Abstract
N-methyl-D-Aspartate receptors (NMDARs) are abundant postsynaptic proteins that are critical for normal synaptic communication. NMDAR channel function is regulated by multiple properties, including phosphorylation. Inhibition of protein phosphatase 1 in hippocampal neurons increases NMDAR activity, an effect abrogated by loss of spinophilin, the major protein phosphatase 1 (PP1)-targeting protein in the postsynaptic density (PSD). However, how spinophilin regulates PP1-dependent NMDAR function is unclear. We hypothesize that spinophilin regulates PP1 binding to the NMDAR to alter NMDAR phosphorylation. Our data demonstrate that spinophilin interacts with the GluN2B subunit of the NMDAR. In HEK293 cells, activation and/or overexpression of protein kinase A increased the association between spinophilin and the GluN2B subunit of the NMDAR. Functionally, we found that spinophilin overexpression decreased PP1 binding to the GluN2B subunit of the NMDAR and attenuated the PP1-dependent dephosphorylation of GluN2B at Ser-1284. Moreover, in P28 hippocampal lysates isolated from spinophilin KO compared to WT mice, there was increased binding of GluN2B to PP1, decreased phosphorylation of GluN2B at Ser-1284, and altered GluN2B protein interactions with PSD-enriched proteins. Together, our data demonstrate that spinophilin decreases PP1 binding to GluN2B and concomitantly enhances the phosphorylation of GluN2B at Ser-1284. The putative consequences of these spinophilin-dependent alterations in GluN2B phosphorylation and interactions on synaptic GluN2B localization and function are discussed.
Keywords: Protein Phosphorylation, Protein Phosphatase 1, Postsynaptic, Signaling, Protein-protein interactions, Proteomics
Introduction
Normal signaling in neurons requires a proper balance between kinases and phosphatases. While serine/threonine (ser/thr) kinases use protein interactions for dynamic localization of specific substrates (Welch et al. 2010), substrate specificity is obtained by consensus sequences located around the phosphorylation site (Brinkworth et al. 2003). In contrast to ser/thr kinases, ser/thr phosphatases are more promiscuous. These differences in promiscuity are due, in part, to a ~6-8-fold greater number of ser/thr kinases compared to phosphatases and a lack of easily defined consensus dephosphorylation sites (Cohen 2002; Morrison et al. 2000). The greater promiscuity and lower number of phosphatases require phosphatases such as protein phosphatase 1 (PP1) to rely more heavily on targeting and regulatory proteins for substrate specificity (Esteves et al. 2012).
The most abundant PP1 targeting protein in the postsynaptic density (PSD) is spinophilin (Colbran et al. 1997). Spinophilin associates with and bundles F-actin (Grossman et al. 2004; Hsieh-Wilson et al. 2003; Satoh et al. 1998) as well as interacts with multiple different synaptic proteins (Baucum et al. 2010; Sarrouilhe et al. 2006; Baucum et al. 2013; Baucum et al. 2012; Muhammad et al. 2015; Smith et al. 1999; Bielas et al. 2007). Specifically, spinophilin interacts with and/or regulates the function of multiple classes of glutamate receptors (Allen et al. 2006; Feng et al. 2000; Yan et al. 1999; Morris et al. 2018; Di Sebastiano et al. 2016), synaptic proteins that are critical mediators of normal synaptic communication and underlie processes such as long-term potentiation (LTP) and long-term depression (LTD) (Bear & Malenka 1994; Malenka & Bear 2004). Whereas spinophilin co-immunoprecipitates with the N-methyl, D-Aspartate receptor (NMDAR) (Baucum et al. 2013; Hiday et al. 2017) to modulate NMDAR channel properties (Allen et al. 2006), the mechanisms and consequences of this regulation are unknown.
We used a heterologous expression system to control interacting partners and show that spinophilin associates with the GluN2B subunit of NMDARs. Mechanistically, the spinophilin interaction with GluN2B is enhanced by protein kinase A (PKA) expression and activity. Consequences of regulating the spinophilin/NMDAR interaction may be to regulate PP1 binding to GluN2B as we found that spinophilin displaces PP1 from GluN2B and that this displacement occurs concurrently with enhanced GluN2B phosphorylation at Ser-1284. Whereas the above studies were performed in heterologous cells, we have identified spinophilin as a critical in vivo regulator of PP1 binding to, and dephosphorylation of, GluN2B. Specifically, spinophilin KO animals had increased association of PP1 with GluN2B, decreased GluN2B Ser-1284 phosphorylation, and enhanced interaction of GluN2B with specific PSD-enriched proteins. Understanding mechanisms that regulate NMDAR phosphorylation at Ser-1284 may have implications in multiple pathologies as this site is decreased under ischemic conditions, increased following reperfusion, and enhanced by acute stress (Lu et al. 2015; Ai et al. 2017). Moreover, mechanisms that regulate GluN2B interactions may modulate GluN2B subcellular localization. This is important as GluN2B localization at synaptic and extrasynaptic sites is linked to pro-survival and proapoptotic processes, respectively (Hardingham & Bading 2010).
Materials and methods:
Materials.
All custom materials will be shared upon reasonable request. Experiments were approved by the institutional biosafety committee (IBC-1594 and IN-1000). cDNAs: Templates used for generation of expression vectors were: human PP1α (PBC004482, Transomic Technologies, Huntsville, AL), rat PP1γ1 (Carmody et al. 2008), human spinophilin (Hiday et al. 2017), human GluN2B (BC113618; Transomic Technologies), human PKAc - pDONR223-PRKACA, (PKAc was a gift from William Hahn & David Root (Johannessen et al. 2010) (Plasmid RRIDs: Addgene_23495). Transfection Reagent: PolyJet (SignaGen Laboratories, Rockville, MD) was used for transfections. Antibodies: Antibodies used for IPs and/or primary blotting: goat polyclonal anti-Neurabin II (spinophilin) (A-20, SC14774, RRID:AB_2169477, Santa Cruz Biotechnology, Dallas, TX), rabbit monoclonal anti-NMDAR2B (GluN2B) (D15B3, RRID:AB_2112463 or D8E10, RRID:AB_2798506, 4212 or 14544, Cell Signaling Technology), Rabbit Phospho-NMDA Receptor 2B (GluN2B) (Ser-1284) (RRID:AB_10922589), goat polyclonal anti-V5 tag (A190-119A, RRID:AB_67317, Bethyl Laboratories, Montgomery, TX), goat polyclonal anti-HA tag (A190-107A, RRID:AB_66970, Bethyl Laboratories), goat polyclonal anti-Myc tag (A190-104A, RRID:AB_66864, Bethyl Laboratories), goat polyclonal anti-PP1γ (sc-6108, RRID:AB_2168091, Santa Cruz Biotechnology), mouse monoclonal anti-PP1 (E-9, sc-7482, RRID:AB_628177, Santa Cruz Biotechnology), rabbit polyclonal anti-V5 (G-14, sc-83849, RRID:AB_2019670, Santa Cruz Biotechnology) and mouse monoclonal anti-Myc (9E10, sc-40, RRID:AB_627268, Santa Cruz Biotechnology). Secondary antibodies used were: Alexa Fluor 790-conjugated AffiniPure Donkey Anti-Mouse IgG (715-655-150, RRID:AB_2340870, Jackson ImmunoResearch Laboratories, West Grove, PA), Alexa Fluor 790-conjugated AffiniPure Donkey Anti-Rabbit IgG (711-655-152, RRID:AB_2340628, Jackson ImmunoResearch Laboratories), Alexa Fluor 790-conjugated AffiniPure Donkey Anti-Goat IgG (705-655-147, RRID:AB_2340441, Jackson ImmunoResearch Laboratories), Alexa Fluor 680-conjugated donkey anti-Goat (A-21084, RRID:AB_2535741, Thermo-Fisher Scientific) and Alexa Fluor 680-conjugated donkey anti-Rabbit (A10043, RRID:AB_2534018, Thermo-Fisher Scientific).
Mutagenesis.
Mutagenesis reactions were performed using QuikChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA). Reactions were carried out using Q5 DNA buffer and 1 μl DNA polymerase in the presence of 5 μM DNTPs and 10 ng of template DNA. The following reaction protocol was performed: 1) an initial denaturation of 98°C for 2 minutes, 2) a 45-second denaturation at 98°C, 3) a 1-minute annealing reaction at a primer-specific temperature, 4) a 15-minute elongation at 68°C. Steps 2-4 were repeated 18 times. To eliminate template DNA, 10 μl of each reaction mixture was digested using 1 μL of DpnI for ~2 hours at 37°C. PCR product (1 μl) was transformed in competent DH5α E. coli. Vectors were then sequence verified (GENEWIZ, Inc South Plainfield, NJ.) for the mutations.
Mammalian Protein Expression.
Human embryonic kidney 293 FT cells (HEK293; Thermo-Fisher Scientific, Waltham MA RRID:CVCL_6911) were used for mammalian protein expression. Cells were purchased, split to passage 7, and frozen down. HEK293 cells are not listed by the International Cell Line Authentication Committee and have not been authenticated after purchase. Cells were used only to passage 23. Cell incubation and growth was performed in Dulbecco’s modified Eagle’s medium (DMEM) that contained 10% FBS, 584 mg/L L-glutamine, 1 mM Sodium Pyruvate, 100 U/mL penicillin and 100 μg/mL streptomycin. 25 mm2 culture flasks were incubated at a constant 37°C and 5% CO2 (Panasonic Healthcare; Secaucus, NJ). Cells were counted and approximately 1,000,000 cells were plated into 25 mm2 flat-bottomed culture flasks and left for overnight growth. Typically, cells were transfected the next day at ~70-80% confluency. Confluency was measured by estimating cell coverage on the bottom of the flask. DNA (0.5 - 5 μg per DNA vector) was added to 250 μL of serum-free DMEM in a 1.7 mL microcentrifuge tube. In a separate microfuge tube, transfection reagent was added to 250 μL of serum-free DMEM. Polyjet was used in a 3:1 volume: mass ratio (e.g. 18 μL of Polyjet was used with 6 μg DNA). For each experiment, DNA concentrations were equalized using an empty DNA vector, so that each condition in the same experiment had an equal mass of DNA and equal amount of transfection reagent. The transfection reagent containing mixture was then added to the tube containing DNA and incubated at room temperature for 15 minutes. The entire mixture was then added to the proper flask and cells were incubated overnight. Following overnight incubation, DMEM was aspirated off and cells were washed with 6 mL of cold 1X phosphate-buffered saline (PBS). PBS was aspirated and cells were lysed in 1.5 mL KCl lysis buffer (150 mM KCl, 1 mM DTT, 2 mM EDTA, 50 mM Tris-HCl pH 7.5, 1% (v/v) Triton X-100, 20 mM sodium fluoride, 20 mM β-glycerophosphate, 20 mM sodium orthovandate, 10 mM sodium pyrophosphate, 1X protease inhibitor cocktail; Thermo-Fisher Scientific or Bimake, Houston TX) then transferred into 2 mL microcentrifuge tubes. If a high percentage of cells were unattached, they were re-suspended in DMEM, then transferred to 15 mL centrifuge tubes and centrifuged at 250 x g for 5 minutes. After aspiration of media, 6 ml of cold PBS was added to cells and the pellet was triturated, which was followed by an additional centrifugation. PBS was then aspirated and cells were lysed in KCl lysis buffer. Cells were sonicated at 25% amplitude for 15 seconds at 4°C using a probe sonicator (Thermo-Fisher Scientific) and centrifuged (4°C for 10 minutes at 16,900 x g). Cell lysates were then used for IPs.
Mice.
Experiments were approved by the School of Science Institutional Animal Care and Use Committee (SC229R, SC239R, SC270R) and performed in accordance with the Guide for the Care and Use of Laboratory Animals and under the oversight of the Indiana University-Purdue University, Indianapolis (IUPUI). Animals were provided food and water ad libitum. Spinophilin KO mice were initially purchased from Jackson Laboratories (Bar Harbor, ME; Stock #018609; RRID:MMRRC_049172-UCD) and a breeding colony has been maintained at IUPUI. Male or female, WT, C57Bl6, (Jackson laboratories) or spinophilin knockout mouse brains were dissected at Postnatal day 28-30 (P28). Animals were group housed and WT and KO littermates were used (WT and KO animals were from heterozygote x heterozygote breeding pairs). Animals were weaned ~P21. 12 total animals were used and animals were euthanized in a randomized order. Animals were euthanized by decapitation without anesthesia. Euthanasia was performed in the afternoon.
Tissue homogenization.
Hippocampal or cortical tissue was flash-frozen in liquid nitrogen. Each brain region was homogenized in 2 mL of low ionic buffer containing 1% Triton X-100 buffer (2 mM Tris pH 7.5, 2 mM EDTA, 1 mM DTT, 1X protease inhibitor, 20 mM sodium fluoride, 20 mM sodium orthovanadate, 10mM sodium pyrophosphate, 20 mM β-Glycerophosphate) using fifteen up-and-down movements of a pestle in a 2 mL tight-fitting glass homogenizer. Tissue homogenates were then transferred to a 2 mL microcentrifuge tube and were sonicated at 25% amplitude for 15 seconds at 4°C using a 505 probe sonicator with 0.3 cm diameter probe (Thermo-Fisher Scientific) and centrifuged (4°C for 10 minutes at 16,900 x g) to reduce non-specific binding. Immunoprecipitations (IPs) were then performed as described below.
IPs.
HEK293 cell lysate or brain homogenates were transferred to a microcentrifuge tube for IPs (400-500 μL) or for a total input (75 μl). For the input, 25 μL of 4X sample buffer (0.2 M Tris HCl pH 6.8, 40% glycerol, 0.1 M DTT, 8% SDS w/v, 0.04% bromophenol blue w/v in water) was added to 75 μL of each input sample, vortexed and stored at −20°C. For the IPs, the appropriate IP antibody (1 – 3 μg) was added to the lysates and incubated at 4°C for approximately 1 hour. After 1-hour incubation of IP antibodies with samples, 25 μL of protein G magnetic beads (Dynabeads 10009D, Invitrogen) that had been previously washed in IP buffer (50 mM Tris HCl, 150 mM NaCl, 0.5% Triton X-100) was added to each sample and incubated rotating overnight at 4°C.
Following incubation, samples were magnetically separated and washed three times with IP wash buffer. Then 40 μL of 2x sample buffer (4x buffer diluted 1:2 with Milli-Q water) was added to each of the samples, vortexed and stored at −20°C until they were analyzed by immunoblot.
Immunoblotting.
Protein IPs or cell lysates were used for western blotting. All samples including IPs and inputs were heated at 70°C for 10 minutes, then IP samples were placed on a magnet prior to loading on the gel to separate magnetic beads out of suspension. 10 μL of input or 10 μL of IP sample were loaded onto a 1-1.5 mm hand-cast 10% polyacrylamide gel or, 26-well, pre-cast Criterion 4-15% polyacrylamide gradient gel (Bio-Rad Laboratories, Hercules, CA) or a 15 well 4-15% Mini-Protein TGX polyacrylamide gradient gel (Bio-Rad Laboratories). The precast gels were typically electrophoresed at 165 V for 1 hour and hand-cast gels were generally electrophoresed at 75 V for 15 minutes and 165 V for approximately 1 hour.
Proteins were transferred to nitrocellulose membranes using either a wet transfer or the Trans-Blot Turbo (Bio-Rad Laboratories). For wet transfer, proteins were transferred to a nitrocellulose membrane using an N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) transfer buffer (10% MeOH, 0.01 M CAPS pH 11). The transfer was performed in a transfer tank attached to a cooling unit set at 4°C and transfer was operated at a constant 1.0 Amps for 1.5 hours. For Trans-Blot Turbo, gels were transferred to nitrocellulose membrane using cold TransBlot Turbo transfer buffer with 20% ethanol. The transfer was performed at 9 V for 30 minutes.
Membranes were stained with a 2 mg/ml Ponceau S stain dissolved in 10% Trichloroacetic acid for 5 minutes to normalize inputs for equal loading where applicable. Following Ponceau staining, membranes were scanned and subsequently washed with deionized water. Membranes were blocked in Tris-buffered saline with Tween (TBST; 50 mM Tris pH 7.5, 150 mM NaCl, 0.1% (v/v) Tween-20) containing 5% (w/v) nonfat dry milk in. Blocking was performed 3 times, 10 minutes each, for a total of 30 minutes. Membrane was incubated with primary antibodies diluted in 5% milk in TBST overnight at 4°C with gentle shaking. Primary antibodies for Myc-tag (1:1000-1:10,000), HA-tag (1:1000-1:10,000), PP1 (1:1000-1:2000), GluN1 (1:1000-1:2000), spinophilin (1:1000-1:2000), GluN2B (1:1000-1:2000), and Ser-1284 (1:1000) were the same as used above for IP. After incubation, membranes were washed 3 times for 10 minutes per wash with TBST containing 5% milk. Appropriate secondary antibodies in TBST containing 5% milk were added to the membranes following the washes. Jackson ImmunoResearch antibodies were typically diluted 1:50000 and Invitrogen antibodies were generally diluted 1:10000. Secondary antibodies were incubated with membranes for one hour at room temperature shaking in darkness. Membranes were washed three times with Tris-Buffered saline without Tween for 10 minutes for each wash. Fluorescence scans were performed using the Odyssey imaging system (LiCor, Lincoln, NE) and data analysis was done using Image Studio software (LiCor). We have previously shown linearity of fluorescence intensity using these conditions for multiple proteins and antibody pairs (Edler et al. 2018; Morris et al. 2018).
Mass Spectrometry.
Samples collected from SDS-PAGE were de-stained with 25 mM ammonium bicarbonate in 50 % acetonitrile (ACN). For all digestion steps, a volume sufficient to cover the gel pieces were used. Next, 10 mM DTT in 25 mM ammonium bicarbonate was added to reduce disulfides. 25 mM iodoacetamide was then added to alkylate free sulfhydryl groups. After addition of iodoacetamide, the reaction was incubated in the dark for 45 minutes. Gel pieces were incubated in 25 mM ammonium bicarbonate. The gel pieces were then dehydrated with 25 mM ammonium bicarbonate in 50% ACN. The samples were then placed in a rotary vacuum and centrifuged until dry and subsequently digested with 12.5 ng/μl trypsin in 25 mM ammonium bicarbonate at 37°C overnight. The supernatant was collected from all samples. The remaining gel pieces were washed with 5% formic acid in 50% ACN and were vortexed and sonicated for 5 minutes. The supernatants were collected and pooled with previous supernatants and were submitted to the Indiana University Proteomics Core Facility for analysis. Digested peptides were loaded onto an Acclaim PepMap C18 trapping column and eluted on a PepMap C18 analytical column with a linear gradient from 3 to 35% acetonitrile (in water with 0.1% formic acid) over 120 minutes in-line with an Orbitrap Velos Pro or Qexactive plus mass spectrometer (Thermo-Fisher Scientific). Raw files generated from the run were analyzed using Thermo-Fisher Proteome Discoverer (PD) 2.2. SEQUEST HT (as a node in PD 2.2) was utilized to perform database searches as previously described (Smith-Kinnaman et al. 2014) with a few modifications: trypsin digestion, 2 maximum missed cleavages, precursor mass tolerance of 10 ppm, fragment mass tolerance of 0.8 Da, a fixed modification of +57.021 Da on cysteine, and a variable modification of +15.995 Da on methionine. The spectral false discovery rate (FDR) was set to ≤ 1% as previously described (Mosley et al. 2011). The FASTA database used was a mouse proteome downloaded from Uniprot on January 9, 2017 with addition of 72 common contaminants. Data was further analyzed in Scaffold Q+ (Proteome Software, Portland, OR) using an FDR cutoff of ≤ 1% for both proteins and peptides. MS/MS spectra of tryptic fragments matching specific phosphorylation sites were validated and the area under the curve (AUC) of the extracted ion chromatogram (XIC) was calculated for both the phosphorylated and non-phosphorylated peptide as previously described (Baucum et al. 2015; Hiday et al. 2017). The XIC reflects the abundance of a specific peptide that has a specific mass. The AUCs of the XICs of the phosphorylated peptide were normalized to the non-phosphorylated peptide AUC of the XIC to create a phosphorylation ratio. The generated ratios were compared across different groups.
Statistical Analyses.
Image Studio software was used for quantification of the integrated fluorescence intensities detected in the western blots. To measure changes in expression, we normalized inputs to total ponceau stain a well-validated approach (Fosang & Colbran 2015). To calculate associations, we divided the integrated fluorescence intensity for the co-immunoprecipitated protein by the integrated fluorescence intensity for the immunoprecipitated protein. In order to normalize for any differences in protein expression, we took the above normalized value and divided it by the input value for the co-immunoprecipitated protein. To compare different conditions across gels, we normalized the above ratio from the experimental condition by the ratio generated on the same gel for the control condition. The formula for this ratio is EXPERIMENTAL((Intensity co-IP proteinPrecipitate/Intensity IP proteinPrecipitate)/(Intensity co-IP proteininput))/CONTROL((Intensity co-IP proteinPrecipitate/Intensity IP proteinPrecipitate)/(Intensity co-IP proteininput)) as we have previously utilized (Edler et al. 2018; Hiday et al. 2017; Morris et al. 2018). If gels were run on different days, this ratio was averaged across multiple transfections, with each transfection corresponding to a unique biological replicate (transfections performed on different days). The N values for each individual experiment correspond to the number of unique biological replicates. To compare between groups, a one-column t-test was performed to compare the experimental condition to a theoretical value of 1. If data were all analyzed on the same gel, a t-test was used. If more than two groups were compared, a one-way ANOVA was used to determine significance. Where appropriate, a two-way ANOVA was used to compare across two different conditions. ANOVA’s were followed by a 1-column t-test for normalized values or a Tukey’s multiple comparison test. All graphs were generated in Prism (Version 8, GraphPad Software, La Jolla, CA). All graphs show the mean +/− standard deviation and all individual data points. All text values show mean +/− standard error of the mean. No inclusion criteria were predetermined. For exclusion, if an outlier was detected by a Grubb’s outlier test, a single outlier was removed from a group. No animals were excluded from the studies. Normality was not evaluated.
Randomization, Preregistration, and Blinding.
Given the study design using cell lines and non-treated animals, the study was not pre-registered, study groups were not randomized, and no blinding was performed. No sample calculation was performed.
Results:
Spinophilin associates with GluN2B subunit of NMDARs.
We immunoprecipitated spinophilin, or GluN2B from P28 WT and spinophilin KO cortical lysates. Whereas GluN2B (Figure 1A) associated with spinophilin in WT animals, no GluN2B co-immunoprecipitated with the spinophilin antibody in the KO lysates where spinophilin was not present. For all figures, the immunoblots for either the inputs or the immunoprecipitates for each protein were performed on the same gel and are shown at the same signal intensity. These data suggest a specific interaction between spinophilin and GluN2B.
Figure 1: Spinophilin and NMDAR interaction:
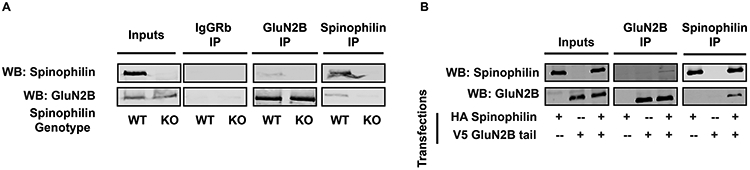
Spinophilin interacts with the GluN2B subunit of NMDARs in brain and HEK293 cells. (A) GluN2B is present in spinophilin immunoprecipitates from P28 cortical lysates of WT, but not spinophilin KO mice. Conversely, spinophilin is detected in GluN2B immunoprecipitates isolated from WT, but not spinophilin KO mice. No signal was detected in the IgG IPs. (B) HEK293 cells were transfected with HA-spinophilin and V5-GluN2BTail. Western blot results show an association between spinophilin andGluN2BTail. Images are representative of 3 animals (A) or 3 independent cell culture preparations (B). Immunoblots for inputs or immunoprecipitates for each protein were taken from the same gel.
As the NMDAR is a heterotetramer with an obligate GluN1 subunit, we utilized a heterologous expression system to delineate if spinophilin binds specifically to the GluN2B subunit of the NMDAR. As full-length GluN2B is trapped in the ER in the absence of GluN1 (Das et al. 1998); we expressed the C-terminal, cytosolic tail of GluN2B (GluN2BTail; amino acids 839-1484) which is not trapped in the ER and is localized to the cytosol. HA-tagged spinophilin and V5-tagged GluN2BTail were overexpressed in HEK293 cells and subsequently immunoprecipitated. We detected GluN2BTail in spinophilin immunoprecipitates and spinophilin in GluN2BTail immunoprecipitates (Figure 1B). As above, this interaction was specific as spinophilin was not detected in GluN2BTail immunoprecipitates when GluN2BTail was absent and GluN2BTail was not detected in spinophilin immunoprecipitates when spinophilin was absent.
Overexpression of the catalytic subunit of PKA (PKAc) in HEK293 cells increases spinophilin and GluN2B interaction.
PKA phosphorylates both spinophilin and NMDA receptor subunits (Hsieh-Wilson et al. 2003; Murphy et al. 2014) and our previous studies have found that PKA can regulate spinophilin interactions (Hiday et al. 2017). V5-tagged GluN2BTail and HA-tagged spinophilin were co-expressed in HEK293 cells alone or alongside a Myc-tagged form of the catalytic subunit of protein kinase A (PKAc). Overexpression of PKAc led to the appearance of multiple bands in the GluN2B input blot, suggesting multiple GluN2B species (most likely differentially phosphorylated forms) (Figure 2A). PKAc overexpression increased the association of spinophilin and GluN2BTail. Quantified data show a PKAc-dependent increase of GluN2BTail in the spinophilin IP (Figure 2B; 2.142 ± 0.3329, **p=0.0089,) and spinophilin in the GluN2BTail IP (Figure 2C) (10.6 ± 1.513, ***p=0.0002).
Figure 2: PKA activity enhances spinophilin-NMDAR interaction.
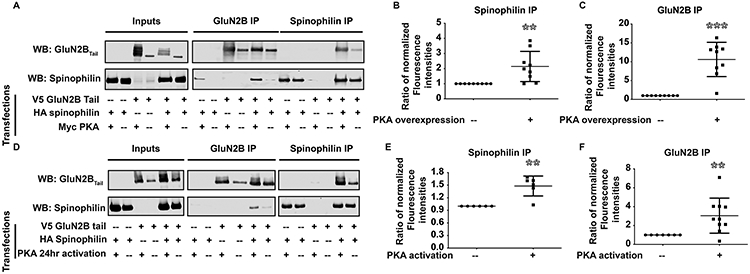
Overexpression of the catalytic subunit of PKA (PKAc) and long-term activation of PKA enhances the spinophilin-NMDAR interaction. A) HEK293 cells were transfected with HA-spinophilin and V5-GluN2BTail with/without overexpression of Myc-PKA and immunoprecipitated and immunoblotted. B-C) There is a significant increase in the spinophilin- GluN2BTail interaction when PKAc is overexpressed (n=10). D) HEK293 cells were transfected with HA-spinophilin and V5-GluN2BTail and endogenous PKA was activated using forskolin and IBMX for 24 hours. E-F) There is a significant increase in the spinophilin- GluN2BTail interaction when PKAc is overexpressed (n=6 independent cell culture preparations). Graphs show the mean +/− the standard deviation. **p<0.01, ***p<0.001.
Activation of endogenous PKA increases the interaction between spinophilin and the GluN2B subunit.
While overexpression of PKAc enhanced the spinophilin-GluN2B interaction, we wanted to determine if PKA activity is responsible for this effect. IBMX and forskolin were used to pharmacologically increase intracellular levels of cAMP to activate endogenous PKA. V5-tagged GluN2BTail was transfected alone or together with HA-tagged spinophilin and cells were incubated for 24 hours. Subsequently, cells were treated with IBMX/forskolin or vehicle alone for 16-20 hours to activate PKA. Long-term activation of endogenous PKA increased the association of spinophilin with GluN2BTail (Figure 2D). Quantitatively, GluN2BTail and spinophilin levels were significantly increased in the HA (1.48 ± 0.09710, **p=0.0043) and V5 IPs (3.046 ± 0.5874, **p=0.0069,), respectively (Figure 2E, 2F) compared to a theoretical value of 1.
Spinophilin-GluN2B interaction domains.
To determine the region on GluN2BTail that interacts with spinophilin, HEK293 cells were transfected with V5-tagged GluN2BTail fragments containing amino acids 839-1088, 1038-1484 and 1268-1484 (Figure 3A), along with HA-tagged spinophilin. We also transfected Myc-tagged PKA in all the conditions to enhance the interaction between spinophilin and the GluN2BTail fragments. Data demonstrate an interaction of HA-tagged spinophilin with the first fragment of GluN2BTail containing amino acids 839-1088 (Figure 3B). This suggests an interaction of spinophilin with the first 250 amino acids of GluN2BTail.
Figure 3: Spinophilin and GluN2BTail interacting domains.
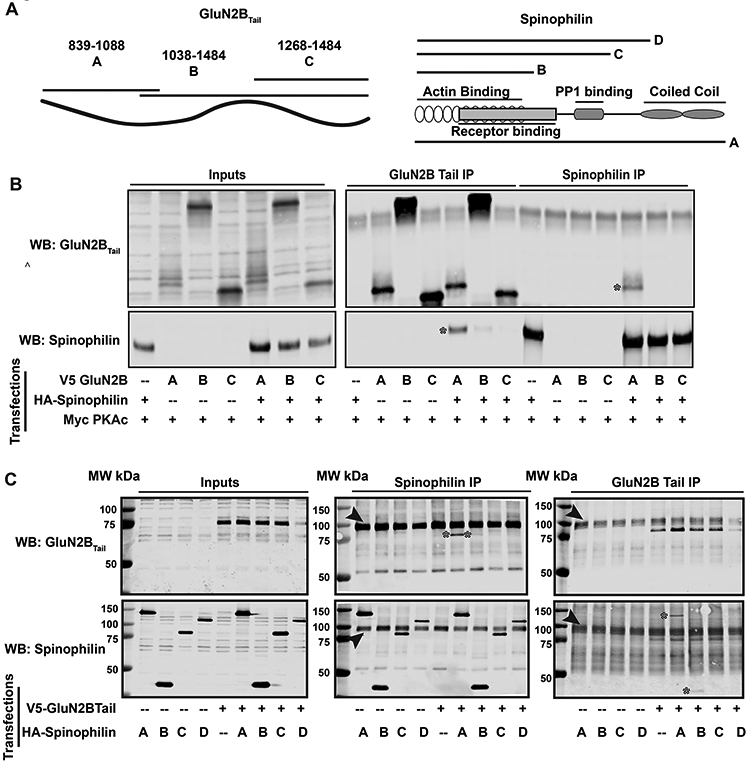
HEK293 cells were transfected with V5-tagged GluN2BTail fragments in the absence or presence of HA-tagged spinophilin. A) Schema of fragments for GluN2BTail (left) and spinophilin (right) used for the co-IP experiments. B) Immunoblots of GluN2BTail fragments using a V5- antibody and spinophilin using an HA antibody. C) Immunoblots of GluN2BTail and spinophilin fragment inputs and immunoprecipitations. Both images are representative of 5 independent cell culture preparations. * are used to highlight co-precipitated protein bands. Arrows are pointing to nonspecific band representing IgG heavy chain.
To determine the domain on spinophilin on which GluN2B binds, we generated three spinophilin truncation fragments with deletions in the C-terminus of the protein (Morris et al. 2018): amino acids 1-817 (full length A), 1-301 (fragment B), 1-460 (fragment C), and 1-670 (fragment D) on spinophilin (Figure 3A). We co-transfected HEK293 cells with V5-tagged GluN2BTail and HA-tagged spinophilin fragments. When full-length or spinophilin fragments are immunoprecipitated (Figure 3C middle panel), we detect GluN2BTail in immunoprecipitates containing full-length spinophilin (* in lane 7 (A fragment)). There is little association of GluN2BTail with the 1-301 fragment (* in lane 8 (B fragment)) and no detectable binding to the 1-460 or 1-670 spinophilin fragments (Lanes 9 and 10 (C and D fragments)) (Figure 3C middle panel). Conversely, when we immunoprecipitated GluN2BTail we observed full-length spinophilin (* in lane 7 (A fragment)). There was some 1-301 fragment (* in lane 8 (B fragment)) and no detectable 1-460 or 1-670 spinophilin fragments (Lanes 9 and 10 (C and D fragments)) in the GluN2BTail immunoprecipitates (Figure 3C far right panel). The major band marked by the arrowhead is predicted to be a non-specific IgG heavy chain. These data suggest that the C-terminal coiled-coiled region (amino acids 665-817) of spinophilin is required for complete association of spinophilin with GluN2B. However, when we expressed only the coiled-coil region, we did not detect consistent binding of spinophilin with GluN2BTail, suggesting it is not sufficient for an interaction (data not shown).
Spinophilin decreases the association of PP1 with GluN2BTail.
As spinophilin is critical in modulating PP1 targeting and PP1 activity, we wanted to determine if spinophilin targets PP1 to GluN2BTail. Both PP1α and PP1γ1 associated with GluN2BTail in the absence of spinophilin, suggesting that both isoforms of PP1 can associate with GluN2BTail directly or via a HEK-cell expressed targeting protein (Figure 4A).
Figure 4: Spinophilin sequesters PP1 away from GluN2BTail.
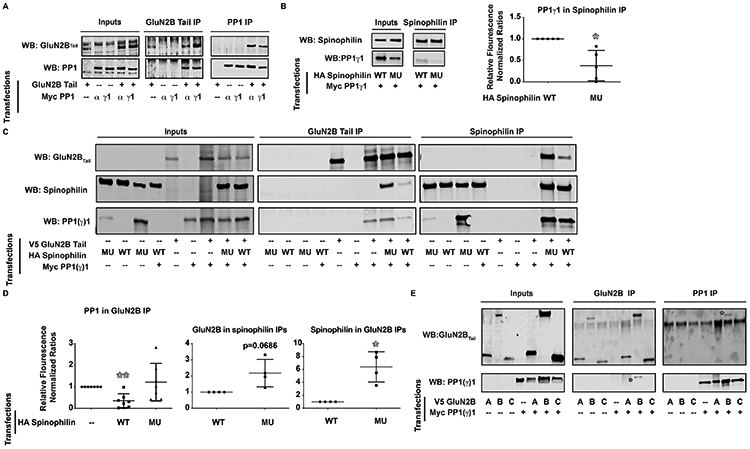
HEK293 cells were transfected with PP1γ1 and/or PP1α and GluN2BTail (Myc and V5 tags respectively). A) Immunoblots of GluN2BTail and PP1 in inputs, GluN2BTail immunoprecipitates, and PP1 immunoprecipitates and the experiment is representative of 4 independent experiments. B) F451A mutant spinophilin has abrogated binding to overexpressed PP1γ1 (n=5). C). GluN2BTail, PP1γ1, WT, and/or F451A (MU) spinophilin were co-transfected in HEK293 cells, immunoprecipitated for GluN2BTail or spinophilin, and immunoblotted. D) WT, but not F451A mutant, spinophilin overexpression decreased the association of PP1γ1 with GluN2BTail (n=7). E) PP1 precipitated with the GluN2B fragment containing residues 1038-1484 and the experiment is representative of 3 independent cell culture preparations. Graphs show the mean +/− the standard deviation. *p<0.05, **p<0.01.
As the major PP1-targeting protein in PSDs, we investigated whether spinophilin overexpression enhances PP1 co-IPs with GluN2BTail. V5-GluN2BTail along with Myc-PP1γ1 was transfected in the presence and absence of WT and a mutant (F451A) HA-spinophilin that has reduced PP1 binding (Ragusa et al. 2010) (Figure 4B; 0.3786 ± 0.1574, *p=0.0168). There was a significant difference across groups for these data (Figure 4D; F(2, 18)=4.899, *p=0.0200). Surprisingly, overexpression of spinophilin decreased the abundance of PP1γ1 bound to GluN2BTail (Figure 4C,D; 0.3564 ± 0.1228, **p=0.0019). Conversely, the F451A mutant spinophilin had no effect on the association of PP1γ1 with GluN2BTail (Figure 4C,D; 1.218 ± 0.3283, p=0.5318).
Given the unexpected effect of spinophilin on PP1γ1-GluN2B interaction, we investigated whether spinophilin and PP1γ1 are competing for the same site on GluN2BTail. (Spinophilin immunoprecipitates: 2.178 ± 0.4227, p=0.0686; GluN2BTail immunoprecipitates: 6.402 ± 1.171, *p=0.0192). Moreover, when we co-expressed GluN2BTail fragments containing amino acids 839-1088, 1038-1484 and 1268-1484 along with PP1γ1, we observed PP1γ1 predominantly binding the second fragment of GluN2B (amino acids 1038-1484; * in Figure 4E). These data suggest that PP1 and spinophilin are binding to different regions on GluN2B, with spinophilin predominantly binding amino acids 839-1088 and PP1 predominantly binding amino acids 1038-1268. Together, these data suggest that spinophilin is neither targeting PP1 to GluN2B, nor is it directly displacing PP1 from, GluN2B.
Spinophilin rescues PP1-dependent dephosphorylation of Ser-1284 on GluN2BTail.
To determine functional implications of decreasing PP1 targeting to GluN2BTail, we transfected V5-tagged GluN2BTail along with Myc-tagged PP1γ1 with and without HA-tagged spinophilin in the presence of Myc-tagged PKAc to increase the interaction between spinophilin and GluN2BTail. Using mass spectrometry on the GluN2BTail immunoprecipitates separated by SDS-PAGE, several PKA phosphorylation sites on GluN2BTail were observed. Phosphorylation was detected at Ser- 929/930 (Figure 5A), Ser-940 (Figure 5B), Ser-1050 (Figure 5C), Ser-1303 (Figure 5D), and Ser-1284 (Figure 5E). Ser-929/930, Ser-940, and Ser-1050 were only observed in the presence of overexpressed PKA. In contrast, Ser-1284 and Ser-1303 were present under all conditions. Of note, Ser-1284 phosphorylation was regulated by spinophilin (F(4, 10)= 139.8, ****p<0.0001). Interestingly, overexpression of PP1γ1 decreased Ser-1284 phosphorylation on GluN2BTail (Figure 5E; GluN2BTail (20.39% ± 0.2537%) vs. GluN2BTail+PP1 (3.447% ± 0.2049%), ****p <0.0001). Overexpression of PKA alone or along with PP1γ1 did not increase Ser-1284 phosphorylation, suggesting that this is not a PKA sensitive site, consistent with other data suggesting it is a CDK5 site (Lu et al. 2015). Co-expression of spinophilin along with PKA and PP1γ1 attenuated the PP1-dependent decrease in Ser-1284 phosphorylation (Figure 5E; GluN2BTail+PP1+PKA (4.948% ± 0.3837%) vs GluN2BTail+PP1+PKA+Spinophilin (14.24% ± 0.7482%), ****p<0.0001). To determine the role of spinophilin in regulating Ser-1284 phosphorylation in the absence of PKA, we overexpressed PP1α or PP1γ1 with GluN2BTail in the absence or presence of spinophilin without overexpressing PKA in the cells. Consistent with the above spinophilin-dependent regulation of GluN2B phosphorylation at Ser-1284, there was a significant effect of spinophilin co-expression on phosphorylation of Ser-1284 (Figure 5F) (F(1,10) = 8.717, *p=0.0145) but no significant individual differences.
Figure 5: Spinophilin and PKA regulate GluN2B phosphorylation.
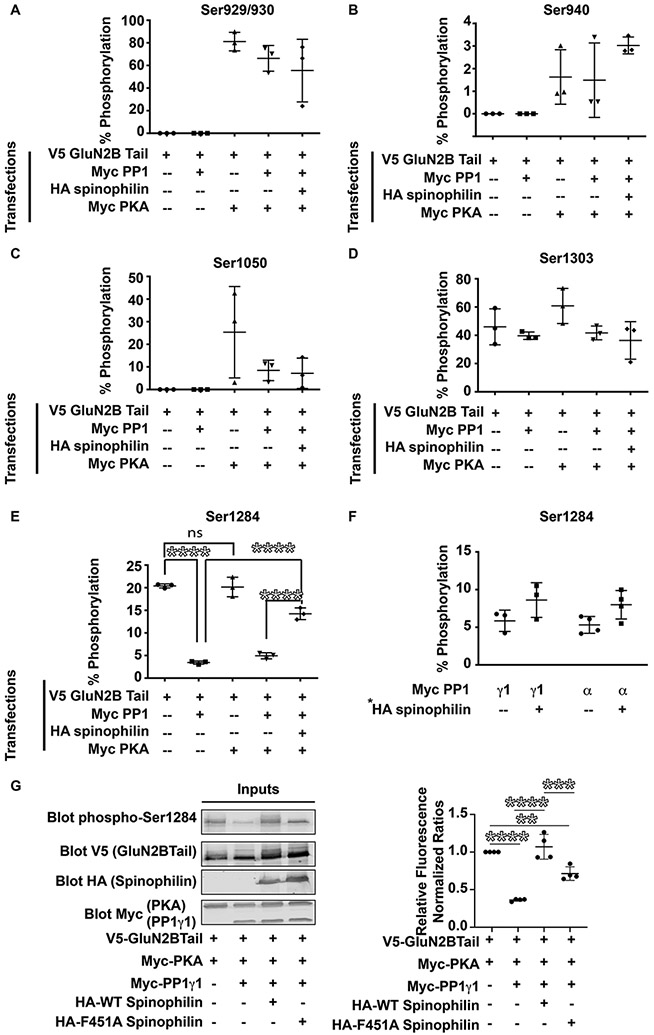
HEK293 cells were transfected with V5-GluN2BTail in presence or absence of Myc-PKA, HA-spinophilin, and/or Myc-PP1γ1. Immunoprecipitations were performed with V5-tag antibodies. Area under the curve of extracted ion chromatograms matching GluN2BTail phosphorylation sites at (A) Ser929/930, (B) Ser940, (C) Ser1050, (D) Ser1303, and (E) Ser 1284. (n=3). (F) GluN2BTail and either PP1γ1 or PP1α were transfected in the absence or presence of overexpressed spinophilin and Ser-1284 phosphorylation was quantified. There was a significant effect (p<0.05) of spinophilin overexpression on the phosphorylation of Ser-1284. (n=3-4 cell culture preparations). (G) HEK293 cells were transfected with GluN2BTail, PKA, PP1γ1, and/or WT or F451A spinophilin and immunoblotted for protein expression and Ser-1284 phosphorylation (n=4 cell culture flasks per group from 2 cell culture preparations). Graphs show the mean +/− the standard deviation. **p<0.01, ***p<0.001, ****p<0.0001.
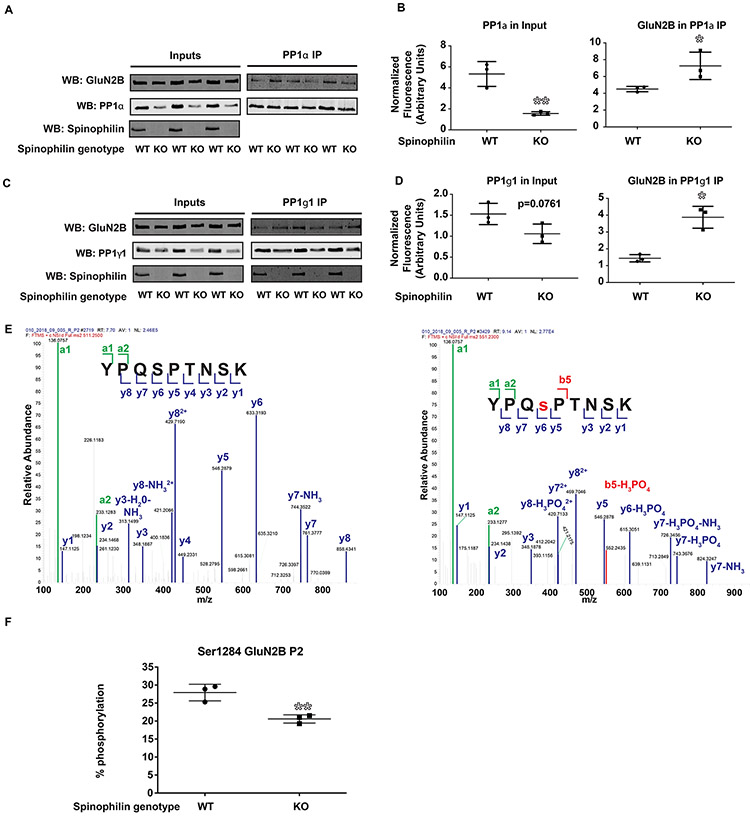
To validate mass spectrometric detection of the Ser-1284 site, we used a S1284A mutant GluN2BTail. We were able to quantify (Figure S1A) and validate by MS/MS the non-phosphorylated (Figure S1B) and phosphorylated (Figure S1C) peaks in the WT condition. We did not detect a peak matching the YPQSPTNSK peptide (phosphorylated or nonphosphorylated; Figure S1D) in the S1284A mutant. However, we did detect a strong peak matching the mass of the YPQAPTNSK tryptic peptide (Figure S1E). Moreover, we validated the MS/MS of this tryptic peptide (Figure S1F).
In addition to validation of the mass spectrometry, we utilized a Ser-1284 antibody to measure phosphorylation under different transfection conditions (F3, 12 = 46.55, P<0.0001). PP1 overexpression decreased phosphorylation of GluN2B by 63.8% (Figure 5G). Moreover, WT spinophilin abrogated the PP1-dependent decrease in phosphorylation (70.7% increase over PP1 condition) whereas the F451A mutant only attenuated this effect (35.1% increase over PP1 condition) (Figure 5G).
Spinophilin KO mice have enhanced GluN2B-PP1 interaction and decreased Ser-1284 phosphorylation in P28 mouse hippocampus.
While the above data suggest that spinophilin can regulate GluN2B phosphorylation in a heterologous cell system, we next wanted to determine if loss of spinophilin regulates the PP1-GluN2B interaction and GluN2B phosphorylation at Ser-1284 in vivo. To achieve this, we used hippocampus dissected from postnatal day 28-30 (P28) wildtype (WT) and global spinophilin KO mice. P28 animals were used since spinophilin expression peaks between P21 and P26 (Allen et al. 1997) and GluN2B expression wanes somewhat into adulthood (McKay et al. 2018). Furthermore, since we are using global, constitutive spinophilin KO animals, we wanted to reduce the period of time that animals lack spinophilin to mitigate potential compensatory changes. As previously observed in striatal lysates (Allen et al. 2006), there were decreases in PP1 expression in hippocampal lysates isolated from the spinophilin KO animals. Specifically, PP1α expression was significantly decreased (WT (4.254 ± 0.5317) vs KO (2.128 ± .08013), **p=0.0051) (Figure 6A-B) whereas PP1γ1 expression trended towards a significant decrease (WT (1.531 ± 0.1461) vs KO (1.059 ± 0.1346), p=0.0761) (Figures 6C-D). Furthermore, PP1α and PP1γ1 were immunoprecipitated and PP1 immunoprecipitates were immunoblotted for GluN2B and spinophilin. When normalized to PP1 in the immunoprecipitate and GluN2B in the input, there was a greater association of GluN2B and PP1α (WT (4.505 ± 0.1826) vs KO (7.265 ± 0.9420), *p=0.0451) (Figure 6A-B) and between GluN2B and PP1γ1 (Figure 6C-D) (WT (1.680 ± 0.2916) vs KO (4.015 ± 0.4585), *p=0.0127) in PP1 IPs isolated from KO compared to WT hippocampus
Figure 6: GluN2B-PP1 interaction is enhanced and Ser-1284 phosphorylation is diminished in P28 spinophilin KO hippocampus.
PP1 and GluN2B were immunoprecipitated from hippocampal lysates generated from P28 WT and spinophilin KO animals. A-B) Western blotting results show that the levels of PP1α are decreased and the amount of GluN2B in PP1α IP are increased in spinophilin KO animals. C-D) There was a trend for lower levels of PP1γ1 and a significantly increased PP1γ1-GluN2B interaction in spinophilin KO hipopocampi. E) MS/MS spectra showing the fragmented ions matching the non-phosphorylated (left) and phosphorylated (right) tryptic peptide YPQSPTNSK. F) Ser-1284 phosphorylation is diminished in spinophilin KO hippocampus. One cohort of an n of 3 mice per group were used for the PP1 IPs and a separate N of 3 mice per group were used for the Ser-1284 phosphorylation. Graphs show the mean +/− the standard deviation. *p<0.05, **p<0.01.
To determine if loss of spinophilin and enhanced PP1 binding correlate with alterations in GluN2B phosphorylation, GluN2B immunoprecipitates were separated by SDS-PAGE, Coomassie stained, and the GluN2B bands were digested and Ser-1284-containing tryptic peptides were analyzed by mass spectrometry (Figure 6E) for Ser-1284 phosphorylation on GluN2B. Consistent with overexpression of spinophilin in HEK293 cells increasing Ser-1284 phosphorylation on GluN2B, when normalized to the non-phosphorylated form, Ser-1284 phosphorylation was decreased in spinophilin KO hippocampus (WT (27.93% ± 1.332%) vs KO (20.60% ± 0.6658%), **p=0.0079) (Figure 6F), suggesting that spinophilin acts to limit Ser-1284 dephosphorylation in vivo.
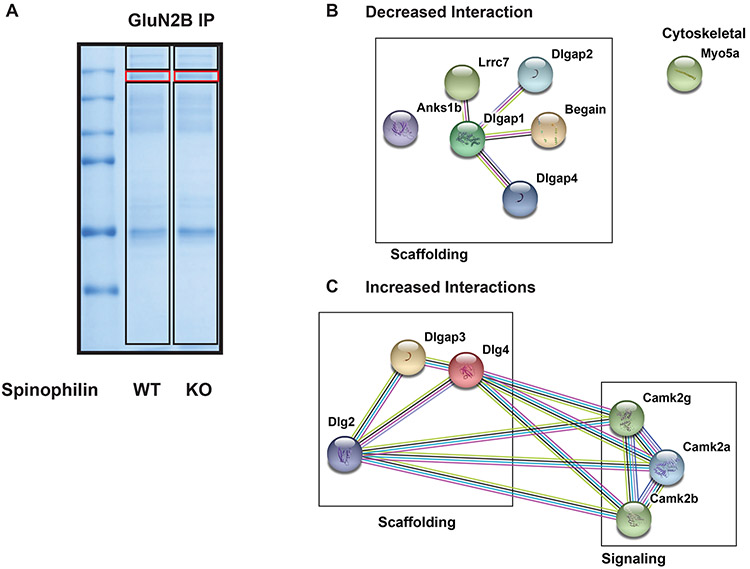
Spinophilin KO modulates the P28 GluN2B interactome.
To determine how spinophilin dependent changes in GluN2B phosphorylation and PP1 interaction regulate GluN2B interactions in brain, GluN2B interacting proteins isolated from WT and spinophilin KO P28 GluN2B immunoprecipitates were separated by SDS-PAGE and Coomassie stained. The GluN2B band (Figure 7 red box) and the other gel regions (black boxes) were excised, digested with trypsin and analyzed by mass spectrometry. We identified multiple proteins in the GluN2B IP (Table S1). To begin to delineate a role for spinophilin in modulating the GluN2B interactome, we combined data from both the GluN2B band and interacting band (Figure 7 red and black boxes on the gel; Table S1). To normalize for GluN2B abundance in the IP, the total number of spectral counts for each interacting protein was divided by the total number of spectral counts matching GluN2B from the same samples. Next, the normalized spectral counts from the WT and spinophilin KO hippocampus immunoprecipitates were summed and a ratio was generated by dividing the KO value by the WT value. To address non-specific interactions, we searched our identified interacting proteins against the CRAPome database (Mellacheruvu et al. 2013). Table 1 shows the 7 proteins that have a total decreased ratio of ≤ −0.5 and the 6 proteins that have an increased ratio of ≥ +0.5. This table only show proteins detected in at least 3 samples and having passed filtration via the CRAPome database. It is important to note that this is an initial screen of potential changes in the GluN2B interactome and follow-up immunoblotting or more quantitative mass spectrometry studies will need to confirm these initial interaction changes.
Figure 7: Spinophilin modulates the P28 mouse hippocampal GluN2B interactome.
GluN2B immunoprecipitates isolated from P28 WT and spinophilin KO hippocampal lysates were separated by SDS-PAGE and Coomassie stained (A). Spectral counts of proteins in GluN2B IPs from WT and spinophilin KO mice were normalized to spectral counts matching GluN2B. A ratio (KO/WT) of normalized spectral counts matching GluN2B interacting proteins was generated. B) The 8 proteins that had a decreased interaction with GluN2B in the KO mice were input into the string db and assigned to categories based on their function. C) The 7 proteins that had an increased interaction with GluN2B in the KO mice were input into the string db and assigned to categories based on their function. n = 3 mice per group (same mice used in Figure 6F).
Table 1.
Proteins having decreased and increased interactions with GluN2B in P28 spinophilin KO/WT mice
| Protein | Gene | Total PSMs | KO/WT Log2 |
|---|---|---|---|
| SAPAP4 | Dlgap4 | 37 | −1.59 |
| Ankyrin repeat and sterile alpha motif domain-containing protein 1b | Anks1b | 20 | −1.33 |
| SAPAP2 | Dlgap2 | 31 | −1.22 |
| SAPAP1/GKAP | Dlgap1 | 48 | −1.04 |
| Myosin Va | Myo5a | 34 | −0.95 |
| Brain-enriched guanylate kinase-associated protein | Begain | 14 | −0.71 |
| Leucine-rich repeat-containing protein 7 | Lrrc7 | 48 | −0.68 |
| SAPAP3 | Dlgap3 | 57 | 1.52 |
| CaMKII beta | Camk2b | 138 | 0.92 |
| CaMKII alpha | Camk2a | 273 | 0.75 |
| CaMKII gamma | CaMK2g | 88 | 0.75 |
| PSD-93 | Dlg2 | 153 | 0.72 |
| PSD-95 | Dlg4 | 414 | 0.59 |
Using the string-db program, we input the 7 decreased (Figure 7B) and 6 increased (Figure 7C) interacting proteins and divided them into scaffolding, signaling, and cytoskeletal categories. Interestingly, loss of spinophilin increased the interaction of GluN2B with CaMKII isoforms, SAPAP3, PSD-95, and PSD-93 whereas it decreased the interaction of GluN2B with the other SAPAP isoforms (1,2,4) and myosin-Va.
Discussion:
Mechanisms regulating the Spinophilin-GluN2B interaction.
While spinophilin has been identified in NMDA receptor complexes (Baucum et al. 2013; Swartzwelder et al. 2016), our current data demonstrate that spinophilin interacts specifically with the GluN2B subunit. While expression of the individual NMDAR subunit suggests a direct interaction, we cannot rule out that a HEK cell-expressed protein is bridging this interaction. Moreover, spinophilin may interact with other NMDAR subunits within a triheteromeric receptor (e.g. GluN1 and GluN2A).
Spinophilin has been previously shown to target PP1 to specific substrates (Allen et al. 1997; Ragusa et al. 2010). Spinophilin binding to GluN2BTail was increased in the presence of overexpressed PKAc. Moreover, activation of endogenous PKA using IBMX, a phosphodiesterase inhibitor (Francis et al. 2001) and forskolin, an adenylyl cyclase activator (Seamon & Daly 1981) enhanced spinophilin binding to NMDARs. Furthermore the intensity of GluN2BTail protein band in the both PKA activated and/or overexpressed blots is higher compared to control condition. This suggests that activation and/or overexpression of PKAc may either increase GluN2BTail protein expression or stabilize the protein. However, when we normalize for the increased expression, we still observe a significant increase in the GluN2BTail-spinophilin interaction when PKAc is activated or overexpressed. Together, these data suggest that PKA activity is regulating the spinophilin NMDAR interaction; however, future studies will need to determine if this is due to phosphorylation of spinophilin, the NMDAR, or an additional protein.
To inform future studies, we input the coiled-coil region of spinophilin, a region critical for the spinophilin/GluN2B interaction, into the NetPhos 3.1 algorithm (Blom et al. 1999; Blom et al. 2004). Three putative PKA sites on spinophilin were detected with this algorithm: Ser-694, Ser-758, and Ser-814. When we added PKA to our samples, we observed multiple GluN2B species that migrated differently on the gel, suggesting that the GluN2BTail is phosphorylated at multiple sites. The mass spectrometry results revealed PKA phosphorylation sites on Ser-929/930, Ser-940 and Ser-1050 on GluN2B, that were within the spinophilin binding region. In addition, Ser-882, was determined as a predicted PKA site using the NetPhos prediction algorithm (Blom et al. 1999; Blom et al. 2004). Future studies will need to detail if phosphorylation of any PKA site(s) on spinophilin and/or GluN2B regulate the interaction of these two proteins.
Spinophilin attenuates PP1 targeting to GluN2B.
Inhibition of PP1 in the presence, but not absence, of spinophilin increases NMDAR currents (Feng et al. 2000). Spinophilin directly binds PP1 (Allen et al. 1997; Colbran et al. 1997) and can alter the phosphorylation state of various proteins by either targeting (Terry-Lorenzo et al. 2002; Grossman et al. 2004; Ragusa et al. 2010) or inhibiting PP1 activity (Bollen et al. 2010; Ragusa et al. 2010). PP1 associates with ~200 targeting/inhibitor proteins and it is not thought to exist in an unbound form; therefore, PP1 catalytic activity is exclusively dependent upon these associated proteins (Bollen et al. 2010; Cohen 2002; Virshup & Shenolikar 2009). We found that spinophilin overexpression decreases PP1γ1 association with GluN2BTail in heterologous cells. Our results suggest that this effect of spinophilin is not due to competition between PP1 and spinophilin interaction domains. Therefore, we posit that decreased PP1 binding to GluN2B is a result of greater affinity of PP1 for spinophilin compared to GluN2B such that when spinophilin is present, it sequesters PP1 away from GluN2B. However, when spinophilin is low or absent, unbound PP1 will find a binding partner (e.g. GluN2B). Spinophilin-dependent sequestration of PP1 is not an overexpression artifact as we observed greater association of GluN2B with PP1 in spinophilin KO compared to WT mice, suggesting an in vivo role of spinophilin. Of note, we have previously found that spinophilin binding to PP1 is regulatable by kinases, such as CDK5 (Edler et al. 2018). Therefore, modulation of the spinophilin/PP1 interaction may regulate PP1 binding to, or activity at, synaptic substrates.
Spinophilin enhances GluN2B phosphorylation at Ser-1284.
In order to determine the functional consequences of the spinophilin/GluN2B interaction and/or spinophilin expression on GluN2B phosphorylation, we utilized MS/MS-based approaches to identify and ratiometrically quantify various phosphorylation sites on GluN2BTail in the absence or presence of overexpressed PP1γ1 and spinophilin. Interestingly, our mass spectrometry and immunoblotting results show a significant decrease in GluN2B Ser-1284 phosphorylation caused by overexpression of PP1γ1. This decrease was partially rescued when spinophilin was mutually overexpressed along with PP1γ1. Moreover, PKA overexpression enhanced the association of spinophilin with GluN2BTail and also led to a greater attenuation of the PP1-induced decrease in Ser-1284 phosphorylation in HEK293 cells both by mass spectrometry and using a Ser-1284 antibody. Of note, the F451A mutant spinophilin which has attenuated binding to PP1γ1 and does not displace PP1 from GluN2B, had a less robust attenuation of the PP1-dependent dephosphorylation of Ser-1284. Together these data suggest that in the absence of spinophilin, PP1 binds GluN2B and dephosphorylates it at Ser-1284. However, when spinophilin is present it displaces PP1 from GluN2B. Moreover, the greater attenuation of Ser-1284 by spinophilin in the presence of PKA suggests that even though the spinophilin-PP1 complex may be bound to GluN2B, in this spinophilin bound state, PP1 cannot dephosphorylate Ser-1284 as readily. Moreover, less Ser-1284 phosphorylation in spinophilin KO compared to WT P28 animals suggest a similar in vivo role for spinophilin. Together, these data suggest that spinophilin acts as an inhibitor of PP1 binding to GluN2B to maintain Ser-1284 phosphorylation (Figure 8). These data are of interest as phosphorylation at this site is altered in ischemic conditions as well as acute stress conditions (Lu et al. 2015). However, the roles of Ser-1284 on regulating GluN2B-containing NMDAR channel activity and localization need to be further evaluated.
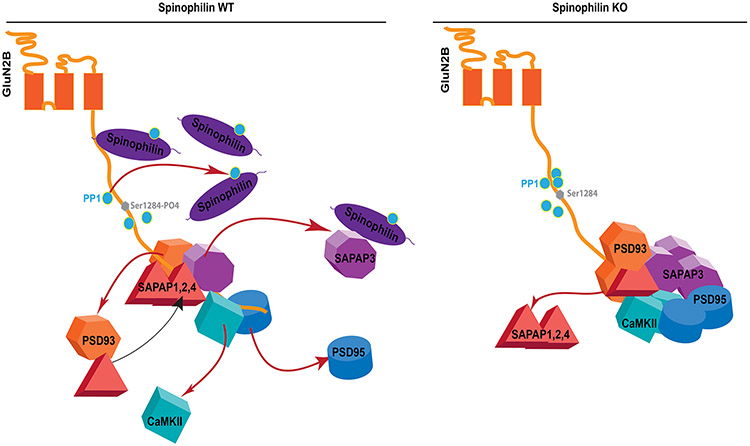
Figure 8: Schema showing spinophilin regulation of GluN2B protein interactions and GluN2B phosphorylation at Ser-1284.
In WT conditions, spinophilin limits PP1 binding to GluN2B thereby maintaining GluN2B phosphorylation at Ser-1284 in a high state. Moreover, spinophilin limits the association of PSD-enriched proteins such as PSD95, CaMKII, and SAPAP3. In spinophilin knockout mice spinophilin no longer limits PP1 binding and therefore, there is a greater interaction of PP1 with GluN2BTail that occurs concurrently with decreased phosphorylation of GluN2B at Ser-1284. Moreover, spinophilin no longer limits the association of proteins such as PSD95, CaMKII, and SAPAP3, which increase their association with GluN2B while concomitantly decreasing the association of the other SAPAP proteins.
Loss of spinophilin alters GluN2B phosphorylation and the GluN2B interactome in vivo.
Spinophilin expression and/or regulation of GluN2B phosphorylation may impact its protein interactions. Using an initial proteomics approach, we found that spinophilin KO mice had altered GluN2B protein interactions. Gel-based proteomics studies may be limited in their ability to detect lowly expressed or lowly interacting proteins and we did not observe PP1 or spinophilin in our dataset possibly be due to low levels of interaction. However, we did observe increased GluN2B interaction with synaptic proteins such as CaMKII isoforms, PSD-95, PSD-93 and SAPAP3, but decreased association with different scaffolding proteins such as SAPAP1, SAPAP2, and SAPAP4 and the cytoskeletal protein, myosin Va. SAPAP3 interacts with spinophilin (Morris et al. 2018) and here we report that the association of SAPAP3 with GluN2B is greater in spinophilin KO compared to WT animals. All four SAPAPs are expressed in hippocampus (Kindler et al. 2004) and therefore spinophilin may normally act to sequester SAPAP3 away from GluN2B to allow for binding of other SAPAP isoforms (Figure 8).
We have observed spinophilin-dependent changes in protein binding to GluN2B that are in the range of 1.5- to 3-fold. It is important to note that these changes in interactions may be due to alterations in protein expression, changes in protein interaction, or both. Moreover, as spinophilin is the major PP1 targeting protein in the PSD, it may be surprising that these changes are not higher. However, compensatory changes, such as decreased PP1 expression in spinophilin KO mice may mitigate these changes. Furthermore, the spinophilin homolog, neurabin, is also highly abundant in the PSD which may limit the effects of spinophilin KO.
The mechanisms and functional consequences of spinophilin-dependent regulation of GluN2B protein interactions are unclear. As stated above, spinophilin KO animals have decreases in Ser-1284 phosphorylation and Ser-1284 phosphorylated GluN2B may be higher in PSD fractions of hippocampal lysates isolated from adult mice (Lu et al. 2015). Functionally, previous studies report that disturbing the CaMKII-GluN2B interaction protects primary hippocampal cultures against ischemic-dependent cell death (Vieira et al. 2016); therefore, regulation of this interaction may have effects on neurotoxicity. Of note, synaptic, compared to extrasynaptic, expression of the NMDAR is associated with neuronal survival (Hardingham & Bading 2010). Pathologically, the GluN2B interactome is implicated in various conditions such as hypoxia-ischemia (Lu et al. 2018). Given the important role of GluN2B interaction with synaptic proteins and GluN2B phosphorylation in ischemic neurotoxicity (Vieira et al. 2016; Lu et al. 2015), future studies will need to delineate if spinophilin-dependent regulation of GluN2B is an important modulator of ischemic neurotoxicity in brain.
Summary
Taken together our data demonstrate that spinophilin attenuates PP1 binding to GluN2B and that this decreased binding enhances Ser-1284 phosphorylation on GluN2B. Moreover, loss of spinophilin enhances GluN2B interactions with PSD-enriched proteins such as PSD-95 and CaMKII.
Supplementary Material
Table S1. Non-normalized and normalized GluN2B interactome isolated from P28 spinophilin WT and KO mouse hippocampus.
Figure S1. Validation of MS/MS spectra for Ser-1284 non-phosphorylated and phosphorylated tryptic peptide. A. Chromatograms matching the total ion current, base peak, non-phosphorylated peptide YPQSPTNSK, and phosphorylated Ser-1284 peptide (YPQsPTNSK) are shown. MS/MS spectra matching the non-phosphorylated peptide YPQSPTNSK (B), and phosphorylated Ser-1284 peptide (YPQsPTNSK; C). Chromatograms showing no proper peak for either the non-phosphorylated peptide YPQSPTNSK or phosphorylated Ser-1284 peptide (YPQsPTNSK) in a sample containing mutant S1284A GluN2BTail. E) Chromatogram showing the S1284A mutant tryptic peptide (YPQAPTNSK). F) MS/MS matching the S1284A mutant tryptic peptide (YPQAPTNSK).
Acknowledgments:
Mass spectrometry was provided by the Indiana University School of Medicine Proteomics Core Facility. We thank Dr. Yvonne Lai (Indiana University and Anagin, Inc) and Dr. Andy Hudmon (Purdue University) for critical evaluation of the manuscript. We also thank Victor Olafusi and Cameron Morris for the help in optimizing/finalizing experiments.
Funding: Acquisition of the IUSM Proteomics core instrumentation used for this project was provided by the Indiana University Precision Health Initiative. Grant support for the studies was from NIH (K01NS073700, R21/R33DA041876 to AJB), Department of Biology Start-up funds and Bridge Funding (to AJB).
Abbreviations:
- NMDA
N-Methyl, D-Aspartate
- NMDAR
N-Methyl, D-Aspartate receptor
- PKAc
Catalytic subunit of Protein kinase A
- IBMX
3-isobutyl-1-methylxanthine
- PP1
Protein Phosphatase 1
- PKA
Protein Kinase A
- CDK5
Cyclin-dependent kinase 5
- PSD
Postsynaptic density
- RRID
Research Resource Identifier
Footnotes
Competing interests. The authors have no competing interests to declare.
References
- Ai H, Shi XF, Hu XP, Fang WQ, Zhang B and Lu W (2017) Acute stress regulates phosphorylation of N-methyl-d-aspartate receptor GluN2B at S1284 in hippocampus. Neuroscience 351, 24–35. [DOI] [PubMed] [Google Scholar]
- Allen PB, Ouimet CC and Greengard P (1997) Spinophilin, a novel protein phosphatase 1 binding protein localized to dendritic spines. Proc Natl Acad Sci U S A 94, 9956–9961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen PB, Zachariou V, Svenningsson P et al. (2006) Distinct roles for spinophilin and neurabin in dopamine-mediated plasticity. Neuroscience 140, 897–911. [DOI] [PubMed] [Google Scholar]
- Baucum AJ 2nd, Brown AM and Colbran RJ (2013) Differential association of postsynaptic signaling protein complexes in striatum and hippocampus. J Neurochem 124, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucum AJ 2nd, Jalan-Sakrikar N, Jiao Y, Gustin RM, Carmody LC, Tabb DL, Ham AJ and Colbran RJ (2010) Identification and validation of novel spinophilin-associated proteins in rodent striatum using an enhanced ex vivo shotgun proteomics approach. Mol Cell Proteomics 9, 1243–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucum AJ 2nd, Shonesy BC, Rose KL and Colbran RJ (2015) Quantitative proteomics analysis of CaMKII phosphorylation and the CaMKII interactome in the mouse forebrain. ACS Chem Neurosci 6, 615–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucum AJ 2nd, Strack S and Colbran RJ (2012) Age-dependent targeting of protein phosphatase 1 to Ca2+/calmodulin-dependent protein kinase II by spinophilin in mouse striatum. PLoS One 7, e31554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear MF and Malenka RC (1994) Synaptic plasticity: LTP and LTD. Curr Opin Neurobiol 4, 389–399. [DOI] [PubMed] [Google Scholar]
- Bielas SL, Serneo FF, Chechlacz M, Deerinck TJ, Perkins GA, Allen PB, Ellisman MH and Gleeson JG (2007) Spinophilin facilitates dephosphorylation of doublecortin by PP1 to mediate microtubule bundling at the axonal wrist. Cell 129, 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom N, Gammeltoft S and Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294, 1351–1362. [DOI] [PubMed] [Google Scholar]
- Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S and Brunak S (2004) Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 4, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Bollen M, Peti W, Ragusa MJ and Beullens M (2010) The extended PP1 toolkit: designed to create specificity. Trends in biochemical sciences 35, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkworth RI, Breinl RA and Kobe B (2003) Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc Natl Acad Sci U S A 100, 74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody LC, Baucum AJ, Bass MA and Colbran RJ (2008) Selective targeting of the γ1 isoform of protein phosphatase 1 to F-actin in intact cells requires multiple domains in spinophilin and neurabin. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology 22, 1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen PT (2002) Protein phosphatase 1--targeted in many directions. Journal of cell science 115, 241–256. [DOI] [PubMed] [Google Scholar]
- Colbran RJ, Bass MA, McNeill RB, Bollen M, Zhao S, Wadzinski BE and Strack S (1997) Association of brain protein phosphatase 1 with cytoskeletal targeting/regulatory subunits. J Neurochem 69, 920–929. [DOI] [PubMed] [Google Scholar]
- Das S, Sasaki YF, Rothe T et al. (1998) Increased NMDA current and spine density in mice lacking the NMDA receptor subunit NR3A. Nature 393, 377–381. [DOI] [PubMed] [Google Scholar]
- Di Sebastiano AR, Fahim S, Dunn HA, Walther C, Ribeiro FM, Cregan SP, Angers S, Schmid S and Ferguson SS (2016) Role of Spinophilin in Group I Metabotropic Glutamate Receptor Endocytosis, Signaling, and Synaptic Plasticity. The Journal of biological chemistry 291, 17602–17615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edler MC, Salek AB, Watkins DS, Kaur H, Morris CW, Yamamoto BK and Baucum AJ 2nd (2018) Mechanisms Regulating the Association of Protein Phosphatase 1 with Spinophilin and Neurabin. ACS Chem Neurosci 9, 2701–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteves SL, Domingues SC, da Cruz e Silva OA, Fardilha M and da Cruz e Silva EF (2012) Protein phosphatase 1alpha interacting proteins in the human brain. OMICS 16, 3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Yan Z, Ferreira A, Tomizawa K, Liauw JA, Zhuo M, Allen PB, Ouimet CC and Greengard P (2000) Spinophilin regulates the formation and function of dendritic spines. Proc Natl Acad Sci U S A 97, 9287–9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosang AJ and Colbran RJ (2015) Transparency Is the Key to Quality. The Journal of biological chemistry 290, 29692–29694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis SH, Turko IV and Corbin JD (2001) Cyclic nucleotide phosphodiesterases: relating structure and function. Progress in nucleic acid research and molecular biology 65, 1–52. [DOI] [PubMed] [Google Scholar]
- Grossman SD, Futter M, Snyder GL, Allen PB, Nairn AC, Greengard P and Hsieh-Wilson LC (2004) Spinophilin is phosphorylated by Ca2+/calmodulin-dependent protein kinase II resulting in regulation of its binding to F-actin. J Neurochem 90, 317–324. [DOI] [PubMed] [Google Scholar]
- Hardingham GE and Bading H (2010) Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci 11, 682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiday AC, Edler MC, Salek AB, Morris CW, Thang M, Rentz TJ, Rose KL, Jones LM and Baucum AJ 2nd (2017) Mechanisms and Consequences of Dopamine Depletion-Induced Attenuation of the Spinophilin/Neurofilament Medium Interaction. Neural Plast 2017, 4153076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Wilson LC, Benfenati F, Snyder GL, Allen PB, Nairn AC and Greengard P (2003) Phosphorylation of spinophilin modulates its interaction with actin filaments. The Journal of biological chemistry 278, 1186–1194. [DOI] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY et al. (2010) COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature 468, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler S, Rehbein M, Classen B, Richter D and Bockers TM (2004) Distinct spatiotemporal expression of SAPAP transcripts in the developing rat brain: a novel dendritically localized mRNA. Brain Res Mol Brain Res 126, 14–21. [DOI] [PubMed] [Google Scholar]
- Lu F, Shao G, Wang Y et al. (2018) Hypoxia-ischemia modifies postsynaptic GluN2B-containing NMDA receptor complexes in the neonatal mouse brain. Exp Neurol 299, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W, Ai H, Peng L, Wang JJ, Zhang B, Liu X and Luo JH (2015) A novel phosphorylation site of N-methyl-D-aspartate receptor GluN2B at S1284 is regulated by Cdk5 in neuronal ischemia. Experimental neurology 271, 251–258. [DOI] [PubMed] [Google Scholar]
- Malenka RC and Bear MF (2004) LTP and LTD: an embarrassment of riches. Neuron 44, 5–21. [DOI] [PubMed] [Google Scholar]
- McKay S, Ryan TJ, McQueen J et al. (2018) The Developmental Shift of NMDA Receptor Composition Proceeds Independently of GluN2 Subunit-Specific GluN2 C-Terminal Sequences. Cell Rep 25, 841–851 e844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellacheruvu D, Wright Z, Couzens AL et al. (2013) The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nature methods 10, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CW, Watkins DS, Salek AB, Edler MC and Baucum AJ 2nd (2018) The association of spinophilin with disks large-associated protein 3 (SAPAP3) is regulated by metabotropic glutamate receptor (mGluR) 5. Molecular and cellular neurosciences 90, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison DK, Murakami MS and Cleghon V (2000) Protein kinases and phosphatases in the Drosophila genome. The Journal of cell biology 150, F57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Sardiu ME, Pattenden SG, Workman JL, Florens L and Washburn MP (2011) Highly reproducible label free quantitative proteomic analysis of RNA polymerase complexes. Mol Cell Proteomics 10, M110 000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad K, Reddy-Alla S, Driller JH et al. (2015) Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function. Nat Commun 6, 8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JA, Stein IS, Lau CG et al. (2014) Phosphorylation of Ser1166 on GluN2B by PKA is critical to synaptic NMDA receptor function and Ca2+ signaling in spines. J Neurosci 34, 869–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragusa MJ, Dancheck B, Critton DA, Nairn AC, Page R and Peti W (2010) Spinophilin directs protein phosphatase 1 specificity by blocking substrate binding sites. Nat Struct Mol Biol 17, 459–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrouilhe D, di Tommaso A, Metaye T and Ladeveze V (2006) Spinophilin: from partners to functions. Biochimie 88, 1099–1113. [DOI] [PubMed] [Google Scholar]
- Satoh A, Nakanishi H, Obaishi H et al. (1998) Neurabin-II/spinophilin. An actin filament-binding protein with one pdz domain localized at cadherin-based cell-cell adhesion sites. The Journal of biological chemistry 273, 3470–3475. [DOI] [PubMed] [Google Scholar]
- Seamon KB and Daly JW (1981) Forskolin: a unique diterpene activator of cyclic AMP-generating systems. Journal of cyclic nucleotide research 7, 201–224. [PubMed] [Google Scholar]
- Smith FD, Oxford GS and Milgram SL (1999) Association of the D2 dopamine receptor third cytoplasmic loop with spinophilin, a protein phosphatase-1-interacting protein. The Journal of biological chemistry 274, 19894–19900. [DOI] [PubMed] [Google Scholar]
- Smith-Kinnaman WR, Berna MJ, Hunter GO, True JD, Hsu P, Cabello GI, Fox MJ, Varani G and Mosley AL (2014) The interactome of the atypical phosphatase Rtr1 in Saccharomyces cerevisiae. Mol Biosyst 10, 1730–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartzwelder HS, Risher ML, Miller KM, Colbran RJ, Winder DG and Wills TA (2016) Changes in the Adult GluN2B Associated Proteome following Adolescent Intermittent Ethanol Exposure. PLoS One 11, e0155951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry-Lorenzo RT, Elliot E, Weiser DC, Prickett TD, Brautigan DL and Shenolikar S (2002) Neurabins recruit protein phosphatase-1 and inhibitor-2 to the actin cytoskeleton. The Journal of biological chemistry 277, 46535–46543. [DOI] [PubMed] [Google Scholar]
- Vieira MM, Schmidt J, Ferreira JS et al. (2016) Multiple domains in the C-terminus of NMDA receptor GluN2B subunit contribute to neuronal death following in vitro ischemia. Neurobiol Dis 89, 223–234. [DOI] [PubMed] [Google Scholar]
- Virshup DM and Shenolikar S (2009) From promiscuity to precision: protein phosphatases get a makeover. Mol Cell 33, 537–545. [DOI] [PubMed] [Google Scholar]
- Welch EJ, Jones BW and Scott JD (2010) Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol Interv 10, 86–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Hsieh-Wilson L, Feng J, Tomizawa K, Allen PB, Fienberg AA, Nairn AC and Greengard P (1999) Protein phosphatase 1 modulation of neostriatal AMPA channels: regulation by DARPP-32 and spinophilin. Nat Neurosci 2, 13–17. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Non-normalized and normalized GluN2B interactome isolated from P28 spinophilin WT and KO mouse hippocampus.
Figure S1. Validation of MS/MS spectra for Ser-1284 non-phosphorylated and phosphorylated tryptic peptide. A. Chromatograms matching the total ion current, base peak, non-phosphorylated peptide YPQSPTNSK, and phosphorylated Ser-1284 peptide (YPQsPTNSK) are shown. MS/MS spectra matching the non-phosphorylated peptide YPQSPTNSK (B), and phosphorylated Ser-1284 peptide (YPQsPTNSK; C). Chromatograms showing no proper peak for either the non-phosphorylated peptide YPQSPTNSK or phosphorylated Ser-1284 peptide (YPQsPTNSK) in a sample containing mutant S1284A GluN2BTail. E) Chromatogram showing the S1284A mutant tryptic peptide (YPQAPTNSK). F) MS/MS matching the S1284A mutant tryptic peptide (YPQAPTNSK).