Abstract
Pharmacogenomics (PGx) is a key area of precision medicine, which is already being implemented in some health systems and may help guide clinicians toward effective therapies for individual patients. Over the last 2 decades, the Pharmacogenomics Knowledgebase (PharmGKB) has built a unique repository of PGx knowledge, including annotations of clinical guideline and regulator‐approved drug labels in addition to evidence‐based drug pathways and annotations of the scientific literature. All of this knowledge is freely accessible on the PharmGKB website. In the first of a series of PharmGKB tutorials, we introduce the PharmGKB coronavirus disease 2019 (COVID‐19) portal and, using examples of drugs found in the portal, demonstrate some of the main features of PharmGKB. This paper is intended as a resource to help users become quickly acquainted with the wealth of information stored in PharmGKB.
The coronavirus disease 2019 (COVID‐19) pandemic is a rapidly evolving global emergency, which looks set to become a long‐term challenge for society. Patients who are hospitalized with COVID‐19 may be treated with multiple drugs, including experimental treatments and adjuvant therapies, in addition to any concomitant medications already prescribed to the patient. As such, it is vital for clinicians to be able to quickly and easily integrate as much information as possible, particularly about patient‐specific factors, such as genetic variants, to optimize treatment.
The Pharmacogenomics Knowledgebase (PharmGKB) has curated pharmacogenomic (PGx) knowledge since 2000. 1 This wealth of information about the impact of genetic variation on drug response collected from research publications, regulator‐approved drug labels, clinical guidelines, and other sources is freely available to all through the PharmGKB website (https://www.pharmgkb.org). Although the majority of PharmGKB content is collected and curated manually by expert scientific curators, automated annotations of PubMed abstracts and full‐text articles in PubMed Central can also be accessed by users. 2
In response to this pandemic, PharmGKB has produced a COVID‐19 portal to bring together relevant PGx resources from across the site as well as some links to external materials. The use of PGx information should only form part of the prescribing decision making process and must be considered alongside other factors, including concomitant medications and drug‐drug interactions. This tutorial paper will guide users through the information presented in the COVID‐19 portal and provide an introduction to navigating the PharmGKB website.
PHARMGKB COVID‐19 PORTAL
The PharmGKB COVID‐19 portal can be accessed at https://www.pharmgkb.org/page/COVID. It is currently also available via the red button displayed on the PharmGKB homepage or users can search for COVID‐19 in the search box and follow the link given on the COVID‐19 disease page to the portal. The portal is updated on a regular basis and provides links to PGx resources for relevant drugs in the following categories:
Resources for research of COVID‐19 PGx. This section includes links to some external resources, including the Liverpool COVID‐19 Drug Interactions site 3 and clinical trials of potential COVID‐19 treatments.
Drugs associated with long QT syndrome and torsades de pointes.
Adjuvant therapies in COVID‐19 treatment.
Antidepressant PGx.
Based on information from ClinicalTrials.gov, drugs undergoing clinical trials as possible experimental therapies for COVID‐19 are listed in the “Resources for research of COVID‐19 PGx” section. All drugs included in the COVID‐19 portal are linked to their drug page on PharmGKB. Drug pages on PharmGKB serve as landing pages, which bring together all PharmGKB resources specifically linked to that drug. Users are able to instantly see what kind of resources are available for each drug by the presence of a blue dot next to a particular resource in the list on the left side of the page. Similar pages are available for genes, variants, and phenotypes.
For example, after clicking on the link to the fluvoxamine page, users will see that there are a number of resources available for fluvoxamine, including prescribing information and drug label annotations (Figure 1 ). Clicking on each resource name takes the user to an overview of all of the annotations of that type.
Figure 1.
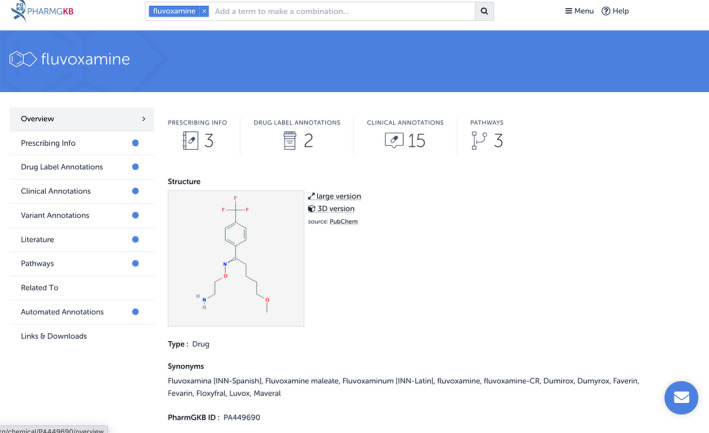
Image of the Pharmacogenomics Knowledgebase (PharmGKB) drug page for fluvoxamine. The tabs on the left side of the page list the different resources provided by PharmGKB. The existence of resources specific to fluvoxamine is indicated by a blue dot next to the resource name.
Some experimental COVID‐19 treatments do not currently have any PGx information available, either because they are awaiting approval from the US Food and Drug Administration (FDA) or because PGx studies of these drugs have not yet been carried out. A dagger symbol next to a drug’s name indicates that there is currently no PGx information available for that drug.
DRUG LABELS
Regulator‐approved drug labels can be important sources of clinical PGx information. To date, PharmGKB has annotated over 750 labels containing PGx information from a number of international regulators, including the FDA. All drug label annotations contain a short summary followed by excerpts of relevant PGx information taken from the label text. Some drug label annotations also have a prescribing section where genotype‐based prescribing guidance from the label is recorded.
Drug label annotations are assigned a “PGx level” by PharmGKB curators. These levels were created by PharmGKB and serve as an indication of the type of PGx information found in the drug label. Definitions of each level are given below and further details can be found on the PharmGKB website. 4
Testing required
The label states or implies that some sort of gene, protein, or chromosomal testing should be conducted before using this drug. Labels that state the variant is an indication for the drug or that a test “should be” performed are also included in this category.
Testing recommended
The label states or implies that some sort of gene, protein, or chromosomal testing is recommended before using this drug. PharmGKB considers labels that say testing “should be considered” or “consider genotyping or phenotyping” to be recommending testing.
Actionable PGx
The label may contain information about changes in efficacy, dosage, metabolism, or toxicity due to variants or phenotypes (e.g., “poor metabolizers”). Alternatively, the label may mention contraindication of the drug in a particular subset of patients with particular variants. However, the label does not require or recommend gene, protein, or chromosomal testing.
Informative PGx
The label contains information stating that particular variants or phenotypes do not affect a drug’s efficacy, dosage, metabolism, or toxicity or that particular variants or phenotypes affect a drug’s efficacy, dosage, metabolism, or toxicity, but this effect is not “clinically” significant. The Informative PGx level is also used for labels that appear on the FDA Biomarker List but do not meet the requirements to be assigned to any of the other PGx levels.
A table showing all PharmGKB drug label annotations can be found by clicking on the “Drug Label Annotations” button on the PharmGKB homepage. The “Dosing Info,” “Alternate Drug,” and “Prescribing Info” tabs can be used to easily find labels that offer information on dosing changes, use of an alternate drug, or any other relevant information on the label for certain patient genotypes. More information about these tabs can be found with the PGx Level definitions on the PharmGKB website.
As mentioned in the previous section, PharmGKB has drug label annotations available for fluvoxamine, which is currently being considered as a potential treatment for cytokine storms caused by COVID‐19. At the time of writing, there were two drug label annotations for fluvoxamine listed in PharmGKB: one from the FDA and one from the Swiss regulator, Swissmedic. These can be found by clicking on the Drug Label Annotations tab on the fluvoxamine drug page (Figure 2 ). Both have been given the PGx Level “Actionable PGx.” Users can access each annotation via the blue “Read Now” button. The annotation of the FDA label displays information stating that caution should be used when administering fluvoxamine to patients known to have low CYP2D6 activity or patients receiving other medications that inhibit CYP2D6. The annotation of the Swissmedic label details how fluvoxamine metabolism is altered in CYP2D6 poor metabolizers.
Figure 2.
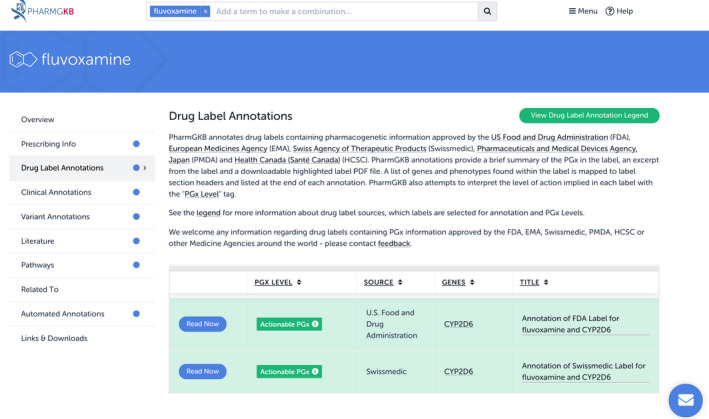
Image of all of the drug label annotations in Pharmacogenomics Knowledgebase (PharmGKB) for fluvoxamine. PGx, pharmacogenomics.
CLINICAL AND VARIANT ANNOTATIONS
Information from the scientific literature is predominantly found in PharmGKB’s variant and clinical annotations. Variant annotations are single sentences that capture individual PGx findings from papers. These sentences are constructed using a standardized structure and vocabularies, allowing them to be easily aggregated and compared. PharmGKB curators can include additional information in a free text section underneath each variant annotation sentence and provide information about the study, such as cohort size or P values, in the study parameters section.
Variant annotations that cover the same combination of drug and variant are collected together as supporting evidence for clinical annotations. Clinical annotations summarize the evidence, both supporting and conflicting, from the curated literature about a variant‐drug pair. A level of evidence is assigned to every clinical annotation to indicate the strength of evidence supporting the association between variant and drug, further details of which can be found on the PharmGKB website. 5 As new evidence is added to clinical annotations, the level of evidence is re‐assessed and can be moved up or down depending on the current strength of the evidence base. Users should be aware that, as PharmGKB clinical annotations are summaries of the published literature, they do not account for other patient‐specific factors, including concomitant medications. Each clinical annotation contains one or more caveats to highlight potential limitations.
The PharmGKB COVID‐19 portal has a list of adjuvant sedatives and analgesics that may potentially be administered to hospitalized patients with COVID‐19. Many of these drugs are linked to at least one clinical annotation in PharmGKB. Clicking on the Clinical Annotations tab on any drug, gene, variant, or phenotype page takes the user to a list of relevant clinical annotations. This tutorial will focus on clinical annotations for fentanyl and clonidine.
At the time of writing, there were 40 clinical annotations on the opioid analgesic fentanyl, including one for fentanyl and the CYP3A4*1 and *1G alleles, which has been assigned a level 2A by PharmGKB curators (PharmGKB ID 1449296086). Users can click on the blue Read Now button to see the annotation in full (Figure 3 ). Level 2A indicates that there is moderate evidence in the scientific literature of an association and that the gene in question, CYP3A4, has been defined as a Very Important Pharmacogene by PharmGKB. Very Important Pharmacogenes are known pharmacogenes, so it is more likely that variants in these genes have a PGx impact. Users should note that fentanyl dosing is not currently recommended to be based on genotype but is monitored clinically; however, the published evidence that fentanyl response may be associated with genotype might be informative to clinicians.
Figure 3.
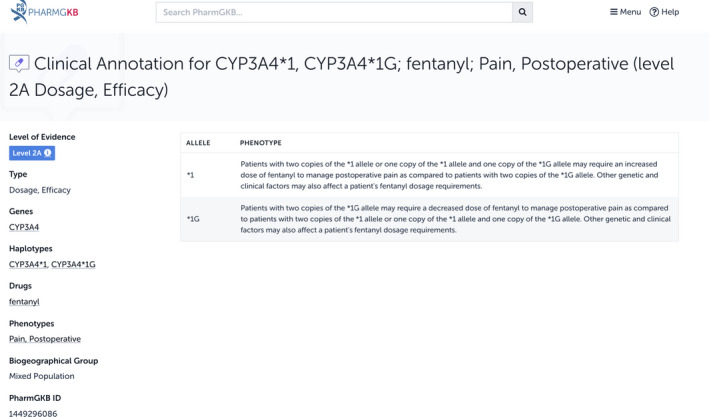
Pharmacogenomics Knowledgebase (PharmGKB) clinical annotation for fentanyl and the CYP3A4*1 and *1G alleles.
At the top of each individual clinical annotation page, is a table showing all the alleles or genotypes covered by the annotation with brief summaries of the nature of the association between that specific allele or genotype and the drug. According to the evidence base for this particular clinical annotation, patients who are homozygous for the *1G allele may require a decreased dose of fentanyl compared with patients who carry at least one *1 allele. Other information about the clinical annotation, such as linked haplotypes and phenotypes, is given next to this table, whereas the variant annotations given as evidence for this clinical annotation are listed further down the page. By looking at the list of variant annotations, users will see that this association has been found in six separate publications.
In contrast to fentanyl, little PGx research has been carried out on the sedative clonidine, which is only linked to two clinical annotations in PharmGKB. Both annotations have been assigned a level 3, indicating that the association may be based on only a single study, for example, the annotation on rs11269124 and clonidine (PharmGKB ID 1183614793). Alternatively, level 3 annotations can show if the variant‐drug pair has been evaluated in multiple studies, but the presence of conflicting evidence means that the nature of the association is not clear. This is the case for the annotation on rs5443 and clonidine, which is linked to two variant annotations that support the association and one variant annotation from a study that failed to find this association.
GUIDELINE ANNOTATIONS
PharmGKB annotates genotype‐based drug prescribing guidelines from Clinical Pharmacogenetics Implementation Consortium (CPIC), the Royal Dutch Association for the Advancement of Pharmacy ‐ Pharmacogenetics Working Group (DPWG), the Canadian Pharmacogenomic Network for Drug Safety (CPNDS), and the French National Network of Pharmacogenetics (RNPGx). PGx guidance from other professional organizations are also annotated as and when curators become aware of them. An overview of all guideline annotations is available via the “Clinical Guideline Annotations” button on the PharmGKB homepage, whereas guideline annotations for specific genes or drugs can be accessed using the Prescribing Info tab on a gene or drug page.
All PharmGKB annotations of clinical guidelines include a short summary of any recommendations in the guideline in addition to a more detailed annotation with excerpts from the guideline itself. Annotations of CPIC guidelines also offer short video summaries of the guideline and an interactive genotype picker, where users can retrieve genotype‐specific recommendations sourced from the guideline.
PGx clinical guidelines are a valuable resource in personalizing drug therapy for patients, an excellent example being the use of such guidelines in psychiatry. The COVID‐19 pandemic, and its health, societal, and economic impacts, is expected to substantially increase the number of patients in need of antidepressant therapy. This increased demand is already being reflected in FDA‐reported shortages of the selective serotonin reuptake inhibitor, sertraline. 6
Establishing an effective antidepressant regime for a patient often involves a “trial and error” approach, which, if not initially successful, can impact on patient well‐being. To assist clinicians in tailoring antidepressant therapy to individual patients, a number of PGx clinical guidelines have been published, including two from the CPIC. 7 , 8 Drugs covered by these CPIC guidelines are displayed in the Antidepressant PGx section of the PharmGKB COVID‐19 portal.
Sertraline is included in the CPIC guideline for selective serotonin reuptake inhibitors and is also mentioned in two guidelines from the DPWG; one with CYP2C19, which gives genotype‐guided prescribing guidance, and one with CYP2D6, which states that there is no interaction between CYP2D6 and sertraline that would affect prescribing.
As mentioned above, the annotation of the CPIC guideline for sertraline includes a short video explaining the guideline and its recommendations as well as the genotype picker. Users can select any combination of alleles from the picker to access specific recommendations for that genotype. For example, selecting the genotype *2/*2 from the picker brings up the recommendations for a CYP2C19 poor metabolizer (Figure 4 ).
Figure 4.

Image of the genotype picker tool on the annotation page for the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for sertraline. Selecting the *2/*2 genotype retrieves the specific recommendations for a CYP2C19 poor metabolizer. PharmGKB, Pharmacogenomics Knowledgebase.
PATHWAYS
Evidence‐based pathway diagrams of a drug’s pharmacokinetics or pharmacodynamics can help researchers to generate hypotheses about potential PGx interactions between a gene and drug. PharmGKB currently has almost 150 freely available, drug‐centered pathways. All pathways are accompanied by a written description and are interactive. Clicking on any drug or gene shown in a pathway takes the user to the PharmGKB page for that object.
Each arrow displayed on a pathway image is supported by primary scientific literature. Users can access a list of publications supporting each arrow via the Components tab, displayed on each pathway page. Pathways can also be downloaded in multiple formats, including as a PDF or in a computable GPML format.
Several drugs that have been, or are currently being, trialed as potential COVID‐19 therapies have previously been associated with an increased risk of serious heart rhythm problems, such as long QT syndrome or torsades de pointes. This issue is exacerbated in patients taking concomitant medications, which are also known to have a QT prolonging effect.
Drugs associated with QT prolongation or torsades de pointes are given in the “Drugs associated with long QT syndrome and torsades de pointes” section of the PharmGKB COVID‐19 portal. One such drug is the antipsychotic haloperidol. Going to the haloperidol drug page shows that three pathways have been linked to this drug on PharmGKB; this tutorial will focus on the haloperidol pharmacokinetics and anti‐arrhythmic pharmacodynamics pathways.
The haloperidol pharmacokinetic pathway (Figure 5 ) illustrates the metabolism of haloperidol in a stylized hepatocyte. Star icons indicate the importance of CYP3A4 in this pathway as the main metabolizing enzyme, whereas the inhibition of CYP2D6 by both haloperidol and its reduced form is also shown. This may be important information for clinicians considering haloperidol for patients with COVID‐19 who are also taking medications primarily metabolized by CYP2D6.
Figure 5.
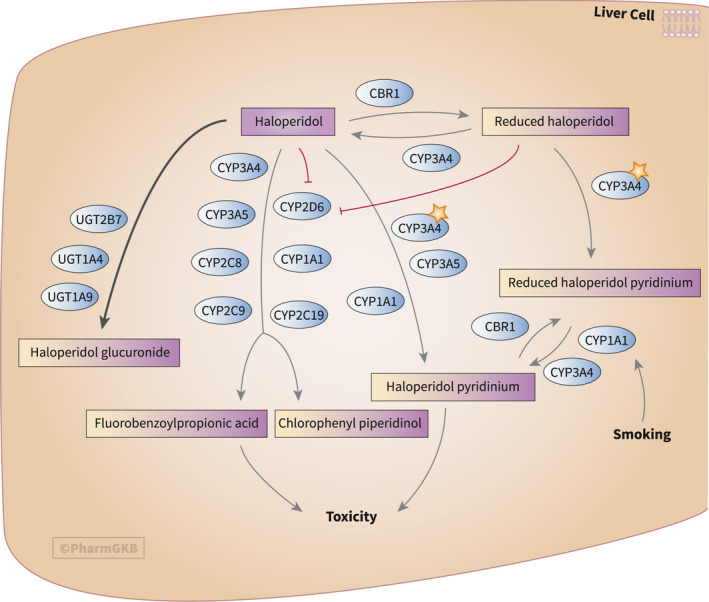
The Pharmacogenomics Knowledgebase (PharmGKB) haloperidol pathway, pharmacokinetics. The original, interactive pathway can be accessed at https://www.pharmgkb.org/pathway/PA166163828. 1
The anti‐arrhythmic pharmacodynamic pathway (Figure 6 ) shows the ion transporters of a cardiomyocyte that may be affected by drug interactions, including QT prolonging drugs. Clicking on each channel brings up a list of constituent subunits, with links to the relevant PharmGKB gene pages. This can provide researchers with a starting list of candidate genes in the search for variants associated with QT prolonging effects of drugs.
Figure 6.
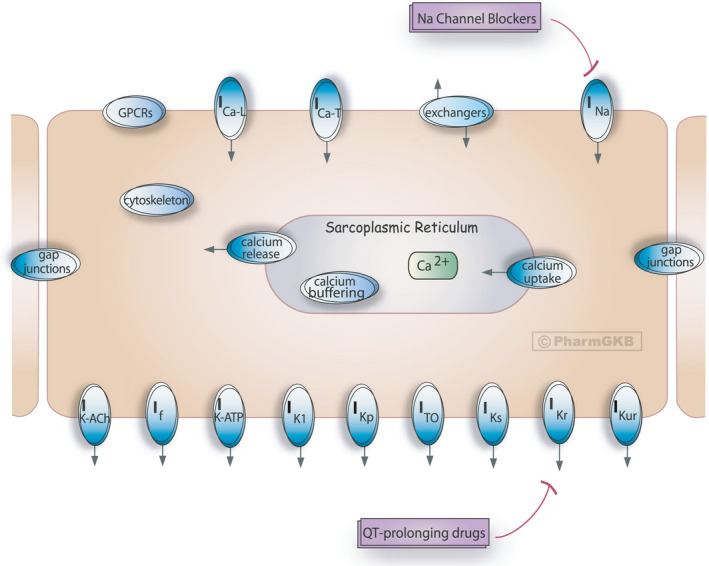
The Pharmacogenomics Knowledgebase (PharmGKB) anti‐arrhythmic pathway, pharmacodynamics. The original, interactive pathway can be accessed at https://www.pharmgkb.org/pathway/PA2033. 1
CONCLUSION
Important information on the impact of genetic variation on a patient’s possible drug response can be drawn from a number of different sources, from drug labels to the scientific literature. Users can access many different PGx resources through the PharmGKB website and the COVID‐19 portal, which will continue to be updated as research into this disease and potential treatments progresses.
FUNDING
PharmGKB is supported by National Institutes of Health (NIH)/National Human Genome Research Institute (NHGRI: U24 HG010615).
CONFLICT OF INTEREST
R.B.A. is a stockholder in Personalis, 23andMe, and Youscript. All other authors declared no competing interests for this work.
[The copyright line for this article was changed on 26 November 2020 after original online publication].
References
- 1. Whirl‐Carrillo, M. et al. Pharmacogenomics knowledge for personalized medicine. Clin. Pharma. Ther. 92, 414–417 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lever, J. et al. PGxMine: text mining for curation of PharmGKB. Pacific Symp. Biocom. 25, 611–622 (2020). [PMC free article] [PubMed] [Google Scholar]
- 3. Back, D. et al. COVID‐19 treatment in patients with comorbidities: awareness of drug‐drug interactions. Br. J. Clin. Pharmacol 10.1111/bcp.14358. [e‐pub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. PharmGKB . Drug label information and legend <https://www.pharmgkb.org/page/drugLabelLegend> (2020). Accessed August 10, 2020.
- 5. PharmGKB . Clinical annotation levels of evidence <https://www.pharmgkb.org/page/clinAnnLevels> (2020). Accessed August 10, 2020.
- 6. US Food and Drug Administration (FDA) . FDA drug shortages: current and resolved drug shortages and discontinuations reported to FDA <https://www.accessdata.fda.gov/scripts/drugshortages/dsp_ActiveIngredientDetails.cfm?AI=Sertraline+Hydrochloride+Tablets&st=c&tab=tabs‐3&i=18&panel=18> (2020). Accessed August 10, 2020.
- 7. Hicks, J.K. et al. Clinical pharmacogenetics implementation consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin. Pharmacol. Ther. 98, 127–134 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hicks, J.K. et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]


