Summary
The current pandemic of severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) has quickly emerged as a global health concern with government bodies worldwide taking drastic control measures. Understanding the virology of SARS‐CoV‐2, its molecular mechanisms, and its pathogenesis are required for a targeted therapeutic approach. In this review, we highlight the current molecular and drug advances that target SARS‐CoV‐2 at the genome level. We also summarize studies that therapeutically target the host angiotensin‐converting enzyme 2 and proteases. Finally, we summarize antibody‐mediated therapeutic approaches, as well as recent trends in vaccine development. Hence, the purpose of this study is to investigate different molecular targets in SARS‐CoV‐2 pathogenesis and their usefulness in developing strategies for drug development.
Keywords: ACE2, antibodies, SARS‐CoV‐2, TMPRSS2, vaccine development, virus
Abbreviations
- ACE2
angiotensin‐converting enzyme 2
- Ang 1–7
angiotensin 1–7
- Ang 1–9
angiotensin 1–9
- Ang I
angiotensin I
- Ang II
angiotensin II
- ARDS
acute respiratory distress syndrome
- AT1
angiotensin II type 1
- AT2
angiotensin II type 2
- CDE
Centre for Drug Evaluation
- Covid‐19
coronavirus disease 2019
- CP
convalescent plasma
- FDA
Food and Drug Administration
- HCoV
human coronavirus
- hrsACE2
human recombinant soluble ACE2
- MERS‐CoV
Middle East respiratory syndrome‐coronavirus
- Mpro
main protease
- NAbs
neutralizing antibodies
- nsps
non‐structural proteins
- PLPro
papain‐like protease
- RAS
renin‐angiotensin system
- RBD
receptor binding domain
- RdRp
RNA‐dependent RNA polymerase
- rhACE2
recombinant ACE2
- SARS‐CoV
severe acute respiratory syndrome coronavirus
- SARS‐CoV‐2
severe acute respiratory syndrome‐coronavirus‐2
- TMPRSS2
transmembrane protease serine 2
- WHO
World Health Organization
1. INTRODUCTION
The current outbreak of coronavirus disease 2019 (Covid‐19) due to severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) was initially reported in December 2019 in Wuhan, China. SARS‐CoV‐2 originated from zoonotic transmission, similar to that of severe acute respiratory syndrome‐coronavirus (SARS‐CoV) and Middle East respiratory syndrome‐coronavirus (MERS‐CoV), but with a severe pandemic potential. 1 , 2 Sustained human–human transmission of SARS‐CoV‐2 is an important characteristic, which facilitates rapid universal spread of Covid‐19. By March 11, 2020, the disease outbreak had spread across 114 countries, infecting more than 118,000 and causing 4292 deaths, so that the World Health Organization (WHO) pronounced it a pandemic disease. 3 The WHO strategized to break human–human transmission by quarantining patients at an earlier stage, recognizing and reducing transmission from animal sources, and promoting research and vaccine development. 4 Severe social and economic impacts were felt amidst the struggle against the deadly virus. Despite the precautionary measures across different countries, SARS‐CoV‐2 had infected more than 13.8 million individuals by July 2020, causing 597,000 deaths worldwide. 5
SARS‐CoV‐2, a 29.8 Kb positive‐sense single‐stranded RNA virus, shares more than 70% sequence homology with SARS‐CoV. 6 Despite its similarity with SARS‐CoV, its transmission competency and complexity are different. Investigations suggest that the contagious SARS‐CoV‐2 virus spread from human to human via respiratory droplet transmission from coughing or sneezing. 7 Recently, the WHO has also announced airborne and aerosol as one possible mode of transmission of SARS‐CoV‐2. 8 , 9 The average incubation period, which is the duration between viral exposure and symptom onset, is estimated to range from 5 to 6 days, however; it can go up to 14 days in some cases. 10 , 11 , 12 Mostly, affected individuals experience mild symptoms, comprised of high body temperature in conjunction with symptoms such as fatigue, cough, diarrhoea, sore throat and headache. 13 Some patients experience pneumonia and acute respiratory distress syndrome (ARDS). Individuals with underlying health complications such as heart disease, chronic pulmonary disease and diabetes develop more severe symptoms. 14
Numerous investigations on SARS‐CoV‐2 focus on delineating potential therapeutic targets, identifying viral inhibitors and re‐purposing antiviral drugs to combat infection. The US Food and Drug Administration (FDA) and other regulatory agencies have granted permission to use drugs like remdesivir, dexamethasone and interferon‐beta to treat severely ill Covid‐19 patients 15 , 16 , 17 Vaccine research for Covid‐19 is also progressing, with several significant studies in the clinical trial phase. With the urgent demand for a vaccine or specific antiviral drugs to combat Covid‐19 pandemic and with the rapid growth of research on novel SAR‐CoV‐2 drug targets and approaches, it is of utmost importance to summarize the newest key findings. Updating findings related to SARS‐CoV‐2 may help understand which contemporary research approaches might provide the best protection until a vaccine can be made available. This review summarizes the recent drug approaches that target SARS‐CoV‐2 at the genome level. We have highlighted the recent therapeutic findings on host molecular targets, ACE2 (angiotensin‐converting enzyme 2), and cell surface and lysosomal proteases, which are essential for the entry and pathogenesis of SARS‐CoV‐2 virus. Lastly, we have summarized antibody‐mediated drug approaches, encompassing the recent trends in SARS‐CoV‐2 vaccine development. Hence, this review investigates the different molecular targets in SARS‐CoV‐2 pathogenesis, and their respective drug approaches, in an attempt to explain the present status of Covid‐19 research.
2. THERAPEUTIC TARGETS IN SARS‐CoV‐2
The genome of SARS‐CoV‐2 codes for non‐structural proteins (nsps), structural proteins and accessory proteins, all of which mediate viral maintenance, replication and life cycle. The ORF 1a and ORF 1b coding for the polyproteins pp1a and pp1ab make up about two‐thirds of the whole genome 18 (Figure 1A). These polyproteins are processed into 16 nsps (nsp1–16) by the main protease (Mpro) and the Papain‐like protease (PLPro). 19 ORFs occupying one‐third of the genome encode structural proteins such as spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins. The ORF3, ORF6, ORF7 and ORF8 genes encode 6–9 accessory proteins. 20 Recent studies employed different strategies to inhibit viral pathogenesis by targeting nsps and structural proteins, combating the progression of viral replication and life cycle progression inside host cells.
FIGURE 1.
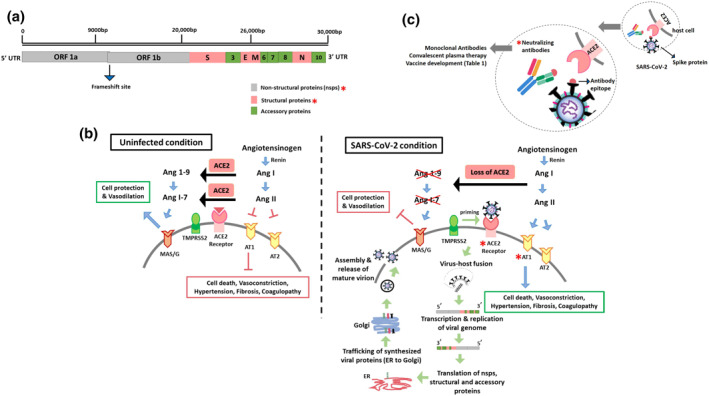
Diverse therapeutic targets to alleviate SARS‐CoV‐2 infection in humans. (A) SARS‐CoV‐2 viral genome. Molecular targets deployed for SARS‐CoV‐2 drug approach studies are marked by red asterisks. (B) Comparison between physiological condition and SARS‐CoV‐2 mediated host receptor invasion. (C) Neutralizing‐antibody‐dependent approach to combat SARS‐CoV‐2 infection
2.1. | Targeting the nsps
Since viral proteases mediate the generation of mature nsps, inhibiting the proteolytic functions of Mpro and PLPro might halt the production of mature nsps. Drugs that target Mpro may decrease the risk of drug resistance, which results from mutations and might exhibit wide spectrum antiviral activity. 21 The SARS‐CoV‐2 Mpro inhibitor 11a showed a high inhibition rate and good pharmacokinetic properties. 19 Recently Jin et al. showed that an FDA approved antineoplastic drug, carmofur, inhibited SARS‐CoV‐2 Mpro‐mediated viral replication in cell cultures. 22 Because HIV protease inhibitors lopinavir and ritonavir were tested on Covid‐19 patients in non‐randomized studies, 23 interim clinical guidelines for treating Covid‐19 patients in Belgium, China, France, India, Italy, South Korea, Spain and Switzerland included lopinavir and ritonavir, either alone or in combination with interferon beta‐1b, along with the standard of care recommended for moderate to severe infections. 24 Preliminary uncontrolled evidence from Chen et al. reported combined benefits of HCV inhibitor danoprevir and ritonavir in the recovery of Covid‐19 patients following 4–12 days of treatment. 25 Most of the nsps (1–16) are critical to the replication of SARS‐CoV‐2 virus. Nsp12, a highly conserved RNA‐dependent RNA polymerase (RdRp), is a potential drug target to alleviate viral replication inside the host. Recently, several molecular docking studies that highlighted the inhibition of RdRp via phytochemicals, antiviral terpenes and phenylpropanoids were published. 26 , 27 Additionally, re‐purposed antiviral RdRp inhibitors remdesivir and favipiravir were clinically assessed for their effectiveness in Covid‐19 patients in many countries. 28 , 29 , 30
2.2. Targeting the structural proteins
The major structural proteins responsible for viral assembly and SARS‐CoV‐2 pathogenesis are spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins. SARS‐CoV‐2 utilizes S protein to bind to the human ACE2 receptor and enables membrane‐mediated fusion and viral entry, thus making this protein a leading target antigen in vaccine development. 31 Xia et al. have identified lipopeptide EK1C4 as a potential inhibitor of the S protein that reduces S protein‐dependent membrane fusion of SARS‐CoV‐2. 32 While studies have seldom focused on the M protein of SARS‐CoV‐2 as a drug target, the E and N proteins have been understood to have critical functions. The E proteins form ion channels, rendering a crucial role in the assembly of viral particles and pathogenesis inside the host. 33 Molecular docking studies report that the antivirals glecaprevir, saquinavir, simeprevir might inhibit SARS‐CoV‐2 and reduce pathogenicity. 34 , 35 Recent studies have targeted the N proteins with pre‐existing FDA approved drugs and re‐purposed anti‐human coronavirus (HCoV) drug combinations such as sirolimus with dactinomycin, mercaptopurine with melatonin, and toremifene with emodin to shorten the viral life cycle inside the host. 36 Due to the evolutionarily conserved nature of the N protein sequence (∼90% with SARS‐CoV) and its contribution to high immunogenicity and abundant expression in infected people, it is one of the vaccine candidates that is highly studied. 37
3. SARS‐CoV‐2 AND ACE2 SIGNALLING
The primary dependency of SARS‐CoV‐2 on the ACE2 receptor has necessitated a detailed understanding of ACE2 downstream functions. Because its discovery over 20 years ago, ACE2 has been recognized as a crucial determinant of the renin‐angiotensin system (RAS). 38 , 39 The harmful effects of the ACE/RAS pathway are counterbalanced via the ACE2/Ang1–7/MAS axis. 40 The ACE/RAS pathway is initiated upon the formation of active renin from an inactive precursor prorenin (Mr 46,000 protein). The renin released in blood cleaves angiotensinogen into Ang I (angiotensin I) (Figure 1B). Physiologically dormant Ang I acts as a precursor of active Ang II (angiotensin II). Binding of Ang II to AT1 and AT2 (Angiotensin Type 1 and Type 2) receptors results in vasoconstriction, endothelial injury, endovascular thrombosis and an elevated blood volume. Additionally, Ang II also promotes hypertension and thrombosis in arterioles. To alleviate the harmful effects of Ang II, ACE2, a monocarboxypeptidase, cleaves Ang I and Ang II into Ang 1–9 (angiotensin 1–9) and Ang 1–7 (angiotensin 1–7), respectively. 41 Activation of the ACE2 receptor impedes the harmful actions of Ang II on the cells, such as fibrosis, thrombosis formation, angiogenesis and cell death. The Ang 1–9 and Ang 1–7 peptides exert beneficial outcomes like vasodilatory and anti‐proliferative and defensive functions by activating the MAS/G receptor. 42 Taken together, these processes demonstrate the objective role of ACE2 receptor in the endogenous counter‐regulatory system (ACE2/Ang1–7/MAS1 axis) that stabilizes the adverse effects of ACE/AngII/AT1 receptor signalling. The overall physiological outcomes of RAS activation elevate vascular tone, body sodium content and total body water.
Together, the affinity of ACE2 receptor to SARS‐CoV‐2 and the impaired endogenous downstream functions of ACE2 contribute to the observed clinical features of ARDS, pulmonary inflammation, myocardial injury, renal failure and increased mortality in the ageing population and people with heart and metabolic diseases. 43 Studies highlight the increased expression of membrane ACE2 receptors and its reduced expression in circulation (soluble form) in uninfected people. 44 Unfortunately, entry of SARS‐CoV‐2 through the ACE2 receptor‐dependent paradigm leads to endocytosis of membrane ACE2 receptors together with SARS‐CoV‐2, and subsequently ACE2 receptor downregulation. 45 This results in the deprivation of the catalytic effect of ACE2 receptors at the membrane surface, explaining unopposed Ang II effects through the ACE/Ang II/AT1 axis. 46 Hence, the combined impact of ACE2 receptor downregulation and AT1 receptor upregulation suggest the use of AT1 receptor blockers, recombinant ACE2, and angiotensin 1–7 as beneficial targets against SARS‐CoV‐2 infection.
3.1. Targeting ACE2 and its receptors
The AT1 receptor antagonist drugs losartan, telmisartan and olmesartan have been widely used in hypertensive patients since the 1990s since their adverse effects are rare. 47 The AT1 receptor antagonists elevate ACE2 expression in rats and humans. Currently, losartan and telmisartan are under phase II clinical trial for SARS‐CoV2 infection. 48 , 49 , 50 While recombinant ACE2 can be an additional target, studies claim that a recombinant ACE2 (rhACE2) lacks the membrane‐anchored domain, thus becoming soluble inside the cell. 51 , 52 , 53 Recently, the Centre for Drug Evaluation (CDE), China, ended a clinical trial for rhACE2 due to its lack of statistically significant effect as a drug treatment approach against SARS‐CoV2. 54 , 55 However, scientists have identified a genetically altered human recombinant soluble ACE2 (hrsACE2), which showed a dose‐dependent effect on SARS‐CoV‐2 viral replication in cell culture studies, and a reduced viral entry by a factor of 1000 to 5000. 56 Despite these benefits, hrsACE2 is not a favoured drug for clinical use because it is a glycosylated protein, and its production will require additional effort, time, and cost, which is not advantageous in drug development. 57
Apart from ATI receptor blockers and recombinant ACE2, molecular docking studies revealed that two selected phytochemicals, 6‐α‐acetoxygedunin and echitamine, showed optimum binding to the human ACE2 receptor, which efficiently blocked the receptor and inhibited receptor‐mediated SAR‐CoV2 entry. 58 This preliminary evidence highlights the need for a comprehensive analysis of phytochemicals in an in vivo setting. Interestingly, a recent study by Minato et al. 59 showed that B38‐CAP, a protein from Paenibacillus, had ACE2‐like enzyme activity and decreased the Ang II levels in mice. Most importantly, recombinant B38‐CAP protein converted Ang II to Ang 1–7 and contributed to the other ACE2 targeted functions. This study highlighted the functional similarity between bacterial B38‐CAP and human ACE2 and B38‐CAP is expected to undergo phase I clinical trial shortly. 57 Taken together, the studies mentioned above offer evidence that the ACE2 receptor is one of the leading potential targets against SARS‐CoV‐2.
Understanding the downsides of ACE2 might minimize the unexpected complications that may arise from ACE2 targeted drugs when used against SARS‐CoV‐2 infection. Stawiski et al. 60 identified variants in ACE2 receptors that modify virus‐host interaction and significantly alter the host's vulnerability to SARS‐CoV‐2. They have deciphered 298 unique variants among 256 codons distributed through 805 amino acid residues of the human ACE2 receptor. 61 The understanding that the natural variations of the ACE2 receptor in human populations may alter ACE2 receptor binding affinity to SARS‐CoV‐2 S‐protein produces divergent evidence regarding virus susceptibility. Additionally, ACE2 expression had a decreasing effect in aged individuals, to a greater extent in males than in females. 62 Diseases like diabetes mellitus reduce ACE2 expression, causing ACE2 deficiency. Though Ang 1–7 has a shielding effect against ARDS, enhanced Ang 1–7 formation from ACE2 in different organs (heart, kidney and vessels) might not render a protective outcome. Patients with diabetes who were treated with pioglitazone, glucagon‐like peptide‐1 agonists and mineralocorticoid inhibitors showed increased endogenous ACE2 levels. 63 Hence, the combined use of these diabetic drugs and ATI receptor blocker/recombinant ACE2/B38‐CAP might cause a decrease in cytosolic pH and an increase in viral load, resulting in exacerbation of SARS‐CoV‐2 infection and mortality. 64 Collective considerations provide essential insights into the ACE2 receptor as potential therapeutic targets; however, the downsides of ACE2 highlights the requirement for additional targets to attenuate SARS‐CoV‐2 infection significantly.
4. TARGETING HOST PROTEASES
Following the recognition of the ACE2 receptor by the S1 receptor binding domain (RBD) of SARS‐CoV‐2, the spike protein undergoes proteolytic activation by host proteases, causing cleavage of S1/S2 and S2, and fusion of viral particles with cellular membranes. 65 Upon entry of SARS‐CoV‐2, activation of host cell surface proteases [e.g., transmembrane protease serine 2 (TMPRSS2)] and lysosomal proteases (e.g., cathepsins L/B) contribute to severe symptoms and fatality in some infected patients. 66 , 67 While these two kinds of proteases were actively investigated recently, furin and other proteases were also shown to contribute to the cumulative action of SARS‐CoV‐2. 65 The findings on TMPRSS2 stem from the earlier studies on influenza H1N1 and SARS‐CoV outbreak, wherein this protease exhibited a similar mechanism to advance disease pathogenesis. 68 Therefore, researchers recommend the existing clinically proven TMPRSS2 inhibitors such as camostat, bromhexine, aprotinin, nafamostat to potentially combat SARS‐CoV‐2 infection. 69 Importantly, camostat has advanced to phase I clinical trial in March 2020, as one of the SARS‐CoV‐2 targets. 70 , 71 Similarly, many pre‐existing cathepsin inhibitors like K11777, oxocarbazate, MDL28170, and E‐64d were studied for their effectiveness against SARS‐CoV‐2. 72 , 73 , 74 , 75 , 76 Dexamethasone, a corticosteroid approved by FDA in 1958 as a broad‐spectrum immunosuppressor, is one of the cathepsin L/B inhibitors that has been approved by WHO to treat critically ill Covid‐19 patients. 77 , 78 One concern about using such inhibitors of host proteases may be the loss of function of innate TMPRSS2 and cathepsin L/B responses, which might result in side effects. Targeting TMPRSS2 and cathepsin L/B host protease at the same time using inhibitors might reduce the entry of the SARS‐CoV‐2 virus into host cells, thereby decreasing the viral load.
5. ANTIBODY‐MEDIATED DRUG THERAPEUTICS
5.1. Neutralizing antibodies
Neutralizing antibodies (NAbs) produced as a humoral response from the adaptive immune system bind to the viral epitope, preventing viral epitope‐human receptor attachment and initiating viral lysis by antibody‐mediated opsonisation or complement activation 79 , 80 (Figure 1C). Interestingly, 95%–100% of SARS‐CoV‐2 infected patients show neutralizing activity 14 days after symptom onset, consisting of IgM, IgG and antibodies that target RBD of SARS‐CoV‐2. 81 , 82 Reports claim a high natural recovery rate of ∼92% from SARS‐CoV‐2 infection; however, the remaining patients experience immunological anomalies like lymphopenia, neutrophilia, elevated inflammatory markers chemokines and cytokines. 83 Especially, the lung injury observed in a few Covid‐19 patients indicates a cytokine storm reaction that progressed to ARDS aggravation, causing multi‐organ failure and death. 84 , 85 Therapeutic deployment of NAbs against SARS‐CoV‐2 is an important area of research that will likely contribute to controlling the pandemic and the potential re‐emergence of viruses in the future.
Among SARS‐CoV‐2 targeted therapies, monoclonal antibodies (mAbs) may be efficient in providing immediate protection. Human monoclonal antibodies were studied against SARS‐CoV during the epidemic in 2002–2004. 86 , 87 Genetic similarity between SARS‐CoV and SARS‐CoV‐2 led the researchers to check for the cross‐reactivity of SARS‐CoV mAbs in SARS‐CoV‐2 infected samples. 88 Notably, the RBD‐specific mAbs, CR3022 and S309, cross‐reacted efficiently with the SARS‐CoV‐2 S1B RBD residues 318–510 and 337–444, respectively, without overlapping with ACE2 receptor binding sites. 89 However, mAbs, which can cross‐react with the crucial SARS‐CoV‐2 S1B RBD residues 460–492, may be considered critical for achieving a therapeutically beneficial outcome. The above‐mentioned critical residues are only 50% conserved between both viruses, entailing the need to develop mAbs that are specific to SARS‐CoV‐2 RBD. 90 , 91 Most importantly, a recent study reported the first human mAbs, 47D11, that neutralizes SARS‐CoV‐2 by binding to S1B RBD, similar to CR3022 and S309, establishing its capacity to cross‐neutralize SARS‐CoV as well. 92 The beneficial effect of mAb 47D11 was demonstrated in HEK‐293T cells, which was independent of receptor‐binding inhibition. 93 AbbVie, an American biotech company, has collaborated with this research team to support further preclinical activities and human trials on mAb 47D11. 94 The major limitation of mAbs is the cost; antibody‐based drug treatment is more expensive than vaccines.
5.2. Convalescent plasma therapy
Convalescent plasma (CP) therapy, an approach of passive immunization by infusion of blood plasma from recovered patients into infected individuals, is also being explored in several countries. CP therapy acquired its popularity because of its use against SARS‐CoV, MERS, H1NI and Ebola viruses in the past. 95 Currently, based on non‐randomized studies, the FDA has given emergency approval to CP therapy as a treatment option, which is mainly deployed in critically ill SARS‐CoV‐2 patients to reduce the viral load and mortality rate. 96 The challenges of CP therapy include the age and state of health of the donor, variations from unit to unit, informed consent from patients, and shortage of plasma. 97 Post‐CP‐therapy complications such as the transmission of other viruses, fever, chills, allergic reactions, acute lung injury, fluid overload and haemolytic events also need to be considered before opting for this treatment option. 98
5.3. Vaccines
Vaccination is defined as an active immunization technique whereby immunity against SARS‐CoV‐2 virus is induced in subjects artificially through exposure to antigens of the virus. 99 On 11 February 2020, the WHO announced that it would take approximately 18 months for a vaccine to become available to the public. 100 Currently, SARS‐CoV‐2 vaccines are under development using approaches like an inactivated virus, antigen presentation through viral vectors, recombinant vaccines, and DNA/mRNA vaccines. 101 As of July 2020, around 219 vaccines are under development worldwide, with 30 vaccines in human trials, including two in phase III clinical trial, two in phase II/III efficacy and dose‐testing trials, two in phase II trials, 10 in phases I–II safety and efficacy trials, and fourteen in phase I trials (Table 1). 102 , 103 , 104 , 105
TABLE 1.
List of vaccines under phase trials for severe acute respiratory syndrome‐coronavirus‐2 infection 102 , 103 , 104 , 105
| Vaccine name | Technology | Industries/Institutes | Clinical trials |
|---|---|---|---|
| mRNA‐1273 | RNA vaccine | Moderna, NIAID, Lonza | Phase III |
| CoronaVac | Inactivated virus | Sinovac, Instituto Butantan | Phase III |
| Bacillus Calmette‐Guerin (BCG) | Live‐attenuated vaccine | University of Melbourne and Murdoch Children's Research Institute, Radboud University Medical Center, Faustman Lab at Massachusetts General Hospital | Phase II/III |
| AZD1222 | Replication‐deficient simian viral vector | The University of Oxford, Astrazeneca | Phase II/III |
| Ad5‐nCoV | Replication‐defecient adenovirus type 5 | Cansino biological Inc., | Phase II |
| RBD‐Dimer | Adjuvant recombinant vaccine | Anhui Zhifei Longcom Biopharmaceutical, Institute of Microbiology of the Chinese Academy of Sciences | Phase II |
| Inactivated vaccine | Inactivated virus | Institute of Medical Biology, Chinese Academy of Medical Sciences | Phase I/II |
| BNT162 | RNA vaccine | BioNTech, Fosun Pharma, Pfizer | Phase I/II |
| T‐COVIDTM | Replication‐deficient adenovirus 5 | Altimmune | Phase I/II |
| GX‐19 | DNA vaccine | Genexine | Phase I/II |
| BBIBP‐CorV | Inactivated virus | Beijing Institute of Biological Products, Sinopharm | Phase I/II |
| Inactivated vaccine | Inactivated virus | Wuhan Institute of Biological Products, Sinopharm | Phase I/II |
| INO‐4800 | DNA vaccine | Inovio Pharmaceuticals, Inc. | Phase I/II |
| NVX‐CoV2373 | Protein subunit | Novavax, Emergent BioSolutions, Praha Vaccines, Serum Institute of India, AGC Biologics | Phase I/II |
| BBV152 (Covaxin) | Inactivated virus | ICMR‐National institute of Virology, Pune, Bharat Biotech | Phase I/II |
| ZyCoV‐D | DNA Vaccine | Zydus Cadila Healthcare | Phase I/II |
| CVnCoV | mRNA vaccine | CureVac | Phase I |
| LNP‐CoVsaRNA | RNA vaccine | Imperial College London, VacEquity Global Health | Phase I |
| SCB‐2019 | Protein subunit | Clover Biopharmaceuticals Inc., GSK, Dynavax | Phase I |
| bacTRL‐S | DNA Vaccine | Symvivo | Phase I |
| aAPC | Modified Lentiviral vector | Shenzhen Geno‐Immune Medical Institute | Phase I |
| LV‐SMENP‐DC | Modified Lentiviral vector | Shenzhen Geno‐Immune Medical Institute | Phase I |
| V‐SARS | Inactivated virus | Immunitor Inc. | Phase I |
| AV‐COVID‐19 | Patient‐specific dendritic cell vaccine | Aivita Biomedical Inc. | Phase I |
| COVAX‐19 | Protein subunit | Vaccine Pty, Flinders University, Oracle | Phase I |
| Molecular Clamp Vaccine | Protein subunit | University of Queensland CSL | Phase I |
| AG0301‐COVID19 | DNA Vaccine | Osaka University, AnGes; Takara Bio | Phase I |
| ARCoV | mRNA Vaccine | PLA Academy of Military Sciences; Walvax Biotechnology | Phase I |
| Gam‐COVID‐Vac | Non‐replicating viral vector | Gamaleya Research Institute | Phase I |
| Plant‐derived VLP vaccine | Adjuvant recombinant vaccine | Medicago Inc., Universite Laval, GSK, Dynavax | Phase I |
6. CONCLUSION
Taken together, specific drugs targeted to the viral genome, human ACE2 receptor, protease inhibitors, and antivirals can potentially aid in alleviating SARS‐CoV‐2 viral infection; however, only antibody‐mediated immune response through vaccine development can hope to bring under control the SARS‐CoV‐2 pandemic. 106
CONFLICT OF INTEREST
The authors have no competing interest.
AUTHORS CONTRIBUTION
Sakthivel Suganya, Suresh Divya and Madasamy Parani wrote the manuscript. Sakthivel Suganya and Suresh Divya contributed equally to the manuscript.
ACKNOWLEDGEMENTS
We acknowledge the funding provided by SRM Institute of Science and Technology, Kattankulathur, Tamil Nadu, India. Sakthivel Suganya and Suresh Divya would like to thank Dr. Ang Chin‐Siang for his valuable support regarding this manuscript and Hsuan Lin for his comments on this manuscript.
References
REFERENCES
- 1. Chan JFW, Yuan S, Kok KH, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person‐to‐person transmission: a study of a family cluster. Lancet. 2020;395(10223):514‐523. 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou P, Lou YangX, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270‐273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. WHO . WHO Director‐General's Opening Remarks at the Media Briefing on COVID‐19–11 March 2020. WHO Dir Gen Speeches; 2020. https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020. [Google Scholar]
- 4. Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5(4):536‐544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Coronavirus cases. Worldometer. 2020:1‐22. https://www.worldometers.info/coronavirus/worldwide-graphs/. [Google Scholar]
- 6. Lim YX, Ng YL, Tam JP, Liu DX. Human coronaviruses: a review of virus‐host interactions. Diseases. 2016;4(3):26. 10.3390/diseases4030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ningthoujam R. COVID 19 can spread through breathing, talking, study estimates. Curr Med Res Pract. 2020;10(3):132‐133. 10.1016/j.cmrp.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. World Health Organization . Transmission of SARS‐CoV‐2 : Implications for Infection Prevention Precautions. 2020. https://www.who.int/publications/i/item/modes‐of‐transmission‐of‐virus‐causing‐covid‐19‐implications‐for‐ipc‐precaution‐recommendations. [Google Scholar]
- 9. Zhang R, Li Y, Zhang AL, Wang Y, Molina MJ. Identifying airborne transmission as the dominant route for the spread of COVID‐19. Proc Natl Acad Sci USA. 2020;117(26):202009637. 10.1073/pnas.2009637117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Jiaye, Liao Xuejiao, Qian Shen, et al. Community Transmission of Severe Acute Respiratory Syndrome Coronavirus 2, Shenzhen, China, 2020. Emerging Infectious Diseases. 2020;26(6):1320‐1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tong ZD, Tang A, Li KF, et al. Potential presymptomatic transmission of SARS‐CoV‐2, Zhejiang province, China, 2020. Emerg Infect Dis. 2020;26(5):1052‐1054. 10.3201/eid2605.200198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (CoVID‐19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020;172(9):577‐582. 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fu L, Wang B, Yuan T, et al. Clinical characteristics of coronavirus disease 2019 (COVID‐19) in China: a systematic review and meta‐analysis. J Infect. 2020;80(6):656‐665. 10.1016/j.jinf.2020.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cen Y, Chen X, Shen Y, et al. Risk factors for disease progression in patients with mild to moderate coronavirus disease 2019—a multi‐centre observational study. Clin Microbiol Infect. 2020;26(9):1242‐1247. 10.1016/j.cmi.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Clinical Evidence Background. 2020. Accessed September 5, 2020. https://www.moh.gov.sg/docs/librariesprovider5/clinical-evidence-summaries/dexamethasone-for-covid-19-(updated-23-july-2020)-(1).pdf. [Google Scholar]
- 16. Clinical Evidence Background. 2020. Accessed September 5, 2020. https://www.clinicaltrials.gov/ct2/show/NCT04344730. [Google Scholar]
- 17. Sallard E, Lescure F‐X, Yazdanpanah Y, Mentre F, Peiffer‐Smadja N. Type 1 Interferons as a Potential Treatment Against COVID‐19. Amsterdam, Netherlands: Elsevier; 2020:178. 10.1016/j.antiviral.2020.104791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silva SJR, Alves da Silva CT, Mendes RPG, Pena L. Role of nonstructural proteins in the pathogenesis of SARS‐CoV‐2. J Med Virol. 2020;92:1427‐1429. 10.1002/jmv.25858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dai W, Zhang B, Su H, et al. Structure‐based design of antiviral drug candidates targeting the SARS‐CoV‐2 main protease. Science. 2020(80):1335. 10.1126/science.abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS‐CoV‐2. Gene Reports. 2020;19:100682. 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Goyal B, Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad‐spectrum therapeutic strategy. ACS Comb Sci. 2020;22(6):297‐305. 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 22. Jin Z, Zhao Y, Sun Y, et al. Structural basis for the inhibition of SARS‐CoV‐2 main protease by antineoplastic drug carmofur. Nat Struct Mol Biol. 2020;27(6):529‐532. 10.1038/s41594-020-0440-6. [DOI] [PubMed] [Google Scholar]
- 23. Mangum EM, Graham KK. Lopinavir‐ritonavir: a new protease inhibitor. Pharmacotherapy. 2001;21(11):1352‐1363. 10.1592/phco.21.17.1352.34419. [DOI] [PubMed] [Google Scholar]
- 24. Clinical evidence background. MOH‐ACE COVID‐19 RAPID Rev. 2020. 10.1101/2020.03.22.20034041v1. [DOI] [Google Scholar]
- 25. Chen H, Zhang Z, Wang L, et al. First clinical study using HCV protease inhibitor danoprevir to treat naïve and experienced COVID‐19 patients. medRxiv 2020. 10.1101/2020.03.22.20034041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Singh S, Sonawane A, Sadhukhan S. Plant‐derived natural polyphenols as potential antiviral drugs against SARS‐CoV‐2 via RNA‐dependent RNA polymerase (RdRp) inhibition: an in‐silico analysis. ChemRxiv. 2020. 10.26434/CHEMRXIV.12312263.V1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kulkarni SA, Nagarajan SK, Ramesh V, Palaniyandi V, Selvam SP, Madhavan T. Computational evaluation of major components from plant essential oils as potent inhibitors of SARS‐CoV‐2 spike protein. J Mol Struct. 2020;1221:1–11. 10.1016/j.molstruc.2020.128823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Götte M. The antiviral compound remdesivir potently inhibits RNA dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020;295(15):4773‐4779. 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin W, Mao C, Luan X, et al. Structural basis for inhibition of the RNA‐dependent RNA polymerase from SARS‐CoV‐2 by remdesivir. Science. 2020;368(6498):eabc1560. 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pachetti M, Marini B, Benedetti F, et al. Emerging SARS‐CoV‐2 mutation hot spots include a novel RNA‐dependent‐RNA polymerase variant. J Transl Med. 2020;18(1):179. 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Satarker S, Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus‐2. Arch Med Res. 2020. 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xia S, Liu M, Wang C, et al. Inhibition of SARS‐CoV‐2 (previously 2019‐nCoV) infection by a highly potent pan‐coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion. Cell Res. 2020;30(4):343‐355. 10.1038/s41422-020-0305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilson L, Mckinlay C, Gage P, Ewart G. SARS coronavirus E protein forms cation‐selective ion channels. Virology. 2004;330(1):322‐331. 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gupta MK, Vemula S, Donde R, Gouda G, Behera L, Vadde R. In‐silico approaches to detect inhibitors of the human severe acute respiratory syndrome coronavirus envelope protein ion channel. J Biomol Struct Dyn. 2020:1‐11. 10.1080/07391102.2020.1751300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anatoly C. Pharmaceutical targeting the envelope protein of SARS‐CoV‐2: the screening for inhibitors in approved drugs. chemRxiv. 2020:1‐13. 10.26434/chemrxiv.12286421.v1. [DOI] [Google Scholar]
- 36. Mukherjee D, Ray U. SARS‐CoV‐2 nucleocapsid assembly inhibitors: repurposing antiviral and antimicrobial drugs targeting nucleocapsid‐RNA interaction. chemRxiv. 2020. 10.26434/CHEMRXIV.12587336.V1. [DOI] [Google Scholar]
- 37. Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS–CoV‐2: a target for vaccine development. J Virol. 2020;94(13):1‐2. 10.1128/jvi.00647-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gheblawi M, Wang K, Viveiros A, et al. Angiotensin‐converting enzyme 2: SARS‐CoV‐2 receptor and regulator of the renin‐angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126(10):1456‐1474. 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Samavati L, Uhal BD. ACE2, much more than just a receptor for SARS‐COV‐2. Front Cell Infect Microbiol. 2020;10:317. 10.3389/fcimb.2020.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. de Kloet AD, Krause EG, Woods SC. The renin angiotensin system and the metabolic syndrome. Physiol Behav. 2010;100(5):525‐534. 10.1016/j.physbeh.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wakahara S, Konoshita T, Mizuno S, et al. Synergistic expression of angiotensin‐converting enzyme (ACE) and ACE2 in human renal tissue and confounding effects of hypertension on the ACE to ACE2 ratio. Endocrinology. 2007;148(5):2453‐2457. 10.1210/en.2006-1287. [DOI] [PubMed] [Google Scholar]
- 42. Santos RAS, Simoes e Silva AC, Maric C, et al. Angiotensin‐(1‐7) is an endogenous ligand for the G protein‐coupled receptor Mas. Proc Natl Acad Sci USA. 2003;100(14):8258‐8263. 10.1073/pnas.1432869100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto K, Ohishi M, Katsuya T, et al. Deletion of angiotensin‐converting enzyme 2 accelerates pressure overload‐induced cardiac dysfunction by increasing local angiotensin II. Hypertens. 1979;47(4):718‐726. 10.1161/01.HYP.0000205833.89478.5b.2006. [DOI] [PubMed] [Google Scholar]
- 44. Behl T, Kaur I, Bungau S, et al. The dual impact of ACE2 in COVID‐19 and ironical actions in geriatrics and pediatrics with possible therapeutic solutions. Life Sci. 2020;257:118075. 10.1016/j.lfs.2020.118075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS‐CoV‐2 infection. Eur J Intern Med. 2020;76:14‐20. 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. D'ardes D, Boccatonda A, Rossi I, et al. COVID‐19 and RAS: unravelling an unclear relationship. Int J Mol Sci. 2020;21(8):1‐8. 10.3390/ijms21083003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gurwitz D. Angiotensin receptor blockers as tentative SARS‐CoV‐2 therapeutics. Drug Dev Res. 2020;81:537‐540. 10.1002/ddr.21656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Losartan for Patients With COVID‐19 Requiring Hospitalization ‐ Full Text View ‐ ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04312009. 2020. Accessed June 28, 2020.
- 49. Rothlin RP, Vetulli HM, Duarte M, Pelorosso FG. Telmisartan as tentative angiotensin receptor blocker therapeutic for COVID‐19. Drug Dev Res. 2020. 10.1002/ddr.21679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Telmisartan for Treatment of COVID‐19 Patients ‐ Full Text View ‐ ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04355936. 2020. Accessed June 28, 2020.
- 51. Batlle D, Wysocki J, Satchell K. Soluble angiotensin‐converting enzyme 2: a potential approach for coronavirus infection therapy? Clin Sci. 2020;134(5):543‐545. 10.1042/CS20200163. [DOI] [PubMed] [Google Scholar]
- 52. Haschke M, Schuster M, Poglitsch M, et al. Pharmacokinetics and pharmacodynamics of recombinant human angiotensin‐converting enzyme 2 in healthy human subjects. Clin Pharmacokinet. 2013;52(9):783‐792. 10.1007/s40262-013-0072-7. [DOI] [PubMed] [Google Scholar]
- 53. Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin‐converting enzyme 2 (ACE2) as a SARS‐CoV‐2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586‐590. 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang X, Li S, Niu S. ACE2 and COVID‐19 and the resulting ARDS. Postgrad Med J. 2020;96:403–407. 10.1136/postgradmedj-2020-137935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Recombinant Human Angiotensin‐converting Enzyme 2 (rhACE2) as a Treatment for Patients With COVID‐19 ‐ Full Text View ‐ ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04287686. 2020. Accessed June 28, 2020.
- 56. Monteil V, Kwon H, Prado P, et al. Inhibition of SARS‐CoV‐2 infections in Engineered human tissues using clinical‐grade soluble human ACE2. Cell. 2020;181(4):905‐913. 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Recombinant Bacterial ACE2 Receptors ‐Like Enzyme of B38‐CAP Could be Promising COVID‐19 Infection‐ and Lung Injury Preventing Drug Better Than Recombinant Human ACE2 ‐ Full Text View ‐ ClinicalTrials.gov. https://clinicaltrials.gov/ct2/show/NCT04375046. 2020. Accessed June 28, 2020.
- 58. Alisha K, Tripti S. Computational screening of phytochemicals from medicinal plants as COVID‐19 inhibitors. chemRxiv. 2020;1. 10.26434/chemrxiv.12320273.v1. [DOI] [Google Scholar]
- 59. Minato T, Nirasawa S, Sato T, et al. B38‐CAP is a bacteria‐derived ACE2‐like enzyme that suppresses hypertension and cardiac dysfunction. Nat Commun. 2020;11(1):1‐12. 10.1038/s41467-020-14867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Stawiski EW, Diwanji D, Suryamohan K, et al. Human ACE2 receptor polymorphisms predict SARS‐CoV‐2 susceptibility. bioRxiv Prepr. 2020. 10.1101/2020.04.07.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Verdecchia P, Cavallini C, Spanevello A, Angeli F. The pivotal link between ACE2 deficiency and SARS‐CoV‐2 infection. Eur J Intern Med. 2020;76:14‐20. 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xudong X, Junzhu C, Xingxiang W, Furong Z, Yanrong L. Age‐ and gender‐related difference of ACE2 expression in rat lung. Life Sci. 2006;78(19):2166‐2171. 10.1016/j.lfs.2005.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cristelo C, Azevedo C, Marques JM, Nunes R, Sarmento B. SARS‐CoV‐2 and diabetes: new challenges for the disease. Diabetes Res Clin Pract. 2020;164:108228. 10.1016/j.diabres.2020.108228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cure E, Cumhur Cure M. Angiotensin‐converting enzyme inhibitors and angiotensin receptor blockers may be harmful in patients with diabetes during COVID‐19 pandemic. Diabetes Metab Syndr Clin Res Rev. 2020;14(4):349‐350. 10.1016/j.dsx.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS‐CoV‐2. Proc Natl Acad Sci USA. 2020;117(21):11727‐11734. 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jean Kaoru Millet GRW. Host Cell Proteases: Critical Determinants of Coronavirus Tropism and Pathogenesis. Elsevier; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie. 2017;142:1‐10. 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Iwata‐Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol. 2019;93(6). 10.1128/jvi.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Azimi A. TMPRSS2 inhibitors, bromhexine, aprotinin, camostat and nafamostat as potential treatments for COVID‐19. Univ Med Sci. 2020. 10.13140/RG.2.2.18254.28484. [DOI] [Google Scholar]
- 70. Stopsack KH, Mucci LA, Antonarakis ES, Nelson PS, Kantoff PW. TMPRSS2 and COVID‐19: serendipity or opportunity for intervention?. Canc Discov. 2020;10(6):779‐782. 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. The Utility of Camostat Mesylate in Patients with COVID‐19 Associated Coagulopathy (CAC) and Cardiovascular Complications ‐ Full Text View ‐ ClinicalTrials.Gov. Accessed July 17, 2020. https://clinicaltrials.gov/ct2/show/NCT04435015. [Google Scholar]
- 72. Ndao M, Nath‐chowdhury M, Sajid M, et al. A cysteine protease inhibitor rescues mice from a Lethal cryptosporidium parvum infection. Antimicrob Agents Chemother. 2013;57(12):6063‐6073. 10.1128/AAC.00734-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Shah PP, Wang T, Kaletsky RL, et al. A small‐molecule oxocarbazate inhibitor of human cathepsin L blocks severe acute respiratory syndrome and Ebola pseudotype virus infection into human Embryonic kidney 293T cells. Mol Pharmacol. 2010;78(2):319‐324. 10.1124/mol.110.064261.to. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang S, Du Q, Zhao K, Li A, Wei D, Chou K. Virtual screening for finding natural inhibitor against cathepsin‐L for SARS therapy. Amino Acids. 2007;33:129‐135. 10.1007/s00726-006-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005;102(33). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Du L, Kao RY, Zhou Y, et al. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem Biophys Res Commun. 2007;359(1):174‐179. 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Theoharides TC, Conti P. Dexamethasone for COVID‐19? Not so fast. J Biol Regul Homeost Agents. 2020;34(3). 10.23812/20-EDITORIAL_1-5. [DOI] [PubMed] [Google Scholar]
- 78. Nguyen‐ba G, Robert S, Dhalluin S, Hornebeck W. Modulatory effect of dexamethasone on ornithine decarboxylase activity and gene Expression: a possible post‐transcriptional regulation by a neutral metalloprotease. Cell Biochem Funct. 1994;12(2):121‐128. 10.1002/cbf.290120207. [DOI] [PubMed] [Google Scholar]
- 79. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS‐CoV‐2 and other human coronaviruses. Trends Immunol. 2020;41(5):355‐359. 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Coughlin MM, Prabhakar BS. Neutralizing human monoclonal antibodies to severe acute respiratory syndrome coronavirus: target, mechanism of action, and therapeutic potential. Rev Med Virol. 2012;22(1):2‐17. 10.1002/rmv.706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhao J, Yuan Q, Wang H, et al. Antibody responses to SARS‐CoV‐2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020; 1–8. Online ahead of print. 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Qu J, Wu C, Li X, et al. Profile of IgG and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2). Clin Infect Dis. 2020;1‐4. Online ahead of print. 10.1093/rfs/hhw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Giamarellos‐Bourboulis EJ, Netea MG, Rovina N, et al. Complex immune dysregulation in COVID‐19 patients with severe respiratory failure. Cell Host Microbe. 2020;27(6):992‐1000. 10.1016/j.chom.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Buonaguro FM, Ascierto PA, Morse GD, et al. Covid‐19: time for a paradigm change. Rev Med Virol. 2020. 10.1002/rmv.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID‐19 cytokine storm; what we know so far. Front Immunol. 2020;11(1446):1–4. 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Zhu Z, Chakraborti S, He Y, et al. Potent cross‐reactive neutralization of SARS coronavirus isolates by human monoclonal antibodies. Proc Natl Acad Sci USA. 2007;104(29):12123‐12128. 10.1073/pnas.0701000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID‐19: immunity, inflammation and intervention. Nat Rev Immunol. 2020. 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565‐574. 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wong SK, Li W, Moore MJ, Choe H, Farzan M. A 193‐amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin‐converting enzyme 2. J Biol Chem. 2004;279(5):3197‐3201. 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tian X, Li C, Huang A, et al. Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus‐specific human monoclonal antibody. Emerg Microbes Infect. 2020;9(1):382‐385. 10.1080/22221751.2020.1729069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Li F, Li W, Farzan M, Harrison SC. Structural biology: structure of SARS coronavirus spike receptor‐binding domain complexed with receptor. Science. 2005;309(5742):1864‐1868. 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 92. Pinto D, Park YJ, Beltramello M, et al. Cross‐neutralization of SARS‐CoV‐2 by a human monoclonal SARS‐CoV antibody. Nature. 2020;583:290‐295. 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 93. Wang C, Li W, Drabek D, et al. A human monoclonal antibody blocking SARS‐CoV‐2 infection. Nat Commun. 2020;11:1‐6. 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Develop Monoclonal Antibody Therapy to Prevent and Treat COVID‐19. 2020. https://news.abbvie.com/news/press-releases/abbvie-harbour-biomed-utrecht-university-and-erasmus-medical-center-announce-collaboration-to-develop-monoclonal-antibody-therapy-to-prevent-and-treat-covid-19.htm.
- 95. Dean CL, Hooper JW, Dye JM, et al. Convalescent plasma to treat COVID‐19 possibilities and challenges. Transfusion. 2020;60(5):1024‐1031. 10.1111/trf.15739. [DOI] [PubMed] [Google Scholar]
- 96. CNA Chinese doctors “using plasma therapy” on COVID‐19 patients. CNA.; 2020. [Google Scholar]
- 97. Zhao Qian, He Yong. Challenges of convalescent plasma therapy on COVID‐19. J Clin Virol J. 2020;127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Maclennan S, Barbara JAJ. Risks and side effects of therapy with plasma and plasma fractions. Best Pract Res Clin Haematol. 2006;19(1):169‐189. 10.1016/j.beha.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 99. Miller E. Controversies and challenges of vaccination: an interview with Elizabeth Miller. BMC Med. 2015;13(1):1‐5. 10.1186/s12916-015-0508-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. WHO . WHO Director‐General's Remarks at the Media Briefing on 2019‐nCoV on 11 February 2020. WHO Director General's Speeches. https://www.who.int/dg/speeches/detail/who-director-general-s-remarks-at-the-media-briefing-on-2019-ncov-on-11-february-2020. 2020. Accessed July 1, 2020. [Google Scholar]
- 101. Engla NEW, Journal ND. Developing covid‐19 vaccines at pandemic speed. N Engl J Med. 2020;382(21):1969‐1973. [DOI] [PubMed] [Google Scholar]
- 102. LSHTM . COVID‐19 Vaccine Development Pipeline. LSHTM Vaccine Cent; 2020:182. https://vac-lshtm.shinyapps.io/ncov_vaccine_landscape/. [Google Scholar]
- 103. Milken Airtable ‐ Milken Institute; 2020. https://airtable.com/shrSAi6t5WFwqo3GM/tblEzPQS5fnc0FHYR/viwDBH7b6FjmIBX5x?blocks=hide. [Google Scholar]
- 104. COVID‐19 Vaccine Tracker. 2020. https://www.raps.org/news-and-articles/news-articles/2020/3/covid-19-vaccine-tracker.
- 105. DRAFT Landscape of COVID‐19 Candidate Vaccines. World Heal Organ; 2020. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines. [Google Scholar]
- 106. Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019‐nCoV). Nat Rev Drug Discov. 2020;19(3):149‐150. 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]


