Abstract
Coronavirus disease 2019 (COVID‐19) has become pandemic since March 11, 2020. Thus, development and integration in clinics of fast and sensitive diagnostic tools are essential. The aim of the study is a development and evaluation of a one‐step quantitative reverse transcription‐polymerase chain reaction (RT‐qPCR) assay (COVID‐19 Amp) for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) detection with an armored positive control and internal controls constructed from synthetic MS2‐phage‐based RNA particles. The COVID‐19 Amp assay limit of detection was 103 copies/ml, the analytical specificity was 100%. A total of 109 biological samples were examined using COVID‐19 Amp and World Health Organization (WHO)‐based assay. Discordance in nine samples was observed (negative by the WHO‐based assay) and discordant samples were retested as positive according to the results obtained from the Vector‐PCRrv‐2019‐nCoV‐RG assay. The developed COVID‐19 Amp assay has high sensitivity and specificity, includes virus particles‐based controls, provides the direct definition of the SARS‐CoV‐2 RdRp gene partial sequence, and is suitable for any hospital and laboratory equipped for RT‐qPCR.
Keywords: COVID‐19, diagnostics, RT‐qPCR, SARS‐CoV‐2
Highlights
One‐step RT‐qPCR assay (COVID‐19 Amp) contains an armored positive control (ARC+) and an armored internal control (ICS) constructed from synthetic MS2‐phage‐based RNA particles.
Displays high specificity and selectivity rendering it a powerful diagnostic test
Suitable for any hospital and laboratory equipped for RT‐qPCR.
1. INTRODUCTION
The novel infectious disease (coronavirus disease 2019 [COVID‐19]) originated from Wuhan, Hubei Province, China, caused by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2; also referred to as novel coronavirus 2019 [nCoV‐19]) has become pandemic since March 11, 2020. 1 Cases of COVID‐19 are now widespread including 188 countries. 2 SARS‐CoV‐2 is the third human‐infecting coronavirus causing severe disease as well as SARS‐CoV and middle east respiratory syndrome‐related coronavirus (MERS‐CoV.) 3 SARS‐CoV‐2 is classified as a member of the subfamily Coronavirinae in the family Coronaviridae and the order Nidovirales, a genera of betacoronavirus. 4 The genome sequence of a novel SARS‐Cov‐2 shows 79.0% and 51.8% identity with SARS‐CoV and MERS‐CoV, respectively, 5 , 6 , 7 as well as the mechanism of cell entry using angiotensin‐converting enzyme 2 receptor. 8 The virus was initially called nCoV‐2019, until the Coronaviridae Study Group of International Committee on Taxonomy of Viruses named the virus SARS‐CoV‐2 based on the phylogenetic analysis, on February 11, 2020. 9 According to the current hypothesis, the first transmission occurred between bats and a yet‐to‐be‐defined intermediate host animal. 10
The most common symptoms related to COVID‐19 are fever and cough 11 , 12 but 22% of patients develop shortness of breath and dyspnea. 13 SARS‐CoV‐2 can be transmitted from human to human. The mode of transmission is through direct contact and droplet spread. 14 In addition, SARS‐CoV‐2 showed the stability in aerosols (<5 μm) for at least up to 3 h and may be more stable on plastic and stainless steel than on copper and cardboard. 15
Accurate diagnosis of COVID‐19 cannot be set based on symptoms due to their nonspecificity. According to the Guan et al. 16 report, 44% of 1099 COVID‐19 patients from China exposed a fever when they entered the hospital and 89% developed a fever while in hospital. Thus, diagnostics can play an important role in the localization of COVID‐19, ensuring the rapid implementation of control measures limiting spread through isolation and contact tracing. The most widely used diagnosing and screening COVID‐19 techniques are nucleic acid testing and computer tomography (CT) scans. 16 , 17 However, molecular techniques are more suitable than syndromic testing targeting specific pathogens when CT scans can only provide an approximate pathology. 18
Real‐time quantitative polymerase chain reaction with reverse transcription (RT‐qPCR) is a widely used technique to detect viral pathogens. As for COVID‐19 diagnostics, RT‐qPCR could target several conserved regions of SARS‐CoV‐2 genome 19 , 20 , 21 , 22 : (1) the RNA‐dependent RNA polymerase gene in the open reading frame ORF1ab region, (2) the envelope protein gene (E), and (3) the nucleocapsid protein gene (N). Both the RdRP and E genes had high analytical sensitivity for detection, whereas the N gene provided poorer analytical sensitivity. 23 The assay also could involve two‐target system detection, where a first primer set detects numerous coronaviruses including SARS‐CoV‐2 and a second primer set is unique for SARS‐CoV‐2. 24
Leastwise 11 nucleic‐acid‐based methods have been approved in China by the National Medical Products Administration for diagnosing SARS‐CoV‐2. 25 Nevertheless, RT‐qPCR is the most generally used method for detecting COVID‐19 in respiratory samples. 26 The United States Centers for Disease Control and Prevention uses an approved one‐step real‐time RT‐qPCR assay to detect the presence of SARS‐CoV‐2. Approximately 11 kits based on RT‐qPCR were approved in the Russian Federation by June 22, 2020 (https://www.roszdravnadzor.ru/). But none of them represents suitable internal and positive controls. Here, we presented a highly sensitive and selective one‐step RT‐qPCR assay for the SARS‐CoV‐2 diagnostics (COVID‐19 Amp), targeting the constant region of RNA‐dependent RNA polymerase gene. The COVID‐19 Amp includes armored recombinant RNA controls and possesses imitation of viral RNA extraction.
2. MATERIALS AND METHODS
2.1. Determination of conservative sites of SARS‐CoV‐2 genome, and primers/probes design for RT‐qPCR
The sequences of the SARS‐CoV‐2 genome available in GenBank in February 2020 were aligned using BLAST software. A 113 bp fragment of RNA‐dependent RNA polymerase (GenBank ID D: NC_045512.2, coordinates 1384–1490) was chosen as a target for amplification using PLOTCON (http://emboss.bioinformatics.nl/cgi‐bin/emboss/plotcon). The primers and probes were designed according to the current guidelines of RT‐PCR techniques. 27 The primers melting temperatures were calculated using the oligonucleotide properties calculator (http://biotools.nubic.northwestern.edu/OligoCalc.html). The analysis of thermodynamic characteristics and secondary structure formation of the probes was conducted using Mfold software (http://unafold.rna.albany.edu/?q=mfold). Probes were modified with attaching of the fluorescent reporter dye rhodamine 6G and black hole quencher 1 at the 5′ and 3′ ends, respectively. The probe CoV_pr was tagged at 5′ end for preventing stable secondary structure formation. The primers and probes were synthesized by DNA‐Synthesis, sequences are presented in Table 1.
Table 1.
Features of the primers and the probes used in the COVID‐19 Amp assay
| Primer/probe | Sequence 5ʹ–3ʹ | Probe type | Gene target | Number of nucleotides | Coordinates on genome (GenBank ID MT457401.1) |
|---|---|---|---|---|---|
| CoV_pr | R6G ‐ TCT TgC CgA ATA CCA TAA TgA ATC Tgg CAA ‐ BHQ1 | TaqMan | RdRp | 30 | 1414–1441 |
| CoV_for | TCA CAA TTC AgA AgT Agg ACC Tg | RdRp | 23 | 1384–1406 | |
| CoV_rev | AgC CTC CAA Agg CAA TAg TgC | RdRp | 21 | 1470–1490 | |
| ICS_pr | FAM ‐ CTA gCT ggg CgT Cag gAA TCC Cag g ‐ BHQ1 | TaqMan | Artificial target | 25 | – |
| ICS_for | CCG GAT TGC GTA TCT CCG GAC T | Artificial target | 22 | – | |
| ICS_rev | CAC GGC GGC ATC TCT ATC ACG A | Artificial target | 22 | – |
Abbreviation: COVID‐19, coronavirus disease 2019.
2.2. Characterization of patients and biological samples
Patients with clinical manifestations of acute respiratory viral infection symptoms were tested for SARS‐CoV‐2 using RT‐qPCR detection. The average age of 109 patients (43 males, 66 females) was 49 years (from 21 to 88), the disease course was mild in 71.6% (n = 78) and moderate in 28.4% (n = 31) cases. All patients received nonspecific treatment at home without hospitalization. No lethal outcomes were registered.
Nasopharyngeal swabs were used as examined biological materials. Samples from patients were collected using commercially available swabs systems at 2–8 days after the onset of COVID‐19. Maximum time storage was estimated as 48 h at 2–8°C with the following shipping on ice pack during no longer than 24 h. Nucleic acid extraction from all 109 samples was performed using the RIBO‐Prep Kit (AmpliSens) with subsequent storage at −70°C.
2.3. Content of reaction mixture and amplification mode
The total volume of reaction mix for the sample was 25 µl containing the following: 1 µl of BioMaster Mix (Biolabmix); 12.5 µl of 2X reaction buffer (Biolabmix); 0.25 µl of each primer and probe (Cov_for, Cov_rev, Cov_pr) with final concentration 0.4 µM for primers and 0.28 µM for probe; 0.25 µl of each primer and probe of the internal control sample (ICS) (ICS_for, ICS_rev, ICS_pr) with the final concentration 0.2 µM for primers and 0.12 µM for probe; and 10 µl of the RNA sample. The amplification regimen was the following: 50°C for 15 min, 95°C for 5 min, and then 40 cycles of 95°C for 10 s and 57°C for 30 s. Fluorescence was observed at 57°C in JOE (for SARS‐CoV‐2) and FAM (for ICS) channels. The reaction was performed using the BioRad CFX96 amplificator. The fluorescence threshold was established as the middle value of the linear increase in the positive‐control fluorescence elevation in the logarithmic units. Amplification results were considered positive if the level of fluorescence crossed the threshold. As controls of the RT‐qPCR, an external positive control for PCR (C+) and an armored recombinant positive control for reverse transcription (ARC+) were applied. The ICS (armored MS2 particles contained artificial RNA sequence) was used to monitor RNA extraction; for this purpose, ICS‐specific primers and probe were added to the reaction mixture. In addition, negative control for extraction (EC−) and PCR (C−) was used to exclude false‐positive results due to possible or unintentional cross‐contamination.
2.4. Positive controls and internal controls preparation
The complementary DNA (cDNA; 113 bp) of SARS‐CoV‐2 RNA‐dependent RNA polymerase gene region that included the primers and probe‐target sequences was constructed using previously developed step‐out amplification. 28 The final PCR product was purified by Zymoclean Gel DNA Recovery Kit (Zymo Research), ligated into the pGEM‐T plasmid vector (Promega), and transformed into Escherichia coli (XL1‐Blue strain). Recombinant plasmids from individual bacterial clones were purified using a Plasmid Miniprep Kit (Axygen). The quality (orientation and nonmutant sequence) of the cloned PCR fragment was estimated by Sanger sequencing (ABI‐Prism 3500 XL, Applied Biosystems). The diluted plasmid of known concentration was used as C+. In addition, the same cDNA fragment was used in ARC+ representing MS2‐phage particles (armored particles [ARP]) contained an artificial target sequence. 29 , 30 The technology of creating ARP used the PCR fragment containing the target region with additional flanking nucleotides that were ligated into a linearized in‐house plasmid vector containing the MS2 coat protein gene. After confirming the correctness by Sanger sequencing, the generated recombinant plasmid was transformed into E. coli (strain B21) with subsequent protein expression induction by isopropyl‐l‐thio‐d‐galactopyranoside. Then, the cells were collected, lysed with combined lysozyme and freeze‐thawing, and treated with DNase I (Life Technologies) and RNase A (Life Technologies). The obtained product was then purified using CsCl gradient centrifugation, measured in concentration, and diluted in RNAlater Stabilization Solution (Life Technologies). The absence of residual DNA in the treated sample was verified using the developed qPCR assay without the reverse transcription step. The C+ and ARC+ concentrations were measured with a QX100 system (BioRad) using a PCR Supermix for Probes Kit (BioRad), a One‐Step ddPCR Supermix for Probes Kit (BioRad), specific primers, and suitable probes as per the manufacturer′s instructions.
To evaluate the efficiency of RNA extraction, an ICS was added to the analyzed samples. The ICS is an artificial RNA sequence (150–170 nt, guanine‐cytosine content 50%; see Table 1), surrounded by an MS2‐derived protective protein coat.
2.5. Limit of detection
The limit of detection (LOD) of the SARS‐CoV‐2 assay was determined using a series of 10‐fold dilutions of ARPs (described above). In particular, 10‐fold RNase‐free water‐diluted ARPs of known concentrations with the final volume of 100 μl were extracted using the RIBO‐Prep Extraction Kit (AmpliSens), in accordance with the manufacturer′s instructions, and then tested using the SARS‐CoV‐2 assay to establish the standard curves and LOD. The LOD was set as the minimal dilution detected in three replicates. 31
2.6. Assay cross‐reactivity
The absence of cross‐reactivity was proved by negative results of the viral panel detection that contained solutions of viral RNA and DNA from seven viral species. The analytical specificity of the COVID‐19 Amp assay was determined as 100%. The summary of RNA or DNA of viruses that were examined in the study is shown in Table 2.
Table 2.
List of viral species used to evaluate the COVID‐19 Amp assay analytical specificity
| Species | Family | Genus | Type of nucleic acid |
|---|---|---|---|
| MERS‐CoV | Coronaviridae | Betacoronavirus | RNA |
| Coronavirus 229E | Coronaviridae | Alphacoronavirus | RNA |
| Coronavirus NL63 | Coronaviridae | Alphacoronavirus | RNA |
| Coronavirus OC43 | Coronaviridae | Betacoronavirus | RNA |
| Adenovirus 3 type | Adenoviridae | Mastadenovirus | DNA |
| RC‐virus | Pneumoviridae | Pneumovirus | RNA |
| Parainfluenza virus 3 type | Paramyxoviridae | Paramyxovirus | RNA |
Abbreviations: COVID‐19, coronavirus disease 2019; MERS‐CoV, middle east respiratory syndrome‐related coronavirus.
2.7. Diagnostic sensitivity and specificity
The COVID‐19 Amp assay diagnostic sensitivity and specificity were determined using the detection of RNA extracted from nasopharyngeal swab samples. All 109 SARS‐CoV‐2 positive samples were tested by the COVID‐19 Amp assay and by the assay based on the World Health Organization (WHO) recommended set of primers and regimens (the WHO‐based assay; Table 3). The 95% confidence interval for a proportion was calculated according to Robert G. Newcombe. 32 In addition, for discordant samples verification, we applied another diagnostic assay Vector‐PCRrv‐2019‐nCoV‐RG (Vector) that is widely used for COVID‐19 diagnostics in Russia.
Table 3.
The list of biological samples from humans used for assessing diagnostic sensitivity of the COVID‐19 Amp assay
| No. | Sex | Age | C t COVID‐19 Amp | C t WHO protocol | No. | Sex | Age | C t COVID‐19 Amp | C t WHO protocol | No. | Sex | Age | C t COVID‐19 Amp | C t WHO protocol |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 63 | 24.04 | 26.03 | 35 | M | 27 | 21.24 | 24.39 | 69 | F | 21 | 24.46 | 27.51 |
| 2 | F | 50 | 19.66 | 21.06 | 36 | M | 29 | 20.11 | 21.73 | 70 | M | 73 | 26.10 | 24.40 |
| 3 | F | 49 | 19.46 | 23.08 | 37 | F | 73 | 21.17 | 23.29 | 71 | F | 82 | 23.49 | 24.61 |
| 4 | F | 62 | 27.14 | 27.36 | 38 | F | 59 | 25.20 | 26.60 | 72 | M | 88 | 20.29 | 23.77 |
| 5 | M | 34 | 18.41 | 21.75 | 39 | F | 41 | 22.79 | 24.53 | 73 | F | 69 | 25.56 | 28.16 |
| 6 | F | 49 | 22.24 | 22.93 | 40 | M | 53 | 24.19 | 24.56 | 74 | F | 67 | 22.04 | 25.22 |
| 7 | F | 37 | 19.74 | 23.23 | 41 | M | 65 | 26.68 | 26.54 | 75 | F | 83 | 23.19 | 24.43 |
| 8 | F | 24 | 25.55 | 35.81 | 42 | F | 61 | 21.18 | 23.07 | 76 | F | 65 | 18.90 | 22.85 |
| 9 | M | 32 | 20.92 | 23.52 | 43 | F | 38 | 19.92 | 22.05 | 77 | M | 31 | 24.93 | 26.69 |
| 10 | M | 56 | 23.07 | 24.62 | 44 | M | 57 | 22.04 | 22.52 | 78 | M | 33 | 23.42 | 27.19 |
| 11 | F | 37 | 26.76 | 28.36 | 45 | M | 27 | 21.51 | 23.30 | 79 | M | 40 | 24.89 | 27.65 |
| 12 | M | 59 | 29.09 | 39.96 | 46 | M | 24 | 23.61 | 27.19 | 80 | M | 58 | 23.21 | 26.34 |
| 13 | F | 23 | 21.63 | 23.72 | 47 | F | 36 | 23.06 | 23.16 | 81 | F | 27 | 26.56 | 27.85 |
| 14 | F | 52 | 26.21 | 27.90 | 48 | F | 51 | 18.53 | 23.04 | 82 | F | 28 | 19.35 | 22.53 |
| 15 | F | 54 | 25.13 | 28.05 | 49 | F | 61 | 25.36 | 27.03 | 83 | F | 63 | 22.96 | 28.22 |
| 16 | F | 59 | 28.73 | 28.29 | 50 | M | 64 | 24.51 | 25.55 | 84 | M | 73 | 19.07 | 22.81 |
| 17 | M | 25 | 22.03 | 24.12 | 51 | F | 23 | 22.63 | 25.44 | 85 | F | 59 | 26.67 | 30.04 |
| 18 | F | 31 | 25.84 | 25.72 | 52 | F | 33 | 28.83 | 30.76 | 86 | M | 23 | 28.09 | 29.52 |
| 19 | M | 56 | 19.27 | 22.80 | 53 | F | 47 | 25.04 | 28.04 | 87 | M | 61 | 28.31 | 30.44 |
| 20 | F | 40 | 20.53 | 23.10 | 54 | F | 50 | 20.48 | 23.49 | 88 | F | 53 | 29.40 | 22.03 |
| 21 | F | 40 | 32.58 | 27.45 | 55 | F | 39 | 21.29 | 22.69 | 89 | F | 47 | 29.43 | 32.60 |
| 22 | M | 43 | 23.19 | 27.01 | 56 | F | 48 | 24.33 | 27.94 | 90 | M | 39 | 26.52 | 28.28 |
| 23 | F | 57 | 22.56 | 23.30 | 57 | M | 30 | 20.59 | 23.37 | 91 | F | 40 | 29.79 | 30.94 |
| 24 | F | 46 | 21.34 | 25.43 | 58 | F | 60 | 21.02 | 23.12 | 92 | F | 33 | 26.37 | 27.23 |
| 25 | F | 30 | 22.23 | 24.94 | 59 | M | 44 | 18.30 | 22.44 | 93 | F | 34 | 26.21 | 29.24 |
| 26 | F | 48 | 20.16 | 23.63 | 60 | M | 51 | 23.79 | 25.80 | 94 | F | 56 | 17.01 | 20.81 |
| 27 | M | 70 | 21.29 | 24.37 | 61 | F | 56 | 24.10 | 25.26 | 95 | F | 55 | 22.42 | 23.79 |
| 28 | M | 23 | 23.77 | 25.47 | 62 | M | 26 | 16.72 | 19.40 | 96 | F | 49 | 28.05 | 28.39 |
| 29 | M | 68 | 22.04 | 26.02 | 63 | F | 71 | 20.64 | 23.78 | 97 | M | 47 | 23.05 | 26.06 |
| 30 | M | 25 | 20.85 | 24.73 | 64 | F | 64 | 19.00 | 22.28 | 98 | F | 58 | 21.78 | 25.30 |
| 31 | M | 40 | 31.32 | 24.58 | 65 | F | 86 | 26.82 | 28.53 | 99 | M | 50 | 24.55 | 28.25 |
| 32 | А | 58 | 23.52 | 26.46 | 66 | F | 41 | 23.85 | 25.73 | 100 | F | 70 | 27.15 | 30.43 |
| 33 | F | 49 | 22.31 | 23.37 | 67 | M | 48 | 23.75 | 26.18 | |||||
| 34 | F | 79 | 18.14 | 22.72 | 68 | M | 31 | 25.21 | 26.37 |
Note: Biological samples swabs from the nasopharynx were applied.
Abbreviations: COVID‐19, coronavirus disease 2019; F, female; M, male; WHO, World Health Organization.
3. RESULTS
Conducted multiple alignments of the sequences of SARS‐CoV‐2 available in the GenBank enabled the identification of highly conserved regions for the designing of the SARS‐CoV‐2‐specific primers and respective probes (Table 1). Chosen oligonucleotide primers and fluorescent probes were designed and synthesized, and the SARS‐CoV‐2 ‐specific assay was invented. The presented assay contained all components required for RT‐qPCR. The proposed assay allows the validation of all diagnostic steps, including extraction, reverse transcription, and PCR. Moreover, the usage of EC− and C− provide the minimization of the risk of false‐positive results due to reduced cross‐contamination. The LOD determined by ARP serial dilutions was 103 copies/ml. Standard detection was linear ranging from 106 (C t = 21.08–22.17) to 10 copies/ml (C t = 38.17–39.16) of the SARS‐CoV‐2 ARPs (R 2 = .97–.99; Figure 1). The cross‐reactivity potential was estimated using the detection of high‐titer RNA or DNA from seven viral species with an absence of positive reaction with the COVID‐19 Amp. Therefore, the evaluated analytical specificity was determined as 100%. To verify the competence of COVID‐19 Amp for SARS‐CoV‐2 diagnostics, we examined the same 109 biological samples previously detected using the WHO‐approved protocol for SARS‐CoV‐2 detection. 19 The C t values of the positive samples analyzed by COVID‐19 Amp ranged from 16.7 to 32.6 cycles, and for WHO protocol from 19.4 to 39.96 (Table 3). The mean C t value difference of COVID‐19 Amp and WHO protocol was 2.2 with less C t for COVID‐19 Amp which determines appropriate relevance of COVID‐19 Amp assay for clinical usage. Discordance in nine samples was observed (negative by the WHO‐based assay and positive by the COVID‐19 Amp assay). However, the discordant samples were retested as positive according to the results obtained from the Vector‐PCRrv‐2019‐nCoV‐RG assay (Table 4). Thus, the COVID‐19 Amp assay was found to be more convenient for diagnostics than the WHO‐based assay.
Figure 1.
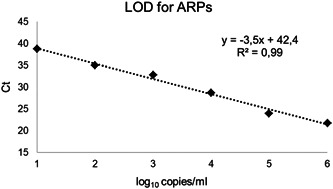
The COVID‐19 Amp assay LOD determined using ARPs. Standard detection was linear ranging from 106 to 10 copies/ml of the SARS‐CoV‐2 ARPs. The LOD was set as the mean minimal dilution detected in three replicates. ARP, armored particle; COVID‐19, coronavirus disease 2019; LOD, limit of detection; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Table 4.
The list of discordant biological samples that were the WHO‐based assay negative
| No. | Sex | Age | Disease course | C t COVID‐19 Amp | C t Vector |
|---|---|---|---|---|---|
| 1 | F | 38 | Mild | 34.21 | 29.56 |
| 2 | F | 68 | Moderate | 26.24 | 30.47 |
| 3 | M | 46 | Mild | 28.91 | 33.1 |
| 4 | F | 71 | Moderate | 29.75 | 25.17 |
| 5 | M | 68 | Moderate | 25.22 | 24.4 |
| 6 | M | 51 | Mild | 25.61 | 28.68 |
| 7 | M | 64 | Moderate | 24.03 | 26.85 |
| 8 | M | 24 | Mild | 25.37 | 24.98 |
| 9 | F | 65 | Moderate | 25.23 | 28.24 |
Note: Biological samples swabs from the nasopharynx were applied.
Abbreviations: COVID‐19, coronavirus disease 2019; F, female; M, male; WHO, World Health Organization.
4. DISCUSSION
Forced by public health needs, there were developed a lot of diagnostic RT‐qPCR Kits for SARS‐CoV‐2 identification. As reported by van Kasteren et al., 33 commercial RT‐PCR Diagnostic Kits for COVID‐19 (Altona Diagnostics, BGI, CerTest Biotec, KH Medical, PrimerDesign, R‐Biopharm AG, and SeeGene) manufactured in several countries (Germany, China, Spain, Korea, England) showed variable sensitivity with the presence of false‐negative results. However, all examined RT‐PCR Kits performed ≥96% PCR efficiency and the estimated LOD varied within a sixfold range between kits. Moreover, not every diagnostic kit contains appropriate controls like MS2‐phage‐based particles to provide adequate nucleic acids’ extraction monitoring 26 that is very important for the contamination screening. Also, RT‐qPCR COVID‐19 diagnostics in several countries (like China, USA, Germany) is based on a two‐target system, where one primer set universally detects numerous coronaviruses including SARS‐CoV‐2 and a second primer set only detects SARS‐CoV‐2. 20 , 25 , 34 One‐target assays can also provide high specificity with decreased primers nonspecific interactions. The proposed COVID‐19 Amp assay is one‐target and assures the precise SARS‐CoV‐2 RdRp gene detection with no cross‐reactivity with other respiratory viruses.
Concerning the rush in the development of RT‐qPCR SARS‐CoV‐2 detection systems in Russia, these kits also possessed analytical specificity and sensitivity issues. Several assays used in Russia and registered up to June 22, 2020 are characterized in Table 5. Some announced assays declare very low LOD, however the most typical sensitivity of viral RT‐qPCR usually is not less than 103 copies/ml. 35 Therefore, there is a lack of diagnostic kits with sufficient characteristics. Developed COVID‐19 Amp assay could reduce the number of false‐positive and false‐negative SARS‐CoV‐2 results and benefit clinical screening of COVID‐19 infection.
Table 5.
Characterization of several SARS‐CoV‐2 RT‐qPCR assays registered in Russia by June 22, 2020
| Assay | Manufacturer | Plexity | Target | Controls | Declared LOD |
|---|---|---|---|---|---|
| Vector‐RT‐PCR‐2019‐nCoV‐RG | Vector, State Research Center of Virology and Biotechnology | SARS‐CoV‐2 | ORF1a | C+—plasmid DNA, ICS—synthetic short RNA, C− | 105 copies/ml of RNA |
| Real‐Best RNA SARS‐CoV‐2 | Vector‐Best JSC | SARS‐CoV‐2 | No data | C+—plasmid DNA, ICS—synthetic short RNA, C− | 103 copies/ml of RNA |
| SARS‐CoV‐2/SARS‐CoV | DNA‐Technology TS LLC | SARS‐CoV‐2, SARS‐CoV‐like | E, N | C+—plasmid DNA, ICS—synthetic short RNA, C− | 500 copies/ml of RNA |
| COVID‐19 OneStep | Genotek | SARS‐CoV‐2 | No data | Positive control sample (PCS)—synthetic short RNA, C− | 102 copies/ml of RNA |
| AmpliPraim SARS‐CoV‐2 DUO | NextBio | SARS‐CoV‐2 | ORF1a, S | PCS—synthetic short RNA, ICS—synthetic short RNA, C−, EC− water | 103 copies/ml of RNA |
Abbreviations: C+, positive control; EC−, extraction; ICS, internal control sample; LOD, limit of detection; ORF, open reading frame; RT‐qPCR, quantitative reverse transcription‐polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
5. CONCLUSION
The ongoing increase in cases of COVID‐19 around the world now is driven by local transmission. 36 Therefore, there is an intense public health necessity for high‐fidelity diagnostic tests for SARS‐CoV‐2 infection. Asymptomatic infection and transmission in patients with COVID‐19 37 greatly increase the demand for screening a massive pool of people. Moreover, the viral load in hospitalized patients could be variable with no disease severity correlation 38 , 39 and infection cannot be excluded after a single negative RT‐qPCR test for SARS‐CoV‐2. Here, we characterized a developed RT‐qPCR assay named COVID‐19 Amp for precise detection of SARS‐CoV‐2 in clinical practice. The major advantages of our COVID‐19 Amp assay are high sensitivity and specificity, presence of virus particle‐based controls (ARCs+ and ICS) for extraction monitoring and DNA positive control (C+) for qPCR monitoring, and direct definition of SARS‐CoV‐2 RdRp gene partial sequence in the clinical sample without genus‐specific primers application. The clinical workflow for using COVID‐19 Amp for COVID‐19 diagnostics is suitable for any hospital and laboratory equipped for RT‐qPCR testing.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Ekaterina A. Goncharova and Vladimir G. Dedkov designed and performed experiments, analyzed data, and co‐wrote the paper. Anna S. Dolgova and Ilia S. Kassirov devised and obtained plasmids used as C+. Marina V. Safonova analyzed data. Yana Voytsekhovskaya provided ARC+ and ICS. Vladimir G. Dedkov and Areg A. Totolian supervised the research.
ETHICS STATEMENT
The study has been evaluated and approved by the Local Ethics Committees of the Pasteur Institute, Saint Petersburg, Russia.
Goncharova EA, Dedkov VG, Dolgova AS, et al. One‐step quantitative RT‐PCR assay with armored RNA controls for detection of SARS‐CoV‐2. J Med Virol. 2021;93:1694–1701. 10.1002/jmv.26540
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1.Coronavirus disease 2019. https://www.who.int/emergencies/diseases/novel-coronavirus-2019. Accessed May 2020.
- 2.Coronavirus COVID‐19 (2019‐nCoV). https://mpsde4.mpslimited.com/DigiEditpro/DigiEditPage.aspx?FileName=14772471348010446663889.xml. Accessed May 20, 2020.
- 3. Epidemiology Working Group for NCIP Epidemic Response, Chinese Center for Disease Control and Prevention . The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID‐19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41:145‐151. 10.46234/CCDCW2020.032 [DOI] [PubMed] [Google Scholar]
- 4. Park SE Epidemiology, virology, and clinical features of severe acute respiratory syndrome‐coronavirus‐2 (SARS‐COV‐2; coronavirus disease‐19). Korean J Pediatr. 2020;63(4):119‐124. 10.3345/cep.2020.00493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ren LL, Wang YM, Wu ZQ, et al. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J. 2020;133:1015‐1024. 10.1097/CM9.0000000000000722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin‐converting enzyme 2 (ACE2) as a SARS‐CoV‐2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586‐590. 10.1007/s00134-020-05985-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5(4):536‐544. 10.1038/s41564-020-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497‐506. 10.1016/S0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus‐infected pneumonia in Wuhan, China. J Am Med Assoc. 2020;323(11):1061‐1069. 10.1001/jama.2020.1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li X, Zai J, Wang X, Li Y Potential of large “first generation” human‐to‐human transmission of 2019‐nCoV. J Med Virol. 2020;92(4):448‐454. 10.1002/jmv.25693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Doremalen N, Bushmaker T, Morris DH, et al. Aerosol and surface stability of SARS‐CoV‐2 as compared with SARS‐CoV‐1. N Engl J Med. 2020;382(16):1564‐1567. 10.1056/NEJMc2004973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guan W, Ni Z, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382(18):1708‐1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee EYP, Ng MY, Khong PL. COVID‐19 pneumonia: what has CT taught us? Lancet Infect Dis. 2020;20(4):384‐385. 10.1016/S1473-3099(20)30134-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang W, Yan F. Patients with RT‐PCR confirmed COVID‐19 and normal chest CT. Radiology. 2020;295(2):E3. 10.1148/radiol.2020200702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller S, Chiu C, Rodino KG, Miller MB Point‐counterpoint: should we be performing metagenomic next‐generation sequencing for infectiousdisease diagnosis in the clinical laboratory? J Clin Microbiol. 2020;58(3):e01739‐19. 10.1128/JCM.01739-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat Biotechnol. 2020;38(4):382‐384. 10.1038/d41587-020-00002-2 [DOI] [PubMed] [Google Scholar]
- 23. Chan JFW, Yip CCY, To KKW, et al. Improved molecular diagnosis of COVID‐19 by the novel, highly sensitive and specific COVID‐19‐RdRp/Hel real‐time reverse transcription‐polymerase chain reaction assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58(5):e00310‐20. 10.1128/JCM.00310-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CDC 2019—Novel Coronavirus (2019‐NCoV). Real‐Time RT‐PCR Diagnostic Panel For Emergency Use Only Instructions for Use; 2019.
- 25. Summary of NMPA approved novel coronavirus 2019‐nCoV test kits 2019‐nCoV test kits . http://english.nmpa.gov.cn/2020-04/03/c_468570.htm. Accessed May 20, 2020.
- 26. Udugama B, Kadhiresan P, Kozlowski HN, et al. Diagnosing COVID‐19: the disease and tools for detection. ACS Nano. 2020;14:3822‐3835. 10.1021/acsnano.0c02624 [DOI] [PubMed] [Google Scholar]
- 27. Taylor SC, Nadeau K, Abbasi M, Lachance C, Nguyen M, Fenrich J. The ultimate qPCR experiment: producing publication quality, reproducible data the first time. Trends Biotechnol. 2019;37(7):761‐774. 10.1016/j.tibtech.2018.12.002 [DOI] [PubMed] [Google Scholar]
- 28. Dedkov VG, Magassouba NF, Safonova MV, et al. Development and evaluation of a real‐time RT‐PCR assay for the detection of Ebola virus (Zaire) during an Ebola outbreak in Guinea in 2014‐2015. J Virol Methods. 2016;228:26‐30. 10.1016/j.jviromet.2015.11.007 [DOI] [PubMed] [Google Scholar]
- 29. Cheng Y, Niu J, Zhang Y, Huang J, Li Q. Preparation of his‐tagged armored RNA phage particles as a control for real‐time reverse transcription‐PCR detection of severe acute respiratory syndrome coronavirus. J Clin Microbiol. 2006;44(10):3557‐3561. 10.1128/JCM.00713-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pasloske BL, Walkerpeach CR, Obermoeller RD, Winkler M, Dubois DB Armored RNA technology for production of ribonuclease‐resistant viral RNA controls and standards. J Clin Microbiol. 1998;36(12):3590‐3594. 10.1128/jcm.36.12.3590-3594.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cherpillod P, Schibler M, Vieille G, et al. Ebola virus disease diagnosis by real‐time RT‐PCR: a comparative study of 11 different procedures. J Clin Virol. 2016;77:9‐14. 10.1016/j.jcv.2016.01.017 [DOI] [PubMed] [Google Scholar]
- 32. Newcombe RG. Two‐sided confidence intervals for the single proportion: comparison of seven methods. Stat Med. 1998;17(8):857‐872. [DOI] [PubMed] [Google Scholar]
- 33. van Kasteren PB, van der Veer B, van den Brink S, et al. Comparison of seven commercial RT‐PCR diagnostic kits for COVID‐19. J Clin Virol. 2020;128:104412. 10.1016/j.jcv.2020.104412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chinese Center for Disease Control and Prevention . Primers and probes for detection 2019‐nCoV. https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2. Accessed May 22, 2020.
- 35. World Health Organization Manual for the Laboratory‐Based Surveillance of Measles, Rubella, and Congenital Rubella Syndrome. WHO. [Google Scholar]
- 36. Liu J, Liao X, Qian S, et al. Community transmission of severe acute respiratory syndrome coronavirus 2, Shenzhen, China, 2020. Emerg Infect Dis. 2020;26(6):1320‐1323. 10.3201/eid2606.200239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bai Y, Yao L, Wei T, et al. Presumed asymptomatic carrier transmission of COVID‐19. J Am Med Assoc. 2020;323(14):1406‐1407. 10.1001/jama.2020.2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS‐CoV‐2 in clinical samples. Lancet Infect Dis. 2020;20(4):411‐412. 10.1016/S1473-3099(20)30113-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holshue ML, DeBolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929‐936. 10.1056/NEJMoa2001191 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


