To the Editor,
We read with interest the study by Chu et al., in which SARS‐CoV‐2 was detected in only 30 of 54 (56%) of healthcare workers (HCW) by real‐time reverse transcription polymerase chain reaction (RT‐PCR) in upper respiratory tract (URT) specimens. 1 At our institution, we similarly had a case of a healthy 33‐year‐old female HCW who developed fevers after working several weeks in clinical assessment areas including the COVID‐19 ward where appropriate personal protective equipment was used. Despite high suspicion for COVID‐19, two nasopharyngeal swabs (NPS) tested negative on days 2 and 4 of illness using a laboratory‐developed assay targeting genes for the envelope (E) protein and RNA‐dependent RNA polymerase. 2 Nevertheless, she continued home self‐isolation given ongoing symptoms.
In the 2nd week of illness, she developed anorexia, nausea, and vomiting but still lacked respiratory symptoms. Due to persisting suspicion for COVID‐19, a third NPS and paired 1 ml saliva specimen was collected on illness day 9. NPS was tested on another platform using the cobas® SARS‐CoV‐2 test (Roche) targeting E and Orf1a genes. Saliva was tested using the LightMix® ModularDx SARS‐CoV (COVID19) E‐gene assay (TIB Molbiol). This third NPS tested “indeterminate”: E gene was positive with a cycle threshold (Ct) value of 41.47, and the Orf1a gene was negative. Saliva tested positive with a Ct of 25.83 for the E gene (Figure 1).
Figure 1.
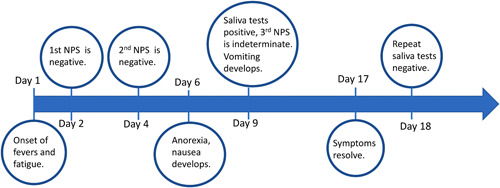
Timeline of illness progression and testing for the healthcare worker
The potentially missed COVID‐19 diagnosis in this HCW highlights several important aspects regarding SARS‐CoV‐2 testing. First, when there is high pretest probability and suspicion for COVID‐19, it is important to maintain isolation measures regardless of an initial negative NPS. The sensitivity of real‐time RT‐PCR of URT specimens may be limited early or late in the disease course with an estimated median false‐negative rate of 38% on the first day of symptom onset. 3 Despite high analytical sensitivity, “false‐negative” results generally arise from preanalytical issues including poor collection technique, nonconducive anatomy, or features of the clinical presentation. 4 , 5 Consequently, saliva for SARS‐CoV‐2 diagnosis is an attractive alternative specimen for ease of collection and less dependence on the collector technique. Reports of cases detected by saliva alone when paired with URT collection are increasing and include prospective studies of 91 inpatients with SARS‐CoV‐2 and 217 quarantine centre outpatients where 11% and 47.5% tested positive only by saliva, respectively. 6 , 7 These findings along with results in Chu et al.'s study of HCWs demonstrate that combining modalities enhances diagnostic yield, especially in cases with high clinical suspicion but negative URT testing. Chu et al. used respiratory imaging in HCWs to enhance diagnosis; however, alternative specimen types like saliva or sera for serological testing, especially beyond the first week of illness, can likewise increase sensitivity and specificity for SARS‐CoV‐2 detection by accounting for limitations of real‐time RT‐PCR of URT samples. 8 , 9
Second, although clinical diagnostic laboratories are accustomed to establishing threshold Ct values for clinical reporting, it is essential to recognize that SARS‐CoV‐2 is a virus of considerable public health importance. Laboratories should carefully review all late Ct values for SARS‐CoV‐2 targets and interpret the results in the clinical context of the patient. Missed diagnoses in HCWs can have substantial consequences including healthcare facility outbreaks. In this case, the third NPS had only one genetic target detected with a very late Ct value, which was reported as “Indeterminate” to indicate a weak positive result. It would have been erroneous to report this as “Negative” given the exposure history and the paired saliva sample, which was clearly positive.
Third, although respiratory symptoms are common in COVID‐19, the extra‐pulmonary disease can exist in isolation. A retrospective study of 651 hospitalized patients found that 28.4% of patients with gastrointestinal symptoms lacked any respiratory symptoms. 10 The HCW's absence of respiratory symptoms, predominant gastrointestinal symptoms, and consistently negative or weakly positive NPS samples likely points toward more gastrointestinal rather than respiratory viral shedding. Clinical judgement must guide testing and interpretation: targeted testing of specimens where symptoms manifest—beyond a single NPS—should be a guiding principle for diagnosing SARS‐CoV‐2 infections and proved to be appropriate and essential here. As the understanding of SARS‐CoV‐2 continues to expand, this case emphasizes that no one diagnostic test fits all.
REFERENCES
- 1. Chu J, Yang N, Wei Y, et al. Clinical characteristics of 54 medical staff with COVID‐19: a retrospective study in a single center in Wuhan, China. J Med Virol. 2020;92(7):807‐813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. Variation in false‐negative rate of reverse transcriptase polymerase chain reaction‐based SARS‐CoV‐2 tests by time since exposure. Ann Intern Med. 2020;173:262‐267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kinloch NN, Ritchie G, Brumme CJ, et al. Suboptimal biological sampling as a probable cause of false‐negative COVID‐19 diagnostic test results. J Infect Dis. 2020;222:899‐902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Piras A, Rizzo D, Uzzau S, De Riu G, Rubino S, Bussu F. Inappropriate nasopharyngeal sampling for SARS‐CoV‐2 detection is a relevant cause of false‐negative reports. Otolaryngol Head Neck Surg. 2020;163:459‐461. [DOI] [PubMed] [Google Scholar]
- 6. Jamal AJ, Mozafarihashjin M, Coomes E, et al. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin Infect Dis. 2020. 10.1093/cid/ciaa848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rao M, Rashid FA, Sabri FSAH, et al. Comparing nasopharyngeal swab and early morning saliva for the identification of SARS‐CoV‐2. Clin Infect Dis. 2020. 10.1093/cid/ciaa1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deeks JJ, Dinnes J, Takwoingi Y, et al. Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Database Syst Rev. 2020;6(6):Cd013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang P. Combination of serological total antibody and RT‐PCR test for detection of SARS‐COV‐2 infections. J Virol Methods. 2020;283:113919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jin X, Lian JS, Hu JH, et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus‐infected disease 2019 (COVID‐19) with gastrointestinal symptoms. Gut. 2020;69(6):1002‐1009. [DOI] [PMC free article] [PubMed] [Google Scholar]


