral shedding on the day of symptom onset (0) and the dotted line indicates the cutoff level of detection. Abbreviations: Covid‐19, coronavirus disease‐2019; H1N1pdm09, 2009 pandemic H1N1 influenza A; MERS, Middle Eastern respiratory syndrome; SARS, severe acute respiratory syndrome coronavirus; URT, upper respiratory tract
Similar to SARS, MERS‐CoV RNA can be detected by RT‐PCR in URT of MERS patients at the time of symptom onset and peaked during the second week after symptom onset, 35 , 36 and higher viral titres were related to the severity of the illness (Figure 1b). 35
4.2. The viral shedding of Covid‐19 and H1N1pdm09 peaked around time of symptom onset
In contrast to SARS and MERS, viral loads of SARS‐CoV‐2 in Covid‐19 patients were detectable in the incubation period and peaked at the time of symptom onset and subsequently declined with time (Figure 1c). 37 , 38 The highest positive rate of viral RNA from the respiratory tract by RT‐PCR was detected in the first week after symptomatic onset and then declined. 37 , 39 The duration of viral shedding in URT ranged 1–53.5 days and the aggregate duration of relative studies was 14.5 days. 40 The dynamics of viral shedding are not affected by disease severity, sex and age groups. 37
H1N1pdm09 infection has a shorter incubation period and viral shedding period compared to Covid‐19, but they share a similar viral shedding dynamic. Viral shedding of H1N1pdm09 starts 1–3 days before symptom onset, peaks at 1–2 days and lasts around 1 week after symptom onset (Figure 1d). 41 , 42 , 43
The cause of differences observed in virus shedding dynamics among these viruses is not completely understood but may relate to the sites of viral replication in the respiratory tract. A detailed study of nine cases of Covid‐19 suggested that the high viral shedding in the URT in the early stage of infection was due to active viral replication in tissues of the URT as evidenced by the detection of viral replicative RNA intermediates in the throat‐swab samples, and sequence‐distinct virus populations in throat and lung samples from the same patient. 44 The active viral replication of SARS‐CoV‐2 and influenza A virus, but not SARS‐CoV and MERS‐CoV in the URT was also demonstrated in animal models. The results from a golden Syrian hamster (GSH) animal model showed that the viral titres in nasal turbinates were higher than the titres in lung tissue in SARS‐CoV‐2 infected GSH, 45 while virus titres in lung tissue were higher than that in nasal turbinate in SARS‐CoV infected animals. 46 By using a non‐human primate cynomolgus macaque model of virus infection, viral shedding can be detected in nasal samples of animals infected by SARS‐CoV‐2, SARS‐CoV or MERS‐CoV‐2. 47 , 48 However, only SARS‐CoV‐2 antigen was detected in ciliated epithelial cells of the nasal mucosae, a feature not seen in SARS‐CoV or MERS‐CoV infected animals, indicating a SARS‐CoV‐2 tropism for the nasal mucosa that enables efficient respiratory transmission. 47 , 48 As SARS‐CoV and SARS‐CoV‐2 engage the same cellular receptor for entry, the factors that contribute this cell tropism difference remain to be investigated. Nasal mucosa tropism was also reported in influenza A virus ferret model in where viral replication in URT was demonstrated to be an important determinant for virus shedding and transmissibility. 49 These observations indicate that the virus shedding temporal dynamic is associated with the sites of viral replication and transmissibility.
The virus shedding of SARS and MERS peaks after 1 week of symptom onset, when patients are hospitalised. This is consistent with predominance of SARS and MERS outbreaks in healthcare settings. 1 , 17 Whereas, the high viral shedding of SARS‐CoV‐2 and influenza during the early stage of infection reflects the highly infectious nature of pre‐symptomatic and mildly symptomatic patients. 50
5. THE PRE‐SYMPTOMATIC TRANSMISSION OF SARS, MERS, Covid‐19 AND H1N1pdm09
Pre‐symptomatic transmission refers to viral transmission by an infected person who is in the incubation period. The investigation of pre‐symptomatic transmission is usually by cluster studies and prospective cohort studies of the close contacts of confirmed cases.
5.1. Pre‐symptomatic transmission of SARS and MERS is rare
There has been no reported instance of SARS transmission by a carrier before symptom onset. 51 A serological investigation in Beijing in 2004 of 363 individuals with a history of close contact with SARS carriers who were in the incubation period showed none became infected. 52
Similar to SARS, there has been no confirmed transmission of MERS by a pre‐symptomatic carrier. However, a study in Korea suggested that a small number of cases might have been infected before their infectors became symptomatic and that infectiousness may begin 0.4 days (95% credible interval [CI]: −1.2 to 2.4) before illness onset. However, other sources of infection could not be excluded. 53 The mean incubation periods of SARS and MERS are 4.7 and 5.8 days respectively, which is much shorter than the mean of serial interval, 8.4 and 12.6 days, suggesting that transmission from pre‐symptomatic infection is unlikely (Table 1).
5.2. Pre‐symptomatic transmission plays roles in Covid‐19 and H1N1pdm09 spread
Due to high levels of viral shedding in the early stage of infection and the shorter serial intervals compared to the incubation period (Table 1), viral transmission of Covid‐19 and H1N1pdm09 by pre‐symptomatic patients was surmised and later supported by evidence, especially as shown by analysis of family clusters that are exemplified in the following.
Several cluster studies of Covid‐19 showed that transmitted secondary cases had symptom onset before or concurrent with index cases, indicating the occurrence of pre‐symptomatic transmission, but the exact time of transmission was not identified. In a familial cluster in Zhengzhou, China, a secondary case had symptom onset a day before the index primary patient and three other secondary cases had symptom onset on the same day as the primary patient. 54 A report of a family cluster in Taiwan identified a husband becoming symptomatic on the same day of his wife who had returned from Wuhan 5 days previously. 55 Another familial cluster in Shanghai, China showed that an 88‐year‐old male had symptoms 5 days earlier than the index patient. 56
Analyses of other clusters pinpointed the time of transmission. A cluster of Covid‐19 in Zhoushan, China showed two people who were infected by contact with an infected traveller, 1 day before he had symptoms. 57 In Singapore, an index patient transmitted Covid‐19 in a singing class 2 days before symptom onset. Covid‐19 transmission in a church involved two index patients who transmitted to three other people 3–4 days before symptom onset. 58 A familial cluster in Zhoushan city, China reported that four family members were infected by SARS‐CoV‐2 from an index patient who left the home and became symptomatic 4 days later. 59 In Xuzhou, China, five members of two households were infected by a pre‐symptomatic index patient 6 days before symptom onset. 60 These cluster data suggest that SARS‐CoV‐2 can be transmitted from an infected individual up to 6 days before symptom onset.
Pre‐symptomatic transmission of SARS‐CoV‐2 in the wider community has been demonstrated. A study in Singapore indicated that 6.4% of locally transmitted Covid‐19 cases by 16 March 2020 involved pre‐symptomatic patients. 58 An analysis of 468 cases in China suggested that 12% of Covid‐19 cases involved pre‐symptomatic viral carriers. 61 Based on the data of serial interval, a model analysis estimated that 44% of secondary cases were infected during the index cases' pre‐symptomatic stage. 37
Regarding H1N1pdm09, a study showed virus shedding and transmission occurred as early as 5 days before symptom onset. Univariate analyses estimated that 69% of children up to 5 years of age, 67% of children 6–15 years of age and 45% of adults (16 years of age or older) showed pre‐symptomatic H1N1pdm09 viral shedding. 62 Investigation of three clusters of H1N1pdm09 infection in Japan demonstrated pre‐symptomatic transmission at least 1 day prior to the index case developing symptoms. 63 A familial cluster in Brazil showed transmission by a pre‐symptomatic patient 1 day before symptomatic onset. 64 However, another study involving party guests showed that none (0 out of 9) of those who had left the party before symptom onset of an index case became infected, compared to 7 out of 17 (41%) who stayed overnight, suggesting that pre‐symptomatic transmission was less effective. 65
In spite of similar viral shedding dynamics in the URT during the incubation period of SARS‐CoV‐2 66 , 67 and H1N1pdm09 infections, 66 , 67 the pre‐symptomatic transmission of Covid‐19 likely plays more important role in virus spread than that of H1N1pdm09 because of a longer incubation period.
6. THE PROPORTION AND TRANSMISSIBILITY OF ASYMPTOMATIC INFECTION OF SARS, MERS, Covid‐19 AND H1N1pdm09
Asymptomatic infections refer to individuals who are infected and carry virus, but lack noticeable symptoms throughout the infection course. The proportions of asymptomatic infection and their roles in SARS, MERS, Covid‐19 and H1N1pdm09 transmission vary significantly.
6.1. Asymptomatic infection of SARS is rare
Serological testing of 80 health workers in Singapore who were exposed to SARS cases prior to strict interdiction control measures showed 45 (56%) positive cases. Of the 45 positive cases, six (13%) were asymptomatic, two (4%) had subclinical illness and 37 (82%) had pneumonic SARS. 68 Of 146 hospital staff in Vietnam who came into contact with SARS patients during the outbreak in 2003, 43 (29.5%) developed SARS, while 16 (11%) were asymptomatic but SARS‐CoV seropositive. 69
Community‐based testing of 1068 asymptomatic close contacts of SARS patients during the 2003 Hong Kong epidemic detected only two (0.19%) individuals with a low titre of anti‐SARS‐CoV IgG antibody. 70 Another community‐based study conducted in Hong Kong 10 months after SARS struck detected 53 (0.44%) of 12,000 people that were IgG antibody positive to the SARS‐CoV nucleocapsid (N) protein. Only seven of these 53 positive sera also reacted with the native N antigen and six of these seven individuals had SARS previously. 71 Thus, only one individual was likely to have had asymptomatic SARS infection. In children, 2 (0.57%) of 353 asymptomatic children from a high‐risk area in China tested positive for SARS‐CoV antibody compared to 0 of 361 from a low‐risk region. 72 Asymptomatic infection was also not detected in SARS case contactors in France 73 and Taiwan. 74
These studies indicate that asymptomatic infection of SARS is uncommon and the role in SARS transmission by asymptomatic infection is negligible.
6.2. Transmission of MERS‐CoV via asymptomatic infection is unconfirmed
In Saudi Arabia, the seroprevalence of MERS‐CoV specific antibody was significantly higher in persons who had occupational exposure to camels compared to the general population (camel shepherds, 2.3%; slaughterhouse workers, 3.6% and the general population 0.2%). 75 A similar study in Abu Dhabi (the United Arab Emirates) detected 17% MERS‐CoV antibody positive people who had occupational contact with camels. 76
A recent systematic review of 10 publications of MERS‐CoV asymptomatic infection found that the extent of asymptomatic MERS infection had increased temporally. 77 In early reports of MERS infections between April 2012 and October 2013, 12.5% were asymptomatic among 144 MERS cases that were confirmed by RT‐PCR. By 2014, the proportion of asymptomatic cases rose to 25.1% among 255 confirmed cases. However, the transmission by asymptomatic infections to close contacts was less than 1%. 77
Among 1125 laboratory‐confirmed MERS‐CoV cases reported to WHO during 1 January 2015 to 13 April 2018, a total of 157 (14%) had an unknown exposure that may have involved transmission by asymptomatic virus carriers. 78 A study in Korea did not detect MERS‐CoV transmission among 82 people that were exposed to an individual with asymptomatic or mild MERS‐CoV infection. 79 In summary, asymptomatic infection of MERS‐CoV is common, but transmission of MERS‐CoV via asymptomatic infection has not been confirmed.
6.3. The role of asymptomatic transmission in H1N1pdm09 spread is not categorical
Prospective studies of H1N1pdm09 outbreak involving households in Canada, Germany and Viet Nam reported the proportion of asymptomatic cases at 10%, 80 14% 81 and 45%, 42 respectively. Retrospective serological studies of the H1N1pdm09 outbreak revealed the proportion of asymptomatic infections were 45% in New Zealand and 84% in Austria. 82 Overall, asymptomatic infection by influenza is common.
The role of asymptomatic transmission in influenza virus spread has not been conclusively demonstrated. One study showed that virus shedding dynamics in symptomatic and asymptomatic cases was similar, 42 while another study indicated that the titre of virus shedding was lower in paucisymptomatic and asymptomatic cases than in symptomatic cases 83 and infectivity of influenza was not associated with the titre of virus shedding. 84
6.4. Asymptomatic infection plays roles in SARS‐CoV‐2 transmission
Familial cluster studies have identified asymptomatic infection of SARS‐CoV‐2. A familial cluster of Covid‐19 in Shenzhen, China showed that five family members had symptomatic infection, while a 10‐year‐old child had only radiological ground‐glass lung opacities but no other symptoms. 85 Another family cluster in Guangzhou, China showed three family members had SARS‐CoV‐2 detected in URT samples but two of them were asymptomatic. 86
A screen of residents in a skilled care nursing facility in Washington identified 13 non‐symptomatic infections among 23 SARS‐CoV‐2 RNA positive cases. Ten of the 13 developed symptoms 1 week later while three remained asymptomatic. 87 In this small cohort, 46% (10 out of 23) of cases were pre‐symptomatic and 13% (3 out of 23) were asymptomatic virus carriers. A modelling analysis of Covid‐19 cases on board the Diamond Princess cruise ship in Japan estimated that the asymptomatic proportion was 17.9% (95% CI:15.5%–20.2%), 67 which overlaps with the estimation of 33.3% (95% CI:8.3%–58.3%) from data of Japanese citizens evacuated from Wuhan. 88
A prospective study of 24 RT‐PCR positive SARS‐CoV‐2 cases that were screened from the close patient‐contacts in Nanjing, China demonstrated seven (29.7%) were asymptomatic. The asymptomatic cases were significantly younger than those who developed symptoms. The period of detectable viral RNA in asymptomatic infections was 2–15 days, with a median of 4 days. 89 A similar study of 78 confirmed SARS‐CoV‐2 infections showed that 33 cases (42.3%) were asymptomatic, while 45 cases (57.7%) were symptomatic. In comparison to symptomatic patients, asymptomatic SARS‐CoV‐2 cases were younger and had a shorter duration of viral shedding. The mean duration of viral shedding in symptomatic and asymptomatic individuals was 19 and 8 days, respectively. 90 In contrast, another study reported longer viral shedding in asymptomatic infection (median duration 19 days) than symptomatic individuals (median duration 14 days). 91
A SARS‐CoV‐2 antibody test used to evaluate 865 community‐based individuals during 13 and 14 April 2020 in Los Angeles County, revealed a 4.06% positive rate. The estimate implies that 36,700 adults in the county had SARS‐CoV‐2 antibodies, which is substantially greater than the 8430 confirmed cases by 10 April in the county. 92 A study of 17,368 individuals in Wuhan and surrounding regions of Hubei province during the period from 9 March 2020 to 10 April 2020 revealed seroprevalence rate of SARS‐CoV‐2 at 3.2% and 3.8%, respectively. 93 A study in Hong Kong revealed a 3.3% (15 out of 452) SARS‐CoV‐2 seroprevalence rate among asymptomatic Hubei returnees. All these studies were conducted without random sampling, so cautious interpretation of these results is required. Nonetheless, they suggest that there is a substantial amount of undiagnosed asymptomatic or mild SARS‐CoV‐2 infections in the epidemic communities.
Transmission of SARS‐CoV‐2 by asymptomatic infections was reported. A prospective study conducted in Nanjing, China found that one asymptomatic infection caused a familial cluster transmission to three other members. 89 Another familial cluster in Anyang, China identified that a 20‐year‐old asymptomatic woman transmitted SARS‐CoV‐2 to her five family members and relatives. 94
A prospective study in Ningbo, China found that 51 symptomatic patients caused 121 new infections, while eight asymptomatically infected individuals caused six new infections, 95 , 96 which is equivalent to an R 0 of 2.37 from symptomatic infection and 0.75 from asymptomatic infection, suggesting that transmissibility of asymptomatic virus carrier is lower than symptomatic carrier but still a significant contributor to community transmission.
The exact role of Covid‐19 transmission by asymptomatic carriers in the pandemic requires further study. 97 In the meantime, a large proportion of mildly symptomatic infections are not easily distinguished from asymptomatic virus carriers, and they are usually young and more active socially. Therefore, asymptomatic and mild infections play important roles in Covid‐19 spread. 98
7. CONCLUSIONS
The combined virological, epidemiological and clinical features of these four viral infections, SARS, MERS, Covid‐19 and H1N1pdm09, determine the patterns of viral spread and the control measures required. For MERS and SARS, low viral shedding and high proportions of symptomatic cases in the early stage of infection, and lack of transmission by pre‐symptomatic and asymptomatic infection enable the symptomatic identification, isolation and quarantine of the transmission sources. Therefore, syndromic surveillance, isolation of patients and quarantine of their contacts were effective measures to contain the spread of SARS‐CoV and MERS‐CoV. 53
In contrast, viral shedding of Covid‐19 and influenza peaks at an early stage of infection when virus carriers show no or only mild symptoms, which results in reduced efficiency of identifying and isolating the transmission source by symptomatic screening. A modelling study showed that airport symptomatic screening was unable to identify 46% of Covid‐19 travellers. 99 Border control measures reduced virus spread between countries, but it did not stop Covid‐19 importations and global spread. 100 Therefore, border closing has been enforced in many countries worldwide. On the other hand, early virus shedding allows identification and isolation of a transmission source by testing for viral RNA, which has become an essential Covid‐19 control measure.
A high proportion of transmission competent, pre‐symptomatic, asymptomatic and mildly symptomatic infection remains a major challenge to controlling Covid‐19 spread in communities so social distancing and public masking are required. The principle of social distancing is to stop a respiratory transmitted pathogen by physically separating individuals in the community, and public masking can set barriers for airborne pathogen release and uptake between individuals, especially when social distance is impossible. 101 These two control measures are effective when most individuals are susceptible to infection, and identification and isolation of the transmission source is difficult. Modelling studies and evidence indicate that these two control measures can effectively cut the transmission chain of SARS‐CoV‐2 and stop Covid‐19 spread in communities. 102 , 103 , 104 , 105 , 106
CONFLICT OF INTEREST
All the authors declare that there is no existing commercial or financial conflict of interest, in any way.
AUTHOR CONTRIBUTIONS
All the authors contributed to the information collection, analysis, writing and approving the final version of the manuscript. Dongsheng Li and Dongsheng Hu also contributed to planning and design.
ACKNOWLEDGEMENTS
Zhonglan Wu is involved in the project of ‘the pathogen and epidemiology study of viral pneumonia in Ningxia, China’ which is supported by the grant (2020BEG01001) of Ningxia Key Research and Development Program and the grant (RQ0126) of Excellent Young Talents Fund Program of Ningxia Province.
Contributor Information
Dongsheng Hu, Email: Dongsheng.Li@qimrberghofer.edu.au.
Dongsheng Li, Email: Dongsheng.Li@qimrberghofer.edu.au.
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analysed during the current study. The data that support the findings of this study are openly available in the cited reference.
References
REFERENCES
- 1. de Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peeri NC, Shrestha N, Rahman MS, et al. The SARS, MERS and novel coronavirus (COVID‐19) epidemics, the newest and biggest global health threats: what lessons have we learned?. Int J Epidemiol. 2020;49(3):717–726. 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO . Middle East respiratory syndrome coronavirus (MERS‐CoV) – Qatar 2020. https://www.who.int/csr/don/12-march-2020-mers-qatar/en/. Accessed 30 March 2020.
- 4. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of Novel coronavirus–Infected pneumonia. N Engl J Med. 2020;382 (13):1199–1207. 10.1056/nejmoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. WHO . Coronavirus disease 2019 (COVID‐19) situation report – 72 [Organization website]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200401-sitrep-72-covid-19.pdf?sfvrsn=3dd8971b_2. Accessed 30 July, 2020.
- 6. WHO . Coronavirus disease (COVID‐19) situation report – 189 [report] 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200727-covid-19-sitrep-189.pdf?sfvrsn=b93a6913_2. Accessed 28 July 2020.
- 7. Fineberg HV. Pandemic preparedness and response‐‐lessons from the H1N1 influenza of 2009. N Engl J Med. 2014;370(14):1335‐1342. [DOI] [PubMed] [Google Scholar]
- 8. Simonsen L, Spreeuwenberg P, Lustig R, et al. Global mortality estimates for the 2009 Influenza Pandemic from the GLaMOR project: a modeling study. PLoS Med. 2013;10(11):e1001558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li W, Shi Z, Yu M, et al. Bats are natural reservoirs of SARS‐like coronaviruses. Science. 2005;310(5748):676‐679. [DOI] [PubMed] [Google Scholar]
- 10. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu A, Peng Y, Huang B, et al. Genome composition and divergence of the novel coronavirus (2019‐nCoV) originating in China. Cell Host Microbe. 2020;27(3):325‐328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4(6):1011‐1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li F. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol. 2016;3(1):237‐261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge XY, Li JL, Yang XL, et al. Isolation and characterization of a bat SARS‐like coronavirus that uses the ACE2 receptor. Nature. 2013;503(7477):535‐538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wrapp D, Wang N, Corbett KS, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lukassen S, Chua RL, Trefzer T, et al. SARS‐CoV‐2 receptor ACE2 and TMPRSS2 are primarily expressed in bronchial transient secretory cells. EMBO J. 2020:e105114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan JF, Lau SK, To KK, Cheng VC, Woo PC, Yuen KY. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS‐like disease. Clin Microbiol Rev. 2015;28(2):465‐522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van Riel D, den Bakker MA, Leijten LM, et al. Seasonal and pandemic human influenza viruses attach better to human upper respiratory tract epithelium than avian influenza viruses. Am J Pathol. 2010;176(4):1614‐1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Neumann G, Kawaoka Y. The first influenza pandemic of the new millennium. Influenza Other Respir Viruses. 2011;5(3):157‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cheng VC, To KK, Tse H, Hung IF, Yuen KY. Two years after pandemic influenza A/2009/H1N1: what have we learned? Clin Microbiol Rev. 2012;25(2):223‐263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nardell EA, Nathavitharana RR. Airborne spread of SARS‐CoV‐2 and a potential role for air disinfection. JAMA. 2020;324(2):141. 10.1001/jama.2020.7603. [DOI] [PubMed] [Google Scholar]
- 22. Tellier R. Aerosol transmission of influenza A virus: a review of new studies. J R Soc Interface. 2009;6(Suppl 6):S783‐S790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N Engl J Med. 2020;382(16):1564‐1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fabian P, McDevitt JJ, DeHaan WH, et al. Influenza virus in human exhaled breath: an observational study. PLoS One. 2008;3(7):e2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lloyd‐Smith JO, Schreiber SJ, Kopp PE, Getz WM. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438(7066):355‐359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Delamater PL, Street EJ, Leslie TF, Yang YT, Jacobsen KH. Complexity of the basic reproduction number (R0). Emerg Infect Dis. 2019;25(1):1‐4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Gayle AA, Wilder‐Smith A, Rocklöv J. The reproductive number of COVID‐19 is higher compared to SARS coronavirus. J Travel Med. 2020;27(2). 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and re‐emerging infection. Clin Microbiol Rev. 2007;20(4):660‐694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Frieden TR, Lee CT. Identifying and interrupting superspreading events‐implications for control of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(6):1059‐1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chowell G, Abdirizak F, Lee S, et al. Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med. 2015;13:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guan W‐J, Ni Z‐Y, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708–1720. 10.1056/nejmoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Poon LL, Chan KH, Wong OK, et al. Early diagnosis of SARS coronavirus infection by real time RT‐PCR. J Clin Virol. 2003;28(3):233‐238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peiris JS, Chu CM, Cheng VC, et al. Clinical progression and viral load in a community outbreak of coronavirus‐associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767‐1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cheng PK, Wong DA, Tong LK, et al. Viral shedding patterns of coronavirus in patients with probable severe acute respiratory syndrome. Lancet. 2004;363(9422):1699‐1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oh MD, Park WB, Choe PG, et al. Viral load kinetics of MERS coronavirus infection. N Engl J Med. 2016;375(13):1303‐1305. [DOI] [PubMed] [Google Scholar]
- 36. Al‐Abdely HM, Midgley CM, Alkhamis AM, et al. Middle East respiratory syndrome coronavirus infection dynamics and antibody responses among clinically diverse patients, Saudi Arabia. Emerg Infect Dis. 2019;25(4):753‐766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He X, Lau EHY, Wu P, et al. Temporal dynamics in viral shedding and transmissibility of COVID‐19. Nat Med. 2020;26(5):672‐675. [DOI] [PubMed] [Google Scholar]
- 38. To KK, Tsang OT, Leung WS, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients with novel coronavirus disease 2019. Clin Infect Dis. 2020. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walsh KA., Jordan K, Clyne B, et al. SARS‐CoV‐2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81(3):357–371. 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. To KK, Chan KH, Li IW, et al. Viral load in patients infected with pandemic H1N1 2009 influenza A virus. J Med Virol. 2010;82(1):1‐7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thai PQ, Mai le Q, Welkers MR, et al. Pandemic H1N1 virus transmission and shedding dynamics in index case households of a prospective Vietnamese cohort. J Infect. 2014;68(6):581‐590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fielding JE, Kelly HA, Mercer GN, Glass K. Systematic review of influenza A(H1N1)pdm09 virus shedding: duration is affected by severity, but not age. Influenza Other Respiratory Viruses. 2014;8 (2):142–150. 10.1111/irv.12216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID‐2019. Nature. 2020;581(7809):465–469. 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 45. Chan JF‐W, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of Coronavirus Disease 2019 (COVID‐19) in golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020. 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roberts A, Vogel L, Guarner J, et al. Severe acute respiratory syndrome coronavirus infection of golden Syrian hamsters. J Virol. 2005;79(1):503‐511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rockx B, Kuiken T, Herfst S, et al. Comparative pathogenesis of COVID‐19, MERS, and SARS in a nonhuman primate model. Science. 2020;29(368):1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kuiken T, Fouchier RA, Schutten M, et al. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet. 2003;362(9380):263‐270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Richard M, van den Brand JMA, Bestebroer TM, et al. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat Commun. 2020;11(1):766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID‐19) outbreak in China: summary of a report of 72314 cases from the Chinese center for disease control and prevention. J Am Med Assoc. 2020;323(13):1239–1242. [DOI] [PubMed] [Google Scholar]
- 51. Peiris JS, Yuen KY, Osterhaus AD, Stohr K. The severe acute respiratory syndrome. N Engl J Med. 2003;349(25):2431‐2441. [DOI] [PubMed] [Google Scholar]
- 52. Zeng G, Xie SY, Li Q, Ou JM. Infectivity of severe acute respiratory syndrome during its incubation period. Biomed Environ Sci. 2009;22(6):502‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cowling BJ, Park M, Fang VJ, Wu P, Leung GM, Wu JT. Preliminary epidemiological assessment of MERS‐CoV outbreak in South Korea, May to June 2015. Euro Surveill. 2015;20(25):7‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Qiu YY, Wang SQ, Wang XL, et al. [Epidemiological analysis on a family cluster of COVID‐19]. Zhonghua Liuxingbingxue Zazhi. 2020;41(4):506‐509. [DOI] [PubMed] [Google Scholar]
- 55. Liu YC, Liao CH, Chang CF, Chou CC, Lin YR. A locally transmitted case of SARS‐CoV‐2 infection in Taiwan. N Engl J Med. 2020;382(11):1070‐1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yu P, Zhu J, Zhang Z, Han Y, Huang L. A familial cluster of infection associated with the 2019 novel coronavirus indicating potential person‐to‐person transmission during the incubation period. J Infect Dis. 2020;221(11):1757–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tong ZD, Tang A, Li KF, et al. Potential presymptomatic transmission of SARS‐CoV‐2, Zhejiang province, China, 2020. Emerg Infect Dis. 2020;26(5):1052–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wei WE, Li Z, Chiew CJ, Yong SE, Toh MP, Lee VJ. Presymptomatic transmission of SARS‐CoV‐2 ‐ Singapore, January 23‐March 16, 2020. MMWR Morb Mortal Wkly Rep. 2020;69(14):411‐415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li P, Fu JB, Li KF, et al. Transmission of COVID‐19 in the terminal stage of incubation period: a familial cluster. Int J Infect Dis. 2020;96:452–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Li C, Ji F, Wang L, et al. Asymptomatic and human‐to‐human transmission of SARS‐CoV‐2 in a 2‐family cluster, Xuzhou, China. Emerg Infect Dis. 2020;26(7):1626–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Du Z, Xu X, Wu Y, Wang L, Cowling BJ, Meyers LA. Serial interval of COVID‐19 among publicly reported confirmed cases. Emerg Infect Dis. 2020;26(6):1341–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ng S, Lopez R, Kuan G, et al. The timeline of influenza virus shedding in children and adults in a household transmission study of influenza in Managua, Nicaragua. Pediatr Infect Dis J. 2016;35(5):583‐586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Gu Y, Komiya N, Kamiya H, Yasui Y, Taniguchi K, Okabe N. Pandemic (H1N1) 2009 transmission during presymptomatic phase, Japan. Emerg Infect Dis. 2011;17(9):1737‐1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Freitas FT, Cabral AP, Barros EN, et al. Pre‐symptomatic transmission of pandemic influenza H1N1 2009: investigation of a family cluster, Brazil. Epidemiol Infect. 2013;141(4):763‐766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hermes J, Bernard H, Buchholz U, et al. Lack of evidence for pre‐symptomatic transmission of pandemic influenza virus A(H1N1) 2009 in an outbreak among teenagers; Germany 2009. Influenza Other Respir Viruses. 2011;5(6):e499‐e503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhou R, Li F, Chen F, et al. Viral dynamics in asymptomatic patients with COVID‐19. Int J Infect Dis. 2020;96:288‐290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Mizumoto K, Kagaya K, Zarebski A, Chowell G. Estimating the asymptomatic proportion of coronavirus disease 2019 (COVID‐19) cases on board the Diamond Princess cruise ship, Yokohama, Japan, 2020. Euro Surveill. 2020;25(10). 10.2807/1560-7917.es.2020.25.10.2000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wilder‐Smith A, Teleman MD, Heng BH, Earnest A, Ling AE, Leo YS. Asymptomatic SARS coronavirus infection among healthcare workers, Singapore. Emerg Infect Dis. 2005;11(7):1142‐1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nishiyama A, Wakasugi N, Kirikae T, et al. Risk factors for SARS infection within hospitals in Hanoi, Vietnam. Jpn J Infect Dis. 2008;61(5):388‐390. [PubMed] [Google Scholar]
- 70. Leung GM, Chung PH, Tsang T, et al. SARS‐CoV antibody prevalence in all Hong Kong patient contacts. Emerg Infect Dis. 2004;10(9):1653‐1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Leung DT, van Maren WW, Chan FK, et al. Extremely low exposure of a community to severe acute respiratory syndrome coronavirus: false seropositivity due to use of bacterially derived antigens. J Virol. 2006;80(18):8920‐8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lee PP, Wong WH, Leung GM, et al. Risk‐stratified seroprevalence of severe acute respiratory syndrome coronavirus among children in Hong Kong. Pediatrics. 2006;117(6):e1156‐e1162. [DOI] [PubMed] [Google Scholar]
- 73. Le Vu S, Yazdanpanah Y, Bitar D, Emmanuelli J, Bonmarin I, Desenclos JC. Absence of infection in asymptomatic contacts of index SARS case in France. Euro Surveill. 2006;11(1):40‐41. [PubMed] [Google Scholar]
- 74. Lee CC, Chen SY, Chang IJ, et al. Seroprevalence of SARS coronavirus antibody in household contacts. Epidemiol Infect. 2005;133(6):1119‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Muller MA, Meyer B, Corman VM, et al. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross‐sectional, serological study. Lancet Infect Dis. 2015;15(5):559‐564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Khudhair A, Killerby ME, Al Mulla M, et al. Risk factors for MERS‐CoV seropositivity among animal market and slaughterhouse workers, Abu Dhabi, United Arab Emirates, 2014‐2017. Emerg Infect Dis. 2019;25(5):927‐935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Al‐Tawfiq JA, Gautret P. Asymptomatic Middle East respiratory syndrome coronavirus (MERS‐CoV) infection: extent and implications for infection control: a systematic review. Travel Med Infect Dis. 2019;27:27‐32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Conzade R, Grant R, Malik MR, et al. Reported direct and indirect contact with dromedary camels among laboratory‐confirmed MERS‐CoV cases. Viruses. 2018;10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Moon SY, Son JS. Infectivity of an asymptomatic patient with Middle East respiratory syndrome coronavirus infection. Clin Infect Dis. 2017;64(10):1457‐1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Papenburg J, Baz M, Hamelin ME, et al. Household transmission of the 2009 pandemic A/H1N1 influenza virus: elevated laboratory‐confirmed secondary attack rates and evidence of asymptomatic infections. Clin Infect Dis. 2010;51(9):1033‐1041. [DOI] [PubMed] [Google Scholar]
- 81. Suess T, Remschmidt C, Schink SB, et al. Comparison of shedding characteristics of seasonal influenza virus (sub)types and influenza A(H1N1)pdm09; Germany, 2007‐2011. PLoS One. 2012;7(12):e51653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Poeppl W, Poeppl G, Hunger M, et al. Pre‐ and post‐pandemic prevalence of antibodies to the 2009 pandemic influenza A (H1N1) virus in Austrian adults. J Med Virol. 2012;84(9):1331‐1334. [DOI] [PubMed] [Google Scholar]
- 83. Ip DK, Lau LL, Leung NH, et al. Viral shedding and transmission potential of asymptomatic and paucisymptomatic influenza virus infections in the community. Clin Infect Dis. 2017;64(6):736‐742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tsang TK, Fang VJ, Chan KH, et al. Individual correlates of infectivity of influenza A virus infections in households. PLoS One. 2016;11(5):e0154418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chan JF, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Pan X, Chen D, Xia Y, et al. Asymptomatic cases in a family cluster with SARS‐CoV‐2 infection. Lancet Infect Dis. 2020;20(4):410‐411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kimball A, Hatfield KM, Arons M, et al. Asymptomatic and presymptomatic SARS‐CoV‐2 infections in residents of a long‐term care skilled nursing facility ‐ king county, Washington, March 2020. MMWR Morb Mortal Wkly Rep. 2020;69(13):377‐381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Nishiura H, Kobayashi T, Suzuki A, et al. Estimation of the asymptomatic ratio of novel coronavirus infections (COVID‐19). Int J Infect Dis. 2020;94:154–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hu Z, Song C, Xu C, et al. Clinical characteristics of 24 asymptomatic infections with COVID‐19 screened among close contacts in Nanjing, China. Sci China Life Sci. 2020;63(5):706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Yang R, Gui X, Xiong Y. Comparison of clinical characteristics of patients with asymptomatic vs symptomatic coronavirus disease 2019 in Wuhan, China. JAMA Netw Open. 2020;3(5):e2010182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat Med. 2020;26(8):1200–1204. [DOI] [PubMed] [Google Scholar]
- 92. Sood N, Simon P, Ebner P, et al. Seroprevalence of SARS‐CoV‐2‐specific antibodies among adults in Los Angeles county, California. J Am Med Assoc. 2020;323(23):2425–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Xu X, Sun J, Nie S, et al. Seroprevalence of immunoglobulin M and G antibodies against SARS‐CoV‐2 in China. Nat Med. 2020;26(8):1193–1195. [DOI] [PubMed] [Google Scholar]
- 94. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. J Am Med Assoc. 2020;323(14):1406–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wu Z. Asymptomatic and pre‐symptomatic cases of COVID‐19 contribution to spreading the epidemic and need for targeted control strategies. Chin J Epidemiol. 2020:41. [DOI] [PubMed] [Google Scholar]
- 96. Chen Yi WA, Yi Bo, Ding Keqin, et al. The epidemiological characteristics of infection in close contacts of COVID‐19 in Ningbo city. Chin J Epidemiol. 2020;41. [DOI] [PubMed] [Google Scholar]
- 97. Gao WJ, Li LM. [Advances on presymptomatic or asymptomatic carrier transmission of COVID‐19]. Zhonghua Liuxingbingxue Zazhi. 2020;41:485‐488. [DOI] [PubMed] [Google Scholar]
- 98. Dong Y, Mo X, Hu Y, et al. Epidemiology of COVID‐19 among children in China. Pediatrics. 2020;145(6):e20200702 [DOI] [PubMed] [Google Scholar]
- 99. Quilty BJ, Clifford S, Flasche S, Eggo RM, Cnw group . Effectiveness of airport screening at detecting travellers infected with novel coronavirus (2019‐nCoV). Euro Surveill. 2020;25(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Wells CR, Sah P, Moghadas SM, et al. Impact of international travel and border control measures on the global spread of the novel 2019 coronavirus outbreak. Proc Natl Acad Sci USA. 2020;117(13):7504‐7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. RDS , Darby S, Giltinan A, Smith N. COVID‐19: in the absence of vaccination ‐ ‘mask‐the‐nation’. Future Microbiol. 2020;15:963–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Sjodin H, Wilder‐Smith A, Osman S, Farooq Z, Rocklov J. Only strict quarantine measures can curb the coronavirus disease (COVID‐19) outbreak in Italy, 2020. Euro Surveill. 2020;25(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Koo JR, Cook AR, Park M, et al. Interventions to mitigate early spread of SARS‐CoV‐2 in Singapore: a modelling study. Lancet Infect Dis. 2020;20(6):678–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang J, Litvinova M, Wang W, et al. Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside Hubei province, China: a descriptive and modelling study. Lancet Infect Dis. 2020;20(7):793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Pan A, Liu L, Wang C, et al. Association of public health interventions with the epidemiology of the COVID‐19 outbreak in Wuhan, China. J Am Med Assoc. 2020;323(19):1915–1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Ruan L, Wen M, Zeng Q, et al. New measures for COVID‐19 response: a lesson from the Wenzhou experience. Clin Infect Dis. 2020;71(15):866–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. WHO . WHO coronavirus disease (COVID‐19) dashboard 2020. https://covid19.who.int/. Accessed 8 July 2020.
- 108. Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181‐192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Mena I, Nelson MI, Quezada‐Monroy F, et al. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Yin Y, Wunderink RG. MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23(2):130‐137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Jiang X, Rayner S, Luo MH. Does SARS‐CoV‐2 has a longer incubation period than SARS and MERS? J Med Virol. 2020;92(5):476‐478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lessler J, Reich NG, Cummings DA, et al. Outbreak of 2009 pandemic influenza A (H1N1) at a New York City school. N Engl J Med. 2009;361(27):2628‐2636. [DOI] [PubMed] [Google Scholar]
- 114. Choi S, Jung E, Choi BY, Hur YJ, Ki M. High reproduction number of Middle East respiratory syndrome coronavirus in nosocomial outbreaks: mathematical modelling in Saudi Arabia and South Korea. J Hosp Infect. 2018;99(2):162‐168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Park JE, Jung S, Kim A, Park JE. MERS transmission and risk factors: a systematic review. BMC Publ Health. 2018;18(1):574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Lipsitch M, Cohen T, Cooper B, et al. Transmission dynamics and control of severe acute respiratory syndrome. Science. 2003;300(5627):1966‐1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Wang Y, Wang Y, Chen Y, Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID‐19) implicate special control measures. J Med Virol. 2020;92(6):568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Echevarria‐Zuno S, Mejia‐Arangure JM. Mar‐Obeso AJ, et al. Infection and death from influenza A H1N1 virus in Mexico: a retrospective analysis. Lancet. 2009;374(9707):2072‐2079. [DOI] [PubMed] [Google Scholar]
- 119. Michaelis M, Doerr HW, Cinatl J, Jr. An influenza A H1N1 virus revival ‐ pandemic H1N1/09 virus. Infection. 2009;37(5):381‐389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the current study. The data that support the findings of this study are openly available in the cited reference.


